Abstract
After cell birth, almost all neurons in the mammalian central nervous system migrate. It is unclear whether and how cell migration is coupled with neurogenesis. Here we report that proneural basic helix-loop-helix (bHLH) transcription factors not only initiate neuronal differentiation but also potentiate cell migration. Mechanistically, proneural bHLH factors regulate the expression of genes critically involved in migration, including down-regulation of RhoA small GTPase and up-regulation of doublecortin and p35, which, in turn, modulate the actin and microtubule cytoskeleton assembly and enable newly generated neurons to migrate. In addition, we report that several DNA-binding-deficient proneural genes that fail to initiate neuronal differentiation still activate migration, whereas a different mutation of a proneural gene that causes a failure in initiating cell migration still leads to robust neuronal differentiation. Collectively, these data suggest that transcription programs for neurogenesis and migration are regulated by bHLH factors through partially distinct mechanisms.
Keywords: cortical migration, doublecortin, neuroD, neurogenin, RhoA
During mammalian cortical neurogenesis, the earliest step involves expression of proneural basic helix-loop-helix (bHLH) genes such as neurogenin 1 and 2 (Ngn1 and 2), which initiates a cascade of bHLH gene activation events that eventually lead to the expression of terminal neuronal differentiation genes (1, 2). However, most neurons do not function in their birth places, and newborn neurons often undergo sometimes quite extensive radial and/or tangential migration to various regions of the nervous system (3). It is well documented that cortical lamination results from multiwave neurogenesis and radial migration (3, 4), whereby later-born neurons migrate and surpass the earlier-born neurons to form the “inside-out” cortical laminar structure. Regulation of both actin and microtubule systems is believed to coordinate for successful neuronal migration (5). Mutations and/or deficiencies in many genes have been found to impair cortical neuronal migration (5, 6). These genes include those that encode a secreted protein (reelin), receptor/membrane proteins (e.g., VLDLR, ApoER2, and α3β1 integrin), signaling molecules (e.g., cyclin-dependent kinase 5, p35, and Disabled), and microtubule or cytoplasmic dynein regulators [e.g., doublecortin (Dcx), LIS1, and NUDEL] (5-8).
Although cell migration and neurogenesis are closely connected temporally, the molecular link between the two biological programs is unknown. Dcx, a gene that regulates microtubule polymerization and is indispensable for cortical migration, has recently been proposed as a marker for neurogenesis (9), making it difficult to distinguish whether Dcx belongs to the neurogenic or the migration program. On the other hand, neurogenesis and cell migration do appear to be two somewhat distinct processes, because specific mouse and human gene mutations exist that affect only one of the two events. For instance, a human disease, periventricular heterotopia (10, 11), which is caused by mutations in the filamin1 gene, involves a population of cortical neurons that differentiate normally but fail to migrate, resulting in the accumulation of postmitotic neurons in the periventricular region. In contrast, in mice lacking cell cycle inhibitors p19Ink4d and p27Kip1, a population of cortical neurons fail to exit the cell cycle but migrate to the cortex (12).
In this study, we explored the potential link between the neurogenic and cell migration processes. We found that proneural bHLH factors not only activate the neurogenic machinery but also regulate genes critically involved in cell migration. Moreover, bHLH genes regulate the neurogenic and migration machineries via partially distinct mechanisms because mutations in bHLH genes were found to specifically affect only the migration or the neurogenic machinery. We conclude that the dual functions of bHLH genes couple migration with neurogenesis, enabling newborn neurons to migrate.
Results
Proneural bHLH Genes Not only Induce Neurogenesis but also Enhance Cell Migration. The developing cerebral cortex contains profound radial migration of newly born cortical pyramidal neurons. Major proneural bHLH genes expressed in the developing cortex include Ngn2, Ngn1, Mash1, and NeuroD (Fig. 1a). Ngn1/2 are primarily expressed in the cortical ventricular zone (VZ), whereas occasional Ngn2 expressing cells can be detected in the intermediate zone (IZ). Although NeuroD is regarded as a proneural gene in the developing retina (13), NeuroD is primarily expressed in postmitotic neurons in the IZ of the developing cortex (14), presumably acting downstream of Ngn1/2. Unlike Ngn1/2, Mash1 does not regulate NeuroD. Mash1 is the primary proneural gene in VZ of the ganglionic eminence, which is critical for striatal neurogenesis, whereas cortical VZ expression of Mash1 (Fig. 1a) is likely to be involved in neurogenesis and oligodendrogliogenesis in the forebrain subventricular zone (SVZ) peri- and postnatally (15).
Fig. 1.
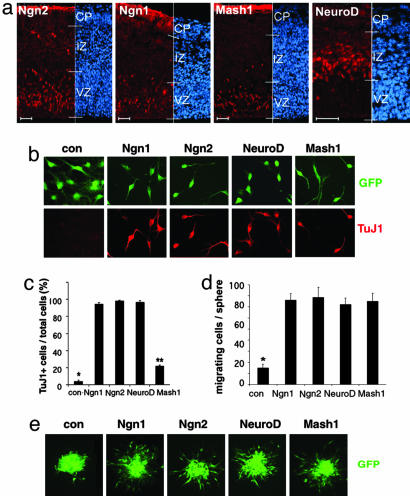
Proneural bHLH factors not only promote neurogenesis but also enhance cell migration. (a) Expression of Ngn2, Ngn1, Mash1, and NeuroD (red) in E14 mouse embryonic cortex. Cells were counterstained with DAPI (blue). (Scale bar: 30 μm.) (b) Expression of TuJ1 (red) in mouse cortical NPCs infected with adenoviruses containing control-, Ngn1-, Ngn2-, NeuroD-, and Mash1-expressing cassettes for 24-48 h. GFP marks infected cells. The quantitative analyses of the experiment are shown in c (*, P < 0.01 compared to the other groups; **, P < 0.01 compared to the other groups; n ≥ 5). (d and e)In cell aggregate migration assay, mouse cortical NPCs infected with adenoviruses at passage 2-3 (P2-3), were fixed after migration from the cell aggregates for 12 h. GFP labels virally infected cells. The quantification of migrating cells is shown in d (*, P < 0.05 compared to the other groups; n ≥ 15).
To explore the potential link between neurogenesis and cortical migration, we introduced replication-deficient adenoviruses carrying either the Ngn1, Ngn2, Mash1, or NeuroD gene expression cassette or a control cassette containing a null mutant Ngn1 (16) into mouse cortical neural stem/progenitor cells (NPCs) at passage 2 [i.e., ≈14 days in vitro upon initial culturing from embryonic day (E) 11/12 mouse cortices in the presence of daily bFGF treatment]. As expected, all four bHLH genes induced neuronal differentiation of NPCs with Mash1 displaying a less-potent neurogenic effect (Fig. 1 b and c). When exogenous bHLH gene expressing NPCs were examined in cell aggregate migration assays, all four bHLH factors were found to enhance cell migration (Fig. 1 d and e), suggesting that neurogenic bHLH factors not only regulate neurogenesis but also enable cortical cells to migrate.
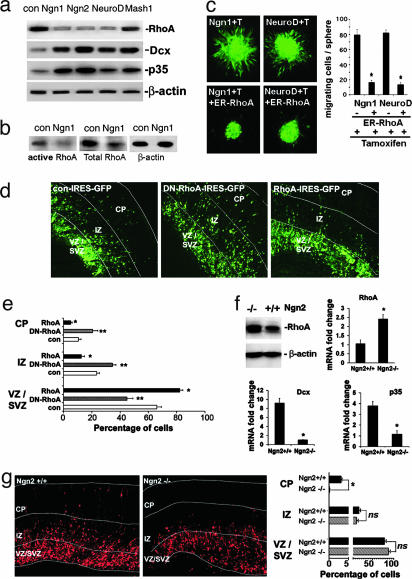
Neurogenic bHLH Factors Negatively and Positively Regulate the Expression of Genes Critically Involved in Cortical Migration. To identify the potential transcription program related to cortical migration, which might be regulated by neurogenic bHLH factors, we took a candidate gene approach. We examined whether neurogenic bHLH factors regulate genes that are known to be critical for cortical migration such as Dcx and p35. In addition, we measured the levels of the Rho family of small GTPases, including RhoA, Rac1, and Cdc42, with or without bHLH gene expression. These small GTPases are well known regulators of cell morphology, adhesion, and motility, which act by modulating the actin cytoskeleton. Our results indicate that neurogenic bHLH factors up-regulate Dcx and p35, and down-regulate RhoA (Fig. 2a; see also Fig. 5, which is published as supporting information on the PNAS web site). Importantly, Ngn1 decreased the overall levels of activated/GTP-bound RhoA in a rhotekin-pull-down assay (17) (Fig. 2b), indicating a reduction in the cellular function of RhoA. Increased expression of Dcx and p35 is expected to promote cortical migration (18), because genetic mutations of these genes lead to profound migration defects in the cortex (7, 19, 20). However, the role of RhoA in cortical migration has not been reported. In situ hybridization of RhoA in the developing cortex indicated that RhoA expression was high in the premigratory cortical ventricular zone and low in the IZ-containing migrating cells (Fig. 6, which is published as supporting information on the PNAS web site). This finding is consistent with the notion that down-regulation of RhoA expression is required for cortical migration. To further determine whether overexpression of RhoA inhibits migration of cortical NPC, we performed NPC aggregate migration assays by using adenovirus carrying a tamoxifen-inducible RhoA or dominant-negative (N19) RhoA (DN-RhoA) expression cassette (Fig. 7 a and b, which is published as supporting information on the PNAS web site), and found that RhoA inhibited, whereas DN-RhoA promoted, migration. Moreover, overexpression of RhoA blocked the migration-inducing effect of Ngn1 and NeuroD as indicated by cell-aggregate and Boyden-chamber migration assays, suggesting that the neurogenic bHLH genes induce cell migration in part by down-regulating RhoA (Fig. 2c; see also Fig. 8, which is published as supporting information on the PNAS web site). To further study cortical migration in a more physiologically relevant setting, we electroporated pCAGGS vector carrying either RhoA or DN-RhoA into E15 mouse cortical VZ followed by cortical slice culture. Our data indicated that ectopic expression of RhoA effectively blocked, whereas DN-RhoA promoted, cortical migration (Fig. 2 d and e).
Fig. 2.
Proneural bHLH factors regulate the expression of RhoA, Dcx, and p35, genes critically involved in cell migration in vitro and in vivo.(a) Western blot of E11 mouse cortical NPCs, infected with control and bHLH gene expression viruses 24 h after infection. β-actin was used as a loading control. (b) Rhotekin-pull-down assay demonstrated that Ngn1 not only decreased total RhoA protein levels but also decreased levels of active/GTP-bound RhoA in cortical NPCs. (c) Cortical NPC aggregate migration assay indicating Tamoxifen-inducible ER-RhoA inhibits cell migration induced by Ngn- and NeuroD-expressing viruses. GFP marks infected cells. Quantification of the experiments is shown in Right (*, P < 0.01 compared to non-ER-RhoA infected group; n ≥10). (d) Ex vivo electroporation of con, DN-RhoA, and RhoA PCAGGS-IRES-GFP constructs into cortical progenitors of wild-type E15 mouse cortices followed by organotypic cortical slice culture for 4 days. Quantification of green cells in each region (VZ/SVZ, IZ, and CP) were shown in e (*, P < 0.05 RhoA compared to control in each zone; **, P < 0.05 DN-RhoA compared to control; n = 15 from total of three independent experiments). (f) Western blot and quantitative RT-PCR of RhoA, Dcx, and P35 in E14 Ngn2-/- and Ngn2 heterozygous mouse cortices. (g) Ex vivo electroporation of red fluorescent protein in cortical progenitors of Ngn2+/+ and Ngn2-/- E15 embryos followed by organotypic cortical slice culture for 3 days. Quantification of red fluorescent protein cells in each zone is shown in Right (*, P < 0.001 compared to the Ngn2-/- group; n ≥ 9 from three independent experiments).
Loss-of-Function Studies Indicate Regulation of RhoA, Dcx, p35, and Cortical Migration by Neurogenic bHLH Genes in Vivo. To determine whether proneural bHLH genes are involved in regulating RhoA, Dcx, and p35 during cortical development in vivo, we used Ngn2-, Ngn1-, and Mash1-knockout mice (21, 22). In situ hybridization analyses demonstrated that the RhoA expressing zone in Ngn1- and Ngn2-double-mutant E14 mouse cortices was expanded. The ratios between RhoA expressing and nonexpressing zones were also higher in Ngn2-/-Ngn1+/-- and Ngn2+/-Ngn1-/--developing cortices as compared to controls (Fig. 9, which is published as supporting information on the PNAS web site). Immunohistochemical analyses indicated that RhoA protein levels were higher in the IZ of Ngn2-/- E14 cortices (Fig. 10, which is published as supporting information on the PNAS web site). In contrast, the expression of p35 and Dcx was substantially decreased in Ngn1-/- and Ngn2-/- cortices (Figs. 9 and 10; ref. 9). More quantitative Western blot and real-time RT-PCR analyses were consistent with the in situ and immunohistochemistry data (Fig. 2f). Mash1 deficiency did not significantly increase RhoA expression in the developing cortex (data not shown), whereas Ngn2 deficiency increased RhoA expression at both the protein and mRNA levels (Fig. 2f), supporting the notion that Ngn2 is a more potent inhibitor for RhoA expression compared to Mash1. To determine whether deregulation of neuronal migration genes due to Ngn2 deficiency leads to defects in cortical migration, we used ex vivo electroporation to label a cohort of cortical VZ/SVZ cells in wild-type (Ngn2+/+) and Ngn2-/- mouse embryos at E15.5 with a red fluorescent protein-expressing plasmid. Three days after cortical slice culturing, some of the labeled cortical cells migrated into the cortical plate in Ngn2+/+, but not in Ngn2-/- cultures, indicating a strong cortical radial migration defect due to Ngn2 deficiency (Fig. 2g).
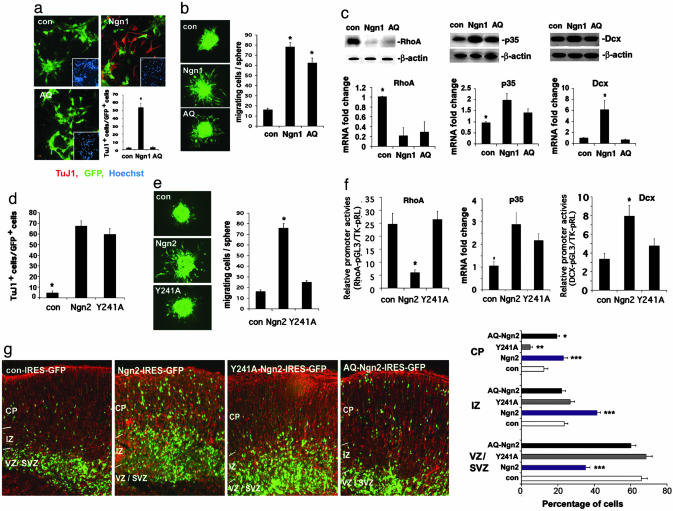
A DNA-Binding Mutant Proneural bHLH Factor Enhances Cortical Cell Migration Without Inducing Neurogenesis. Although it has been reported that Ngn2 knockout embryos do not have any overt early neurogenic phenotype due to compensation by Mash1, it is difficult to assess whether the cell migration defect observed in Ngn2-/- cortices is due to a replacement of the Ngn2-defined dorsal forebrain neurogenic program with a ventral program defined by Mash1 (21) or, alternatively, due to Ngn2 deficiency-induced direct defects in cortical migration. In culture, Ngn1 appears to be able to change cell morphology and reduce cell adhesion without inducing neurogenesis (Fig. 11, which is published as supporting information on the PNAS web site). To address whether proneural bHLH genes may induce cortical migration independent of their ability to trigger neurogenesis, we used a mutant form of Ngn1/2, AQ-Ngn1/2, which harbors two amino acid mutations at the C terminus of its basic domain (16). AQ-Ngn1/2, although localized to the nucleus, cannot bind to E-box elements (i.e., DNA-binding cis elements for proneural bHLH factors) and, therefore, does not activate the canonical Ngn1/2 downstream genes, such as NeuroD, and fails to initiate the neurogenic transcriptional activation cascade (16). When AQ-Ngn1 was introduced into E11 mouse cortical NPCs at passage 3 (P3, ≈21 days in vitro), it did not induce neurogenesis (Fig. 3a). However, both Ngn1 and AQ-Ngn1 induced cell migration in cortical NPC aggregate and Boyden chamber migration assays (23) (Fig. 3b; see also Fig. 12a, which is published as supporting information on the PNAS web site). As expected, unlike Ngn1, AQ-Ngn1 enhanced migration of nestin-positive neural progenitors that do not express neuronal markers (Fig. 12b). Together, these observations suggest that Ngn1 promotes NSC migration independent of its neurogenic function.
Fig. 3.
Regulation of neurogenesis and migration by AQ- and Y241A-Ngn1/2. (a) Infection of mouse E11 NPCs with con, Ngn1, and AQ indicates that AQ-Ngn1 fails to induce neurogenesis when examined 24 h after infection. TuJ1 (red) marks neuronal cells. Hoechst labels nuclei. GFP shows the infected cells.(*, P < 0.01 compared to the other groups; n ≥ 5). (b) GFP-marked infected cells in the cell aggregate migration assay (*, P < 0.05 compared to control; n ≥ 15). (c) RhoA, P35, and Dcx expression in mouse E11 cortical NPCs infected with control (con), Ngn1, or AQ-Ngn1 viruses, 24 h after infection. (c Upper) Western blotting. (c Lower) Quantitative RT-PCR (*, P < 0.01 compared to the rest of the groups; n ≥ 4). (d) Quantification of neurogenesis induced by con, Ngn2, or Y241A infection of P5 mouse E11 NPCs as measured by TuJ1 staining. (*, P < 0.05 compared to the rest of the groups; n = 5). (e) Cell aggregate migration assay in E11 mouse cortical NPCs. Quantification is shown in Right (*, P < 0.05 compared to the other groups; n = 6). (f) Luciferase analysis of RhoA and Dcx promoter activities in mouse E14 primary neuronal cultures infected with Ngn2 and Y241A expressing viruses (*, P < 0.05 compared to the rest of the groups; n = 9), Quantitative RT-PCR indicating less potent effect of Y241A-Ngn2 in inducing p35 expression. (g) Ex vivo electroporation of con, Ngn2, Y241A, and AQ-Ngn2 pCAGGS-IRES-GFP constructs into E15 wild-type mouse cortices followed by organotypic slice culture for 4 days. Cortical slices were double labeled with Nestin (red) and GFP (green). Quantification of green cells in each zone (VZ/SVZ, IZ, and CP) were shown in Right (*, P < 0.05 AQ-Ngn2 compared to control; **, P < 0.05 Y241ANgn2 compared to control; ***, P < 0.05 Ngn2 compared to control; n = 15 from three independent experiments).
To further determine whether AQ-Ngn1, similar to Ngn1, regulates cell migration-related genes, including RhoA, Dcx, and p35, Western blot and quantitative RT-PCR analyses were performed on cortical NPCs overexpressing AQ-Ngn1. Our results indicated that AQ-Ngn1 was capable of down-regulating RhoA and slightly up-regulating p35 and serine 732 phosphorylation of focal adhesion kinase (Figs. 3c and 12c). On the other hand, AQ-Ngn1 did not increase NeuroD or Dcx expression (Fig. 3c and 12d), suggesting that E-box binding is indispensable for Ngn1 to regulate Dcx and NeuroD. E-box binding is partially required for p35 regulation but not required for RhoA regulation by Ngn1 (Fig. 3c).
A Mutant Ngn2 Induces Neurogenesis Without Enhancing Cortical Cell Migration. Although the E-box binding mutant of Ngn1, AQ-Ngn1, is capable of inducing cortical cell migration without promoting neurogenesis, another mutant form of Ngn2, Y241A-Ngn2, was capable of doing the opposite. It has been proposed that tyrosine 241(Y241) of the Ngn2 protein could potentially be phosphorylated and that this posttranslational modification might be involved in the normal function of Ngn2 (24). Mutation of Y241 to alanine (A) blocked cortical cell migration without changing its ability to regulate the NeuroD promoter or initiating the neurogenic program (Fig. 3 d and e; see also Fig. 13a, which is published as supporting information on the PNAS web site). In young cortical neurons, Y241A-Ngn2, unlike Ngn2, did not appear to inhibit the RhoA promoter or decrease endogenous RhoA mRNA levels (Fig. 3f and 13b). Y241A-Ngn2 was also less effective in activating the Dcx promoter or p35 expression at both mRNA and protein levels, as compared to wild-type Ngn2 (Fig. 3f and 13c). When we electroporated control, Ngn2, AQ-Ngn2, and Y241A-Ngn2 overexpression constructs into E15 mouse cortical slices, followed by 4 days of culturing, we found that Ngn2 and AQ-Ngn2 enhanced cortical migration, whereas Y241A-Ngn2 failed to enhance migration (Fig. 3g). Together the data suggest that Ngn1/2 induces cortical migration and neurogenesis through distinct mechanisms.
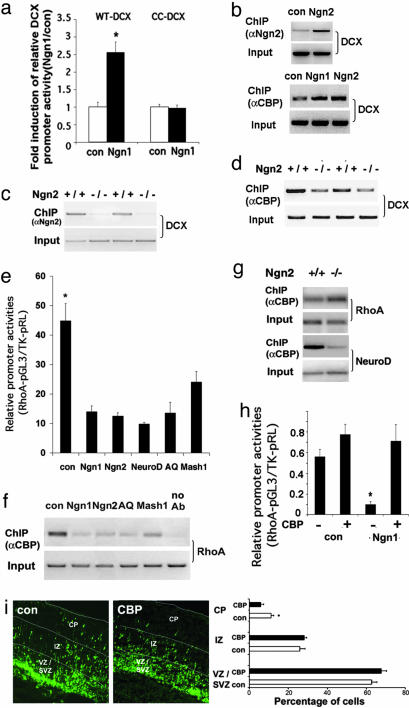
Molecular Mechanisms by Which Ngn1/2 Regulate Dcx, NeuroD, and RhoA. To explore the molecular mechanisms by which proneural bHLH factors regulate genes such as Dcx or p35, we used the genomatrix/matinspector program (Genomatix Software, Munich, Germany) to find that the Dcx promoter contains canonical Ngn1/2-, NeuroD-, and Mash1-binding E-box elements within the 2 kb upstream region from Dcx transcriptional initiation site. We cloned this 2-kb Dcx promoter and constructed a promoter-luciferase reporter plasmid. Like the NeuroD promoter, the 2-kb Dcx promoter was inducible by Ngn1, and mutating the putative E-box element (from -910 bp 5′-CATCTG-3′ -905 bp into -910 bp 5′-CACCCG-3′ -905 bp) within the promoter completely abolished promoter activation by Ngn1 (Fig. 4a). Chromatin immunoprecipiation (ChIP) analyses further demonstrated that Ngn1/2 directly binds to the Dcx promoter both in cultured NPCs and in vivo in the developing mouse cortex (E14) (Fig. 4 b and c). The association of Ngn2 with the promoter helps to recruit a transcriptional coactivator, CBP, which contains histone acetyltransferase activity, to activate the promoter (Fig. 4b). In addition, less CBP was found to associate with the Dcx promoter in E14 Ngn2-/- cortices (Fig. 4 c and d), supporting the in vivo role of Ngn2 in CBP recruitment to the Dcx promoter. Within the 2-kb promoter region of the p35 gene, there are multiple E-box elements, some of which are canonical for Mash1 (5′-CAGGTG-3′). It remains to be determined whether any of the proneural bHLH factors directly bind to the p35 promoter.
Fig. 4.
Proneural bHLH factors regulate RhoA and Dcx, partially by redistributing CBP between RhoA and Dcx promoters. (a) Luciferase analysis of wild-type Dcx promoter (WT-Dcx) and an E-box element mutant (CC-Dcx) Dcx promoter activities in E11 mouse NPC cultures infected with control or Ngn1 virus (*, P < 0.01 compared to the rest of the group; n ≥ 12). (b) ChIP analysis of E11 mouse NPC culture shows greater association of Ngn2 and CBP with the Dcx promoter with exogenous Ngn1/2 expression. (c and d) ChIP analyses of Ngn2-/- E14 mouse cortices indicate decreased Ngn2/CBP association with the Dcx promoter due to Ngn2 deficiency. (e) Luciferase analysis of the RhoA promoter in E11 mouse cortical NPCs 24 h after infection (*, P < 0.05 compared to the rest of the group; n ≥ 12). (f and g) ChIP analysis showing changes of CBP association with RhoA promoter in both E11 NPC cultures with exogenous bHLH gene expression (f), and in Ngn2-/- mouse E14 cortices (g). (h) Luciferase analysis of RhoA promoter in E11 mouse NPC cultures, 24 h after cotransfection of CBP with con or Ngn1 plasmid (*, P < 0.01 compared to the rest of the group; n ≥ 12). (i) Electroporation of CBP into E15 mouse cortices followed by 4-day slice culturing (*, P < 0.05 compared to CBP in CP; n = 15 from three independent experiments).
To understand the potential mechanisms by which proneural bHLH genes negatively regulate RhoA expression, we isolated a 2-kb mouse RhoA promoter (-2,112 bp to + 75 bp) and cloned it into a luciferase reporter construct. When the RhoA promoter luciferase construct was introduced into mouse NPCs, coexpression of Ngn1, Ngn2, NeuroD, or AQ-Ngn1 significantly decreased RhoA promoter activity. Mash1, although less effectively, still significantly decreased RhoA-luciferase activity (Fig. 4e). We have previously shown that Ngn1 and AQ-Ngn1 inhibit astroglial genes by displacing transcription coactivators such as p300/CBP away from the glial promoters onto neuronal differentiation genes (16). We hypothesized that similar mechanisms might also be used by bHLH factors to down-regulate the RhoA promoter. CBP ChIP assays in NPCs indicated that expression of Ngn1/2, AQ-Ngn1, and Mash1 caused decreased association of CBP with the endogenous RhoA promoter (Fig. 4f). To test the in vivo relevance of this finding, we performed ChIP analyses by using Ngn2-knockout and control mouse embryonic cortices. Consistent with the CBP redistribution model, we found more CBP association with the RhoA promoter and less with the Ngn2 target, the NeuroD promoter, in Ngn2-/- E14 mouse cortices as compared to those in Ngn2+/+ cortices (Fig. 4g). To further interrogate the CBP sequestration model, we overexpressed CBP in NPCs and found that CBP expression could reverse the effect of RhoA promoter inhibition by Ngn1 (Fig. 4h). In addition, CBP expression also prevented down-regulation of endogenous RhoA mRNA levels induced by Ngn1 or Ngn2 (Fig. 14a, which is published as supporting information on the PNAS web site). When the CBP overexpression vector was introduced into E15 mouse cortices, cortical migration was inhibited (Fig. 4i). Together these data support the notion that displacement of CBP from the RhoA promoter by bHLH factors down-regulates RhoA, which, in part, is involved in proneural gene-induced cortical cell migration.
Discussion
Coupling of Cell Migration with Neurogenesis by Neurogenic bHLH Genes. In the developing central nervous system, neurogenesis is tightly coordinated with subsequent neuronal migration. However, the molecular mechanisms underlying the coupling of these two events are unknown. Our results indicate that both the neurogenic and the cell migration machineries can be independently regulated by proneural bHLH genes such as Ngn1/2. Specifically, a mutant form of Ngn1/2 (AQ-Ngn1/2) that does not activate neurogenensis still enhances cortical cell migration, whereas another mutant form of Ngn2 (Y241A-Ngn2) that is fully capable of inducing neurogenesis does not promote cell migration. Thus, proneural bHLH genes serve as molecular “linkers” connecting neurogenesis with migration.
Neurogenic bHLH Genes Regulate the Cell Migration Machinery in Vivo. Previous Ngn1/2 knockout studies have demonstrated certain cortical phenotypes that could have resulted from defects in cell migration (22, 25). However, because Ngn1/2 are known to be cell fate determination factors, the anatomical abnormalities observed in knockout mice have been attributed to misspecification of the neuronal fate, which is assumed to lead subsequently to changes in cell migration. Our studies using Ngn1/2 mutations indicate that neurogenic bHLH genes could influence the cell migration property independent of cell fate specification and vice versa, suggesting that cell fate and migration are regulated in parallel by neurogenic bHLH genes (Fig. 15, which is published as supporting information on the PNAS web site).
Both of our gain- and loss-of function studies demonstrate that neurogenic bHLH factors regulate a number of genes related to the cell migration machinery. In addition, bHLH genes modulate the migration machinery through both gene activation and inhibition. We believe that down-regulation of RhoA and up-regulation of p35 and Dcx are merely a representation of a broader spectrum of genes involved in cell migration that are targeted by neurogenic bHLH factors. Because the function of Rho, Rac, and Cdc42 is thought to be regulated primarily through their association with GTP or GDP (26, 27), it is possible that Ngn1/2 may also regulate the function of the Rho family of small GTPases by controlling the expression of GEFs and GAPs. However, suppression of RhoA gene expression provides more-effective long-lasting inhibition of RhoA function as compared to regulation of GEFs and GAPs.
Among the bHLH genes we tested, NeuroD is primarily expressed in the cortical IZ. Thus, we propose that proneural bHLH genes expressed in the VZ including Ngn1/2 are involved in the initial down-regulation of RhoA right before NPCs become post-mitotic, whereas Ngn1/2 target genes, such as NeuroD, are primarily involved in the continuous suppression of RhoA during cortical neuronal migration. In addition to NeuroD, other neurogenic bHLH factors, including Math2, neuroD2, and Nscl1, may also suppress RhoA expression and participate in regulating the migration machinery in postmitotic neurons, which might explain the lack of cortical development phenotype in neuroD-/- mice.
Molecular Mechanisms by Which Neurogenic bHLH Genes Regulate Migration-Related Genes. Functional analyses of AQ-Ngn1/2 suggest that neurogenic bHLH factors regulate migration genes through various mechanisms. In fact, RhoA, p35, and Dcx represent three types of target genes for Ngn1/2. RhoA expression is inhibited by Ngn1/2 independent of their E-box binding ability. Dcx expression is activated directly by Ngn1/2 binding to the E-box element within the Dcx promoter, whereas the positive regulation of p35 expression by bHLH factors depends only partially on E-box binding.
The redistribution of transcription coactivators such as CBP appears to be critical for both activation and inhibition of the migration genes by neurogenic bHLH factors (28). Our CBP ChIP analyses indicated that expression of neurogenic bHLH factors displaced CBP away from the RhoA promoter and onto their direct target genes such as Dcx and NeuroD. This redistribution of transcriptional coactivators allows for simultaneous inhibition and activation of genes. Overexpression of CBP can reverse RhoA suppression by Ngn1/2, further supporting the CBP displacement model. In addition, AQ-Ngn1 binds to CBP but not to the E-box element (Fig. 14b), indicating that AQ-Ngn1 is capable of displacing CBP away from the RhoA promoter and suppressing RhoA expression without activating Dcx or NeuroD.
The Y241A mutation of Ngn2, in contrast to AQ-Ngn2, blocks cell migration without inhibiting neurogenesis. We found that the Y241A mutation of Ngn2 reduced the ability of Ngn2 to associate with CBP (Fig. 14c), which could, in part, explain the decreased ability of Y241A-Ngn2 to inhibit RhoA and activate Dcx and p35. We propose that different Ngn1/2 target genes might have different dependence on CBP. In support of this hypothesis, we found that overexpression of CBP blocked the inhibitory effect of Ngn1 on the RhoA promoter, whereas the effect of Ngn1 on the NeuroD promoter was not further enhanced by CBP overexpression (Fig. 16 a and b, which is published as supporting information on the PNAS web site). Conversely, in the presence of lower amount of CBP small interfering RNA (siRNA), whereas the NeuroD promoter was not significantly affected, the RhoA promoter activity was reduced, suggesting that the RhoA promoter is more sensitive to CBP deficiency than the NeuroD promoter. At a higher concentration, CBP siRNA attenuated Ngn-induced RhoA and NeuroD promoter activation, suggesting that the NeuroD promoter still uses CBP for gene activation (Fig. 16 c-f). It remains to be determined why the NeuroD promoter depends less on CBP as compared to the RhoA promoter. It is possible that additional transcriptional coactivators are used by Ngn1/2 to regulate the NeuroD promoter. Taken together, the low CBP dependence of NeuroD-like neurogenic promoters as compared to that of migration-related genes might partially explain the migration defect without a neurogenic defect caused by Y241A mutation of Ngn2.
Although we tested only CBP redistribution in this study, other transcription cofactors of the proneural bHLH genes including the SWI-SNF complex (29) may also be displaced by bHLH factors in a fashion similar to CBP. Because accumulating evidence suggests that different Ngn1/2 target genes may use different transcriptional activating complexes, our studies on CBP displacement merely provide a mechanistic model whereby redistribution of transcriptional coactivators could be used to coordinate gene inhibition and activation. In addition, our studies suggest that during neuronal fate determination, many genes related to later neuronal differentiation processes are induced by proneural genes and that parallel regulation of different modular transcriptional programs could be a feature common to all cell fate-specification factors.
Methods
Transgenic Mice, Cell Cultures, Transfection, and Viral Infection. Transgenic mice with mutations in Ngn2 and Mash1 were generated as reported in ref. 22. Mouse cortical NPCs from E11.5 cortices were cultured as described in ref. 30. Primary neurons from E14 mouse cortices were cultured in serum containing medium as described in ref. 31. FuGENE 6 (Roche) reagent was used to transfect NPCs as described in ref. 30, and calcium phosphate based method was used for cortical neuronal transfection (31). Replication-deficient adenoviruses carrying various bHLH gene expression cassettes were generated as described in ref. 16. High infection efficiency (>90%) was achieved in NPC cultures 24 h after virus infection (see also Fig. 17, which is published as supporting information on the PNAS web site).
Immunostaining of Cortical Tissue, Cultured Cortical Slice, and NPCs. The fixation and immunostaining procedure, as well as the antibody information, are listed in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. Images were captured by an Olympus fluorescence microscope.
ChIP Analysis, Western Blot and RT-PCR Analyses, and Promoter Assays. Cultured NPCs or minced E14 mouse cortices were cross-linked with 1% formaldehyde for 20 min at room temperature for ChIP analysis as described in ref. 31. Western blot and RT-PCR analyses were performed as described in ref. 32. Detailed methods of promoter assays as well as antibody and primer information are listed in Supporting Materials and Methods.
Cell Aggregate Migration Assay, ex Vivo Electroporation, Cortical Slice Culture, and Migration Analyses. Detailed methods are published in Supporting Materials and Methods.
Statistical Analysis. All statistical analyses in the study were carried out by using one-way ANOVA plus Fisher's post hoc test.
Supplementary Material
Acknowledgments
We thank Dr. Tracy Xie (University of California, Los Angeles) for providing the NVSLM1 motorized vibratome, Dr. Franck Polleux for communicating unpublished observations, and Drs. David Anderson and Jane Johnson (University of Texas Southwestern, Dallas) for providing antibodies. This work is supported by Basil O'Connor Starter Scholar Awards, a Whitehall Foundation Grant, a Beckman Young Investigator Award, and a Sloan Fellowship (to Y.E.S.).
Author contributions: W.G., J.G.G., F.G., and Y.E.S. designed research; W.G., F.H., K.J.K., B.B., V.C., L.N., X.W., J.Z., J.I.-T.H., K.M., J.T., H.W., M.M.S., and Y.E.S. performed research; W.G., F.H., K.J.K., B.B., L.N., X.W., D.C., G.C., J.G.G., M.E.G., F.G., and Y.E.S. contributed new reagents/analytic tools; W.G., F.H., K.J.K., F.G., and Y.E.S. analyzed data; and W.G. and Y.E.S. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: bHLH, basic helix-loop-helix; ChIP, chromatin immunoprecipiation; Dcx, doublecortin; En, embryonic day n; IZ, intermediate zone; NPC, neural stem/progenitor cell; SVZ, subventricular zone; VZ, ventricular zone.
References
- 1.Bertrand, N., Castro, D. S. & Guillemot, F. (2002) Nat. Rev. Neurosci. 3, 517-530. [DOI] [PubMed] [Google Scholar]
- 2.Ma, Q., Sommer, L., Cserjesi, P. & Anderson, D. J. (1997) J. Neurosci. 17, 3644-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer, S. A. & Altman, J. (1991) Neocortical Development (Raven, New York), pp. 116-128.
- 4.Caviness, V. S., Crandall, J. E. & Edwards, M. A. (1988) in Cerebral Cortex, eds. Peters, A. & Jones, E. G. (Plenum, New York), Vol. 7, pp. 59-89. [Google Scholar]
- 5.Lambert de Rouvroit, C. & Goffinet, A. M. (2001) Mech. Dev. 105, 47-56. [DOI] [PubMed] [Google Scholar]
- 6.Nadarajah, B. & Parnavelas, J. G. (2002) Nat. Rev. Neurosci. 3, 423-432. [DOI] [PubMed] [Google Scholar]
- 7.Dhavan, R. & Tsai, L. H. (2001) Nat. Rev. Mol. Cell. Biol. 2, 749-759. [DOI] [PubMed] [Google Scholar]
- 8.Smith, D. S. & Tsai, L. H. (2002) Trends Cell Biol. 12, 28-36. [DOI] [PubMed] [Google Scholar]
- 9.Mattar, P., Britz, O., Johannes, C., Nieto, M., Ma, L., Rebeyka, A., Klenin, N., Polleux, F., Guillemot, F. & Schuurmans, C. (2004) Dev. Biol. 273, 373-389. [DOI] [PubMed] [Google Scholar]
- 10.Fox, J. W., Lamperti, E. D., Eksioglu, Y. Z., Hong, S. E., Feng, Y., Graham, D. A., Scheffer, I. E., Dobyns, W. B., Hirsch, B. A., Radtke, R. A., et al. (1998) Neuron 21, 1315-1325. [DOI] [PubMed] [Google Scholar]
- 11.Fox, J. W. & Walsh, C. A. (1999) Am. J. Hum. Genet. 65, 19-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zindy, F., Cunningham, J. J., Sherr, C. J., Jogal, S., Smeyne, R. J. & Roussel, M. F. (1999) Proc. Natl. Acad. Sci. USA 96, 13462-13467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrow, E. M., Furukawa, T., Lee, J. E. & Cepko, C. L. (1999) Development (Cambridge, U.K.) 126, 23-36. [DOI] [PubMed] [Google Scholar]
- 14.Sommer, L., Ma, Q. & Anderson, D. J. (1996) Mol. Cell. Neurosci. 8, 221-241. [DOI] [PubMed] [Google Scholar]
- 15.Parras, C. M., Galli, R., Britz, O., Soares, S., Galichet, C., Battiste, J., Johnson, J. E., Nakafuku, M., Vescovi, A. & Guillemot, F. (2004) EMBO J. 23, 4495-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun, Y., Nadal-Vicens, M., Misono, S., Lin, M. Z., Zubiaga, A., Hua, X., Fan, G. & Greenberg, M. E. (2001) Cell 104, 365-376. [DOI] [PubMed] [Google Scholar]
- 17.Shamah, S. M., Lin, M. Z., Goldberg, J. L., Estrach, S., Sahin, M., Hu, L., Bazalakova, M., Neve, R. L., Corfas, G., Debant, A. & Greenberg, M. E. (2001) Cell 105, 233-244. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka, T., Serneo, F. F., Higgins, C., Gambello, M. J., Wynshaw-Boris, A. & Gleeson, J. G. (2004) J. Cell Biol. 165, 709-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gleeson, J. G., Allen, K. M., Fox, J. W., Lamperti, E. D., Berkovic, S., Scheffer, I., Cooper, E. C., Dobyns, W. B., Minnerath, S. R., Ross, M. E. & Walsh, C. A. (1998) Cell 92, 63-72. [DOI] [PubMed] [Google Scholar]
- 20.Gleeson, J. G., Lin, P. T., Flanagan, L. A. & Walsh, C. A. (1999) Neuron 23, 257-271. [DOI] [PubMed] [Google Scholar]
- 21.Fode, C., Ma, Q., Casarosa, S., Ang, S. L., Anderson, D. J. & Guillemot, F. (2000) Genes Dev. 14, 67-80. [PMC free article] [PubMed] [Google Scholar]
- 22.Nieto, M., Schuurmans, C., Britz, O. & Guillemot, F. (2001) Neuron 29, 401-413. [DOI] [PubMed] [Google Scholar]
- 23.Mason, H. A., Ito, S. & Corfas, G. (2001) J. Neurosci. 21, 7654-7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hand, R., Bortone, D., Mattar, P., Nguyen, L., Heng, J. I., Guerrier, S., Boutt, E., Peters, E., Barnes, A. P., Parras, C., et al. (2005) Neuron 48, 45-62. [DOI] [PubMed] [Google Scholar]
- 25.Schuurmans, C., Armant, O., Nieto, M., Stenman, J. M., Britz, O., Klenin, N., Brown, C., Langevin, L. M., Seibt, J., Tang, H., Cunningham, J. M., et al. (2004) EMBO J. 23, 2892-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall, A. (1992) Mol. Biol. Cell 3, 475-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raftopoulou, M. & Hall, A. (2004) Dev. Biol. 265, 23-32. [DOI] [PubMed] [Google Scholar]
- 28.Koyano-Nakagawa, N., Wettstein, D. & Kintner, C. (1999) Mol. Cell Neurosci. 14, 327-339. [DOI] [PubMed] [Google Scholar]
- 29.Seo, S., Richardson, G. A. & Kroll, K. L. (2005) Development (Cambridge, U.K.) 132, 105-115. [DOI] [PubMed] [Google Scholar]
- 30.Ge, W., Martinowich, K., Wu, X., He, F., Miyamoto, A., Fan, G., Weinmaster, G. & Sun, Y. E. (2002) J. Neurosci. Res. 69, 848-860. [DOI] [PubMed] [Google Scholar]
- 31.Martinowich, K., Hattori, D., Wu, H., Fouse, S., He, F., Hu, Y., Fan, G. & Sun, Y. E. (2003) Science 302, 890-893. [DOI] [PubMed] [Google Scholar]
- 32.He, F., Ge, W., Martinowich, K., Becker-Catania, S., Coskun, V., Zhu, W., Wu, H., Castro, D., Guillemot, F., Fan, G., et al. (2005) Nat. Neurosci. 8, 616-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.