Abstract
The receptor activator of NF-κB (RANK) and its ligand RANKL are key molecules for differentiation and activation of osteoclasts. RANKL stimulates transcription factors AP-1 through mitogen-activated protein kinase (MAPK) activation, and NF-κB through IκB kinase (IKK) activation. Tumor necrosis factor receptor-associated factor 6 (TRAF6) is essential for activation of these kinases. In the interleukin-1 signaling pathway, TAK1 MAPK kinase kinase (MAPKKK) mediates MAPK and IKK activation via interaction with TRAF6, and TAB2 acts as an adapter linking TAK1 and TRAF6. Here, we demonstrate that TAK1 and TAB2 participate in the RANK signaling pathway. Dominant negative forms of TAK1 and TAB2 inhibit NF-κB activation induced by overexpression of RANK. In 293 cells stably transfected with full-length RANK, RANKL stimulation facilitates the formation of a complex containing RANK, TRAF6, TAB2, and TAK1, leading to the activation of TAK1. Furthermore, in murine monocyte RAW 264.7 cells, dominant negative forms of TAK1 and TAB2 inhibit NF-κB activation induced by RANKL and endogenous TAK1 is activated in response to RANKL stimulation. These results suggest that the formation of the TRAF6-TAB2-TAK1 complex is involved in the RANK signaling pathway and may regulate the development and function of osteoclasts.
Skeletal remodeling is a dynamic and continual process that involves the coupled events of bone formation by osteoblasts and bone resorption by osteoclasts. Osteoclasts are professional bone-resorbing polykaryons derived from hematopoietic cells of the monocyte-macrophage lineage (27, 34). The receptor activator of NF-κB (RANK) is a member of the tumor necrosis factor (TNF) receptor family and is involved in osteoclastogenesis and lymph node development (1, 10). The ligand for RANK, RANKL (also called osteoclast differentiation factor [46], TNF-related activation induced cytokine [44], and osteoprotegerin ligand [21]), is a TNF receptor family ligand that regulates the functions of dendritic cells and osteoclasts. RANKL is expressed on osteoblasts and bone marrow stromal cells, while its receptor RANK is expressed on osteoclast progenitors or mature osteoclasts. RANKL interacts with RANK via direct cell-cell contact, thereby promoting the differentiation, survival, and bone-resorbing capability of osteoclasts (reviewed in references 13 and 35). RANK interacts with members of the family of TNF receptor-associated factors (TRAFs) that mediate activation of NF-κB and c-Jun NH2-terminal kinase (JNK) (8, 11, 17, 43). Furthermore, the RANK cytoplasmic tail associates with c-Src kinase, which is responsible for the activation of Akt/PKB, a factor that has an antiapoptotic effect on osteoclasts (42). However, the proximal molecular components of RANK signal transduction and their interactions are not well understood.
The TRAF family consists of six distinct proteins, each containing a ring and zinc finger motif in their N terminus and C-terminal TRAF domains that are responsible for self-association and protein interaction. The TRAF proteins serve as cytoplasmic adapters that can interact directly with the intracellular domains of cell surface receptors, such as the TNF receptor family, and mediate signaling (2). When overexpressed in cell lines, RANK can interact with TRAF1, -2, -3, -5, and -6. Among these TRAF molecules, TRAF6 has been shown to be a pivotal component in the RANK signaling pathway. TRAF6-deficient mice exhibit severe osteopetrosis and are defective in bone remodeling and tooth eruption caused by impaired osteoclast function (22, 25).
TRAF6 also mediates NF-κB and JNK activation in the interleukin-1 (IL-1) signaling pathway (7). Recent studies have suggested a model by which the IL-1 signaling cascade is regulated. IL-1 signaling is initiated by the formation of a high-affinity complex composed of IL-1, the IL-1 receptor, and the IL-1 receptor accessory protein (12, 16, 20, 41). The intracellular adapter protein MyD88 is then recruited to the complex, where it mediates the association of IL-1 receptor-associated kinase (IRAK) with the receptor. (5, 6, 24, 40). IRAK then dissociates from the receptor complex and interacts with TRAF6, which transduces the IL-1 signal downstream, leading to NF-κB and JNK activation. Thus, TRAF6 links several families of cytokine receptors to NF-κB and JNK activation.
TAK1 is a member of the mitogen-activated protein kinase kinase kinase (MAPKKK) family and is activated by various cytokines, including the family of transforming growth factor-β ligands (45). It was previously demonstrated that TAK1 is also involved in the IL-1 signaling pathway (26). Following exposure of cells to IL-1, endogenous TAK1 is recruited to the TRAF6 complex and activated, whereupon it stimulates both JNK and NF-κB activation. Thus, TAK1 functions at the same point in the IL-1-activated signaling cascade as TRAF6. In previous studies, the yeast two-hybrid system was employed to isolate TAB2, a protein that interacts with TAK1. It was recently shown that TAB2 is also an intermediate in the IL-1 signaling pathway (36, 37). IL-1 stimulates the translocation of TAB2 from the membrane to the cytosol, where it interacts with TRAF6 and mediates its association with TAK1. These results suggest that TAB2 functions as an adapter that links TAK1 and TRAF6 in response to IL-1 and thereby mediates TAK1 activation. These results indicate that IL-1 activation of the NF-κB and JNK cascades involves the formation of a TRAF6-TAB2-TAK1 complex.
In contrast to IL-1 signaling, the mechanism of RANKL-induced signal transduction is not well understood. In this work, we report that TAK1 and TAB2 are involved in the RANK signaling pathway. We show that TRAF6, TAB2, and TAK1 assemble into the RANK complex upon ligand engagement. Furthermore, RANKL stimulation activates endogenous TAK1 activity. These results suggest that the formation of the TRAF6-TAB2-TAK1 complex with RANK is important for the RANK signaling.
MATERIALS AND METHODS
Plasmids.
The full-length human RANK cDNA was amplified by PCR from human leukocyte cDNA (Clontech). The PCR product was verified as RANK cDNA by sequencing. To generate a C-terminal Myc-tagged RANK expression vector, full-length RANK cDNA was subcloned into the HindIII-SacII site of pcDNA3.1/Myc-His(+) mammalian expression vector (Invitrogen). The expression vectors for TAK1, TAK1(K63W), TAB2, TAB2C, MyD88, MyD88C, and hemagglutinin (HA)-JNK were described previously (29, 30, 32, 36, 40). Expression plasmids for mouse TRAF6 and the dominant negative form of TRAF6 (ΔTRAF6) were generously provided by H. Nakano (Juntendo University, Japan).
Cell cultures and transfection.
RAW 264.7 and 293 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum at 37°C in 5% CO2. For the transfection studies, 293 cells were transfected with various expression vectors using Lipofectamine plus reagent (Gibco-BRL) according to the manufacturer's instructions. RAW 264.7 cells were transfected using FuGENE6 Transfection Reagent (Roche Diagnostics).
Generation of cell lines stably expressing RANKs.
To establish stable cell lines that express RANK, 293 cells were transfected with 1 μg of pcDNA3.1/Myc-His-RANK and selected in medium containing 800 μg of G418 (Geneticin; Gibco-BRL) per ml. Resistant colonies were isolated and characterized for RANKL-induced activation of NF-κB by luciferase reporter assay.
Transient-transfection and reporter assay.
Cells were plated into 12-well plates and transfected with the luciferase reporter plasmid pNFκB-Luc (Stratagene) or Ig-κ-luciferase and pRSV-β-gal (kindly provided by M. Tsuda at Toyama Medical and Pharmaceutical University). In some experiments, cells were cotransfected with multiple expression vectors and the total amount of DNA was adjusted with empty vector. After 24 h, cells were treated for 5 h with 1,000 ng of sRANKL (Pepro Tech) per ml or left untreated, and cell extracts were prepared and assayed for luciferase activity with a luciferase assay system (Wako). Relative luciferase activities were normalized to β-galactosidase activity.
Antibodies.
Polyclonal rabbit antibody to RANK (anti-RANK) was produced against bacterially expressed RANK (amino acids 1 to 212). The expression vector for the RANK extracellular domain was generated by inserting human RANK cDNA encoding amino acids 1 to 212 into the SphI-HindIII site of pQE32 (Qiagen). Polyclonal rabbit antibodies to TRAF6 (anti-TRAF6C), TAK1 (anti-TAK1), TAB1 (anti-TAB1), and TAB2 (anti-TAB2) have been described previously (26, 36). Anti-c-Myc monoclonal antibody (9E10), anti-Flag M5 monoclonal antibody (Sigma), anti-T7 monoclonal antibody (Novagen), anti-HA (Y-11) polyclonal antibody, anti-JNK1 (FL) polyclonal antibody, anti-IKKα (H-744) polyclonal antibody, and anti-Xpress polyclonal antibody (Santa Cruz) were used for immunoprecipitation and immunoblotting. Polyclonal rabbit antibody to phospho-specific p38 MAP kinase (New England Biolabs) was used to detect the phosphorylated form of p38. Purified rabbit immunoglobulin G (IgG) (Sigma) were used as control antibodies.
Immunoprecipitation assay.
293-RANK cells plated in 10-cm-diameter dishes were treated with 1,000 ng of RANKL per ml for the indicated times or left untreated. Cells were washed once with ice-cold phosphate-buffered saline and lysed in 0.3 ml of lysis buffer (25 mM HEPES [pH 7.7], 0.3 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.1% Triton X-100, 10 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol (DTT), 10 μg of aprotinin per ml, and 10 μg of leupeptin per ml). Cell lysates were then diluted with an equal volume of dilution buffer (20 mM HEPES [pH 7.7], 2.5 mM MgCl2, 0.1 mM EDTA, 0.05% Triton X-100, 10 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 1 mM DTT, 10 μg of aprotinin per ml, and 10 μg of leupeptin per ml). After centrifugation, lysates were incubated with various antibodies on ice for 1.5 h and rotated with protein G-Sepharose (Amersham Pharmacia). The beads were washed three times with washing buffer (20 mM HEPES [pH 7.7], 0.15 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.05% Triton X-100). The immunoprecipitates or whole-cell lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to Immobilon-P membranes (Millipore). The membranes were immunoblotted with various antibodies, and the primary antibodies were detected with horseradish peroxidase-conjugated antibodies to rabbit IgG (Calbiochem) or mouse IgG (Sigma) and visualized by the ECL system (Amersham Pharmacia).
In vitro kinase assay.
Endogenous TAK1, JNK, or IKK was immunoprecipitated with anti-TAK1, anti-JNK, or anti-IKK antibody, respectively, as described above. Immunoprecipitates were incubated with 30 μl of kinase buffer (20 mM HEPES [pH 7.6], 20 mM MgCl2, 2 mM DTT, 20 μM ATP, 20 mM β-glycerophosphate, 20 mM disodium ρ-nitrophenylphosphate, 0.1 mM sodium orthovanadate, and 2 μCi of [γ-32P]ATP). For the TAK1, JNK, or IKK kinase assay, 2.5 μg of bacterially expressed 6xHis-MKK6, glutathione S-transferase (GST)-c-Jun (1-79), or GST-IκB (1-54) (29) was added in each kinase buffer as a substrate. The reaction mixtures were resolved by SDS-PAGE, followed by autoradiography.
RESULTS
Effects of TAK1 and TAB2 on RANK-mediated signal transduction.
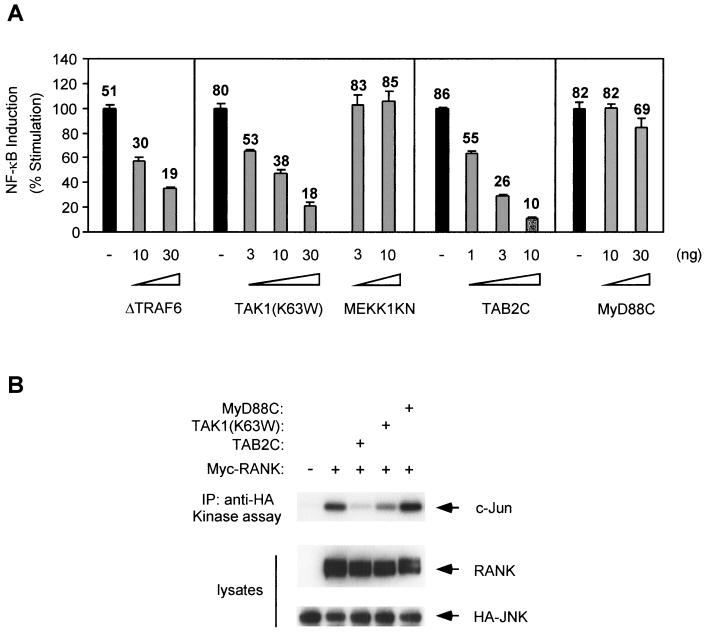
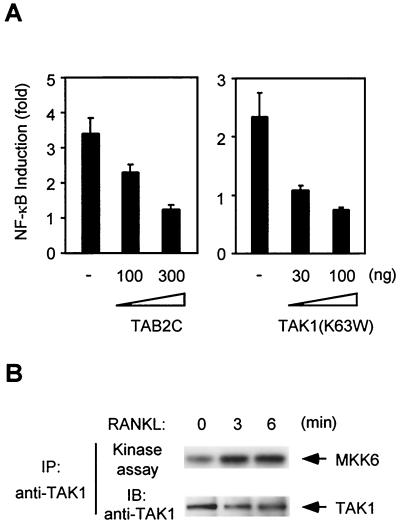
Human embryonic kidney epithelial 293 cells were transfected with a full-length RANK expression plasmid, and NF-κB activity was assayed with an NF-κB-dependent luciferase reporter gene. As reported previously (11, 43), overexpression of RANK causes a 50- to 100-fold activation of NF-κB in the absence of ligand. RANK-mediated NF-κB activation was blocked by coexpression of a truncated version of TRAF6 (ΔTRAF6) (Fig. 1A), which acts as a dominant negative inhibitor of NF-κB activation in the IL-1 pathway (7). In the IL-1 signaling pathway, formation of the TRAF6-TAB2-TAK1 complex is essential for NF-κB activation. Therefore, we investigated whether TAK1 and TAB2 participate in RANK signaling by examining the effects of dominant negative mutants on RANK-induced NF-κB activation. Overexpression of a kinase-negative mutant of TAK1, TAK1(K63W), or a truncated form of TAB2, TAB2C, was previously shown to inhibit IL-1-induced activation of NF-κB (26, 36). Similarly, we found that TAK1(K63W) and TAB2C blocked RANK-induced NF-κB activation in a dose-dependent manner (Fig. 1A). Another member of the MAPKK family, MEKK1, has been shown to be involved in TNF-α-induced NF-κB activation (14). Overexpression of a kinase-negative mutant of MEKK1 (MEKK1KN) had no effect on NF-κB activation by RANK. Moreover, the inhibitory effect was not observed when the truncated form of MyD88 (MyD88C), an inhibitor of NF-κB activation in the IL-1 pathway (40), was cotransfected (Fig. 1A). These results indicate that the dominant negative effects on RANK-induced NF-κB activation are specific for TAK1(K63W) and TAB2C.
FIG. 1.
Effects of dominant negative forms of TAK1 and TAB2 on RANK-induced NF-κB activation. (A) 293 cells were transiently transfected with pNFκB-Luc, pRSV-β-gal, and the RANK expression plasmid along with indicated amounts of ΔTRAF6, TAK1(K63W), MEKK1KN, TAB2C, or MyD88C expression plasmids. Twenty-four hours later the cells were harvested, lysed, and assayed for luciferase expression. Luciferase activity was normalized to β-galactosidase activity resulting from the cotransfected β-galactosidase gene. Data are expressed as percentages of induction. Values shown above columns represent the fold increase relative to cells transfected with vector alone. (B) 293 cells were transfected with HA-JNK and RANK expression plasmid with TAB2C, TAK1(K63W), or MyD88C expression plasmids, as indicated. After 24 h, cell lysates were immunoprecipitated with anti-HA antibody, and immunoprecipitates were subjected to in vitro kinase assays with GST-c-Jun as a substrate (upper panel). Expression levels of Myc-RANK and HA-JNK were determined by immunoblotting with anti-Myc and anti-HA antibodies (middle and lower panels).
Overexpression of RANK also induced activation of JNK (8) (Fig. 1B). Since TAK1 is known to participate in activation of both NF-κB and JNK by IL-1, we next examined whether TAK1(K63W) and TAB2C also block RANK-induced JNK activation by an in vitro kinase assay. Coexpression of TAK1(K63W) and TAB2C reduced activation of JNK induced by RANK (Fig. 1B). However, MyD88C did not block RANK-induced JNK activation (Fig. 1B). These results suggest that TAK1 and TAB2 are involved in the RANK signaling pathway.
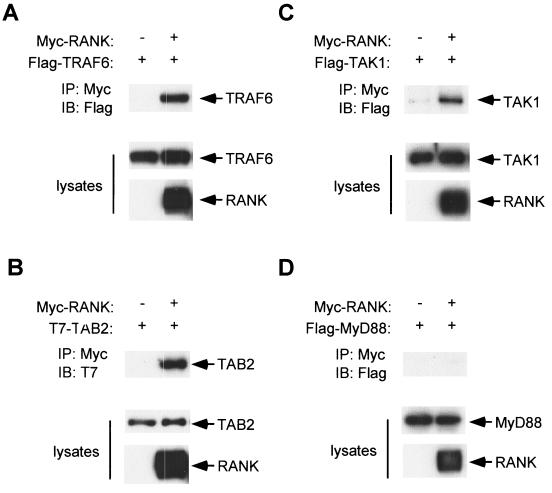
TAB2 and TAK1 interact with RANK.
When overexpressed, TRAF6 binds to RANK (8). We confirmed this interaction by coexpressing Flag-tagged TRAF6 (Flag-TRAF6) and Myc-tagged RANK (Myc-RANK) in 293 cells (Fig. 2A). We tested the potential of TAB2 and TAK1 to associate with RANK by coimmunoprecipitation experiments. Myc-RANK was coexpressed with T7-tagged TAB2 (T7-TAB2) or Flag-tagged TAK1 (Flag-TAK1) in 293 cells and immunoprecipitated with anti-Myc antibody. The immunoprecipitates were subjected to immunoblotting with anti-T7 or anti-Flag antibody, respectively. TAB2 and TAK1 were found to coprecipitate with RANK (Fig. 2B and C). These results indicate that TRAF6, TAB2, and TAK1 constitutively interact with RANK when overexpressed. In contrast to TAK1 and TAB2, Flag-tagged MyD88, which is known to be an adapter protein linking IRAK and IL-1 receptor in IL-1 signaling pathway, did not coprecipitate with RANK under the same conditions (Fig. 2D).
FIG. 2.
Interaction of RANK with TRAF6, TAB2 and TAK1. 293 cells were transiently transfected with expression plasmid encoding Flag-TRAF6 (A), T7-TAB2 (B), Flag-TAK1 (C), or Flag-MyD88 (D) in the presence of either vector plasmid or the expression plasmid for Myc-RANK as indicated. After 24 h, cell lysates were immunoprecipitated with anti-Myc antibody to detect RANK and coprecipitated TRAF6, TAB2, TAK1, or MyD88 was detected by immunoblotting with anti-Flag or anti-T7 antibody, respectively (upper panels). Expression levels of TRAF6, TAB2, TAK1, MyD88, and RANK were determined by immunoblotting (middle and lower panels).
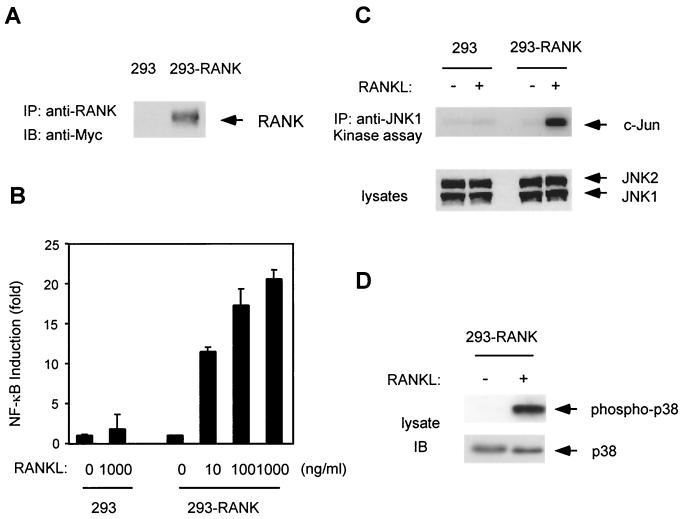
Isolation of 293 cells stably expressing RANK.
293 cells contain only small amounts of RANK and respond weakly to RANKL, as judged by NF-κB activation (11) (Fig. 3B). To elucidate the role of endogenous TAB2 and TAK1 in the RANK signaling pathway, we generated 293 cells stably expressing Myc epitope-tagged RANK (Myc-RANK). 293 cells were transfected with a Myc-RANK expression vector carrying the neomycin resistance gene and selected in medium containing G418. Twenty individual colonies were isolated and expanded. Each clone was transfected with an NF-κB-dependent luciferase reporter gene, stimulated with RANKL, and assayed for luciferase activity. We chose the clone which showed maximum-fold activation of NF-κB by RANKL. RANK expression in this clone, 293-RANK, was analyzed by immunoblotting. Whole-cell extracts were prepared from 293-RANK cells and immunoprecipitated with anti-RANK antibody. The immunoprecipitates were subjected to immunoblotting with a monoclonal antibody to c-Myc. Expression of Myc-RANK was detected in 293-RANK cells but not in parental 293 cells (Fig. 3A). 293-RANK cells also showed a dose-dependent response to RANKL in the NF-κB reporter assay, while 293 cells showed a weak response (Fig. 3B).
FIG. 3.
Characterization of 293-RANK cells. (A) Expression of Myc-tagged RANK in 293-RANK cells. Cell extracts from 293 and 293-RANK cells were immunoprecipitated with anti-RANK antibody. Immunoprecipitates were separated by SDS-PAGE, and immunoblotting was performed with antibody to c-Myc. (B) NF-κB activation by RANKL. 293 and 293-RANK cells were transiently transfected with pNFκB-Luc. After 24 h, cells were left unstimulated or stimulated with indicated concentrations of RANKL for 5 h, and luciferase activity was measured. Values represent luciferase activities relative to unstimulated cells. The 293-RANK cells had 1.8-fold basal activity of 293 cells. (C) JNK activation by RANKL. 293 and 293-RANK cells were left unstimulated or stimulated with 1,000 ng of RANKL per ml for 10 min. Cell lysates were immunoprecipitated with anti-JNK antibody, and immunoprecipitates were subjected to in vitro kinase assays with GST-c-Jun as a substrate (upper panel). Whole-cell extracts were immunoblotted with anti-JNK (lower panel). (D) Activation of p38 MAPK by RANKL. 293-RANK cells were stimulated with 1,000 ng of RANKL per ml for 7 min. Whole-cell extracts were immunoblotted with phospho-specific p38 MAP kinase antibody (upper panel) and p38 MAP kinase antibody (lower panel).
RANKL can induce activation of the JNK and p38 MAPKs (23, 44). When stimulated with RANKL, JNK activation in 293-RANK cells was observed by immunocomplex kinase assay using the GST-c-Jun protein as a substrate (Fig. 3C). The active, phosphorylated form of p38 MAPK was also detected following RANKL stimulation of 293-RANK (Fig. 3D), but not parent 293 cells (data not shown). These results indicate that NF-κB, JNK, and p38 are activated in response to RANKL stimulation in 293-RANK cells and therefore can be used to analyze endogenous signal transduction mechanisms mediated by RANKL.
RANKL induces the formation of a complex containing RANK, TRAF6, TAB2, and TAK1.
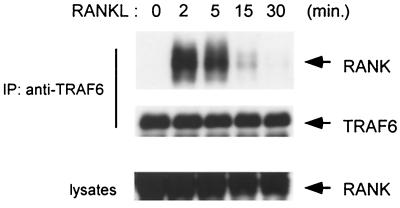
When individually overexpressed in 293 cells, TRAF6, TAB2, and TAK1 interact with RANK, even in the absence of ligand stimulation (Fig. 2). To determine whether endogenous TRAF6, TAB2, and TAK1 form complexes with RANK, we investigated their association in 293-RANK cells stimulated with RANKL. After being stimulated for various lengths of time, endogenous TRAF6 was immunoprecipitated with anti-TRAF6 antibody. The immune complexes were subjected to immunoblotting with anti-Myc antibody to detect coimmunoprecipitated RANK. Association of endogenous TRAF6 with RANK was observed in cells stimulated with RANKL, but not in the unstimulated, control cells. The presence of RANK in TRAF6 immunoprecipitates was observed starting at 2 min after RANKL treatment (Fig. 4). This is consistent with the previous observation that TRAF6 and RANK associate in a RANKL-dependent manner in dendritic cells (42). The amounts of RANK found associated with TRAF6 peaked 2 min after RANKL induction and declined steeply thereafter. These results demonstrate that RANK forms a transient complex with TRAF6 in response to RANKL stimulation.
FIG. 4.
RANKL-induced association between endogenous RANK and TRAF6. 293-RANK cells were treated with 1,000 ng of RANKL per ml for the indicated times. Cell extracts were immunoprecipitated with anti-TRAF6 antibody, and coprecipitated Myc-tagged RANK was detected by immunoblotting with anti-Myc antibody (upper panel). The amounts of TRAF6 in each immune complex were determined by immunoblotting (middle panel). Whole-cell extracts were immunoblotted with anti-Myc antibody (lower panel).
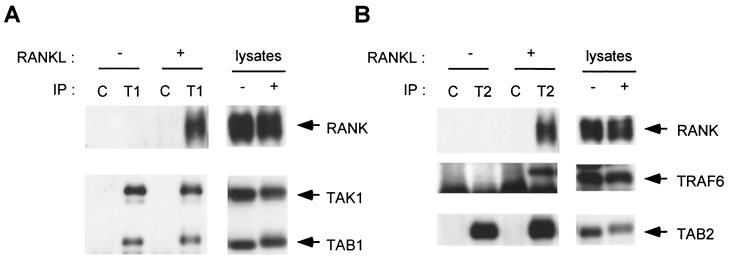
We next analyzed the interaction of endogenous TAK1 and TAB2 with RANK. No association between RANK and TAK1 or TAB2 was detected in unstimulated cells, but association was detected upon addition of RANKL (Fig. 5A and B). The kinetics of association were comparable to those between TRAF6 and RANK (data not shown). Collectively, these results suggest that RANKL transiently induces the formation of a complex composed of RANK with TRAF6, TAB2, and TAK1. TAB1, another TAK1 binding protein which is known to be a specific activator of TAK1 (31), constitutively associated with TAK1 (Fig. 5A), suggesting that TAB1 is also a component of this complex.
FIG. 5.
RANKL induces the formation of a signaling complex containing RANK, TRAF6, TAB2, and TAK1. (A) RANKL-induced association of endogenous TAK1 with RANK. 293-RANK cells were treated with 1,000 ng of RANKL per ml for 5 min or left untreated. Cell extracts were immunoprecipitated with anti-TAK1 antibody (T1) or rabbit IgG (C). Coprecipitated RANK and TAB1 were detected by immunoblotting with anti-Myc and anti-TAB1 antibodies, respectively (left, upper and lower panels). The amounts of immunoprecipitated TAK1 were determined with anti-TAK1 antibody (left, lower panel). Total amounts of RANK, TAB1, and TAK1 were determined by immunoblotting in cell lysates (right panels). (B) RANKL-induced association of endogenous TAB2 with RANK and TRAF6. 293-RANK cells were treated with 1,000 ng of RANKL per ml for 5 min or left untreated. Cell extracts were immunoprecipitated with anti-TAB2 antibody (T2) or rabbit IgG (C). Coprecipitated RANK and TRAF6 were detected by immunoblotting with anti-Myc and anti-TRAF6 antibodies, respectively (left, top and middle panels). The amounts of immunoprecipitated TAB2 were determined with anti-TAB2 antibody (left, bottom panel). Total amounts of RANK, TRAF6, and TAB2 were determined by immunoblotting in cell lysates (right panels).
RANKL-induced interaction of TRAF6 with TAB2 and TAK1.
In the IL-1 signaling pathway, TAB2 facilitates the formation of a TRAF6-TAB2-TAK1 complex by interacting with both TRAF6 and TAK1 (36). The above results raised the possibility that RANKL stimulation might induce the formation of a complex containing TRAF6, TAB2, and TAK1 with RANK in a similar manner. To test this possibility, we examined the interaction between endogenous TRAF6 and TAB2 in response to RANKL. 293-RANK cells were treated with RANKL or left untreated, cell extracts were subjected to immunoprecipitation with anti-TAB2 antibody, and immunocomplexes were probed for the presence of TRAF6 by immunoblot analysis. It was observed that RANKL treatment induced the association of TAB2 with TRAF6 (Fig. 5B).
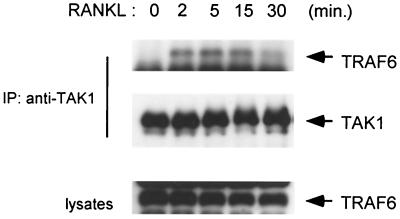
We next examined the effect of RANKL stimulation on the interaction between endogenous TRAF6 and TAK1. Cell extracts from 293-RANK cells treated with RANKL were immunoprecipitated with anti-TAK1 antibody and analyzed by immunoblotting with anti-TRAF6 antibody. Association of endogenous TAK1 with TRAF6 was observed at 2 min after RANKL treatment and subsequently released from TRAF6 at 30 min (Fig. 6). The kinetics of RANKL-induced TRAF6-TAK1 association are similar to those for RANK-TRAF6 association (Fig. 4).
FIG. 6.
RANKL-induced association of endogenous TAK1 with TRAF6. 293-RANK cells were treated with 1,000 ng of RANKL per ml for indicated times. Cell extracts were immunoprecipitated with anti-TAK1 antibody. Coprecipitated TRAF6 was detected by immunoblotting with anti-TRAF6 antibody (top panel). The amounts of TAK1 in each immune complex were determined by immunoblotting (middle panel). Total amounts of TRAF6 were determined by immunoblotting in cell lysates (bottom panel).
RANKL-induced activation of endogenous TAK1 kinase.
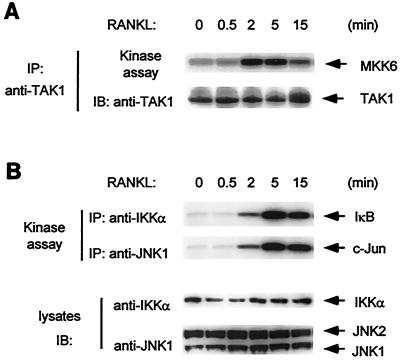
To examine whether TAK1 is involved in RANK signaling, we examined activation of endogenous TAK1 in response to RANKL stimulation. Cell extracts were prepared from 293-RANK cells, either untreated or stimulated with RANKL for various times, and subjected to immunoprecipitation with anti-TAK1 antibody. These TAK1 immunoprecipitates were assayed for TAK1 kinase activity in vitro using bacterially expressed MKK6 as a substrate. RANKL stimulation elicited a rapid but rather short-lived effect, with maximum kinase activity observed at 2 to 5 min after stimulation (Fig. 7A). TAK1 activity decreased after 15 min of treatment. The time course of TAK1 activation by RANK parallels the formation of complexes among RANK, TRAF6, TAB2, and TAK1. These results collectively suggest that RANKL stimulates the transient formation of a signaling complex containing RANK, TRAF6, TAB2, and TAK1, which in turn activates TAK1 kinase activity.
FIG. 7.
Activation of endogenous TAK1, IKK and JNK by RANKL. (A) TAK1 activation by RANKL in 293-RANK cells. 293-RANK cells were left unstimulated or stimulated with 1,000 ng of RANKL per ml for the indicated times. Cell extracts were immunoprecipitated with anti-TAK1 antibody, and immunoprecipitates were subjected to in vitro kinase assays using MKK6 as a substrate (upper panel). The amounts of TAK1 in each immune complex were determined by immunoblotting (lower panel). (B) Activation of IKK and JNK by RANKL. 293-RANK cells were stimulated with 1,000 ng of RANKL per ml for the indicated times. Cell extracts were immunoprecipitated with anti-IKKα or anti-JNK1 antibody. Kinase activities of IKK (top panel) and JNK (second panel) were measured by in vitro kinase assays with GST-IκB or GST-c-Jun as a substrate. Whole-cell extracts were immunoblotted with anti-IKKα and anti-JNK1 antibodies (third and bottom panels).
Since activation of TAK1 was shown to lead to the activation of both IKK and JNK (26), we next examined the kinetics of RANKL-induced activation of these kinases in 293-RANK cells. We observed that endogenous IKK and JNK activities reached their maximums within 5 min of RANKL stimulation (Fig. 7B). Thus, activation of IKK and JNK follows the activation of TAK1, suggesting that TAK1 is an upstream activator of IKK and JNK in the RANK signaling pathway.
TAK1 and TAB2 are involved in the RANK signaling pathway in RAW 264.7 cells.
To confirm the significance of TAK1 and TAB2 in the RANK signaling pathway, we tested the effects of TAK1(K63W) and TAB2C on RANKL-induced activation of NF-κB under physiological conditions. The mouse macrophage cell line, RAW 264.7, was chosen because this cell line is known to express RANK and to differentiate into osteoclast-like cells in response to RANKL treatment (15). RANKL activated NF-κB in RAW 264.7 cells in reporter gene assay (Fig. 8A), as reported previously (39). We found that this RANKL-induced NF-κB activation was blocked by TAK1(K63W) and TAB2C (Fig. 8A), suggesting that TAK1 and TAB2 also play important roles in RANK signaling in osteoclasts. Moreover, endogenous TAK1 was activated within 3 min after addition of RANKL in RAW 264.7 cells (Fig. 8B). This rapid activation of TAK1 was comparable to that seen in 293-RANK cells. Thus, TAK1 activation is linked to RANKL-mediated signaling in RAW 264.7 cells as well as in 293-RANK cells.
FIG. 8.
TAK1 and TAB2 are involved in the RANK signaling pathway in RAW 264.7 cells. (A) Effect of dominant negative forms of TAB2 and TAK1 on NF-κB activation by RANKL. RAW 264.7 cells were transiently transfected with Ig-κ-luciferase and pRSV-β-gal along with indicated amounts of TAB2C or TAK1(K63W) expression plasmids. Twenty-four hours later the cells were left unstimulated or stimulated with 1,000 ng of RANKL per ml for 5 h, and luciferase activity was measured. Luciferase activity was normalized to β-galactosidase activity resulting from the cotransfected β-galactosidase gene. Values represent the fold increases relative to cells unstimulated and transfected with vector alone. (B) TAK1 activation by RANKL in RAW 264.7 cells. RAW 264.7 cells were left unstimulated or stimulated with 1,000 ng of RANKL per ml for the indicated times. Cell extracts were immunoprecipitated with anti-TAK1 antibody, and immunoprecipitates were subjected to in vitro kinase assays using MKK6 as a substrate (upper panel). The amounts of TAK1 in each immune complex were determined by immunoblotting (lower panel).
DISCUSSION
RANK plays an important role in the function of osteoclasts and dendritic cells. The signal transduction pathways activated by RANK include the TRAF family of cytoplasmic adapter proteins. Studies of RANK signaling in model cell lines have demonstrated that multiple TRAF proteins bind to the RANK cytoplasmic domain and mediate activation of the NF-κB and JNK pathways (8, 43). TRAF6 gene knockout mice exhibit severe osteopetrosis and are defective in osteoclast formation (22, 25). Thus, among TRAF family members TRAF6 is the most critical for RANK signaling. However, the molecular mechanisms by which TRAF6 exerts its biological effects in the RANK pathway have not previously been defined. In addition to its role in RANK signaling, TRAF6 has also been shown to be involved in the IL-1 signal transduction pathway linked to NF-κB and JNK activation (7). Previous studies have demonstrated that TRAF6 interacts with the TAK1-associating protein TAB2 in an IL-1-dependent manner, resulting in the formation of a TRAF6-TAB2-TAK1 complex (36). Formation of this complex appears to be required for IL-1-mediated activation of NF-κB and JNK. Thus, TAB2 acts as an adapter that links TRAF6 and TAK1 and thereby mediates the activation of TAK1 in the IL-1 signaling pathway. In this study we show that RANK employs a similar mechanism.
Our evidence suggesting that TAB2 and TAK1 function in the RANKL-RANK signal transduction pathway may be summarized as follows. First, dominant negative forms of TAB2 and TAK1 block activation of NF-κB and JNK induced by overexpression of RANK. Second, in transient transfection experiments, overexpressed TAB2 and TAK1 constitutively associate with RANK. Third, in 293 cells stably expressing RANK (293-RANK), RANKL treatment induces the associations of RANK with TRAF6, RANK with TAB2, RANK with TAK1, TRAF6 with TAB2, and TRAF6 with TAK1, all with similar kinetics. This suggests that RANKL may induce the formation of a TRAF6-TAB2-TAK1 complex with RANK. Fourth, in 293-RANK cells, TAK1 activation occurs rapidly following RANKL stimulation, with kinetics that parallel the observed formation of complexes among RANK, TRAF6, TAB2, and TAK1. It therefore seems reasonable to assume that formation of the TRAF6-TAB2-TAK1 complex constitutes an early event in the activation of TAK1 by RANK. Fifth, dominant negative forms of TAB2 and TAK1 block RANKL-induced NF-κB activation in RAW 264.7 cells, which are able to differentiate into osteoclast-like cells following this stimulation. Finally, RANKL stimulation also induces activation of endogenous TAK1 in RAW 264.7 cells, suggesting that TAK1 and TAB2 are involved in the RANK signaling pathway in osteoclasts. Taken together, these results suggest a model in which RANKL stimulation facilitates the formation of a RANK-TRAF6-TAB2-TAK1 complex, leading to activation of TAK1. In this model, TRAF6 acts as a mediator between RANK and TAK1 and TAB2 functions as an adapter that links TAK1 and TRAF6. However, the intramolecular mechanisms by which TAK1 is activated remain to be elucidated. Other studies have shown that TAK1 is activated by autophosphorylation (18, 28); thus, we speculate that complex formation may cause a conformational change in the catalytic domain of TAK1, which induces its kinase activity leading to autophosphorylation. Further study is required to clarify the role of each component of this large signaling complex in TAK1 activation.
In the case of IL-1 signaling, IRAK, not TRAF6, is recruited to the activated IL-1 receptors. IRAK then mediates the formation of a TRAF6-TAB2-TAK1 complex by interacting with TRAF6 (37). On the other hand, in RANK signaling, TRAF6 is directly recruited to the activated receptor, bypassing the dependence on IRAK seen in the IL-1 pathway. Consistent with this, overexpression of RANK was able to induce NF-κB activation in IRAK-deficient cells (data not shown). Darnay et al. (9) have identified a novel TRAF6 binding motif (basic QXPXEX acidic) in RANK (340-RQMPTEDE-347) (homologous residues are underlined). Indeed, this TRAF6 binding region (positions 340 to 358) is necessary and sufficient for RANK-induced NF-κB activation (9). Interestingly, this TRAF6 binding motif is also present in the C terminus of IRAK (701-RQGPEESD-708) (9). These results suggest that TRAF6 forms a complex with TAB2 and TAK1 on IRAK and RANK in the IL-1 and RANKL signaling pathways, respectively.
Wong et al. (42) have recently demonstrated that RANKL activates the antiapoptotic Akt/PKB pathway through TRAF6 and c-Src kinase. c-Src is constitutively bound to the cytoplasmic tail of RANK and further activated by recruitment of TRAF6 to the receptor complex following RANKL engagement (42). c-Src kinase is ubiquitously expressed and activated by many pathways, including tyrosine kinase growth factor receptors, G-protein-coupled receptors, and integrin cell surface adhesion molecules (4, 38). However, targeted disruption of the c-src gene in mice leads to only one predominant phenotype, osteopetrosis, resulting from an intrinsic functional defect in osteoclast development (33). Osteoclasts from Src-deficient mice failed to form ruffled borders or resorption lacunae (3). These observations, together with our present results, suggest that in osteoclasts, RANKL stimulates the formation of a signaling complex containing c-Src, TAK1, and other molecules at the cytoplasmic tail of RANK, and thereby activates downstream signaling events leading to osteoclast differentiation, cell survival, and cytoskeletal reorganization.
TRAF6-deficient (TRAF6−/−) mice also exhibit severe osteopetrosis and are defective in osteoclast formation due to blockade of the RANKL-RANK pathway (22). Recently, Kobayashi et al. (19) have delineated functional domains of TRAF6 by expressing mutant TRAF6 transgenes at endogenous wild-type levels in TRAF6−/− cells. These authors found that the finger domain of TRAF6 is responsible for the maturation of osteoclasts and activation of TAK1. Therefore, TAK1 may play a key role in osteoclastogenesis and may be an important pharmacological target in the treatment of bone-destructive diseases, such as osteoporosis and rheumatoid arthritis. Further development of a selective TAK1 inhibitor or generation of TAK1 or TAB2 gene knockout mice may provide more information on their roles in osteoclastogenesis.
Acknowledgments
We thank H. Nakano and M. Tsuda for materials, M. Tsuda, N. Sato, and N. Yanaka for helpful discussions, and M. Lamphier for critical reading of the manuscript.
This work was supported by special grants to Advanced Research on Cancer from the Ministry of Education, Culture and Science of Japan (K.M.).
REFERENCES
- 1.Anderson, D. M., E. Maraskovsky, W. L. Billingsley, W. C. Dougall, M. E. Tometsko, E. R. Roux, M. C. Teepe, R. F. DuBose, D. Cosman, and L. Galibert. 1997. A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature 390:175-179. [DOI] [PubMed] [Google Scholar]
- 2.Arch, R. H., R. W. Gedrich, and C. B. Thompson. 1998. Tumor necrosis factor receptor-associated factors (TRAFs); a family of adapter proteins that regulates life and death. Genes Dev. 12:2821-2830. [DOI] [PubMed] [Google Scholar]
- 3.Boyce, B. F., T. Yoneda, C. Lowe, P. Soriano, and G. R. Mundy. 1992. Requirement of pp60c-src expression for osteoclasts to form ruffled borders and resorb bone in mice. J. Clin. Investig. 90:1622-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, M. T., and J. A. Cooper. 1996. Regulation, substrates and functions of src. Biochim. Biophys. Acta 1287:121-149. [DOI] [PubMed] [Google Scholar]
- 5.Burns, K., F. Martinon, C. Esslinger, H. Pahl, P. Schneider, J. L. Bodmer, F. Di Marco, L. French, and J. Tschopp. 1998. MyD88, an adapter protein involved in interleukin-1 signaling. J. Biol. Chem. 273:12203-12209. [DOI] [PubMed] [Google Scholar]
- 6.Cao, Z., W. J. Henzel, and X. Gao. 1996. IRAK: a kinase associated with the interleukin-1 receptor. Science 271:1128-1131. [DOI] [PubMed] [Google Scholar]
- 7.Cao, Z., J. Xiong, M. Takeuchi, T. Kurama, and D. V. Goeddel. 1996. TRAF6 is a signal transducer for interleukin-1. Nature 383:443-446. [DOI] [PubMed] [Google Scholar]
- 8.Darnay, B. G., V. Haridas, J. Ni, P. A. Moore, and B. B. Aggarwal. 1998. Characterization of the intracellular domain of receptor activator of NF-κB (RANK). Interaction with tumor necrosis factor receptor-associated factors and activation of NF-κB and c-Jun N-terminal kinase. J. Biol. Chem. 273:20551-20555. [DOI] [PubMed] [Google Scholar]
- 9.Darnay, B. G., J. Ni, P. A. Moore, and B. B. Aggarwal. 1999. Activation of NF-κB by RANK requires tumor necrosis factor receptor-associated factor (TRAF) 6 and NF-κB-inducing kinase. Identification of a novel TRAF6 interaction motif. J. Biol. Chem. 274:7724-7731. [DOI] [PubMed] [Google Scholar]
- 10.Dougall, W. C., M. Glaccum, K. Charrier, K. Rohrbach, K. Brasel, T. De Smedt, E. Daro, J. Smith, M. E. Tometsko, C. R. Maliszewski, A. Armstrong, V. Shen, S. Bain, D. Cosman, D. Anderson, P. J. Morrissey, J. J. Peschon, and J. Schuh. 1999. RANK is essential for osteoclast and lymph node development. Genes Dev. 13:2412-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galibert, L., M. E. Tometsko, D. M. Anderson, D. Cosman, and W. C. Dougall. 1998. The involvement of multiple tumor necrosis factor receptor (TNFR)-associated factors in the signaling mechanisms of receptor activator of NF-κB, a member of the TNFR superfamily. J. Biol. Chem. 273:34120-34127. [DOI] [PubMed] [Google Scholar]
- 12.Greenfeder, S. A., P. Nunes, L. Kwee, M. Labow, R. A. Chizzonite, and G. Ju. 1995. Molecular cloning and characterization of a second subunit of the interleukin 1 receptor complex. J. Biol. Chem. 270:13757-13765. [DOI] [PubMed] [Google Scholar]
- 13.Hofbauer, L. C., S. Khosla, C. R. Dunstan, D. L. Lacey, W. J. Boyle, and B. L. Riggs. 2000. The roles of osteoprotegerin and osteoprotegerin ligand in the paracrine regulation of bone resorption. J. Bone Miner. Res. 15:2-12. [DOI] [PubMed] [Google Scholar]
- 14.Hirano, M., S. Osada, T. Aoki, S. Hirai, M. Hosaka, J. Inoue, and S. Ohno. 1996. MEK kinase is involved in tumor necrosis factor α-induced NF-κB activation and degradation of IκB-α. J. Biol. Chem. 271:13234-13238. [DOI] [PubMed] [Google Scholar]
- 15.Hsu, H., D. L. Lacey, C. R. Dunstan, I. Solovyev, A. Colombero, E. Timms, H. L. Tan, G. Elliott, M. J. Kelley, I. Sarosi, L. Wang, X. Z. Xia, R. Elliott, L. Chiu, T. Black, S. Scully, C. Capparelli, S. Morony, G. Shimamoto, M. B. Bass, and W. J. Boyle. 1999. Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc. Natl. Acad. Sci. USA 96:3540-3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, J., X. Gao, S. Li, and Z. Cao. 1997. Recruitment of IRAK to the interleukin 1 receptor complex requires interleukin 1 receptor accessory protein. Proc. Natl. Acad. Sci. USA 94:12829-12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, H. H., D. E. Lee, J. N. Shin, Y. S. Lee, Y. M. Jeon, C. H. Chung, J. Ni, B. S. Kwon, and Z. H. Lee. 1999. Receptor activator of NF-κB recruits multiple TRAF family adaptors and activates c-Jun N-terminal kinase. FEBS Lett. 443:297-302. [DOI] [PubMed] [Google Scholar]
- 18.Kishimoto, K., K. Matsumoto, and J. Ninomiya-Tsuji. 2000. TAK1 mitogen-activated protein kinase kinase kinase is activated by autophosphorylation within its activation loop. J. Biol. Chem. 275:7359-7364. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi, N., Y. Kadono, A. Naito, K. Matsumoto, T. Yamamoto, S. Tanaka, and J. Inoue. 2001. Segregation of TRAF6-mediated signaling pathways clarifies its role in osteoclastogenesis. EMBO J. 20:1271-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korherr, C., R. Hofmeister, H. Wesche, and W. Falk. 1997. A critical role for interleukin-1 receptor accessory protein in interleukin-1 signaling. Eur. J. Immunol. 27:262-267. [DOI] [PubMed] [Google Scholar]
- 21.Lacey, D. L., E. Timms, H. L. Tan, M. J. Kelley, C. R. Dunstan, T. Burgess, R. Elliott, A. Colombero, G. Elliott, S. Scully, H. Hsu, J. Sullivan, N. Hawkins, E. Davy, C. Capparelli, A. Eli, Y. X. Qian, S. Kaufman, I. Sarosi, V. Shalhoub, G. Senaldi, J. Guo, J. Delaney, and W. J. Boyle. 1998. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell 93:165-176. [DOI] [PubMed] [Google Scholar]
- 22.Lomaga, M. A., W. C. Yeh, I. Sarosi, G. S. Duncan, C. Furlonger, A. Ho, S. Morony, C. Capparelli, G. Van, S. Kaufman, A. van der Heiden, A. Itie, A. Wakeham, W. Khoo, T. Sasaki, Z. Cao, J. M. Penninger, C. J. Paige, D. L. Lacey, C. R. Dunstan, W. J. Boyle, D. V. Goeddel, and T. W. Mak. 1999. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 13:1015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsumoto, M., T. Sudo, T. Saito, H. Osada, and M. Tsujimoto. 2000. Involvement of p38 mitogen-activated protein kinase signaling pathway in osteoclastogenesis mediated by receptor activator of NF-κB ligand. J. Biol. Chem. 275:31155-31161. [DOI] [PubMed] [Google Scholar]
- 24.Muzio, M., J. Ni, P. Feng, and V. M. Dixit. 1997. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science 278:1612-1615. [DOI] [PubMed] [Google Scholar]
- 25.Naito, A., S. Azuma, S. Tanaka, T. Miyazaki, S. Takaki, K. Takatsu, K. Nakao, K. Nakamura, M. Katsuki, T. Yamamoto, and J. Inoue. 1999. Severe osteopetrosis, defective interleukin-1 signalling and lymph node organogenesis in TRAF6-deficient mice. Genes Cells 4:353-362. [DOI] [PubMed] [Google Scholar]
- 26.Ninomiya-Tsuji, J., K. Kishimoto, A. Hiyama, J. Inoue, Z. Cao, and K. Matsumoto. 1999. The kinase TAK1 can activate the NIK-IκB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature 398:252-256. [DOI] [PubMed] [Google Scholar]
- 27.Roodman, G. D. 1996. Advances in bone biology: the osteoclast. Endocr. Rev. 17:308-332. [DOI] [PubMed] [Google Scholar]
- 28.Sakurai, H., H. Miyoshi, J. Mizukami, and T. Sugita. 2000. Phosphorylation-dependent activation of TAK1 mitogen-activated protein kinase kinase kinase by TAB1. FEBS Lett. 474:141-145. [DOI] [PubMed] [Google Scholar]
- 29.Sakurai, H., H. Miyoshi, W. Toriumi, and T. Sugita. 1999. Functional interactions of transforming growth factor β-activated kinase 1 with IκB kinases to stimulate NF-κB activation. J. Biol. Chem. 274:10641-10648. [DOI] [PubMed] [Google Scholar]
- 30.Sakurai, H., N. Shigemori, K. Hasegawa, and T. Sugita. 1998. TGF-β-activated kinase 1 stimulates NF-κB activation by an NF-κB-inducing kinase-independent mechanism. Biochem. Biophys. Res. Commun. 243:545-549. [DOI] [PubMed] [Google Scholar]
- 31.Shibuya, H., K. Yamaguchi, T. Shirakabe, A. Tonegawa, Y. Gotoh, N. Ueno, K. Irie, E. Nishida, and K. Matsumoto. 1996. TAB1: an activator of the TAK1 MAPKKK in TGF-β signal transduction. Science 272:1179-1182. [DOI] [PubMed] [Google Scholar]
- 32.Shirakabe, K., K. Yamaguchi, H. Shibuya, K. Irie, S. Matsuda, T. Moriguchi, Y. Gotoh, K. Matsumoto, and E. Nishida. 1997. TAK1 mediates the ceramide signaling to stress-activated protein kinase/c-Jun N-terminal kinase. J. Biol. Chem. 272:8141-8144. [DOI] [PubMed] [Google Scholar]
- 33.Soriano, P., C. Montgomery, R. Geske, and A. Bradley. 1991. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell 64:693-702. [DOI] [PubMed] [Google Scholar]
- 34.Suda, T., I. Nakamura, E. Jimi, and N. Takahashi. 1997. Regulation of osteoclast function. J. Bone Miner. Res. 12:869-879. [DOI] [PubMed] [Google Scholar]
- 35.Suda, T., N. Takahashi, N. Udagawa, E. Jimi, M. T. Gillespie, and T. J. Martin. 1999. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr. Rev. 20:345-357. [DOI] [PubMed] [Google Scholar]
- 36.Takaesu, G., S. Kishida, A. Hiyama, K. Yamaguchi, H. Shibuya, K. Irie, J. Ninomiya-Tsuji, and K. Matsumoto. 2000. TAB2, a novel adaptor protein, mediates activation of TAK1 MAPKKK by linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Mol. Cell 5:649-658. [DOI] [PubMed] [Google Scholar]
- 37.Takaesu, G., J. Ninomiya-Tsuji, S. Kishida, X. Li, G. R. Stark, and K. Matsumoto. 2001. Interleukin-1 (IL-1) receptor-associated kinase leads to activation of TAK1 by inducing TAB2 translocation in the IL-1 signaling pathway. Mol. Cell. Biol. 21:2475-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas, S. M., and J. S. Brugge. 1997. Cellular functions regulated by Src family kinases. Annu. Rev. Cell. Dev. Biol. 13:513-609. [DOI] [PubMed] [Google Scholar]
- 39.Wei, S., S. L. Teitelbaum, M. W. Wang, and F. P. Ross. 2001. Receptor activator of nuclear factor-κB ligand activates nuclear factor-κB in osteoclast. Endocrinology 142:1290-1295. [DOI] [PubMed] [Google Scholar]
- 40.Wesche, H., W. J. Henzel, W. Shillinglaw, S. Li, and Z. Cao. 1997. MyD88: an adapter that recruits IRAK to the IL-1 receptor complex. Immunity 7:837-847. [DOI] [PubMed] [Google Scholar]
- 41.Wesche, H., C. Korherr, M. Kracht, W. Falk, K. Resch, and M. U. Martin. 1997. The interleukin-1 receptor accessory protein (IL-1RAcP) is essential for IL-1-induced activation of interleukin-1 receptor-associated kinase (IRAK) and stress-activated protein kinases (SAP kinases). J. Biol. Chem. 272:7727-7731. [DOI] [PubMed] [Google Scholar]
- 42.Wong, B. R., D. Besser, N. Kim, J. R. Arron, M. Vologodskaia, H. Hanafusa, and Y. Choi. 1999. TRANCE, a TNF family member, activates Akt/PKB through a signaling complex involving TRAF6 and c-Src. Mol. Cell 4:1041-1049. [DOI] [PubMed] [Google Scholar]
- 43.Wong, B. R., R. Josien, S. Y. Lee, M. Vologodskaia, R. M. Steinman, and Y. Choi. 1998. The TRAF family of signal transducers mediates NF-κB activation by the TRANCE receptor. J. Biol. Chem. 273:28355-28359. [DOI] [PubMed] [Google Scholar]
- 44.Wong, B. R., J. Rho, J. Arron, E. Robinson, J. Orlinick, M. Chao, S. Kalachikov, E. Cayani, F. S. Bartlett, W. N. Frankel, S. Y. Lee, and Y. Choi. 1997. TRANCE is a novel ligand of the tumor necrosis factor receptor family that activates c-Jun N-terminal kinase in T cells. J. Biol. Chem. 272:25190-25194. [DOI] [PubMed] [Google Scholar]
- 45.Yamaguchi, K., K. Shirakabe, H. Shibuya, K. Irie, I. Oishi, N. Ueno, T. Taniguchi, E. Nishida, and K. Matsumoto. 1995. Identification of a member of the MAPKKK family as a potential mediator of TGF-β signal transduction. Science 270:2008-2011. [DOI] [PubMed] [Google Scholar]
- 46.Yasuda, H., N. Shima, N. Nakagawa, K. Yamaguchi, M. Kinosaki, S. Mochizuki, A. Tomoyasu, K. Yano, M. Goto, A. Murakami, E. Tsuda, T. Morinaga, K. Higashio, N. Udagawa, N. Takahashi, and T. Suda. 1998. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc. Natl. Acad. Sci. USA 95:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]