Abstract
We previously showed that thioredoxins are required for dithiothreitol (DTT) tolerance, suggesting they maintain redox homeostasis in response to both oxidative and reductive stress conditions. In this present study, we screened the complete set of viable deletion strains in Saccharomyces cerevisiae for sensitivity to DTT to identify cell functions involved in resistance to reductive stress. We identified 195 mutants, whose gene products are localized throughout the cell. DTT-sensitive mutants were distributed among most major biological processes, but they particularly affected gene expression, metabolism, and the secretory pathway. Strikingly, a mutant lacking TSA1, encoding a peroxiredoxin, showed a similar sensitivity to DTT as a thioredoxin mutant. Epistasis analysis indicated that thioredoxins function upstream of Tsa1 in providing tolerance to DTT. Our data show that the chaperone function of Tsa1, rather than its peroxidase function, is required for this activity. Cells lacking TSA1 were found to accumulate aggregated proteins, and this was exacerbated by exposure to DTT. Analysis of the protein aggregates revealed that they are predominantly composed of ribosomal proteins. Furthermore, aggregation was found to correlate with an inhibition of translation initiation. We propose that Tsa1 normally functions to chaperone misassembled ribosomal proteins, preventing the toxicity that arises from their aggregation.
INTRODUCTION
Cells originally evolved in a reducing environment. The accumulation of oxygen in the atmosphere then made possible the efficient energy generation process of respiration. Cells have therefore evolved to survive both oxidative and reductive conditions. Although much is now known regarding the damaging effects of oxidation, little is known regarding the molecular responses to a reducing environment, which is the subject of this present study.
Glutathione is the most abundant low-molecular-weight thiol in eukaryotic cells and has been widely used as an indicator of the cellular redox state (Schafer and Buettner, 2001). The cytoplasm is generally very reducing; for example, the ratio of reduced glutathione (GSH) to oxidized glutathione (GSSG) in yeast is in the range of ∼70-190:1 (Grant et al., 1998; Garrido and Grant, 2002). In contrast, the endoplasmic reticulum is more oxidizing and the GSH:GSSG ratio has been measured in the range of 1:1-3:1 (Hwang et al., 1992; Bass et al., 2004). Disulfide bonds are essential for the folding and stability of proteins that are secreted or localized through the secretory pathway. In eukaryotic cells, disulfide bonds are formed in nascent proteins within the lumen of the endoplasmic reticulum (ER). Genetic evidence has identified a pathway for disulfide bond formation where oxidizing equivalents are transferred between thiol-containing proteins and secretory proteins (Frand et al., 2000; Gross et al., 2002). Thus, oxidative protein folding proceeds primarily by protein-protein-based relays. Glutathione provides a buffer against hyperoxidizing conditions in the ER and functions in the reductive pathway for the formation of native disulfide bonds (Cuozzo and Kaiser, 1999; Jessop and Bulleid, 2004). However, GSH also competes for oxidizing equivalents in the ER, because elevated GSH disrupts ER function and activates the unfolded protein response (UPR) owing to an accumulation of misfolded proteins (Cuozzo and Kaiser, 1999). Compartmentalization of the oxidative chemistry required for disulfide formation is important to protect cells from being exposed to nonspecific oxidation or reduction events.
DTT is a strong reducing agent that is often used to promote reductive stress. It can cross membranes and prevent disulfide bond formation. For example, DTT reversibly blocks the folding and transport from the ER of vacuolar carboxypeptidase Y (CPY), which contains five disulfides in its native form (Simons et al., 1995). A reductive stress induced by DTT treatment leads to an accumulation of misfolded proteins in the ER, which signals the UPR through the action of the Ire1 transmembrane kinase/nuclease (Cox et al., 1993; Kohno et al., 1993). Microarray analysis has elucidated the transcriptional scope of the UPR and shown that it affects multiple ER and secretory pathway functions that normally function to counteract ER stress (Travers et al., 2000). Interestingly, many genes are up-regulated in response to DTT treatment whose functions seem unrelated to the ER and do not form part of the UPR (Gasch et al., 2000). These gene products are likely to function in protection against reductive stress.
A reductive stress can occur in response to any conditions that shift the redox balance of important biological redox couples, such as the NAD+/NADH, NADP+/NADPH, and GSSG/2GSH couples, to a more reducing state (Schafer and Buettner, 2001). These couples are often thermodynamically linked, and the redox environment of a cell is a reflection of the state of these couples. There is also increasing evidence that alterations in the redox state of protein-sulfhydryl groups can affect many protein functions and activities (Rietsch and Beckwith, 1998). Thus, any reductive or oxidative challenges that alter the cellular redox environment will influence many cellular processes. Not surprisingly, therefore, a strong correlation has been observed between the redox environment of the cell and cellular growth and differentiation; for example, a more reducing environment is linked with the proliferation of some tumor cells (Schafer and Buettner, 2001; Nkabyo et al., 2002; Yeh et al., 2005). Excess reductants may also disrupt signaling pathways that rely on low physiologically relevant concentrations of reactive oxygen species (ROS) as well as redox-sensitive transcription factors (Sen, 2000). A reductive stress can also arise from an imbalance in the levels of key metabolites. For example, the inability to oxidize NAD+ results in an NADH-reductive stress that impairs yeast cell growth (Valadi et al., 2004). NAD is required for numerous biological processes, and many human diseases are associated with changes in NAD+ levels and/or the NAD+:NADH ratio (Lin and Guarente, 2003).
We have previously tested whether known redox regulatory systems are required for protection against a reductive stress and shown that thioredoxin mutants (trx1 trx2) are sensitive to DTT (Trotter and Grant, 2002). Thioredoxins are small, highly conserved oxidoreductases that have been best characterized for their role in protection against ROS (Holmgren, 1989). Sensitivity to a reducing agent is somewhat surprising, given their accepted role as antioxidants. This sensitivity to reducing conditions is not a general property of mutants affected in redox control because mutants lacking components of the GSH/glutaredoxin system are unaffected. Further evidence that thioredoxins form part of the cellular response to a reductive challenge came from the finding that TRX2 gene expression is induced in response to DTT treatment and does not depend on the presence of IRE1 (Trotter and Grant, 2002). Loss of thioredoxins is known to result in elevated glutathione levels, indicating a link between the thioredoxin system and glutathione metabolism in the cell (reviewed in Carmel-Harel and Storz, 2000; Wheeler and Grant, 2004). We proposed that the combination of exogenous DTT and high intracellular GSH results in an additive reductive stress that is toxic to thioredoxin mutants (Trotter and Grant, 2002). However, subsequent analysis of the cellular redox potential of a trx1 trx2 mutant revealed that it is comparable with that of a wild-type strain because the mutant also contains elevated levels of GSSG (Trotter and Grant, 2003). Thus, there does not seem to be a correlation between the cellular redox state and DTT sensitivity.
In this present study, we have a performed a genome-wide screen to identify mutants that are sensitive to DTT. This analysis shows that a wide range of gene functions are required to protect against DTT stress, including a number of factors that affect the thioredoxin system. We show that the role of thioredoxins in protection against reductive stress is dependent on their activity as a cofactor for the Tsa1 peroxiredoxin (Prx). In particular, the chaperone activity of Tsa1 is required to protect against DTT-mediated protein aggregation. Surprisingly, ribosomal proteins were found to be particularly susceptible to aggregation in response to reductive stress. This finding has important implications in the regulation of protein synthesis in response to changes in redox status.
MATERIALS AND METHODS
Yeast Strains and Plasmids
The Saccharomyces cerevisiae strains used in this study were isogenic derivatives of CY4 (MATa ura3-52 leu2-3 leu2-112 trp1-1 ade2-1 his3-11 can1-100) (Grant et al., 1996b). Strains deleted for thioredoxins (trx1::TRP1 trx2::URA3) and YAP1 (yap1::HIS3) have been described previously (Grant et al., 1996a; Draculic et al., 2000). Strain CY399, which is deleted for GPX3 (gpx3::HIS3); strain CY947, which is deleted for SKN7 (skn7::LEU2); and strain CY954, which is deleted for TSA1 (tsa1::LEU2) were constructed using a one-step PCR amplification protocol (Baudin et al., 1993). Multicopy plasmids containing TRX1, TRX2, TSA1 (Garrido and Grant, 2002), TSA1 containing mutations in the active site cysteine residues (Cys47 and Cys170) (Wong et al., 2004), ERO1 (Haynes et al., 2004), HAC1, and a HAC1 gene fusion with the activation domain of the Gal4 transcriptional activator [mcGAL4(AD)-HAC1] (Mori et al., 2000) have all been described previously.
Growth Conditions
Strains were grown in rich YEPD medium [2% (wt/vol) glucose, 2% (wt/vol) bactopeptone, and 1% (wt/vol) yeast extract) or minimal SD medium (0.17% (wt/vol) yeast nitrogen base without amino acids, 5% (wt/vol) ammonium sulfate, 2% (wt/vol) glucose supplemented with appropriate amino acids and bases (Sherman et al., 1974) at 30°C and 180 rpm. Media were solidified by the addition of 2% (wt/vol) agar. SGal plates contained 2% (wt/vol) galactose. For anaerobic growth conditions, media was supplemented with 0.1% (vol/vol) Tween 80 and 30 mg/l ergosterol, and plates were maintained in an anaerobic jar (Oxoid, Basingstoke, Hampshire, England) containing a gas generating kit (Anaerobic system BR38; Oxoid). Stress sensitivity was determined by growing cells to stationary phase and spotting onto agar plates containing various concentrations of DTT, cysteine, H2O2, or diamide.
Genome-Wide Screening for Mutations Causing Sensitivity to DTT
The S.. cerevisiae strains used in this study were derivatives of BY4743 (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 lys2Δ0/LYS2 ura3Δ0/ura3Δ0), which are homozygous for the relevant gene deletion. The construction of the yeast genome deletion library by the European S. cerevisiae Archive for Functional Analysis (EUROSCARF) has been described previously (Winzeler, 1999). Cells were grown to stationary phase in YEPD medium in static 96 well-plates. After resuspension, cells were plated onto YEPD containing 32 mM DTT using a 96 pin replicator. Plates were incubated at 30°C for 3-4 d before scoring growth. The screen was repeated twice, and mutants were identified which were reproducibly sensitive to DTT.
Measurement of Intracellular Oxidation
Oxidized glutathione levels were determined as described previously (Grant et al., 1998). Briefly, cells were grown to exponential phase in YEPD medium, treated with DTT for 1 h, and harvested by centrifugation. Cells were washed twice with phosphate-buffered saline (PBS), pH 7.4, to remove any traces of growth medium and resuspended in ice-cold 8 mM HCl, 1.3% (wt/vol) 5-sulfosalicyclic acid. Cells were broken with glass beads using a Minibead beater (Biospec Products, Bartlesville, OK) for 40 s at 4°C. Cell debris and precipitated proteins were pelleted in a microcentrifuge for 15 min (13,000 rpm; 4°C). The collected supernatants were pretreated with 5% (vol/vol) 2-vinylpyridine for 1 h at room temperature before analysis. GSSG levels are expressed as nanomoles per absorbance at 600 nm of cells.
Intracellular ROS were measured using the oxidant-sensitive probe 2′,7′-dichlorofluorescin diacetate (DCFH-DA). Cells growing exponentially in SD medium were treated with DTT for 15 min. After treatment, cells were harvested, resuspended in PBS containing 10 μM DCFH-DA, and incubated for 1 h at 28°C. Cells were broken with glass beads using a Minibead beater, and fluorescence was measured using a SynegyHT plate reader (Bio-Tek Instruments, Winooski, VT) with excitation and emission wavelengths of 485 and 528 nm, respectively. Extracts from untreated cells that had been incubated without DCFH-DA were used as blanks for background fluorescence.
Measurement of Protein Aggregation
The aggregation of soluble proteins was analyzed essentially as described by Jang et al. (2004). Cells were grown to exponential phase in YEPD medium and treated with DTT for 1 h. Equivalent cell numbers (10 A600 units) were harvested by centrifugation, washed, and resuspended in 300 μl of lysis buffer [50 mM potassium phosphate buffer, pH 7, 1 mM EDTA, 5% (vol/vol) glycerol, 1 mM phenylmethylsulfonyl fluoride, and Complete Mini protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN)]. The cells were lysed by freezing and thawing followed by an incubation (30°C for 30 min) with 100 μl of lyticase (9830 U/ml) (Sigma-Aldrich, St. Louis, MO). Cells were disrupted with glass beads using a Minibead beater for 3 × 1 min at 4°C. Each beating was followed by a 1-min cooling at 4°C. Intact cells were removed by centrifugation at 3000 × g for 15 min. The membrane and aggregated proteins were isolated by centrifugation at 15,000 × g for 20 min. The membrane proteins were removed by washing twice with 320 μl of lysis buffer and 80 μl of 10% Igepal CA-630 (NP-40) (Sigma-Aldrich), centrifuging at 15,000 × g for 20 min each time. The final aggregated protein extract was resuspended in 100 μl of lysis buffer and analyzed by 12% reducing SDS-PAGE. Proteins were visualized by silver staining with the Bio-Rad (Hercules, CA) silver stain plus kit.
For protein identification, gels were stained using colloidal Coomassie blue (Sigma-Aldrich) and dried using a Bio-Rad Gelair drier. Proteins of interest were excised from gels and peptide mass fingerprints were generated using matrix-assisted laser desorption ionization/time of flight (MALDI-TOF). Proteins were identified using the Mascot mass fingerprinting program (www.matrixscience.com) to search the NCBInr and Swissprot databases.
Western Blot Analysis
Protein extracts were electrophoresed under reducing conditions on 12% SDS-PAGE minigels (αRps3) or 16% tricine Novex minigels (αTrx1,2) and electroblotted onto polyvinylidene difluoride membrane (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). Bound antibody was visualized by chemiluminescence (GE Healthcare) after incubation of the blot in donkey anti-rabbit immunoglobulin-horseradish peroxidase conjugate (GE Healthcare).
Measurement of Protein Synthesis Rate
To measure the rate of protein synthesis, exponential phase cells were treated with DTT for 1 h and pulse labeled for the last 5 min with 85 μM l-[35S]cysteine/methionine. Aliquots of cells were lysed by boiling them in 20% (wt/vol) trichloroacetic acid (TCA) for 10 min and then placing them on ice for 10 min. Proteins were precipitated onto GF/C glass microfiber filters (Whatman, Maidstone, United Kingdom) and washed using cold (4°C) 20% (wt/vol) TCA. Radioactive incorporation was measured by scintillation counting radioactivity retained on the filters and expressed as cpm per minute per absorbance at 600 nm.
Analysis of Ribosome Distribution on Sucrose Density Gradients
Yeast cultures were grown to exponential phase and treated with DTT as appropriate. Extracts were prepared in 100 μg of cycloheximide/ml, and these were layered onto 15-50% sucrose gradients. The gradients were sedimented via centrifugation at 40,000 rpm in a Beckman ultracentrifuge for 2.5 h, and the A254 measured continuously to give the traces shown (Ashe et al., 2000).
RESULTS
Identification of DTT-sensitive Strains
The homozygous diploid deletion mutant collection (EURO-SCARF) was screened to identify nonessential gene products that are required for resistance to DTT. Strains were grown in YEPD media in microtiter plates and spotted onto YEPD plates containing 32 mM DTT. This concentration of DTT was chosen because it allowed for normal growth of the wild-type strain but prevented growth of a hac1 mutant that is known to be DTT sensitive (our unpublished data). The screen was repeated twice with 708 sensitive mutants identified from the first screen and 616 mutants identified from the second screen. Mutants were scored as sensitive if they showed reduced growth after 3 d of incubation on media containing DTT. Common mutants (204) were identified as DTT sensitive in both screens. These mutants were rescreened for DTT sensitivity, resulting in a total of 195 mutants identified as DTT sensitive from the 4711 mutants tested (Table 1).
Table 1.
Genes required for DTT resistance
| Gene | ORF | Function |
|---|---|---|
| Protein synthesis | ||
| RPL31A | YDL075W | 60S large subunit ribosomal protein L31.e |
| RPP1B | YDL130W | 60S large subunit acidic ribosomal protein L44 prime |
| RPL35A | YDL191W | 60S large subunit ribosomal protein |
| RPL1B | YGL135W | 60S large subunit ribosomal protein |
| RPL27A | YHR010W | 60S large subunit ribosomal protein L27.e |
| RPL39 | YJL189W | 60S large subunit ribosomal protein L39.e |
| RPL13B | YMR142C | 60S large subunit ribosomal protein |
| RPS27B | YHR021C | Ribosomal protein S27.e |
| RPS12 | YOR369C | 40S small subunit acidic ribosomal protein S12 |
| RPS6A | YPL090C | Ribosomal protein S6.e |
| MRPL23 | YOR150W | Mitochondrial ribosomal protein, large subunit |
| IMG1 | YCR046C | Mitochondrial ribosomal protein, large subunit |
| MRPS28 | YDR337W | Mitochondrial ribosomal protein, small subunit |
| MSR1 | YHR091C | Arginyl-tRNA synthetase, mitochondrial |
| ARX1 | YDR101C | Required for 60S pre-ribosome formation |
| FUN12 | YAL035W | General translation factor, IF2 homolog |
| SWS2 | YNL081C | Mitochondrial ribosomal protein of the small subunit |
| YER087W | Putative prolyl-tRNA synthetase | |
| Transcription/RNA processing | ||
| SRB5 | YGR104C | DNA-directed RNA polymerase II holoenzyme |
| GAL11 | YOL051W | DNA-directed RNA polymerase II holoenzyme |
| RPB9 | YGL070C | DNA-directed RNA polymerase II, 14.2-kDa subunit |
| PAF1 | YBR279W | DNA-directed RNA polymerase II regulator |
| RRN10 | YBL025W | RNA polymerase I-specific transcription initiation factor |
| SSN8 | YNL025C | DNA-directed RNA polymerase II holoenzyme |
| NUT1 | YGL151W | Negative transcription regulator from artificial reporters |
| TUP1 | YCR084C | General transcription repressor |
| HFI1 | YPL254W | Transcriptional coactivator |
| CYC8 | YBR112C | General repressor of transcription |
| HHO1 | YPL127C | Histone H1 protein |
| MOT2 | YER068W | Transcriptional repressor |
| NOT5 | YPR072W | Component of the NOT protein complex |
| ROX3 | YBL093C | Transcription factor |
| SPT3 | YDR392W | General transcriptional adaptor or coactivator |
| NAM2 | YLR382C | Leucine-tRNA ligase precursor, mitochondrial |
| CBP2 | YHL038C | Apo-cytochrome b pre-mRNA processing protein 2 |
| DBP7 | YKR024C | RNA helicase required for 60S ribosomal subunit assembly |
| MRM2 | YGL136C | Mitochondrial rRNA methyltransferase |
| PRP18 | YGR006W | U5 snRNA-associated protein |
| RAI1 | YGL246C | Required for pre-rRNA processing |
| SIN4 | YNL236W | Global regulator protein |
| HPR1 | YDR138W | Hyperrecombination protein related to Top1p |
| PGD1 | YGL025C | Mediator complex subunit |
| Secretory pathway | ||
| Vacuolar protein sorting | ||
| VPS5 | YOR069W | Involved in Golgi retention and vacuolar sorting |
| VPS16 | YPL045W | Vacuolar sorting protein |
| VPS17 | YOR132W | Vacuolar protein sorting-associated protein |
| SNF7 | YLR025W | Class E Vps protein |
| VPS34 | YLR240W | Phosphatidylinositol 3-kinase |
| VPS20 | YMR077C | Vacuolar protein sorting (putative) |
| VPS33 | YLR396C | Vacuolar sorting protein |
| VPS45 | YGL095C | Vacuolar protein sorting-associated protein |
| VPS54 | YDR027C | Subunit of VP51-54 complex |
| PEP8 | YJL053W | Vacuolar protein sorting/targeting protein |
| PEP3 | YLR148W | Vacuolar membrane protein |
| PEP7 | YDR323C | Vacuolar segregation protein |
| FAB1 | YFR019W | Phosphatidylinositol 3-phosphate 5-kinase |
| VPS27 | YNR006W | Vacuolar protein sorting-associated protein |
| PEP12 | YOR036W | Syntaxin (t-SNARE), vacuolar |
| VPS1 | YKR001C | Member of the dynamin family of GTPases |
| Glycosylation | ||
| ANP1 | YEL036C | Required for protein glycosylation in the Golgi |
| MNN9 | YPL050C | Required for complex N-glycosylation |
| OCH1 | YGL038C | α-1,6-Mannosyltransferase |
| OST3 | YOR085W | Oligosaccharyltransferase γ subunit |
| LDB7 | YBL006C | Mannosylphosphorylation of cell wall mannoproteins |
| Vesicular transport (Golgi network, etc.) | ||
| AKR1 | YDR264C | Ankyrin repeat-containing protein |
| AFR1 | YDL192W | Small GTP-binding protein of the ARF family |
| CHC1 | YGL206C | Clathrin heavy chain |
| CLC1 | YGR167W | Clathrin light chain |
| SWA2 | YDR320C | Clathrin-binding protein |
| GYP1 | YOR070C | GTPase activating protein for Ypt1p and Sec4p |
| TRS33 | YOR115C | TRAPP subunit of 33 kDa |
| SAC6 | YDR129C | Actin filament bundling protein, fimbrin |
| SAC1 | YKL212W | Recessive suppressor of secretory defect |
| SIW14 | YNL032W | Protein involved in actin filament organization |
| YDJ1 | YNL064C | Mitochondrial and ER import protein |
| Cell cycle and differentiation | ||
| TPD3 | YAL016W | Ser/Thr protein phosphatase 2A, regulatory chain A |
| YVH1 | YIR026C | Protein tyrosine phosphatase |
| SLK19 | YOR195W | Involved in control of spindle dynamics together with kar3p |
| SFP1 | YLR403W | Zinc finger protein |
| BUB3 | YOR026W | Cell cycle arrest protein |
| DOC1 | YGL240W | Component of the anaphase promoting complex |
| RAD6 | YGL058W | E2 ubiquitin-conjugating enzyme |
| RAD50 | YNL250W | DNA repair protein |
| PHR1 | YOR386W | Deoxyribodipyrimidine photolyase |
| IWR1 | YDL115C | Interacts with RNA polymerase II |
| GLO3 | YER122C | Zinc finger protein |
| SET3 | YKR029C | Meiotic-specific repressor of the sporulation gene program |
| SPO19 | YPL130W | Putative GPI-anchored spore wall protein |
| HTL1 | YCR020W-B | High-temperature lethal, chromosome stability and fertility |
| SNT1 | YCR033W | Component of meiotic-specific repressor of the sporulation |
| SAC7 | YDR389W | Suppressor of actin mutation |
| FYV6 | YNL133C | Protein required for survival upon exposure to K1 killer toxin |
| BUD19 | YJL188C | Questionable protein |
| BUD22 | YMR014W | Protein involved in bud-site selection |
| BUD23 | YCR047C | Protein involved in bud-site selection |
| BUD25 | YER014C-A | Questionable protein |
| BUD32 | YGR262C | Protein involved in bud-site selection |
| BUD7 | YOR299W | Involved in bud-site selection |
| BUD30 | YDL151C | Involved in bipolar bud site selection |
| BEM1 | YBR200W | Bud emergence mediator |
| Metabolism | ||
| Carbon metabolism | ||
| PFK2 | YMR205C | 6-Phosphofructokinase |
| GCR2 | YNL199C | Glycolytic genes transcriptional activator |
| ADH1 | YOL086C | Alcohol dehydrogenase |
| GND1 | YHR183W | 6-Phosphogluconate dehydrogenase |
| RPE1 | YJL121C | d-Ribulose-5-phosphate 3-epimerase |
| REG1 | YDR028C | Regulatory subunit for protein phosphatase Glc7p |
| SHP1 | YBL058W | Potential regulatory subunit for Glc7p |
| SNF1 | YDR477W | Carbon catabolite derepressing Ser/Thr protein kinase |
| TPS1 | YBR126C | α,α-Trehalose-phosphate synthase |
| GRR1 | YJR090C | Required for glucose repression/glucose and cation transport |
| RBK1 | YCR036W | Ribokinase |
| Amino acid metabolism | ||
| ARO2 | YGL148W | Chorismate synthase |
| PRO1 | YDR300C | Glutamate 5-kinase |
| PRS3 | YHL011C | Ribose-phosphate pyrophosphokinase |
| AAT2 | YLR027C | Aspartate aminotransferase, cytosolic |
| URE2 | YNL229C | Nitrogen catabolite repression regulator |
| PHO2 | YDL106C | Homeodomain protein |
| NPR2 | YEL062W | Nitrogen permease regulator |
| Lipid metabolism | ||
| ERG2 | YMR202W | C-8 sterol isomerase |
| ERG6 | YML008C | S-Adenosyl-methionine delta-24-sterol-c-methyltransferase |
| ERG24 | YNL280C | C-14 sterol reductase |
| ARV1 | YLR242C | Sterol uptake and distribution into the plasma membrane |
| CRD1 | YDL142C | Cardiolipin synthase |
| Nucleotide metabolism | ||
| NPT1 | YOR209C | Nicotinate phosphoribosyltransferase |
| RIB1 | YBL033C | GTP cyclohydrolase II |
| ADK1 | YDR226W | Adenylate kinase, cytosolic |
| IRA2 | YOL081W | GTPase-activating protein for RAS proteins |
| RNR4 | YGR180C | Ribonucleotide reductase small subunit |
| RNR1 | YER070W | Ribonucleoside-diphosphate reductase, large subunit |
| Energy | ||
| ATP2 | YJR121W | F1F0-ATPase complex, F1 β subunit |
| ATP12 | YJL180C | F1F0-ATPase complex assembly protein |
| COX14 | YML129C | Cytochrome c oxidase assembly protein |
| Cell rescue and defense | ||
| PKR1 | YMR123W | Resistance against Pichia farinosa killer toxin (SMK toxin) |
| HYR1 (GPX3) | YIR037W | Glutathione peroxidase |
| GON7 | YJL184W | Hypothetical protein |
| TSA1 | YML028W | Thiol-specific antioxidant |
| SSE1 | YPL106C | Heat-shock protein of Hsp70 family |
| HIT1 | YJR055W | Required for growth at high temperature |
| ARR4 | YDL100C | Similarity to Escherichia coli arsenical pump-driving ATPase |
| SKN7 | YHR206W | Transcription factor with similarity to Hsf1p |
| CRZ1 | YNL027W | Calcineurin-responsive zinc finger |
| SSQ1 | YLR369W | Mitochondrial Hsp70 |
| UB14 | YLL039C | Ubiquitin |
| DOA1 | YKL213C | Involved in ubiquitin-dependent proteolysis |
| DOA4 | YDR069C | Ubiquitin-specific protease |
| MAP1 | YLR244C | Methionine aminopeptidase |
| IRE1 | YHR079C | Protein kinase |
| HAC1 | YFL031W | Transcription factor |
| Ion homeostasis | ||
| CUP5 | YEL027W | H+-ATPase V0 domain 17-kDa subunit, vacuolar |
| PPA1 | YHR026W | H+-ATPase 23-kDa subunit, vacuolar |
| VMA8 | YEL051W | H+-ATP synthase V1 domain 32-kDa subunit, vacuolar |
| VMA7 | YGR020C | H+-ATPase V1 domain 14-kDa subunit, vacuolar |
| VMA13 | YPR036W | H+-ATPase V1 domain 54-kDa subunit, vacuolar |
| VMA2 | YBR127C | H+-ATPase V1 domain 60-kDa subunit, vacuolar |
| VMA10 | YHR039C-B | H+-transporting ATPase V0 domain 13-kDa subunit, vacuolar |
| VMA22 | YHR060W | Vacuolar ATPase assembly protein |
| VPH2 | YKL119C | H+-ATPase assembly protein |
| CSG2 | YBR036C | Calcium-dependent regulatory protein |
| VMA21 | YGR105W | ATPase assembly integral membrane protein |
| PHO88 | YBR106W | Involved in phosphate transport |
| CNB1 | YKL190W | Calcineurin B, regulatory subunit |
| PER1 | YCR044C | Involved in manganese homeostasis |
| KSP1 | YHR082C | Ser/Thr protein kinase |
| IMP2′ | YIL154C | Involved in maintenance of ion homeostasis |
| SPF1 | YEL031W | P-type ATPase |
| Cellular transport | ||
| SSH1 | YBR283C | Involved in cotranslational pathway of protein transport |
| TOM70 | YNL121C | Mitochondrial outer membrane specialized import receptor |
| NUP84 | YDL116W | Nuclear pore protein |
| NUP133 | YKR082W | Nuclear pore protein |
| MON2 | YNL297C | Role in endocytosis and vacuole integrity |
| FEN2 | YCR028C | Pantothenate permease |
| Miscellaneous/unclassified | ||
| MDM39 | YGL020C | Protein involved in determination of mitochondrial structure |
| APQ13 | YJL075C | Questionable protein |
| IES6 | YEL044W | INO80 chromatin remodeling complex subunit 6 |
| ARP8 | YOR141C | Actin-related protein |
| KRE25 | YNL296W | Questionable ORF |
| FYV5 | YCL058C | Required for survival upon exposure to K1 killer toxin |
| YLR193C | Protein of unknown function localized to mitochondria | |
| YLR358C | Protein of unknown function | |
| YLR426W | Putative 3-oxoacyl-[acyl-carrier-protein] reductase | |
| YML036W | Similarity to Caenorhabditis elegans hypothetical protein CELW03F8 | |
| YDR048C | Questionable ORF | |
| YDR049W | Found in mitochondrial proteome | |
| YDR532C | Protein of unknown function localized to spindle pole body | |
| YEL033W | Questionable protein | |
| YFL032W | Questionable ORF | |
| YHR100C | Hypothetical ORF | |
| YJL028W | Hypothetical protein | |
| YJL120W | Questionable ORF | |
| YKL118W | Questionable ORF | |
| YNL140C | Protein of unknown function | |
| YNL115C | Protein of unknown function localized to vacuole | |
| YOR304C-A | Protein of unknown function localized to cytoplasm and bud | |
| YOR331C | Questionable protein | |
| YBR025C | Protein of unknown function localized to cytoplasm | |
| YJL070C | Putative AMP deaminases |
ORF, open reading frame.
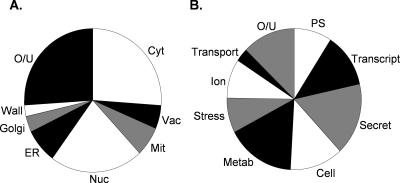
The reductive stress caused by DTT has primarily been studied because of its effects on ER function and protein processing in the secretory pathway. However, grouping the genes according to their cellular component revealed that DTT-sensitive mutants are localized throughout the cell (Figure 1A). As predicted, a large number of gene products that localize to the ER, Golgi, and vacuole were identified. In addition, gene products that localize to other cellular compartments, including the cytoplasm, mitochondria, nucleus, and cell wall were required for DTT resistance. Gene products were grouped into functional categories according to the MIPS functional database (http://mips.gsf.de/genre/proj/yeast/index.jsp) and Saccharomyces Genome Database GO term mapper (http://db.yeastgenome.org/cgi-bin/SGD/GO/goTermMapper). DTT-sensitive mutants were distributed among most major biological processes but particularly affected gene expression (transcription and protein synthesis), metabolism, and the secretory pathway (Figure 1B and Table 1). Approximately 30% of the mutants are defective for growth on nonfermentable carbon sources (http://www.yeastgenome.org/). However, there does not seem to be any link between respiratory ability and DTT tolerance because a respiratory incompetent petite strain shows a similar level of resistance to DTT as a wild-type strain (our unpublished data).
Figure 1.
Functional grouping of deletion mutant sensitivity data. (A) Localization was assigned based on the “Component” term of the Saccharomyces Genome Database GO term mapper. Cyt, cytoplasm; Vac, vacuole; Mit, mitochondrion; Nuc, nucleus; Golgi, Golgi apparatus; Wall, cell wall; O/U, other/unknown. (B) Gene products were grouped into functional categories according to the MIPS functional database and Saccharomyces Genome Database, combined with visual inspection. More detailed functional data are available in Table 1. PS, protein synthesis; Transcript, transcription; Secret, secretory pathway; Cell, cell cycle and differentiation; Metab, metabolism; Stress, cell rescue and defense; ion, ion homeostasis; Transport, cellular transport; O/U, other/unknown.
As expected, a major class of mutants included those that are defective in the secretory pathway, including vacuolar function, glycosylation, and vesicular transport. In addition, a number of mutants predicted to influence ion homeostasis are DTT sensitive, including several mutants in the vacuolar membrane H+-ATPase. A diverse range of mutants affecting metabolism are DTT sensitive. These could be subdivided into groups involved in carbon, amino acid, lipid, nucleotide, and energy metabolism. Together, these data indicate that active metabolism may be required to protect against reductive stress. A number of gene functions involved in transcription and translation are required for DTT tolerance, indicating that gene expression may be required in response to a reductive stress. Previous analysis of mutants that are required for general oxidative stress resistance has indicated that a number of similar gene functions are commonly required for protection against ROS (Thorpe et al., 2004). Unique to reductive stress, a number of mutants affecting ribosomal proteins are sensitive to DTT. These include components of the small subunit, large subunit, mitochondrial ribosome, and factors that are involved in ribosome export and biogenesis.
Relatively few mutants affected in cell rescue and defense were identified from this screen. They include genes that are generally required under conditions that generate denatured proteins, confirming that DTT can increase the cellular levels of misfolded proteins. Examples include members of the heat-shock protein (Hsp) 70 family, chaperones, and genes involved in ubiquitin-mediated protein degradation. As might be expected, loss of IRE1 and HAC1, which are essential for the induction of UPR-regulated genes, resulted in DTT sensitivity (Cox et al., 1993; Mori et al., 1993). Interestingly, strains deleted for GPX3, encoding a phospholipid hydroperoxide glutathione peroxidase (PHGPx), and TSA1, encoding a Prx, were sensitive to DTT. In addition, a strain deleted for SKN7, encoding a transcription factor involved in the oxidative stress response was found to be DTT sensitive. Given our original observation that thioredoxins are required for protection against DTT stress (Trotter and Grant, 2002), these mutants were chosen for further investigation.
Mutants Affecting the Thioredoxin System Are Sensitive to Reductive Stress
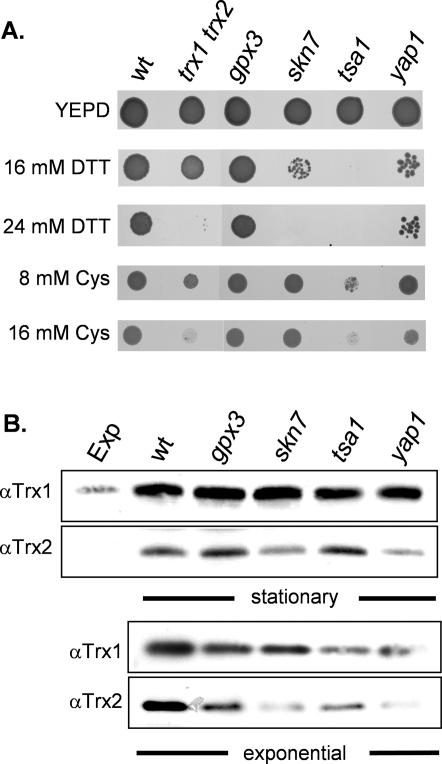
Isogenic haploid deletion strains were constructed to confirm the sensitivity of selected mutants to reductive stress. Strains were grown to stationary phase and spotted onto plates containing various concentrations of DTT (Figure 2A). The tsa1 mutant was the most sensitive and showed no growth on 16 mM DTT. Yeast contains five thioredoxin peroxidase isoforms, including cytoplasmic (Tsa1, Tsa2, and Ahp1), mitochondrial (Prx1/mTpx), and nuclear (Dot5/nTpx) isoforms (Park et al., 2000). However, only a strain deleted for the TSA1 isoform is sensitive to DTT (our unpublished data). Strains deleted for SKN7 were also sensitive to 16 mM DTT. Faster growing revertants were generated at this concentration of DTT, whereas no growth was detected on 24 mM DTT. The gpx3 mutant was the least sensitive and was able to grow on plates containing 24 mM DTT. Growth of the gpx3 mutant could be prevented by 32 mM DTT (our unpublished data). Yeast contains three PHGPxs encoded by GPX1-3 (Inoue et al., 1999; Avery and Avery, 2001). However, only a strain deleted for the GPX3 isoform is sensitive to DTT (our unpublished data). We next examined the sensitivity of these mutants to a reductive stress induced by the naturally occurring low-molecular-weight thiol cysteine (Figure 2A). The trx1 trx2 and tsa1 mutants were sensitive and showed poor growth on plates containing 16 mM cysteine.
Figure 2.
Mutants affecting the thioredoxin system are sensitive to reductive stress. (A) Stress sensitivity was determined by spotting strains onto YEPD plates containing DTT or SD plates containing cysteine (Cys). Cultures of wild-type, trx1 trx2, gpx3, skn7, tsa1, and yap1 strains were grown into stationary phase and adjusted to an A600 of 1.0 before spotting onto appropriate plates. Plates were incubated at 30°C for 3 d before scoring growth. (B) Western blot analysis of thioredoxin protein levels. Thioredoxin protein concentrations were measured in wild-type, trx1 trx2, gpx3, skn7, tsa1, and yap1 mutant strains grown to exponential or stationary phases in YEPD media. Blots were analyzed with antibodies specific for Trx1 or Trx2 as indicated.
We examined the DTT sensitivity of a yap1 mutant because Gpx3, Skn7, and Tsa1 are all known to affect the activity of the Yap1 transcription factor. Yap1 is a redoxsensitive bZip-transcription factor that regulates the expression of many antioxidant genes, including TRX2 and TRR1 (Toledano et al., 2004). The yap1 mutant was sensitive and showed poor growth on 16 mM DTT (Figure 2A). Given that Gpx3, Skn7, Tsa1, and Yap1 can all affect thioredoxin gene expression, we next examined the levels of thioredoxins in these mutants to determine whether DTT sensitivity could be explained by affects on thioredoxin gene expression.
Loss of SKN7 and YAP1 Affects the Basal Levels of Trx2
We examined the concentrations of Trx1 and Trx2 in the various mutants by Western blot analysis with the use of specific antibodies. Yeast contains two gene pairs encoding cytoplasmic thioredoxins (TRX1, TRX2), which seem to have redundant activities as antioxidants (Garrido and Grant, 2002). TRX2 plays the predominant role and its expression is regulated by both oxidative and reductive stress conditions. The cellular concentrations of Trx1 and Trx2 were elevated in stationary phase compared with exponential growth (Figure 2B) in agreement with previous transcriptional studies (Garrido and Grant, 2002). The amount of stationary phase Trx1 was unaffected by the loss of GPX3, SKN7, TSA1, or YAP1 compared with the wild-type strain. In contrast, the levels of Trx2 were somewhat lower in the skn7 and yap1 mutants compared with the wild-type, gpx3, and tsa1 mutant strains. Similarly, the concentrations of Trx2 in exponential phase cells were lower in the skn7 and yap1 mutants compared with the other strains. Lowered levels of Trx2 may therefore account for the DTT sensitivity of skn7 and yap1 mutants. In addition, all the mutants contained somewhat lower levels of Trx1 and Trx2 compared with the wild-type strain in exponential phase cells. Thus, there does not seem to be any strong correlation between the basal levels of Trx1-2 in these mutants and DTT sensitivity. The tsa1 mutant did not significantly affect thioredoxin expression, yet showed the greatest sensitivity to DTT, and so it was chosen for further investigation.
Thioredoxins Require Tsa1 for Reductive Stress Resistance
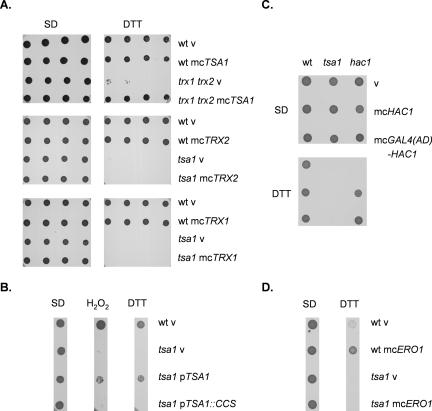
Tsa1 may influence thioredoxin activity through an effect on thioredoxin gene expression as described above, or as a thioredoxin-dependent enzyme. To differentiate between these two possibilities, we examined the epistasis of TSA1 and TRX1-2 in the response to reductive stress. Overexpression of TRX1 or TRX2 was unable to rescue the DTT sensitivity of a tsa1 mutant, indicating that thioredoxins require Tsa1 to promote DTT tolerance (Figure 3A). We have previously confirmed that thioredoxins are overexpressed in these strains and can promote resistance to hydrogen peroxide (Garrido and Grant, 2002). In contrast, overexpression of TSA1 rescued the growth of a trx1 trx2 mutant on DTT media, indicating that Tsa1 does not require thioredoxins to promote DTT tolerance. Thus, Tsa1 can act independently of Trx1-2 and must use an alternative source of reducing equivalents under these conditions. In agreement with this idea, previous studies have shown that GSH and DTT can act as alternative electron donors for peroxiredoxins (Park et al., 2000). Together, these data indicate that thioredoxins act upstream of Tsa1 in the thioredoxin-dependent DTT resistance pathway.
Figure 3.
Role of Trx1-2, Tsa1, and the UPR in DTT tolerance. (A) Overexpression of TSA1 increases the resistance of thioredoxin mutants to DTT, whereas overexpression of thioredoxins does not affect the tsa1 mutant. The wild-type, trx1 trx2 and tsa1 mutant strains containing empty vector (V), mcTRX1, mcTRX2, or mcTSA1, as indicated, were grown into stationary phase and adjusted to an A600 of 1.0, 0.5, 0.25, or 0.125 before spotting onto SD plates containing 2-4 mM DTT. (B) The active site cysteine residues of Tsa1 are required for resistance to DTT. The wild-type and tsa1 mutant strains containing empty vector (V), pTSA1 expressing wild-type Tsa1 or pTSA1::CCS expressing the active-site mutant of Tsa1p, in which cysteine 47 and 170 have been replaced by serines, were grown into stationary phase and adjusted to an A600 of 1.0 before spotting onto SGal plates containing 1 mM H2O2 or 8 mM DTT. (C) Overexpression of HAC1 does not rescue the DTT sensitivity of a tsa1 mutant. The wild-type, tsa1, and hac1 mutant strains containing empty vector (v), mcHAC1, or mcGAL4(AD)-HAC1 were grown into stationary phase and adjusted to an A600 of 1.0 before spotting onto SD plates containing 2 mM DTT. (D) Overexpression of ERO1 does not rescue the DTT sensitivity of a tsa1 mutant. The wild-type and tsa1 mutant strains containing empty vector (V) or mcERO1 were grown into stationary phase and adjusted to an A600 of 1.0 before spotting onto SD plates containing 4 mM DTT. Plates were incubated at 30°C for 3 d before scoring growth.
In common with all typical Prxs, Tsa1 contains two redox active Cys residues that are directly involved in enzyme activity (Wood et al., 2003). We therefore determined whether an active site mutant of Tsa1, in which Cys47 and Cys170 were substituted with serines (Wong et al., 2004), could promote resistance to DTT. Tsa1 lacking the active site cysteine residues was unable to rescue the DTT sensitivity of a tsa1 mutant, indicating that peroxiredoxin enzyme activity is required (Figure 3B). Tsa1 has mainly been characterized as an antioxidant in the detoxification of hydroperoxides, but more recently it has been shown to act as molecular chaperone that promotes resistance to heat shock (Jang et al., 2004). The active site Cys residues are required for these activities and Tsa1 may therefore protect against reductive stress via its peroxidase or chaperone activities.
Activating the UPR in a tsa1 Mutant Does Not Rescue DTT Sensitivity
Because DTT can disrupt disulfide bond formation, most studies of DTT stress have focused on protein folding in the ER and the UPR. We therefore tested whether there is any link between the DTT sensitivity of the tsa1 mutant and the UPR. The magnitude of the UPR, measured using a reporter construct under the control of the UPR element (Sidrauski et al., 1996), is comparable in the wild-type and tsa1 mutant strains (our unpublished data). When unfolded proteins accumulate in the ER, the UPR is activated and the Hac1 transcription factor recognizes the UPR element present in target genes. We therefore tested whether overexpression of HAC1 could rescue the growth of a tsa1 mutant on DTT. HAC1 overexpression was achieved using a multicopy vector containing wild-type HAC1 (YEp-HAC1) or HAC1 fused to the Gal4 activation domain at its N terminus [YEp-GAL4(AD)-HAC1] (Mori et al., 2000). An mRNA-splicing event is normally required to fuse the HAC1 DNA-binding domain to its activation domain, and the presence of the Gal4 activation domain results in constitutively active Hac1 in this construct. However, overexpression of Hac1 or the Gal4-fusion construct was unable to rescue DTT sensitivity of the tsa1 mutant (Figure 3C). In comparison, both constructs could rescue the DTT sensitivity of a hac1 mutant. Ero1 is a flavoenzyme that is required for oxidative protein folding in the ER. It is a target of the UPR and overexpression has been show to confer DTT resistance (Frand and Kaiser, 1998; Pollard et al., 1998). We therefore tested whether overexpression of ERO1 could rescue the DTT sensitivity of the tsa1 mutant. ERO1 increased the tolerance of the wild-type strain to DTT, but it was unable to rescue the DTT sensitivity of the tsa1 mutant (Figure 3D). Together, these data indicate that the DTT sensitivity of the tsa1 mutant does not seem to result from an inability to prevent protein misfolding in the ER and is not related to the UPR.
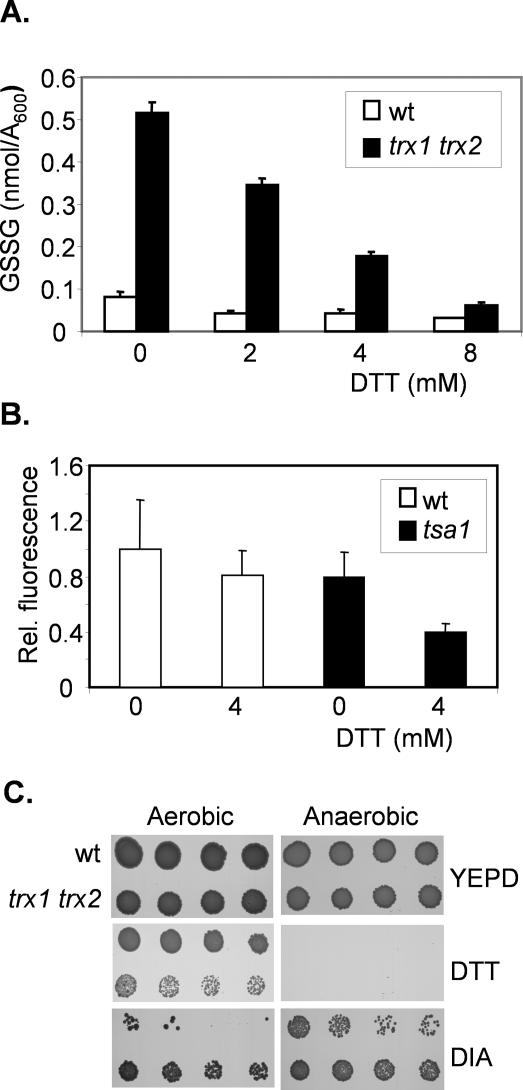
DTT Toxicity Is Not Mediated by Reactive Oxygen Species
Although DTT is well known as an ER reducing agent, it is possible that it causes cellular oxidation through an indirect mechanism. For example, recent reports have shown that ER stress is linked with the generation of ROS (Harding et al., 2003; Haynes et al., 2004; Tu and Weissman, 2004). We therefore tested whether DTT exposure generates ROS using two different approaches. First, we determined whether the levels of GSSG are increased in response to DTT. Glutathione is the major cellular redox buffer and the redox state of the glutathione system is often taken as an indicator of the cellular redox environment (Meister and Anderson, 1983; Schafer and Buettner, 2001). Indeed, increases in yeast GSSG of sixfold are observed during conditions promoting ROS formation (Grant et al., 1998). The cellular concentrations of GSSG can therefore be used as an indicator of ROS formation. Exposure of wild-type cells to DTT resulted in lowered levels of GSSG (Figure 4A). This effect was most apparent in a trx1 trx2 mutant, which contains higher basal levels of GSSG (Muller, 1996; Garrido and Grant, 2002). DTT resulted in a dose-dependent decrease in GSSG levels in the trx1 trx2 mutant (Figure 4A). Second, to more directly examine ROS levels, we used DCFH-DA. This is a cell-permeable fluorogenic probe that is widely used to estimate ROS formation (Davidson et al., 1996; Izawa et al., 1999). Fluorescence was decreased upon exposure to 4 mM DTT in both a wild-type and tsa1 mutant (Figure 4B). These data indicate that rather than generating oxidizing conditions, DTT shifts the intracellular redox environment to a more reducing state. As a further test of whether DTT might act through an oxidative mechanism, we examined DTT toxicity under anaerobic conditions (Figure 4C). Anaerobic growth exacerbated the reductive stress and prevented the growth of a wild-type and trx1 trx2 mutant on plates containing DTT. In comparison, anaerobic conditions rescued the growth of a wild-type strain on the thiol oxidant diamide. Thus, DTT toxicity does not arise through a mechanism that involves ROS formation, making it unlikely that the peroxidase activity of Tsa1 is required to protect against DTT.
Figure 4.
DTT toxicity is not caused by the formation of reactive oxygen species. (A) DTT treatment lowers the cellular levels of oxidized GSSG. The wild-type and thioredoxin mutant (trx1 trx2) were grown to exponential phase (A600 = 1.0) in YEPD media and treated with DTT for 1 h. GSSG concentrations shown are the means of at least three independent determinations and are given in nanomoles per milliliter per A600. (B) DTT treatment lowers intracellular ROS levels. ROS were measured in the wild-type and tsa1 mutant strain grown in SD media using the cell-permeable fluorogenic probe DCFH-DA after treatment with 4 mM DTT for 15 min. Data are means ± SD from three independent experiments. (C) Anaerobic growth conditions do not prevent DTT toxicity. The wild-type and thioredoxin mutant strains (trx1 trx2) were spotted onto YEPD plates containing DTT or diamide (DIA) and grown under aerobic or anaerobic conditions.
DTT Induces Protein Aggregation in Thioredoxin Mutants
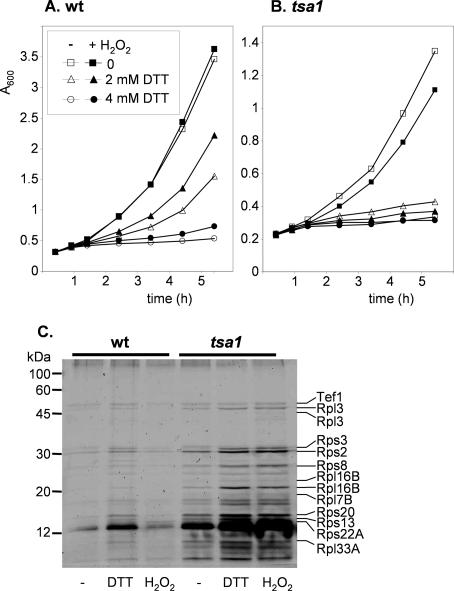
Oxidative stress conditions induced by exposure to low levels of H2O2 have been shown to convert Tsa1 to a high-molecular-weight complex chaperone form (Jang et al., 2004). This chaperone form can enhance yeast resistance to heat shock, and so we determined whether it could also protect against a reductive stress. Wild-type cells were pretreated with 50 mM H2O2 before exposure to toxic levels of DTT. Treatments with 2 or 4 mM DTT decreased the growth rate of wild-type cells, but growth could be improved by pretreatment with H2O2 (Figure 5A). This adaptive response requires Tsa1 because it is absent in the tsa1 mutant (Figure 5B).
Figure 5.
DTT stress induces protein aggregation. Induction of DTT resistance by oxidative stress. Wild-type (A) and tsa1 (B) mutant cells grown in YEPD media were pretreated with 50 μM H2 O2 for 1 h before growth in the presence of 2 or 4 mM DTT. Growth was monitored by absorbance at 600 nm. (C) Aggregation of ribosomal proteins in the tsa1 mutant and in response to DTT. Insoluble proteins were extracted from the wild-type and tsa1 mutant strains grown in YEPD media after treatment with 16 DTT or 4 mM H2 O2 for 1 h. Proteins were separated by SDS-PAGE and detected by silver staining. The indicated proteins were identified via MALDI-TOF mass spectrometry.
Exposure to heat stress has been shown to promote protein aggregation in the tsa1 mutant, and so we examined whether DTT has a similar effect. Protein aggregation was measured using a technique that solubilizes membrane proteins and separates them from aggregated proteins, effectively reducing the background in aggregate fractions (Tomoyasu et al., 2001; Jang et al., 2004). Relatively few aggregated proteins were detected in the wild-type strain during normal growth conditions (Figure 5C). Exposure to DTT increased the levels of several proteins in the aggregate fraction. The tsa1 mutant contained high basal levels of protein aggregates, comparable with the levels detected in the wild-type strain after DTT stress. Interestingly, the concentrations of these aggregated proteins increased further after exposure to DTT. These data indicate that the sensitivity of a tsa1 mutant to reductive stress may be caused by an inability to protect against the toxic effects of protein aggregates. We compared the levels of aggregated proteins formed following a reductive stress with those formed during an oxidative stress generated by exposure to hydrogen peroxide. Whereas H2 O2 had relatively little effect on the wild-type strain, it increased protein aggregation in the tsa1 mutant to a similar level to that caused by DTT (Figure 5C).
Thioredoxin Mutants Contain Elevated Levels of Aggregated Ribosomal Proteins
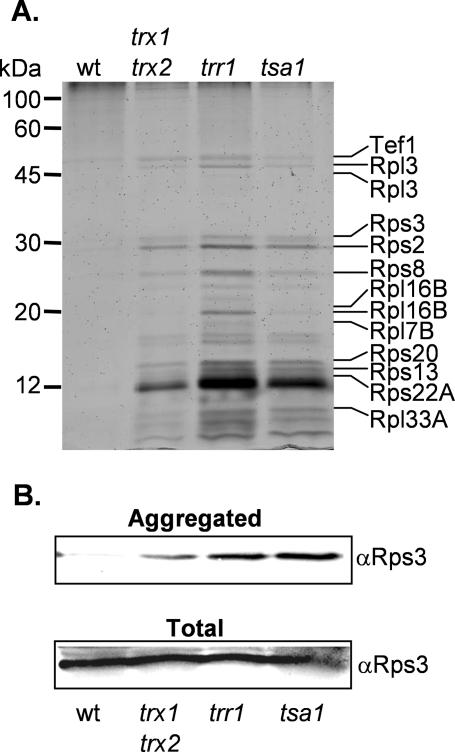
Given that protein aggregation may account for the toxicity of DTT, we analyzed the pattern of proteins that aggregate in a tsa1 mutant. The major proteins were identified by mass spectrometry and are indicated on Figure 5C. Remarkably, the identified proteins were all ribosomal proteins and a ribosome associated protein (Tef1). These include proteins of both the small and large ribosomal subunits. We examined whether other thioredoxin pathway mutants affect ribosomal protein aggregation. High basal levels of aggregated proteins were detected in the trx1 trx2 and trr1 mutants, and were particularly elevated in the trr1 mutant (Figure 6A).
Figure 6.
Thioredoxin system mutants contain elevated levels of aggregated ribosomal proteins. (A) Analysis of insoluble proteins from the wild-type, trx1 trx2, trr1, and tsa1 mutant strains as for Figure 5B. (B) Western blot analysis of Rps3 in the aggregated protein fraction and in a total cell extract from the wild-type, trx1 trx2, trr1, and tsa1 mutant strains.
To check that there are no differences in the basal levels of ribosomal proteins in thioredoxin mutants, Western blot analysis was used to examine the levels of Rps3. Similar levels of Rps3 were detected in total cell extracts prepared from thioredoxin mutant strains (Figure 6B). In contrast, Rps3 is elevated in the aggregated protein preparations from the tsa1, trr1, and trx1 trx2 mutants compared with the wild-type strain (Figure 6B). Probing the Western blots with an antibody against glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which is a cytoplasmic enzyme, confirmed that no GAPDH is present in the aggregate fraction, whereas it is present in total cell extracts (our unpublished data). These data indicate that a fraction of the ribosomal proteins present in thioredoxin mutants form insoluble aggregates.
Reductive Stress Inhibits Protein Synthesis in a tsa1 Mutant
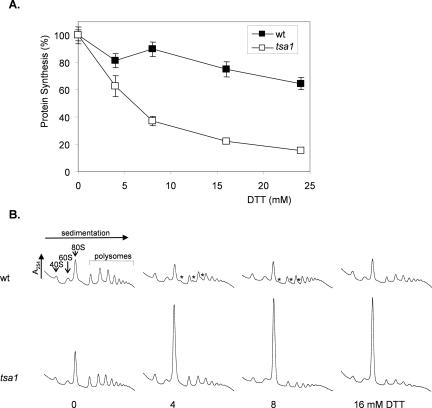
Given that reductive stress particularly affects ribosomal proteins, we examined whether there is any effect on protein synthesis. The rate of protein synthesis was measured as the incorporation of [35S]cysteine/methionine into proteins during a 5-min labeling period. DTT inhibited protein synthesis in the wild-type strain with a treatment of 24 mM DTT for 1 h reducing the rate of protein synthesis by ∼35% (Figure 7A). Protein synthesis was much more sensitive to DTT in the tsa1 mutant; for example, 4 mM DTT reduced the rate of protein synthesis by a similar amount to that seen for 24 mM DTT in the wild-type strain.
Figure 7.
Reductive stress inhibits protein synthesis. (A) Cultures of the wild-type and tsa1 mutant strain were grown to exponential phase in minimal SD media. Aliquots of cell culture were used to measure the rate of protein synthesis by pulse labeling with [35S]cysteine/methionine for 5 min. Data are shown for untreated cultures (100%) and after treatment with DTT for 1 h. (B) DTT treatment specifically inhibits translation initiation. Polyribosome traces are shown from the wild-type and tsa1 mutant strains. Yeast were grown in SD medium and treated with the indicated concentrations of DTT for 1 h. Polyribosomes were analyzed as described in Materials and Methods. The peaks that contain the small ribosomal subunit (40S), the large ribosomal subunit (60S), and both subunits (80S) are indicated by arrows. The polysome peaks generated by 2, 3, 4, 5, etc., 80S ribosomes on a single mRNA are bracketed. Asterisks indicate an accumulation of halfmer polysomes.
The inhibition of protein synthesis caused by DTT prompted us to analyze translational activity by examining the distribution of polysomes. Polysomes are ribosomes that are actively translating mRNAs, and they can be separated on sucrose density gradient and quantified by measuring absorbance at 254 nm. Extracts prepared from the wild-type and tsa1 mutant strains exhibited normal profiles, including peaks corresponding to 40S and 60S ribosomal subunits, monosomes (80S ribosomes), and polysomes (Figure 7B). Interestingly, treatment of the wild-type strain with lower concentrations of DTT (4 and 8 mM) caused an accumulation of halfmer polysomes. This effect was lost at a higher concentration of 16 mM DTT. In the tsa1 mutant, there was a dramatic shift of ribosomes from the polysomal region into the monosome or 80S peak after treatment with DTT (4-16 mM). The accumulation of ribosomes in the 80S peak of a sucrose gradient is indicative of decreased translation initiation. These data indicate that a reductive stress caused by DTT affects translation initiation in wild-type and tsa1 mutant strains.
DISCUSSION
We previously showed that thioredoxins are required for DTT tolerance suggesting that thioredoxins maintain redox homeostasis in response to both oxidative and reductive stress conditions (Trotter and Grant, 2002). The data in the current study indicate that the requirement for thioredoxins during DTT stress is dependent on the chaperone activity of Tsa1. Tsa1 is a typical 2-Cys peroxiredoxin that has primarily been characterized as an antioxidant (Chae et al., 1994; Park et al., 2000). More recently, a chaperone function has been demonstrated for Tsa1, which enhances yeast resistance to heat shock (Jang et al., 2004). Studies of several typical 2-Cys Prxs have revealed changes in oligomeric state that are linked to changes in redox state (Wood et al., 2003). In the yeast Tsa1 protein, the chaperone function has been shown to predominate in the higher molecular weight complex forms (Jang et al., 2004). The switch between thioredoxin-dependent peroxidase and chaperone functions is linked to oxidation of the peroxidatic Cys47 residue. In this current study, we have shown that Tsa1 is required to protect against DTT-induced protein aggregation. This activity is enhanced by prior treatment with hydrogen peroxide, which shifts the pool of Tsa1 to the higher molecular weight complex chaperone form (Jang et al., 2004). Analysis of the proteins that aggregate in a tsa1 mutant identified ribosomal proteins as likely substrates for Tsa1 chaperone function. The resulting defect in protein synthesis seems to underlie the toxicity of reductive stress in thioredoxin mutants.
Genome-wide studies have indicated that there is a common regulatory program, called the environmental stress response (ESR), that is activated in response to diverse environmental stress conditions (Gasch et al., 2000; Causton et al., 2001). In addition to these general responses, each particular stress can invoke a unique pattern of transcriptional regulation that seems to be specialized for the specific conditions. The initial response to DTT includes the induction of ER chaperones consistent with the idea that DTT disrupts protein folding in the ER (Gasch et al., 2000; Travers et al., 2000). Prolonged exposure to DTT activates the ESR, along with the induction of genes that are involved in the response to cell wall damage, indicating that cell wall perturbation may eventually trigger the ESR in response to DTT (Gasch et al., 2000). However, the analysis of genome-wide responses to DNA-damaging agents and oxidants has indicated that relatively few of the genes induced after exposure to these agents are necessary to protect the viability of the cell against these agents (Birrell et al., 2002; Thorpe et al., 2004). Similarly, of the 195 deletion mutants identified in this study as being sensitive to DTT, only 28 are induced within 60 min after DTT stress (Travers et al., 2000). The induced genes are distributed throughout the cell and do not cluster into any particular functional category (our unpublished data). The remaining genes presumably represent constitutive functions that are required for DTT tolerance.
Extensive analysis of gene functions that are required during exposure to different ROS has revealed that distinct gene classes maintain protection against particular oxidants (Thorpe et al., 2004). Additionally, core gene functions are required for tolerance against all forms of ROS, including gene expression, vacuolar protein sorting, and cell wall biosynthesis and maintenance. These core functional classes are also required for DTT tolerance and may represent gene functions that are generally required during stress conditions. Mutants affecting the secretory pathway are overrepresented in the DTT-sensitive strains, compared with ROS-sensitive strains, presumably reflecting the effect of DTT on protein folding and secretion. A strikingly large class of DTT-sensitive mutants are defective in ribosomal proteins and ribosome export and processing. This may indicate that perturbing ribosome production results in DTT sensitivity, consistent with the idea that aggregation of ribosomal proteins is the cause of DTT toxicity in thioredoxin mutants.
Yeast, like most eukaryotes, contains a complete cytoplasmic thioredoxin system, including two thioredoxins (TRX1-2) and a thioredoxin reductase (TRR1), which functions in protection against oxidative stress (reviewed in Carmel-Harel and Storz, 2000; Wheeler and Grant, 2004). Trx1 and Trx2 are active as antioxidants and play key roles in protection against oxidative stress induced by various ROS. Trx2 seems to play the predominant role because mutants lacking TRX2 are hypersensitive to hydroperoxides and mutants containing TRX2, in the absence of TRX1, are resistant to these oxidants (Garrido and Grant, 2002). Additionally, five thioredoxin peroxidases have been identified in yeast, although their exact intracellular roles and targets remain unclear (Park et al., 2000). Components of the thioredoxin system are similarly regulated, with the expression of TRX2, TRR1, TSA1, TSA2, and PRX1 regulated by the cooperative action of the Yap1 and Skn7 transcription factors (Charizanis et al., 1999; Lee et al., 1999; Park et al., 2000). Tsa1 is essential for the transcriptional induction of TRX2 and TRR1, and Gpx3 plays the role of a hydroperoxide sensor and transducer that signals the activation of Yap1 (Kuge and Jones, 1994; Morgan et al., 1997; Ross et al., 2000; Delaunay et al., 2002). Strains lacking TSA1, SKN7, GPX3 or YAP1 were all identified as being DTT sensitive in this study, which may therefore result from lowered levels of thioredoxins in these mutants. Although this seemed to be the case for skn7 and yap1 mutants, alterations in thioredoxin levels do not seem to account for the DTT sensitivity of the gpx3 mutant. We have previously shown that TRX2 expression is regulated by reductive stress (Trotter and Grant, 2002), and it is possible that GPX3, or the other genes identified in this study, are required for this regulation. Because Tsa1 represents the most downstream component of the thioredoxin system identified in this screen, we focused on characterizing the DTT sensitivity of a tsa1 mutant in this study.
DTT stress was found to disrupt ribosome function and to inhibit translation initiation. Eukaryotic translation initiation is a complex highly regulated process involving more than 30 polypeptide factors interacting with ribosomal subunits, Met-tRNAmeti, and mRNAs (McCarthy, 1998). The initiation phase of protein synthesis is rate limiting and is a target of regulation. Four mammalian protein kinases (HRI, PERK, PKR, and GCN2) have been identified that regulate translation initiation by phosphorylating eukaryotic initiation factor-2 (eIF2) in response to cellular stress (Wek, 1994; Dever, 2002). In yeast, Gcn2 is the sole eIF2 kinase and phosphorylates eIF2a in response to diverse stress conditions, including amino acid, purine, and glucose starvation, and sodium and rapamycin exposure. This causes a global inhibition of protein synthesis as well as gene-specific translational activation of GCN4, which encodes a transcriptional activator protein (Hinnebusch, 1994). More recently, Gcn4 has been found to play an essential role in the UPR: Gcn4 and Gcn2 are required for the induction many UPR target genes during ER stress. Both Hac1 and Gcn4 bind target gene promoters to stimulate transcriptional induction (Patil et al., 2004).
PERK has been found in all multicellular eukaryotes and is a component of the unfolded protein response. Consistent with its central role in the ER stress response, cells lacking PERK fail to phosphorylate eIF2α and do not down-regulate protein synthesis during ER stress conditions (Kaufman, 1999; Bertolotti et al., 2000). Yeast does not contain PERK and hence DTT stress has not previously been shown to inhibit protein synthesis. Our data show that DTT-stress causes an accumulation of halfmers in a wild-type strain, which are polyribosomes that have an extra 40S or 43S ribosomal subunit attached to the mRNA (Tuite et al., 1999). They have previously been described for yeast strains deficient in translation factors, including Rpl16 and Gcd2, and are thought to reflect a reduced ability of the 60S subunit to join the 43S complex on mRNA (Rotenberg et al., 1988; Foiani et al., 1991). DTT resulted in a more pronounced defect in translation initiation in the tsa1 mutant, caused ribosome aggregation and strongly inhibited protein synthesis. This block in protein synthesis correlated with the aggregation of ribosomal proteins caused by DTT exposure. These data indicate that some aspect of ribosomal production/function is sensitive to reductive stress conditions.
The production of ribosomes is a complex, highly regulated energy-requiring process (Warner, 1999). It must be able to respond to environmental conditions and several conserved signaling pathways (TOR, RAS/protein kinase A, and protein kinase C) are known to regulate ribosome biosynthesis (Jorgensen et al., 2004; Schawalder et al., 2004). As yet, it is unclear whether DTT disrupts existing ribosomes or affects the assembly and processing of new ribosomes. Interestingly, ARX1, DBP7, SFP1, and SSQ1, which are all implicated in ribosome export and biogenesis were identified in the screen for DTT-sensitive mutants (Table 1). Sfp1 is a transcription factor that regulates ribosomal protein gene expression in response to nutrients and stress conditions (Fingerman et al., 2003; Marion et al., 2004). In response to diverse environmental conditions, including DTT-stress, it down-regulates transcription of ribosomal protein genes (Marion et al., 2004). Arx1 has been identified as a protein associated with the ribosomal export complex (Nissan et al., 2002). Dbp7 is a putative ATP-dependent RNA helicase that is required for 60S ribosomal subunit assembly (Daugeron and Linder, 1998). Ssq1 is a mitochondrial Hsp70-type molecular chaperone required for the assembly of iron/sulfur clusters into proteins (Muhlenhoff et al., 2003). It may have an indirect effect on ribosomal processing and production because the biogenesis of ribosomes requires the essential iron/sulfur protein Rli1 (Kispal et al., 2005; Yarunin et al., 2005). The finding that several mutants affected in ribosome production as well as mutants lacking individual ribosomal proteins are DTT sensitive may indicate that it is the production of new ribosomes that is DTT sensitive. Identification of the exact step targeted by DTT will require further experimentation.
Like other molecular chaperons, Tsa1 has been shown to suppress thermal aggregation in vitro by binding to unfolded proteins via hydrophobic protein sites (Jang et al., 2004). Additionally, the absence of TSA1 led to an accumulation of heat-shock-induced protein aggregates in yeast cells, although the aggregated proteins were not identified (Jang et al., 2004). We have now identified ribosomal proteins as a major target for Tsa1 chaperone activity. Intact ribosomes require numerous protein-protein and protein-RNA interactions, and so it is perhaps, not surprising, that disrupting their normal structure results in the formation of nonspecific aggregates. Binding of any misassembled ribosomal proteins by Tsa1 would prevent their aggregation, before recycling or degradation. Given the dramatic effect on protein synthesis of disrupting this process it seems to represent a previously unrecognized stress-protective mechanism.
Acknowledgments
We thank Dong-Yan Jin (University of Hong Kong, China), Martin Poole (University of Manchester, United Kingdom), Antony Cooper (University of Missouri-Kansas City, Kansas City, MO) and Kazutoshi Mori (Kyoto University, Kyoto, Japan) for the kind gift of plasmids and antibodies. We are grateful to Kathleen Carroll (University of Manchester) for mass spectrometry analysis, and we thank Mark Ashe (University of Manchester) for critical reading of the manuscript and advice on polysome analysis. We thank Gabriel Perrone (University of New South Wales, Sydney, Australia) for advice and providing data before publication. This work was supported by the Biotechnology and Biological Sciences Research Council.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-06-0520) on October 26, 2005.
References
- Ashe, M. P., De Long, S. K., and Sachs, A. B. (2000). Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell 11, 833-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery, A. M., and Avery, S. V. (2001). Saccharomyces cerevisiae expresses three phospholipid hydroperoxide glutathione peroxidases. J. Biol. Chem. 276, 33730-33735. [DOI] [PubMed] [Google Scholar]
- Bass, R., Ruddock, L. W., Klappa, P., and Freedman, R. B. (2004). A major fraction of endoplasmic reticulum-located glutathione is present as mixed disulfides with protein. J. Biol. Chem. 279, 5257-5262. [DOI] [PubMed] [Google Scholar]
- Baudin, A., Ozier-Kalogeropoulos, O., Danouel, A., Lacroute, F., and Cullin, C. (1993). A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21, 3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti, A., Zhang, Y., Hendershot, L. M., Harding, H. P., and Ron, D. (2000). Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2, 326-332. [DOI] [PubMed] [Google Scholar]
- Birrell, G. W., Brown, J. A., Wu, H., Giaever, G., Chu, A. M., Davis, R. W., and Brown, J. M. (2002). Transcriptional response of Saccharomyces cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents. Proc. Natl. Acad. Sci. USA 99, 8778-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmel-Harel, O., and Storz, G. (2000). Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and saccharomyces cerevisiae responses to oxidative stress. Annu. Rev. Microbiol. 54, 439-461. [DOI] [PubMed] [Google Scholar]
- Causton, H. C., Ren, B., Koh, S. S., Harbison, C. T., Kanin, E. J., Jennings, E. G., Lee, T. I., True, H. L., Lander, E. S., and Young, R. A. (2001). Remodelling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12, 323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae, H. Z., Chung, S. J., and Rhee, S. G. (1994). Thioredoxin-dependent peroxide reductase from yeast. J. Biol. Chem. 269, 27670-27678. [PubMed] [Google Scholar]
- Charizanis, C., Juhnke, H., Krems, B., and Entian, K.-D. (1999). The oxidative stress response mediated via Pos9/Skn7 is negatively regulated by the Ras/PKA pathway in Saccharomyces cerevisiae. Mol. Gen. Genet. 261, 740-752. [DOI] [PubMed] [Google Scholar]
- Cox, J. S., Shamu, C. E., and Walter, P. (1993). Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73, 1197-1206. [DOI] [PubMed] [Google Scholar]
- Cuozzo, J. W., and Kaiser, C. A. (1999). Competition between glutathione and protein thiols for disulphide-bond formation. Nat. Cell Biol. 1, 130-135. [DOI] [PubMed] [Google Scholar]
- Daugeron, M. C., and Linder, P. (1998). Dbp7p, a putative ATP-dependent RNA helicase from Saccharomyces cerevisiae, is required for 60S ribosomal subunit assembly. RNA 4, 566-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, J. F., Whyte, B., Bissinger, P. H., and Schiestl, R. H. (1996). Oxidative stress is involved in heat-induced cell death in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93, 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay, A., Pflieger, D., Barrault, M. B., Vinh, J., and Toledano, M. B. (2002). A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111, 471-481. [DOI] [PubMed] [Google Scholar]
- Dever, T. E. (2002). Gene-specific regulation by general translation factors. Cell 108, 545-556. [DOI] [PubMed] [Google Scholar]
- Draculic, T., Dawes, I. W., and Grant, C. M. (2000). A single glutaredoxin or thioredoxin is essential for viability in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 36, 1167-1174. [DOI] [PubMed] [Google Scholar]
- Fingerman, I., Nagaraj, V., Norris, D., and Vershon, A. K. (2003). Sfp1 plays a key role in yeast ribosome biogenesis. Eukaryot. Cell 2, 1061-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani, M., Cigan, A. M., Paddon, C. J., Harashima, S., and Hinnebusch, A. G. (1991). GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 3203-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frand, A. R., Cuozzo, J. W., and Kaiser, C. A. (2000). Pathways for protein disulphide bond formation. Trends Cell Biol. 10, 203-210. [DOI] [PubMed] [Google Scholar]
- Frand, A. R., and Kaiser, C. A. (1998). The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol. Cell 1, 161-170. [DOI] [PubMed] [Google Scholar]
- Garrido, E. O., and Grant, C. M. (2002). Role of thioredoxins in the response of Saccharomyces cerevisiae to oxidative stress induced by hydroperoxides. Mol. Microbiol. 43, 993-1003. [DOI] [PubMed] [Google Scholar]
- Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., Botstein, D., and Brown, P. O. (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, C. M., Collinson, L. P., Roe, J.-H., and Dawes, I. W. (1996a). Yeast glutathione reductase is required for protection against oxidative stress and is a target gene for yAP-1 transcriptional regulation. Mol. Microbiol. 21, 171-179. [DOI] [PubMed] [Google Scholar]
- Grant, C. M., MacIver, F. H., and Dawes, I. W. (1996b). Glutathione is an essential metabolite required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. Curr. Genet. 29, 511-515. [DOI] [PubMed] [Google Scholar]
- Grant, C. M., Perrone, G., and Dawes, I. W. (1998). Glutathione and catalase provide overlapping defenses for protection against hydrogen peroxide in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Co. 253, 893-898. [DOI] [PubMed] [Google Scholar]
- Gross, E., Sevier, C. S., Vala, A., Kaiser, C. A., and Fass, D. (2002). A new FAD-binding fold and intersubunit disulfide shuttle in the thiol oxidase Erv2p. Nat. Struct. Biol. 9, 61-67. [DOI] [PubMed] [Google Scholar]
- Harding, H. P., et al. (2003). An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619-633. [DOI] [PubMed] [Google Scholar]
- Haynes, C. M., Titus, E. A., and Cooper, A. A. (2004). Degradation of misfolded proteins prevents ER-derived oxidative stress and cell death. Mol. Cell 15, 767-776. [DOI] [PubMed] [Google Scholar]
- Hinnebusch, A. G. (1994). Translational control of GCN4:an in vivo barometer of initiation factor activity. Trends Biochem. Sci. 19, 409-414. [DOI] [PubMed] [Google Scholar]
- Holmgren, A. (1989). Thioredoxin and Glutaredoxin systems. J. Biol. Chem. 264, 13963-13966. [PubMed] [Google Scholar]
- Hwang, C., Sinskey, A. J., and Lodish, H. F. (1992). Oxidized redox state of glutathione in the endoplasmic reticulum. Science 257, 1496-1502. [DOI] [PubMed] [Google Scholar]
- Inoue, Y., Matsuda, T., Sugiyama, K.-I., Izawa, S., and Kimura, A. (1999). Genetic analysis of glutathione peroxidase in oxidative stress response of Saccharomyces cerevisiae. J. Biol. Chem. 274, 27002-27009. [DOI] [PubMed] [Google Scholar]
- Izawa, S., Maeda, K., Sugiyama, K.-I., Mano, J., Inoue, Y., and Kimura, A. (1999). Thioredoxin deficiency causes the constitutive activation of Yap1, an AP-1-like transcription factor in Saccharomyces cerevisiae. J. Biol. Chem. 274, 28459-28565. [DOI] [PubMed] [Google Scholar]
- Jang, H. H., et al. (2004). Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell 117, 625-635. [DOI] [PubMed] [Google Scholar]
- Jessop, C. E., and Bulleid, N. J. (2004). Glutathione directly reduces an oxidoreductase in the endoplasmic reticulum of mammalian cells. J. Biol. Chem. 279, 55341-55347. [DOI] [PubMed] [Google Scholar]
- Jorgensen, P., Rupes, I., Sharom, J. R., Schneper, L., Broach, J. R., and Tyers, M. (2004). A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18, 2491-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman, R. J. (1999). Stress signalling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13, 1211-1233. [DOI] [PubMed] [Google Scholar]
- Kispal, G., et al. (2005). Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria. EMBO J. 24, 589-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno, K., Normington, K., Sambrook, J., Gething, M.-J., and Mori, K. (1993). The promoter region of the yeast KAR2 (Bip) gene contains a regulatory region domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Cell 13, 877-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge, S., and Jones, N. (1994). YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 13, 655-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J., Godon, C., Lagniel, G., Spector, D., Garin, J., Labarre, J., and Toledano, M. B. (1999). Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 274, 16040-16046. [DOI] [PubMed] [Google Scholar]
- Lin, S. J., and Guarente, L. (2003). Nicotinamide adenine dinucleotide, a metabolic regulator of transcription, longevity and disease. Curr. Opin. Cell Biol. 15, 241-246. [DOI] [PubMed] [Google Scholar]
- Marion, R. M., Regev, A., Segal, E., Barash, Y., Koller, D., Friedman, N., and O'Shea, E. K. (2004). Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl. Acad. Sci. USA 101, 14315-14322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, J.E.G. (1998). Posttranscriptional control of gene expression in yeast. Microbiol. Mol. Biol. Rev. 62, 1492-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister, A., and Anderson, M. E. (1983). Glutathione. Annu. Rev. Biochem. 52, 711-760. [DOI] [PubMed] [Google Scholar]
- Morgan, B. A., Banks, G. R., Toone, W. M., Raitt, D., Kuge, S., and Johnston, L. H. (1997). The Skn7 response regulator controls gene expression in the oxidative stress response of the yeast Saccharomyces cerevisiae. EMBO J. 16, 1035-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, K., Ma, W., Gething, M. J., and Sambrook, J. (1993). A transmemebrane protein with cdc2+/CDC28-related kinase activity is required for signalling from the ER to the nucleus. Cell 74, 743-756. [DOI] [PubMed] [Google Scholar]
- Mori, K., Ogawa, N., Kawahara, T., Yanagi, H., and Yura, T. (2000). mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc. Natl. Acad. Sci. USA 97, 4660-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlenhoff, U., Gerber, J., Richhardt, N., and Lill, R. (2003). Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p. EMBO J. 22, 4815-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, E.G.D. (1996). A glutathione reductase mutant of yeast accumulates high levels of oxidized glutathione and requires thioredoxin for growth. Mol. Biol. Cell 7, 1805-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan, T. A., Bassler, J., Petfalski, E., Tollervey, D., and Hurt, E. (2002). 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21, 5539-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkabyo, Y. S., Ziegler, T. R., Gu, L. H., Watson, W. H., and Jones, D. P. (2002). Glutathione and thioredoxin redox during differentiation in human colon epithelial (Caco-2) cells. Am. J. Physiol. 283, G1352-G1359. [DOI] [PubMed] [Google Scholar]
- Park, S. G., Cha, M.-K., Jeong, W., and Kim, I.-H. (2000). Distinct physiological functions of thiol peroxidase isoenzymes in Saccharomyces cerevisiae. J. Biol. Chem. 275, 5723-5732. [DOI] [PubMed] [Google Scholar]
- Patil, C. K., Li, H., and Walter, P. (2004). Gcn4p and novel upstream activating sequences regulate targets of the unfolded protein response. PLoS Biol. 2, E246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, M. G., Travers, K. J., and Weissman, J. S. (1998). Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol. Cell 1, 171-182. [DOI] [PubMed] [Google Scholar]
- Rietsch, A., and Beckwith, J. (1998). The genetics of disulfide bond metabolism. Annu. Rev. Genet. 32, 163-184. [DOI] [PubMed] [Google Scholar]
- Ross, S. J., Findlay, V. J., Malakasi, P., and Morgan, B. A. (2000). Thioredoxin peroxidase is required for the transcriptional response to oxidative stress in budding yeast. Mol. Biol. Cell 11, 2631-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotenberg, M. O., Moritz, M., and Woolford, J.L.J. (1988). Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of halfmer polyribosomes. Genes Dev. 12, 160-172. [DOI] [PubMed] [Google Scholar]
- Schafer, F. Q., and Buettner, G. R. (2001). Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 30, 1191-1212. [DOI] [PubMed] [Google Scholar]
- Schawalder, S. B., Kabani, M., Howald, I., Choudhury, U., Werner, M., and Shore, D. (2004). Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature 432, 1058-1061. [DOI] [PubMed] [Google Scholar]
- Sen, C. K. (2000). Cellular thiols and redox-regulated signal transduction. Curr. Top. Cell Regul. 36, 1-30. [DOI] [PubMed] [Google Scholar]
- Sherman, F., Fink, G. R., and Lawrence, C. W. (1974). Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Sidrauski, C., Cox, J. S., and Walter, P. (1996). tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell 87, 405-413. [DOI] [PubMed] [Google Scholar]
- Simons, J. F., S., F.-N., Rose, M. D., and Helenius, A. (1995). Bip/Kar2p serves as a molecular chaperone during carboxypeptidase Y folding in yeast. J. Cell Biol. 130, 41-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe, G. W., Fong, C. S., Alic, N., Higgins, V. J., and Dawes, I. W. (2004). Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc. Natl. Acad. Sci. USA 10, 6564-6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledano, M. B., Delaunay, A., Monceau, L., and Tacnet, F. (2004). Microbial H2O2 sensors as archetypical redox signaling modules. Trends Biochem. Sci. 29, 351-357. [DOI] [PubMed] [Google Scholar]
- Tomoyasu, T., Mogk, A., Langen, H., Goloubinoff, P., and Bukau, B. (2001). Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol. Microbiol. 40, 397-413. [DOI] [PubMed] [Google Scholar]
- Travers, K. J., Patil, C. K., Woodicka, L., Lockhart, D. J., Weissman, J. S., and Walter, P. (2000). Functional and genomic analysis reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101, 249-258. [DOI] [PubMed] [Google Scholar]
- Trotter, E. W., and Grant, C. M. (2002). Thioredoxins are required for protection against a reductive stress in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 46, 869-878. [DOI] [PubMed] [Google Scholar]
- Trotter, E. W., and Grant, C. M. (2003). Non-reciprocal regulation of the redox state of the glutathione/glutaredoxin and thioredoxin systems. EMBO Rep. 4, 184-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, B. P., and Weissman, J. S. (2004). Oxidative protein folding in eukaryotes: mechanisms and consequences. J. Cell Biol. 164, 341-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite, M. F., Stansfield, I., and Planta, R. J. (1999). Identifying genes encoding components of the protein synthesis machinery of the yeast Saccharomyces cerevisiae. In: Yeast Gene Analysis, ed. A. J. Brown and M. F. Tuite, London: Academic Press.
- Valadi, H., Valadi, A., Ansell, R., Gustafsson, L., Adler, L., Norbeck, J., and Blomberg, A. (2004). NADH-reductive stress in Saccharomyces cerevisiae induces the expression of the minor isoform of glyceraldehyde-3-phosphate dehydrogenase (TDH1). Curr. Genet. 45, 90-95. [DOI] [PubMed] [Google Scholar]
- Warner, J. R. (1999). The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24, 437-440. [DOI] [PubMed] [Google Scholar]
- Wek, R. C. (1994). eIF-2 kinases: regulators of general and gene-specific translation initiation. Trends Biochem. Sci. 19, 491-496. [DOI] [PubMed] [Google Scholar]
- Wheeler, G. L., and Grant, C. M. (2004). Regulation of redox homeostasis in the yeast Saccharomyces cerevisiae. Physiol. Plant 120, 12-20. [DOI] [PubMed] [Google Scholar]
- Winzeler, E. A., et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901-906. [DOI] [PubMed] [Google Scholar]
- Wong, C. M., Siu, K. L., and Jin, D. Y. (2004). Peroxiredoxin-null yeast cells are hypersensitive to oxidative stress and are genomically unstable. J. Biol. Chem. 22, 23207-23213. [DOI] [PubMed] [Google Scholar]
- Wood, Z. A., Schroder, E., Harris, J. R., and Poole, L. B. (2003). Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 28, 32-40. [DOI] [PubMed] [Google Scholar]
- Yarunin, A., Panse, V. G., Petfalski, E., Dez, C., Tollervey, D., and Hurt, E. C. (2005). Functional link between ribosome formation and biogenesis of ironsulfur proteins. EMBO J. 24, 580-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, C. C., Hou, M. F., Wu, S. H., Tsai, S. M., Lin, S. K., Hou, L. A., Ma, H., and Tsai, L. Y. (2005). A study of glutathione status in the blood and tissues of patients with breast cancer. Cell Biochem. Funct. (in press). [DOI] [PubMed]