Abstract
We investigated the binding regions of components of the origin recognition complex (ORC) in the human genome. For this purpose, we performed chromatin immunoprecipitation assays with antibodies against human Orc1 and Orc2 proteins. We identified a binding region for human Orc proteins 1 and 2 in a <1-kbp segment between two divergently transcribed human genes. The region is characterized by CpG tracts and a central sequence rich in AT base pairs. Both, Orc1 and Orc2 proteins are found at the intergenic region in the G1 phase, but S-phase chromatin contains only Orc2 protein, supporting the notion that Orc1p dissociates from its binding site in the S phase. Sequences corresponding to the intergenic region are highly abundant in a fraction of nascent DNA strands, strongly suggesting that this region not only harbors the binding sites for Orc1 protein and Orc2 protein but also serves as an origin of bidirectional DNA replication.
The origin recognition complex (ORC) was first identified in Saccharomyces cerevisiae as a multiprotein factor that binds to yeast origins of replication in an ATP-dependent manner (4). ORC is composed of six protein subunits, Orc1p to Orc6p, encoded by essential yeast genes whose deletions result in lethality. Components of ORC interact with the Cdc6 protein as an early step in the assembly of prereplicative complexes that are subsequently completed by the loading of Mcm protein hexamers (3, 18, 43, 64).
The binding sites for ORC in yeast DNA are known as ARS (for autonomously replicating sequences) because they direct the extrachromosomal replication of ARS-bearing plasmids. Chromosomal ARS elements serve as origins of bidirectional DNA replication in yeast and consist of about 150 bp of DNA with an essential 11-bp AT-rich ARS consensus sequence A and two or three short stimulatory sequences, B1 to B3, which are functionally important but divergent in sequence (36, 40). The ORC binding site in yeast origins is a bipartite DNA sequence that includes the consensus A element and the adjacent B1 element (4, 47, 54). The B3 element is the binding site for a transcription factor, Abf1 (19). Protein-DNA cross-linking studies show that subunits Orc1p, Orc2p, Orc4p, and Orc5p contact a DNA strand in the major groove of the ARS binding site and may determine the specificity of the reaction, whereas subunits Orc3p and Orc6p appear to form protein-protein contacts (33). Bidirectional DNA replication is initiated in the immediate neighborhood of the ARS B1 element (8), suggesting that DNA-bound ORC determines the start points for DNA replication in yeast.
Proteins homologous to the yeast ORC, as well as to other proteins required for prereplicative complex formation, have been detected in all eukaryotes examined, suggesting that the manner of prereplicative complex formation may be highly conserved. Experiments with Xenopus egg extracts have clearly shown that chromatin-bound ORC serves as a landing pad for the subsequent binding of Cdc6 which, together with the Cdt1 protein, is required for the recruitment of Mcm proteins in a chain of reactions that leads to replication-competent chromatin (9, 10, 34, 35, 51, 52). The order of events appears to be similar in mammalian cells since studies with synchronously proliferating cultured mammalian cells show a stepwise binding of the Cdc6 protein and Mcm proteins to chromatin-bound ORC during the G1 phase of the cell cycle (28, 30, 38, 44, 55). In fact, human homologues of all six ORC subunits have been identified by sequence similarity searches (17, 21, 45, 46, 62) and shown to form specific complexes when extracted from chromatin of HeLa cells or when simultaneously expressed in baculovirus-infected insect cells (16, 63). When prepared from HeLa chromatin, ORC complexes appear to be composed of a core of proteins Orc2p, Orc3p, Orc4p, and Orc5p which together associate with Orc1p in some but not in all cases (63). Protein hOrc6 is absent from endogenous ORC but binds to the ORC core when Orc proteins are expressed in the baculovirus system (16, 63). However, it is not known whether mammalian ORC proteins, like their yeast counterparts, assemble at specific DNA sites and whether these sites coincide with origins of bidirectional DNA replication.
Much recent research has focused on the identification of replication origins in mammalian systems (7, 15, 24, 58). Since ARS assays in cultured mammalian cells are inefficient and frequently quite inconclusive, different mapping techniques have been developed (14), including the isolation and analysis of newly synthesized (nascent) DNA strands. These techniques have led to the identification of a number of mammalian origins. A major conclusion of these experiments is that mammalian origins are extremely heterogeneous in size and sequence, and it is therefore quite possible that replication start sites in mammalian genomes are determined rather more by features of chromatin organization than by base pair sequence or composition. Well-mapped mammalian origins are frequently located in the vicinity of actively transcribed genes. These areas are defined by a more open, decondensed chromatin conformation, allowing origin binding proteins to associate with their target sites and to establish replication forks. For example, the first human replication origin that has been characterized at the single nucleotide level is located within a relatively short region downstream of the lamin B2 gene (LMNB2) and upstream of the closely adjacent PPV1 gene (1, 23). Furthermore, a random collection of nascent DNA strands from human and hamster cells was found to be highly enriched for sequences corresponding to CpG islands, important regulatory elements in the vicinity of ca. 50% of all mammalian genes (13), and an assessment of putative replication origins in a 500 kb-region of the human X chromosome suggests that the usage of replication initiation sites depends on the transcription of adjacent genes (50).
In order to identify ORC binding sites in the human genome, we used a modified chromatin immunoprecipitation (CHIP) procedure (25) that allows a high purification of specific in vivo cross-linked nucleoprotein fragments under carefully controlled conditions. The DNA components were recovered from nucleoprotein precipitated by antibodies against the human ORC proteins, hOrc1p and hOrc2p (referred to here as hOrc1p and hOrc2p antibodies). We describe below how hOrc1p and hOrc2p are cross-linked to a specific DNA region between the divergently transcribed human genes MCM4 and PRKDC (11, 31). We propose that this region contains the in vivo binding sites for human ORC. Thus, components of ORC can be assigned to a specific region of the human genome. In addition, we also show that the same MCM4 upstream promoter sequence is highly abundant among nascent DNA strands, suggesting that the hOrc1p and hOrc2p binding region serves as a replication origin.
MATERIALS AND METHODS
Cells.
Human HeLa S3 cells were grown to semiconfluency on plastic dishes in Dulbecco modified Eagle medium plus 5% fetal calf serum. For S-phase synchronization, cells were first subjected to a double-thymidine block (29, 49) and then released for 4 h. G1-phase cells were prepared in the following manner: they were first arrested by a single thymidine block, released for 9 h, and then arrested again by a short treatment (2 h) with nocodazole. Mitotic cells were collected and, after removal of nocodazole, released for 4 h into G1 phase.
In vivo cross-linking.
Formaldehyde was diluted to 1% in prewarmed medium (37°C) and added to monolayers of ca. 108 cells for 4 min if not stated otherwise (25). After removal of the medium, cells were washed three times on plates with cold phosphate-buffered saline (PBS) (20), scraped off, washed again twice in cold PBS, and then resuspended in hypotonic RSB buffer (10 mM Tris-HCl, 3 mM MgCl2; pH 8.0). All buffers contained 10 mM sodium bisulfite as a protease inhibitor.
After 10 min on ice, the swollen cells were disrupted by Dounce homogenization. Nuclear material was collected and washed twice in RSB buffer and once in high-salt NSB buffer (1 M NaCl, 10 mM Tris-HCl, 0.1% NP-40, 1 mM EDTA; pH 8.0). Nuclear material was then resuspended in low-salt NSB buffer (0.1 M NaCl) and loaded onto a step gradient made up of 1.75 g of CsCl/ml, 1.5 g of CsCl/ml, and 1.3 g of CsCl/ml in 20 mM Tris-HCl-0.5% sarcosyl-1 mM EDTA (pH 8). Nucleoprotein complexes were collected after centrifugation (37,000 rpm, 24 h, 18°C) and dialyzed overnight against TE (10 mM Tris-HCl, 1 mM EDTA; pH 7.4) containing 10 mM sodium bisulfite. Nucleoprotein was then briefly sonicated on ice and digested with micrococcal nuclease in TE with 3 mM CaCl2 (1 U of micrococcal nuclease/100μg of nucleoprotein for 15 min at 37°C).
CHIP.
Affinity-purified antibodies against the human ORC proteins, hOrc1p and hOrc2p (30), and against Mcm proteins (49) have been described. The control antibodies were nonspecific rabbit immunoglobulin G (IgG) antibodies (Sigma).
Immunoprecipitations were performed in NET buffer (50 mM Tris, pH 7.4; 150 mM NaCl; 5 mM EDTA; 0.5% NP-40). Nuclease-digested nucleoprotein was centrifuged for 10 min at 15,000 × g for clarification. The supernatants (1 mg of nucleoprotein) were incubated with antibodies (15 μg of hOrc1p antibodies, 10 μg of hOrc2p antibodies, 10 μg of hMcm3p antibodies, or 15 μg of control IgG) for 2 h at 20°C. Then, 50 μl of protein A-Sepharose (Amersham Pharmacia Biotech) was added for an additional 2 h. Immunocomplexes were washed eight times with RIPA (50 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 0.5% sodium desoxycholate, 0.1% sodium dodecyl sulfate [SDS]; pH 8), three times in Lithium Buffer (10 mM Tris-HCl, 250 mM LiCl2, 0.5% NP-40, 0.5% sodium desoxycholate, 1 mM EDTA; pH 8), and five times in TE buffer. The washed precipitates were divided for Western blotting and DNA extraction. For Western blot analyses, proteins were eluted from cross-linked chromatin and processed as previously described (49). For DNA extraction, the immunoprecipitates were first washed again as described above to maximize the signal-to-noise ratio. The final pellet was then resuspended in TE with 1% SDS and incubated overnight at 37°C with 200 μg of proteinase K/ml. The DNA was purified by standard phenol-chloroform extraction and ethanol precipitation. The precipitated DNA (usually between 5 and 10 ng) was disolved in 40 μl of TE. One-fifth of the sample was used for conventional PCR assays, and one-twentieth was used for real-time quantitative PCR in the Light Cycler instrument (Roche Diagnostics).
Isolation of nascent DNA strands.
Nascent DNA strands were prepared from 108 exponentially growing HeLa S3 cells essentially as described by Giacca et al. (22). Total genomic DNA was prepared from isolated nuclei and denatured in TE buffer at 85°C for 10 min, followed by rapid cooling on ice. The denatured DNA was loaded on a 5 to 30% neutral sucrose gradient (in TE plus 0.1 M NaCl) and centrifuged in a Beckman SW28 rotor for 20 h at 26,000 rpm. DNA size markers (1-kb ladder; MBI Fermentas) were centrifuged in a parallel tube. The distribution of DNA size markers were determined by agarose gel electrophoresis of samples from the gradient fractions. DNA fragments corresponding to an average of 1 kb were collected from the gradient with denatured genomic DNA and concentrated by ethanol precipitation. The quality of the DNA strands was tested by PCR with specific primers for the origin region of the human lamin B2 locus, as well as for adjacent control regions (23) (see below).
PCR analyses.
Conventional PCR assays were performed in 50-μl volumes containing 25 pmol of the respective primers (Table 1) and 0.2 mM concentrations of nucleoside triphosphates with 2.5 U of RedTaq polymerase (Sigma) and total genomic HeLa cell DNA (200 ng) or DNA from the immunoprecipitates (ca. 0.5 ng) as the templates. The template DNA was denatured at 90°C before PCR. Thirty PCR cycles were performed; each cycle consisted of denaturation for 1 min at 90°C, annealing for 1 min (see Table 1), and extension for 1 min at 72°C. Light Cycler PCR reactions were performed exactly as suggested in the manufacturer's manual (FastStart DNA Master SYBR Green I; Roche Diagnostics). Annealing temperatures for each primer pair are given in Table 1.
TABLE 1.
Sequences and amplification conditions for primers
| GenBank accession no. and primer | Sequence (5′ to 3′) | Map positions (bp) | Length (bp) | Annealing temp (°C)
|
|
|---|---|---|---|---|---|
| Standard PCR | Quantitative PCR | ||||
| U63630a | |||||
| EX9-F | ATGTCTTCCGGAGACTCCTGAAGC | 6342-6365 | |||
| EX9-R | GGCCTCCTATTCTCAGAATCATGC | 6705-6728 | 387 | 52 | 57 |
| EX7-F | TAATCCGTCACCTTGACTACCACC | 8901-8924 | |||
| EX7-R | ACAGCACGTGCATGATTCTGTAGG | 9277-9300 | 400 | 66 | |
| EX6-F | TACCTGTGGGTAAGAGATGAGTTG | 10691-10704 | |||
| EX6-R | TGCCTGTTCCCAAATGCTATATGC | 10998-11021 | 331 | 65 | |
| EX2-F | TCTGCACTCCGTTCAGCTCCTCTG | 11894-11917 | |||
| EX2-R | GAGTGAGGATGCCAGGTCATCTCC | 12191-12214 | 321 | 68 | |
| UPR-F | AAACCAGAAGTAGGCCTCGCTCGG | 12946-12969 | |||
| UPR-R1 | GGCCAGTAAGCGCGCCTCTTTGG | 13460-13482 | 537 | 62 | |
| UPR-R2 | GTCTGACCTGCGGAGGTAGTTTGG | 13405-13428 | 483 | 66 | |
| IN1-F | ATCTCGCCTAATCCCACCAGTACC | 14364-14387 | |||
| IN2-R | CATATTCACTACTAGACCCTCCGG | 14633-14656 | 293 | 62 | |
| IN6-F | GACATTCTGCTTCCATAGATGTGG | 19943-19966 | |||
| IN6-R | GTTGGGAAAGATGTCATCATCAGG | 20265-20288 | 346 | 55 | |
| IN7-F | GAGGAATGCCAGAATTTCCAGAGG | 26412-26435 | |||
| IN7-R | TTCCATCTGGAATGAGATCCCAGC | 26118-26741 | 327 | 58 | |
| M94363b | |||||
| LB2C1-F | GTTAACAGTCAGGCGCATGGGCC | 1-23 | |||
| LB2C1-R | CCATCAGGGTCACCTCTGGTTCC | 217-240 | 240 | 64 | |
| LB2-F | GGCTGGCATGGACTTTCATTTCAG | 3839-3862 | |||
| LB2-R | GTGGAGGGATCTTTCTTAGACATC | 4047-4070 | 232 | 66 | |
| LB2C2-F | CACAGCATGCGGCTGCTGATCTG | 6648-6670 | |||
| LB2C2-R | CCTGGTGCGTCCCATCTGCCTGC | 6910-6932 | 285 | 68 | |
RESULTS
In vivo cross-linking of ORC proteins.
To identify binding sites for ORC proteins, we covalently linked proteins to their target DNA in vivo by a formaldehyde-mediated cross-linking procedure. This technique is widely used in eukaryotic biochemistry to assess the interaction of proteins with DNA (12, 42). The procedure requires a brief treatment of proliferating cells with formaldehyde and the purification of cross-linked nucleoprotein complexes by repeated wash steps and CsCl equilibrium centrifugation to separate nucleoprotein from free protein and other material that is not covalently bound to DNA (see Materials and Methods).
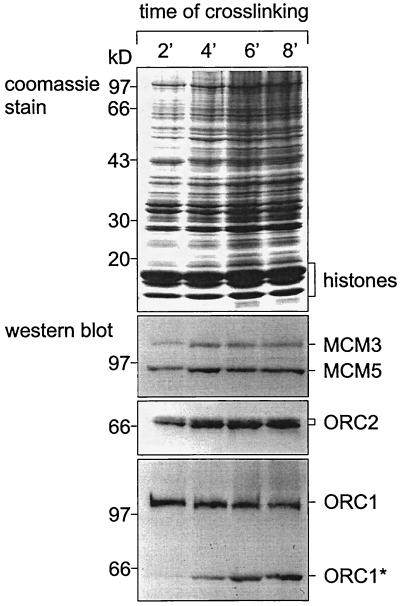
In the experiment shown in Fig. 1,we recovered cross-linked nucleoprotein complexes from CsCl equilibrium gradients (at buoyant densities of between 1.3 and 1.5 g/ml) and analyzed them by Western blotting for the presence of human Orc1p (hOrc1p) and hOrc2p. As Coomassie blue staining shows, core histones and a variety of nonhistone proteins were covalently linked to DNA at cross-linking times as short as 2 min, while longer treatment with formaldehyde increased the fraction of associated nonhistone proteins. Among the early cross-linking proteins were human Mcm proteins (49), as well as hOrc1p and hOrc2p. In most experiments, degradation products of hOrc1p were detected at longer cross-linking times (Fig. 1), suggesting either that hOrc1p was degraded as a consequence of formaldehyde treatment or that endogenous partially degraded hOrc1p requires longer exposure to formaldehyde for cross-linking. In either case, the results (Fig. 1) show that 2 to 4 min of exposure to formaldehyde are sufficient to covalently link ORC proteins to DNA.
FIG. 1.
Short treatment with formaldehyde suffices to cross-link Orc proteins to DNA. HeLa cells were treated with formaldehyde for 2 to 8 min as indicated. Nucleoprotein was prepared by repeated wash steps and CsCl purification (Materials and Methods). Aliquots (100 μg of DNA each) were suspended in Laemmli buffer and analyzed by denaturing polyacrylamide gel electrophoresis (32). We show Coomassie blue-stained gels and immunoblots (Western blot) (60) with polyclonal monospecific antibodies against human proteins Mcm3p and Mcm5p (49), as well as hOrc1p and hOrc2p (30) as indicated. The asterisk indicates a degradation product of hOrc1p.
CHIP.
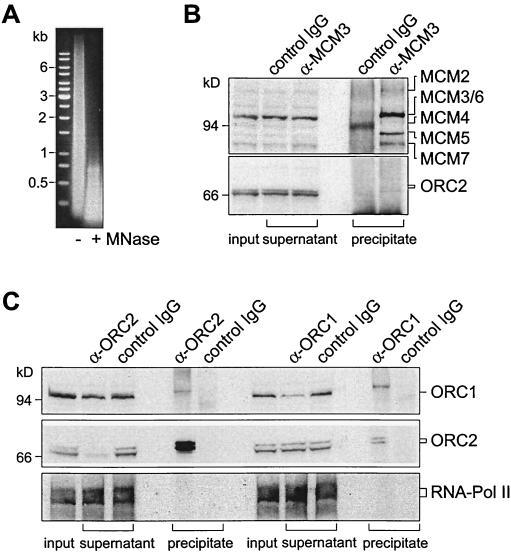
Next, we analyzed whether hOrc1p and hOrc2p were accessible to antibodies after covalent linkage. CsCl gradient-purified nucleoprotein contained a wide spectrum of DNA fragments ranging in lengths from a few hundred to up to ten-thousand base pairs. To obtain chromatin fragments with DNA sizes suitable for further investigations, we digested the original nucleoprotein preparation with micrococcal nuclease (or restriction endonucleases [see below]), reducing the lengths of DNA fragments to smaller than 1 kb (Fig. 2A).As a control, nuclease-treated chromatin was first incubated with Mcm3-specific antibodies in comparison to unspecific antibodies. It has been shown previously that the Mcm3-specific antibodies immunoprecipitate chromatin bearing cross-linked Mcm proteins (49). We show in Fig. 2B that Mcm3-specific antibodies, but not control antibodies, precipitated chromatin fragments that carried the six human Mcm proteins. As described and discussed earlier (49), only a small fraction of hOrc2p coimmunoprecipitated with Mcm proteins (Fig. 2B).
FIG. 2.
Cross-linked chromatin is efficiently immunoprecipitated. (A) DNA in cross-linked nucleoprotein before (−) and after (+) treatment with micrococcal nuclease (MNase). (B) Immunoprecipitation of cross-linked, micrococcal nuclease-treated chromatin with antibodies against hMcm3p (α-MCM3) and with unspecific control antibodies (IgG). Input (chromatin sample before immunoprecipitation) and supernatants (remaining chromatin after treatment with antibodies and protein A-Sepharose) are analyzed in Western blots with α-MCM3. Immunoprecipitates are analyzed with DEFD antibodies that recognize a polypeptide common to all human Mcm proteins (49) and with hOrc2p-specific antibodies. (C) Immunoprecipitation of cross-linked, micrococcal nuclease-treated chromatin with hOrc2p-specific (α-ORC2) and hOrc1p-specific (α-ORC1) antibodies as indicated. The input, supernatant, and precipitated samples were Western blotted and analyzed with hOrc1p and hOrc2p antibodies, as well as with antibodies against a large subunit of RNA polymerase II.
We next used hOrc2p and hOrc1p antibodies for CHIP. As shown in Fig. 2C, hOrc2 antibodies specifically and efficiently precipitated chromatin carrying hOrc2p. The immunoprecipitate also contained some hOrc1p that was apparently linked to the hOrc2p-bearing chromatin fragments. Likewise, hOrc1 antibodies precipitated chromatin carrying bound hOrc1p, as well as bound hOrc2p. However, only a fraction of hOrc1p in cross-linked chromatin appeared to be accessible to antibodies because two- to threefold-higher amounts of hOrc1p antibodies failed to precipitate all hOrc1p-bearing nucleoprotein (not shown). To exclude unspecific protein aggregations, we used antibodies against an RNA polymerase II subunit (Santa Cruz Laboratories) and detected the protein on cross-linked nucleoprotein. However, neither hOrc1p nor hOrc2p antibodies precipitated nucleoprotein with covalently linked RNA polymerase II (Fig. 2C), indicating that the ORC proteins and RNA polymerase II did not occur on the same chromatin fragment. Another control experiment involved the p60 subunit of the chromatin assembly factor CAF-1 (37), which was efficiently cross-linked to DNA after exposure to formaldehyde. Cross-linked nucleoprotein carrying the p60 protein was immunoprecipitated with p60-specific antibodies but not with hOrc2p-specific antibodies (not shown).
As the experiment of Fig. 2C indicates, only minor fractions of hOrc1p and hOrc2p coimmunoprecipitated and were therefore linked to the same chromatin fragments. Rather, hOrc2 antibodies precipitated essentially all chromatin-bound hOrc2p but only a small fraction of chromatin-bound hOrc1p and, conversely, hOrc1 antibodies precipitated a significant fraction of chromatin-bound hOrc1p but only a minor fraction of hOrc2p. Thus, hOrc1p and hOrc2p are frequently linked independently of each other to chromatin. This observation does not exclude the possibility that both hOrc1p and hOrc2p could be components of a multisubunit complex in vivo (63). It could indicate that short formaldehyde treatments stochastically cause the covalent linkage of hOrc1p in some complexes and of hOrc2p in other complexes. Furthermore, hOrc1p may dissociate from chromatin (and may be degraded) during S phase, while hOrc2p remains bound to chromatin during all phases of the cell cycle (30) (see below). Therefore, chromatin preparations from asynchronously proliferating cells are expected to always contain more hOrc2p than hOrc1p.
In conclusion, Fig. 2 shows that our hOrc1p and hOrc2p antibodies are suitable for the immunoprecipitation of chromatin fragments with covalently linked ORC proteins.
DNA in immunoprecipitated nucleoprotein.
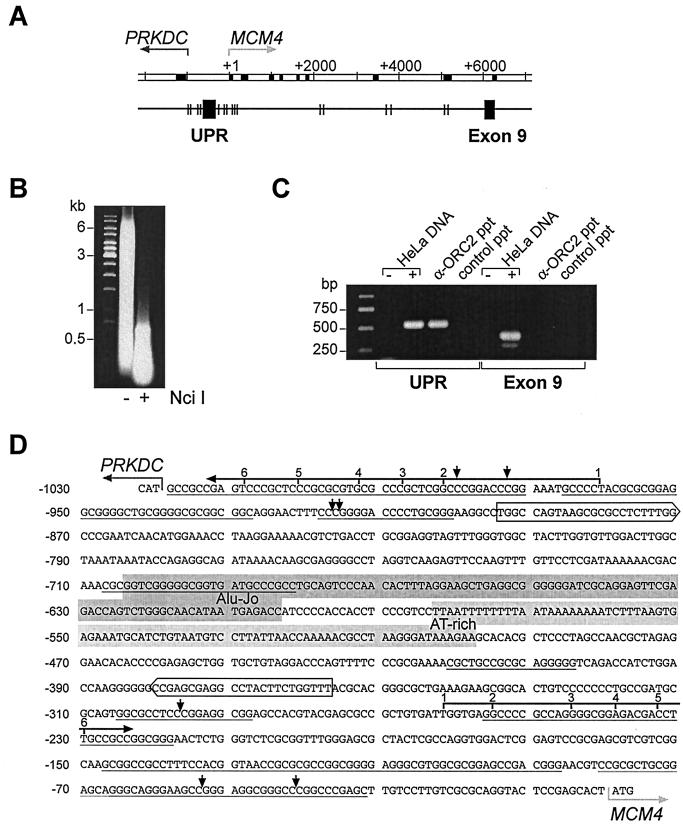
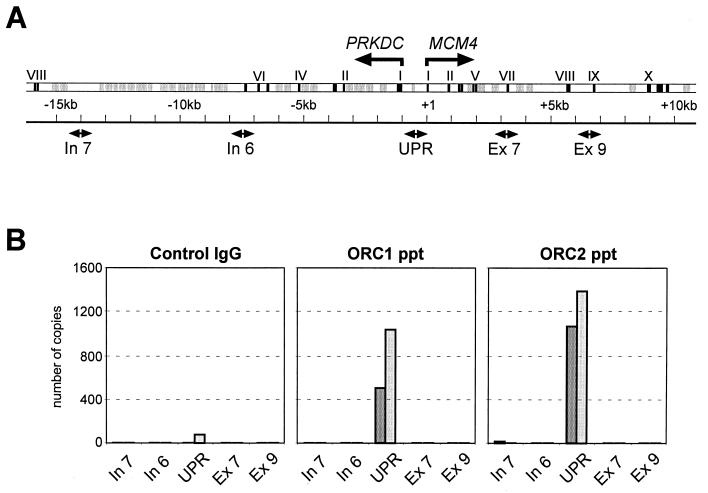
We characterized the DNA sequences which are associated with immunoprecipitated ORC proteins. For this purpose, we cloned and analyzed ca. 20 DNA fragments of 1 to 2 kbp extracted from immunoprecipitated chromatin. We found no apparent sequence similarities except that more than one-half of the fragments had a higher-than-average GC content (>50%) with many CpG dinucleotides, as characteristically described for CpG islands (data not shown). Since such DNA elements frequently occur upstream of many mammalian genes, we looked for well-characterized upstream gene regions in immunoprecipitated chromatin and detected among others (not shown) the genomic region between the divergently transcribed genes MCM4, encoding the human Mcm4 protein, and PRKDC, which encodes the catalytic subunit of the DNA-dependent protein kinase (11, 31) (Fig. 3A).As described in detail below, this intergenic region was enriched in immunoprecipitated chromatin treated with either micrococcal nuclease or the restriction endonuclease NciI (Fig. 3B).
FIG. 3.
hOrc2p is specifically associated with the UPR of gene MCM4. (A) Genomic organization. In the double-line diagram, exons are shown as black boxes, and arrows indicate the starts and directions of transcription. In the single-line diagram, boxes show regions amplified in PCRs, and the vertical lines show NciI restriction sites. (B) PCR assays. UPR, primers complementary to the MCM4 upstream region; Exon 9, primers complementary to exon IX; HeLa DNA, PCR assays with genomic DNA as template (+) or without template DNA (−); α-ORC2 ppt, template DNA extracted from cross-linked chromatin immunoprecipitated with anti hOrc2p antibodies; control ppt, DNA extracted from mock immunoprecipitates with control antibodies. (C) DNA in cross-linked material before (−) and after (+) digestion with NciI. (D) Intergenic region. The translational start codons of the two divergent genes are marked by gray horizontal arrows. Multiple transcriptional start sites (black horizontal arrows with numbers) have been mapped by Conelly et al. (11) for the gene PRKDC and for the gene MCM4. Short vertical arrows indicate NciI restriction sites. Boxed sequences show complementary to PCR primers. Gray backgrounds indicate portions of the Alu-Jo repetitive element and the central AT-rich sequence.
We show in Fig. 3C the results of conventional PCR assays with two sets of oligonucleotide primers; one primer pair corresponded to sequences in the MCM4 upstream promoter region (UPR), and the second pair was complementary to exon IX sequences. The control shows that PCR assays with both primer pairs amplified sections in genomic DNA with equal efficiencies. However, only the upstream region primers, but not the exon IX primers, gave positive PCR signals with template DNA extracted from chromatin that had been immunoprecipitated with hOrc2p antibodies (Fig. 3C).
We conclude that hOrc2p was specifically cross-linked to DNA in the region between the two divergent genes and not to DNA in the exon IX coding region of the MCM4 gene.
Quantitative PCR.
In previous studies, competitive PCR has been used for the quantitative assessment of specific DNA sequences (23). In the present study we used a Light Cycler (Roche Diagnostics) for quantitative real-time PCR, which has the advantages that specifically designed competing DNA sequences are not necessary and that several different amplicons can be accurately quantitated with high sensitivity. The FastStart DNA Master SYBR Green I system allows for the real-time detection of PCR products because amplification products are visualized by the fluorescent dye SYBR Green I that binds to the minor groove in double-stranded DNA. For quantitative analyses, genomic DNA samples of known concentrations were serially diluted and amplified by using the oligonucleotide primers corresponding to the MCM4 upstream region in one assay and the exon VII primers (or other primers, see below) in parallel assays. After the completion of the PCR, the Light Cycler software calculates the copy number in target molecules by plotting the logarithm of fluorescence versus cycle number and setting a baseline x axis. The x-axis crossing point of each standard sample was determined and plotted against the logarithm of concentration to produce a standard curve. Identical PCR assays were performed with the DNA from immunoprecipitated chromatin as a template. The concentration of specific DNA sequences was determined by extrapolation to the standard curve.
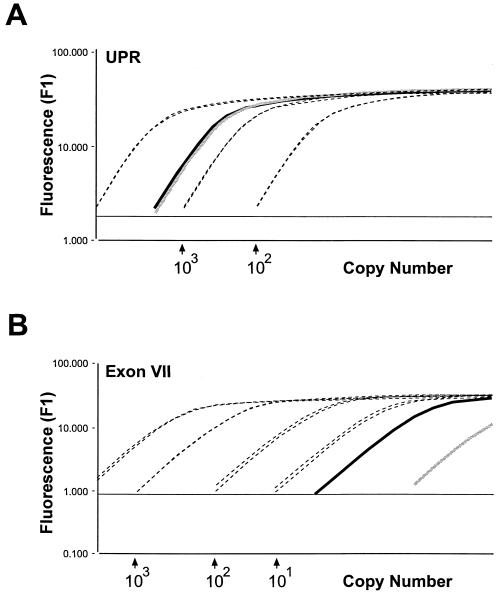
First, we quantitated by real-time PCR ca. 0.1 ng of the DNA from immunoprecipitated chromatin by using the upstream promoter primer set (UPR; Fig. 4A)and an exon VII primer set (Fig. 4B). We detected ca. 2,000 copies of the upstream promoter sequence and <10 copies of the exon VII sequence in the particular DNA sample examined. In a control experiment, the same procedure was used for the determination of DNA sequences prepared from cross-linked chromatin prior to nuclease digestion and immunoprecipitation. In this case, we detected no excess of the upstream promoter region over exon VII sequences (not shown). Thus, immunoprecipitation resulted in a high enrichment of hOrc2p-bearing chromatin fragments.
FIG. 4.
Quantitative PCR. Broken lines, two dilution series of genomic HeLa cell DNA. Dilutions are based on the estimate that 30 ng of DNA correspond to 104 genome equivalents. The x axis as calculated by the Light Cycler software cuts parallel running calibration curves. The y axis corresponds to fluorescence in arbitrary units. Solid lines show template DNA extracted from immunoprecipitated chromatin (gray line, Orc1 precipitated; black line, Orc2 precipitated). (A) PCR primers corresponding to the UPR (see Fig. 3C and Table 1). (B) PCR primers corresponding to MCM4 exon VII (Table 1).
The results described thus far were obtained with DNA prepared from chromatin immunoprecipitated by hOrc2p-specific antibodies. We next performed experiments to determine whether hOrc1p antibodies also result in a specific immunoprecipitation of chromatin containing the MCM4 upstream promoter region. In these PCR experiments, we used the upstream promoter primers as well as primers complementary to several sequences within the adjacent genes. A calibration curve was constructed with serially diluted genomic DNA for each one of the additional primer pairs as explained in Fig. 4.
The results of two independent experiments are shown in Fig. 5and demonstrate that, in spite of some variations between individual experiments, hOrc1p antibodies just like hOrc2p antibodies always preferentially precipitate chromatin containing MCM4 upstream region DNA.
FIG. 5.
hOrc1p- and hOrc2p-bearing MCM4 upstream promoter DNA is enriched in chromatin immunoprecipitates. (A) Overview over the genomic region encompassing genes PRKDC and MCM4. Black boxes, exons; gray boxes, repeat elements; divergent arrow heads, sequences complementary to the PCR primers used (see Table 1). (B) Quantitative real-time PCR with DNA templates extracted from chromatin precipitated with control antibodies and with antibodies against hOrc1p and hOrc2p. We show the results of two independent experiments.
We conclude that hOrc1p and hOrc2p were specifically cross-linked to a region between the two divergently transcribed genes MCM4 and PRKDC.
Variations during S phase.
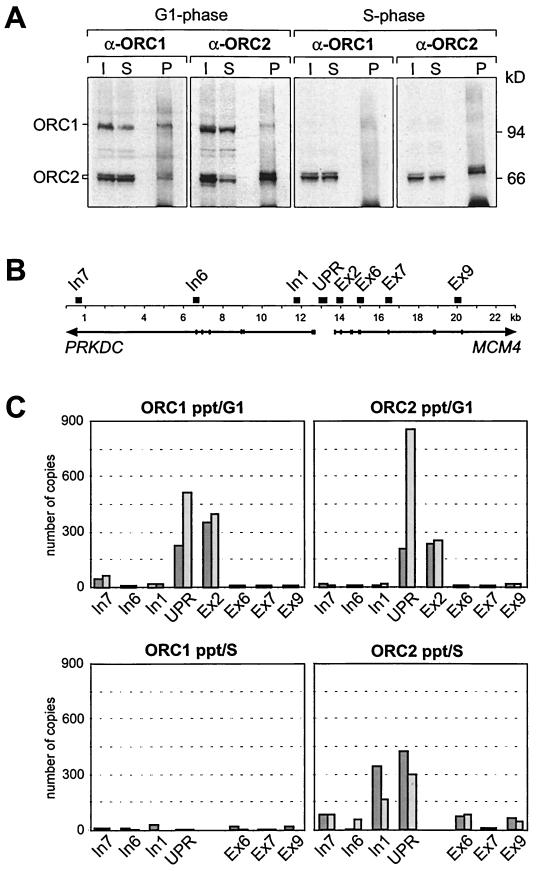
The experiments reported so far were performed with asynchronously proliferating HeLa cells. However, we know from previous work that the composition of ORC may change during S phase because hOrc1p most likely dissociates from chromatin (30). It was therefore of interest to compare chromatin from cells in G1 phase and cells in S phase. Cross-linked chromatin from G1-phase cells and from S-phase cells was prepared and immunoprecipitated with hOrc1p and with hOrc2p antibodies as described above.
We could precipitate both hOrc1p and hOrc2p associated with G1-phase chromatin but failed to precipitate hOrc1p on chromatin from S-phase cells. In contrast, hOrc2p was present in immunoprecipitates of G1-phase chromatin and of S-phase chromatin (Fig. 6A).
FIG. 6.
S-phase chromatin contains no hOrc1p. (A) Cross-linked chromatin from G1-phase cells and S-phase cells was immunoprecipitated with hOrc1p antibodies (α-ORC1) or hOrc2p antibodies (α-ORC2). Input (I), supernatants (S), and precipitates (P) of the α-ORC1- and α-ORC2-treated samples were analyzed with hOrc1 and with hOrc2p antibodies on Western blots. (B) Schematic representation of the genomic region analyzed in this study. The positions of the segments amplified by PCR are indicated by black boxes. (C) DNA extracted from chromatin immunoprecipitates of G1-phase cells (upper panels) and of S-phase cells (lower panels) were PCR amplified as in Fig. 5. We show the results of two independent experiments.
Next, we analyzed the DNA extracted from immunoprecipitated chromatin by quantitative real-time PCR. In these experiments, we used eight rather than only five primer pairs as in Fig. 5. Four of the eight primer sets correspond to sequences close to or within the promoter region and allow for a more precise determination of the binding region (Fig. 6B). The results of two independent experiments showed that both hOrc1p and hOrc2p were bound to upstream promoter DNA sequences in G1-phase chromatin, whereas only hOrc2p appeared on upstream promoter DNA in S-phase chromatin (Fig. 6C).
In the PCR experiment of Fig. 6C, we used the primer pairs Ex2 and In1 (Table 1, Fig. 6B) for the amplification of DNA regions that are located at a short distance (0.7 to 1 kb) on both sides of the UPR site. It is interesting that hOrc1p and hOrc2p antibodies precipitated chromatin fragments with UPR and Ex2 sequences from G1-phase cells but that hOrc2p antibodies precipitated chromatin fragments with UPR and In1 sequences from S-phase cells (Fig. 6C). Future studies will resolve whether these findings reflect cell cycle-dependent variations in the chromatin structure around the MCM4 gene promoter.
Control experiments were performed with cross-linked chromatin precipitated with irrelevant antibodies. No amplifiable DNA could be extracted from these control precipitates (data not shown; see also Fig. 3 and 5).
Abundance of nascent DNA strands.
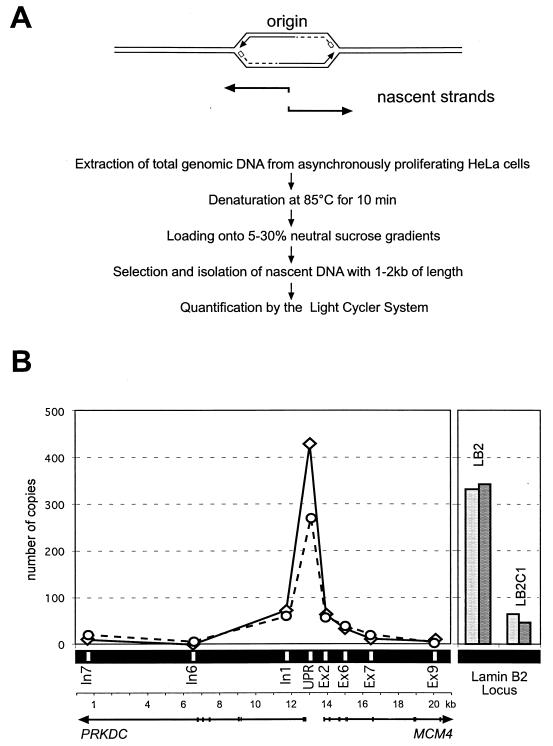
In budding yeast, ORC marks the origin of bidirectional DNA replication in chromatin (see the introduction). It was therefore of interest to determine whether the hOrc1p and/or hOrc2p-binding region upstream of gene MCM4 coincides with a replication origin.
To investigate this possibility, we used the nascent strand abundance assay (22, 23). This assay is based on the assumption that DNA strands of ca. 1 kb preferentially occur in the vicinity of replication start sites because leading strands at more advanced replication forks are much longer and lagging strands consist of Okazaki fragments of 100 to 200 bp (Fig. 7A).As demonstrated by Giacca et al. (23), sequences corresponding to known origin DNA are highly enriched in the 1-kb fraction of denatured DNA from asynchronously proliferating cells.
FIG. 7.
The UPR of gene MCM4 is highly abundant among nascent DNA strands. (A) Rationale and procedure (see the text). (B) In the left panel, quantitation of the DNA sequences in the 1- to 2-kbp fraction of nascent DNA is shown. The Primers UPR, In7, In6, In1, Ex2, Ex6, Ex7, and Ex9 (Table 1) were used in two independent PCRs (solid and broken line). In the right panel, LB2 corresponds to a primer pair with sequences of the known and well characterized lamin B2 replication origin (23). LB2C1 is a control primer pair complementary to a genomic region in a 3.5-kbp distance upstream from the lamin B2 origin (see Table 1).
Accordingly, we prepared genomic DNA from proliferating HeLa cells. The DNA was heat denatured and fractionated by sucrose gradient centrifugation. DNA in the 1- to 2-kbp fraction was used as a template for quantitative PCR. We first performed control reactions with a primer pair corresponding to the well-characterized lamin B2 origin downstream of the LMNB2 gene (23) and a second primer pair complementary to a noncoding region 3.5 kbp upstream of the origin (see Table 1). Our results clearly demonstrate a greater abundance of the LMNB2 origin sequence in the 1-kbp fraction of denatured DNA fragments (Fig. 7), exactly as expected from the published work of Giacca et al. (23). As an additional control for this assay, we detected no significant enrichment of these sequences when nascent DNA strands were isolated from HeLa cells that were blocked in late G1 phase by mimosine (data not shown).
We then used oligonucleotide primers corresponding to the MCM4 upstream promoter region, as well as primers complementary to neighboring intragenic regions, for quantitative PCR assays. We determined a much higher abundance of the UPR than of intragenic sequences in the 1-kbp fraction of denatured DNA (Fig. 7).
We conclude that a start point for DNA replication is located in the same genomic region that also contains binding sites for hOrc1p and hOrc2p.
Association of lamin B2 origin sequences with ORC proteins.
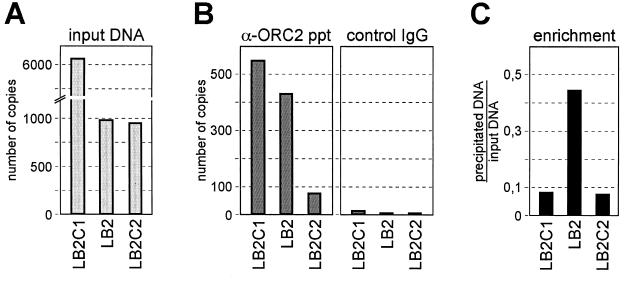
The question arises as to whether the procedure that we used to characterize the ORC binding site in the MCM4 gene promoter could also be used to identify ORC binding sites elsewhere in the genome. For this purpose, we tried to to demonstrate that other mapped human origin sequences are also marked by the binding of ORC. The lamin B2 origin has been mapped with the high precision (1, 23) that is required for the design of specific PCR primers.
Accordingly, we applied the CHIP assay on the lamin B2 locus. For quantitative analyses, we designed a primer set corresponding to the mapped replicative start sites of the lamin B2 locus (LB2) and two adjacent primer pairs at a 3- to 4-kbp distance as controls (LB2C1 and LB2C2; see Table 1). We first determined the copy number of nuclease-resistant DNA in the cross-linked chromatin sample prior to immunoprecipitation and found that fragments containing the LB2C1 sequence was six to eight times more abundant than fragments containing the other two regions investigated (Fig. 8A),suggesting that the chromatin region with the LB2C1 sequence is more resistant to nuclease attack than the other investigated regions.
FIG. 8.
Sequences of the replication start site of the lamin B2 origin are enriched in the ORC immunoprecipitate. (A) Quantitative real-time PCR was used to determine the copy numbers of sequences in cross-linked, nuclease-treated input chromatin. (B) The input chromatin sample was immunoprecipitated with hOrc2p-specific antibodies (α-ORC2 ppt). The copy numbers of amplifiable sequences were determined in the precipitates. Also shown are control immunoprecipitation experiments performed under identical conditions with nonspecific antibodies (control IgG). (C) Enrichment of sequences expressed as the ratio of precipitated over input DNA.
However, treatment with hOrc2p-specific antibodies resulted in the precipitation of a higher percentage of chromatin fragments with the mapped lamin B2 origin (amplifiable by the LB2 primers) than of the adjacent control regions (amplifiable by the LB2C1 and LB2C2 primers; Fig. 8C). Thus, even though sequences containing the lamin B2 origin region were not more abundant than the adjacent region in the immunoprecipitated chromatin fraction, they were certainly more enriched relative to the input DNA. The conclusion is that the CHIP approach does not appear to be a general method suited for the specific identification of human ORC binding sites. Rather, the technique is useful for determining ORC binding regions in sections of the genome with known nucleotide sequences. In these cases specific primer sets can be designed to evaluate the enrichment of DNA regions in specifically immunoprecipitated chromatin.
DISCUSSION
A CHIP procedure was used to identify sequences carrying bound hOrc1p and hOrc2p. We could show that a binding site is located in a region between the two divergently transcribed genes MCM4 and PRKDC, a region that also appears to contain an origin of bidirectional DNA replication. We achieved this result by closely adhering to several technical details which we shall discuss first.
Cross-linking and immunoprecipitation.
CHIP has been successfully used for the study of various aspects of protein-DNA interaction in eukaryotes (12, 42), including the interaction of transcription factors with specific DNA sequences in yeast (48). A similar approach for the investigation of mammalian transcription factor binding sites could be problematic since intergenic repeating DNA sequences are in a huge excess over transcription units in mammalian genomes. The situation could be less demanding for the determination of ORC binding sites because mammalian chromatin may possess many thousands of sites carrying ORC proteins, if we assume that ORCs are located at replication origins which are spaced at distances of roughly 105 bp in chromatin (14, 26).
We found, however, that several technical points are important for a successful application of the CHIP technology to the investigation of ORC binding regions in the human genome. These points include the length of the formaldehyde treatment, the size of the DNA fragments in cross-linked chromatin, and the quality of the antibodies. A crucial step of the procedure is an extensive washing of immunoprecipitates in buffers of various compositions to improve the signal-to-noise ratio for the high enrichment of specific DNA sequences in immunoprecipitated chromatin.
However, it must be added that the CHIP procedure as it stands now seems not to be suitable for the specific identification of human ORC binding sites in general. Rather, it is useful for the determination of ORC sites in genomic regions of known nucleotide sequences which allow the design of PCR primer sets to assay the quantities of specifically immunoprecipitated DNA.
Finally, we found the Light Cycler instrument to be highly useful for a rather convenient and reliable PCR-based estimate of the number of DNA copies in a given sample.
hOrc1p and hOrc2p on isolated chromatin.
We used hOrc1p and hOrc2p antibodies to precipitate chromatin with specific genomic DNA sequences. In most experiments, hOrc2p antibodies precipitated the majority of hOrc2p-bearing chromatin fragments, whereas hOrc1p antibodies usually were only able to precipitate a third or maximally one-half of the hOrc1p-bearing chromatin fragments. Since the hOrc1p antibodies have previously been shown to effectively precipitate soluble hOrc1p (30), we assume that much of the cross-linked hOrc1p is either not accessible to antibodies or structurally altered as a consequence of the formaldehyde treatment. Furthermore, since Orc1p and Orc2p have been found together in ORC after release from HeLa chromatin (63), it is reasonable to assume that hOrc1p and hOrc2p are components of a multisubunit human ORC in vivo. It is therefore likely that hOrc1p and hOrc2p do not occur independently of each other on chromatin but are components of chromatin-bound ORC. In CHIP assays, however, only a minority of chromatin fragments that are precipitated by hOrc1p antibodies also contain hOrc2p and vice versa. We assume that the reason for this is the stochastic manner by which formaldehyde links various components of ORC to DNA during the short cross-linking times chosen. The possibility that hOrc1p and hOrc2p are components of chromatin-bound ORC is supported by the results described above showing that antibodies to hOrc1p and hOrc2p precipitate chromatin with identical DNA sequences.
Earlier experiments had shown that ORC complexes are not static during the mammalian cell cycle, in contrast to the situation in yeast, where ORC remains intact during the entire cell cycle. In fact, the contacts of Orc1p to other ORC components may change during the S phase in animal cells (27, 39, 53, 59), and hOrc1p most likely dissociates from chromatin during the S phase of proliferating HeLa cells (30). This behavior of hOrc1p is reflected in our experiments with chromatin from cells investigated at 4 h after release from a double-thymidine block. Chromatin fragments prepared from cells in the middle or late S phase cannot be immunoprecipitated with hOrc1p antibodies and consequently do not carry hOrc1p. However, hOrc2p remained bound to its binding region during S phase. Thus, the S-phase-dependent dissociation of hOrc1p does not seem to promote a major change in the localization of hOrc2p (and possibly other components of ORC) during the cell cycle.
A hOrc1p- or hOrc2p-binding region.
We have found that chromatin immunoprecipitated by hOrc1p and hOrc2p antibodies contains a high fraction of DNA sequences that are unusually rich in GC base pairs, including runs of CpG dinucleotides (not shown). CpG dinucleotides are relatively rare in the human genome but occur in the upstream promoter regions of many human genes, where they form CpG islands as important regulatory elements for the expression of constitutive genes (2). This prompted us to search for the presence of upstream regions of known human genes among the DNA sequences extracted from immunoprecipitated chromatin. By using conventional and quantitative real-time PCR, we indeed found an enrichment of sequences corresponding to the region between the divergently transcribed genes MCM4 and PRKDC (and among other sequences [results not shown]). The sequence of the hOrc1p- and hOrc2p-carrying region is interesting since it includes several tracts with CpG dinucleotides bracketing an AT-rich element (see Fig. 3D), as well as several transcription factor binding sites, including an E2F-binding site that may be responsible for stimulating the transcription of gene MCM4 at the beginning of the S phase (41, 56, 61). This molecular architecture is reminiscent of the simian virus 40 origin of replication, which combines on a DNA stretch of ca. 100 bp several GC-rich elements (as a binding site for the viral replication initiator, T antigen), in addition to an essential run of AT base pairs and transcription factor binding sites that greatly stimulate the viral origin function (57). It will be a challenge for future research to determine the functions of the corresponding sequence elements in the MCM4 upstream region. A first step could be a precise determination of the ORC binding site by in vivo or in vitro footprinting methods.
We have performed biochemical tests to determine whether ORC, extracted from HeLa chromatin (63), specifically binds to sites in the UPR. We found that isolated ORC does bind to DNA, although in a sequence-nonspecific manner (16; unpublished data), suggesting that factors in addition to ORC are necessary for specific DNA binding.
Presently, we note that the region that cross-links to hOrc1p and hOrc2p overlaps or is closely adjacent to transcriptional control elements. This is a sequence context believed to favor the formation of mammalian replication origins (see the introduction). In fact, Delgado et al. (13) have shown that many early replicating DNA sequences contain runs of CpG dinucleotides such as those found in the upstream regions of many transcriptional start sites. Consequently, we determined by the nascent strand abundance assay that sequences of the MCM4 upstream region occur in high copy numbers in the fraction of nascent DNA strands. Thus, it is quite possible that bound ORC proteins determine the functions of origins in the human genome just as yeast ORC proteins appear to determine the initiation of replication at yeast ARS elements (8).
Conclusion.
We were able to show that components of the human ORC specifically cross-link to a genomic region close to or overlapping with the genetic control elements of two divergently transcribed human genes. We also showed that hOrc2p cross-links to the well-mapped origin in the lamin B2 region. It is thus quite likely that these regions include binding sites for human ORC. Corresponding sequences are highly abundant in preparations of nascent DNA strands. This indicates that, at least in these particular regions, ORC could mark the start site for DNA replication in the human genome.
Acknowledgments
We thank Sandra Kreitz and Torsten Krude for their comments on the manuscript.
This work was supported by the Bundesministerium für Bildung und Forschung (BMBF) and by the Deutsche Forschungsgemeinschaft (DFG).
REFERENCES
- 1.Abdurashidova, G., M. Deganuto, R. Klima, S. Riva, G. Biamonti, M. Giacca, and A. Falaschi. 2000. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287:2023-2026. [DOI] [PubMed] [Google Scholar]
- 2.Antequera, F., and A. Bird. 1993. Number of CpG islands and genes in human and mouse. Proc. Natl. Acad. Sci. USA 90:11995-11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aparicio, O. M., D. M. Weinstein, and S. P. Bell. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91:59-69. [DOI] [PubMed] [Google Scholar]
- 4.Bell, S. P., and B. Stillman. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357:128-134. [DOI] [PubMed] [Google Scholar]
- 5.Biamonti, G., M. Giacca, G. Perini, G. Contreas, L. Zentilin, F. Weighardt, M. Guerra, G. Della Valle, S. Saccone, S. Riva, et al. 1992. The gene for a novel human lamin maps at a highly transcribed locus of chromosome 19 which replicates at the onset of S phase. Mol. Cell. Biol. 12:3499-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biamonti, G., G. Perini, F. Weighardt, S. Riva, M. Giacca, P. Norio, L. Zentilin, S. Diviacco, D. Dimitrova, and A. Falaschi. 1992. A human DNA replication origin: localization and transcriptional characterization. Chromosoma 102:S24-S31. [DOI] [PubMed] [Google Scholar]
- 7.Bielinsky, A. K. 2001. Where it all starts: eukaryotic origins of DNA replication. J. Cell Sci. 114:643-651. [DOI] [PubMed] [Google Scholar]
- 8.Bielinsky, A. K., and S. A. Gerbi. 1998. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science 279:95-98. [DOI] [PubMed] [Google Scholar]
- 9.Chong, J. P., H. M. Mahbubani, C. Y. Khoo, and J. J. Blow. 1995. Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature 375:418-421. [DOI] [PubMed] [Google Scholar]
- 10.Coleman, T. R., P. B. Carpenter, and W. G. Dunphy. 1996. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell 87:53-63. [DOI] [PubMed] [Google Scholar]
- 11.Connelly, M. A., H. Zhang, J. Kieleczawa, and C. W. Anderson. 1998. The promoters for human DNA-PKcs (PRKDC) and MCM4: divergently transcribed genes located at chromosome 8 band q11. Genomics 47:71-83. [DOI] [PubMed] [Google Scholar]
- 12.Crane-Robinson, C., F. A. Myers, T. R. Hebbes, A. L. Clayton, and A. W. Thorne. 1999. Chromatin immunoprecipitation assays in acetylation mapping of higher eukaryotes. Methods Enzymol. 304:533-547. [DOI] [PubMed] [Google Scholar]
- 13.Delgado, S., M. Gomez, A. Bird, and F. Antequera. 1998. Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J. 17:2426-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DePamphilis, M. L. 1993. Eukaryotic DNA replication: anatomy of an origin. Annu. Rev. Biochem. 62:29-63. [DOI] [PubMed] [Google Scholar]
- 15.DePamphilis, M. L. 1999. Replication origins in metazoan chromosomes: fact or fiction? Bioessays 21:5-16. [DOI] [PubMed] [Google Scholar]
- 16.Dhar, S. K., L. Delmolino, and A. Dutta. 2001. Architecture of the human origin recognition complex. J. Biol. Chem. 6:6.. [DOI] [PubMed] [Google Scholar]
- 17.Dhar, S. K., and A. Dutta 2000. Identification and characterization of the human ORC6 homolog. J. Biol. Chem. 275:34983-34988. [DOI] [PubMed] [Google Scholar]
- 18.Diffley, J. F., J. H. Cocker, S. J. Dowell, and A. Rowley 1994. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell 78:303-316. [DOI] [PubMed] [Google Scholar]
- 19.Diffley, J. F., and B. Stillman. 1988. Purification of a yeast protein that binds to origins of DNA replication and a transcriptional silencer. Proc. Natl. Acad. Sci. USA 85:2120-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dulbecco, R. L., and M. Vogt. 1954. Plaque formation and isolation of pure lines with poliomyelitis virus. J. Exp. Med. 99:167-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gavin, K. A., M. Hidaka, and B. Stillman. 1995. Conserved initiator proteins in eukaryotes. Science 270:1667-1671. [DOI] [PubMed] [Google Scholar]
- 22.Giacca, M., C. Pelizon, and A. Falaschi. 1997. Mapping replication origins by quantifying relative abundance of nascent DNA strands using competitive polymerase chain reaction. Methods 13:301-312. [DOI] [PubMed] [Google Scholar]
- 23.Giacca, M., L. Zentilin, P. Norio, S. Diviacco, D. Dimitrova, G. Contreas, G. Biamonti, G. Perini, F. Weighardt, S. Riva, et al. 1994. Fine mapping of a replication origin of human DNA. Proc. Natl. Acad. Sci. USA 91:7119-7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert, D. M. 1998. Replication origins in yeast versus metazoa: separation of the haves and the have nots. Curr. Opin. Genet. Dev. 8:194-199. [DOI] [PubMed] [Google Scholar]
- 25.Göhring, F., and F. O. Fackelmayer. 1997. The scaffold/matrix attachment region binding protein hnRNP-U (SAF-A) is directly bound to chromosomal DNA in vivo: a chemical cross-linking study. Biochemistry 36:8276-8283. [DOI] [PubMed] [Google Scholar]
- 26.Hand, R. 1978. Eucaryotic DNA: organization of the genome for replication. Cell 15:317-25. [DOI] [PubMed] [Google Scholar]
- 27.Hua, X. H., H. Yan, and J. Newport. 1997. A role for Cdk2 kinase in negatively regulating DNA replication during S phase of the cell cycle. J. Cell Biol. 137:183-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang, W., N. J. Wells, and T. Hunter. 1999. Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc. Natl. Acad. Sci. USA 96:6193-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, R. T., C. S. Downes, and R. E. Meyns. 1993. The synchronization of mammalian cells, p. 1-24. In P. Fantes, and R. Brooks (ed.), The cell cycle: a practical approach. IRL Press/Oxford University Press, New York, N.Y.
- 30.Kreitz, S., M. Ritzi, M. Baack, and R. Knippers. 2001. The human origin recognition complex protein 1 dissociates from chromatin during S phase in HeLa Cells. J. Biol. Chem. 276:6337-6342. [DOI] [PubMed] [Google Scholar]
- 31.Ladenburger, E. M., F. O. Fackelmayer, H. Hameister, and R. Knippers. 1997. MCM4 and PRKDC, human genes encoding proteins MCM4 and DNA-PKcs, are close neighbours located on chromosome 8q12-q13. Cytogenet. Cell Genet. 77:268-270. [DOI] [PubMed] [Google Scholar]
- 32.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 33.Lee, D. G., and S. P. Bell. 1997. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol. Cell. Biol. 17:7159-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madine, M. A., C. Y. Khoo, A. D. Mills, and R. A. Laskey 1995. MCM3 complex required for cell cycle regulation of DNA replication in vertebrate cells. Nature 375:421-424. [DOI] [PubMed] [Google Scholar]
- 35.Maiorano, D., J. Moreau, and M. Mechali. 2000. XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature 404:622-625. [DOI] [PubMed] [Google Scholar]
- 36.Marahrens, Y., and B. Stillman. 1992. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255:817-823. [DOI] [PubMed] [Google Scholar]
- 37.Marheineke, K., and T. Krude. 1998. Nucleosome assembly activity and intracellular localization of human CAF-1 changes during the cell division cycle. J. Biol. Chem. 273:15279-15286. [DOI] [PubMed] [Google Scholar]
- 38.Mendez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Natale, D. A., C. J. Li, W. H. Sun, and M. L. DePamphilis. 2000. Selective instability of Orc1 protein accounts for the absence of functional origin recognition complexes during the M-G1 transition in mammals. EMBO J. 19:2728-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newlon, C. S. 1996. DNA replication in yeast, p. 873-914. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 41.Ohtani, K., R. Iwanaga, M. Nakamura, M. Ikeda, N. Yabuta, H. Tsuruga, and H. Nojima. 1999. Cell growth-regulated expression of mammalian MCM5 and MCM6 genes mediated by the transcription factor E2F. Oncogene 18:2299-2309. [DOI] [PubMed] [Google Scholar]
- 42.Orlando, V. 2000. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 25:99-104. [DOI] [PubMed] [Google Scholar]
- 43.Perkins, G., and J. F. Diffley. 1998. Nucleotide-dependent prereplicative complex assembly by Cdc6p, a homolog of eukaryotic and prokaryotic clamp-loaders. Mol. Cell 2:23-32. [DOI] [PubMed] [Google Scholar]
- 44.Quintana, D. G., and A. Dutta. 1999. The metazoan origin recognition complex. Front. Biosci. 4:D805-D815. [DOI] [PubMed] [Google Scholar]
- 45.Quintana, D. G., Z. Hou, K. C. Thome, M. Hendricks, P. Saha, and A. Dutta. 1997. Identification of HsORC4, a member of the human origin of replication recognition complex. J. Biol. Chem. 272:28247-28251. [DOI] [PubMed] [Google Scholar]
- 46.Quintana, D. G., K. C. Thome, Z. H. Hou, A. H. Ligon, C. C. Morton, and A. Dutta. 1998. ORC5L, a new member of the human origin recognition complex, is deleted in uterine leiomyomas and malignant myeloid diseases. J. Biol. Chem. 273:27137-27145. [DOI] [PubMed] [Google Scholar]
- 47.Rao, H., and B. Stillman. 1995. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc. Natl. Acad. Sci. USA 92:2224-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren, B., F. Robert, J. J. Wyrick, O. Aparicio, E. G. Jennings, I. Simon, J. Zeitlinger, J. Schreiber, N. Hannett, E. Kanin, T. L. Volkert, C. J. Wilson, S. P. Bell, and R. A. Young. 2000. Genome-wide location and function of DNA binding proteins. Science 290:2306-2309. [DOI] [PubMed] [Google Scholar]
- 49.Ritzi, M., M. Baack, C. Musahl, P. Romanowski, R. A. Laskey, and R. Knippers. 1998. Human minichromosome maintenance proteins and human origin recognition complex 2 protein on chromatin. J. Biol. Chem. 273:24543-24549. [DOI] [PubMed] [Google Scholar]
- 50.Rivella, S., B. Palermo, C. Pelizon, C. Sala, G. Arrigo, and D. Toniolo. 1999. Selection and mapping of replication origins from a 500-kb region of the human X chromosome and their relationship to gene expression. Genomics 62:11-20. [DOI] [PubMed] [Google Scholar]
- 51.Romanowski, P., M. A. Madine, A. Rowles, J. J. Blow, and R. A. Laskey. 1996. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr. Biol. 6:1416-1425. [DOI] [PubMed] [Google Scholar]
- 52.Rowles, A., J. P. Chong, L. Brown, M. Howell, G. I. Evan, and J. J. Blow. 1996. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell 87:287-296. [DOI] [PubMed] [Google Scholar]
- 53.Rowles, A., S. Tada, and J. J. Blow. 1999. Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J. Cell Sci. 112:2011-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rowley, A., J. H. Cocker, J. Harwood, and J. F. Diffley. 1995. Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J. 14:2631-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saha, P., J. Chen, K. C. Thome, S. J. Lawlis, Z. H. Hou, M. Hendricks, J. D. Parvin, and A. Dutta. 1998. Human CDC6/Cdc18 associates with Orc1 and cyclin-cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol. Cell. Biol 18:2758-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schulte, D., R. Burkhart, C. Musahl, B. Hu, C. Schlatterer, H. Hameister, and R. Knippers. 1995. Expression, phosphorylation and nuclear localization of the human P1 protein, a homologue of the yeast Mcm3 replication protein. J. Cell Sci. 108:1381-1389. [DOI] [PubMed] [Google Scholar]
- 57.Simmons, D. T. 2000. SV40 large T antigen functions in DNA replication and transformation. Adv. Virus Res. 55:75-134. [DOI] [PubMed] [Google Scholar]
- 58.Spradling, A. C. 1999. ORC binding, gene amplification, and the nature of metazoan replication origins. Genes Dev. 13:2619-2623. [DOI] [PubMed] [Google Scholar]
- 59.Sun, W., M. Hola, K. Pedley, S. Tada, J. J. Blow, I. T. Todorov, S. E. Kearsey, and R. F. Brooks. 2000. The replication capacity of intact mammalian nuclei in Xenopus egg extracts declines with quiescence, but the residual DNA synthesis is independent of Xenopus MCM proteins. J. Cell Sci. 113:683-695. [DOI] [PubMed] [Google Scholar]
- 60.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsuruga, H., N. Yabuta, K. Hashizume, M. Ikeda, Y. Endo, and H. Nojima. 1997. Expression, nuclear localization and interactions of human MCM/P1 proteins. Biochem. Biophys. Res. Commun. 236:118-125. [DOI] [PubMed] [Google Scholar]
- 62.Tugal, T., X. H. Zou-Yang, K. Gavin, D. Pappin, B. Canas, R. Kobayashi, T. Hunt, and B. Stillman. 1998. The Orc4p and Orc5p subunits of the Xenopus and human origin recognition complex are related to Orc1p and Cdc6p. J. Biol. Chem. 273:32421-32429. [DOI] [PubMed] [Google Scholar]
- 63.Vashee, S., P. Simancek, M. D. Challberg, and T. J. Kelly. 2001. Assembly of the human origin recognition complex. J. Biol. Chem. 276:26666-26673. [DOI] [PubMed] [Google Scholar]
- 64.Weinreich, M., C. Liang, and B. Stillman. 1999. The Cdc6p nucleotide-binding motif is required for loading Mcm proteins onto chromatin. Proc. Natl. Acad. Sci. USA 96:441-446. [DOI] [PMC free article] [PubMed] [Google Scholar]