Abstract
Mathematical modeling was performed to test the extent to which cytopathic and noncytopathic T cell effector functions contribute to resolution of hepatitis B virus (HBV) infection in three acutely infected chimpanzees. Simulations based exclusively on cytopathic functions show a poor fit to the data and would require the destruction and regeneration of ≈11 livers for clearance to occur. In contrast, a simulation based on a combination of cytopathic and noncytopathic functions provided a significantly better fit to the data (P < 0.001) and required as much as 5-fold less destruction to clear the virus from the liver. The best fit simulation supports the notion that during the early phase of HBV clearance, noncytopathic T cell effector mechanisms inhibit viral replication and greatly shorten the half-life of the long lived covalently closed circular viral DNA transcriptional template, thereby limiting the extent to which cytopathic T cell effector functions and tissue destruction are required to terminate acute HBV infection.
Keywords: mathematical modeling, pathogenesis, covalently closed circular DNA
The hepatitis B virus (HBV) is a noncytopathic, hepatotropic, DNA virus (hepadnavirus) that causes acute and chronic hepatitis and hepatocellular carcinoma (1). Although it is generally agreed that viral clearance and disease pathogenesis during HBV infection are mediated by the immune response (2-6), the extent to which cytopathic and noncytopathic CD8+ T cell antiviral functions contribute to these events is a matter of some debate (7-11). On the one hand, it has been amply demonstrated that termination of acute hepadnavirus infections in the woodchuck (7, 12) and chimpanzee (9-11) systems is associated with the destruction of infected cells and hepatocellular regeneration (7-10, 12-15). On the other hand, it is equally clear from chimpanzee (9-11) and transgenic mouse (16, 17) experiments, that HBV replication is strongly inhibited by the antiviral effects of inflammatory cytokines produced by HBV-specific CD8+ T cells when they recognize antigen in the infected or transgenic liver. Furthermore, it has been shown that viral clearance in acutely infected chimpanzees is heralded by a sharp drop in the HBV DNA content of the liver, which depends on the influx of IFN-γ-producing CD8+ T cells into the liver (10), but which precedes and is out of proportion to the amount of liver disease (10). Moreover, in the transgenic mouse model, adoptively transferred HBV-specific CD8+ T cells have been shown to inhibit HBV replication by an IFN-γ-dependent mechanism (16), even when they cannot cause hepatitis because their cytolytic effector functions have been abrogated genetically (16). On the other hand, HBV replication proceeds unabated when IFN-γ-deficient CD8+ T cells are injected into HBV transgenic mice despite the induction of liver disease.
Although noncytolytic inhibition of HBV replication by CD8+ T cell-derived antiviral cytokines is sufficient to prevent the production of new virus particles and, therefore, viral spread, complete viral clearance from the liver, requires elimination of the covalently closed circular (ccc) HBV DNA nuclear episome that serves as the transcriptional template of the virus in infected cells (18). Although the half-life of the cccDNA episome has never been accurately determined, the available evidence suggests that it is a long-lived species (19-22). If that assumption is correct, either the infected cells must be destroyed or the half-life of the cccDNA must be shortened by noncytolytic mechanisms, or both, for viral clearance to occur. Because HBV cccDNA is not readily produced by HBV transgenic mice (23), it is not possible to ascertain the impact of antiviral cytokines on cccDNA half-life in that model. In a recent study, however, the cccDNA content of the liver was quantitated at weekly intervals in three acutely infected chimpanzees (9), and the results were compared with the dynamic changes in many other HBV-specific virological and disease parameters (9, 10). In that study, it appeared that the cccDNA content of the infected liver decreased before and out of proportion to the amount of liver disease in these animals following the onset of a CD8+ T cell response. Nonetheless, it was difficult to be certain that the cccDNA was controlled by noncytolytic mechanisms in those animals because the cytopathic events were superimposed and because termination of the infection in all of the animals coincided with peak liver disease (9).
Thus, in the current report, we assess the relative contributions of cytolytic and noncytolytic processes to viral clearance during HBV infection by comparing the observed viral and disease dynamics in three acutely infected chimpanzees with simulations of those dynamics by using a mathematical model that includes all of the underlying infection components and uses the simplest assumptions to interpret the basis for changes in each component according to currently accepted principles of CD8+ T cell-based viral immunopathology. In each simulation, we examined the likely relative contributions of cytopathic versus noncytopathic T cell effector mechanisms in the clearance of the virus, total viral DNA, and viral cccDNA from the liver. Models based exclusively on cytopathic T cell effector mechanisms failed to reproduce the observations and required an exceedingly large amount of liver disease and regeneration to terminate the infection. A model that incorporates both cytopathic and noncytopathic mechanisms, including noncytopathic shortening of the cccDNA half-life, provides a significantly better fit to the data without requiring massive liver destruction and regeneration. We conclude that T cell-mediated noncytopathic antiviral effector functions limit viral spread and reduce the viral cccDNA template during HBV infection, thereby minimizing the amount of infected tissue that must be destroyed to terminate the infection.
Materials and Methods
Mathematical Model. We assume newly infected cells arise from infection of uninfected hepatocytes, H, by free virus, V. The rate at which infection occurs is proportional not only to the size of each of these components but where infection saturates relative to the number of uninfected hepatocytes kHV/(Kh + H). This Michaelis-Menten form is typical of systems such as predatorprey problems where the number of prey, e.g., uninfected hepatocytes, can be much larger than predators, e.g., HBV, such as early in infection. New infection produces a cell with a single copy of cccDNA, I1. The infected cell moves from the component with one cccDNA copy, I1, to cells with two copies, I2, at rate a. Cells are assumed to increase their cccDNA copy number to a maximum of N copies, giving rise to the components Ii, i = 1,..., N of infected cells with i cccDNA copies. The rate at which infected cells move to a higher copy number is proportional to a but decreases in a linear fashion so that there is zero increase from the highest copy number N.
The total amount of pregenomic RNA (pgRNA) in the liver is denoted by R, whereas the number of DNA containing capsids is given by D. Each cccDNA template produces M pgRNA molecules within the cell per day, and these mature through encapsidation and conversion to single- and double-stranded HBV DNA capsids at rate α. Inhibition of this maturation (as well as of amplification of the cccDNA number within infected hepatocytes) is modeled by a factor u that is 1 when a cytokine effect is absent (t < tc) and a lower value when it is present [u = 1 - eff, t ≥ tc + 7, with a linear change between the values on tc, tc + 7, and time in days). HBV DNA capsids are exported from the cell at rate β to produce free virus V. Free virus is removed at rate c by mechanisms including the immune response.
Clearance of infection arises through death of infected cells. The amount of death per day is assumed proportional (with proportionality constant b) to A = max(ALT - 40,0), and alanine transaminase (ALT) levels above the upper limit of normal, taken as 40. Cell death at any time is distributed among the Ii, i = 1,..., N in proportion to their number compared to total infected hepatocytes, I. Killed cells are assumed to be immediately replaced by uninfected cells by requiring H = 100 - I, where all cell numbers have been rescaled to percentages of hepatocytes. An infected cell that is killed is assumed to be replaced through cell division of another hepatocyte. The dividing cell is chosen randomly from the remaining infected and uninfected cells. If the dividing cell is also infected, then we assume it produces two uninfected daughter cells. In this situation, two infected cells are replaced with the death of one cell, whereas if the dividing cell is uninfected, one infected cell is replaced with the death of one cell. This process is captured in the term (1 + (I/100)) by multiplying the expressions for cell death.
The differential equation description of this model is given by
 |
[1] |
Simulations including degradation of cccDNA are modeled by the inclusion of terms -i(1 - u)μIi for i =2,..., N on the right hand side of the equations for dIi/dt, where log(2)/μ is the half-life of cccDNA during this time. We excluded this term from dIi/dt because we use the sum of Ii as a measure of HBc+ hepatocytes, and cells can be HBc+ while all cccDNA have been degraded. We model this possible discrepancy between HBc and cccDNA positivity by keeping all infected cells at least cccDNA+ via I1. Hence, these latter simulations will provide an upper bound of hepatocyte death. Variable descriptions are collected in Table 1, which is published as supporting information on the PNAS web site, and parameter descriptions are shown in Table 2, which is published as supporting information on the PNAS web site. Assumptions used in the construction of this model, justifications for their choice, and alternative assumptions are listed in Table 3, which is published as supporting information on the PNAS web site.
Three scenarios of infection clearance were investigated: Scenario 1, removal of infected cells through cell division and cell death (linearly scaled from observed ALT above upper limit of normal) so u ≡ 1 for all time; Scenario 2, cell division and death plus a cytokine effect blocking formation of pgRNA-containing capsids and, therefore, a block in HBV DNA containing capsids, u < 1 after tc with μ ≡ 0; and Scenario 3, cell division and death, block of formation of pgRNA containing capsids, plus decay of cccDNA through either an inherently short lifespan or destabilization via cytokines u < 1 after tc with μ > 0.
To obtain the best performance possible within each scenario, a maximum likelihood estimate (MLE) method was used to find the best fit of model simulations, searching over the space of all parameter values, to log10 data of HBcAg positive hepatocyte percentage, cccDNA per hepatocyte, pgRNA% of maximum, HBV DNA in liver, and HBV DNA in serum for Ch 1615 and Ch 1627 simultaneously. Simulation values below levels of detection were reset to that level. A sequential quadratic programming method was used to find MLE parameter values. The optimal parameter values were then used in the simulations of each scenario and for each chimpanzee. Model parameters were either required to be the same, but chosen optimally, for all animals [k, Kh, a, β, M (and μ for Scenario 3)], or animal specific [b, c, eff, where eff registers the effectiveness of cytokines in inhibiting pgRNA maturation and possibly depletion of cccDNA (u = 1 - eff, t ≥ tc + 7)], also chosen optimally. Variability in the animal-specific parameters was required to accommodate the different levels of cell death reflected in ALT and immune effectiveness. Any cytokine effects were assumed to commence at time tc taken to be 1 week after peak infection levels (week 8, Ch 1615 and Ch 1627; week 10, Ch 1620). Maximum cccDNA copies per infected cell N were set at peak cccDNA copies per hepatocyte. Because only relative levels of pgRNA were observed, the value for α, the rate of maturation to HBV DNA within an infected cell, could not be determined. Because this dynamic should be similar to the export rate of HBV DNA, we set α = β. The MLE parameter values were then applied to the data for Ch 1620, where once again the animal-specific parameters b, c, and eff were chosen as MLEs. The significance of fit between different models was determined by using the likelihood ratio test, which takes into consideration the advantages of a model with extra parameters. Ninety-five percent confidence intervals were calculated in a similar fashion (see Supporting Text, which is published as supporting information on the PNAS web site). Simulations were performed in matlab, version 6.5, (MathWorks, Natick, MA).
Supporting Information. The infectious inocula, chimpanzees, and assay and statistical methods are described in the Supporting Text.
Results
Course of HBV Infection in Acutely Infected Chimpanzees. Ch1627, Ch1615, and Ch1620 were inoculated with 0.5 ml of HBV-transgenic mouse serum containing 1 × 108 genome equivalents of HBV DNA (10). As shown in Fig. 1, all three animals became infected with similar kinetics of viral spread reaching maximal serum HBV DNA levels between 4.3 × 108 and 3.4 × 109 genome equivalents (GE)/ml during weeks 7-8 after inoculation (Fig. 1 A). For the non-CD8-depleted animals Ch1615 and Ch1627, intrahepatic HBV DNA, cccDNA, and pgRNA reached maximal levels by 1 week later (Fig. 1B; refs. 9 and 10). Furthermore, between 86% and 99% of the hepatocytes were HBcAg positive at the peak of infection (Fig. 1B) in these animals, indicating that virtually the entire liver became infected (9, 10).
Fig. 1.
Course of acute HBV infection in chimpanzees after experimental inoculation with HBV in the presence or absence of CD4+ and CD8+ cells. All animals were inoculated with 108 genome equivalents (GEs) of HBV intravenously on week 0, and a CD4- or CD8-specific monoclonal antibody was injected at week 6 in Ch1615 and Ch1620, respectively. Ch1627 was injected with irrelevant control antibody. (A) Serum HBV DNA (green line) in all animals is presented as log10 values of GE/ml, and serum alanine transaminase activity (ALT, shaded area) is expressed in units per liter (U/L). (B) Total HBV DNA (green line) and cccDNA (red line) in the liver are expressed as copies per hepatocyte and were calculated as described in Materials and Methods. The intrahepatic level of HBV pregenomic-(pg)RNA (blue line) is represented as a percentage of the corresponding peak pgRNA level in each animal. Hepatitis B core antigen positive (HBcAg+) hepatocytes (shaded area) are expressed as percentage of the total number of hepatocytes. Short upward arrows indicate partial restoration of the corresponding T cell population, and the longer upward arrow indicates that CD8+ T cells returned to baseline levels. Total RNA isolated from liver biopsies was analyzed for the expression of CD3, IFN-γ, and L32 by an RNase protection assay. The L32 signals reflect the amount of RNA used in the assay.
As described in refs. 9 and 10, the clearance phase in the control Ch1627 and the CD4-depleted Ch1615 started almost immediately after peak infection at week 8, whereas depletion of CD8 cells in Ch1620 (Fig. 1B, solid line) prolonged the phase of maximal infection for all viral parameters (Fig. 1B; refs. 9 and 10) until as late as week 12 (peak cccDNA), when CD8 cells gradually reappeared (Fig. 1B, upward arrrow). The beginning of the viral clearance phase in all three animals coincided with the influx of T cells (Fig. 1B, CD3 mRNA) into the liver and intrahepatic IFN-γ production (Fig. 1B, IFN-γ) as shown by RNase protection analysis of total liver RNA (Fig. 1B; refs. 9-11). Importantly, during the first 4 to 6 weeks of viral clearance, all viral nucleic acid species decreased between 8- and 50-fold. This decrease outpaced the observed reduction in the percentage of HBcAg-positive hepatocytes (Fig. 1B; refs. 9 and 10) and it occurred before serum ALT activity reached maximal levels (Fig. 1 A; refs. 9-11). As suggested in refs. 9-11, the kinetics of these changes implies that HBV replication within infected hepatocytes is reduced by noncytopathic mechanisms, probably mediated by the antiviral effects of IFN-γ (Fig. 1B; refs. 9-11). Later in the course of infection in all animals, peak serum ALT activity corresponded to losses of between 7% and 20% of HBcAg-positive hepatocytes and increased removal rates of viral nucleic acids, reflecting the cytolytic activity of the virus-specific CD8 T cell response (10).
The importance of CD8+ T cells was further demonstrated by CD8 depletion in Ch1620, which changed the kinetics of viral clearance and liver disease (Fig. 1), both of which depended on the reappearance of CD8 cells (Fig. 1B, upward arrows; refs. 9 and 10). As shown in Fig. 1B, all viral parameters in Ch1620 remained at or near peak levels and serum ALT activity was normal during the period of complete CD8 depletion (Fig. 1B, weeks 6-11). Because CD8+ T cells reappeared from weeks 12 to 17, albeit at subnormal levels (data not shown), all viral parameters fell significantly, except the number of HBcAg positive hepatocytes, similar to the other two animals, and serum ALT activity rose. The failure of the HBcAg-positive hepatocytes to fall significantly during this period suggests that the extent of liver cell destruction was minimal, despite the elevated serum ALT activity. Indeed, because 97% of HBV DNA in serum and liver and 87% of cccDNA disappeared during this interval, whereas HBcAg positive hepatocytes were reduced by only 17%, the noncytolytic activity of the CD8+ T cells was dominant at that point. All viral parameters disappeared, and serum ALT activity spiked when CD8+ T cell levels surged to baseline levels on week 29 (Fig. 1B, upward arrow), suggesting that the cytolytic function of the CD8+ T cells was required for complete viral clearance to occur (9-11). Notably, the cccDNA was the slowest viral element to disappear because they were detectable for as long as 73, 111, and 111 weeks for Ch 1615, 1620, and 1627, respectively. Nonetheless, at the late time points shown in Fig. 1B, total liver HBV DNA, inclusive of cccDNA, is no higher than the cccDNA levels and the pgRNA levels are below detection, suggesting that the remaining cccDNA molecules are long-lived but not highly transcriptionally active.
Transition Between Infection Components. The processes from infection of a cell to virion production follow the order: a cell is infected (I), a cccDNA molecule is established in the nucleus (C), HBV RNA molecules including pgRNA are transcribed (R), pgRNA-containing capsids are formed and these mature to single- and double-stranded DNA-containing capsids (D), and some of these capsids are transported to the nucleus to amplify cccDNA numbers, whereas the majority are exported to plasma as virions (V). We assessed immune system interference in these processes, especially with regard to IFN-γ, by comparing the ratios of a downstream component with its upstream parent. If a particular process is affected, then the ratios in the clearance phase should be lower than the ratios at the end of the infection phase. Here we are particularly interested in the role of noncytopathic processes and have represented the clearance phase by the period of elevated IFN-γ expression (Fig. 1B) and from the time of the first measured liver HBV DNA after peak levels so that relevant ratios can be calculated (Ch 1615, weeks 11-15; Ch 1620, weeks 13-30; Ch 1627, weeks 11-14). The ratios of weekly interpolated values are shown in Fig. 2. For ease of comparison, time is represented relative to the end of the infection period, with negative times representing the infection phase and positive times representing the clearance phase.
Fig. 2.
Changes in transition between components of infection for Ch 1615 (blue circles), 1620 (green squares), and 1627 (red triangles). (A) Total intracellular HBV DNA per cell per pgRNA% of maximum (D/R%). (B) cccDNA copies per infected hepatocyte (C/I). (C) pgRNA% of maximum per cccDNA copy per cell (R%/C). (D) HBV DNA in serum per total intracellular HBV DNA in liver (V/D). Ratios were calculated by using linear interpolants for missing weekly time points. Time in weeks is represented relative to the end of the infection period (Ch 1615, week 10; Ch 1620, week 12; Ch 1627, week 10).
The ratios D/R% (Fig. 2 A) drop considerably from the start of clearance. Values of this ratio during the clearance phase are significantly lower than earlier values (P ≤ 0.03 for each animal, Wilcoxon rank sum test). Reduced levels of this ratio indicate an immune component specifically inhibiting HBV replication post-transcriptionally possibly by preventing the formation of pgRNA-containing capsids and, hence, the production of single- and double-stranded DNA-containing capsids.
Values for the C/I ratio (Fig. 2B) for the clearance phase are also lower than earlier values (Ch 1615, P = 0.13; Ch 1620, P = 0.001; Ch 1627, P = 0.03). This decreased ratio can be explained by depletion of cccDNA molecules within infected cells, or removal of infected cells with high cccDNA complement and replacement by infected cells with fewer cccDNA copies.
The ratios R%/C (Fig. 2C) do not differ significantly between the clearance and infection phases (Ch 1615, P = 0.79; Ch 1620, P = 0.08; Ch 1627, P = 0.34). Hence, the immune system has little effect on pgRNA production from the cccDNA template. Similarly, the ratio V/D (Fig. 2D) is not significantly different between periods (Ch 1615, P = 0.08; Ch 1620, P = 0.12; Ch 1627, P = 0.69). This lack of difference implies little immune inhibition of virion export and no change in virion clearance in plasma.
Model of Cytokine and Cytolytic Effects. To accurately investigate the impact of the various immune effects on the time course of HBV infection in these chimpanzees, we constructed a mathematical model containing elements of infection and clearance (Materials and Methods). We specifically investigated three scenarios for clearance of the infection: Scenario 1, cell division and death (scaled from observed ALT above the upper limit of normal); Scenario 2, cell division and death plus a cytokine effect blocking formation of pgRNA-containing capsids; and Scenario 3, cell division and death, block of formation of pgRNA containing capsids, plus decay of cccDNA through either an inherently short lifespan or destabilization via cytokines.
We developed the scenario models by first determining best estimates jointly for two of the animals and then tested the results further by applying these estimates to the remaining animal. Therefore, we determined MLE parameter values for each scenario by fitting simulations to all viral data for the non-CD8-depleted Ch1627 and Ch1615 simultaneously. For the additional test for the suitability of these scenarios, we then applied these common MLE parameter values to Ch 1620 and determined MLE values for the remaining animal specific parameters (Table 2). These model simulations are displayed against the data for Ch1627, Ch1615, and Ch1620 in Figs. 3, 4, 5.
Fig. 3.
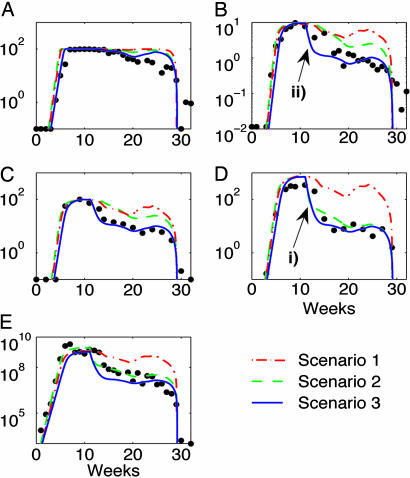
Data (dots) and model simulations (lines) for Ch 1627 by using maximum likelihood estimates for parameters for each of the three scenarios. (A) Percentage of HBcAg-positive hepatocytes. (B) Average copy number of cccDNA per hepatocyte. (C) pgRNA as a percentage of maximal pgRNA levels. (D) Average copy number of HBV DNA replicative intermediates per hepatocyte. (E) HBV genome equivalents per milliliter of serum. Scenario 1 (red dash-dot line), total simulated hepatocyte death 516%; Scenario 2 (green dashed line), total simulated hepatocyte death 199%; Scenario 3 (blue solid line), total simulated hepatocyte death 199%. Red dash-dot line, Scenario 1, cell division and death only; green dashed line, Scenario 2, cell division and death plus blocking of pgRNA maturation; blue solid line, Scenario 3, cell division and death plus blocking of pgRNA maturation plus decay of cccDNA. Time is measured in weeks from inoculation. Drop in HBV DNA/cell indicated by i) here and in Figs. 4 and 5 is due to continuous export and cytokine mediated block in production of DNA containing capsids. Decay of cccDNA per hepatocyte through either direct destabilization by cytokines or an inherently short lifespan indicated by ii) in B here and in Figs. 4 and 5.
Fig. 4.
Data (dots) and model simulations (lines) for Ch 1615 by using maximum likelihood estimates for parameters for each of the three scenarios. (A) Percentage of HBcAg-positive hepatocytes. (B) Average copy number of cccDNA per hepatocyte. (C) pgRNA as a percentage of maximal pgRNA levels. (D) Average copy number of HBV DNA replicative intermediates per hepatocyte. (E) HBV genome equivalents per milliliter of serum. Scenario 1 (red dash-dot line), total simulated hepatocyte death 772%; Scenario 2 (green dashed line), total simulated hepatocyte death 183%; Scenario (blue solid line), total simulated hepatocyte death 143%. For symbols, see Fig. 3.
Fig. 5.
Data (dots) and model simulations (lines) for Ch 1620 by using maximum likelihood estimates for parameters for each of the three scenarios. (A) Percentage of HBcAg-positive hepatocytes. (B) Average copy number of cccDNA per hepatocyte. (C) pgRNA as a percentage of maximal pgRNA levels. (D) Average copy number of HBV DNA replicative intermediates per hepatocyte. (E) HBV genome equivalents per milliliter of serum. Scenario 1 (red dash-dot line), total simulated hepatocyte death 1,174%; Scenario 2 (green dashed line), total simulated hepatocyte death 347%; Scenario 3 (blue solid line), total simulated hepatocyte death, 278%. For symbols, see Fig. 3.
The likelihood ratio test allowed comparison of the scenarios at both the model developmental stage, with respect to data for Ch 1627 and 1615, and at the testing stage, with respect to data for Ch 1620. Scenario 1 (red dash-dot lines) was significantly worse at reproducing data in both stages (P < 0.001), and Scenario 3 (blue solid lines) was significantly better than each of the other scenarios (P < 0.001 against each scenario in the development stage, and P < 0.001 compared to Scenario 1 and P = 0.012 against Scenario 2 in the testing stage).
The differences in quality of fit essentially arise because of two processes evident in the data. In D of each figure, i) indicates where Scenarios 2 and 3 both give consistency with data, but Scenario 1 fails. This section of the data is indicative of loss of HBV DNA in hepatocytes relative to pgRNA, because simulations in C (Figs. 3, 4, 5) for Scenario 2 do not exhibit any corresponding decay. The slope of the decline in i) and for the corresponding time of its downstream component of HBV DNA in serum (E), is determined by the export rate of HBV DNA, β = 0.46. This slope predicts an export half-life of 1.5 days (log(2)/β). The extent of decline is determined by the effectiveness of cytokines blocking the process of maturation of pgRNA. For both Ch 1627 and the CD4-depleted Ch 1615, there is no delay in this decrease with effectiveness of this block in pgRNA maturation being on average 88% for Scenarios 2 and 3.
The other section of data that distinguishes the accuracy of the scenario simulations occurs in B of each figure and is indicated by ii). It shows a fast first-phase decay in cccDNA followed by slower removal, and this cccDNA decay also induces a similar profile in the downstream component of pgRNA in C. The removal of cccDNA molecules within infected cells causes the fast decline in this viral component that is evident in the data and is reproduced by Scenario 3 but not Scenarios 1 or 2. The slope of this decline is determined by the decay rate of cccDNA μ = 0.24, in combination with the effectiveness of blocking HBV DNA repopulating cccDNA. This slope predicts a half-life of cccDNA, at least when cytokines are active, of ≈3 days. The 95% confidence interval for the half-life of cccDNA during this clearance phase is 0.6-8 days.
Discussion
Quantification of all intracellular components of HBV infection allowed us to accurately determine the dynamics of infection and clearance in acutely infected chimpanzees. To assess the interaction of various hypothesized processes of HBV clearance in this complicated dynamical system, we developed a mathematical model that contained these processes and ran it over three scenarios. The comparison between any two scenarios involved the likelihood ratio test, which compares twice the difference between the maximized log-likelihoods to a χ2 distribution with degrees of freedom given by the number of extra parameters of one scenario over the other. Therefore, P values <0.05 imply that the scenario with more parameters is a better description of the data regardless of the extra parameters. Only Scenario 3, which included a cytokine effect that blocks conversion of pgRNA into DNA-containing capsids and is associated with a fast decay of cccDNA molecules, accurately simulated all components of infection (blue solid lines, Figs. 3, 4, 5), with this scenario significantly better than Scenario 1, which only used cell division and death (P < 0.001), and also significantly better in comparison to Scenario 2, which did not allow for the loss of cccDNA (P ≤ 0.012).
The profile of a fast decline of HBV DNA in hepatocytes after peak infection, Figs. 3, 4, 5D, i), indicates the rate of export of HBV DNA from cells and the effectiveness of the block in pgRNA maturation. Calculations suggest that ≈90% of pgRNA is blocked from maturation to HBV DNA in the presence of these cytokines, consistent with the significant change in ratios of intracellular HBV DNA and pgRNA (P ≤ 0.03,D/R%, Fig. 2A).
Additionally, a specific effect of cccDNA removal within cells was observed through the fast decline of cccDNA (Figs. 3, 4, 5B, ii). Calculations from Ch 1627 and 1615 suggest that during the block in replenishment of the intracellular HBV DNA pool and possibly under direct cytokine effects that cccDNA molecules have a half-life of ≈3 days, with a 95% confidence interval of between 0.6 and 8 days.
If cell death in conjunction with cell division were the only mechanisms responsible for clearance of HBV infection, these simulations indicate that very large amounts of death and replacement would be required to clear the infection (516%, Ch 1627; 772%, Ch 1615; 1,174%, Ch 1620), assuming that any uninfected cell that serves as a replacement is infected at the same rate as in the initial infection period. On the other hand, scenarios incorporating the cytokine effects described above require significantly less hepatocyte death (199%, Ch 1627; 143%, Ch 1615; 278%, Ch 1620).
HBV pgRNA levels had dropped below detection limits within 20 weeks after inoculation for Ch 1615 and Ch 1627 and 32 weeks for Ch 1620, and the number of HBV DNA molecules per hepatocyte (which includes cccDNA) fell to levels indistinguishable from the cccDNA copy number per hepatocyte. HBV DNA in the serum decreased >6 orders of magnitude. Yet, cccDNA was still detectable as late as 111 weeks after inoculation (9). The lack of mRNA and markedly less reinfection due to negligible detectable virus implies that these cccDNA viral templates are not transcriptionally active, in which case their continued presence would indicate that cccDNA molecules are inherently stable and long-lived. Hence, the very short half-life of 3 days calculated for these molecules during active immune clearance and the presence of a strong cytokine effect would imply that it is the direct effects of cytokines that remove cccDNA rather than the block in replenishment of this pool uncovering an underlying fast half-life.
Mathematical modeling inherently simplifies a problem. That is its aim while still retaining sufficient complexity to uncover major forces shaping outcomes. Our model in Eq. 1 has simplified HBV infection to a considerable extent, yet it contains the essential features of the infection. It assumes that cytokines inhibit the conversion of pgRNA into DNA-containing capsids and destabilize cccDNA and that cell death can be scaled directly from observed ALT levels. Based on these assumptions, it has faithfully reproduced the complicated dynamic profiles of five viral components (Figs. 3, 4, 5, blue solid lines) in three genetically unrelated animals of different ages, sizes, and sex. Importantly, scenarios that do not incorporate noncytopathic antiviral effects of these cytokines fail in this regard to a statistically significant level (green dashed and red dash-dot lines). At a more basic level, Scenario 1 (Figs. 3, 4, 5, red dash-dot lines), will also be inconsistent with the observed reduction in ratios of intracellular HBV DNA and pgRNA (P ≤ 0.03, D/R%, Fig. 2A), because cell death and/or division should not affect one of these components more than the other. We conclude from our results and mathematical modeling that processes whereby CD8+ T cells not only kill infected cells, but also induce a noncytopathic effect that blocks conversion of pgRNA to DNA-containing capsids and removes cccDNA, can provide a statistically significant description of clearance of acute HBV infection.
Supplementary Material
Acknowledgments
We thank Dr. Alan Perelson for discussions on HBV modeling and Dr. William Mason for helpful comments and suggestions. This work was supported by National Institutes of Health Grant AI20001 (to F.V.C.). J.M.M. was assisted by a University of NSW Goldstar Award. This work is manuscript 17260-MEM from The Scripps Research Institute.
Author contributions: J.M.M., S.F.W., R.H.P., and F.V.C. designed research; J.M.M. and S.F.W. performed research; J.M.M., S.F.W., and F.V.C. analyzed data; J.M.M. and S.F.W. wrote the paper; and F.V.C. edited the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: ALT, alanine transaminase; ccc, covalently closed circular; HBV, hepatitis B virus; MLE, maximum likelihood estimate; pgRNA, pregenomic RNA.
References
- 1.Chisari, F. V. & Ferrari, C. (1995) Annu. Rev. Immunol. 13, 29-60. [DOI] [PubMed] [Google Scholar]
- 2.Ferrari, C., Bertoletti, A., Penna, A., Cavalli, A., Valli, A., Missale, G., Pilli, M., Fowler, P., Giuberti, T., Chisari, F. V., et al. (1991) J. Clin. Invest. 88, 214-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertoletti, A., Ferrari, C., Fiaccadori, F., Penna, A., Margolskee, R., Schlicht, H. J., Fowler, P., Guilhot, S. & Chisari, F. V. (1991) Proc. Natl. Acad. Sci. USA 88, 10445-10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nayersina, R., Fowler, P., Guilhot, S., Missale, G., Cerny, A., Schlicht, H. J., Vitiello, A., Chesnut, R., Person, J. L., Redeker, A. G., et al. (1993) J. Immunol. 150, 4659-4671. [PubMed] [Google Scholar]
- 5.Missale, G., Redeker, A., Person, J., Fowler, P., Guilhot, S., Schlicht, H. J., Ferrari, C. & Chisari, F. V. (1993) J. Exp. Med. 177, 751-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rehermann, B., Fowler, P., Sidney, J., Person, J., Redeker, A., Brown, M., Moss, B., Sette, A. & Chisari, F. V. (1995) J. Exp. Med. 181, 1047-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo, J. T., Zhou, H., Liu, C., Aldrich, C., Saputelli, J., Whitaker, T., Barrasa, M. I., Mason, W. S. & Seeger, C. (2000) J. Virol. 74, 1495-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Summers, J., Jilbert, A. R., Yang, W., Aldrich, C. E., Saputelli, J., Litwin, S., Toll, E. & Mason, W. S. (2003) Proc. Natl. Acad. Sci. USA 100, 11652-11659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wieland, S. F., Spangenberg, H. C., Thimme, R., Purcell, R. H. & Chisari, F. V. (2004) Proc. Natl. Acad. Sci. USA 101, 2129-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thimme, R., Wieland, S., Steiger, C., Ghrayeb, J., Reimann, K. A., Purcell, R. H. & Chisari, F. V. (2003) J. Virol. 77, 68-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guidotti, L. G., Rochford, R., Chung, J., Shapiro, M., Purcell, R. & Chisari, F. V. (1999) Science 284, 825-829. [DOI] [PubMed] [Google Scholar]
- 12.Kajino, K., Jilbert, A. R., Saputelli, J., Aldrich, C. E., Cullen, J. & Mason, W. S. (1994) J. Virol. 68, 5792-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jilbert, A. R., Wu, T. T., England, J. M., Hall, P. M., Carp, N. Z., O'Connell, A. P. & Mason, W. S. (1992) J. Virol. 66, 1377-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jilbert, A. R. & Kotlarski, I. (2000) Dev. Comp. Immunol. 24, 285-302. [DOI] [PubMed] [Google Scholar]
- 15.Mason, W. S., Jilbert, A. R. & Summers, J. (2005) Proc. Natl. Acad. Sci. USA 102, 1139-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guidotti, L. G., Ishikawa, T., Hobbs, M. V., Matzke, B., Schreiber, R. & Chisari, F. V. (1996) Immunity 4, 25-36. [DOI] [PubMed] [Google Scholar]
- 17.Guidotti, L. G., Ando, K., Hobbs, M. V., Ishikawa, T., Runkel, L., Schreiber, R. D. & Chisari, F. V. (1994) Proc. Natl. Acad. Sci. USA 91, 3764-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Summers, J. & Mason, W. S. (1982) Cell 29, 403-415. [DOI] [PubMed] [Google Scholar]
- 19.Zhu, Y., Yamamoto, T., Cullen, J., Saputelli, J., Aldrich, C. E., Miller, D. S., Litwin, S., Furman, P. A., Jilbert, A. R. & Mason, W. S. (2001) J. Virol. 75, 311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fourel, I., Cullen, J. M., Saputelli, J., Aldrich, C. E., Schaffer, P., Averett, D. R., Pugh, J. & Mason, W. S. (1994) J. Virol. 68, 8321-8330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luscombe, C., Pedersen, J., Uren, E. & Locarnini, S. (1996) Hepatology 24, 766-773. [DOI] [PubMed] [Google Scholar]
- 22.Zhang, Y. Y. & Summers, J. (2000) J. Virol. 74, 5257-5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guidotti, L. G., Matzke, B., Schaller, H. & Chisari, F. V. (1995) J. Virol. 69, 6158-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.