Abstract
Overexpression of the c-myc oncogene contributes to the development of a significant number of human cancers. In response to deregulated Myc activity, the p53 tumor suppressor is activated to promote apoptosis and inhibit tumor formation. Here we demonstrate that p53 induction in response to Myc overexpression requires the ataxia-telangiectasia mutated (ATM) kinase, a major regulator of the cellular response to DNA double-strand breaks. In a transgenic mouse model overexpressing Myc in squamous epithelial tissues, inactivation of Atm suppresses apoptosis and accelerates tumorigenesis. Deregulated Myc expression induces DNA damage in primary transgenic keratinocytes and the formation of γH2AX and phospho-SMC1 foci in transgenic tissue. These findings suggest that Myc overexpression causes DNA damage in vivo and that the ATM-dependent response to this damage is critical for p53 activation, apoptosis, and the suppression of tumor development.
Keywords: p53, DNA damage
The c-myc oncogene is overexpressed in a large percentage of human tumors, including cancers of lymphoid, mesenchymal, and epithelial origin. Increased Myc activity contributes to tumorigenesis by promoting proliferation and making cells refractory to some antimitogenic signals. Myc is a transcription factor that regulates the expression of a number of genes involved in cell cycle control and metabolism (1, 2). It has also been suggested that deregulated Myc expression leads to DNA damage and genomic instability and in this way also contributes to cancer development (3-5).
The p53 tumor suppressor limits cell proliferation and tumor development in response to increased Myc activity by promoting apoptosis (6-8). One mechanism by which Myc overexpression is signaled to p53 is through the ARF (p14ARF in humans and p19Arf in mice) tumor suppressor (9). ARF regulates p53 by binding to and inhibiting the action of Mdm2, a negative regulator of p53 (10). Like p53 loss, the inactivation of Arf suppresses apoptosis and promotes tumorigenesis in response to Myc overexpression (9, 11-13).
The activity of p53 is also regulated in response to DNA damage and other stresses by posttranslational modifications, including phosphorylation (14). Phosphorylation of p53 at N-terminal residues is especially critical because these modifications can inhibit Mdm2 binding, increase p53 transcriptional activation capacity, and promote additional posttranslational modifications that regulate DNA binding. The ataxia-telangiectasia mutated (ATM) and ATM-and Rad3-related (ATR) kinases directly phosphorylate p53 on serine-15 (15-17). In addition, ATM indirectly regulates other p53 phosphorylation events by phosphorylating and activating additional kinases such as Chk2, Chk1, and Plk3 (18-24). Other proteins phosphorylated by ATM as part of the DNA damage response include Mdm2, BRCA1, SMC1, NBS1, and E2F1 (25-32). It is thought that ATM responds primarily to DNA double-strand breaks whereas ATR responds to UV radiation-induced DNA damage and blocks in transcription (33).
Recent reports have demonstrated that the ATM DNA damage response pathway is activated early during the formation of several types of human tumors (34, 35). This finding is consistent with findings from cell culture experiments showing that a number of oncogenic factors, such as E2F1, cyclin E, and Myc, stimulate the phosphorylation of p53 and some other ATM targets (3, 12, 34, 36-38). It has been suggested that the activation of this checkpoint response by oncogenic stresses inhibits the formation of cancer. In the present study, a transgenic mouse model overexpressing Myc in squamous epithelial tissues is used to demonstrate that ATM plays a critical role in activating p53, inducing apoptosis, and suppressing tumorigenesis in response to Myc.
Results
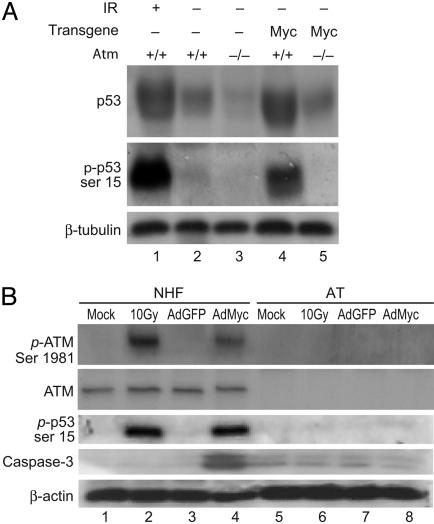
ATM Is Required for p53 Accumulation and Phosphorylation in Response to Myc. K5 Myc-transgenic mice display hyperproliferative epidermis and spontaneously develop tumors in the skin and oral epithelium (39, 40). K5 Myc mice also exhibit aberrant apoptosis in their epidermis that depends largely on functional p53 (40). Consistent with these findings, K5 Myc-transgenic epidermis contains elevated levels of p53 protein compared with nontransgenic epidermis (Fig. 1A). To determine whether this elevated p53 in K5 Myc tissue is phosphorylated, antisera specific for the serine-18 phosphorylated form of murine p53 was used in a Western blot of epidermal protein isolated from K5 Myc mice. As a positive control for phosphorylated p53, epidermal protein from a nontransgenic mouse irradiated with 10 Gy of ionizing radiation (IR) was used (Fig. 1 A, lane 1). K5 Myc mice contained elevated levels of total and phosphorylated p53 that were similar to those of IR-treated mice whereas phosphorylated p53 was barely detectable in epidermal extracts from an untreated, nontransgenic sibling mouse (Fig. 1 A). This finding that Myc overexpression leads to p53 phosphorylation in K5 Myc tissue in vivo agrees with previous results by others on the ability of Myc to stimulate p53 phosphorylation in cultured fibroblasts (3, 36).
Fig. 1.
ATM-dependent accumulation and phosphorylation of p53 in response to Myc. (A) Western blot analysis was performed on epidermal protein lysates from nontransgenic (lanes 1-3) and K5 Myc-transgenic (lanes 4 and 5) mice that were wild-type (lanes 1, 2, and 4) or nullizygous (lanes 3 and 5) for Atm. The wild-type mouse in lane 1 was treated with 10 Gy of IR 30 min before it was killed. Antisera specific to total p53 (Top), phospho-serine-15 p53 (Middle), or β-tubulin (Bottom) were used as indicated. (B) NHFs (lanes 1-4) or primary fibroblasts from an AT patient (AT, lanes 5-8) were mock-treated (lanes 1 and 5), exposed to 10 Gy of IR (lanes 2 and 6), or infected with AdGFP (lanes 3 and 7) or AdMyc (lanes 4 and 8) at a multiplicity of infection of 100. Cells were harvested for protein extract 1 h after IR or 24 h after infection, and Western blot analysis was performed by using antisera or antibody specific for phospho-serine-1981 ATM, total ATM, phospho-serine-15 p53, activated caspase-3, or β-actin as indicated.
Human p53 serine-15 and the homologous murine p53 serine-18 are phosphorylated by several members of the phosphatidylinositol 3-kinase family, including ATM and ATR (15-17). To determine whether ATM is involved in p53 stabilization and serine-18 phosphorylation in the K5 Myc model, these mice were bred into an Atm-null background. The total level of p53 protein in K5 Myc tissue was significantly reduced and serine-18 phosphorylation was undetectable in the absence of ATM (Fig. 1 A).
To further examine the requirement for ATM in mediating p53 accumulation and phosphorylation induced by Myc, normal human fibroblasts (NHFs) and primary fibroblasts from a patient with ataxia-telangiectasia (AT) were obtained. These cells were infected with a recombinant adenovirus expressing Myc (AdMyc) or a control adenovirus expressing GFP (AdGFP). As a positive control for ATM-dependent phosphorylation, these cell cultures were also irradiated with IR. Exposure to IR induced the phosphorylation of ATM on serine-1981, a marker of ATM activation (41). Phosphorylation of ATM also occurred in NHFs infected with AdMyc but not AdGFP (Fig. 1B). As expected, IR induced p53 phosphorylation at serine-15 in NHFs, and this was significantly reduced in the AT cells (Fig. 1B). Phosphorylation of p53 also occurred in NHFs infected with AdMyc but not AdGFP. In AT cells, however, overexpression of Myc did not induce significant levels of phosphorylated p53 (Fig. 1B). Thus, ATM is required for p53 accumulation and phosphorylation in response to Myc overexpression in both primary transgenic epidermal tissue and cultured primary human fibroblasts.
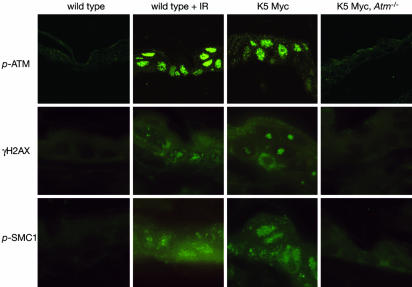
Myc Induces γH2AX and Phospho-SMC1 Foci in Vivo. Consistent with the Western blot data from cells infected with the AdMyc virus, ATM phosphorylated at serine-1981 could also be detected in primary tissue from K5 Myc mice by immunofluorescence (Fig. 2). Phosphorylated ATM localizes to subnuclear foci that are sites of DNA double-strand breaks (41). The staining pattern of phospho-ATM in Myc-transgenic epidermis was similar to the focal pattern of staining in mouse epidermis exposed to IR (Fig. 2). This finding suggests that transgenic expression of Myc may cause DNA double-strand breaks as the mechanism for ATM activation. One of the most commonly used and sensitive markers for detecting DNA double-strand breaks is the formation of subnuclear foci containing the phosphorylated form of the histone variant H2AX, termed γH2AX (42, 43). As expected, exposure of wild-type mice to IR led to the formation of subnuclear γH2AX foci (Fig. 2). In unirradiated K5 Myc epidermis, γH2AX foci were also observed in most nuclei, and this depended on the presence of ATM. (Fig. 2). K5 Myc epidermis also stained positive for other markers of DNA double-strand breaks, including phospho-SMC1 (Fig. 2). SMC1 is phosphorylated by ATM and, like γH2AX, localizes to sites of DNA double-strand breaks (26, 27, 44). As with γH2AX, phospho-SMC1 displayed a focal pattern of staining in K5 Myc epidermis, similar to the pattern observed in IR-treated epidermis, and this depended on the presence of ATM (Fig. 2). Thus, transgenic expression of Myc in the epidermis results in the ATM-dependent formation of γH2AX and phospho-SMC1 foci that may represent sites of double-strand breaks.
Fig. 2.
K5 Myc-transgenic tissue stains for markers of DNA double-strand breaks. Immunofluorescent staining was performed on skin sections from untreated wild-type mice, wild-type mice exposed to 3 Gy of IR 20 min before killing, K5 Myc mice, and K5 Myc, Atm-/- mice by using antibodies specific for ATM phosphorylated at serine-1981 (p-ATM), γH2AX, or SMC1 phosphorylated at serine-957 (p-SMC1).
Myc Induces DNA Damage in Vivo. To confirm that transgenic expression of Myc caused DNA damage, primary keratinocytes were isolated from K5 Myc mice and immediately used in the single-cell gel electrophoresis (comet) assay. Keratinocytes isolated from mice exposed to IR were used as a positive control for DNA damage. In the absence of IR, only a few wild-type keratinocytes had a detectable comet tail (Table 1). After IR treatment, the majority of keratinocytes had comet tails, indicative of DNA damage. Many keratinocytes isolated from K5 Myc mice also had detectable comet tails, although the number of positive cells and the length of the comet tails was lower than in the IR-treated keratinocytes. Nonetheless, measurement of the comet tail moment confirmed that K5 Myc keratinocytes had significantly more DNA damage compared with untreated wild-type keratinocytes (Table 1). Taken together, these findings suggest that transgenic expression of Myc induces DNA damage that is then recognized by the ATM response pathway.
Table 1. DNA damage in primary keratinocytes as measured by comet assay.
| Cells | % with tails | Average olive moment |
|---|---|---|
| Untreated | 10.3 | 2.8 ± 0.5 |
| 5-Gy IR | 71.1 | 14.3 ± 2.1* |
| K5 Myc | 41.8 | 9.7 ± 1.2* |
, P < 0.001, as compared with untreated wild-type samples
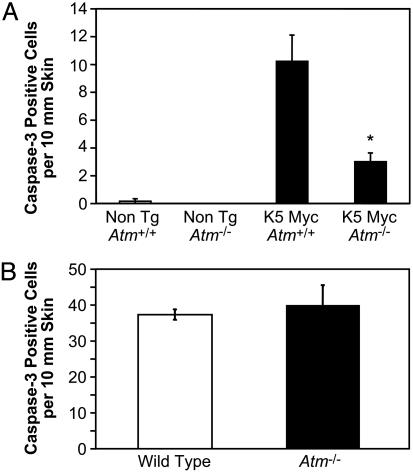
ATM Promotes Apoptosis Induced by Myc. Activation of p53 in response to Myc overexpression leads to the induction of apoptosis. Consistent with this, infection of NHFs with AdMyc, but not AdGFP, resulted in the formation of the activated, cleaved form of caspase-3 (Fig. 1B). In contrast, Myc overexpression did not increase the level of activated caspase-3 in AT cells. This finding suggests that ATM participates in Myc-induced apoptosis. To determine whether this is the case in vivo, the effect of Atm status on apoptosis in K5 Myc mice was examined by measuring the number of epidermal keratinocytes staining for the activated form of caspase-3. Inactivation of Atm resulted in a significant decrease in the number of apoptotic cells observed in K5 Myc-transgenic epidermis (Fig. 3A). In contrast to the impaired apoptosis observed in response to Myc overexpression, Atm-/- mice had normal levels of apoptosis in the epidermis after exposure to UV radiation (Fig. 3B). This finding demonstrates that the absence of ATM does not cause a general impairment of apoptosis in mouse keratinocytes, but rather inactivation of Atm specifically inhibits apoptosis in response to Myc overexpression.
Fig. 3.
Inactivation of Atm reduces apoptosis in K5 Myc-transgenic mice. (A) Skin sections taken from mice with the indicated genotypes were immunohistochemically stained with an antibody specific for the activated form of caspase-3. The average number of positive epidermal cells per 10 mm of skin was determined microscopically from at least four independent mice in each group. The number of caspase-3-positive cells in K5 Myc-transgenic mice null for Atm is statistically different from the number in K5 Myc-transgenic mice wild-type for Atm by unpaired t test (P = 0.0175). (B) Three wild-type and three Atm-null mice were treated with 200 mJ/cm2 UVB, and skin sections were taken 24 h later. Skin sections were immunohistochemically stained for activated caspase-3, and the average number of positive cells per 10 mm of epidermis was determined microscopically for each group.
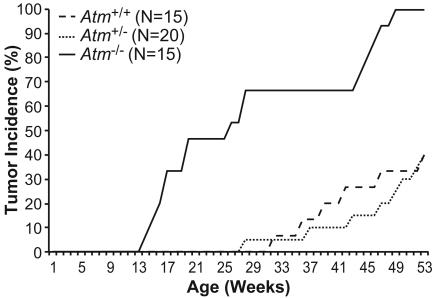
ATM Inactivation Cooperates with Myc Overexpression in Tumorigenesis. Suppressing Myc-induced apoptosis, through the inactivation of p53 or Arf or the overexpression of Bcl-2, has been shown to accelerate tumorigenesis in a number of experimental systems (7, 8, 13, 45, 46). To determine whether Atm inactivation would also promote Myc-driven tumor development, K5 Myc mice that were wild-type, hemizygous, or nullizygous for Atm were generated and maintained. K5 Myc mice develop spontaneous tumors in several K5-expressing tissues, including the skin and oral epithelium, starting at ≈30 weeks of age (39, 40). Atm-/- mice normally develop lymphomas by 6 months of age but very rarely develop tumors in epithelial tissues (47-49). Consistent with our previous results, no K5 Myc mouse wild-type for Atm developed a tumor before 30 weeks of age (Fig. 4). Inactivation of one Atm allele had no significant effect on the rate of tumor development in K5 Myc mice. In contrast, 67% of K5 Myc mice null for Atm developed an epithelial tumor by 30 weeks of age (Fig. 4).
Fig. 4.
Inactivation of Atm accelerates epithelial tumorigenesis in K5 Myc-transgenic mice. K5 Myc-transgenic mice wild-type (+/+), hemizygous (+/-), or nullizygous (-/-) for Atm were monitored for spontaneous tumor development for 1 year. Only tumors from squamous epithelial tissues are included. Tumor incidence in K5 Myc-transgenic mice that are Atm-/- is statistically different from transgenic mice that are Atm+/+ or Atm+/- by univariate ANOVA (P < 0.05).
Similar to Atm-/- mice, more than half of the K5 Myc mice null for Atm (15 of 26) developed a lymphoma before or concurrent with the development of an epithelial tumor. However, all mice that developed a lymphoma before developing an epithelial tumor were removed from the study shown in Fig. 4 and Table 2. By 49 weeks of age, 100% of K5 Myc, Atm-/- mice that did not develop a lymphoma had developed an epithelial tumor. In contrast, only 40% of the transgenic mice wild-type or hemizygous for Atm developed a tumor by 1 year of age (Fig. 4 and Table 2). All of the epithelial tumors that arose in the K5 Myc mice were in the skin or oral epithelium (Table 2).
Table 2. Epithelial tumors in K5 Myc-transgenic mice.
| Genotype | n | Tumor incidence at 1 year, % | Average age of onset, weeks | Tumor types |
|---|---|---|---|---|
| Atm+/+ | 15 (8 F, 7 M) | 40 | 42.0 | Two squamous papillomas and four SCC of skin |
| Atm+/– | 20 (12 F, 8 M) | 40 | 44.8 | Three squamous papillomas, one SCC of skin, and four SCC of oral cavity |
| Atm–/– | 15 (7 F, 8 M) | 100 | 29.2 | Four squamous papillomas, three SCC of skin, and nine SCC of oral cavity |
F, female; M, male; SCC, squamous cell carcinoma.
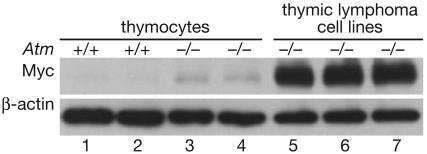
These findings demonstrate that the absence of ATM, like the absence of p53, cooperates with Myc overexpression to promote the development of at least some types of epithelial cancers. To determine whether a similar cooperation might also occur in lymphoma development, Myc expression was examined in cell lines derived from lymphomas that had arisen in Atm-/- mice lacking the K5 Myc transgene. In all three Atm-/- thymic lymphoma cell lines examined, Myc was highly overexpressed compared with the levels of Myc observed in primary T cells from wild-type or Atm-/- mice (Fig. 5). This result is consistent with the observation that c-myc gene amplification is a common event in lymphomas from Atm-/- mice (50). Together, these findings suggest that loss of ATM function also cooperates with Myc overexpression in the development of the spontaneous thymic lymphomas that arise in Atm-/- mice.
Fig. 5.
Thymic lymphoma cell lines from Atm knockout mice overexpress Myc. Western blot analysis for Myc and β-actin was performed by using extracts from primary thymocytes isolated from 4-week-old wild-type (lanes 1 and 2) or Atm-null (lanes 3 and 4) mice or from three independent thymic lymphoma cell lines derived from Atm-/- mice (lanes 5-7).
Discussion
Phosphorylation of several DNA damage response proteins, including ATM, Chk2, p53, and H2AX, can be observed in precursor stage cancers of the breast, colon, lung, skin, testes, and urinary bladder (34, 35, 51). This has led to the suggestion that, in many cancers, DNA damage occurs during the earliest stages of tumor development, before genomic instability and the loss of wild-type p53 function. In agreement with this idea, forced expression of oncogenic growth factors, cyclin E, cdc25A, or E2F1 in cultured cells can induce the ATM signaling pathway (34, 35, 37, 38). Overexpression of Myc was also shown to induce DNA damage in vitro (3, 5), but whether this occurs in vivo has been a matter of debate (52). It has been reported that deregulated expression of Myc in the liver of transgenic mice leads to chromosomal damage, but only when coexpressed with a TGFα transgene (4).
Here we show that transgenic expression of Myc on its own induces the ATM DNA damage response pathway, as evidenced by ATM and p53 phosphorylation and the formation of γH2AX and phospho-SMC1 foci. An increase in DNA damage over background levels could also be detected in primary keratinocytes isolated from K5 Myc mice. This finding indicates that Myc overexpression, a common event in human cancers, causes DNA damage in vivo and could contribute to the activation of the ATM signaling pathway that is observed in early-stage clinical specimens.
In addition to promoting cell proliferation, Myc also induces apoptosis, and this functions to suppress tumor development. A number of mediators have been shown to be important for Myc-induced apoptosis, with one of the most critical being p53 (6). It has been proposed that oncogenes such as Myc and DNA damage activate p53 through distinct mechanisms. Specifically, ARF is believed to be involved in the activation of p53 in response to oncogenes whereas kinases such as ATM are thought to be important for p53 activation in response to DNA damage (53, 54). Indeed, in the K5 Myc mice used in this study, inactivation of Arf reduced the level of apoptosis observed in the epidermis (12).
However, the findings presented here demonstrate that ATM is also critical for p53 activation and the induction of apoptosis in response to Myc. In the absence of ATM, p53 accumulation and phosphorylation in response to Myc are greatly reduced. Moreover, the spontaneous apoptosis observed in the epidermis of K5 Myc mice is significantly decreased by Atm inactivation. Atm inactivation reduced apoptosis in K5 Myc-transgenic tissue to a level that is similar to that observed when Arf was inactivated, but not to the near-complete inhibition of apoptosis observed in the absence of p53 (12, 39). This finding suggests that, in at least some cases, the ARF and ATM pathways cooperate to activate p53 and promote apoptosis. ATM phosphorylates additional targets, including Mdm2 and Chk2, that could also potentially contribute to p53 activation and apoptosis. Whether Myc promotes the ATM-dependent phosphorylation of these other targets remains to be determined.
Inhibition of Myc-induced apoptosis has been shown to accelerate tumor development in a number of experimental systems (7, 8, 40, 45, 46). Consistent with this finding, Atm inactivation not only impairs apoptosis but also accelerates epithelial tumor development in K5 Myc mice. This finding provides experimental evidence to support the idea that the ATM DNA damage response pathway functions to suppress the emergence of tumors from precancerous lesions. In addition to impairing apoptosis, the absence of ATM may also promote tumorigenesis by causing genetic instability. Cells from humans and mice lacking ATM have defective telomere maintenance, end-to-end chromosome fusions, and an increased frequency of spontaneous translocations that could contribute to cancer development (50, 55-58).
A previous study using a different transgenic mouse model demonstrated that p53 induction and apoptosis induced by another type of oncogenic stress were unaffected by the absence of ATM (59). In this transgenic model, a fragment of SV40 T antigen, which binds and inactivates Rb family members but not p53, is expressed in the brain choriod plexus epithelium. Transgenic expression of this T antigen fragment induces aberrant proliferation and apoptosis that depends on p53 and E2F1 (60, 61). The discrepancy in the ATM requirement for apoptosis in our K5 Myc model and this T antigen model may be due to differences in the oncogenic signal or to tissue-specific differences between the choriod plexus and the epidermis. Interestingly, inactivation of Atm did not accelerate tumor development in the T antigen transgenic mouse model (59). Thus, an oncogenic stress that does not require ATM to activate p53 and induce apoptosis does not cooperate with the absence of ATM to promote tumor development. This finding suggests that it is the ability of ATM to promote apoptosis in response to Myc that is critical for suppressing tumorigenesis in K5 Myc mice, although other activities of ATM, such as maintaining genomic stability, may also play a role.
Materials and Methods
Mice. The generation of K5 Myc-transgenic mice is described in ref. 40. Briefly, the transgene contains the bovine K5 promoter (62), the rabbit β-globin intron 2, the simian virus 40 polyadenylation signal, and a murine c-myc genomic fragment. K5 Myc-transgenic mice (line MM5) were bred to mice containing an inactivated Atm allele (47) to generate K5 Myc mice heterozygous for Atm. Male K5 Myc mice heterozygous for Atm were then bred to Atm heterozygous female mice to generate K5 Myc and nontransgenic mice that were homozygous, heterozygous, or nullizygous for Atm. K5 Myc mice were originally in the SSIN strain background. Atm+/- mice were obtained from The Jackson Laboratory and were in the C57BL/6J strain background. The genetic background of mice in this study was, therefore, a mixture of SSIN and C57BL/6J strains. Sibling mice were used for comparisons in all experimental procedures. All experiments with mice were performed in accordance with national guidelines and regulations and approved by the Institutional Animal Care and Use Committee.
Cells and Viruses. NHFs and primary AT fibroblasts (Coriell Cell Repositories, Camden, NJ) were maintained in MEM with 2 mM glutamine, nonessential amino acids, and 15% FBS. Cells from age-, sex-, and ethnicity-matched individuals GM08399 (wild type) and GM02052 (from an AT patient) were used. Recombinant adenoviruses expressing human c-myc and GFP are described in ref. 37.
Immunoblotting and Antibodies. Epidermal protein lysate was collected by scraping dorsal skin and resuspending epidermal tissue in modified RIPA lysis buffer (50 mM Hepes, pH 7.4/1% IGEPAL/0.25% Na deoxycholate/150 mM NaCl/1 mM EDTA/1 mM PMSF/1 mM NaF/1 mM Na3VO4) with 6 M urea containing protease (Sigma, P-8340) and phosphatase (Sigma, P-2850) inhibitor mixtures. Protein extract was prepared by freeze-thawing followed by supernatant collection after ultracentrifugation (135,000 × g, Beckman TL-100). Cells were harvested and lysed in RIPA lysis buffer with protease and phosphatase inhibitors, and proteins were extracted as described above.
Protein samples (50 μg) were separated on 6-10% SDS/PAGE gels depending on the protein size and transferred to a poly(vinylidene difluoride) membrane. After transferring, the membranes were blocked in 5% milk in TBST (TBS and 0.01% Tween 20) for 30 min and probed with primary antisera or antibodies for 1 h in 5% milk in TBST). The following rabbit polyclonal antisera were acquired: p53 pS15 (catalog no. 9284; 1:1,000), p53 (catalog no. 9282; 1:1,000), and cleaved caspase-3 (catalog no. 9661; 1:1,000) from Cell Signaling Technology (Beverly, MA) and c-Myc (catalog no. sc-788; 1:1,000) and β-tubulin (catalog no. sc-9104; 1:2,000) from Santa Cruz Biotechnology. The following mouse monoclonal antibodies were obtained: ATM pS1981 (catalog no. 200-301-400; 1:1,000) from Rockland, ATM (catalog no. GTX70103; 1:1,000) from Genetex, and β-actin (catalog no. sc-8342; 1:2,000) from Santa Cruz Biotechnology. Bands were visualized by using enhanced chemiluminescence reagent (Amersham Pharmacia).
Single-Cell Gel Electrophoresis (Comet) Assay. Primary keratinocytes were isolated from adult mouse epidermis as described in ref. 63. Briefly, dorsal skin samples were incubated in trypsin for 2 h at 32°C, and then the epidermal layer was separated from the dermis. The scraped epidermis was stirred at 100 rpm on a magnetic stirrer for 20 min at room temperature. The cells were then pelleted and resuspended in PBS. Primary keratinocytes at a concentration of 2 × 105 cells per ml were then embedded in low-melting-point agarose on a glass slide by using the CometAssay kit and the manufacturer's protocol (Trevigen, catalog no. 4250-050-K). Briefly, the embedded cells were incubated overnight at 4°C in lysis solution (2.5 M NaCl/100 mM EDTA, pH 10/10 mM Tris/1% sodium lauryl sarcosinate/0.01% Triton X-100) followed by incubation in alkaline solution (300 mM NaOH/EDTA 1 mM) for 40 min to denature the DNA. The samples were then washed twice in TBE buffer and electrophoresed at 19 V for 10 min in 1× TBE buffer. The samples were stained with 50 μl of SYBR Green, and nuclei were visualized by fluorescent microscopy. Tail length and olive tail moment of 70 nuclei per slide were scored by using cometscore software (TriTek). Student's t test was used to derive P values. Tail moment is the product of the tail length and the fraction of total DNA in the tail, which gives a measure of the extent of DNA damage.
Phospho-ATM, γH2AX, and Phospho-SMC1 Immunofluorescence. Formalin-fixed, paraffin-embedded mouse skin sections were deparaffinized, boiled in 10 mM sodium citrate for 10 min, and blocked in 50% goat serum for 30 min. Sections were then incubated overnight at 4°C with antibodies against histone H2AX pS139 (γH2AX) (catalog no. 05-636, Upstate Biotechnology; 1:300), ATM pS1981 (catalog no. 600-401-400, Rockland; 1:200) and SMC1 pS957 (catalog no. 200-301-397, Rockland; 1:300). Alexa Fluor 488-conjugated goat anti-rabbit or anti-mouse secondary antibodies from Molecular Probes were used, and the sections were imaged by using an Olympus laser confocal microscope or an Olympus BX60 fluorescent microscope.
Activated Caspase-3 Immunohistochemistry. Formalin-fixed, paraffin-embedded skin sections were immunohistochemically stained with an antibody specific for the activated form of caspase-3 (R & D Systems, 1:2,000 dilution) by using the Histostain-Plus kit (Zymed). The stained slides were examined microscopically to determine the average number of positive epidermal keratinocytes per 10 mm of linear skin. For UV treatment, 6- to 8-week-old wild-type and Atm-/- knockout mice were irradiated with 200 mJ/cm2 UVB from 12 Westinghouse FS20 sunlamps and killed 24 h later. Skin samples were immunohistochemically stained and analyzed for activated caspase-3 as above.
Acknowledgments
We thank Jen Smith and Pam Blau for technical assistance; Dale Weiss, Lezlee Coghlan, and coworkers for animal care; Irma Gimenez-Conti and coworkers for histology; Chris Brown and Joi Holcomb for artwork; Dennis Johnston and Howard Thames for statistical analysis; and Shawnda Sanders for preparation of the manuscript. We also thank Thomas Berton, Hideaki Kaneto, Toru Kawamoto, Qingyi Wei, Ping Xiong, and Rebecca Morris for training and advice. This work was supported by National Institutes of Health Grants CA098601 (to D.G.J.), ES07784, and CA16672 and a grant from the Longevity Foundation of Austin (to P.K.W.). J.T.P. was supported by National Institutes of Health Training Grant T32 ES07247.
Author contributions: P.K.W. and D.G.J. designed research; and R.V.P., R.J.R., S.H., J.T.P., M.Y., K.K., and M.J.M. performed research.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IR, ionizing radiation; NHF, normal human fibroblast; AT, ataxia-telangiectasia; AdMyc, adenovirus expressing Myc; AdGFP, adenovirus expressing GFP.
References
- 1.Pelengaris, S., Khan, M. & Evan, G. (2002) Nat. Rev. Cancer 2, 764-776. [DOI] [PubMed] [Google Scholar]
- 2.Dang, C. V. (1999) Mol. Cell. Biol. 19, 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vafa, O., Wade, M., Kern, S., Beeche, M., Pandita, T. K., Hampton, G. M. & Wahl, G. M. (2002) Mol. Cell 9, 1031-1044. [DOI] [PubMed] [Google Scholar]
- 4.Sargent, L. M., Sanderson, N. D. & Thorgeirsson, S. S. (1996) Cancer Res. 56, 2137-2142. [PubMed] [Google Scholar]
- 5.Felsher, D. W. & Bishop, J. M. (1999) Proc. Natl. Acad. Sci. USA 96, 3940-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermeking, H. & Eick, D. (1994) Science 265, 2091-2093. [DOI] [PubMed] [Google Scholar]
- 7.Elson, A., Deng, C., Campos-Torres, J., Donehower, L. A. & Leder, P. (1995) Oncogene 11, 181-190. [PubMed] [Google Scholar]
- 8.Blyth, K., Terry, A., O'Hara, M., Baxter, E. W., Campbell, M., Stewart, M., Donehower, L. A., Onions, D. E., Neil, J. C. & Cameron, E. R. (1995) Oncogene 10, 1717-1723. [PubMed] [Google Scholar]
- 9.Zindy, F., Eischen, C. M., Randle, D. H., Kamijo, T., Cleveland, J. L., Sherr, C. J. & Roussel, M. F. (1998) Genes Dev. 12, 2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sherr, C. J. (2001) Nat. Rev. Mol. Cell Biol. 2, 731-737. [DOI] [PubMed] [Google Scholar]
- 11.Schmitt, C. A., McCurrach, M. E., de Stanchina, E., Wallace-Brodeur, R. R. & Lowe, S. W. (1999) Genes Dev. 13, 2670-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russell, J. L., Powers, J. T., Rounbehler, R. J., Rogers, P. M., Conti, C. J. & Johnson, D. G. (2002) Mol. Cell. Biol. 22, 1360-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eischen, C. M., Weber, J. D., Roussel, M. F., Sherr, C. J. & Cleveland, J. L. (1999) Genes Dev. 13, 2658-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giaccia, A. J. & Kastan, M. B. (1998) Genes Dev. 12, 2973-2983. [DOI] [PubMed] [Google Scholar]
- 15.Banin, S., Moyal, L., Shieh, S., Taya, Y., Anderson, C. W., Chessa, L., Smorodinsky, N. I., Prives, C., Reiss, Y., Shiloh, Y. & Ziv, Y. (1998) Science 281, 1674-1677. [DOI] [PubMed] [Google Scholar]
- 16.Canman, C. E., Lim, D. S., Cimprich, K. A., Taya, Y., Tamai, K., Sakaguchi, K., Appella, E., Kastan, M. B. & Siliciano, J. D. (1998) Science 281, 1677-1679. [DOI] [PubMed] [Google Scholar]
- 17.Tibbetts, R. S., Brumbaugh, K. M., Williams, J. M., Sarkaria, J. N., Cliby, W. A., Shieh, S. Y., Taya, Y., Prives, C. & Abraham, R. T. (1999) Genes Dev. 13, 152-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gatei, M., Sloper, K., Sorensen, C., Syljuasen, R., Falck, J., Hobson, K., Savage, K., Lukas, J., Zhou, B. B., Bartek, J. & Khanna, K. K. (2003) J. Biol. Chem. 278, 14806-14811. [DOI] [PubMed] [Google Scholar]
- 19.Ahn, J. Y., Schwarz, J. K., Piwnica-Worms, H. & Canman, C. E. (2000) Cancer Res. 60, 5934-5936. [PubMed] [Google Scholar]
- 20.Chaturvedi, P., Eng, W. K., Zhu, Y., Mattern, M. R., Mishra, R., Hurle, M. R., Zhang, X., Annan, R. S., Lu, Q., Faucette, L. F., et al. (1999) Oncogene 18, 4047-4054. [DOI] [PubMed] [Google Scholar]
- 21.Chehab, N. H., Malikzay, A., Appel, M. & Halazonetis, T. D. (2000) Genes Dev. 14, 278-288. [PMC free article] [PubMed] [Google Scholar]
- 22.Hirao, A., Kong, Y. Y., Matsuoka, S., Wakeham, A., Ruland, J., Yoshida, H., Liu, D., Elledge, S. J. & Mak, T. W. (2000) Science 287, 1824-1827. [DOI] [PubMed] [Google Scholar]
- 23.Shieh, S. Y., Ahn, J., Tamai, K., Taya, Y. & Prives, C. (2000) Genes Dev. 14, 289-300. [PMC free article] [PubMed] [Google Scholar]
- 24.Xie, S., Wu, H., Wang, Q., Cogswell, J. P., Husain, I., Conn, C., Stambrook, P., Jhanwar-Uniyal, M. & Dai, W. (2001) J. Biol. Chem. 276, 43305-43312. [DOI] [PubMed] [Google Scholar]
- 25.Maya, R., Balass, M., Kim, S. T., Shkedy, D., Leal, J. F., Shifman, O., Moas, M., Buschmann, T., Ronai, Z., Shiloh, Y., et al. (2001) Genes Dev. 15, 1067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, S. T., Xu, B. & Kastan, M. B. (2002) Genes Dev. 16, 560-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yazdi, P. T., Wang, Y., Zhao, S., Patel, N., Lee, E. Y. & Qin, J. (2002) Genes Dev. 16, 571-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cortez, D., Wang, Y., Qin, J. & Elledge, S. J. (1999) Science 286, 1162-1166. [DOI] [PubMed] [Google Scholar]
- 29.Wu, X., Ranganathan, V., Weisman, D. S., Heine, W. F., Ciccone, D. N., O'Neill, T. B., Crick, K. E., Pierce, K. A., Lane, W. S., Rathbun, G., et al. (2000) Nature 405, 477-482. [DOI] [PubMed] [Google Scholar]
- 30.Zhao, S., Weng, Y. C., Yuan, S. S., Lin, Y. T., Hsu, H. C., Lin, S. C., Gerbino, E., Song, M. H., Zdzienicka, M. Z., Gatti, R. A., et al. (2000) Nature 405, 473-477. [DOI] [PubMed] [Google Scholar]
- 31.Lim, D. S., Kim, S. T., Xu, B., Maser, R. S., Lin, J., Petrini, J. H. & Kastan, M. B. (2000) Nature 404, 613-617. [DOI] [PubMed] [Google Scholar]
- 32.Lin, W. C., Lin, F. T. & Nevins, J. R. (2001) Genes Dev. 15, 1833-1844. [PMC free article] [PubMed] [Google Scholar]
- 33.Shiloh, Y. (2001) Curr. Opin. Genet. Dev. 11, 71-77. [DOI] [PubMed] [Google Scholar]
- 34.Bartkova, J., Horejsi, Z., Koed, K., Kramer, A., Tort, F., Zieger, K., Guldberg, P., Sehested, M., Nesland, J. M., Lukas, C., et al. (2005) Nature 434, 864-870. [DOI] [PubMed] [Google Scholar]
- 35.Gorgoulis, V. G., Vassiliou, L. V., Karakaidos, P., Zacharatos, P., Kotsinas, A., Liloglou, T., Venere, M., Ditullio, R. A., Jr., Kastrinakis, N. G., Levy, B., et al. (2005) Nature 434, 907-913. [DOI] [PubMed] [Google Scholar]
- 36.Lindstrom, M. S. & Wiman, K. G. (2003) Oncogene 22, 4993-5005. [DOI] [PubMed] [Google Scholar]
- 37.Powers, J. T., Hong, S., Mayhew, C. N., Rogers, P. M., Knudsen, E. S. & Johnson, D. G. (2004) Mol. Cancer Res. 2, 203-214. [PubMed] [Google Scholar]
- 38.Rogoff, H. A., Pickering, M. T., Frame, F. M., Debatis, M. E., Sanchez, Y., Jones, S. & Kowalik, T. F. (2004) Mol. Cell. Biol. 24, 2968-2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rounbehler, R. J., Rogers, P. M., Conti, C. J. & Johnson, D. G. (2002) Cancer Res. 62, 3276-3281. [PubMed] [Google Scholar]
- 40.Rounbehler, R. J., Schneider-Broussard, R., Conti, C. J. & Johnson, D. G. (2001) Oncogene 20, 5341-5349. [DOI] [PubMed] [Google Scholar]
- 41.Bakkenist, C. J. & Kastan, M. B. (2003) Nature 421, 499-506. [DOI] [PubMed] [Google Scholar]
- 42.Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S. & Bonner, W. M. (1998) J. Biol. Chem. 273, 5858-5868. [DOI] [PubMed] [Google Scholar]
- 43.Paull, T. T., Rogakou, E. P., Yamazaki, V., Kirchgessner, C. U., Gellert, M. & Bonner, W. M. (2000) Curr. Biol. 10, 886-895. [DOI] [PubMed] [Google Scholar]
- 44.Kitagawa, R., Bakkenist, C. J., McKinnon, P. J. & Kastan, M. B. (2004) Genes Dev. 18, 1423-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fanidi, A., Harrington, E. A. & Evan, G. I. (1992) Nature 359, 554-556. [DOI] [PubMed] [Google Scholar]
- 46.Schmitt, C. A., Fridman, J. S., Yang, M., Baranov, E., Hoffman, R. M. & Lowe, S. W. (2002) Cancer Cell 1, 289-298. [DOI] [PubMed] [Google Scholar]
- 47.Barlow, C., Hirotsune, S., Paylor, R., Liyanage, M., Eckhaus, M., Collins, F., Shiloh, Y., Crawley, J. N., Ried, T., Tagle, D. & Wynshaw-Boris, A. (1996) Cell 86, 159-171. [DOI] [PubMed] [Google Scholar]
- 48.Elson, A., Wang, Y., Daugherty, C. J., Morton, C. C., Zhou, F., Campos-Torres, J. & Leder, P. (1996) Proc. Natl. Acad. Sci. USA 93, 13084-13089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu, Y., Ashley, T., Brainerd, E. E., Bronson, R. T., Meyn, M. S. & Baltimore, D. (1996) Genes Dev. 10, 2411-2422. [DOI] [PubMed] [Google Scholar]
- 50.Liyanage, M., Weaver, Z., Barlow, C., Coleman, A., Pankratz, D. G., Anderson, S., Wynshaw-Boris, A. & Ried, T. (2000) Blood 96, 1940-1946. [PubMed] [Google Scholar]
- 51.Bartkova, J., Bakkenist, C. J., Rajpert-De Meyts, E., Skakkebaek, N. E., Sehested, M., Lukas, J., Kastan, M. B. & Bartek, J. (2005) Cell Cycle 4, 838-845. [DOI] [PubMed] [Google Scholar]
- 52.Soucek, L. & Evan, G. (2002) Cancer Cell 1, 406-408. [DOI] [PubMed] [Google Scholar]
- 53.de Stanchina, E., McCurrach, M. E., Zindy, F., Shieh, S. Y., Ferbeyre, G., Samuelson, A. V., Prives, C., Roussel, M. F., Sherr, C. J. & Lowe, S. W. (1998) Genes Dev. 12, 2434-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harris, S. L. & Levine, A. J. (2005) Oncogene 24, 2899-2908. [DOI] [PubMed] [Google Scholar]
- 55.Undarmaa, B., Kodama, S., Suzuki, K., Niwa, O. & Watanabe, M. (2004) Biochem. Biophys. Res. Commun. 315, 51-58. [DOI] [PubMed] [Google Scholar]
- 56.Hande, M. P., Balajee, A. S., Tchirkov, A., Wynshaw-Boris, A. & Lansdorp, P. M. (2001) Hum. Mol. Genet. 10, 519-528. [DOI] [PubMed] [Google Scholar]
- 57.Smilenov, L. B., Morgan, S. E., Mellado, W., Sawant, S. G., Kastan, M. B. & Pandita, T. K. (1997) Oncogene 15, 2659-2665. [DOI] [PubMed] [Google Scholar]
- 58.Stumm, M., Neubauer, S., Keindorff, S., Wegner, R. D., Wieacker, P. & Sauer, R. (2001) Cytogenet. Cell Genet. 92, 186-191. [DOI] [PubMed] [Google Scholar]
- 59.Liao, M. J., Yin, C., Barlow, C., Wynshaw-Boris, A. & van Dyke, T. (1999) Mol. Cell. Biol. 19, 3095-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Symonds, H., Krall, L., Remington, L., Saenz-Robles, M., Lowe, S., Jacks, T. & Van Dyke, T. (1994) Cell 78, 703-711. [DOI] [PubMed] [Google Scholar]
- 61.Pan, H., Yin, C., Dyson, N. J., Harlow, E., Yamasaki, L. & Dyke, T. V. (1998) Mol. Cell. Biol. 2, 283-292. [DOI] [PubMed] [Google Scholar]
- 62.Ramirez, A., Bravo, A., Jorcano, J. L. & Vidal, M. (1994) Differentiation 58, 53-64. [DOI] [PubMed] [Google Scholar]
- 63.Wu, W. Y. & Morris, R. J. (2005) Methods Mol. Biol. 289, 79-86. [DOI] [PubMed] [Google Scholar]