Abstract
Current theory indicates that mitochondria were obtained 1.5 billion years ago from an ancient prokaryote. The mitochondria provided the capacity for aerobic respiration, the creation of the eukaryotic cell, and eventually complex multicellular organisms. Recent reports have found that mitochondria play essential roles in aging and determining lifespan. A variety of heritable and acquired diseases are linked to mitochondrial dysfunction. We report here that mitochondria are more dynamic than previously considered: mitochondria or mtDNA can move between cells. The active transfer from adult stem cells and somatic cells can rescue aerobic respiration in mammalian cells with nonfunctional mitochondria.
Keywords: human bone marrow, nonhematopoietic, stem/progenitor cells, ischemia
Mitochondria are essential organelles in plant and animal cells that are from a prokaryotic ancestor and play a key role in processes such as oxidative phosphorylation, aerobic metabolism of glucose and fat, calcium signaling, and apoptosis (1, 2). The human mitochondrial genome is 16,568 bp and encodes a limited number of mitochondria-specific proteins, rRNAs, and tRNAs (3). All other mitochondrial proteins are encoded in the nucleus. The mitochondrial genome is maternally inherited and undergoes a high rate of mutation because mtDNA is not protected by histones, is inefficiently repaired (4), and is exposed to oxygen radicals generated by oxidative phosphorylation (1).
Nearly every tissue contains stem-like progenitor cells that repair tissues after damage (5), but local repair can be supplemented by stem-like progenitor cells from the bone marrow. Stem/progenitor cells repair tissues by differentiating to replace lost cells, providing cytokines and growth factors, and by cell fusion with endogenous cells.
In this article, we ask whether stem/progenitor cells or other somatic cells can repair cells with nonfunctional mitochondria by transfer of functional mitochondria or mtDNA. We used cells that were pretreated with ethidium bromide so that the mtDNA became mutated and depleted and the cells became incapable of aerobic respiration and growth (A549 ρ° cells), except in a permissive medium containing uridine and pyruvate to supplement anaerobic glycolysis (6, 7). The A549 ρ° cells were cocultured with either adult nonhematopoietic stem/progenitor cells from human bone marrow (hMSCs) or with skin fibroblasts. The cocultures produced clones of rescued A549 ρ° cells with functional mitochondria.
Results
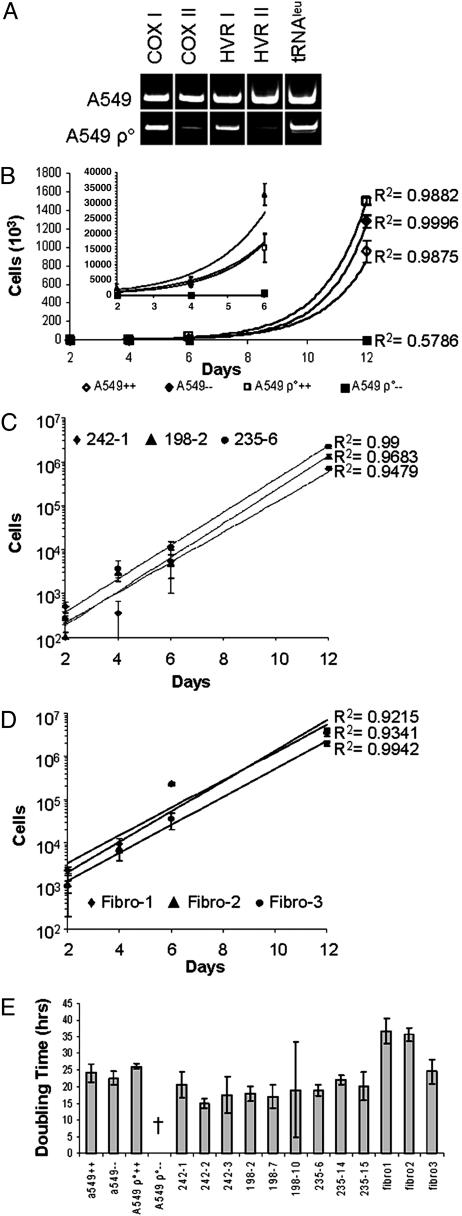
Rescue of A549 ρ° Cells by Coculture with hMSCs. PCR assays indicated that a number of mitochondrial genes and DNA sequences were absent or depleted in A549 ρ° cells: the genes for cytochrome oxidase I and II, the gene for tRNA leucine, and hypervariable regions I and II of the D loop (Fig. 1A). Some of the PCR products may have been generated by nonfunctional nuclear pseudogenes in the A549 ρ° cells that are only detectable after mtDNA depletion (8). Sequencing of the PCR products demonstrated additional extensive damage and mutations of A549 ρ° mtDNA (data not shown). The A549 ρ° cells expanded in a permissive medium that contained pyruvate and uridine to stimulate anaerobic glycolysis (Fig. 1B). In contrast, some A549 ρ° cells persist but do not expand when observed for up to 2 months in the restrictive medium lacking uridine and pyruvate.
Fig. 1.
Exponential growth of rescued A549 ρ° clones. (A) PCR analysis of 100 ng of DNA reveals depletion in mtDNA from A549 ρ° cells for COX I and COX II, the D-loop hypervariable regions I and II (HVR I and HVR II), and the leucine transfer RNA (tRNAleu). (B) Growth of the parental A549 cell line and the mtDNA-depleted cell line A549 ρ° in permissive medium containing pyruvate and uridine (⋄, A549++; □, A549 ρ°++) and in restrictive medium lacking supplements (♦, A549–; ▪, A549 ρ°–). (Inset) Expanded scale (0 to 40,000 cells) to indicate growth of cells early in the incubation period. (C) Exponential growth is restored in three representative rescued A549 ρ° clones after transfer of hMSC mitochondria. Each clone is from a different human donor and from a separate coculture experiment (♦, 242-1; ▴, 198-2; •, 235-6). (D) Exponential growth of the rescued A549 ρ° clones derived from fibroblast mitochondria transfer (♦, Fibro-1; ▴, Fibro-2; •, Fibro-3). (E) Doubling times of the parental A549 cell line, A549 ρ° cells, and rescued A549 ρ° clones. (++, growth in permissive medium; ––, growth in restrictive medium). Note that some of the rescued clones have shorter doubling times than the original A549 cells. †, negative value indicating no growth. Error bars represent one standard deviation from the mean (B–D) or the greatest variation of the maximum or minimum doubling times from average between three replicates at day 6 and day 12 (E).
For the coculture experiments, A549 ρ° cells were grown to confluency in permissive medium, GFP-labeled hMSCs were added, and the cultures continued in the permissive medium. hMSCs added to the culture joined the A549 ρ° monolayer and flattened into a fibroblast-like morphology. The cultures became very active, rapidly acidifying the medium and causing mainly A549 ρ° cell death. After 4 days, the medium was then replaced with restrictive medium lacking pyruvate and uridine. Microscopic examination of the cultures revealed GFP-negative colonies of small, round, morphologically distinct A549 cells that formed over the next 10 days (not shown). In control cultures, hMSCs grew in the restrictive medium; however, they became senescent in cocultures, apparently because of contact inhibition with A549 ρ° cells. Similar results were obtained with fibroblasts.
In coculture experiments using hMSCs from six different individuals, we were able to isolate over a hundred single cell-derived, A549 colonies that grew in restrictive medium. The rescued A549 colonies were easily distinguished by the lack of GFP and their distinct A549-like morphology compared with hMSCs. All of the rescued clones recovered from cocultures grew exponentially in the restrictive medium (Fig. 1C). Similar results were obtained with rescued clones isolated from cocultures of humanized GFP (hrGFP)+ human skin fibroblasts and A549 ρ° cells (Fig. 1D). Doubling times of the rescued clones were comparable with the doubling times of A549 cells (Fig. 1E). For reasons that were not apparent, a few of the hMSC-rescued clones grew more rapidly than did the control A549 cells (clones 242-2, 198-2, and 198-7; Fig. 1E).
To assess the frequency of rescue of A549 ρ° cells, we performed a limiting dilution analysis of the cocultures. We repeated the coculture experiments with decreasing numbers of hrGFP+ hMSCs from three different donors (d202, d224, and d235) and counted colonies of GFP-negative A549 cells that grew in restrictive medium. The results (See Table 2, which is published as supporting information on the PNAS web site) indicated that the addition of as few as 100 hMSCs was sufficient to generate one rescued A549 ρ° cell colony in 10 days.
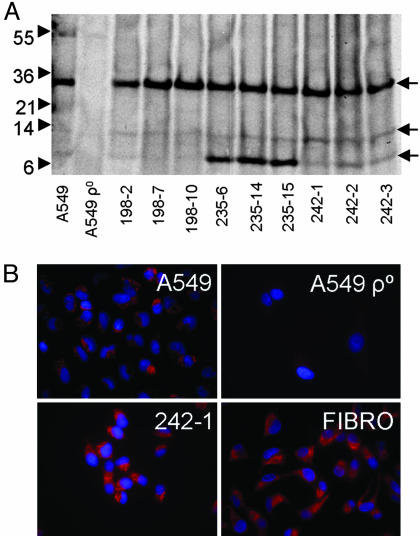
Protein Expression from Mitochondria in Rescued Clones. Mitochondria encode a number of their own proteins, and translation occurs in the mitochondrial matrix along the inner membrane. To demonstrate the functionality of mitochondria in the rescued cells, clones from three cocultures were incubated with [35S]methionine in the presence of emetine to inhibit cytoplasmic translation. The parent A549 cells generated 35S-labeled bands of newly synthesized protein (Fig. 2A). As expected, no 35S-labeled proteins were synthesized by the A549 ρ° cells. Distinct bands of proteins similar to those synthesized by A549 cells were generated by the rescued clones from hMSC cocultures. Interestingly, three clones that were rescued with hMSCs from one donor (d235) synthesized increased amounts of an 8-kDa band, indicating a distinct pattern of mitochondrial protein expression not present in the parental A549 cells.
Fig. 2.
Translation of mitochondrial proteins in rescued A549 ρ° clones. (A) Metabolic labeling of A549 cells, A549 ρ° cells, and rescued A549 ρ° clones was for 5 h with [35S]methionine in methionine-free medium in the presence of emetine to inhibit translation of proteins encoded in the nucleus. Mitochondria were isolated by differential centrifugation, and 10 μg of mitochondria protein was analyzed per sample. A549 ρ° cells did not translate mitochondria proteins, whereas the rescued A549 ρ° clones translate proteins of similar molecular weights to those produced by the parental cell line (arrows). Note that the clones derived from mitochondria transfer from donor 235 have higher levels of an ≈8-kDa protein relative to the parental cell line. (B) Immunocytochemistry of COX II indicating the presence and correct localization of COX II in rescued clones. FIBRO, fibroblast.
Furthermore, staining with antibodies to cytochrome c oxidase (COX) subunit II demonstrated restoration of the mitochondrial network in rescued clones (Fig. 2B). Therefore, the depleted COX II gene found in A549 ρ° cells (Fig. 1A) was rescued.
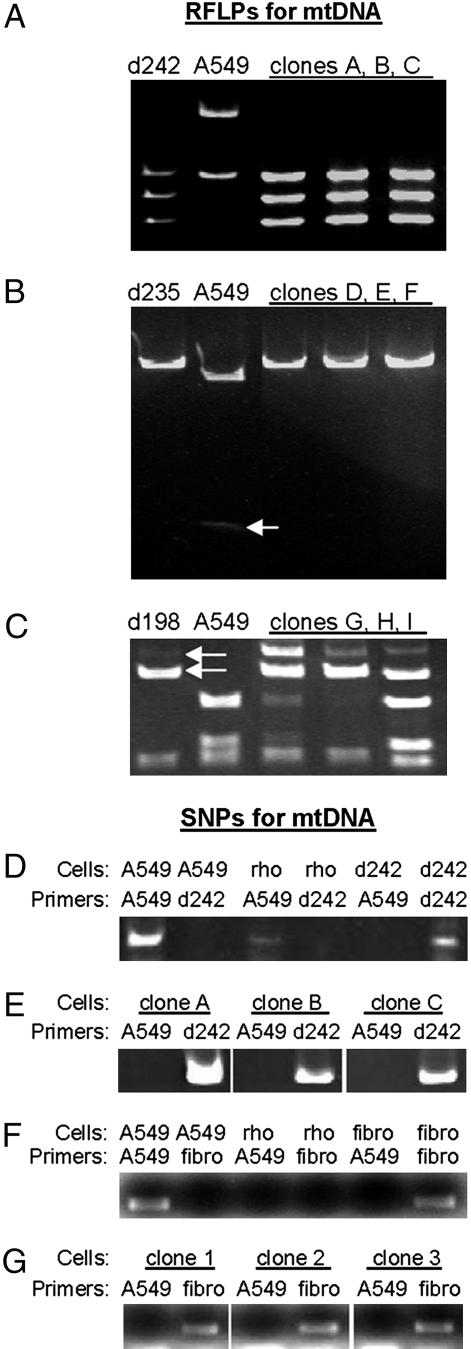
Genetic Analysis of Rescued A549 ρ° Clones. To establish that mitochondria were transferred from hMSCs and fibroblasts to A549 ρ° cells without cell fusion, we assayed both mtDNA and nuclear DNA from rescued clones (Fig. 3 and Table 1). Sequencing of the mtDNAs from the parental A549 cell line and the hMSC donors revealed polymorphic sites that could be used to distinguish the different mtDNAs by restriction fragment length polymorphisms (RFLPs) and SNPs (Fig. 3). RFLP analysis of rescued clones demonstrated the presence of mtDNA from hMSC in 69 clones assayed (Fig. 3 A–C and Table 1). In one experiment (d198 in Fig. 3C and Table 1), heteroplasmy was seen because residual or repaired mtDNA from A549 cells was present. SNP analysis by PCR confirmed the RFLP results in that 76 of 79 rescued clones contained detectable mtDNA from the hMSCs (Fig. 3 D and E). SNP-PCR analysis demonstrated similar results for clones derived from A549 ρ° cells cocultured with human skin fibroblasts: all of the clones contained mtDNA derived from the fibroblast donor cells (Fig. 3 F and G and Table 1).
Fig. 3.
Assays for RFLPs and SNPs for mitochondria transfer from hMSCs and fibroblasts. (A) HpyCH4V restriction enzyme digestion of the PCR products of the hypervariable region I PCR product (donor 242). (B) ApaLI restriction enzyme digestion of PCR products of the hypervariable region II PCR product (donor 235). Arrow, faint second band in digestion of A549 cells that is not present in digestion products from d235 or rescued clones D, E, and F. (C) BfaI restriction enzyme digestion of PCR products of the COX I PCR product (donor 198). In one coculture experiment, some clones contained mtDNA derived from both donor and target cells, implying persistence of the original mtDNA. Arrows, two top bands indicate presence of d198 mtDNA in rescued clones G, H, and I. (D and E) SNP-PCR assays to identify A549-derived mitochondria and hMSC donor-derived mitochondria in rescued clones. Note that clones A, B, and C are the same as those analyzed by in A. (F and G) SNP-PCR assays to identify A549-derived mitochondria and fibroblast (fibro) donor-derived mitochondria in rescued clones. D is the control for E; F is the control for G.
Table 1. Summary data for mitochondrial and nuclear genotypes of rescued A549 ρ° clones.
| Mitochondrial genome
|
Nuclear genome
|
|||||||
|---|---|---|---|---|---|---|---|---|
| RFLP
|
SNP
|
Col1A2
|
STR
|
|||||
| Donor cells | A549 | Donor | A549 | Donor | A549 | Donor | A549 | Donor |
| d198 | 18/24 | 21/24 | 0/24 | 21/24 | 14/14 | 0/14 | 3/3 | 0/3 |
| d242 | 0/24 | 24/24 | 0/31 | 31/31 | 10/10 | 0/10 | 3/3 | 0/3 |
| d235 | 0/24 | 24/24 | 0/24 | 24/24 | 6/6 | 0/6 | 3/3 | 0/3 |
| fibro | 0/24 | 24/24 | 0/23 | 23/23 | N/A | N/A | 3/3 | 0/3 |
The mitochondrial genotype was assayed by RFLPs and SNP. Nuclear genotypes of rescued clones from several coculture experiments were assayed by a polymorphism in the intron downstream of exon 28 in the type 1 collagen (COL1A2 exon 28 IVS + 15) and by analysis of nine different microsatellite markers on different chromosomes (STR). The observed mitochondria genotypes indicate transfer of mitochondria from three different hMSC donors (d198, d242, and d235) and one fibroblast donor (fibro). The nuclear genotypes of rescued clones indicate that in all cases the nucleus was derived from the original A549 cell line, excluding cell fusion as the primary mechanism for mitochondria transfer. N/A, not assayed.
To exclude cell fusion as an explanation for the rescue of the clones, we assayed nuclear DNA by polymorphisms found in the COL1A2 gene (See Fig. 6, which is published as supporting information on the PNAS web site) that could distinguish donor and A549 nuclei and by analysis of short tandem repeats (Table 1; see also Table 3, which is published as supporting information on the PNAS web site). The results indicated that the COL1A2 gene was exclusively from A549 cells in 30 of 30 clones assayed. Analysis of nine short tandem repeats, which provided a probability of erroneous identify of ≈1 × 10–11 (Profiler Plus, Applied Biosystems), confirmed that the nuclear genome in 12 of 12 clones was from A549 cells and that there was no evidence of nuclear DNA from the donor cells (See Table 3). Therefore, the observations did not provide any evidence of cell fusion as the mechanism for mitochondrial transfer.
Lack of Passive Transfer of Mitochondria. Experiments were performed to test for passive transfer of mitochondria. In one series of experiments, we cocultured A549 ρ° cells with human platelets as the source of membrane-bound functional mitochondria. Platelets were isolated from plasma from three individuals and used to prepare cocultures in which 150,000 to 500,000 platelets were added to confluent plates of ≈6 million A549 ρ° cells in restrictive medium. The cocultures were observed for up to 30 days without detection of proliferating colonies of A549 ρ° cells. In additional experiments, we added mitochondria isolated by a differential centrifugation from 1 × 106 hMSCs derived from three individuals to cultures of A549 ρ° cells. Again, no A549 ρ° cells were rescued for up to 30 days.
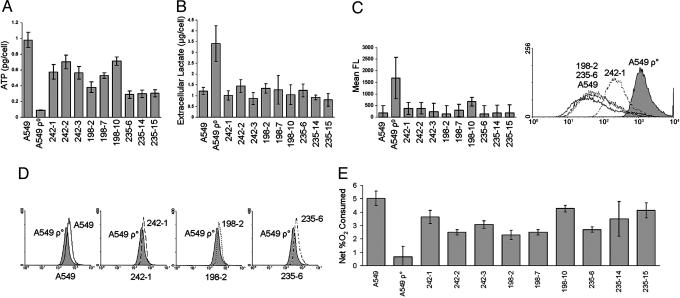
Functional Measurements of Mitochondrial Activity in Rescued Clones. A series of assays for mitochondrial function were carried out on the rescued clones of A549 ρ° cells (9). Because aerobic respiration produces 9-fold more ATP than glycolysis, the levels of intracellular ATP reflect the metabolic activity of the mitochondria. Intracellular ATP was measured by using a luciferase-based bioluminescent assay. All of the rescued clones tested exhibited at least a 3-fold increase in ATP levels compared with A549 ρ° cells (Fig. 4A). In several rescued clones, the ATP levels approached 70% of the ATP levels measured in parental A549 cells.
Fig. 4.
Recovery of mitochondrial function in rescued clones. A549 cells, A549 ρ° cells, and rescued clones were analyzed for mitochondrial function by several assays. (A) Without exception, the rescued clones had significantly increased intracellular ATP compared with A549 ρ° cells. (B) Extracellular lactate was significantly reduced in the rescued clones when normalized for cell number, reaching levels comparable to parental A549 cells. (C) There was a decrease in reactive oxygen species compared with A549 ρ° cells in all of the rescued clones. Values (Left) are relative levels of mean fluorescence (FL) derived from the histogram shown (Right). (D) There was an increase in mitochondrial membrane potential in all rescued clones compared with A549 ρ° cells (242-1, 198-2, and 235-6). (E) Rescued clones consumed almost as much O2 as the parental A549 cells, indicating aerobic respiration. Error bars represent one standard deviation from the mean of three individual cultures (A–D) or three measurements of a single culture (E). Two-tailed Student t tests for A, B, and E indicated P values <0.05.
Because of their nonfunctional mitochondria, A549 ρ° cells produce large amounts of extracellular lactate through anaerobic glycolysis. We measured lactate indirectly with an assay in which lactate and NAD+ were converted to pyruvate and NADH by lactate dehydrogenase. After normalizing for cell number, the results indicated that all of the rescued clones had significantly reduced extracellular lactate production compared with A549 ρ° cells (Fig. 4B).
A549 ρ° cells have higher levels of intracellular reactive oxygen species because of a decrease in antioxidants such as glutathione and a decrease in glutathione reductase and other enzymes that remove free radicals (10). We used a reactive fluorescent dye (5′,6′-chloromethyl-2′,7′dichlorodihydrofluorescein diacetate) and flow cytometry to measure the relative levels of reactive oxygen species in the cells. There was a decrease in reactive oxygen species in all of the rescued clones compared with A549 ρ° cells (Fig. 4C).
The membrane potential in mitochondria is created by the electron transport chain as it pumps H+ ions into the intermembrane space, creating an electrochemical gradient that drives ATP production. By assaying a membrane potential-sensitive fluorescent dye (tetramethylrhodamine ethyl ester) with flow cytometry, we demonstrated an increase in membrane potentials in all rescued clones compared to A549 ρ° cells (Fig. 4D).
The last step of the electron transport chain uses oxygen as an electron acceptor, creating H2O as a final product. Without a functional electron transport chain, A549 ρ° cells consume little or no O2. To measure aerobic respiration of the clones, we incubated the cells in a sealed, constantly agitated container for 20 h and then assayed dissolved O2 with an oxygen probe. The rescued clones consumed 3- to 7-fold more O2 than the original A549 ρ° cells (Fig. 4E).
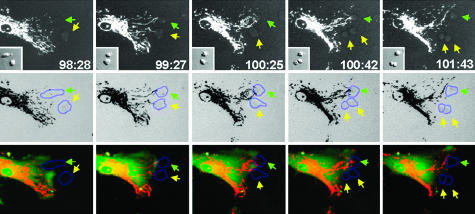
Interactions Between hMSCs and A549 ρ° Cells. To demonstrate visually the transfer of mitochondria, we transduced hMSCs with lentiviral vectors encoding hrGFP and the red fluorescent protein DsRed2 fused to a mitochondrial localizing sequence. We then followed the cocultures by time-lapse microscopy (Fig. 5). Mitochondrial segments moved as fast as 20 μm/min, continually fusing to and pinching off from the larger mitochondrial network. Also, the mitochondria streamed forward and backward through cytoplasmic extensions of the cells. Upon coculture with A549 ρ° cells in the absence of pyruvate and uridine, we observed that the hMSCs directed cytoplasm toward A549 ρ° cells either as cytoplasmic extensions (Fig. 5; see also Movie 1, which is published as supporting information on the PNAS web site) or broad areas of cellular contact (data not shown). At areas of cellular contact, mitochondria streamed toward the A549 ρ° cells (Fig. 5; see also Movie 1). Shortly after contact with an hMSC, some of the A549 ρ° cells were seen to divide (Fig. 5, yellow arrows), an observation consistent with mitochondrial rescue. It was not apparent whether mitochondria had been transferred through direct cytoplasmic transfer or as discrete vesicles.
Fig. 5.
Microscopy of cocultures. hMSCs were transduced with lentiviral vectors encoding the gene for DsRed2 with a mitochondrial localization signal and the gene for hrGFP. They were then added to cocultures with A549 ρ° cells, and time lapse microscopy of cocultures in restrictive medium was recorded. Images were captured every minute for 8 h, starting on the fourth day of coculture. The numbers are time elapsed in hours and min from the initiation of the coculture. (Top) The sequence records an interaction between an hMSC and two A549 ρ° cells in restrictive medium. (Middle) The images are inverse images together with digitally added outlines of the target A549 ρ° cells by differential interference contrast microscopy. (Bottom) The images are composite overlays of red and green fluorescence and digitally added outlines. The hMSC mitochondria localized to two focal areas of contact with the A549 ρ° cells. In the third frame, the hMSC mitochondria became concentrated near the upper A549 ρ° cell. Shortly afterward, the bottom A549 ρ° cell divided, indicating restored aerobic respiration. Green arrow, upper of two A549 ρ° cells; yellow arrow, lower A549 ρ° cell that divided in the fourth frame (Left to Right).
We also examined control cultures of transduced hMSCs that were unsorted and in which ≈50% of the cells expressed the DsRed2-mito transgene. Some hMSCs deposited small, membrane-bound vesicles onto the tissue culture plates that fluoresced red, implying that the vesicles contained functioning mitochondria (see Fig. 7A, which is published as supporting information on the PNAS web site). Nearby nontransduced hMSCs in the same cultures acquired these extracellular membrane-bound mitochondria on their plasma membrane, but it was unclear whether they were internalized (Fig. 7B).
Discussion
The results presented here demonstrated that, after hMSCs or human skin fibroblasts were cocultured with A549 ρ° cells that have defective or deleted mtDNA, some of the A549 ρ° cells acquired functional mitochondria. The rescued cells were able to propagate exponentially in a restrictive medium similar to the parental cell line with functional mitochondria. As few as 100 hMSCs were sufficient to generate a colony of rescued cells. Genetic analysis of rescued clones from the cocultures demonstrated that 99 of 102 (97%) of the clones contained mtDNA from the donor cells. The genomic DNA was from the A549 ρ° cells as demonstrated both by a polymorphism in the COL2A1 gene and by the automated assay of multiple short tandem repeats widely used to identify individual genomes. The results do not fully exclude the possibility that the cells underwent fusion followed by selective loss of the donor cell nuclei, but this explanation is unlikely because we never detected transfer of any of 10 genetic markers spread throughout the genome in any clone. The rescue of mitochondrial function was demonstrated by assays of exponential growth kinetics, translation and localization of mitochondrial proteins, and several parameters of mitochondrial activity: decreased production of extracellular lactate, decreased levels of reactive oxygen species, increased intracellular ATP, increased membrane potential, and increased oxygen consumption. Time-lapse microscopy demonstrated that hMSCs in the cocultures developed cytoplasmic projections directed toward the target cells. They made contact with the target cells either through long, thin extensions or through broad areas of the cell surfaces. Mitochondria streamed through the extended cytoplasmic projections, and subsequently, some of the target cells began to divide in the restrictive medium, an observation consistent with mitochondrial rescue.
The results did not establish whether mitochondria were transferred to the target cells directly through structures such as tunneling nanotubes (11–13) or through uptake of vesicles containing mitochondria that budded off from the donor cells (see Fig. 8, which is published as supporting information on the PNAS web site). Some of the time-lapse frames suggested direct transfer from the cytoplasm of the donor cells to the target cells, but they did not exclude vesicular transfer, as occurs with melanosomes (14, 15) and exosomes (16). The possibility of vesicular transfer was supported by the observation that hMSCs were seen to deposit vesicles in the culture dishes that apparently contained mitochondria because they contained the mitochondria-targeted red fluorescent protein. We were not able to demonstrate transfer of mitochondria from cocultured platelets or isolated mitochondria. Therefore, the transfer of mitochondria appeared to involve an active cellular process rather than passive uptake of cellular fragments or organelles. The results did not exclude the possibility that the target cells were rescued by a transfer of mtDNA without intact mitochondria, but the simplest explanation of the data are that functional mitochondria were transferred and then propagated in the target cells.
A large number of heritable diseases are caused by mutations that are found in mitochondrial and nuclear genes encoding mitochondrial proteins and that produce heritable skeletal or cardiac myopathies (1, 3, 17, 18). In addition, environmentally induced mutations in mtDNA have been implicated in many common acquired disorders, including ischemic diseases of the heart and brain, neurodegenerative diseases, some liver diseases, and some cancers (17). The data presented here indicate that genetic defects in mtDNA can be rescued by transfer of normally functioning mitochondria from wild-type cells ex vivo. The frequency of such transfers in vivo is difficult to estimate. However, they may explain some of the beneficial effects observed when large numbers of hMSCs or other progenitor cells are administered in animal models for diseases such as spinal cord injury (19, 20), stroke (21), and heart disease (22, 23). Injuries to mitochondria are early events in most of the animal models, and transfers of mitochondria may well explain why functional improvements were observed, but few of the donor cells engrafted long-term.
Materials and Methods
Cell Culture. Frozen vials of passage 2 or 3 hMSCs (24) were obtained from the Tulane Center for Preparation and Distribution of Adult Stem Cells. Vials of ≈106 cells were plated on a 150-cm2 culture dish (Nunc) in complete culture medium consisting of α-MEM (Gibco) with 20% FBS (lot selected for rapid growth of hMSCs; Atlanta Biologicals, Norcross, GA)/2 mM l-glutamine/100 units/ml penicillin/100 μg/ml streptomycin (Gibco). The medium was replaced after 1 day, and the viable adherent cells were lifted with 0.25% trypsin/1 mM EDTA for 2 to 5 min at 37°C. The cells were expanded by plating at 50 cells/cm2, incubated for 8 to 10 days, and harvested with trypsin/EDTA at 70% confluence. Cells were then lifted by 0.25% trypsin/1 mM EDTA and frozen at 1 × 106 cells per ml per vial.
Passage 5 or 7 normal adult human skin fibroblasts were obtained from the Cell Repository of the Tulane Center for Gene Therapy and cultured to 60% confluence in complete culture medium. A549 ρ° cells were provided by Ian Holt (Dunn Human Nutrition Unit, Medical Research Council, Cambridge, U.K.).
Coculture Experiments. A549 ρ° cells in three T175 flasks (Nunc) were cultured in permissive medium consisting of DMEM (Gibco) containing 10% FBS (Atlanta Biologicals), 2 mM l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, 110 μg/ml sodium pyruvate, and 50 μg/ml uridine. After the cells reached confluency, 150,000 or 500,000 hrGFP+ hMSCs or hrGFP+ fibroblasts were added as mitochondrial donor cells. The cocultures were incubated in permissive medium for 4 days with no medium change. On the 4th day, the medium was replaced with restrictive medium that consisted of the same medium lacking sodium pyruvate and uridine. After 10 days, clones were isolated by placing sterile trypsin/EDTA-saturated paper disks (Scienceware, Research Products International) over discrete colonies for 5 min. The disks were placed into 24-well plates (Nunc) with restrictive medium and allowed to grow for 1 week before seeding into T25 flasks. Alternatively, the cells in the cocultures were lifted with 0.25% trypsin/1 mM EDTA and flow-sorted for A549 ρ° cells (GFP-negative). The sorted A549 ρ° cells were expanded in restrictive medium, and clones were isolated by paper disks as above. After propagation in restrictive medium, the rescued A549 ρ°-derived clones were lifted by 0.25% trypsin/1 mM EDTA and frozen for subsequent isolation of DNA and for assays of growth and mitochondrial function. For additional procedures, see also Supporting Methods, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
Lentiviral constructs were created from the pWPT-GFP construct donated by the Tronolab (Swiss Institute of Technology Lausanne, Lausanne, Switzerland). This work was supported in part by National Institutes of Health Grants AR-48323, HL-073755, HL-073252, and P01 HL-07561; Columbia/HCA Healthcare Corp.; and the Louisiana Gene Therapy Research Consortium.
Author contributions: J.L.S., S.D.O., and D.J.P. designed research; J.L.S., S.D.O., and M.J.W. performed research; J.L.S., S.D.O., M.J.W., and D.J.P. analyzed data; and J.L.S., S.D.O., M.J.W., and D.J.P. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: hMSCs, adult nonhematopoietic stem/progenitor cells from human bone marrow; hrGFP, humanized GFP; COX, cytochrome c oxidase; RFLPs, restriction fragment length polymorphisms.
References
- 1.Wallace, D. C. (1994) Proc. Natl. Acad. Sci. USA 91, 8739–8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dyall, S. D., Brown, M. T. & Johnson, P. J. (2004) Science 304, 253–257. [DOI] [PubMed] [Google Scholar]
- 3.Brandon, M. C., Lott, M. T., Nguyen, K. C., Spolim, S., Navathe, S. B., Baldi, P. & Wallace, D. C. (2005) Nucleic Acids Res. 33, D611–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mason, P. A., Matheson, E. C., Hall, A. G. & Lightowlers, R. N. (2003) Nucleic Acids Res. 31, 1052–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prockop, D. J., Gregory, C. A. & Spees, J. L. (2003) Proc. Natl. Acad. Sci. USA 100, Suppl. 1, 11917–11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King, M. P. & Attardi, G. (1989) Science 246, 500–503. [DOI] [PubMed] [Google Scholar]
- 7.Bodnar, A. G., Cooper, J. M., Holt, I. J., Leonard, J. V. & Schapira, A. H. (1993) Am. J. Hum. Genet. 53, 663–669. [PMC free article] [PubMed] [Google Scholar]
- 8.Wallace, D. C., Stugard, C., Murdock, D., Schurr, T. & Brown, M. D. (1997) Proc. Natl. Acad. Sci. USA 94, 14900–14905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pallotti, F., Baracca, A., Hernandez-Rosa, E., Walker, W. F., Solaini, G., Lenaz, G., Melzi d'Eril, G. V., Dimauro, S., Schon, E. A. & Davidson, M. M. (2004) Biochem. J. 384, 287–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vergani, L., Floreani, M., Russell, A., Ceccon, M., Napoli, E., Cabrelle, A., Valente, L., Bragantini, F., Leger, B. & Dabbeni-Sala, F. (2004) Eur. J. Biochem. 271, 3646–3656. [DOI] [PubMed] [Google Scholar]
- 11.Rustom, A., Saffrich, R., Markovic, I., Walther, P. & Gerdes, H. H. (2004) Science 303, 1007–1010. [DOI] [PubMed] [Google Scholar]
- 12.Önfelt, B. & Davis, D. M. (2004) Biochem. Soc. Trans. 32, 676–678. [DOI] [PubMed] [Google Scholar]
- 13.Önfelt, B., Nedvetski, S., Yanagi, K. & Davis, D. M. (2004) J. Immunol. 173, 1511–1513. [DOI] [PubMed] [Google Scholar]
- 14.Scott, G., Leopardi, S., Printup, S. & Madden, B. C. (2002) J. Cell Sci. 115, 1441–1451. [DOI] [PubMed] [Google Scholar]
- 15.Sugden, D., Davidson, K., Hough, K. A. & Teh, M. T. (2004) Pigm. Cell. Res. 17, 454–460. [DOI] [PubMed] [Google Scholar]
- 16.Denzer, K., Kleijmeer, M. J., Heijnen, H. F., Stoorvogel, W. & Geuze, H. J. (2000) J. Cell Sci. 113, 3365–3374. [DOI] [PubMed] [Google Scholar]
- 17.Wallace, D. C. (1999) Science 283, 1482–1488. [DOI] [PubMed] [Google Scholar]
- 18.Green, D. R. & Kroemer, G. (2004) Science 305, 626–629. [DOI] [PubMed] [Google Scholar]
- 19.Chopp, M., Zhang, X. H., Li, Y., Wang, L., Chen, J., Lu, D., Lu, M. & Rosenblum, M. (2000) NeuroReport 11, 3001–3005. [DOI] [PubMed] [Google Scholar]
- 20.Hofstetter, C. P., Schwarz, E. J., Hess, D., Widenfalk, J., El Manira, A., Prockop, D. J. & Olson, L. (2002) Proc. Natl. Acac. Sci. USA 99, 2199–21204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen, J., Li, Y., Katakowski, M., Chen, X., Wang, L., Lu, D., Lu, M., Gautam, S. C. & Chopp, M. (2003) J. Neurosci. Res. 73, 778–786. [DOI] [PubMed] [Google Scholar]
- 22.Pittenger, M. F. & Martin, B. J. (2004) Circ. Res. 95, 9–20. [DOI] [PubMed] [Google Scholar]
- 23.Gnecchi, M., He, H., Liang, O. D., Melo, L. G., Morello, F., Mu, H., Noiseux, N., Zhang, L., Pratt, R. E., Ingwall, J. S. & Dzau, V. J. (2005) Nat. Med. 11, 367–368. [DOI] [PubMed] [Google Scholar]
- 24.Sekiya, I., Larson, B. L., Smith, J. R., Pochampally, R., Cui, J. G. & Prockop, D. J. (2002) Stem Cells (Dayton) 20, 530–541. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.