Abstract
Saccharomyces cerevisiae Spo11 protein (Spo11p) is thought to generate the DNA double-strand breaks (DSBs) that initiate homologous recombination during meiosis. Spo11p is related to a subunit of archaebacterial topoisomerase VI and appears to cleave DNA through a topoisomerase-like transesterase mechanism. In this work, we used the crystal structure of a fragment of topoisomerase VI to model the Spo11p structure and to identify amino acid residues in yeast Spo11p potentially involved in DSB catalysis and/or DNA binding. These residues were mutated to determine which are critical for Spo11p function in vivo. Mutation of Glu-233 or Asp-288, which lie in a conserved structural motif called the Toprim domain, abolished meiotic recombination. These Toprim domain residues have been implicated in binding a metal ion cofactor in topoisomerases and bacterial primases, supporting the idea that DNA cleavage by Spo11p is Mg2+ dependent. Mutations at an invariant arginine (Arg-131) within a second conserved structural motif known as the 5Y-CAP domain, as well as three other mutations (E235A, F260R, and D290A), caused marked changes in the DSB pattern at a recombination hotspot, suggesting that Spo11p contributes directly to the choice of DNA cleavage site. Finally, certain DSB-defective mutant alleles generated in this study conferred a semidominant negative phenotype but only when Spo11p activity was partially compromised by the presence of an epitope tag. These results are consistent with a multimeric structure for Spo11p in vivo but may also indicate that the amount of Spo11 protein is not a limiting factor for DSB formation in normal cells.
Homologous recombination during meiosis in Saccharomyces cerevisiae proceeds via the formation and subsequent repair of DNA double-strand breaks (DSBs) (reviewed in references 24 and 38). Formation of these DSBs requires the products of at least 10 genes, including SPO11. The Spo11 protein (Spo11p) was found covalently linked to the 5"-strand termini of DSBs in certain mutants (e.g., rad50S) that are defective for the normal 5"- to-3" nucleolytic processing of DSB ends (23), and Spo11p shares sequence similarity with a subunit of an archaebacterial topoisomerase (6). Based on these observations, it is thought that Spo11p is the catalytic subunit of the meiotic DNA cleaving activity and that it cuts DNA by a topoisomerase-like transesterification reaction. The role of Spo11p in promoting meiotic recombination initiation is widely conserved, as functional homologs have been characterized in fungi, plants, and animals (3, 10, 11, 16, 18, 26, 30, 31, 35, 43).
It is likely that all of the archaebacterial Spo11p homologs function as type II topoisomerases, but topoisomerase activity has been directly demonstrated only for the enzyme from Sulfolobus shibatae (5, 8). Because the amino acid sequence of this topoisomerase is unlike the previously known eukaryotic and prokaryotic type II enzymes, it was named topoisomerase VI to distinguish it from these proteins (6). Topoisomerase VI is an A2B2 heterotetramer, of which the smaller subunit (called Top6A) shares similarity with Spo11p. The Top6A subunit can bind DNA nonspecifically (32) but does not catalyze efficient DNA cleavage by itself (8). The Top6B subunit contains an ATP-binding domain found in other type II topoisomerases, as well as in the Hsp90 family of heat shock proteins and the MutL class of mismatch repair proteins (6).
Crystallographic studies identified two domains in the type II topoisomerases of bacteria and eukaryotes (referred to as type IIA enzymes) that are also found in archaeal Top6A (referred to as type IIB) (32) and in topoisomerase I from Escherichia coli (a type IA enzyme) (4, 25). These shared folds are embedded within significantly different tertiary and quaternary structures of the proteins and appear to be diagnostic of enzymes that generate 5"-phosphodiester linkages because they are not found in the type I topoisomerases of vaccinia virus or humans (type IB enzymes), which cleave DNA to form a 3"-phosphodiester linkage (14).
The first of these structural domains is an α-helical fold similar to the E. coli catabolite gene activator protein (CAP) DNA binding domain (36). This domain contains the catalytic tyrosine residue (Tyr-135 in S. cerevisiae Spo11p) (6) and is termed the 5Y-CAP motif because it is common to all topoisomerases that generate a 5"-tyrosyl phosphodiester (32). The second domain is an abbreviated Rossmann fold, consisting of a four-stranded parallel β sheet sandwiched between two pairs of α-helices (4, 32). This domain shows modest sequence similarity among different families of topoisomerases and corresponds to a sequence motif identified using an iterative database search seeded with the sequence of E. coli primase (2). This motif has been termed the Toprim domain (for topoisomerases and primases). Only 3 residues—a glutamate and two aspartates—are conserved in nearly all Toprim-motif containing proteins. The function of these residues is not known, but they coordinate metal ions in both the Top6A and primase structures (20, 32). Divalent metal ions are known to be important for the activities of a number of Toprim-containing enzymes (discussed further below).
In the work described here, we exploited the sequence similarity of Spo11p with Top6A to identify regions of yeast Spo11p that are likely to contribute to DNA binding and to catalysis of strand cleavage. We then tested whether conserved residues in these regions were critical for meiotic DSB formation in vivo using site-directed mutagenesis. The effects of these mutations provide insight on several aspects of Spo11p activity, including the functional significance of conserved structural motifs in the Spo11p/Top6A family, homotypic interactions critical for DSB formation, the site specificity for DNA cleavage, and mechanisms that control Spo11p activity.
MATERIALS AND METHODS
Yeast strains and plasmids.
All yeast strains used in this study are isogenic diploid derivatives of SK1 (19) and were derived from strains originally provided by N. Kleckner, Harvard University. Unless otherwise indicated, all are MATa/MATα and his4X::LEU2/his4B::LEU2 and are homozygous for ho::LYS2, lys2, ura3, and leu2. SKY10 is also homozygous for spo11Δ. SKY103 (used for DSB analysis) is homozygous for his4B::LEU2, spo11Δ, trp1::hisG, and rad50(K81I)::ura3. Except for SKY10 and SKY103, all other strains are homozygous for nuc1Δ::LEU2 and carry alleles of SPO11 as indicated in text and tables. Yeast transformations were by the lithium acetate/polyethylene glycol method (17).
The plasmids used in this study use the pRS316 vector backbone unless otherwise indicated (ARS, CEN, and URA3) (37). The spo11-HA3His6::kanMX4 allele carries a 3" fusion of the SPO11 coding sequence to a sequence encoding three copies of the HA epitope (YPYDVPDYA) and six histidines. This allele confers phenotypes that are recessive to the wild type (see below and reference 21), so it is designated in lowercase. The kanMX4 drug resistance cassette (39) was inserted into a unique SwaI site in the SPO11 3" untranslated region and is transcribed in the same direction as SPO11. Details for the generation of this construct are provided elsewhere (21). The SPO11::kanMX4 allele is identical except that it lacks the epitope tag sequence. Point mutations were introduced into the spo11-HA3His6 coding sequence using the QuickChange method (Stratagene) according to the manufacturer's instructions. Sequences of the mutagenic primers are available on request. All mutated constructs were confirmed by sequencing of the entire open reading frame. To replace the genomic SPO11 locus with specific mutant alleles, cells were transformed to G418 resistance with appropriate restriction fragments. Correct targeting was confirmed by PCR and Southern blotting of genomic DNA, and retention of the desired point mutations was confirmed by direct sequencing of PCR-amplified genomic DNA.
Culture methods.
Synchronous meiotic cultures were prepared essentially as described previously (1, 34) with slight modification to select for retention of plasmids. Briefly, fresh transformants were grown to saturation in liquid yeast extract-Peptone-dextrose (1% yeast extract, 2% Bacto Peptone, and 2% dextrose) containing 200 μg of G418/ml. These cultures were used to inoculate liquid yeast extract-Peptone-acetate (1% yeast extract, 2% Bacto Peptone, and 1% potassium acetate) containing 200 μg of G418/ml and were grown for 13 to 14 h at 30°C with vigorous shaking (G418 was omitted when constructs were stably integrated). Cells were then harvested, washed, and resuspended in SPM (0.3% potassium acetate and 0.02% raffinose) prewarmed to the desired temperature. For experiments with 2μm-based vectors, transformants were grown in liquid uracil dropout medium prior to the yeast extract-Peptone-acetate step.
Spore viabilities were determined by dissection of four-spored asci produced in liquid SPM. Where indicated, the statistical significance of observed differences in spore viability was evaluated by the Z test, using a Microsoft Excel spreadsheet calculator written by M. F. F. Abdullah (laboratory of R. Borts, Oxford University). For measurements of meiotic intragenic recombination frequencies, strains with his4::LEU2 heteroalleles (9) were induced to enter meiosis as described above, and samples were collected at 0 h (to measure the frequency of spontaneous prototroph formation) and at either 8 h (for sporulation at 30°C) or 13 h (for sporulation at 16°C). Samples were sonicated, diluted in sterile distilled water, and plated on yeast extract-Peptone-dextrose to measure total viable cells and on histidine dropout medium to measure His+ prototroph formation. Colonies were scored after 2 or 3 days at 30°C.
DSB measurements.
Genomic DNA was purified from synchronized cultures as described earlier (7, 9) and was quantitated using a DynaQuant DNA fluorometer (Hoeffer). DNA (1 μg) was digested with PstI, electrophoresed on 0.8% agarose gels in 1× Tris-acetate-EDTA, and transferred to Hybond N+ membranes (Amersham) by positive pressure blotting (Posiblot; Stratagene). For higher-resolution analysis, 5 μg of genomic DNA was digested with EcoRI, electrophoresed on 5% polyacrylamide gels in 1× Tris-borate-EDTA, and electrophoretically transferred to Hybond N membranes. DSBs at the his4::LEU2 hotspot were detected using 32P-labeled probe 291 or 207 as described previously (22). Blots were visualized by autoradiography, and DSBs were quantified as a fraction of total hybridizing DNA using a Fuji BAS-2500 phosphorimager system.
RESULTS
Selection of amino acid residues for targeted mutagenesis.
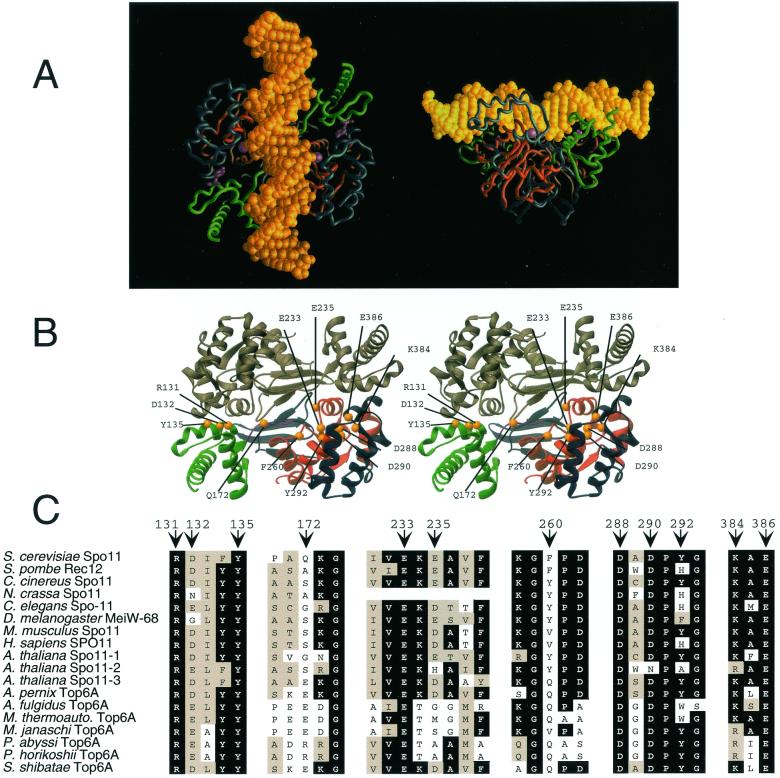
The overall sequence identity among Spo11p/Top6A family members is only 20 to 30% in pairwise combinations, but several blocks of much higher conservation are apparent (6, 23, 24). These blocks correspond in many cases to core structural motifs identified in the Methanococcus jannaschii Top6A crystal structure (32). To identify residues in yeast Spo11p that might contribute directly to DNA cleavage, we first looked for conserved residues within the 5Y-CAP and Toprim domains whose side chains in M. jannaschii Top6A lie near the catalytic tyrosine residue, near the acidic metal binding pocket or in the space between these putative components of the active site (Fig. 1A and B).We targeted the equivalent of eight such residues in S. cerevisiae Spo11p for mutagenesis. We also chose three residues in Top6A whose side chains contribute prominently to the putative DNA binding surface, on the premise that these might be involved in DNA binding. These residues are not as highly conserved, but likely equivalent residues in Spo11p could be identified from sequence alignments and from homology-based structural modeling (Fig. 1C and data not shown).
FIG. 1.
(A) Orthogonal views of the M. jannaschii Top6A crystal structure, with B-form DNA (yellow) modeled into the putative DNA binding channel. The Top6A monomers are colored to indicate the 5Y-CAP (green) and Toprim (red) domains; the bound Mg2+ ions are represented by magenta spheres. (B) Stereo view of the M. jannaschii Top6A crystal structure, showing the positions of residues equivalent to those mutated in yeast Spo11p in this study (yellow spheres). One monomer is shaded gray; the other is colored as in panel A. The residues and numbering are according to the yeast Spo11p sequence. (C) Alignment of eukaryotic Spo11p and archaeal Top6A amino acid sequences. Portions of the sequences around the residues mutated in this study are shown (see reference 24 for a more detailed alignment). Conserved residues are highlighted in black; conservative substitutions are highlighted in gray. The positions of the mutated S. cerevisiae residues are indicated by arrows.
A total of 18 point mutations at 11 positions were generated and analyzed for their effect on Spo11p activity in vivo. The mutant proteins were tagged at their C termini with three repeats of the hemagglutinin (HA) epitope plus a hexahistidine sequence and were expressed under the control of the SPO11 promoter on an ARS/CEN vector in diploid spo11Δ/spo11Δ strains. Spo11p activity was assessed by measuring meiotic DSBs at the his4::LEU2 hotspot in both RAD50 and rad50S backgrounds, his4::LEU2 heteroallele recombination frequencies, and spore viability (an indirect measure of total recombination in the genome). DSB Southern blots for a representative subset of the mutants are presented in Fig. 2A,and the phenotypic analyses are summarized in Table 1. The spore viability and recombination frequency with a plasmid-borne wild-type SPO11 allele were slightly lower than with chromosome-borne alleles (compare Table 1 with Table 2, below), most likely because plasmid loss renders a fraction of the cells spo11. For those cells that retained the plasmid, however, the copy number was at least two per cell on average, based on analysis of plasmid segregation in four-spore-viable tetrads (data not shown). For epitope-tagged proteins, all of the point mutants were expressed at levels comparable to wild-type levels, measured by Western blotting of whole-cell extracts (Fig. 2B). For the purpose of this analysis, we defined a residue as being essential for Spo11p function if mutation caused a ≥100-fold decrease in recombination frequency.
FIG. 2.
(A) Effects of selected spo11 mutations on DSB formation. DSBs that were formed at the his4::LEU2 locus during meiosis in a spo11Δ rad50S strain (SKY103) carrying each of the indicated spo11-HA3His6 alleles on an ARS/CEN vector were analyzed by Southern blotting. Results are shown for a representative subset of the mutations in this study. The positions of the parental (unbroken) DNA and of the fragments generated by cleavage at either of the two prominent DSB sites in this region are indicated. The band marked with the asterisk derives from variable cross-hybridization of the probe to the vector carrying the spo11 alleles. (B) Steady-state levels of Spo11-HA3His6p. Whole-cell extracts were prepared 4 h after transfer to sporulation conditions from cultures of SKY10 (spo11Δ) carrying the indicated spo11-HA3His6 alleles on ARS/CEN vectors. Levels of Spo11-HA3His6p were determined by Western blotting with an anti-HA antibody. Stripping and reprobing the same blot with antitubulin antibodies controlled for loading. A representative experiment is shown. The small apparent variations in steady-state levels were not reproducible (data not shown), so they are presumably due to small differences in the synchronization and/or sporulation efficiency of individual cultures.
TABLE 1.
Analysis of mutations in SPO11
| Location of plasmid-borne allelesa | Frequency of His+ (103)b
|
% Viable sporesc for
|
% DSB (rad50S) | ||||
|---|---|---|---|---|---|---|---|
| HA3His6-tagged alleles at: | Untagged alleles at:
|
HA3His6-tagged alleles at:
|
Untagged alleles at:
|
HA3His6-tagged alleles at:
|
|||
| 30°C | 30°C | 16°C | 30°C | 30°C | 16°C | 30°C | |
| Wild type | 6.3 ± 1.9 | 6.7 ± 1.1 | 5.5 ± 0.51 | 90.9 | 92.8 | 86.5 | 7.6 |
| No plasmid | 0.006 ± 0.006 | 0.50 | <0.1d | ||||
| spo11Δ | 0.012 ± 0.010 | <0.25e | |||||
| 5Y-CAP | |||||||
| R131K | 2.8 ± 1.7 | 8.0 ± 1.3 | 70.0 | 87.3 | 0.35 | ||
| R131A | 0.018 ± 0.011 | 0.005 ± 0.002 | <0.25e | 0.11 | |||
| D132A | 6.9 ± 1.3 | 78.7 | 2.0 | ||||
| Toprim | |||||||
| E233A | 0.003 ± 0.001 | 0.10 ± 0.15 | <0.2e | 0.25 | <0.1d | ||
| E233Q | 0.015 ± 0.014 | 0.50 | <0.1d | ||||
| D288A | 0.025 ± 0.022 | 0.009 ± 0.004 | 0.50 | 2.5 | <0.1d | ||
| D288N | 0.023 ± 0.022 | 0.50 | <0.1d | ||||
| D290A | 1.9 ± 0.53 | 8.1 ± 0.60 | 0.92 ± 0.55 | 19.3 | 87.0 | 2.0 | 0.62 |
| D290N | 6.4 ± 2.2 | 81.1 | 2.2 | ||||
| E235A | 2.2 ± 1.0 | 7.2 ± 1.2 | 0.43 ± 0.17 | 53.2 | 88.0 | 1.0 | 0.56 |
| K384A | 0.055 ± 0.044 | 6.1 ± 0.83 | 1.5 ± 0.62 | 0.50 | 90.3 | 22.5 | <0.1d |
| E386A | 2.4 ± 0.75 | 4.2 ± 0.25 | 67.0 | 92.0 | 3.0 | ||
| DNA binding surface | |||||||
| Q172A | 8.6 ± 1.4 | 92.2 | 3.6 | ||||
| Q172R | 10.3 ± 4.2 | 82.2 | 2.7 | ||||
| F260A | 1.1 ± 0.45 | 3.2 ± 0.32 | 0.37 ± 0.14 | 50.3 | 84.0 | <0.25e | 1.6 |
| F260R | 8.2 ± 6.2 | 91.3 | 4.3 | ||||
| Y292A | 4.9 ± 1.3 | 88.8 | 5.7 | ||||
| Y292R | 0.035 ± 0.03 | 0.002 ± 0.001 | 1.0 | <0.1d | |||
SKY10 (spo11Δ/spo11Δ) was transformed with ARS/CEN plasmids carrying the indicated SPO11 alleles.
Intragenic recombination was measured as the His+ prototroph frequency (per viable cell) after 8 h (30°C) or 13 h (16°C) in SPM. Each value is the mean ± standard deviation of at least three determinations. The premeiotic prototroph frequencies have not been subtracted.
At least 100 four-spored asci were dissected for each determination.
DSB signal (if any) was below the detection limit for this assay.
No viable spores were recovered.
TABLE 2.
Semidominance of SPO11 alleles
| Test allele | Results for configurationa
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A (Test allele with SPO11)
|
B (Test allele with SPO11 [both tagged])
|
C (Test allele with SPO11 [test allele tagged])
|
D (Test allele with SPO11 [SPO11 tagged])
|
||||||||||
| Frequency of His+b | Viability (%) (n)c | Strain | Frequency of His+ | Viability (%) (n) | Strain | Frequency of His+ | Viability (%) (n) | Strain | Frequency of His+ | Viability (%) (n) | Strain | ||
| Wild type | 18.9 ± 3.6 | 98.0 (100) | SKY534 | 11.8 ± 5.8 | 97.1 (140) | SKY334 | |||||||
| spo11Δ | 10.4 ± 6.8 | 96.3 (100) | SKY486 | 8.4 ± 2.5 | 94.5 (160) | SKY344 | |||||||
| Y135F | 9.9 ± 3.9 | 94.7 (99) | SKY546 | 4.4 ± 2.2 | 51.1 (162) | SKY335 | 14.2 ± 4.0 | 95.8 (100) | SKY533 | 0.12 ± 0.046 | 2.7 (100) | SKY545 | |
| E233A | 9.6 ± 5.0 | 95.0 (100) | SKY487 | 11.1 ± 5.4 | 86.0 (100) | SKY587 | 13.8 ± 6.6 | 98.0 (100) | SKY588 | 0.16 ± 0.045 | 0.50 (100) | SKY458 | |
| D288N | 8.7 ± 0.75 | 97.3 (100) | SKY488 | 1.1 ± 1.0 | 44.0 (138) | SKY337 | 7.5 ± 2.3 | 99.0 (100) | SKY529 | 0.18 ± 0.11 | 2.5 (100) | SKY459 | |
| D290A | 13.1 ± 2.2 | 98.5 (100) | SKY538 | 7.1 ± 1.9 | 70.0 (109) | SKY336 | 7.8 ± 2.5 | 97.0 (100) | SKY530 | 7.03 ± 1.3 | 94.8 (100) | SKY539 | |
Each of the indicated test alleles (with or without the HA3His6 tag) was crossed to wild-type SPO11 (with or without the tag), as indicated (configuration).
Meiotic intragenic recombination at his4::LEU2 was measured as the frequency of His+ prototrophs per viable cell after 8 h in SPM at 30°C (frequency × 103; mean ± standard deviation of at least three determinations).
Spore viability was assayed by dissection of the indicated number (n) of four-spored asci produced after ≥24 h in SPM at 30°C.
Potential active site residues in the 5Y-CAP domain.
Tyr-135 is the only tyrosine absolutely conserved in all Spo11p/Top6A family members (Fig. 1C) and is thus thought to be the catalytic residue that carries out a nucleophilic attack on the DNA backbone, becoming covalently attached to the DNA. Consistent with this interpretation, Tyr-135 was previously shown to be essential for Spo11p activity in vivo (6). We obtained similar results with an independently generated phenylalanine substitution at this position (12; data not shown).
The arginine at position 131 is conserved in all Spo11p/Top6A family members (Fig. 1C), and an arginine is found at a similar position relative to the catalytic tyrosine in three-dimensional space in all 5Y-CAP topoisomerases (4, 32). Replacing it with alanine at this position severely compromised Spo11p activity in vivo (R131A, Table 1). Interestingly, a conservative substitution of lysine at this position (R131K) supported high levels of meiotic intragenic recombination (44% of wild type) and spore viability (76% of wild type), but DSBs were more severely reduced: 5% of wild type in rad50S (Table 1) and <10% of wild type (i.e., undetectable) in RAD50 (data not shown). This apparent discrepancy between DSB levels and recombination frequencies appears to reflect effects of the mutation on the distribution of cleavage events at the recombination hotspot (see below).
Most Spo11p/Top6A family members have an acidic residue at the position equivalent to Asp-132 in Spo11p (Fig. 1C). Nevertheless, substituting alanine for Asp-132 had little, if any, effect on the ability of the mutant allele to complement the defects of a spo11 null (Table 1), indicating that this residue is not critical for Spo11p catalytic activity in vivo. This result is consistent with the fact that this residue is not conserved in Neurospora crassa Spo11p (asparagine) or Drosophila melanogaster MeiW-68 (glycine).
Potential active site residues in the Toprim domain.
The three conserved acidic residues which coordinate a Mg2+ ion in the Top6A structure were each mutated to alanine or to glutamine or asparagine (E233A, E233Q, D288A, D288N, D290A, and D290N). Of these mutants, E233A, E233Q, D288A, and D288N were indistinguishable from a null mutation (Table 1). In contrast, substitutions at Asp-290 had less effect: D290A showed 10 to 30% of wild-type levels for the parameters assayed, while D290N was nearly normal.
The position equivalent to Spo11p Glu-235 contains an acidic residue in nearly all of the eukaryotic Spo11p homologs but is not conserved in the archaebacterial Top6A proteins (Fig. 1C). Replacement with alanine at Glu-235 resulted in a relatively modest defect in intragenic recombination (35% of wild type) and spore viability (60% of wild type). However, as in the case of R131K (above), DSB frequency appeared to be more severely compromised (7% of wild type) and the distribution of cleavage sites at the his4::LEU2 hot spot was altered (see below).
The C-terminal regions of members of the Spo11p/Top6A family are poorly conserved, but one block of conserved sequence includes two residues (equivalent to Lys-384 and Glu-386 of S. cerevisiae Spo11p) that lie in or near the putative active site in M. jannaschii Top6A (Fig. 1). Replacement by alanine at Glu-386 caused a modest defect (38 to 75% of wild type for the parameters assayed), whereas the K384A allele was severely compromised.
Mutations in residues potentially involved in binding DNA.
Mutants were also generated that affected each of three Spo11p residues equivalent to M. jannaschii Top6A residues which protrude into the putative DNA binding channel and which might be expected to interact directly with DNA. The first of these (equivalent to Gln-172 in yeast Spo11p) is glutamate in most of the archaeal proteins and serine, glycine, or alanine in most of the other eukaryotic proteins (Fig. 1C). Mutation of this residue in yeast Spo11p (Q172A and Q172R) had little if any effect on the parameters examined (Table 1). The second such residue (equivalent to Phe-260) is a glutamine in most of the archaeal proteins but is a bulky hydrophobic residue in the eukaryotic proteins. Replacement by alanine at this position significantly decreased Spo11p activity in vivo (F260A). A charged amino acid substitution at this position supported normal amounts of recombination (F260R, Table 1) but caused an alteration in DSB site specificity (see below). A third residue, Tyr-292, is more widely conserved between eukaryotes and archaebacteria; it is tyrosine, phenylalanine, or tryptophan in three-quarters of the members of the Spo11p/Top6A family and histidine in most of the remainder. Placing alanine at this position (Y292A) had little effect on Spo11p activity in vivo, but replacement with arginine (Y292R) inactivated Spo11p (Table 1). A possible reason for this mutational effect is discussed below.
Synergistic defects from combining certain point mutations with the epitope tag.
The epitope-tagged version of Spo11p used for this analysis (Spo11-HA3His6p) supports high levels of recombination at 30°C. However, while this analysis was in progress, we discovered that the tag renders Spo11p slightly cold sensitive, indicating that the protein is not completely normal (21). To address the concern that the phenotypes described above might be affected by the presence of the tag, a subset of the mutations was reengineered in an untagged form and analyzed as described above. For the E233A and D288A mutations in the Toprim domain and for R131A in the 5Y-CAP domain, the untagged mutant alleles were still severely compromised (Table 1). These residues thus appear to be essential for Spo11p activity.
In contrast, a set of mutations that conferred a partial loss-of-function phenotype when epitope tagged (D290A, E235A, R131K, E386A, and F260A) supported significantly higher recombination levels when the tag was removed, in many cases indistinguishable from wild-type levels (Table 1). The K384A mutant was even more striking: for this allele, intragenic recombination and spore viability were <1% of wild-type levels for the tagged form but were indistinguishable from wild-type levels for the untagged form. It thus appears that the D290A, E235A, R131K, K384A, E386A, and F260A mutations confer slight defects which are only revealed when Spo11p is sensitized by the modest defect caused by the C-terminal HA3His6 epitope tag.
Because the epitope tag caused a weak cold-sensitive phenotype by itself, we also asked whether conditional defects were associated with the point mutations that acted synergistically with the tag. None of the untagged point mutants was noticeably deficient relative to the wild type at elevated temperature (data not shown), but D290A, E235A, and F260A showed roughly 10-fold-lower intragenic recombination frequencies and spore viability at 16°C than at 30°C and K384A was reduced roughly three- to fourfold (Table 1).
Alterations of DSB site preference.
A subset of the mutants that retained recombination-promoting activity (R131K, E235A, F260R, and D290A) generated noticeably unusual DSB patterns at site I of the his4::LEU2 hotspot in Southern blots of agarose gels (Fig. 2A and 3A; data not shown). In higher-resolution analysis on polyacrylamide gels, DSBs at site I were spread over a roughly 100- to 150-bp region in SPO11/SPO11 rad50S/rad50S cells, as previously reported (41) (Fig. 3B). At this resolution, the epitope tag had little if any effect on the distribution of DSBs (Fig. 3B and data not shown), although the total DSB frequency was somewhat diminished, consistent with intragenic recombination frequencies measured with integrated constructs (see below). The four mutations that gave aberrant patterns on agarose gels gave even more dramatically altered patterns in this higher-resolution analysis (Fig. 3B). There were slight variations in DSB distribution (in addition to the previously established differences in DSB frequency; see above) between tagged and untagged alleles, but the major effects of these point mutations on DSB site preference appeared to be largely independent of the epitope tag. Untagged D290A and E235A gave cleavage patterns similar to one another, whereas the patterns for R131K and F260R were distinctly different.
FIG. 3.
Alteration of the distribution of cleavage events at a recombination hotspot. kbp, kilobase pairs. ARS/CEN vectors carrying the indicated SPO11 alleles (with or without the HA3His6 tag, as indicated) were introduced into SKY103 (spo11Δ rad50S). DNA was prepared from meiotic cultures 6 h after transfer to sporulation medium, and DSBs at the his4::LEU2 hotspot were analyzed by Southern blotting after restriction enzyme digestion and electrophoresis on 0.8% agarose (A) or 5% polyacrylamide (B). The restriction-enzyme-and-probe combination shown in panel B detects DSBs only at site I.
Semidominance of DSB-defective alleles.
Two Spo11p monomers must act in concert to cleave both strands of the DNA duplex (15, 23, 24). In the Top6A structure, the catalytic tyrosine of each monomer lies closer to the Toprim metal binding pocket of its dimer partner than it does to its own Toprim domain (Fig. 1B). This arrangement suggests that the dimer forms two hybrid active sites, each responsible for cleaving a single strand of the DNA duplex (32). Extrapolating this dimeric structure to Spo11p predicts that catalytically inactive mutants should confer a semidominant-negative phenotype when coexpressed with the wild type, if Spo11p is limiting in vivo. To test this prediction, we replaced the normal SPO11 locus with each of several mutant alleles and tested whether single copies of the mutants were recessive, dominant, or semidominant relative to the wild type. We examined three severely compromised mutants (Y135F, E233A, and D288N) and one partial loss-of-function mutant (D290A). Because of the effects of the epitope tag on Spo11p activity, the mutations were examined in four different configurations. Each configuration consisted of a test allele (wild-type or mutant) with or without the HA3His6 tag in combination with a wild-type SPO11 allele, also with or without the tag. All alleles were marked with the kanMX4 selectable marker inserted downstream of the SPO11 coding sequence. Intragenic recombination frequency at his4::LEU2 and spore viability were assessed for each mutation in each configuration (Table 2).
When neither allele was tagged (configuration A in Table 2), the mutations had little or no effect on the parameters assayed. The spo11(Y135F)/SPO11 strain (SKY546) was indistinguishable from a matched hemizygous control (SKY486, SPO11/spo11Δ), with a modest decrease in the intragenic recombination frequency relative to a wild-type control and no significant change in spore viability (P > 0.1). Similar results were obtained with the E233A (SKY487) and D288N (SKY488) mutations. These results indicate that DSB-defective alleles of SPO11 are completely or nearly completely recessive when present in single copy along with a wild-type allele. The spo11(D290A)/SPO11 strain (SKY538, Table 2) was essentially indistinguishable from the wild type, consistent with the fact that the untagged D290A allele supported nearly wild-type recombination levels on its own (Table 1).
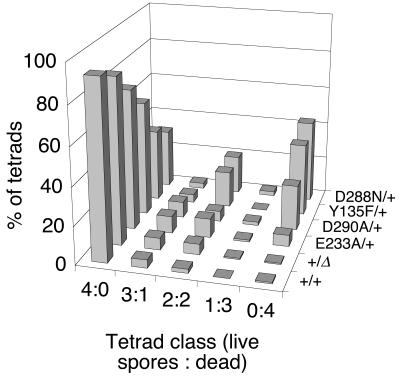
If the mutant and wild-type proteins can dimerize with one another, then the cell should contain roughly one-fourth the normal amount of wild-type Spo11p homodimer, with the remainder consisting of heterodimers and mutant homodimers, neither of which should be able to catalyze DSB formation. One explanation for the recessive nature of the DSB-defective alleles in configuration A would be that Spo11p activity is not limiting, such that a fourfold reduction in the amount of active protein has little effect on recombination frequencies. We therefore also tested the DSB-defective alleles under conditions in which Spo11p activity was slightly compromised, i.e., when the protein was tagged with an HA3His6 epitope tag (configuration B in Table 2). Strikingly, the Y135F and D288N mutations were semidominant in this configuration, with intragenic recombination frequencies reduced to roughly 10 to 50% of the levels in matched homozygous and hemizygous controls and with spore viability reduced to roughly 50 to 60% of normal (SKY335 and SKY337, respectively). In contrast, the E233A mutation was at best only weakly semidominant: a strain heterozygous for this mutation (SKY587) had no detectable defect in recombination at his4::LEU2 but had a small but statistically significant decrease in spore viability (P < 0.01). The D290A mutation, which was partially defective for recombination initiation when tagged (see above), conferred an intermediate semidominant phenotype (SKY336, Table 2). The decreased spore viability in these strains resulted mostly from an increase in the frequency of two- and zero-spore-viable tetrads, with little or no increase in one- and three-spore-viable tetrads, as expected for an increase in meiosis I nondisjunction caused by a recombination initiation defect (Fig. 4).
FIG. 4.
Spore viability patterns indicative of meiosis I nondisjunction in strains carrying semidominant configurations of SPO11 mutant alleles. Tetrads were dissected from strains with the indicated epitope-tagged alleles integrated at the SPO11 locus (configuration B in Table 2), and the frequencies of four-spore-viable, three-spore-viable, etc., tetrads were determined.
We also examined configurations in which only one of the SPO11 alleles was tagged. When the point mutant allele carried the tag, the mutation was recessive (configuration C, Table 2). In contrast, if the mutant allele was untagged, the Y135F, E233A, and D288N mutations were dominant over a tagged wild-type allele (configuration D, Table 2). Intragenic recombination frequency and spore viability were reduced to roughly 1 to 3% of the levels seen in a matched SPO11/spo11Δ control. The spo11(D290A)/spo11-HA3His6 heterozygote in configuration D was essentially normal, as expected because the untagged D290A mutant itself behaved normally.
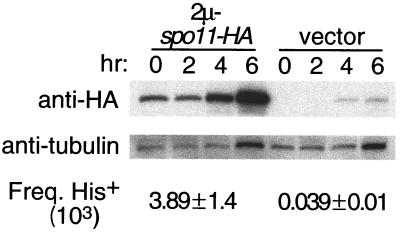
A simple explanation for these dominance patterns would be that the tag causes a decrease in the steady-state level of the protein. Unfortunately, polyclonal anti-Spo11p antibodies that are sufficiently specific to detect the protein in crude lysates are presently unavailable to us, so it has not been possible to directly measure relative levels of tagged and untagged proteins. However, this hypothesis predicts that increasing the expression of a tagged allele should at least partially overcome its recessivity. We therefore introduced spo11-HA3His6 on a high-copy-number 2μm plasmid into SKY545 [spo11(Y135F)/spo11-HA3His6], resulting in a roughly 20- to 40-fold increase in the amount of tagged Spo11p. The increased Spo11-HA3His6p expression gave significant rescue of the recombination defect, with the recombination frequency at his4::LEU2 rising ca. 100-fold (Fig. 5).
FIG. 5.
Overexpression partially rescues the recessivity of spo11-HA3His6. Freq., frequency of; hr, hours. High-copy-number 2μm (2μ) vectors pDA7 (spo11-HA3His6) or pRS426 (vector control) were introduced into strain SKY545 [heterozygous for untagged spo11(Y135F) and tagged spo11-HA3His6]. Whole-cell extracts were prepared at the indicated times after transfer to sporulation medium and were analyzed for Spo11-HA3His6p levels by Western blotting with an anti-HA antibody. Stripping and reprobing the same blot with antitubulin antibodies controlled for loading. Cells were also collected at 8 h and plated to determine the frequency of intragenic recombination between his4::LEU2 heteroalleles (mean ± standard deviation of three independent cultures).
DISCUSSION
To better understand the mechanism of DNA cleavage by yeast Spo11p, we mutagenized conserved residues predicted to lie in or near regions thought to constitute the protein's active site and/or DNA binding surface, based on extrapolation from the crystal structure of the homologous Top6A protein from M. jannaschii. Our findings are in good agreement with an independent study by Nicolas and colleagues (P.C. Varoutas, B. de Massy, C. Mezard, and A. Nicolas, personal communication). Of the residues analyzed thus far, only Tyr-135, Glu-233, and Asp-288 were intolerant of substitution, while another, Arg-131, could tolerate only a conservative change. None of these mutations affected steady-state levels of the epitope-tagged versions of the protein, so the null recombination phenotypes were not caused by destabilization of the protein. These residues thus appear to be essential for Spo11p catalytic activity. Based on studies of equivalent residues in the structurally related proteins yeast topoisomerase II and E. coli topoisomerase I (discussed further below), it seems likely that these residues in yeast Spo11p contribute directly to DNA cleavage. However, it is difficult to attribute phenotypic endpoints to specific biochemical defects, so we cannot rule out the possibility that the mutations cause defects in other functions of Spo11p instead of or in addition to DNA cleavage, such as DNA binding or protein-protein interactions.
Several studies mapping Spo11p-dependent DSBs at nucleotide resolution have shown that cleavage occurs across fairly large regions within hotspots (on the order of 100 or more bp) (15, 27, 40, 41). Breaks are distributed nonrandomly across these regions, but no clear rules have emerged for the nucleotide-level specificity of DNA cleavage, nor have any of the meiosis-specific components of the DSB-forming machinery been implicated in controlling this specificity. At least four of the mutations analyzed in this work (R131K, E235A, F260R, and D290A) significantly affected the distribution of cleavage events at the his4::LEU2 hotspot (Fig. 3), suggesting that Spo11p itself contributes to the choice of site for DSB formation. The side chains of all four residues are predicted to lie on the surface of the putative DNA binding channel (Fig. 1), so we favor the idea that these residues contact the DNA and contribute directly to an inherent cleavage site bias of Spo11p.
Three of these mutations (R131K, E235A, and D290A) gave reduced recombination levels when combined with the HA3His6 epitope tag (Table 1). In each case, the effect on DSB formation as measured directly by Southern blotting of genomic DNA was three- to fivefold more severe than that measured by intragenic recombination assays. It is likely that this discrepancy reflects a quantification artifact in the Southern blot analysis caused by the simultaneous reduction in DSB frequency and change in the distribution of DSBs. Consistent with this idea, good agreement was seen between DSB measurements and intragenic recombination frequencies for mutations that affected DSB frequency without affecting cleavage site distribution (e.g., E386A) or affected DSB distribution without affecting the frequency of cleavage (e.g., F260R).
Several mutations showed synergistic defects when combined with an HA3His6 epitope tag (Table 1). By itself, this tag renders Spo11p slightly cold sensitive (21), and overexpression studies are consistent with the idea that the tag causes a decrease in the steady-state amount of tagged Spo11p (Fig. 5). The synergistically acting point mutations did not further reduce steady-state protein levels, so it appears that these mutations cause subtle defects in Spo11p function that are only revealed in a sensitized system. All of these mutations also rendered recombination cold sensitive when present in an untagged form (Table 1), perhaps implying that Spo11p function is especially sensitive to perturbations in the assembly or disassembly of higher order protein-protein and/or protein-DNA complexes.
Mutations affecting a putative Mg2+ binding pocket.
The role of the three conserved Toprim acidic residues in phosphoryl transfer reactions is not known, but they have been proposed to contribute to catalytic activity through general acid/base chemistry or by providing a binding pocket for a divalent cation cofactor. Consistent with the latter role, these residues in crystals of M. jannaschii Top6A and E. coli DNA primase (DnaG) can coordinate a divalent cation (20, 32) and they are required for normal Mg2+ binding by E. coli topoisomerase I (45). We show here that two of these residues in yeast Spo11p (Glu-233 and Asp-288) are essential for DSB formation, whereas the third (Asp-290) is not.
The Top6A crystal structure provides a plausible structural explanation for this distinction: the carboxylate side chains of the Top6A residues equivalent to Glu-233 and Asp-288 directly coordinate the Mg2+ ion, but the Asp-290 equivalent makes indirect contacts through two bridging water molecules (32). If a similar arrangement occurs in Spo11p, then mutations affecting Glu-233 or Asp-288 would be expected to severely affect Mg2+ binding, whereas mutations affecting Asp-290 would have less effect. In particular, changing Asp-290 to asparagine should still allow hydrogen bonding to the bridging waters and provide a relatively normal coordination sphere for the bound metal, thus accounting for the almost wild-type behavior of this mutant. One implication of this interpretation is that DNA cleavage by Spo11p requires Mg2+. Interestingly, these three acidic residues are absolutely conserved in all known Spo11p/Top6A family members except Arabidopsis thaliana SPO11-3, in which the residue equivalent to Asp-290 is asparagine (24). This suggests that the detailed architecture of this metal binding pocket is conserved throughout the Spo11p/Top6A family.
Another mutation generated in this study may also impinge on Mg2+ binding: the side chain of the Top6A residue equivalent to Spo11p Tyr-292 lies in the putative DNA binding channel near the Mg2+ binding site. Replacement by an arginine at this position would be expected to place a positively charged guanidinium group very near the Mg2+ binding site, perhaps interfering with metal binding. This could account for the observed defect for the Y292R mutant (Table 1).
These studies reveal interesting similarities and differences between Spo11p and other Toprim domain-containing transesterases. For the Glu-449 residue in yeast topoisomerase II (equivalent to Spo11p Glu-233), replacement by alanine completely abolished DNA cleavage activity, similar to the case for Spo11p and consistent with the fact that topoisomerase II requires a divalent metal cofactor for DNA cleavage (29). Similarly, a glutamine substitution for Glu-214 in bacteriophage P4 α protein abolished primase activity in vitro and in vivo (46). In contrast, mutations at Glu-9 of E. coli topoisomerase I behaved somewhat differently. Replacement by alanine almost completely eliminated DNA cleavage, but glutamine supported high levels of DNA cleavage (although DNA relaxation activity was still severely compromised) (13, 44, 45). Moreover, replacement by alanine of Asp-111 (equivalent to Spo11p Asp-288) supported high levels of DNA cleavage and relaxation activity, whereas substitution at Asp-113 (equivalent to Asp-290 of Spo11p) caused a roughly 5- to 10-fold decrease in relaxation and a modest defect in DNA cleavage. These differences between Spo11p and E. coli topoisomerase I probably reflect in part that Mg2+ is required for DNA relaxation but not for DNA cleavage by topoisomerase I. They may also indicate that, despite the sequence and structural conservation of these Toprim acidic residues, the precise manner in which they interact with Mg2+ and participate in phosphoryl transfer is different in these distinct enzyme families.
Essential residues in the 5Y-CAP domain.
The 5Y-CAP structural motif is found in type IA, type IIA, and type IIB topoisomerases. In all of these enzymes, a tyrosine side chain provides the nucleophilic center that attacks the DNA phosphodiester backbone, forming a 5"-tyrosyl phosphodiester linkage. Structural alignments place this tyrosine in a similar position in three-dimensional space for these enzymes, even though the residue lies on different secondary structural elements within the 5Y-CAP domain of each class of protein (4, 32). Not surprisingly, mutation of this tyrosine abolished DNA cleavage activity for all of these enzymes (e.g., 6, 13, 33, 42).
In type IA and type IIA topoisomerases, there is an invariant arginine residue near the catalytic tyrosine on a loop between two β strands, and in the type IIB (Spo11p/Top6A) family, an invariant arginine is encoded 4 residues upstream, within the same α-helix. Despite these different primary and secondary structure positions, the arginines in all cases occupy similar positions relative to the catalytic tyrosine in three-dimensional space (4, 32). Replacements by alanine at this residue in yeast topoisomerase II or E. coli topoisomerase I reduced but did not abolish DNA cleavage activity (13, 28), whereas lysine substitutions were well tolerated in E. coli topoisomerase I and human topoisomerase IIα (13, 33). For Spo11p, DSB formation was severely compromised by an alanine substitution at Arg-131, but lysine substitution had less effect (Table 1), suggesting that this structurally conserved residue plays similar roles in all three classes of topoisomerase. Chen and Wang (13) proposed that this residue may interact with an oxygen of the scissile phosphate of the DNA backbone or that it contributes slightly to deprotonation of the catalytic tyrosine. Our finding that replacement by lysine at this position affects the site preference for DSB formation (Fig. 3) seems to support the idea that this residue makes direct contacts with DNA.
Implications of semidominance of spo11 point mutations.
As discussed above, the arrangement of the 5Y-CAP and Toprim domains in the M. jannaschii Top6A structure suggests that the dimer forms two hybrid active sites. Similar hybrid active sites are thought to form in type IIA topoisomerases (4). Consistent with this idea, yeast topoisomerase II heterodimers formed between mutant and wild-type protomers or between two different mutant protomers (e.g., one carrying a mutation in the 5Y-CAP domain and the other a mutation in the Toprim domain) assemble one mutant active site and one fully wild-type active site and are capable of nicking DNA (28, 29).
Based on these observations, we expected that catalytically inactive mutants of Spo11p would confer a semidominant negative phenotype if Spo11p formed a similar dimeric structure and if Spo11 protein was limiting for recombination initiation. We found that this prediction was met for several DSB-defective mutations (Y135F, D288N, and D290A) if both the mutant allele and the wild-type allele were epitope tagged (Table 2). Because we also found that epitope-tagged spo11 alleles were always recessive to untagged alleles and that introducing extra copies of a tagged allele on a 2μm vector could partially overcome this recessivity, it is possible that the epitope tag causes a reduction in the steady-state level of Spo11p relative to an untagged form. Taken together, these observations lead to two main conclusions. First, these findings are consistent with an oligomeric structure for Spo11p in vivo and suggest that homotypic interactions are critical for Spo11p function. Notably, these semidominant phenotypes were observed with single-copy mutant alleles, not overexpression constructs. Thus, we consider it unlikely that the semidominance is due to titration of other essential factors. Second, because the same mutations are recessive when all of the Spo11 protein in the cell is in an untagged form, we conclude that the amount of Spo11p competent to make DSBs is not a limiting factor for recombination initiation in normal cells. This idea rules out an earlier proposal, namely, that a suicide mechanism of DSB formation by Spo11p might serve to control the number of DSBs made (23).
We were interested to find that the E233A allele was only very weakly semidominant under these conditions (Table 2), even though by itself it is just as recombination defective as Y135F or D288N (Table 1). The reason for this difference is not known, but it could indicate that the E233A mutation causes defects in dimer formation or that it causes a defect in DNA binding that leads to an inability of heterodimers or mutant homodimers to compete effectively with wild-type homodimers. The latter possibility is consistent with the observation that replacement by alanine at this position in Top6A eliminates DNA binding in vitro (32).
The results presented here provide information about the functional significance of conserved domains in the Spo11p/Top6A family and the mechanism of DNA cleavage during meiotic recombination initiation. These studies also provide a battery of reagents with which to titrate Spo11p activity in vivo; this “recombination rheostat” will provide a valuable tool for further analysis of the roles of recombination in meiosis and of the mechanisms by which it is controlled and coordinated with other events in the cell.
Acknowledgments
We thank Julie Pui Yuen for constructing many of the untagged SPO11 alleles and Jennifer Giordano for her help in characterizing the altered DSB cleavage patterns. We also thank Bernard de Massy and Paul-Christophe Varoutas for discussions and sharing information prior to publication and Jerry Hurwitz, Ken Marians, and Stewart Shuman for critical comments on the manuscript.
This work was supported in part by grants to S.K. from the NIH (GM58673) and from the New York City Council Speaker's Fund for Biomedical Research.
REFERENCES
- 1.Alani, E., R. Padmore, and N. Kleckner. 1990. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61:419-436. [DOI] [PubMed] [Google Scholar]
- 2.Aravind, L., D. D. Leipe, and E. V. Koonin. 1998. Toprim—a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 26:4205-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baudat, F., K. Manova, J. P. Yuen, M. Jasin, and S. Keeney. 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6:989-998. [DOI] [PubMed] [Google Scholar]
- 4.Berger, J. M., D. Fass, J. C. Wang, and S. C. Harrison. 1998. Structural similarities between topoisomerases that cleave one or both DNA strands. Proc. Natl. Acad. Sci. USA 95:7876-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergerat, A., D. Gadelle, and P. Forterre. 1994. Purification of a DNA topoisomerase II from the hyperthermophilic archaeon Sulfolobus shibatae. A thermostable enzyme with both bacterial and eucaryal features. J. Biol. Chem. 269:27663-27669. [PubMed] [Google Scholar]
- 6.Bergerat, A., B. de Massy, D. Gadelle, P. C. Varoutas, A. Nicolas, and P. Forterre. 1997. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386:414-417. [DOI] [PubMed] [Google Scholar]
- 7.Bishop, D. K., D. Park, L. Xu, and N. Kleckner. 1992. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69:439-456. [DOI] [PubMed] [Google Scholar]
- 8.Buhler, C., D. Gadelle, P. Forterre, J. C. Wang, and A. Bergerat. 1998. Reconstitution of DNA topoisomerase VI of the thermophilic archaeon Sulfolobus shibatae from subunits separately overexpressed in Escherichia coli. Nucleic Acids Res. 26:5157-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao, L., E. Alani, and N. Kleckner. 1990. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61:1089-1101. [DOI] [PubMed] [Google Scholar]
- 10.Celerin, M., S. T. Merino, J. E. Stone, A. M. Menzie, and M. E. Zolan. 2000. Multiple roles of Spo11 in meiotic chromosome behavior. EMBO J. 19:2739-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cervantes, M. D., J. A. Farah, and G. R. Smith. 2000. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell 5:883-888. [DOI] [PubMed] [Google Scholar]
- 12.Cha, R. S., B. M. Weiner, S. Keeney, J. Dekker, and N. Kleckner. 2000. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev. 14:493-503. [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, S. J., and J. C. Wang. 1998. Identification of active site residues in Escherichia coli DNA topoisomerase I. J. Biol. Chem. 273:6050-6056. [DOI] [PubMed] [Google Scholar]
- 14.Cheng, C., P. Kussie, N. Pavletich, and S. Shuman. 1998. Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell 92:841-850. [DOI] [PubMed] [Google Scholar]
- 15.de Massy, B., V. Rocco, and A. Nicolas. 1995. The nucleotide mapping of DNA double-strand breaks at the CYS3 initiation site of meiotic recombination in Saccharomyces cerevisiae. EMBO J. 14:4589-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dernburg, A. F., K. McDonald, G. Moulder, R. Barstead, M. Dresser, and A. M. Villeneuve. 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94:387-398. [DOI] [PubMed] [Google Scholar]
- 17.Gietz, R. D., and R. A. Woods. 1998. Transformation of yeast by the lithium acetate/single-stranded carrier DNA/PEG method. Methods Microbiol. 26:53-66. [Google Scholar]
- 18.Grelon, M., D. Vezon, G. Gendrot, and G. Pelletier. 2001. AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 20:589-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kane, S., and R. Roth. 1974. Carbohydrate metabolism during ascospore development in yeast. J. Bacteriol. 118:8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keck, J. L., D. D. Roche, A. S. Lynch, and J. M. Berger. 2000. Structure of the RNA polymerase domain of E. coli primase. Science 287:2482-2486. [DOI] [PubMed] [Google Scholar]
- 21.Kee, K., and S. Keeney. Functional interactions between SPO11 and REC102 during initiation of meiotic recombination in Saccharomyces cerevisiae. Genetics, in press. [DOI] [PMC free article] [PubMed]
- 22.Keeney, S., and N. Kleckner. 1995. Covalent protein-DNA complexes at the 5" strand termini of meiosis-specific double-strand breaks in yeast. Proc. Natl. Acad. Sci. USA 92:11274-11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keeney, S., C. N. Giroux, and N. Kleckner. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88:375-384. [DOI] [PubMed] [Google Scholar]
- 24.Keeney, S. 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52:1-53. [DOI] [PubMed] [Google Scholar]
- 25.Lima, C. D., J. C. Wang, and A. Mondragon. 1994. Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature 367:138-146. [DOI] [PubMed] [Google Scholar]
- 26.Lin, Y., and G. R. Smith. 1994. Transient, meiosis-induced expression of the rec6 and rec12 genes of Schizosaccharomyces pombe. Genetics 136:769-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, J., T.-C. Wu, and M. Lichten. 1995. The location and structure of double-strand DNA breaks induced during yeast meiosis: evidence for a covalently linked DNA-protein intermediate. EMBO J. 14:4599-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, Q., and J. C. Wang. 1998. Identification of active site residues in the “GyrA” half of yeast DNA topoisomerase II. J. Biol. Chem. 273:20252-20260. [DOI] [PubMed] [Google Scholar]
- 29.Liu, Q., and J. C. Wang. 1999. Similarity in the catalysis of DNA breakage and rejoining by type IA and IIA DNA topoisomerases. Proc. Natl. Acad. Sci. USA 96:881-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahadevaiah, S. K., J. M. A. Turner, F. Baudat, E. P. Rogakou, P. de Boer, J. Blanco-Rodriguez, M. Jasin, S. Keeney, W. M. Bonner, and P. S. Burgoyne. 2001. Recombinational DNA double strand breaks in mice precede synapsis. Nat. Genet. 27:271-276. [DOI] [PubMed] [Google Scholar]
- 31.McKim, K. S., and A. Hayashi-Hagihara. 1998. mei-W68 in Drosophila melanogaster encodes a Spo11 homolog: evidence that the mechanism for initiating meiotic recombination is conserved. Genes Dev. 12:2932-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nichols, M. D., K. DeAngelis, J. L. Keck, and J. M. Berger. 1999. Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11. EMBO J. 18:6177-6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okada, Y., Y. Ito, A. Kikuchi, Y. Nimura, S. Yoshida, and M. Suzuki. 2000. Assignment of functional amino acids around the active site of human DNA topoisomerase IIα. J. Biol. Chem. 275:24630-24638. [DOI] [PubMed] [Google Scholar]
- 34.Padmore, R., L. Cao, and N. Kleckner. 1991. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell 66:1239-1256. [DOI] [PubMed] [Google Scholar]
- 35.Romanienko, P. J., and R. D. Camerini-Otero. 2000. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6:975-987. [DOI] [PubMed] [Google Scholar]
- 36.Schultz, S. C., G. C. Shields, and T. A. Steitz. 1991. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science 253:1001-1007. [DOI] [PubMed] [Google Scholar]
- 37.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, K. N., and A. Nicolas. 1998. Recombination at work for meiosis. Curr. Opin. Genet. Dev. 8:200-211. [DOI] [PubMed] [Google Scholar]
- 39.Wach, A., A. Brachat, C. Rebischung, S. Steiner, K. Pokorni, S. te Heesen, and P. Philippsen. 1998. PCR-based gene targeting in Saccharomyces cerevisiae. Methods Microbiol. 26:67-81. [Google Scholar]
- 40.Xu, F., and T. D. Petes. 1996. Fine-structure mapping of meiosis-specific double-strand DNA breaks at a recombination hotspot associated with an insertion of telomeric sequences upstream of the HIS4 locus in yeast. Genetics 143:1115-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu, L., and N. Kleckner. 1995. Sequence non-specific double-strand breaks and interhomolog interactions prior to double-strand break formation at a meiotic recombination hot spot in yeast. EMBO J. 14:5115-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yokochi, T., J. Kato, and H. Ikeda. 1996. DNA nicking by Escherichia coli topoisomerase IV with a substitution mutation from tyrosine to histidine at the active site. Genes Cells 1:1069-1075. [DOI] [PubMed] [Google Scholar]
- 43.Zenvirth, D., and G. Simchen. 2000. Meiotic double-strand breaks in Schizosaccharomyces pombe. Curr. Genet. 38:33-38. [DOI] [PubMed] [Google Scholar]
- 44.Zhu, C. X., C. J. Roche, N. Papanicolaou, A. DiPietrantonio, and Y. C. Tse-Dinh. 1998. Site-directed mutagenesis of conserved aspartates, glutamates and arginines in the active site region of Escherichia coli DNA topoisomerase I. J. Biol. Chem. 273:8783-8789. [DOI] [PubMed] [Google Scholar]
- 45.Zhu, C. X., and Y. C. Tse-Dinh. 2000. The acidic triad conserved in type IA DNA topoisomerases is required for binding of Mg(II) and subsequent conformational change. J. Biol. Chem. 275:5318-5322. [DOI] [PubMed] [Google Scholar]
- 46.Ziegelin, G., N. A. Linderoth, R. Calendar, and E. Lanka. 1995. Domain structure of phage P4 α protein deduced by mutational analysis. J. Bacteriol. 177:4333-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]