Abstract
In vivo induction of the Escherichia coli lactose operon as a function of inducer concentration generates a sigmoidal curve, indicating a non-linear response. Suggested explanations for this dependence include a 2:1 inducer–repressor stoichiometry of induction, which is the currently accepted view. It is, however, known for decades that, in vitro, operator binding as a function of inducer concentration is not sigmoidal. This discrepancy between in vivo and in vitro data has so far not been resolved. We demonstrate that the in vivo non-linearity of induction is due to cooperative repression of the wild-type lac operon through DNA loop formation. In the absence of DNA loops, in vivo induction curves are hyperbolic. In the light of this result, we re-address the question of functional molecular inducer–repressor stoichiometry in induction of the lac operon.
INTRODUCTION
The lactose operon of Escherichia coli is, together with phage lambda (1), probably the best analysed model system for transcriptional regulation (2). The interactions of Lac repressor with lac operator and inducer have been the subject of intensive studies for about five decades. The culmination so far of these efforts has been the solution of X-ray crystal structures of repressor bound to lac operator, non-operator DNA or to the gratuitous inducer isopropyl-β,d-thiogalactoside (IPTG) (3–5). Still, important features of regulation in the lac operon have yet to be fully elucidated.
It had early been noticed that in vivo expression of the lac operon as a function of inducer concentration does not follow a simple hyperbolic saturation function but instead yields a distinctively sigmoidal curve (6). This observation has prompted over the years several different explanations: cooperative inducer binding, a two-step mechanism of induction and the suggestion that two molecules of inducer are necessary to abolish operator binding of Lac repressor (7–11). The latter assumption prevailed and became the accepted view (12,13), despite the fact that in vitro binding studies of Lac repressor binding to lac operator, later tacitly ignored, failed to show the non-linearity of in vivo induction (14). It should be pointed out that most of these studies have been performed before it was generally realized that Lac repressor can bind to two operator sequences at the same time (15,16).
Lac repressor is a homo-tetramer that can be thought of as consisting of two DNA-binding dimers (9,17) which aggregate into a dimer of dimers by means of a 4-helix bundle (3,5,18,19). While a Lac repressor dimer is needed for specific DNA binding, each monomer binds with equal affinity to one molecule of inducer (20,21).
Lac repressor is the negative regulator of the lactose operon (22). The repressor prevents initiation of transcription of the lac messenger RNA by binding with high affinity to the first lac operator, O1 (23), which lies immediately downstream of the lac promoter. Occupancy of O1 by Lac repressor is cooperatively increased through DNA loop formation (24) by binding of the other dimer of the homotetrameric Lac repressor to either of the auxiliary operators, O2 (25), which lies downstream of the lac promoter within the coding sequence of lacZ, or O3 (26), which lies upstream of the lac promoter.
Upon binding of inducer, the conformation of Lac repressor changes such that the DNA-binding headpieces alter their orientation relative to the repressor core and relative to each other (5). As a result, affinity to operator drops ∼1000-fold (27), and transcription of the lac messenger RNA increases accordingly. The induction of the lac operon as a function of inducer concentration can be followed in vivo through the expression of β-galactosidase, which is encoded by the first gene (lacZ) of the tricistronic lac mRNA. These measurements are done in a Lac permease negative background (lacY−), where the cellular concentration of the gratuitous inducer IPTG is close to that of the medium (28).
Our earlier finding of cooperativity in repression of the lac operon (24) prompted us to look into the difference between in vivo and in vitro induction curves and to re-evaluate the current model for induction of the lac operon.
MATERIALS AND METHODS
Strains, lambda phages and plasmids
E.coli strain BMH 8117 is Δ(lac, proAB). E.coli strains BMH 8117(λEwt100) and BMH 8117(λEwt123) are derivatives of BMH 8117 which carry lambda prophages bearing the β-galactosidase gene under control of the indicated combination of lac operators (24). Wild-type tetrameric Lac repressor and dimeric active Lac repressor were expressed from plasmids pSO1010-P1 and pSO331Stop (carrying an iadi allele), (24,29), respectively, for in vivo repression measurements. Constitutive expression was determined in the presence of plasmid pSO1000ΔA (24), bearing an i− allele. Plasmid pWB1000 (30) was used for over-expression of wt tetrameric Lac repressor for in vitro experiments.
Plasmid pBlueOid was generated by cloning the ideal lac operator, Oid, which has an ∼10-fold higher affinity to Lac repressor than O1 (31,32), from pWB300 (33) as a XbaI fragment into the SpeI site of pBluescript SK(+) (Stratagene). Plasmid pBCOid(−2) was constructed by cloning the polylinker, containing the ideal lac operator, from pBlueOid as a KpnI–SacI fragment into the respective sites of pBC KS(+), followed by deletion of 4 bp (blunting of the PstI site) and insertion of 2 bp (fill-in of the ClaI site).
β-Galactosidase assays
Specific β-galactosidase activities were determined as described (24,34). Each data point is the mean from two experiments. Constitutive expression is the mean of at least five independent cultures.
Band shift assays
Binding of wt Lac repressor to a single lac operator in vitro as a function of IPTG concentration was determined by band shifts, which were performed as described (35). Binding reactions with a 257 bp radiolabelled DNA fragment carrying a single ideal lac operator at 5 × 10−11M and with 2 × 10−10M Lac repressor dimer had a volume of 20 µl. The DNA fragment was generated by PCR, using primers PCRfor1 (5′-GTTGTAAAACGACGGCC-3′), PCRrev1 (5′-CAGGAAACAGCTATGACC-3′) and plasmid pBlueOid as template. Non-bindable DNA was estimated as unbound DNA at 2.2 × 10−9 M Lac repressor dimer and corrected for.
Band shifts of wt Lac repressor binding to a DNA fragment with two lac operators (ideal operator Oid and wt O1) were as above, with the following modifications: a 298 bp DNA fragment was generated using primers PCRfor1 and PCRrev0 (5′-GCTCGTATGTTGTGTGG-3′) and plasmid pBCOid(−2) as template. Binding reactions were separated on 4% acrylamide gels (acrylamide to bis-acrylamide, 79:1).
Lac repressor was partially purified with a 40–50% ammonium sulfate cut from sonication extracts in CSB (36) of BMH 8117 cells carrying pWB1000. Concentration of active repressor was determined by stoichiometric titration with 5 × 10−10 M ideal lac operator fragment. Dried gels were analysed with a Storm 840 phosphoimager (Molecular Dynamics). Phosphoimager data were quantified with ImageQuant 5.2 (Molecular Dynamics). Data points are the means of at least five experiments.
Data analysis
To derive operator binding data from specific β-galactosidase activities, we substituted:
[Ot] (total operator) with Ac,
[Of] (free operator) with A,
[Ooc] (occupied operator) with Ac − A,
where Ac = constitutive specific β-galactosidase activity and A = specific β-galactosidase activity at a given concentration of IPTG.
Regression analyses were performed with the program DataFit 7.0 (Engeneered Software, PA).
RESULTS
Induction of the lac promoter in the presence and in the absence of DNA loops
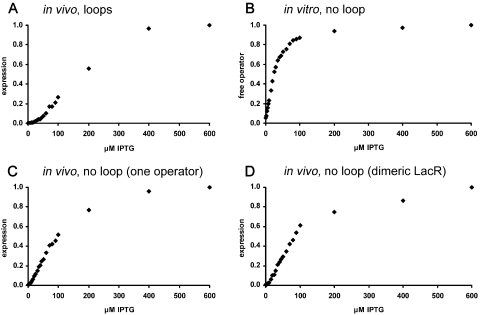
We first compared the in vivo induction curve of the wild-type (wt) configuration of the lac operon (the lac promoter is controlled through DNA loops by all three wt lac operators and wt tetrameric Lac repressor) with an in vitro induction curve of wt Lac repressor binding to a DNA fragment containing a single lac operator (Figure 1A and B). The graphs reflect the known discrepancy: The in vivo curve is sigmoidal, the in vitro curve is hyperbolic.
Figure 1.
Induction of cooperative and non-cooperative lac systems. (A) Relative β-galactosidase activities (corresponding to a plot of [Of]/[Ot]) of strain BMH 8117 (lacY−), harbouring a plasmid expressing wt Lac repressor and carrying on a λ prophage a lacZ gene driven by the wt lac promoter with all three operators, are plotted against the IPTG concentration in the growth medium. (B) Complex of wt Lac repressor with a DNA fragment carrying a single lac operator. The fraction ([Of]/[Ot]) of free DNA fragment (total fragment concentration is 5 × 10−11 M) in the presence of 2 × 10−10 M wt Lac repressor is plotted as a function of IPTG concentration. (C) As in (A), but in the presence of the first lac operator only. (D) As in (A), but harbouring a plasmid expressing dimeric active Lac repressor instead of wt tetrameric Lac repressor. To normalize the curves, values at 600 µM IPTG are set to 1.
We then determined two in vivo induction curves of lac promoters controlled without DNA loops. Figure 1C: in the presence of wt tetrameric Lac repressor but only the first lac operator. Figure 1D: in the presence of all three lac operators but repressed by a mutant dimeric active Lac repressor. Neither combination allows DNA loop formation (24). Both curves are hyperbolic at inducer concentrations above the dissociation constant (∼5 × 10−6 M) of the repressor–IPTG complex (37,38), demonstrating that the sigmoidality of the induction curve of the wt system reflects cooperative repression through DNA loop formation. Consequently, induction data of the wt lac promoter do not allow straightforward inference as to the functional stoichiometry of inducer–Lac repressor dimer interaction. To address the question of how many molecules of inducer are required to abolish operator binding of a Lac repressor dimer, systems without DNA loops have to be analysed.
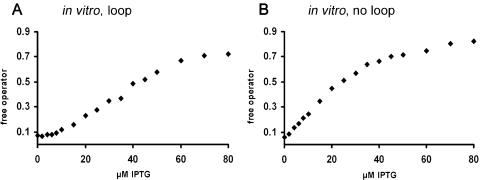
Figure 2 demonstrates that, also in vitro, template binding of Lac repressor engaged in a DNA loop, plotted as a function of inducer concentration, yields a sigmoidal induction curve. Lac repressor forms DNA loops in vitro with a DNA template carrying two suitably spaced operators, here O1 and the ideal lac operator Oid, with the centres of symmetry separated by 168 bp, corresponding to 16 helical turns. Loop complexes with a linear template are less stable than those that can be achieved with supercoiled templates (39). Also, in contrast to the in vitro conditions, loops in vivo appear to be additionally stabilized by architectural DNA-binding proteins which increase the flexibility of the DNA (40). Cooperative binding of Lac repressor in vitro is accordingly weaker than in vivo and the sigmoidality of the induction curve restricted to its initial part (Figure 2A).
Figure 2.
Induction of a DNA loop in vitro. The fraction ([Of]/[Ot]) of free DNA fragment (total fragment concentration is 5 × 10−11 M) in the presence of 2 × 10−10 M wt Lac repressor is plotted as a function of IPTG concentration. Note the different scale compared with Figure 1. (A) Loop complex of wt Lac repressor with a DNA fragment carrying two lac operators separated by 16 helical turns. (B) Control: complex of wt Lac repressor with a DNA fragment carrying a single lac operator. To normalize the curves, values at 600 µM IPTG are set to 1.
The stoichiometry of lac induction
In an experimental system devoid of DNA loops, three possible forms of the Lac repressor dimer, D (free dimer), DI (complex of dimer with one molecule of inducer) and DI2 (complex of dimer with two molecules of inducer) can react with lac operator. In our experiments, inducer is in a large excess over repressor, and two identical and independent inducer-binding sites can be assumed for a Lac repressor dimer (20,21). Expressing the respective Lac repressor–inducer complexes as the fraction of total repressor [Dt], the occupancy R (defined as the quotient [Ooc]/[Of] of operator Ooc, occupied by any one of the three forms of the repressor dimer, and free operator Of) of O1, can therefore be described by the following equation.
| 1 |
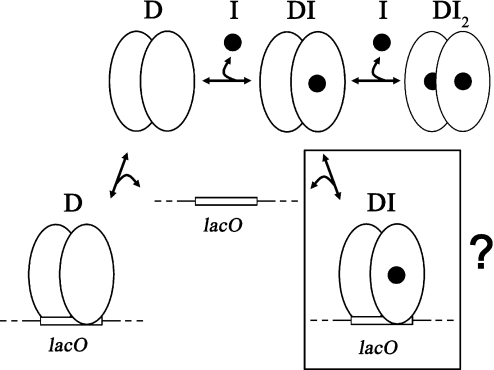
where KoX (X = 1 for the complex D–O1, X = 2 for the complex DI–O1 and X = 3 for the complex DI2–O1) is the equilibrium dissociation constant of the respective lac operator–Lac repressor complexes and Ki is the repressor monomer–inducer equilibrium dissociation constant. Figure 3 gives a schematic overview of the relevant equilibria.
Figure 3.
Equilibria of inducer and operator binding of Lac repressor dimers. While it is known that free repressor (D) binds strongly to lac operator, binding of the complex of the repressor dimer and two molecules of inducer (DI2) is too weak to play a role at low inducer concentrations. Operator binding of the complex of a Lac repressor dimer with one molecule of inducer (DI) cannot be directly measured and is therefore boxed and labelled with a question mark. Closed circles symbolize inducer, open ovals monomers of Lac repressor and the open box lac operator (lacO).
The contribution of species DI2 to repression is negligible at low experimental concentrations of inducer. The operator affinity of the Lac repressor dimer saturated with IPTG (DI2) is ∼1000-fold lower than that of Lac repressor in the absence of inducer (27). For low concentrations of inducer, Equation 1 consequently reduces to the following equation.
| 2 |
Depending on the functional stoichiometry of induction, this equation simplifies further in one of two ways. Model 1: if the functional inducer–repressor stoichiometry of induction is 1:1, only D contributes to repression and DI is as inefficient in operator binding as is DI2. In this case, Equation 2 simplifies further to the following equation.
| 3 |
Model 2: if the functional stoichiometry is 2:1, only DI2 loses affinity to operator DNA, and D and DI have the same high affinity for operator (Ko1 = Ko2 = Ko). This is the current view (19). In that case, Equation 2 simplifies to the following equation.
| 4 |
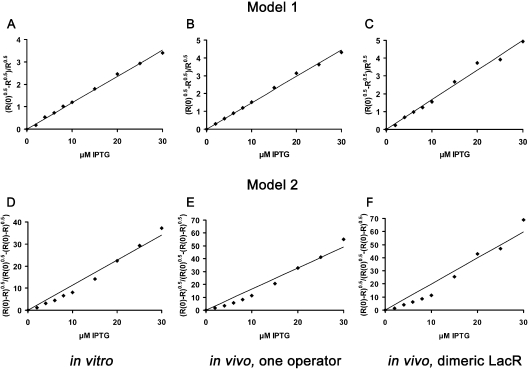
Using these two equations, we performed non-linear regression on the non-cooperative in vitro binding data and the two sets of induction data without DNA loops at low inducer concentrations. Taking the unexplained variance as a measure of the fit, we find that model 1 describes the data substantially better. In all three cases, the unexplained variance for model 2 is at least twice that for model 1 (in vitro, wt repressor: 1.2% versus 0.6%; in vivo, wt repressor: 0.7% versus 0.01%; in vivo, dimeric repressor: 1.9% versus 0.7%).
Linear transformation of the alternative Equations 3 and 4 demonstrates that this difference is due to the fact that the best fit of model 2 exhibits a systematic deviation from the data (Figure 4D–F), while the data are well approximated by model 1 (Figure 4A–C). Model 2 is apparently not an appropriate description of the induction data. Both in vivo and in vitro data can thus be explained assuming a functional inducer–repressor dimer stoichiometry of 1:1. Most of the operator affinity of the Lac repressor dimer is lost upon binding to one molecule of inducer. Operator binding by DI can be neglected at low inducer concentrations and at the resolution the induction analysis has.
Figure 4.
The functional inducer-binding stoichiometry of Lac repressor. Substituting [Dt]/Ko with R(0), the experimental data are transformed using linearizing rearrangements of Equation 3 (Model 1: (R(0)0.5 − R0.5)/R0.5 = ) and 4 (Model 2: (R(0) − R)0.5/(R(0)0.5 − (R(0) − R)0.5) = ). Best fit straight lines were derived by linear regression. All plots are of non-cooperative systems. (A) In vitro, model 1. (B) In vivo, model 1. The lac promoter is controlled by wt tetrameric Lac repressor and the first lac operator only. (C) In vivo, model 1. The lac promoter is controlled by dimeric active Lac repressor and all three lac operators. (D) In vitro, model 2. (E) In vivo, model 2. The lac promoter is controlled by wt tetrameric Lac repressor and the first lac operator. (F) In vivo, model 2. The lac promoter is controlled by dimeric active Lac repressor and all three lac operators.
Table 1 gives the respective inducer and operator binding equilibrium constants of Lac repressor, as determined by non-linear regression analysis using model 1.
Table 1.
Equilibrium dissociation constants (±SE) for operator binding (Ko) and inducer (IPTG) binding (Ki) of wt tetrameric Lac repressor and the dimeric active mutant 331Stop
| Ko (M) | Ki (M) | |
|---|---|---|
| In vitro (Oid)/tet. LacR | 1.26 (±0.05) × 10−11 | 8.2 (±0.4) × 10−6 |
| In vivo (O1)/tet. LacR | 4.39 (±0.02) × 10−10 | 6.7 (±0.1) × 10−6 |
| In vivo (O1)/dim. LacR | 2.44 (±0.06) × 10−10 | 6.4 (±0.4) × 10−6 |
DISCUSSION
We demonstrate that the sigmoidality of the induction curve of the wt lac operon in vivo is a consequence of the relief of cooperative repression through DNA loops during induction. In the absence of DNA loops in vivo, induction curves are hyperbolic. There are mainly two reasons why cooperative operator binding (and therefore sigmoidality of induction curves) by Lac repressor had not been seen previously in vitro. Firstly, DNA loops are not detectable by filter binding, the most commonly used assay for Lac repressor–DNA interactions for many years (35). Secondly, while DNA loops can be shown with gel-shift assays, they do not form readily on linear lac operon DNA as template (41,42).
We therefore used a synthetic construct, carrying two suitably spaced lac operators (16 helical turns, assuming a helix repeat of 10.5) with high affinity to Lac repressor (O1 and Oid) for in vitro measurements of Lac repressor binding to a DNA template in a loop complex as a function of inducer concentration. While, as expected for a linear template in vitro, less pronounced than in vivo, the resulting curve is clearly sigmoidal. This confirms our conclusion that sigmoidality of induction of the wt lac operon is the consequence of DNA loop formation. Abolishing DNA loops in vivo converts a sigmoidal induction curve into a hyperbolic one, and introducing a DNA loop in vitro converts a hyperbolic induction curve into a sigmoidal one.
The reconciliation of in vivo and in vitro observations reopens at the same time the question of functional stoichiometry of Lac repressor–inducer interaction, which had been answered using in vivo induction data of the wt lac promoter.
It is a commonly held belief that a plot of log {[r − r(0)]/r(0)}, where r is [Of]/[Ooc] and r(0) is r at [Inducer] = 0, against log [Inducer] yields the molecular functional induction stoichiometry of inducible systems as the slope of the resulting straight line (13). A slope of ∼2 has been reported before for the wt lac operon (11). Using this plot on our in vivo data on the wt lac system, we find a slope of 1.98 (±0.06), consistent with the previous findings. Our experiments show, however, that this apparent stoichiometry of 2 is coincidental. It is a consequence of cooperative repression through DNA loop formation. For the non-cooperative lac systems, in vivo and in vitro, the slope of this plot is ∼1.4–1.5 and thus not compatible with simple stoichiometry, reflecting the simplification of setting Dt = D + DIn (where operator affinity is lost in the transition from DIn−1 to DIn). We therefore avoided this approximation here.
Our analysis of lac induction in the absence of DNA loops is compatible with the suggestion that most of the operator binding of a Lac repressor dimer is lost upon binding of one molecule of inducer. This finding is consistent with what is structurally known about Lac repressor. Operator binding should be weakened as soon as the headpieces of a repressor dimer are out of alignment. This is already the case when one repressor monomer in a dimer undergoes inducer caused allosteric change.
The subsequent binding of Lac repressor dimers to an additional molecule of inducer contributes to induction of the lac operon by shifting the equilibrium towards the inactivated form of the repressor dimer (Figure 3).
One could imagine a repressor dimer that exhibits strong negative inducer-binding cooperativity, upon inducer binding of one monomer essentially precluding the second monomer from binding to inducer. Binding to inducer is here described by the following equation.
| 5 |
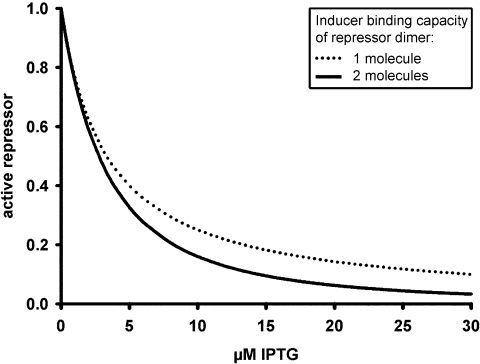
It is obvious that inactivation of this hypothetical repressor would lag considerably behind that of wt Lac repressor (Figure 5). Thus, the formation of DI2 has an important thermodynamical role that leads to inactivation of more Lac repressor at lower concentrations of inducer.
Figure 5.
Induction as a function of the inducer-binding capacity of a Lac repressor dimer. Two calculated curves using Ki and Ko from Table 1 (in vivo, tet. LacR) are given. Dotted line, inactivation of a (hypothetic) repressor dimer that can only bind to one molecule of inducer (Equation 5). Solid line, inactivation of wt Lac repressor which binds to two molecules of inducer (Equation 3).
The analysis of lac induction in the absence of DNA loops yields estimates for in vitro and in vivo inducer and operator binding constants of Lac repressor, which are comparable with affinities reported in the literature (14,20,29,31,32,43–45).
Interestingly, analysis of inactivation of the E.coli Cyt repressor, which is a member of the LacI family of bacterial repressors (46), reveals similarities of the induction process (47). Here, specific binding of the dimeric repressor protein to its operator depends on protein–protein contacts of the repressor monomers to one molecule each of the CAP proteins, which are bound to two CAP binding sites flanking the cyt operator. In this system, binding of inducer (cytidine) to the repressor does not directly affect its affinity to operator. Instead, the protein–protein contacts to the CAP proteins are weakened, which then leads to dissociation of the repressor–operator complex. Despite these important differences, also here, binding of one monomer of the repressor dimer to inducer (and, consequently, loss of contact to only one of the two CAP molecules) appears to be sufficient for efficient induction.
It seems not unreasonable to assume that the other members of the family of Lac repressor related proteins, as, for example, the Gal repressor (48), exhibit equivalent mechanisms of induction as the Lac repressor. Future analyses are required to show if this assumption is true.
Acknowledgments
We thank Regina Alex and Alexandros Kiupakis for helpful discussions. This work was supported by a grant of Deutsche Forschungsgemeinschaft to B.M.H. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ptashne M. A Genetic Switch: Phage Lambda Revisited. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2004. [Google Scholar]
- 2.Müller-Hill B. The lac Operon: A Short History of a Genetic Paradigm. Berlin, NY: Walter de Gruyter; 1996. [Google Scholar]
- 3.Friedman A.M., Fischmann T.O., Steitz T.A. Crystal structure of lac repressor core tetramer and its implications for DNA looping. Science. 1995;268:1721–1727. doi: 10.1126/science.7792597. [DOI] [PubMed] [Google Scholar]
- 4.Bell C.E., Lewis M. Crystallographic analysis of Lac repressor bound to natural operator O1. J. Mol. Biol. 2001;312:921–926. doi: 10.1006/jmbi.2001.5024. [DOI] [PubMed] [Google Scholar]
- 5.Lewis M., Chang G., Horton N.C., Kercher M.A., Pace H.C., Schumacher M.A., Brennan R.G., Lu P. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science. 1996;271:1247–1254. doi: 10.1126/science.271.5253.1247. [DOI] [PubMed] [Google Scholar]
- 6.Herzenberg L.A. Studies on the induction of beta-galactosidase in a cryptic strain of Escherichia coli. Biochim. Biophys. Acta. 1959;31:525–538. doi: 10.1016/0006-3002(59)90029-0. [DOI] [PubMed] [Google Scholar]
- 7.Boezi J.A., Cowie D.B. Kinetic studies of beta-galactosidase induction. Biophys. J. 1961;1:639–647. doi: 10.1016/s0006-3495(61)86913-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark D.J., Marr A.G. Studies on the repression of beta-galactosidase in Escherichia coli. Biochim. Biophys. Acta. 1964;92:85–94. doi: 10.1016/0926-6569(64)90272-x. [DOI] [PubMed] [Google Scholar]
- 9.Miller J.H. The lacI gene: its role in lac operon control and its use as a genetic system. In: Miller J.H., Reznikoff W.S., editors. The Operon. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1978. pp. 31–88. [Google Scholar]
- 10.Zubay G., Chambers D.A., Cheong L.C. Cell-free studies on the regulation of the lac operon. In: Beckwith J.R., Zipser D., editors. The Lactose Operon. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1970. pp. 375–391. [Google Scholar]
- 11.Yagil G., Yagil E. On the relation between effector concentration and the rate of induced enzyme synthesis. Biophys. J. 1971;11:11–27. doi: 10.1016/S0006-3495(71)86192-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sasse-Dwight S., Gralla J.D. Probing co-operative DNA-binding in vivo. The lac O1:O3 interaction. J. Mol. Biol. 1988;202:107–119. doi: 10.1016/0022-2836(88)90523-2. [DOI] [PubMed] [Google Scholar]
- 13.Yagil G. Enzyme induction. In: Segel L.A., editor. Biological Kinetics. Cambridge: Cambridge University Press; 1991. pp. 57–73. [Google Scholar]
- 14.Barkley M.D., Bourgeois S. Repressor recognition of operator and effectors. In: Beckwith J.R., Zipser D., editors. The Lactose Operon. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1970. pp. 177–220. [Google Scholar]
- 15.Kania J., Müller-Hill B. Construction, isolation and implications of repressor-galactosidase—beta-galactosidase hybrid molecules. Eur. J. Biochem. 1977;79:381–386. doi: 10.1111/j.1432-1033.1977.tb11819.x. [DOI] [PubMed] [Google Scholar]
- 16.O'Gorman R.B., Dunaway M., Matthews K.S. DNA binding characteristics of lactose repressor and the trypsin-resistant core repressor. J. Biol. Chem. 1980;255:10100–10106. [PubMed] [Google Scholar]
- 17.Kania J., Brown D.T. The functional repressor parts of a tetrameric lac repressor-beta-galactosidase chimaera are organized as dimers. Proc. Natl Acad. Sci. USA. 1976;73:3529–3533. doi: 10.1073/pnas.73.10.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alberti S., Oehler S., von Wilcken-Bergmann B., Krämer H., Müller-Hill B. Dimer-to-tetramer assembly of Lac repressor involves a leucine heptad repeat. New Biol. 1991;3:57–62. [PubMed] [Google Scholar]
- 19.Alberti S., Oehler S., von Wilcken-Bergmann B., Müller-Hill B. Genetic analysis of the leucine heptad repeats of Lac repressor: evidence for a 4-helical bundle. EMBO J. 1993;12:3227–3236. doi: 10.1002/j.1460-2075.1993.tb05992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohshima Y., Mizokoshi T., Horiuchi T. Binding of an inducer to the lac repressor. J. Mol. Biol. 1974;89:127–136. doi: 10.1016/0022-2836(74)90166-1. [DOI] [PubMed] [Google Scholar]
- 21.Schmitz A., Schmeissner U., Miller J.H. Mutations affecting the quaternary structure of the lac repressor. J. Biol. Chem. 1976;251:3359–3366. [PubMed] [Google Scholar]
- 22.Jacob F., Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 23.Schlax P.J., Capp M.W., Record M.T., Jr Inhibition of transcription initiation by lac repressor. J. Mol. Biol. 1995;245:331–350. doi: 10.1006/jmbi.1994.0028. [DOI] [PubMed] [Google Scholar]
- 24.Oehler S., Eismann E.R., Krämer H., Müller-Hill B. The three operators of the lac operon cooperate in repression. EMBO J. 1990;9:973–979. doi: 10.1002/j.1460-2075.1990.tb08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reznikoff W.S., Winter R.B., Hurley C.K. The location of the repressor binding sites in the lac operon. Proc. Natl Acad. Sci. USA. 1974;71:2314–2318. doi: 10.1073/pnas.71.6.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert W., Gralla J., Majors J., Maxam A. Lactose operator sequences and the action of lac repressor. In: Sund H., Blauer G., editors. Symposium on Protein–Ligand Interactions. Berlin: Walter de Gruyter; 1975. pp. 193–206. [Google Scholar]
- 27.Barkley M.D., Riggs A.D., Jobe A., Burgeois S. Interaction of effecting ligands with lac repressor and repressor–operator complex. Biochemistry. 1975;14:1700–1712. doi: 10.1021/bi00679a024. [DOI] [PubMed] [Google Scholar]
- 28.Kepes A. Kinetic studies on galactoside permease of Escherichia coli. Biochim. Biophys. Acta. 1960;40:70–84. doi: 10.1016/0006-3002(60)91316-0. [DOI] [PubMed] [Google Scholar]
- 29.Oehler S., Amouyal M., Kolkhof P., von Wilcken-Bergmann B., Müller-Hill B. Quality and position of the three lac operators of E.coli define efficiency of repression. EMBO J. 1994;13:3348–3355. doi: 10.1002/j.1460-2075.1994.tb06637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehming N., Sartorius J., Oehler S., von Wilcken-Bergmann B., Müller-Hill B. Recognition helices of lac and lambda repressor are oriented in opposite directions and recognize similar DNA sequences. Proc. Natl Acad. Sci. USA. 1988;85:7947–7951. doi: 10.1073/pnas.85.21.7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadler J.R., Sasmor H., Betz J.L. A perfectly symmetric lac operator binds the lac repressor very tightly. Proc. Natl Acad. Sci. USA. 1983;80:6785–6789. doi: 10.1073/pnas.80.22.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simons A., Tils D., von Wilcken-Bergmann B., Müller-Hill B. Possible ideal lac operator: Escherichia coli lac operator-like sequences from eukaryotic genomes lack the central G X C pair. Proc. Natl Acad. Sci. USA. 1984;81:1624–1628. doi: 10.1073/pnas.81.6.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehming N., Sartorius J., Niemöller M., Genenger G., v Wilcken-Bergmann B., Müller-Hill B. The interaction of the recognition helix of lac repressor with lac operator. EMBO J. 1987;6:3145–3153. doi: 10.1002/j.1460-2075.1987.tb02625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller J.H. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 35.Oehler S., Alex R., Barker A. Is nitrocellulose filter binding really a universal assay for protein-DNA interactions? Anal. Biochem. 1999;268:330–336. doi: 10.1006/abio.1998.3056. [DOI] [PubMed] [Google Scholar]
- 36.Rosenberg J.M., Khallai O.B., Kopka M.L., Dickerson R.E., Riggs A.D. Lac repressor purification without inactivation of DNA binding activity. Nucleic Acids Res. 1977;4:567–572. doi: 10.1093/nar/4.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Gorman R.B., Rosenberg J.M., Kallai O.B., Dickerson R.E., Itakura K., Riggs A.D., Matthews K.S. Equilibrium binding of inducer to lac repressor.operator DNA complex. J. Biol. Chem. 1980;255:10107–10114. [PubMed] [Google Scholar]
- 38.Donner J., Caruthers M.H., Gill S.J. A calorimetric investigation of the interaction of the lac repressor with inducer. J. Biol. Chem. 1982;257:14826–14829. [PubMed] [Google Scholar]
- 39.Krämer H., Amouyal M., Nordheim A., Müller-Hill B. DNA supercoiling changes the spacing requirement of two lac operators for DNA loop formation with lac repressor. EMBO J. 1988;7:547–556. doi: 10.1002/j.1460-2075.1988.tb02844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Becker N.A., Kahn J.D., Maher L.J., III Bacterial repression loops require enhanced DNA flexibility. J. Mol. Biol. 2005;349:716–730. doi: 10.1016/j.jmb.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 41.Borowiec J.A., Zhang L., Sasse-Dwight S., Gralla J.D. DNA supercoiling promotes formation of a bent repression loop in lac DNA. J. Mol. Biol. 1987;196:101–111. doi: 10.1016/0022-2836(87)90513-4. [DOI] [PubMed] [Google Scholar]
- 42.Eismann E.R., Müller-Hill B. lac repressor forms stable loops in vitro with supercoiled wild-type lac DNA containing all three natural lac operators. J. Mol. Biol. 1990;213:763–775. doi: 10.1016/S0022-2836(05)80262-1. [DOI] [PubMed] [Google Scholar]
- 43.Friedman B.E., Olson J.S., Matthews K.S. Interaction of lac repressor with inducer, kinetic and equilibrium measurements. J. Mol. Biol. 1977;111:27–39. doi: 10.1016/s0022-2836(77)80129-0. [DOI] [PubMed] [Google Scholar]
- 44.Gilbert W., Müller-Hill B. The lac operator is DNA. Proc. Natl Acad. Sci. USA. 1967;58:2415–2421. doi: 10.1073/pnas.58.6.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riggs A.D., Bourgeois S., Newby R.F., Cohn M. DNA binding of the lac repressor. J. Mol. Biol. 1968;34:365–368. doi: 10.1016/0022-2836(68)90261-1. [DOI] [PubMed] [Google Scholar]
- 46.Weickert M.J., Adhya S. A family of bacterial regulators homologous to Gal and Lac repressors. J. Biol. Chem. 1992;267:15869–15874. [PubMed] [Google Scholar]
- 47.Barbier C.S., Short S.A., Senear D.F. Allosteric mechanism of induction of CytR-regulated gene expression. Cytr repressor-cytidine interaction. J. Biol. Chem. 1997;272:16962–16971. doi: 10.1074/jbc.272.27.16962. [DOI] [PubMed] [Google Scholar]
- 48.von Wilcken-Bergmann B., Müller-Hill B. Sequence of galR gene indicates a common evolutionary origin of lac and gal repressor in Escherichia coli. Proc. Natl Acad. Sci. USA. 1982;79:2427–2431. doi: 10.1073/pnas.79.8.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]