Abstract
Regulated intramembrane proteolysis of the factors SREBP and ATF6 represents a central control mechanism in sterol homeostasis and stress response within the endoplasmic reticulum. Here, we compare localization of ATF6-related bZip factors CREB4, CREB-H, Luman, and OASIS. These factors contain the defining features of a bZip domain, a predicted transmembrane domain and an adjacent cleavage site for the Golgi protease S1P, with conserved features which indicate that it represents a specific subclass of S1P sites. Each factor localizes to the endoplasmic reticulum (ER), but a population of CREB4 was also observed in the Golgi. Deletion of the transmembrane domain in CREB4 resulted in efficient nuclear accumulation. An N-terminal variant of CREB4 containing the BZIp domain potently activated expression from a target gene containing ATF6 binding sites and from the promoter for the ER chaperone GRP78/BIP. CREB4 was cleaved in a site-specific manner in response to brefeldin A disruption of the Golgi or by coexpression with S1P but only after deletion or substitution of its C-terminal lumenal domain. Thus, S1P cleavage of CREB4 may be suppressed by a determinant in the C-terminal region. Dithiothreitol induced more complete transport of CREB4 to the Golgi, but not cleavage. Together, the data identify at least one additional bZip factor whose localization responds to ER stress, and we propose a model based on these results that indicates additional levels of control of this novel class of transcription factors.
INTRODUCTION
Several key metabolic processes, including sterol and fatty acid homeostasis, and stress signaling within the endoplasmic reticulum (ER) are controlled by a mechanism known as regulated intramembrane proteolysis (Brown and Goldstein, 1997; Yoshida et al., 1998; Haze et al., 1999; Kaufman, 1999; Ye et al., 2000; Sakai and Rawson, 2001). The central components in these systems comprise a distinct class of transcription factors that contain one or more transmembrane domains, anchoring partners that localize the factors to a cellular compartment, and proteases that are located in a different compartment. Generally, the transcription factors are inserted into ER membranes, with the domain involved in DNA binding and transcriptional regulation oriented toward the cytosolic face of the membrane (Sato et al., 1994; Hua et al., 1995; Haze et al., 1999). The main step in controlling the activity of these factors resides in their regulated release from the ER in response to specific cues, to traffic to another site, usually a Golgi-associated compartment. Resident proteases at this site cleave the factor in a stepwise manner, resulting in release of the cytosolic domain that then traffics to the nucleus to effect transcription of specific target genes (Brown and Goldstein, 1997; Rawson et al., 1997; Sakai et al., 1998b; Kaufman, 1999; Seidah et al., 1999).
The prototypical membrane-bound transcription factors are the sterol regulatory element binding proteins SREBP 1 and 2, which regulate genes involved in cholesterol and fatty acid homeostasis (Brown and Goldstein, 1999). SREBP contains an N-terminal helix-loop-helix DNA binding and activation domain, a central section containing two transmembrane regions, and a C-terminal regulatory domain (Hua et al., 1993; Yokoyama et al., 1993; Brown and Goldstein, 1997). Newly synthesized SREBP is anchored in the ER via its two transmembrane domains (Sato et al., 1994; Brown and Goldstein, 1997), and when cholesterol levels in the membrane are high, SREBP is retained in the ER in a complex with a polytopic transmembrane protein SREBP cleavage activating protein (SCAP) (Sakai et al., 1998a,b; Yang et al., 2000; Matsuda et al., 2001). Further interaction between SCAP and a transmembrane protein termed INSIG seems to be involved in ER retention of this multiprotein complex (Yang et al., 2002). When cholesterol levels decrease, an SREBP-SCAP complex is released and is transported to the Golgi where it is subject to sequential cleavage at two sites (Hua et al., 1996; Sakai et al., 1996). The lumenal loop is first cleaved at site 1, resulting in a membrane-anchored intermediate. In SREBP2, this site encompasses the sequence P4-RSVL-P1, and cleavage is catalyzed by the site 1 protease S1P (Duncan et al., 1997; Sakai et al., 1998b). The second cleavage is at site 2, at a conserved Leu-Cys bond within the first transmembrane domain of SREBP, and it is catalyzed by the site 2 metalloprotease S2P (Sakai et al., 1996). Current evidence indicates that SCAP is required in the complex with SREBP for the latter's recognition and cleavage by S1P. Cleavage by S2P is not thought to be regulated directly by sterols, but it is dependent upon prior cleavage of SREBP by S1P.
An interesting convergence has recently been demonstrated between these processes and other stress responses. ATF6, a bZip transcription factor, was shown to be synthesized as a single transmembrane protein that was also activated by proteolysis, in this case in response to ER stress (Yoshida et al., 1998; Haze et al., 1999; Li et al., 2000; Wang et al., 2000). It was subsequently shown that ATF6 also possesses an RxxL motif within its lumenal domain and that the protein is indeed subject to cleavage by the same proteases S1P and S2P that cleave SREBP (Ye et al., 2000). Cleavage of ATF6 is not, however, regulated by cholesterol. Recent results indicate that ATF6 is retained in the ER through interaction with the ER chaperone BiP/GRP78 (Chen et al., 2002; Shen et al., 2002). Loss of BiP binding by ER stress seems to unmask Golgi localization signals, allowing ATF6 to be transported to the site of active proteases in the Golgi (Shen et al., 2002). Subsequent nuclear transport of the active N-terminal domain of ATF6 results in the induction of molecular chaperones, including BiP itself as part of a coordinated response, which involves additional factors besides ATF6.
We previously identified an additional member of this group of membrane-bound transcription factors, Luman/CREB3 (Lu et al., 1997), and we demonstrated that it is also subject to proteolysis by the same proteases that cleave SREBP and ATF6 (Raggo et al., 2002). Primary cleavage of Luman likely occurs in the Golgi because brefeldin A redistribution of Golgi components to the ER caused efficient processing of the protein to yield an N-terminal product (Lu et al., 1997b; Raggo et al., 2002). Furthermore, coexpression of Luman with an ER-localized form of S1P resulted in the appearance of a specific N-terminal cleavage product (Raggo et al., 2002). Although Luman contains an RxxL consensus cleavage S1P, we identified an adjacent site, RQLR, as the S1P cleavage motif. However, unlike ATF6, induction of the unfolded protein response did not result in relocation of Luman from the ER, and the signals that induce Luman trafficking to the Golgi remain to be identified.
On the basis of the conservation of a bZip domain followed by a putative transmembrane domain, we identify a family of proteins that are clearly related to ATF6 and Luman. These factors, CREB4, OASIS, and CREBH, were cloned and subjected to direct comparative analysis, and CREB4 was analyzed in greater detail. All of these factors localized in the ER, although a population of CREB4 was also present in the Golgi. Deletion of the CREB4 transmembrane domain resulted in nuclear accumulation, and a truncated N-terminal form of CREB4 also accumulated in the nucleus and potently activated expression of test target genes. Although CREB4 could be cleaved by S1P, this was only after removal of the C-terminal region of the protein. Analysis of deletion constructs and chimeras indicated the possibility of additional factors determining CREB4 cleavage. Moreover, dithiothreitol (DTT) induced more complete transport of CREB4 to the Golgi, but not cleavage. Together, our results indicate that CREB4 is a member of the class of proteins whose localization is regulated by the unfolded protein response and that it can be a substrate for S1P. They also indicate the possibility of additional forms of regulation of S1P cleavage or additional types of ER stress, as discussed.
MATERIALS AND METHODS
Cell Culture
COS-1 cells, HeLa, and Vero cells were maintained in DMEM (Invitrogen, Carlsbad, CA) under standard conditions. Transfections were carried out with 1-2 μg of the appropriate vectors as described previously (Chen and Okayama, 1987; Raggo et al., 2002). Where indicated, brefeldin A (final concentration 1 μg/ml) was added to cells at ∼24 h after transfection, and cultures were further incubated for 4-8 h. For studies examining the effect of the induction of the unfolded protein response, DTT was added in normal medium with the times and concentrations indicated in the figure legends.
Plasmids
The construction of a vector (pJS6), containing the coding sequence of Luman (CREB3), flanked by an SV5 epitope tag at its N terminus and a hemagglutinin (HA) tag at its C terminus, has been described previously (Raggo et al., 2002). The coding sequence of CREB4 was amplified from a HeLa cDNA library (Marathon Ready cDNA; BD Biosciences Clontech, Palo Alto, CA), using the primers AGATCTATGGATCTCGGAATCCCTGACCC and CTCGAGCATCTCATCTG CATGCAGCACGG, in which a BglII and XhoI site were incorporated onto the 5′ and 3′ primers, respectively, to facilitate subsequent cloning steps. The underlined ATG represents the initiator methionine, and the 3′ primer is designed so that the last coding triplet will read in-frame to a C-terminal tag after the subsequent cloning steps. The amplified fragment was then digested with BglII and XhoI, and inserted into the BamHI and XhoI sites of pJS6, creating a construct that expressed intact CREB4 (pJS23), tagged at the N terminus and C terminus. Using a native BamHI site just downstream of the CREB4 start site, a BamHI-XhoI fragment was also cloned between the BamHI and XhoI sites of pJS6 creating a vector that expressed a version of CREB4 lacking the first 22 residues (pJS24; CREB.ΔN1).
A chimera consisting of the N-terminal domain of CREB4 (1-275) fused to the transmembrane and C-terminal domains of Luman was constructed by digestion of pJS23 with BlpI, a natural unique site just before the putative transmembrane domain; treatment with T4 polymerase to remove the 3′ extensions; and digestion with XhoI. This removed the transmembrane and C-terminal domain of CREB4 (residues 276-395). A fragment (EcoRV-XhoI) from pJS6 containing the transmembrane and C-terminal tail of Luman (residues 220-371) was then inserted to create pJS42 (CH2). A chimera consisting of the N-terminal domain and transmembrane region of CREB4 (1-324) fused to the C-terminal lumenal domain of CREB3 (258-371) was created by amplifying the N terminus and transmembrane domain of CREB4 using the primers AGATCTATGGATCTCGGAATCCCTGACCC and AGATCTAGACCCAGCTTCTGGTCGAC. This was cloned between the BamHI at the N terminus and the internal BlpI site of pJS6 to create a vector pJS43 (CH3). An additional chimera was constructed (pJS66, CH6) that contained the N terminus of CREB4, including the transmembrane and S1P sites, linked to the lumenal domain of Luman. For this, a C-terminal Luman fragment was amplified using the primers CTCGAGGCCCTCCCCAGTGAGGACCCTTAC and GATCCTCGAG GCCTGAGTATC TGTCCAG and cloned into the XhoI site of pJS60 to create a chimera composed of the N-terminal 344 residues of CREB4 followed by residues 269-371 of Luman. Finally a reciprocal chimera containing the N terminus of CREB3 with the transmembrane domain and C terminus of CREB4 (pJS58, CH4) was constructed by replacing an EcoRV-XhoI fragment of pJS6 with a BlpI-XhoI fragment from pJS23.
For deletion mutants, CREB4.ΔTM (pJS32) was otherwise full length but contained an internal deletion to remove the putative transmembrane domain (residues 281-313). Other deletion constructs contained the N-terminal portion of the protein, CREB.ΔTMC1, truncated at residue 276 and lacking the transmembrane and C-terminal region; CREB4.ΔC1, truncated at residue 319, and including the transmembrane region but lacking the putative S1P site at position 335-338; CREB4.ΔC2, truncated at residue 344 and containing the N terminus plus the transmembrane domain and putative S1P site. Residues 1-344 were amplified using the primers AGATCTATGGATCTCGGAATCCCTGACCC and CTCGAGTGTTACGTCCTTGTGGGTCAGG, and the resulting cDNA was cloned between the BamHI and XhoI sites of pJS23. CREB4.ΔC1 was made by digesting pJS23 with SalI and XhoI, treating with Klenow and religating. This contains residues 1-319 of CREB4 and includes seven residues (HKGEPAS) before new stop codon. All constructs were confirmed by sequencing and are their structures are summarized in the appropriate figures.
Fusions of CREB 4 to GAL4 DNA binding domain were constructed using the GAL4 DNA binding domain pM1 (Sadowski et al., 1992). CREB4 was amplified from pJS23 using the primers GAATTCATGGATCTCGGAATCCCTGAC and CTCGAGTTACAGGCTGGCATAGTCAGGC. This product, containing full-length CREB4 with the addition of an HA tag at the C terminus followed by a termination codon was digested with EcoRI and XhoI and inserted between the EcoRI and SalI sites of pM1 to produce pJS55, expressing the GAL4 DNA binding domain fused to full-length CREB4. To produce a GAL4 fusion to the N-terminal region only of CREB4, pJS44 (CREB.ΔTMC1) was digested with HindIII and XbaI to isolate the N-terminal residues 1-276, which were cloned between the HindIII and XbaI sites of pJS55 to produce pJS62.
A vector expressing a green fluorescent protein (GFP)-CREB4 fusion protein was constructed. By inserting a fragment (NdeI-BamHI) containing the coding sequence for GFP from pEGFP-C1 (BD Biosciences Clontech) into pJS23 to create a vector that expresses a fusion protein of CREB4.ΔN1 with GFP at the N terminus (pJS51). The BglII-BlpI fragment from intact CREB4 was then transferred into the BamHI and BlpI sites of pJS51 expressing the intact GFP-CREB4 fusion protein with GFP at the N terminus (pJS52).
To construct an expression vector for OASIS, cDNA (IMAGE ID 4856946) was digested with BamHI. The cDNA clone contains a native BamHI site at residue 17 of OASIS and a BamHI in the plasmid vector. The 1.5-kb BamHI fragment encompassing residues 17 to the C-terminal residue (519) was then transferred to pJS6 to create the intermediate vector pJS6OAS1. To construct the expression vector such that OASIS contained the same C-terminal HA tag as the other constructs, primers GGTCACCAACAAGATCTCCAGACC containing a BstEII site and CTCGAGGGAGAGTTTGATGGTGG containing an XhoI site were used to amplify the 3′ end of the coding region (residues 354-519), so that the region would read in frame when inserted into pJS6OAS1. The cDNA fragment digested with BstEII and XhoI was then inserted between the BstEII (in Oasis) and XhoI (Vector) sites of pJS6OAS1 to create a second intermediate vector, pJS6OAS2. To complete the cloning, the 5′ end of the OASIS coding region was then amplified from the Image cDNA clone using the primers (AGATCTATGGACGCCGTCTTGG) and (TCCGGACTCTCTTCAAGGCCTTC). This 874-bp fragment was digested with BglII and BspEI and cloned between the BamHI and the native BspEI sites of pJS6OAS2 to produce a construct that expresses intact OASIS with the SV5 epitope at the N terminus and the HA epitope at the C terminus (pJS22).
The strategy to construct an expression vector for CREB-H was the same as that described above for CREB4 from a HeLa cDNA library using the primers CGCGGATCCATGAATACGGATTTAGCTGC and ACGCGTCGACCAGCTCGTCTCCCGCCGCCT. The fragment BamHI-SalI was cloned into the BamHI-XhoI sites of pJS6 (pSM20). The construct expressing ATF6 (p3XFLAG-CMV-ATF6) and the N-terminal variant 1-373 were kindly supplied by Dr. Ron Prywes (Department of Biological Sciences, Columbia University, New York, NY). Expression vectors for S1P were kindly supplied by Dr. Joseph Goldstein (Department of Molecular Genetics, University of Texas, Southwestern Medical Center, Dallas, TX).
Mutagenesis
Mutation of the S1P cleavage site within CREB4 was carried out using QuikChange kit (Stratagene, La Jolla, CA). Within CREB4, the sequence 335-RNIL-338 is located ∼20 residues downstream of the transmembrane region. A mutant version of either CREB4 (pJS50) or the truncated CREB4.ΔC2 (pJS67) that contained a single amino acid substitution (R335G) within this motif was created using the primers GGAGTGACTTCCGGAAATATCCTGACCCACAAGG and CCTTGTGGGTCAGGATATTTCCGGAAGTCACTCC. The parental plasmids pJS24 or pJS60 were denatured and incubated with the primers according to the manufacturer's instructions. After completion of the reaction, the sample was digested with DpnI to remove parental plasmid template and then transformed. Insertion of the mutation in this construct as for all constructs was confirmed by plasmid sequencing.
Immunofluorescence
Cells grown on coverslips were transfected with 2 μg of the appropriate expression vector. At ∼30 h after transfection, the cells were washed in phosphate-buffered saline (PBS) and fixed for 15 min in methanol at -20°C. After blocking in PBS containing 10% fetal calf serum, primary antibodies diluted in blocking solution were applied. These included the monoclonal antibodies SV5 (1/10000) for the SV5 tag and anti-FLAG M2 (1/1000; Stratagene) and polyclonal anti-GFP (1/500; Research Diagnostics, Flanders, NJ), anti-calreticulin (1/500, Calbiochem, San Diego, CA), and anti-β-Cop (1/100; Affinity Bioreagents, Golden, CO). After washing in several changes of PBS, secondary antibodies conjugated with Alexa 488 or Alexa 543 (Invitrogen) were applied at a dilution of 1/200. After incubation for 20 min, the slides were again washed and mounted in Mowiol (Sigma Chemical, Poole, Dorset, United Kingdom). For localization studies, CREB4 was also cotransfected with a plasmid (enhanced yellow fluorescent protein [EYFP]-Golgi; BD Biosciences Clontech), which expresses the Golgi-targeting signal of β-galactosyl transferase fused to yellow fluorescent protein. Fluorescence images were acquired on a Zeiss LSM410 laser scanning confocal microscope. Images were routinely acquired using a Plan-apochromat 63× oil immersion objective lens, numerical aperture 1.4, and zoom factors ranging from 1 to 8 of the LSM 410 acquisition software.
Luciferase Assays
COS cells were transfected with the different expression and target vectors, and cell lysates prepared 24 or 48 h after transfection by addition of Glo lysis buffer (Promega, Madison, WI). Firefly luciferase activity was determined using the Bright-Glo assay system as described by the manufacturer (Promega).
For GAL4 fusion experiments, 2 μg of the commercial target vector containing five GAL4 binding sites, pFR-LUC (Stratagene), was cotransfected with various amounts (0, 0.25, 1, or 4 μg) of pM1 (empty vector expressing the GAL4 DNA binding domain alone), pJS55 (GAL4-CREB4), or pJS62 (GAL4-CREB4.ΔTMC1).
In other activation assays, activity of native CREB4 and variants was compared with ATF6 using the vectors pJS23 (CREB4), pJS44 (CREB4.ΔTMC1, residues 1-276), p3XFLAG-CMV-ATF6, and pCGNATF6 (1-373). These latter two vectors were kindly supplied by Dr. Ron Prywes and have been described previously (Wang et al., 2000). These were cotransfected with target vectors containing multiple ATF6 binding sites, 5XATF6-GL3 (2 μg), or vectors containing the native promoters for ER chaperones such GRP78, pGL3-GRP78 promoter (0.5 μg). This vector, kindly supplied by Dr. Mori (HSP Research Institute, Kyoto Research Park, Kyoto, Japan), has been described previously (Yoshida et al., 1998).
Western Blotting
Cultures of transfected cells (1-2 × 105 cells) were washed in cold PBS, and total lysates were prepared by adding 200 μl of SDS loading buffer. Samples were briefly sonicated and boiled before electrophoresis. Equal amounts of total proteins were fractionated by SDS-PAGE and transferred onto Hybond-C membranes (GE healthcare, Little Chalfont, Buckinghamshire, United Kingdom). Membranes were blocked with PBS containing 5% nonfat milk and 0.1% Tween 20. Primary antibodies diluted in blocking solution were anti-SV5 (1/10000) and anti-FLAG M5 (1/1000; Sigma Chemical). Protein bands were detected by standard procedures for chemiluminescence detection
RESULTS
Analysis of a Conserved Multisection Domain in Subset bZIP Transcription Factors
The transcription factors SREBP 1 and 2 are key components in the homeostatic control of sterol and fatty acid metabolism (Brown and Goldstein, 1997; Horton et al., 2002). These proteins are regulated by compartmentalization, trafficking, and subsequent cleavage by Golgi-resident proteases. It was subsequently shown that ATF6 is also a member of this class of factors, subject to cleavage by the same proteases that cleave SREBP 1 and 2 (Haze et al., 1999; Li et al., 2000; Wang et al., 2000; Ye et al., 2000; Yoshida et al., 2000), in this case as a consequence of the accumulation of unfolded proteins, rather than sterol or fatty acid fluctuation. Recently, we demonstrated that Luman(CREB3) was a new member of the ER-associated bZIP transcription factors (Lu et al., 1997b; Raggo et al., 2002), also located in the ER, and subject to cleavage by S1P (Raggo et al., 2002). However, unlike ATF6, treatments such as tunicamycin or DTT, which induce the unfolded protein response, did not induce trafficking of Luman. The environmental cues that induce Luman transport and cleavage remain to be identified.
To examine whether there were additional members of this distinct class of transcription factors, we searched protein databases for the defining features conserved between ATF6 and Luman, i.e., a bZip domain in proximity to a hydrophobic transmembrane region possessing particular features (see below). This search revealed three human proteins with strong similarity to ATF6 and Luman within a central domain (Figure 1a). The protein termed CREB4 was originally identified as part of a sequencing analysis of human cDNAs (Cao et al., 2002). The same protein termed AIbZIP was identified in a screen for genes increased in androgen-treated prostate cancer cell lines (Qi et al., 2002), although we adopt the name CREB4 here. The factor termed OASIS (old astrocyte specifically induced substance) was originally identified based on its increase in long-term versus short-term cultured astrocytes (Nikaido et al., 2001; Omori et al., 2002). From random sequencing of cDNA clones from the hepatoma cell line HepG2, the factor termed CREB-H was identified (Omori et al., 2001).
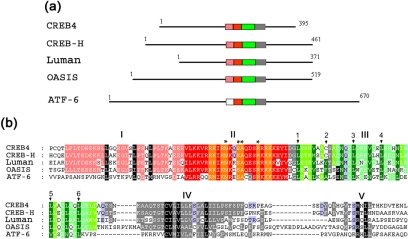
Figure 1.
Comparison of bZIP transcription factors containing transmembrane domains. (a) Schematic illustration of the relative sizes and organization of the central conserved domain in Luman, CREB4, OASIS, CREBH, and ATF-6. The total length of the proteins is indicated, and the core region exhibiting a high degree of sequence similarity is divided into subregions and colored for ease of reference as discussed in the text. (b) Amino acid sequence alignments of the conserved domains of Luman (AAB69652), CREB4 (AAH38962), OASIS (BAC01278), CREB-H (NP_115996), and ATF-6 (AAB64434). Numbers indicate the positions of the first residues in the block for each protein. The subregions discussed in the text are colored as follows: region I (pink); region II, the basic domain (red); region III, the leucine zipper (green); and the putative transmembrane domains (gray). The blocks indicate regions with identity or similarity in four of the five sequences, but shading has also been used to highlight features of interest. For example, although region I is not well conserved in ATF6, certain residues (black background) are conserved in all the proteins. Within region III, shading has been used to highlight similarities and differences. In region IV, there is a consensus motif conforming to C∅∅∅∅∅∅n∅∅∅, as discussed in the text. This is colored in gray with certain resides in black for ease of reference.
Although the presence of bZip region in these proteins was noted in their original identification, consideration of specific features within this region are informative in the broader context of regulated intramembrane proteolysis (RIP) and for our experimental analysis (Figure 1). The transcription factors are of a generally similar size, whereas ATF6 is significantly larger. They exhibit short pockets of limited homology within the N-terminal and C-terminal regions but the most notable feature is clearly the presence of a highly conserved section, approximately in the middle of each of the proteins. This central region (150 residues) comprises five distinct subregions, which for ease of comparison are indicated by colored boxes, and shown in detail in corresponding shading in Figure 1b.
We have labeled the basic domain and leucine zipper homologies as domains II and III, respectively. These regions are extremely well conserved. Each of the factors contains the invariant asparagine and adjacent hydrophobic residues within the core basic domain (Figure 1b, asterisks), together with the conserved leucine residues at the appropriate spacing (Figure 1b, numbered), which are the defining features of bZip transcription factors (Vinson et al., 1989). Interestingly, immediately adjacent to the N-terminal end of the bZip region, there is an additional region of ∼30 residues (Figure 1b, region I), which is conserved in the proteins CREB4, Luman, CREB-H, and OASIS but lacking in ATF6. This latter region is not part of the typical consensus bZip DNA binding domain and therefore may provide a distinct function in these factors. Luman and ATF6 have been shown to be anchored in the ER through the possession of a hydrophobic domain (Haze et al., 1999; Raggo et al., 2002). This region (IV) is positioned just C terminal of the leucine zipper, is clearly well conserved in all the factors, and notably contains a completely conserved cysteine residue followed by a run of exclusively hydrophobic residues (see below). Adjacent to this region on the C-terminal side is the next conserved region (region V). The motif for SIP cleavage in SREBP was defined as RSVLS, where SIP cleaves between the L and S (Duncan et al., 1997). An RxxL motif is conserved in ATF6 and is the site for cleavage for SIP (Ye et al., 2000). Although Luman possesses an RXXL motif just adjacent to the transmembrane region (Figure 1b), we showed that the motif SRQLR located ∼10 residues away was in fact the site of S1P cleavage (Raggo et al., 2002). Alignment of the additional members of this subfamily now demonstrates that the Luman site lies within a conserved motif with the refined consensus S R xL/I x, where R is the invariant P4 position and P1 shows a strong preference for L. Interestingly, with the exception of ATF6, this family retains a serine residue immediately adjacent to the invariant R residue. This alignment together with experimental examination of the site in the context of CREB4 (see below) is relevant to the broader consideration (see Discussion) of the target sites for S1P cleavage and potential regulation of site selection (Brown and Goldstein, 1999; Espenshade et al., 1999; Toure et al., 2000; Elagoz et al., 2002).
Localization of bZIP Transcription Factors
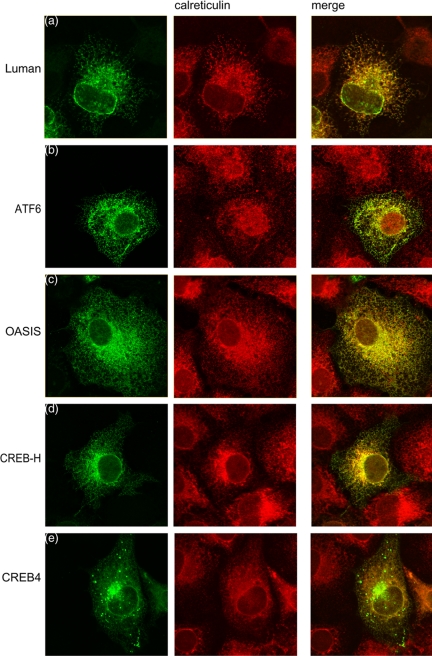
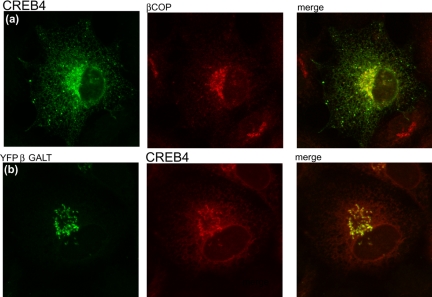
To investigate the subcellular localization of CREB4, OASIS, and CREB-H, the corresponding cDNAs were isolated by reverse transcription-PCR, and vectors were constructed that contained epitope-tagged versions of the proteins to facilitate direct comparative analyses. Expression and localization of the factors were then examined in parallel with that of Luman and ATF6. We have previously shown that Luman is located in the ER (Raggo et al., 2002), and a typical cell showing Luman in the cytoplasm in reticular structures that costain for calreticulin is shown in Figure 2a. A similar pattern was observed for ATF6 (Figure 2b), OASIS (Figure 2b) and CREB-H (Figure 2d), each being found solely in the cytoplasm where they colocalized with calreticulin in the ER. CREB4 was also present in a reticular pattern within the cytoplasm, with no detectable nuclear material (Figure 2e). A population of CREB4 exhibited a distinct pattern of accumulation in a perinuclear region resembling Golgi localization and to varying extents in punctate vesicular structures within the cytoplasm, which possibly represented compartments of the endosomal/lysomal system; see below). Colocalization of a population of CREB4 in the Golgi was confirmed using the marker β-Cop (Figure 3a). Alternatively, the cells were cotransfected with pEYFP-Golgi (BD Biosciences Clontech), which encodes a fusion protein consisting of EYFP and the Golgi-targeting N-terminal 81 amino acids of human β1,4-galactosyl transferase. Again a population of CREB4 colocalized with the marker protein (Figure 3b). This localization was noted for CREB4 when originally identified as AIbZip (Qi et al., 2002). Although characterization of the compartmentalization and processing of CREBH and OASIS are ongoing, the remainder of this work concerns analysis of CREB4.
Figure 2.
Localization of CREB4, Luman, ATF6, OASIS, and CREBH. Cells grown on coverslips were transfected with 2 μg of the appropriate expression vector as indicated. At 24 h posttransfection, cells were fixed in methanol and stained with a monoclonal antibody that recognized the N-terminal epitope tag on each of the proteins (green channel), together with an antibody to the ER protein calreticulin (red channel).
Figure 3.
A population of CREB4 localizes to the Golgi. Transfectection and processing were as for Figure 3. In a, cells were costained for CREB4 (green channel) or the Golgi-associated protein β-Cop. In b, CREB4 was cotransfected with the commercial expression vector for a Golgi-associated yellow fluorescent protein, YFP-β-galactosyl transferase, (YFP-B GALT). In this case, CREB4 localization is in the red channel, and YFP-B GALT is in the green channel. By both methods, a population of CREB4 clearly associated with the Golgi apparatus.
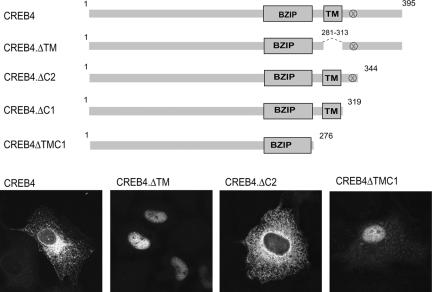
Deletion of the Predicted Transmembrane Domains Results in Nuclear Accumulation
Consistent with the alignment of CREB4, prediction algorithms (http://www.ch.embnet.org/software/TMPRED_form.html) indicated that CREB4 is a type II membrane protein with a transmembrane domain (TM) located around residues 294-313. To demonstrate that this region was responsible for the ER and Golgi localization of the protein, we deleted it, removing residues 281-313 in the context of an otherwise full-length protein, in the construct CREB4.ΔTM (Figure 4, top). The result was clear and striking. Removal of this region from CREB4 resulted in quantitative relocalization of the protein from the cytoplasm to an exclusively nuclear localization in all cells, a typical example of which is shown in Figure 4, bottom. We also constructed a version of CREB4, CREB4.ΔTMC1, which contained only residues 1-276, encompassing the N terminus and bZip domains. We predicted that this version would approximate the active cleaved form of CREB4, which would be released from membrane localization. This version was also found to exhibit virtually exclusive nuclear localization (Figure 4, bottom). On the other hand, variants containing residues to position 319, or 344 (Figure 4, top), encompassing the predicted transmembrane region IV, were found predominantly in the cytoplasm (Figure 4, bottom; our unpublished data). These results are consistent with the prediction that the conserved region IV was indeed responsible for membrane localization of CREB4 and that deletion of this region resulted in an N-terminal product that efficiently localized in the nucleus. For variants within the nucleus, no obvious subnuclear localization was observed although both forms showed typical nucleolar sparing.
Figure 4.
CREB4 localizes to the ER and Golgi dependent on the putative transmembrane domain. Top, schematic summary of CREB4 and variants. Bottom, cells were transfected with were transfected with 2 μg of the appropriate expression vector as indicated and processed as in Materials and Methods. Removal of the TM domain in the context of the full-length protein, or deletion of the TM domain and the C-terminal region, resulted in compete relocation of the protein to the nucleus.
Promoter Activation by CREB4
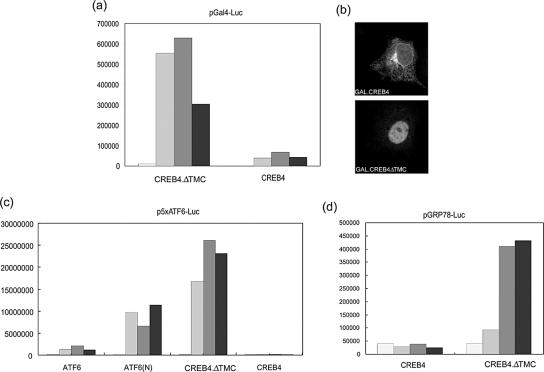
We next wanted to examine whether CREB4, particularly the predicted N-terminal cleavage product, could act as a transcriptional activator. We examined activity in the context of GAL4 fusion constructs. Intact CREB4, or the variant CREB4.ΔTMC, was fused to the GAL4 DNA binding domain. These constructs or a control vector expressing the GAL4 DNA binding domain alone (pM1), were cotransfected in various amounts with a commercial target luciferase vector (pFR-Luc) containing GAL4 DNA binding sites linked to a basal promoter. The control vector pM1 gave no detectable activation above background levels of promoter activity (our unpublished data). By comparison, very potent activation by GAL4.CREB4.ΔTMC was observed, with limited, but above background, activation by GAL4.CREB4 (Figure 5a). Control experiments demonstrated that these proteins were synthesized in approximately equal amounts. The efficient activation presumably reflects both the possession of an activation domain within the N terminus of the protein and the observation, as expected from the results mentioned above, that GAL4-CREB4.ΔTMC was almost completely nuclear, whereas the fusion to the intact protein was mainly in the cytoplasm (Figure 5b).
Figure 5.
Promoter activation by a GAL4-CREB4 chimeric protein. (a) Cells were transfected with 2 μg of the GAL4 UAS target vector pFR-LUC together with increasing amounts of GAL4-CREB4 or GAL4-CREB4. ΔTMC1 expression vectors. The increasing shading represents 0, 0.25, 1, or 4 μg of either vector. In each case, total DNA was equalized with the appropriate amount of pUC19 plasmid DNA. Cell lysates were prepared 24 h after transfection by addition of Glo lysis buffer (Promega), and firefly luciferase activity was determined using the Bright-Glo luciferase assay system (Promega). (b) Typical localization patterns of GAL4-CREB4 or GAL4-CREB4.ΔTMC 1. Cells were fixed and processed as described in Figure 2. (c) Activation by the native N-terminal product of CREB4. A target vector containing multimerized ATF6 sites, 5XATF6-GL3 (2 μg), was transfected with 0, 0.5, 1, or 4 μg of p3XFLAG-CMV-ATF6 (intact ATF6), pCGNATF6 (ATF6, 1-373), CREB4 (pJS23), or CREB4.ΔTMC 1 (pJS44) as indicated by the increasing gray shading. Cells were harvested and assayed 24 h later. Compared with the activated levels, basal expression from this promoter was extremely low. Very potent activation was seen for CREB4.ΔTMC 1 and ATF6, 1-373. (d) Activation of the native BiP promoter by CREB4.ΔTMC 1. A target vector containing the native BiP/GRP 78 promoter (0.5 μg) was transfected with 0, 0.5, 1, or 4 μg of CREB4 (pJS23) or CREB4.ΔTMC 1 (pJS44) as indicated. Cells were harvested and assayed 24 h later.
We next wanted to confirm whether native CREB4 could activate expression from a target promoter and first compared its activity with that of ATF6 on a target promoter containing ATF6 binding sites. In parallel, we examined activation by ATF6 itself or an N-terminal derivative of ATF6 (ATF6N, residues 1-373), which lacks the transmembrane and C-terminal regions in a corresponding manner to CREB4.ΔTMC. The results (Figure 5c) demonstrated that ATF6 induced expression from the target promoter, with overall increase ranging up to 30-fold. Consistent with previous results (Wang et al., 2000), the truncated form of ATF6 was considerably more active than the intact protein, with increases of up to several 100-fold. Although intact CREB4 gave very minor activity (3- to 4-fold increases), the N-terminal variant of CREB4 induced extremely potent activation of the target promoter, resulting in increases in activity even greater (by 3- to 5-fold) than that induced by the N-terminal variant of ATF6. Activation by CREB4.ΔTMC was dependent upon the ATF6 sites in the promoter because essentially no activation was observed on a control promoter lacking these sites (our unpublished data). The activity of intact ATF6 observed in this and other studies is presumably because of a level of constitutive cleavage releasing the N-terminal active form, although the appearance of this product can be variable and difficult to detect (Wang et al., 2000). Given the observation of very potent activation by CREB4.ΔTMC, the very low activity of intact CREB4 likely reflects very low constitutive cleavage.
We also examined whether CREB4ΔTMC could activate expression from a native stress-responsive promoter, that of GRP78/BIP (Figure 5d). Again, although intact CREB4 was virtually inactive, significant induction (15-fold) was observed by the N-terminal CREB4 protein. This activation, although lower than on the multimerized ATF6 site, is typical of what has been observed previously on this native promoter. Although this analysis was not intended to compare specific activity of ATF6 and CREB4 (the proteins have different epitope tags and their levels cannot be directly compared), these results do supply robust evidence that CREB4 can act as a transcriptional activator, that the intact protein exhibits little intrinsic activity (due to retention in the cytoplasm), and that the N-terminal fragment comprises in a potent activator located within the nucleus and is able to transactivate promoter activity of ER chaperones.
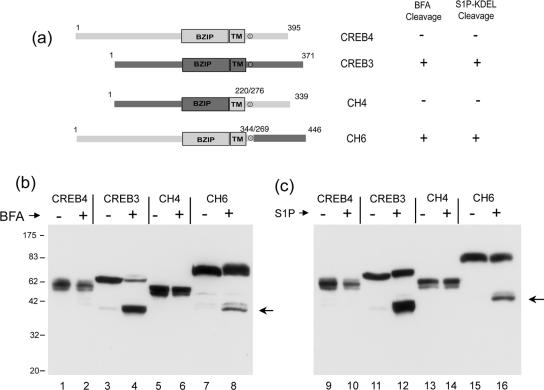
Comparison of Brefeldin A Treatment on Luman and CREB4 Cleavage
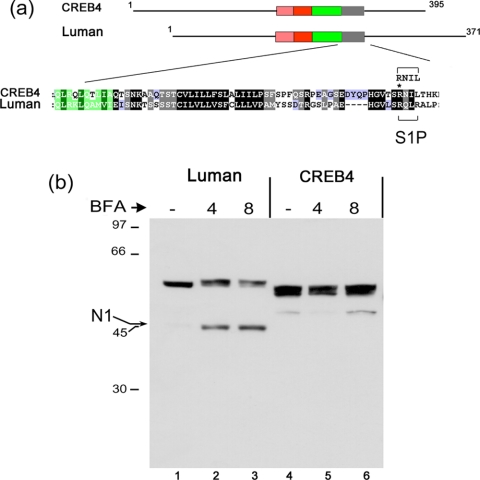
One of the first demonstrations that ATF6 was a target for S1P came from the use of brefeldin A to relocalize Golgi enzymes, resulting in ATF6 cleavage (Ye et al., 2000). We demonstrated previously (Raggo et al., 2002) that Luman was also subject to cleavage induced by brefeldin A relocalization of S1P and that cleavage was at the specific motif within region V. The schematic diagram in Figure 6 illustrates a specific comparison between Luman and CREB4. The enlarged section indicates the high degree of sequence conservation within the transmembrane domains and highlights the position of the S1P cleavage motif in Luman, RQLR (residues 264-267). CREB4 also has a consensus cleavage site, RNIL (residues 335-338), which is exactly in the appropriate position and is actually closer to the sequence of the SREBP cleavage site RSVL than is the Luman motif.
Figure 6.
CREB4 resistance to brefeldin A-induced cleavage. (a) Selective comparison of Luman and CREB4 and alignment of regions IV and V as discussed in the text. The position of a single R>G substitution (see Figure 9) is indicated by an asterisk. (b) Cells were transfected with expression vectors for Luman, CREB4, or CREB4.ΔN1, and 24 h later they were treated with brefeldin A (1 μg/ml) 4 or 8 h, or left untreated (-). The cells were then harvested and samples separated by SDS-PAGE and analyzed by Western blot analysis using the anti-epitope SV5 antibody. An arrow indicates the position of the induced cleavage product of Luman (N1). Very minor low levels of an N-terminal product of CREB4 are indicated, but these were not reproducibly observed (see, e.g., Figures 8 and 9) and importantly were not affected by brefeldin A treatment.
To compare the susceptibility of Luman and CREB4 to SIP cleavage, we first examined the effect of brefeldin A treatment. Cells were transfected with expression vectors for Luman and CREB4 and 24 h later were treated with brefeldin A for 4 or 8 h. Note that Luman migrates with a molecular weight of approximately 55 K, larger than the value of 41 K predicted from its amino acid sequence (Figure 7b, lane 1), due at least partially to glycosylation (Raggo et al., 2002). Similarly CREB4 migrated with a molecular weight of approximately 53 K, larger than its predicted size of 43 K (Figure 6b, lane 4). CREB4 migrated as a doublet indicative of post-translational modification such as glycosylation, and/or phosphorylation.
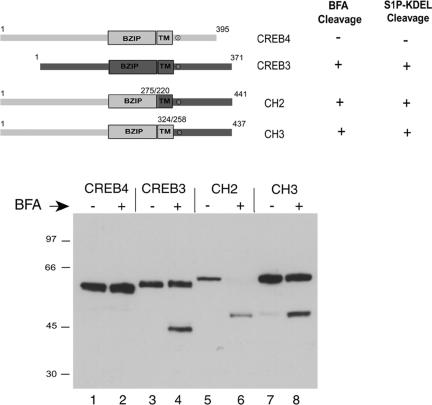
Figure 7.
Cleavage of chimeric CREB4 variants. Top, summary of the chimeric proteins in which the C-terminal region of CREB4 was replaced with the C-terminal region of Luman/CREB3. The fusion junction was either before the CREB4 TM domain (CH2) or after the TM domain but upstream of the S1P site (CH3). Bottom, cells were transfected, treated with brefeldin A (1 μg/ml; 8 h), and processed as in Figure 6.
As expected, brefeldin A induced the specific accumulation of a 45-K N-terminal Luman product (Figure 6b, lanes 1-3). For CREB4 in the absence of brefeldin A treatment, a minor and variable population of an N-terminal product was occasionally observed (Figure 6b, lane 4), which we believe is due to nonspecific cleavage. However, in contrast to Luman, there was no effect of brefeldin A treatment, evidenced by the accumulation of any N-terminal product (Figure 6b, lanes 4-6). These results indicated that despite its possession of a consensus S1P cleavage site in the appropriate position, CREB4 was either not a substrate for S1P, or its cleavage was somehow controlled in a manner different from that for Luman, and ATF6. Results below indicate that the latter explanation may be the case.
Brefeldin A-induced Cleavage of CREB4 Chimeric Proteins
Considering the sequence similarity between Luman and CREB4, we next wanted to address possible mechanisms for the apparent resistance of CREB4. We reasoned that the C-terminal regions of these factors, encompassing the transmembrane domain, the predicted S1P sites, and the lumenal region, were likely to influence cleavage. To approach this, we constructed chimeric proteins consisting of the N-terminal region of CREB4 fused to the C-terminal region of Luman to examine whether cleavage susceptibility could be transferred to CREB4. Two chimera were constructed: CH2 in which the Luman TM domain and lumenal region were transferred to CREB4 and CH3 in which only the lumenal domain, encompassing the S1P site, was transferred. These constructs including the TM domains and the S1P sites (x) are summarized in Figure 7, top. Each of the constructs was transfected as described above, and the cells were treated with brefeldin A or mock treated. The results were clear and consistent. In contrast to the parental CREB4, both of the chimeric proteins were susceptible to brefeldin A-induced cleavage, resulting in the appearance of a specific N-terminal cleavage product (Figure 7, bottom, cf. lanes 2, 6, and 8). As expected, the N-terminal product of the CREB4/Luman chimera migrated with a slower mobility than the product of Luman (Figure 7, cf. lanes 4, 6, and 8) due to the larger size of the N terminus of CREB4.
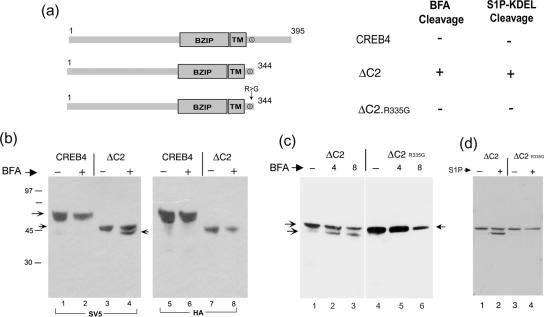
Considering the comparison between CREB4 and CH3, these results indicate that the C-terminal domain of CREB4 seems to determine resistance to brefeldin A-induced cleavage. To examine the possibility that determinants within the C-terminal region of CREB4 somehow conferred a resistance to cleavage at the S1P site, which was otherwise susceptible, we next constructed CREB4.ΔC2, which terminates at residue 344, i.e., six residues downstream of the consensus S1P site, and thus lacks the bulk of the CREB4 lumenal domain (Figure 8). Each of the constructs contained both N-terminal and C-terminal epitope tags to allow for the discrimination of cleavage and the provenance of any products. Cells transfected with the parental or ΔC2 constructs were treated with brefeldin A, and cleavage assessed as described above. In contrast to parental CREB4 (lanes 1 and 2), brefeldin A induced ΔC2 cleavage and the appearance of a single, specific cleavage product, migrating just below full-length ΔC2 (Figure 8b, lanes 3 and 4). To confirm that this product represented cleavage, the samples were probed with the antibody to the C-terminal tag (Figure 8c, lanes 5-8). Although the full-length product was detected with both antibody probes, the smaller brefeldin A-induced product lacked the C-terminal epitope (cf. Figure 8b, lanes 4 and 8). To demonstrate that this cleavage indeed represented cleavage at the specific S1P site, we tested a variant of ΔC2 in which a single amino acid, R335, was mutated to glycine. This residue represents the invariant P4 position of the consensus S1P site (Figures 1 and 4). No cleavage was observed for the version containing the single substitution R335G (Figure 8c, lanes 4-6), providing strong evidence that cleavage was indeed due to S1P. To confirm this, we cotransfected ΔC2 or the mutant variant with the ER-localized SIP-KDEL. The results (Figure 8d) demonstrated that S1P-KDEL induced the cleavage of CREB4ΔC2 and the appearance of the truncated product and that this was dependent upon R335 because substitution abrogated cleavage.
Figure 8.
The lumenal domain of CREB4 blocks S1P cleavage. (a) Summary of the deletion variant (ΔC2) in which the C-terminal region of CREB4 (from residue 345 to the C terminus) was deleted, either with the natural S1P site or with a single substitution R>335G as indicated. (b) Cells were transfected with vectors for CREB4 or ΔC2, treated with brefeldin A (1 μg/ml; 8 h), and processed as described in Figure 7 with antibody (SV5) to detect the N-terminal end of the proteins or with anti-HA to detect the C-terminal end. The BFA-induced product was both smaller than ΔC2, and lacked the C-terminal tag, demonstrating BFA induced cleavage (c). Cells were transfected with vectors for ΔC2 or ΔC2.R335G, treated with brefeldin A (1 μg/ml; 8 h), and processed with antibody (SV5) to detect the N-terminal end of the proteins. (d) Cells were transfected with vectors for ΔC2 or ΔC2.R335G, with or without cotransfection of S1P-KDEL and processed with antibody (SV5) to detect the N-terminal end of the proteins. The single R335G substitution completely abrogated BFA induced cleavage or S1P-KDEL-induced cleavage of ΔC2.
Finally, to further examine resistance to cleavage determined by the C-terminal region of CREB4, we transferred this region to the N terminus of CREB3 (Figure 9a, CH4). Consistent with expectation, this chimera was resistant to both brefeldin A- (Figure 9b, lanes 5 and 6) and S1P (Figure 9c, lanes 13 and 14)-induced cleavage, whereas the parental CREB3 was readily cleaved (lanes 3 and 4 and 10 and 11).
Figure 9.
(a) Summary of the additional CREB4/CREB3 chimeric proteins. CH4 contains the C-terminal region of CREB4 fused to the N-terminal region of CREB3. The fusion junction was before the CREB4 TM domain. CH6 contained the lumenal domain of CREB3 fused to the N-terminal region of CREB4. The fusion junction was just after the S1P site of CREB4. (b) Cells were transfected, treated with brefeldin A (1 μg/ml; 8 h), or (d) contransfected with S1P and processed as described in legends to Figures 7 and 8.
We also examined a chimera (CH6) that is similar to CH2 and CH3 but in which only the C-terminal luminal domain of CREB3 was transferred to CREB4 (Figure 9a). Consistent with the cleavage of CH2 and CH3 (Figure 7), CH6 was also sensitive to both brefeldin A-induced (lanes 7 and 8) and S1P-induced cleavage (lanes 15 and 16).
Together, the results demonstrate first that CREB4 contains a S1P consensus site, second that this site can be cleaved either by brefeldin A redistribution of Golgi enzymes, or by introduction of S1P-KDEL, and third that cleavage is dependent upon a specific residue within the consensus The results provide convincing evidence that the C-terminal domain of CREB4 somehow confers resistance to cleavage by S1P, which can be released either by removal of the region or physiologically by some regulatory signal (see Discussion).
ER Stress-induced Relocalization of CREB4
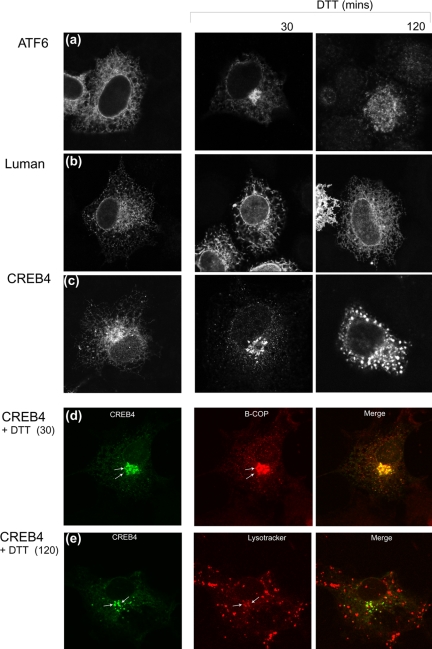
Although we detected a population of CREB4 in the Golgi in the absence of exogenous stress (see Discussion), we further examined whether CREB4 localization was modified by ER stress and whether there was any evidence of an alteration in the ER population. We used DTT, which has been reported to induce a more robust unfolded trafficking response for ATF6 (Haze et al., 1999; Ye et al., 2000; Chen et al., 2002; Shen et al., 2002).
Cells were transfected with expression vectors for CREB4, ATF6, or Luman and examined 30 and 120 min after the addition of DTT. Fields of cells typifying the results are shown in Figure 10. In untreated cells, the localization of each of the proteins was as described above, with both ATF6 (Figure 10a) and Luman (Figure 10b) localized in the ER, whereas CREB4 showed ER localization and a Golgi population (Figure 10c). For ATF6, there was accumulation of the protein in a Golgi localization within 30 min (Figure 10a, ATF6, DTT 30 min), consistent with previous results (Chen et al., 2002). At later times after DTT treatment (120 min), we observed translocation of ATF6 to the nucleus, reflecting cleavage by S1P and S2P within the Golgi (Chen et al., 2002). DTT had no discernible effect on the localization of Luman (Figure 10b), consistent with previous results (Chen et al., 2002; Raggo et al., 2002).
Figure 10.
ER stress induced CREB4 relocalization. Cells were transfected with expression vectors as indicated. At 24 h posttransfection, DTT (5 mM) was added, and incubation was continued for 30 or 120 min. The cells were then fixed in methanol and processed as described in text. In a-c, the location of each of the factors before and after DTT treatment is shown. (d) To confirm colocalization of CREB4 with the Golgi, DTT-treated cells (30 min) expressing CREB4 (green channel) were costained with antibody to β-Cop (red channel). (e) Cells transfected with a GFP-CREB4 fusion protein (green channel) were treated with DTT for 120 min, and live cells were incubated with Lyso-Tracker (red channel) for the last 30 min of incubation. The vesicular population of CREB4 colocalized with the LysoTracker label. Overall, the results indicate that like ATF6, CREB4 localization clearly responded to DTT, which induced a much more restricted Golgi distribution and reduced ER localization.
CREB4 localization responded to DTT treatment. Within 30 min, DTT induced a clear accumulation of CREB4 in a Golgi-like localization with concomitant loss of the ER population (Figure 10c). Golgi associated localization was confirmed as determined by colocalization with the marker β-Cop (Figure 10d). The results indicate that CREB4 localization is regulated in response to ER stress. However, at later times after DTT addition, CREB4 accumulated in a distinct punctate vesicular pattern in the cytoplasm, which could be observed either together with the Golgi localization or exclusively vesicular (Figure 10c, DTT 120 min). Using a GFP-CREB4 fusion protein in conjunction with LysoTracker (BD Biosciences Clontech) labeling of live cells, this punctuate labeling seen later after DTT treatment was consistent with lysosomal compartmentalization of a least a population of CREB4 (Figure 10e). Unlike ATF6, CREB4 was not observed to accumulate in the nucleus.
Together, these results provide strong evidence for the proposal that the localization of CREB4 is responsive to ER stress, which promotes its forward trafficking to the Golgi. However, CREB4 seemed not to be released from the Golgi, accumulating instead in a vesicular compartment. The results indicated that although DTT induced the trafficking of CREB4, the protein may not have been cleaved by the Golgi associated S1P and/or S2P. Consistent with this, we were unable to demonstrate DTT induced cleavage products of CREB4 (our unpublished data).
In conclusion, from consideration of (I) the very striking conservation of bZIP, transmembrane, and S1P motifs, (II) results demonstrating that a truncated CREB4 is nuclear and a potent activator of gene expression, (III) the fact that the protein can be subject to Brefeldin A or S1P cleavage in a site specific manner, and (IV) the altered localization induced by DTT, we believe that CREB4 is an ER associated transcription factor whose localization is regulated in response to ER stress, but that it is also subject to cleavage in a distinct manner.
DISCUSSION
To date, a small set of transmembrane transcription factors, including SREBP1 and 2, and ATF6 have been shown to be subject to RIP, whereby their movement from the ER to the Golgi is regulated by environmental cues (Brown and Goldstein, 1997; Yoshida et al., 1998; Haze et al., 1999; Kaufman, 1999; Li et al., 2000; Wang et al., 2000; Ye et al., 2000; Sakai and Rawson, 2001). Here, we compare localization of the additional proteins Luman, OASIS, CREBH, and CREB4, which contain a conserved central domain related to that in ATF6, and examine further CREB4.
Our analysis of the central region of homology reveals several features worth specific consideration. The general organization comprises five subsections (Figure 1). Although overall, the 150-residue region is very well conserved between all the factors, CREB4, Luman, CREBH, and OASIS seem to make up a more closely related group compared with ATF6. The basic domain and leucine zipper (regions II and III, respectively) are highly related, but these factors contain a region (region I), just adjacent to and N-terminal of the basic domain, not present in ATF6. The conservation of this region might reflect a common feature in the DNA binding specificity and/or potential cofactor interaction in these factors, for example, affecting promoter selectivity or cell type specificity. The next region (region IV) is the putative transmembrane domain and has a completely conserved core including a cysteine residue, C∅∅∅∅∅∅n∅∅∅, where ∅ represents the hydrophobic residues I, L, M, V, or A. Residues flanking this core are again somewhat better conserved in CREB4, OASIS, CREBH, and Luman than in ATF6. Within SREBP, the transmembrane region, which encompasses the S2P cleavage site, conforms very closely to this sequence and also contains a cysteine residue toward the N-terminal end of the domain (Rawson et al., 1997; Nohturfft et al., 1998). Following the transmembrane domain is the fifth element of this conserved core, region V. This element has the completely conserved motif RxL, and, with the exception of OASIS, which seems to have a short insertion, it is always located 35-39 residues from the invariant cysteine noted above in region IV. The conservation of this region is further discussed below in relation to S1P cleavage.
CREB4 Localization
Each of the factors was localized to the ER, but CREB4 showed additional localization to the Golgi. The possibility of CREB4 localization to the Golgi was noted in the original identification of AIbZIP (Qi et al., 2002). It is possible that overexpression of CREB4 itself induced stress in the ER either from the accumulation of a population of CREB4 in an unfolded form (although generally unfolded proteins are retained in the ER) or general induction of an unfolded response, with the result that a population of CREB4 trafficked to the Golgi. ATF6 is actively retained in the ER at least in part by virtue of a stable association with BIP. Less stable interaction with a chaperone, or reduced levels of a specific chaperone, might allow some forward transport of CREB4. Furthermore, certain variants of CREB4 with deletions in the N terminus (our unpublished data) are more efficiently retained in the ER, indicating potential masking of ER retention signals. It is noteworthy that in ATF6 cytoplasmic determinants may influence ER retention and trafficking (Chen et al., 2002).
CREB4 Cleavage by S1P
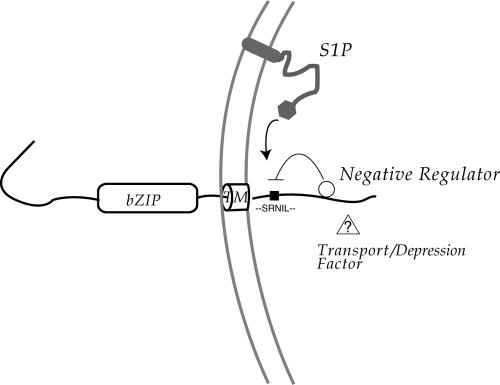
We found little evidence for any constitutive cleavage of CREB4, nor did we observe brefeldin A-induced cleavage, a treatment that has been shown to cause redistribution of S1P from the Golgi and cleavage of ATF6 or Luman (DeBose-Boyd et al., 1999; Raggo et al., 2002). However, we show that the S1P site in CREB4 is indeed subject to cleavage by brefeldin A treatment or coexpression of S1P but that the C-terminal lumenal tail seems to block cleavage. The cleavage was specific and was abolished by a single R-to-G mutation in the conserved S1P site at position 335. These results, together with the striking conservation of the central domain, provide compelling evidence that CREB4 is, at least under certain circumstances, a target for S1P. In Figure 11, we propose a model whereby the CREB4 lumenal domain is bound by a negative regulator of cleavage. This factor could be involved both in transport and cleavage, and as part of the response regulating CREB4, would have to be displaced or modified to relieve repression. It is noteworthy in this regard that cleavage of other substrates, i.e., SREBP and ATF6 is not a default activity when these factors reach the Golgi. Cleavage of SREBP requires copresentation with SCAP. In ATF6, results from analysis of deletion mutants indicates the requirement for C-terminal determinant(s) specifically involved in cleavage in the Golgi and not for transport to the Golgi (Chen et al., 2002; Shen et al., 2002). In this regard, it has been recently shown that ATF6 binds the ER chaperone BiP stably in the ER (Shen et al., 2005) and that active recognition and induced release of BiP is involved in forward transport. However C-terminal deletion variants of ATF6, such as variants 1-475, or 1-550 bind and release BiP and are transported to the Golgi in response to stress (Chen et al., 2002; Shen et al., 2002), but nevertheless they do not seem to be activated, i.e., cleaved by S1P. The basis for this is unclear but indicates that certain other components/cofactors may be important, and in principle these could be selective for different bZip factors. It is therefore readily possible that CREB4 binds BiP or indeed another ER factor that blocks S1P cleavage and/or that CREB4 requires a positive cofactor for cleavage.
Figure 11.
Summary and proposal for CREB4 localization The model as discussed in the text indicates negative regulation of CREB4 cleavage in the Golgi, by association of a negative factor/masking factor with the CREB4 lumenal tail. It could be either that the lumenal tail itself blocks access to the S1P site by an intramolecular interaction or that association of a negative regulator with the tail blocks access or activity of S1P. In a further variant of this proposal, it may be that the CREB4 S1P site is itself directly subject to regulation, for example, phosphorylation at the conserved serine, and this selective modification or context of the site contributes to masking or interaction with the C-terminal determinant.
The CREB4 S1P Cleavage Site
S1P-KDEL cleaved CREB4.ΔC2, and this cleavage was abolished by substitution of R335 within the conserved motif. In SREBP-2, S1P cleaves after the leucine of the sequence RSVL (Duncan et al., 1997), with a strong preference for R at the P4 position, and L/K at the P1 position. Analysis of S1P autocatalytic processing, and cleavage of additional substrates indicates that the S1P consensus may be defined as (R/K)x∅z, where ∅ represents a hydrophobic residue, and z any residue, preferentially L or T, and not V, P, or E (Elagoz et al., 2002). The conserved motif in the CREB4, Luman, CREBH, OASIS family, including ATF6, conforms completely to this latter sequence (R/K)x∅z. We note further, however, that residues adjacent to this motif are also very well conserved, including a serine residue immediately adjacent to R, and a hydrophobic residue at position -3 relative to the R. This serine residue is completely conserved in CREB4, Luman, CREBH, and OASIS (but absent in ATF6), making the overall consensus for these factors ∅xSRx(L/I)x. Thus, the site in this family may be a subset of S1P sites and could be subject to additional regulation, including cell typespecific regulation. For example, it is possible that phosphorylation at this serine residue could affect cleavage efficiency adding an additional dimension to control of processing and activity. We note that because CREB4 was first shown to be expressed in prostate cancer cells, we have attempted (although unsuccessfully) to demonstrate CREB4 cleavage in this cell type in response to ER stress. The possibility of cell type-specific regulation of cleavage will be the subject of further analysis.
Trafficking of CREB4 in Response to ER Stress
DTT induced the efficient accumulation of CREB4 in the Golgi. For ATF6, recent results indicate that the lumenal domain contains elements required both for cleavage at the S1P site and separate motifs for forward transport from the ER to the Golgi (Chen et al., 2002; Shen et al., 2002, 2005). The region involved in forward transport to the Golgi is thought to contain two positively acting motifs (as opposed to a retention signal, which is masked), mapping within residues 467-475 and 475-500 (Chen et al., 2002). More recently, it has been shown that multiple regions, without a recognizable consensus motif, within the lumenal domain of ATF6 bind the ER chaperone BiP. Release from BiP binding combined with the separable positive Golgi localization motifs are together involved in regulating ATF6 forward transport to the Golgi in response to stress (Shen et al., 2002). CREB4, Luman, CREBH, and OASIS have generally shorter regions C terminal of the conserved regions IV and V (i.e., lumenal domains), and we find no obvious homology within this region of ATF6 and CREB4. Future deletion and mutational analysis should help delineate the signal(s) within CREB4 and allow comparisons with those in ATF6.
Whatever the precise signals involved in regulating movement and cleavage, we demonstrate that the N-terminal domain of CREB4 acts as a very potent transactivator. Not only did CREB4 activate a multimerized ATF6 promoter target (and was at least as potent as ATF6 itself) but also significant activation was observed with a native promoter of an ER chaperone, that from BIP/GRP78. Although further work is necessary to compare specific DNA binding activities and profiles of target genes, these results add strong weight to the proposal that CREB4 is an activator of gene expression and additional component in the overall ER stress response.
In conclusion, we present refined analysis of features of a five-section domain conserved in a family of proteins, including CREB4. The conservation within this bZip family, of the putative transmembrane domain and S1P cleavage site, strongly indicates that each of these factors will be subject to transmembrane localization and regulated intramembrane proteolysis by S1P, or a related protease with similar specificity. We show that CREB4 is localized to both the ER and Golgi and that deletion of the transmembrane results in complete nuclear accumulation. Although the significance of the Golgi population remains to be established (i.e., whether this represents a physiologically relevant stress in culture), we provide results that indicate that CREB4 localization is regulated by ER stress and that it is a new substrate for S1P. Future work should help elucidate how the C-terminal domain blocks S1P cleavage and the nature of additional cell type-specific or regulatory features that allow S1P cleavage of CREB4 in the Golgi, and this is integrated together with ATF6 and other components in the unfolded protein response.
Acknowledgments
We are grateful to Joseph Goldstein, Michael Brown, Kazutoshi Mori, and Ron Prywes for generous provision of plasmids. This work is funded by Marie Cure Cancer Care.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-06-0500) on October 19, 2005.
References
- Brown, M. S., and Goldstein, J. L. (1997). The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell, 89, 331-340. [DOI] [PubMed] [Google Scholar]
- Brown, M. S., and Goldstein, J. L. (1999). A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl. Acad. Sci. USA 96, 11041-11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, G., Ni, X., Jiang, M., Ma, Y., Cheng, H., Guo, L., Ji, C., Gu, S., Xie, Y., and Mao, Y. (2002). Molecular cloning and characterization of a novel human cAMP response element-binding (CREB) gene (CREB4). J. Hum. Genet. 47, 373-376. [DOI] [PubMed] [Google Scholar]
- Chen, C., and Okayama, H. (1987). High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7, 2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Shen, J., and Prywes, R. (2002). The luminal domain of ATF6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the Golgi. J. Biol. Chem. 277, 13045-13052. [DOI] [PubMed] [Google Scholar]
- DeBose-Boyd, R.A., Brown, M. S., Li, W. P., Nohturfft, A., Goldstein, J. L., and Espenshade, P. J. (1999). Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell 99, 703-712. [DOI] [PubMed] [Google Scholar]
- Duncan, E. A., Brown, M. S., Goldstein, J. L., and Sakai, J. (1997). Cleavage site for sterol-regulated protease localized to a Leu-Ser bond in the lumenal loop of sterol regulatory element-binding protein-2. J. Biol. Chem. 272, 12778-12785. [DOI] [PubMed] [Google Scholar]
- Elagoz, A., Benjannet, S., Mammarbassi, A., Wickham, L., and Seidah, N. G. (2002). Biosynthesis and cellular trafficking of the convertase SKI-1/S1P: ectodomain shedding requires SKI-1 activity. J. Biol. Chem. 277, 11265-11275. [DOI] [PubMed] [Google Scholar]
- Espenshade, P. J., Cheng, D., Goldstein, J. L., and Brown, M. S. (1999). Autocatalytic processing of site-1 protease removes propeptide and permits cleavage of sterol regulatory element-binding proteins. J. Biol. Chem. 274, 22795-22804. [DOI] [PubMed] [Google Scholar]
- Haze, K., Yoshida, H., Yanagi, H., Yura, T., and Mori, K. (1999). Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10, 3787-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton, J. D., Goldstein, J. L., and Brown, M. S. (2002). SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Investig. 109, 1125-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, X., Sakai, J., Brown, M. S., and Goldstein, J. L. (1996). Regulated cleavage of sterol regulatory element binding proteins requires sequences on both sides of the endoplasmic reticulum membrane. J. Biol. Chem. 271, 10379-10384. [DOI] [PubMed] [Google Scholar]
- Hua, X., Sakai, J., Ho, Y. K., Goldstein, J. L., and Brown, M. S. (1995). Hairpin orientation of sterol regulatory element-binding protein-2 in cell membranes as determined by protease protection. J. Biol. Chem. 270, 29422-29427. [DOI] [PubMed] [Google Scholar]
- Hua, X., Yokoyama, C., Wu, J., Briggs, M. R., Brown, M. S., Goldstein, J. L., and Wang, X. (1993). SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc. Natl. Acad. Sci. USA 90, 11603-11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman, R. J. (1999). Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13, 1211-1233. [DOI] [PubMed] [Google Scholar]
- Li, M., Baumeister, P., Roy, B., Phan, T., Foti, D., Luo, S., and Lee, A. S. (2000). ATF6 as a transcription activator of the endoplasmic reticulum stress element: thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol. Cell. Biol. 20, 5096-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R., Yang, P., O'Hare, P., and Misra, V. (1997). Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol. Cell. Biol. 17, 5117-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda, M., Korn, B. S., Hammer, R. E., Moon, Y. A., Komuro, R., Horton, J. D., Goldstein, J. L., Brown, M. S., and Shimomura, I. (2001). SREBP cleavage-activating protein (SCAP) is required for increased lipid synthesis in liver induced by cholesterol deprivation and insulin elevation. Genes Dev. 15, 1206-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido, T., Yokoya, S., Mori, T., Hagino, S., Iseki, K., Zhang, Y., Takeuchi, M., Takaki, H., Kikuchi, S., and Wanaka, A. (2001). Expression of the novel transcription factor OASIS, which belongs to the CREB/ATF family, in mouse embryo with special reference to bone development. Histochem. Cell Biol. 116, 141-148. [DOI] [PubMed] [Google Scholar]
- Nohturfft, A., Brown, M. S., and Goldstein, J. L. (1998). Topology of SREBP cleavage-activating protein, a polytopic membrane protein with a sterolsensing domain. J. Biol. Chem. 273, 17243-17250. [DOI] [PubMed] [Google Scholar]
- Omori, Y., Imai, J., Suzuki, Y., Watanabe, S., Tanigami, A., and Sugano, S. (2002). OASIS is a transcriptional activator of CREB/ATF family with a transmembrane domain. Biochem. Biophys. Res. Commun. 293, 470-477. [DOI] [PubMed] [Google Scholar]
- Omori, Y., Imai, J., Watanabe, M., Komatsu, T., Suzuki, Y., Kataoka, K., Watanabe, S., Tanigami, A., and Sugano, S. (2001). CREB-H: a novel mammalian transcription factor belonging to the CREB/ATF family and functioning via the box-B element with a liver-specific expression. Nucleic Acids Res. 29, 2154-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, H., Fillion, C., Labrie, Y., Grenier, J., Fournier, A., Berger, L., El-Alfy, M., and Labrie, C. (2002). AIbZIP, a novel bZIP gene located on chromosome 1q21.3 that is highly expressed in prostate tumors and of which the expression is up-regulated by androgens in LNCaP human prostate cancer cells. Cancer Res. 62, 721-733. [PubMed] [Google Scholar]
- Raggo, C., Rapin, N., Stirling, J., Gobeil, P., Smith-Windsor, E., O'Hare, P., and Misra, V. (2002). Luman, the cellular counterpart of herpes simplex virus VP16, is processed by regulated intramembrane proteolysis. Mol. Cell. Biol. 22, 5639-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson, R. B., Zelenski, N. G., Nijhawan, D., Ye, J., Sakai, J., Hasan, M. T., Chang, T. Y., Brown, M. S., and Goldstein, J. L. (1997). Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol. Cell 1, 47-57. [DOI] [PubMed] [Google Scholar]
- Sadowski, I., Bell, B., Broad, P., and Hollis, M. (1992). GAL4 fusion vectors for expression in yeast or mammalian cells. Gene 118, 137-141. [DOI] [PubMed] [Google Scholar]
- Sakai, J., Duncan, E. A., Rawson, R. B., Hua, X., Brown, M. S., and Goldstein, J. L. (1996). Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell 85, 1037-1046. [DOI] [PubMed] [Google Scholar]
- Sakai, J., Nohturfft, A., Goldstein, J. L., and Brown, M. S. (1998a). Cleavage of sterol regulatory element-binding proteins (SREBPs) at site-1 requires interaction with SREBP cleavage-activating protein. Evidence from in vivo competition studies. J. Biol. Chem. 273, 5785-5793. [DOI] [PubMed] [Google Scholar]
- Sakai, J., and Rawson, R. B. (2001). The sterol regulatory element-binding protein pathway: control of lipid homeostasis through regulated intracellular transport. Curr. Opin. Lipidol. 12, 261-266. [DOI] [PubMed] [Google Scholar]
- Sakai, J., Rawson, R. B., Espenshade, P. J., Cheng, D., Seegmiller, A. C., Goldstein, J. L., and Brown, M. S. (1998b). Molecular identification of the sterol-regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol. Cell 2, 505-514. [DOI] [PubMed] [Google Scholar]
- Sato, R., Yang, J., Wang, X., Evans, M. J., Ho, Y. K., Goldstein, J. L., and Brown, M. S. (1994). Assignment of the membrane attachment, DNA binding, and transcriptional activation domains of sterol regulatory element-binding protein-1 (SREBP-1). J. Biol. Chem. 269, 17267-17273. [PubMed] [Google Scholar]
- Seidah, N. G., et al. (1999). Mammalian subtilisin/kexin isozyme SKI-1, a widely expressed proprotein convertase with a unique cleavage specificity and cellular localization. Proc. Natl. Acad. Sci. USA, 96, 1321-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, J., Chen, X., Hendershot, L., and Prywes, R. (2002). ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 3, 99-111. [DOI] [PubMed] [Google Scholar]
- Shen, J., Snapp, E. L., Lippincott-Schwartz, J., and Prywes, R. (2005). Stable binding of ATF6 to BiP in the endoplasmic reticulum stress response. Mol. Cell Biol. 25, 921-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toure, B. B., Munzer, J. S., Basak, A., Benjannet, S., Rochemont, J., Lazure, C., Chretien, M., and Seidah, N. G. (2000). Biosynthesis and enzymatic characterization of human SKI-1/S1P and the processing of its inhibitory prosegment. J. Biol. Chem. 275, 2349-2358. [DOI] [PubMed] [Google Scholar]
- Vinson, C. R., Sigler, P. B., and McKnight, S. L. (1989). Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science 246, 911-916. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Shen, J., Arenzana, N., Tirasophon, W., Kaufman, R. J., and Prywes, R. (2000). Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J. Biol. Chem. 275, 27013-27020. [DOI] [PubMed] [Google Scholar]
- Yang, T., Espenshade, P. J., Wright, M. E., Yabe, D., Gong, Y., Aebersold, R., Goldstein, J. L., and Brown, M. S. (2002). Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 110, 489-500. [DOI] [PubMed] [Google Scholar]
- Yang, T., Goldstein, J. L., and Brown, M. S. (2000). Overexpression of membrane domain of SCAP prevents sterols from inhibiting SCAP.SREBP exit from endoplasmic reticulum. J. Biol. Chem. 275, 29881-29886. [DOI] [PubMed] [Google Scholar]
- Ye, J., Rawson, R. B., Komuro, R., Chen, X., Dave, U. P., Prywes, R., Brown, M. S., and Goldstein, J. L. (2000). ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6, 1355-1364. [DOI] [PubMed] [Google Scholar]
- Yokoyama, C., Wang, X., Briggs, M. R., Admon, A., Wu, J., Hua, X., Goldstein, J. L., and Brown, M. S. (1993). SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell 75, 187-197. [PubMed] [Google Scholar]
- Yoshida, H., Haze, K., Yanagi, H., Yura, T., and Mori, K. (1998). Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J. Biol. Chem. 273, 33741-33749. [DOI] [PubMed] [Google Scholar]
- Yoshida, H., Okada, T., Haze, K., Yanagi, H., Yura, T., Negishi, M., and Mori, K. (2000). ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 20, 6755-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]