Abstract
Chironomus tentans-repressor splicing factor (Ct-RSF) represses the activation of splicing by SR proteins in vitro. Ct-RSF colocalizes with the Ser-Arg-rich (SR) protein hrp45 in interchromatin granule clusters and coimmunoprecipitates with hrp45 in nuclear extracts. Ct-RSF and hrp45 can also interact directly in vitro. Ct-RSF and hrp45 are recruited together to transcribing genes and associate with growing pre-mRNAs. Ct-RSF and hrp45 colocalize at a large number of gene loci. Injection of anti-Ct-RSF antibodies into nuclei of living cells blocks association of both Ct-RSF and hrp45 with the growing pre-mRNA, whereas binding of U2 small nuclear ribonucleoprotein particle (snRNP) to the pre-mRNA is unaffected. On the intron-rich Balbiani ring (BR) 3 pre-mRNA, hrp45 as well as U1 and U2 snRNPs bind extensively, whereas relatively little Ct-RSF is present. In contrast, the BR1 and BR2 pre-mRNAs, dominated by exon sequences, bind relatively much Ct-RSF compared with hrp45 and snRNPs. Our data suggest that Ct-RSF represses SR protein function at exons and that the assembly of spliceosomes at authentic splice sites displaces Ct-RSF locally.
INTRODUCTION
Most eukaryotic genes contain introns that interrupt the exons. The introns are removed constitutively or alternatively during the splicing reaction. Precise recognition of exon–intron borders is essential for gene expression. Assembly of the splicing machinery, the spliceosome, at each intron is guided by conserved 5′ splice site-, 3′ splice site-, and branch point sequences in the pre-mRNA. A multitude of dynamic interactions between the spliceosomal small nuclear RNAs (snRNAs) themselves and between the snRNAs and the pre-mRNA as well as extensive protein–protein and protein–pre-mRNA interactions are essential (Collins and Guthrie, 2000). The same basic splicing machinery is involved both in constitutive and alternative splicing. Recognition of splice sites must therefore be robust enough to distinguish similar, but false splice sites from correct splice sites and at the same time be flexible enough to allow recognition of alternative splice sites.
Assembly of the spliceosome, and to a considerable extent the splicing reaction, are cotranscriptional events in vivo. Evidence has come from visualization (Beyer and Osheim, 1988) and direct isolation (Baurén and Wieslander, 1994) of spliced nascent transcripts and from the kinetics of intron excision from very long transcripts (Tennyson et al., 1995). Spliceosomal components are recruited from presumed storage sites in interchromatin granule clusters (IGCs) to the nascent transcripts on transcribing genes (Misteli et al., 1997). Three-dimensional reconstruction of a transcribing gene in situ has shown that spliceosomal components, the pre-messenger ribonucleoprotein particle (mRNP), and RNA polymerase II are all present in a large RNA–protein complex during transcription elongation (Wetterberg et al., 2001). Splicing is influenced by other pre-mRNA processing events, for example, capping and 3′ end cleavage/polyadenylation (reviewed by Proudfoot et al., 2002). All pre-mRNA processing events are coupled to RNA transcription by RNA polymerase II. The C-terminal domain of the largest subunit of RNA polymerase II plays a role in coordinating transcription with processing events (Bentley, 2002). Chromatin immunoprecipitation experiments in yeast show that different spliceosomal components come to and leave the growing transcript with different kinetics (Kotovic et al., 2003), in accordance with data from in vitro analyses of splicing. It has also been shown that preassembled spliceosomal complexes are present in yeast (Stevens et al., 2002). It is still not settled how spliceosomes are assembled in vivo. It is essential to better understand how spliceosomal assembly takes place on the nascent transcripts to understand both constitutive and alternative splicing.
A conserved protein family, the Ser-Arg-rich (SR) protein family, is essential for splicing. The SR proteins have one or two RNA-binding domains (RBD) at their amino terminus, coupled to a domain rich in Ser-Arg dipeptides, the SR domain (Sanford et al., 2003). The SR proteins function through protein–protein interactions (reviewed in Graveley, 2000; Sanford et al., 2005) and through interactions with the pre-mRNA itself (Shen and Green, 2004). SR proteins can stimulate splice site recognition independent of pre-mRNA sequences. SR proteins can also bind exonic splicing enhancer sequences (ESEs) and recruit and stabilize binding of other splicing components during the splicing reaction. SR proteins therefore perform several important functions during spliceosomal assembly and splicing (Graveley, 2000). The SR proteins can have redundant functions in constitutive splicing. The concentration of SR proteins may influence the choice of alternative splice sites. It has been shown that SR proteins have several other functions during gene expression. They can act as export adapters (Huang et al., 2003) and some SR proteins accompany the mRNP to the cytoplasm. ASF/SF2 has been shown to regulate the stability of mRNA (Lemaire et al., 2002) and to stimulate translation (Sanford et al., 2004). In addition, SR proteins may be involved in mRNA surveillance (Zhang and Krainer, 2004) and may be involved in maintenance of genomic stability (Li and Manley, 2005).
SR proteins are targets for the regulation of splicing. They are subjected to cycles of phosphorylation/dephosphorylation. The degree of phosphorylation influences the function of the proteins. It is believed that hyperphosphorylated SR proteins are recruited to intron-containing nascent pre-mRNAs from the IGCs. The splicing reaction requires dephosporylation of SR proteins (Mermoud et al., 1994); and subsequent to splicing, hypophosphorylated SR proteins bind the export receptor TAP (Huang et al., 2004; Lai and Tarn, 2004). Shutdown of endogenous gene expression during virus infection is partly achieved through dephosphorylation of SR proteins (Kanopka et al., 1998; Huang et al., 2002). The dephosphorylated SRp38 is involved in down-regulating general gene expression during M phase in the cell cycle (Shin and Manley, 2002) and during heat shock (Shin et al., 2004). Some proteins, such as heterogeneous nuclear RNP (hnRNP) proteins, are known to influence SR protein binding to pre-mRNA and thereby repress splicing (Zhu et al., 2001; Wollerton et al., 2001). Furthermore, proteins that are similar to SR proteins, such as SRrp86 in mammals (Barnard and Patton, 2000) and RSF1 in Drosophila melanogaster (Labourier et al., 1999a) have been shown to repress splicing by antagonizing the function of SR proteins.
Here, we have identified a protein, Chironomus tentans-repressor splicing factor (Ct-RSF) in the dipteran C. tentans that inhibits splicing in vitro. Ct-RSF has an RBD that is similar to RBDs in SR proteins, coupled to a glycine-arginine-serine-rich domain. Ct-RSF is similar to the D. melanogaster RSF1 protein. Our data suggest that Ct-RSF is associated with the SR protein hrp45 in IGCs and is recruited to nascent transcripts together with hrp45. Ct-RSF remains bound to exon-rich transcripts together with hrp45, but Ct-RSF is displaced by assembly of spliceosomes on intron-rich transcripts. Our data propose that Ct-RSF plays a role in avoiding spliceosomal assembly at incorrect splice sites.
MATERIALS AND METHODS
Animals and Cells
C. tentans was cultured as described by Meyer et al., (1983). Where indicated, larvae were treated with galactose for 8 d in water containing 0.5 g of galactose per 100 ml. A C. tentans embryonic epithelial cell line was cultured as described by Wyss (1982). Certain C. tentans cells were incubated in standard medium to which the transcription inhibitors 5,6-dichloro-1-β-d-ribofuranosyl benzimidazole (DRB) (90 μM) or actinomycin D (5 μg/ml) were added. New synthesis of proteins was inhibited by addition of cycloheximide (20 μg/ml).
Antibodies
Polyclonal antibodies against Ct-RSF were raised in rabbits. Antibodies that were specific to the protein were purified by chromatography on CNBr-activated Sepharose 4B columns (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). The monoclonal antibody (mAb) 2E4 (Kiseleva et al., 1994; Wurtz et al., 1996), which recognizes the C. tentans SR protein hrp45; the mAb 11:1D3, recognizing the hrp23/Ct-RSF protein (Sun et al., 1998); and the mAb 4F9, which recognizes the C. tentans hnRNP protein hrp36 (Visa et al., 1996) were gifts from Dr. B. Daneholt (Karolinska Institutet, Stockholm, Sweden). The mAb 4G3, which recognizes the U2 snRNP-specific B″ protein, was obtained from Euro-Diagnostics (Melmo, Sweden). The 3C5 mAb, detecting a phosphoepitope in SR proteins (Turner et al., 1985), was a gift from Dr. B. M. Turner (University of Birmingham, Birmingham, United Kingdom). The human anti-70K antibodies (Welin Henriksson and Pettersson, 1996) were a gift from Drs. I. Lundberg and E. Hedfors (Huddinge Hospital, Huddinge, Sweden) and E. Welin Henriksson (Karolinska Institutet). Anti-U1 70K-specific antibodies were affinity purified by binding them to a D. melanogaster U1 70K fusion protein (Welin Henriksson and Pettersson, 1997) blotted to a filter, as described in Sambrook and Russell (2001). The secondary antibodies used for immunocytology were rabbit anti-mouse Ig fluorescein isothiocyanate (FITC) (DakoCytomation Denmark A/S, Glostrup, Denmark), swine anti-rabbit Ig FITC (DakoCytomation Denmark A/S), donkey anti-mouse IgG Texas Red (Jackson ImmunoResearch Laboratories, West Grove, PA), goat anti-rabbit IgG Cy-5 (GE Healthcare), and donkey anti-mouse IgG Cy-5 (GE Healthcare), all diluted 1:100. The secondary antibodies used for Western blots were swine anti-rabbit IG HRP (DakoCytomation Denmark A/S) and goat anti-mouse Ig horseradish peroxidase (HRP) (Dako-Cytomation Denmark A/S), both diluted 1:3000.
Expression of Fusion Proteins
Ct-RSF was expressed as a His-tagged fusion protein from the vector pET15B (Novagen, Madison, WI). Expression in BL21 bacteria was induced by bacteriophage CE6 infection, and the fusion protein was purified on a Ni-NTA resin affinity column (QIAGEN, Valencia, CA). The SR protein ASF/SF2 (expression plasmid was a gift from Dr. J. L. Manley, Columbia University, New York, NY) was expressed as a His-tagged protein in BL21 bacteria and purified on a Ni-NTA resin affinity column. ASF/SF2 was dialyzed into buffer D (Dignam et al., 1983), containing 0.5 M guanidine hydrochloride. Before use, the protein was diluted 10 times in buffer D. The His-hrp36 and the His-Ct-Y14 proteins were expressed in BL21 bacteria and purified on Ni-NTA resin affinity columns. The CLK/STY protein kinase (expression plasmid was a gift from Dr. J. L. Manley) was produced as glutathione S-transferase (GST)-fusion protein in the bacterial host JM101 as described by Xiao and Manley (1997). GST-Ct-RSF and GST-hrp45 fusion proteins were produced using the pGEX-4T-1 vector and purified on glutathione-Sepharose 4B (GE Healthcare). His-hrp45 was expressed from the pET21d(+) vector in Escherichia coli BL21 (Novagen). The pET21d-hrp45 clone was a gift from Dr. B. Daneholt (Karolinska Institutet). His-hrp45 was purified on a HiPrep 16/10 QFF column (GE Healthcare).
Protein Preparation and Western Blotting
SR proteins from C. tentans tissue culture cells were prepared as described by Zahler et al. (1992). Proteins to be analyzed by Western blotting were boiled in sample buffer (62.5 mM Tris-HCl at pH 6.8, 10% glycerol, 2.3% SDS, 5% mercaptoethanol, and 0.02% bromphenol blue) and separated on 12% or 15% SDS-polyacrylamide gels. The separated proteins were transferred to polyvinylidene diflouride (PVDF)-filters by semidry electrophoresis. HRP-labeled secondary antibodies were detected by the enhanced chemiluminescence method (GE Healthcare).
Immunoprecipitation
Immunoprecipitation from nuclear extracts of C. tentans tissue culture cells was performed essentially as described previously (Sun et al., 1998). In brief, mAb 11:1D3 or mAb 2E4 was added to nuclear extract in the presence of 0.1% NP-40 and incubated for 90 min at 4°C with gentle rotation. Rabbit anti-mouse immunoglobulins (DakoCytomation Denmark A/S), covalently coupled to protein G-Sepharose beads (Zymed Laboratories, South San Francisco, CA) were added, and the rotation was continued for 90 min at 4°C. The Sepharose beads were washed three times with phosphate-buffered saline (PBS) containing 0.1% NP-40 and once with PBS. The immunoprecipitated complexes were eluted with 0.5% SDS at room temperature. The proteins were precipitated with acetone and subsequently analyzed by Western blotting. The extracts were either immunoprecipitated directly or after treatment with RNase A (50 μg/ml) and RNase T1 (200 U) for 20 min at room temperature.
Protein Phosphorylation
The recombinant Ct-RSF or hrp45 proteins were incubated in phosphorylation buffer (20 mM HEPES, pH 7.6, 5 mM MgCl2, 1 mM dithiothreitol [DTT], 20 μCi of [γ32P]ATP, and 1 mM ATP), and the recombinant GST-CLK/STY protein at 30°C for 1 h.
In Vitro Splicing
Nuclear extracts and S100 extracts from HeLa cells and from C. tentans cells were prepared essentially according to Dignam et al. (1983) with slight modifications as described by Mühlemann and Akusjärvi (1998). A PCR fragment corresponding to part of the β-globin gene (Hornig et al., 1986), containing 174 base pairs of exon 1, the complete intron 1 of length 126 base pairs, and 53 base pairs of exon 2 with a built-in T7 promoter at its 5′ end, was transcribed in vitro with T7 polymerase in the presence of [α-32P]CTP. An adenovirus E1A transcript was prepared by cleaving the plasmid Sp4 (Popielarz et al., 1993) at the HindIII site and carrying out transcription in vitro with the SP6 polymerase in the presence of [α-32P]CTP. The plasmid pGEM2-V61, a gift from Dr. D. Rio (University of California, Berkeley, Berkeley, CA) was transcribed by T7 polymerase to produce the Fushi tarazu pre-mRNA (Rio, 1988). The plasmid pTA10 (Ghosh and Garcia-Blanco, 2000) was transcribed by T7 polymerase to produce an AdML transcript. All transcripts were purified on 4% sequencing polyacrylamide gels. Splicing reactions were carried out essentially as described by Mühlemann and Akusjärvi (1998). The reactions were performed in 25 μl containing 10 μl of nuclear extract or 10 μl of S100 extract, with a final concentration of 3.2 mM MgCl2, 2 mM ATP, 20 mM creatine phosphate, 2.6% polyvinyl alcohol, 5–10 fmol of RNA substrate, 12 mM HEPES, pH 7.9, 60 mM KCl, 0.12 mM EDTA, 0.3 mM DTT, 0.04 mM phenylmethylsulfonyl fluoride, and 3% glycerol. The reactions were incubated at 30°C for 90 min or the time indicated. Splicing products were treated with proteinase-K, extracted by phenol, precipitated by ethanol, and analyzed on 4 or 6% polyacrylamide gels containing 8 M urea. For analysis of spliceosomal complexes, HeLa cell nuclear extract was incubated at room temperature for 30 min in the presence of 2 mM glucose to deplete ATP. After incubation with the labeled pre-mRNA at 30°C, formation of E complex was analyzed on 1.5% agarose gels using 50 mM Tris-glycine as running buffer, as described previously (Das and Reed, 1999). Formation of A complex was analyzed on 2% agarose gels (Das and Reed, 1999) or on 4% nondenaturing polyacrylamide gels (Konarska and Sharp, 1988).
Immunocytological Localization
Cultured Cells. Cultured C. tentans diploid cells were prepared and stained essentially as described by Baurén et al. (1996), with the modification of using 0.5% Triton X-100 instead of SDS.
Isolated Chromosomes. Polytene chromosomes from the salivary glands of C. tentans were isolated and immunostained as described previously (Zhao et al., 2002). Some isolated chromosomes were treated with 100 μg/ml RNase A for 60 min at room temperature and rinsed in TKM buffer (10 mM triethanolamine-HCl, pH 7.0, 100 mM KCl, and 1 mM MgCl2) before fixation and antibody staining.
Microscopy
All preparations were mounted in antifade medium VectaShield (Vector Laboratories, Burlingame, CA). Preparations were viewed and photographed in a Zeiss Axiovert 100 M, equipped with a Zeiss confocal LSM 510 laser-scanning module or in an Axioplan II microscope (Carl Zeiss, Jena, Germany).
Microinjection
Salivary glands were dissected from C. tentans fourth instars and placed in a drop of ZO medium (Wyss, 1982) surrounded by paraffin oil. Anti-Ct-RSF antibodies, 10 mg/ml control antibodies, 10 mg/ml anti-Ct-RBD1 antibodies (Björk et al., 2002), or PBS was injected into individual nuclei (AIS MicroSystems; Carl Zeiss). The glands were incubated in hemolymph for 90 min. Immunostaining was performed essentially as described by Baurén et al. (1996).
Far-Western Analyses
GST-Ct-RSF and GST-hrp45 were labeled by CLK/STY kinase as described under Protein Phosphorylation. Labeled proteins were purified by binding to glutathione-Sepharose beads (GE Healthcare). After washing, the proteins were eluted in 20 mM glutathione (Sigma-Aldrich, St. Louis, MO). Purified proteins (1 μg each) or C. tentans SR proteins (∼1 μg) were separated on a 12% SDS-PAGE and transferred to PVDF filters. U1 snRNP was a gift from Dr. R. Lührmann (Max-Planck-Institute of Biophysical Chemistry, Göttingen, Germany). The proteins were renatured and probed with 10 mg of labeled GST-Ct-RSF or GST-hrp45 in 10 ml of binding buffer as described previously (Kohtz et al., 1994). After washing in binding buffer lacking bovine serum albumin and milk powder, the filters were dried and exposed to x-ray films.
RESULTS
Ct-RSF Colocalizes with the SR Protein hrp45 in Nuclear Speckles
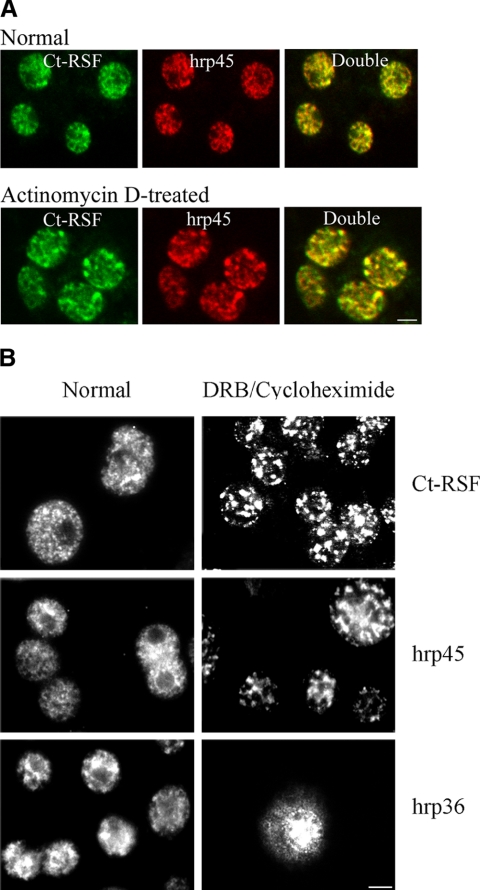
Ct-RSF-specific antibodies stained diploid cell nuclei from C. tentans in a diffuse pattern, apparently excluding the nucleoli, combined with a dominating, more intense speckled pattern (Figure 1A, Normal). The speckled pattern is characteristic for the localization of splicing factors, including SR proteins, in IGCs. Double staining of nuclei with Ct-RSF antibodies and a mAb against the SR protein hrp45 showed that the two proteins colocalized in the speckles (Figure 1A, top).
Figure 1.
Ct-RSF is restricted to the nucleus and colocalizes with the SR protein hrp45 in nuclear speckles. (A) C. tentans diploid cells were stained with anti-Ct-RSF and anti-hrp45 antibodies. In cells grown in normal medium (top), the staining patterns for the two proteins overlapped extensively. In cells treated with actinomycin D (bottom), both Ct-RSF and hrp45 were redistributed into the same larger and more intensively stained speckles. (B) Ct-RSF and hrp45 were restricted to the nucleus and shifted into large speckles also upon treatment with DRB and cycloheximide. In contrast, the hnRNP protein hrp36 did not accumulate in the same speckles. To some extent, it accumulated in the cytoplasm. Bars, 2 μm.
It has previously been shown that upon transcription shutdown, SR proteins redistribute into fewer and more brightly stained speckles. After treatment of the C. tentans diploid cells with actinomycin D at a concentration that inhibits RNA polymerase II transcription, both Ct-RSF and hrp45 behaved in the same manner and colocalized in large and more brightly stained speckles (Figure 1A, bottom). In Figure 1B, it is shown that Ct-RSF also behaved as hrp45 upon inhibition of transcription by treatment of the cells with DRB. As before, the two proteins concentrated in fewer, brightly stained speckles. It is also shown that Ct-RSF was retained in the nucleus during these conditions, as was hrp45. In contrast, the hnRNP protein hrp36 behaved differently. It did not accumulate in the large speckles, and this protein partly accumulated in the cytoplasm, indicating that it is a shuttling protein. Ct-RSF does not shuttle in a transcription-dependent manner detectable in this assay. We conclude that a substantial amount of Ct-RSF colocalizes with the SR protein hrp45 in nuclear regions corresponding to IGCs.
Ct-RSF and hrp45 Interact through Protein–Protein Contacts
To investigate more in detail whether Ct-RSF and hrp45 are associated in the nucleus, we used nuclear extracts for immunoprecipitation experiments. The extracts were prepared to mainly contain soluble, nonchromatin-associated material. In Figure 2, it is shown that Ct-RSF was coimmunoprecipitated with hrp45 and vice versa (Figure 2, A and B, lanes 2). The coimmunoprecipitations were not affected by RNase treatment of the nuclear extracts (Figure 2, A and B, lanes 5). In contrast, coimmunoprecipitation of the Y14 and Mago proteins was abolished by the RNase treatment (our unpublished data). In agreement with the colocalization of Ct-RSF and hrp45 in the cell nucleus (Figure 1), the two proteins interact directly or indirectly in vivo, and this interaction is mediated by protein–protein contacts.
Figure 2.
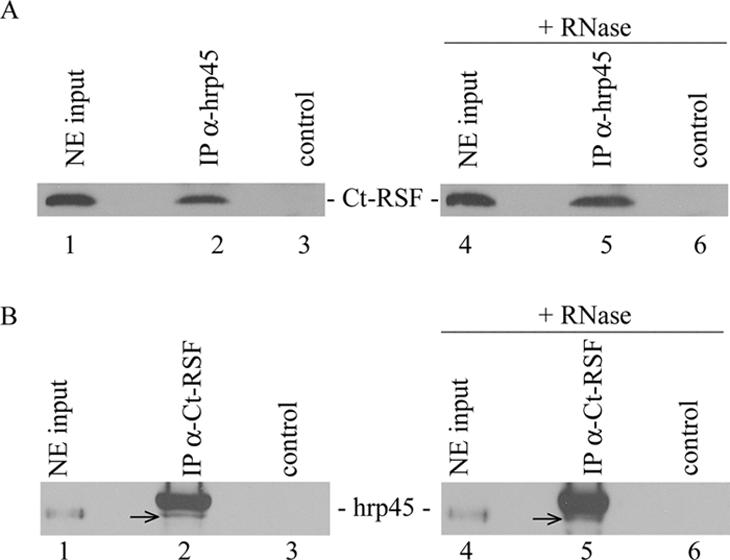
Ct-RSF and hrp45 are coimmunoprecipitated. Nuclear extract from C. tentans cells was immunoprecipitated with anti-hrp45 antibodies (A) or anti-Ct-RSF antibodies (B). Ct-RSF coimmunoprecipitated with hrp45 and vice versa (A and B, lanes 2). The hrp45 signal in B (arrows) is located just below the position of the antibody used in the experiment. Controls represent immunoprecipitations in the absence of specific antibodies. RNase treatment of the nuclear extracts before immunoprecipitation did not affect the signal (A and B, lanes 5).
Ct-RSF and hrp45 Interact Directly In Vitro
To investigate whether Ct-RSF and hrp45 can bind directly to each other, we performed Far-Western experiments. Purified Ct-RSF and hrp45 were 32P-labeled with the kinase CLK/STY, and a series of proteins were probed (Figure 3). In Figure 3A, it is shown that Ct-RSF bound to hrp45 under these conditions. It also bound to several other C. tentans SR proteins and to itself. Ct-RSF did not bind to GST, the hnRNP protein hrp36, or Ct-Y14. We also found that hrp45 bound to itself, to other SR proteins, and to Ct-RSF (Figure 3B). The interaction with Ct-RSF involved the C-terminal domain, rich in glycine, serine, and arginine residues. We also tested binding to the U1 snRNP-specific protein U1 70K that has an SR-like domain. Both Ct-RSF and hrp45 bound to the U1 70K protein.
Figure 3.
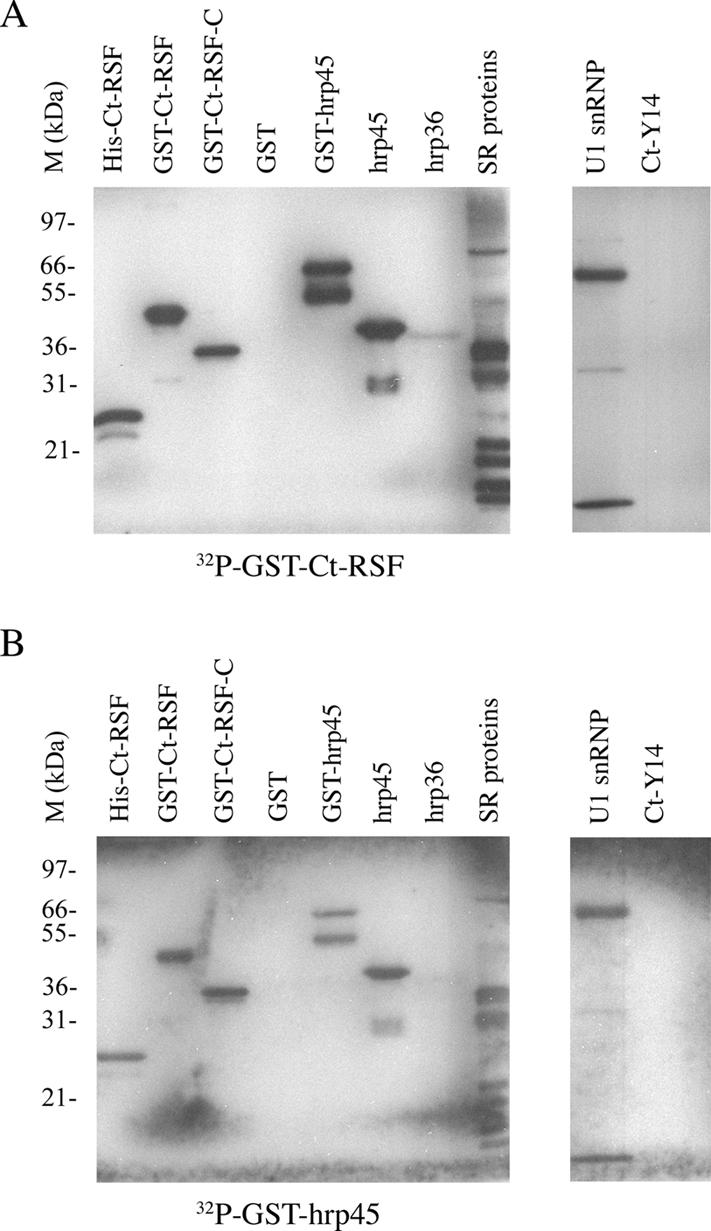
Ct-RSF and hrp45 bind directly to each other in vitro. Far-Western experiment in which the indicated proteins were separated on polyacrylamide gels, blotted to a nylon filter, and refolded. The proteins were probed with labeled Ct-RSF (A) or labeled hrp45 (B). SR proteins were purified from C. tentans cells. U1 snRNP represents purified mammalian U1 snRNP. The positions of size marker proteins are shown to the left of each filter.
Ct-RSF and hrp45 Are Recruited to Growing Pre-mRNAs
The nuclear distribution of Ct-RSF outside the speckles seen in Figure 1 may partly reflect Ct-RSF binding to mRNA in transit from the gene loci to the nuclear pore complexes and partly Ct-RSF binding to nascent pre-mRNA at many active gene loci. It has been shown that nascent transcripts on polytene chromosomes are immediately associated with RNA-binding proteins, including splicing factors during transcription (Matunis et al., 1993; Kiseleva et al., 1994; Baurén et al., 1996). We therefore focused specifically on the binding of Ct-RSF and hrp45 to growing transcripts.
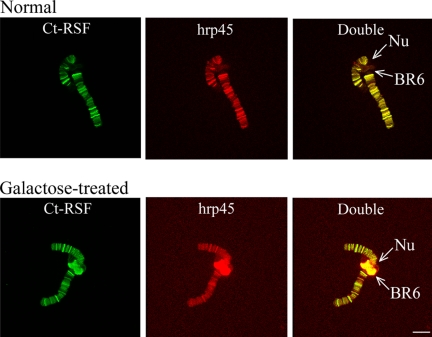
Using C. tentans salivary gland cells containing polytene chromosomes, we could demonstrate that both Ct-RSF and hrp45 are recruited to gene loci upon activation of transcription. When C. tentans larvae are treated with galactose, transcription of salivary gland-specific genes are either turned off (the BR2 genes) or turned on (the BR6 genes). As shown in Figure 4, Ct-RSF and hrp45 were present only at the transcribing BR6 gene and not at the nontranscribing gene (compare Figure 4, A and B). The immunofluorescence signal was sensitive to RNase treatment, demonstrating that the proteins were bound to the pre-mRNA (Figures 7B and 8C).
Figure 4.
t-RSF and hrp45 are recruited to transcriptionally active gene loci. Polytene chromosome III was isolated from C. tentans salivary gland cells and double-stained for Ct-RSF and hrp45. In A, the larvae were grown under normal conditions, and in B, in the presence of galactose. The BR6 gene locus (BR6), located close to the nucleolus (Nu) on chromosome III, is transcriptionally inactive under normal conditions. In the presence of galactose, the BR6 gene is transcribed and forms a large puff. Bar, 10 μm.
Figure 7.
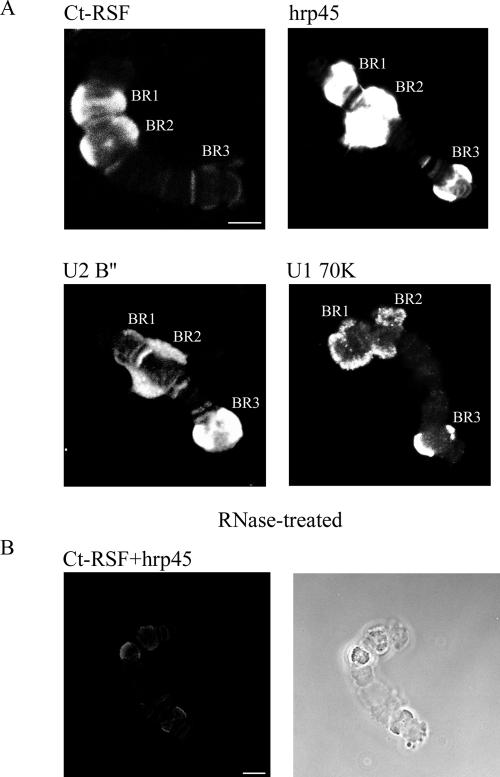
Ct-RSF binds preferentially to pre-mRNAs that have few introns and long exons. Immunostaining of isolated polytene chromosome IV. Individual chromosomes were stained with antibodies directed against Ct-RSF, hrp45, U2 B″, or U1 70K as indicated (A). The highly transcribed BR genes (BR1, BR2, and BR3) were stained as well as additional gene loci. The intensity of staining for U2 B″ and U1 70K was reproducibly highest for the BR3 gene locus that contains 38 introns. In contrast, the staining of the BR3 gene locus was weak for Ct-RSF. RNase treatment (B) abolished immunostaining for Ct-RSF and hrp45, immunostaining (left), phase contrast (right). Compare the RNase treated and double stained chromosome in B (left) with the chromosomes in A. Bars, 10 μm.
Figure 8.
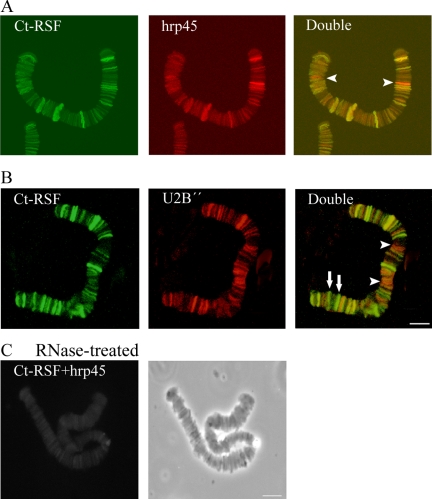
Ct-RSF colocalizes extensively with hrp45 and U2 snRNP on pre-mRNAs. Polytene chromosome I from salivary gland cells was isolated and stained with antibodies directed against Ct-RSF, hrp45, and U2 B″. In A, chromosome I was double-stained for Ct-RSF and hrp45. All stained gene loci were immunolabeled for both proteins. In some cases, staining for hrp45 was considerably stronger than for Ct-RSF (arrowheads). In B, chromosome I was stained for Ct-RSF and U2 B″. Most stained gene loci were labeled with both antibodies. In some cases, staining for Ct-RSF was relatively much stronger than for U2 B″ (arrows), and for others, staining for U2 B″ was relatively stronger (arrowheads). In C, RNase treatment abolished staining for Ct-RSF and hrp45, double staining (left), phase contrast (right). Bars, 10 μm.
Ct-RSF Inhibits Splicing and SR Protein Activation of Splicing
Based on the recruitment of Ct-RSF to growing pre-mRNA and the relationship of Ct-RSF to hrp45 and other SR proteins, we studied whether Ct-RSF is involved in splicing. We therefore analyzed the possible influence of Ct-RSF on splicing, using extracts from both HeLa and C. tentans cells and different RNA substrates.
Ct-RSF could not activate splicing in S100 extract from C. tentans cells (Figure 5A, lane 4), in contrast to the individual SR protein hrp45 (Figure 5A, lane 3) or SR protein preparations from C. tentans (Figure 5A, lane 2). Furthermore, Ct-RSF repressed the activating effect of SR proteins (Figure 5A, compare lanes 2 and 5) and of hrp45 (Figure 5A, compare lanes 3 and 6). Ct-RSF could not activate splicing of an E1A transcript in HeLa S100 extract (Figure 5B) or a β-globin transcript in HeLa S100 extract (our unpublished data).
Figure 5.
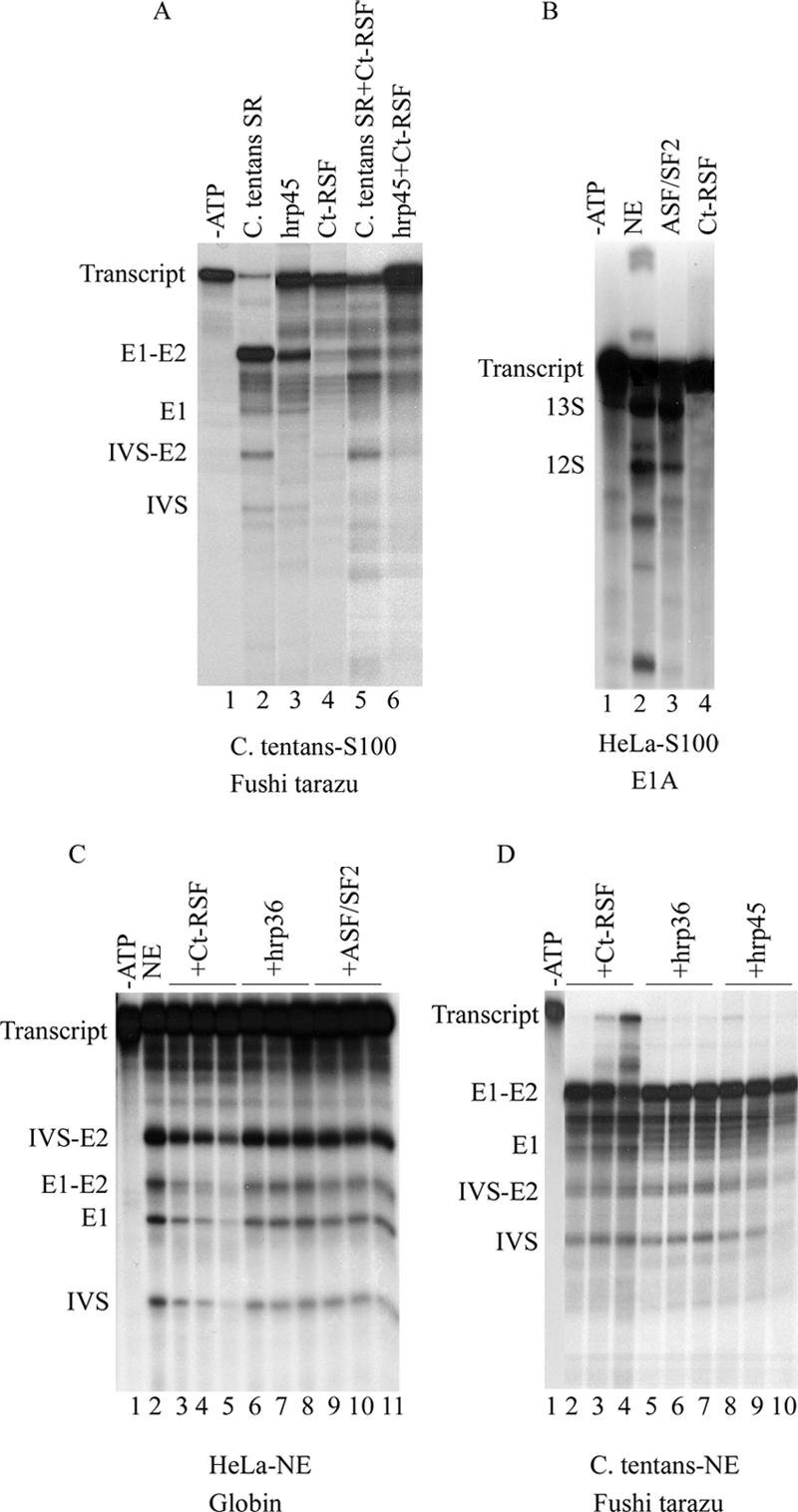
Ct-RSF cannot activate splicing in S100 extracts and represses splicing in nuclear extracts. In vitro splicing of a Fushi tarazu transcript in S100 extract from C. tentans cells (A). Lanes 1 and 2, control reactions after addition of C. tentans SR proteins (0.5 μg). ATP was left out in lane 1. Splicing was activated by hrp45 (0.5 μg) (lane 3) but not by Ct-RSF (0.5μg) (lane 4). Ct-RSF (0.5 μg) repressed the activation of splicing by C. tentans SR proteins (0.5 μg) (lane 5) and by hrp45 (0.5 μg) (lane 6). In vitro splicing of an E1A transcript (B) in S100 extract from HeLa cells. Lane 1, control reaction supplemented with nuclear extract but lacking ATP. Lane 2, control reaction supplemented with nuclear extract. Addition of ASF/SF2 (0.5 μg) (lane 3) activated splicing. Addition of Ct-RSF (0.5 μg) did not activate splicing (lane 4). In vitro splicing of β-globin pre-mRNA in HeLa nuclear extract (C). Lane 1, control reaction in the absence of ATP. Lane 2, control reaction with ATP. Lanes 3–5, increasing amounts of Ct-RSF (0.5, 1, and 1.5 μg) repressed splicing. No repression was seen after addition of hrp36 (lanes 6–8, 0.5, 1, and 1.5 μg) or ASF/SF2 (lanes 9 –11, 0.5, 1, and 1.5 μg). In vitro splicing of Fushi tarazu pre-mRNA in C. tentans nuclear extract (D). Lane 1, control reaction supplemented with nuclear extract, but lacking ATP. Increasing amounts of Ct-RSF (0.5, 1, and 1.5 μg) (lanes 2– 4) repressed splicing, but this was not seen after addition of hrp36 (0.5, 1, and 1.5 μg) (lanes 5–7) or hrp45 (0.5, 1, and 1.5 μg) (lanes 8 –10). Positions of the pre-mRNAs, splicing intermediates and products are indicated to the left of each figure.
In addition, Ct-RSF specifically inhibited splicing in nuclear extracts from HeLa cells (Figure 5C, lanes 3–5) and from C. tentans cells (Figure 5D, lanes 2– 4), using the β-globin and the Fushi tarazu (Ftz) transcripts, respectively. At the same concentrations, ASF/SF2 (Figure 5C, lanes 9 –11) and hrp45 (Figure 5D, lanes 8 –10) did not inhibit the splicing reaction and neither did the hnRNP protein hrp36 (Figures 5C, lanes 6–8, and 6D, lanes 5–7). We note that Ct-RSF repression of splicing is the same in extracts from both HeLa and C. tentans cells. This shows that Ct-RSF repression is not only seen in a heterologous system and in addition that the mechanism of repression is most likely the same in both mammalian and insect systems.
Figure 6.

Ct-RSF represses formation of spliceosomal E complex. HeLa cell nuclear extract was depleted for ATP and incubated with labeled AdML pre-mRNA at 30°C for the indicated times. Complexes were analyzed on 1.5% agarose gels. Lanes 3 and 4, increasing amounts (1 and 1.5 μg) of Ct-RSF repressed E complex formation. Positions of E and H complexes are shown to the right.
Splicing of the E1A transcript was also highly influenced by Ct-RSF. ASF/SF2 shifted 5′ splice site use to proximal 5′ splice sites as described previously (Ge and Manley, 1990; Krainer et al., 1990), whereas Ct-RSF had a considerably stronger effect (our unpublished data).
Finally, we investigated if Ct-RSF inhibited the formation of a specific spliceosomal complex. Figure 6 (lanes 3 and 4) shows that Ct-RSF repressed formation of E complex in HeLa nuclear extract using an AdML transcript. In agreement, repression by Ct-RSF of A complex formation was also observed for the AdML, E1A, and β-globin transcripts (our unpublished data).
Ct-RSF Colocalizes with hrp45 on Exons of a Large Number of Nascent Pre-mRNAs
Next, we analyzed the association of Ct-RSF and hrp45 with nascent transcripts that have known exon–intron structure. Immunostaining for the U1 snRNP-specific 70K and U2 snRNP-specific B″ were included as markers for spliceosomal assembly. We took advantage of the characterized gene structures of the BR1, BR2, and BR3 genes on chromosome IV in C. tentans salivary gland cells (Wieslander, 1994). The BR1 and BR2 genes both contain four introns, however, these genes are dominated by an ∼35-kb exon. It has previously been shown that hrp45 binds along most of the 35-kb exon (Alzhanova-Ericsson et al., 1996), whereas snRNPs bind at the beginning and end of the transcripts where the introns are located (Kiseleva et al., 1994). In contrast to the BR1 and BR2 genes, ∼50% of the BR3 pre-mRNA is made up of 38 introns, evenly spaced throughout the transcript. A majority of these introns are cotranscriptionally removed in the splicing reaction (Wetterberg et al., 1996). In Figure 7A, it is shown that hrp45 was associated with nascent transcripts on the BR1, BR2, and BR3 genes. These transcripts also bound the U1 and U2 snRNPs as shown by detecting U1 70K and the U2 B″. Strikingly, only the BR1 and BR2 loci bound substantial amounts of Ct-RSF. In the BR3 locus, only a small amount of Ct-RSF was detected (Figure 7A). The immunostaining was sensitive to RNase treatment, showing that both Ct-RSF and hrp45 are associated with the pre-mRNA on the active genes (Figure 7B).
Importantly, although the relative amount of Ct-RSF on the BR3 nascent transcript was small compared with the BR1 and BR2 loci, the relative staining intensity for hrp45 was approximately the same for the BR1, BR2, and BR3 genes. These results show that Ct-RSF was much less associated with an intron-rich transcript than with transcripts dominated by exon sequences. In contrast, the U1 and U2 snRNPs were more associated with the intron-rich transcript and hrp45 present equally much with both transcripts.
We stained chromosomes I–III to get an overview of the entire genome. Along each of the three chromosomes, ∼30 loci were strongly stained with each of the antibodies. In addition, a large number of weakly stained chromosomal loci were observed. RNase treatment abolished the staining, showing that binding was to nascent transcripts (Figure 8C). A comparison of a large number of chromosomes showed that the staining patterns for U2 snRNP B″ and hrp45 antibody overlapped largely, if not completely. Double staining for Ct-RSF and hrp45 (Figure 8A) or for Ct-RSF and U2 B″ (Figure 8B) showed that all strongly stained chromosomal loci that contained U2 snRNP and hrp45 also contained Ct-RSF, although the staining intensity for Ct-RSF varied and at some loci was considerably weaker than staining for the two splicing factors (Figure 8, arrowheads). Close inspection of the many weakly stained loci revealed that this pattern was also true for them. We did not find any locus stained only for Ct-RSF. In a few cases, staining for Ct-RSF and hrp45 was about equally intense, whereas the staining for U2 snRNP was considerably weaker (Figure 8B, arrows). We conclude that Ct-RSF colocalized with hrp45 and U2 snRNP on nascent transcripts throughout the genome, but the relative staining intensity for the three proteins varied. Based on the staining patterns for the characterized BR1–3 gene loci, it is likely that the specific patterns observed throughout the genome reflect different exon–intron structures in the different gene loci.
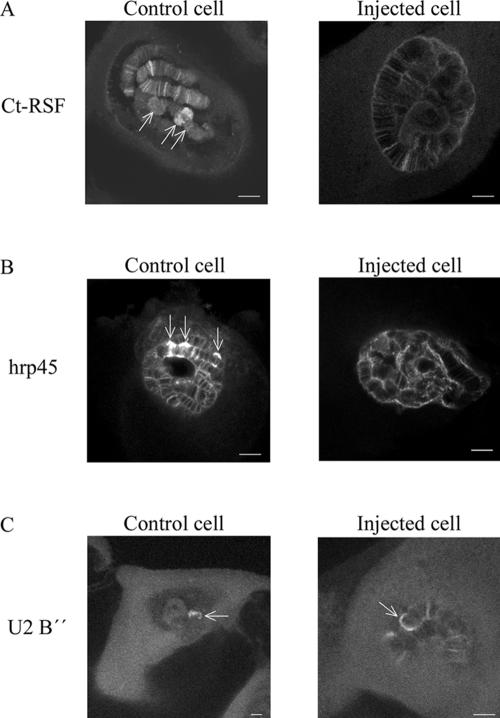
Ct-RSF-specific Antibodies Interfere with the Association of Both Ct-RSF and hrp45 with Nascent Pre-mRNAs In Vivo
To learn more about the function of Ct-RSF in the cell nucleus, we injected anti-Ct-RSF antibodies into nuclei of living salivary gland cells (Figure 9). On injection of the antibodies, the staining for Ct-RSF as well as for hrp45 decreased considerably in the active BR1, 2, and 3 gene loci and also in other gene loci (Figure 9, A and B). The staining for U2 snRNP B″ in the BR gene loci was unaffected (Figure 9C). Injection of an unrelated antibody did not produce these effects (control cells). These results show that the Ct-RSF-specific antibodies inhibited the localization of both Ct-RSF and hrp45 at active gene loci. Therefore, it is likely that the two proteins are associated with each other on nascent transcripts. It is possible that the antibodies influenced the proteins already bound to the pre-mRNAs. Another explanation could be that the antibodies inhibited the recruitment of the two proteins to the active gene loci. If so, Ct-RSF and hrp45 is recruited as a complex to the nascent transcripts.
Figure 9.
Anti-Ct-RSF antibodies interfere with the association of Ct-RSF and hrp45 with nascent pre-mRNAs. Individual salivary gland cells were injected with anti-Ct-RSF antibodies. After incubation for 90 min in hemolymph, the glands were fixed and immunostained for Ct-RSF, hrp45, or U2 B″ as indicated. Each picture shows a confocal section through a cell nucleus. Arrows point out the transcriptionally highly active BR genes on chromosome IV. Controls cells were injected with an unrelated antibody. Bars, 20 μm.
DISCUSSION
Ct-RSF Is Structurally Related to SR Proteins and Influences Their Function
Ct-RSF has previously been described as hrp23 (Sun et al., 1998). We have chosen to call it Ct-RSF because of its structural and functional similarities to RSF1 in D. melanogaster. One striking property of Ct-RSF is the similarity of its RBD to the RBDs found in SR proteins. This suggests that Ct-RSF can bind to similar sites as SR proteins, as shown for RSF1 (Labourier et al., 1999b). Ct-RSF does not have a typical RS-domain. Instead, it has a C-terminal domain rich in glycine, arginine, and serine residues, including eight scattered RS-dipeptides. Furthermore, Ct-RSF can be phosphorylated by the kinase CLK/STY that also phosphorylates SR proteins, and the phosphoepitope in Ct-RSF is recognized by the mAb 3C5 (Turner et al., 1985) that specifically binds to phosphoepitopes in SR proteins (our unpublished data). We also found that Ct-RSF is copurified with SR proteins in the two-step salt precipitation procedure for SR proteins (our unpublished data). This copurification is in agreement with our data showing colocalization in the nucleus and direct interaction between Ct-RSF and the SR protein hrp45. A functional relationship is indicated by the observation that Ct-RSF specifically represses the splicing activation property of SR proteins. Our results indicate that Ct-RSF is structurally related and functionally coupled to SR proteins.
We have shown that Ct-RSF inhibits splicing in vitro. Less is known about the mechanisms of inhibition of splicing than about activation of splicing and spliceosomal assembly. During mitosis (Shin and Manley, 2002) and during heat shock (Shin et al., 2004), there is a large-scale repression of gene expression and a general shutdown of pre-mRNA splicing. The splicing machinery remains intact and the repression is in both conditions believed to be mediated by a dephosphorylated form of the SR protein SRp38. The dephosporylated SRp38 seems to repress splicing by binding to the U1 snRNP-specific U1 70K protein and thereby prevents SR protein-mediated recruitment of U1 snRNP to the 5′ splice site. In cells infected by adenovirus (Kanopka et al., 1998) and vaccinia virus (Huang et al., 2002), host cell gene expression is down-regulated at several levels, including pre-mRNA splicing, and again, SR proteins are involved. In the infected cells, SR proteins are functionally inactivated by a virus-induced dephosphorylation.
A crucial problem in pre-mRNA splicing is how exon– intron borders are accurately defined. It is conceivable that correct splice site recognition and regulation of splice site use is achieved not only by proteins that promote splicing but also by specific proteins that repress splicing. Among these are SR proteins. Apart from SRp38, the SR protein ASF/SF2 can inhibit splicing if it binds to intron sequences (Kanopka et al., 1996). SR-like proteins may repress splicing by direct interference with SR protein function. SRrp86 in mammals represses activation of splicing of some SR proteins, but not of others (Barnard and Patton, 2000). RSF1 in D. melanogaster represses U1 snRNP binding to 5′ splice sites (Labourier et al., 1999a). Other proteins that can interfere with splicing are hnRNP proteins. The hnRNP A/B proteins can bind to exonic splicing silencers (ESSs) and block the activation effect of SR proteins (Zhu et al., 2001), supporting a model in which one function for ESEs is to antagonize the effect of neighboring ESSs (Blencowe, 2000). Splicing repression by competitive binding to the pre-mRNA has also been described for the Sex-lethal protein in D. melanogaster (Valcárcel et al., 1993) and the PTB in mammals (also called hnRNP I) (Wollerton et al., 2001). PTB binds to polypyrimidine sequences at many 3′ splice sites and may thereby compete out factors essential for 3′ splice site recognition (Singh et al., 1995).
It is evident that SR proteins are essential for both down-regulation and activation of splicing. Either the phosphorylation level of SR proteins is important or other proteins can interfere with the action of SR proteins. In line with this theme, our data indicate that Ct-RSF functions through interaction with SR proteins.
Ct-RSF Is Functionally Coupled to hrp45
Ct-RSF and RSF1 have similar N-terminal RBDs. The C-terminal domains cannot be aligned, but they share the property of containing many glycine, arginine, and serine residues. As for Ct-RSF, RSF1 interacts with SR proteins and inhibits splicing in HeLa extracts at an early step in spliceosomal assembly. Apparently, RSF1 represses the cooperation between ASF/SF2 and U1 snRNP in binding to the 5′ splice site (Labourier et al., 1999a). We demonstrate that in spite of the differences in the C-terminal domains, the function of Ct-RSF is similar to the function of RSF1. In this report, we further analyze the relationship of Ct-RSF to nascent pre-mRNAs and to the specific SR protein hrp45 in intact cells. The C. tentans hrp45 corresponds to the previously characterized D. melanogaster B52 (Champlin et al., 1991) and the mammalian SRp55 (Sanford et al., 2003). SRp55 binds to ESEs and is involved in regulation of alternative splicing (Nagel et al., 1998; Tran and Roesser, 2003). A B52 null mutant is lethal at early larval stages in D. melanogaster; furthermore, B52 is important for splicing of a subset of pre-mRNAs (Ring and Lis, 1994; Kim et al., 2003). Overexpression of B52 results in developmental defects. These defects are rescued by over expression of RSF1 (Labourier et al., 1999a). The results from our coimmunoprecipitation, localization, and microinjection experiments provide further evidence that Ct-RSF/RSF1 and hrp45/B52 are functionally coupled in vivo.
Splicing Inhibition and In Vivo Behavior of Ct-RSF
How does Ct-RSF repress splicing in vitro? Our data suggest that Ct-RSF interferes with the function of SR proteins. It can bind directly to SR proteins and the repressing activity of Ct-RSF on splicing can be titrated out by an excess of SR proteins (our unpublished data). The C-terminal domain of Ct-RSF is sufficient for binding to SR proteins, but the C-terminal domain itself does not repress splicing (our unpublished data), similar to what has been described for RSF1 (Labourier et al., 1999a). This suggests that the complete protein is required, possibly because its binding to the pre-mRNA is necessary for splicing repression. SR proteins can function through protein–protein interactions with spliceosomal components (reviewed in Graveley, 2000). It has also been shown recently that the RS domain of SR proteins directly contacts the 5′ splice site and the branch point in the pre-mRNA (Shen and Green, 2004). Both kinds of interactions might be blocked by Ct-RSF at cryptic splice sites, when bound to SR proteins. Both RSF1 and the dephosphorylated SRp38 inhibit SR protein stimulated U1 snRNP association at 5′ splice sites. Based on the observations that Ct-RSF can bind U1 70K, that it represses E complex formation, and its similarity to RSF1, it is likely that Ct-RSF also interferes with U1 snRNP association at 5′ splice sites. The dipteran Ct-RSF and RSF1 and the mammalian SRp38 may therefore be functionally related.
Ct-RSF is present in considerable amounts in the cell nucleus; but in contrast to the in vitro situation, it obviously does not in general inhibit splicing at correct splice sites. The conditions in the cell must therefore be different from in the in vitro splicing reactions. Our data are consistent with the following view. The majority of Ct-RSF is present in IGCs, known to contain stored splicing factors, including SR proteins (Misteli et al., 1997; Lamond and Spector, 2003). The facts that Ct-RSF colocalizes with hrp45 in IGCs, coimmunoprecipitates with hrp45 in nuclear extracts, and can bind directly to hrp45 in vitro suggest that Ct-RSF interacts with hrp45 in the IGCs. We have clearly demonstrated that Ct-RSF and hrp45 are present at transcriptionally active, but not at inactive, gene loci and colocalize on the same nascent pre-mRNAs. Therefore, both proteins are likely to be recruited to nascent pre-mRNAs from IGCs. We have shown that Ct-RSF and hrp45 colocalize on growing transcripts but that the relative amount of Ct-RSF compared with hrp45 is much lower at some gene loci. At such loci, exemplified by the BR3 gene, hrp45 and U2 snRNP (and U1 snRNP) are abundant. Based on the correlation between the presence of a large number of introns, 38 in the BR3 transcript, and relatively low amounts of Ct-RSF on the one hand, and between a large amount of Ct-RSF and transcripts dominated by exon sequences, 35-kb exons in the BR1 and BR2 transcripts, on the other hand, we propose that Ct-RSF colocalizes with hrp45 on exon sequences.
Injection of Ct-RSF-specific antibodies into nuclei interfered with the association of both Ct-RSF and hrp45 but not with U2 snRNP to nascent pre-mRNAs. Analyzes of constitutive splicing of the BR1 gene transcript showed that the efficiency of excision of intron 3 was not significantly changed after antibody injection (our unpublished data), but we could not analyze whether use of cryptic splice sites was changed. It is possible that small amounts of hrp45 remained bound to the transcript to support splicing or that some other SR protein supported splicing. Notably, association of hrp45 to the BR3 nascent pre-mRNA was also inhibited after antibody injection in spite of that only a small amount of Ct-RSF is present on this transcript. It should be pointed out that in the experiments, 90 min elapsed between antibody injection and immunostaining of the cells. The known transcription time for the BR1 and BR2 genes is 15–20 min and for the BR3 gene it is presumably about one-third of that time. Therefore, the population of nascent transcripts on the genes completely turned over several times during the experiment. Although it is possible that the antibodies released Ct-RSF and hrp45 that were already present on the nascent transcripts, we believe that it is more likely that the antibodies inhibited the recruitment of the two proteins to the transcripts. According to this interpretation, Ct-RSF and hrp45 are recruited to the growing pre-mRNA from the IGCs as a complex. At the nascent transcript, both proteins bind to exon sequences. It has previously been demonstrated that hrp45 binds along most of the 35-kb exons in the BR1 and BR2 nascent transcripts (Alzhanova-Ericsson et al., 1996). Both RSF1 (Labourier et al., 1999b) and B52/SRp55 (Nagel et al., 1998) have been shown to bind to exonic sequences. On the BR3 pre-mRNA, hrp45 is present in relatively higher amounts than Ct-RSF. Therefore, it is possible that spliceosomal assembly at the many correct splice sites along the BR3 nascent transcript displaces Ct-RSF but allows hrp45 to remain. It is also possible that hrp45 in addition can be recruited alone to exon sequences located close to correct splice sites. A small amount of Ct-RSF was detected on the BR3 pre-mRNA presumably bound to the short exon sequences. Ct-RSF is likely to bind similar sequences as RSF1 (Labourier et al., 1999b). The number of such identified so called A and B sequences per kilobase is approximately similar in the exons in BR1, BR2, and BR3 transcripts, but the total number of such sequences is 5–7 times higher in the BR1 and BR2 transcripts due to the total length of their exons. Our immunostaining data are in agreement with binding of Ct-RSF to exon sequences in BR1, 2, and 3 (Figure 7). Bound to the exons, Ct-RSF may locally repress SR protein stimulated initial steps in spliceosome assembly. U1 snRNP binding to cryptic splice sites in exons may be such a step.
It has been shown that Ct-RSF (Sun et al., 1998) and hrp45 (Alzhanova-Ericsson et al., 1996) are associated with BR1 and BR2 mRNPs until the mRNPs reach the nuclear pore complexes. Ct-RSF would then continue to repress any spliceosomal assembly on exon sequences during mRNP transport in the nucleus. This view of the function of Ct-RSF in vivo is in accordance with the model put forward for the D. melanogaster RSF1 (Labourier et al., 1999a). In agreement with this model, Ct-RSF together with hrp45 binds to exon sequences. At positions of introns, Ct-RSF is displaced locally, possibly by many cooperating interactions during spliceosomal assembly. Our finding that the staining for Ct-RSF, hrp45 and U2 snRNP varied for different gene loci would also agree with this model. For example, some loci bound relatively little U2 snRNP but considerably more Ct-RSF (and hrp45). Such transcripts would contain very few or no introns. Other loci were associated with relatively little Ct-RSF but considerably more hrp45 and U2 snRNP. Such transcripts would then contain many introns. We propose that Ct-RSF is involved in regulating spliceosomal assembly such that it inhibits spliceosomal early assembly at incorrect exonic splice sites.
Acknowledgments
We thank Kerstin Bernholm and Mona Tibäck for excellent technical assistance. We also thank Drs. Göran Akusjärvi, Oliver Mühlemann, and Jan-Peter Kreivi for technical advice and for providing material for in vitro splicing, and Drs. R. Reed and D. Black for technical advice. This work was supported by grants from the Swedish Research Fund and Magnus Bergvalls Stiftelse.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–04–0339) on October 19, 2005.
References
- Alzhanova-Ericsson, A. T., Sun, X., Visa, N., Kiseleva, E., Wurtz, T., and Daneholt, B. (1996). A protein of the SR family of splicing factors binds extensively to exonic Balbiani ring pre-mRNA and accompanies the RNA from the gene to the nuclear pore. Genes Dev. 10, 2881–2893. [DOI] [PubMed] [Google Scholar]
- Barnard, D. C., and Patton, J. G. (2000). Identification and characterization of a novel serine-arginine-rich splicing regulatory protein. Mol. Cell. Biol. 20, 3049 –3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baurén, G., Jiang, W., Bernholm, K., Gu, F., and Wieslander, L. (1996). Demonstration of a dynamic transcription dependent organization of splicing factors in polytene nuclei. J. Cell Biol. 133, 929 –941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baurén, G., and Wieslander, L. (1994). Splicing of Balbiani ring 1 pre-mRNA occurs simultaneously with transcription. Cell 76, 183–192. [DOI] [PubMed] [Google Scholar]
- Bentley, D. (2002). The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 14, 336 –342. [DOI] [PubMed] [Google Scholar]
- Beyer, A. L., and Osheim, Y. N. (1988). Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 2, 336 –342. [DOI] [PubMed] [Google Scholar]
- Björk, P., Baurén, G., Jin, S., Tong, Y.-G., Bürglin, T. R., Hellman, U., and Wieslander, L. (2002). A novel conserved RNA-binding domain protein, RBD-1, is essential for ribosome biogenesis. Mol. Biol. Cell 13, 3683–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe, B. J. (2000). Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25, 106–110. [DOI] [PubMed] [Google Scholar]
- Champlin, D. T., Frasch, M., Saumweber, H., and Lis, J. T. (1991). Characterization of a Drosophila protein associated with boundaries of transcriptionally active chromatin. Genes Dev. 5, 1611–1621. [DOI] [PubMed] [Google Scholar]
- Collins, C. A., and Guthrie, C. (2000). The question remains: is the spliceosome a ribozyme? Nat. Struct. Biol. 7, 850–854. [DOI] [PubMed] [Google Scholar]
- Das, R., and Reed, R. (1999). Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA 5, 1504 –1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam, J. D., Lebovitz, R. M., and Roeder, R. G. (1983). Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, H., and Manley, J. L. (1990). A protein factor, ASF, controls cell specific alternative splicing of SV40 early pre-mRNA in vitro. Cell 62, 25–34. [DOI] [PubMed] [Google Scholar]
- Ghosh, S., and Garcia-Blanco, M. A. 2000. Coupled in vitro synthesis and splicing of RNA polymerase II transcripts. RNA 6, 1325–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley, B. R. (2000). Sorting out the complexity of SR protein functions. RNA 6, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig, H., Aebi, M., and Weissmann, C. (1986). Effect of mutations at the lariat branch acceptor site on b– globin pre-mRNA splicing in vitro. Nature 324, 589 –591. [DOI] [PubMed] [Google Scholar]
- Huang, T. S., Nilsson, C. E., Punga, T., and Akusjärvi, G. (2002). Functional inactivation of the SR family of splicing factors during a vaccinia virus infection. EMBO Rep. 3, 1088 –1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y., Gattoni, R., Stévenin, J., and Steitz, J. A. (2003). SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell 11, 837– 843. [DOI] [PubMed] [Google Scholar]
- Huang, Y., Yario, T. A., and Steitz, J. A. (2004). A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl. Acad. Sci. USA 101, 9666 –9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanopka, A., Mühlemann, O., and Akusjärvi, G. (1996). Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature 381, 535– 538. [DOI] [PubMed] [Google Scholar]
- Kanopka, A., Mühlemann, O., Petersen-Mahrt, S., Estmer, C., Öhrmalm, C., and Akusjärvi, G. (1998). Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature 393, 185–187. [DOI] [PubMed] [Google Scholar]
- Kohtz, J. D., Jamison, S. F., Will, C. L., Lührmann, R., Garcia-Blanco, M. A., and Manley, J. L. (1994). Protein-protein interactions and 5′ splice site recognition in mammalian mRNA precursors. Nature 368, 119 –124. [DOI] [PubMed] [Google Scholar]
- Kim, S., Shi, H., Lee, D. K., and Lis, J. T. (2003). Specific SR protein-dependent splicing substrates identified through genomic SELEX. Nucleic Acids Res. 31, 1955–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva, E., Wurtz, T., Visa, N., and Daneholt, B. (1994). Assembly and disassembly of spliceosomes along a specific pre-messenger RNP fiber. EMBO J. 15, 6052– 6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska, M. M., and Sharp, P. A. (1988). Interaction between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell 49, 763–774. [DOI] [PubMed] [Google Scholar]
- Kotovic, K. M., Lockshon, D., Boric, L., and Neugebauer, K. M. (2003). Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol. Cell. Biol. 23, 5768 –5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer, A. R., Conway, G. C., and Kozak, D. (1990). The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell 62, 35– 42. [DOI] [PubMed] [Google Scholar]
- Labourier, E., Allemand, E., Brand, S., Fostier, M., Tazi, J., and Bourbon, H.-M. (1999b). Recognition of exonic splicing enhancer sequences by the Drosophila splicing repressor RSF1. Nucleic Acids Res. 27, 2377–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labourier, E., Bourbon, H.-M., Gallouzi, I.-E., Fostier, M., Allemand, E., and Tazi, J. (1999a). Antagonism between RSF1 and SR proteins for both splice-site recognition in vitro and Drosophila development. Genes Dev. 13, 740 –753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, M.-C. and Tarn, W.-Y. (2004). Hypophosphorylated ASF/SF2 binds TAP and is present in messenger ribonucleoproteins. J. Biol. Chem. 279, 31745– 31749. [DOI] [PubMed] [Google Scholar]
- Lamond, A. I., and Spector, D. L. (2003). Nuclear speckles: a model for nuclear organelles. Nature Rev. Mol. Cell. Biol. 4, 605– 612. [DOI] [PubMed] [Google Scholar]
- Lemaire, R., Prasad, J., Kashima, T., Gustafson, J., Manley, J. L., and Lafyatis, R. (2002). Stability of PKCI-1-related mRNA is controlled by the splicing factor ASF/SF 2, s novel function for SR proteins. Genes Dev. 16, 594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. and Manley, J. L. 2005. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122, 365–378. [DOI] [PubMed] [Google Scholar]
- Matunis, E. L., M. J. Matunis, and Dreyfuss, G. (1993). Association of individual hnRNP proteins and snRNPs with nascent transcripts. J. Cell Biol. 1211, 219 –228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermoud, J. E., Cohen, P.T.W., and Lamond, A. I. (1994). Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO J. 13, 5679 –5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, B., Mähr, R., Eppenberger, H. M., and Lezzi, M. (1983). The activity of Balbiani rings 1 and 2 in salivary glands of Chironomus tentans larvae under different modes of development and after pilocarpine treatment. Dev. Biol. 98, 265–277. [DOI] [PubMed] [Google Scholar]
- Misteli, T., Cáceres, J. F., and Spector, D. L. (1997). The dynamics of a pre-mRNA splicing factor in living cells. Nature 29, 523–527. [DOI] [PubMed] [Google Scholar]
- Mühlemann, O., and Akusjärvi, G. (1998). Preparation of splicing competent nuclear extracts from adenovirus infected cells. In: Methods in Molecular Medicine: Adenovirus Methods and Protocols, Vol. 21, ed. W.S.M. Wold, Totowa, NJ: Humana Press, 203–216. [Google Scholar]
- Nagel, R. J., Lancaster, A. M., and Zahler, A. M. (1998). Specific binding of an exonic splicing enhancer by the pre-mRNA splicing factor SRp55. RNA 4, 11–23. [PMC free article] [PubMed] [Google Scholar]
- Popielarz, M., Gattoni, R., and Stevenin, J. (1993). Contrasted cis-acting effects of downstream 5′ splice sites on the splicing of a retained intron: the adenoviral E1A pre-mRNA model. Nucleic Acids Res. 21, 5144 –5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot, N. J., Furger, A., and Dye, M. J. (2002). Integrating mRNA processing with transcription. Cell 108, 501–512. [DOI] [PubMed] [Google Scholar]
- Ring, H. Z., and Lis, J. T. (1994). The SR protein B52/SRp55 is essential for Drosophila development. Mol. Cell. Biol. 14, 7499 –7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio, D. C. (1988). Accurate and efficient pre-mRNA splicing in Drosophila cell-free extracts. Proc. Natl. Acad. Sci. USA 85, 2904 –2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford, J. R., Ellis, J., and Cáceres, J. F. 2005. Multiple roles of arginine/serine splicing factors in RNA processing. Biochem. Soc. Trans. 33, 443– 446. [DOI] [PubMed] [Google Scholar]
- Sanford, J. R., Gray, N. K., Beckmann, K., and Cáceres, J. F. (2004). A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 18, 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford, R., Longman, D., and Cáceres, J. F. (2003). Multiple roles of the SR protein family in splicing regulation. Prog. Mol. Subcell. Biol. 31, 33–58. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D. W. 2001. Molecular Cloning: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Shen, H., and Green, M. R. (2004). A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol. Cell 16, 362–373. [DOI] [PubMed] [Google Scholar]
- Shin, C., and Manley, J. L. (2002). The SR protein SRp38 represses splicing in M phase cells. Cell 111, 407– 417. [DOI] [PubMed] [Google Scholar]
- Shin, C., Feng, Y., and Manley, J. L. (2004). Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature 427, 553–558. [DOI] [PubMed] [Google Scholar]
- Singh, R., Valcárcel, J., and Green, M. R. (1995). Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 26, 1173–1176. [DOI] [PubMed] [Google Scholar]
- Stevens, S. W., Ryan, D. E., Ge, H. Y., Moore, R. E., Young, M. K., Lee, T. D., and Abelson, J. (2002). Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell 9, 31– 44. [DOI] [PubMed] [Google Scholar]
- Sun, X., Alzhanova-Ericsson, A., Visa, N., Aissouni, Y., Zhao, J., and Daneholt, B. (1998). The hrp23 protein in the Balbiani ring pre-mRNP particle is released just before or at the binding of the particle to the nuclear pore complex. J. Cell Biol. 142, 1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennyson, C. N., Klamut, H. J., and Worton, R. G. (1995). The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nat. Genet. 9, 184 –190. [DOI] [PubMed] [Google Scholar]
- Tran, Q., and Roesser, J. R. (2003). SRp55 is a regulator of calcitonin/CGRP alternative RNA splicing. Biochemistry 42, 951–957. [DOI] [PubMed] [Google Scholar]
- Turner, B. M., Davies, S., and Whitfield, G. F. (1985). Characterization of a family of nuclear and chromosomal proteins identified by a mAb. Eur. J. Cell Biol. 38, 344 –352. [PubMed] [Google Scholar]
- Valcárcel, J., Singh, R., Zamore, P. D., and Green, M. R. (1993). The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature 362, 171–175. [DOI] [PubMed] [Google Scholar]
- Visa, N., Alzhanova-Ericsson, A., Sun, X., Kiseleva, E., Björkroth, B., Wurtz, T., and Daneholt, B. (1996). A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell 84, 253–264. [DOI] [PubMed] [Google Scholar]
- Welin Henriksson, E., and Pettersson, I. (1996). Human anti-RNP sera contain both human-specific and cross-reactive anti-70K autoantibodies. J. Autoimmun. 9, 551–559. [DOI] [PubMed] [Google Scholar]
- Welin Henriksson, E., and Pettersson, I. (1997). Autoepitope-mapping of the U1–70K protein with human-Drosophila chimeric proteins. J. Autoimmun. 10, 559 –568. [DOI] [PubMed] [Google Scholar]
- Wetterberg, I., Baurén, G., and Wieslander, L. (1996). The intranuclear site of excision of each intron in the Balbiani ring 3 pre-mRNA is influenced by the time remaining to transcription termination and different excision efficiencies for the various introns. RNA 2, 641– 651. [PMC free article] [PubMed] [Google Scholar]
- Wetterberg, I., Zhao, J., Masich, S., Wieslander, L., and Skoglund, U. (2001). In situ transcription and splicing in the Balbiani ring 3 gene. EMBO J. 20, 2564 –2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieslander, L. (1994). The Balbiani ring multigene family: coding repetitive sequences and evolution of a tissue specific cell function. Prog. Nucleic Acid Res. Mol. Biol. 48, 275–313. [DOI] [PubMed] [Google Scholar]
- Wollerton, M. C., Gooding, C., Robinson, F., Brown, E. C., Jackson, R. J., and Smith, C. W. (2001). Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein (PTB). RNA 7, 819–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtz, T., Kiseleva, E., Nacheva, A., Alzhanova-Ericsson, A., Rosen, A., and Daneholt, B. (1996). Identification of two RNA-binding proteins in Balbiani ring pre-messenger RNP granules and the presence of these proteins in specific subsets of nuclear ribonucleoprotein particles. Mol. Cell. Biol. 139, 297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss, C. (1982). Chironomus tentans epithelial cell lines sensitive to ecdysteroids, juvenile hormone, insulin and heat shock. Exp. Cell Res. 139, 309 –319. [DOI] [PubMed] [Google Scholar]
- Xiao, S.-H. and Manley, J. L. (1997). Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 11, 334 –344. [DOI] [PubMed] [Google Scholar]
- Zahler, A. M., Lane, W. S., Stolk, J. A., and Roth, M. B. (1992). SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 6, 837– 847. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., and Krainer, A. R. (2004). Involvement of SR proteins in mRNA surveillance. Mol. Cell 19, 597– 607. [DOI] [PubMed] [Google Scholar]
- Zhao, J., Jin, S.-B., Björkroth, B., Wieslander, L., and Daneholt, B. (2002). The mRNA export factor Dbp5 is associated with Balbiani ring mRNP from gene to cytoplasm. EMBO J. 21, 1177–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., Mayeda, A., and Krainer, A. R. (2001). Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell 8, 1351–1361. [DOI] [PubMed] [Google Scholar]