Abstract
The mechanisms that govern intracellular transport of sterols in eukaryotic cells are not well understood. Saccharomyces cerevisiae is a facultative anaerobic organism that becomes auxotroph for sterols and unsaturated fatty acids in the absence of oxygen. To identify pathways that are required for uptake and transport of sterols, we performed a systematic screen of the yeast deletion mutant collection for genes that are required for growth under anaerobic conditions. Of the ∼4800 nonessential genes represented in the deletion collection, 37 were essential for growth under anaerobic conditions. These affect a wide range of cellular functions, including biosynthetic pathways for certain amino acids and cofactors, reprogramming of transcription and translation, mitochondrial function and biogenesis, and membrane trafficking. Thirty-three of these mutants failed to grow on lipid-supplemented media when combined with a mutation in HEM1, which mimics anaerobic conditions in the presence of oxygen. Uptake assays with radio- and fluorescently labeled cholesterol revealed that 17 of the 33 mutants strongly affect uptake and/or esterification of exogenously supplied cholesterol. Examination of the subcellular distribution of sterols in these uptake mutants by cell fractionation and fluorescence microscopy indicates that some of the mutants block incorporation of cholesterol into the plasma membrane, a presumably early step in sterol uptake. Unexpectedly, the largest class of uptake mutants is affected in mitochondrial functions, and many of the uptake mutants show electron-dense mitochondrial inclusions. These results indicate that a hitherto uncharacterized mitochondrial function is required for sterol uptake and/or transport under anaerobic conditions and are discussed in light of the fact that mitochondrial import of cholesterol is required for steroidogenesis in vertebrate cells.
INTRODUCTION
Sterols are important lipid components of eukaryotic membranes that determine different membrane characteristics important for membrane transport and sorting in animal, plant, and fungal cells (Liscum and Dahl, 1992; Maxfield and Wustner, 2002; Soccio and Breslow, 2004).
Either lack or excess of cellular sterols is detrimental, necessitating mechanisms that control sterol homeostasis at different levels, from its synthesis, storage in form of steryl esters, to its release and appropriate distribution between subcellular membranes. Cholesterol is synthesized in the endoplasmic reticulum (ER) and transported to the plasma membrane, which harbors ∼90% of the free sterol pool of the cell (Lange et al., 1989). ER-to-plasma membrane transport of cholesterol is temperature and energy dependent but only partially sensitive to brefeldin A, suggesting that both vesicular and nonvesicular transport pathways may be operating (Kaplan and Simoni, 1985; Urbani and Simoni, 1990; Puglielli et al., 1995; Smart et al., 1996; Field et al., 1998; Heino et al., 2000).
Fungal cells synthesize ergosterol instead of cholesterol as their main sterol and many aspects of sterol homeostasis are conserved between yeast and human (Sturley, 2000). Under aerobic conditions, ergosterol is synthesized in the ER membrane and is greatly enriched at the yeast plasma membrane (Schneiter et al., 1999). Under these conditions, cells do not take up exogenous sterols, a phenomenon known as “aerobic sterol exclusion.” Under anaerobic growth conditions or in mutants that lack heme, however, yeast becomes auxotrophic for sterols and unsaturated fatty acids, because the synthesis of these lipids requires molecular oxygen (Andreasen and Stier, 1953; Lewis et al., 1985). The fact that Saccharomyces cerevisiae is a facultative anaerobic organism that displays robust growth when appropriately supplemented indicates that lipid uptake is efficient. The cellular processes that are required for sterol uptake and the mechanisms that regulate sterol homeostasis under conditions where de novo synthesis is bypassed, however, are not well understood (Lorenz et al., 1986).
Under aerobic conditions, yeast cells bypass the aerobic sterol exclusion if they bear a hypermorphic allele of the transcription factor UPC2 or in its homologue ECM22, or if they overexpress the transcriptional regulator Sut1p (Lewis et al., 1988; Bourot and Karst, 1995; Crowley et al., 1998). These conditions result in the up-regulation of two ATP-binding cassette (ABC) transporters, Aus1p and Pdr11p, and a putative cell wall mannoprotein, Dan1 (Wilcox et al., 2002; Alimardani et al., 2004). Consistent with a role of Aus1p and Pdr11p in sterol uptake, an aus1Δ pdr11Δ double mutant does not grow under anaerobic conditions and has a greatly reduced rate of steryl ester formation (Wilcox et al., 2002; Li and Prinz, 2004). A defect in sterol uptake and impaired growth under anaerobic conditions was also observed in cells that lack ARV1, a gene that is required for viability of cells that lack steryl esters (Tinkelenberg et al., 2000). ARV1 encodes a conserved protein with six putative transmembrane domains and localizes to the ER and Golgi membranes (Swain et al., 2002).
To identify cellular processes that are important for sterol uptake, transport, and homeostatic regulation in the absence of its de novo synthesis, we screened the yeast deletion mutant collection for genes that are required for growth under anaerobic conditions. Here, we report the identification and preliminary characterization of a novel set of potential sterol uptake/transport mutants. Further characterization of the pathways that are affected in these mutants is likely to provide important new insights into the mechanisms that regulate sterol uptake, transport, and homeostasis in yeast and possibly also in mammalian cells.
MATERIALS AND METHODS
Yeast Strains and Growth Conditions
The yeast strains used in this study are shown in Table 1. The haploid gene deletion collections in the BY4742 background (MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0) and BY4741 background (MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0) were obtained from EUROSCARF (Winzeler et al., 1999). Strains were cultivated in YPD rich media (1% Bacto yeast extract, 2% Bacto peptone [US Biological, Swampscott, MA], and 2% glucose) or minimal media (0.67% yeast nitrogen base without amino acids [US Biological], 2% glucose, and 2 g/l amino acids). Media supplemented with sterols and fatty acids contained 5 mg/ml Tween 80 and 20 μg/ml ergosterol or cholesterol (Sigma-Aldrich, St. Louis, MO). hem1Δ mutant cells were supplemented with 10 μg/ml δ-aminolevulinic acid (ALA). The colony forming capacity of hem1Δ double mutant cells was determined by plating cells after 24, 48, or 72 h of growth on sterol-supplemented YPD plates containing ALA or Tween 80 and cholesterol. Cell viability was determined by staining cells with 0.1% methylene blue. Selection for the kanMX4 marker was on media containing 200 μg/ml G418 (Invitrogen, Carlsbad, CA). Anaerobic conditions were maintained using an anaerobic jar containing an AnaeroGen sachet (Oxoid, Basingstoke, Hampshire, England). Yeast was transformed by lithium acetate (Ito et al., 1983). For DNA cloning and propagation of plasmids, Escherichia coli strain XL1-Blue (Stratagene, La Jolla, CA) was used.
Table 1.
S. cerevisiae strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| BY4742 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 | EUROSCARF; Winzeler et al. (1999) |
| MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 xyz::kanMX4 | EUROSCARF; Winzeler et al. (1999); xyz indicates a deletion in a nonessential gene used in this study | |
| MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 xyz::kanMX4 hem1::LEU2 | This study, heme-deficient xyz-double mutant strains | |
| YRS1707 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 hem1::LEU2 | This study |
| YRS1945 | MAT? his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 aus1::kanMX4 pdr11::kanMX4 | This study |
| YRS1962 | MAT? his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 aus1::kanMX4 pdr11::kanMX4 hem1::LEU2 | This study |
| YRS1851 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 are1::kanMX4 are2::HIS3MX6 hem1::LEU2 | This study |
| YRS2135 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 atf2::kanMX4 hem1::LEU2 | This study |
| YRS2114 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 hem1::LEU2 ERG6-RFP-kanMX6 | This study |
Plasmid Constructions and DNA Manipulations
The plasmid pHEM1-LEU2 containing the hem1::LEU2 disruption cassette (kindly provided by I. Hapala, Slovak Academy of Sciences, Bratislava, Slovak Republic) was cut with BamHI/HindIII to release the disruption cassette, and yeast transformants were selected on minimal media without leucine but supplemented with ALA. Correct insertion of the disruption cassette at the HEM1 locus was confirmed by phenotypic analysis of the transformants, i.e., growth on ALA-supplemented media but no growth on nonsupplemented media. Plasmids expressing the functional, green fluorescent protein (GFP)-tagged versions of Aus1p (pW1220) and Pdr11p (pWP1251) were kindly provided by W. Prinz (NIH, Bethesda, MD) (Li and Prinz, 2004). The plasmid for overexpression of Aus1p (pNEV-AUS1) was kindly provided by T. Berges (University Poitiers, Poitiers, France) (Alimardani et al., 2004).
Cholesterol Uptake
Uptake of [14C]cholesterol was performed essentially as described previously (Crowley et al., 1998). hem1Δ mutant cells were cultured in ALA-containing media, washed, and then diluted in minimal media supplemented with Tween 80 (5 mg/ml), cholesterol (20 μg/ml), and 0.025 μCi/ml [14C]cholesterol (American Radiolabeled Chemicals, St. Louis, MO) and cultivated for 24 or 48 h at 24°C. Equal optical density (OD) units of cells were collected and washed with 0.5% tergitol. [3H]Palmitic acid was added to the cell pellet as internal standard. Cells were disrupted with glass beads in the presence of chloroform/methanol (1:1), and lipids were extracted into the organic phase. Lipids were separated by thin-layer chromatography (TLC) (Merck, Darmstadt, Germany) with the solvent system petroleum ether/diethyl ether/acetic acid (70:30:2; per volume) and free cholesterol and cholesteryl esters were quantified by scanning with a Tracemaster 40 automatic TLC-linear analyzer (Berthold Technologies, Bad Wildbad, Germany). TLC plates were then exposed to a phosphorimager screen and visualized using a PhosphorImager (Bio-Rad, Hercules, CA). Lipid extracts containing 25-{N-[(7-nitrobenz-2-oxa-1,3-diazol-4-yl)-methyl]amino}-27-norcholesterol (NBD-cholesterol) were visualized using a FluorChem 8900 imaging system (Alpha Innotech, San Leandro, CA).
Fluorescence Microscopy
To visualize uptake of NBD-cholesterol (Avanti Polar Lipids, Alabaster, AL) hem1Δ mutant cells were incubated with a 1:1 mixture of cholesterol and NBD-cholesterol (20 μg/ml) in Tween 80 (5 mg/ml). Cells were collected after 24 h of incubation at 24°C, washed with minimal media containing 1 mg/ml p-phenylenediamine as antibleach, and examined by fluorescence microscopy using a Plan-Neofluor objective (100×, numerical aperture 1.3) on a Zeiss Axioplan 2 (Carl Zeiss, Oberkochen, Germany) equipped with an AxioCam charge-coupled device camera and AxioVision 4.2 software. Intracellular lipid particles were stained with Nile red. Vacuoles were stained with FM4-64. Pictures were recorded with equal exposure times, processed to maintain intensity differences, and mounted using Adobe Photoshop CS (Adobe Systems, Mountain View, CA).
Electron Microscopy
For ultrastructural examination, heme-deficient mutant cells were cultivated in media containing ALA and then diluted into Tween 80 and cholesterol-supplemented media. Cells were cultivated for 24 h at 24°C and then fixed in 1.5% aqueous solution of KMnO4 for 30 min at room temperature. Fixed cells were washed several times in distilled water and incubated in 0.5% aqueous uranyl acetate overnight with shaking at 4°C. Samples were dehydrated in a graded series of ethanol (50–100%) and embedded in Spurr resin. Ultrathin sections (80 nm) were stained with lead citrate and viewed with a Philips CM 10 electron microscope.
Subcellular Fractionation and Western Blot Analysis
To determine the sterol distribution in subcellular membranes, heme-deficient mutant cells were cultivated in minimal media supplemented with Tween 80 (5 mg/ml), cholesterol (20 μg/ml), and 0.25 μCi/ml [14C]cholesterol for 24 h at 24°C. Cells were converted to spheroplasts, and the homogenate was fractionated on an Accudenz density gradient (Accurate Chemical & Scientific, Oslo, Norway) as described previously (Cowles et al., 1997). Twelve fractions were collected from the top of the gradient; lipids were extracted and analyzed by TLC, proteins were precipitated with 10% trichloroacetic acid, and marker proteins were detected by Western blot analysis. Protein concentration was determined by the method of Lowry, using bovine serum albumin as standard. Radiolabeled cholesterol was determined by scintillation counting. Western blot analyses to determine the distribution of marker proteins in Accudenz gradient fractions were performed with the following rabbit antibodies: anti-Sec61p (1:5000; R. Schekman, University of California, Berkeley, Berkeley, CA), anti-Gas1p (1:5000; A. Conzelmann, University of Fribourg, Fribourg, Switzerland), anti-porin (1:10,000; G. Daum, Graz University of Technology, Graz, Austria), anti-Erg6p (1:10,000; G. Daum, Graz University of Technology), anti-Tlg1p (1:5000; H. Pelham, MRC Laboratory of Molecular Biology, Cambridge, United Kingdom), anti-Pma1p (1:10,000), or anti-CPY (1:10,000). Rabbit antibodies against GFP (1:5000; Torrey Pines Biolabs, Houston, TX) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:10,000) were used to examine the expression of the GFP-tagged ABC transporters in the sterol uptake mutants.
RESULTS
To identify genes that are required for the uptake and intracellular transport of sterols, we screened the haploid yeast deletion mutant collection containing ∼4800 strains for mutants that fail to grow under anaerobic conditions on YPD media supplemented with the fungal-specific sterol ergosterol or the mammalian sterol cholesterol and Tween 80 as a source of unsaturated fatty acids (Winzeler et al., 1999). The maintenance of oxygen-limiting conditions during these growth tests was assessed by ensuring that wild-type cells did not grow on media lacking the lipid supplementation (Figure 1). Candidate mutants that displayed weak growth in the first round of screening were retested four to five times to confirm their growth phenotype under anaerobic conditions. Linkage of the phenotype to the gene disruption was confirmed by examining mutant strains from the deletion mutant collection of the opposite mating type and by PCR analysis with gene-specific primers (our unpublished data). Only mutants that displayed the anaerobic growth phenotype in both mating types remained in the mutant collection.
Figure 1.
Identification of genes that are essential for growth under anaerobic conditions. Cells bearing a deletion of the gene indicated were serially diluted 10-fold and spotted on YPD media containing cholesterol (Chol/Tw) or ergosterol (Erg/Tw) and Tween 80 or on nonsupplemented YPD media (Non-suppl.). Plates were incubated under anaerobic or aerobic conditions for 4 d at 30°C. Anaerobic growth of wild-type (BY4742) and nongrowth of an aus1Δ pdr11Δ (YRS1945) double mutant strain is shown for comparison on the top two lines.
Ergosterol supports a number of different cellular processes, including cell cycle progression (known as the sparking function of ergosterol), cell polarization, membrane fusion during mating, endocytosis and vacuolar fusion, and sorting of the tryptophan permease along the secretory pathway (Rodriguez and Parks, 1983; Rodriguez et al., 1985; Munn et al., 1999; Kato and Wickner, 2001; Bagnat and Simons, 2002; Umebayashi and Nakano, 2003). Under the growth conditions used here, these processes did not become growth limiting as indicated by the fact that wild-type cells grew equally well on media supplemented with cholesterol or ergosterol. However, eight of the mutants identified in this screen (aro2Δ, aro7Δ, tkl1Δ, adh1Δ, rib4Δ, drs2Δ, pho85Δ, and ypk1Δ) displayed reduced growth on media containing cholesterol compared with ergosterol, suggesting that these mutants have a more stringent requirement for ergosterol in a growth limiting process that is not satisfied by cholesterol (Figure 1). The aus1Δ pdr11Δ double mutant was included in this and all subsequent assays as a control, known to be defective in sterol uptake (Wilcox et al., 2002; Li and Prinz, 2004).
The 37 mutants that showed a growth defect under anaerobic conditions were classified according to the function of the gene defective in the respective strain. As shown in Table 2, these mutants affect various cellular functions: nine bear deficiencies in the metabolism of amino acids, vitamins, and cofactors, five affect transcription and translation, three affect vesicular transport, and three bear defects in protein kinases. The largest class of mutants with 11 members, however, affects mitochondrial functions; five of these directly impair the F1 subunit of the ATP synthase. Under anaerobic conditions, F1-catalyzed ATP hydrolysis is required to maintain an electrochemical potential across the inner mitochondrial membrane, which is essential for mitochondrial biogenesis (Lefebvre-Legendre et al., 2003). Mutations in GUP1 and NUP84 affect glycerol transport and nuclear import, respectively, and SHE1 affects cell growth when overexpressed (Espinet et al., 1995). Three of the mutants bear deletions of functionally uncharacterized open reading frames, all of which affect growth on nonfermentable carbon sources and hence are required for respiration (YBL100, YJR120, and YLR202) (Entian et al., 1999; Dimmer et al., 2002). Together, the results of these analyses indicate that various cellular processes are differentially regulated under anaerobic conditions and become essential under oxygen-limiting conditions.
Table 2.
Genes required for growth under anaerobic conditions
| Functional class | ORF | Gene | Function |
|---|---|---|---|
| Amino acid metabolism | YGL148 | ARO2 | Chorismate synthase; required for biosynthesis of aromatic amino acids |
| YPR060 | ARO7 | Chorismate mutase; required for biosynthesis of tyrosine and phenylalanine | |
| YJR139 | HOM6 | Homoserine dehydrogenase; required for biosynthesis of methionine and threonine | |
| YOR184 | SER1 | Phosphoserine transaminase; required for biosynthesis of l-serine | |
| YPR074 | TKL1 | Transketolase I; required for the pentose phosphate pathway and for biosynthesis of aromatic amino acids | |
| Fermentation of glucose and metabolism of cofactors | YOL086 | ADH1 | Alcohol dehydrogenase; required for glucose fermentation and catabolism of tryptophan and phenylalanine |
| YOR209 | NPT1 | Nicotinate phosphoribosyltransferase; required for salvage pathway of NAD+ biosynthesis, affects silencing | |
| YBL033 | RIB1 | GTP cyclohydrolase II; required for riboflavin, FMN, and FAD biosynthesis | |
| YOL143 | RIB4 | Riboflavine synthase; required for riboflavin, FMN, and FAD biosynthesis | |
| Regulation of transcription and translation | YKL139 | CTK1 | RNA polymerase II C-terminal domain kinase |
| YML112 | CTK3 | RNA polymerase II C-terminal domain kinase | |
| YNL139 | RLR1 | RNA polymerase II transcription elongation, mRNA export | |
| YBL093 | ROX3 | RNA polymerase II holoenzyme component | |
| YGL135 | RPL1B | Structural constituent of the large (60S) ribosomal subunit | |
| Vesicular transport | YAL026 | DRS2 | P-type ATPase, potential aminophospholipid translocase |
| YNL243 | SLA2 | Protein involved in membrane cytoskeleton assembly, required for cell polarization and endocytosis | |
| YLR240 | VPS34 | Phosphatidylinositol 3-kinase, required for vacuolar protein sorting | |
| Protein kinases | YLR226 | BUR2 | Cyclin-dependent protein kinase regulator |
| YPL031 | PHO85 | Cyclin-dependent protein kinase, regulates glycogen and phosphate metabolism | |
| YKL126 | YPK1 | Serine/threonine protein kinase required for endocytosis | |
| Mitochondrial function and biogenesis | YLR304 | ACO1 | Aconitase; required for TCA cycle, mutation leads to glutamate auxotrophy |
| YBL099 | ATP1 | α-Subunit of F1 sector of F1F0 ATP synthase | |
| YJR121 | ATP2 | β-Subunit of F1 sector of F1F0 ATP synthase | |
| YBR039 | ATP3 | γ-Subunit of F1 sector of F1F0 ATP synthase | |
| YNL315 | ATP11 | Molecular chaperone required for assembly of F1 sector of the F1F0 ATP synthase | |
| YJL180 | ATP12 | Molecular chaperone required for assembly of F1 sector of the F1F0 ATP synthase | |
| YOR125 | CAT5 | May encode a protein involved in one or more mono-oxygenase or hydroxylase steps of ubiquinone biosynthesis | |
| YJR144 | MGM101 | Involved in mitochondrial genome maintenance | |
| YLR369 | SSQ1 | Chaperon involved in the synthesis and assembly of iron/sulfur clusters into proteins | |
| YDR470 | UGO1 | Outer membrane protein required for mitochondrial fusion | |
| YPR024 | YME1 | Mitochondrial inner membrane peptidase | |
| Miscellaneous | YGL084 | GUP1 | Multimembrane-spanning protein and putative glycerol transporter that is essential for proton symport of glycerol |
| YDL116 | NUP84 | Subunit of the nuclear pore complex | |
| YBL031 | SHE1 | Cytoskeletal protein of unknown function; overexpression causes growth arrest | |
| Unknown function | YBL100 | Dubious open reading frame | |
| YJR120 | Hypothetical open reading frame | ||
| YLR202 | Dubious open reading frame |
Characterization of the Anaerobic Nonviable Mutants in a Heme-deficient Background
Deficiency in heme biosynthesis mimics anaerobic conditions in the presence of oxygen (Gollub et al., 1977). We thus examined whether disruption of HEM1 in the anaerobic nonviable mutants results in growth arrest on sterol-supplemented media as would be predicted for mutants that fail to take up sterols. The lack of HEM1 can be overcome either by supplementing cells with sterols and unsaturated fatty acids or it can be bypassed by providing the enzymatic product of the Hem1p-catalyzed first step in heme biosynthesis, ALA (Gollub et al., 1977). Using a disruption cassette for HEM1,33 of the 37 mutants could be obtained in a heme-deficient background. For rib1Δ, rlr1Δ, sla2Δ, and atp3Δ double mutants with hem1Δ could not be obtained despite repeated attempts.
Growth analysis of the heme-deficient mutants under aerobic conditions on sterol-supplemented media revealed that most of the mutants were unable to grow when supplemented with either ergosterol or cholesterol and Tween 80, but they were rescued by ALA (Figure 2). This phenotype is thus consistent with a possible defect in sterol uptake/transport in these mutants. Moreover, the observation that the mutants were not viable in a heme-deficient background suggests that their growth defect under anaerobic conditions is due to a deficiency of a heme-dependent reaction. hom6Δ, ypk1Δ, and aco1Δ mutants displayed some growth on lipid-supplemented media, but they did not reach the growth capacity of wild-type cells and thus remained in the collection for further analysis of a possible lipid uptake defect. Other mutants, such as aro2Δ, aro7Δ, tkl1Δ, and rib4Δ grew significantly better on ergosterol-than on cholesterol-supplemented media and hence maintained their preference for ergosterol over cholesterol even in the heme-deficient background.
Figure 2.
Characterization of anaerobic nonviable mutants in a heme-deficient background. Heme-deficient cells of the indicated genotype were serially diluted 10-fold and spotted on YPD media containing cholesterol (Chol/Tw) or ergosterol (Erg/Tw) and Tween 80, YPD media containing ALA (ALA), and on nonsupplemented YPD media (Non-suppl.). Plates were incubated under aerobic conditions for 4 d at 30°C. Growth of a hem1Δ (YRS1707) and nongrowth of an aus1Δ pdr11Δ hem1Δ (YRS1962) triple mutant strain is shown for comparison on the top two lines.
Growth Characteristics of Heme-deficient Mutants
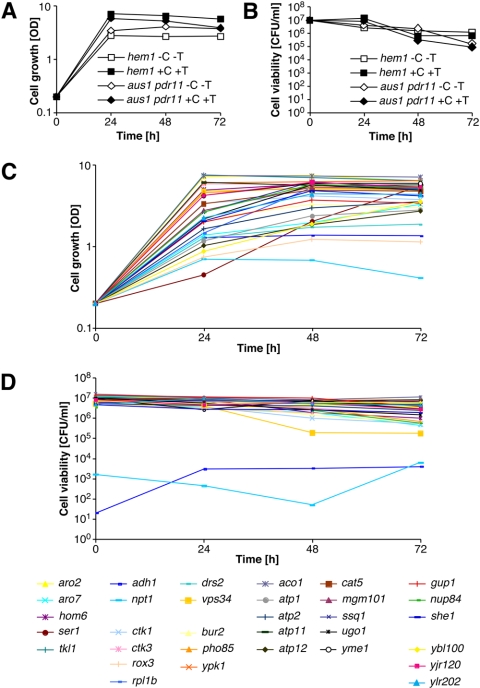
To examine a possible defect of these mutants in sterol uptake/transport in liquid media, we examined growth and viability of the heme-deficient mutants in liquid media that either contained or lacked cholesterol. Therefore, cells were grown in ALA-supplemented minimal media and then diluted into media with or without cholesterol. Under these conditions, the hem1Δ mutant and even an uptake-deficient aus1Δ pdr11Δ hem1Δ triple mutant continued to grow and reached stationary phase within 24 h of growth, irrespective of whether the media contained cholesterol (Figure 3A). Plating the cells on rich media containing ALA revealed that the uptake deficient aus1Δ pdr11Δ hem1Δ triple mutant begins to loose viability after 48 h (Figure 3B). Examination of the growth properties of the strains from the mutant collection revealed that the majority of these heme-deficient double mutants reached the stationary phase, albeit only within 48 h. Only the strains lacking NPT1, ROX3, ADH1, or DRS2 grew more slowly and did not reach an OD above 2 (Figure 3C). When plated from the liquid culture on solid media supplemented with ALA, the majority of mutants retained their capacity to grow colonies from single cells. Only npt1Δ, and adh1Δ, and to a much lesser degree vps34Δ, lost viability in the cholesterol media (Figure 3D). Because of their slow growth and low viability, npt1Δ and adh1Δ were excluded from the collection and not further analyzed. The estimates of cell viability based on plating correlated well with those obtained by staining cells with a vital dye, methylene blue (our unpublished data). Moreover, ATP levels in the 33 heme-deficient mutants grown for 48 h in cholesterol media were comparable with that of a heme-deficient wild-type (our unpublished data). Together, these data indicate that the heme-deficient mutants, despite their possible defect in lipid uptake, reached stationary phase within 48 h of growth in cholesterol media and that they remained energized and fully viable during this time.
Figure 3.
Growth and viability of heme-deficient mutants in cholesterol-supplemented media. (A) Growth of hem1Δ (YRS1707) and an aus1Δ pdr11Δ hem1Δ (YRS1962) mutant in media with or without cholesterol. Cells of the indicated genotype were pre-grown in minimal media supplemented with ALA, washed, and then diluted into media with (+C +T) or without cholesterol and Tween 80 (–C –T). Cell growth was monitored by reading absorbance at 600 nm. (B) Viability of heme-deficient wild-type and aus1Δ pdr11Δ sterol uptake mutant. Cells of the indicated genotype were grown as described for A. Aliquots were diluted and plated on solid rich media containing ALA. The number of colony-forming units per OD unit of culture over time is shown. (C) Growth of the anaerobic nonviable mutants. Heme-deficient cells of the indicated genotype were cultivated as described for panel A and cell growth was monitored. (D) Viability of the anaerobic nonviable mutants. Heme-deficient cells of the indicated genotype were cultivated as described for A; aliquots were diluted and plated on solid rich media containing cholesterol. The number of colony-forming units per OD unit of culture over time is shown. Mutants are grouped into functional classes.
Biochemical Characterization of a Sterol Uptake Defect
To directly examine cholesterol uptake and/or transport, heme-deficient wild-type cells were grown in the presence of [14C]cholesterol for 48 h, and uptake and esterification of the cholesterol were quantified by TLC analysis of radiolabeled lipids. Under these conditions, the radiolabeled cholesterol was efficiently taken up and incorporated into steryl esters (Figure 4). Only little uptake and esterification of [14C]cholesterol was seen if the cells were cultivated with ALA, consistent with exclusion of sterols under aerobic conditions. Sterol uptake was dependent on the ABC transporters Aus1p and Pdr11p, because it was greatly reduced in an aus1Δ pdr11Δ hem1Δ triple mutant (Wilcox et al., 2002; Li and Prinz, 2004) (Figure 4). Esterification of the radiolabeled cholesterol was dependent on the two ER-localized acyl CoA:cholesterol acyltransferases (ACATs), Are1p and Are2p, because no steryl esters were formed in an are1Δ are2Δ hem1Δ triple mutant (Yang et al., 1996; Yu et al., 1996) (Figure 4). These experiments thus indicate that sterol uptake under heme deficiency was dependent on the same factors that affect sterol uptake in cells expressing the hypermorphic allele of UPC2 or in cells overexpressing SUT1 (Wilcox et al., 2002; Alimardani et al., 2004; Li and Prinz, 2004).
Figure 4.
Uptake and esterification of [14C]cholesterol in heme-deficient cells. Heme-deficient cells of the indicated genotype were grown in the presence of [14C]cholesterol and Tween 80 for 48 h at 24°C. Equal OD units of cells were harvested, lipids were extracted, separated by TLC, and [14C]cholesterol in the free and esterified sterol fraction was quantified by radioscanning. Values represent mean and SD of two independent experiments. (A) Example of a TLC plate showing uptake of [14C]cholesterol and its incorporation into steryl esters in hem1Δ (YRS1707) grown in the presence (+ALA) or absence of ALA (–ALA), in aus1Δ pdr11Δ hem1Δ (YRS1962), and in are1Δ are2Δ hem1Δ (YRS1851) triple mutant cells. The position of [14C]cholesterol in the free and esterified sterol fraction is indicated to the right. Chol, free cholesterol; STE, steryl ester. (B) Quantification of [14C]cholesterol uptake (sterol) and incorporation into steryl esters (STE) in the heme-deficient cells shown in A.
Next, we examined sterol uptake by the anaerobic nonviable mutants. This analysis revealed greatly reduced levels of both free and esterified [14C]cholesterol in drs2Δ, bur2Δ, atp1Δ, gup1Δ, and yjr120Δ mutant cells, indicating that these mutants are strongly deficient in uptake and esterification of the radiolabeled cholesterol (Figure 5). Mutations in HOM6, TKL1, RPL1b, PHO85, ACO1, ATP2, ATP11, ATP12, CAT5, MGM101, and UGO1, in contrast, resulted in intermediate levels of free cholesterol but reduced levels of esterified sterols (Figure 5). On the contrary, a deletion of ROX3 resulted in significantly elevated levels of esterified sterols. Together, the sterol uptake analyses indicate that 17 of the 30 mutants tested in this in vivo cholesterol uptake assay displayed defects in sterol homeostasis under auxotrophic conditions.
Figure 5.
Uptake and esterification of [14C]cholesterol in the anaerobic nonviable mutants. Heme-deficient cells of the indicated genotype were grown in the presence of [14C]cholesterol and Tween 80 for 48 h at 24°C. Equal OD units of cells were harvested, lipids were extracted, separated by TLC, and [14C]cholesterol in the free and esterified sterol fraction (STE) was quantified by radioscanning. Values are expressed as percentage relative to heme-deficient wild-type cells. They represent mean and standard deviations of two independent experiments. Mutants are grouped into functional classes as shown in Table 2.
Visualization of Cholesterol Uptake Defects with NBD-Cholesterol
To visualize a defect in cholesterol uptake/trafficking of the mutants under heme-deficient conditions and to identify possible transport phenotypes, the intracellular distribution of cholesterol was examined using a fluorescently labeled cholesterol derivative, NBD-cholesterol (Mukherjee and Chattopadhyay, 1996; Sparrow et al., 1999). Localization studies using fluorescently labeled lipid analogues are hampered by the fact that the lipid is chemically modified and thus may behave aberrantly. However, in our hands, the NBD-labeled cholesterol represented the biochemically characterized sterol distribution pattern more uniformly, faithfully, and reproducibly than did staining with filipin, a fluorescent polyene. To stain cells with NBD-cholesterol, heme-deficient mutants were cultivated in media containing a 1:1 mixture of unlabeled cholesterol and NBD-cholesterol for 24 h. Under these conditions, NBD-cholesterol supported growth even when added without the unlabeled cholesterol, indicating that it is not toxic to the cells. Uptake of NBD-cholesterol paralleled that of [14C]cholesterol over a 24-h period. As observed in mammalian cells, the NBD-cholesterol was even metabolized to steryl esters as indicated by the appearance of a more hydrophobic fluorescent derivative that was absent in an esterification deficient are1Δ are2Δ hem1Δ triple mutant (Sparrow et al., 1999) (Figure 6A). The band labeled by an asterisk in Figure 6A was identified as an acetylated form of NBD-cholesterol, because it is absent in a mutant lacking ATF2, which encodes an alcohol acetyltransferase that has been shown to acetylate the steroid pregnenolone (Cauet et al., 1999). Although an aus1Δ pdr11Δ hem1Δ triple mutant displayed levels of free NBD-cholesterol that were comparable with wild-type cells, it failed to metabolize the fluorescent cholesterol, indicating that the free NBD-cholesterol observed in the lipid extract of these cells may be because of adherence of the label to the cells rather than due to bona fide uptake (Figure 6A).
Figure 6.
Uptake, metabolic conversion, and subcellular distribution of NBD-cholesterol in heme-deficient cells. (A) Uptake and esterification of [14C]cholesterol compared with NBD-cholesterol. hem1Δ (YRS1707), are1Δ are2Δ hem1Δ (YRS1851), and aus1 pdr11 hem1Δ (YRS1962) mutant cells were cultivated in presence of either [14C]cholesterol or NBD-cholesterol and aliquots were removed after 0 or 24 h of incubation. Lipids were separated by TLC and visualized by phosphorimaging or fluorescence. The position of free (Chol) and esterified sterol (STE) is indicated to the right. The asterisk indicates the position of acetylated NBD-cholesterol, which is absent in an atf2Δ hem1Δ mutant (YRS2135). (B) Subcellular distribution of NBD-cholesterol. hem1Δ (YRS1707) mutant cells were incubated with NBD-cholesterol for 24 h either in the absence or presence of ALA and examined by fluorescence microscopy. Plasma membrane and lipid particle staining is indicated by arrows and arrowheads, respectively. The distribution of NBD-cholesterol in an uptake deficient aus1Δ pdr11Δ hem1Δ (YRS1962), esterification deficient are1Δ are2Δ hem1Δ (YRS1851), or acetylation deficient atf2Δ hem1Δ (YRS2135) mutant is shown to the right. All pictures were recorded with the same exposure settings and processed to maintain differences in intensity using Photoshop. Nomarski view of the same visual fields is shown below. Bar, 5 μm. (C) Lipid particle staining by NBD-cholesterol. A hem1Δ mutant expressing an RFP-tagged Erg6p (YRS2114) as marker for lipid particles was incubated with NBD-cholesterol for 24 h. The distribution of NBD-cholesterol and that of Erg6p-RFP were recorded by fluorescence microscopy. Colocalization of NBD-cholesterol and Erg6p-RFP in intracellular lipid particles is indicated by arrowheads. Nomarski view of the same visual field is shown to the right. Bar, 5 μm.
When examined by fluorescence microscopy, the heme-deficient cells cultivated in the presence of NBD-cholesterol for 24 h exhibited uniform ring staining at the cell periphery, corresponding to plasma membrane, and staining of intracellular punctuate structures (Figure 6B). Staining by NBD-cholesterol was dependent on the heme deficiency because it was absent when cells were supplemented with ALA. Uptake of NBD-cholesterol was dependent on Aus1p Pdr11p, because an aus1Δ pdr11Δ hem1Δ triple mutant displayed only very weak plasma membrane staining, indicating that the NBD-cholesterol that adheres to these cells does not reach a hydrophobic environment, which is important to induce a large Stoke's shift in NBD fluorescence. The esterification-deficient are1Δ are2Δ hem1Δ triple mutant, in contrast, displayed normal staining of the plasma membrane but no staining of intracellular punctuate structures (Figure 6B). The fact that the NBD-cholesterol localization in an atf2Δ mutant is the same as that in wild-type cells indicates that the acetylated NBD-cholesterol does not significantly perturb the staining in wild-type cells. The punctuate structures that were stained with NBD-cholesterol in wild-type cells corresponded to lipid particles as they colocalized with Erg6p-red fluorescent protein (RFP), a marker protein for lipid particles (Figure 6C). These results thus indicate that NBD-cholesterol was efficiently taken up, converted to steryl esters, and incorporated into lipid particles in heme-deficient cells, indicating that it mimics unlabeled cholesterol and thus could serve as a valuable tool to study sterol uptake and transport in yeast.
Examination of the distribution of NBD-cholesterol in the heme-deficient mutant collection revealed low levels of plasma membrane and lipid particle staining in drs2Δ, bur2Δ, gup1Δ, and yjr120Δ (Figure 7). Mutations in HOM6, TKL1, RPL1b, PHO85, ACO1, ATP1, ATP2, ATP11, ATP12, CAT5, MGM101, and UGO1, in contrast, resulted in intermediate levels of plasma membrane staining but only weak staining of lipid particles. Mutants that displayed no or weak staining of lipid particles had apparently normal lipid particles when stained with Nile red, a vital dye that specifically stains neutral lipids (our unpublished data), indicating that the biogenesis of lipid particles was not generally affected in these mutants. rox3Δ, in contrast, exhibited stronger staining of the plasma membrane and lipid particles, consistent with the elevated uptake and esterification of the radiolabeled cholesterol observed in this mutant (Figure 7). mgm101Δ displayed staining of a single large punctuate structure whose identity is unknown. Together, the analysis of the uptake and intracellular distribution of the fluorescent NBD-cholesterol indicate that drs2Δ, bur2Δ, gup1Δ, and yjr120Δ are as strongly affected in sterol uptake as is an aus1Δ pdr11Δ mutant and that these mutations seem to affect the incorporation of NBD-cholesterol into the plasma membrane.
Figure 7.
Subcellular localization of NBD-cholesterol in the anaerobic nonviable mutants. Heme-deficient cells of the indicated genotype were grown in the presence of NBD-cholesterol for 24 h at 24°C, and NBD-cholesterol localization was examined by fluorescence microscopy using identical exposure settings. Bar, 5 μm.
Morphological Analysis of Heme-deficient Mutants
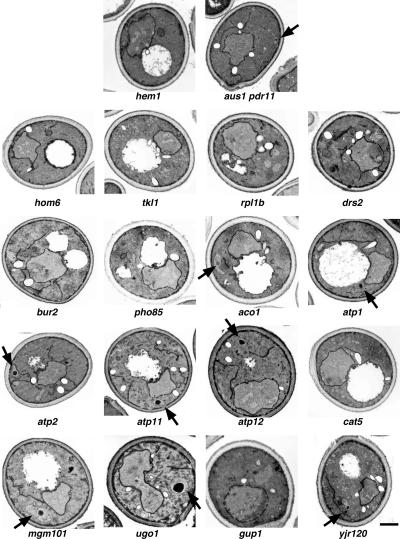
To identify possible morphological alterations that may accompany the defect in sterol uptake in some of the mutants, heme-deficient cells were grown in the presence of cholesterol for 24 h and then fixed and prepared for examination by transmission electron microscopy. This analysis revealed unusual, membrane enclosed, electron-dense structures in aus1Δ pdr11Δ, aco1Δ, atp1Δ, atp2Δ, atp11Δ, atp12Δ, mgm101Δ, ugo1Δ, and yjr120Δ (Figure 8). Similar structures have been observed previously in cells lacking a mitochondrial matrix protease and may represent aggregated mitochondrial proteins that accumulate in the absence of intramitochondrial proteolysis (Suzuki et al., 1994).
Figure 8.
Morphological analysis of the anaerobic nonviable mutants. Heme-deficient cells of the indicated genotype were grown in the presence of cholesterol and Tween 80 for 24 h at 24°C, fixed, and examined by transmission electron microscopy. The presence and position of unusual structures is indicated by arrows. Bar, 1 μm.
Analysis of the Subcellular Distribution of Cholesterol in Heme-deficient Uptake Mutants
To examine whether the sterol uptake mutants already block uptake of the radiolabeled sterol at the plasma membrane, as suggested by the NBD-cholesterol distribution, heme-deficient cells were grown in the presence of [14C]cholesterol for 24 h, and the subcellular distribution of the radiolabeled cholesterol was analyzed by fractionation on an Accudenz density gradient. Lipids from individual gradient fractions were extracted, separated on TLC, and levels of free [14C]cholesterol were quantified. In heme-deficient wild-type cells, the radiolabeled free cholesterol was greatly enriched in a peak of the gradient, centering around fraction 8 (Figure 9A). The same fraction was enriched in two plasma membrane markers, the proton pumping ATPase Pma1p and the GPI-anchored Gas1p, but it also contained significant levels of the ER marker Sec61p and the mitochondrial porin. The profile of the [14C]cholesterol distribution, however, closely matched that of the plasma membrane markers, indicating that the sterol peak is largely due to plasma membrane-localized free cholesterol (Figure 9B).
Figure 9.
Biochemical characterization of the subcellular distribution of free [14C]cholesterol. (A) hem1Δ (YRS1707) and hem1Δ aus1 pdr11Δ (YRS1962) mutant cells were grown in the presence of [14C]cholesterol for 24 h at 24°C. Equal OD units of cells were harvested, and membranes were separated on an Accudenz density gradient. Fractions were collected, lipids were extracted and separated by TLC, and free sterols were quantified by radioscanning. (B) Distribution of marker proteins. Membranes from hem1Δ (YRS1707) mutant cells grown in cholesterol were fractionated on an Accudenz density gradient, and the distribution of marker proteins was examined by Western blotting with antibodies against the proteins indicated. Organelles are indicated as follows: PM, plasma membrane; LP, lipid particles; VAC, vacuole; and Mito, mitochondria. (C) Distribution of free cholesterol in anaerobic nonviable mutants. Strains of the indicated genotype were cultivated as in A, membranes were fractionated on an Accudenz gradient, lipids were extracted and separated by TLC, and the distribution of free [14C]cholesterol was determined by radioscanning.
In contrast to the heme-deficient wild-type, the aus1Δ pdr11Δ mutant incorporated only very little radiolabeled cholesterol into its membranes with a small peak in fraction 9 of the gradient, indicating that this mutant is deficient already in the uptake step of cholesterol at the plasma membrane (Figure 9A).
Fractionation of some of the heme-deficient uptake mutants revealed essentially two classes of profiles. bur2Δ, drs2Δ, and gup1Δ showed low levels of uptake with a flat distribution over the gradient fractions, suggesting that these mutants affect an early step in uptake, resulting in low levels of free sterols in the plasma membrane (Figure 9C). pho85Δ, mgm101Δ, and hom6Δ, in contrast, displayed higher levels of uptake with a discernible enrichment of the free sterol in fractions 7–9 of the gradient, suggesting that these mutants may be compromised in the efficiency of the uptake process or in the distribution of the sterol that has been taken up.
Sterol Uptake Mutants Affect the Expression and Localization of the ABC Transporters Aus1p and Pdr11p
Because Aus1p and Pdr11p are two components known to be required for sterol uptake under anaerobic conditions, we examined whether the uptake mutants that we have isolated affect sterol uptake through Aus1p or Pdr11p (Wilcox et al., 2002; Alimardani et al., 2004). Therefore, we first examined the subcellular localization of functional GFP-tagged versions of these two ABC transporters in our sterol uptake mutants (Figure 10A). This analysis revealed that many of the mutants, particularly tkl1Δ, rpl1bΔ, drs2Δ, bur2Δ, pho85Δ, aco1Δ, atp2Δ, atp12Δ, cat5Δ, mgm101, ugo1, gup1Δ, and yjr120Δ displayed an increased GFP fluorescence in internal structures, which were identifiable as vacuoles. The increased staining of internal structures was frequently accompanied by a reduced staining of the plasma membrane, indicating that the ABC transporters in these mutants might either be unstable upon arrival at the cell surface or mistargeted to the vacuole during surface transport. The steady-state levels of these proteins were then examined more quantitatively by Western blot analysis (Figure 10B). This analysis revealed large differences in the levels of Aus1p-GFP, which migrated as multiple bands above 180 kDa, in the different mutants. The identity of the different bands is unknown, but it is possible that they are due to the formation of complexes that partially resist dissociation by SDS or represent polyubiquitin conjugates. Aus1p-GFP levels were strongly elevated in drs2Δ, pho85Δ, aco1Δ, and atp12Δ, i.e., in mutants that also showed internal GFP staining. hom6Δ, bur2Δ, atp1Δ, atp2Δ, atp11Δ, and cat5Δ, in contrast, displayed reduced levels of Aus1p-GFP compared with the hem1Δ mutant. Levels of Pdr11p-GFP were generally lower than that of Aus1p-GFP, but the full-length protein was detectable in all of the mutants. These data thus indicate that many of the sterol uptake mutants affect steady-state levels and localization of at least one of the two ABC transporters, Aus1p and Pdr11p. To examine whether a reduced expression of Aus1p could account for the sterol uptake defect, we tested whether overexpression of Aus1p would rescue growth of the sterol uptake mutants on media containing cholesterol. Therefore, the heme-deficient mutants were transformed with a high-copy number plasmid that confers expression of AUS1 from a strong constitutive PMA1 promoter (Wilcox et al., 2002; Alimardani et al., 2004). Growth analysis of the resulting strains on media containing cholesterol, however, revealed that none of the mutants was rescued by the overexpression of Aus1p (our unpublished data). These results thus suggest that the sterol uptake defect of these mutants is independent of Aus1p and Pdr11p, either of which is sufficient for sterol uptake to proceed normally (Wilcox et al., 2002; Alimardani et al., 2004).
Figure 10.
Subcellular localization and steady-state levels of the two ABC transporters Aus1p and Pdr11p in the sterol uptake mutants. (A) Sterol uptake mutants expressing GFP-tagged versions of Aus1p and Pdr11p were cultivated for 24 h in cholesterol containing media, and the subcellular distribution of the two ABC transporters was analyzed by fluorescent microscopy. Bar, 5 μm. (B) Sterol uptake mutants were cultivated as described under A. Equal OD units of cells were harvested, and levels of Aus1p and Pdr11p were examined by Western blots, which were also probed for GAPDH.
DISCUSSION
This study was aimed at the identification of novel components required for sterol uptake by yeast. Using a genomewide screening strategy, we first identified genes that are essential under anaerobic conditions, i.e., under conditions where yeast is auxotroph for sterols. To facilitate the subsequent analysis of the anaerobic nonviable mutants, they were rendered heme-deficient, which allowed us to examine uptake and esterification of radiolabeled cholesterol under more practicable aerobic conditions. Consistent with the growth defect seen under anaerobic conditions, the heme-deficient mutants failed to grow on sterol-supplemented media but were viable when supplemented with ALA. More direct analysis of sterol uptake/transport using radiolabeled cholesterol revealed that 17 of the 30 mutants tested were deficient in at least one of the steps along a putative sterol uptake/transport pathway. The uptake defect of these mutants seemed to be specific for sterols because radiolabeled fatty acids were still taken up and incorporated into lipids (our unpublished data).
Sterol Transport through Equilibration?
Sterols may be transported by proteins, by membranes, or a combination of the two. A putative sterol uptake and transport pathway is likely to be composed of separate steps, including uptake of the free sterol at the plasma membrane, its internalization, and possibly recycling to the plasma membrane, followed by the transport back to the ER, esterification, and deposition into lipid particles. This back transport pathway may or may not be mechanistically related to the forward transport of sterols from the ER to the plasma membrane that operates under aerobic conditions. The volume and possibly also the rate of the back transport, however, may be comparable with that of the forward transport because anaerobic cells grow efficiently and have approximately equal levels of plasma membrane sterols as they have steryl esters, which is also the case for aerobically grown cells. Two recent reports have shown that transport of sterols from the ER to the plasma membrane in yeast cannot efficiently be blocked by mutations in the classical secretory pathway (Baumann et al., 2005; Schnabl et al., 2005). It has thus been proposed that sterol transport relies on a “nonvesicular equilibration” of the free sterol concentration between the ER and the plasma membrane (Baumann et al., 2005). In this model, free sterol would be trapped at the plasma membrane through its condensation with sphingolipids and hence removed from the equilibrium of the free, nonsphingolipid-associated sterol pool (Baumann et al., 2005). Under anaerobic conditions, the back transport, in contrast, would have to act against the high sphingolipid concentration in the plasma membrane or would first need to saturate the binding capacity of the plasma membrane sphingolipids before an equilibrium with the ER could be established. At the ER, however, the free sterol may again be removed from the equilibrium through its esterification. Thus, in principle, a similar equilibration model as proposed for the forward transport could also account for the back transport. Such a model would be consistent with the observation that back transport of sterols to the ER requires steryl ester synthesis in the ER and that the rate of esterification is inversely proportional to the propensity of different sterols to associate with plasma membrane lipid rafts (Li and Prinz, 2004). Thus, in both models, the free sterol would efficiently be removed from the equilibrium by the target membrane, either through a direct chemical modification, i.e., esterification in the ER, or through the condensation into a lipid–lipid complex, i.e., raft formation in the plasma membrane. Our observation that the esterification deficient are1Δ are2Δ double mutant grows normally under anaerobic or heme-depleted conditions together with the fact that the mutant takes up normal levels of free sterols indicates that esterification is not required for sterol incorporation into the plasma membrane, as has been proposed by Li and Prinz (2004). The equilibration model is complicated by the fact that under aerobic conditions, both sterols and ceramides are synthesized in the ER, one would thus assume that they condense already at their site of synthesis, in which case surface delivery of sterols should be sensitive to a block in secretion, because such a block efficiently inhibits the transport-dependent maturation of sphingolipids (Puoti et al., 1991). Regardless, equilibration of the sterol pool between different membranes could be mediated by close apposition of donor and target membranes, i.e., membrane contacts (Levine, 2004) or through a protein-mediated transport of the sterols through the aqueous phase. Based on structural considerations, members of the oxysterol-binding protein (OSPB) family are good candidates for mediating such a transport (Im et al., 2005). Whether these OSPBs indeed transport sterols between membranes in vivo, however, remains to be established.
Three Classes of Sterol Uptake/Transport Mutants
The sterol transport mutants that we isolated in this screen fell into three classes. First, defects in BUR2, DRS2, GUP1, ATP1, and YJR120 resulted in the strongest defect in sterol uptake. The uptake defect in these mutants is in fact as strong as that of the aus1Δ pdr11Δ double mutant. Similar to an aus1Δ pdr11Δ mutant, defects in BUR2, DRS2, GUP1, ATP1, and YJR120 resulted in low levels of both free and esterified sterols, indicating a defect early in the pathway, most likely already in the uptake step at the plasma membrane. Consistent with this proposition, bur2Δ, drs2Δ, and gup1Δ mutants incorporated little NBD-cholesterol into the plasma membrane and had low levels of radiolabeled cholesterol incorporated into the plasma membrane, as assessed by subcellular fractionation, thereby, again resembling the aus1Δ pdr11Δ double mutant. These observations indicate that these mutants affect sterol incorporation into the plasma membrane, a presumably early step in sterol uptake. Three of these mutants, bur2Δ, drs2Δ, and gup1Δ, are particularly interesting, because their block in sterol uptake does not seem to be related to mitochondrial function.
BUR2 encodes a divergent cyclin that is involved in transcriptional regulation through histone methylation and thus is likely to act indirectly on the expression of factors involved in sterol transport or the regulation of membrane lipid composition (Yao et al., 2000; Laribee et al., 2005).
DRS2, in contrast, may more directly be involved in sterol transport because it encodes a candidate aminophospholipid translocase. These lipid translocases form a class of integral membrane proteins that flip fluorescent- or spin-labeled derivatives of phosphatidylserine and phosphatidylethanolamine from the external leaflet of a membrane bilayer to the cytosolic leaflet and thereby maintain the lipid asymmetry of a cell membrane. Drs2p has been localized to the trans-Golgi complex and a lack of Drs2p affects protein transport from that organelle (Chen et al., 1999; Gall et al., 2002). Remarkably, translocation of phosphatidylserine in the Golgi membrane is required for Drs2p-dependent protein transport; the precise in vivo substrate of Drs2p, however, remains to be identified (Natarajan et al., 2004). The strong sterol uptake defect that we observe in a drs2Δ mutant could thus be because of the protein transport defect of that mutant, as exemplified by the mistargeting of Aus1p-GFP in drs2Δ (Figure 10), or, alternatively, could more directly be related to an impaired lipid asymmetry of drs2Δ mutant membranes that could, for example, be incompatible with the efficient absorption of sterols at the plasma membrane.
GUP1, finally, encodes a polytopic integral membrane protein that localizes to the ER and has originally been identified as being required for glycerol uptake (Holst et al., 2000; Neves et al., 2004a). However, cells lacking GUP1 display multiple phenotypes, apparently unrelated to glycerol uptake, such as defects in bipolar bud site selection, defects in protein sorting to the vacuole, and changes in telomere length (Ni and Snyder, 2001; Bonangelino et al., 2002; Askree et al., 2004). The fact that a bona fide glycerol symporter, which localizes to the plasma membrane has recently been identified suggests that the defect of gup1Δ in glycerol uptake may be more indirect (Ferreira et al., 2005). Interestingly, however, Gup1p belongs to the superfamily of membrane-bound O-acyl transferases, which also include Are1p and Are2p, and as such may be more directly involved in lipid metabolism or protein lipidation (Hofmann, 2000; Neves et al., 2004b).
The second class of mutants displayed intermediate levels of free sterols, i.e., between 30 and 70% of that typically found in wild-type cells, and low levels of steryl esters. This class contains hom6Δ, tkl1Δ, rpl1bΔ, pho85Δ, and the majority of the mitochondrial mutants, i.e., aco1Δ, atp2Δ, atp11Δ, atp12Δ, cat5Δ, mgm101Δ, and ugo1Δ. These mutants had normal looking lipid particles when stained with Nile red and wild-type levels of ACAT activity when assayed in vivo or in vitro (our unpublished data) (Yang et al., 1996; Chang et al., 1997), suggesting that cholesterol availability in the ER membrane, rather than a reduced activity of the acyl CoA:cholesterol acyltransferase itself, is likely to account for the low levels of steryl esters in these cells. Consistent with this proposition, pho85Δ, mgm101Δ, and hom6Δ had reduced levels of plasma membrane sterols, but a normal looking profile, when fractionated on a density gradient.
The third class of mutants that can be distinguished based on the levels of cholesterol they take up and esterify is represented by rox3Δ. ROX3 mutants had elevated levels of esterified sterols. Rox3p is part of the RNA polymerase II transcription mediator complex, suggesting that the effects of Rox3p on the cellular level of free and esterified sterols are more indirect (Myers et al., 1998).
Although the identity of the mutants isolated in this screen does not yet reveal any molecular mechanism of the sterol uptake and transport pathway that is operating under anaerobic conditions, they clearly indicate that this pathway is directly or indirectly affected by a mitochondrial function.
A Function of Mitochondria in Sterol Uptake/Transport
The largest class of mutants that we isolated in this screen contains genes known to be required for mitochondrial functions. Of 10 anaerobic nonviable mutants in this group, eight displayed defects in sterol uptake/transport. Four of these, atp1Δ, atp2Δ, atp11Δ, and atp12Δ, directly affect the biogenesis of the soluble F1 sector of the F1F0 ATP synthase. Mutations in the F1 ATPase are known to result in an anaerobic nonviable phenotype, and these mutants are termed “petite negative,” i.e., they are lethal in a respiratory deficient background even when grown on glucose, indicating that the lack of the ATPase can only be tolerated if the mitochondrial electron transport chain is functional. The petite negative phenotype of these ATPase mutants has been rationalized by the hypothesis that the ATPase activity of F1 is required to maintain an essential electrochemical gradient across the inner mitochondrial membrane by converting ATP4– to ADP3– + Pi (Lefebvre-Legendre et al., 2003). Although this would explain why the ATPase mutants are nonviable under anaerobic conditions or in a heme-deficient background, it does not account for the deficiency of these mutants to take up sterols.
Yme1, an ATP- and metal-dependent protease associated with the mitochondrial inner membrane, is also required for viability in a respiratory-deficient background (Thorsness et al., 1993). Deletion of YME1, however, does not affect sterol uptake, indicating that the sterol uptake defect of the mitochondrial mutants is independent of their petite negative phenotype. Similarly, strains with deletions or such that completely lack the mitochondrial genome (rho– and rho°) have no growth defect under anaerobic conditions (our unpublished data), indicating that sterol uptake is independent of the presence of a mitochondrial genome and hence is not affected by the lack of F0 subunits of the ATPase, some of which are mitochondrially encoded.
A deficient electron transport chain, as caused by the heme mutation, combined with an inability to generate an alternative membrane potential, because a mutation in the ATPase, is thus likely to deenergize the mitochondria and hence arrest its functions and biogenesis. Energy depletion of mitochondria has been reported to affect both transport of ER-synthesized ergosterol to the plasma membrane and uptake of exogenously supplied cholesterol (Hunakova et al., 1997). In addition, energy depletion may induce the collapse of the matrix and formation of electron-dense inclusions. These ultrastructural changes, however, are unlikely to account for the block in sterol uptake, because we found that pim1Δ/lon1Δ mutant cells are viable under anaerobic conditions (our unpublished data).
The observation, that electron-dense structures are also apparent in an aus1Δ pdr11Δ mutant, which lacks two ABC transporters that have been localized to the plasma membrane and thus have no obvious relation to mitochondrial functions (Li and Prinz, 2004), rather suggests that the aberrant mitochondria could be because of a defect of these cells to take up sterols. The inner mitochondrial membrane is relatively rich in ergosterol, but the total and the mitochondrial ergosterol contents drop up to fourfold in anaerobic cells, even when supplemented with ergosterol (Jollow et al., 1968; Paltauf and Schatz, 1969; Schneiter et al., 1999). Such a reduced mitochondrial sterol content is known to affect the activity of the adenine nucleotide transporter in the inner mitochondrial membrane, leading to decreased intramitochondrial ATP levels and a decreased mitochondrial macromolecular synthesis (Haslam et al., 1977). Reduced ATP levels, in turn, are likely to inhibit the ATP-dependent mitochondrial protease Pim1/Lon1 and thereby might result in a phenotype characteristic for the protease deficient mutant (van Dijl et al., 1998). Thus, it seems reasonable to suggest that the electron-dense mitochondrial inclusions are a consequence rather than a cause of the reduced sterol uptake in these mutants. In this case, however, these structures should be visible in all the uptake mutants and may even be diagnostic for a sterol uptake defect, unless some of the mutants arrest at a stage that precedes the mitochondrial collapse. Thus, a direct role of membrane sterols in mitochondrial ATP import would explain the altered mitochondrial structures that are observed in many of the sterol uptake mutants, but it does not explain why the presumably deenergized mitochondria affect sterol uptake.
Even under anaerobic conditions, mitochondrial dysfunctions are likely to affect many different cellular functions, such as calcium and iron homeostasis, or modify the expression of nuclear genes, each of which could directly or indirectly affect sterol transport (Bianchi et al., 2004; Butow and Avadhani, 2004; Lill and Mühlenhoff, 2005). Alternatively, in vertebrate and insect cells, mitochondria themselves are known to be important for sterol metabolism, because they harbor the cytochrome P450 side chain cleavage enzyme that converts cholesterol to pregnenolone, which is then further metabolized to steroids (Miller, 1995). Remarkably, heterologous expression of P450 side chain cleavage activity in yeast mitochondria results in the conversion of endogenously synthesized sterols to pregnenolone, thus indicating that sterols are accessible and thus transported to the P450 system in the matrix site of the inner mitochondrial membrane (Duport et al., 1998). Transport routes for sterols from the ER or the plasma membrane to mitochondria thus seem to have evolved at some point. Our results would indicate that such routes may already be present in yeast and that they could be important for sterol uptake under anaerobic conditions.
Acknowledgments
We thank A. Conzelmann for helpful discussions during the course of this study, A. Conzelmann and I. Hapala for comments on the manuscript, Linda Corbino for expert technical assistance, Alexandre Toulmay for help with the revision, and T. Berges and W. Prinz for plasmids. This work was supported by grants from the Austrian Science Foundation (P15210), the “Novartis Stiftung für Medizinisch-Biologische Forschung” (02C62), and by the Swiss National Science Foundation (631-065925).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–06–0515) on October 26, 2005.
Abbreviations used: ABC, ATP-binding cassette; ACAT, acyl CoA: cholesterol acyltransferase; ALA, δ-aminolevulinic acid; NBD-cholesterol, 25-{N-[(7-nitrobenz-2-oxa-1,3-diazol-4-yl)-methyl]amino}-27-norcholesterol; TLC, thin-layer chromatography.
References
- Alimardani, P., Regnacq, M., Moreau-Vauzelle, C., Ferreira, T., Rossignol, T., Blondin, B., and Berges, T. (2004). SUT1-promoted sterol uptake involves the ABC transporter Aus1 and the mannoprotein Dan1 whose synergistic action is sufficient for this process. Biochem. J. 381, 195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen, A., and Stier, T.J.B. (1953). Anaerobic nutrition of Saccharomyces cerevisiae, I. Ergosterol requirement for growth in defined medium. J. Cell. Comp. Physiol. 41, 23–36. [DOI] [PubMed] [Google Scholar]
- Askree, S. H., Yehuda, T., Smolikov, S., Gurevich, R., Hawk, J., Coker, C., Krauskopf, A., Kupiec, M., and McEachern, M. J. (2004). A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl. Acad. Sci. USA 101, 8658–8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat, M., and Simons, K. (2002). Cell surface polarization during yeast mating. Proc. Natl. Acad. Sci. USA 99, 14183–14188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann, N. A., Sullivan, D. P., Ohvo-Rekila, H., Simonot, C., Pottekat, A., Klaassen, Z., Beh, C. T., and Menon, A. K. (2005). Transport of newly synthesized sterol to the sterol-enriched plasma membrane occurs via nonvesicular equilibration. Biochemistry 44, 5816–5826. [DOI] [PubMed] [Google Scholar]
- Bianchi, K., Rimessi, A., Prandini, A., Szabadkai, G., and Rizzuto, R. (2004). Calcium and mitochondria: mechanisms and functions of a troubled relationship. Biochim. Biophys. Acta 1742, 119–131. [DOI] [PubMed] [Google Scholar]
- Bonangelino, C. J., Chavez, E. M., and Bonifacino, J. S. (2002). Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 2486–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourot, S., and Karst, F. (1995). Isolation and characterization of the Saccharomyces cerevisiae SUT1 gene involved in sterol uptake. Gene 165, 97–102. [DOI] [PubMed] [Google Scholar]
- Butow, R. A., and Avadhani, N. G. (2004). Mitochondrial signaling: the retrograde response. Mol. Cell 14, 1–15. [DOI] [PubMed] [Google Scholar]
- Cauet, G., Degryse, E., Ledoux, C., Spagnoli, R., and Achstetter, T. (1999). Pregnenolone esterification in Saccharomyces cerevisiae. A potential detoxification mechanism. Eur. J. Biochem. 261, 317–324. [DOI] [PubMed] [Google Scholar]
- Chang, T. Y., Chang, C. C., and Cheng, D. (1997). Acyl-coenzyme A:cholesterol acyltransferase. Annu. Rev. Biochem. 66, 613–638. [DOI] [PubMed] [Google Scholar]
- Chen, C. Y., Ingram, M. F., Rosal, P. H., and Graham, T. R. (1999). Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J. Cell Biol. 147, 1223–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles, C. R., Odorizzi, G., Payne, G. S., and Emr, S. D. (1997). The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell 91, 109–118. [DOI] [PubMed] [Google Scholar]
- Crowley, J. H., Leak, F. W., Jr., Shianna, K. V., Tove, S., and Parks, L. W. (1998). A mutation in a purported regulatory gene affects control of sterol uptake in Saccharomyces cerevisiae. J. Bacteriol. 180, 4177–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmer, K. S., Fritz, S., Fuchs, F., Messerschmitt, M., Weinbach, N., Neupert, W., and Westermann, B. (2002). Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 847–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duport, C., Spagnoli, R., Degryse, E., and Pompon, D. (1998). Self-sufficient biosynthesis of pregnenolone and progesterone in engineered yeast. Nat. Biotech. 16, 186–189. [DOI] [PubMed] [Google Scholar]
- Entian, K. D., et al. (1999). Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol. Gen. Genet. 262, 683–702. [DOI] [PubMed] [Google Scholar]
- Espinet, C., de la Torre, M. A., Aldea, M., and Herrero, E. (1995). An efficient method to isolate yeast genes causing overexpression-mediated growth arrest. Yeast 11, 25–32. [DOI] [PubMed] [Google Scholar]
- Ferreira, C., van Voorst, F., Martins, A., Neves, L., Oliveira, R., Kielland-Brandt, M. C., Lucas, C., and Brandt, A. (2005). A member of the sugar transporter family, Stl1p is the glycerol/H+ symporter in Saccharomyces cerevisiae. Mol. Biol. Cell 16, 2068–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field, F. J., Born, E., Murthy, S., and Mathur, S. N. (1998). Transport of cholesterol from the endoplasmic reticulum to the plasma membrane is constitutive in CaCo-2 cells and differs from the transport of plasma membrane cholesterol to the endoplasmic reticulum. J. Lipid Res. 39, 333–343. [PubMed] [Google Scholar]
- Gall, W. E., Geething, N. C., Hua, Z., Ingram, M. F., Liu, K., Chen, S. I., and Graham, T. R. (2002). Drs2p-dependent formation of exocytic clathrin-coated vesicles in vivo. Curr. Biol. 12, 1623–1627. [DOI] [PubMed] [Google Scholar]
- Gollub, E. G., Liu, K. P., Dayan, J., Adlersberg, M., and Sprinson, D. B. (1977). Yeast mutants deficient in heme biosynthesis and a heme mutant additionally blocked in cyclization of 2,3-oxidosqualene. J. Biol. Chem. 252, 2846–2854. [PubMed] [Google Scholar]
- Haslam, J. M., Astin, A. M., and Nichols, W. W. (1977). The effects of altered sterol composition on the mitochondrial adenine nucleotide transporter of Saccharomyces cerevisiae. Biochem. J. 166, 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heino, S., Lusa, S., Somerharju, P., Ehnholm, C., Olkkonen, V. M., and Ikonen, E. (2000). Dissecting the role of the Golgi complex and lipid rafts in biosynthetic transport of cholesterol to the cell surface. Proc. Natl. Acad. Sci. USA 97, 8375–8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann, K. (2000). A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling. Trends Biochem. Sci. 25, 111–112. [DOI] [PubMed] [Google Scholar]
- Holst, B., Lunde, C., Lages, F., Oliveira, R., Lucas, C., and Kielland-Brandt, M. C. (2000). GUP1 and its close homologue GUP2, encoding multimembrane-spanning proteins involved in active glycerol uptake in Saccharomyces cerevisiae. Mol. Microbiol. 37, 108–124. [DOI] [PubMed] [Google Scholar]
- Hunakova, A., Daum, G., and Hapala, I. (1997). Changes in cellular ergosterol distribution in intramitochondrial energy-depleted Saccharomyces cerevisiae cells. Folia Microbiol. 42, 229–231. [DOI] [PubMed] [Google Scholar]
- Im, Y. J., Raychaudhuri, S., Prinz, W. A., and Hurley, J. H. (2005). Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature 437, 154–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, H., Fukada, Y., Murata, K., and Kimura, A. (1983). Transformation of intact cells with alkali cations. J. Bacteriol. 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jollow, D., Kellerman, G. M., and Linnane, A. W. (1968). The biogenesis of mitochondria. 3. The lipid composition of aerobically and anaerobically grown Saccharomyces cerevisiae as related to the membrane systems of the cells. J. Cell Biol. 37, 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, M. R., and Simoni, R. D. (1985). Transport of cholesterol from the endoplasmic reticulum to the plasma membrane. J. Cell Biol. 101, 446–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, M., and Wickner, W. (2001). Ergosterol is required for the Sec18/ATP-dependent priming step of homotypic vacuole fusion. EMBO J. 20, 4035–4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, Y., Swaisgood, M. H., Ramos, B. V., and Steck, T .L. (1989). Plasma membranes contain half the phospholipid and 90% of the cholesterol and sphingomyelin in cultured human fibroblasts. J. Biol. Chem. 264, 3786–3793. [PubMed] [Google Scholar]
- Laribee, R. N., Krogan, N. J., Xiao, T., Shibata, Y., Hughes, T. R., Greenblatt, J. F., and Strahl, B. D. (2005). BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr. Biol. 15, 1487–1493. [DOI] [PubMed] [Google Scholar]
- Lefebvre-Legendre, L., Balguerie, A., Duvezin-Caubet, S., Giraud, M. F., Slonimski, P. P., and Di Rago, J. P. (2003). F1-catalysed ATP hydrolysis is required for mitochondrial biogenesis in Saccharomyces cerevisiae growing under conditions where it cannot respire. Mol. Microbiol. 47, 1329–1339. [DOI] [PubMed] [Google Scholar]
- Levine, T. (2004). Short-range intracellular trafficking of small molecules across endoplasmic reticulum junctions. Trends Cell Biol. 14, 483–490. [DOI] [PubMed] [Google Scholar]
- Lewis, T. A., Taylor, F. R., and Parks, L. W. (1985). Involvement of heme biosynthesis in control of sterol uptake by Saccharomyces cerevisiae. J. Bacteriol. 163, 199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, T. L., Keesler, G. A., Fenner, G. P., and Parks, L. W. (1988). Pleiotropic mutations in Saccharomyces cerevisiae affecting sterol uptake and metabolism. Yeast 4, 93–106. [DOI] [PubMed] [Google Scholar]
- Li, Y., and Prinz, W. A. (2004). ATP-binding cassette (ABC) transporters mediate nonvesicular, raft-modulated sterol movement from the plasma membrane to the endoplasmic reticulum. J. Biol. Chem. 279, 45226–45234. [DOI] [PubMed] [Google Scholar]
- Lill, R., and Mühlenhoff, U. (2005). Iron-sulfur-protein biogenesis in eukaryotes. Trends Biochem. Sciences 30, 133–141. [DOI] [PubMed] [Google Scholar]
- Liscum, L., and Dahl, N. K. (1992). Intracellular cholesterol transport. J. Lipid Res. 33, 1239–1254. [PubMed] [Google Scholar]
- Lorenz, R. T., Rodriguez, R. J., Lewis, T. A., and Parks, L. W. (1986). Characteristics of sterol uptake in Saccharomyces cerevisiae. J. Bacteriol. 167, 981–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield, F. R., and Wustner, D. (2002). Intracellular cholesterol transport. J. Clin. Investig. 110, 891–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, W. L. (1995). Mitochondrial specificity of the early steps in steroidogenesis. J. Steroid Biochem. Mol. Biol. 55, 607–616. [DOI] [PubMed] [Google Scholar]
- Mukherjee, S., and Chattopadhyay, A. (1996). Membrane organization at low cholesterol concentrations: a study using 7-nitrobenz-2-oxa-1,3-diazol-4-yl-labeled cholesterol. Biochemistry 35, 1311–1322. [DOI] [PubMed] [Google Scholar]
- Munn, A. L., HeesePeck, A., Stevenson, B. J., Pichler, H., and Riezman, H. (1999). Specific sterols required for the internalization step of endocytosis in yeast. Mol. Biol. Cell 10, 3943–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, L. C., Gustafsson, C. M., Bushnell, D. A., Lui, M., Erdjument-Bromage, H., Tempst, P., and Kornberg, R. D. (1998). The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 12, 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan, P., Wang, J., Hua, Z., and Graham, T. R. (2004). Drs2p-coupled aminophospholipid translocase activity in yeast Golgi membranes and relationship to in vivo function. Proc. Natl. Acad. Sci. USA 101, 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves, L., Lages, F., and Lucas, C. (2004a). New insights on glycerol transport in Saccharomyces cerevisiae. FEBS Lett. 565, 160–162. [DOI] [PubMed] [Google Scholar]
- Neves, L., Oliveira, R., and Lucas, C. (2004b). Yeast orthologues associated with glycerol transport and metabolism. FEMS Yeast Res. 5, 51–62. [DOI] [PubMed] [Google Scholar]
- Ni, L., and Snyder, M. (2001). A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell 12, 2147–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paltauf, F., and Schatz, G. (1969). Promitochondria of anaerobicallly grown yeast. II. Lipid composition. Biochemistry 8, 335–339. [DOI] [PubMed] [Google Scholar]
- Puglielli, L., Rigotti, A., Greco, A. V., Santos, M. J., and Nervi, F. (1995). Sterol carrier protein-2 is involved in cholesterol transfer from the endoplasmic reticulum to the plasma membrane in human fibroblasts. J. Biol. Chem. 270, 18723–18726. [DOI] [PubMed] [Google Scholar]
- Puoti, A., Desponds, C., and Conzelmann, A. (1991). Biosynthesis of mannosylinositol-phosphoceramide in Saccharomyces cerevisiae is dependent on genes controlling the flow of secretory vesicles from the endoplasmic reticulum to the Golgi. J. Cell Biol. 113, 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, R. J., Low, C., Bottema, C. D., and Parks, L. W. (1985). Multiple functions for sterols in Saccharomyces cerevisiae. Biochim. Biophys. Acta 837, 336–343. [DOI] [PubMed] [Google Scholar]
- Rodriguez, R. J., and Parks, L. W. (1983). Structural and physiological features of sterols necessary to satisfy bulk membrane and sparking requirements in yeast sterol auxotrophs. Arch. Biochem. Biophys. 225, 861–871. [DOI] [PubMed] [Google Scholar]
- Schnabl, M., Daum, G., and Pichler, H. (2005). Multiple lipid transport pathways to the plasma membrane in yeast. Biochim. Biophys. Acta 1687, 130–140. [DOI] [PubMed] [Google Scholar]
- Schneiter, R., et al. (1999). Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J. Cell Biol. 146, 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart, E. J., Ying, Y., Donzell, W. C., and Anderson, R. G. (1996). A role for caveolin in transport of cholesterol from endoplasmic reticulum to plasma membrane. J. Biol. Chem. 271, 29427–29435. [DOI] [PubMed] [Google Scholar]
- Soccio, R. E., and Breslow, J. L. (2004). Intracellular cholesterol transport. Arterioscler. Thromb. Vasc. Biol. 24, 1150–1160. [DOI] [PubMed] [Google Scholar]
- Sparrow, C. P., Patel, S., Baffic, J., Chao, Y. S., Hernandez, M., Lam, M. H., Montenegro, J., Wright, S. D., and Detmers, P. A. (1999). A fluorescent cholesterol analog traces cholesterol absorption in hamsters and is esterified in vivo and in vitro. J. Lipid Res. 40, 1747–1757. [PubMed] [Google Scholar]
- Sturley, S. L. (2000). Conservation of eukaryotic sterol homeostasis: new insights from studies in budding yeast. Biochim. Biophys. Acta 1529, 155–163. [DOI] [PubMed] [Google Scholar]
- Suzuki, C. K., Suda, K., Wang, N., and Schatz, G. (1994). Requirement for the yeast gene LON in intramitochondrial proteolysis and maintenance of respiration. Science 264, 273–276. [DOI] [PubMed] [Google Scholar]
- Swain, E., Stukey, J., McDonough, V., Germann, M., Liu, Y., Sturley, S. L., and Nickels, J. T., Jr. (2002). Yeast cells lacking the ARV1 gene harbor defects in sphingolipid metabolism. Complementation by human ARV1. J. Biol. Chem. 277, 36152–36160. [DOI] [PubMed] [Google Scholar]
- Thorsness, P. E., White, K. H., and Fox, T. D. (1993). Inactivation of YME1, a member of the ftsH-SEC18-PAS1-CDC48 family of putative ATPase-encoding genes, causes increased escape of DNA from mitochondria in Saccharomyces cerevisiae. Mol. Cell. Biol. 13, 5418–5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinkelenberg, A. H., Liu, Y., Alcantara, F., Khan, S., Guo, Z., Bard, M., and Sturley, S. L. (2000). Mutations in yeast ARV1 alter intracellular sterol distribution and are complemented by human ARV1. J. Biol. Chem. 275, 40667–40670. [DOI] [PubMed] [Google Scholar]
- Umebayashi, K., and Nakano, A. (2003). Ergosterol is required for targeting of tryptophan permease to the yeast plasma membrane. J. Cell Biol. 161, 1117–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbani, L., and Simoni, R. D. (1990). Cholesterol and vesicular stomatitis virus G protein take separate routes from the endoplasmic reticulum to the plasma membrane. J. Biol. Chem. 265, 1919–1923. [PubMed] [Google Scholar]
- van Dijl, J. M., Kutejova, E., Suda, K., Perecko, D., Schatz, G., and Suzuki, C. K. (1998). The ATPase and protease domains of yeast mitochondrial Lon: roles in proteolysis and respiration-dependent growth. Proc. Natl. Acad. Sci. USA 95, 10584–10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox, L. J., Balderes, D. A., Wharton, B., Tinkelenberg, A. H., Rao, G., and Sturley, S. L. (2002). Transcriptional profiling identifies two members of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast. J. Biol. Chem. 277, 32466–32472. [DOI] [PubMed] [Google Scholar]
- Winzeler, E. A., et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906. [DOI] [PubMed] [Google Scholar]
- Yang, H., Bard, M., Bruner, D. A., Gleeson, A., Deckelbaum, R. J., Aljinovic, G., Pohl, T. M., Rothstein, R., and Sturley, S. L. (1996). Sterol esterification in yeast: a two-gene process. Science 272, 1353–1356. [DOI] [PubMed] [Google Scholar]
- Yao, S., Neiman, A., and Prelich, G. (2000). BUR1 and BUR2 encode a divergent cyclin-dependent kinase-cyclin complex important for transcription in vivo. Mol. Cell. Biol. 20, 7080–7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, C., Kennedy, N. J., Chang, C. C., and Rothblatt, J. A. (1996). Molecular cloning and characterization of two isoforms of Saccharomyces cerevisiae acyl-CoA:sterol acyltransferase. J. Biol. Chem. 271, 24157–24163. [DOI] [PubMed] [Google Scholar]