Abstract
The ErbB1 and ErbB2 receptors are oncogenes with therapeutic significance in human cancer, whereas the transforming potential of the related ErbB4 receptor has remained controversial. Here, we have addressed whether four alternatively spliced ErbB4 isoforms differ in regulating cellular responses relevant for tumor growth. We show that the two tumor necrosis factor-α converting enzyme (TACE)-cleavable ErbB4 isoforms (the juxtamembrane [JM]-a isoforms) were overexpressed in a subset of primary human breast cancers together with TACE. The overexpression of the JM-a cytoplasmic (CYT)-2 ErbB4 isoform promoted ErbB4 phosphorylation, survival of interleukin-3-dependent cells, and proliferation of breast cancer cells even in the absence of ligand stimulation, whereas activation of the other three ErbB4 isoforms required ligand stimulation. Ligand-independent cellular responses to ErbB4 JM-a CYT-2 overexpression were regulated by both tyrosine kinase activity and a two-step proteolytic generation of an intracellular receptor fragment involving first a TACE-like proteinase, followed by γ-secretase activity. These data suggest a novel transforming mechanism for the ErbB4 receptor in human breast cancer that is 1) specific for a single receptor isoform and 2) depends on proteinase cleavage and kinase activity but not ligand activation of the receptor.
INTRODUCTION
ErbB receptors constitute the epidermal growth factor receptor (EGFR) subfamily of receptor tyrosine kinases (RTKs). Members of the family include ErbB1 (also known as EGFR or HER1), ErbB2 (c-Neu, HER2), ErbB3 (HER3), and ErbB4 (HER4). ErbB receptors regulate cellular functions in response to activation by epidermal growth factor (EGF)-like growth factors (Yarden and Sliwkowski, 2001). Ligand-dependent formation of homo- or heterodimeric receptor complexes initiates multiple downstream signaling cascades, such as the Ras/Raf/mitogen-activated protein kinase kinase/extracellular signal-regulated kinase pathway, that indirectly convey the signal to the nucleus. Recently, an alternative direct signaling mechanism has been proposed for one of the ErbB receptors, ErbB4 (Ni et al., 2001; Lee et al., 2002). This mechanism, reminiscent to that used by amyloid precursor protein and the Notch receptors, is based on proteolytic generation of a soluble intracellular domain (ICD) of ErbB4 that is translocated to the nucleus and may possess transcriptional activity (Ni et al., 2001; Lee et al., 2002; Komuro et al., 2003; Williams et al., 2004).
ErbB1 and ErbB2 are well-characterized cancer drug targets (Gschwind et al., 2004). In contrast, the oncogenic potential and clinical significance of ErbB4 is poorly understood. Clinical reports have indicated high expression levels of ErbB4 in neoplasias, including thyroid (Haugen et al., 1996), breast (Srinivasan et al., 2000), ovarian (Furger et al., 1998), endometrial (Srinivasan et al., 1999), and oral squamous cell cancer (Bei et al., 2001) as well as medulloblastoma (Gilbertson et al., 1997), ependymoma (Gilbertson et al., 2002), and osteosarcoma (Hughes et al., 2004). However, ErbB4 expression is down-regulated or lost in prostate (Lyne et al., 1997), renal (Thomasson et al., 2004), and local pancreatic cancer (Graber et al., 1999). Furthermore, there are contradictory reports about the significance of ErbB4 expression levels for clinical outcome (Gullick, 2003).
Inconsistent conclusions about the transforming potential of ErbB4 have also been drawn from experimental studies. There are observations that support oncogenicity of ErbB4: 1) Rodent fibroblast transfectants overexpressing ErbB4 together with any other ErbB and an activating ligand grow in soft agar and in nude mice (Cohen et al., 1996). Also, 2) analyses of human breast cancer cell lines suggest that overexpression of exogenous ErbB4 promotes growth (Junttila etal., 2005), and down-regulation of endogenous ErbB4 blocks tumor formation both in vitro and in vivo (Tang et al., 1999). In contrast, there are also observations that support an antioncogenic or differentiation-inducing role for ErbB4: 1) ErbB4 ligands and activating ErbB4 antibodies stimulate differentiation and suppress proliferation of human breast cancer cell lines in vitro (Peles et al., 1992; Chen et al., 1996; Sartor et al., 2001). In addition, 2) a constitutively active mutant of ErbB4 fails to promote anchorage-independent growth of rodent fibroblasts (Penington et al., 2002).
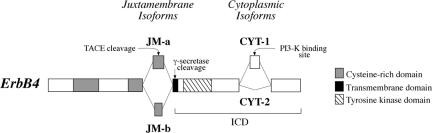
We have identified naturally occurring isoforms of ErbB4 that differ from each other both structurally and functionally (Elenius et al., 1997, 1999; Junttila et al., 2000; Kainulainen et al., 2000). These isoforms, generated by tissue-specific alternative splicing (Junttila et al., 2003), are characterized by variant extracellular juxtamembrane (JM) domains and intracellular cytoplasmic (CYT) domains. The JM domain of type a (JM-a) includes 23 amino acids that confer the receptor a proteinase cleavage site that is missing from the alternative 13 amino acids in the JM domain of type b (JM-b) (Elenius et al., 1997). One proteinase that has been shown to regulate ErbB4 ectodomain shedding in a JM-a-specific manner is tumor necrosis factor-α converting enzyme (TACE) (Rio et al., 2000). The CYT isoforms differ by having (CYT-1) or not having (CYT-2) a sequence of 16 amino acids within their cytoplasmic tails (Elenius et al., 1999). These 16 amino acids can serve as a binding site for phosphoinositide 3-kinase (PI3-K) (Elenius et al., 1999). When overexpressed in rodent fibroblasts, an ErbB4 isoform with CYT-1 tail, but not an isoform with CYT-2 tail, can mediate stimulation of PI3-K activity (Elenius et al., 1999), and PI3-K-dependent cellular responses such as activation of Akt kinase, survival, and chemotaxis (Kainulainen et al., 2000).
Some of the controversy about the oncogenic potential of ErbB4 could be attributed to the existence of functionally dissimilar and differentially regulated isoforms. However, analyses of ErbB4 function in cancer have almost exclusively been carried out using tools that do not differentiate between the individual isoforms. In support of differential tumorigenic potential of cleavable and noncleavable ErbB4 isoforms, we recently reported that breast cancer patients with localization of an ErbB4 ICD epitope in the cancer cell nuclei have worse prognosis compared with patients with ErbB4 immunoreactivity at cell surfaces (Junttila et al., 2005). Here, we have analyzed the growth-promoting potential and signaling mechanisms of the major ErbB4 isoforms simultaneously in cells devoid of endogenous ErbB background as well as in human breast cancer cells. The relative expression levels of ErbB4 as well as the ErbB4-cleaving enzyme TACE were also assessed using samples representing normal mammary gland and breast cancer tissue. Our findings indicate that a specific ErbB4 isoform has more oncogenic potential than the other isoforms. The results also suggest that strategies that specifically target ErbB4 JM-a CYT-2 may generate better candidates for cancer prognostics and therapy than strategies that do not differentiate between the ErbB4 isoforms.
MATERIALS AND METHODS
Immunohistochemistry
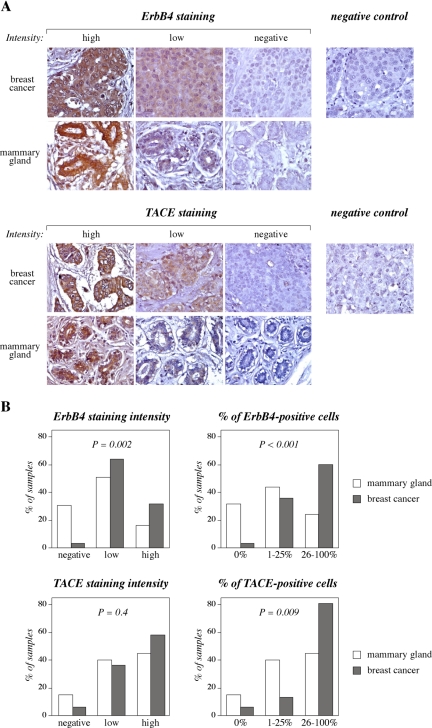
Paraffin sections of breast cancer tissue (n = 53) and histologically nonmalignant mammary gland tissue (n = 25) were obtained from the University Hospitals of Turku and Tampere, Finland. Cancer samples were from patients (median age 55; range 33–89) with ductal (76%), lobular (15%), or other (9%) breast cancer histology of grade 1 (17%), grade 2 (44%), or grade 3 (39%). Seventy-one percent of the tumors were determined estrogen receptor positive, and 53% of the tumors were progesterone receptor positive. Nonmalignant samples consisted of eight normal samples from patients undergoing reduction mammoplasty, nine histologically normal peripheral samples from patients with breast cancer, seven samples from patients with fibrocystic disease, and one sample from a patient with benign hyperplasia. Immunohistochemistry was carried out using 1:50 dilutions of the primary antibodies against ErbB4 (monoclonal HFR-1; Neomarkers, Fremont, CA) or TACE (polyclonal C-15; sc-6416; Santa Cruz Biotechnology, Santa Cruz, CA) as described previously (Junttila et al., 2003). As negative controls, primary antibodies were omitted, replaced by a nonspecific mouse IgG (sc-2025; Santa Cruz Biotechnology) (Figure 1A, top), or competed with a blocking peptide according to manufacturer's instructions (sc-6416P; Santa Cruz Biotechnology) (Figure 1A, bottom). The sections stained for ErbB4 and TACE were mostly adjacent sections from the same tissue blocks. The intensity of immunostaining was classified (negative, weak, or strong) (Figure 1A), and the percentage of positive cancer or epithelial cells was also scored under microscope. The scoring was carried out independently by two investigators. For statistical analyses, chi-square and Fisher's exact tests were performed using SAS software version 8.2 (SAS Institute, Cary, NC).
Figure 1.
ErbB4 and an ErbB4 cleaving enzyme, TACE, are overexpressed in breast cancer. (A) ErbB4 and TACE expression was analyzed by immunohistochemistry from 53 breast cancer samples and 25 samples representing nonmalignant mammary gland. Staining with a nonspecific mouse IgG and the effect of a blocking peptide are shown as negative controls for ErbB4 and TACE staining, respectively. (B) Positive signal was scored based on intensity (no staining, weak or strong) and on percentage of positive epithelial cells (0, 1–25, and 26–100%). All epithelial cells from paraffin sections (5–200 mm2) were scored.
Real-Time Reverse Transcription-PCR Analysis
ErbB4 isoform and TACE mRNA expression levels were determined by real-time quantitative reverse transcription (RT)-PCR (TaqMan; Applied Biosystems, Foster City, CA), as described previously (Junttila et al., 2003). For TACE mRNA analysis, primers 5′-GAGGATGGGTTTGAGAAGGACC-3′ and 5′-GGTCCGTGAGATCCTCAAATGA-3′, and probe 5′-6-Fam-TTCCCAAATAGCAGCACAGCTGCCAA-Tamra-3′ were used. Total RNA was extracted using RNAzol B reagent (Tel-Test, Friendswood, TX). Samples were analyzed in triplicate, and in each measurement, SD of the threshold cycle (CT) values was <5% of the mean. ErbB expression was presented as the percentage of ErbB mRNA expression relative to the mRNA expression of the internal control gene β-actin.
Cell Culture
MCF-7 human breast cancer cells (Soule et al., 1973) and interleukin (IL-3)-dependent murine 32D cells (Pierce et al., 1988) were maintained in RPMI 1640 medium (Invitrogen, Carlsbad, CA), 10% fetal calf serum (FCS) (Bioclear or Promocell), 50 μg/ml streptomycin sulfate, and 100 IU/ml penicillin. The culture medium for MCF-7 cells was further supplemented with 1 nM 17-β-estradiol (Sigma-Aldrich, St. Louis, MO), and the medium for 32D cells with 5% conditioned RPMI 1640 medium of WEHI cells as a source of IL-3.
Expression Vectors
To generate vectors for stable expression of ErbB4 JM-a CYT-1, ErbB4 JM-b CYT-1, and ErbB4 JM-b CYT-2, cDNA fragments encoding full reading frames of the respective isoforms were digested from previously prepared pCDM8ErbB4JM-aCYT-1 (cH4M2) (Elenius et al., 1997), pCDM8ErbB4JM-bCYT-1 (cH4M2 JM-b) (Elenius et al., 1997), and pCDM8ErbB4JM-bCYT-2 (Elenius et al., 1999) with SmaI and XbaI. The inserts were ligated into pcDNA3.1 (–) vector (Invitrogen) after digestion of the vector multiple cloning site with EcoRV and XbaI. The resulting vectors were named pcDNA3.1ErbB4JM-aCYT-1, pcDNA3.1ErbB4JM-bCYT-1, and pcDNA3.1ErbB4JM-bCYT-2. To generate pcDNA3.1ErbB4JM-aCYT-2, a 2219-base pair KpnI fragment of pcDNA3.1JM-bCYT-2, including CYT-2 sequence, was swapped to replace a corresponding 2244-base pair KpnI fragment in pcDNA3.1ErbB4JM-aCYT-1. Generation of pcDNA3.1ErbB4JM-aCYT-1HA and pcDNA3.1JM-aCYT-2Myc encoding hemagglutinin (HA) and Myc epitope tags in the carboxy terminus of ErbB4 JM-a CYT-1 and JM-a CYT-2, respectively, will be described elsewhere (Peri, Sundvall, Määttä, Tvorogov, and Elenius, unpublished data). For episomal expression of ErbB4 isoforms JM-a CYT-2 and JM-b CYT-2, the respective inserts were cloned into pCEP4 vector (Invitrogen) to generate pCEP4ErbB4JM-aCYT-2 and pCEP4ErbB4JM-bCYT-2, as described previously (Junttila et al., 2005).
Generation of Cell Lines Overexpressing ErbB4 Isoforms
MCF-7 cells were transfected with pCEP4-based episomal vectors, as described previously (Egeblad and Jaattela, 2000), and resistant pools were maintained in medium containing 150 μg/ml hygromycin B (Roche Diagnostics, Mannheim, Germany).
32D cells were transfected with PvuI-linearized pcDNA3.1 vectors by electroporation. Stable transfectants were selected in the presence of 400 μg/ml G418 (Geneticin; Calbiochem, San Diego, CA). Cells expressing ErbB4 were enriched by magnet-assisted cell sorting using a mouse monoclonal anti-ErbB4 (H4.77.16; Neomarkers) and anti-mouse IgG magnetic beads (Dynal Biotech, Oslo, Norway). Clonal lines were established by limiting dilution.
Western Blot Analysis of ErbB Protein Expression Levels
To prepare Triton-soluble cellular fractions mostly representing (≥95%) membrane-anchored ErbB4 forms, MCF-7 cells were grown to 90% confluence and 32D cells into a density of 500,000 cells/ml. Cells were lysed for 15 min on ice in lysis buffer (1% Triton X-100, 10 mM Tris-HCl, pH 7.4, 1 mM EDTA, 2 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM sodium orthovanadate, 10 mM sodium fluoride, and 10 mM sodium pyrophosphate), the lysates were centrifuged (12,000 × g; 4°C; 15 min), and the supernatants were used for analyses. To prepare cellular fractions mostly representing Triton-soluble cytosolic proteins, 5 × 107 32D cells/sample were fractionated following a protocol described previously (Vecchi et al., 1996). Briefly, cells were lysed to hypotonic buffer (20 mM HEPES, pH 7.4, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 1 mM PMSF, and 1 mM sodium orthovanadate), and the lysate was homogenized with a Dounce homogenizer. NaCl was added to final concentration of 100 mM. Nuclei and unbroken cells were removed by centrifuging twice for 5 min at 800 × g. The supernatants were ultracentrifuged for 50 min at 100,000 × g to remove unsoluble material. Triton X-100 was added to supernatants (final concentration 1%) representing soluble cytosolic proteins. Samples equivalent to 75 μg of total protein were analyzed by Western blotting using an anti-ErbB4 antibody (sc-283; Santa Cruz Biotechnology), as described previously (Kainulainen et al., 2000).
Analysis of ErbB4 Tyrosine Phosphorylation
MCF-7 transfectants were maintained without serum overnight in RPMI 1640 medium supplemented with 1 nM 17-β-estradiol and stimulated for 10 min at 37°C with or without neuregulin (NRG-1) (50 ng/ml; R&D Systems, Minneapolis, MN). 32D cells were seeded at density of 200,000 cells/ml. Next day, 32D cells were centrifuged, washed three times with phosphate-buffered saline (PBS), maintained for an additional 1 h at 37°C in RPMI supplemented with 10% FCS and stimulated for 10 min at 37°C with or without NRG-1 (50 ng/ml). After NRG-1 stimulation, both MCF-7 and 32D cells were washed with ice-cold phosphate-buffered saline (PBS) and lysed, as described previously (Kainulainen et al., 2000).
Samples equivalent to 1 mg of total protein were immunoprecipitated with an anti-ErbB4 antibody (sc-283), separated in SDS-PAGE gels, and analyzed by Western blotting using an anti-phosphotyrosine antibody (4G10; Upstate Biotechnology, Lake Placid, NY). The intensity of tyrosine phosphorylation in 32D cells was quantitated by densitometry using MCID Image Analyzer (Imaging Research, St. Catharines, ON, Canada).
Inhibition of ErbB4 Expression by Small Interfering RNAs
Synthetic single-stranded small interfering RNA (siRNA) oligonucleotides designed to block all ErbB4 isoforms (5′-ACUGAGCUCUCUCUCUGACTT-3′ and 5′-GUCAGAGAGAGAGCUCAGUTT-3′) or to selectively block ErbB4 JM-b (negative control siRNA for cells not expressing this isoform; 5′-GUAUUGAAGACUGCAUCGGTT-3′ and 5′-CCGAUGCAGUCUUCAAUACTT-3′) were purchased from Eurogentec, Seraing, Belgium) annealed, and used according to manufacturer's instructions. Double-stranded siRNAs were introduced into MCF-7 transfectants using TransIT-TKO transfection reagent (Mirus Bio, Madison, WI) according to manufacturer's instructions. ErbB4 protein expression was analyzed 2 d after siRNA transfection by Western blotting.
Effect of Inhibitors on Cell Proliferation
For siRNA experiments, MCF-7 transfectants were grown in full culture medium on 24-well plates and transfected with siRNA oligonucleotides. Triplicate wells of each treatment were counted 2, 4, and 5 d after transfection using a hemocytometer.
For experiments with tyrosine kinase and γ-secretase inhibitors, equal numbers of MCF-7 transfectants were plated on 24-well plates and grown in full culture medium in the presence of AG 1478 (10 μM; Calbiochem) and/or γ-secretase inhibitor compound IX (10 μM; GSI IX, DAPT; Calbiohem). Equal volume of solvent was added to each well. Triplicate wells of each treatment were counted by hemocytometer 2 and 5 d after plating and addition of inhibitors.
Confocal Microscopy
MCF-7 cells were transiently transfected with ErbB4 constructs using Fu-GENE 6 Transfection Reagent (Roche Diagnostics) according to manufacturer's instructions. Twenty-four hours after transfection cells were treated for 10 h with or without GSI IX (10 μM) in the presence or absence of leptomycin B (20 ng/ml; Sigma-Aldrich). When NRG-1 (50 ng/ml) was used, it was added for 30 min together with leptomycin B. Cells were washed with PBS and fixed with methanol at –20°C. Fixed cells were stained with mouse monoclonal anti-ErbB4 antibody (HFR-1; Neomarkers) recognizing the carboxy terminus of the receptor, mouse anti-c-Myc (Zymed Laboratories, South San Francisco, CA), or rat anti-HA antibody (Roche Diagnostics). The nuclei were visualized with 4,6-diamidino-2-phenylindole (DAPI) (0.5 μg/ml; Sigma-Aldrich). Localization of ErbB4 and nuclei was analyzed by confocal microscopy (Axiovert 200M with LSM 510 Meta; Carl Zeiss, Jena, Germany). Representative images of single 0.7-μm sections are shown.
3-(4,5-Dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium Assay
32D cells were washed three times with PBS, resuspended in RPMI 1640 medium supplemented with 10% FCS, and dispensed into 96-well plates at a density of 25,000 cells/200 μl/well in the presence or absence of 100 ng/ml NRG-1, 5% WEHI-conditioned medium as a source of IL-3, and tyrosine kinase or γ-secretase inhibitors. The number of viable cells was determined at time points from three parallel wells by measuring mitochondrial activity using 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) staining assays (Promega, Madison, WI), following manufacturer's recommendations. Briefly, 20 μl of mitochondrial MTS staining reagent was added into each well for 3 h, and absorbance at 492 nm was measured spectrophotometrically.
Analysis of ErbB4 Processing
To stimulate ErbB4 shedding with phorbol ester, 32D cells were cultured in full medium supplemented with 100 ng/ml phorbol 13-myristate 12-acetate (PMA; Sigma-Aldrich) (Vecchi et al., 1996; Elenius et al., 1997).
To inhibit TACE activity, the original tumor necrosis-α peptide inhibitor (TAPI-0) (40 μM; Peptides International, Louisville, KY) was used as a 30-min pretreatment before addition of GSI IX, or lysing the cells for tyrosine phosphorylation analysis.
To inhibit proteosomal degradation, cells were cultured for 3 h in full medium supplemented with N-acetyl-l-leucyl-l-leucyl-l-norleucinal (50 μM; ALLN; Calbiochem) or lactacystin (10 μM; Calbiochem). To assess the effect of proteasome inhibition on PMA-stimulated ErbB4 cleavage, cells were treated for 3 h with ALLN followed by 40-min treatment with a combination of ALLN and PMA.
To inhibit γ-secretase activity in 32D cells, GSI IX (10 μM) or DFK167 (10 μM; Enzyme System Products, Livermore, CA) were used as 30-min pretreatments before addition of PMA or lysing the cells for tyrosine phosphorylation analysis, or added to culture media of cells assessed for proliferation. To analyze the effect of γ-secretase inhibition on ErbB4 cleavage in MCF-7 cells, cells were grown in the presence of GSI IX (10 μM) for 4 d.
Analysis of ErbB4 Kinase Activity
For in vitro kinase assay, 32D cells were maintained in full medium for 6 h in the presence of ALLN (200 μM) and GSI IX (10 μM). Cells were lysed in lysis buffer not including EDTA or orthovanadate. Glycoproteins interacting with concanavalin A (Con A) were separated from 1.25 mg of the total lysate using Con A-Sepharose beads (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). Material not bound to concanavalin A and 1 mg of total lysate were immunoprecipitated at 4°C with anti-ErbB4 antibody (sc-283) and washed three times with lysis buffer and twice with kinase buffer (50 mM NaCl, 20 mM HEPES, pH 7.3, 3 mM MnCl2, 20 mM MgCl2, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 mM orthovanadate). Washed samples were incubated in kinase buffer in the presence or absence of ATP (25 μM; Roche Diagnostics) or [γ-32P]ATP (2 μCi/sample; MP Biomedicals, Irvine, CA) for 30 min. Phosphorylated samples were separated in SDS-PAGE gels, and visualized by anti-phosphotyrosine Western blotting or autoradiography. To inhibit ErbB4 kinase activity, a tyrosine kinase inhibitor AG 1478 (Calbiochem), reported to inhibit ErbB4 tyrosine phosphorylation (Egeblad et al., 2001), was used. MCF-7 transfectants were grown for 4 d in the presence of AG 1478 (10 μM; Calbiochem), a tyrosine kinase inhibitor reported to inhibit ErbB4 tyrosine phosphorylation (Egeblad et al., 2001), before tyrosine phosphorylation analysis. For 32D transfectants, AG 1478 (10 μM) was used as 30-min pretreatments before addition of PMA, GSI IX, or inhibitors of proteasomal degradation, or lysing the cells for tyrosine phosphorylation analysis. For 32D MTS assays 10 μM AG 1478 was added to the culture medium.
RESULTS
Cleavable ErbB4 Isoforms (JM-a CYT-1 and JM-a CYT-2) and an ErbB4-Cleaving Enzyme (TACE) Are Overexpressed in a Subset of Primary Human Breast Cancers
ErbB4 and TACE protein expression was analyzed by immunohistochemistry from 53 breast cancer samples and 25 samples representing histologically normal mammary gland (Figure 1A). Statistically significant ErbB4 overexpression in cancer samples was observed when either intensity of immunoreactivity (p = 0.002) or percentage of ErbB4-positive epithelial cells (p < 0.001) was scored (Figure 1B). TACE expression was also observed in a significantly greater percentage of cancer cells compared with normal epithelial cells (p = 0.009), but differences in TACE staining intensity per cell did not reach statistical significance (p = 0.4) (Figure 1B).
Structurally and functionally different ErbB4 isoforms are generated as a result of tissue-specific alternative splicing of extracellular JM and intracellular CYT domains (Figure 2). Real-time RT-PCR analyses demonstrated that transcripts encoding the JM-a domain were present in the majority of breast cancer samples together with both CYT-1 and CYT-2 types of cytoplasmic domains, whereas no signal for ErbB4 JM-b mRNA was detected (Junttila et al., 2005). TACE mRNA was also present in the majority of breast cancer cases (our unpublished data). Together, the immunohistochemical and RT-PCR data suggest that both the cleavable ErbB4 JM-a isoforms and TACE, an enzyme capable of cleaving ErbB4 at the JM-a domain, are overexpressed in some primary breast cancers in vivo.
Figure 2.
Schematic presentation of juxtamembrane (JM-a and JM-b) and cytoplasmic (CYT-1 and CYT-2) ErbB4 isoforms. JM-a isoforms, but not JM-b isoforms, can be proteolytically cleaved by TACE, resulting in shedding of the extracellular domain. Only ErbB4 first cleaved by TACE is a substrate for γ-secretase that releases a soluble ICD from the cell membrane. CYT-1 isoforms, but not CYT-2 isoforms, include an additional module that may interact with intracellular signaling molecules, such as PI3-K. Putative TACE and γ-secretase cleavage sites and a PI3-K binding site are indicated.
ErbB4 JM-a CYT-2 Overexpressed in Breast Cancer Cells Is Constitutively Cleaved and Phosphorylated, and Promotes Proliferation
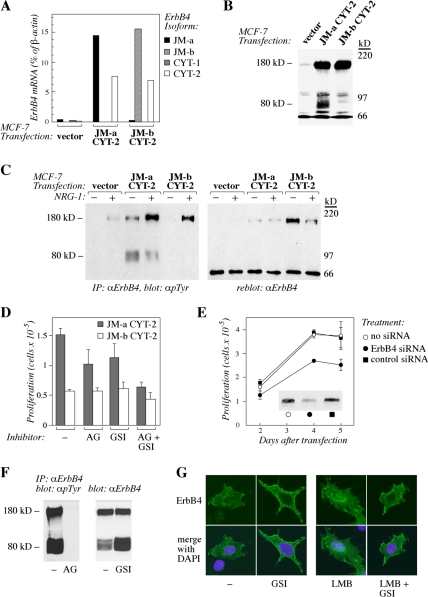
To assess whether ErbB4 JM-a isoforms support signaling unique for cancer-associated receptor variants, MCF-7 cells overexpressing ErbB4 JM-a CYT-2 or an isoform not present in breast cancer, ErbB4 JM-b CYT-2, were generated by transfection. ErbB4 mRNA expression in transfectants (Figure 3A) reached similar levels, as observed in breast cancer patients with high ErbB4 mRNA levels (Junttila et al., 2005). Western analysis indicated that both ErbB4 transfectants expressed full-length 180-kDa ErbB4 proteins to similar extent (Figure 3B). Consistent with earlier findings that ErbB4 JM-a, but not ErbB4 JM-b, can be proteolytically cleaved (Elenius et al., 1997), an additional 80-kDa band was detected selectively in cells expressing JM-a CYT-2 using an antibody against the carboxy terminus of ErbB4 (Figure 3B). The 80-kDa protein probably represented basal generation of a membrane-anchored fragment of ErbB4, because treatment with an inhibitor of TACE (TAPI-0) resulted in complete disappearance of the band (our unpublished data), and treatment with an inhibitor of intramembraneous proteolysis (GSI IX) resulted in accumulation of the 80-kDa band (Figure 3F; see also Figure 2 for location of proteinase cleavage sites in ErbB4). Both full-length 180-kDa ErbB4 isoforms responded to ligand stimulation with NRG-1 by phosphorylation, but, unexpectedly, ErbB4 JM-a CYT-2 also demonstrated basal ligand-independent phosphorylation (Figure 3C). Moreover, the 80-kDa ErbB4 JM-a CYT-2 was also constitutively phosphorylated (Figure 3C).
Figure 3.
Overexpression of ErbB4 JM-a CYT-2 promotes ligand-independent breast cancer cell growth. (A) Real-time RT-PCR analysis of ErbB4 isoform mRNA expression in MCF-7 cells transfected with an empty vector or vectors encoding ErbB4 JM-a CYT-2 or JM-b CYT-2. (B) Western analysis of ErbB4 protein expression in MCF-7 transfectants. The light band around 90 kDa on lane 3 may represent an amino-terminally truncated form of JM-b CYT-2 variant, because it was not observed in vector control cells (lane 1). (C) MCF-7 transfectants were stimulated for 10 min with or without 50 ng/ml NRG-1 and analyzed for ErbB4 tyrosine phosphorylation by immunoprecipitation with an anti-ErbB4 antibody followed by Western blotting with an anti-phosphotyrosine antibody. To control loading, the membranes were reblotted with the anti-ErbB4 antibody. (D) MCF-7 transfectants overexpressing ErbB4 JM-a CYT-2 or ErbB4 JM-b CYT-2 were cultured for 5 d in the presence or absence of AG 1478 (AG) and GSI IX (GSI), and proliferation was assessed by cell counting. (E) Proliferation assay of MCF-7 transfectants overexpressing ErbB4 JM-a CYT-2 after transfection with siRNAs designed to target ErbB4 or with control siRNAs. The effects of the siRNAs on ErbB4 expression are shown by Western blotting. (F) MCF-7 transfectants overexpressing ErbB4 JM-a CYT-2 were cultured for 4 d in the presence or absence of AG 1478 (AG) and GSI IX (GSI), and the effects on ErbB4 tyrosine phosphorylation and accumulation of membrane-anchored 80-kDa ErbB4 fragment, respectively, were determined. (G) MCF-7 cells were transiently transfected with a plasmid encoding ErbB4 JM-a CYT-2. The effect of a 10-h treatment with GSI IX (GSI) on subcellular localization of an intracellular ErbB4 epitope was determined by confocal microscopy both in the presence and absence of leptomycin B (LMB), an inhibitor or nuclear export. Green color, ErbB4 staining; blue color, nuclei stained with DAPI.
To study whether the ligand-independent ErbB4 JM-a CYT-2 phosphorylation or cleavage associated with cellular responses, growth of MCF-7 transfectants was assessed. Both proliferation and DNA synthesis rate were enhanced in cells overexpressing the cleavable ErbB4 JM-a isoform compared with cells overexpressing the noncleavable JM-b isoform or vector control cells (Figure 3D; our unpublished data), suggesting that overexpression of a cleavable ErbB4 JM-a CYT-2 was functionally significant. The causal role of ErbB4 JM-a CYT-2 expression in enhanced proliferation was further controlled by ErbB4-specific siRNAs that specifically down-regulated growth of cells transfected with this isoform (Figure 3E) but not of vector control cells (our unpublished data). These data demonstrate that overexpression of JM-a CYT-2, an ErbB4 isoform also overexpressed in breast cancer cells in vivo, specifically promotes breast cancer cell proliferation.
The Mechanism of Enhanced Breast Cancer Cell Proliferation by ErbB4 JM-a CYT-2 Overexpression Involves Both ErbB4 Tyrosine Kinase Activity and γ-Secretase Activity
To analyze whether the observed constitutive receptor phosphorylation or proteolytic cleavage by γ-secretase (generating a soluble receptor fragment putatively capable of signaling) have a role in ErbB4 JM-a CYT-2-stimulated MCF-7 proliferation, an inhibitor of ErbB kinase activity (AG 1478) and GSI IX were tested. Both AG 1478 and GSI IX clearly inhibited proliferation of MCF-7 cells overexpressing ErbB4 JM-a CYT-2 when administered alone and a combination of agents resulted in an additive effect (Figure 3D). No similar effect was seen with cells expressing ErbB4 JM-b CYT-2 when AG 1478 or GSI IX were analyzed separately, but the combination had a small but reproducible effect also on these cells (Figure 3D) that make low levels of endogenous ErbB4 JM-a isoforms (Figure 3, A and B). The continuous effects of the used 10 μM AG 1478 and GSI IX concentrations over 4 d were controlled by ErbB4 tyrosine phosphorylation analyses and Western analyses of 80-kDa ErbB4 accumulation (Figure 3F). The effect of γ-secretase inhibition on ErbB4 JM-a CYT-2 was also shown by confocal microscopy demonstrating that GSI IX reduced perinuclear and nuclear localization while enhancing cell surface-associated localization of a carboxy-terminal epitope of ErbB4 (Figure 3G). In addition, inhibiting ErbB4 cleavage using ilomastat, a metalloproteinase inhibitor that blocks TACE activity, suppressed proliferation of MCF-7 cells overexpressing ErbB4 JM-a CYT-2 (Supplemental Figure S1A). Combining GSI IX did not add to the inhibitory effect of ilomastat (Supplemental Figure S1A), consistent with both inhibitors blocking the release of a soluble ErbB4 ICD (see Figure 2 for TACE- and γ-secretase cleavage sites). Data obtained using chemical inhibitors were further controlled by demonstrating similar growth-suppressing effects of siRNAs down-regulating either TACE or presenilin-1, a critical protein subunit of γ-secretase complex (Supplemental Figure S1B). These data suggest that the mechanism by which ErbB4 JM-a CYT-2 promotes breast cancer cell proliferation involves both ErbB4 kinase activity and proteolytic cleavage.
Overexpression of ErbB4 JM-a CYT-2 Promotes Ligand-independent Survival in a Cellular Context with no ErbB Background
MCF-7 cells expressed endogenous ErbB4, and similar to other adherent cell lines, also other members of the ErbB receptor family (Figure 3, A and B; Supplemental Figure S2A; our unpublished data) with potential to form ErbB heterodimers and mediate signaling. To further characterize the function of ErbB4 isoforms in a more defined cellular context without endogenous ErbB background, myeloid 32D cells that did not express detectable levels of endogenous ErbB1, ErbB2, ErbB3, or ErbB4 protein (Supplemental Figure S2A), nor mRNA encoding any of the ErbB4 isoforms (Supplemental Figure S2B), were chosen for further experiments.
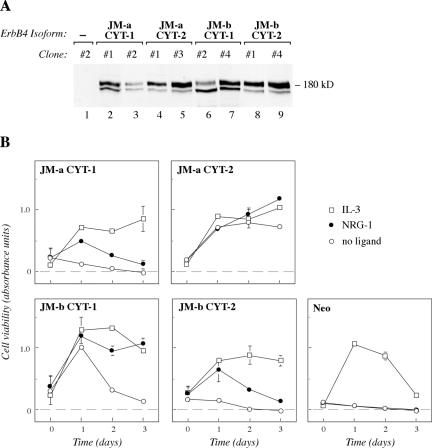
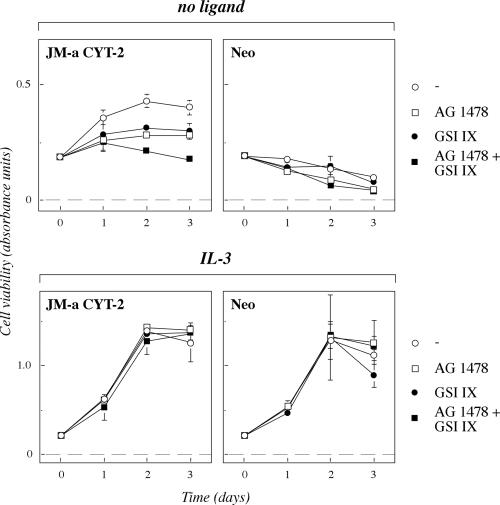
Expression plasmids encoding ErbB4 isoforms JM-a CYT-1, JM-a CYT-2, JM-b CYT-1, or JM-b CYT-2 (Figure 2) were constructed and transfected into 32D cells (Figure 4A). The functionality of all four ErbB4 isoforms was demonstrated by a response of the transfectants to stimulation with NRG-1 in MTS mitochondrial staining assays measuring the number of viable cells (Figure 4B). As expected, all clones of the IL-3-dependent 32D cells also survived in the presence of IL-3 supplied in the form of 5% conditioned medium of IL-3-producing WEHI cells. However, clones expressing ErbB4 JM-a CYT-2 survived for 72 h also in the absence of both IL-3 and NRG-1. Overexpression of other ErbB4 isoforms did not improve survival (JM-a CYT-1 and JM-b CYT-2) or promoted it only for the first 24 h (JM-b CYT-1) (Figure 4B). The signaling potential of the sole ErbB4 ICD of CYT-2 type was tested in transiently transfected COS-7 cells by analyzing cell cycle progression by fluorescence-activated cell sorting, and survival by assessing nuclear morphology after DAPI staining. Overexpression of CYT-2 ICD and the full-length JM-a CYT-2, but not of JM-b CYT-2, was sufficient to stimulate cell cycle progression and survival in COS-7 cells (our unpublished data). These data indicate that homodimers of one specific ErbB4 isoform, ErbB4 JM-a CYT-2, are capable of promoting 32D cell survival in a ligand-independent manner.
Figure 4.
ErbB4 JM-a CYT-2 promotes 32D cell survival in a ligand-independent manner. (A) Western analysis of ErbB4 expression in independent 32D clones transfected with plasmids encoding different ErbB4 isoforms or only the neomycin resistance gene (–). (B) MTS survival assay of 32D clones expressing ErbB4 isoforms JM-a CYT-1 (clone #1), JM-a CYT-2 (clone #3), JM-b CYT-1 (clone #4), or ErbB4 JM-b CYT-2 (clone #4) and of a control clone transfected with an empty vector (Neo; clone #2). Cells were cultured in the absence or presence of NRG-1 or 5% WEHI cell conditioned medium (IL-3). Similar results were obtained with independent clones.
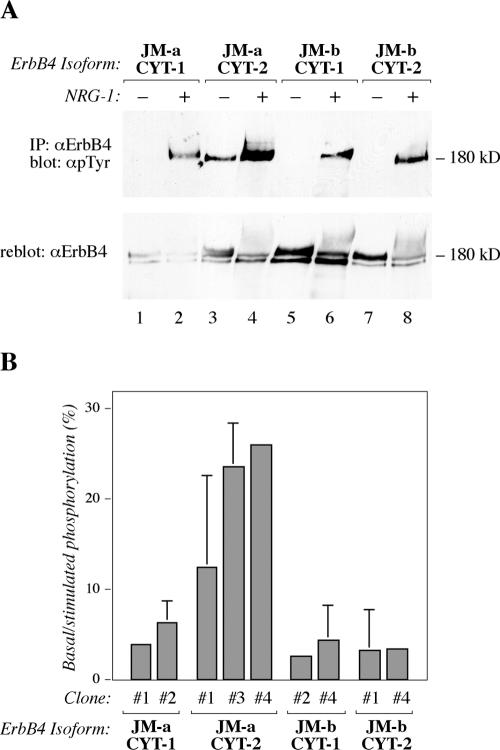
ErbB4 JM-a CYT-2 Homodimers Are Constitutively Phosphorylated
To assess the mechanism underlying constitutive ErbB4 JM-a CYT-2 activity in 32D cells, tyrosine phosphorylation of each of the isoforms was analyzed. All four isoforms responded to NRG-1 by enhanced tyrosine phosphorylation (Figure 5A, top). Analysis with an anti-ErbB4 antibody demonstrated that all isoforms were also down-regulated after ligand stimulation (Figure 5A, bottom). However, ErbB4 JM-a CYT-2 isoform consistently demonstrated more basal tyrosine phosphorylation than the other isoforms in the absence of exogenous ErbB4 ligands (Figure 5A, top). This ligand-independent activity was also evident when independent 32D transfectant clones were quantitated for the ratio of basal to NRG-1-stimulated tyrosine phosphorylation by densitometry (Figure 5B). Clones expressing ErbB4 JM-a CYT-2 had basal ErbB4 tyrosine phosphorylation levels at 15–25% of levels maximally reached by ligand stimulation, compared with ≈5% in the case of other isoforms. These data demonstrate that homodimers of ErbB4 JM-a CYT-2 can be tyrosine phosphorylated in a ligand-independent manner.
Figure 5.
ErbB4 JM-a CYT-2 is constitutively phosphorylated. (A) ErbB4 tyrosine phosphorylation analysis of 32D cells expressing different ErbB4 isoforms. Cells were stimulated for 10 min with or without 50 ng/ml NRG-1 and analyzed for ErbB4 tyrosine phosphorylation as in Figure 3C. (B) Densitometric quantitation of ErbB4 phosphorylation analyses of independent 32D clones expressing different ErbB4 isoforms. Each isoform was analyzed from 2 to 3 clones in three to six assays.
Constitutive Cleavage of ErbB4 JM-a CYT-2 Generates a Stable Membrane-anchored 80-kDa Carboxy-terminal Fragment and a Soluble ICD
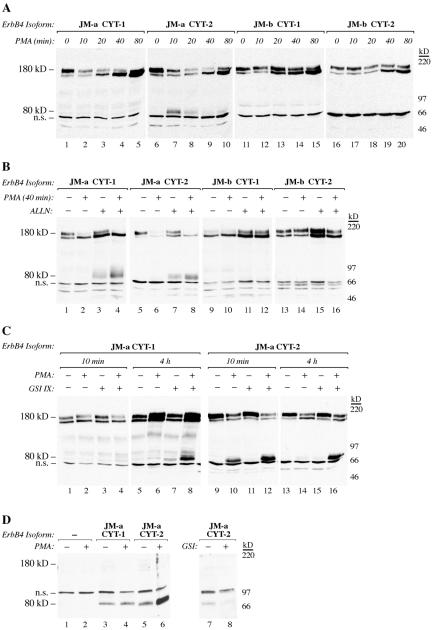
The effect of γ-secretase inhibition on the growth of MCF-7 transfectants (Figure 3D; Supplemental Figure S1B) indicated that ErbB4 may signal by releasing a proteolytic fragment. To further elucidate the molecular mechanisms of ligand-independent signaling by ErbB4 JM-a CYT-2, the potential of the phorbol ester PMA to stimulate cleavage of ErbB4 isoforms was analyzed. Both isoforms with JM-a type of juxtamembrane domains responded to PMA within 10 min by cleavage, as indicated by reduction of the full-length 180-kDa ErbB4 (Figure 6A). However, neither of the isoforms of the JM-b type was cleaved by PMA. In all transfectants, a 160-kDa ErbB4 band was also up-regulated by PMA in 40–80 min. The identity of the 160-kDa ErbB4 band is not known but may represent a less glycosylated biosynthetic intermediate of the a mature 180-kDa ErbB4, because both 160- and 180-kDa proteins were reduced into a single 140-kDa core protein by inhibiting N-glycosylation (Supplemental Figure S3A). Furthermore, the 160-kDa band disappeared before 180-kDa band after blocking protein synthesis (Supplemental Figure S3B), and only the 180-kDa band was phosphorylated upon stimulation with an extracellular ligand (Figure 5A). The 160-kDa ErbB4 may thus not be accessible for PMA-stimulated cleavage at cell surface but reflect an independent effect of PMA on ErbB4 turnover.
Figure 6.
ErbB4 JM-a CYT-2 is constitutively cleaved to an 80-kDa carboxy-terminal fragment that is resistant to proteasomal degradation and processed by γ-secretase. Proteins recognized with an antibody against the carboxy terminus of ErbB4 (sc-283) were visualized from 32D cells expressing the indicated ErbB4 isoforms by Western blotting. (A–C) Cell fractions mostly representing membrane-anchored ErbB4 were analyzed after treating the cells with PMA for indicated periods of time (A), after pretreating the cells with the proteasome inhibitor ALLN for 3 h before addition of PMA for a further 40 min (B), and after pretreating the cells for 30 min with GSI IX before addition of PMA for a further 10 min or 4 h (C). (D) Cell fractions representing cytosolic proteins were analyzed after treating the cells for 10 min with or without PMA, or 30 min with or without GSI IX. n.s., nonspecific band.
Together, the findings suggest that both 180-kDa JM-a isoforms of ErbB4 are cleaved in response to PMA.
PMA-stimulated cleavage of 180-kDa ErbB4 JM-a CYT-2 was also reflected in the appearance of an 80-kDa carboxy-terminal fragment in the cellular fraction primarily (≥95%) containing membrane-anchored ErbB4. Interestingly, however, no 80-kDa ErbB4 protein was observed in cells expressing JM-a CYT-1 (Figure 6A). To address the possibility that 80-kDa carboxy-terminal fragments were also generated from JM-a CYT-1 but rapidly degraded, 32D transfectants were treated with the proteasome inhibitor ALLN before a 40-min stimulation with PMA (Figure 6B). Under these conditions, an 80-kDa fragment was generated from both ErbB4 JM-a isoforms both with and without PMA stimulation (Figure 6B). Similar results were obtained when another proteasome inhibitor, lactacystin, was used (our unpublished data). Constitutive and similar cleavage of both 180-kDa JM-a isoforms was also indicated by similar accumulation of 120-kDa ErbB4 ectodomains into culture media of 32D transfectants expressing either JM-a CYT-1 or JM-a CYT-2 (Supplemental Figure S4). Basal ALLN- or GSI IX-induced accumulation of membrane-anchored 80-kDa ErbB4 was blocked by peptide inhibitors of TACE (Supplemental Figure S5), consistent with previous observations of ErbB4 cleavage by TACE in other cellular contexts (Vecchi and Carpenter, 1997; Rio et al., 2000). These data indicate that both JM-a isoforms are constitutively cleaved in 32D cells by TACE-like activity but that the membrane-anchored 80-kDa CYT-1 is more readily degraded in proteasomes.
Absence of the 80 kDa CYT-1 from cellular fractions representing mostly membrane-anchored ErbB4 (Figure 6A) could also result from an efficient release of soluble 80-kDa CYT-1 ICD from cell membrane by γ-secretase activity. However, pretreatment of 32D transfectants with GSI IX before 10-min PMA stimulation clearly promoted accumulation of the membrane-anchored 80-kDa CYT-2, but accumulation of 80-kDa CYT-1 required a 4-h treatment with PMA in the continuos presence of GSI IX (Figure 6C). Moreover, PMA treatment induced γ-secretase-dependent appearance of a soluble 80-kDa CYT-2 ICD into purified cytosolic cell fractions (not including membrane-anchored proteins) to a significantly greater extent than soluble 80-kDa CYT-1 ICD (Figure 6D). However, very little if any 80-kDa ErbB4 protein was detected in purified nuclear fractions (our unpublished data).
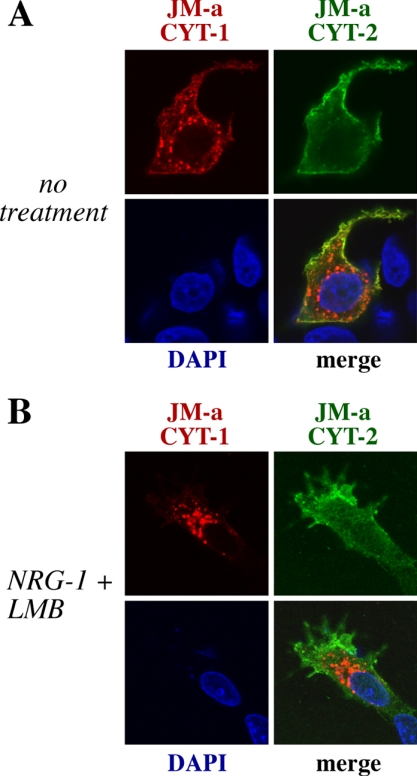
Because some nuclear staining was observed in MCF-7 background (Figure 3G), subcellular targeting of JM-a CYT-1 and JM-a CYT-2 was also compared by confocal immunofluorescence microscopy of transiently transfected MCF-7 cells. JM-a CYT-2 but not JM-a CYT-1 was localized to nuclei in some cells (Figure 7). The nuclear signal was sensitive to GSI IX (Figure 3G), suggesting it originated from ICD. Interestingly, JM-a CYT-1, but not JM-a CYT-2, was localized to intracellular vesicles (Figure 7). Some of these vesicles possibly represented early endosomes as they were also stained for the endosomal marker Rab-5 (our unpublished observations). The signal in intracellular vesicles was not reduced by GSI IX (our unpublished data), suggesting that the vesicles included membrane-anchored receptor forms. Only a small fraction of cytosolic ErbB4 staining colocalized with the marker of Golgi compartment Golgin-97 (our unpublished data). Together, these observation suggest that both types of ErbB4 JM-a isoforms are constitutively cleaved but that the generated 80-kDa fragments are differentially targeted.
Figure 7.
ErbB4 JM-a CYT-1 and JM-a CYT-2 have differential subcellular localization. MCF-7 cells were transiently transfected with plasmids encoding HA-tagged ErbB4 JM-a CYT-1 and Myc-tagged ErbB4 JM-a CYT-2. Cells were left untreated (A) or treated for 10 h with leptomycin B (LMB), an inhibitor of nuclear export, followed by addition of NRG-1 for 30 min (B). Subcellular localization was determined by confocal microscopy. Red, ErbB4 JM-a CYT-1; green, ErbB4 JM-a CYT-2; blue, nuclei stained with DAPI.
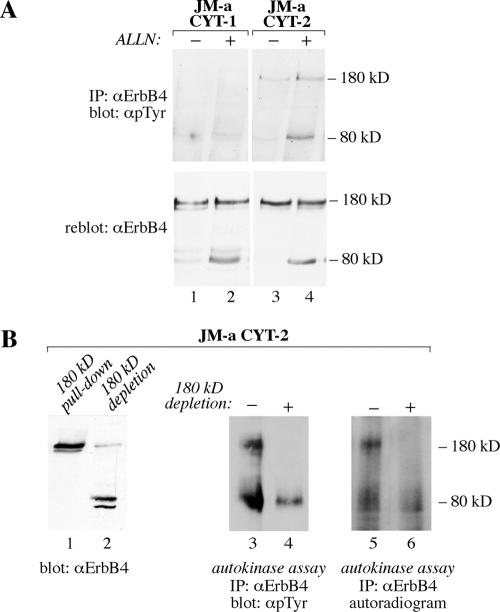
Constitutive Tyrosine Phosphorylation and Generation of an 80-kDa Proteolytic Fragment Are Two Independent Signaling Mechanisms of ErbB4 JM-a CYT-2
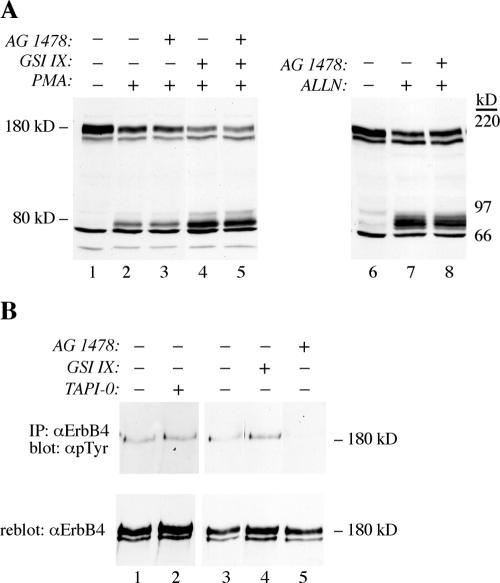
Tyrosine phosphorylation analyses and in vitro kinase assays demonstrated that both the 180-kDa ErbB4 JM-a CYT-2 as well as its 80-kDa carboxy-terminal fragment were constitutively phosphorylated (Figure 8A) and possessed autokinase activity (Figure 8B). Interestingly, neither the 180-nor the 80-kDa JM-a CYT-1 proteins were significantly phosphorylated, even when accumulated with ALLN (Figure 8A). The ErbB4 isoform JM-a CYT-2 also demonstrated ligand-independent proteolytic processing into putative signaling fragments, and ErbB4 JM-a cleavage was stimulated by PMA (Figure 6), a known activator of protein kinase C (PKC). Because kinase-active ErbB4 itself could activate PKC via phospholipase C-γ pathway (Vecchi et al., 1996), the possibility that constitutive ErbB4 tyrosine kinase activity was necessary for both basal tyrosine phosphorylation and cleavage, was addressed. 32D cells expressing ErbB4 JM-a CYT-2 were treated with AG 1478, and accumulation of membrane-anchored 80-kDa ErbB4 was induced by PMA, GSI IX, or ALLN (Figure 9A). Although AG 1478 efficiently blocked constitutive tyrosine phosphorylation of 180-kDa ErbB4 JM-a CYT-2 (Figure 9B), it did not affect accumulation of the 80-kDa fragment stimulated by any of the treatments (Figure 9A). The finding that an ErbB kinase inhibitor does not affect PMA-stimulated ErbB4 cleavage has also been reported previously (Cheng et al., 2003). Moreover, two different chemical inhibitors of PKC did not affect ALLN-stimulated 80-kDa fragment accumulation (our unpublished data).
Figure 8.
The 80-kDa ErbB4 JM-a CYT-2 fragment is tyrosine phosphorylated and has autokinase activity. (A) ErbB4 tyrosine phosphorylation analysis of the 80-kDa ErbB4 carboxy-terminal fragments. 32D cells expressing ErbB4 JM-a CYT-1 or ErbB4 JM-a CYT-2 were treated for 3 h with ALLN, and analyzed as in Figure 3C. (B) In vitro kinase assay of the 80-kDa CYT-2. 32D cells expressing ErbB4 JM-a CYT-2 were treated with ALLN and GSI IX for 3 h, and full-length glycosylated 180-kDa receptor was separated from the nonglycosylated 80-kDa fragment by pull-down using concanavalin A-Sepharose beads, as shown by Western blotting (lanes 1 and 2). Samples depleted for 180-kDa ErbB4, and total cell lysates were immunoprecipitated with an anti-ErbB4 antibody and analyzed for autotyrosine kinase activity by a 30-min incubation in the presence of ATP followed by ErbB4 phosphotyrosine analysis (lanes 3 and 4), as in Figure 3C. No signal was detected in parallel analysis carried without incubation with ATP using similar exposure time (our unpublished data). Alternatively, a radioactive kinase-assay was performed using [32P]ATP and autoradiography (lanes 5 and 6).
Figure 9.
Generation of an 80-kDa proteolytic fragment and basal tyrosine phosphorylation of the full-length receptor are two independent signaling events of ErbB4 JM-a CYT-2. (A) Western blot analysis with an anti-ErbB4 (sc-283) antibody. 32D cells expressing ErbB4 JM-a CYT-2 were pretreated for 30 min with AG 1478 or GSI IX before addition of PMA for a further 10 min, or ALLN for a further 3 h. (B) ErbB4 tyrosine phosphorylation analysis. 32D cells expressing ErbB4 JM-a CYT-2 were treated for 30 min with AG 1478, GSI IX, or TAPI-0, and analyzed as in Figure 3C.
Generation of a proteolytic carboxy-terminal RTK fragment with some kinase activity could theoretically also associate with and stimulate phosphorylation of the full-length receptor. To test whether constitutive ErbB4 JM-a CYT-2 cleavage was the primary event necessary for subsequent stimulation of receptor phosphorylation, 32D cells expressing ErbB4 JM-a CYT-2 were treated with a peptide inhibitor of TACE, TAPI-0, or GSI IX and analyzed for basal ErbB4 tyrosine phosphorylation (Figure 9B). However, no inhibitory effect of either TAPI-0 or GSI IX was observed. Together, these data indicate that constitutive tyrosine phosphorylation and generation of proteolytic fragments are two independent signaling mechanisms of ErbB4 JM-a CYT-2.
Both ErbB4 Tyrosine Kinase Activity and γ-Secretase Activity Contribute to Ligand-Independent Survival Promoted by ErbB4 JM-a CYT-2
The findings about basal tyrosine kinase activity (Figures 5 and 8), constitutive generation of 80-kDa fragments (Figure 6), and the observation that the two events were not interdependent (Figure 9) provide two molecular explanations for the ligand-independent survival signaling exclusively promoted by homodimers of ErbB4 JM-a CYT-2 (Figure 4B). To measure the relative contribution of the two pathways for survival, 32D transfectants were analyzed using MTS assays in the absence and presence of AG 1478 and GSI IX (Figure 10). Both AG 1478 and GSI IX alone reduced the number of viable cells expressing ErbB4 JM-a CYT-2 in the absence of exogenous ligands. Similar to observations with MCF-7 cells expressing ErbB4 JM-a CYT-2 (Figure 3D), the combinatory effect of both AG 1478 and GSI IX was additive (Figure 9). Similar results were obtained when GSI IX was replaced by another γ-secretase inhibitor, DFK167 (our unpublished data). However, the inhibitors did not significantly affect the viability of the vector control cells in the absence of a ligand, or the response of either cells expressing ErbB4 JM-a CYT-2 or vector control cells to IL-3, demonstrating specificity of the effect. The effect of GSI IX on ErbB signaling in general was further controlled by demonstrating a lack of effect on EGF-stimulated survival of 32D cells overexpressing ErbB1 (our unpublished data). These data support the hypothesis that the two signaling routes are parallel pathways that both are necessary for efficient ligand-independent survival stimulated by ErbB4 JM-a CYT-2. Together with data about inhibitory effects of AG 1478 and GSI IX also on the growth of breast cancer transfectants (Figure 3D), these observations suggest that overexpression of ErbB4 isoform JM-a CYT-2 promotes cellular growth by a mechanism depending on both ErbB4 kinase activity and proteolytic generation of signaling receptor fragments.
Figure 10.
Ligand-independent survival stimulated by ErbB4 JM-a CYT-2 is dependent on both ErbB4 tyrosine kinase activity and γ-secretase activity. MTS survival assay of 32D clones expressing ErbB4 JM-a CYT-2 (clone #1) or Neo control cells (clone #2) cultured in the absence (no ligand) or presence (IL-3) of 5% WEHI cell conditioned medium. The culture media were supplemented with or without AG 1478 and GSI IX.
DISCUSSION
In spite of extensive research on the role of ErbB receptors in cancer, the significance of ErbB4 as an oncogenic factor has remained controversial (Junttila et al., 2000; Gullick, 2003). Here, we demonstrate that alternatively spliced ErbB4 isoforms have critically different signaling capabilities and that one of the isoforms, ErbB4 JM-a CYT-2, has more oncogenic potential than the others. This isoform was overexpressed in a subset of breast cancer cases in vivo, and overexpression of the isoform also specifically promoted growth of breast cancer cells in vitro. Analyses using both MCF-7 cells and ErbB4 isoform-transfected myeloid cells devoid of endogenous ErbB background, demonstrated that the growth-promoting mechanism unique for the single ErbB4 isoform was dependent on ligand-independent constitutive tyrosine phosphorylation and proteolytic generation of an 80-kDa soluble receptor fragment. The described experiments represent the first analysis in which signaling properties of all the four predominant isoforms of human ErbB4 have been assessed.
Data suggesting that proteinase activity was necessary for full ligand-independent signaling potential of ErbB4 JM-a CYT-2, together with the fact that the noncleavable JM-b isoforms did not demonstrate ligand-independent activity, indicated that proteolytic processing of the receptor was involved in the growth-stimulating mechanism of ErbB4 JM-a CYT-2 overexpression. This was supported by the finding that transient expression of just the ICD of JM-a CYT-2 promoted cell cycle progression of transiently transfected COS-7 cells. Interestingly, both ErbB4 JM-a isoforms expressed in 32D cells were constitutively cleaved in two consecutive steps by a TACE-like proteinase and γ-secretase activity, known to generate a soluble receptor ICD with putative transcriptional activity (Ni et al., 2001; Komuro et al., 2003; Williams et al., 2004). However, a mechanism solely dependent on ErbB4 proteolysis would not account for differential signaling between the two cleavable JM-a isoforms, JM-a CYT-1 and JM-a CYT-2, that only differ by a 16-amino acid insert present in the cytoplasmic tail of CYT-1 but not in CYT-2 (Figure 2). The membrane-anchored 80-kDa fragment of ErbB4 JM-a CYT-1 had a shorter half-life, compared with ErbB4 JM-a CYT-2, in the absence of proteasome inhibitors, indicating that 80 kDa CYT-1 was more efficiently degraded. Furthermore, findings that more soluble CYT-2 ICD was found in cytosolic and nuclear compartments, and experiments with inhibitors of γ-secretase that block release of ICDs from cell membranes, indicated that more soluble CYT-2 ICD was generated. Together, these findings indicate that the two types of membrane-anchored ErbB4 80-kDa fragments are differentially targeted with a greater proportion of 80-kDa CYT-1 being degraded, and a greater proportion of 80-kDa CYT-2 being solubilized with γ-secretase. Moreover, the results suggest that the relatively efficient generation of soluble ICD is critical for the ligand-independent growth-promotion by ErbB4 JM-a CYT-2.
Differential proteasomal targeting of the two 80-kDa ErbB4 JM-a fragments could result from the coupling of ErbB4 CYT-1, but not of ErbB4 CYT-2, with PI3-K signaling (Elenius et al., 1999). Indeed, domains interacting with PI3-K have been shown to target RTKs, such as platelet-derived growth factor receptor-β to degradation, whereas receptors with mutated PI3-K binding sites recycle back to cell surface (Joly et al., 1995). Inhibition of PI3-K activity by chemical inhibitors, however, did not affect the production, stability or phosphorylation of the 80-kDa CYT-1 in 32D cells (our unpublished data), suggesting that the 16-amino acid stretch specific for ErbB4 CYT-1 isoform regulates its targeting in a mechanism independent of PI3-K activity. These results imply that there may be other interaction partners that differentially regulate the targeting and signaling of the 80-kDa fragments of ErbB4 JM-a CYT-1 and ErbB4 JM-a CYT-2.
Constitutive phosphorylation of the full-length receptor, also unique for ErbB4 JM-a CYT-2, provides another mechanistic explanation for the ligand-independent activity of the isoform. Our experiments with transfectants and various normal and cancer tissues suggest that the constitutive phosphorylation requires expression above the levels found in normal cells. This phosphorylation could be both a cause and an outcome of the generation of a signaling 80-kDa fragment: an activated receptor could stimulate its own cleavage via signaling molecules such as PKC (Vecchi et al., 1996), or deletion of the ectodomain from an RTK could activate the kinase in the remaining 80-kDa protein (Cabrera et al., 1996; Boerner et al., 2003). However, our data with both inhibitors of ErbB4 kinase and ErbB4 cleavage support the conclusions that these two phenomena are independent parallel signaling steps that both contribute to constitutive ErbB4 JM-a CYT-2 activity. This is in accordance with the findings that using combinations of the two types of inhibitors resulted in additive effects in assays measuring ErbB4 JM-a CYT-2-dependent cell growth.
Consistent with a role of ErbB4 JM-a CYT-2 as a human oncogene, this isoform, along with the other cleavable ErbB4 isoform JM-a CYT-1, was selectively overexpressed in a subset of human breast cancer patients. Also, the growth of a breast cancer cell line T-47D endogenously expressing JM-a isoforms, is suppressed by ErbB4 down-regulation with either ribozymes (Tang et al., 1999) or siRNAs (Junttila et al., 2005). Similar ErbB4 isoform expression pattern has been described previously for a subset of ependymoma (Gilbertson et al., 2002) and bladder cancer (Junttila et al., 2003) patients. Overexpression of nondefined ErbB4 isoforms in breast cancer have been reported previously (Srinivasan et al., 2000; Bieche et al., 2003), although the significance of the overexpression for outcome of patients has remained controversial (Bieche et al., 2003; Gullick, 2003; Lodge et al., 2003; Witton et al., 2003). Interestingly, immunohistochemical analyses of clinical breast cancer series suggest that nuclear localization of carboxy-terminal ErbB4 epitope associates with an invasive phenotype (Srinivasan et al., 2000) and poor survival in the estrogen receptor-positive subset of patients (Junttila et al., 2005). These observations suggest that cleavage of ErbB4 JM-a isoforms in breast cancer cells in vivo may be relevant for tumor biology. Some of the breast cancer tissue samples analyzed here also demonstrated overexpression of TACE, a proteinase capable of cleaving ErbB4. There was also a significant association of ErbB4 and TACE immunoreactivity when either the staining intensity or the percentage of positive cells was scored (p = 0.016 and 0.011 by chi-square and Fishers exact tests, respectively). Therefore, one could speculate that the normal regulation of ErbB4 JM-a CYT-2 processing is lost in tumors with enhanced TACE activity, resulting in enhanced ligand-independent signaling via proteolytic ErbB4 fragments.
Together, the results indicate that one specific ErbB4 isoform, ErbB4 JM-a CYT-2, promotes ligand-independent signaling by a mechanism involving both ligand-independent tyrosine phosphorylation and proteolytic generation of a soluble ICD. Together with findings demonstrating overexpression of ErbB4 JM-a isoforms in breast cancer, these data suggest that ErbB4 JM-a CYT-2 has more oncogenic potential than other ErbB4 isoforms. ErbB4 JM-a CYT-2 may thus represent a more specific and effective cancer drug target than nonspecific targeting of all ErbB4 gene products. The unique signaling mechanism exploited by ErbB4 JM-a CYT-2 may also provide opportunities to block signaling by reagents, such as metalloproteinase or γ-secretase inhibitors, that interfere with ErbB4 processing.
Supplementary Material
Acknowledgments
We thank Tero Vahlberg for statistical analyses and Maria Tuominen for excellent technical assistance. This work has been supported by the Academy of Finland, Emil Aaltonen Foundation, Sigrid Juselius Foundation, Turku University Central Hospital, Finnish Cancer Organizations, Turku University Foundation, Foundation for the Finnish Cancer Institute, Ida Montin Foundation, Finnish Medical Society Duodecim, Paulo Foundation, K. Albin Johansson Foundation, and Aurator Biomed Fund.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–05–0402) on October 26, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Bei, R., Pompa, G., Vitolo, D., Moriconi, E., Ciocci, L., Quaranta, M., Frati, L., Kraus, M. H., and Muraro, R. (2001). Co-localization of multiple ErbB receptors in stratified epithelium of oral squamous cell carcinoma. J. Pathol. 195, 343–348. [DOI] [PubMed] [Google Scholar]
- Bieche, I., Onody, P., Tozlu, S., Driouch, K., Vidaud, M., and Lidereau, R. (2003). Prognostic value of ERBB family mRNA expression in breast carcinomas. Int. J. Cancer 106, 758–765. [DOI] [PubMed] [Google Scholar]
- Boerner, J. L., Danielsen, A., and Maihle, N. J. (2003). Ligand-independent oncogenic signaling by the epidermal growth factor receptor: v-ErbB as a paradigm. Exp. Cell Res. 284, 111–121. [DOI] [PubMed] [Google Scholar]
- Cabrera, N., Diaz-Rodriguez, E., Becker, E., Martin-Zanca, D., and Pandiella, A. (1996). TrkA receptor ectodomain cleavage generates a tyrosine-phosphorylated cell-associated fragment. J. Cell Biol. 132, 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Levkowitz, G., Tzahar, E., Karunagaran, D., Lavi, S., Ben-Baruch, N., Leitner, O., Ratzkin, B. J., Bacus, S S., and Yarden, Y. (1996). An immunological approach reveals biological differences between the two NDF/heregulin receptors, ErbB-3 and ErbB-4. J. Biol. Chem. 271, 7620–7629. [PubMed] [Google Scholar]
- Cheng, Q. C., Tikhomirov, O., Zhou, W., and Carpenter, G. (2003). Ectodomain cleavage of ErbB-4, characterization of the cleavage site and m80 fragment. J. Biol. Chem. 278, 38421–38427. [DOI] [PubMed] [Google Scholar]
- Cohen, B. D., Kiener, P. A., Green, J. M., Foy, L., Fell, H. P., and Zhang, K. (1996). The relationship between human epidermal growth-like factor receptor expression and cellular transformation in NIH3T3 cells. J. Biol. Chem. 271, 30897–30903. [DOI] [PubMed] [Google Scholar]
- Egeblad, M., and Jaattela, M. (2000). Cell death induced by TNF or serum starvation is independent of ErbB receptor signaling in MCF-7 breast carcinoma cells. Int. J. Cancer 86, 617–625. [DOI] [PubMed] [Google Scholar]
- Egeblad, M., Mortensen, O. H., van Kempen, L. C., and Jaattela, M. (2001). BIBX1382BS, but not AG1478 or PD153035, inhibits the ErbB kinases at different concentrations in intact cells. Biochem. Biophys. Res. Commun. 281, 25–31. [DOI] [PubMed] [Google Scholar]
- Elenius, K., Corfas, G., Paul, S., Choi, C. J., Rio, C., Plowman, G. D., and Klagsbrun, M. (1997). A novel juxtamembrane domain isoform of HER4/ErbB4. Isoform-specific tissue distribution and differential processing in response to phorbol ester. J. Biol. Chem. 272, 26761–26768. [DOI] [PubMed] [Google Scholar]
- Elenius, K., Choi, C. J., Paul, S., Santiestevan, E., Nishi, E., and Klagsbrun, M. (1999). Characterization of a naturally occurring ErbB4 isoform that does not bind or activate phosphatidyl inositol 3-kinase. Oncogene 18, 2607–2615. [DOI] [PubMed] [Google Scholar]
- Furger, C., Fiddes, R. J., Quinn, D. I., Bova, R. J., Daly, R. J., and Sutherland, R. L. (1998). Granulosa cell tumors express erbB4 and are sensitive to the cytotoxic action of heregulin-beta2/PE40. Cancer Res. 58, 1773–1778. [PubMed] [Google Scholar]
- Gilbertson, R. J., et al. (2002). ERBB receptor signaling promotes ependymoma cell proliferation and represents a potential novel therapeutic target for this disease. Clin. Cancer Res. 8, 3054–3064. [PubMed] [Google Scholar]
- Gilbertson, R. J., Perry, R. H., Kelly, P. J., Pearson, A. D., and Lunec, J. (1997). Prognostic significance of HER2 and HER4 coexpression in childhood medulloblastoma. Cancer Res. 57, 3272–3280. [PubMed] [Google Scholar]
- Graber, H. U., Friess, H., Kaufmann, B., Willi, D., Zimmermann, A., Korc, M., and Buchler, M. W. (1999). ErbB-4 mRNA expression is decreased in nonmetastatic pancreatic cancer. Int. J. Cancer 84, 24–27. [DOI] [PubMed] [Google Scholar]
- Gschwind, A., Fischer, O. M., and Ullrich, A. (2004). The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat. Rev. Cancer 4, 361–370. [DOI] [PubMed] [Google Scholar]
- Gullick, W. J. (2003). c-erbB-4/HER 4, friend or foe? J. Pathol. 200, 279–281. [DOI] [PubMed] [Google Scholar]
- Haugen, D. R., Akslen, L. A., Varhaug, J. E., and Lillehaug, J. R. (1996). Expression of c-erbB-3 and c-erbB-4 proteins in papillary thyroid carcinomas. Cancer Res. 56, 1184–1188. [PubMed] [Google Scholar]
- Hughes, D. P., Thomas, D. G., Giordano, T. J., Baker, L. H., and McDonagh, K. T. (2004). Cell surface expression of epidermal growth factor receptor and Her-2 with nuclear expression of Her-4 in primary osteosarcoma. Cancer Res. 64, 2047–2053. [DOI] [PubMed] [Google Scholar]
- Joly, M., Kazlauskas, A., and Corvera, S. (1995). Phosphatidylinositol 3-kinase activity is required at a postendocytic step in PDGF receptor trafficking. J. Biol. Chem. 270, 13225–13230. [DOI] [PubMed] [Google Scholar]
- Junttila, T. T., Laato, M., Vahlberg, T., Söderström, K.-O., Visakorpi, T., Isola, J., and Elenius, K. (2003). Identification of patients with transitional cell carcinoma of the bladder overexpressing ErbB2, ErbB3 or ErbB4 isoforms. Quantitative RT-PCR analysis in estimation of ErbB receptor status from cancer patients. Clin. Cancer Res. 9, 5346–5357. [PubMed] [Google Scholar]
- Junttila, T. T., Sundvall, M., Lundin, M., Lundin, J., Tanner, M., Harkonen, P., Joensuu, H., Isola, J., and Elenius, K. (2005). Cleavable ErbB4 isoform in estrogen receptor-regulated growth of breast cancer cells. Cancer Res. 65, 1384–1393. [DOI] [PubMed] [Google Scholar]
- Junttila, T. T., Sundvall, M., Määttä, J. A., and Elenius, K. (2000). ErbB4 and its isoforms: selective regulation of growth factor responses by naturally occurring receptor variants. Trends Cardiovasc. Med. 10, 304–310. [DOI] [PubMed] [Google Scholar]
- Kainulainen, V., Sundvall, M., Määttä, J. A., Santiestevan, E., Klagsbrun, M., and Elenius, K. (2000). A natural ErbB4 isoform that does not activate phosphoinositide 3-kinase mediates proliferation but not survival or chemotaxis. J. Biol. Chem. 275, 8641–8649. [DOI] [PubMed] [Google Scholar]
- Komuro, A., Nagai, M., Navin, N. E., and Sudol, M. (2003). WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxy-terminal fragment of ErbB-4 that translocates to the nucleus. J. Biol. Chem. 278, 33334–33341. [DOI] [PubMed] [Google Scholar]
- Lee, H. J., Jung, K. M., Huang, Y. Z., Bennett, L. B., Lee, J. S., Mei, L., and Kim, T. W. (2002). Presenilin-dependent gamma-secretase-like intramembrane cleavage of ErbB4. J. Biol. Chem. 277, 6318–6323. [DOI] [PubMed] [Google Scholar]
- Lodge, A. J., Anderson, J. J., Gullick, W. J., Haugk, B., Leonard, R. C., and Angus, B. (2003). Type 1 growth factor receptor expression in node positive breast cancer: adverse prognostic significance of c-erbB-4. J. Clin. Pathol. 56, 300–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyne, J. C., Melhem, M. F., Finley, G. G., Wen, D., Liu, N., Deng, D. H., and Salup, R. (1997). Tissue expression of neu differentiation factor/heregulin and its receptor complex in prostate cancer and its biologic effects on prostate cancer cells in vitro. Cancer J. Sci. Am. 3, 21–30. [PubMed] [Google Scholar]
- Ni, C. Y., Murphy, M. P., Golde, T. E., and Carpenter, G. (2001). gamma-Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science 294, 2179–2181. [DOI] [PubMed] [Google Scholar]
- Peles, E., Bacus, S. S., Koski, R. A., Lu, H. S., Wen, D., Ogden, S. G., Levy, R. B., and Yarden, Y. (1992). Isolation of the neu/HER-2 stimulatory ligand: a 44 kd glycoprotein that induces differentiation of mammary tumor cells. Cell 69, 205–216. [DOI] [PubMed] [Google Scholar]
- Penington, D. J., Bryant, I., and Riese, D. J., 2nd (2002). Constitutively active ErbB4 and ErbB2 mutants exhibit distinct biological activities. Cell Growth Differ. 13, 247–256. [PubMed] [Google Scholar]
- Pierce, J. H., Ruggiero, M., Fleming, T. P., Di Fiore, P. P., Greenberger, J. S., Varticovski, L., Schlessinger, J., Rovera, G., and Aaronson, S. A. (1988). Signal transduction through the EGF receptor transfected in IL-3-dependent hematopoietic cells. Science 239, 628–631. [DOI] [PubMed] [Google Scholar]
- Rio, C., Buxbaum, J. D., Peschon, J. J., and Corfas, G. (2000). Tumor necrosis factor-alpha-converting enzyme is required for cleavage of erbB4/HER4. J. Biol. Chem. 275, 10379–10387. [DOI] [PubMed] [Google Scholar]
- Sartor, C. I., et al. (2001). Her4 mediates ligand-dependent antiproliferative and differentiation responses in human breast cancer cells. Mol. Cell Biol. 21, 4265–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soule, H. D., Vazguez, J., Long, A., Albert, S., and Brennan, M. (1973). A human cell line from a pleural effusion derived from a breast carcinoma. J. Natl. Cancer Inst. 51, 1409–1416. [DOI] [PubMed] [Google Scholar]
- Srinivasan, R., Benton, E., McCormick, F., Thomas, H., and Gullick, W. J. (1999). Expression of the c-erbB-3/HER-3 and c-erbB-4/HER-4 growth factor receptors and their ligands, neuregulin-1 alpha, neuregulin-1 beta, and beta-cellulin, in normal endometrium and endometrial cancer. Clin. Cancer Res. 5, 2877–2883. [PubMed] [Google Scholar]
- Srinivasan, R., Gillett, C. E., Barnes, D. M., and Gullick, W. J. (2000). Nuclear expression of the c-erbB-4/HER-4 growth factor receptor in invasive breast cancers. Cancer Res. 60, 1483–1487. [PubMed] [Google Scholar]
- Tang, C. K., Concepcion, X. Z., Milan, M., Gong, X., Montgomery, E., and Lippman, M. E. (1999). Ribozyme-mediated down-regulation of ErbB-4 in estrogen receptor-positive breast cancer cells inhibits proliferation both in vitro and in vivo. Cancer Res. 59, 5315–5322. [PubMed] [Google Scholar]
- Thomasson, M., Hedman, H., Junttila, T. T., Elenius, K., Ljungberg, B., and Henriksson, R. (2004). ErbB4 is downregulated in renal cell carcinoma: a quantitative RT-PCR and immunohistochemical analysis of the epidermal growth factor receptor family. Acta Oncol. 43, 453–459. [DOI] [PubMed] [Google Scholar]
- Vecchi, M., Baulida, J., and Carpenter, G. (1996). Selective cleavage of the heregulin receptor ErbB-4 by PKC activation. J. Biol. Chem. 271, 18989–18995. [DOI] [PubMed] [Google Scholar]
- Vecchi, M., and Carpenter, G. (1997). Constitutive proteolysis of the ErbB-4 receptor tyrosine kinase by a unique, sequential mechanism. J. Cell Biol. 139, 995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, C. C., Allison, J. G., Vidal, G. A., Burow, M. E., Beckman, B. S., Marrero, L., and Jones, F. E. (2004). The ERBB4/HER4 receptor tyrosine kinase regulates gene expression by functioning as a STAT5A nuclear chaperone. J. Cell Biol. 167, 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witton, C. J., Reeves, J. R., Going, J. J., Cooke, T. G., and Bartlett, J. M. (2003). Expression of the HER1–4 family of receptor tyrosine kinases in breast cancer. J. Pathol. 200, 290–297. [DOI] [PubMed] [Google Scholar]
- Yarden, Y., and Sliwkowski, M. X. (2001). Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2, 127–137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.