Abstract
PTEN (phosphatase and tensin homolog deleted on chromosome 10) is a potent tumor suppressor gene frequently mutated in human prostate cancers. Deletion of Pten in a murine model of prostate cancer recapitulates the disease progression seen in humans. Using defined cell lineage markers, we demonstrate that PTEN negatively regulates p63-positive prostatic basal cell proliferation without blocking differentiation. Concomitant with basal cell proliferation is the expansion of a prostate stem/progenitor-like subpopulation as evidenced by the progressive increase of stem cell antigen-1 (Sca-1)- and BCL-2-positive cells. This observation provides strong evidence that basal cell proliferation can be an initiating event for precancerous lesions. Sca-1+ and BCL-2+ progenitors may serve as cancer-initiating cells in this model.
Keywords: animal model, prostate cancer, Pten conditional knockout, stem cell proliferation and differentiation, tumor progression
Prostate cancer is the most common malignancy in men and the second leading cause of male cancer-related deaths in the Western world (1). The development of human prostate cancer proceeds through a series of defined states, initiating with prostatic intraepithelial neoplasia (PIN) and progressing to invasive hormone-dependent then -independent cancers (2, 3). The normal human prostatic epithelium consists of basal cells, transit amplifying/intermediate cells (TA/IC), secretory luminal cells, and neuroendocrine cells (4-6). Most human prostate cancers are adenocarcinomas and express markers associated with luminal epithelial cells. Because prostate cancer arises as a result of unbalanced cell proliferation, cell differentiation, and cell death, a central goal in prostate cancer biology is to identify the tumor precursor or tumor-initiating cells that are capable of maintaining tumor growth.
The prostate is an androgen-dependent organ that undergoes involution after androgen withdrawal but can completely regenerate upon androgen restoration. Results from androgen ablation/restoration cycling experiments on rodents suggest the existence of a population of long-lived, self-renewable stem cells (7, 8) that are responsible for maintaining the homeostasis of the prostate gland. These stem cells are proposed to be present predominantly in the basal cell layer of the human prostate gland (4, 9). Using the kidney capsule reconstitution system initially developed by Cunha and Lung (10), Richardson et al. (11) demonstrated that human prostatic epithelial cells positive for CD133 have stem cell activity. Similarly, we found that the regenerative capacity of prostatic stem/progenitor cells resides in a unique subpopulation of murine epithelial cells that express cell surface markers such as Sca-1 and that are sensitive for oncogenic transformation (12). Consistent with BrdUrd label-retaining stem/progenitor cells, the Sca-1+ population is enriched in the proximal regions of the prostate gland (12, 13).
Here, we report that altered stem/progenitor cell proliferation is directly associated with prostate tumor initiation and progression in the Pten-null murine model. PTEN negatively regulates basal cell proliferation, and Pten deletion in the basal cells leads to expansion of Sca-1+ and BCL-2+ stem/progenitor cells with concomitant differentiation. Collectively, our results illustrate that basal cell proliferation is an important event for prostate cancer initiation and early progression.
Results
Definition of Cell Types Associated with Murine Prostatic Epithelium Compartment. To identify the cell of origin for prostate cancer, we first defined the cell types associated with the normal prostatic epithelium compartment. We chose the proximal region of the dorsolateral lobe to analyze because this region is enriched for the slow cycling, BrdUrd label-retaining, and Sca-1+ stem/progenitor cells (8, 12, 13).
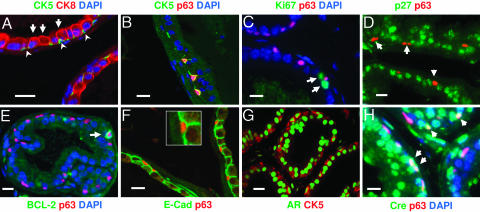
To simultaneously colocalize the expression of multiple cellular markers, we performed dual or triple immunofluorescence analyses, using markers that have been used to identify subpopulations of human and murine prostatic epithelial cells (4, 14-16). Our analysis showed that the normal mouse prostate epithelium consists of two distinct cell populations: the CK5+ basal cells that are directly attached to the basement membrane (Fig. 1A, arrowheads) and the CK8+ secretory cells that face the lumen (Fig. 1 A, arrows). In the WT prostate epithelium, all CK5+ cells are positive for p63 (Fig. 1B) and occasionally positive for BCL-2 (indicated by the arrow in Fig. 1E) but are negative for the cell proliferation marker Ki67 (Fig. 1C) and the cell cycle inhibitor p27kip1 (Fig. 1D).
Fig. 1.
Characterization of basal and luminal epithelial cells in the normal murine prostate, and localization of Cre expression. Immunofluorescence analyses were carried out by using sections from the proximal region of dorsolateral lobes of WT adult mice. Each section was double-stained by two independent cell markers, and some sections were counterstained with DAPI to visualize the nuclei. Only overlaid results are shown here. The WT adult prostatic acini are composed of two major cell types. The luminal cells are immunoreactive to antibodies against CK8 (A, arrows), p27 (D), epithelial cadherin (E-cad) (F), and androgen receptor (AR) (G). By contrast, the basal cells form a discontinuous cell layer scattered along the basement membrane and are positive for cellular markers CK5 (A, arrowheads), p63 (B), AR (G), and occasionally BCL-2 (E) but are negative for p27 (D), E-cad (F), and Ki67 (C). Cre is expressed in both basal (H) and luminal epithelial cell compartments (shown in green). (Inset) A high-power image of F. (Scale bar, 20 μm.)
The cadherin family is known to be a critical mediator of cell-cell adhesion and stem cell-niche interaction. Strong epithelial cadherin (E-cad) staining can be detected at the cell-cell junction between luminal cells and basal-luminal cells (Fig. 1F). However, different from Drosophila germ stem cells in which E-cad serves as a glue to anchor germ stem cells to their niche (17, 18), no E-cad staining can be detected at the interface of p63+ basal cells and the basement membrane (Fig. 1F). Thus, the murine prostatic basal cells can be defined as CK5+;CK8-;p63+;Ki67-;p27kip1- and are associated with the basement membrane. In contrast, the luminal secretory cells are defined as CK5-;CK8+;p63-;p27kip1+.
Cre Recombinase Is Expressed in both Basal and Luminal Epithelial Cells. In the Pten-null prostate cancer model, the tumor-initiation event commences with the expression of the Cre recombinase, resulting in Pten homozygous deletion. We achieved prostate epithelium-specific Pten deletion by crossing Pten conditional knockout mice (floxed) with transgenic mice in which the Cre expression was driven by the ARR2 probasin promoter, a modified rat prostate-specific probasin promoter known to be regulated by androgen (19). We found that androgen receptor (AR) is expressed in both CK5+ mouse basal cells and CK5- luminal cells (Fig. 1G). By using the same condition, CK5+;AR+ basal cells can be detected also in human prostate tissue sections (Fig. 6i, which is published as supporting information on the PNAS web site). When we conducted immunofluorescence analyses to identify Cre-expressing cells in Ptenloxp/+ mice carrying the probasin-Cre4 transgene, we consistently found that ≈50% of p63+ basal cells (Fig. 1H) and close to 85% of the p63- luminal cells (data not shown) express significant levels of the Cre recombinase. Thus, Pten deletion can occur in both basal and luminal epithelial cell compartments, and, therefore, both cell types could potentially serve as tumor-initiating cells.
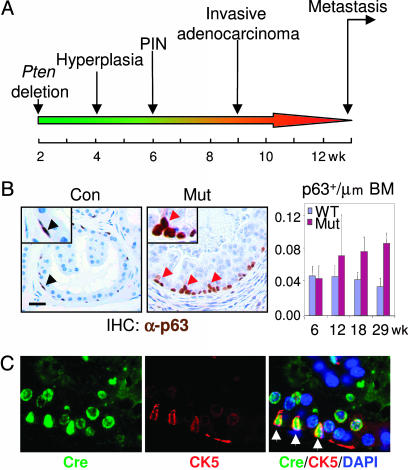
Pten Deletion Leads to Increased Basal Cell Density, Altered Basal Cell Morphology, and Localization. Mice with prostatic-specific Pten deletion develop prostate cancer with pathological changes similar to human disease with the following well defined kinetics: at 4 weeks, hyperplasia; at 6 weeks, PIN; at 9 weeks, invasive adenocarcinoma; and at 12 weeks, metastasis (Fig. 2A). During the course of prostate cancer initiation and progression, we observed a remarkable increase of CK5+;p63+ cells along the basement membrane in the proximal region of the dorsolateral lobe (Figs. 2B and 4B Left). In addition, basal cells in the Pten-null prostate glands display distinct morphological change. The Cre+;CK5+ cells have a larger cell size and elongated nuclei and are often perpendicular to the basement membrane (Fig. 2C). This morphology is in direct contrast with the WT p63+ cells, which lie flat against the basement membrane (Fig. 2B Left, arrowheads). Additionally, a fraction of the p63+ cells appear to “bud-out” from the basal cell layer that is directly attached to the basement membrane (Fig. 2B Center, red arrowheads), and those “budding” p63+ cells often show a diminished p63 level, perhaps indicative of their differentiation status (see below).
Fig. 2.
Increased p63+ cell number and altered morphology along with tumor initiation and progression. (A) A schematic illustration of the kinetics of prostate cancer development caused by Pten conditional knockout. (B) Increased p63+ cell number and altered morphology in the Pten-null prostate epithelium. Immunohistochemical analysis was performed with anti-p63 antibody. Note that scattered p63+ cells lie flat on the basement membrane of WT epithelium (Left). p63+ cells in Pten-null prostate show altered morphology, including bigger cell size, changed polarity, and altered localization (Center). Most p63+ cells are organized into individual small clusters. Red arrowheads point to p63+ cells that appear to “bud-out” from the basal cell layer through asymmetric-like cell division. (Scale bar, 20 μm, 40 μm for Insets.) (Right) Quantification of p63+ cells. p63-stained sections at each time point, with five animals per point, were quantified for the density of p63+ cells along the basement membrane (BM). The values are presented as mean ± SD. (C) Cre-mediated Pten deletion in CK5+ basal cells leads to change in cell morphology. Arrows (Right) show Cre and CK5 double-positive Pten-null cells.
Fig. 4.
Pten deletion leads to increased basal and TA/IC populations without blocking cell differentiation. (A) Immunofluorescence analysis illustrates asymmetric-like cell division events in Pten-null prostate glands. (Scale bar, 20 μm.) (B) Pten deletion does not perturb cell differentiation. From basal layer (BC) to terminally differentiated luminal cell layer (LC), the cancerous acini are composed of localized p63+;CK5+ basal cells (Left), TA/IC cells that coexpress both CK5 basal and CK8 luminal cell markers, and CK8 single-positive luminal cells (second panel from the left). Most of the proliferative Ki67+ cells are AR- and BCL-2-positive in the basal and TA/IC regions. (Scale bar, 50 μm.)
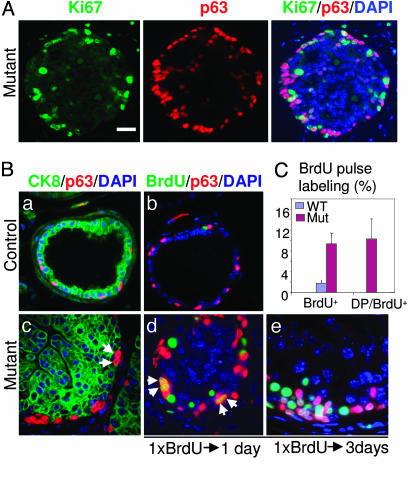
Pten Deletion Leads to Increased Cell Proliferation Near the Basal Cell Compartment. Cancer can originate from either a self-renewable stem cell population (20) or its differentiated progeny that have acquired self-renewal capacity owing to accumulated mutations (21-24). Unlike normal, replication-quiescent stem cells, tumor-initiating cells or their immediate progenitors are competent for continuous proliferation to maintain tumor growth (25). When colocalizing Ki67, a cell proliferation marker, with the luminal and basal cell markers, we observed that most of the Ki67+ cells are localized within or immediately above the basal cell compartment but are rarely associated with terminally differentiated luminal cells that are localized in the center of the hyperplastic duct (Fig. 3A).
Fig. 3.
Pten deletion in the basal cell compartment leads to increased cell proliferation. (A) Immunofluorescence analysis demonstrates that most Ki67+ proliferating cells are associated with or near the basal cell compartment: an overlaid image of Ki67+p63+ cells in white or light yellow is shown (Right). (Scale bar, 50 μm.) (B) a and c show the localization of CK8+ (green) and p63+ cells (red). Arrows in C point to two adjacent p63+ cells along the basement membrane. b, d, and e illustrate BrdUrd+ cells (green) after 24-h (b and d) and 72-h (e) chases. (a and b) Control prostate. (c-e) Mutant prostate. Arrows in d point to BrdUrd-labeled p63+ cells undergoing symmetric cell division. (Scale bar, 20 μm.) (C) Quantification of BrdUrd+ and BrdUrd/p63 double-positive cells after a 24-h chase. The number of each cell type was counted from five microscopic fields of dorsolateral prostate from each of two individual mice, and the data are presented as mean ± SD.
To directly evaluate the rate of cell proliferation, we gave a single dose of BrdUrd to 16-week-old mice and then quantified the percentage of BrdUrd-labeled cells 24 and 72 h later. The BrdUrd+ cells were further defined by luminal and basal cell markers. As shown in Fig. 3Bd and quantified in Fig. 3C, there is a 5-fold increase in BrdUrd pulse-labeled cells (green) in the Pten-null prostate gland as compared with the WT controls (Fig. 3Bb). About 10% of these BrdUrd+ cells are p63+ basal cells, in contrast to <0.01% in the WT controls (Fig. 3Bd, double-positive cells are shown in yellow). Importantly, basal cells with “symmetric-like” cell divisions, in which two p63+ nuclei are closely adjacent to each other (Fig. 3Bc, arrows) and with equal intensity of BrdUrd labeling (Fig. 3Bd, arrows), are clearly evident in the mutant ducts but absent in the controls (Fig. 3B a and b). Three days after initial labeling, these BrdUrd+ cells began to form small clusters, which were closely associated with the basal cell compartment (Fig. 3Be). On the basis of their cell proliferation potential, basal cells and cells immediately adjacent to the basal cell compartment, but not the luminal cell population, likely serve as tumor-initiating cells in the Pten-null prostate cancer model.
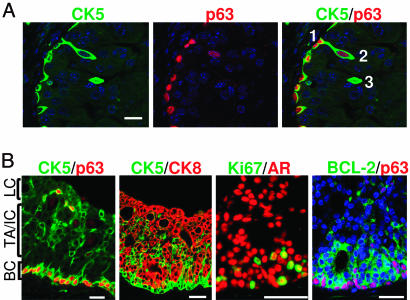
PTEN Loss Leads to Increased Stem/Progenitor Cell Populations Without Blocking Differentiation. Recent work suggests that p63 is required to maintain the commitment of the urogenital sinus endoderm to the prostate cell lineage and that p63+ basal progenitor cells are the precursors of luminal secretory cells (26). To further study this hierarchical relationship and to test whether PTEN affects p63+ basal cell differentiation, we performed dual immunohistochemical analysis using p63 to localize basal cell nuclei and CK5 to mark the cell boundary. Fig. 4A shows the events associated with initial cell differentiation in the Pten-null prostatic epithelium: the segregation of p63+ basal cell and its p63low;CK5+ progeny. Confocal images clearly show the association of two cell types (marked 1 and 2). Cell 1 is anchored on the basement membrane. Cell 2 exhibits an enlarged and elongated cell body perpendicular to the basement membrane (Fig. 4A Left) and possesses weaker p63 staining (Fig. 4A Center), similar to the “budding cells” observed in Fig. 2B. Other cells (such as cell 3) have completely lost association with the basement membrane and p63 expression but have retained CK5 expression, suggesting that the separation of p63 and CK5 basal cell markers signals the initial differentiation event.
In contrast to the WT prostatic epithelium shown in Fig. 1, three distinct subpopulations could be easily identified within the multilayer epithelium of PIN lesions, depending upon the expression of basal and luminal cell markers: p63 and CK5 double-positive cells are predominantly restricted to the basal layer (Fig. 4B Left), and CK8+ terminally differentiated secretory cells face the luminal space (Fig. 4B, second panel from left). Clusters of CK5 and CK8 double-positive cells (Fig. 4B, second panel from left) are found between the basal and luminal cell layers, similar to the transient TA/IC zone described in human prostate specimens (14). Most of the cycling cells are localized in these TA/IC clusters (Fig. 4B, third panel from left). Increased p27 (data not shown) and BCL-2 expression can also be detected in this region (Fig. 4B Right). Thus, we can define three major subpopulations of epithelial cells that are present at the prostate cancer-initiation stage: basal cells (CK5+;CK8-;p63+;Ki67+/-;AR+;p27kip1-), TA/IC cells (CK5+;CK8+;p63-;Ki67+;AR+;p27kip1+;BCL-2+), and the luminal secretory cells (CK5-;CK8+;p63-;AR+;p27kip1+;BCL-2-;Ki67-). Pten deletion dramatically perturbs the basal and TA/IC compartments, leading to increased basal cell density and expansion of the TA/IC cell population. This phenotype is further extended from PIN to invasive adenocarcinoma lesions in which CK5+ single-positive basal cell and CK5+;CK8+ double-positive TA/IC cell numbers are consistently increased, and in many cases, mislocalized (Fig. 7, which is published as supporting information on the PNAS web site).
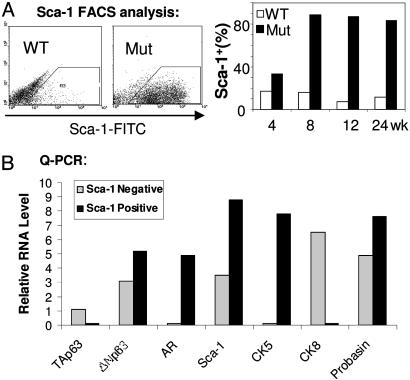
Sca-1 Cell Surface Marker-Positive Cells Are Enriched in Pten-Null Prostate Cancers. Recent work identified Sca-1 as a cell surface antigen that marks a subpopulation within the prostatic epithelial compartment with enhanced regenerative capacity and increased susceptibility to oncogenic transformation (12). Sca-1+ cells are enriched near the proximal region of the murine prostate ducts, and the Sca-1 cell surface antigen can be used for identifying prostate stem/progenitor cells (12, 13).
To determine whether Sca-1+ cells are increased in the Pten-null prostate gland, we examined Sca-1+ cells at different stages of tumor development. Starting at 4 weeks, at the hyperplastic stage (Fig. 5A), a significant increase in the Sca-1+ population was found in the Pten-null prostate glands. The Sca-1+ population reaches its peak at 8 weeks, corresponding to the PIN lesion stage, with a 10-fold difference between mutants and WT controls (Fig. 5A Right). Expression of a constitutively activated form of AKT/PKB serine/threonine kinase in WT epithelial cells gave a similar level of increase in the Sca-1+ population and pathological progression in regenerated prostate cancer tissues (12).
Fig. 5.
PTEN loss leads to an increase in Sca-1+ stem/progenitor subpopulations. (A) FACS analysis demonstrates increased Sca-1+ populations in Pten-null prostate gland, as compared with their age- and genetic background-matched littermate controls (n = 3). (B) Quantitative PCR analysis on mRNA samples prepared from Sca-1+ and Sca-1- populations. Relative expression levels compared with a G3PDH control are illustrated. The experiment was repeated three times and one representative result is shown.
To further define the Sca-1+ cell population, we separated the Sca-1+ from the Sca-1- cell population by FACS sorting. Quantitative PCR analysis demonstrated that some of the basal cell-associated genes, such as ΔNp63 (27) and CK5 (Fig. 5B), are highly expressed in the Sca-1+ population. Western blot analysis further confirmed increased Sca-1 protein levels in the Pten-null prostate glands, starting at 8 weeks, accompanied by increased p63 protein levels (Fig. 8, which is published as supporting information on the PNAS web site). Thus, the number of stem/progenitor cells, identified either in situ by cell lineage markers or by expression of the Sca-1 cell surface antigen, increases progressively with time in Pten-null prostate glands.
Discussion
In this study, we used both cell lineage and cell surface markers to demonstrate the dynamic cellular changes associated with prostate cancer initiation and progression in the Pten-null murine model. Our study demonstrated that PTEN loss in prostatic basal cells has two major consequences: (i) it promotes basal cell division, leading to increased basal cell density, and (ii) it enhances cell differentiation, resulting in the production of more differentiated progeny corresponding to the TA/IC cell population. Our results indicate that alterations in the basal and TA/IC cell compartments are associated with prostate cancer initiation. These cells may serve as tumor-initiating cells in our Pten-null prostate cancer model.
p63 is expressed in the regenerative epithelial compartment of several organs, including the prostate (27-29). p63 knockout mice fail to develop multiple epithelial compartments such as the prostate (27). Recent work suggests that p63 is required to maintain the commitment of the urogenital sinus endoderm to the prostate cell lineage and that the p63+ basal progenitor cell is the cell of origin for luminal secretory cells (26). Our study suggests that prostate cancer initiation in Pten-null mice is associated with increased p63+ cell numbers and change in cell morphology and localization. In addition, PTEN loss leads to the expansion of the basal-derived TA/IC cell population and a significant increase in the Sca-1+ population whose gene expression profile overlaps with that of TA/IC. The TA/IC cell has been suggested to be the cell of origin for human prostate adenocarcinomas (4, 9, 14). Consistent with this idea, previous studies demonstrated that PSCA, a marker associated with late TA/IC prostate epithelial cells and highly expressed in human prostate cancers (30), is up-regulated in Pten-null prostate cancers (31), and the percentage of PSCA-positive cells is increased in the prostates of Pten-deleted animals (32). Of note, an increased Sca-1+ population has been associated with mammary tumor stem cell activity (33), although the functional significance of this increase remains to be tested.
The molecular mechanisms by which PTEN controls stem cell behavior are currently unknown. We have previously shown that PTEN loss leads to increased neural stem cell self-renewal and proliferation (34), at least in part, by modulating the G0 to G1 cell cycle transition, a critical step for stem cell self-renewal and differentiation (35). How PTEN controls prostatic stem cell self-renewal and modulates the reciprocal interactions between niche and stem cells, stem cells and progenitors, and progenitors and terminally differentiated cell types needs to be investigated. Although an increase in the TA/IC population is a common event in human and murine prostate cancer initiation, the role of TA/IC cells as cancer stem cells is a subject of debate. Two scenarios have been proposed. In the first scenario, a slight change in stem cell number leads to a dramatic expansion of the TA population, which serves as a precursor for luminal epithelial cells and is capable of rapid cell division. Although the amplification of TA cells is a common event observed during prostate cancer development, the real tumor-initiating cells are their parents, the mutated stem cells. Alternatively, the second scenario suggests that oncogenic events may not take place in the stem cell population. Instead, TA cells with limited replicative capacity can be transformed by acquiring self-renewal potential (23). Our current study cannot distinguish between these two possibilities. Future studies with lineage-specific labeled transgenic mice may provide additional information toward answering this fundamental question.
There are both significant similarities and differences in the gross and microscopic anatomy of the rodent and the human prostate. These need to be considered when comparing results derived from murine models with results derived from human prostate (36). Unlike the single lobular structure of the human prostate gland, the mouse prostate gland has four paired lobes (36). Besides this anatomic difference, our detailed analysis indicates several differences between human and murine prostatic epithelium: e.g., TA/IC cells, marked by their concomitant expression of both CK5 and CK8 markers, are below our threshold of detection in the WT murine prostate gland (Fig. 1 A). In addition, human basal cells are positive for p27 and BCL-2 (4), whereas murine basal cells do not express or only occasionally express p27 and BCL-2 (Fig. 1 D and E, arrowheads). Our study also suggests that prostate cancer initiation in Pten-null mice is associated with an increased p63+ cell number and a change in cell morphology and localization. These results seem to contradict reports on human prostate cancers, in which the presence of p63+ cells is negatively correlated with progression of adenocarcinoma (37, 38). This discrepancy may be due to the different anatomies of human and murine prostates (36). Because our study concentrated on the proximal region of the dorsolateral prostate lobe, an area highly enriched for the stem/progenitor cell population (12, 13), we may have had a unique opportunity to observe a significant association of stem/progenitor cells with tumor initiation and progression. Of note, we observed that murine tumors located distal from the urethra often lack p63+ cells. Further study is needed to determine whether there are stem cell-rich areas in the human prostate gland that contribute to prostate cancer formation.
Materials and Methods
Histology and Immunochemistry Analysis. Tissue samples were prepared as described by Wang et al. (31). To visualize fluorescence, signals were amplified by using a PerkinElmer Life Sciences (Boston) TSA-Plus Cy3/Cy5/FITC System. Slides were incubated for 1 h at room temperature or 4°C overnight with the following antibodies diluted according to manufacturers' instructions: anti-p63 (BD Pharmingen, catalog no. 559951), anti-CK-5 and anti-CK-8 (Covance Research Products, Denver, PA; catalog nos. AF 138 and HK-8), anti-Ki67 polyclonal antibody (Novocastra, Newcastle, U.K.; catalog no. NCL-Ki67p), anti-AR (Upstate Biotechnology, Lake Placid, NY; catalog no. 06-680), anti-BCL-2 (BD Biosciences, Franklin Lakes, NJ; catalog no. 610538), and anti-BrdUrd (BD Biosciences, catalog no. 551321). Signals were amplified by using the biotin-streptavidin-based detection system from BioGenex Laboratories (San Ramon, CA). Briefly, slides were first incubated with biotinylated anti-mouse IgG (or anti-rabbit IgG for rabbit polyclonal antibody) and then incubated with streptavidin-conjugated peroxidase (BioGenex Laboratories) for 30 min at room temperature. Tyramide signal amplification reagent (PerkinElmer Life Sciences, catalog no. NEL741) with the appropriate fluorescent dye was added to the slides for 5 min, and a cover slide was mounted with antifading fluorescent mounting media containing DAPI (Vector Laboratories, catalog no. H1200). Samples were visualized with an Olympus (Melville, NY) BX60 fluorescence microscope or a Leica (Bannockburn, IL) TCS-SP confocal microscope.
FACS Analysis and Quantitative PCR. Dissociated prostate cells were stained with FITC-conjugated anti-Sca-1 antibody or FITC-conjugated rat IgG2a isotype control as described in ref. 12 (Pharmingen, 1 μM final concentration). For quantitative PCR, total RNAs were extracted from sorted Sca-1+ and Sca-1- cells of mutant mice with an RNeasy Mini Kit (Qiagen, Valencia, CA). RNAs were reverse-transcribed by oligo(dT) primer by using an Advantage RT-for-PCR kit from BD Biosciences, according to the manufacturer's instructions. The primer sets and PCR protocol used were as described in ref. 39. Results were analyzed by the relative quantification method and expressed as relative RNA levels (ΔCT, difference of cycling threshold). ΔCT values represent CT[gene] - CT[G3PDH]; thus, higher values indicate relatively lower expression levels. The relative RNA level was then calculated by normalizing the ΔCT of each gene to the least abundant RNA species, which was arbitrarily set to 8. The relative RNA level of gene x is thus equal to 8 - ΔCT(x).
BrdUrd Pulse Labeling. Two mice at each time point from each genotype group were injected with a single dose of BrdUrd (i.p., dissolved in PBS to a final concentration of 10 mg/ml) at 100 mg/kg body weight. Prostates were harvested along with small intestine, which served as a positive control, 24 or 72 h after injection.
Quantification of p63+, CK5+, Ki67+, and BrdUrd+ Cells. Prostate sections were stained with antibody against p63, CK5, BrdUrd, and Ki67 and visualized by using either the BioGenex Laboratories DAB system or PerkinElmer Life Sciences TSA reagent. After counterstaining and mounting, 300-400 epithelial cell nuclei in the proximal regions of the dorsolateral lobes were marked for each mouse prostate, and nuclei positive for p63, CK5, Ki67, and BrdUrd were counted. The number of p63-positive nuclei was reported either as a percentage (per 100 nuclei) or as a density (per micrometer of basement membrane). All quantification and measurements were done by using imagepro software (Media Cybernetics, Silver Spring, MD). Similar measurements were done for CK-5-, BrdUrd-, and Ki67-positive nuclei.
Supplementary Material
Acknowledgments
We thank Drs. Sawyers, Chow, Herschman, Reiter, Ellwood, and Thomas and members of our laboratories for helpful comments. O.N.W. is an Investigator of the Howard Hughes Medical Institute. S.W. and A.J.G. are partially supported by National Institutes of Health Grant P50 CA86306 and National Cancer Institute Minority Supplement Award R01 CA107166:01S1 (to H.W.), respectively. D.A.L. is supported by National Institutes of Health Tumor Cell Biology Training Grant PHS T32 CA09056. This work was partially supported by National Institutes of Health Grants U01 CA84128, P50 CA092131 (to H.W. and O.N.W.), and R01 CA107166; Department of Defense Grant PC031130 (to H.W.); and Prostate Cancer Foundation grants (to H.W. and O.N.W.).
Author contributions: S.W., O.N.W., and H.W. designed research; S.W., A.J.G., M.W., and D.A.L. performed research; S.W., A.J.G., M.W., D.A.L., O.N.W., and H.W. analyzed data; and S.W., A.J.G., D.A.L., O.N.W., and H.W. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: AR, androgen receptor; E-cad, epithelial cadherin; PIN, prostatic intraepithelial neoplasia; PTEN, phosphatase and tensin homolog deleted on chromosome 10; Sca-1, stem cell antigen-1; TA/IC, transit amplifying/intermediate cell.
References
- 1.Greenlee, R. T., Murray, T., Bolden, S. & Wingo, P. A. (2000) CA Cancer J. Clin. 50, 7-33. [DOI] [PubMed] [Google Scholar]
- 2.Feldman, B. J. & Feldman, D. (2001) Nat. Rev. Cancer 1, 34-45. [DOI] [PubMed] [Google Scholar]
- 3.Isaacs, W., De Marzo, A. & Nelson, W. G. (2002) Cancer Cell 2, 113-116. [DOI] [PubMed] [Google Scholar]
- 4.De Marzo, A. M., Nelson, W. G., Meeker, A. K. & Coffey, D. S. (1998) J. Urol. 160, 2381-2392. [DOI] [PubMed] [Google Scholar]
- 5.Bui, M. & Reiter, R. E. (1998) Cancer Metastasis Rev. 17, 391-399. [DOI] [PubMed] [Google Scholar]
- 6.Isaacs, J. T. (1999) Urol. Clin. North Am. 26, 263-273. [DOI] [PubMed] [Google Scholar]
- 7.English, H. F., Santen, R. J. & Isaacs, J. T. (1987) Prostate 11, 229-242. [DOI] [PubMed] [Google Scholar]
- 8.Tsujimura, A., Koikawa, Y., Salm, S., Takao, T., Coetzee, S., Moscatelli, D., Shapiro, E., Lepor, H., Sun, T. T. & Wilson, E. L. (2002) J. Cell Biol. 157, 1257-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hudson, D. L. (2004) Prostate Cancer Prostatic Dis. 7, 188-194. [DOI] [PubMed] [Google Scholar]
- 10.Cunha, G. R. & Lung, B. (1978) J. Exp. Zool. 205, 181-193. [DOI] [PubMed] [Google Scholar]
- 11.Richardson, G. D., Robson, C. N., Lang, S. H., Neal, D. E., Maitland, N. J. & Collins, A. T. (2004) J. Cell Sci. 117, 3539-3545. [DOI] [PubMed] [Google Scholar]
- 12.Xin, L., Lawson, D. A. & Witte, O. N. (2005) Proc. Natl. Acad. Sci. USA 102, 6942-6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burger, P. E., Xiong, X., Coetzee, S., Salm, S. N., Moscatelli, D., Goto, K. & Wilson, E. L. (2005) Proc. Natl. Acad. Sci. USA 102, 7180-7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schalken, J. A. & van Leenders, G. (2003) Urology 62, 11-20. [DOI] [PubMed] [Google Scholar]
- 15.Wang, Y., Hayward, S., Cao, M., Thayer, K. & Cunha, G. (2001) Differentiation 68, 270-279. [DOI] [PubMed] [Google Scholar]
- 16.De Marzo, A. M., Meeker, A. K., Epstein, J. I. & Coffey, D. S. (1998) Am. J. Pathol. 153, 911-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song, X. & Xie, T. (2002) Proc. Natl. Acad. Sci. USA 99, 14813-14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song, X., Zhu, C. H., Doan, C. & Xie, T. (2002) Science 296, 1855-1857. [DOI] [PubMed] [Google Scholar]
- 19.Wu, X., Wu, J., Huang, J., Powell, W. C., Zhang, J., Matusik, R. J., Sangiorgi, F. O., Maxson, R. E., Sucov, H. M. & Roy-Burman, P. (2001) Mech. Dev. 101, 61-69. [DOI] [PubMed] [Google Scholar]
- 20.Passegue, E., Wagner, E. F. & Weissman, I. L. (2004) Cell 119, 431-443. [DOI] [PubMed] [Google Scholar]
- 21.Huntly, B. J., Shigematsu, H., Deguchi, K., Lee, B. H., Mizuno, S., Duclos, N., Rowan, R., Amaral, S., Curley, D., Williams, I. R., Akashi, K. & Gilliland, D. G. (2004) Cancer Cell 6, 587-596. [DOI] [PubMed] [Google Scholar]
- 22.Daley, G. Q. (2004) Cell 119, 314-316. [DOI] [PubMed] [Google Scholar]
- 23.Jamieson, C. H., Ailles, L. E., Dylla, S. J., Muijtjens, M., Jones, C., Zehnder, J. L., Gotlib, J., Li, K., Manz, M. G., Keating, A., Sawyers, C. L. & Weissman, I. L. (2004) N. Engl. J. Med. 351, 657-667. [DOI] [PubMed] [Google Scholar]
- 24.Jamieson, C. H., Weissman, I. L. & Passegue, E. (2004) Cancer Cell 6, 531-533. [DOI] [PubMed] [Google Scholar]
- 25.Al-Hajj, M., Becker, M. W., Wicha, M., Weissman, I. & Clarke, M. F. (2004) Curr. Opin. Genet. Dev. 14, 43-47. [DOI] [PubMed] [Google Scholar]
- 26.Signoretti, S., Pires, M. M., Lindauer, M., Horner, J. W., Grisanzio, C., Dhar, S., Majumder, P., McKeon, F., Kantoff, P. W., Sellers, W. R. & Loda, M. (2005) Proc. Natl. Acad. Sci. USA 102, 11355-11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Signoretti, S., Waltregny, D., Dilks, J., Isaac, B., Lin, D., Garraway, L., Yang, A., Montironi, R., McKeon, F. & Loda, M. (2000) Am. J. Pathol. 157, 1769-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang, A., Kaghad, M., Wang, Y., Gillett, E., Fleming, M. D., Dotsch, V., Andrews, N. C., Caput, D. & McKeon, F. (1998) Mol. Cell 2, 305-316. [DOI] [PubMed] [Google Scholar]
- 29.Levrero, M., De Laurenzi, V., Costanzo, A., Gong, J., Wang, J. Y. & Melino, G. (2000) J. Cell Sci. 113, 1661-1670. [DOI] [PubMed] [Google Scholar]
- 30.Tran, C. P., Lin, C., Yamashiro, J. & Reiter, R. E. (2002) Mol. Cancer Res. 1, 113-121. [PubMed] [Google Scholar]
- 31.Wang, S., Gao, J., Lei, Q., Rozengurt, N., Pritchard, C., Jiao, J., Thomas, G. V., Li, G., Roy-Burman, P., Nelson, P. S., Liu, X. & Wu, H. (2003) Cancer Cell 4, 209-221. [DOI] [PubMed] [Google Scholar]
- 32.Dubey, P., Wu, H., Reiter, R. E. & Witte, O. N. (2001) Cancer Res. 61, 3256-3261. [PubMed] [Google Scholar]
- 33.Li, Y., Welm, B., Podsypanina, K., Huang, S., Chamorro, M., Zhang, X., Rowlands, T., Egeblad, M., Cowin, P., Werb, Z., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 15853-15858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Groszer, M., Erickson, R., Scripture-Adams, D. D., Lesche, R., Trumpp, A., Zack, J. A., Kornblum, H. I., Liu, X. & Wu, H. (2001) Science 294, 2186-2189. [DOI] [PubMed] [Google Scholar]
- 35.Groszer, M., Erickson, R., Scripture-Adams, D. D., Dougherty, J., LeBelle, J., Zack, J., Geschwind, D., Liu, X., Kornblum, H. & Wu, H. (2006) Proc. Natl. Acad. Sci. USA 103, 111-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy-Burman, P., Wu, H., Powell, W. C., Hagenkord, J. & Cohen, M. B. (2004) Endocr. Relat. Cancer 11, 225-254. [DOI] [PubMed] [Google Scholar]
- 37.Parsons, J. K., Gage, W. R., Nelson, W. G. & De Marzo, A. M. (2001) Urology 58, 619-624. [DOI] [PubMed] [Google Scholar]
- 38.Shah, R. B., Zhou, M., LeBlanc, M., Snyder, M. & Rubin, M. A. (2002) Am. J. Surg. Pathol. 26, 1161-1168. [DOI] [PubMed] [Google Scholar]
- 39.Xin, L., Ide, H., Kim, Y., Dubey, P. & Witte, O. N. (2003) Proc. Natl. Acad. Sci. USA 100, Suppl. 1, 11896-11903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.