Abstract
The major problem of using somatic mutations as markers of malignancy is that the clinical samples are frequently containing a trace amounts of mutant allele in a large excess of wild-type DNA. Most methods developed thus far for the purpose of tickling this difficult problem require multiple procedural steps that are laborious. We report herein the development of a rapid and simple protocol for detecting a trace amounts of mutant K-ras in a single tube, one-step format. In a capillary PCR, a 17mer peptide nucleic acid (PNA) complementary to the wild-type sequence and spanning codons 12 and 13 of the K-ras oncogene was used to clamp-PCR for wild-type, but not mutant alleles. The designated PNA was labeled with a fluorescent dye for use as a sensor probe, which differentiated all 12 possible mutations from the wild-type by a melting temperature (Tm) shift in a range of 9 to 16°C. An extension temperature of 60°C and an opposite primer 97 nt away from the PNA were required to obtain full suppression of wild-type PCR. After optimization, the reaction detected mutant templates in a ratio of 1:10 000 wild-type alleles. Using this newly devised protocol, we have been able to detect 19 mutants in a group of 24 serum samples obtained from patients with pancreatic cancer. Taken together, our data suggest that this newly devised protocol can serve as an useful tool for cancer screening as well as in the detection of rare mutation in many diseases.
INTRODUCTION
Somatic mutations are useful markers for the early detection of cancer (1,2). For example, the K-ras mutation in codons 12 and 13 occurs in 80–90% of pancreatic cancer and 35–50% of colorectal cancer (3–5). However, detecting such mutations for population screening purposes presents a challenge for the clinical laboratory because of the large excess of wild-type DNA usually found in clinical samples, especially in body fluids or stool. This excess of wild-type DNA can exhaust essential reagents during PCR, and tends to mask the mutant signal during detection assays. The general strategy used to date to overcome this difficulty was to employ suppression of the wild-type allele or enrichment of the mutant allele during amplification, and followed by using a detection procedure that provided a sufficient resolution to reveal the mutant signals. Methods used to enrich mutant template level include allele-specific amplification (6), restriction enzyme digestion of wild-type DNA (4,7,8) and sequence-specific ligation (9). Methods used to detect mutant signal include: (i) distinguishing the conformational or length differences by gel electrophoresis (10–12) or denaturing high-performance liquid chromatography (13); (ii) detecting short sequences by mass spectrometry (14,15); and (iii) detecting nucleotide sequence changes by melting curve analysis (16), endonuclease V reaction (17) or hybridization on a microarray chip (18). However, most of these methods are not convenient for use in clinical laboratories owing to multiple procedural manipulations that are time-consuming and cost ineffective. Most importantly, the risk of contamination during multiple transfers was increased.
Recently, the peptide nucleic acid (PNA)-based PCR procedure has been developed for the enrichment of mutant alleles (19). PNA is a synthetic DNA analog in which the normal phosphodiester backbone is replaced with a 2-aminoethylglycine chain. Its nucleobases complement DNA or RNA in the normal A-T and G-C geometry (20–22). Two important features make PNA a superior PCR clamp for specific alleles. PNA cannot serve as a primer for polymerization, nor can it be a substrate for exonuclease activities of Taq polymerase. In addition, the melting temperature (Tm) of a perfectly matched PNA–DNA duplex is higher than that of DNA–DNA of the same length, but a single mismatch destabilizes the PNA–DNA hybrids, causing a Tm shift of 10–18°C (23). Therefore, PNA can specifically block primer annealing or chain elongation on a perfectly matched template without interfering with these reactions on templates with mismatched bases (15,24,25). In addition, the large Tm difference between perfectly matched and mismatched hybrids makes PNA a good sensor of point mutations. For example, a PNA sensor probe has been used to detect GNAS mutations after PCR (26).
The use of melting curve analysis in combination with fluorescent probes provides a powerful tool for the detection of single base alterations. The hybridization probe system is most widely used for this purpose. This system usually comprises a pair of oligonucleotides—the anchor and the sensor—each labeled with a different fluorescent dye, such that fluorescence energy transfer occurs between the two when they anneal adjacent sites of a complementary PCR strand (27). The melting curve profile of the sensor probe (designed to anneal to the variable region), allows for homogeneous genotyping in a closed tube (27). Recently, hybridization probes was combined with PNA-mediated PCR clamping for detection of variant bcr-abl allele in leukemia (28), and of K-ras mutation in pancreatic cancer (29,30).
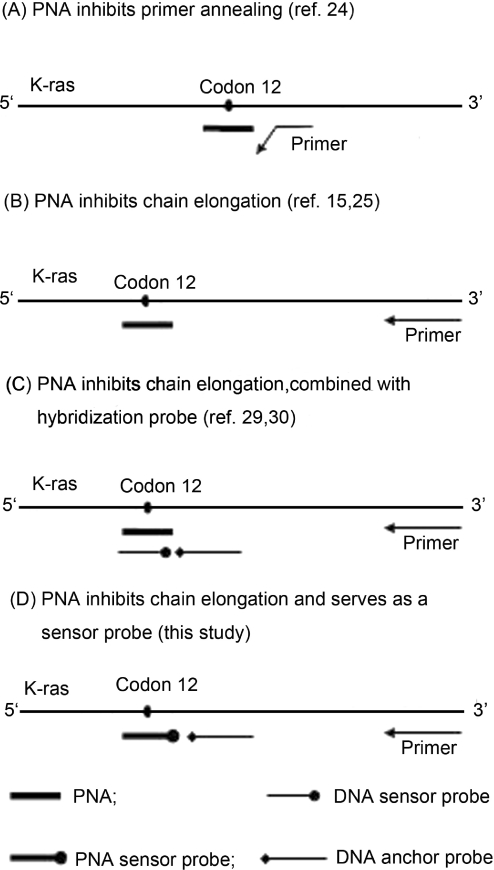
In this report we devised a convenient PCR procedure for the detection of a trace amounts of mutant K-ras in a large excess of wild-type DNA. The key feature of this procedure is that a PNA oligomer serves as both PCR clamp and sensor probe, which allows for differentiation from the wild-type of sequence alterations in codons 12 and 13. The principle of this design, as well as other methods for PNA-mediated PCR clamping, is depicted in Figure 1.
Figure 1.
The schematic concepts using PNA-mediated PCR for detection of mutant K-ras. For simplification, only one template strand is shown.
MATERIALS AND METHODS
Primers and probes
Three forward primers (F1, F2 and F3) and a reverse primer (R) were designed to amplify K-ras fragments in exon 1. The sensor probe covering the variable region is a 17mer PNA labeled with fluorescein at the N-terminus (equivalent to the 5′-end of a DNA oligomer) The anchor probe is a 44mer DNA labeled with fluorescent dye LC-Red 640 at the 3′-end. PCR primers and the anchored probe were provided by TIB MOLBIOL (Berlin, Germany). PNA was provided by Applied Biosystems (Forster City, CA, USA). Sequences of primers and probes used in this study are listed in Table 1.
Table 1.
Primers and probes used in this study
| Name | Sequence (5′–3′ for DNA or N to C for PNA) | Positiona |
|---|---|---|
| Primers | ||
| F1 | atgactgaatataaacttgtggta | 1 ∼ 24 |
| F2 | attaaccttatgtgtgacat | −70 ∼ −51 |
| F3 | tactggtggagtatttgata | −94 ∼ −75 |
| R | caagatttacctctattgtt | 121 ∼ 102 |
| Probes | ||
| Sensor (PNA) | (FIuorescein)-cctacgccaccagctcc | 44 ∼ 28 |
| Anchor | gtccacaaaatgattctgaattagctgtatcgtcaaggcactct-(640) | 90 ∼ 47 |
| Primers for site-directed mutagenesis | ||
| M1Xb | atgactgaatataaacttgtggtagttggagctXgtggcgta | 1 ∼ 42 |
| M2X | atgactgaatataaacttgtggtagttggagctgXtggcgta | 1 ∼ 42 |
| M3X | atgactgaatataaacttgtggtagttggagctggtXgcgta | 1 ∼ 42 |
| M4X | atgactgaatataaacttgtggtagttggagctggtgXcgta | 1 ∼ 42 |
aA of the ATG start codon is designated as position 1.
bX represents either A, T or C. The number represents the position of four guanines in codons 12 and 13. Therefore, ‘M1C’ would indicate a G to C change at the first guanine.
Templates
Wild-type genomic DNA was purified from the cultured human leukemia cell line K-562 (BCRC60007; from Resource Collection and Research Center, Hsinchu, Taiwan) using a QIAamp DNA-blood-mini kit (Qiagen, Hilden, Germany). Purified DNA was quantitated by ultraviolet (UV) spectrophotometry and stored at −20°C until use. Mutant templates were either purified from cell line SW480 (BCRC60249) by the QIAamp kit or synthesized by PCR-based site-directed mutagenesis. SW480 cells harbor a G to T mutation at the second base of codon 12 in the K-ras gene. The PCR-based site-directed mutagenesis was performed using one of 12 different primers complementary to the variable region of the K-ras gene but bearing a mismatch at either the first or second position of codon 12 or 13 (Table 1). Sequences of all the synthesized mutant templates were verified by an autosequencer. The synthesized templates were purified with QIAquick PCR purification kit (Qiagen) and diluted with 10 mM Tris–HCl (pH 8.0) containing 1 µg/ml salmon sperm DNA and stored at −20°C before use.
PCR analysis
PCR was performed in a 20 µl reaction mixture containing 1× reaction buffer [50 mM Tris (pH 8.5), 3 mM MgCl2, 500 µg/ml BSA, 200 µM each deoxyribonucleoside triphosphate], 0.5 µM forward and reverse primers, 0.25 µM PNA, 0.5 U Platinum Taq (Invitrogen, Carlsbad, CA, USA), and templates. The amplification was performed on a LightCycler (Roche Diagnostics, Mannheim, Germany) starting with a 2 min denaturation at 94°C, then running for 50 cycles as follows: 94°C held for 0 s for denaturation; 70°C held for 5 s for PNA binding, 56°C held for 0 s for primer annealing and 10 s at various temperatures for extension. Melting analysis was performed after a 20 s denaturation at 95°C and then decreasing the temperature to 45°C at ramp rate 0.7°C/s. Detection was in channel F2 for the LC-Red 640 labeled probes.
Detection of K-ras mutation in patients' sera
Serum samples were collected from 24 pancreatic cancer patients in Chang Gung Memorial Hospital, Taiwan. Control samples were collected from 10 healthy volunteers. DNA was extracted from 200 µl aliquots of serum using a QIAamp DNA-blood-mini kit (Qiagen). One-fourth of the eluted DNA was used as PCR template. PCR was performed using F2 as the forward primer and 60°C (clamp condition) or 72°C (non-clamp condition) as extension temperature. Presumptive mutants were identified as those samples having melting peaks close to 60°C. To confirm results and determine specific mutation types, PCR products were separated on a 2% agarose gel, eluted, and then sequenced by an automated DNA sequencer.
RESULTS
The 17mer PNA probe differentiated wild-type K-ras from mutants
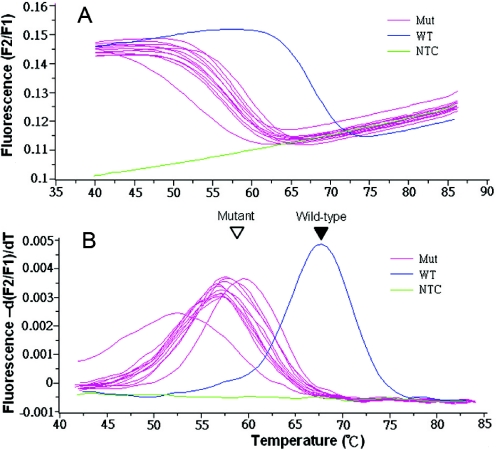
To determine whether our hybridization probes could differentiate wild-type from mutant K-ras, we made 12 mutant templates by PCR-based site-directed mutagenesis. These 12 mutants cover all possible single nucleotide mutations in codons 12 and 13 that result in amino acid changes. In the regular PCR conditions, all of these mutant templates as well as the wild-type template can be amplified. Melting curve analysis revealed that the Tm of the PNA probe bound to wild-type K-ras is 69°C, but varies between 53 and 60°C when bound to the different mutant templates (Figure 2). This indicates that a single nucleotide change causes a Tm shift of 9–16°C for the 17mer PNA probe.
Figure 2.
The PNA sensor probe differentiated the 12 possible K-ras mutations in codons 12 and 13 from the wild-type. Mutant K-ras were generated by PCR-mediated site-directed mutagenesis and were used as PCR templates to test the resolution of the PNA sensor probe. All the PCR were performed under non-clamping conditions, using primer F1, and 72°C for extension. Melting curves (A) and melting peaks (B) were plotted after PCR. Filled and open arrowheads indicate wild-type and mutant melting peaks, respectively. Mut, mutant; WT, wild-type; NTC, no template control.
PNA suppresses PCR of wild-type but not mutant templates at a lower extension temperature
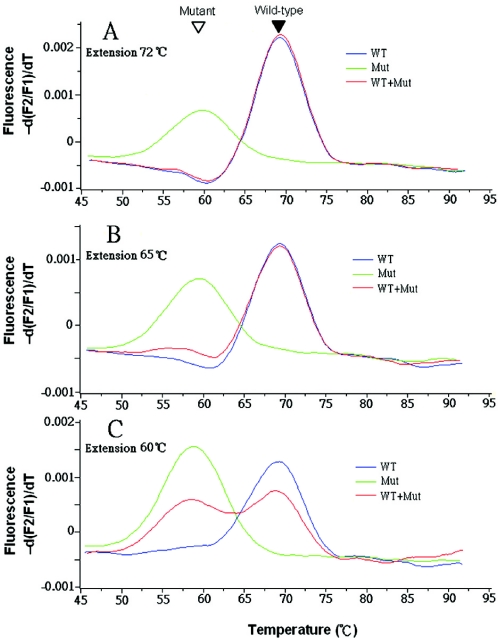
To determine whether the conventional PCR extension step is conducted at a temperature that is too high for the PNA to inhibit elongation through the wild-type template, a mixture of 1 ng of mutant genomic DNA and a 100-fold excess of wild-type DNA was used as template for real-time PCR performed using different extension temperatures. When doing extension at 72°C or 65°C, only the wild-type melting curve was seen, indicating that no obvious clamp occurred (Figure 3A and B). After lowering the extension temperature to 60°C, which is around the Tm of mutants and 9°C lower than the wild-type Tm, the mutant peak started to appear (Figure 3C). Further decreases the extension temperature, to 55°C or 50°C, slowed PCR amplification of both wild-type and mutants (data not shown).
Figure 3.
Extension temperature affected the efficiency of the PNA-mediated PCR clamp. PCR were performed using either 100 ng wild-type (WT), 1 ng mutant (Mut) or a mix containing 100 ng wild-type and 1 ng mutant genomic DNA (WT + Mut) as templates and F1 as the forward primer. Extension temperatures were 72 (A), 65 (B) or 60°C (C). Filled and open arrowheads indicate wild-type and mutant melting peaks, respectively.
Distant primers enhance PCR clamping of wild-type templates
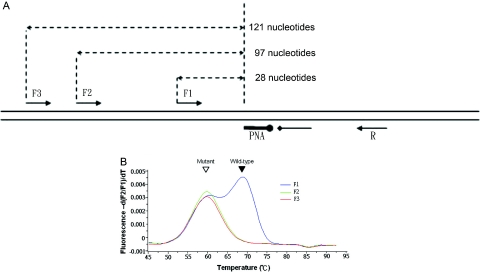
In addition to extension temperature, we demonstrated that the position of opposite primers also influences the PNA-mediated PCR clamping. When using primer F1, which is 28 nt away from the PNA binding site (Figure 4A), wild-type amplification could not be completely suppressed (Figures 3C and 4B). However, when using primers F2 and F3, which are more than 97 nt from the PNA binding site, wild-type amplification was successfully inhibited, and only the mutant was amplified (Figure 4B).
Figure 4.
Primer position affects the clamp efficiency of PNA-mediated PCR clamp. (A) Relative positions and orientations of PCR primers and probes. (B) Melting peaks after clamp-PCR using 100 ng wild-type plus 1 ng mutant genomic DNA as templates and either F1, F2 or F3 as the forward primer. Filled and open arrowheads indicate wild-type and mutant melting peaks, respectively.
The assay detects rare mutants in a large excess of wild-type DNA
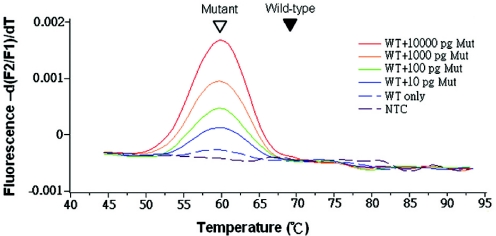
To determine the assay's limits for detecting rare mutants, we used different amounts of mutant genomic DNA mixed with 100 ng of wild-type genomic DNA as templates. When performing PCR under optimal conditions, using primer F2 and doing extension at 60°C, as little as 10 pg of mutant DNA (about three genomes, or 1:10 000) was detected by melting curve analysis (Figure 5).
Figure 5.
Assay sensitivity for detection of K-ras mutants in a large excess of wild-type DNA. 100 ng wild-type genomic DNA plus various amounts of mutant genomic DNA were used as templates for clamp-PCR. Under the optimal condition using F2 as the forward primer and 60°C as extension temperature, the assay detected the signal from as few as 10 pg mutant DNA without interference from the wild-type DNA. Filled and open arrowheads indicate wild-type and mutant melting peaks, respectively.
The assay detects K-ras mutations in serum DNA
Of 24 purified serum DNA samples from pancreatic cancer patients, 19 (79%) had melting peaks close to 60°C and were presumed to be mutants; the other five samples without obvious peaks were presumed to be wild-type. Sequence analysis confirmed that each of the 19 ‘expected mutants’ had a point mutation in codon 12 (Table 2). Note that all the ‘expected wild-type’ could not be amplified during clamp-PCR: these samples were subjected to PCR under non-clamp condition to ensure that they contained amplifiable DNA (data not shown). In addition, using our assay, all samples from the 10 healthy controls were determined to be wild-type (data not shown).
Table 2.
Type of K-ras mutations found in serum DNA from pancreatic cancer patients
| (Codon) mutationa (amino acid change) | Number | Mutant melting peak |
|---|---|---|
| (12) GGT → GTT (Gly → Val) | 16 | Yes |
| (12) GGT → AGT (Gly → Ser) | 1 | Yes |
| (12) GGT → GAT (Gly → Asp) | 1 | Yes |
| (12) GGT → GTT, GATb (Gly → Val, Asp) | 1 | Yes |
| Wild-type | 5 | No |
| Total | 24 |
aAltered bases are underlined.
bTwo mutations co-exist in the patient.
DISCUSSION
The essence of this study was the development of a PCR procedure for the detection of trace K-ras mutations using PNA as a PCR clamp as well as a sensor probe. The uniqueness of our newly developed method is that PCR amplification, mutant enrichment and mutation detection can be accomplished in a single tube on a LightCycler without having to go through several laborious procedures including electrophoresis, hybridization and enzymatic reaction. Next, the Tm difference between perfectly matched templates and mismatched templates is larger for PNA probes than oligonucleotide probes. For this reason, all alleles with single base changes can be readily distinguished from wild-type by melting peak analysis. Most importantly, only one pair of primers and probes is required to detect all 12 possible mutations from the wild-type in codons 12 and 13 of K-ras. All these advantages greatly simplifies the manipulating procedure and thus can be potentially useful in multiplex assays. Finally, our procedure can detect as low as 10 pg of mutant genomic DNA against a background of 100 ng of wild-type DNA (a 1:10 000 ratio).
Using PNA as both sensor probe and PCR clamp is an important feature of our method. Association and dissociation of PNA and its complementary DNA is revealed by melting curve analysis. This allows for easy optimization of thermal conditions for PCR clamping of wild-type, but not mutant templates. The Tm of the PNA-wild-type templates is 69°C: at 72°C, the extension temperature used in conventional PCR, less than half of these templates are associated with PNA, leading us to hypothesize that the PNA clamp would not be efficient under this temperature. As indicated in the Figure, we found that in order to inhibit the wild-type amplification to a full extent, an extension temperature of 60°C, 9°C lower than the wild-type Tm, should be used. Conversely, an extension temperature of 60°C has a minimal effect on mutant amplification. Since the Tm's of all the mutants studied fall between 53 and 60°C, we can conclude that at a temperature above Tm, Taq polymerase can readily repel a PNA block and continue the chain elongation.
The fact that primer position also affected the efficiency of clamping is somewhat surprising. Our explanation is that primer position determines the running off distance from the PNA binding site. At the extension temperature (60°C) used in PCR during this experiment, the progression of Taq polymerase on the wild-type template is hindered because of PNA binding. When reaction temperature ramps from 60 (extension) to 94°C (denaturation phase of the next cycle), PNA leaves the wild-type DNA at some temperature close to its Tm (69°C) and polymerization resumes. This polymerization will occur over a very short time, however, because the polymerase will quickly dissociate from the template as the temperature continue to rise. Given this narrow ‘window of opportunity’, whether or not the polymerization can be finished depending on the distance to run off. If this distance is too long, the polymerase does not have sufficient time to run off the template. The ‘chain reaction’ of PCR is therefore abolished, because the truncated products, lacking a primer-binding site, can no longer serve as templates during the next cycle. Our results also suggest that the ramp rate of a thermal cycler may also influence the efficiency of a PNA clamp. We have found that the slower the ramp rate, the longer is the distance required between PNA and primer for successful clamping (C.C. Chiou, unpublished data).
Applying the optimal conditions resulted in the successful detection of mutant alleles in serum DNA from patients with pancreatic cancer, indicating that the procedure has potential for use in screening for malignant diseases in clinical laboratories. Although the assay cannot identify specific mutation types, the PCR products can be subjected to further analysis, such as sequencing, to confirm preliminary findings; this also makes the procedure useful for research purposes. Note that although mutations occurring in codons 12 and 13 are more likely to be found in cancer patients, other mutations occurring in the flanking region covered by the PNA can also be differentiated by the assay. The mutation spectrum shown in Table 2 reveals that the majority of K-ras mutations in our samples are GGT to GTT in codon 12, which is consistent with a previous study indicating that this mutation accounts for 94.5% of pancreatic cancer in the Taiwanese population (31).
PNA in combination with oligonucleotide hybridization probes was used for rapid detection of K-ras mutations in two previous studies. In both studies, a pair of oligonucleotide hybridization probes was used to detect mutations, and a 17mer PNA was used to suppress PCR of the wild-type allele (see Figure 1C and references 29 and 30). Because the PNA bound to DNA so tightly, not only did it suppress PCR, but also it competed with the sensor probe for binding to K-ras templates. As a compromise, the investigators designed mutant-specific sensor probes. The perfectly matched mutant, with its high Tm (70.6°C), had the largest Tm difference from wild-type (66.3°C). Other mutants had Tm's closer to that of wild-type and were therefore less easily differentiated. The design of these studies and ours look similar but utilize very different underlying logic. In these other studies, in addition to use of mutant-specific probes, 72°C was used as the extension temperature during PCR, leading to inefficient clamping of wild-type amplification. However, the assay was also sensitive (1:10 000) because the PNA inhibited probe binding to wild-type products, so only the mutant signal was revealed on the melting curve. A possible problem with this design is that competition always exists between the PNA and the probe. The extend to which the signal is affected by this competition when a mutant is mismatched to both PNA and probe is not clear.
Recently, another DNA analogue, namely locked nucleic acid (LNA), was introduced and used in molecular detection assays (32). PNA and LNA probes have been used in combination to detect genetic heterogeneity of epidermal growth factor receptor (EGFR) in non-small cell lung cancer (33). In that study, mutant-specific LNA probes were used in a real-time PCR to generate amplification curve, and PNA was used to clamp wild-type amplification. The LNA probes resembled TaqMan probes, with a fluorophore at one end and a quencher at the other end. An amplification curve was generated when the probes were cleaved by Taq DNA polymerase during PCR. By analysis of the second derivative of the amplification curve, mutants containing a point mutation or a deletion in the EGFR gene were detected in 100 to 1000-fold excess of wild-type alleles. This study suggests that combining different DNA analogues can be used to develop powerful tools for detecting gene alterations.
Our assay may prove to have additional advantages. For example, mutant detection can be quantitative if samples with standard concentrations are assayed in parallel with clinical specimens, which may be useful in some situations for evaluation of the severity of disease. In addition, the PCR products of our assay can be used for further sequencing analysis, or in other enzymatic reactions, after a simple purification step, without interference by the PNA. Furthermore, multiplex assays can be performed in a single tube as long as the real-time PCR machine can differentiate fluorescent signals between different probes.
In summary, our study has provided a simple method to detect a trace amounts of K-ras mutants in large excess of wild-type DNA. This method has great potential for use in cancer screening, and could be adapted for detection of trace mutants pertinent to other diseases. Our study has defined important factors affecting the efficiency of PNA-mediated PCR clamping. These findings will facilitate further development of the role of PNA in molecular diagnosis.
Acknowledgments
The authors thank Dr Tsan-Zon Liu for editorial assistance and comments on the manuscript. This work was supported in part by a grant (NSC 92-2622-B-182-001-CC3) from National Science Council, ROC. Funding to pay the Open Access publication charges for this article was provided by Chang Gung University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Srivastava S., Verma M., Henson D.E. Biomarkers for early detection of colon cancer. Clin. Cancer Res. 2001;7:1118–1126. [PubMed] [Google Scholar]
- 2.Hirsch F.R., Franklin W.A., Gazdar A.F., Bunn P.A., Jr Early detection of lung cancer: clinical perspectives of recent advances in biology and radiology. Clin. Cancer Res. 2001;7:5–22. [PubMed] [Google Scholar]
- 3.Motojima K., Urano T., Nagata Y., Shiku H., Tsunoda T., Kanematsu T. Mutations in the Kirsten-ras oncogene are common but lack correlation with prognosis and tumor stage in human pancreatic carcinoma. Am. J. Gastroenterol. 1991;86:1784–1788. [PubMed] [Google Scholar]
- 4.Dieterle C.P., Conzelmann M., Linnemann U., Berger M.R. Detection of isolated tumor cells by polymerase chain reaction-restriction fragment length polymorphism for K-ras mutations in tissue samples of 199 colorectal cancer patients. Clin. Cancer Res. 2004;10:641–650. doi: 10.1158/1078-0432.ccr-1355-02. [DOI] [PubMed] [Google Scholar]
- 5.Anker P., Lefort F., Vasioukhin V., Lyautey J., Lederrey C., Chen X.Q., Stroun M., Mulcahy H.E., Farthing M.J. K-ras mutations are found in DNA extracted from the plasma of patients with colorectal cancer. Gastroenterology. 1997;112:1114–1120. doi: 10.1016/s0016-5085(97)70121-5. [DOI] [PubMed] [Google Scholar]
- 6.Iinuma H., Okinaga K., Adachi M., Suda K., Sekine T., Sakagawa K., Baba Y., Tamura J., Kumagai H., Ida A. Detection of tumor cells in blood using CD45 magnetic cell separation followed by nested mutant allele-specific amplification of p53 and K-ras genes in patients with colorectal cancer. Int. J. Cancer. 2000;89:337–344. doi: 10.1002/1097-0215(20000720)89:4<337::aid-ijc4>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 7.Jacobson D.R., Mills N.E. A highly sensitive assay for mutant ras genes and its application to the study of presentation and relapse genotypes in acute leukemia. Oncogene. 1994;9:553–563. [PubMed] [Google Scholar]
- 8.Norheim Andersen S., Breivik J., Lovig T., Meling G.I., Gaudernack G., Clausen O.P., Schjolberg A., Fausa O., Langmark F., Lund E., et al. K-ras mutations and HLA-DR expression in large bowel adenomas. Br. J. Cancer. 1996;74:99–108. doi: 10.1038/bjc.1996.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nickerson D.A., Kaiser R., Lappin S., Stewart J., Hood L., Landegren U. Automated DNA diagnostics using an ELISA-based oligonucleotide ligation assay. Proc. Natl. Acad. Sci. USA. 1990;87:8923–8927. doi: 10.1073/pnas.87.22.8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishikawa T., Maemura K., Hirata I., Matsuse R., Morikawa H., Toshina K., Murano M., Hashimoto K., Nakagawa Y., Saitoh O., et al. A simple method of detecting K-ras point mutations in stool samples for colorectal cancer screening using one-step polymerase chain reaction/restriction fragment length polymorphism analysis. Clin. Chim. Acta. 2002;318:107–112. doi: 10.1016/s0009-8981(01)00806-3. [DOI] [PubMed] [Google Scholar]
- 11.Toyooka S., Tsukuda K., Ouchida M., Tanino M., Inaki Y., Kobayashi K., Yano M., Soh J., Kobatake T., Shimizu N., et al. Detection of codon 61 point mutations of the K-ras gene in lung and colorectal cancers by enriched PCR. Oncol. Rep. 2003;10:1455–1459. doi: 10.3892/or.10.5.1455. [DOI] [PubMed] [Google Scholar]
- 12.Imai M., Hoshi T., Ogawa K. K-ras codon 12 mutations in biliary tract tumors detected by polymerase chain reaction denaturing gradient gel electrophoresis. Cancer. 1994;73:2727–2733. doi: 10.1002/1097-0142(19940601)73:11<2727::aid-cncr2820731113>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 13.Lilleberg S.L., Durocher J., Sanders C., Walters K., Culver K. High sensitivity scanning of colorectal tumors and matched plasma DNA for mutations in APC, TP53, K-RAS, and BRAF genes with a novel DHPLC fluorescence detection platform. Ann. N Y Acad. Sci. 2004;1022:250–256. doi: 10.1196/annals.1318.039. [DOI] [PubMed] [Google Scholar]
- 14.Lleonart M.E., Ramon y Cajal S., Groopman J.D., Friesen M.D. Sensitive and specific detection of K-ras mutations in colon tumors by short oligonucleotide mass analysis. Nucleic Acids Res. 2004;32:e53. doi: 10.1093/nar/gnh051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun X., Hung K., Wu L., Sidransky D., Guo B. Detection of tumor mutations in the presence of excess amounts of normal DNA. Nat. Biotechnol. 2002;20:186–189. doi: 10.1038/nbt0202-186. [DOI] [PubMed] [Google Scholar]
- 16.Nakao M., Janssen J.W., Seriu T., Bartram C.R. Rapid and reliable detection of N-ras mutations in acute lymphoblastic leukemia by melting curve analysis using LightCycler technology. Leukemia. 2000;14:312–315. doi: 10.1038/sj.leu.2401645. [DOI] [PubMed] [Google Scholar]
- 17.Pincas H., Pingle M.R., Huang J., Lao K., Paty P.B., Friedman A.M., Barany F. High sensitivity EndoV mutation scanning through real-time ligase proofreading. Nucleic Acids Res. 2004;32:e148. doi: 10.1093/nar/gnh150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maekawa M., Nagaoka T., Taniguchi T., Higashi H., Sugimura H., Sugano K., Yonekawa H., Satoh T., Horii T., Shirai N., et al. Three-dimensional microarray compared with PCR-single-strand conformation polymorphism analysis/DNA sequencing for mutation analysis of K-ras codons 12 and 13. Clin. Chem. 2004;50:1322–1327. doi: 10.1373/clinchem.2004.032060. [DOI] [PubMed] [Google Scholar]
- 19.Demers D.B., Curry E.T., Egholm M., Sozer A.C. Enhanced PCR amplification of VNTR locus D1S80 using peptide nucleic acid (PNA) Nucleic Acids Res. 1995;23:3050–3055. doi: 10.1093/nar/23.15.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen P.E., Egholm M., Berg R.H., Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 21.Hanvey J.C., Peffer N.J., Bisi J.E., Thomson S.A., Cadilla R., Josey J.A., Ricca D.J., Hassman C.F., Bonham M.A., Au K.G., et al. Antisense and antigene properties of peptide nucleic acids. Science. 1992;258:1481–1485. doi: 10.1126/science.1279811. [DOI] [PubMed] [Google Scholar]
- 22.Egholm M., Buchardt O., Christensen L., Behrens C., Freier S.M., Driver D.A., Berg R.H., Kim S.K., Norden B., Nielsen P.E. PNA hybridizes to complementary oligonucleotides obeying the Watson–Crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 23.Kyger E.M., Krevolin M.D., Powell M.J. Detection of the hereditary hemochromatosis gene mutation by real-time fluorescence polymerase chain reaction and peptide nucleic acid clamping. Anal. Biochem. 1998;260:142–148. doi: 10.1006/abio.1998.2687. [DOI] [PubMed] [Google Scholar]
- 24.Thiede C., Bayerdorffer E., Blasczyk R., Wittig B., Neubauer A. Simple and sensitive detection of mutations in the ras proto-oncogenes using PNA-mediated PCR clamping. Nucleic Acids Res. 1996;24:983–984. doi: 10.1093/nar/24.5.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taback B., Bilchik A.J., Saha S., Nakayama T., Wiese D.A., Turner R.R., Kuo C.T., Hoon D.S. Peptide nucleic acid clamp PCR: a novel K-ras mutation detection assay for colorectal cancer micrometastases in lymph nodes. Int. J. Cancer. 2004;111:409–414. doi: 10.1002/ijc.20268. [DOI] [PubMed] [Google Scholar]
- 26.Karadag A., Riminucci M., Bianco P., Cherman N., Kuznetsov S.A., Nguyen N., Collins M.T., Robey P.G., Fisher L.W. A novel technique based on a PNA hybridization probe and FRET principle for quantification of mutant genotype in fibrous dysplasia/McCune-Albright syndrome. Nucleic Acids Res. 2004;32:e63. doi: 10.1093/nar/gnh059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernard P.S., Ajioka R.S., Kushner J.P., Wittwer C.T. Homogeneous multiplex genotyping of hemochromatosis mutations with fluorescent hybridization probes. Am. J. Pathol. 1998;153:1055–1061. doi: 10.1016/s0002-9440(10)65650-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kreuzer K.A., Le Coutre P., Landt O., Na I.K., Schwarz M., Schultheis K., Hochhaus A., Dorken B. Preexistence and evolution of imatinib mesylate-resistant clones in chronic myelogenous leukemia detected by a PNA-based PCR clamping technique. Ann. Hematol. 2003;82:284–289. doi: 10.1007/s00277-003-0644-y. [DOI] [PubMed] [Google Scholar]
- 29.Dabritz J., Hanfler J., Preston R., Stieler J., Oettle H. Detection of Ki-ras mutations in tissue and plasma samples of patients with pancreatic cancer using PNA-mediated PCR clamping and hybridisation probes. Br. J. Cancer. 2005;92:405–412. doi: 10.1038/sj.bjc.6602319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen C.Y., Shiesh S.C., Wu S.J. Rapid detection of K-ras mutations in bile by peptide nucleic acid-mediated PCR clamping and melting curve analysis: comparison with restriction fragment length polymorphism analysis. Clin. Chem. 2004;50:481–489. doi: 10.1373/clinchem.2003.024505. [DOI] [PubMed] [Google Scholar]
- 31.Wang J.Y., Lian S.T., Chen Y.F., Yang Y.C., Chen L.T., Lee K.T., Huang T.J., Lin S.R. Unique K-ras mutational pattern in pancreatic adenocarcinoma from Taiwanese patients. Cancer Lett. 2002;180:153–158. doi: 10.1016/s0304-3835(01)00818-7. [DOI] [PubMed] [Google Scholar]
- 32.Braasch D.A., Corey D.R. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem. Biol. 2001;8:1–7. doi: 10.1016/s1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- 33.Nagai Y., Miyazawa H., Huqun, Tanaka T., Udagawa K., Kato M., Fukuyama S., Yokote A., Kobayashi K., Kanazawa M., et al. Genetic heterogeneity of the epidermal growth factor receptor in non-small cell lung cancer cell lines revealed by a rapid and sensitive detection system, the peptide nucleic acid-locked nucleic acid PCR clamp. Cancer Res. 2005;65:7276–7282. doi: 10.1158/0008-5472.CAN-05-0331. [DOI] [PubMed] [Google Scholar]