Abstract
Morphogenesis of filamentous ascomycetes includes continuously elongating hyphae, frequently emerging lateral branches, and, under certain circumstances, symmetrically dividing hyphal tips. We identified the formin AgBni1p of the model fungus Ashbya gossypii as an essential factor in these processes. AgBni1p is an essential protein apparently lacking functional overlaps with the two additional A. gossypii formins that are nonessential. Agbni1 null mutants fail to develop hyphae and instead expand to potato-shaped giant cells, which lack actin cables and thus tip-directed transport of secretory vesicles. Consistent with the essential role in hyphal development, AgBni1p locates to tips, but not to septa. The presence of a diaphanous autoregulatory domain (DAD) indicates that the activation of AgBni1p depends on Rho-type GTPases. Deletion of this domain, which should render AgBni1p constitutively active, completely changes the branching pattern of young hyphae. New axes of polarity are no longer established subapically (lateral branching) but by symmetric divisions of hyphal tips (tip splitting). In wild-type hyphae, tip splitting is induced much later and only at much higher elongation speed. When GTP-locked Rho-type GTPases were tested, only the young hyphae with mutated AgCdc42p split at their tips, similar to the DAD deletion mutant. Two-hybrid experiments confirmed that AgBni1p interacts with GTP-bound AgCdc42p. These data suggest a pathway for transforming one axis into two new axes of polar growth, in which an increased activation of AgBni1p by a pulse of activated AgCdc42p stimulates additional actin cable formation and tip-directed vesicle transport, thus enlarging and ultimately splitting the polarity site.
INTRODUCTION
Elongated cells, such as neurites, pollen tubes, and root hair cells, are generated when polar growth is maintained for extended time periods. An extreme case of polar growth has evolved in filamentous fungi, which are able to extend the tips of their tubelike cells, called hyphae, for unlimited time, provided nutrients are available (Gow, 1995; Momany, 2002; Harris et al., 2005). Hyphae not only very efficiently elongate but regularly establish new axes of polarity along their cortex, thus forming lateral branches, which themselves again generate lateral branches. This results in a fast spreading network of hyphae and the typical appearance of a fungal mycelium. In most filamentous fungi the initial hyphal tip elongation speed can increase by an order of magnitude or even more. Some filamentous fungi display hyphal tip splitting, the unique ability to simultaneously generate at tips of fast growing hyphae two sister hyphae.
The ascomycete Ashbya gossypii shows all the hallmarks of fungal filamentous growth, including tip splitting, although its recently completed genome sequence reveals an evolutionary relation with the Saccharomyces cerevisiae genome (Dietrich et al., 2004). A. gossypii is amenable to functional genome analysis using gene targeting methods or autonomously replicating plasmids (Wright and Philippsen, 1991; Steiner and Philippsen, 1994; Steiner et al., 1995; Wendland et al., 2000), which have promoted functional analyses of polarity genes in this fungus. Polar growth in A. gossypii starts from an isotropically growing germ bubble. The first steps in polarity establishment involve the A. gossypii proteins AgCdc24p and AgCdc42p (Wendland and Philippsen, 2001). Once the first germ tube has emerged hyphal growth of A. gossypii proceeds with frequent lateral branching and a steadily increasing elongation speed from initially 5 μm/h up to a maximum of 170 μm/h (Knechtle et al., 2003). For this process of hyphal maturation AgBem2p, AgRho3p, AgCla4p, AgSpa2p, and AgRsr1p are important (Ayad-Durieux et al., 2000; Wendland and Philippsen, 2000, 2001; Knechtle et al., 2003; Bauer et al., 2004). Although AgBem2p, AgRho3p and AgRsr1p are responsible for maintenance of polarity, both AgCla4p and AgSpa2p are necessary to reach maximal growth speed.
An important late step in the development to a fast spreading A. gossypii mycelium is the splitting of hyphal tips which, under optimal growth conditions, begins 12–14 h after emergence of the first hypha (Ayad-Durieux et al., 2000). A study with a GFP-labeled AgSpa2p showed that during tip splitting the existing polarity control center divides into two new centers of polarity, yielding two hyphae that elongate, after a short lag phase, with a speed similar to that before tip splitting (Knechtle et al., 2003). So far, the molecular basis for the initiation of hyphal tip splitting is unknown. We assumed that the apparent duplication of polar growth capacity depends on an approximately two-fold increase in secretory vesicle transport at or shortly after tip splitting and that, before this increase, additional tip-located actin cables had to form. Given the conserved role of formins in nucleating actin cables (Pruyne et al., 2002; Sagot et al., 2002b), we therefore hypothesized that a formin homolog could play an important role for the regulation of tip splitting.
Formins are common to all eukaryotic species and participate in many different processes, from cell polarization to embryonic development (see Wallar and Alberts (2003) and Evangelista et al. (2003) for reviews). Except for some cases in higher cells where a formin is involved in signaling (Habas et al., 2001), most formins participate in the organization of the actin cytoskeleton. Recently, the ability of formins to nucleate actin at the barbed end of actin filaments was described for different organisms (Pruyne et al., 2002; Sagot et al., 2002b; Kobielak et al., 2003; Kovar et al., 2003; Li and Higgs, 2003). Actin nucleation is mediated by the conserved formin homology domain FH2. As found by analyzing the crystal structures of the mouse formin, mDIA, and the yeast formin, ScBni1p, the core of the FH2 domain seems to have actin binding capacity, whereas adjacent amino acids are necessary for oligomerization and gain of polymerization capability (Shimada et al., 2004; Xu et al., 2004). A subclass of formins, the so-called diaphanous related formins (DRFs), are defined by two properties: First, they are activated when a GTP-bound Rho-type protein interacts with their amino terminus (Kohno et al., 1996; Evangelista et al., 1997; Imamura et al., 1997; Watanabe et al., 1997; Habas et al., 2001). Second, they posses a carboxy-terminal diaphanous autoregulatory domain (DAD), which binds to the amino-terminus in the inactive state of these formins (Watanabe et al., 1999; Alberts 2001).
In fungi, six formins have been studied to date. In S. cerevisiae, the two formins Bni1p and Bnr1p are required for cell polarity and cytokinesis with some overlapping and some different functions (Zahner et al., 1996; Evangelista et al., 1997; Imamura et al., 1997; Kamei et al., 1998; Pruyne et al., 2004). In Schizosaccharomyces pombe, three formins exist which are also involved in cell polarity and cytokinesis. The protein SpCdc12p is involved in cytokinesis (Chang et al., 1997; Kovar et al., 2003), SpFus1p in cell fusion (Petersen et al., 1998b) and SpFor3p in cell polarity control via regulation of the actin and microtubule network (Feierbach and Chang, 2001; Nakano et al., 2002). The only formin family member described so far in a filamentous fungus is the SepA protein of Aspergillus nidulans. SepA is an essential protein that locates to growing hyphal tips and to sites of cytokinesis (septation). Some SepA mutants still grow in a filamentous manner although they can no longer form septa (Harris et al., 1997; Sharpless and Harris, 2002).
In this article we first document by videomicroscopy the distinct differences in polar growth of budding yeast and A. gossypii and compare the domain compositions of the three A. gossypii formins with the two S. cerevisiae formins. Then we show that mutants lacking the AgBnr1p or AgBnr2p formin develop like wild type and that mutants lacking both formins are unable to grow. We next provide evidence that the AgBni1p formin is essential for hyphal emergence and elongation, that it localizes at hyphal tips, and that it is essential for organization of actin cables and thus tip-directed transport of secretory vesicles. In addition, we demonstrate that constitutively active AgBni1p leads to premature tip splitting and that this is most likely triggered by AgCdc42p-GTP.
MATERIALS AND METHODS
A. gossypii Strains and Growth Conditions
All strains were constructed by PCR-based gene targeting according to the method described by Wendland et al. (2000). Either pGEN3 (Wendland et al., 2000), pGUG (Knechtle et al., 2003), or pUC19NATPS (D. Hoepfner, personal communication) were used as templates to generate gene-targeting cassettes encoding geneticin-resistance, GFP plus geneticin-resistance, and ClonNAT-resistance, respectively. Strains and names of the oligonucleotides and templates used are given in Table 1. The strains were verified by PCR using a PTC 100 thermocycler (MJ Research, Waltham, MA). All oligonucleotides are listed in Table 2. Strains were grown at 30°C in AFM (Ashbya Full Medium) with or without geneticin or ClonNAT or at room temperature on synthetic medium for live cell imaging (Knechtle et al., 2003).
Table 1.
Ashbya gossypii strains and details of construction
| Strain | Genotype | Oligo-nucleotides | Template | Reference |
|---|---|---|---|---|
| ATCC10895 | Wild type | — | — | Ashby and Nowell (1926) |
| ΔlΔt | Agleu2Δ Agthr4Δ | — | — | Altmann-Johl and Philippsen (1996) |
| Agbni1Δ2 | Agbni1Δ::GEN3, Agleu2Δ Agthr4Δ | AgBNI1-S1 AgBNI1-S2 | pGEN3 | This study |
| AgBNI1ΔD | AgBNI1Δ5308-5757::GEN3 | AgBNI1dD-S1 AgBNI1dD-S2 | pGEN3 | This study |
| AgBNI1-GFP | AgBNI1-GFP-GEN3, Agleu2Δ Agthr4Δ | AgBNI1-GS1 AgBNI1-GS2 | pGUG | This study |
| AgSPA2-GFP | AgSPA2-GFP-GEN3, Agleu2Δ Agthr4Δ | — | — | Knechtle et al. (2003) |
| Agbni1Δ AgSPA2-GFP | Agbni1Δ::NAT1, AgSPA2-GFP-GEN3, Agleu2Δ Agthr4Δ | AgBNI1-NS1 AgBNI1-NS2 | pUC19NATPS | This study |
| Agbni1S1642G | Agbni1T4924G, C4925G-GEN3, Agleu2Δ Agthr4Δ | See text for details | This study | |
| AgbnilΔ1530G, K1620R | Agbni1A4589G, A4859G-GEN3, Agleu2Δ Agthr4Δ | See text for details | This study | |
| Agbnr1Δ-GEN3 | Agbnr1Δ::GEN3, Agleu2Δ Agthr4Δ | AgBNR1-S1 AgBNR1-S2 | pGEN3 | This study |
| Agbnr1Δ-NAT1 | Agbnr1Δ::NAT1, Agleu2Δ Agthr4Δ | AgBNR1-NS1 AgBNR1-NS2 | pUC19NATPS | This study |
| Agbnr2Δ | Agbnr2Δ::GEN3, Agleu2Δ Agthr4Δ | AgBNR2-S1 AgBNR2-S2 | pGEN3 | This study |
| Agbnr1,2ΔGL | Agbnr1Δ::GEN3, Agbnr2Δ::LEU2, Agleu2Δ Agthr4Δ | AgBNR2-S1 AgBNR2-S2 | pScLEU2 | This study |
| Agbnr1,2ΔGN | Agbnr1Δ::GEN3, Agbnr2Δ::NAT1 | AgBNR1-NS1 AgBNR1-NS2 | pUC19NATPS | This study |
| Agcdc42* | Agcdc42G183C-GEN3, Agleu2Δ Agthr4Δ | See text for details | This study | |
| AgBNI1ΔD-GFP | AgBNI1ΔD-GFP-GEN3, Agleu2Δ Agthr4Δ | AgBNI1dD-F1 AgBNI1dD-F2 | pAGT141 | This study |
Table 2.
Oligonucleotides used in this study
| Name | Sequencea | Purpose |
|---|---|---|
| AgBNI1-S1 | CTCGACCGTAGAGTCGATAGCGAGTTCTGCTCTAGCAAAATCCCCGCTAGGGATAACAGGGTAAT | Deletion |
| AgBNI1-S2 | GAGTTGTTCGATGGAGATCTTGACCACCTGAAGTACAGGTTCAAGAGGCATGCAAGCTTAGATCT | Deletion |
| AgBNI1-G1 | CCCGACCACCGTCTTGGACC | Verification |
| AgBNI1-G2 | GGGCATTTGCGCCGGGAGAG | Verification |
| AgBNI1-GS1 | CGGTGAACACCCGGAGTCTCGCAAGTCAATGCTCGATGAGCACAAGGGTGCAGGCGCTGGAGCTG | GFP-tagging |
| AgBNI1-GS2 | GCGGCGCGGCGGCGACATCGCGCTGGTCTATCAGTTTCTTGGTGCGGCAGGGACCTGGCACGGAGC | GFP-tagging |
| AgBNI1-11 | CAGATCGGGCCTGTGTTACC | Verification |
| AgBNR2-S1 | GATAAAGAACATCGCTAGTTAATTTTGCTAATAAAGAAGAGACCTTTAAgctagggataacagggtaat | Deletion |
| AgBNR2-S2 | GGCCGATAAGTGTGGAATCGGATAAGTGTTGAATAGGGTAACGGAAGAGGCATGCAAGCTTAGATCT | Deletion |
| AgBNR2-I1 | CGCACTCCGCACCGTGGACC | Verification |
| BNR2G1neu | CTGTCGAGGAATTAACTTTAAG | Verification |
| BNR2G2neu | GCCAGCGCATTTTGCACCGG | Verification |
| AgBNR1-S2 | GCCCGAGCGCCGAAGGCGAAGGGATGCGAAAAGTATATAGATGCAAGTTGaggcatgcaagcttagatct | Deletion |
| AgBNR1-S1 | GCGGGAGCAGTAGATTTGTGCCTGGGCCCCTCCCCCGCGAGTGCGCAATGgctagggataacagggtaat | Deletion |
| AgBNR1-I1 | CAGGCTGTGCGGCGCGCGCG | Verification |
| BNR1G1neu | GAGCAGCTGGTCGCTGGGCATCTCGTGG | Verification |
| BNR1G4neu | GCCTCGAATCACCAATCCCC | Verification |
| AgBNR1-NS1 | GGCGGGAGCAGTAGATTTGTGCCTGGGCCCCTCCCCCGCGAGTGCGCAccagtgaattcgagctcgg | Deletion |
| AgBNR1-NS2 | GCCCGAGCGCCGAAGGCGAAGGGATGCGAAAAGTATATAGATGCAAGTTG tacgccaagcttgcatgcct | Deletion |
| V2PDC1P | GAACAAACCCAAATCTGATTGCAAGGAGAGTGAAAGAGCCTT | Verification |
| V3PDC1T | GACCAGACAAGAAGTTGCCGACAGTCTGTTGAATTGGCCTG | Verification |
| AgBNI1dD-S1 | TCTGGAGTACAAGCGCGCGCAGGAGTTTAACCGCAAGATCTAAGCTAGGGATAACAGGGTAAT | Deletion |
| AgBNI1dD-S2 | GCTGGTCTATCAGTTTCTTGGTGCGGCGCTGGCGAACCTTAGGCATGCAAGCTTAGATCT | Deletion |
| Agbnil-12 C1 | CTTGAACTACGTGGAACGCATCGTCAGCCAGAATTATCCAgagctcgttttcgacactgg | pCORE integration |
| Agbnil-12 C2 | CCTGAAGTACAGGTTCAAGTTCCTGGAGAAAGCTGTTGAAtccttaccattaagttgatc | pCORE integration |
| Agbnil-12 IRO1 | ATGACTTTCTTGAACTACGTGGAACGCATCGTCAGCCAGAATTATCCAGGGTTCAACAGC | Mutagenesis |
| Agbnil-12 IRO2 | TTGACCACCTGAAGTACAGGTTCAAGTTCCTGGAGAAAGCTGTTGAACCCTGGATAATTC | Mutagenesis |
| Agbnil-11.1 C1 | TAAGTCTCCGGAAAAGGATCCAAGCGAGCTTCAGCGATCAgagctcgttttcgacactgg | pCORE integration |
| Agbnil-11.1 C2 | AGTAGCTTTGAAGATTAACAATGAGGTTCAAGAATAACTGtccttaccattaagttgatc | pCORE integration |
| Agbnil-11.1 IRO1 | GAGGATGCTAAGTCTCCGGAAAAGGATCCAAGCGAGCTTCAGCGATCAGGCCAGTTATTC | Mutagenesis |
| Agbnil-11.1 IRO2 | TGAGCTCCAGTAGCTTTGAAGATTAACAATGAGGTTCAAGAATAACTGGCCTGATCGCTG | Mutagenesis |
| Agbnil-11.2 C1 | GGGGTTCAAGTTGAGTACATTGCAGAGGCTCACCTTCATTgagctcgttttcgacactgg | pCORE integration |
| Agbnil-11.2 C2 | GTTCCACGTAGTTCAAGAAAGTCATGCTGTTTTTCTCGTCtccttaccattaagttgatc | pCORE integration |
| Agbnil-11.2 IRO1 | GCACAGGGGTTCAAGTTGAGTACATTGCAGAGGCTCACCTTCATTAGAGACGAGAAAAAC | Mutagenesis |
| Agbnil-11.2 IRO2 | CTGACGATGCGTTCCACGTAGTTCAAGAAAGTCATGCTGTTTTTCTCGTCTCTAATGAAG | Mutagenesis |
| AgBNRCFP-N | CCAGCATGTCAACCACTATATTGATCACCGATATATGGACTTCCACACCAACTAGacgcggccgccagctgaagc | In vivo recombination |
| AgBNI1-GS2 | GCGGCGCGGCGGCGACATCGCGCTGGTCTATCAGTTTCTTGGTGCGGCagggacctggcacggagc | In vivo recombination |
| AgRHO1a-ATG | GAGATCGAATTCATGGCGTACCAGACAGGCGGCA | Cloning |
| AgRHO1aG204C | GCCGGTCGTAGTCCTCcTGGCCCGCTGTATCCC | Mutagenesis |
| AgRHO1a-TAG | CGACGGATCCCTTTCTTCTTCTTCTTGTCACCGT | Cloning |
| AgRHO1b-ATG | GATCGAATTCATGTCTCAGCAAATGCATAAC | Cloning |
| AgRHO1bG207C | GCCTGTCGTAATCCTCGTGCCCAGCCGTATCC | Mutagenesis |
| AgRHO1b-TAG | CGACGGATCCCCTTTTTCTTCTTCTTCTTGTCAGAC | Cloning |
| AgRHO2-ATG | GAGATCGAATTCATGACGGTCAACGTTGTGAGAC | Cloning |
| AgRHO2G195C | CAGACGCTCGTATTCTTCGTGACCAGCAGTATCC | Mutagenesis |
| AgRHO2-TAG | CGACGGATCCCGCCTTGACCCGGCTCCTTGTTCAC | Cloning |
| AgRHO3-ATG | GATCGAATTCATGCCTCTGTGTGGGTCGAG | Cloning |
| AgRHO3G219C | GCAACCGGTCAAACTCCTCGTGCCCAGCAGTGTCCCACAGG | Mutagenesis |
| AgRHO3-TAG | GACGGATCCCACTGCTGCTTTCGGCCTCGG | Cloning |
| AgRHO4-ATG | GAGATCGAATTCATGAGCGCAGGGCCGTTGCAAG | Cloning |
| AgRHO4G333C | CAGCCGGCTGTACTCCTCGTGCCCAGCAGTGTCCCACAGC | Mutagenesis |
| AgRHO4-TAG | GACGGATCCCGCGGTGCTTGCGCACCCGCTTG | Cloning |
| AgRHO5-ATG | GATCGAATTCATGTGTTTTTCGCAGAGCGGCAG | Cloning |
| AgRHO5G255neu | CCGCAACCGGTCGTACTCCTCGTGCCCCGCCGTGTCCCACAG | Mutagenesis |
| AgRHO5-TAG | CGACGGATCCCTCTAGATCTCTTCCTCCTCTTTTTC | Cloning |
| AgCDC42-ATG | GATCGAATTCATGCAGACATTGAAGTGCGTGGTC | Cloning |
| AgCDC42-TAG | CGACGGATCCCCTTCTTGCTCTTCTTGATG | Cloning |
| AgCDC42G183C | GCCGCAACCTGTCGTAGTCCTCGTGGCCGGCAGTGTCGAACAAGC | Mutagenesis |
| u-40 | GTTTTCCCAGTCACGAC | Mutagenesis |
| Reverse | CAGGAAACAGCTATGACCATG | Mutagenesis |
| Sec4-ECO | cgcgaattcTCGGGGCTAAGAACGGTGTC | Cloning |
| Sec4P-BAM | ccggatccCATTATAGCTACTGTACTGC | Cloning |
| Sec4P-HIND | ggaagcttCTACTACGCCTGAGCGCCGC | Cloning |
| Sec4-SPE | caccaactagtTGGGCGGCCTGTGCTGCAAAG | Cloning |
| GFP-BAM | cgggatccAGTAAAGGAGAAGAACTTTTCAC | Cloning |
| GFP-ECO | cgcgaattcTTTGTATAGTTCATCCATGCC | Cloning |
| AgBNI1-ATG2Hy | GTAGTAACAAAGGTCAAAGACAGTTGACTGTATCGCCGGAATTCatgaagaagtccacgcactcgaac | In vivo recombination |
| AgBNI1-TAG2Hy | CATAAGAAATTCGCCCGGAATTAGCTTGGCTGCAGGTCGACGGATCCttacttgtgctcatcgagcattg | In vivo recombination |
| AgBNI1-P_HindIII | gcgcgcaagcttCTGGGACGACGACAAGCCTAC | Cloning |
| AgBNI1-T_Smal | gcgccccgggCCTGCACCGAAATCCGCCGA | Cloning |
| AgBNI1-GS1 | CGGTGAACACCCGGAGTCTCGCAAGTCAATGCTCGATGAGCACAAGggtgcaggcgctggagctg | In vivo recombination |
| AgBNI1-GS2 | GCGGCGCGGCGGCGACATCGCGCTGGTCTATCAGTTTCTTGGTGCGGCagggacctggcacggagc | In vivo recombination |
| AgBNI1-NS1 | GGTATAGTGGCAGCGTCGGCGGCTGGGCACATATGCAAGGccagtgaattcgagctcgg | Deletion |
| AgBNI1-NS2 | GCTGGTCTATCAGTTTCTTGGTGCGGCGCTGGCGAACCTTtacgccaagcttgcatgcct | Deletion |
| Gal4-ad | GTTTGGAATCACTACAGGG | sequencing |
| Gal4-bd | GATTGGCTTCAGTGGAG | sequencing |
| Gad-term | GAGATGGTGCACGATGCACAGTTG | sequencing |
| Gbt-term | CGTTTTAAAACCTAAGAGTCAC | sequencing |
| AgCdc42_c1 | GGACGAGCCGTACACGTTGGGCTTGTTCGACACTGCCGGGgagctcgttttcgacactgg | pCORE integration |
| AgCdc42_c2 | GTCGACGGGTACGACAACGGCCGCAACCTGTCGTAGTCCTCtccttaccattaagttgatc | pCORE integration |
| AgCdc42_Q-H-antisense | cacgtccgtcgacgggtacgacaacggccgcaacctgtcgtagtcctcgtgcccggcagtg | Mutagenesis |
| AgCdc42_Q-H-sense | gtgatgatcggggacgagccgtacacgttgggcttgttcgacactgccgggcacgaggac | Mutagenesis |
| INT_CDC42*_S1 | ACCAAGTGCCAGCTCTAGCGCCAGGACCCGCGCGAGACCTGCTAGGGATAACAGGGTAAT | Mutagenesis |
| INT_CDC42*_S2 | TTGCCGATTTTTGGGCAGGGCGCAAAGACAGATATAGCAGAGGCATGCAAGCTTAGATCT | Mutagenesis |
| AgCDC42-ATG | GATCGAATTCATGCAGACATTGAAGTGCGTGGTC | Verification |
| AgCDC42-TAG | CGACGGATCCCCTTCTTGCTCTTCTTGATG | Verification |
| AgBNI1dD-F1 | GACTTTATTCTGGAGTACAAGCGCGCGCAGGAGTTTAACCGCAAGATCaaaacgacggccagtgaattcg | In vivo recombination |
| AgBNI1dD-F2 | GACATCGCGCTGGTCTATCAGTTTCTTGGTGCGGCGCTGGCGAACCTTaccatgattacgccaagcttgc | In vivo recombination |
Underscore indicates nucleotides exchanged for mutagenesis.
DNA Manipulations and Sequencing
All DNA manipulations were carried out according to Sambrook et al. (2001) with DH5αF′ as host (Hanahan, 1983). Sequencing was done using an ABI prism 377 DNA sequencer (PE Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. Plasmids were isolated from yeast using the High Pure Plasmid Purification Kit (Roche Diagnostics, Mannheim, Germany) according to the instruction manual for plasmid preparation from Escherichia coli but with the following modifications: 5 ml of a yeast culture was collected by centrifugation and the supernatant was discarded. The cells were resuspended in solution 1 and 0.2 g of 0.5-mm glass beads were added. Cells were lysed by vortexing for 8 min at 4°C. From here on the instructions of the manufacturer were followed, only the elution volume was decreased to 20 μl. Ten microliters of the elute were transformed into E. coli for plasmid amplification.
Plasmids and Constructs
All plasmids are listed in Table 3. All constructs carrying genes for Rho-type GTPases were constructed using the same scheme. Each gene was amplified from A. gossypii genomic DNA using the corresponding primers from Table 2 for PCR. All primers were designed to amplify the open reading frame (ORF) from the start codon to the end, excluding the nucleotides coding for the CAAX motif at the carboxy terminus, thus avoiding lipid modification (primers AgRHOx-ATG, AgRHOx-TAG and AgCDC42-ATG, and AgCDC42-TAG). To facilitate cloning, EcoRI and BamHI restriction sites were added to the oligonucleotides. The fragments were purified, cut, and ligated into pUC19 (Vieira and Messing, 1991). Activated alleles of all Rho-GTPases were constructed using the method by Boles and Miosga (1995) with primers named AgRHOx and AgCDC42 plus the corresponding nucleotide exchange. DNA of wild-type and mutant alleles was cloned into pGBT9 (Bartel et al., 1993) using again EcoRI and BamHI. The resulting plasmids were verified by sequencing. AgBNI1 was amplified by PCR from genomic A. gossypii DNA using primers AgBNI1-ATG2Hy and AgBNI1-TAG2Hy, which add on both sides of the amplified AgBNI1 45 base pairs homologous to pGBT9 for in vivo cloning. This PCR product was cotransformed with EcoRI- and BamHI-digested pGBT9 into the yeast strain DHD5. Transformants were restreaked on selective medium, and recombinant plasmids were isolated from the yeast cells and amplified in E. coli. This resulted in pGBT9AgBNI1. From this, a 2.5-kb EcoRI-BglII fragment was cut out and ligated into the EcoRI-BamHI sites of pGAD424 (Bartel et al., 1993). Again the plasmid was first verified by digestion and sequencing.
Table 3.
Plasmids
| Name | Insert | Reference |
|---|---|---|
| pUC19 | — | Vieira and Messing (1991) |
| pGBT9 | — | Bartel et al. (1993) |
| pGAD424 | — | Bartel et al. (1993) |
| YCPlac111 | — | Gietz and Sugino (1988) |
| pRS415 | — | Sikorski and Hieter (1989) |
| pUC19NATPS | — | D. Hoepfner, personal communication |
| pAGT141 | pUC19 carrying the PstI fragment from pGEN and the SacI fragment from pGUG | A. Kaufmann, personal communication |
| pCORE | — | Storici et al. (2001) |
| pUC19AgRHO1a | A. gossypii RHO1a from nucleotide 1-615 | This study |
| pUC19AgRHO1b | A. gossypii RHO1b from nucleotide 1-609 | This study |
| pUC19AgRHO2 | A. gossypii RHO2 from nucleotide 1-546 | This study |
| pUC19AgRHO3 | A. gossypii RHO3 from nucleotide 1-660 | This study |
| pUC19AgRHO4 | A. gossypii RHO4 from nucleotide 1-765 | This study |
| pUC19AgRHO5 | A. gossypii RHO5 from nucleotide 1-870 | This study |
| pUC19AgCDC42 | A. gossypii CDC42 from nucleotide 1-561 | This study |
| pGBTAgRHO1a | A. gossypii RHO1a from nucleotide 1-615 | This study |
| pGBTAgRHO1b | A. gossypii RHO1b from nucleotide 1-609 | This study |
| pGBTAgRHO2 | A. gossypii RHO2 from nucleotide 1-546 | This study |
| pGBTAgRHO3 | A. gossypii RHO3 from nucleotide 1-660 | This study |
| pGBTAgRHO4 | A. gossypii RHO4 from nucleotide 1-765 | This study |
| pGBTAgRHO5 | A. gossypii RHO5 from nucleotide 1-870 | This study |
| pGBTAgCDC42 | A. gossypii CDC42 from nucleotide 1-561 | This study |
| pGBTAgRHO1a* | A. gossypii RHO1a G204C from nucleotide 1-615 | This study |
| pGBTAgRHO1b* | A. gossypii RHO1b G207C from nucleotide 1-609 | This study |
| pGBTAgRHO2* | A. gossypii RHO2 G195C from nucleotide 1-546 | This study |
| pGBTAgRHO3* | A. gossypii RHO3 G219C from nucleotide 1-660 | This study |
| pGBTAgRHO4* | A. gossypii RHO4 G333C from nucleotide 1-765 | This study |
| pGBTAgRHO5* | A. gossypii RHO5 G255C from nucleotide 1-870 | This study |
| pGBTAgCDC42* | A. gossypii CDC42 from nucleotide 1-561 | This study |
| pGBTAgBNI1-N | A. gossypii BNI1 from nucleotide 1-3152 | This study |
| pGADAgBNI1-N | A. gossypii BNI1 from nucleotide 1-3152 | This study |
| YCp111GFPSEC4 | N-terminal Fusion of GFP to A. gossypii SEC4 | This study |
| pAMK1 | A. gossypii BNI1 from nucleotide -682 to +523 | This study |
| pCDC42 | AgCDC42 including promoter and terminator | This study |
| pCDC42cas | Agcdc42 with codon CAG starting at position 181 replaced by pCORE | This study |
| pcdc42cons | pCDC42 with codon CAG starting at position 181 altered to CAC | This study |
| pcdc42kanr | pcdc42cons with integrated GEN3 cassette behind ORF | This study |
| pUC19cdc42cons | 3.1 kb BamHI/PstI fragment from pcdc42kanr with pUC19 backbone | This study |
| pSEC4NAT | PstI fragment from pUC19NATPS carrying ClonNAT cassette ligated into PstI site of YCP111GFPSEC4. Orientation reverse | This study |
The plasmid carrying AgBNI1 (pAMK1) was constructed by ligation of an HindIII/SmaI fragment generated by PCR using primers AgBNI1-P_HindIII and AgBNI1-T_SmaI from genomic A. gossypii DNA. GFP was fused to the gene by in vivo recombination in S. cerevisiae strain DHD5 with a PCR fragment derived from pGUG with primers AgBNI1-GS1 and AgBNI1-GS2. The fusion was verified by sequencing.
An amino-terminal fusion of AgSEC4 to GFP was constructed by a sequential series of PCR amplification and cloning steps. First, the AgSEC4 promoter was amplified by PCR out of the genome using primers Sec4P-Hind and Sec4P-Bam and cloned into YCPlac111 using HindIII and BamHI. The resulting plasmid was cut with BamHI and EcoRI to allow fusion of the SEC4 promoter to GFP that was amplified by PCR from pGUG with primers GFP-Eco and GFP-Bam. In a final step the AgSEC4 ORF and terminator were fused to the GFP-ORF using the enzymes EcoRI and SpeI and a fragment generated from genomic DNA using primers Sec4-Eco and Sec4-Spe.
The S1642G allele of AgBNI1 was generated using the method described by Storici et al. (2001) using primers Agbni1–12C1, Agbni1–12C2, Agbni1–12IRO1, and Agbni1–12IRO2 and S. cerevisiae strain CEN/PK2 (http://www.rz.uni-frankfurt.de/FB/fb16/mikro/euroscarf/) transformed with pAMK1. The resulting plasmid pAMK1ts12 was isolated from yeast digested with AscI and dephosphorylated. The linearized vector was cotransformed together with a PCR product generated with primers AgBNRCFP-N and AgBNI1-GS2 from pGUG for in vivo recombination to integrate the URA3 terminator from S. cerevisiae behind the mutagenized Agbni1 gene. The resulting vector was isolated and verified by sequencing of the altered regions. An XbaI fragment containing the altered region of AgBNI1, the URA3 terminator and the G418 resistance cassette was cloned into pUC19. The resulting plasmid pHPS232 was cut again with XbaI and transformed into the A. gossypii ΔlΔt strain. Construction of the D1530R,K1620R allele of AgBNI1 was identical to the S1642G allele except for a sequential round of mutagenesis first using primers Agbni1–11.1C1, Agbni1–11.1C2, Agbni1–11.1IRO1 and Agbni1–11.1IRO2 and Agbni1–11.2C1, Agbni1–11.2C2, Agbni1–11.2IRO1 and Agbni1–11.2IRO2 respectively.
For construction of the Q61H allele of AgCDC42 the gene was first PCR-amplified from genomic A. gossypii DNA using primers cdc42genom_sense and cdc42genom_antisense. The resulting product was cut with EcoRV, and PstI was ligated into pRS415 cut with SmaI and PstI. In the resulting plasmid pCDC42 the codon CAG starting at position 181 was deleted by integration of the pCORE-cassette (Storici et al., 2001) using primers AgCdc42_Q-H-sense and AgCdc42_Q-H-antisense. This resulted in plasmid pCDC42cas. By homologous recombination in yeast with annealed primers and counterselection, the pCORE-cassette was replaced by the mutagenized codon, resulting in pcdc42cons. The Gen3 resistance cassette was amplified from pGEN3 using primers INT_CDC42*_S1 and INT_CDC42*_S2. The resulting PCR product was integrated behind the cdc42 gene in the vector pcdc42cons yielding pcdc42kanr. The 3.1-kb BamHI/PstI fragment from pcdc42kanr carrying the mutagenized cdc42 and the resistance cassette was cloned into pUC19, resulting in pUC19cdc42cons. DNA of this vector was again cut with BamHI/PstI and transformed into A. gossypii. Transformants resistant against G418 were sporulated and spores were separated to get homokaryotic Geneticin (G418) resistant mycelium. From this mycelium the CDC42 gene was amplified by PCR using primers AgCDC42-ATG and AgCDC42-TAG. The resulting PCR product was sequenced to verify the presence of the desired mutation.
Image Acquisition and Processing
The microscopy setup used was the same as described in Knechtle et al. (2003). Actin staining was done according to Knechtle et al. (2003). The illumination time and light intensity for standard phase contrast, DIC, or fluorescence acquisitions was chosen to reach minimally 25% of the maximal intensity. For multiple exposures of the same sample bleaching of the sample had to be taken into account. The Z-distance between two planes in stack acquisitions was set between 0.1 and 0.5 μm. Phase contrast, DIC, and single-plane fluorescence images were processed using the “scale image” and “unsharp mask” feature in MetaMorph (Universal Imaging, West Chester, PA). Stacks were deblurred with MetaMorph's “remove haze,” flattened by maximum projection with “stack arithmetics” and scaled as mentioned above. Fluorescence and phase-contrast images were colored and overlaid using MetaMorph's “overlay images” tool. For time-lapse acquisition spores were cultured on a slide with a cavity (time-lapse slide) that was filled with solid medium (Ashbya Full Medium). Spores were incubated in a humid chamber without coverslip until they reached the required developmental stage. Then a coverslip was applied. For Supplementary Movies S1 to S7 cells or spores were cultured on Petri plates with AFM agar, cultivated in a humid chamber, and placed for image acquisitions under a light microscope. The acquisition frequency varied between 1 and 0.2 min–1. The time-lapse picture series were exported from MetaMorph as 8-bit TIFF files converted to a QuickTime (Apple Computer, Redmond, WA) or AVI movie with Adobe Premiere 4.2 (Adobe Systems, San Jose, CA).
Two-hybrid Experiments
For two-hybrid experiments prey and bait plasmids were cotransformed into S. cerevisiae strain PJ69-4a (James et al., 1996) and selected on minimal medium lacking leucine and tryptophan but containing a fourfold concentration (80 mg/l) of adenine. Transformants were grown in the same liquid medium to an OD600 of ∼1. Of these, 5 μl were spotted on plates lacking in addition to leucine and tryptophan either histidine or adenine to monitor activity of the reporter genes.
RESULTS
Filamentous Growth of A. gossypii Compared with Unicellular Growth of S. cerevisiae
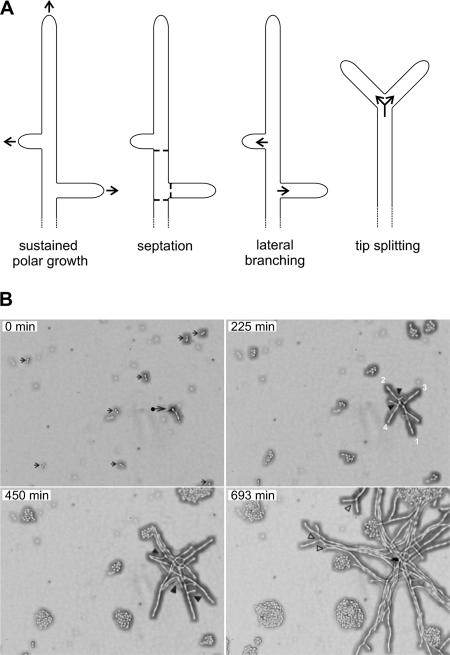
A. gossypii grows like a filamentous ascomycete (Momany, 2002) and the main differences to yeastlike growth are shown in Figure 1A. Sustained polar growth results in continuous tip extensions of hyphae and lateral branches. Thus, growth of the hyphal surface is restricted to the tips, and isotropic growth phases are absent. In a yeast cell, a short phase of polar growth of the emerging bud is followed by isotropic growth of the bud. Then a septum grows at the junction between the mother cell and the bud and both cells separate. Septation in A. gossypii like in other filamentous ascomycetes occurs in the growing hyphae and at the base of branches but separation of hyphal segments does not occur. Lateral branching leads to the successive establishment of several novel axes of polarity at the cortex of growing hyphae, in A. gossypii usually in an about right angle from the growth axis of the mother hyphae. In budding yeast, only one novel axis of polarity (site of bud emergence) is established in each cell cycle. Finally, the process of dividing the polarity-axis at the tip of mature hyphae (tip splitting) is restricted to a subset of filamentous ascomycetes including A. gossypii. Budding yeasts do not simultaneously generate two buds (two axes of polarity).
Figure 1.
Differences in polar growth between A. gossypii and S. cerevisiae. (A) Schematic presentation of landmarks of hyphal growth of A. gossypii: Sustained polar growth (accelerating growth along a selected polarity-axis by continuous surface expansion at the tip), septation (growth of chitin-containing open rings at 30–50-μm distances at the cortex of hyphae and at the base of branches), lateral branching (successive selection of new axes of polarity at the cortex of elongating hyphae), and tip splitting (simultaneous generation of two new axes of polarity at fast growing tips). (B) Simultaneous monitoring of A. gossypii and S. cerevisiae growth. A. gossypii spores and S. cerevisiae cells were mixed and plated on AFM agar. After 4 h the agar surface was screened under the microscope. An area was selected for the start of the video (top left panel) showing eight S. cerevisiae cells after their first division (small arrows) and one germinated A. gossypii spore with the first emerged hypha (•). Growth in this area was monitored for 15 h at 25°C with pictures taken every 3 min (Supplementary Movie S1). After 225 min, the S. cerevisiae mini-colonies consist of four to eight cells, whereas the A. gossypii young mycelium has already developed nine extending hyphae. Two emerged, in addition to the first hypha, from the germinated spore and six originated from the initiation of lateral branches at the cortex of growing hyphae (two branches are marked by black arrow heads). At 450 min, the S. cerevisiae minicolonies have 12–32 cells and the A. gossypii young mycelium now consists of 23 hyphae due to 14 newly emerged lateral branches (black arrow heads). At 690 min the fastest hyphae, marked with open arrowheads, have started to symmetrically divide at their tips. Ten additional tip-splitting events were observed until the end of the movie at 897 min. The elongation speeds of the main hyphae during early mycelial development are ∼10 μm/h (0 min), 15 μm/h (225 min), and 25 μm/h (450 min), as determined from the elongation of the four hyphae marked in frame 225 min, which were measured every four frames until 501 min. For technical reasons 21 frames were not recorded during the early period between the frames representing 132 and 135 min. To determine hyphal speeds at tip splitting hyphal elongations were measured during 18 frames before and after tip splitting. This mode of branching starts at elongation speeds of 80 μm/h. The highest speed observed at the end of Supplementary Movie S1 before a tip splitting was 140 μm/h. Four cases of consecutive tip splitting events were observed with an average distance of 190 μm and an average time interval of 140 min. Scale bar, 20 μm.
To visualize the differences in growth dynamics of S. cerevisiae and A. gossypii, we simultaneously monitored, by time-lapse video microscopy, growth of both fungi on AFM agar (Movie S1). Four representative frames were selected to show the development of S. cerevisiae mini colonies and of an A. gossypii mycelium (Figure 1B). Arrows in the zero minute frame point to eight S. cerevisiae cells, which have just finished the first division, and to one germinated A. gossypii spore from which the first hypha has emerged. After 225 min, the S. cerevisiae cells have undergone only one or two additional divisions whereas the A. gossypii young mycelium has already developed nine growing hyphae. At 450 min, the S. cerevisiae cells have divided one or two more times, and the A. gossypii young mycelium is now spreading from 23 hyphal tips. After nearly 700 min, the fastest hyphae (marked with open arrow heads) have started to symmetrically divide at their tips, the common mode of branching when hyphal tip speeds have reached 80 μm/h or more. This switch in branching pattern allows optimal radial spreading of an A. gossypii mycelium. The apparently symmetric division of the polar growth machinery leads only to a transient decrease of tip elongation speeds by ∼20% as determined for seven tip splitting events in Supplementary Movie S1 (unpublished data). Using similar conditions, Knechtle et al. (2003) presented the dynamics of successive tip splitting events every 90–120 min at the edge of a radially expanding mature A. gossypii mycelium.
How can these differences in morphogenesis be explained when both organisms carry a very similar set of proteins? One possible answer is that homologous polar growth components may exert not only common functions but also distinct functions in the cellular networks of both organisms. One example for shared and specific functions is AgSpa2p, the first component identified in A. gossypii to permanently localize at hyphal tips, where it plays an essential role to reach maximal tip speeds (Knechtle et al., 2003). These are properties specific for A. gossypii, because ScSpa2p, the homolog in S. cerevisiae, does not localize permanently but only transiently to the bud tip and because a cellular process, controlling maximal bud tip speed, is unknown in morphogenesis of S. cerevisiae (Sheu et al., 1998). Because ScSpa2p is part of the polarisome, we speculated that A. gossypii homologues of other components of the polarisome complex, in particular the formin homology protein ScBni1p (Evangelista et al., 2002; Sagot et al., 2002a), could fulfill, in addition to common functions, also specific functions in sustained hyphal tip growth and tip splitting. Therefore, we searched for homologues in A. gossypii to ScBni1p and the related formin ScBnr1p and investigated to which degree the homologues would contribute to sustained hyphal tip growth and tip splitting.
The A. gossypii Formin Family Members
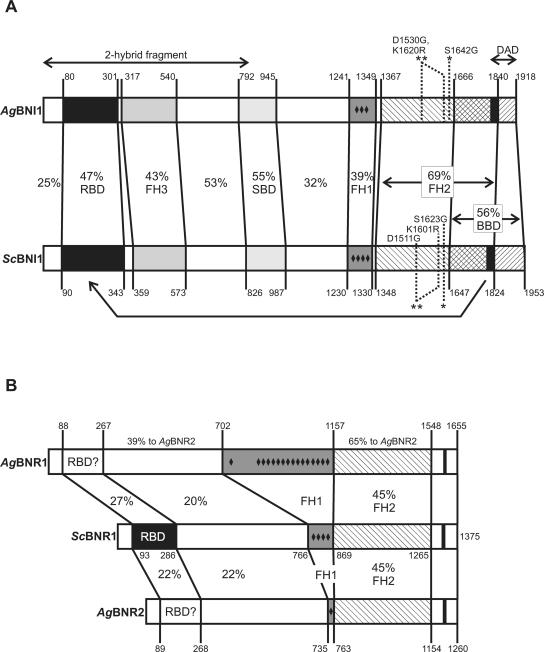
Using FASTA database searches (Pearson, 1990), we identified three formin homology members in the genome of A. gossypii (Dietrich et al., 2004). By combining information from homology searches with synteny, these proteins can be categorized as follows: one syntenic homolog to the S. cerevisiae BNI1 gene (AFR669W, in the following referred to as AgBNI1) and one syntenic and one nonsyntenic, telomere-located homolog to the S. cerevisiae BNR1 gene (AFR301C and AGL364C, referred to as AgBNR1 and AgBNR2, respectively). The formins ScBni1p and AgBni1p have almost the same size and domain composition (Figure 2A) using the domain definition from Evangelista et al. (2002). The two homologues to ScBnr1p differ much more in size and, interestingly, these size differences are located exclusively within the FH1 domains leading to differences in the number of short polyproline stretches (Figure 2B).
Figure 2.
Domain comparisons between formin homologues of S. cerevisiae and A. gossypii. (A) Comparison of AgBni1p and ScBni1p. The domain structure of ScBni1p is based on the one published by Evangelista et al. (2002). Domain borders were first determined using the gap program of the GCG Wisconsin Package (Accelrys), which uses an Needleman-Wunsch algorithm (Needleman and Wunsch, 1970). Each domain was realigned and the identity values were taken from the output. All distances are drawn to a relative scale. RBD, Rho-binding domain; FH1–3, Formin Homology domain 1–3; SPD, Spa2-binding domain; BBD, Bud6-binding domain; DAD, diaphanous autoregulatory domain, which is drawn as black bar within BBD. The point mutations and the DAD deletion used in this work are indicated as well as the boundaries of the amino-terminal fragment used in two-hybrid studies. (B) Comparison of AgBnr1p and AgBnr2p with ScBnr1p. The domain structure of ScBnr1p is based on that of ScBni1p. Domain borders were determined and marked as described in A. No clear indication of an RBD was found in the two A. gossypii homologues. Small black diamonds indicate positions of short stretches of two or more prolines that are not separated by more than one other residue.
It was shown that the FH1 domain of formins can mediate interactions with profilin, an abundant actin-binding protein, thereby sequestering actin monomers (Pruyne et al., 2002; Sagot et al., 2002b; Moseley et al., 2004). The FH2 domain provides the activity for nucleation of actin cables (Pruyne et al., 2002; Sagot et al., 2002a; Kovar et al., 2003; Harris et al., 2004; Moseley et al., 2004). This domain is the most conserved and best characterized domain within formins and was used as a basis for the phylogenetic classification of over a hundred formins (Higgs and Peterson, 2005). The FH3 or Formin Homology 3 domain is only loosely conserved among formins and is defined by its role, in S. cerevisiae, to help locating ScBni1p to the bud tip (Ozaki-Kuroda et al., 2001).
Other domains shown in Figure 2 are known to be involved in diverse protein interactions in S. cerevisiae. The Spa2-binding domain (SBD) and to a lesser degree the Bud6-binding domain (BBD) are important for localization of ScBni1p to the growing bud tip (Ozaki-Kuroda et al., 2001; Sagot et al., 2002a). The Rho-binding domain (RBD) interacts with small GTPases of the Rho-type and also undergoes an intramolecular association with a short carboxy-terminal domain called diaphanous autoregulatory domain (DAD). This association, indicated by the arrow in Figure 2A, prevents actin cable assembly unless an activated GTPase binds to this domain (Alberts, 2001). A short sequence similar to the known functional DAD residues of ScBni1p is also found in all members of the Bnr1 group (small black bars in Figure 2B) but its function remains unclear. Importantly, in contrast to ScBnr1p, am RBD could not be identified in the A. gossypii homologues searching for this domain (DRF_GBD) in PFAM version 17.0. This result is consistent with our finding that neither full-length nor amino-terminal parts of AgBnr1p and AgBnr2p bind to wild-type or activated forms of all A. gossypii Rho-GTPases in a two-hybrid test (unpublished data).
Because the two S. cerevisiae formins have partially overlapping functions allowing, for example, single deletions to grow, we expected, based on the domain comparisons, also to find overlapping functions in the three formins of A. gossypii.
Single Deletions of Agbnr1 and Agbnr2 Are Viable But Not the Double Deletion
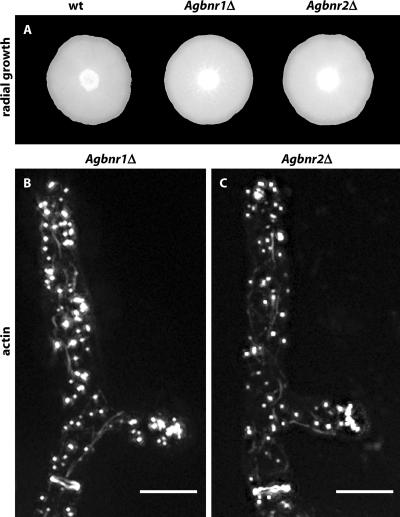
To investigate the cellular function of the two homologues of ScBnr1p, we first deleted the entire ORFs of AgBNR1 and AgBNR2 by PCR-based gene targeting using a cassette coding for resistance against geneticin (G418). Young mycelium with 20–40 nuclei was transformed to geneticin-resistance. These primary transformants are heterokaryotic because their haploid nuclei carry either the wild-type or the deletion allele. Homokaryotic deletion transformants were obtained from spores that contain single nuclei and that were dissected on selective agar medium. Growing homokaryotic transformants were rechecked for the loss of the wild-type and presence of the deletion allele and were phenotypically characterized.
As can be seen in Figure 3A, the mycelial development of both single deletions is not perturbed. Consistent with this wild type-like growth, all actin structures can be observed by Alexa-phalloidin staining as shown in Figure 3, B and C. Staining with calcofluor white revealed an increased number of incomplete chitin rings in the Agbnr2 deletion, and a fusion with GFP revealed that at least a fraction of the AgBnr2p-GFP proteins locates at septa, whereas no fluorescence was detected at the tips (unpublished data). Several attempts to locate an AgBnr1-GFP fusion remained unsuccessful.
Figure 3.
Wild type-like growth of Agbnr1 and Agbnr2 deletions. (A) Mycelial colonies from A. gossypii wild-type and single deletion strains. Small aliquots of mycelia were transferred to the middle of full medium plates and grown for6dat 30°C. Bar, 1.0 cm. (B and C) Visualization of actin patches, actin cables, and actin rings by staining young mycelium of the respective deletion strains with Alexa-phalloidin (Knechtle et al., 2003). In both panels a growing tip is at the top and an actin ring is seen at the bottom below an emerged lateral branch. Bars, 5 μm.
The viability of both deletions indicates that the two AgBnrp homologues probably encode overlapping functions. The genomic arrangement of the A. gossypii BNR genes indeed suggests that AgBNR2 most likely originates from a duplication of an ancient AgBNR1 gene involving transposition of one copy to the telomere region of chromosome VI. We therefore constructed and tested double deletions of both homologues. Because of the lack of a functional mating system in A. gossypii, we had to delete the AgBNR2 gene in a strain already deleted for the AgBNR1 gene. This resulted in transformants that are homokaryotic for the Agbnr1 deletion (ClonNAT resistance marker) and heterokaryotic for the Agbnr2 deletion (geneticin resistance marker). PCR-verified heterokaryotic transformants were sporulated and spores were dissected on different selection media. Spores were viable on ClonNAT, selecting for the Agbnr1 deletion, but nonviable on geneticin, selecting for the Agbnr1/Agbnr2 double deletion (n > 100). To avoid the possibility that the Agbnr1 deletion strain used to construct the double deletion carried an unknown mutation, which is colethal with the introduced Agbnr2 deletion, we repeated the experiment with a newly constructed Agbnr1 transformant as the recipient strain, now marked by geneticin resistance. Then we used leucine auxotrophy as selection for the Agbnr2 deletion. Again, we were unable to isolate any mature mycelium carrying a homokaryotic Agbnr1/Agbnr2 double deletion. To see if the nonviable spores showed residual growth, we looked for germination structures under the microscope. None of the spores formed a germ bubble (n > 100), indicating a common function of AgBnr1p and AgBnr2p essential very early in germination.
Deletion of AgBNI1 Is Lethal
To investigate the functions of AgBNI1, we first deleted the entire ORF. More than 100 spores obtained from the primary heterokaryotic mycelium were dissected and placed on selective medium. None of these spores gave rise to a mature mycelium. Thus, deletion of AgBNI1 is lethal despite the presence of the two other formin genes, AgBNR1 and AgBNR2, indicating an essential role of the formin AgBni1p. In S. cerevisiae, mutants with deletions of either one of the two formin genes, ScBNI1 and ScBNR1, are viable and only deletion of both genes is lethal (Imamura et al., 1997).
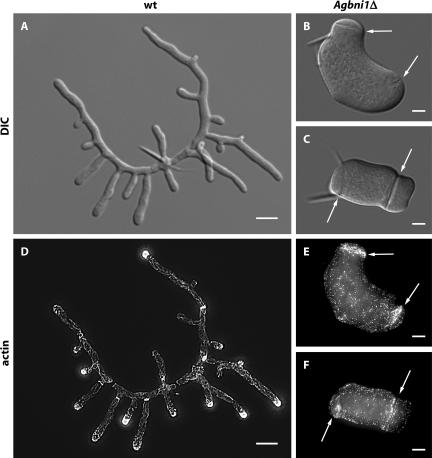
Microscopic observation of Agbni1Δ (Figure 4) revealed that spores started swelling like wild type, but were not able to initiate hyphal growth. Instead, irregular isotropic growth proceeded for at least 20 h, generating giant, potatolike cells containing over 100 nuclei. Because formins were reported to catalyze actin cable polymerization (Pruyne et al., 2002; Sagot et al., 2002b), we investigated the actin cytoskeleton of Agbni1Δ stained with Alexa-phalloidin. Long, polarized actin cables, reaching the tips, have been reported for A. gossypii wild type (Knechtle et al. (2003) and Figure 4D). The images in Figure 4, E and F, show that Agbni1 deletion mutants do not contain polarized actin cables and that actin patches are distributed all over the cell cortex. This is in contrast to wild-type hyphae, where patches are concentrated at the tips The only concentration of actin patches is observed along structures which, according to the DIC images in Figure 4, B and C, most likely mark sites of septum formation as described before (Knechtle et al., 2003). Thus, AgBNI1 might not be essential for septum formation but is required for actin cable formation and hyphal morphology.
Figure 4.
Lack of mycelial development after deletion of the single formin gene AgBNI1. Spores from wild type and from heterokaryotic Agbni1Δ transformants were grown over night at 30°C in AFM and AFM-Geneticin medium, respectively. DIC images are shown for wild-type (A) and Agbni1Δ (B and C), and fluorescence images for rhodamine-stained wild-type (D) and Alexa-phalloidin-stained Agbni1Δ (E and F). Arrows mark giant septumlike structures in Agbni1Δ that are visible by DIC and by actin staining. The needlelike structures attached to the middle of the young mycelium (A) and to one pole of the giant cells (B and C) originate from the spores, which have the shape of ∼20-μm-long needles. Bars, (A and D) 10 μm and 5 μm for all other images.
The loss of actin cable formation in the Agbni1 deletion raises an important question. Why is this loss not compensated, as in S. cerevisiae, by one of the two other formin homology members? The domain comparisons of Figure 2 do not reveal an answer because there is no significant difference between formin homologues of both organisms with respect to the domains responsible for actin cable formation. Therefore we hypothesized that AgBni1p is essential because its essential function may be connected to its location.
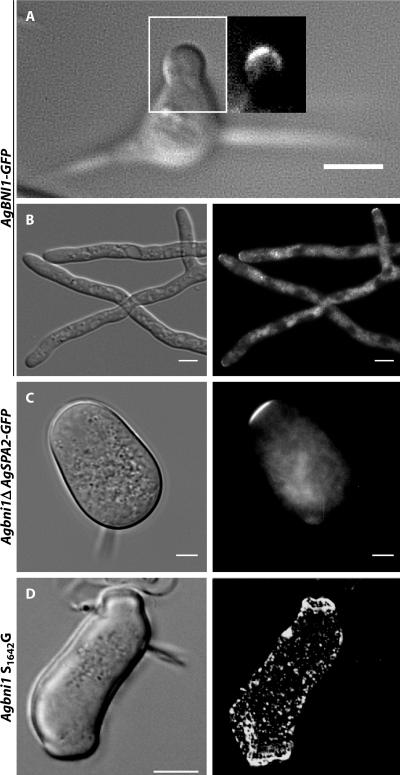
AgBni1p Locates to Hyphal Tips
To determine the location of AgBni1p, we constructed a GFP fusion at the genomic locus of AgBNI1 and first tested whether the fusion protein is functional. Spores expressing AgBni1p-GFP developed to a young mycelium with actin cables, actin patches, and actin rings indistinguishable from wild type. Mature hyphae displayed regular tip splitting, only their maximal elongation speed was slightly slower than wild type (unpublished data). Microscopic images revealed a very weak GFP signal, always located at the tip of hyphae, which was detectable in both, young mycelium (Figure 5A) and mature hyphae (Figure 5B). Interestingly, no signal could be detected at sites of septation in over hundred apical regions inspected. This is in contrast to S. cerevisiae where Bni1p-GFP localizes, in addition to polarized bud tips, also as ringlike structures at sites of septation (Ozaki-Kuroda et al., 2001), where it is most likely involved in cell separation, a process that does not occur in growing A. gossypii hyphae. Therefore the observed localization is consistent with an exclusive role for AgBni1p in tip growth.
Figure 5.
Localization of AgBni1p-GFP at hyphal tips. (A) Brightfield and fluorescence image of an A. gossypii germling carrying one copy of AgBNI1-GFP expressed from the endogenous BNI1 promoter. Because of high autofluorescence of spores, the fluorescence image is only shown for the area corresponding to the white rectangle. Scale bar, 5 μm. (B) Brightfield and fluorescence image of a mature hypha expressing AgBni1p-GFP from a genomic fusion gene. Scale bar, 5 μm. (C) Localization of AgSpa2p-GFP in Agbni1Δ. DIC image and fluorescence images of Agbni1Δ cells carrying a SPA2-GFP fusion at the AgSPA2 locus. Spores from heterokaryotic transformants were grown in AFM-Geneticin medium over night at 30°C. Scale bar, 5 μm. (D) Phenotype of the single S1642G substitution in the FH2 domain of AgBni1p. Spores from heterokaryotic transformants were grown over night in liquid AFM-Geneticin medium at 30°C and stained for actin with Alexa phalloidin. The fluorescence image (top) and the DIC image (bottom) were taken from the same cell. Scale bar, 10 μm.
Localization of the Polarisome Component AgSpa2p in the Agbni1Δ Mutant
One possible essential process AgBni1p that could be involved in at hyphal tips is the assembly or stabilization of a polarisome-like structure, which acts as a polarity control complex. For example, Bauer et al. (2004) could recently demonstrate that the GTPase AgBud1p is important to stably maintain the polarisome marker AgSpa2p-GFP at hyphal tips and that transient loss of this tip marker in an Agbud1 deletion causes an immediate arrest of polar growth. To test a possible essential role for AgBni1p in the assembly or stabilization of the “polarisome complex,” we deleted AgBNI1 in a strain expressing GFP-labeled AgSpa2p. AgSpa2p permanently localizes to the tips of elongating hyphae (Knechtle et al., 2003). In the giant Agbni1Δ cells AgSpa2p-GFP is always found at the cortex of the slowly expanding zone opposite from the spore needle (Figure 5C), indicating that AgBni1p does not play an essential role in polarizing AgSpa2p. The observed polarization may be the reason why giant Agbni1Δ cells form potato-shaped structures instead of large round cells, like mutants of Agcdc24 and Agcdc42, which are blocked in early steps of polarity establishment (Wendland and Philippsen, 2001).
Mutation of a Single Amino Acid in the FH2 Domain of AgBni1p Is Lethal
The experiments presented so far and the domain comparison of Bni1p from A. gossypii and S. cerevisiae discussed in Figure 2 suggest similar molecular properties of both proteins. One important property of AgBni1p and ScBni1p is the nucleation of actin cables, which resides within the FH2 domain. We asked whether the characteristic phenotype of the AgBNI1 deletion is only due to a tip-located loss of actin polymerization capability and subsequent loss of actin cables rather than loss of other functions. To test this, we constructed point mutations in the FH2 domain of AgBni1p, leading to the amino acid substitutions S1642G in one and D1530G, K1620R in a second mutant. Especially the residues mutated in the second mutant are highly conserved throughout all formin proteins (Higgs and Peterson, 2005). In S. cerevisiae, both homologous substitutions S1623G and D1511G, K1601R lead to temperature-sensitive strains in a background deleted for ScBNR1, the second formin gene (Evangelista et al., 2002). We expected to find temperature-sensitive phenotypes for both A. gossypii mutants. Surprisingly, the analogous substitutions in AgBni1p were lethal, even at 16°C. Like the complete deletion, these point mutants were able to form giant cells with many actin patches but without actin cables (Figure 5D). These experiments indicate that the tip-located assembly of actin cables represents the essential function of the AgBni1p formin, which emphasizes the importance of actin cables for hyphal growth.
Actin Cable-based Vesicle Transport Is Defect in Agbni1D
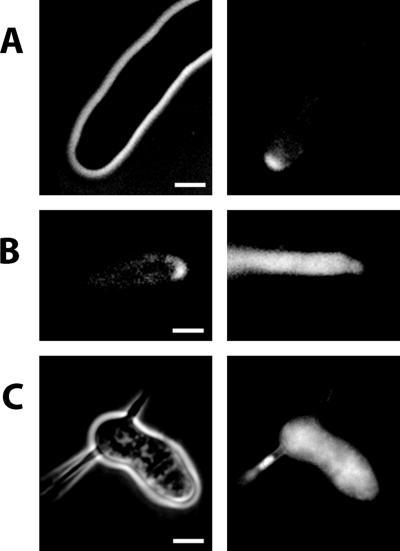
The loss of actin cables should severely impede delivery of secretory vesicles to hyphal tips. To confirm that AgBni1p is needed for secretory vesicle transport, we isolated the AgSEC4 gene by homology to the S. cerevisiae SEC4 gene and constructed a GFP-fusion. In budding yeast, a GFP-Sec4p fusion protein localizes to secretory vesicles and moves in a directed manner along actin cables toward the bud tip (Schott et al., 2002). We transformed A. gossypii with a plasmid expressing an amino-terminal GFP-AgSec4p fusion under control of the native AgSEC4 promoter. As shown in Figure 6A, the fusion product localizes mainly to the tip. Addition of latrunculin A, which disrupts actin structures, abolishes vesicular movement and apical localization, giving rise to a uniform fluorescence within hyphae (Figure 6B). To observe the localization of vesicles in the absence of AgBni1p, we transformed a heterokaryotic Agbni1Δ mycelium with the plasmid expressing the GFP-AgSec4p fusion protein. Spores of these transformants were grown under conditions selective for both, the Agbni1 deletion and the plasmid. In all growing giant cells the GFP fluorescence was evenly distributed (Figure 6C). This verifies that continuous tip-directed transport of secretory vesicles via actin cables is essential for hyphal growth.
Figure 6.
Distribution of GFP-Sec4p in wild type and Agbni1Δ. (A) Brightfield (left) and GFP-fluorescence (right) micrographs of a single wild-type hypha carrying a GFP-SEC4 fusion. The main fluorescence localizes to the tip. (B) Localization of GFP-Sec4p prior (left) and after (right) treatment with 200 μM latrunculin A for 1.5 min. (C) Brightfield (left) and GFP-fluorescence (right) images of Agbni1Δ carrying a GFP-SEC4 fusion gene. All strains were grown in liquid selective medium at 30°C over night. Scale bars, 5 μm.
Hyphal Morphogenesis in the Presence of Constitutively Active AgBni1p (AgBNI1ΔD)
We wanted to know whether an overactivation of the AgBni1 protein would increase hyphal elongation rates. We speculated that such a mutant should assemble more actin cables emanating from the tips, thus enhancing the transport rate of secretory vesicles toward the tip. To test this hypothesis, we deleted in AgBNI1 the coding sequence of the carboxy-terminal DAD (see Figure 2), thereby eliminating in the expressed protein the inactivating interaction of the DAD domain with the amino terminus.
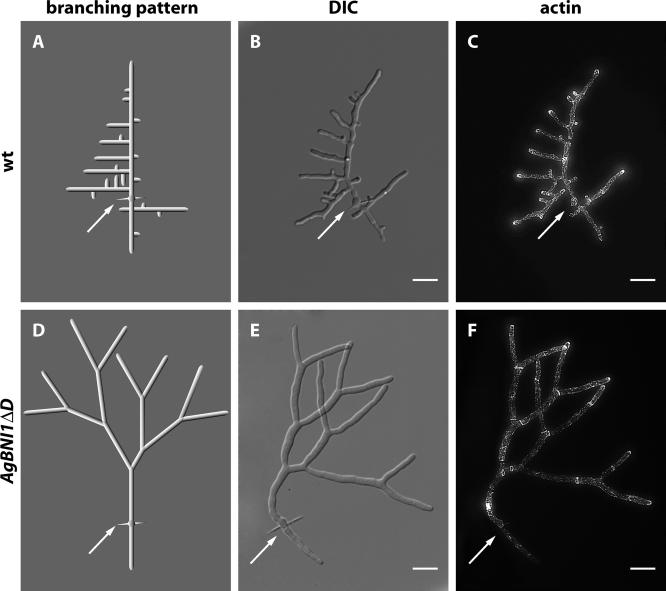
Several independent mutants were isolated. All show a complete change in branching pattern. Although morphogenesis of young wild-type mycelium exclusively displays lateral branching (Figure 7, A–C), the overactivation of the AgBni1p formin suppresses this branching mode and induces successive and symmetric hyphal splitting at tips (Figure 7, D–F). Lateral branching events occur only late and very rarely in this mutant, as concluded from videomicroscopy of 10 developing young mycelia. Examples are documented in Supplementary Movies S2-S4 for wild type and Supplementary Movies S5-S7 for AgBNI1ΔD (see Supplementary Material).
Figure 7.
Altered morphogenesis of AgBNI1ΔD mycelium. Wild-type and AgBNI1ΔD spores were incubated in liquid AFM medium at 30°C. After 18 h aliquots were stained with Alexa-phalloidin and mounted for microscopy. (A) Idealized branching scheme of the young wild-type mycelium shown as DIC image in B and as fluorescence image in C. The white arrows mark the needle of the spore from which two main hyphae developed in opposite directions. New branches always emerged distant from the tip and in an about right angle from the polarity-axis of the main hyphae, leading to a typical “Christmas tree” appearance. (D) Idealized tip splitting pattern of the young AgBNI1ΔD mycelium shown as DIC image in E and as fluorescence image in F. The white arrows mark the original position of the spore. Scale bar, 10 μm.
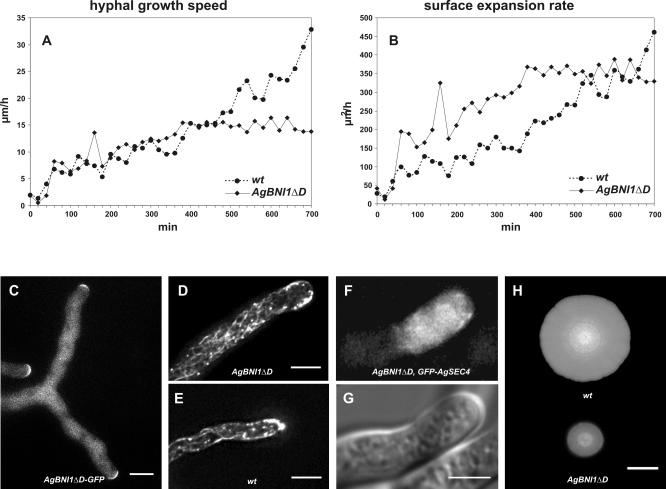
Quantitative evaluation of the movies reveals similar initial tip extension speeds for wild type and AgBNI1ΔD (Figure 8A). However, the average diameter of young hyphae of the mutant has almost doubled compared with wild type based on 300 measurements of three hyphae each (4.63 ± 0.46 μm for wild type and 7.52 ± 1.1 μm for mutant hyphae). These parameters allowed us to calculate for each time point the surface expansion rate, which, during the first 4 h, is up to three times higher in the mutant compared with wild type (Figure 8B). This increase in surface growth strongly indicates that an overactivation of AgBni1p can enhance the traffic of tip-directed secretory vesicles. This idea is supported by three experimental results. First, constitutively activated AgBni1ΔD proteins still locate to the tips (20/20) of hyphae (Figure 8C). Second, a two to three times higher number of actin cables can be observed in the apical region of mutant hyphae (Figure 8D) compared with wild-type hyphae (Figure 8E). And third, more vesicles are transported to the tip region as evident from the larger area of accumulated secretory vesicles in mutant hyphae (Figure 8, F and G) compared with wild-type hyphae (Figure 6A).
Figure 8.
Increase in surface expansion and number of actin cables in AgBNI1ΔD. (A) Elongation speeds in μm/h of wild-type hyphae (•) and AgBNI1ΔD hyphae (♦) during the first 700 min of hyphal development. The data points represent the average from two hyphae followed in three young mycelia each, starting shortly after the first hypha has emerged from a germinated spore. (B) Surface expansion rates at hyphal tips of wild type (•) and AgBNI1ΔD (♦) were calculated from the elongation speeds determined in A, and the hyphal diameters were measured every 10 μm. (C) Localization of AgBni1ΔD-GFP fusion proteins at hyphal tips. (D) Visualization by epifluorescence microscopy of rhodamine-phalloidin-stained F-actin in AgBNI1ΔD. For several hyphae a stack of 16 planes taken at 0.4-μm distances was analyzed. Because of the high density of actin cables along the cortex only a maximum projection of the top third of the hypha (three planes) is shown. Four to five actin cables can be seen per cross section, which represents about one-third of the density of cortical actin cables in the hypha. (E) Rhodamine-phalloidin stained F-actin in wild-type A. gossypii. The stack of 16 planes was processed as described for D, and only the top third of the hypha is shown. Only one to two cortical actin cables can be seen per cross section. (F and G) Visualization of GFP-AgSEC4-labeled vesicles in the tip region of an AgBNI1ΔD hypha and the corresponding DIC image. (H) Mycelial colonies of wild type and AgBNI1ΔD after growth on AFM agar at 30°C for 5 d. Bars, (C–F) 5 μm, (H) 2 cm.
The nonregulatable, overactive AgBni1ΔD formin leads to a growth advantage during the early mycelial development but has a negative effect on the further development to a fast spreading colony (Figure 8H). Already after 6 to 7 h the elongation rate of mutant hyphae and with that the surface expansion rate reaches a plateau, whereas the elongation of wild-type hyphae continues to accelerate (Figure 8, A and B). This difference in growth rates may be due to the high frequency of premature tip splitting events in the mutant, which impedes hyphal speed acceleration and thus leads to a slower growing mycelium compared with wild type.
AgBni1p Is an Effector of AgCdc42p
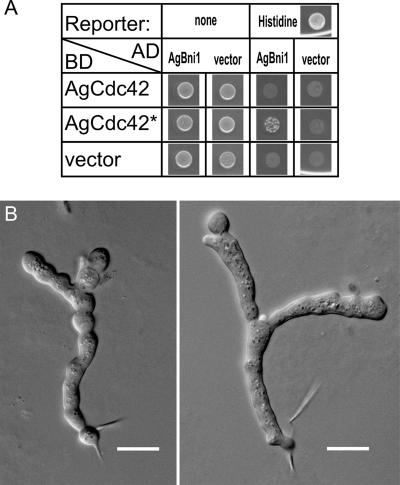
The domain composition of AgBni1p predicts that it is activated when a GTP-bound Rho-type GTPase interacts with the amino-terminal RBD (Figure 2). We searched for candidates of such activator among A. gossypii homologues of Rho-type GTPases using two criteria: Its GTP-bound form should interact with the amino-terminus of AgBni1p, and a GTP-locked form of this protein should induce tip splitting in young hyphae due to a permanent activation of AgBni1p, similarly as observed for young hyphae of the AgBNI1ΔD mutant. By systematic two-hybrid analysis of the AgBni1p amino-terminus against all A. gossypii Rho-type GTPases and by videomicroscopy of GTP-locked mutants of the three Rho-type GTPases, which scored positive in the first test, we identified AgCdc42p as the only candidate. The two-hybrid result is summarized in Figure 9A and two microscopic images showing tip splitting in young hyphae expressing GTP-locked AgCdc42p are presented in Figure 9B. These hyphae carry only the mutated AgCDC42 allele and show growth defects preventing the young mycelia from further development. This is most likely due the multitude of effector proteins that are regulated by AgCdc42p.
Figure 9.
AgCdc42p regulates tip splitting. (A) Two-hybrid assay of AgCdc42p-GTP and the amino-terminal half of AgBni1p. Each spot corresponds to 5 μl of a yeast culture of strain PJ69–4a with an OD600 of 1, transformed with plasmids encoding fusions of the indicated proteins to either the Gal4-activation domain (AD) or to the Gal4-DNA-binding domain (BD) or, as control, transformed with the vector plasmid. The cells were spotted on a medium allowing to select for the plasmids and were grown overnight at 30°C to screen for protein-protein interactions by growth in the absence of histidine. (B) Altered morphogenesis of A. gossypii expressing GTP-locked AgCdc42p. A heterokaryotic strain of A. gossypii was constructed carrying the point mutation AgCdc42-Q61H that encodes a protein mimicking the GTP-bound and therefore activated state of the GTPase. Spores were dissected by micromanipulation and allowed to germinate under conditions selecting only for spores carrying the GTP-locked AgCDC42 allele. The two microscopic images show examples of tip splitting soon after the first hypha emerged from the germling. These hyphae carry only the mutated AgCDC42 allele and show growth defects preventing the young mycelia from further development. This is most likely due the multitude of effector proteins that are regulated by AgCdc42p. Scale bar, 10 μm.
DISCUSSION
The filamentous fungus A. gossypii and the budding yeast S. cerevisiae have different life styles (see Supplementary Movie S1) despite very similar gene contents and conserved domain compositions of gene products (Dietrich et al., 2004). Both organisms can establish polar growth but in S. cerevisiae periods of growth alternate with cell divisions, whereas A. gossypii cells (hyphae) grow for unlimited periods without undergoing divisions. In addition, A. gossypii hyphae can establish multiple new axes of polarity, e.g., during lateral branching or tip splitting. In contrast, wild-type yeast cells never form more than one new bud at the same time (Caviston et al., 2002). We describe here a role for the AgBni1p formin in two of these processes: the establishment of polarity (formation of hyphae) and the controlled division of one axis of polarity into two new polarity axes (tip splitting), a novel function for this protein class.
Redundance of Formins in A. gossypii
The lethality of the Agbni1 deletion was surprising because A. gossypii contains two additional formin genes, AgBNR1 and AgBNR2, and because all three formin genes encode proteins with similar domains, notably the actin nucleation domain FH2. The deletion of either one of the AgBNR genes is fully viable indicating overlapping functions in the AgBnr formins. These results made us at first conclude that the lethality of Agbni1Δ is due to distinct functions of members of the AgBni1p and the AgBnr proteins. This idea was supported by comparison to the more distantly related fission yeast S. pombe, which expresses three members of the formin family with clear separation of functions. For3 is involved in cell polarity (Feierbach and Chang, 2001), Cdc12 is involved in cytokinesis (Chang et al., 1997) and Fus1 is necessary for cell fusion during mating (Petersen et al., 1998a). Although S. pombe for3Δ had no visible actin cables (Feierbach and Chang, 2001) or the cables were shorter in size and reduced in number (Nakano et al., 2002), cells were still able to grow in a polarized manner unlike a deletion of Agbni1.
Different functions were also described for the baker's yeast formins ScBni1p and ScBnr1p (Pruyne et al., 2004). ScBni1p localizes mainly to the bud tip where it forms actin cables. It is also found at the septum in a very late phase of bud formation, when the direction of growth polarity has already switched. ScBnr1p in contrast localizes exclusively and already during early phases of bud formation to the forming septum. However, budding yeast cells can still survive in the absence of ScBni1p because the septum-associated ScBnr1p is also able to nucleate actin cables, thereby providing the close-by growing bud with secretory vesicles. This short distance between the septum and the growing bud tip, e.g., during axial budding, is probably of key importance for the viability of cells lacking the ScBni1p formin. At increasing distances between the septum and the growing tip, formin deletion experiments may give different results. For example, assuming similar actin nucleating functions for the formins in A. gossypii but distinct protein localization requirements, the lethality of the Agbni1 deletion could be explained by the large distance between the growing tip and the nearest septum, preventing septum-associated formin functions to complement the loss of such functions at hyphal tips. This alternative interpretation is supported by the fact that we were able to visualize AgBni1p-GFP at hyphal tips, but not at septa, and AgBnr2p-GFP at septa but not at hyphal tips.
The fact that AgBni1p functions in hyphal emergence and elongation, but probably not in septum formation, indicates that it is also functionally different from SepA, the only other formin described in a filamentous fungus so far. SepA from Aspergillus nidulans localizes to the tip of hyphae and to septa, and it is essential for morphogenesis. Interestingly, some SepA mutants, defective in septum formation, are still able to form hyphae (Sharpless and Harris, 2002). Therefore, mutant formins can probably still induce actin cables at hyphal tips, or another, so far unidentified formin gene, is present in the genome of A. nidulans.
AgBni1p and the Actin Cytoskeleton
The phenotype observed for the AgBNI1 deletion, lack of actin cables and hyphal growth, indicated that an important function of AgBni1p might be the regulation of vesicle transport via actin cables. A. gossypii hyphae are capable to grow with very high growth speeds of up to 170 μm/h (Knechtle et al., 2003). This speed obviously requires a constant and very efficient transport of growth material toward the elongating tips. Our results show that single amino acid changes in the FH2 domain of AgBni1p, which is responsible for actin cable assembly, can only expand to potato-shaped cells, which lack polarized actin cables like the deletion mutant. Therefore, it might be the defect of actin cable-based transport, which leads to the phenotype observed when AgBNI1 is deleted. It is known from several studies, that actin cables can serve as tracks for different cargos such as secretory vesicles (Johnston et al., 1991) and several organelles (Simon et al., 1995; Hill et al., 1996; Hoepfner et al., 2001; Rossanese et al., 2001) as well as mRNA (Bobola et al., 1996; Sil and Herskowitz, 1996; Takizawa et al., 2000). Using the AgSec4p fused to GFP, we were able to show that secretory vesicles use actin cables for accumulation at the hyphal tips. They no longer do so in AgBNI1-deleted hyphae even though the polarisome component AgSpa2p localizes correctly to the cortex at the slowly expanding growth zone, which explains the potatolike shape of the giant mutant cells.
Regulation of Tip Splitting by AgBni1p
Tip splitting, the symmetric division of the polar growth zone of a single growing cell, is to our knowledge limited to filamentous fungi and to dendrites of neuronal cells. Hyphae of A. gossypii usually start tip splitting under optimal conditions 12–14 h after the first hyphae emerged from germinated spores (Ayad-Durieux et al., 2000). Premature tip splitting, already after 6–8 h of elongation and before any lateral branching, as found for the constitutively active AgBni1p protein, was not described before. In an Agcla4 deletion strain, premature tip splitting was previously observed but after normal development of a young mycelium, including lateral branching (Ayad-Durieux et al., 2000). AgCla4p is a homologue of the PAK21 kinase Cla4p from S. cerevisiae, which is involved in polarization of the cytoskeleton during the cell cycle (Holly and Blumer, 1999). Evidence for an involvement of actin in tip splitting was reported in studies of other filamentous fungi in which tip splitting was not observed in wild type but was seen in different mutants as a neomorphic phenotype. For example, the ramosa-1 mutant of Aspergillus niger can divide at hyphal tips when shifted to the restrictive temperature (Reynaga-Pena and Bartnicki-Garcia, 1997), and the authors suggested that tip splitting might be triggered by a transient alteration in the cytoskeletal organization. Similarly, viable mutations in the formin SepA in A. nidulans show tip splitting (Sharpless and Harris, 2002). In Neurospora crassa, several mutants are known that lead to hyphal tip splitting (Sone and Griffiths, 1999; Bok et al., 2001; Seiler and Plamann, 2003; Virag and Griffiths, 2004). It was suggested that a calcium gradient controls the transport of vesicles. This control mechanism also involves actin and determines whether a tip divides or not. The increased surface expansion rate in A. gossypii hyphae with an activated allele of AgBNI1, reported here, is most likely caused by the increase in actin cables and thus vesicle transport. This overstimulation in vesicle transport also leads to premature tip splitting. The fact that GTP-bound AgCdc42p interacts with AgBni1p in a two-hybrid assay and that mutants expressing the activated allele of AgCDC42 also show premature tip splitting suggest that this event is regulated by an AgBni1p branch of the AgCdc42p signaling network. The observed growth problems of the activated Agcdc42 mutant compared with the activated Agbni1 mutant can be explained by the fact that changes in the activity state of AgCdc42p will influence the interaction with multiple effector proteins (Nelson et al., 2003).
The involvement of the Cdc42 GTPase in tip splitting is also supported by mutants described for the CDC24 homolog of N. crassa (Seiler and Plamann, 2003) because Cdc24p is a GDP-GTP exchange factor for the small GTPase Cdc42p.
On the basis of the data discussed here, we suggest a molecular model for tip splitting regulation. AgCdc42p activates AgBni1p which in turn increases the number of actin cables emanating from the tip. This leads than to a stimulation of actin cable-based vesicle transport, which first enlarges and finally divides the polar growth site into two new sites. These data, when combined, suggest that altering the activities of formin molecules by Cdc42p can lead to dramatically different cell shapes. Incorporation of future experiments into this model should help to increase our understanding of this unique process like e.g., its growth speed dependence or it might help us to understand in more detail how tip splitting is triggered.
Supplementary Material
Acknowledgments
We thank Sophie Brachat and Anne-Laure Vitte for their preliminary work on Agbni1Δ; Dominic Hoepfner and Anne L'Hernault for providing pUC19NatPS before publication; and Amy Gladfelter, Anja Lorberg, and Tom Bickle for critical reading of the manuscript. This work was supported by the University of Basel and the Swiss National Science Foundation Grant 3100A0-100734.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–06–0479) on October 19, 2005.
Note added in proof. After submission of this manuscript, two other fungal formins, CaBni1p and CaBnr1p, were shown to play an important but nonessential role in yeast and hyphal growth of Candida albicans (Li et al., 2005).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Alberts, A. S. (2001). Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J. Biol. Chem. 276, 2824–2830. [DOI] [PubMed] [Google Scholar]
- Altmann-Johl, R., and Philippsen, P. (1996). AgTHR4, a new selection marker for transformation of the filamentous fungus Ashbya gossypii, maps in a four-gene cluster that is conserved between A. gossypii and Saccharomyces cerevisiae. Mol. Gen. Genet. 250, 69–80. [DOI] [PubMed] [Google Scholar]
- Ashby, S. F., and Nowell, W. (1926). The fungi of stigmatomycosis. Ann. Botany 40, 69–86. [Google Scholar]
- Ayad-Durieux, Y., Knechtle, P., Goff, S., Dietrich, F., and Philippsen, P. (2000). A PAK-like protein kinase is required for maturation of young hyphae and septation in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 113 (Pt 24), 4563–4575. [DOI] [PubMed] [Google Scholar]
- Bartel, P. L., Chien, C.-T., Sternglanz, R., and Fields, S. (1993). Using the two-hybrid system to detect protein-protein interactions. In: Cellular Interactions in Development: A Practical Approach, ed. D. A. Hartley, Oxford: Oxford University Press, 153–179.
- Bauer, Y., Knechtle, P., Wendland, J., Helfer, H., and Philippsen, P. (2004). A Ras-like GTPase is involved in hyphal growth guidance in the filamentous fungus Ashbya gossypii. Mol. Biol. Cell 15, 4622–4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobola, N., Jansen, R. P., Shin, T. H., and Nasmyth, K. (1996). Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell 84, 699–709. [DOI] [PubMed] [Google Scholar]
- Bok, J. W., Sone, T., Silverman-Gavrila, L. B., Lew, R. R., Bowring, F. J., Catcheside, D. E., and Griffiths, A. J. (2001). Structure and function analysis of the calcium-related gene spray in Neurospora crassa. Fungal Genet. Biol. 32, 145–158. [DOI] [PubMed] [Google Scholar]
- Boles, E., and Miosga, T. (1995). A rapid and highly efficient method for PCR-based site-directed mutagenesis using only one new primer. Curr. Genet. 28, 197–198. [DOI] [PubMed] [Google Scholar]
- Caviston, J. P., Tcheperegine, S. E., and Bi, E. (2002). Singularity in budding: a role for the evolutionarily conserved small GTPase Cdc42p. Proc. Natl. Acad. Sci. USA 99, 12185–12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, F., Drubin, D., and Nurse, P. (1997). cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J. Cell Biol. 137, 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, F. S. et al. (2004). The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304, 304–307. [DOI] [PubMed] [Google Scholar]
- Evangelista, M., Blundell, K., Longtine, M. S., Chow, C. J., Adames, N., Pringle, J. R., Peter, M., and Boone, C. (1997). Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 276, 118–122. [DOI] [PubMed] [Google Scholar]
- Evangelista, M., Pruyne, D., Amberg, D. C., Boone, C., and Bretscher, A. (2002). Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 4, 260–269. [DOI] [PubMed] [Google Scholar]
- Evangelista, M., Zigmond, S., and Boone, C. (2003). Formins: signaling effectors for assembly and polarization of actin filaments. J. Cell Sci. 116, 2603–2611. [DOI] [PubMed] [Google Scholar]
- Feierbach, B., and Chang, F. (2001). Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr. Biol. 11, 1656–1665. [DOI] [PubMed] [Google Scholar]
- Gietz, R. D., and Sugino, A. (1988). New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74, 527–534. [DOI] [PubMed] [Google Scholar]
- Gow, N.A.R. (1995). Tip Growth and Polarity. In: The Growing Fungus, ed. N.A.R. Gow, G. M. Gadd, London: Chapman and Hall, 277–299.
- Habas, R., Kato, Y., and He, X. (2001). Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell 107, 843–854. [DOI] [PubMed] [Google Scholar]
- Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. [DOI] [PubMed] [Google Scholar]
- Harris, E. S., Li, F., and Higgs, H. N. (2004). The mouse formin, FRLalpha, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J. Biol. Chem. 279, 20076–20087. [DOI] [PubMed] [Google Scholar]
- Harris, S. D., Read, N. D., Roberson, R. W., Shaw, B., Seiler, S., Plamann, M., and Momany, M. (2005). Polarisome meets spitzenkorper: microscopy, genetics, and genomics converge. Eukaryot. Cell 4, 225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S. D., Hamer, L., Sharpless, K. E., and Hamer, J. E. (1997). The Aspergillus nidulans sepA gene encodes an FH1/2 protein involved in cytokinesis and the maintenance of cellular polarity. EMBO J. 16, 3474–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs, H. N., and Peterson, K. J. (2005). Phylogenetic analysis of the formin homology 2 domain. Mol. Biol. Cell 16, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, K. L., Catlett, N. L., and Weisman, L. S. (1996). Actin and myosin function in directed vacuole movement during cell division in Saccharomyces cerevisiae. J. Cell Biol. 135, 1535–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner, D., van den Berg, M., Philippsen, P., Tabak, H. F., and Hettema, E. H. (2001). A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 155, 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holly, S. P., and Blumer, K. J. (1999). PAK-family kinases regulate cell and actin polarization throughout the cell cycle of Saccharomyces cerevisiae. J. Cell Biol. 147, 845–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura, H., Tanaka, K., Hihara, T., Umikawa, M., Kamei, T., Takahashi, K., Sasaki, T., and Takai, Y. (1997). Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 16, 2745–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E. A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, G. C., Prendergast, J. A., and Singer, R. A. (1991). The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J. Cell Biol. 113, 539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei, T., Tanaka, K., Hihara, T., Umikawa, M., Imamura, H., Kikyo, M., Ozaki, K., and Takai, Y. (1998). Interaction of Bnr1p with a novel Src homology 3 domain-containing Hof1p. Implication in cytokinesis in Saccharomyces cerevisiae. J. Biol. Chem. 273, 28341–28345. [DOI] [PubMed] [Google Scholar]
- Knechtle, P., Dietrich, F., and Philippsen, P. (2003). Maximal polar growth potential depends on the polarisome component AgSpa2 in the filamentous fungus Ashbya gossypii. Mol. Biol. Cell 14, 4140–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobielak, A., Pasolli, H. A., and Fuchs, E. (2003). Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nat. Cell Biol. 6, 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno, H. et al. (1996). Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 15, 6060–6068. [PMC free article] [PubMed] [Google Scholar]
- Kovar, D. R., Kuhn, J. R., Tichy, A. L., and Pollard, T. D. (2003). The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J. Cell Biol. 161, 875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. R., Wang, Y. M., DeZheng, X., Liang, H. Y., Tang, J. C., and Wang, Y. (2005). The formin family protein CaBni1p has a role in cell polarity control during both yeast and hyphal growth in Candida albicans. J. Cell. Sci. 118, 2637–2648. [DOI] [PubMed] [Google Scholar]
- Li, F., and Higgs, H. N. (2003). The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 13, 1335–1340. [DOI] [PubMed] [Google Scholar]
- Momany, M. (2002). Polarity in filamentous fungi: establishment, maintenance and new axes. Curr. Opin. Microbiol. 5, 580–585. [DOI] [PubMed] [Google Scholar]
- Moseley, J. B., Sagot, I., Manning, A. L., Xu, Y., Eck, M. J., Pellman, D., and Goode, B. L. (2004). A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol. Biol. Cell 15, 896–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, K., Imai, J., Arai, R., Toh, E. A., Matsui, Y., and Mabuchi, I. (2002). The small GTPase Rho3 and the diaphanous/formin For3 function in polarized cell growth in fission yeast. J. Cell Sci. 115, 4629–4639. [DOI] [PubMed] [Google Scholar]
- Needleman, S. B., and Wunsch, C. D. (1970). A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48, 443–453. [DOI] [PubMed] [Google Scholar]
- Nelson, B. et al. (2003).. RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol. Biol. Cell 14, 3782–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki-Kuroda, K., Yamamoto, Y., Nohara, H., Kinoshita, M., Fujiwara, T., Irie, K., and Takai, Y. (2001). Dynamic localization and function of Bni1p at the sites of directed growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, W. R. (1990). Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183, 63–98. [DOI] [PubMed] [Google Scholar]
- Petersen, J., Nielsen, O., Egel, R., and Hagan, I. M. (1998a). F-actin distribution and function during sexual differentiation in Schizosaccharomyces pombe. J. Cell Sci. 111 (Pt 7), 867–876. [DOI] [PubMed] [Google Scholar]
- Petersen, J., Nielsen, O., Egel, R., and Hagan, I. M. (1998b). FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J. Cell Biol. 141, 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne, D., Evangelista, M., Yang, C., Bi, E., Zigmond, S., Bretscher, A., and Boone, C. (2002). Role of formins in actin assembly: nucleation and barbed-end association. Science 297, 612–615. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., Gao, L., Bi, E., and Bretscher, A. (2004). Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol. Biol. Cell 15, 4971–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaga-Pena, C. G., and Bartnicki-Garcia, S. (1997). Apical branching in a temperature sensitive mutant of Aspergillus niger. Fungal Genet. Biol. 22, 153–167. [DOI] [PubMed] [Google Scholar]
- Rossanese, O. W., Reinke, C. A., Bevis, B. J., Hammond, A. T., Sears, I. B., O'Connor, J., and Glick, B. S. (2001). A role for actin, Cdc1p, and Myo2p in the inheritance of late Golgi elements in Saccharomyces cerevisiae. J. Cell Biol. 153, 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagot, I., Klee, S. K., and Pellman, D. (2002a). Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 4, 42–50. [DOI] [PubMed] [Google Scholar]
- Sagot, I., Rodal, A. A., Moseley, J., Goode, B. L., and Pellman, D. (2002b). An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 4, 626–631. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Russel, D. W., and Sambrook, J. (2001). Molecular Cloning: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Schott, D. H., Collins, R. N., and Bretscher, A. (2002). Secretory vesicle transport velocity in living cells depends on the myosin-V lever arm length. J. Cell Biol. 156, 35–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler, S., and Plamann, M. (2003). The genetic basis of cellular morphogenesis in the filamentous fungus Neurospora crassa. Mol. Biol. Cell 14, 4352–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless, K. E., and Harris, S. D. (2002). Functional characterization and localization of the Aspergillus nidulans formin SEPA. Mol. Biol. Cell 13, 469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu, Y. J., Santos, B., Fortin, N., Costigan, C., and Snyder, M. (1998). Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol. Cell. Biol. 18, 4053–4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, A., Nyitrai, M., Vetter, I. R., Kuhlmann, D., Bugyi, B., Narumiya, S., Geeves, M. A., and Wittinghofer, A. (2004). The core FH2 domain of diaphanous-related formins is an elongated actin binding protein that inhibits polymerization. Mol. Cell 13, 511–522. [DOI] [PubMed] [Google Scholar]
- Sikorski, R. S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sil, A., and Herskowitz, I. (1996). Identification of asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell 84, 711–722. [DOI] [PubMed] [Google Scholar]
- Simon, V. R., Swayne, T. C., and Pon, L. A. (1995). Actin-dependent mitochondrial motility in mitotic yeast and cell-free systems: identification of a motor activity on the mitochondrial surface. J. Cell Biol. 130, 345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone, T., and Griffiths, A. J. (1999). The frost gene of Neurospora crassa is a homolog of yeast cdc1 and affects hyphal branching via manganese homeostasis. Fungal Genet. Biol. 28, 227–237. [DOI] [PubMed] [Google Scholar]
- Steiner, S., and Philippsen, P. (1994). Sequence and promoter analysis of the highly expressed TEF gene of the filamentous fungus Ashbya gossypii. Mol. Gen. Genet. 242, 263–271. [DOI] [PubMed] [Google Scholar]
- Steiner, S., Wendland, J., Wright, M. C., and Philippsen, P. (1995). Homologous recombination as the main mechanism for DNA integration and cause of rearrangements in the filamentous ascomycete Ashbya gossypii. Genetics 140, 973–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici, F., Lewis, L. K., and Resnick, M. A. (2001). In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 19, 773–776. [DOI] [PubMed] [Google Scholar]
- Takizawa, P. A., DeRisi, J. L., Wilhelm, J. E., and Vale, R. D. (2000). Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 290, 341–344. [DOI] [PubMed] [Google Scholar]
- Vieira, J., and Messing, J. (1991). New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100, 189–194. [DOI] [PubMed] [Google Scholar]
- Virag, A., and Griffiths, A. J. (2004). A mutation in the Neurospora crassa actin gene results in multiple defects in tip growth and branching. Fungal Genet. Biol. 41, 213–225. [DOI] [PubMed] [Google Scholar]
- Wallar, B. J., and Alberts, A. S. (2003). The formins: active scaffolds that remodel the cytoskeleton. Trends Cell Biol. 13, 435–446. [DOI] [PubMed] [Google Scholar]
- Watanabe, N., Kato, T., Fujita, A., Ishizaki, T., and Narumiya, S. (1999). Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat. Cell Biol. 1, 136–143. [DOI] [PubMed] [Google Scholar]
- Watanabe, N., Madaule, P., Reid, T., Ishizaki, T., Watanabe, G., Kakizuka, A., Saito, Y., Nakao, K., Jockusch, B. M., and Narumiya, S. (1997). p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 16, 3044–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland, J., Ayad-Durieux, Y., Knechtle, P., Rebischung, C., and Philippsen, P. (2000). PCR-based gene targeting in the filamentous fungus Ashbya gossypii. Gene 242, 381–391. [DOI] [PubMed] [Google Scholar]
- Wendland, J., and Philippsen, P. (2000). Determination of cell polarity in germinated spores and hyphal tips of the filamentous ascomycete Ashbya gossypii requires a rhoGAP homolog. J. Cell Sci. 113(Pt 9), 1611–1621. [DOI] [PubMed] [Google Scholar]
- Wendland, J., and Philippsen, P. (2001). Cell polarity and hyphal morphogenesis are controlled by multiple rho-protein modules in the filamentous ascomycete Ashbya gossypii. Genetics 157, 601–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, M. C., and Philippsen, P. (1991). Replicative transformation of the filamentous fungus Ashbya gossypii with plasmids containing Saccharomyces cerevisiae ARS elements. Gene 109, 99–105. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Moseley, J. B., Sagot, I., Poy, F., Pellman, D., Goode, B. L., and Eck, M. J. (2004). Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell 116, 711–723. [DOI] [PubMed] [Google Scholar]
- Zahner, J. E., Harkins, H. A., and Pringle, J. R. (1996). Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 1857–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.