Abstract
Background
Experimental populations of Escherichia coli have evolved for 20,000 generations in a uniform environment. Their rate of improvement, as measured in competitions with the ancestor in that environment, has declined substantially over this period. This deceleration has been interpreted as the bacteria approaching a peak or plateau in a fitness landscape. Alternatively, this deceleration might be caused by non-transitive competitive interactions, in particular such that the measured advantage of later genotypes relative to earlier ones would be greater if they competed directly.
Results
To distinguish these two hypotheses, we performed a large set of competitions using one of the evolved lines. Twenty-one samples obtained at 1,000-generation intervals each competed against five genetically marked clones isolated at 5,000-generation intervals, with three-fold replication. The pattern of relative fitness values for these 315 pairwise competitions was compared with expectations under transitive and non-transitive models, the latter structured to produce the observed deceleration in fitness relative to the ancestor. In general, the relative fitness of later and earlier generations measured by direct competition agrees well with the fitness inferred from separately competing each against the ancestor. These data thus support the transitive model.
Conclusion
Non-transitive competitive interactions were not a major feature of evolution in this population. Instead, the pronounced deceleration in its rate of fitness improvement indicates that the population early on incorporated most of those mutations that provided the greatest gains, and subsequently relied on beneficial mutations that were fewer in number, smaller in effect, or both.
Background
Twelve populations of Escherichia coli B were founded from a common ancestor, and they have evolved for 20,000 generations in a uniform environment with glucose as the density-limiting resource [1-3]. During that period, the populations adapted to this environment by the substitution of spontaneous beneficial mutations. Figure 1 shows the overall fitness trajectory of the 12 populations, measured in competition with the common ancestor. As shown by the excellent fit of the hyperbolic curve, the rate of fitness gain relative to the ancestor has strongly decelerated over time.
Figure 1.

Long-term E. coli populations show decelerating improvement. Each point shows the mean fitness (averaged over 12 replicate populations) measured relative to the common ancestor. Error bars are 95% confidence intervals calculated from the 12 population estimates. The hyperbolic curve was fit to the data. Data from Cooper and Lenski [3].
In this paper, we seek to test two hypotheses that could, in principle, account for this deceleration. (H1) The observed deceleration indicates that the rate of evolutionary adaptation is truly slowing down. This occurs as the evolving population approaches a fitness peak or plateau because the remaining number of beneficial mutations, their marginal effect, or both become progressively smaller. This hypothesis assumes that fitness values of the chronologically ordered population samples are both qualitatively and quantitatively transitive; i.e., they follow a strict competitive hierarchy, such that the cumulative fitness improvement relative to the ancestor could be predicted from the incremental gains over the constituent time intervals. (H2) The rate of adaptation continues at the initial rapid pace, and the apparent deceleration is an artifact of using the ancestor as the common yardstick to measure adaptation. Under this hypothesis, fitness values are non-transitive. Consequently, the total fitness improvement relative to the ancestor cannot be predicted from the incremental gains over smaller intervals.
Several studies have documented non-transitive interactions. Paquin and Adams [4] observed unexpected declines in fitness in populations of Saccharomyces cerevisiae evolving in glucose-limited chemostats, when fitness was based on competition with the ancestral strain. However, when a series of chronologically ordered isolates was each competed against its immediate predecessor, the later isolate was invariably more fit. The precise mechanism responsible for this non-transitivity was not identified, but presumably involved the accumulation of metabolic by-products in the culture media, which affected competition. Rainey and Travisano [5] observed non-transitive fitness interactions between three different variants of Pseudomonas fluorescens that evolved in cultures without physical shaking, which allowed gradients or other heterogeneities to develop. The non-transitive effects were based on competitive trade-offs between niche specialists, which provided an advantage to each type when it was rare relative to one or both other types in the overall heterogeneous environment. Non-transitive interactions sometimes also result from cyclical competitive hierarchies that resemble the "rock-paper-scissors" game, in which rock crushes scissors, scissors cut paper, and paper covers rock [6]. Biological examples of such cyclical hierarchies include: (i) competition among toxin producers, resistant types, and sensitive non-producers in species that use allelopathy to compete, such as coral-reef dwelling invertebrates [7] and various microbes [8,9]; (ii) competition between three morphologically different types with distinct reproductive strategies in side-blotched lizards [10]; and (iii) other situations with more than two limiting resources that can lead to complex dynamics in plankton communities [11].
In this study, we seek to test whether non-transitive interactions could account for the observed deceleration in fitness gains, measured relative to the ancestral strain, in the long-term E. coli evolution experiment. To that end, we first develop transitive and non-transitive models that can explain – both equally well – the measured trajectory of fitness relative to the ancestor, but which make quantitatively different predictions about the fitness values that would result from competing evolved samples taken at different time points. We then perform experiments to test which model is better.
Results and Discussion
Transitive and non-transitive models
Here we present two theoretical models that could, in principle, account for the mean fitness trajectory previously measured relative to the ancestor (Figure 1). The main features of the two models are shown in Figure 2 and described below. Some further details are provided in Additional file 1. For both models, let A, B, C ... represent a sequence of bacteria sampled at equal temporal intervals, starting with the ancestor, and define W(j:i) as the fitness of j relative to i.
Figure 2.

Transitive and non-transitive models. Both models could, in principle, account for the declining rate of fitness improvement relative to the ancestor. In competition with the ancestor, both models yield identical results, and were parameterized to match the data in Figure 1 (see Additional file 1). However, the two models make different predictions about fitness measured relative to clones isolated from later generations. (A) Transitive model. (B) Transitive model, ln-transformed. (C) Non-transitive model. (D) Non-transitive model, ln-transformed.
According to the transitive model, the fitness of two samples relative to one another can be calculated from the fitness of each sample relative to their common ancestor: for example, W(C:B) = W(C:A) / W(B:A). If the same samples were all to compete against a more fit clone isolated from later in the experiment (instead of against the ancestor), then their relative fitness values should shift down by a constant proportion, as shown in Figure 2A. When these same values are ln-transformed, the trajectories against a set of progressively more and more fit competitors form a series of parallel lines (Figure 2B). Also, the fitness trajectories against a set of competitors isolated at equal intervals from the evolution experiment should be more closely packed against the later competitors, which are progressively more similar in their fitness relative to one another.
In general, non-transitive competition models can take many different forms. All such models will have the property that ln-transformed fitness trajectories measured against a series of competitors isolated from different times deviate from parallelism. More specifically, we are interested in the possibility that non-transitive interactions might produce the observed deceleration in fitness relative to the ancestor, yet be consistent with a constant rate of on-going adaptive change. Figure 2C shows the non-transitive model that uniquely fulfils both these conditions. A ln-transformation of this model does not produce parallel fitness trajectories, nor do these trajectories become more closely packed when competitions are performed against later competitors (Figure 2D). The transformation does show, however, that all of the trajectories have equal and maximum slopes, which correspond to samples that are temporally adjacent.
Perhaps the most compelling way to capture the difference between the two models is by contrasting their cumulative fitness gains: W(B:A) × W(C:B) × ... Figure 3 shows that the ln-transformed cumulative gains for the non-transitive model follow a straight line, indicating no deceleration in the rate of adaptation, despite the deceleration when fitness is measured against the ancestor. By contrast, the ln-transformed cumulative gains for the transitive model follow precisely the same decelerating trajectory as ln-fitness measured against the ancestral type.
Figure 3.

Cumulative fitness gains under two models. The two models depicted in Figure 2 differ most strikingly in their cumulative fitness gains, here shown ln-transformed. The transitive model shows cumulative gains that are identical to fitness measured relative to the common ancestor. The non-transitive model predicts cumulative gains that increase indefinitely at a constant rate.
Fitness trajectories measured against five different competitors
To distinguish between the transitive and non-transitive models, we performed 315 relative fitness assays, all involving population samples and clones from the Ara-1 line of the long-term experiment. We use the term "population sample" to refer to the heterogeneous mixture of clones that existed in the population when the sample was obtained, whereas a "clone" refers to a pure culture that was derived from a single randomly chosen cell. Twenty-one evenly spaced samples (generations 0, 1,000, 2,000, ..., 20,000) were each tested in competition against five evenly spaced clones (from generations 0, 5,000, 10,000, 15,000 and 20,000), all bearing a neutral genetic marker, with three-fold replication. Figure 4A shows all the data as five trajectories for relative fitness, and Figure 4B shows the same data ln-transformed.
Figure 4.

Fitness trajectory measured relative to clones from five time-points. (A) Each line shows the fitness trajectory of the Ara-1 population measured relative to one of five clones bearing a neutral marker. The clones are the ancestor and isolates from generations 5,000, 10,000, 15,000 and 20,000. Each competition was replicated three-fold, and error bars are standard errors. (B) The same data ln-transformed.
Several important features of these data are immediately apparent. First, the fitness trajectories against all five competitors decelerate over time, confirming the findings reported previously based on competitions against the common ancestor only [1-3]. Second, the five ln-transformed fitness trajectories are nearly parallel to one another. Third, the trajectories are much more closely packed against the clonal competitors from later generations. All these features are consistent with the transitive model, but not with the non-transitive model. To formalize this conclusion, we perform two rigorous statistical tests in the next section.
Statistical tests support transitive model, reject non-transitive model
Our first test is a two-way analysis of variance (ANOVA) using the ln-transformed fitness data. Here again, "sample" refers to the 21 different population samples and "clone" indicates the five different clones used as competitors against each population sample. For the purposes of the ANOVA, both variables are categorical. The sample-by-clone interaction includes any non-parallelism, and thus the ANOVA tests all forms of non-transitivity (not only the particular form that was developed earlier and that especially interests us). Table 1 presents the full ANOVA. Both of the main effects, sample and clone, are highly significant (both P < 0.0001). Together they explain > 93% of the variation in the ln-transformed fitness data. By contrast, their interaction does not even approach statistical significance (P > 0.5). The interaction term accounts for < 2% of the overall variation, while the residual error accounts for about 5%. One cannot invoke insufficient statistical power as being responsible for the non-significant interaction, because the interaction term has more degrees of freedom than the two main effects, which are both highly significant. Thus, the ANOVA provides no evidence for any non-transitive fitness effects among the 21 population samples and 5 marked clones tested in all pairwise combinations from this population. Because this analysis might appear superficially to be dominated by the initial period of rapid fitness change, we also performed an ANOVA that excluded fitness data involving all samples and clones from before 5,000 generations. The overall conclusions were not substantively affected: the main effects of sample and clone were both still highly significant (P < 0.0001), and their interaction remained non-significant (P > 0.5).
Table 1.
ANOVA testing for non-transitive competitive interactions. Sample refers to the 21 population samples taken from generations 0, 1,000, 2,000, ..., and 20,000. Clone refers to the 5 neutrally marked clones from generations 0, 5,000, 10,000, 15,000, and 20,000. Each sample competed against each clone with 3-fold replication. The sample-by-clone interaction is used to test for any deviations from the transitive model.
| Source | DF | SS | MS | F | P |
| Sample | 20 | 5.095 | 0.2548 | 49.50 | < 0.0001 |
| Clone | 4 | 14.37 | 3.593 | 698.1 | < 0.0001 |
| Interaction | 80 | 0.3290 | 0.004112 | 0.7990 | 0.8763 |
| Residual | 210 | 1.081 | 0.005147 | --- | --- |
Our second test addresses the pronounced difference in cumulative fitness gains that is predicted (Figure 3) under the particular non-transitive model in which the rate of adaptation has not slowed, and the transitive model where the measured deceleration in fitness against the ancestor also indicates a slowing pace of adaptation. Using ln-transformed fitness gains, Figure 5 shows that the non-transitive model is decisively rejected, whereas this analysis is again consistent with transitivity. Bar A shows the mean ln-transformed fitness of the 20,000-generation sample relative to the ancestor. This value, although it was itself measured, represents precisely the cumulative gain predicted by the transitive model. Bar B shows the cumulative gain measured from the competitions of the 5,000-generation population sample against the ancestor, the 10,000-generation sample with the 5,000-generation clone, the 15,000-generation sample with the 10,000-generation clone, and the 20,000-generation sample against the 15,000-generation clone, along with corresponding values obtained by competing earlier samples against later clones. Notice that the 95% confidence intervals for both A and B reciprocally overlap the A and B means, therefore indicating that the prediction of the transitive model is consistent with the independently estimated cumulative gains. Bar C shows the fitness gain measured over the first 5,000 generations extrapolated to the full 20,000 generations, which corresponds to the prediction of this particular form of non-transitive model. The 95% confidence intervals for B and C do not come close to overlapping one another, which shows that this non-transitive model prediction is strongly rejected by the observed cumulative gains.
Figure 5.
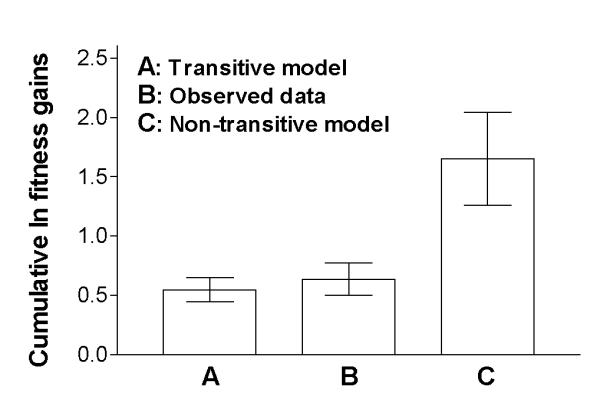
Cumulative fitness gains measured and compared with two models. Test of the two model predictions shown in Figure 3 using cumulative ln-transformed fitness gains, as explained in the text. Error bars are 95% confidence intervals. A: Cumulative gains predicted by the transitive model. B: Cumulative gains observed over the four 5,000-generation intervals. C: Cumulative gains predicted by the non-transitive model. Both the A and B confidence intervals reciprocally overlap the A and B means, indicating that the prediction of the transitive model is consistent with the independently measured cumulative gains. By contrast, neither the B nor C confidence interval includes the reciprocal mean, and thus the non-transitive model is strongly rejected by the data.
Conclusions
We tested two hypotheses for the observed deceleration of fitness improvement in an experimental population of evolving E. coli. One hypothesis assumes competitive interactions are transitive, and that the deceleration indicates an actual decline in the rate of adaptation – owing to fewer beneficial mutations, mutations having smaller benefits, or both – which is expected when a population approaches a fitness peak or plateau. The other hypothesis assumes that the instantaneous rate of adaptation is as fast after 20,000 generations as was the initial rate, and that the observed deceleration is an artifact resulting from non-transitive interactions. By measuring the fitness of samples obtained from the population over the course of 20,000 generations relative to five clones from different time points, we found absolutely no evidence for non-transitive interactions using two statistical tests. First, an analysis of variance showed that the fitness trajectories relative to clones from different time points were almost perfectly parallel, as predicted by the transitive model, whereas the non-transitive model predicts strongly non-parallel trajectories. The interaction term in this analysis tested for any possible deviation from transitivity, and not only the particular form that could have accounted for the deceleration in fitness gains measured relative to the ancestor. Second, the cumulative fitness increase after 20,000 generations, relative to the ancestor, was consistent with the incremental gains measured across a series of temporal intervals, as predicted by the transitive model but significantly contrary to the non-transitive model.
Our purpose in presenting these findings is not to argue that non-transitivity is either uninteresting or unimportant in some other systems. As reviewed in the Background, non-transitive interactions have been well documented in several systems. Also, the features of the environment used in the E. coli long-term experiment (including fairly low glucose concentration and the absence of physical structure) were chosen, in part, to minimize this potential complication, in order to focus on the most fundamental dynamics of evolution. We do not even claim that all forms of frequency-dependent interaction (of which non-transitivity is only one type) are completely absent from the long-term evolution experiment with E. coli. In fact, it has been shown elsewhere that another population than the one we have studied here has a balanced polymorphism that is maintained by a strong frequency-dependent interaction [12], and other populations contain clones with much weaker advantages when rare [13].
What we do assert, based on our new data and analyses, is that non-transitive effects are either absent or much too weak to account for the pronounced overall deceleration in the rate of fitness gains measured relative to the ancestor. Consequently, this deceleration indicates that the pace of genetic adaptation has indeed slowed, although it has not completely stopped [3], in the long-term E. coli evolution experiment. Therefore, this and the other evolving populations must have fairly quickly incorporated mutations that provided the greatest gains, and their slower subsequent adaptation reflects beneficial mutations that are fewer in number, smaller in effect, or both [1,14].
Methods
Bacteria and culture conditions
The bacteria used in this study were all derived from a single population, designated Ara-1, that is part of the long-term evolution experiment [1-3]. Details concerning the founding strain of E. coli B, culture conditions, and so on are given elsewhere [1]. In brief, this population was propagated serially by 1:100 daily transfers for 20,000 generations (= 3,000 days) in Davis minimal (DM) salts medium supplemented with glucose at 25 μg/mL and incubated at 37°C. In this study, we use population samples from 1,000-generation intervals, which were stored at -80°C with glycerol as a cryo-protectant. Each sample contains millions of cells and includes essentially all of the genetic diversity present in the population at the time of storage.
The Ara-1 population and its ancestor are unable to grow on the sugar L-arabinose. However, an Ara+ variant of the ancestor has been generated, and it has previously served as the common competitor for samples from the evolving Ara-1 population. Numerous experiments have shown that this Ara+ marker is selectively neutral under the culture conditions used in the long-term evolution experiment. However, this marker allows evolved and ancestral competitors to be distinguished and enumerated by plating on tetrazolium-arabinose (TA) indicator agar [1].
For this study, we needed Ara+ clones derived from the Ara-1 population at generations 5,000, 10,000, 15,000 and 20,000 in addition to the ancestral Ara+ clone. We used an Ara+ mutant clone that was isolated previously from a clone sampled at generation 10,000 from Ara-1 [15]. For generations 5,000, 15,000 and 20,000 we randomly chose single clones from each of the corresponding samples taken from Ara-1. Each clone was grown to high density, concentrated by centrifugation, and plated on minimal-arabinose agar medium [16] to find spontaneous Ara+ mutants. Three Ara+ mutants were isolated from each clone, and each mutant was assayed in preliminary competitions against the population sample from which it came. We retained for this study the single Ara+ mutant whose estimated fitness relative to its source sample was closest to unity (and, in all cases, not significantly different from 1). To summarize, this study uses 21 population samples of Ara- cells taken at 0, 1,000, 2,000, ..., 20,000 generations from the evolving Ara-1 population, and 5 neutrally marked Ara+ mutant clones derived from generations 0, 5,000, 10,000, 15,000 and 20,000 of the same population.
Measuring relative fitness
We performed a total of 315 competitions to measure the fitness of each population sample relative to each marked clone, with three fold-replication. The competitions followed the same protocol described in detail elsewhere [1]. To summarize briefly, the competitions were performed under the exact same culture conditions as in the long-term evolution experiment. To ensure that any two competitors were comparably acclimated to the competition environment, they were simultaneously removed from the freezer, grown separately in a nutrient-rich broth for one day, and then acclimated for another day to the competition environment. The competitors were then mixed at a 1:1 volumetric ratio, diluted 1:100 into fresh DM and incubated at 37°C for one day. Appropriate dilutions were plated on TA agar at the start of the competition and again after 24 h, to estimate the initial and final numbers of each competitor. In a few cases, plate counts were less than 50 total or less than 15 for one competitor. In these cases, we repeated the competition experiments to obtain more accurate estimates of relative fitness based on higher counts. The relative fitness of two competitors was calculated simply as the ratio of their realized population growth rates during competition [1]. Let N(0) and N(1) denote initial and final population densities, respectively, and let subscripts i and j indicate the two competitors. Then relative fitness is calculated as:
W(j:i) = ln [Nj(1) / Nj(0)] / ln [Ni(1) / Ni(0)].
Relative fitness is dimensionless because the same time units are used in calculating both competitors' realized population growth rates.
Statistical methods
Standard statistical analyses were used throughout, with one exception. The jackknife method [[17], pp 795–799] was used to calculate the confidence interval around the mean value in Figure 5B. In particular, the mean cumulative gain observed over four 5,000-generation intervals was calculated as the product of four successive means, each based on six measured values (three from a later sample competed against an earlier clone, and three from a later clone competed against an earlier sample). Owing to the non-linear dependence of a multiplicative product of means on the measured values, one cannot calculate a confidence interval using standard methods. The jackknife method yields a confidence interval that accurately reflects variation in the 24 underlying measurements. The jackknife method was not needed to compute the confidence intervals around the means in Figure 5A and 5C, because both of these means are simple arithmetic averages and hence do not involve any non-linearities.
Authors' contributions
JAGMdV performed the particular experiments reported here, while REL developed the long-term evolution experiment. Both authors contributed substantially to the experimental design, statistical analyses, and writing this paper. Both authors read and approved the final manuscript.
Supplementary Material
Fitness values and defining features of the transitive and non-transitive models The values correspond to the trajectories shown in Figure 2. The values are colour-coded to show how the two models were constructed from previous data.
Acknowledgments
Acknowledgements
The authors thank T. Czárán and R. Hoekstra for helpful discussions and two anonymous reviewers for suggestions that improved the manuscript. JAGMdV's research is supported by the Netherlands Organization for Scientific Research. REL's long-term evolution experiment is funded by the United States National Science Foundation (DEB-9981397).
Contributor Information
J Arjan GM de Visser, Email: arjan.devisser@wur.nl.
Richard E Lenski, Email: lenski@msu.edu.
References
- Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat. 1991;138:1315–1341. doi: 10.1086/285289. [DOI] [Google Scholar]
- Lenski RE, Travisano M. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc Natl Acad Sci USA. 1994;91:6808–6814. doi: 10.1073/pnas.91.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper VS, Lenski RE. The population genetics of ecological specialization in evolving E. coli populations. Nature. 2000;407:736–739. doi: 10.1038/35037572. [DOI] [PubMed] [Google Scholar]
- Paquin CE, Adams J. Relative fitness can decrease in evolving asexual populations of S. cerevisiae. Nature. 1983;306:368–371. doi: 10.1038/306368a0. [DOI] [PubMed] [Google Scholar]
- Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J. Evolution and the Theory of Games. Cambridge University Press. 1982.
- Jackson JBC, Buss L. Allelopathy and spatial competition among coral reef invertebrates. Proc Natl Acad Sci USA. 1975;72:5160–5163. doi: 10.1073/pnas.72.12.5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czárán T, Hoekstra RF, Pagie L. Chemical warfare between microbes romotes biodiversity. Proc Natl Acad Sci USA. 2002;99:786–790. doi: 10.1073/pnas.012399899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr B, Riley M, Feldman M, Bohannan BJM. Local dispersal promotes biodiversity in a real life game of rock-paper-scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- Sinervo B, Lively CM. The rock-paper-scissors game and the evolution of alternative male strategies. Nature. 1996;380:240–243. doi: 10.1038/380240a0. [DOI] [Google Scholar]
- Huisman J, Weissing FJ. Biodiversity of plankton by species oscillations and chaos. Nature. 1999;402:407–410. doi: 10.1038/46540. [DOI] [Google Scholar]
- Rozen DE, Lenski RE. Long-term experimental evolution in Escherichia coli. VIII. Dynamics of a balanced polymorphism. Am Nat. 2000;155:24–35. doi: 10.1086/303299. [DOI] [PubMed] [Google Scholar]
- Elena SF, Lenski RE. Long-term experimental evolution in Escherichia coli. VII. Mechanisms maintaining genetic variability within populations. Evolution. 1997;51:1058–1067. doi: 10.1111/j.1558-5646.1997.tb03953.x. [DOI] [PubMed] [Google Scholar]
- Gerrish PJ, Lenski RE. The fate of competing beneficial mutations in an asexual population. Genetica. 1998;102/103:127–144. doi: 10.1023/A:1017067816551. [DOI] [PubMed] [Google Scholar]
- Elena SF, Ekunwe L, Hajela N, Oden SA, Lenski RE. Distribution of fitness effects caused by random insertion mutations in Escherichia coli. Genetica. 1998;102/103:349–358. doi: 10.1023/A:1017031008316. [DOI] [PubMed] [Google Scholar]
- Lenski RE. Experimental studies of pleiotropy and epistasis in Escherichia coli. I. Variation in competitive fitness among mutants resistant to virus T4. Evolution. 1988;42:425–432. doi: 10.1111/j.1558-5646.1988.tb04149.x. [DOI] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry. New York, Freeman. 1981.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fitness values and defining features of the transitive and non-transitive models The values correspond to the trajectories shown in Figure 2. The values are colour-coded to show how the two models were constructed from previous data.


