Abstract
The human immunodeficiency virus type 1 (HIV-1) Tat protein activates transcription elongation by stimulating the Tat-activated kinase (TAK/p-TEFb), a protein kinase composed of CDK9 and its cyclin partner, cyclin T1. CDK9 is able to hyperphosphorylate the carboxyl-terminal domain (CTD) of the large subunit of RNA polymerase during elongation. In addition to TAK, the transcription elongation factor Spt5 is required for the efficient activation of transcriptional elongation by Tat. To study the role of Spt5 in HIV transcription in more detail, we have developed a three-stage Tat-dependent transcription assay that permits the isolation of active preinitiation complexes, early-stage elongation complexes, and Tat-activated elongation complexes. Spt5 is recruited in the transcription complex shortly after initiation. After recruitment of Tat during elongation through the transactivation response element RNA, CDK9 is activated and induces hyperphosphorylation of Spt5 in parallel to the hyperphosphorylation of the CTD of RNA polymerase II. However, immunodepletion experiments demonstrate that Spt5 is not required for Tat-dependent activation of the kinase. Chase experiments using the Spt5-depleted extracts demonstrate that Spt5 is not required for early elongation. However, Spt5 plays an important role in late elongation by preventing the premature dissociation of RNA from the transcription complex at terminator sequences and reducing the amount of polymerase pausing at arrest sites, including bent DNA sequences. This novel biochemical function of Spt5 is analogous to the function of NusG, an elongation factor found in Escherichia coli that enhances RNA polymerase stability on templates and shows sequence similarity to Spt5.
Transcription from the human immunodeficiency virus (HIV) promoter is regulated both at the level of initiation and elongation (for recent reviews, see references 14, 26, 48, and 54). In activated T cells and other permissive cell types, initiation of HIV transcription is very efficient, but production of full-length transcripts requires the viral regulatory protein Tat. In the absence of Tat, the majority of RNA polymerase II complexes that initiate transcription at the HIV promoter disengage from the template near to the promoter (24, 35). Tat strongly activates RNA polymerase processivity and permits the synthesis of full-length HIV transcripts both in vivo and in cell-free transcription systems (11, 17, 27, 28, 39, 40, 50, 53).
Tat is an RNA-binding protein that is recruited to early HIV transcription complexes by binding to the transactivation response element (TAR), an RNA stem-loop structure that is located at the 5" end of HIV transcripts (5, 12, 28, 51, 52). The interaction between Tat and TAR involves not only the binding of Tat to TAR RNA but also its association with the Tat-activated kinase (TAK) (20, 38, 69), a protein kinase that is able to phosphorylate the carboxyl-terminal domain (CTD) of the large subunit of RNA polymerase II (21, 43, 65).
TAK is composed of a kinase subunit, CDK9, and its cyclin partner, cyclin T1 (CycT1) (38, 60, 69). In addition to regulating CDK9, CycT1 is able to increase the affinity of Tat for TAR RNA (60). In the absence of CycT1, the recognition sequence of Tat on TAR is restricted to the region surrounding a trinucleotide bulge located near the apex of the TAR RNA stem-loop structure. In the presence of CycT1, the recognition sequence on TAR RNA is extended to include the apical loop sequence (16, 60).
Formation of the quaternary complex between CDK9, CycT1, Tat, and TAR RNA induces conformational changes in the enzyme complex that result in the constitutive activation of the CDK9 kinase (6, 8, 13, 16, 23, 34). The CDK9 kinase is then able to hyperphosphorylate the CTD of the RNA polymerase, resulting in the formation of a novel and unusually highly phosphorylated form of the polymerase which we have called RNA polymerase IIo* (21). Although experimental evidence demonstrating that RNA polymerase processivity is regulated by CTD phosphorylation has not yet been presented, it seems likely that CTD modification has a direct influence on elongation.
Modification of the transcription complex by Tat and TAK to make it more processive also requires specific cofactors that have been identified using partially reconstituted cell-free transcription systems (61, 67, 68). One of the best characterized of these factors is Spt5, a subunit of the 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) sensitivity-inducing factor (DSIF). In normal transcription, DSIF works in concert with the negative elongation factor (NELF) to induce polymerase pausing near promoter start sites (57-59, 62, 63). This pausing event can be enhanced by DRB and reversed by a CDK9-dependent phosphorylation event. In addition to playing a role in early transcription, Spt5 may act as a generalized elongation factor. In an important experiment demonstrating a direct role for Spt5 in HIV transcription, Gaynor and coworkers (22, 61) showed that immunodepletion of Spt5 from transcription extracts strongly inhibited Tat-activated transcription but had only minimal effects on basal transcription. A further link between Tat and Spt5 comes from very recent studies demonstrating that Spt5 can be phosphorylated by CDK9 (22, 31, 47).
Although the evidence that Spt5 is needed as a Tat cofactor is compelling, one limitation of the published studies is that they did not distinguish between the possibilities that Spt5 and CDK9 might play distinct biochemical roles in promoting the stability and/or processivity of Tat-activated transcription complexes or, alternatively, whether Spt5 is required for the activation or regulation of CDK9. To examine the roles of Spt5, CDK9, and Tat in HIV transcription in greater detail, we used an immobilized DNA template assay (21, 28) to isolate transcription complexes paused during three distinct phases of the transcription cycle. The results demonstrate that Spt5 plays a distinct role in transcription elongation by promoting the stability of transcription complexes at terminator sequences. This biochemical function is analogous to the role of the Escherichia coli RNA polymerase-binding protein NusG, a prokaryotic elongation factor that is able to stabilize prokaryotic RNA polymerase during elongation (18, 45).
MATERIALS AND METHODS
Template DNAs.
The templates contain the entire HIV long terminal repeat (LTR) from HIVNL-4-3 (1). PW1 carries the wild-type (wt) TAR region. Mutations in the Tat-binding site (G26·C39 to C·G; mGC) or in the loop (G32-34 to UUU; mLG) were described previously (11). pH3.3, which contains two tandem bent DNA arrest sequences (4, 30), was constructed by inserting the sequence 5"-CTAGCATTTTTAAAAGAGGGACGTTTTTTTCCCTTTTTTGGAGAGGCGGAAACTTGATGT-3" between the NheI and AccI sites flanking the stem-loop terminator sequence of pW1. The control plasmid was created by replacing the terminator sequence by a DNA sequence of equivalent length from the corresponding region of the HIV genome (5"-GAGCGTCGGTATTAAGCGGGGGAGAATTAGATAAGT-3").
Biotinylation of DNA and binding to streptavidin beads.
After cleavage with XbaI, template DNAs were 5" and 3" end labeled with biotin-16-dUTP (Roche) using Klenow enzyme (Roche). The free biotin-dUTP was removed from the biotinylated DNA by gel filtration using Sephadex G-50 (Pharmacia). Biotin-DNA was absorbed onto 50 μl of streptavidin magnetic beads (Dynabeads M280 streptavidin; Dynal). The beads were prewashed in EBCD wash buffer (50 mM Tris-HCl [pH 8.0], 120 mM NaCl, 0.5% NP-40, 5 mM dithiothreitol [DTT], 4 mM MgCl2). DNA was incubated on ice for 5 min before being washed in EBCD to remove unbound DNA. The final amount of DNA bound on the beads was 0.5 μg.
Cell-free full transcription reactions.
Streptavidin-bound DNA was incubated in a 40-μl reaction containing final concentrations (including the nuclear extract) of 20 mM HEPES (pH 7.9), 3 mM DTT, 11.25 μM ZnSO4, 4 mM MgCl2, 75 mM KCl, 50 μM ATP, 50 μM CTP, 50 μM GTP, 5 μM UTP, 2.5 μg of creatine kinase per ml, 10 μM creatine phosphate, and 1 μg of poly(dI-dC) (Roche), in the presence of 20 μl of HeLa nuclear extract and 5 to 10 μCi of [α-32P]UTP. Recombinant Tat (20 ng) and Lac repressor protein (100 ng) were added where indicated. The reaction mixtures were incubated for 20 min at 30°C and then washed twice with 250 μl of EBCD buffer containing 0.1% Sarkosyl. The immobilized templates were separated by magnetic concentration before being resuspended in 50 μl of 10 mM Tris-HCl (pH 7.4) and 1 mM EDTA. The bound and free fractions collected after magnetic separation were treated by the same procedure, and equivalent amounts of the two fractions were analyzed.
To analyze RNA transcripts, 50 μl of stop buffer (300 mM sodium acetate, 1% SDS, 20 mM EDTA, 0.1 μg of tRNA per μl) was added to each sample and the samples were subjected to phenol extraction and ethanol precipitation. The precipitated nucleic acids were dissolved in 10 μl of RNA loading buffer (80% [vol/vol] formamide, 10 mM Tris-HCl [pH 8.0], 0.1 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol), denatured at 95°C for 5 min, and analyzed by electrophoresis on a 6% polyacrylamide gel containing 7 M urea and TBE buffer (90 mM Tris base, 89 mM boric acid, 2.5 mM EDTA).
To analyze the protein constituents of the transcription complexes, washed beads were mixed with sodium dodecyl sulfate (SDS) loading buffer (50 mM Tris-HCl [pH 6.8], 2.5% SDS, 100 mM β-mercaptoethanol, 10% glycerol, 0.02% bromophenol blue), heated to 95°C for 5 min, and loaded onto appropriate precast protein gels (Novex/Invitrogen).
Three-stage transcription reactions.
For the three-stage transcription assays, the HeLa nuclear extract was first treated with 20 U of hexokinase (HK) per ml and 25 mM glucose for 5 min at 30°C to remove endogenous ATP. Preinitiation complexes (PIC) were prepared by incubating 0.5 μg of bound DNA for 10 min at 30°C in 40-μl reaction mixtures containing 20 mM HEPES (pH 7.9), 3 mM DTT, 11.25 μM ZnSO4, 4 mM MgCl2, 50 mM KCl, 1 μg of poly(dI-dC), and 20 μl of HK- and glucose-treated HeLa nuclear extract. To permit transcription initiation in the absence of ATP, 50 μM dATP was added as a substitute phosphate donor.
After formation of the preinitiation complexes, early transcription complexes paused at the uridine residue at +14 were prepared by adding 10 μCi of [α-32P]UTP, 50 μM CTP, 50 μM GTP, 2.5 μg of creatine kinase per ml, and 10 μM creatine phosphate to the PIC reaction mixtures and incubating them for a further 10 min at 30°C. Under these labeling conditions, we routinely observed paused polymerases between +1 and +14 which arose due to the low levels of UTP. To circumvent this problem, we followed the initial reaction by a 2-min incubation in the presence of 125 μM UTP. At the end of the elongation reaction, the beads carrying complexes at +14 were washed once with 250 μl of EBCD buffer.
For elongation from +14 to the end of the template, the beads were resuspended in 40 μl of buffer containing 20 mM HEPES (pH 7.9), 3 mM DTT, 11.25μM ZnSO4, 4 mM MgCl2, 100 mM KCl, 2.5 μg of creatine kinase per ml, 10 μM creatine phosphate, 5 μM UTP, and 50 μM each ATP, CTP, and GTP in the absence or presence of 10 μl of fresh nuclear extract and incubated for 15 min at 30°C. Where indicated, Tat (20 ng) was added during this chase.
Aliquots corresponding to one-fifth of the reaction mixture were removed following each step (PIC, +14, or elongation) for analysis of RNA and protein. The beads were washed twice with EBCD buffer, and RNA and protein analysis was carried out as described above.
Preparation of recombinant p65 protein.
A recombinant baculovirus expressing polyhistidine-tagged p65 was provided by J Hiscott (36). A 350-ml volume of Sf9 cells (1.2 × 106/ml) was infected at a final concentration of 1% first-passage virus and incubated for 3 days at 27°C. The cell pellet was resuspended in 10 ml of homogenization buffer (10 mM Tris-HCl [pH 7.5], 500 mM NaCl, 0.1% NP-40, 15 mM imidazole, 2 mM 2-mercaptoethanol, 2 mM phenylmethylsulfonyl fluoride [PMSF], 10% glycerol, complete protease inhibitor cocktail [Boehringer-Mannheim]), and the cells were then lysed using a Dounce homogenizer (A pestle). After centrifugation at 15,000 rpm in an SS-34 rotor for 10 min, the cleared cell lysate was applied to a Quick 300 Ni-nitrilotriacetic acid syringe column (Novagen). The column was washed twice with wash buffer (10 mM Tris-HCl [pH 7.5], 200 mM NaCl, 0.2% NP-40, 15 mM imidazole, 2 mM 2-mercaptoethanol, 2 mM PMSF, 10% glycerol), and the recombinant protein was then eluted with 1 ml of elution buffer (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 0.1% NP-40, 250 mM imidazole, 2 mM 2-mercaptoethanol, 2 mM PMSF, 10% glycerol). The eluted protein was then dialyzed against NDBZ buffer (20 mM HEPES [pH 7.9], 100 mM KCl, 2 mM MgCl2, 10μM ZnSO4, 10% glycerol, 2 mM DTT) and stored in liquid nitrogen.
Dephosphorylation of elongation complexes.
Elongation complexes bound on magnetic beads were obtained as described in the previous section and purified by washing the beads twice with EBCD buffer containing 0.1% Sarkosyl. Complexes were resuspended in 10 mM HEPES (pH 7.9), 2 mM DTT, 6.25 μM ZnSO4, and 4 mM MgCl2 and incubated with 2.5 U of protein phosphatase 1 for 1 h at 30°C, with frequent mixing.
Antibodies and Western blot analysis.
Protein samples were loaded onto Novex precast gels. For the RNA polymerase II and Spt5 analysis, we used 3 to 8% Tris-acetate gels. For the other proteins, we used either 10 or 4 to 12% Bis-Tris gels. After SDS-polyacrylamide gel electrophoresis, proteins were transferred to nitrocellulose membranes (Schleicher & Schuell) and probed with appropriate antibodies. Spt5 was detected using either a polyclonal antibody directed against the terminal 852 to 1,087 amino acids of the protein (kindly supplied by R. Gaynor) or a monoclonal antibody directed against amino acids 866 to 985 (anti-DSIF-p160; BD Transduction Laboratories). The LacR monoclonal antibody (NL1.1B2.10) has been described previously (28, 29). Finally, the following polyclonal antibodies were supplied by Santa Cruz Biotechnology: RNA polymerase II (N-20), CDK9 (H-169), cyclin T1 (C-20), CDK7 (C-19), cyclin H (D-10), TAFII250 (6B3), and RAP74 (N-16).
Immunodepletion of nuclear extracts.
Protein A-Sepharose beads were washed twice with phosphate-buffered saline (PBS) (125 mM NaCl, 10 mM Na2PO4 [pH 7.0]), saturated with bovine serum albumin for 30 min, and then washed twice again with PBS before being incubated with antibodies for 1 h at 4°C with rolling mixing. Between 20 and 40 μg of antibodies was bound to 100 μl of beads. The antibodies used were polyclonal anti-CDK7 (30 μg of C-19), a mixture of equal amounts of polyclonal anti-CDK9 (20 μg of H-169) and affinity-purified polyclonal anti-CycT1 (20 μg), monoclonal anti-DSIF-p160 (20 μg) or control rabbit immunoglobulin G (20 μg to 40 μg [Santa Cruz Biotechnology]). For Spt5 and TAK codepletion, 20 μg of each antibody (Spt5, CDK9, and CycT1) was used per 100 μl of beads. The beads were washed twice with PBS buffer to remove the free antibodies.
To deplete 100 μl of nuclear extract, four sequential depletions using 25 μl of antibody bound to beads were performed. The initial depletion mixture was incubated for 2 h, and the subsequent depletions were incubated for 1 h each. After the final round of depletion, the nuclear extract was collected and incubated for 20 min with 10 to 20 μl of bovine serum albumin-saturated protein A-Sepharose beads to remove remaining free antibodies from the extract. Beads were collected by centrifugation, and the nuclear extract was used immediately in the transcription assays or frozen in liquid nitrogen. The efficiency and specificity of the depletions were checked by analysis of the nuclear extract by Western blotting with various antibodies.
RESULTS
Experimental design.
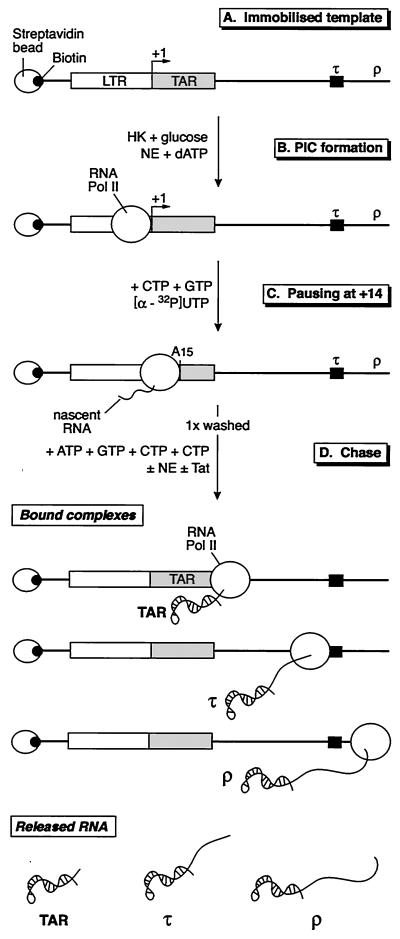
In the experiments described below, we employed a three-stage transcription system that permits the sequential analysis of preinitiation complexes, complexes paused during early elongation, and complexes paused after elongation through TAR (Fig. 1). This experimental system is based on our previous studies using immobilized DNA templates to purify preinitiation and elongation transcription complexes in vitro and to analyze their functional characteristics (21, 28).
FIG. 1.
Three-stage Tat-dependent transcription reactions. (A) Immobilized template. The DNA template, containing the HIV LTR and an artificial terminator (τ), was biotinylated on both ends using Klenow DNA polymerase and bound to streptavidin-coated magnetic beads. (B) PIC formation. PIC are obtained following incubation of immobilized templates with a nuclear extract (NE) depleted of ATP by a treatment with HK and glucose. Phosphorylation of the CTD of RNA polymerase is allowed by adding dATP in the reaction mixture. Protein composition of preinitiation complexes can be analyzed by Western blotting. (C) Pausing at −14. Early elongation complexes were formed by incubation of the preinitiation complexes in the absence of ATP and in the presence of CTP, GTP, and [α-32P]UTP. This allows the RNA polymerase to carry out the transcription of the first 14 nt of the RNA chain and to pause before the first A residue located at +15 on the DNA template. (D) Chase. Elongation complexes were obtained by allowing the +14 complexes to extend following addition of all the nucleotides in the presence or absence of Tat. The population of elongation complexes, which have reached various points on the template, can be purified by washing the beads and analyzed for their RNA and/or protein composition. Released transcripts can be analyzed by purifying the RNA present in the supernatant collected after magnetic separation. The major stop sites correspond to the end of TAR, the terminator (τ), and the end of the template (ρ).
In the first stage of the reaction, preinitiation complexes that contained only nonphosphorylated RNA polymerase II were assembled using nuclear extracts that were depleted of ATP by treatment with hexokinase and glucose (21). Phosphorylation of the RNA polymerase II CTD was then initiated by adding dATP. The second stage of the reaction creates transcription complexes that are paused at +14, a stage that is shortly after initiation but before complete clearance of the promoter. These complexes were obtained by chasing the preinitiation complexes in the presence of all the ribonucleotides except ATP, using dATP as a phosphate donor for the phosphorylation of the RNA polymerase II CTD (44, 46, 47). Because there is no ATP remaining in the extract, elongation stops at +14, immediately before the first adenine on the template, which is at position +15 (Fig. 1). Addition of ATP to the reaction mixture permits further elongation. Since no additional label is added during the chase, the only products detected are the result of elongation of the arrested transcripts and are not due to new initiation events (21, 28).
The majority of the template DNAs used in these experiments carried a synthetic terminator sequence (τ) placed between +290 and +326, downstream of the start of transcription. The terminator contains a stable RNA stem-loop structure followed by a tract of 9 uridine residues (17, 50). Transcription complexes reaching the terminator either dissociate from the template at the end of the U tract or remain bound to the template at the end of the stem-loop structure. Alternatively, templates carried arrest sites created by bent DNA sequences (4, 30). Synthesis of full-length transcripts from these templates is greatly enhanced in the presence of Tat and Spt5. To measure the effects of Spt5 on transcript release, we also took advantage of the immobilized template system to separate transcription complexes that remained tightly bound to the template from the transcripts that had been released due to termination activity.
Tat activation of transcription complexes paused during early elongation.
As shown in Fig. 2A, the staged transcription reactions permit efficient chases from the early elongation complexes arrested at +14 to late-stage elongation complexes. The +14 transcripts were synthesized with equal efficiency in the presence and absence of Tat (data not shown). All of the complexes paused at +14 resume elongation following the addition of a full complement of nucleotides. However, efficient transcription elongation requires the addition of nuclear extract during the chase. In the absence of added extract, the majority of the transcription complexes disengage from the template shortly after TAR (+59) and short RNA transcripts appear in the free fraction, suggesting that general elongation factors present in the extract are required to read through TAR. There is also significant release of transcripts at the end of the second major stem-loop structure in the HIV leader sequence (stem-loop II) at +104, and only a limited number of transcription complexes reach the terminator (τ).
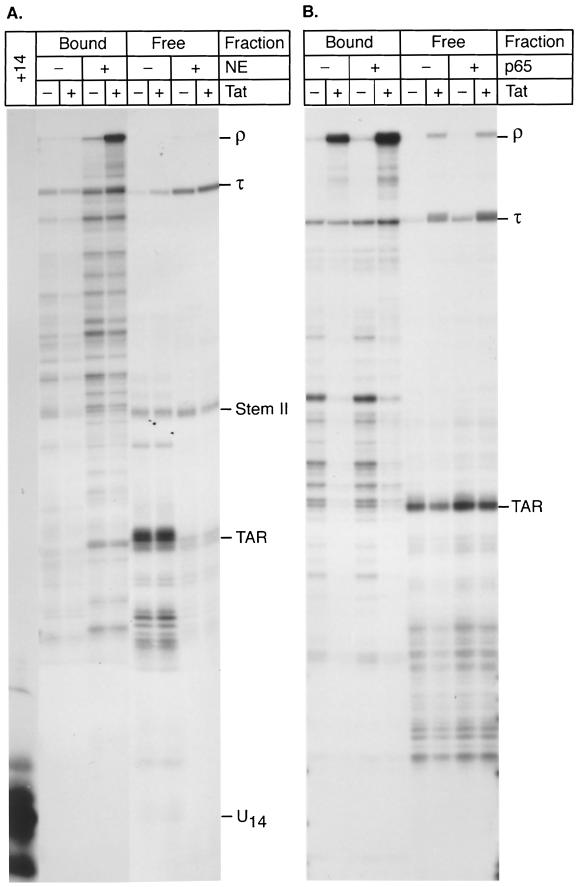
FIG. 2.
Efficient elongation from transcription complexes paused at +14 requires the addition of nuclear extract. (A) Preinitiation complexes were set up on immobilized templates as described in the legend to Fig. 1 and paused at +14 by elongation in the presence of CTP, GTP, and [α-32P]UTP. The +14 products were then washed and chased by addition of unlabeled nucleotides. Reactions were performed in the absence (−) or presence (+) of 10 μl of fresh nuclear extract (NE) and of 20 ng of Tat protein. RNA products tightly associated with the elongation complexes (bound) or prematurely released from the template (free) were analyzed: Abbreviations: TAR, paused transcription complexes or transcripts released at +59, at the end of the TAR stem-loop structure; Stem II, complexes released at +104, at the end of stem-loop II in the HIV sequence; τ, paused transcription complexes or transcripts released at the terminator; ρ, transcripts reaching the end of the template. (B) Transcription in the presence of NF-κB. Recombinant NFκB p65 protein (300 ng) purified from baculoviruses (+) or the corresponding storage buffer (NDBZ buffer; see Materials and Methods) (−) was added prior to the nuclear extract to set up the preinitiation complex. Transcription was then carried out to pause the complexes at+14, and beads were washed and then chased in the presence of fresh nuclear extract and the absence (−) or presence (+) of 20 ng of Tat. Bound or released (free) RNA products were analyzed.
By contrast, in the presence of nuclear extract, the transcription complexes are stabilized on the template and comparatively few transcripts dissociate from the template after TAR and stem-loop II. However, a significant fraction of the transcription complexes dissociate from the template at the artificial terminator sequence (τ). Efficient synthesis of the runoff transcript (518 nt) is strongly dependent on the presence of the Tat protein in the reaction (Fig. 2A). Therefore, the net effect of Tat is to increase the proportion of transcripts that are able to read through the terminator sequence and remain tightly associated with the template.
The ability of Tat to promote readthrough of the terminator sequence cannot be mimicked by general activators such as the transcription factor NF-κB p65 (Fig. 2B). Addition of p65 stimulated basal transcription 2.7-fold but did not alter the distribution of transcript lengths or the ability of Tat to further stimulate elongation. For example, addition of Tat to the reaction mixture in the absence of p65 resulted in a reduction of short transcripts and a 16.9-fold increase in the amount of runoff product. Similarly, in the presence of p65, Tat increased the amount of runoff transcripts 10.3-fold.
Similar strategies for studying transcription complexes during various stages of initiation and elongation have been employed by other groups studying HIV transcription (37, 44, 47, 65). However, in all of these studies the Tat response was lost or severely inhibited. The experimental system that we developed is unique because we have been able to retain a Tat response during a chase from complexes paused during early elongation that is nearly equivalent to that observed in uninterrupted transcription reactions. In addition to its experimental utility, the system provides evidence that all the elongation factors required for Tat activation are present at each stage in the reaction.
Three technical features of the system deserve mention. First, we have found that preinitiation complexes after phosphorylation by dATP are extremely fragile and that critical factors that cannot be subsequently replaced are lost when these complexes are washed with isotonic buffers. Second, in the chase experiments that follow, Tat is added to complexes that have reached +14. This demonstrates that Tat is fully functional even when it is added after initiation, in agreement with a previous report (37). Finally, it should be noted that since no new label is introduced during the chase, the results provide a direct comparison of transcription elongation in the presence and absence of Tat. By contrast, under continuous labeling conditions, the proportion of long transcripts is exaggerated because of their increased specific activity.
Spt5 prevents premature release of transcripts at terminator sequences.
We next examined the role of Spt5 during various stages of elongation. To selectively remove Spt5 from the transcription system, without removing other transcription elongation factors, nuclear extracts were immunodepleted using a mouse monoclonal antibody raised against Spt5. As a control, nuclear extracts were also depleted using equivalent amounts of rabbit immunoglobulin G (mock depleted).
The results of transcription experiments performed with the mock-depleted and Spt5-depleted extracts are shown in Fig. 3.In the mock-depleted extracts, addition of recombinant Tat protein to the reaction stimulated the production of runoff product (ρ) remaining bound to the template by more than 30-fold. Removal of Spt5 from the extracts by immunodepletion reduced the amount of runoff product in the bound fraction by sevenfold but did not reduce the amount of products found at the terminator. Thus, Spt5 is required for maximal readthrough of the terminator sequence.
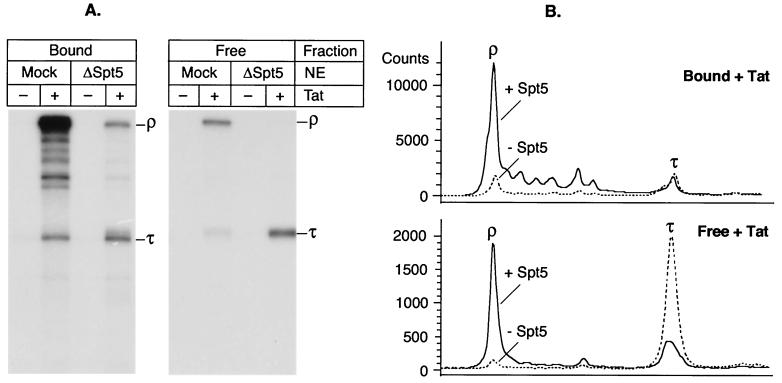
FIG. 3.
Spt5 prevents release of transcripts at terminator sequences. (A) Standard transcription reactions using an immobilized wild-type HIV-1 LTR template with either a mock-depleted (Mock) or Spt5-depleted (ΔSpt5) nuclear extract (NE), in the absence (−) or presence (+) of 20 ng of Tat. RNA products present in elongation complexes (bound fraction) as well as released transcripts (free fraction) were analyzed. (B) Densitometry of the gels shown in panel A. Continuous line, mock-depleted extracts in the presence of Tat; broken line, Spt5-depleted extracts in the presence of Tat.
Examination of transcripts released from the templates shows that the reduction in the runoff product retained on the template in Spt5-depleted extracts is due to an increase in transcript release at the terminator sequence, τ. Densitometry of the gels (Fig. 3B) shows that in the absence of Spt5 there is a 3.3-fold increase in the amount of RNA released at the terminator and a concomitant 13.4-fold decrease in the amount of RNA released from the end of the template.
Spt5 does not promote transcript release at arrest sites.
To test whether Spt5 can help to overcome blocks imposed by strong arrest sites, we also prepared templates carrying arrest sites present in the human histone H3.3 gene intron (Fig. 4). These sequences contain two tandem polypyrimidine tracts flanked by purines. The effect of these sequences is to bend the DNA and force polymerase pausing (4, 30). As a control, we also included a template that did not carry either the artificial terminator or the strong arrest sites.
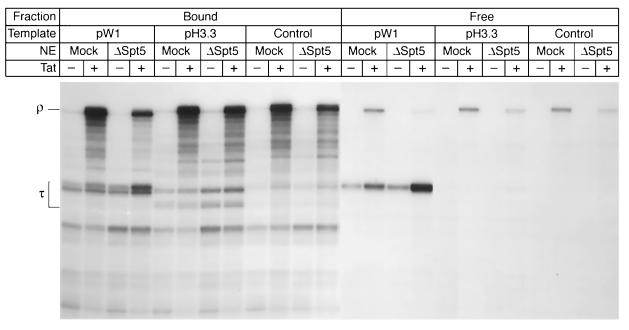
FIG. 4.
Spt5-enhanced readthrough of transcription arrest sites. Three different templates were used in this experiment: the wild-type HIV-1 LTR (pW1), which contains the artificial stem-loop terminator sequence; constructs in which the terminator sequence has been replaced by an arrest sequence present in the histone H3.3 gene (H3.3); and a control sequence (control). Standard transcription reactions using the three different immobilized templates with either a mock-depleted (Mock) or a Spt5-depleted (ΔSpt5) nuclear extract (NE), in the absence (−) or presence (+) of 20 ng of Tat, were performed. RNA products present in elongation complexes (bound fraction) as well as released transcripts (free fraction) are shown.
As shown in Fig. 4, the stem-loop terminator sequence induced pausing at +290 (τ) whereas the histone H3.3 arrest sequence introduced two new stop sites of the expected sizes that were absent at the equivalent positions on the control template. During the transcription reaction, using mock-depleted extracts, Tat increased the synthesis of long transcripts on each of the templates by more than 15-fold and greatly enhanced readthrough of both the terminator sequence and the histone arrest sites. Consistent with the results shown in Fig. 3, when the same reactions were performed using Spt5-depleted extracts, there was a three- to fourfold increase in the amount of transcripts released at the terminator site present in pW1.
By contrast, there was no transcript release using the pH3.3 templates. However, an approximately twofold increase in the amount of paused transcription complexes was found at the arrest sites when transcription was performed in the absence of Spt5. Similarly, increased pausing at strong arrest sites found in the HIV sequence was observed when the control template was transcribed in the absence of Spt5.
We interpret these results as indicating that Spt5 is able to relieve pausing at a wide variety of sites ranging from the naturally occurring pause sites on the control templates to the strong arrest sites on the H3.3 template and the strong terminator sequence present in pW1. Thus, using the pW1 template, full-length transcript levels were reduced 2.9-fold when Spt5 was removed. Similarly, full-length transcript levels were reduced 1.6- and 2.4-fold in the absence of Spt5 when the pH3.3 and control templates were used.
TAK and Spt5 play distinct and complementary roles during elongation.
Tat activation of elongation requires TAK (p-TEFb), a kinase that is able to phosphorylate the RNA polymerase CTD as well as to relieve blocks imposed during early elongation by Spt5 (DSIF). Immunodepletion of transcription reactions of the CDK9 subunit of TAK has been reported previously to severely inhibit Tat-dependent elongation and reduce basal transcription (9, 66), but the relative contributions of TAK and Spt5 to the elongation reaction have not been measured.
In the experiment in Fig. 5,we took advantage of the observation that early-stage elongation complexes paused at +14 could be efficiently prepared using extracts in which both TAK (CDK9 and CycT1) and Spt5 were depleted. The immunoblots shown in Fig. 5B demonstrate that CDK9, CycT1, and Spt5 can be removed from the extracts both individually and without significantly depleting the extracts for other transcription factors, including TAFII250, RNA polymerase II, CDK7, and RAP74.
FIG. 5.
Complementary activities of Spt5 and TAK during elongation. (A) Elongation complexes arrested at +14 were prepared using a nuclear extract depleted of both TAK and Spt5. The +14 complexes were washed and subsequently chased in the presence of either mock-depleted (Mock), TAK-depleted (ΔTAK), or Spt5-depleted (ΔSpt5) nuclear extracts (NE). Each chase was performed in the absence (−) or presence (+) of 20 ng of Tat protein. RNA products present in elongation complexes (bound fraction) as well as released transcripts (free fraction) were analyzed. (B) Immunoblot showing the protein composition of depleted nuclear extracts. A corresponding amount of each nuclear extract was analyzed by immunoblotting using antibodies directed against the following proteins (from top to bottom): TAFII250, the core of RNA polymerase II (Pol II) (N-20), Spt5 (anti-DSIF-p160), cyclin T1, CDK9, CDK7, and RAP74.
It is important to note that depletion of Spt5 in this system does not impair the synthesis of the +14 transcripts (Fig. 5; also see Fig. 8). These observations are consistent with earlier reports showing that NELF and DSIF have only minimal effects on early-stage elongation reactions (62). Thus, Spt5 is not required for initiation or early elongation. The paused transcription complexes prepared in the absence of TAK and Spt5 could then be chased in the presence and absence of mock-depleted, TAK-depleted, and Spt5-depleted nuclear extracts. Under these conditions, efficient elongation and Tat responses were obtained using the mock-depleted extracts in the chase (Fig. 5A). However, there was a severe restriction in the formation of long transcripts when TAK was depleted. As expected, in the absence of TAK, Tat was also unable to stimulate elongation. By contrast, Tat was able to stimulate elongation in the absence of Spt5, but the overall levels of long transcripts were reduced due to enhanced pausing and RNA chain release at the terminator sequence. Thus, the complementary activities of TAK, Tat, and Spt5 are evident at the terminator sequence. Tat and TAK were able to stimulate the production of runoff templates, but the total amount of these products was strongly reduced in the absence of Spt5 due to the increased release of transcripts at the terminator sequence.
FIG. 8.
Spt5 recruitment is independent of RNA polymerase II phosphorylation. (A) PIC and early elongation complexes (+14) were assembled as described in the legend to Fig. 1, using mock-depleted (Mock) or Spt5-depleted (ΔSpt5) nuclear extract (NE) pretreated with HK and glucose, and in the presence of increasing levels of dATP (5, 10, or 25 μM). The protein compositions of the different transcription complexes were analyzed by Western blotting with core RNA polymerase II (N-20)- or Spt5 (provided by R. Gaynor)-specific antibodies. (B) Transcription gel showing the corresponding +14 RNA products. Note that RNA polymerase can be extensively phosphorylated under conditions that do not permit formation of the +14 products. Spt5 is recruited proportionally to the amount of the +14 product.
In addition to transcript release at the terminator, a significant amount of transcripts of approximately 60 nucleotides (nt) was released at the end of the TAR RNA sequence and after the end of stem-loop II. It is notable that equivalent levels of released TAR were observed when mock-depleted, TAK-depleted, and Spt5-depleted extracts were used. Thus, neither Spt5 nor TAK plays a significant role in stabilizing transcription complexes at TAR.
Tat-dependent hyperphosphorylation of Spt5 in late-stage elongation complexes.
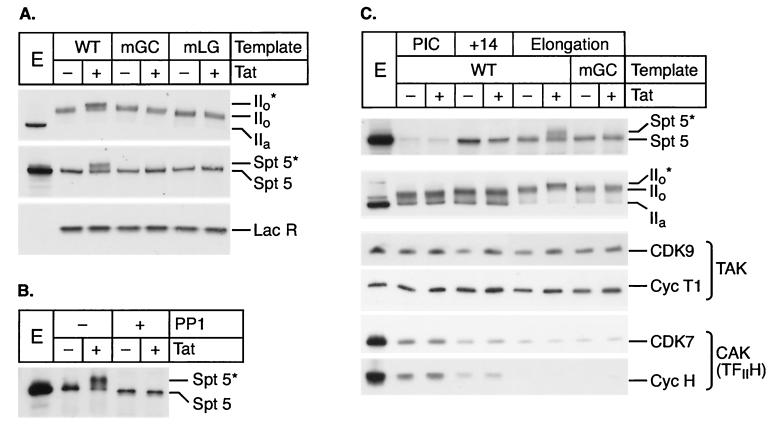
We have reported previously (21) that RNA polymerase, which is mainly unphosphorylated in the nuclear extract (IIa), becomes phosphorylated during early elongation (IIo) and hyperphosphorylated in the presence of Tat after transcription through TAR (IIo*) (Fig. 6A). Immunoblotting of the elongation complexes with Spt5-specific antibodies (Fig. 6A) demonstrated that Spt5 also becomes phosphorylated in parallel to the RNA polymerase II CTD. Addition of Tat to the reaction mixture induced the appearance of a second band migrating above Spt5, which we have called Spt5*. This shift in the migration of Spt5 is dependent on a functional TAR element, as shown with templates carrying a point mutation either in the Tat-binding site (mGC) or in the cyclin T1-binding site (mLG).
FIG. 6.
Spt5 is recruited shortly after initiation and hyperphosphorylated during elongation in the presence of Tat. (A) Immunoblot showing the protein composition of elongation complexes. Standard transcription reactions were performed using templates carrying either wild-type (WT), bulge (mGC), or loop (mLG) mutants of the TAR element, in the presence of 100 ng of LacR and in the absence (−) or presence (+) of 20 ng of Tat. Elongation complexes were purified and analyzed with antibodies directed (from top to bottom) against the core of RNA polymerase II (N-20), Spt5 (provided by R. Gaynor) or LacR (NL1.1B2.10). Tat induces hyperphosphorylation of both RNA polymerase (IIo*) and Spt5 (Spt5*). A 1-μl volume of nuclear extract (E) was used as a control. (B) Phosphatase treatment of elongation complexes. Elongation complexes purified as in panel A were incubated in the absence (−) or presence (+) of protein phosphatase 1 (PP1) and analyzed by immunoblotting with Spt5 antibodies. (C) Immunoblot showing the protein composition of the different transcription complexes. PIC and +14 complexes were assembled on wild-type immobilized template and purified as described in the legend to Fig. 1. Elongation complexes were purified from parallel standard transcription reactions, using templates carrying wild-type or mutant (mGC) TAR. Tat was added (+) or not (−) at different times to monitor its effect in different complexes. Purified complexes were loaded on precast gels of appropriate concentration for analysis of different proteins and probed with the corresponding antibodies (from top to bottom): Spt5 (provided by R. Gaynor), the core of RNA polymerase II (N-20), CDK9 (H-169), cyclin T1 (C-20), CDK7 (C-19), and cyclin H (D-10).
Treatment of the elongation complexes with protein phosphatase 1 results in the complete disappearance of the band corresponding to the Spt5* protein (Fig. 6B), demonstrating that Spt5* is a form of Spt5 which is modified by phosphorylation. It should also be noted that the migration of the Spt5 protein present in the elongation complexes in the absence of Tat was slightly decreased compared to that of the dephosphorylated protein. This suggests that Spt5 is partially phosphorylated in these complexes and that Tat induces a further phosphorylation of the protein after its binding to the TAR element.
Reports published when these experiments were in progress have also demonstrated that Spt5 can become phosphorylated during transcription elongation (31, 47). However, these experiments were performed by adding limiting amounts of 32P-labeled nucleotides to arrested transcription complexes and consequently underestimated the extent of phosphorylation that Spt5 undergoes during elongation in the presence of Tat. As a result, the Spt5* form was not detected in these earlier experiments. In agreement with our results, a highly phosphorylated form of Spt5, which may correspond to Spt5*, was detected by Garber et al. (15) after the in vitro phosphorylation of Spt5 by CDK9.
Recruitment of Spt5 to early-stage transcription elongation complexes.
The elongation factor Spt5 has been identified previously as necessary for Tat-dependent transactivation of the transcription from the LTR promoter (61). To measure the dynamics of Spt5 association with transcription complexes during various stages of the transcription cycle, preinitiation, +14, and elongation complexes were analyzed by immunoblotting. Antibodies against TAK (CDK9 and cyclin T1) and the CTD-associated kinase (CDK7 and cyclin H) were used as controls (Fig. 6).
Surprisingly only background levels of Spt5 were observed in preinitiation complexes (Fig. 6C). However, in agreement with the observations of Ping and Rana (47), we found that Spt5 was recruited shortly after initiation and was present at high levels in the transcription complexes paused at +14. By contrast, control experiments show that the CTD-associated kinase subunits CDK7 and cyclin H were present at high levels in preinitiation complexes but were then recycled during transcription. These factors are found at significantly reduced levels in the complexes paused at +14 and are essentially absent from complexes that have extended beyond +14 (33, 64, 65). Both CDK9 and its associated cyclin T1 were present in the complex assembled on the DNA template during initiation, and no change in their levels were observed during subsequent elongation.
Spt5 is not required for activation of TAK by Tat.
The observation that there was still a significant increase in transcription elongation on addition of Tat to Spt5-depleted extracts (Fig. 3 to 5) suggests that Tat and Spt5 are operating through distinct biochemical mechanisms but show complementary functions. In support of this hypothesis, the immunoblotting experiment in Fig. 7demonstrates that although Spt5 is phosphorylated by CDK9 during elongation, it is not required for the activation of CDK9 by Tat.
FIG. 7.
Spt5 is not required for Tat-dependent hyperphosphorylation of RNA polymerase II. (A) Elongation complexes were assembled using mock-depleted extracts (Mock) or immunodepleted extracts with anti-CDK7 antibodies (ΔCDK7) or a combination of antibodies against CDK9 and cyclin T1 (ΔTAK). The complexes were analyzed by Western blotting using antibodies directed against (from top to bottom) Spt5 (provided by R. Gaynor), the core of RNA polymerase II (N-20), CDK7 (C-19), CDK9 (H-169), and cyclin T1 (C-20). (B) Elongation complexes were assembled using mock-depleted extracts or extracts immunodepleted of Spt5 and analyzed with antibodies directed against (from top to bottom) the core of RNA polymerase II (N-20), Spt5 and CDK9 (H-169). NE, nuclear extract.
Figure 7A shows the effects of immunodepletion of CDK9 and cyclin T1 on protein phosphorylation. In these extracts, transcription initiation was normal and there was no reduction in the levels of transcripts reaching +14. However, removal of CDK9 and cyclin T1 strongly inhibited Tat-activated elongation in the subsequent chase (data not shown). As shown in Fig. 7A, immunodepletion of CDK9 and cyclin T1 also resulted in a complete absence of both the RNA polymerase IIo* and the Spt5* proteins from elongation complexes prepared in the presence of Tat. Thus, both RNA polymerase II and Spt5 are substrates for CDK9 during Tat-activated transcription.
In contrast, depletion of CDK7 resulted in a severe inhibition of transcription initiation (data not shown). Under these conditions, there was a strong reduction in the levels of RNA polymerase IIo and polymerase IIo∗, with approximately 50% of the template-associated RNA polymerase remaining hypophosphorylated (IIa), and a corresponding reduction in Spt5 levels. However, due to some residual initiation events, some of the Spt5 that was recruited to the promoter became phosphorylated by CDK9 during elongation.
Significantly, in the Spt5-depleted extracts (Fig. 7B), hyperphosphorylation of the RNA polymerase CTD took place with the same efficiency as in mock-depleted extracts. Thus, the defects in elongation observed in the absence of Spt5 cannot be attributed to changes in the phosphorylation pattern of RNA polymerase. Furthermore, the experiment shows that although Spt5 is itself a substrate for the Tat-activated CDK9, it is not required for the phosphorylation of the CTD by CDK9 and does not appear to act as a regulator of CDK9 activity.
Spt5 is not required for initiation or early elongation.
To examine whether Spt5 is required for initiation or early elongation, we also examined the synthesis of the +14 transcripts using mock-depleted and Spt5-depleted extracts. The reactions in Fig. 8 were performed using three different concentrations of dATP to regulate the level of CDK7-dependent phosphorylation in the preinitiation complexes. At 5 and 10 μM dATP, synthesis of the +14 complex was strongly inhibited compared to the levels obtained using 25 μM dATP.
At dATP concentrations below 25 μM, the phosphorylation of the RNA polymerase CTD in the preinitiation complex was restricted and the majority of the RNA polymerase remained unphosphorylated. During the subsequent chase there was additional phosphorylation of the RNA polymerase CTD, and the amount of the RNA polymerase IIo form that was produced was equivalent to that seen in the presence of 25μM dATP. We attribute the additional CTD phosphorylation to the use of GTP in the chase mixture by CDK7 and/or conformational changes that are induced in the RNA polymerase during the synthesis of the first residues of the nascent transcript. However, at dATP concentrations below 25 μM, the formation of the +14 complex was strongly inhibited. This result is consistent with studies showing that although the CTD kinase activity of TFIIH enhances initiation rates, it is not obligatory for initiation and early elongation (55). The dATP-dependent increase in early elongation most probably reflects the increasing ability of the XPB subunit of TFIIH to use dATP as a substrate for its ATP-dependent helicase activity, which is the main enzymatic activity required for initiation (7, 55).
As noted above, Spt5 is not present in the preinitiation complexes but is recruited to early-stage elongation complexes. As shown in Fig. 8, there is a strict correlation between the amount of Spt5 recruitment and the amount of +14 RNA synthesis. However, there is no correlation between Spt5 recruitment and the extent of RNA polymerase CTD phosphorylation. It therefore seems likely that Spt5 is recruited to early elongation complexes because of conformational changes in the polymerase rather than because of recognition of the phosphorylated form of the CTD.
DISCUSSION
Positive and negative roles for Spt5 during transcription elongation.
The human homologue of Spt5 was first identified as one of the subunits of DSIF, a factor that could mediate the inhibition of transcriptional elongation in the presence of the protein kinase inhibitor DRB (59). DSIF induces promoter-proximal pausing when CDK9 kinase activity is absent, such as in extracts that have been immunodepleted of CDK9 or in extracts that have been treated with DRB (58, 59). This elongation block is relieved by addition of purified p-TEFb (58) together with a second factor called NELF (negative elongation factor) (62).
In an independent study pointing to a positive role for Spt5 in transcription elongation, Wu-Baer et al. (61) discovered that Spt5 is an essential cofactor for Tat activation of HIV transcription elongation. Studies of the Drosophila heat shock promoter also indicate that Spt5 can play a role in general elongation (2). Spt5 is present at the promoter in the absence of heat shock, suggesting that it might be associated with the paused transcription complex or that it might play a role in maintaining the chromatin structure under an open conformation. Following heat shock, Spt5 is redistributed to occupy both the 5" and 3" ends of heat shock genes, where it colocalizes with RNA polymerase and p-TEFb (2). This association with active transcription complexes suggests that Spt5 can also play a positive role in transcription elongation. Consistent with this hypothesis, Drosophila Spt5 colocalizes extensively with the actively elongating IIo form of RNA polymerase and with p-TEFb on polytene chromosomes (25).
Spt5 is a general elongation factor that stabilizes RNA polymerase at terminator sequences.
In this study we used immobilized DNA template to analyze the role of Spt5 during the transactivation of the HIV-1 LTR transcription in detail. Using a three-stage transcription assay, we demonstrated that Spt5 is not present in preinitiation complexes but that it is recruited immediately after initiation, independently of Tat. Our studies have also shown that Spt5 plays a positive role in transcription elongation by enhancing the stability of transcription complexes at terminator sequences and by promoting readthrough of strong arrest sites. In the absence of Spt5, release of transcripts at terminator sites is increased over threefold, while polymerase pausing at bent DNA sequences is increased by as much as twofold.
This novel biochemical function of Spt5 is reminiscent of that activity of the E. coli NusG protein, with which it has a region of sequence homology. NusG is an essential E. coli protein that regulates Rho-dependent termination by increasing the rate of transcription of elongation of E. coli RNA polymerase in the absence of Rho. This renders the transcription complex more susceptible to the termination activity of Rho and leads to early chain release (45). NusG selectively reduces transcription pausing at class II terminators, a set of pause sites where less stable RNA-DNA hybrids induce backtracking of RNA polymerase (3). In the experiments reported here, the τ terminator sequence, which consists of a stable stem-loop structure followed by a tract of nine U residues, has a structure and activity similar to those of the class II terminators. Moreover, release of RNA from the template in the absence of Spt5 occurs preferentially at the end of the poly(U) tract, where the RNA-DNA hybrid is expected to be particularly unstable.
Genetic and biochemical studies have shown that Spt5, like NusG, interacts directly with RNA polymerase II (19, 59, 61, 62). The interactions between Spt5 and RNA polymerase II are mediated by four KOW domains, which are also found in NusG (18). Significantly, deletion of one of the two N-terminal KOW motifs also resulted in loss of Spt5 activity. It is not yet known whether, by analogy to NusG, Spt5 complements the function of a Rho-like termination factor or whether it acts directly on the RNA polymerase to stabilize it at terminator sequences.
Spt5 does not induce transcript release after TAR.
In the original experiments used to characterize DSIF function, Wada et al. (58, 59) found that addition of DSIF to partially reconstituted transcription systems resulted in promoter-proximal pausing, creating a series of early termination products of 20 to 30 nt. This negative effect seems to be a specific feature of the partially purified transcription system and specific reaction conditions used in these experiments. Subsequent studies by Yamaguchi et al. (62) have shown that blocks to elongation imposed by DSIF and NELF become apparent only during the later stages of elongation, when RNA polymerase has transcribed at least 60 nt. These effects of DSIF and NELF are largely kinetic, and over time only full-length transcripts are produced even in the absence of DSIF (49).
In HIV, the TAR RNA sequence is located within the first 59 nt of the transcript and forms a strong kinetic barrier to elongation (41, 42). In our experiments, using HeLa cell nuclear extracts depleted of either Spt5 or CDK9, we could find no evidence that Spt5 moderated the promoter-proximal pausing of transcription release at the TAR RNA sequence. By contrast, we have found evidence that transcription elongation factors other than Spt5 are required for early stability of elongation, particularly after stem-loop structures such as TAR RNA and stem-loop II. When +14 complexes are chased in the absence of added nuclear extract, the majority of the RNA polymerases disengage from the template near the end of the TAR RNA. This block to elongation can be relieved by the addition of nuclear extracts, including extracts that have been depleted of Spt5. Although the identity of these factors has not yet been established, it seems very likely that the general elongation factors TFIIF and TFIIS, which are known to stimulate transcription from the HIV LTR in cell-free systems, are also able to contribute to polymerase stability during HIV elongation through TAR (27).
Regulation of Spt5 by phosphorylation.
It is now well established that DSIF activity is regulated by phosphorylation. For example, addition of CDK9 to depleted extracts is able to restore DRB-independent inhibition of transcription by DSIF and NELF (57-59, 62).
Spt5 contains two separate domains named CTR1 and CTR2, which show some sequence similarity to the RNA polymerase II CTD (63). The CTR1 motif is well conserved and contains nine repeats of the consensus sequence GS(R/Q)TPXY. The CTR2 motif is less well conserved and has 10 repeats of the consensus P(T/S)PSP(Q/A)(S/G)Y. Spt5 is a known substrate for phosphorylation by CDK9 in vitro (15, 22). This phosphorylation is restricted mainly to CTR1; however, the functional significance of the various phosphorylation sites has not been unambiguously established. In one study, DRB-sensitive transcription was unaffected by deletion of the C-terminal region (63); however, a second report suggested that CTR1 conferred DRB sensitivity (22).
Here we have provided additional evidence that phosphorylation of Spt5 by CDK9 is important for its activity. We have shown that Spt5 becomes highly phosphorylated in elongation complexes that have been activated by Tat. This results in the appearance of a modified form of the protein with markedly reduced mobility in SDS-polyacrylamide gels (Spt5∗). Earlier experiments showing that Spt5 can become phosphorylated during transcription elongation (31, 47) were performed under limiting nucleotide concentrations and underestimated the extent to which the protein is phosphorylated. The hyperphosphorylation of Spt5 does not occur when CDK9 and cyclin T1 are immunodepleted from the nuclear extracts or when Tat is absent from the system, suggesting strongly that Tat-activated CDK9 is the kinase responsible for Spt5 hyperphosphorylation.
Phosphatase treatment showed also that Spt5 is partially phosphorylated in early elongation complexes that have not been exposed to Tat or transcribed through TAR RNA. Significantly, this form of the protein can also be detected in the absence of CDK9, implying that another kinase is able to phosphorylate Spt5 when it is recruited to early elongation complexes. In keeping with these observations, Ivanov et al. (22) reported that CDK7 is able to phosphorylate Spt5 carrying mutations in the CTR1 domain. This suggests that additional phosphorylation sites located outside the CTR regions could be subject to modification by protein kinases other than CDK9.
Recruitment of Spt5 to early-stage elongation complexes.
Spt5 is not present in preinitiation complexes but is recruited shortly after initiation. Although it was originally proposed that Spt5 could interact only with the nonphosphorylated form of RNA polymerase II (58), our results suggest strongly that Spt5 is recruited following CTD phosphorylation and transcription initiation. A clear demonstration of the separation between CTD phosphorylation and Spt5 recruitment is found in the experiment in Fig. 8. In this experiment, preinitiation complexes were formed under conditions that limit initiation due to restricted levels of dATP. In subsequent chases in the absence of additional ATP, the RNA polymerase CTD becomes fully phosphorylated due to CDK7 activity but initiation is compromised and only a small fraction of the polymerases are able to transcribe up to +14. Under these conditions, there is a strict correlation between the amount of Spt5 that is recruited and the levels of +14 transcripts but there is no recruitment of Spt5 to the phosphorylated RNA polymerases remaining at the promoter. Thus, it appears that Spt5 is recruited to early elongation complexes because of conformational changes in the polymerase and/or its release from initiation factors that permits Spt5 binding rather than recognition of the phosphorylation state of the CTD. In keeping with this hypothesis, we have also found that dephosphorylation of the +14 transcription complexes does not result in release of Spt5 (data not shown).
Complementary roles of Spt5 and TAK during HIV-1 LTR transcription.
How do Spt5 and TAK act in concert to promote efficient elongation? In both prokaryotes and eukaryotes, transcription elongation is discontinuous and is characterized by multiple and specific pausing events. The duration of these pauses is a function of the specific DNA sequences being transcribed as well as of interactions with regulatory proteins.
In this study we examined the effects of Spt5 and TAK on transcription through a variety of blocks to elongation. The first transcription block that we studied was an RNA stem-loop structure followed by 9 uridine residues. This artificial sequence, which closely resembles bacterial termination signals, causes RNA polymerase to stop elongation and to release transcripts in cell-free transcription systems. The second transcription block was an arrest site normally found in the histone H3.3 gene. This site carries structural elements that bend DNA and causes polymerase pausing but not the release of the nascent transcripts in cell-free systems and in vivo (4, 30). Bent DNA sequences are also potent arrest sites in yeast (10, 32).
The heterologous terminator and arrest sequences that we have used in these experiments provide effective tools for measuring small changes in RNA polymerase processivity. However, the HIV genome does not carry any specific termination signals. Instead, there are numerous sites where pausing can occur. The cumulative effect of each of these pauses is to prevent the RNA polymerase from transcribing the entire HIV genome unless Tat and Spt5 are present. Enhancement of elongation therefore involves not only increasing the intrinsic processivity of the polymerase but also ensuring that the activated polymerase remains tightly bound to the template (56). The results presented in this paper strongly suggest that Spt5 helps to stabilize RNA polymerase during elongation and thereby functions as a cofactor for Tat.
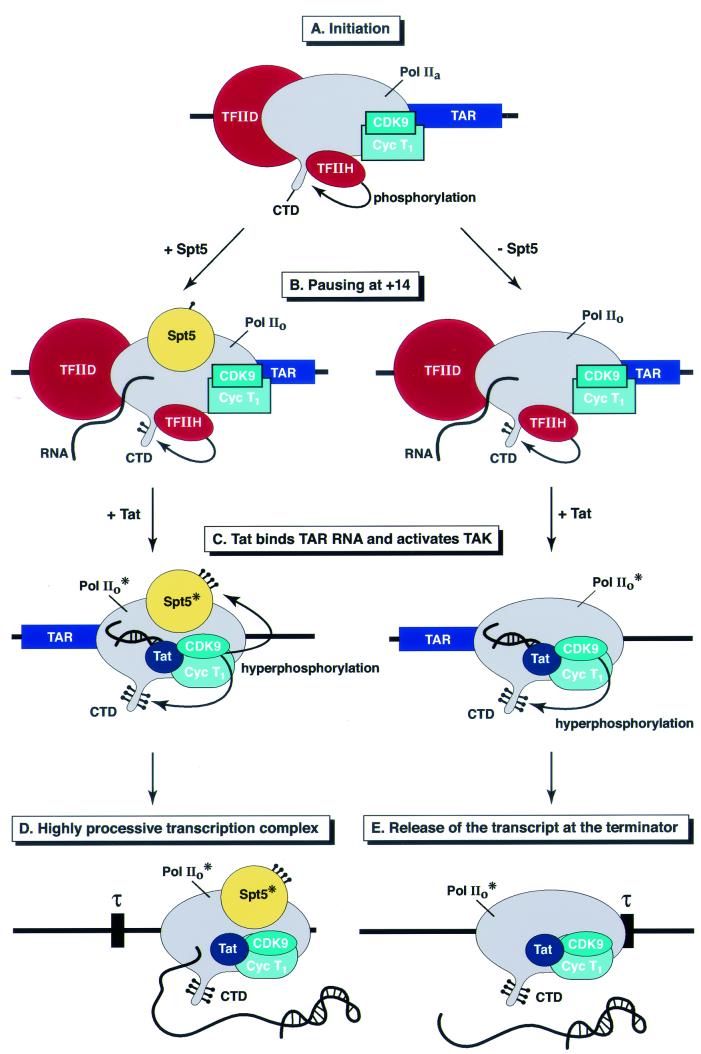
As shown in the model presented in Fig. 9,it seems likely that phosphorylation of the RNA polymerase CTD and Spt5 by Tat-activated CDK9 are two separate activation events. In agreement with our previous results (21), we have shown that CDK9 and cyclin T1 are present at equivalent levels in the preinitiation complexes and transcription complex during various stages of elongation. However, immunodepletion experiments show that CDK9 is required only for late elongation, not for HIV-1 transcription initiation or early elongation. CDK9 activity does not appear to be required to enhance polymerase stability on templates. Depletion of extracts of CDK9 and CycT1 blocks Tat-activated transcription but does not result in premature release of transcripts, suggesting that TAK is required to enhance polymerase processivity directly. Consistent with this hypothesis, we have found in experiments to be presented elsewhere (Y. K. Kim et al., unpublished data) that CTD phosphorylation can enhance the ability of RNA polymerase to read through terminator sequences.
FIG. 9.
Model for the activity of Spt5 in the Tat-mediated transactivation of transcription at the HIV-1 LTR promoter. (A) Initiation. The RNA polymerase II (Pol II) complex, containing all the general transcription factors assembles on the promoter (only TFIID and TFIIH are represented). CDK9 and cyclin T1 are also present in the complex but are inactive (blue boxes). During initiation, the CTD of RNA polymerase is extensively phosphorylated by the CDK7 subunit of TFIIH. (B) Pausing at +14. Shortly after initiation, as measured by the +14 complexes, Spt5 is recruited to the transcription complex. Assembly of the +14 complex and phosphorylation of the CTD by TFIIH are equivalent in the presence and absence of Spt5. (C) Tat binds TAR RNA and activates TAK. Shortly after promoter clearance, the general transcription factors, including TFIIH, dissociate from the transcription complex. After the transcription of the TAR element, Tat is recruited and activates TAK (the CDK9 and CycT1 complex represented by blue circles). The active kinase complex phosphorylates both the CTD of RNA polymerase II (II0* form) and Spt5 (Spt5*). (D) Highly processive transcription complex. In the presence of Spt5, the highly processive transcription complex formed after Tat-mediated activation is stabilized and can carry out the transcription of the full HIV genome. (E) Release of the transcript at the terminator. In the absence of Spt5, the transcription complex containing a highly phosphorylated RNA polymerase II is less stable than complexes containing Spt5. As a result, there is an increased rate of release of transcripts at pause sites, such as the terminator sequence (τ).
The role of Spt5 appears to be complementary to but distinct from the activation of elongation by CTD phosphorylation. Like CDK9, Spt5 is not required for initiation or during the early stages of elongation and can be recruited to complexes paused at +14. Furthermore, in the HIV system, Spt5 is not required for basal transcription from the HIV LTR and there is no evidence for an increase of short transcripts in Spt5-depleted extracts. However, as shown here, Spt5 is required to prevent premature release of RNA chains from specific terminator sequences and minimize arrest at bent DNA sequences. It is this novel biochemical function of Spt5 that permits it to play a critical role in promoting Tat-dependent transcriptional elongation.
Acknowledgments
We thank our colleague Marc Bailey for helpful discussions, and we thank A. D. Lowe for preparation of the transcription cell extracts. We are grateful to Richard Gaynor, Jim Hiscott, and Tsafi Pe'ery for gifts of reagents.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and non-human cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrulis, E. D., E. Guzmán, P. Döring, J. Werner, and J. T. Lis. 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14:2635-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Artsimovitch, I., and R. Landick. 2001. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc. Natl. Acad. Sci. USA 97:7090-7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Awrey, D. E., R. G. Weilbaecher, S. A. Hemming, S. M. Orlicky, C. M. Kane, and A. M. Edwards. 1997. Transcription elongation through DNA arrest sites: a multistep process involving both RNA polymerase II subunit RPB9 and TFIIS. J. Biol. Chem. 272:14747-14754. [DOI] [PubMed] [Google Scholar]
- 5.Berkhout, B., R. H. Silverman, and K.-T. Jeang. 1989. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 59:273-282. [DOI] [PubMed] [Google Scholar]
- 6.Bieniasz, P. D., T. A. Grdina, H. P. Bogerd, and B. R. Cullen. 1998. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 17:7056-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradsher, J., F. Coin, and J.-M. Egly. 2000. Distinct roles for the helicases of TFIIH in transcript initiation and promoter escape. J. Biol. Chem. 275:2532-2538. [DOI] [PubMed] [Google Scholar]
- 8.Chen, D., Y. Fong, and Q. Zhou. 1999. Specific interaction of Tat with the human but not rodent pTEFb complex mediates the species-specific Tat activation of HIV-1 transcription. Proc. Natl. Acad. Sci. USA 96:2728-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, D., and Q. Zhou. 1999. Tat activates human immunodeficiency virus type 1 transcriptional elongation independent of TFIIH kinase. Mol. Cell. Biol. 19:2863-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christie, K. R., D. E. Awrey, A. M. Edwards, and C. M. Kane. 1994. Purified yeast RNA polymerase II reads through intrinsic blocks to elongation in response to the yeast TFIIS analogue, p37. J. Biol. Chem. 269:936-943. [PubMed] [Google Scholar]
- 11.Churcher, M. J., A. D. Lowe, M. J. Gait, and J. Karn. 1995. The RNA element encoded by the trans-activation-responsive region of human immunodeficiency virus type 1 is functional when displaced downstream of the start of transcription. Proc. Natl. Acad. Sci. USA 92:2408-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dingwall, C., I. Ernberg, M. J. Gait, S. M. Green, S. Heaphy, J. Karn, A. D. Lowe, M. Singh, M. A. Skinner, and R. Valerio. 1989. Human immunodeficiency virus 1 Tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc. Natl. Acad. Sci. USA 86:6925-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujinaga, K., R. Taube, J. Wimmer, T. P. Cujec, and B. M. Peterlin. 1999. Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc. Natl. Acad. Sci. USA 96:1285-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garber, M. E., and K. A. Jones. 1999. HIV-1 Tat: coping with negative elongation factors. Curr. Opin. Immunol. 11:460-465. [DOI] [PubMed] [Google Scholar]
- 15.Garber, M. E., T. P. Mayall, E. M. Suess, J. Meisenhelder, N. E. Thompson, and K. A. Jones. 2000. CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 Tat-P-TEFb complex to TAR RNA. Mol. Cell Biol. 20:6958-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garber, M. E., P. Wei, V. N. KewelRamani, T. P. Mayall, C. H. Herrmann, A. P. Rice, D. R. Littman, and K. A. Jones. 1998. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 12:3512-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graeble, M. A., M. J. Churcher, A. D. Lowe, M. J. Gait, and J. Karn. 1993. Human immunodeficiency virus type 1 trans-activator protein Tat, stimulates transcriptional read-through of distal terminator sequences in vitro. Proc. Natl. Acad. Sci. USA 90:6184-6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenblatt, J., J. R. Nodwell, and S. W. Mason. 1993. Transcriptional antitermination. Nature 364:401-406. [DOI] [PubMed] [Google Scholar]
- 19.Hartzog, G. A., T. Wada, H. Handa, and F. Winston. 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12:357-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrmann, C. H., and A. P. Rice. 1995. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J. Virol. 69:1612-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isel, C., and J. Karn. 1999. Direct evidence that HIV-1 Tat activates the Tat-associated kinase (TAK) during transcriptional elongation. J. Mol. Biol. 290:929-941. [DOI] [PubMed] [Google Scholar]
- 22.Ivanov, D., Y. T. Kwak, J. Guo, and R. B. Gaynor. 2000. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell. Biol. 20:2970-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanov, D., Y. T. Kwak, E. Nee, J. Guo, L. F. Garcia-Martinez, and R. B. Gaynor. 1999. Cyclin T domains involved in complex formation with Tat and TAR RNA are critical for Tat-activation. J. Mol. Biol. 288:41-56. [DOI] [PubMed] [Google Scholar]
- 24.Kao, S.-Y., A. F. Calman, P. A. Luciw, and B. M. Peterlin. 1987. Anti-termination of transcription within the long terminal repeat of HIV-1 by Tat gene product. Nature 330:489-493. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan, C. D., J. R. Morris, C.-T. Wu, and F. Winston. 2000. Spt5 and Spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 14:2623-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karn, J. 1999. Tackling Tat. J. Mol. Biol. 293:235-254. [DOI] [PubMed] [Google Scholar]
- 27.Kato, H., H. Sumimoto, P. Pognonec, C.-H. Chen, C. A. Rosen, and R. G. Roeder. 1992. HIV-1 Tat acts as a processivity factor in vitro in conjunction with cellular elongation factors. Genes Dev. 6:655-666. [DOI] [PubMed] [Google Scholar]
- 28.Keen, N. J., M. J. Churcher, and J. Karn. 1997. Transfer of Tat and release of TAR RNA during the activation of the human immunodeficiency virus type-1 transcription elongation complex. EMBO J. 16:5260-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keen, N. J., M. J. Gait, and J. Karn. 1996. Human immunodeficiency virus type-1 Tat is an integral component of the activated transcription-elongation complex. Proc. Natl. Acad. Sci. USA 93:2505-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerppola, T. K., and C. M. Kane. 1990. Analysis of signals for transcription termination by purified RNA polymerase II. Biochemistry 29:269-278. [DOI] [PubMed] [Google Scholar]
- 31.Kim, J. B., and P. A. Sharp. 2001. Positive transcription elongation factor b phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J. Biol. Chem. 276:12317-12323. [DOI] [PubMed] [Google Scholar]
- 32.Kulish, D., and K. Struhl. 2001. TFIIS enhances transcriptional elongation through an artificial arrest site in vivo. Mol. Cell. Biol. 21:4162-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar, K. P., S. Akoulitchev, and D. Reinberg. 1998. Promoter-proximal stalling results from the inability to recruit transcription factor IIH to the transcription complex and is a regulated event. Proc. Natl. Acad. Sci. USA 95:9767-9772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwak, Y. T., D. Ivanov, J. Guo, E. Nee, and R. B. Gaynor. 1999. Role of the human and murine cyclin T proteins in regulating HIV-1 Tat-activation. J. Mol. Biol. 288:57-69. [DOI] [PubMed] [Google Scholar]
- 35.Laspia, M. F., A. P. Rice, and M. B. Mathews. 1989. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell 59:283-292. [DOI] [PubMed] [Google Scholar]
- 36.Lin, R., D. Gewert, and J. Hiscott. 1999. Differential transcriptional activation in vitro by NF-κB/Rel proteins. J. Biol. Chem. 270:3123-3131. [DOI] [PubMed] [Google Scholar]
- 37.Liu, Y., C. Suñé, and M. A. Garcia-Blanco. 1999. Human immunodeficiency virus type 1 Tat-dependent activation of an arrested RNA polymerase II elongation complex. Virology 255:337-346. [DOI] [PubMed] [Google Scholar]
- 38.Mancebo, H. S. Y., G. Lee, J. Flygare, J. Tomassini, P. Luu, Y. Zhu, J. Peng, C. Blau, D. Hazuda, D. Price, and O. Flores. 1997. p-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 11:2633-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marciniak, R. A., B. J. Calnan, A. D. Frankel, and P. A. Sharp. 1990. HIV-1 Tat protein trans-activates transcription in vitro. Cell 63:791-802. [DOI] [PubMed] [Google Scholar]
- 40.Marciniak, R. A., and P. A. Sharp. 1991. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 10:4189-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palangat, M., and R. Landick. 2001. Roles of RNA:DNA hybrid stability, RNA structure, and active site conformation in pausing by human RNA polymerase II. J. Mol. Biol. 311:265-282. [DOI] [PubMed] [Google Scholar]
- 42.Palangat, M., T. I. Meier, R. G. Keene, and R. Landick. 1998. Transcriptional pausing at +62 of the HIV-1 nascent RNA modulates formation of the TAR RNA structure. Mol. Cell 1:1033-1042. [DOI] [PubMed] [Google Scholar]
- 43.Parada, C. A., and R. G. Roeder. 1996. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature 384:375-378. [DOI] [PubMed] [Google Scholar]
- 44.Parada, C. A., J.-B. Yoon, and R. G. Roeder. 1995. A novel LBP-1-mediated restriction of HIV-1 transcription at the level of elongation in vitro. J. Biol. Chem. 270:2274-2283. [DOI] [PubMed] [Google Scholar]
- 45.Pasman, Z., and P. H. von Hippel. 2000. Regulation of Rho-dependent transcription termination by NusG is specific to the Escherichia coli elongation complex. Biochemistry 39:5573-5585. [DOI] [PubMed] [Google Scholar]
- 46.Ping, Y.-H., and T. M. Rana. 1999. Tat-associated kinase (P-TEF-b): a component of transcription preinitiation and elongation complexes. J. Biol. Chem. 274:7399-7404. [DOI] [PubMed] [Google Scholar]
- 47.Ping, Y. H., and T. M. Rana. 2001. DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates pTEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. J. Biol. Chem. 276:12951-12958. [DOI] [PubMed] [Google Scholar]
- 48.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Renner, D. B., Y. Yamaguchi, T. Wada, H. Handa, and D. H. Price. 2001. A highly purified RNA polymerase II elongation control system. J. Biol. Chem. 276:42601-42609. [DOI] [PubMed] [Google Scholar]
- 50.Rittner, K., M. J. Churcher, M. J. Gait, and J. Karn. 1995. The human immunodeficiency virus long terminal repeat includes a specialised initiator element which is required for Tat-responsive transcription. J. Mol. Biol. 248:562-580. [DOI] [PubMed] [Google Scholar]
- 51.Selby, M. J., E. S. Bain, P. Luciw, and B. M. Peterlin. 1989. Structure, sequence and position of the stem-loop in TAR determine transcriptional elongation by Tat through the HIV-1 long terminal repeat. Genes Dev. 3:547-558. [DOI] [PubMed] [Google Scholar]
- 52.Sodroski, J., R. Patarca, C. Rosen, F. Wong-Staal, and W. A. Haseltine. 1985. Location of the trans-acting region on the genome of human T-cell lymphotropic virus type III. Science 229:74-77. [DOI] [PubMed] [Google Scholar]
- 53.Suñé, C., and M. A. Garcia-Blanco. 1995. Transcriptional trans-activation by human immunodeficiency virus type 1 Tat requires specific coactivators that are not basal factors. J. Virol. 69:3098-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taube, R., K. Fujinaga, J. Wimmer, M. Barboric, and B. M. Peterlin. 1999. Tat transactivation: a model for the regulation of eukaryotic transcriptional elongation. Virology 264:245-253. [DOI] [PubMed] [Google Scholar]
- 55.Tirode, F., D. Busso, F. Coin, and J.-M. Egly. 1999. Reconstitution of the transcription factor TFIIH: assignment of functions for the three subunits XPB, XPD, and cdk7. Mol. Cell 3:87-95. [DOI] [PubMed] [Google Scholar]
- 56.von Hippel, P. H. 1998. An integrated model of the transcription complex in elongation, termination and editing. Science 281:660-665. [DOI] [PubMed] [Google Scholar]
- 57.Wada, T., G. Orphanides, J. Hasegawa, D.-K. Kim, D. Shima, Y. Yamaguchi, A. Fukada, K. Hisatake, O. Sangtaek, D. Reinberg, and H. Handa. 2000. FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between p-TEFb and TFIIH. Mol. Cell 5:1067-1072. [DOI] [PubMed] [Google Scholar]
- 58.Wada, T., T. Takagai, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that pTEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei, P., M. E. Garber, S.-M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel cdk9-associated c-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 61.Wu-Baer, F., W. S. Lane, and R. B. Gaynor. 1998. Role of the human homolog of the yeast transcription factor SPT5 in HIV-1 Tat-activation. J. Mol. Biol. 277:179-197. [DOI] [PubMed] [Google Scholar]
- 62.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41-51. [DOI] [PubMed] [Google Scholar]
- 63.Yamaguchi, Y., T. Wada, D. Watanabe, T. Takagi, J. Hasegawa, and H. Handa. 1999. Structure and function of the human transcription elongation factor DSIF. J. Biol. Chem. 274:8085-8092. [DOI] [PubMed] [Google Scholar]
- 64.Zawel, L., P. Kumar, and D. Reinberg. 1995. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 9:1479-1490. [DOI] [PubMed] [Google Scholar]
- 65.Zhou, M., M. A. Halanski, M. F. Radonovich, F. Kashanchi, J. Peng, D. H. Price, and J. N. Brady. 2000. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 20:5077-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou, Q., D. Chen, E. Pierstorff, and K. Luo. 1998. Transcription elongation factor p-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 17:3681-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou, Q., and P. A. Sharp. 1995. Novel mechanism and factor for regulation by HIV-1 Tat. EMBO J. 14:321-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou, Q., and P. A. Sharp. 1996. Tat-SF1: cofactor for stimulation of transcriptional elongation by HIV-1 Tat. Science 274:605-610. [DOI] [PubMed] [Google Scholar]
- 69.Zhu, Y., T. Pe'ery, J. Peng, Y. Ramanathan, N. Marshall, T. Marshall, B. Amendt, M. B. Mathews, and D. H. Price. 1997. Transcription elongation factor p-TEFb is required for HIV-1 Tat trans-activation in vitro. Genes Dev. 11:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]