Abstract
We have used fluorescence microscopy and the technique of rapamycin-regulated protein heterodimerization to examine the dynamics of the subcellular localizations of fluorescent proteins fused to lipid-modified protein sequences and to wild-type and mutated forms of full-length K-ras4B. Singly prenylated or myristoylated fluorescent protein derivatives lacking a “second signal” to direct them to specific subcellular destinations, but incorporating a rapamycin-dependent heterodimerization module, rapidly translocate to mitochondria upon rapamycin addition to bind to a mitochondrial outer membrane protein incorporating a complementary heterodimerization module. Under the same conditions analogous constructs anchored to the plasma membrane by multiply lipid-modified sequences, or by a transmembrane helix, show very slow or no transfer to mitochondria, respectively. Interestingly, however, fluorescent protein constructs incorporating either full-length K-ras4B or its plasma membrane-targeting sequence alone undergo rapamycin-induced transfer from the plasma membrane to mitochondria on a time scale of minutes, demonstrating the rapidly reversible nature of K-ras4B binding to the plasma membrane. The dynamic nature of the plasma membrane targeting of K-ras4B could contribute to K-ras4B function by facilitating redistribution of the protein between subcellular compartments under particular conditions.
INTRODUCTION
A variety of cellular proteins are modified with single iso-prenyl groups attached to carboxy-terminal cysteine residues, which are typically also carboxy-methylated as well (Zhang and Casey, 1996; Fu and Casey, 1999; Sinensky, 2000; Roskoski, 2003). After initial farnesylation or geranylgeranylation mediated by cytoplasmic protein prenyltransferases, singly prenylated proteins undergo further processing at the endoplasmic reticulum (ER), where the terminal three amino acid residues of the -CAAX prenylation motif are endoproteolytically removed and the carboxyl group of the now-terminal prenylated cysteine residue is methylated (Gutierrez et al., 1989; Dai et al., 1998; Romano et al., 1998; Schmidt et al., 1998; Otto et al., 1999). After completing these common processing steps at the ER, different singly prenylated proteins diverge markedly in their subsequent trafficking to their final intracellular destinations (Choy et al., 1999; Apolloni et al., 2000; Michaelson et al., 2001; Silvius, 2002). H- and N-ras, for example, are transported to the plasma membrane in association with vesicles of the classical secretory pathway, whereas K-ras4B reaches the same membrane by a distinct, apparently nonvesicular pathway (Choy et al., 1999; Apolloni et al., 2000), and the closely related rap proteins localize primarily to the Golgi (Beranger et al., 1991; Pizon et al., 1994; Nomura et al., 2004).
Correct final subcellular targeting of the different ras species and of other singly prenylated proteins has been shown to require both the prenylated carboxy-terminal cysteine residue and a “second signal,” such as one or more nearby S-acylation (“palmitoylation”) sites (as in H- and N-ras) or a polybasic sequence (as in K-ras4B) located near the prenylated carboxy-terminus (Hancock et al., 1990, 1991; Willumsen et al., 1996; Apolloni et al., 2000; Michaelson et al., 2001). For mammalian H- and N-ras and for Ras2p in yeast, plasma membrane targeting has been shown to be mediated by S-acylation of the prenylated carboxy-terminal sequence, which leads to kinetic trapping of these proteins at the plasma membrane (Dong et al., 2003; Goodwin et al., 2005; Rocks et al., 2005; Roy et al., 2005). The mechanism of plasma membrane localization targeting of proteins such as K-ras4B, with a prenylated and polybasic targeting sequence, remains unclear. To date it has not been clearly established whether plasma membrane targeting of K-ras4B is dynamic (i.e., rapidly reversible) or instead might rest on some form of kinetic-trapping mechanism.
In this study we have combined fluorescence microscopy and the technique of rapamycin-induced protein heterodimerization to examine the dynamics of membrane localization of prenylated or myristoylated protein constructs that lack a second targeting signal or that additionally carry either S-acylation sites or the polybasic plasma membrane-targeting signal of K-ras4B. In the absence of a functional second signal, prenylated and myristoylated proteins distribute among different cellular membranes in a highly dynamic manner, whereas prenylated or myristoylated proteins that are also S-acylated are kinetically trapped on the plasma membrane, as recently demonstrated for H- and N-ras (Goodwin et al., 2005; Rocks et al., 2005). Interestingly, however, plasma membrane targeting of fluorescent protein constructs incorporating full-length K-ras4B or its farnesylated, polybasic carboxy-terminal sequence is also dynamic, with a halftime for membrane desorption on the order of minutes, indicating that plasma membrane targeting of K-ras4B rests on an equilibrium-binding rather than a kinetic-trapping mechanism.
MATERIALS AND METHODS
Materials
Rapamycin was obtained from Sigma/Aldrich Canada (Oakville, Ontario), and the rapamycin analogue AP29167 was provided by Ariad Pharmaceuticals (Cambridge, MA).
Plasmids
The plasmids pC4EN-F1, pC4M-F2E, and pC4-RHE were provided by Ariad Pharmaceuticals. XbaI/SpeI digestion fragments from these plasmids were ligated into the XbaI sites of the parent plasmids to produce sequences comprising three repeats of an 11-kDa rapamycin-binding domain (FKBP) from human FKBP12 or two repeats of the rapamycin-binding domain (FRB) from FRAP/mTOR, including a mutation that allows selective binding to the mutated FRB domain of the rapamycin analogue AP21967, which unlike rapamycin does not inhibit the mTOR/FRAP kinase. These tandem sequences were subcloned into XbaI/SpeI-digested pC4EN-F1 or pC4-RHE to produce the plasmids pC4EN-F3 and pC4-2RHE, respectively.
PCR fragments containing the entire coding sequences of enhanced cyan and yellow fluorescent proteins flanked by a 5′-XbaI site and 3′-SpeI site plus short linker sequences were prepared from pECFP-N1 or pEYFP-N1 (BD Biosciences/Clontech, San Jose, CA), using the 5′-primer TCTAGAGGAGCTGGTGCAGTGAGCAAGGGCGAGAGCT and the 3′-primer ACTAGTGAGTCCGGACTTGTACAGCTCGTCCATGCC. A PCR fragment containing the coding sequence of monomeric red fluorescent protein (mRFP) and the same flanking sequences was prepared from a plasmid containing the complete mRFP coding sequence (generously provided by Dr. Robert Nabi, University of British Columbia, and used with the kind permission of Dr. Richard Tsien, University of California at San Diego) and the 5′- and 3′-primers TCTAGAGGAGCTGGTGCAATGGCCTCCCTCCGAGGACGTC and ACTAGTGAGCCGGGAGGCGCCGGTGGAGTGGCGG. The XbaI/SpeI-digested PCR products were ligated into a XbaI/SpeI-digested modified form of pC4EN-F1, in which the sequence between the EcoRI and XbaI sites had been replaced by that of pC4-RHE, to produce the plasmids pC4-CFP, pC4-YFP, and pC4-mRFP. EcoRI/BamHI fragments from the first two plasmids were subcloned into pALTER-1, mutated using the Altered Sites procedure (Promega, Madison, WI) to produce the Cerulean and monomeric Citrine variants, respectively (Zacharias et al., 2002; Rizzo et al., 2004), and then subcloned back into the parent plasmids to yield pC4-Cer and pC4-mCit.
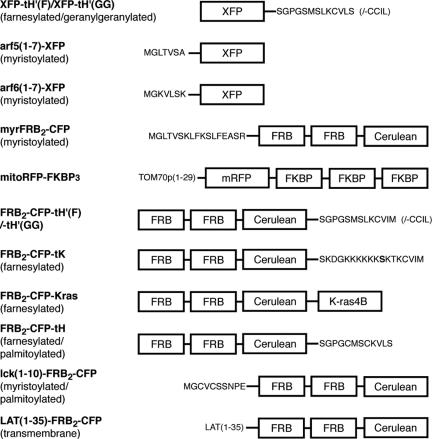
Plasmids encoding constructs linking fluorescent proteins to prenylated carboxy-terminal sequences were constructed from pC4-Cer or pC4-mCit by replacing the sequences between the SpeI and BamHI sites with the following sequences (flanking restriction sites underlined): ACTAGTAGCGGCCCCGGCTCCATGAGCCTGAAGTGTGTGTCCTAAGGATCC for–tH′(F) constructs, ACTAGTAGCGGCCCCGGCTCCATGAGCCTGAAGTGTTGCATCCTGGGATCC for–tH′(GG) constructs, ACTAGTAGCGGCCCCGGCTGCATGAGCTGCAAGTGTGTGCTCTCCTAAGGATCC for -tH constructs and ACTAGTAAAGATGGTAAAAAGAAGAAAAAGAAGTCAAAGACAAAGTGTGTAAATATGTAAGGATCC for -tK constructs (replacing the highlighted TCA codon in the -tK sequence by GCA for -tK(S181A) constructs). A plasmid encoding the myr-CFP construct was prepared using PCR to replace the sequence of pC4-Cer between the EcoRI and XbaI sites with the sequence GAATTCACCATGGGCCTGACCGTGAGCAAGCTGTTCAAGTCCCTGTTTGAAGCTTCTAGA. PCR was also used to replace the sequence of pC4-Cer between the EcoRI and XbaI sites with sequences encoding amino acids 1–10 of human p56lck or amino acids 1–35 of human LAT. A XbaI/SpeI restriction fragment from pC4-2RHE was ligated into the XbaI site of the above plasmids to obtain sequences encoding the corresponding FRB2-CFP-module-containing proteins (see Figure 1). pcDNA3-based plasmids encoding Arf5(1–7)- or Arf6(1–7)-fluorescent protein constructs were prepared as described previously for the analogous derivative of enhanced green fluorescent protein (Roy et al., 2000).
Figure 1.
Structures of fluorescent protein constructs created for this study. The construct FRB2-CFP-tK(S181A) was generated by mutating to alanine the serine residue shown in boldface in the structure of FRB2-CFP-tK.
PCR fragments containing the complete coding sequences of K-ras4B and its GTPase-defective G12V mutant (including the initial methionine and the final stop codon), flanked by 5′ SpeI and 3′-BamHI sites, were prepared using template plasmids generously provided by Dr. Adrienne Cox (University of North Carolina) and ligated between the SpeI and BamHI sites of the plasmid encoding the FRB2-CFP-tK construct to prepare plasmids encoding the FRB2-CFP-Kras and FRB2-CFP-Kras(G12V) constructs. The related FRB2-CFP-Kras(S17N) construct was prepared by mutagenesis as described above, replacing codons 17–18 of K-ras4B (AGTGCC) by AATGCA to introduce the indicated mutation as well as an NsiI restriction site to facilitate screening for mutated clones.
A PCR fragment encoding the mitochondrial outer membrane-targeting sequence (residues 1–29) of yeast TOM70p flanked by EcoRI and XbaI sites was prepared from the plasmid pSP(pOMD29; Li and Shore, 1992) and ligated into EcoRI/XbaI-digested pC4-mRFP to prepare the plasmid pC4-T70-mRFP. A XbaI/SpeI restriction fragment from pC4EN-F3 was ligated into the SpeI site of the latter vector to obtain a plasmid encoding the mitoRFP-FKBP3 construct. A plasmid encoding the pOCT(1–37)-CFP construct was prepared by ligating between the KpnI and XbaI sites of pcDNA3 a PCR fragment consisting of a KpnI restriction site, the coding sequence of amino acid residues 1–37 of human preornithine carbamyltransferase, and a linker sequence TACTCTAGAGGATCC followed by the complete coding sequence of ECFP (less the initiator ATG codon).
Cell Transfections and Fluorescence Microscopy
CV-1 and COS-1 monkey kidney fibroblasts were seeded on glass coverslips in six-well culture dishes and transfected 16–24 h later using the calcium phosphate procedure as described previously (Roy et al., 2000). Transiently transfected cells were imaged live 48–72 h posttransfection using a Zeiss LSM 5 Pascal confocal microscope (Thornwood, NY; using a pinhole setting of 1.5 μm) or a Nikon Eclipse TE300 epifluorescence microscope (Melville, NY) fitted with a Photometrics Quantix digital camera (Tucson, AZ). Cells expressing the lowest levels of fluorescent protein suitable for photographic imaging were chosen for analysis. We did not observe any noteworthy differences in the distributions of fluorescence in cells expressing different levels of a given protein construct, with the exception that cells expressing very high levels of some lipidated proteins exhibited diffuse cytoplasmic and nuclear fluorescence, suggestive of incomplete protein processing. Rapamycin-induced translocation of FRB domain-incorporating fluorescent protein constructs to mitochondria in cells coexpressing the mitoRFP-FKBP3 construct was assessed by adding medium containing rapamycin or AP29167 (0.5 or 1.0 μM, respectively) to the cells at 22°C and imaging by conventional fluorescence microscopy as described above. Average pixel intensities were determined for defined regions of the cell images as a function of time after rapamycin addition, and the intensity data, corrected for photobleaching effects using standard curves determined from parallel exposure series for untreated cells, were plotted versus time and analyzed by curve-fitting to an equation of the form I(t) = A + B·exp(–kt), where I(t) is the average pixel intensity, t is the time after rapamycin addition, and k is the rate constant for redistribution of fluorescence.
RESULTS
In this study we adapted the technique of rapamycin-induced protein heterodimerization to examine the dynamics of association of lipid-modified proteins with the cytoplasmic faces of cellular membranes. As demonstrated previously (Chen et al., 1995; Choi et al., 1996; Stockwell and Schreiber, 1998; Castellano et al., 1999), upon addition of rapamycin to intact cells, an expressed protein containing one or more repeats of the rapamycin-binding domain of mTOR (FRB) can heterodimerize rapidly and efficiently with a coexpressed protein incorporating multiple repeats of an 11-kDa rapamycin-binding domain (FKBP) from the human FKBP12 protein. If the two proteins are normally localized to different cellular membranes, such heterodimerization will be possible only if at least one of the proteins can transfer at an observable rate to the membrane where the other resides. For this study we prepared an FKBP domain-containing protein construct designated mitoRFP-FKBP3 (Figure 1), comprising three FKBP domains and an mRFP fluorescent marker stably anchored to the mitochondrial outer membrane via the transmembrane sequence of human Tom70p. This construct was coexpressed with various lipid-modified protein constructs, incorporating two FRB domains and a CFP fluorescent marker (Figure 1), which in the absence of rapamycin associate with different nonmitochondrial membrane compartments. Because most cellular membrane compartments do not traffic materials to mitochondria via vesicular pathways, addition of rapamycin to cells coexpressing a pair of protein constructs as described above is predicted (and shown below) to cause redistribution of the lipidated FRB domain-containing protein to mitochondria if (and only if) the lipidated species can desorb at a finite rate from its normal membrane environment. Under these conditions, the rate of rapamycin-induced transfer of the lipidated species to mitochondria can provide a measure of the rate of membrane dissociation of this species.
Kinetics of Redistribution of Singly Lipid-modified Protein Constructs
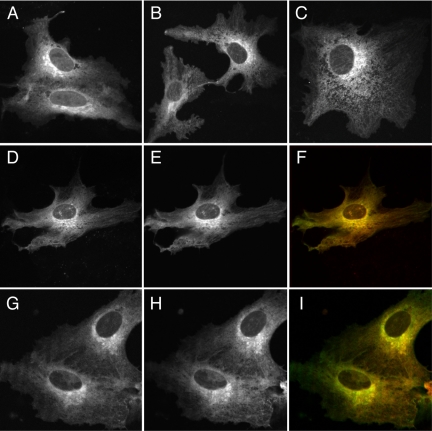
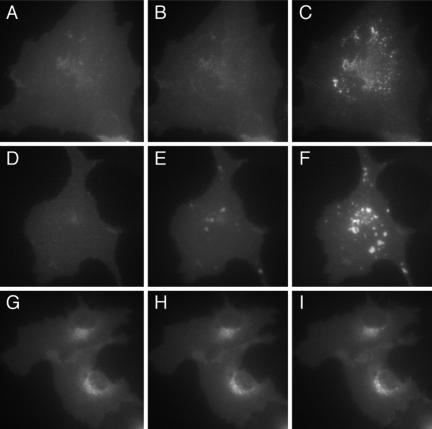
We have previously shown (Silvius and l'Heureux, 1994; Schroeder et al., 1997; Leventis and Silvius, 1998) that prenylated or myristoylated peptides bound to lipid bilayers (lipid vesicles) can transfer between different bilayers through the aqueous phase on time scales of a few minutes or less. To investigate whether singly lipid-anchored proteins can show comparable rates of intermembrane transfer in living cells, we used the approach outlined above to examine the rates of intermembrane transfer of prenylated or myristoylated proteins lacking a second signal (e.g., a nearby polybasic domain or S-acylation site(s)) to specify targeting to particular cellular membranes. Previous studies of prenylated proteins lacking second signals have observed that these proteins are prominently associated with the ER and perinuclear structures (Hancock et al., 1990, 1991; Choy et al., 1999; Calero et al., 2003; Gomes et al., 2003). As shown in Figure 2, A and B, this was true also for protein constructs linking two FRB domains and a CFP module (FRB2-CFP-) to modified forms of the H-ras carboxy-terminus, which are farnesylated (-tH′(F)) or geranylated (-tH′(GG)) but in which the S-acylation sites that comprise the second signal for H-ras targeting (Hancock et al., 1990, 1991; Willumsen et al., 1996) were mutated. A very similar subcellular distribution was observed for an analogous myristoylated construct (myr-FRB2-CFP), as illustrated in Figure 2C. The localizations of these constructs closely matched those of constructs fusing YFP to the same lipid-modified sequences, as illustrated for YFP–tH′(GG) and FRB2-CFP-tH′(GG) in Figure 2, D–F, indicating that the presence of FRB domains did not alter the subcellular distributions of these species. Interestingly, as illustrated in Figure 2, G–I, the subcellular localization of the YFP–tH′(GG) protein closely resembled that of coexpressed proteins linking CFP to minimal myristoylation sequences derived from either human Arf6, a plasma membrane-associating protein (Donaldson, 2003), or (unpublished data) Arf5, a Golgi-associating protein (Haun et al., 1993). This result suggests that the subcellular distributions of prenylated proteins lacking second signals may not depend on any distinctive structural features of their hydrophobic modifications.
Figure 2.
Confocal microscopic images showing the subcellular distributions of prenylated and myristoylated fluorescent proteins in CV-1 cells. (A–C) Images of (A) FRB2-CFP-tH′(F), (B) FRB2-CFP-tH′(GG), and (C) myr-FRB2-CFP. (D and E) Images of FRB2-CFP-tH′(GG) and YFP-tH′(GG), respectively; (F) merged image (green is YFP-tH′(GG)). (G and H) Images of Arf6(1–7)-CFP and YFP-tH′(GG), respectively; (I) merged image (green is YFP-tH′(GG)).
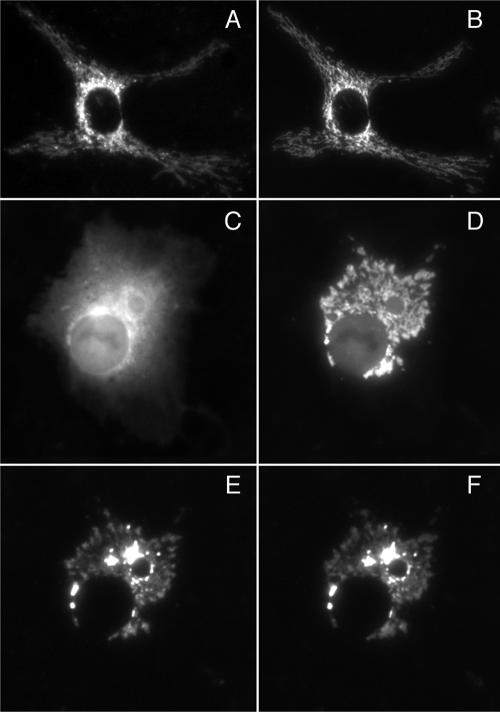
As illustrated in Figure 3, A and B, when expressed in COS-1 or CV-1 cells the mitoRFP-FKBP3 construct is targeted exclusively to mitochondria, as demonstrated by its colocalization with pOCT(1–37)-CFP, a derivative of CFP that is targeted to the mitochondrial matrix by fusion to the matrix-targeting sequence of human preornithine carbamoyltransferase. When rapamycin is added to cells coexpressing mitoRFP-FKBP3 and the farnesylated FRB2-CFP-tH′(F) construct, the farnesylated protein rapidly redistributes to mitochondria to colocalize with mitoRFP-FKBP3 (Figure 3, C, D, and F). Rapamycin-induced redistribution of the farnesylated protein is essentially complete within 5 min, whereas as expected, the distribution of the mitoRFP-FKBP3 construct remains unchanged upon rapamycin addition (Figure 3, E and F), indicating that the drug does not perturb mitochondrial morphology or the integrity of the mitochondrial membrane compartment. Similarly rapid translocation was observed for the geranylgeranylated FRB2-CFP-tH′(GG) and myristoylated myrFRB2-CFP constructs upon addition of rapamycin to cells coexpressing these proteins with mitoRFP-FKBP3 (Figure 4, A–C and D–F).
Figure 3.
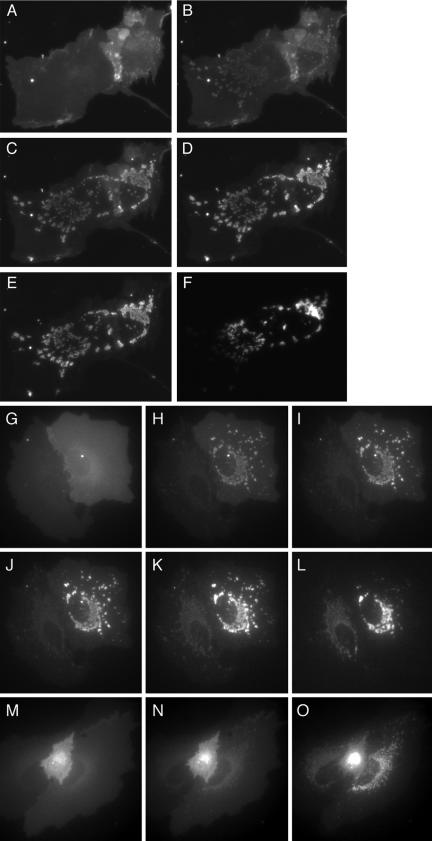
Rapamycin-induced redistribution to mitochondria of farnesylated FRB2-CFP-tH′(F) in cells transiently expressing the protein together with mitochondrially anchored mitoRFP-FKBP3. Limited resolution in some images is due to the use of short exposure times to minimize potential photobleaching. (A and B) Fluorescence images of mitoRFP-FKBP3 and of a mitochondrial matrix-targeted CFP derivative, pOCT(1–37), respectively, in a COS-1 cell transiently expressing the two proteins. (C and D) Fluorescence images of FRB2-CFP-tH′(F) before (C) and 5 min after (D) addition of rapamycin (0.5 μM). (E and F) Fluorescence images of mitoRFP-FKBP3 before (E) and 5 min after (F) addition of rapamycin to the same cell as shown in C and D.
Figure 4.
Rapamycin-induced redistribution of lipid-modified, FRB-domain-incorporating fluorescent proteins. (A–C) Subcellular distribution of FRB2-CFP-tH′(GG) at 0 (A) and 5 min after (B) addition of rapamycin to a CV-1 cell coexpressing the protein together with mitoRFP-FKBP3, whose distribution 5 min after rapamycin addition is shown in C. (D–F) Subcellular distribution of myrFRB2-CFP at 0 (D) and 5 min after (E) addition of rapamycin to CV-1 cells coexpressing the protein together with mitoRFP-FKBP3, whose distribution 5 min after rapamycin addition is shown in F. (G and H) Subcellular distribution of FRB2-CFP-tH′(GG) at 0 (G) and 5 min after (H) addition of rapamycin to a CV-1 cell not coexpressing the mitoRFP-FKBP3 construct.
Several control experiments were carried out to validate the above approach to monitor intercompartmental transfer of membrane-associating proteins. No significant redistribution of fluorescence was observed when rapamycin was added to cells expressing only the FRB2-CFP-tH′(GG) construct (Figure 4, G and H) or (unpublished data) the FRB2-CFP-tH′(F) construct. Similarly, rapamycin caused no redistribution of the FRB domain-lacking CFP-tH′(F) or CFP-tH′(GG) constructs in cells in which these proteins were coexpressed with the mitoRFP-FKBP3 protein (unpublished data). Finally, under the photographic conditions used for the above experiments, photobleaching of the lipidated fluorescent protein constructs was minor (<1% between consecutive exposures), as assessed by quantitative image analysis (see below) for control experiments like those just described. The rapid disappearance of fluorescence from nonmitochondrial membranes in the experiments shown in Figures 3 (C–F) and 4 (A–F) thus can be attributed essentially completely to rapid intermembrane translocation of the lipidated fluorescent proteins, with a minimal contribution from photobleaching.
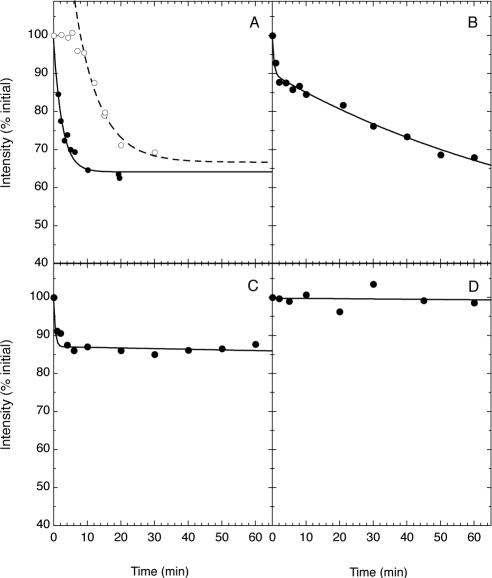
To obtain more quantitative information about the time courses of intermembrane translocation of lipid-anchored FRB2-CFP-constructs, we measured the mean fluorescence intensities for defined regions of cells like those shown in Figures 3 and 4 as a function of time after rapamycin addition. We then analyzed the resulting intensity-time profiles as described in Materials and Methods to extract the effective rate constants for redistribution of different protein constructs. Analyses of this type are illustrated in Figure 5, and the transfer rate constants thereby estimated are summarized in Table 1. Very similar estimates of rate constants for rapamycin-induced redistribution of the fluorescent constructs were obtained by measuring depletion of fluorescence from regions of the cell lacking mitochondria or accumulation in regions rich in mitochondria (unpublished data). As illustrated in Figure 5A for geranylgeranylated FRB2-CFP-tH′(GG), singly lipid-modified constructs lacking second signals show halftimes for transfer to mitochondria of the order of 1–2 min or less. It is possible that for such rapidly translocating species the kinetics of rapamycin permeation into cells (or the intrinsic rate constants for rapamycin-mediated protein heterodimerization) are at least partly rate-limiting for the observed redistribution to mitochondria. If so, the intrinsic rate constants for intermembrane transfer of these species may in fact be still faster than those measured for their transfer to mitochondria in the above experiments, and may vary to a greater extent between species with different (single) lipid chains than do the apparent rate constants given in Table 1.
Figure 5.
Quantitative analysis, carried out as described in the text, of the kinetics of rapamycin- or rapalog-induced translocation to mitochondria of protein constructs coexpressed with mitoRFP-FKBP3 in CV-1 cells. (A) Time courses of redistribution of FRB2-CFP-tH′(GG) upon addition of 0.5 μM rapamycin (•) or 1 μM rapalog AP21967 (○). The complete time courses determined for rapamycin-treated cells, and the rapid (post-lag) phases of those determined for AP29167-treated cells, were fit as illustrated to an equation of the form I = A + B·exp(–kt), where I is the mean pixel intensity determined for a fixed region of the cell image, A and B are adjustable scaling parameters, and k is the rate constant for the redistribution process. Values of k determined in this manner are summarized in Table 1. (B–D) Time courses of rapamycin-induced redistribution of (B) FRB2-CFP-tH, (C) lck(1–10)-FRB2-CFP, and (D) LAT(1–35)-FRB2-CFP in CV-1 cells under conditions similar to those described for A. Data shown in B and C are fit with a biexponential decay curve and those in D to a monoexponential decay curve (in the latter case yielding a rate constant << 10–4 min–1, indicating negligible transfer from the plasma membrane). The values of k summarized in Table 1 for translocation of FRB2-CFP-tH, and lck(1–10)-FRB2-CFP correspond to the rate constants estimated for the slow phase of fluorescence redistribution. Because of the nature of the image analysis and the deliberate use of overly conservative image-background corrections in data quantification, the magnitudes of the intensity decays shown (expressed as a percentage of the initial intensity) should not be directly equated to the percentages of translocation of the protein constructs, which will typically be somewhat larger and can be better assessed from original images like those shown in Figures 3, 4, and 6.
Table 1.
Estimated rate constants for rapamycin-induced intracellular redistribution of FRB-domain-containing Cerulean constructs fused to different lipidation sequences
| Construct | Redistribution rate constant (min-1)a | Redistribution halftime (min)b |
|---|---|---|
| FRB2-CFP-tH′(GG) | 0.69 ± 0.19 | 1.72 ± 0.33 (11) |
| FRB2-CFP-tH′(F) | 0.54 ± 0.18 | 1.17 ± 0.67 (5) |
| myrAmphi-FRB2-CFP | 1.19 + 0.75 | 1.95 + 1.23 (5) |
| FRB2-CFP-tK | 10.6 ± 1.7 × 10-2 | 9.3 ± 1.2 (17) |
| FRB2-CFP-tK(S181A) | 8.4 ± 1.1 × 10-2 | 10.1 ± 1.2 (19) |
| FRB2-CFP-Kras | 13.7 ± 2.2 × 10-2 | 6.9 ± 1.1 (14) |
| FRB2-CFP-Kras(G12V) | 10.2 ± 1.3 × 10-2 | 10.6 ± 1.8 (25) |
| FRB2-CFP-Kras(S17N) | 13.6 ± 2.1 × 10-2 | 6.3 ± 0.7 (14) |
| FRB2-CFP-tH | 8.8 ± 1.8 × 10-3 | 137 ± 58 (7) |
| Lck(1-10)-FRB2-CFP | 7.2 ± 1.4 × 10-4 | 1250 ± 370 (5) |
| Rapalog | ||
| FRB2-CFP-tH′(GG) | 8.3 ± 1.2 × 10-2 | 8.9 ± 1.3 (5) |
| FRB2-CFP-tK | 1.8 ± 0.2 × 10-2 | 46 ± 10 (10) |
| FRB2-CFP-Kras | 3.4 ± 0.5 × 10-2 | 32 ± 5 (22) |
Rate constants for intracellular redistribution of the indicated protein constructs in CV-1 cells coexpressing the mitoRFP-FKBP3 construct were determined by curve-fitting analysis as discussed in Materials and Methods and as illustrated in Figures 6 and 9. Values represent the mean ± SEM of n separate determinations.
Redistribution halftimes (t1/2) were estimated from each time course using the formula t1/2 = ln(2)/k, where k is the estimated redistribution rate constant. Average halftime values shown were calculated from these individual determinations of t1/2, not from the average rate-constant values shown. Values represent the mean ± SEM of n separate determinations, with n in parentheses.
To ensure that the above translocation phenomena were not linked to the ability of rapamycin to inhibit the protein kinase mTOR/FRAP (Huang et al., 2003), we repeated the above experiments using the rapamycin analogue (“rapalog”) AP21967, which does not inhibit mTOR/FRAP but can still induce heterodimerization of FRB- and FKBP-domain-containing protein constructs. As illustrated in Figure 5A and summarized in Table 1, at a concentration of 1 μM the rapalog also induces mitochondrial translocation of the FRB2-CFP-tH′(GG) construct in cells coexpressing this protein and mitoRFP-FKBP3, albeit with somewhat slower kinetics and with a lag that is not observed using rapamycin (0.5 μM) under the same conditions. This lag may reflect a slower rate of cell permeation, or a need to attain a higher intracellular concentration in order to promote efficient heterodimerization of FRB and FKBP domains, for the rapalog compared with rapamycin itself. Consistent with these possibilities, the rapalog-induced translocation of the FRB2-CFP-tH′(GG) construct was significantly accelerated by increasing the rapalog concentration (unpublished data). However, in order to ensure complete and consistent solubilization the rapalog was routinely used at a concentration of 1 μM.
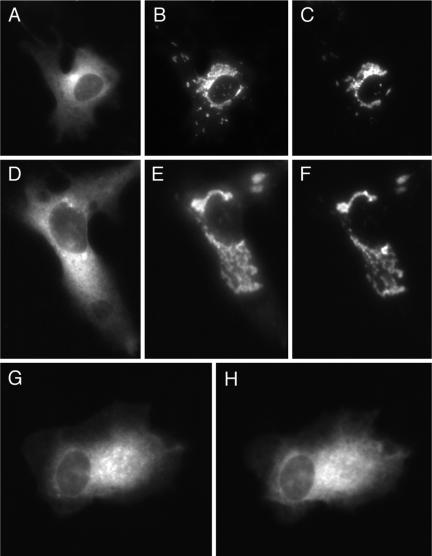
To compare with the above observations for singly lipidmodified protein constructs, we also examined the effects of rapamycin (in cells coexpressing mitoRFP-FKBP3) on the localization of constructs comprising an FRB2-CFP module anchored to the plasma membrane by multiply lipidated sequences or a transmembrane helix. As illustrated in Figures 5B and 6, A–C, for cells expressing an FRB2-CFP construct linked to the plasma membrane-targeting sequence of H-ras (-tH), which incorporates two S-acylation sites as well as a farnesylated carboxy-terminus, addition of rapamycin induces rapid redistribution of a small fraction of the construct to mitochondria, after which the remaining fraction redistributes much more slowly. A construct linking the FRB2-CFP module via its amino terminus to amino acid residues 1–10 of human p56lck, a myristoylated sequence incorporating two S-acylation sites, also shows a biphasic loss of plasma membrane-associated fluorescence, with a markedly slower second phase of redistribution than that observed for the -tH construct, upon addition of rapamycin to cells coexpressing this protein and mitoRFP-FKBP3 (Figures 5C and 6, D–F). The estimated halftimes for the slow phases of the redistribution process for the FRB2-CFP-tH and lck(1–10)-FRB2-CFP constructs (Table 1) are comparable to or longer than the previously reported halftimes for turnover of individual covalently bound S-acyl residues in H-ras and p56lck, respectively (Paige et al., 1993; Lu and Hofmann, 1995; Baker et al., 2000, 2003). This result is consistent with the proposal that the slow phase of translocation of these constructs represents release of their de-S-acylated forms from the plasma membrane. A construct fusing the FBP2-CFP module to the transmembrane domain of the Linker in Activation of T-cells (LAT) protein showed no rapamycininduced redistribution of fluorescence from the plasma membrane in cells coexpressing this protein with mitoRFP-FKBP3 (Figures 5D and 6, G–I).
Figure 6.
Effects of rapamycin on the subcellular distribution (in CV-1 cells coexpressing mitoRFP-FKBP3) of fluorescent FRB domain-containing protein constructs anchored to membranes via multiply lipid-modified or transmembrane sequences. (A–C) Cell expressing FRB2-CFP-tH imaged before (A) and 5 min (B) or 60 min (C) after addition of rapamycin (0.5 μM). (D–I) Cells expressing lck(1–10)-FRB2-CFP (D–F) and LAT(1–35)-FRB2-CFP (G–I), imaged at the same times as indicated for A–C.
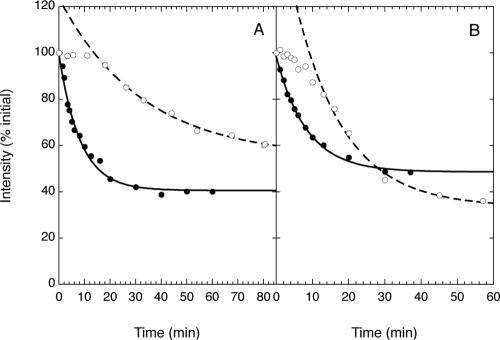
Having established that the experimental approach described here can detect intermembrane diffusion of membrane-associated proteins on a wide range of time scales, we examined the kinetics of dissociation from the plasma membrane of constructs in which FRB2-CFP modules were linked either to the carboxy-terminus of K-ras4B (-tK), which targets K-ras4B and heterologous proteins to the plasma membrane (Hancock et al., 1990, 1991) or to full-length K-ras4B. Both the FRB2-CFP-tK and the FRB2-CFP-Kras constructs are targeted specifically to the plasma membrane (Figure 7, A, G, and M), as confirmed by confocal microscopy (unpublished data). As illustrated in Figures 7, A–F, and 8A, rapamycin-induced translocation of the FRB2-CFP-tK construct to mitochondria (in cells coexpressing the mitoRFP-FKBP3 construct) could be detected within 1–2 min after rapamycin addition and proceeded over a time course of a few tens of minutes. Quantitative analysis as described above showed that the halftime for translocation of the FRB2-CFP-tK construct was significantly longer than that observed for the FRB2-CFP-tH′(F) construct, which is also farnesylated but lacks a polybasic sequence. Nonetheless, the halftime for intermembrane transfer of the tK-anchored construct is still only of the order of 10 min at 22°C and is likely even shorter at 37°C. Addition of the rapalog AP21967 (1 μM) also induced mitochondrial translocation of this construct, at a rate roughly fourfold slower than that observed using 0.5 μM rapamycin, similar to the results of analogous experiments described above for FRB2-CFP-tH′(GG) (Table 1). The construct FRB2-CFP-tK(S181A), in which serine-181 of the K-ras4B targeting sequence is mutated to an alanine residue to block potential phosphorylation, exhibits rapamycin-induced translocation to mitochondria in cells coexpressing mitoRFP-FKBP3 with kinetics very similar to those observed for the FRB2-CFP-tK construct under the same conditions (Table 1).
Figure 7.
Rapamycin- or rapalog-induced redistribution to mitochondria of fluorescent FRB-domain-containing constructs incorporating the K-ras4B-targeting sequence or fulllength K-ras4B in CV-1 cells coexpressing the mitoRFP-FKBP3 construct. (A–F) Subcellular distribution of FRB2-CFP-tK at 0 (A), 1 (B), 5 (C), 20 (D), and 60 min (E) after addition of rapamycin; (F) distribution of mitoRFP-FKBP3 in the same cells 60 min after rapamycin addition. The distribution of mitoRFP-FKBP3 at 0 min (unpublished data) was very similar. (G–L) Subcellular distribution of FRB2-CFP-Kras at 0 (G), 2 (H), 5 (I), 10 (J), and 45 min (K) after rapamycin addition; (L) distribution of mitoRFP-FKBP3 45 min after rapamycin addition. (M–O) Distribution of FRB2-CFP-Kras at 0 (M), 10 (N), and 45 min (O) after addition of rapalog AP29167.
Figure 8.
Kinetic analysis of rapamycin-induced intracellular redistribution of FRB2-CFP-tK and FRB2-CFP-Kras upon addition of rapamycin or rapalog AP29167 to CV-1 cells coexpressing the mitoRFP-FKBP3 protein. (A) Time courses of redistribution of FRB2-CFP-tK upon addition of 0.5 μM rapamycin (•)or1 μM rapalog (○). (B) Redistribution of FRB2-CFP-Kras upon addition of rapamycin (•) or rapalog (○).
Protein constructs fusing an FRB2-CFP module to fulllength K-ras4B also readily transfer from the plasma membrane to mitochondria upon addition of rapamycin or rapalog to cells coexpressing these constructs with mitoRFP-FKBP3, as illustrated for FRB2-CFP-Kras in Figure 7, G–O. As shown in Figure 8B and Table 1, the rate of translocation of the K-ras4B fusion protein is comparable to but slightly faster than that observed with the FRB2-CFP-tK construct. A fusion protein incorporating the GDP/GTP exchange-defective, and hence constitutively deactivated, S17N mutant of K-ras4B exhibited a rate of intermembrane redistribution very similar to that observed for the wild-type K-ras4B fusion construct. An analogous fusion protein incorporating the constitutively active (GTPase-defective) form of K-ras4B appears to show a modestly slower rate of transfer from the plasma membrane than do the equivalent wild-type and S17N constructs, a difference that approaches the level of statistical significance (p < 0.10 by Student's t test based on the calculated rate constants, and p < 0.05 based on the calculated half-times, for intermembrane transfer). This result suggests that the inactive (GDP-bound) form of the protein may exhibit a somewhat faster rate of desorption from the plasma membrane than the active (GTP-bound) form.
DISCUSSION
Our present findings agree with recent reports that farnesylated H-ras in its unpalmitoylated form associates dynamically with cellular membranes (Goodwin et al., 2005; Rocks et al., 2005) and with the results of previous biophysical studies showing that singly farnesylated, geranylgeranylated, and myristoylated peptides can rapidly transfer between different lipid bilayers (Silvius and l'Heureux, 1994; J. Silvius, unpublished observations). Our conclusion that the lipid-modified FRB2-CFP module-containing proteins studied here redistribute between membranes (here, to mitochondria) by diffusion through the cytoplasm, rather than by membrane trafficking processes, is based on several considerations. First, no pathway is known to exist for transfer of membrane-bound proteins from the plasma membrane, or from the ER/Golgi or other endomembrane compartments, to the mitochondrial outer membrane. Although transfer of newly synthesized phosphatidylserine between the ER and mitochondrial membranes may occur at zones of apposition between the two membrane compartments (Vance and Shiao, 1996; Daum and Vance, 1997; Shiao et al., 1998), this process is highly specific for phosphatidylserine over other lipids (Shiao et al., 1998) and does not appear to represent a general pathway for transfer of membrane material between the two compartments. Second, the rapamycin-induced redistribution to mitochondria of singly lipid-modified protein constructs is very rapid, with halftimes for transfer of ca. 10 min or less (and in some cases <1–2 min) at 22°C, whereas vesicular membrane trafficking processes are typically considerably slower at this temperature (Saraste et al., 1986; Kuismanen and Saraste, 1989). Third, a fluorescent protein construct stably anchored to the plasma membrane via the transmembrane sequence of LAT shows no detectable translocation to mitochondria under our experimental conditions. Finally, we observe that upon rapamycin addition singly lipid-modified protein constructs can be rapidly depleted from diverse cellular membranes. This result is exactly that expected if these constructs can readily diffuse between different cellular membranes, but it is much more difficult to explain as a consequence of the working of specific (and, given the different initial localizations of various prenylated species, presumably multiple) membrane trafficking pathways.
Our present findings do not necessarily imply that single lipid-modified proteins diffuse through the cytoplasm as monomeric entities. Soluble proteins such as Rho-GDI (Michaelson et al., 2001) and phosphodiesterase δ-subunit (Nancy et al., 2002) can bind prenylated proteins with varying degrees of specificity, and it is quite possible that one or more of these species can serve at least to enhance the rate of intermembrane transfer of a given prenylated protein. However, we note again that fast rates of intermembrane transfer, similar to those we observe in the intracellular milieu for prenylated protein constructs lacking additional lipid groups, are also observed for prenylated or myristoylated peptides in lipid model membrane systems (Silvius and l'Heureux, 1994; Leventis and Silvius, 1998; J. Silvius, unpublished results).
Previous studies of the dynamics of plasma membrane association of K-ras4B have reached divergent conclusions. Yokoe and Meyer (1996) concluded that the rate of exchange of a GFP-K-ras4B fusion protein between the plasma membrane and cytoplasm is very rapid, with a halftime of 1.5 s. By contrast, Niv et al. (1999), from an extensive analysis of photobleaching-recovery kinetics for a similar fusion construct, concluded that the halftime for desorption of the activated G12V mutant of K-ras4B from the plasma membrane must be considerably greater, although they were unable to estimate a precise value for the desorption rate. Our present findings are compatible with the conclusions of Niv et al., but they indicate that desorption of the FRB2-CFP-Kras fusion protein from the plasma membrane is nonetheless relatively rapid, with a halftime of the order of several minutes at 22°C, and likely even less at 37°C. Fusion constructs incorporating wild-type, constitutively activated (GTPase-defective) or constitutively deactivated (GDP/GTP exchange-defective) forms of K-ras4B desorb from the plasma membrane at similar rates, although the activated form may desorb at a modestly slower rate than does the unactivated species. This latter observation is reminiscent of previous reports that when expressed at low levels, fluorescent protein-tagged derivatives of constitutively activated mutants of H- and K-ras show greater restriction of their lateral diffusion in the plasma membrane than do analogous fluorescent derivatives of the constitutively deactivated S17N mutant (Niv et al., 2002; Lommerse et al., 2005).
Our present results strongly support the proposal that plasma membrane targeting mediated by the K-ras4B targeting sequence rests on a high equilibrium binding affinity for the plasma membrane vis-à-vis other cellular membranes, rather than on a “kinetic trapping” mechanism (Shahinian and Silvius, 1995) as is the case for S-acylated proteins such as H- and N-ras (Goodwin et al., 2005; Rocks et al., 2005). This conclusion is consistent with previous observations that in contrast to H- and N-ras, K-ras4B reaches the plasma membrane by a nonvesicular, possibly diffusional mechanism (Choy et al., 1999; Apolloni et al., 2000). It will be of interest to determine whether the dynamic plasma membrane association observed for K-ras4B is also characteristic of other proteins, such as pp60src and various species of Rho proteins, whose plasma membrane targeting sequences also combine a single lipid modification with an adjacent polybasic sequence (Michaelson et al., 2001).
The functional implications of the dynamic nature of the K-ras4B/plasma membrane interaction remains to be fully assessed, but at least two possibilities can be readily suggested from our current knowledge of the properties of K-ras4B and other ras proteins. First, an activated form of H-ras, but not of K-ras4B, has been shown to accumulate in endosomes when the rate of endocytosis is enhanced vis-à-vis that of endosomal recycling using an activated mutant of rab5 (Roy et al., 2002). It has been suggested on the basis of such results that K-ras4B (but not H-ras) may be retrieved from the endosomal compartment to the plasma membrane by simple diffusion rather than through endosomal recycling (Roy et al., 2002). This suggestion is fully consistent with our finding that membrane association of K-ras4B is dynamic, whereas that of H-ras is much more slowly reversible (Goodwin et al., 2005; Rocks et al., 2005; note also our present findings for the FRB2-CFP-tH protein construct). Second, Hand N-ras have been shown to mediate some of their biological functions in cellular compartments other than the plasma membrane (Chiu et al., 2002; Bivona et al., 2003; Caloca et al., 2003; Perez de Castro et al., 2004; Peyker et al., 2005), and it is possible that the same may be true for K-ras4B as well. If so, the dynamic nature of the K-ras4B/plasma membrane interaction could facilitate rapid recruitment of K-ras4B to (or sequestration in) cellular compartments other than the plasma membrane under particular signaling conditions. This possibility is supported by very recent reports of calcium/calmodulin- or phosphorylation-induced relocation of K-ras4B from the plasma membrane to the Golgi or to mitochondria, respectively (Fivaz and Meyer, 2005; Philips, 2005).
Acknowledgments
This research was supported by a grant from the Canadian Institutes of Health Research (CIHR) to J.R.S. and by scholarship support to P.B., from the CIHR Strategic Training Initiative in Chemical Biology, and to D.T., from the CIHR-funded Strategic Training Initiative of the McGill Cancer Research Consortium.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–05–0408) on October 19, 2005.
Abbreviations used: CFP, Cerulean variant of cyan fluorescent protein; ER, endoplasmic reticulum; FKBP, rapamycin-binding heterodimerization domain of human FKBP12; FRB, rapamycin-binding heterodimerization domain of mTOR/FRAP; (m)RFP, monomeric red fluorescent protein; YFP, monomeric Citrine variant of yellow fluorescent protein.
References
- Apolloni, A., Prior, I. A., Lindsay, M., Parton, R. G., and Hancock, J. F. (2000). H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol. Cel. Biol. 20, 2475–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, T. L., Booden, M. A., and Buss, J. E. (2000). S-Nitrosocysteine increases palmitate turnover on Ha-Ras in NIH 3T3 cells. J. Biol. Chem. 275, 22037–22047. [DOI] [PubMed] [Google Scholar]
- Baker, T. L., Zheng, H., Walker, J., Coloff, J. L., and Buss, J. E. (2003). Distinct rates of palmitate turnover on membrane-bound cellular and oncogenic Hras. J. Biol. Chem. 278, 19292–19300. [DOI] [PubMed] [Google Scholar]
- Beranger, F., Goud, B., Tavitian, A., and de Gunzburg, J. (1991). Association of the Ras-antagonistic Rap1/Krev-1 proteins with the Golgi complex. Proc. Natl. Acad. Sci. USA 88, 1606–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivona, T. G., Perez De Castro, I., Ahearn, I. M., Grana, T. M., Chiu, V. K., Lockyer, P. J., Cullen, P. J., Pellicer, A., Cox, A. D., and Philips, M. R. (2003). Phospholipase Cγ activates Ras on the Golgi apparatus by means of RasGRP1. Nature 424, 694–698. [DOI] [PubMed] [Google Scholar]
- Calero, M., Chen, C. Z., Zhu, W., Winand, N., Havas, K. A., Gilbert, P. M., Burd, C. G., and Collins, R. N. (2003). Dual prenylation is required for Rab protein localization and function. Mol. Biol. Cell 14, 1852–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caloca, M. J., Zugaza, J. L., and Bustelo, X. R. (2003). Exchange factors of the RasGRP family mediate Ras activation in the Golgi. J. Biol. Chem. 278, 33465–33473. [DOI] [PubMed] [Google Scholar]
- Castellano, F., Montcourrier, P., Guillemot, J.-C., Gouin, E., Machevsky, L., Cossart, P., and Chavrier, P. (1999). Inducible recruitment of Cdc424 or WASP to a cell-surface receptor triggers actin polymerization and filopodium formation. Curr. Biol. 9, 351–360. [DOI] [PubMed] [Google Scholar]
- Chen, J., Zheng, X. F., Brown, E. J., and Schreiber, S. L. (1995). Identification of an 11–kDa rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc. Natl. Acad. Sci. USA 92, 4947–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, V. K., Bivona, T., Hach, A., Sajous, J. B., Silletti, J., Wiener, H., Johnson, R. L., 2nd, Cox, A. D., and Philips, M. R. (2002). Ras signalling on the endoplasmic reticulum and the Golgi. Nat. Cell Biol. 4, 343–350. [DOI] [PubMed] [Google Scholar]
- Choi, J., Chen, J., Schreiber, S. L., and Clardy, J. (1996). Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science 273, 239–242. [DOI] [PubMed] [Google Scholar]
- Choy, E., Chiu, V. K., Silletti, J., Feoktistov, M., Morimoto, T., Michaelson, D., Ivanov, I. E., and Philips, M. R. (1999). Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell 98, 69–80. [DOI] [PubMed] [Google Scholar]
- Dai, Q., Choy, E., Chiu, V., Romano, J., Slivka, S. R., Steitz, S. A., Michaelis, S., and Philips, M. R. (1998). Mammalian prenylcysteine carboxyl methyltransferase is in the endoplasmic reticulum. J. Biol. Chem. 273, 15030–15034. [DOI] [PubMed] [Google Scholar]
- Daum, G., and Vance, J. E. (1997). Import of lipids into mitochondria. Prog. Lipid Res. 36, 103–130. [DOI] [PubMed] [Google Scholar]
- Donaldson, J. G. (2003). Multiple roles for Arf6, sorting, structuring, and signaling at the plasma membrane. J. Biol. Chem. 278, 41573–41576. [DOI] [PubMed] [Google Scholar]
- Dong, X., Mitchell, D. A., Lobo, S., Zhao, L., Bartels, D. J., and Deschenes, R. J. (2003). Palmitoylation and plasma membrane localization of Ras2p by a nonclassical trafficking pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 23, 6574–6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fivaz, M., and Meyer, T. (2005). Reversible intracellular translocation of KRas but not HRas in hippocampal neurons regulated by Ca2+/calmodulin. J. Cell Biol. 170, 429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H. W., and Casey, P. J. (1999). Enzymology and biology of CaaX protein prenylation. Rec. Prog. Hormone Res. 54, 315–342. [PubMed] [Google Scholar]
- Gomes, A. Q., Ali, B. R., Ramalho, J. S., Godfrey, R. F., Barral, D. C., Hume, A. N., and Seabra, M. C. (2003). Membrane targeting of Rab GTPases is influenced by the prenylation motif. Mol. Biol. Cell 14, 1882–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin, J. S., Drake, K. R., Rogers, C., Wright, L., Lippincott-Schwartz, J., Philips, M. R., and Kenworthy, A. K. (2005). Depalmitoylated Ras traffics to and from the Golgi complex via a nonvesicular pathway. J. Cell Biol. 170, 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez, L., Magee, A. I., Marshall, C. J., and Hancock, J. F. (1989). Posttranslational processing of p21ras is two-step and involves carboxyl-methylation and carboxy-terminal proteolysis. EMBO J. 8, 1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock, J. F., Paterson, H., and Marshall, C. J. (1990). A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 63, 133–139. [DOI] [PubMed] [Google Scholar]
- Hancock, J. F., Cadwallader, K., Paterson, H., and Marshall, C. J. (1991). A CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targeting of ras proteins. EMBO J. 10, 4033–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haun, R. S., Tsai, S. C., Adamik, R., Moss, J., and Vaughan, M. (1993). Effect of myristoylation on GTP-dependent binding of ADP-ribosylation factor to Golgi. J. Biol. Chem. 268, 7064–7068. [PubMed] [Google Scholar]
- Huang, S., Bjornsti, M. A., and Houghton, P. J. (2003). Rapamycins: mechanism of action and cellular resistance. Cancer Biol. Ther. 2, 222–232. [DOI] [PubMed] [Google Scholar]
- Kuismanen, E., and Saraste, J. (1989). Low temperature-induced transport blocks as tools to manipulate membrane traffic. Methods Cell Biol. 32, 257–274. [DOI] [PubMed] [Google Scholar]
- Leventis, R., and Silvius, J. R. (1998). Lipid-binding characteristics of the polybasic carboxy-terminal sequence of K-ras4B. Biochemistry 37, 7640–7648. [DOI] [PubMed] [Google Scholar]
- Li, J. M., and Shore, G. C. (1992). Protein sorting between mitochondrial outer and inner membranes. Insertion of an outer membrane protein into the inner membrane. Biochim. Biophys. Acta 1106, 233–241. [DOI] [PubMed] [Google Scholar]
- Lommerse, P. H., Snaar-Jagalska, B. E., Spaink, H. P., and Schmidt, T. (2005). Single-molecule diffusion measurements of H-Ras at the plasma membrane of live cells reveal microdomain localization upon activation. J. Cell Sci. 118, 1799–1809. [DOI] [PubMed] [Google Scholar]
- Lu, J. Y., and Hofmann, S. L. (1995). Depalmitoylation of CAAX motif proteins. Protein structural determinants of palmitate turnover rate. J. Biol. Chem. 270, 7251–7256. [DOI] [PubMed] [Google Scholar]
- Michaelson, D., Silletti, J., Murphy, G., D'Eustachio, P., Rush, M., and Philips, M. R. (2001). Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J. Cell Biol. 152, 111–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nancy, V., Callebaut, I., El Marjou, A., and de Gunzburg, J. (2002). The delta subunit of retinal rod cGMP phosphodiesterase regulates the membrane association of Ras and Rap GTPases. J. Biol. Chem. 277, 15076–15084. [DOI] [PubMed] [Google Scholar]
- Niv, H., Gutman, O., Henis, Y., and Kloog, Y. (1999). Membrane interactions of a constitutively active GFP-Ki-ras 4B and their role in signaling. Evidence from lateral mobility studies. J. Biol. Chem. 274, 1606–1613. [DOI] [PubMed] [Google Scholar]
- Niv, H., Gutman, O., Kloog, Y., and Henis, Y. I. (2002). Activated K-Ras and H-Ras display different interactions with saturable nonraft sites at the surface of live cells. J. Cell Biol. 157, 865–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, K., Kanemura, H., Satoh, T., and Kataoka, T. (2004). Identification of a novel domain of Ras and Rap1 that directs their differential subcellular localizations. J. Biol. Chem. 279, 22664–22673. [DOI] [PubMed] [Google Scholar]
- Otto, J. C., Kim, E., Young, S. G., and Casey, P. J. (1999). Cloning and characterization of a mammalian prenyl protein-specific protease. J. Biol. Chem. 274, 8379–8382. [DOI] [PubMed] [Google Scholar]
- Paige, L. A., Nadler, M. J., Harrison, M. L., Cassady, J. M., and Geahlen, R. L. (1993). Reversible palmitoylation of the protein-tyrosine kinase p56lck. J. Biol. Chem. 268, 8669–8674. [PubMed] [Google Scholar]
- Perez de Castro, I., Bivona, T. G., Philips, M. R., and Pellicer, A. (2004). Ras activation in Jurkat T cells following low-grade stimulation of the T-cell receptor is specific to N-Ras and occurs only on the Golgi apparatus. Mol. Cell. Biol. 24, 3485–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyker, A., Rocks, O., and Bastiaens, P. I. (2005). Imaging activation of two ras isoforms simultaneously in a single cell. ChemBioChem 6, 78–85. [DOI] [PubMed] [Google Scholar]
- Philips, M. R. (2005). Compartmentalized signalling of Ras. Biochem. Soc. Trans. 33, 657–661. [DOI] [PubMed] [Google Scholar]
- Pizon, V., Desjardins, M., Bucci, C., Parton, R. G., and Zerial, M. (1994). Association of Rap1a and Rap1b proteins with late endocytic/phagocytic compartments and Rap2a with the Golgi complex. J. Cell Sci. 107, 1661–1670. [DOI] [PubMed] [Google Scholar]
- Rizzo, M. A., Springer, G. H., Granada, B., and Piston, D. W. (2004). An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 22, 445–449. [DOI] [PubMed] [Google Scholar]
- Romano, J. D., Schmidt, W. K., and Michaelis, S. (1998). The Saccharomyces cerevisiae prenylcysteine carboxyl methyltransferase Ste14p is in the endoplasmic reticulum membrane. Mol. Biol. Cell 9, 2231–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocks, O., Peyker, A., Kahms, M., Verveer, P. J., Koerner, C., Lumbierres, M., Kuhlman, J., Waldmann, H., Wittinghofer, A., and Baestiaens, P.I.H. (2005). An acylation cycle regulates localization and activity of palmitoylated ras isoforms. Science 307, 1746–1752. [DOI] [PubMed] [Google Scholar]
- Roskoski, R., Jr. (2003). Protein prenylation: a pivotal posttranslational process. Biochem. Biophys. Res. Commun. 303, 1–7. [DOI] [PubMed] [Google Scholar]
- Roy, M.-O., Leventis, R., and Silvius, J. R. (2000). Mutational and biochemical analysis of plasma membrane targeting mediated by the farnesylated, polybasic carboxy terminus of K-ras4B. Biochemistry 39, 8298–8307. [DOI] [PubMed] [Google Scholar]
- Roy, S., Wyse, B., and Hancock, J. F. (2002). H-Ras signaling and K-Ras signaling are differentially dependent on endocytosis. Mol. Cell. Biol. 22, 5128–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, S., Plowman, S., Rotblat, B., Prior, I. A., Muncke, C., Grainger, S., Parton, R. G., Henis, Y. I., Kloog, Y., and Hancock, J. F. (2005). Individual palmitoyl residues serve distinct roles in H-ras trafficking, microlocalization, and signaling. Mol. Cell. Biol. 25, 6722–6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste, J., Palade, G. E., and Farquhar, M. G. (1986). Temperature-sensitive steps in the transport of secretory proteins through the Golgi complex in exocrine pancreatic cells. Proc. Natl. Acad. Sci. USA 83, 6425–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, W. K., Tam, A., Fujimura-Kamada, K., and Michaelis, S. (1998). Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage. Proc. Natl. Acad. Sci. USA 95, 11175–11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, H., Leventis, R., Rex, S., Schelhaas, M., Nägele, E., Waldmann, H., and Silvius, J. R. (1997). S-Acylation and plasma membrane targeting of the farnesylated carboxyl-terminal peptide of N-ras in mammalian fibroblasts. Biochemistry 36, 13102–13109. [DOI] [PubMed] [Google Scholar]
- Shahinian, S., and Silvius, J. R. (1995). Doubly-lipid-modified protein sequence motifs exhibit long-lived anchorage to lipid bilayer membranes. Biochemistry 34, 3813–3822. [DOI] [PubMed] [Google Scholar]
- Shiao, Y. J., Balcerzak, B., and Vance, J. E. (1998). A mitochondrial membrane protein is required for translocation of phosphatidylserine from mitochondria-associated membranes to mitochondria. Biochem. J. 331, 217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvius, J. R. (2002). Mechanisms of Ras protein targeting in mammalian cells. J. Membr. Biol. 190, 83–92. [DOI] [PubMed] [Google Scholar]
- Silvius, J. R., and l'Heureux, F. (1994). Fluorimetric evaluation of the affinities of isoprenylated peptides for lipid bilayers. Biochemistry 33, 3014–3022. [DOI] [PubMed] [Google Scholar]
- Sinensky, M. (2000). Recent advances in the study of prenylated proteins. Biochim. Biophys. Acta 1484, 93–106. [DOI] [PubMed] [Google Scholar]
- Stockwell, B. R., and Schreiber, S. L. (1998). Probing the role of homomeric and heteromeric receptor interactions in TGF-β signaling using small molecule dimerizers. Curr. Biol. 8, 761–770. [DOI] [PubMed] [Google Scholar]
- Vance, J. E., and Shiao, Y. J. (1996). Intracellular trafficking of phospholipids: import of phosphatidylserine into mitochondria. Anticancer Res. 16, 1333–1339. [PubMed] [Google Scholar]
- Willumsen, B. M., Cox, A. D., Solski, P. A., Der, C. J., and Buss, J. E. (1996). Novel determinants of H-Ras plasma membrane localization and transformation. Oncogene 13, 1901–1909. [PubMed] [Google Scholar]
- Yokoe, H., and Meyer, T. (1996). Spatial dynamics of GFP-tagged proteins investigated by local fluorescence enhancement. Nat. Biotechnol. 14, 1252–1256. [DOI] [PubMed] [Google Scholar]
- Zacharias, D. A., Violin, J. D., Newton, A. C., and Tsien, R. Y. (2002). Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296, 913–916. [DOI] [PubMed] [Google Scholar]
- Zhang, F. L., and Casey, P. J. (1996). Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 65, 241–269. [DOI] [PubMed] [Google Scholar]