Abstract
The trans-Golgi matrix consists of a group of proteins dynamically associated with the trans-Golgi and thought to be involved in anterograde and retrograde Golgi traffic, as well as interactions with the cytoskeleton and maintenance of the Golgi structure. GMx33 is localized to the cytoplasmic face of the trans-Golgi and is also present in a large cytoplasmic pool. Here we demonstrate that GMx33 is dynamically associated with the trans-Golgi matrix, associating and dissociating with the Golgi in seconds. GMx33 can be locked onto the trans-Golgi matrix by GTPγS, indicating that its association is regulated in a GTP-dependent manner like several other Golgi matrix proteins. Using live-cell imaging we show that GMx33 exits the Golgi associated with tubules and within these tubules GMx33 segregates from transmembrane proteins followed by fragmentation of the tubules into smaller tubules and vesicles. Within vesicles produced by an in vitro budding reaction, GMx33 remains segregated in a matrixlike tail region that sometimes contains Golgin-245. This trans-matrix often links a few vesicles together. Together these data suggest that GMx33 is a member of the trans-Golgi matrix and offer clues regarding the role of the trans-Golgi matrix in sorting and exit from the Golgi.
INTRODUCTION
The Golgi matrix was described biochemically in 1994 as a detergent-insoluble network (Slusarewicz et al., 1994), which was hypothesized to function in maintenance of Golgi structure and to promote the formation of new stacked cisternae after mitosis (Barr et al., 1997; Shorter and Warren, 1999; Shorter et al., 1999; Seemann et al., 2000). A “matrixlike material” between and around Golgi cisternae was observed in all electron microscopy (EM) studies and was emphasized by early microscopists (Mollenhauer, 1965; Amos and Grimstone, 1968). In the intervening years many Golgi-matrix proteins have been identified and localized to the cis-, medial- and trans-regions of the Golgi ribbon (Barr and Short, 2003; Gillingham and Munro, 2003). Many Golgi matrix proteins have no transmembrane domain and associate with the cytosolic face of Golgi membranes, whereas a few are transmembrane proteins. The unifying structural feature of Golgi matrix proteins is their large coiled-coil domains and primarily based on this feature it has been suggested that most Golgi matrix proteins be included in the Golgin family, proteins originally identified as Golgi-localized autoantigens (Fritzler et al., 1984; Barr and Short, 2003; Nozawa et al., 2005).
It is becoming increasingly apparent that many Golgi matrix proteins are dynamic, associating and dissociating with membranes in seconds (Ward et al., 2001; Brandon et al., 2003) and are more diverse in function than first predicted. In addition to their role in cisternal stacking, the Golgi matrix has been implicated in vesicle tethering and ER to Golgi transport (reviewed in Lupashin and Sztul, 2005), as well as exit from the trans-Golgi and regulation of endosome-to-Golgi transport (Brown et al., 2001; Yoshino et al., 2003; Kakinuma et al., 2004; Lu et al., 2004; Yoshino et al., 2005). One cis-Golgi matrix protein, GM130, has been described as a docking site for signaling molecules (Preisinger et al., 2004) and another, p115, may play a role in initiating Golgi fragmentation during apoptosis (Chiu et al., 2002). Recently, we have shown that several cis- and medial Golgi matrix proteins move associated with tubules to ER exit sites, adding to the evidence that these proteins interact with membrane structures other than stacked cisternae (Mardones et al., 2005). An even broader functional heterogeneity for Golgi matrix proteins is anticipated as more data become available and suggests our current definition of the Golgi matrix might need to be broadened.
The trans-Golgi matrix proteins and their functions are less well defined than those of the cis-medial Golgi. Currently, the trans-Golgi matrix consists of GRIP family proteins, p230/Golgin-245, Golgin97, GCC88, and GCC185, and Bicaudal-D 1 and 2 (Kjer-Nielsen et al., 1999; Munro and Nichols, 1999; Matanis et al., 2002; Luke et al., 2003). Bicaudal D 1 and 2 have been shown to interact with the dynein-dynactin complex and participate in COPI-independent retrograde traffic (Hoogenraad et al., 2001; Matanis et al., 2002; Hoogenraad et al., 2003). The GRIP-containing proteins, on the other hand, have been implicated in exit from the Golgi as well as retrograde traffic from endosomes to the Golgi (Gleeson et al., 1996; Brown et al., 2001; Kakinuma et al., 2004; Lu et al., 2004; Yoshino et al., 2005). Each GRIP protein is thought to have distinct properties (Derby et al., 2004). For all known GRIP proteins, the GRIP domain alone is sufficient for Golgi targeting (Kjer-Nielsen et al., 1999; Munro and Nichols, 1999; McConville et al., 2002). However, GRIP protein recruitment may be regulated in different ways (Derby et al., 2004). In the best characterized interaction leading to Golgi targeting, the GRIP domain interacts directly with Arl-1 in a GTP-regulated and brefeldin A (BFA)-sensitive manner and Arl-1 targets the complex to the Golgi (Lu and Hong, 2003; Panic et al., 2003a, 2003b; Wu et al., 2004). One GRIP domain-containing protein, Golgin-245 (p230, tGolgin-1), has been found associated with vesicles budding from the trans-Golgi and implicated in exit from the Golgi and transport to the Golgi from endosomes, perhaps through its interactions with MACF, a large molecule thought to bridge microtubules and actin (Erlich et al., 1996; Gleeson et al., 1996; Kakinuma et al., 2004; Yoshino et al., 2005).
GMx33 was identified through a proteomic analysis of detergent-insoluble proteins in an isolated Golgi fraction (Wu et al., 2000). GMx33 is conserved from yeast to mammals and there are two known forms, α and β, which are very homologous to each other and similarly localized. GMx33 is highly posttranslationally modified, peripherally associated with the trans-Golgi and present in a cytoplasmic pool. The human orthologues of GMx33α and β (GPP34 and GPP34R, respectively) were simultaneously identified through a separate proteomic analysis and described as a Golgi-localized coatlike protein (Bell et al., 2001). Unlike most Golgi matrix proteins, GMx33 is relatively small (∼33 kDa) and lacks an extensive coiled-coil structure, although it is predicted to have three coiled domains. Almost nothing is known about the function of GMx33. Its yeast homologue, Vps74p, has been implicated in an interaction with Vps26p, a member of the yeast retromer complex (Bonangelino et al., 2002). Deletion of vps74 results in a modest mis-sorting of the vacuolar protein carboxypeptidase Y.
Further characterization suggests that GMx33α is the predominant form expressed in HeLa and NRK cells. Fluorescence recovery after photobleaching (FRAP) data show that GMx33α is highly dynamic, rapidly moving between membrane and cytosolic pools. GMx33α is associated with tubules exiting the trans-Golgi and appears to sort into distinct domains along the length of these tubules before the fission of the tubules from the Golgi. Finally, biochemistry and immunoelectron microscopy reveals that GMx33 localizes to matrixlike structures associated with and between budded vesicles and tubulovesicular structures. Our data suggest that GMx33 is a part of the trans-Golgi matrix despite its unique properties.
MATERIALS AND METHODS
Cell Lines and DNA Constructs
NRK cells were used for all transfection and immunofluorescence experiments described. A GFP-GMx33α construct was described previously (Wu et al., 2000). GFP-GRASP55 and GFP-GRASP65 (Barr et al., 1998; Shorter et al., 1999) were kindly provided by Francis Barr (Max-Planck Institute of Biochemistry, Martinsreid, Germany).
Cell Culture and Transfection
NRK cells were grown in DMEM supplemented with 10% fetal calf serum (FCS; Hyclone, Logan, UT), penicillin and streptomycin (GIBCO-Invitrogen, Grand Island, NY). BFA (5 μg/ml final) or ammonium chloride (50 mM final) were added directly to the culture media and incubated for the indicated lengths of time. For inhibition of endocytosis, cells were either incubated for 2 h in complete media supplemented with glacial acetic acid to a final pH of 5 (∼40 mM glacial acetic acid from Fisher Scientific (Pittsburgh, PA; Cosson et al., 1989) or were potassium depleted essentially as described (Larkin et al., 1983; Salazar and Gonzalez, 2002). Briefly, cells were rinsed two times in buffer A (20 mM HEPES, pH 7.4, 140 mM NaCl, 1 mM CaCl2, 1 mM MgCl2) and then incubated in hypotonic media (buffer A diluted 1:1 in water) for 5 min. Cells were then rinsed once more in buffer A and then incubated in buffer A for 2 h before imaging.
Transient and stable transfections were performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Transiently transfected cells were typically visualized 6-8 h after transfection. Stable cell lines were produced by selection for Geneticin resistance at 500 μg/ml (GIBCO-Invitrogen) and were maintained in Geneticin throughout the culture period.
RNA Interference
SMARTpools of small interference RNA (siRNA) specific for GMx33α, GMx33β, and lamin were purchased from Dharmacon (Lafayette, CO) and suspended at 2 μM according to the manufacturer's instructions. NRK cells were grown to confluency in a 24-well plate and transfected with 100 nM of each siRNA using DharmaFECT 1 according to the manufacturer's instructions (Dharmacon) and OptiMEM (GIBCO-Invitrogen). Cells were incubated in a total transfection volume of 200 μl for 2 h before complete media was added. After 24 h, the cells were split 1:4 onto coverslips in a new 24-well plate and cultured for another 72 h before fixation and preparation for immunofluorescence as described below.
Immunofluorescence
Immunofluorescence microscopy was carried out using cells grown on coverslips overnight and fixed by a 3-min incubation in 100% methanol at room temperature. Fixed cells were rinsed in phosphate-buffered saline (PBS) and blocked for 30 min at room temperature or overnight at 4°C with a solution of 0.2% BSA and 0.1% gelatin in PBS. The following monoclonal antibodies were used to localize specific molecules: 2F7 (anti-TGN38; Horn and Banting, 1994), GM10 (anti-rat LAMP-I; Reaves et al., 1996) kindly provided by John Hutton (University of Colorado), and H68.4 (anti-transferrin receptor, Zymed, South San Francisco, CA). Polyclonal human autoimmune sera that recognizes Golgin-245 (G4 anti-p230, courtesy of Alfonso Gonzalez) and polyclonal rabbit anti-GMx33 antisera have been described elsewhere (Kreitzer et al., 2000; Wu et al., 2000). Primary antibodies were detected with either anti-mouse or anti-rabbit secondary antibodies conjugated to either Alexa Fluor 488 or Alexa Fluor 594 (Invitrogen, Carlsbad, CA). Primary and secondary incubations were carried out at 37°C for 20-30 min. Cells were washed in 100-200 ml PBS between and after antibody incubations. During the secondary staining, nuclei were stained with 0.3 μM DAPI (Invitrogen). Coverslips were immersed (cells down) in Fluoromount G (Electron Microscopy Systems, Washington, PA) on glass slides and baked for 15 min at 65°C. Images were obtained with an inverted Zeiss Axiovert 200M deconvolution microscope (Thornwood, NY) with a 63× 1.4 NA Zeiss PlanApochromat oil immersion objective and processed using Slidebook software (Intelligent Imaging Innovations, Denver, CO).
Time-Lapse Imaging
Live cells were held at 37°C using a microscope stage heater (Warner Instruments, Hamden, CT). One of three microscopes was used for imaging. Time-lapse images of tubules were captured using an inverted Olympus IX81 spinning disk microscope and a 60× 1.4 NA Olympus oil immersion objective (Olympus America, Melville, NY) with an attached Hamamatsu ORCA IIER monochromatic CCD camera (Hamamatsu, Bridgewater, NJ). Cell surface images were captured using an inverted Olympus IX81 Total Internal Reflection Fluorescence Microscope and a 60× 1.45 NA Olympus TIRFM objective (Olympus America). Photobleaching was performed on a Zeiss LSM 510 NLO laser scanning confocal microscope using a PlanApochromat 63× NA 1.4 oil immersion objective (Carl Zeiss International).
Images acquired on Olympus microscopes were acquired and processed using Slidebook software as above. Quicktime movies were produced directly from slidebook software and single frames were processed with Adobe Photoshop 7 (Adobe Systems, Mountain View, CA). Images acquired on the Zeiss 510 were acquired using Zeiss LSM Image Examiner and processed with the same software or NIH ImageJ software (Rasband, 1997-2005).
Isolation of Membrane and Cytosolic Fractions
Rat liver stacked Golgi and cytosolic fractions were prepared as described (Taylor et al., 1997) from control animals; i.e., rats were not treated with cyclohexamide. For analysis of experiments in NRK cells, whole cell lysates from control and GFP-GMx33α transfected cells were prepared from duplicate wells of a six-well plate. Cells were lysed in 1 ml cold lysis buffer (0.1 M KH2PO4, 0.1 M K2HPO4, 5 mM MgCl2, 1% TX-100, pH 7.2) containing a cocktail of protease inhibitors including: 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1 μg/ml each of chymotrypsin, leupeptin, antipain, and pepstatin. The lysed cells were mixed and then centrifuged at 16,100 × g for 10 min at 4°C and the supernatant was collected. For isolation of membranes and cytosol from the same cell lines, cells from four wells of a six-well plate were scraped with a rubber policeman in 1 ml of homogenization buffer (0.1 M KH2PO4, 0.1 M K2HPO4, 5 mM MgCl2, 0.25 M sucrose, pH 7.2) and protease inhibitors as above. The scraped cells were passed 15 times through a precooled ball bearing cell cracker and the homogenate was spun at 100,000 × g for 15 min at 4°C. The supernatant (cytosol) and pellet (membranes) were collected. For direct analysis, membranes were solubilized in 1 ml of lysis buffer and spun at 16,100 × g for 10 min at 4°C to separate solubilized membrane proteins (supernatant) from insoluble proteins (pellet).
Cytosolic Association and Membrane Budding Assays
Rat liver Golgi fraction and cytosol were maintained on ice or at 4°C throughout the manipulations except where indicated. Golgi fractions were pelleted and resuspended in the appropriate buffer, typically cytosol buffer (20 mM HEPES, pH 7.4, 50 mM potassium acetate, 2 mM EDTA) containing 1 mM PMSF. For high pH stripping, Golgi fractions were suspended in 200 mM sodium carbonate and incubated on ice for 20 min before being washed in cytosol buffer. For salt stripping, an equal volume of 2 M KCl or 4 M NaCl (final of 1 and 2 M, respectively) was added to the resuspended Golgi fraction, which were then incubated for 1 h on ice before being pelleted at 16,100 × g for 20 min and washed in cytosol buffer. To test the association of cytosolic GMx33 with the Golgi fraction, salt-stripped Golgi fractions were resuspended in cytosol buffer containing 1 mM PMSF at 300 μg/ml (15 μg of membrane protein was used for each experimental group in 50 μl of buffer). Rat liver cytosol was precleared by ultracentrifugation (100,000 × g for 20 min in a Beckman TL-100 ultracentrifuge using a TLA100.3 fixed angle rotor; Beckman Coulter, Fullerton, CA). Typically, 50 μl (800 μg) of precleared cytosol was added to each experimental tube. For the indicated samples, GTPγS was added at a final concentration of 100 μM (from 100× stock) and ATP was added to a final concentration of 1 mM (from 100× stock). Whenever ATP was added to a sample, an ATP-regenerating system was also added to a final concentration of 8 mM creatine phosphate and 43 μg/ml creatine kinase (from 25× stock), except when ATP was specifically depleted. For ATP depletion, ATP was added as above and a depleting system of glucose (5 mM final concentration) and hexokinase (200 μg/ml final concentration) replaced the regenerating system. Samples were brought to a final volume of 120 μl per tube using cytosol buffer and were typically incubated for 30 min at 37°C in an Eppendorf Thermomixer R (Eppendorf, Westbury, NY). After incubation, samples were briefly cooled on ice and spun at 4°C at 16,100 × g for 20 min through a 0.5 M sucrose cushion to pellet the Golgi fractions and separate them from cytosol. Pelleted fractions were washed 1× in cytosol buffer and frozen until gel electrophoresis was performed. Typically one half of each sample (7.5 μg of starting membrane) was run on 10% SDS-polyacrylamide gels.Golgi budding assays were performed essentially as described (de Almeida et al., 1993; Heimann et al., 1999). Briefly, 75 μg of rat liver Golgi fraction was suspended in HKM buffer (25 mM HEPES, pH 7.4, 20 mM KCl, 2.5 mM magnesium acetate) and samples were maintained in 1 mM PMSF throughout the assay. Buffer alone, ATP, and the ATP regenerating system with or without GTPγS were added to 4.5 mg of precleared cytosol in separate tubes and maintained on ice until being added to the Golgi fractions. For GTPγS treatment, the Golgi fractions were incubated at 37°C with 100 μM GTPγS for 5 min before the addition of cytosol. Control and ATP-treated Golgi fractions were incubated at 37°C for the same length of time, but in the absence of any nucleotides. Cytosol mixtures containing nucleotides were then added to each sample and the mixture was incubated at 37°C for 30 min as above. All final concentrations were the same as above. After 30 min, samples were cooled on ice for 10 min and remnant Golgi was separated from the budded fraction and cytosol by a low-speed spin of 16,100 × g for 20 min at 4°C. The supernatants were then layered on top of a discontinuous sucrose gradient consisting of 2 M sucrose and 20% sucrose (diluted in HKM) and the samples were spun at 100,000 × g for 90 min in a TLS55, swinging bucket rotor. The budded fractions were recovered at the interface of the 2 M and 20% sucrose layers. Typically one half of the budded fraction and one fifth of the remnant Golgi was resolved using 10% SDS-PAGE gels. For detergent and salt extractions of budded fractions or remnant Golgi fractions, samples derived as above were incubated in HKM buffer containing: 1% Triton X-100 (Tx100) alone, 1% Tx100 + 150 mM NaCl, or 150 mM NaCl alone for 30 min on ice. The unextracted material was then pelleted (100,000 × g for the budded fraction, 16,100 × g for the remnant Golgi). All of the budded fraction and one fifth of the remnant Golgi was used to resolve proteins on SDS-PAGE gels. For immunoblotting, proteins were transferred to nitrocellulose and blocked in 5% defatted milk either overnight at 4°C or for 1 h at room temperature. Monoclonal antibodies M3A5 (anti-β-COP; Allan and Kreis, 1986) and 166 (anti-pIgAR; Kuhn and Kraehenbuhl, 1983) and polyclonal antibodies against p115 (kindly provided by Elizabeth Sztul, University of Alabama, Birmingham AL) and p24 (kindly provided by John Bergeron, McGill University, Montreal, Quebec, Canada) were used to probe membranes after dilution in 1% milk. Primary antibodies were incubated for 1 h at room temperature or overnight at 4°C. Antibodies were detected by enhanced chemiluminescence (ECL) using HRP-conjugated anti-rabbit or anti-mouse antibodies (both from Rockland, Gilbertsville, PA). ECL signal was detected with ECL Western Blotting detection reagents and Hyperfilm ECL (both from GE Health Care, Little Chalfont, Buckinghamshire, United Kingdom). Densitometry of these signals was performed using NIH Image J software (Rasband, 1997-2005). Alternatively, the Typhoon 9400 Variable Mode Imager was used to detect fluorescent signal from anti-rabbit Alexa Fluor 633 or anti-mouse Alexa Fluor 488.
Electron Microscopy and Immunogold Labeling
The budded fractions from 150 μg of rat liver Golgi were prepared as above and suspended in 50 μl of HKM buffer. Ten microliters of sample was placed onto the coated side of a formvar-coated/carbon-coated/glow-discharged 100-mesh copper-rhodium grid and allowed to settle (without drying) for ∼10 min. After settling, paraformaldehyde (Electron Microscopy Sciences, Washington, PA) was diluted in HKM buffer to 4% and 10 μl was placed on each grid to fix the samples. Grids were rinsed three times for 5 min each by inversion over drops of PBS and samples were blocked with 10% FCS (Hyclone) in PBS for 30 min at room temperature. Antibodies for labeling were diluted in 5% FCS in PBS. The primary antibodies (anti-GMx33 and human anti-Golgin-245) and gold-labeled secondary antibodies were incubated sequentially on the grids for 1.5 h at room temperature. After each incubation, grids were rinsed four times for 10 min each by inversion onto drops of PBS. After the final wash, grids were rinsed twice with ddH2O and negativestained with a solution of 1% uranyl acetate and 1% methylcellulose for 30 min. The grids were air-dried and then exposed to vapors from 2% OsO4 for 10 min. Samples were observed with a Tecnai-12 electron microscope (FEI, Hillsboro, OR) running at 80 keV. Images recorded digitally with a Gatan 2k × 2k CCD camera using DigitalMicrograph software (Gatan, Pleasanton, CA). For tomographic imaging and 3D reconstruction, samples were prepared on grids as above. The grid was placed in a Model 925 Double-Tilt Rotation holder (Gatan) and viewed with an FEI Tecnai TF-20 FEG electron microscope operating at 200 keV. The sample was tilted ±60° and images were recorded at 1° intervals with a pixel size corresponding to 0.45 nm. The grid was then rotated and a similar tilt-series acquired about the orthogonal axis. Tilt-series were acquired automatically using Serial-EM software. Tomograms of individual labeled vesicles were calculated and analyzed with the IMOD software package on a Macintosh G4 computer.
RESULTS
GFP-GMx33α Is Phenotypically Indistinguishable from Endogenous GMx33
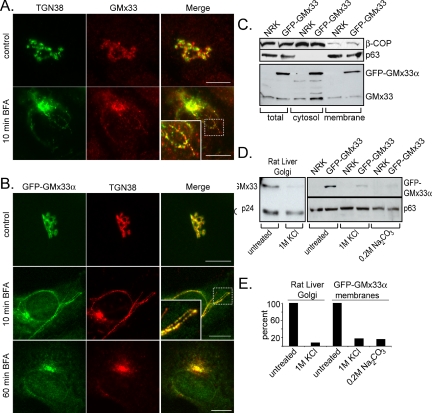
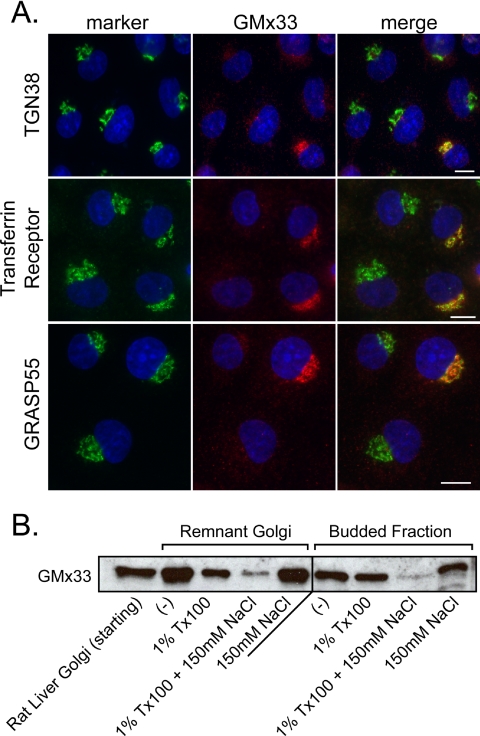
GMx33 is peripherally associated with the trans-Golgi (Wu et al., 2000; Bell et al., 2001). To study GMx33 in more detail, a GFP-GMx33α fusion construct was produced and a stably transfected NRK cell line was established. GFP-GMx33α is morphologically and biochemically indistinguishable from the endogenous protein (Figure 1). In both nontransfected, control NRK cells and stably transfected cells, GMx33 appears to almost colocalize with TGN38, a trans-Golgi marker, but is slightly more peripheral than TGN38 (Figure 1, A and B, top panels). The addition of 5 μg/ml BFA for 10 min results in the appearance of TGN38-positive tubules that are also positive for either endogenous GMx33 (Figure 1A, bottom panels) or GFP-GMx33α (Figure 1B, middle panels). A closer examination of the tubules reveals that GMx33 and TGN38 do not precisely overlap, but rather a portion of each resides in distinct puncta along the length of the tubule, whereas another portion appears to overlap (Figure 1, A and B, insets). After 1 h of BFA treatment, both TGN38 and GFP-GMx33α relocate to the region of the microtubule organizing center, the so-called “BFA dot,” as previously shown for endogenous GMx33 (Figure 1B, bottom panels; Wu et al., 2000). Immunoblot analysis of GMx33 in transfected and nontransfected cells reveals the existence of a large (∼50%) cytosolic pool of both GFP-GMx33α and endogenous GMx33 (Figure 1C). Finally, to demonstrate that GFP-GMx33α was indeed peripherally associated with the Golgi, high salt washes were performed. Isolated rat-liver Golgi fraction and a total membrane fraction from GFP-GMx33α-transfected cells were treated with 1 M KCl, resulting in ∼80-90% of GMx33 being stripped from the membrane of both samples (Figure 1D and quantitation in Figure 1E). Similar results were obtained after treatment of Golgi fractions with 2 M NaCl (see Figure 5B). High pH also stripped GFP-GMx33α as expected (Figure 1, D and E).
Figure 1.
Endogenous GMx33 and GFP-GMx33α are peripherally localized to the TGN. (A) NRK cells were left untreated (top panels) and treated with 5 μg/ml brefeldin A for 10 min (bottom panels), fixed, and stained with antibodies specific for either TGN38 or GMx33. Our GMx33-specific antibody recognizes both α and β forms of GMx33. Primary antibodies were detected with either anti-mouse or anti-rabbit secondary antibodies conjugated to either Alexa Fluor 488 or Alexa Fluor 594. The inset is an enlarged image from the area indicated by the dotted box. (B) NRK cells stably expressing GFP-GMx33α were left untreated (top panel) or treated with brefeldin A for 10 or 60 min (middle and bottom panels). Fixed cells were stained for TGN38. The inset is an enlarged image from the area indicated by the dotted box. (C) NRK cells stably expressing GFP-GMx33α were lysed in detergent (total) or used to prepare membrane and cytosol fractions as described in Materials and Methods. Protein (50 μg per lane) was analyzed by immunoblot with antibodies specific for the indicated proteins. (D) Fifteen micrograms of rat liver Golgi fraction were left untreated or stripped with 1 M KCl for 1 h on ice and washed (left panel). One hundred micrograms of total membrane fractions from wild-type NRK cells or those expressing GFP-GMx33α (right panel) were left untreated or stripped with 1 M KCl as above or a high pH wash, 0.2 M sodium carbonate for 15 min on ice. Half of each sample was subjected to SDS-PAGE and analyzed by immunoblot with antibodies specific for the indicated proteins. p24 and p63 proteins were followed as loading controls. (E) Densitometry was performed on the immunoblots in D using the NIH Image J software. The amount of p24 or p63 signal in each lane was used to normalize slight differences in loading. Bars, 10 μm.
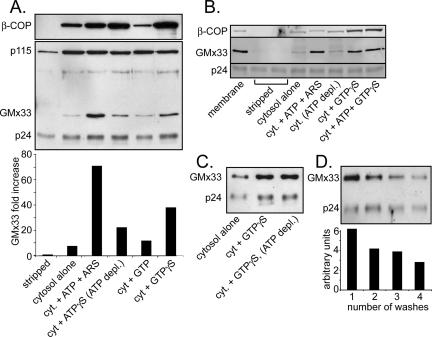
Figure 5.
Cytosolic GMx33 associates with stripped membranes in a GTPor ATP-dependent manner. (A) Rat liver Golgi fraction was stripped with 1 M KCl, split into 15 μg aliquots, and incubated with cytosol (800 μg per aliquot) for 30 min with the indicated treatment as described in Materials and Methods. Half of each sample was analyzed by immunoblot with antibodies against the following proteins: β-COP, p115, GMx33, and p24. For densitometry, the GMx33 signal was standardized to the signal for the Golgi-transmembrane protein, p24, and the data are in the bottom part of the panel. (B) Rat liver Golgi fraction was stripped with 2 M NaCl. Cytosolic proteins were incubated with the stripped membrane in the presence of the indicated nucleotides and samples were analyzed as in A. (C) Rat liver Golgi fraction was stripped as in A and treated as indicated with the indicated nucleotides. (D) Rat liver Golgi fraction was not stripped and was subjected to the indicated number of washes at 4°C. Between each wash the fraction was incubated in cytosol buffer for 10 min at 37°C. The final membrane pellets were immunoblotted with antibodies specific for GMx33 and p24. Quantitation was performed as in A and is provided below the blot.
Cytosolic GMx33α Rapidly Exchanges with Membrane-bound Protein
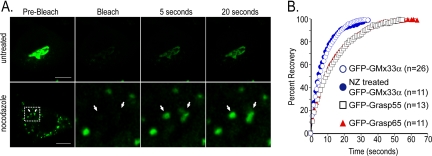
Like many Golgi matrix proteins, there is a large cytosolic pool of GMx33 (∼50% of total protein, Figure 1C). To determine whether this pool exchanges with the membrane-bound pool, the FRAP of GFP-GMx33α transfected cells was measured. As shown in Figure 2, after bleaching the entire Golgi region, GFP-GMx33α fluorescence rapidly returned to the membrane with a t1/2 of ∼5.4 s. Maximal recovery was achieved within 20-30 s. The recovery of GFP-GMx33α fluorescence was not dependent on microtubules because treatment of the cells with 33.3 μM nocodazole for 2 h resulted in no significant change in the recovery half-time on Golgi ministacks (Figure 2, A and B). Correlative experiments in which the cytosol, but not the Golgi, was bleached repeatedly, ultimately led to fluorescence loss at the Golgi, confirming that Golgi-localized GMx33α exchanges rapidly with its cytosolic pool (our unpublished data). As a comparison, NRK cells were transfected with GFP-GRASP55 or GFP-GRASP65, two Golgi matrix proteins with cytosolic pools, one of which, GRASP65, has been previously shown to rapidly exchange between Golgi-bound and cytosolic pools (Ward et al., 2001). The fluorescence recovery of GFP-GRASP65 and GFP-GRASP55 was slightly slower than that of GFP-GMx33α in our hands, with t1/2s of 9.3 and 8.7 s, respectively. As a control, cells were also transfected with GFP-Golgin84, a transmembrane Golgi matrix protein with no cytosolic pool, and bleached as above. As expected, these cells showed little fluorescence recovery after photobleaching, even after 1 h (our unpublished data). It is important to note that the dynamics of the GFP-GMx33α membrane association and dissociation may not precisely mimic those of the endogenous protein, especially because the transfected protein may have to compete with the endogenous protein for Golgi binding. However, these data clearly demonstrate the general phenomenon that GMx33α is extremely dynamic, rapidly cycling between membrane-bound and cytosolic pools.
Figure 2.
GFP-GMx33α rapidly cycles between membrane and cytosolic pools. (A) Cells stably or transiently expressing GFP-GMx33α were left untreated (top panels) or were treated with 33.3 μM nocodazole for 2 h at 37°C (bottom panels). The whole Golgi region or the indicated Golgi-stacklets were bleached with the 488-nm laser set to 100%. Approximately one image was captured each second after bleaching and the fluorescence recovery at 5 and 20 s is shown. The bottom leftmost panel shows the entire cell. The region indicated by the white dotted box is enlarged in the right three panels. (B) The indicated number of cells transfected with GFP-GMx33α, GFP-GRASP55, or GFP-GRASP65 were bleached and the average fluorescence recovery after bleaching was graphed. The actual fluorescence intensity for each cell over time was converted to a percentage of the total recovery. Fluorescence values obtained immediately after bleaching were used as 0%. One hundred percent recovery corresponds to the point for each cell at which overall fluorescence no longer increased over time. Because of the rapid exchange of membrane and cytosolic forms of GFP-GMx33α, the cytosolic pool was bleached as it exchanged on the membrane during the bleaching period. Therefore, the total fluorescence never recovered to the initial prebleach levels, and recovery level was inversely proportional to the time of bleaching. Bars, 10 μm.
GMx33α Traffics to Other Membranes in the Cell
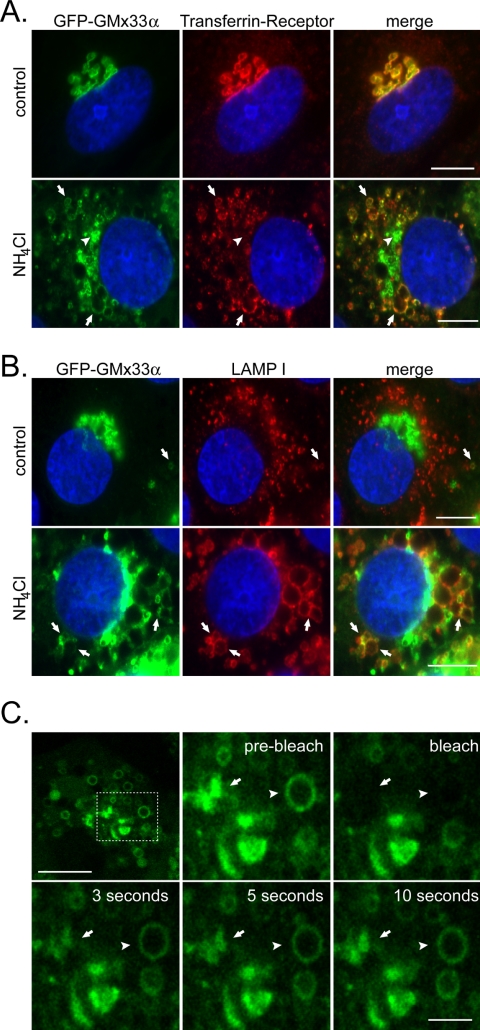
When examining GFP-GMx33α by microscopy, we periodically observed small GFP-GMx33α-positive structures near the periphery of the cell that were clearly separate from the Golgi. In some cases, these structures colocalized with endosomal/lysosomal markers such as LAMPI (Figure 3B, top panels). However, such colocalization was rare in control cells. To determine if GFP-GMx33α could be trapped and accumulated at any other membrane compartment besides the Golgi, cells were first treated with inhibitors of endosomal acidification for 1 h. As shown in Figure 3, A and B, ammonium chloride results in a dramatic expansion of transferrin receptor-positive structures that normally reside very close to the Golgi in NRK cells (Figure 3A, bottom panels) and LAMPI-positive structures that are normally distinct from the Golgi in NRK cells (Figure 3B, bottom panels). The response of GFP-GMx33α to this treatment was somewhat different. Nearly all of the transferrin receptor-positive structures and many, but not all, of the LAMPI-positive structures, even when significantly removed from the peri-nuclear region (Figure 3, A and B, arrows) also contained GFP-GMx33α. These structures frequently stained positive for TGN-38 as well, which has been previously shown to migrate to endosomes after similar treatment (Chapman et al., 1994; Reaves and Banting, 1994; and our unpublished data). However, a portion of GFP-GMx33α also remained in the peri-nuclear region and clearly no longer colocalized with the transferrin receptor (arrowheads in Figure 3A, best seen in the merged image where no transferrin receptor staining can be seen). This portion of GFP-GMx33α always colocalized with other Golgi markers such as GM130, indicating that a portion of GMx33α was retained at the Golgi under these conditions (our unpublished data). A similar phenotype was observed in nontransfected cells and also after treatment with alternative acidification inhibitors monensin and chloroquine (our unpublished data). The fact that the transferrin-receptor-positive and LAMPI-positive structures are clearly distinct from the remnant Golgi indicates that a pool of GFP-GMx33α is being recruited to these peripheral structures and away from the Golgi.
Figure 3.
GMx33 can be “trapped” at endosomal membranes. (A) NRK cells stably transfected with GFP-GMx33α were left untreated or treated with ammonium chloride for 2 h before fixation and staining. Cells were stained with an antibody specific for the transferrin receptor and DAPI. Arrows indicate enlarged endosomal membranes positive for both GFP-GMx33α and the transferrin receptor. Arrowheads indicate GFP-GMx33α that is still associated with the Golgi. Bar, 10 μm. (B) Cells were treated as in A and stained with an antibody specific for LAMP I and DAPI. Arrows indicate LAMP I-positive membranes that are also positive for GFP-GMx33α in both control cells (top panels) and ammonium chloride treated (bottom panels) cells. Bar, 10 μm. (C) NRK cells stably transfected with GFP-GMx33α were treated with ammonium chloride as in A and B. An enlarged endosome (arrowhead) and an edge of the Golgi (arrow) were both bleached as in Figure 2. The dotted box in the leftmost top panel indicates the region enlarged in the other panels. Bar in top leftmost panel, 10 μm. Bar in bottom right panel, 3 μm.
To determine whether GMx33α or its binding partner(s) were trapped in these structures, cells expressing GFP-GMx33α were treated with ammonium chloride as above and analyzed by FRAP. As shown in Figure 3C, after simultaneous photobleaching of both compartments, GFP-GMx33α associated rapidly with enlarged endosomelike structures (arrowhead) and remaining Golgi (arrow). These data demonstrate that GMx33α interacting with two different compartments continued to exchange with the cytosol and implies that GMx33α can follow a binding partner or partners to endosomal compartments.
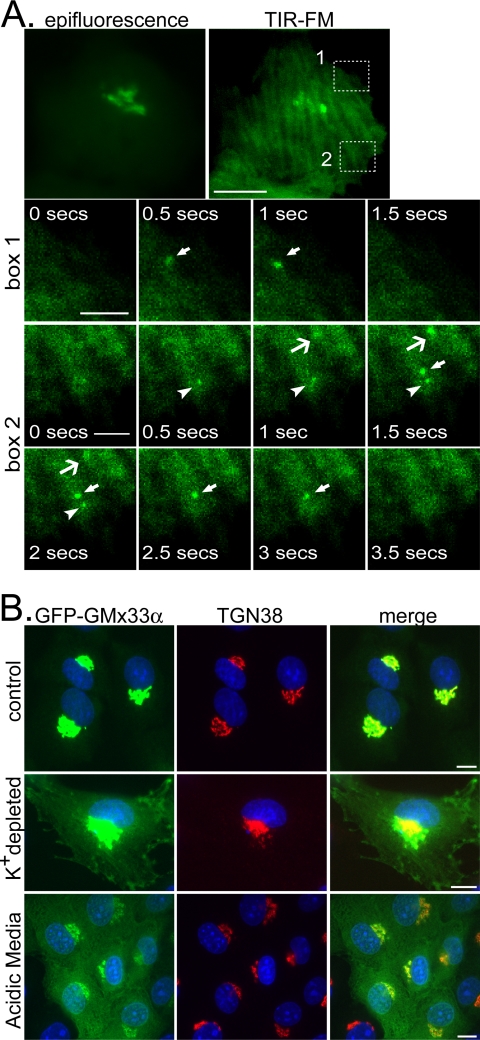
To determine whether GMx33α localized to the plasma membrane as well, total internal reflection fluorescence microscopy (TIR-FM) was used in control cells. Because there is so much GFP-GMx33α in the cytosol, considerable background fluorescence was observed just beneath the plasma membrane by TIR-FM. Nevertheless, as shown in Figure 4A and Supplementary Video 1, GFP-positive vesicles could be observed frequently at the plasma membrane. These vesicles appeared at the plasma membrane and rapidly disappeared, typically within 0.5-1.5 s. It is unclear whether these are exocytic or endocytic events because we were never able to clearly observe the GFP-GMx33α diffusing into the plane of the plasma membrane. However, we often saw the GFP-GMx33α-positive vesicles moving laterally just before disappearing.
Figure 4.
GFP-GMx33α can be found at the plasma membrane by TIR-FM and can be trapped at the plasma membrane by inhibitors of endocytosis. (A) NRK cells stably transfected with GFP-GMx33α were maintained at 37°C and visualized by TIR-FM over time. The epifluorescent image of the entire cell is also shown (top left panel). The bottom three rows show enlarged images of the boxed areas in the top right-hand panel. Images were captured every 0.5 s. Arrows and arrowheads point to GFP-GMx33α-positive events at the plasma membrane. This localization is transient. These events are seen more clearly in Supplementary Video 1. Bar in top panels, 10 μm; bars in bottom panels, 3 μm. (B) NRK cells stably transfected with GFP-GMx33α were left untreated or were cultured in the presence of a K+-depleting buffer or acidic (pH 5) media. Cells were fixed and stained with antibodies specific for TGN38 before imaging. Bars, 10 μm.
To determine whether GFP-GMx33α could be accumulated at the plasma membrane, we used inhibitors of endocytosis. As shown in Figure 4B, potassium depletion and hypotonic shock (middle panels) as well as cytosol acidification (bottom panels), both induced the accumulation of GFP-GMx33α associated with the plasma membrane. Together these data indicate that GMx33α does indeed traffic to the plasma membrane as well as to endosomes.
GMx33 Association with Golgi Can Be Regulated by Either ATP or GTP
To determine the biochemical properties governing GMx33 association with the Golgi membrane, the rat liver Golgi fraction was stripped with high salt (either 1 M KCl or 2 M NaCl) and incubated with precleared cytosol in the presence or absence of various nucleotides. Cytosol alone resulted in binding of a negligible amount of GMx33 to the stripped Golgi membranes. However, the presence of ATP and an ATP-regenerating system resulted in binding of ∼10-fold more GMx33 (Figure 5A), which was negated by the presence of an ATP-depleting system (Figure 5B). A nonhydrolyzable analog of ATP (ATPγS) induced much less GMx33 binding (about threefold). Although GTP had no effect on GMx33 binding (Figure 5A), GTPγS induced an intermediate increase in GMx33 binding (about fivefold, Figure 5A). This effect of GTPγS was reproducible even in the presence of an ATP-depleting system, indicating that the effect of GTPγS was independent of ATP (Figure 5C). A cis-Golgi matrix protein, p115, showed no response to any nucleotide tested (Figure 5A), whereas β-COP was extremely responsive to GTPγS, as originally reported (Donaldson et al., 1991a; Figure 5, A and B).
To determine the amount of cytosol necessary for GMx33 association with stripped rat liver Golgi fraction, cytosol was titrated in the absence or presence of either GTPγS or ATP. At least 400 μg of cytosol was required to detect a significant amount of GMx33 binding over background (Supplementary Figure 1). In comparison, significant β-COP association with stripped Golgi membranes was detectable with ∼100 μg of cytosol in the same assay. Finally, to determine whether GMx33 spontaneously disassociates from membranes, Golgi fractions were suspended in cytosol buffer without salt and incubated for 10-min intervals at 37°C. The Golgi fractions were pelleted at 4°C between each incubation. As shown in Figure 5D, GMx33 dissociated slowly under these conditions. After four washes, ∼50% of the original GMx33 signal remained associated with the Golgi membranes. Because these dissociation assays were performed in the absence of cytosol, we would not necessarily expect the rate of dissociation observed here to be identical to that seen in the in vivo FRAP experiments (Figure 2), especially if a cytosolic component is essential to rapidly remove GMx33 from the membrane. However, given that our antibody recognizes both GMx33α and β, it is also possible that these two isoforms behave with different kinetics, making the dissociation of GMx33 as observed by immunoblot of rat liver fractions, which contain both isoforms, slower than that of GMx33α alone as observed in transfected NRK cells.
GMx33α Exits the Golgi in Tubules
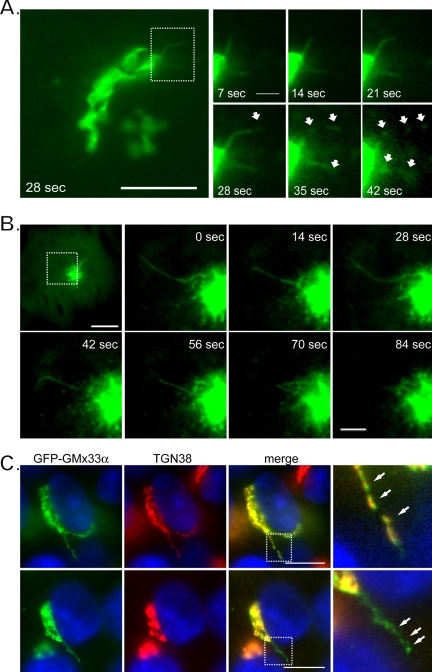
To further observe the dynamic nature of GMx33, live cell imaging was performed. Short tubules emerged from the Golgi in nearly every cell (Figure 6A) and in all cases, after a few seconds GFP-GMx33α segregated into puncta along the length of the tubule. In some instances these puncta separated dramatically, appearing to break off into shorter tubular or vesicular structures before moving throughout the cell (Figure 6A and Supplementary Video 2). The tubules moved relatively rapidly, emerging and separating in less than 1 min. Similar structures also could be observed extending from the “BFA dot” after 1 h of BFA treatment. These data indicate that Arfs, Arls, and coatomer are not required for GMx33 tubule formation, because BFA has been shown to dissociate all three from the Golgi membrane (Figure 6B and Supplementary Video 3).
Figure 6.
GFP-GMx33α exits the Golgi in tubules. (A) NRK cells stably expressing GFP-GMx33α were held at 37°C and visualized by time-lapse imaging every 7 s. Shown is a pair of tubules extending from the Golgi that fragment into short tubules or vesicles between 28 and 42 s. The left panel shows the whole Golgi; the right panels are enlarged images of the region indicated by a dashed box. Bar in leftmost panel, 10 μm; in enlarged images, 2 μm. (B) NRK cells stably expressing GFP-GMx33α were treated with brefeldin A for 1 h and visualized by time-lapse imaging as in A. Shown is a whole cell just before tubule formation and enlarged images of the region outlined by the dotted box in the top leftmost panel. Bar in the leftmost top panel, 10 μm; in the enlarged images, 3μm. (C) NRK cells stably transfected with GFP-GMx33α were fixed and stained with antibodies specific for TGN38. Two cells with tubules extending from the Golgi are shown. Bars, 10 μm.
In fixed cells the GFP-GMx33α-positive tubules were observed and were frequently (Figure 6C, top panels), but not always (bottom panels) positive for TGN38. As in BFAtreated cells (Figure 1, A and B), the GFP-GMx33α and TGN38 appeared to segregate into puncta along the length of the tubules that only partially overlapped (Figure 6A, rightmost panels). Similar results were observed in nontransfected cells. These data suggest that GMx33 exits the Golgi associated with tubules.
GMx33 Associates with Tethers Attached to Budded Vesicles
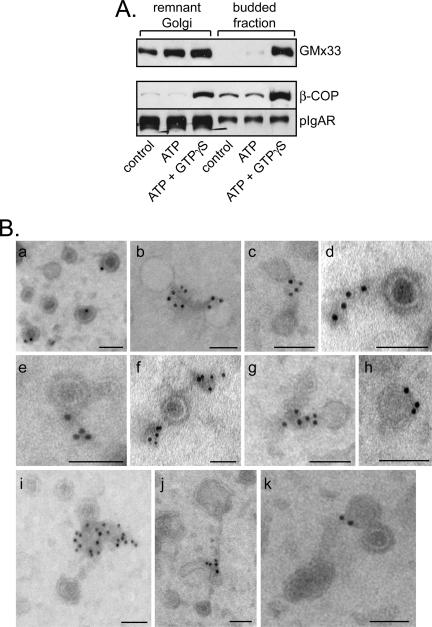
Seeing GMx33-positive tubules leaving the Golgi was surprising, because GMx33 did not move to a budded membrane fraction in our standard budding assay (Wu et al., 2000). However, only ATP, not GTPγS, was used in the original experiments. Because our data indicate that GMx33 can associate with Golgi fraction in the presence of GTPγS (Figure 5), it is possible that, like coatomer (Donaldson et al., 1991a, 1991b), GMx33 dissociates from budded vesicles in a GTP-dependent manner. To test this, a budding assay was performed with rat liver Golgi fraction in the presence or absence of GTPγS. As expected, budded membranes recovered in the presence of cytosol alone, or cytosol with ATP and an ATP-regenerating system had no detectable, or extremely small amounts of GMx33 associated (Figure 7A). Strikingly however, in the presence of both ATP and GTPγS we easily detected GMx33 in the budded fraction. Therefore, GMx33 associates with budding membranes in a GTP-dependent manner and GTPγS stabilizes this interaction.
Figure 7.
GMx33 associates in a GTP-dependent manner with matrixlike structures attached to vesicles in a budded vesicle fraction. (A) Rat liver Golgi fraction (75 μg) was incubated with 4.5 mg cytosol and the indicated nucleotides for 30 min at 37°C and remnant Golgi and budded fractions were separated as described in Materials and Methods. Protein was analyzed by immunoblot with antibodies specific for GMx33, β-COP, and pIgA receptor (a cargo molecule moving to the plasma membrane). (B) Budded fractions were prepared as in A in the presence of ATP and GTPγS, placed on an EM grid, and immunolabeled with antibodies specific for GMx33. Shown are 11 different examples of budded vesicles associated with electron dense (osmophilic) “tails” that are positive for GMx33. Some of these tails appear to connect adjacent vesicles (b, c, g, j, and k). The gold particles lie almost exclusively within the tail material and are distinct from the vesicles' membranes. Bars, 100 nm.
If GMx33 is associating with the budded fraction in the presence of GTPγS, this could explain the fact that GTPγS did not promote as much GMx33 association with stripped membranes as ATP (Figure 5A). This budded fraction was not recovered in the original assay because of the low-speed centrifugation used to pellet the stacked Golgi fraction. To test this, rat liver Golgi membranes were stripped as before and the association of GMx33 from the cytosol was measured over time. Both ATP and GTPγS induced rapid association with stripped Golgi membranes to an approximately equal degree. However, although the presence of ATP caused GMx33 to remain associated with Golgi for the entire 30-min time course, the total amount of GMx33 associated with recovered Golgi steadily decreased in the presence of GTPγS, indicating that the GMx33 that had bound to the Golgi was leaving over time on the exiting tubules or vesicles (Supplementary Figure 2).
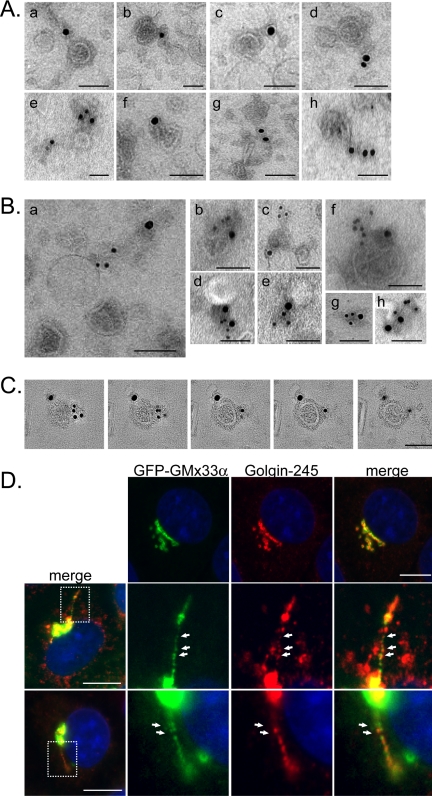
To identify what GMx33 was associating with in these experiments, the budded fraction was examined by EM after immunolabeling for GMx33. In the presence of GTPγS, much more GMx33 associated with the budded fraction (our unpublished data), confirming the immunoblot results. GMx33 was localized to the outside of budded vesicles (Figure 7B), which, by morphology, were either surrounded by a dense, coatlike structure or were uncoated. Strikingly however, the primary structure labeled by the GMx33 specific antibodies was a “matrixlike,” osmophilic structure associated with the vesicles. This matrix was often connecting two or more vesicles and labeled with multiple GMx33 antibody-gold complexes (Figure 7B). In addition, many of these matrixlike structures were present independent of vesicles (Figure 8B, panel g). 3-D tomography revealed no discernable membranes within these matrixlike structures, which appeared to be contiguous with the coats surrounding the vesicle membrane (Supplementary Videos 4 and 5). Morphometric analysis showed that GMx33 almost exclusively (∼97%) associated with these matrixlike structures (Supplementary Figure 3). In rare instances when the gold label looked to be directly associated with a vesicle, there typically was a small matrix-piece that surrounded or attached to the same or another part of the vesicle (Figure 7B, a, h, and k). As an antigen is localized within 15-18 nm of the gold particle (Griffiths, 1993), because of the length of the two IgG molecules, it is difficult to determine where the coat ends and matrixlike structures begin.
Figure 8.
GMx33 associates with the trans- Golgi matrix and colocalizes with p230/Golgin-245 in tubules exiting the Golgi. (A) Budded fractions were immunolabeled on EM grids as in Figure 7 with antibodies specific for p230/Golgin-245. Shown are eight different examples of Golgin-245-positive electron dense (osmophilic) “tails” associated with the budded vesicles. Bars, 100 nm. (B) Budded fractions were prepared as in A and probed with antibodies specific for p230/Golgin-245 (15 nm gold) and GMx33 (10 nm gold). Shown are eight examples of structures, both vesicles and matrix alone (g) that colabeled for both proteins. Bars, 100 nm. (C) A vesicle that colabeled with antibodies for GMx33 and Golgin-245 was analyzed by 3-D tomography. Shown are five tomographic slices (0.45 nm) through the volume of the vesicle. Gold particles corresponding to the two antigens are localized to the electron-dense matrix material surrounding the vesicle and both are distinct from the vesicle membrane itself. Bar, 100 nm. (D) NRK cells stably transfected with GFP-GMx33α were fixed and labeled with antibodies specific for p230/Golgin-245 and fluorophore-conjugated secondary antibodies. The top panels show that there is good, but not perfect colocalization between GMx33α and Golgin-245. The bottom two panels show a tubule extending from different cells. The boxed regions in the leftmost panels depict the areas enlarged in the subsequent panels. Bars, 10 μm.
GMx33 Associates with the Golgi Matrix
The structures that labeled with the GMx33-specific antisera are very similar to those previously described to contain p230/Golgin-245, a GRIP-domain containing Golgi matrix protein (Gleeson et al., 1996). To investigate whether these were the same structures, a budded vesicle fraction was colabeled with GMx33 and Golgin-245-specific antisera and both antibodies labeled nearly identical structures (Figure 8A). The same structures were sometimes double-labeled (Figure 8, B and C) and often the label was on two different matrix-pieces associated with the same vesicle as shown both in negative stain and 3-D tomography (Figure 8B, c and f, and 8C and Supplementary Video 6). These data indicate that GMx33 is associated with the trans-Golgi matrix on budded vesicles.
To verify that these two proteins could be found on tubules exiting the Golgi, fixed cells were stained with p230/Golgin-245-specific antisera. Both GFP-GMx33α and Golgin-245 could be found on the same tubules and both segregated into puncta along the length of the tubule (Figure 8D). Although the two proteins showed some regions of overlap on the tubules, there were also regions of unique localization. Because GMx33 and Golgin-245 sort into these puncta along tubules (Figures 6 and 8) and do not associate with the membranes of budded vesicles, but rather the matrixlike “tails” as observed by EM (Figures 7 and 8) and because GMx33 and TGN38 often appear to be in distinct puncta along naturally occurring tubules (Figure 6C) and those formed by BFA treatment (Figure 1), our data suggest that matrix sorts away from transmembrane proteins within tubules.
GMx33 Is an Atypical Golgi Matrix Protein
Inhibition of GRIP protein function by specific antibodies (Lu et al., 2004) or overexpression of either a GRIP domain alone or the GRIP domain protein GCC88 has been shown to disrupt the trans-Golgi structure (Luke et al., 2003; Yoshino et al., 2003). To determine whether depletion of GMx33 would also lead to Golgi fragmentation, we treated NRK cells with siRNA specific for GMx33α and GMx33β individually or together. GMx33α-specific siRNA depleted the majority of GMx33 expression (Figure 9A). By comparison GMx33β-specific or lamin-specific siRNA had no detectable effect on the overall GMx33 signal by immunofluorescence or immunoblot (our unpublished data). Our antisera recognizes both GMx33 α and β, indicating that GMx33α is the predominant form expressed by NRK and HeLa cells. Even when all detectable GMx33 has been depleted, no major changes in the Golgi structure were obvious by immunofluorescence (Figure 9A). These data indicate that although GMx33 associates with the trans-Golgi matrix in a GTP-dependent manner, Golgi structure is not disrupted in a significant way and therefore GMx33 may not be a “classical” trans-Golgi matrix protein.
Figure 9.
Depletion of GMx33 does not visibly disrupt the Golgi structure even though GMx33 has the biochemical properties of the Golgi matrix. (A) NRK cells were transfected with siRNA specific for GMx33α, incubated for 96 h, and stained with antibodies specific for GMx33 and the indicated marker protein. Each image includes cells that continued to express GMx33 for comparison. Bar, 10 μm. (B) Budded fractions were isolated from 75 μg rat liver Golgi fraction in the presence of ATP and GTPγS. After isolation, the pelleted budded fractions or remnant Golgi fractions were analyzed directly (-) subjected to extraction with 1% Tx100 alone, 1% Tx100 + 150 mM NaCl, or 150 mM NaCl alone for 30 min on ice. Each sample was then pelleted again and protein was analyzed by immunoblot with antibodies specific for GMx33.
Because the Golgi matrix was originally defined as insoluble in Tx100 and partially resistant to extraction by Tx100 plus 150 mM NaCl, both the budded fraction and the remaining Golgi were submitted to these conditions, and the resultant fractions wee pelleted and analyzed by immunoblot. In both fractions GMx33 was predominantly insoluble in 1% Tx100, whereas greater that 75% of the GMx33 was extracted with 1% Tx100 plus 150 mM NaCl. These biochemical properties are hallmarks of Golgi matrix proteins.
DISCUSSION
GMx33 is dynamically associated with the trans-Golgi matrix, associating and dissociating with the Golgi in seconds. GMx33 can be locked onto the trans-Golgi matrix by GTPγS, indicating that its association is regulated in a GTP-dependent manner like many other Golgi matrix proteins (Barr and Short, 2003; Jackson, 2003; Gleeson et al., 2004). Using live-cell imaging, we show that GFP-GMx33α exits the Golgi in tubules and within the tubules the GFP-GMx33α appears to segregate from transmembrane proteins followed by fragmentation of the tubules into smaller tubules and vesicles. Within vesicles produced in an in vitro budding reaction, GMx33 remains segregated in a matrixlike tail region that also contains Golgin-245, and this trans-matrix often links a few vesicles together. Together this data suggests that GMx33 is a member of the trans-Golgi matrix and may participate in sorting and exit from the Golgi.
GMx33α and β were identified as unknown proteins in an effort to define the Golgi proteome and characterized to be detergent insoluble, predominately localized to the trans- Golgi but having a large cytoplasmic pool (Wu et al., 2000). The Golgi bound and cytosolic pools are differentially post-translationally modified. The human orthologues of GMx33, GPP34, and GPP34R were identified in an independent proteomic study of the Golgi and were shown to associate with the cytoplasmic face of the trans-Golgi (Bell et al., 2001). Neither isoform contains strong homology to previously known domains. GMx33α has limited similarity to GRIP domains, but mutagenesis studies have indicated that GMx33α is not a GRIP protein since the putative GRIP domain is not sufficient for Golgi localization (our unpublished data). Despite significant homology between the α and β isoforms, there are regions that are not shared and these will be useful for teasing apart the individual function of each. Our data suggest that GMx33α is the dominant isoform expressed in NRK and HeLa cells because alphaspecific siRNA reduced almost all detectable GMx33 expression, whereas beta-specific siRNA had no detectable effect (our unpublished data).
The homologous proteins to GMx33 have been deleted from yeast (vps74; Bonangelino et al., 2002) and Drosophila (l(2)s5379; Spradling et al., 1999) and depleted from Caenorhabditis elegans (Y47G6A; Fraser et al., 2000), in broad deletion studies. Although the absence of GMx33 in Drosophila and C. elegans offers few clues as to its function, in yeast, where there is only a single form, Vps74p, deletion resulted in a modest mis-sorting and secretion of the vacuolar protein carboxypeptidase Y. Additionally, an interaction between Vps74p and Vps26p, a member of the yeast retromer complex, was reported (Bonangelino et al., 2002), which is interesting in light of our evidence that GMx33 can be localized to and trapped on both endosomal and the plasma membranes. Because there is a large cytosolic pool of GMx33, it is possible that GMx33 from the cytosol is simply binding to various membranes directly from the cytosol as its binding partners or ligands traffic. This idea is supported by our finding that even after treatment of cells with inhibitors of endosomal acidification, GFP-GMx33α continues to cycle rapidly between the membrane-bound and cytoplasmic pools. Similar experiments have not been carried out on other Golgi matrix proteins, so it is possible they also associate dynamically with membrane compartments other than the Golgi. Our preliminary results suggest that Golgin-245 does not follow the same pathway as GMx33.
GMx33 is highly posttranslationally modified (Wu et al., 2000). Phosphorylation is known to regulate several Golgi matrix proteins and membrane bound GMx33 is phosphorylated, whereas cytosolic GMx33 seems to lack the phosphorylated modifications (Barr et al., 1997; Nakamura et al., 1997; Lowe et al., 1998; Dirac-Svejstrup et al., 2000; Lowe et al., 2000; Wu et al., 2000; Brandon et al., 2003; Wang et al., 2005). ATP promotes GMx33's Golgi association and it is likely that this association involves phosphorylation. The role of GTP in regulating the Golgi matrix as a whole is clearly multifactorial and Rab, Arf, and Arl GTPases are known to function in the regulation of Golgi matrix membrane association and tethering and perhaps the regulation of conformation to govern the specificity of interactions (Barr and Short, 2003; Jackson, 2003; Gleeson et al., 2004). Given this, it is not surprising that GTP plays a role in regulating GMx33's Golgi association, even though we were not able to detect similar regulation of the cis-Golgi matrix protein p115 with these conditions (Figure 5A). GMx33 itself is not predicted to bind to GTP and its Golgi association is not disrupted by BFA, ostensibly eliminating Arl1 and Arf1 as possible binding partners. Therefore, it is likely that GMx33 is regulated by a Rab GTPase and GTPγS is simply locking GMx33 onto the trans-Golgi matrix. At this time, we cannot rule out the possibility that there are two pools of membrane bound GMx33, one regulated by ATP and phosphorylation and a second regulated by GTP, especially because there are two isoforms of the protein, α and β, and each isoform could be regulated differently.
Golgi matrix proteins are considered to be important for maintaining the structure of the Golgi. The GRASP proteins have been shown to stack Golgi cisternae in vitro (Barr et al., 1997; Shorter et al., 1999). Cleavage of Golgi matrix proteins by caspases is essential for Golgi fragmentation during apoptosis (Mancini et al., 2000; Chiu et al., 2002; Lane et al., 2002). Inhibition of Golgin-97 by specific antibodies results in Golgi fragmentation (Lu et al., 2004), whereas overexpression of GCC88 resulted in accumulation of large matrix-rich domains, termed “cauliflowers” in the trans-Golgi region (Luke et al., 2003) and overexpression of the GRIP domain of p230/Golgin-245 resulted in perturbations in the trans-Golgi (Yoshino et al., 2003). These effects may be indirect however because depletion of Golgin-97 by RNA interference did not result in any obvious structural changes in the Golgi by light microscopy (Lu et al., 2004). These data and our findings that depletion of GMx33 does not appear to affect the structure of the Golgi by light microscopy suggest a subtle role for these proteins in maintenance of the trans-Golgi. It is interesting to note that although some bacteria may have proteins related to GMx33 as defined by low (10-9 and 10-11) E values in a BLAST search, plants do not seem to have any proteins related to GMx33, supporting the idea that it is not required for maintaining Golgi structure.
The presence of GMx33 on tubules exiting the Golgi is striking. All GMx33-positive tubules observed show distinct puncta of GMx33 along the length of the tubule and these puncta do not precisely colocalize with TGN38 in fixed cells. Only one other trans-Golgi matrix protein, p230/Golgin-245, has been shown to associate with vesicles and tubules exiting the Golgi (Brown et al., 2001; Gleeson et al., 2004). Both GMx33 and Golgin-245 appear in puncta along tubules, and both proteins associate with similar matrix structures after a standard budding assay from an isolated Golgi fraction. Our hypothesis is that during tubulation, transmembrane proteins are sorted from matrix within the tubules. Our EM data further suggests that the Golgi matrix may tether these sorted vesicles together for some time along the length of tubules, before fission. This suggests an interesting role for the trans-Golgi matrix proteins in tubulation and sorting in exit from the Golgi.
The Golgi matrix has been classically defined as a detergent-insoluble and salt-resistant group of proteins that bind to resident Golgi transmembrane proteins and regulate the structure of the Golgi stack. GM130, the prototypical Golgi matrix protein, is ∼80% insoluble in Tx100, and ∼35% of the protein is resistant to extraction by 1% Tx100 in 150 mM NaCl (Slusarewicz et al., 1994). Similarly, GMx33 is also mostly resistant to solubilization in 1% Tx100, especially when associated with the Golgi matrix in a budded fraction (Figure 9B). The addition of 150 mM NaCl to the 1% Tx100 increases the efficiency of the GMx33 extraction from the matrix, but GMx33 is clearly salt sensitive (Figure 1D), so this is not surprising.
As more Golgi matrix proteins have been discovered, long coiled-coil domains have been defined as a common structural feature. Many of these proteins are targets of autoantibodies and have been termed Golgins (Nozawa et al., 2005). GMx33 is not predicted to have an extensive coiled-coil structure, although it may have three coiled domains, and by this definition it is not a Golgin (Barr and Short, 2003). However, GRASP65 and GRASP55, two matrix proteins involved in maintaining the Golgi stack (Barr et al., 1997; Shorter et al., 1999), are also not predicted to be highly coiled (Gillingham and Munro, 2003), but are sometimes categorized as Golgins (Barr and Short, 2003).
The functions of the trans-Golgi matrix are poorly understood. Much of what we know has come from studies characterizing the individual matrix proteins and that data have been used to predict functions. These predicted functions include a role in both anterograde and retrograde trafficking as well as interactions with the cytoskeleton and maintenance of Golgi structure (Erlich et al., 1996; Matanis et al., 2002; Luke et al., 2003; Yoshino et al., 2003; Kakinuma et al., 2004; Lu et al., 2004; Yoshino et al., 2005). As other proteins are characterized, the predicted functions will be modified to include the new data. One such protein is GMx33, which has the biochemical properties of a matrix protein and associates dynamically with the trans-Golgi matrix. Its presence on tubules exiting the Golgi and its localization within segregated puncta on these tubules, as well as its localization to endosomes and the plasma membrane offer clues about the role that GMx33, and perhaps other trans-Golgi matrix proteins, play in several basic sorting processes.
Supplementary Material
Acknowledgments
We thank Francis Barr, John Bergeron, Alfonso Gonzalez, and Elizabeth Sztul for kindly providing antibodies. The live cell imaging and photobleachingwas carried out at the UCHSC Light Microscopy Facility, and we thank Steven Fadul for his help. We also thank Rytis Prekeris for the use of the Zeiss Axiovert 200. This work was supported by Grants GM42629 and PO1 GM61306 from the National Institutes of Health, awarded to K.E.H. and a postdoctoral fellowship from the NIH, F32 GM072236-01, awarded to C.M.S.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-07-0682) on October 19, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Allan, V. J., and Kreis, T. E. (1986). A microtubule-binding protein associated with membranes of the Golgi apparatus. J. Cell Biol. 103, 2229-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos, W. B., and Grimstone, A. V. (1968). Intercisternal material in the Golgi body of Trichomonas. J. Cell Biol. 38, 466-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr, F. A., Nakamura, N., and Warren, G. (1998). Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae. EMBO J. 17, 3258-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr, F. A., Puype, M., Vandekerckhove, J., and Warren, G. (1997). GRASP65, a protein involved in the stacking of Golgi cisternae. Cell 91, 253-262. [DOI] [PubMed] [Google Scholar]
- Barr, F. A., and Short, B. (2003). Golgins in the structure and dynamics of the Golgi apparatus. Curr. Opin. Cell Biol. 15, 405-413. [DOI] [PubMed] [Google Scholar]
- Bell, A. W. et al. (2001). Proteomics characterization of abundant Golgi membrane proteins. J. Biol. Chem. 276, 5152-5165. [DOI] [PubMed] [Google Scholar]
- Bonangelino, C. J., Chavez, E. M., and Bonifacino, J. S. (2002). Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 2486-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon, E., Gao, Y., Garcia-Mata, R., Alvarez, C., and Sztul, E. (2003). Membrane targeting of p115 phosphorylation mutants and their effects on Golgi integrity and secretory traffic. Eur. J. Cell Biol. 82, 411-420. [DOI] [PubMed] [Google Scholar]
- Brown, D. L., Heimann, K., Lock, J., Kjer-Nielsen, L., van Vliet, C., Stow, J. L., and Gleeson, P. A. (2001). The GRIP domain is a specific targeting sequence for a population of trans-Golgi network derived tubulo-vesicular carriers. Traffic 2, 336-344. [DOI] [PubMed] [Google Scholar]
- Chapman, R. E., Munro, S., Reaves, B., and Banting, G. (1994). Retrieval of TGN proteins from the cell surface requires endosomal acidification. Vacuolar ATPase inactivation blocks recycling to the trans-Golgi network from the plasma membrane. EMBO J. 13, 2305-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, R., Novikov, L., Mukherjee, S., and Shields, D. (2002). A caspase cleavage fragment of p115 induces fragmentation of the Golgi apparatus and apoptosis. J. Cell Biol. 159, 637-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson, P., de Curtis, I., Pouyssegur, J., Griffiths, G., and Davoust, J. (1989). Low cytoplasmic pH inhibits endocytosis and transport from the trans-Golgi network to the cell surface. J. Cell Biol. 108, 377-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida, J. B., Doherty, J., Ausiello, D. A., and Stow, J. L. (1993). Binding of the cytosolic p200 protein to Golgi membranes is regulated by heterotrimeric G proteins. J. Cell Sci. 106(Pt 4), 1239-1248. [DOI] [PubMed] [Google Scholar]
- Derby, M. C., van Vliet, C., Brown, D., Luke, M. R., Lu, L., Hong, W., Stow, J. L., and Gleeson, P. A. (2004). Mammalian GRIP domain proteins differ in their membrane binding properties and are recruited to distinct domains of the TGN. J. Cell Sci. 117, 5865-5874. [DOI] [PubMed] [Google Scholar]
- Dirac-Svejstrup, A. B., Shorter, J., Waters, M. G., and Warren, G. (2000). Phosphorylation of the vesicle-tethering protein p115 by a casein kinase II-like enzyme is required for Golgi reassembly from isolated mitotic fragments. J. Cell Biol. 150, 475-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson, J. G., Kahn, R. A., Lippincott-Schwartz, J., and Klausner, R. D. (1991a). Binding of ARF and beta-COP to Golgi membranes: possible regulation by a trimeric G protein. Science 254, 1197-1199. [DOI] [PubMed] [Google Scholar]
- Donaldson, J. G., Lippincott-Schwartz, J., and Klausner, R. D. (1991b). Guanine nucleotides modulate the effects of brefeldin A in semipermeable cells: regulation of the association of a 110-kD peripheral membrane protein with the Golgi apparatus. J. Cell Biol. 112, 579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich, R., Gleeson, P. A., Campbell, P., Dietzsch, E., and Toh, B. H. (1996). Molecular characterization of trans-Golgi p230. A human peripheral membrane protein encoded by a gene on chromosome 6p12-22 contains extensive coiled-coil alpha-helical domains and a granin motif. J. Biol. Chem. 271, 8328-8337. [DOI] [PubMed] [Google Scholar]
- Fraser, A. G., Kamath, R. S., Zipperlen, P., Martinez-Campos, M., Sohrmann, M., and Ahringer, J. (2000). Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325-330. [DOI] [PubMed] [Google Scholar]
- Fritzler, M. J., Etherington, J., Sokoluk, C., Kinsella, T. D., and Valencia, D. W. (1984). Antibodies from patients with autoimmune disease react with a cytoplasmic antigen in the Golgi apparatus. J. Immunol. 132, 2904-2908. [PubMed] [Google Scholar]
- Gillingham, A. K., and Munro, S. (2003). Long coiled-coil proteins and membrane traffic. Biochim. Biophys. Acta 1641, 71-85. [DOI] [PubMed] [Google Scholar]
- Gleeson, P.A., Anderson, T. J., Stow, J. L., Griffiths, G., Toh, B. H., and Matheson, F. (1996). p230 is associated with vesicles budding from the trans-Golgi network. J. Cell Sci. 109(Pt 12), 2811-2821. [DOI] [PubMed] [Google Scholar]
- Gleeson, P. A., Lock, J. G., Luke, M. R., and Stow, J. L. (2004). Domains of the TGN: coats, tethers and G proteins. Traffic 5, 315-326. [DOI] [PubMed] [Google Scholar]
- Griffiths, G. (1993). Fine Structure Immuno-cytochemistry, Heidelberg: Springer-Verlag.
- Heimann, K., Percival, J. M., Weinberger, R., Gunning, P., and Stow, J. L. (1999). Specific isoforms of actin-binding proteins on distinct populations of Golgi-derived vesicles. J. Biol. Chem. 274, 10743-10750. [DOI] [PubMed] [Google Scholar]
- Hoogenraad, C. C., Akhmanova, A., Howell, S. A., Dortland, B. R., De Zeeuw, C. I., Willemsen, R., Visser, P., Grosveld, F., and Galjart, N. (2001). Mammalian Golgi-associated Bicaudal-D2 functions in the dynein-dynactin pathway by interacting with these complexes. EMBO J. 20, 4041-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenraad, C. C., Wulf, P., Schiefermeier, N., Stepanova, T., Galjart, N., Small, J. V., Grosveld, F., de Zeeuw, C. I., and Akhmanova, A. (2003). Bicaudal D induces selective dynein-mediated microtubule minus end-directed transport. EMBO J. 22, 6004-6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn, M., and Banting, G. (1994). Okadaic acid treatment leads to a fragmentation of the trans-Golgi network and an increase in expression of TGN38 at the cell surface. Biochem. J. 301(Pt 1), 69-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, C. L. (2003). Membrane traffic: Arl GTPases get a GRIP on the Golgi. Curr. Biol. 13, R174-R176. [DOI] [PubMed] [Google Scholar]
- Kakinuma, T., Ichikawa, H., Tsukada, Y., Nakamura, T., and Toh, B. H. (2004). Interaction between p230 and MACF1 is associated with transport of a glycosyl phosphatidyl inositol-anchored protein from the Golgi to the cell periphery. Exp. Cell Res. 298, 388-398. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen, L., Teasdale, R. D., van Vliet, C., and Gleeson, P. A. (1999). A novel Golgi-localisation domain shared by a class of coiled-coil peripheral membrane proteins. Curr. Biol. 9, 385-388. [DOI] [PubMed] [Google Scholar]
- Kreitzer, G., Marmorstein, A., Okamoto, P., Vallee, R., and Rodriguez-Boulan, E. (2000). Kinesin and dynamin are required for post-Golgi transport of a plasma-membrane protein. Nat. Cell Biol. 2, 125-127. [DOI] [PubMed] [Google Scholar]
- Kuhn, L. C., and Kraehenbuhl, J. P. (1983). Monoclonal antibodies recognizing the secreted and membrane domains of the IgA dimer receptor. Ann. NY Acad. Sci. 409, 751-759. [DOI] [PubMed] [Google Scholar]
- Lane, J. D., Lucocq, J., Pryde, J., Barr, F. A., Woodman, P. G., Allan, V. J., and Lowe, M. (2002). Caspase-mediated cleavage of the stacking protein GRASP65 is required for Golgi fragmentation during apoptosis. J. Cell Biol. 156, 495-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, J. M., Brown, M. S., Goldstein, J. L., and Anderson, R. G. (1983). Depletion of intracellular potassium arrests coated pit formation and receptor-mediated endocytosis in fibroblasts. Cell 33, 273-285. [DOI] [PubMed] [Google Scholar]
- Lowe, M., Gonatas, N. K., and Warren, G. (2000). The mitotic phosphorylation cycle of the cis-Golgi matrix protein GM130. J. Cell Biol. 149, 341-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, M., Rabouille, C., Nakamura, N., Watson, R., Jackman, M., Jamsa, E., Rahman, D., Pappin, D. J., and Warren, G. (1998). Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell 94, 783-793. [DOI] [PubMed] [Google Scholar]
- Lu, L., and Hong, W. (2003). Interaction of Arl1-GTP with GRIP domains recruits autoantigens Golgin-97 and Golgin-245/p230 onto the Golgi. Mol. Biol. Cell 14, 3767-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, L., Tai, G., and Hong, W. (2004). Autoantigen Golgin-97, an effector of Arl1 GTPase, participates in traffic from the endosome to the trans-Golgi network. Mol. Biol. Cell 15, 4426-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke, M. R., Kjer-Nielsen, L., Brown, D. L., Stow, J. L., and Gleeson, P. A. (2003). GRIP domain-mediated targeting of two new coiled-coil proteins, GCC88 and GCC185, to subcompartments of the trans-Golgi network. J. Biol. Chem. 278, 4216-4226. [DOI] [PubMed] [Google Scholar]
- Lupashin, V., and Sztul, E. (2005). Golgi tethering factors. Biochim. Biophys. Acta 1744, 325-339. [DOI] [PubMed] [Google Scholar]
- Mancini, M., Machamer, C. E., Roy, S., Nicholson, D. W., Thornberry, N. A., Casciola-Rosen, L. A., and Rosen, A. (2000). Caspase-2 is localized at the Golgi complex and cleaves golgin-160 during apoptosis. J. Cell Biol. 149, 603-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardones, G., Snyder, C. M., and Howell, K. E. (2005). cis-Golgi matrix proteins move directly to endoplasmic reticulum exit sites by association with tubules. Mol. Biol. Cell 17, 525-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matanis, T. et al. (2002). Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dynein-dynactin motor complex. Nat. Cell Biol. 4, 986-992. [DOI] [PubMed] [Google Scholar]
- McConville, M. J., Ilgoutz, S. C., Teasdale, R. D., Foth, B. J., Matthews, A., Mullin, K. A., and Gleeson, P. A. (2002). Targeting of the GRIP domain to the trans-Golgi network is conserved from protists to animals. Eur J. Cell Biol. 81, 485-495. [DOI] [PubMed] [Google Scholar]
- Mollenhauer, H. H. (1965). An intercisternal structure in the Golgi apparatus. J. Cell Biol. 24, 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro, S., and Nichols, B. J. (1999). The GRIP domain—a novel Golgi-targeting domain found in several coiled-coil proteins. Curr. Biol. 9, 377-380. [DOI] [PubMed] [Google Scholar]
- Nakamura, N., Lowe, M., Levine, T. P., Rabouille, C., and Warren, G. (1997). The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell 89, 445-455. [DOI] [PubMed] [Google Scholar]
- Nozawa, K., Fritzler, M. J., and Chan, E. K. (2005). Unique and shared features of Golgi complex autoantigens. Autoimmun. Rev. 4, 35-41. [DOI] [PubMed] [Google Scholar]
- Panic, B., Perisic, O., Veprintsev, D. B., Williams, R. L., and Munro, S. (2003a). Structural basis for Arl1-dependent targeting of homodimeric GRIP domains to the Golgi apparatus. Mol. Cell 12, 863-874. [DOI] [PubMed] [Google Scholar]
- Panic, B., Whyte, J. R., and Munro, S. (2003b). The ARF-like GTPases Arl1p and Arl3p act in a pathway that interacts with vesicle-tethering factors at the Golgi apparatus. Curr. Biol. 13, 405-410. [DOI] [PubMed] [Google Scholar]
- Preisinger, C., Short, B., De Corte, V., Bruyneel, E., Haas, A., Kopajtich, R., Gettemans, J., and Barr, F. A. (2004). YSK1 is activated by the Golgi matrix protein GM130 and plays a role in cell migration through its substrate 14-3-3zeta. J. Cell Biol. 164, 1009-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband, W. S. (1997-2005). IMAGE J: U.S. National Institutes of Health, Bethesda, MD.
- Reaves, B., and Banting, G. (1994). Vacuolar ATPase inactivation blocks recycling to the trans-Golgi network from the plasma membrane. FEBS Lett. 345, 61-66. [DOI] [PubMed] [Google Scholar]
- Reaves, B. J., Bright, N. A., Mullock, B. M., and Luzio, J. P. (1996). The effect of wortmannin on the localisation of lysosomal type I integral membrane glycoproteins suggests a role for phosphoinositide 3-kinase activity in regulating membrane traffic late in the endocytic pathway. J. Cell Sci. 109(Pt 4), 749-762. [DOI] [PubMed] [Google Scholar]
- Salazar, G., and Gonzalez, A. (2002). Novel mechanism for regulation of epidermal growth factor receptor endocytosis revealed by protein kinase A inhibition. Mol. Biol. Cell 13, 1677-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann, J., Jokitalo, E., Pypaert, M., and Warren, G. (2000). Matrix proteins can generate the higher order architecture of the Golgi apparatus. Nature 407, 1022-1026. [DOI] [PubMed] [Google Scholar]
- Shorter, J., and Warren, G. (1999). A role for the vesicle tethering protein, p115, in the post-mitotic stacking of reassembling Golgi cisternae in a cell-free system. J. Cell Biol. 146, 57-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter, J., Watson, R., Giannakou, M. E., Clarke, M., Warren, G., and Barr, F. A. (1999). GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 18, 4949-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slusarewicz, P., Nilsson, T., Hui, N., Watson, R., and Warren, G. (1994). Isolation of a matrix that binds medial Golgi enzymes. J. Cell Biol. 124, 405-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling, A. C., Stern, D., Beaton, A., Rhem, E. J., Laverty, T., Mozden, N., Misra, S., and Rubin, G. M. (1999). The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153, 135-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, R. S., Jones, S. M., Dahl, R. H., Nordeen, M. H., and Howell, K. E. (1997). Characterization of the Golgi complex cleared of proteins in transit and examination of calcium uptake activities. Mol. Biol. Cell 8, 1911-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Satoh, A., and Warren, G. (2005). Mapping the functional domains of the Golgi stacking factor GRASP65. J. Biol. Chem. 280, 4921-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, T. H., Polishchuk, R. S., Caplan, S., Hirschberg, K., and Lippincott-Schwartz, J. (2001). Maintenance of Golgi structure and function depends on the integrity of ER export. J. Cell Biol. 155, 557-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. C., Taylor, R. S., Lane, D. R., Ladinsky, M. S., Weisz, J. A., and Howell, K. E. (2000). GMx33, a novel family of trans-Golgi proteins identified by proteomics. Traffic 1, 963-975. [PubMed] [Google Scholar]
- Wu, M., Lu, L., Hong, W., and Song, H. (2004). Structural basis for recruitment of GRIP domain golgin-245 by small GTPase Arl1. Nat. Struct. Mol. Biol. 11, 86-94. [DOI] [PubMed] [Google Scholar]
- Yoshino, A., Bieler, B. M., Harper, D. C., Cowan, D. A., Sutterwala, S., Gay, D. M., Cole, N. B., McCaffery, J. M., and Marks, M. S. (2003). A role for GRIP domain proteins and/or their ligands in structure and function of the trans Golgi network. J. Cell Sci. 116, 4441-4454. [DOI] [PubMed] [Google Scholar]
- Yoshino, A. et al. (2005). tGolgin-1 (p230, golgin-245) modulates Shiga-toxin transport to the Golgi and Golgi motility towards the microtubule-organizing centre. J. Cell Sci. 118, 2279-2293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.