Abstract
In order to revisit the architecture of the catalytic center of the antigenomic hepatitis delta virus (HDV) ribozyme we developed an unbiased in vitro selection procedure that efficiently selected novel variants from a relatively small set of sequences. Using this procedure we examined all possible variants from a pool of HDV ribozymes that had been randomized at 25 positions (425). The isolated set of sequences shows more variability than do the natural variants. Nucleotide variations were found at all randomized positions, even at positions when the general belief was that the specific base was absolutely required for catalytic activity. Covariation analysis supports the presence of several base pairs, although it failed to propose any new tertiary contacts. HDV ribozyme appears to possess a greater number of constraints, in terms of sequences capable of supporting the catalysed cleavage, than do other catalytic RNAs. This supports the idea that the appearance of this catalytic RNA structure has a low probability (i.e. is a rare event), which may explain why to date it has been found in nature only in the HDV. These contrasts with the hammerhead self-cleaving motif that is proposed to have multiple origins, and that is widespread among different organisms. Thus, just because a self-cleaving RNA motif is small does not imply that it occurs easily.
INTRODUCTION
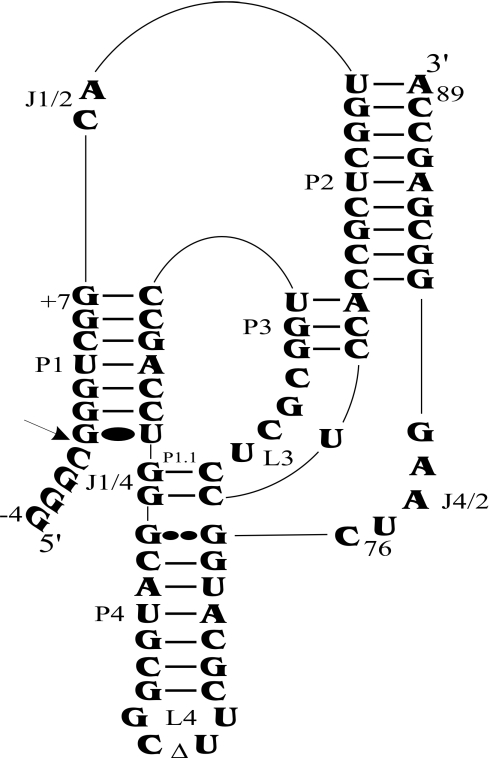
Both the genomic and antigenomic hepatitis delta virus (HDV) RNA strands include a self-cleaving motif essential for their replication (1,2). The HDV self-cleaving motif folds into a double-pseudoknot model secondary structure composed of one stem (P1 stem), two pseudoknots (P1.1 and P2 stems), two stem–loops (P3–L3 and P4–L4) and three single-stranded junctions (J1/2, J1/4 and J4/2) (Figure 1). The catalytic core takes the form of two coaxial helices formed by the stacking of the P1–P1.1–P4 stems and of the P2–P3 stems. The use of X-ray diffraction and nuclear magnetic resonance has provided high-resolution definition of the ternary structure of this catalytic RNA (3–5), and both approaches have contributed to the elucidation of the network of interactions that take place within the catalytic center. However, the number of attempts to confirm the existence of these interactions in solution remains limited. For example, chemically synthesized ribozymes with site-specific functional modifications in the sugar provided relatively good support for the identification of the important 2′-hydroxyl groups of the ribose moieties, although some minor discrepancies were also identified (6,7). The latter experiments also identified several other important groups, although without identifying the chemical residue with which they were interacting. Clearly, much more work is required in order to be able to picture the architecture of the catalytic center in solution.
Figure 1.
Secondary structure of antigenomic self-cleaving HDV RNA. The numbering system used is that of Shih and Been (2). The triangle in the L4 loop indicates the P4 deletion (as compared with the natural variants). The homopurine base pair at the top of the P4 stem is represented by two large dots (G••G), while the Wobble base pair is represented by a single large dot (G•U). The arrow indicates the cleavage site.
The best means of identifying interactions between the bases of an RNA species is the in vitro selection of RNA molecules from a pool of randomized sequences (8). At its fundamental level this method consists of sequentially repeating a process that includes the selection of a specific activity (e.g. the self-cleaving sequence) coupled to an amplification of the RNA molecules with this activity. Selection experiments have been carried out to isolate ribozymes that catalyze a variety of chemical transformations, as well as to investigate the repertoire of functional sequences for a known ribozyme [e.g. Refs (9–12)]. Applying this approach to the study of various versions of HDV cis- and trans-acting ribozymes has permitted the isolation of only a very limited number of sequence variants that were, most of time, reminiscent of either the natural variants or mutations reported previously (13–16). We hypothesized that the low rate of variability was due to the fact that the sequences composing the catalytic core were only partially randomized. If a tertiary interaction involves a non-randomized nucleotide, it creates a bias in favour of variants highly homologous to the wild-type HDV sequence. Recently, it has been demonstrated that the base pairs located at the middle of the P1 stem are important for both P1 stem formation and for the subsequent steps of the cleavage pathway (17). Cross-linking experiments have shown that a thiouridine inserted in position +4 of the P1 stem ends up in close proximity to both C22 and U17 at the junction of the P3–L3 stem–loop (18). This may explain the limited sequence variation observed previously.
In order to revisit the architecture of the catalytic center of the antigenomic HDV ribozyme, we developed an in vitro selection procedure that resulted in a pool of HDV sequences randomized for 25 nt, and then examined all possible sequence variants. The set of sequences selected shows significantly more variability than that of the sequence variants reported in nature. However, HDV ribozyme appears to possess a significantly greater number of constraints, in terms of sequences capable of supporting the catalyzed cleavage, than do other catalytic RNAs of similar size.
MATERIALS AND METHODS
Generation of random libraries
The sequence used as the starting point for all randomized oligonucleotides consisted of modified version of the antigenomic cis-acting HDV sequence (5′GTTTGTTTGTTTGTTGAGGTGGCTCGCCCTTAGCCATGCGAAGCCGCATGCCCAGGTCGGACCGCGAGGAGGTGGCGAGCCATGCCGACCCTTTTTTTTTTTCCCTATAGTGAGTCGTATTAG-3′). Oligonucleotides with randomized positions were synthesized using a manual protocol that ensured the obtention of an equal distribution of each of the 4 nt at each position (19) (Medicorp Inc.). All oligonucleotides serving as templates were desalted, purified by denaturing PAGE (19:1 ratio of acrylamide to bisacrylamide in buffer containing 45 mM Tris–borate, pH 7.5, 7 M urea and 1 mM EDTA, pH 8.0) and then the bands of the correct sizes cut out and the DNA recovered. The library of DNA oligonucleotide randomized in 25 positions was amplified by five cycles of PCR using the sense primer T7polyA (5′-CTAATACGACTCACTATAGGGAAAAAAAAAAGG-3′) and antisense primer OP2 5′-GTTTGTTTGTTTGTTGAGGTGGCTCGC-3′).
PCRs
All PCRs were performed in a final volume of 100 µl containing 10 mM Tris–HCl, pH 8.3, 50 mM KCl, 2 mM MgCl2, 0.001% gelatin, 1.25 mM of each dNTP, 2.5 U Taq DNA polymerase and 1 mM of each primer. Except if mentioned in the text, all PCRs included 30 amplification cycles of 1 min at 94°C, 1 min at 55°C and 1 min at 72°C. The sequences of the various oligonucleotides (Invitrogen) used as primers, as well as for other aspects of this study, are shown in the Supplementary Table 1.
In vitro selection
The SELEX cycle was initiated by the run-off transcription of the PCR products. Briefly, transcriptions were performed in the presence of purified T7 RNA polymerase (10 µg), RNA Guard (24 U; Amersham Biosciences), pyrophosphatase (0.01 U; Roche Diagnostics) and either linearized plasmid DNA (5 µg) or PCR product (2–5 µM) in a buffer containing 80 mM HEPES–KOH, pH 7.5, 24 mM MgCl2, 2 mM spermidine, 40 mM DTT, 5 mM of each NTP and either with or without 50 µCi [α-32P]GTP (3000 Ci/mmol; New England Nuclear) in a final volume of 100 µl at 37°C for 2 h. Upon completion, the reaction mixtures were treated with DNase RQ1 (Amersham Biosciences) at 37°C for 20 min. The RNA was then purified by phenol/chloroform extraction and ethanol precipitation, and was then fractionated by denaturing 8% PAGE. The bands corresponding to the correct sizes for the self-cleaving RNA species were cut out, the transcripts eluted and then ethanol precipitated.
The primer extension reactions were performed using SuperScript II reverse transcriptase for 1 h at 45°C either in the presence or absence of [α-32P]dCTP according to the manufacturer's protocol (Invitrogen). The samples were then incubated for 10 min at 37°C in the presence of 10 µg of RNase A before being fractionated on 8% denaturing PAGE gels. The bands corresponding to 91 nt were recovered and the cDNA extracted and ethanol precipitated. The 3′ end extension reactions were then performed in a total volume of 20 µl containing 25 mM Tris–HCl, pH 6.6, 1.5 mM CoCl2, 200 mM potassium cacodylate, 0.25 ng BSA, 6.25 µM of either dTTP (even cycles) or dATP (odd cycles), 400 U of terminal transferase (Invitrogen). After an incubation of 20 min at 37°C, the terminal transferase was inactivated by heating the mixtures at 75°C for 15 min followed by cooling on ice for 3 min. Then, 20 nmol of dNTP and 100 pmol of either T7polyA or T7polyT primers were added. The mixtures were heated at 65°C for 3 min and chilled on ice and then 5 U of T7 DNA polymerase added prior incubation at 37°C for 15 min. The mixtures were purified by passage through Sephadex G25 columns and the nucleic acids ethanol precipitated. The resulting double-stranded DNAs (dsDNAs) were used as PCR templates for the subsequent selection step.
In general, all of the products of each step were used as template for the subsequent steps, with two exceptions. Firstly, in the case of the PCR amplification, only half of the resulting mixtures were used for the subsequent transcription reactions. Secondly, when the active RNA species dominated a population, only 32 nmol were used as template for the reverse transcription reactions.
Cloning, sequencing and self-cleavage assays
Aliquots of the PCR products were cloned into pGem T vector (Promega). Screening of positive clones was performed by enzymatic hydrolysis using both the SacI and SacII restriction sites. Positive colonies produced an insert of ∼175 bp, while negative ones yielded an insert of only 55 bp. The positive clones inserts were sequenced using the T7 sequencing kit (GE Healthcare). The self-cleavage activity of each variant was verified by in vitro transcription. Briefly, the inserts were PCR amplified using either T7polyA or T7polyT as sense primer and OP2 as antisense primer. After purification of the PCR products on agarose gels, in vitro transcriptions were performed in the presence of [α-32P]GTP and the resulting transcripts analysed on denaturing 6% PAGE gels. Non-cleaved transcripts of several selection cycles were also tested for self-cleavage activity. In these cases, transcripts isolated after gel extraction were resuspended in buffer containing 50 mM Tris–HCl (pH 7.5) and 10 mM MgCl2, incubated at 37°C for 1–4 h and analysed on denaturing 6% PAGE gels. No cleavage activity was detected, confirming that all of the active variants self-cleaved during the transcription step.
RESULTS AND DISCUSSION
Development of an in vitro selection procedure
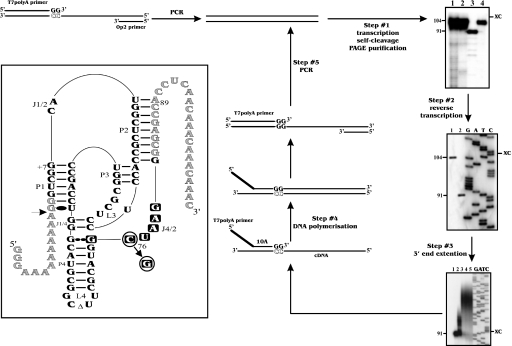
An in vitro selection procedure permitting the use of the largest possible pool of sequences without any bias was developed. All steps described below were optimized using modified versions of the antigenomic cis-acting HDV sequence (Figure 2, inset). The modifications used were included in order to favour the in vitro selection, and were shown to have no influence on the level of self-cleavage of the active sequence. For example, the nucleotides upstream to the cleavage site were replaced by adenosines, a process that should increase the cleavage activity (20). An inactive version with a guanosine, rather than a cytosine in position 76 (G76), served as a negative control that mimicked RNA molecules deprived of self-cleavage activity (Figure 2).
Figure 2.
Strategy of in vitro selection and autoradiograms of 6% PAGE gels. Step #1, run-off transcription: lanes 1 and 2 are ladders produced by alkaline and ribonuclease T1 hydrolysis, respectively, of the inactive G76 RNA strand. Lanes 3 and 4 are the transcription products from the active (C76) and inactive (G76) RNA strands, respectively. The sizes of the RNA species in nucleotides are indicated beside the gel. XC indicates xylene cyanol. Step #2, reverse transcription: lanes 1 and 2 are the primer extension products from the inactive (G76) and active (C76) RNA strands, respectively. Lanes 3–6 are the G, A, T and C sequencing reactions performed using plasmid pGEMT. Step #3, 3′ end extension: lanes 1–5 are aliquots recovered after 0, 1, 5, 10 and 30 min. Lanes 6–9 are the G, A, T and C sequencing reactions performed using plasmid pGEMT. Inset: secondary structure and sequence of the self-cleaving RNA species used. Nucleotides in grey correspond to the binding sequences of the PCR primers. The active and inactive RNA species harbour a C76 and a G76, respectively. The squared nucleotides were randomized in the J4/2 junction experiment. The arrow indicates the cleavage site.
Prior to the first cycle of selection, a PCR was performed using an oligonucleotide corresponding to the T7 RNA promoter followed by the entire HDV antigenomic self-cleaving sequence as template (Figure 2). In addition to producing dsDNA minigenes, five PCR cycles amplified the starting population 12-fold. The run-off transcription was then performed. Since this reaction is performed in the presence of 25 mM MgCl2, the active RNA molecules self-cleaved, producing 91 nt transcripts (Figure 2). After PAGE gel purification, we estimated that, starting from each copy of DNA template, 200 copies of full-length transcripts were produced from each copy of DNA template. Subsequently, a reverse transcription reaction was performed using a 27mer oligonucleotide complementary to both the P2 stem and the adjacent 3′ end 19 nt (Figure 2, inset). When the experiment was performed in the presence of [α-32P]dCTP and analysed on a PAGE gel beside sequencing reactions of the corresponding DNA version, only the bands with the desired lengths were detected (91 and 104 nt; Figure 2). Approximately 95% of the RNA molecules were estimated to be reverse transcribed. After ribonuclease hydrolysis (to remove all traces of template) and gel purification, the 3′ end of the cDNAs were extended using terminal transferase in the presence of dTTP. More than 90% of the molecules were extended during the first 5 min of the reaction by an average of 30–35 adenine residues (Figure 2). Longer incubations did not significantly increase the length of the poly(A) tail; however, the quantity of extended cDNA was found to be slightly larger. Finally, the cycle was completed by a T7 DNA polymerization coupled to a PCR (Figure 2). If the T7 DNA polymerization step was omitted, the PCR was not productive. As shown above, this step significantly contributes to the amplification factor of the cycle of selection. Therefore, the only steps that did not contribute to amplifying the population of molecules appeared to be the reverse transcription and the cDNA extension (i.e. steps 2 and 3). However, the yields of these reactions are largely compensated by both the transcription and PCR amplification (i.e. steps 1 and 4). The global yield per cycle was estimated to be up to 2000 copies [i.e. 12 (PCR amplification in the first cycle) × 200 (transcription) × 0.95 (reverse transcription) ×0.90 (cDNA extension)]. This means that every active ribozyme in the library will have the possibility to be amplified at least 2000 times per selection cycle. All steps in the subsequent selection cycles were similar to those of the first, except that the PCRs now involved 30 cycles.
Randomization of the J4/2 junction (46 variants)
In order to verify the design of the selection procedure we initially performed a test using a partially randomized population of molecules. The positions corresponding to the guanosine residue at the top of the P4 stem, which participates in the formation of the homopurine base pair, and to the five consecutive nucleotides forming the J4/2 junction were randomized (i.e. positions 75–80; Figure 2, inset). Subsequent to the preliminary PCR step an aliquot of the dsDNA was cloned in pGEM T vector, and >50 of the resulting colonies were sequenced. No identical sequences were found, and a perfect ratio of 25% for each base at each position was observed, supporting the idea that the oligonucleotides were successfully randomized without bias.
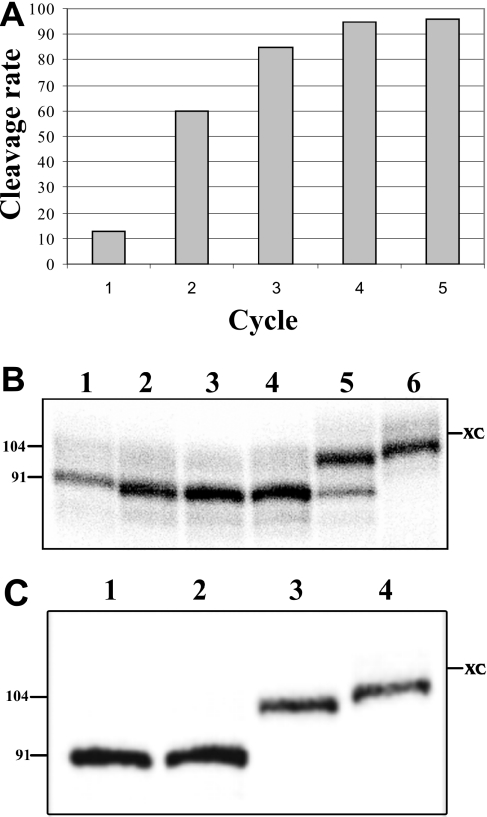
Starting with a concentration of 2 nmol, five cycles of selection were performed. The fraction of self-cleaving sequences increased from 13% after the first cycle to >90% after the fourth (Figure 3A). Run-off transcriptions of several isolated clones were performed, and the percentage of self-cleaving transcripts determined. A typical autoradiogram for the sequence variants obtained after the fourth cycle is presented in Figure 3B. Three sequence variants self-cleaved to near completion, while another self-cleaved only at a 10% level (compare lanes 2–4 to lane 5), indicating that the selection process was not restricted to the most active clones. More generally, we noted that the more frequently a given clone was found, the higher the likelihood that it self-cleaved efficiently. Since our goal was to investigate the sequence variability, rather than isolate the most catalytically active sequences, aliquots of the PCR products from each cycle were cloned and a total of 76 colonies were sequenced (Supplementary Table 2). Forty-five clones were isolated only once, while the remaining sequences were either found more than once in the same cycle (i.e. mainly in the fourth cycle) or in different cycles. Two clones appeared to dominate. The more frequent variant corresponded to the wild-type sequence, suggesting that the wild-type sequence is catalytically more advantageous that any other in the initial population (i.e. J4/2-WT with the sequence G75CUAAG80 was retrieved eight times). The other frequent variant was retrieved four times (i.e. C75GCGAA80). This latter sequence possessed a guanosine residue in position 76, which is unexpected based on the demonstration that this position is normally occupied by the catalytic cytosine. In fact, this variation has never been reported. Examination of all variants showed that 15 other sequences possess a residue other than a cytosine at position 76. One possible explanation for this discrepancy is if the first nucleotide (i.e. the one prior to the homopurine base pair) forms a mismatch, the G76 would consequently be involved in the homopurine base pair and the C77 will act as the catalytic cytosine. If this was indeed the case, then the J4/2 region would include only four single-stranded nucleotides (C60GAA63). This is shorter than what is found in nature, but is also observed to occur in other variants. In order to verify this hypothesis two mutants derived from the original HDV sequence were produced: one possessed the sequence G75G76CAAG80, and the other G75G76GAAG80 (i.e. mutated nucleotides are underlined). The mutant with the C77 self-cleaved as efficient as the wild-type sequence (i.e. Figure 3C, compare lane 2 to lane 1). Conversely, the mutant with the G77 did not exhibit any self-cleavage activity (lane 3). These results support the hypothesis that the cytosine is the catalytic residue, and, consequently, that these self-cleaving RNAs possess both a mismatch adjacent to the homopurine base pair and a shorter J4/2 junction. More importantly, this shows that our in vitro selection procedure allowed us to isolate sequence variants never reported previously.
Figure 3.
Results from the randomization of the J4/2 junction. (A) Profile of the selection performed with the library randomized for the J4/2 junction. (B) Analysis of the self-cleavage activity on a 6% PAGE gel. Lanes 1 and 6 are active (C76) and inactive (G76) control RNA species, respectively. Lanes 2–5 are various representative sequence variants (J4/2N23, J4/2N34, J4/2N27 and J4/2N110, respectively; for the detailed sequences see Supplementary Table 2). The sizes of the RNA species in nucleotides are indicated beside the gel. XC indicates xylene cyanol. (C) Autoradiogram of a 6% PAGE of the self-cleavage assay performed in order to verify the hypothesis of a mismatch adjacent to the homopurine base pair. Lanes 1 and 4 were run-off transcriptions of the active (C76) and inactive (G76) control RNA species, respectively. Lanes 2 and 3 are transcriptions of the G75G76CAAG80 and G75G76GAAG80 mutants.
The nucleotides present at each position of both types of self-cleaving species (i.e. either with or without the mismatch adjacent to the homopurine base pair) were analysed. Considering the correction required for the second type (i.e. G76C77), the consensus sequence appeared to be G/A75CNNNN80. This sequence respects the requirements of the catalysis: a homopurine at the top of the P4 step (positions 41 and 75) and an adjacent catalytic cytosine (position 76). Most of the sequence variations observed were reported previously, supporting the idea of the presence of a bias caused by the conserved sequences of the catalytic core when only a small domain is randomized.
Selection from a larger pool of sequences (425 variants)
We estimated that, starting with 11.2 nmol of oligonucleotide randomized for 25 positions would yield six copies of each possible ribozyme (i.e. the number of copies of each possibility = 11.2 × 10−9 mol × 6.023 × 1023/425 possibilities). The randomizing of any additional position would compromise the objective of having all possible sequences represented in the starting pools. The schematic representation of the HDV sequence randomization is illustrated in Figure 4A. The selection of the positions to be randomized was based mainly on the crystal structures of the genomic HDV ribozyme (3,5). In summary, all of the nucleotides kept intact possess a structural role (e.g. the P1.1, P2 and P4 stems), except for the catalytic C76 that is essential for the chemistry of the reaction; while all of the nucleotides forming the catalytic center, as well as those susceptible to participating in tertiary interactions (e.g. the middle of the P1 stem, the P3–L3 stem–loop and the J4/2 junction), were randomized.
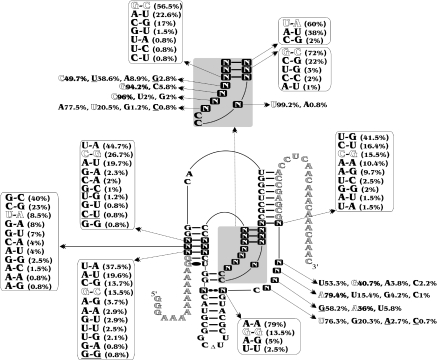
Figure 4.
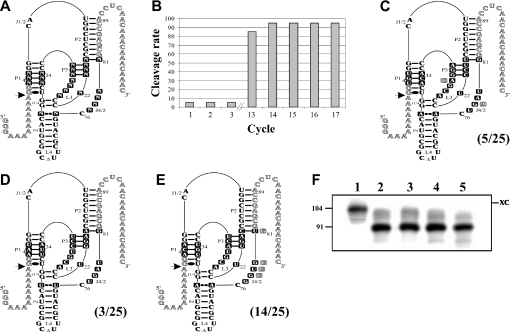
Results from the randomization of 25 positions. (A) Secondary structure and nucleotides of the HDV species. The squared nucleotides were randomized. N indicates the presence of 25% of each residue (A, C, G and U). (B) Evolution of the selection based on the percentage of active RNA species as a function of the cycle number. (C–E) Variants from the three most abundant families. The nucleotide sequences depicted are those of the most active and abundant species of each family. The mutations found in the other members of these families are illustrated. The frequency of the sequence families among the 25 clones is indicated in parentheses. (F) Self-cleavage assays of the three representative RNA species. Lane 1 is the inactive (G76) control RNA species. Lanes 2–4 are variants of the ACA, UGU and AAU families, respectively. Lane 5 is the active (C76) control RNA species.
All experiments were performed using an initial concentration of 5.6 nmol of randomized oligonucleotides. Initially, six cycles of the designed selection procedures were performed. In order to work with all possible sequences in the library during the first cycle, the amplification step was performed using 200 PCRs, and the resulting dsDNA were pooled prior to performing the run-off transcription reaction in a final volume of 15 ml. After the sixth cycle, a band representing ∼50% of the transcription products and possessing the same electrophoretic mobility as the active RNA species was observed. An aliquot of the corresponding PCR product was cloned, and the DNA from several colonies sequenced. Only RNA species with deleted sequences were obtained. The deletions were internal, and occurred at various positions. These 91 nt RNA species did not exhibit any self-cleavage activity. The mechanism responsible for the deletions remains unknown; the most reasonable hypotheses are recombination, reverse transcriptase skipping or template switching during the in vitro selection (21,22). In this form, the protocol used allowed the selection of RNA species of 91 nt, but did not discriminate between inactive and active molecules.
In addition to the deletions within the sequence, the inactive RNA species conserved their 5′ end poly(A) region, and, therefore, the corresponding cDNA possessed two poly(A) regions. Conversely, the cDNA resulting from an active self-cleaving species harboured only one poly(A) tail. In order to avoid amplification of the inactive RNA species, the strategy was modified so as to include an alternation of two sense primers (i.e. the T7polyT and T7polyA primers; Supplementary Figure 1). The 3′ end extension of the cDNA was alternatively performed in the presence of either dATP or dTTP. As a result, the primers can only bind to the newly added tail during the DNA filling step, exclusively yielding a PCR product of 122 bp. Consequently, the 122 bp PCR products corresponding to non-deleted sequences, can produce transcriptional products of 104 nt that will self-cleave into 91 nt transcripts. Conversely, the 122 bp PCR products corresponding to the deleted sequences will only lead to transcripts of 104 nt. This strategy was demonstrated to be an efficient means of discriminating between the deleted and non-deleted sequences (Supplementary Figure 1).
Following this strategy the experiment was repeated from the beginning. During the 12 first cycles, very little to no RNA species in the neighbourhood of 91 nt in size were detected (Figure 4B). The percentage of active sequences then increased drastically to near completion after cycle 13. Using the PCR products from cycle 12, the cycle was repeated in independent experiments under carefully controlled conditions so as to avoid any potential contamination. The almost spontaneous enrichment of the self-cleaving sequence in cycle 13 was consistently observed. Aliquots of the PCR products from cycle 13 were cloned and 25 colonies sequenced. Three families of sequence variants, characterized by the residues in positions 3–5 of the P1 stem that varied from ACA to UGU to AAU, dominated (Figure 4C–E and Supplementary Table 3). Other sequences forming the P1 stem were obtained (e.g. the wild-type sequence); however, only in amounts insufficient to be considered as a family. The self-cleavage activity of each sequence variant was assessed by run-off transcription. A typical autoradiogram of a subsequent PAGE purification showed that a representative variant of each of the three families exhibited a self-cleavage activity equivalent to that of the wild-type sequence (Figure 4F). Finally, aliquots of the PCR products from cycles 13 to 17 were also cloned and sequenced. Only sequence variants belonging to the three families were isolated (Supplementary Table 3).
In order to avoid the domination of these three families, a second PCR amplification strategy was designed using oligonucleotides possessing three additional residues at the 3′ end. These three positions were randomized in order to obtain a population of 64 different sequences as sense primers (i.e. 43). The rationale behind this design was that when the members of the ACA, UGU and AUU families have consumed their respective primer molecules during the amplification step, the possibility of amplifying the other 61 sub-populations remains, if indeed such a population existed in the pool. Cycles 14 and 15 were repeated using these randomized oligonucleotides. The fraction of self-cleaving variants was found to be smaller in cycle 14, and to then increase by 65% in cycle 15. This result was expected because it is the less active sequence variants that are amplified under these conditions. When the same strategy was applied after cycle 12, i.e. to say before the exponential enrichment, the drastic increase in the self-cleaving fraction was not observed. The fractions of self-cleaving RNA species increased gradually up to 80% after cycle 17. Clearly, this result supports the idea that the randomized oligonucleotides in the PCR permitted the appearance of the less abundant sequence variants. After each cycle an aliquot of the PCR products was cloned, and a total of 255 colonies were sequenced (i.e. 55 after cycle 13, 54 after cycle 14, 50 after cycle 15, 46 after cycle 16 and 50 after cycle 17; Supplementary Table 3). These sequences included the wild-type version, members of the three families characterized previously and several new variants for a total of 45.
Sequence variation in the catalytic center
Together, the different experiments performed with the library randomized in 25 positions resulted in the sequencing of 330 clones. All sequence variants were compiled in a database (Supplementary Table 3), and a summary is presented in Figure 5. The selected ribozymes exhibited different relative rates of cleavage. For example, some variants self-cleaved as efficiently as the wild-type ribozyme (e.g. variants 25N1311, 25N1317 and 25N1316), while others exhibited only minimal or residual self-cleavage activity (e.g. <5% for variants 25N1519, 25N1622 and 25N1310). However, most of the variants possessed a relative self-cleavage rate located somewhere between these two limits. Nucleotide variations were found at all randomized positions, even when the general belief was that a specific base was an absolute requirement for catalytic activity (i.e. U22 and G27). In these specific cases independent experiments revealed that these sequence variants exhibited only residual self-cleavage activity. In addition, variants with mutations in the base pairs comprising the P3–L3 stem–loop were also retrieved, even if they were previously thought to be highly conserved. This is considerably more variation than is observed in the 124 natural antigenomic HDV species in the daily, up-to-date, subviral RNA database (23). In fact, only eight natural substitutions were found at the positions corresponding to those that were randomized (see the underlined nucleotides in Figure 5). The higher number of mutations obtained from the in vitro selection procedure suggests to us that either other selection pressures occur in nature, thereby providing a net advantage for the wild-type sequence, or that the sequence variability potential has not yet been explored in the cellular environment. The latter possibility appears to be improbable, based on the fact that natural HDV species show a relatively large number of mutations in both the P2 and P4 stems.
Figure 5.
Compilation of sequence variations. All sequence variations retrieved from the library of the oligonucleotides randomized in 25 positions (i.e. identified by a squared letter N). The mutations also retrieved in natural species are underlined. Shadowed nucleotides correspond to the wild-type sequence.
Some of the sequence variations observed by in vitro selection have been reported previously in studies with either cis- or trans-acting antigenomic HDV ribozyme (1,24), while others have yet to be reported. For example, the variation of the P3 stem observed here is in good agreement with that seen previously (24). The size of the P3 stem (i.e. 3 bp) is critical, and there is a preference for either an AU or an UA bp at the top of the P3 stem and a GC or a CG bp for the other positions. Second, it has been shown that the presence of a homopurine bp at the top of the P4 stem is mandatory (24). In natural species this is always a GG bp, while we observed a large dominance of AA bp. This supports the idea that differences exist between the selection pressures taking place in nature versus in the laboratory. However, the symmetry in the nature of the bases was almost perfect, that is to say either two As or two Gs were present. Third, the in vitro selection process yields a relatively limited set of base pairs for the middle of the P1 stem even though there is no known restriction on these base pairs. Moreover, the presence of mismatches was tolerated at these positions, although only in a limited number of variants. Lastly, the C18G81 bp at the bottom of the P2 stem was considered to be a conserved Watson–Crick base pair (24). According to the in vitro selection results, many different nucleotides, either forming a Watson–Crick base pair or not, may be tolerated in these positions. Clearly, a lot more biochemical experiments are required in order to establish the effect of each specific mutation and to decipher the role of each residue.
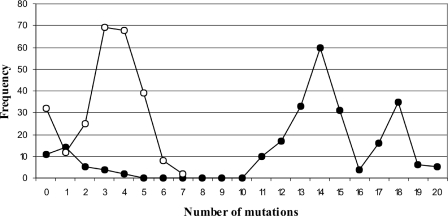
The collection of sequences was analysed not only on an individual basis to find new mutations, but also together. For example, the 255 sequences from the last selection (i.e. in order to avoid any bias observed previously) were used to study the number of mutations observed per variant (i.e. the Hamming distance from the wild-type sequence) as a function of the frequency among the 255 clones (Figure 6). When all of the sequence variation was considered, 86% of the clones included at least 11 mutations as compared with the wild-type sequences. The highest frequency was found for the clones of the sequences with 14 mutations. This showed the large variation among the sequence variants retrieved. There were even two variants that possessed 20 out of 25 possible mutations (i.e. 25N1622 and 25N1310). This analysis was repeated using only the single-stranded positions (i.e. removing both the P1 and P3 stems, as well as the homopurine base pair at the top of the P4 stem). In that case, 82% of the clones include at least three mutations. The highest frequency of mutations was observed in the variants possessing five mutations, and the clones with the largest number of mutations possessed 9 out of 11 mutated nucleotides. This is in agreement with an independent study showing that changing nearly half of the HDV ribozyme may lead to the observation of variants with relatively efficient self-cleavage activities (depending the position of the mutations) (25). More importantly, this shows that the collection of sequences exhibited a lot of variability, and should be ideal for nucleotide covariation analysis.
Figure 6.
Graph of the number of mutations observed per sequence variant as a function of frequency amongst the 255 clones. The closed circles represent data obtained when considering the 25 randomized positions, while the open circles indicate that obtained when considering a subset of only 11 randomized positions (see the text).
Analysis of nucleotide covariation was performed using various bioinformatic software packages. Covariation was observed for the Watson–Crick base pairs forming both the P1 and the P3 stems. Moreover, covariation analysis supports the presence of the homopurine base pair at the top of the P4 stem. However, no covariation that would support the presence of a tertiary contact was detected. This might be due to the fact that the number of different variant was relatively small (i.e. 45 variants). However, it is not impossible that additional sequencing efforts may lead to the observation of covariation.
HDV self-cleaving RNA motif is highly constrained
In vitro selection experiments using an unbiased sample of random sequences to find self-cleaving motifs showed that, under near-physiological conditions, the hammerhead ribozyme motif was the most common RNA structure capable of self-cleavage (12). Since selection and amplification should favour the simplest motifs, and hammerheads were the most common motifs found, the latter appears to be simplest than HDV. If we accept the premise that the more complex the active tertiary structure of a given RNA species, the more difficult its generation will be either in nature or by in vitro selection, then clearly the HDV self-cleaving motif has to be more complex than that of the hammerhead. Compared with the hammerhead ribozyme, the architecture of the catalytic center of the HDV self-cleaving motif is most likely highly constrained including several tertiary contacts that are not of the base–base type. Physical evidence also supports the idea that HDV possesses a compact catalytic center. For example, it retains activity at temperatures of about 80°C and in buffer containing either 5 M urea or 18 M formamide (26).
The in vitro selection experiment using an unbiased sample also led to the proposition that the hammerhead self-cleaving motif most likely has multiple origins (12). This may explain why the hammerhead self-cleaving motif is widespread among different organisms, including viroids, satellite RNA of plant viruses, newt, schistosomes, insects and plants (27). Conversely, to date the HDV self-cleaving motif has been found (in nature) only in the HDV, which may support the idea that, due to its complexity, its appearance has a lower probability (i.e. is a rare event). Just because a self-cleaving RNA motif is small does not imply that it appeared easily. The complexity of the structure appears to be the more important selective pressure.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We are grateful to Dominique Lévesque for technical assistance. This work was supported by grants from the Canadian Institute of Health Research (CIHR; grant number MOP-44002 and EOP-38322) to J.P.P. The RNA group is supported by grants from Université de Sherbrooke. A.N. was the recipient of a pre-doctoral fellowship from Ministère de l'Enseignement Supérieur de la Tunisie. J.P.P. holds the Canada Research Chair in Genomics and Catalytic RNA. Funding to pay the Open Access publication charges for this article was provided by CIHR grant number MOP-44002.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bergeron L.J., Ouellet J., Perreault J.P. Ribozyme-based gene-inactivation systems require a fine comprehension of their substrate specificities; the case of delta ribozyme. Curr. Med. Chem. 2003;10:2589–2597. doi: 10.2174/0929867033456486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shih I.H., Been M.D. Catalytic strategies of the hepatitis delta virus ribozymes. Annu. Rev. Biochem. 2002;71:887–917. doi: 10.1146/annurev.biochem.71.110601.135349. [DOI] [PubMed] [Google Scholar]
- 3.Ferré D'Amaré A.R., Zhou K., Doudna J.A. Crystal structure of a hepatitis δ virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka Y., Tagaya M., Hori T., Sakamoto T., Kurihara Y., Uesugi S. Imino proton NMR analysis of HDV ribozyme: nested double pseudoknot structure and Mg2+ ion-binding site close to the catalytic core in solution. Nucleic Acids Res. 2002;30:766–774. doi: 10.1093/nar/30.3.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ke A., Zhou K., Ding F., Cate J., Doudna J.A. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 6.Fiola K., Perreault J.P. Kinetic and binding analysis of the catalytic involvement of ribose moieties of a trans-acting δ ribozyme. J Biol. Chem. 2002;277:26508–26516. doi: 10.1074/jbc.M203468200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishikawa F., Shirai M., Nishikawa S. Site-specific modification of functional groups in genomic hepatitis delta virus (HDV) ribozyme. Eur. J. Biochem. 2002;269:5792–5803. doi: 10.1046/j.1432-1033.2002.03280.x. [DOI] [PubMed] [Google Scholar]
- 8.Wilson D.S., Szostak J.W. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 9.Johnston W.K., Unrau P.J., Lawrence M.S., Glasner M.E., Bartel D.P. RNA-catalyzed RNA polymerization: accurate and general RNA-templated primer extension. Science. 2001;292:1319–1325. doi: 10.1126/science.1060786. [DOI] [PubMed] [Google Scholar]
- 10.Curtis E.A., Bartel D.P. New catalytic structures from an existing ribozyme. Nature Struct. Mol. Biol. 2005;12:994–1000. doi: 10.1038/nsmb1003. [DOI] [PubMed] [Google Scholar]
- 11.Li Y., Sen D. A catalytic DNA for porphyrin metallation. Nature Struct. Biol. 1996;3:743–747. doi: 10.1038/nsb0996-743. [DOI] [PubMed] [Google Scholar]
- 12.Salehl-Ashtiani K., Szostak J.W. In vitro evolution suggests multiple origins for the hammerhead ribozyme. Nature. 2001;414:82–84. doi: 10.1038/35102081. [DOI] [PubMed] [Google Scholar]
- 13.Suh Y.A., Kumar P.K.R., Kawakami J., Nishikawa F., Taira K., Nishikawa S. Systematic substitution of individual bases in two important single-stranded regions of the HDV ribozyme for evaluation of the role of specific bases. FEBS Lett. 1993;326:158–162. doi: 10.1016/0014-5793(93)81782-u. [DOI] [PubMed] [Google Scholar]
- 14.Kawakami J., Kumar P.K.R., Suh Y.A., Nishikawa F., Kawakami K., Taira K., Othsuka E., Nishikawa S. Identification of important bases in a single-stranded region (SSrC) of the hepatitis delta (δ) virus ribozyme. Eur. J. Biochem. 1993;217:29–36. doi: 10.1111/j.1432-1033.1993.tb18214.x. [DOI] [PubMed] [Google Scholar]
- 15.Branch A.D., Polaskova J.A. 3-D models of the antigenomic ribozyme of the hepatitis delta agent with eight new contacts suggested by sequence analysis of 188 cDNA clones. Nucleic Acids Res. 1995;23:4180–4189. doi: 10.1093/nar/23.20.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishikawa F., Kawakami J., Chiba A., Shirai M., Kumar P.K.R., Nishikawa S. Selection in vitro of trans-acting genomic human hepatitis delta virus (HDV) ribozymes. Eur. J. Biochem. 1996;237:712–718. doi: 10.1111/j.1432-1033.1996.0712p.x. [DOI] [PubMed] [Google Scholar]
- 17.Ananvoranich S., Perreault J.P. Substrate specificity of delta ribozyme cleavage. J. Biol. Chem. 1998;273:13182–13188. doi: 10.1074/jbc.273.21.13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ouellet J., Perreault J.P. Cross-linking experiments reveal the presence of novel structural features between a hepatitis delta virus ribozyme and its substrate. RNA. 2004;10:1059–1072. doi: 10.1261/rna.7230604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy M., Ellington A.D. In vitro selection of a deoxyribozyme that can utilize multiple substrates. J. Mol. Evol. 2002;54:180–190. doi: 10.1007/s00239-001-0066-1. [DOI] [PubMed] [Google Scholar]
- 20.Deschênes P., Lafontaine D.A., Charland S., Perreault J.P. Nucleotides -1 to -4 of hepatitis delta ribozyme substrate increase the specificity of ribozyme cleavage. Antisense Nucleic Acid Drug Dev. 2000;10:53–61. doi: 10.1089/oli.1.2000.10.53. [DOI] [PubMed] [Google Scholar]
- 21.Svarovskaia E.S., Cheslock S.R., Zhang W.H., Hu W.S., Pathak V.K. Retroviral mutation rates and reverse transcriptase fidelity. Front. Biosci. 2003;8:117–134. doi: 10.2741/957. [DOI] [PubMed] [Google Scholar]
- 22.Lehman N., Unrau P.J. Recombination during in vitro evolution. J. Mol. Evol. 2005;61:245–252. doi: 10.1007/s00239-004-0373-4. [DOI] [PubMed] [Google Scholar]
- 23.Pelchat M., Rocheleau L., Perreault J., Perreault J.P. Subviral RNA: a database of the smallest known auto-replicable RNA species. Nucleic Acids Res. 2003;31:444–445. doi: 10.1093/nar/gkg026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Been M.D., Wickham G.S. Self-cleaving ribozymes of hepatitis delta virus RNA. Eur. J. Biochem. 1997;247:741–753. doi: 10.1111/j.1432-1033.1997.00741.x. [DOI] [PubMed] [Google Scholar]
- 25.Schultes E.A., Bartel D.P. One sequence, two ribozymes: implications for the emergence of new ribozyme folds. Science. 2000;289:448–452. doi: 10.1126/science.289.5478.448. [DOI] [PubMed] [Google Scholar]
- 26.Doherty E.A., Doudna J.A. Ribozyme structure and mechanism. Annu. Rev. Biochem. 2000;69:597–615. doi: 10.1146/annurev.biochem.69.1.597. [DOI] [PubMed] [Google Scholar]
- 27.Pzybilski R., Graf S., Lescoute A., Nellen W., Westhof E., Steger G., Hammann C. Functional hammerhead ribozymes naturally encoded in the genome of Arabidopsis thaliana. Plant Cell. 2005;17:1877–1885. doi: 10.1105/tpc.105.032730. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.