Abstract
By microinjecting purified glutathione S-transferase linked to all or parts of herpes simplex virus type 1 US11 protein into either the nucleus or the cytoplasm, we have demonstrated that this nucleolar protein exhibits a new type of localization signal controlling both retention in nucleoli and export to the cytoplasm. Saturated mutagenesis combined with computer modeling allowed us to draw the fine-structure map of this domain, revealing a new proline-rich motif harboring both activities, which are temperature dependent and regulated by phosphorylation. Finally, crossing the nuclear pore complex from the cytoplasm to the nucleus is an energy-dependent process for US11 protein, while getting to nucleoli through the nucleoplasm is energy independent.
Nucleoli are mainly the seat of ribosome biogenesis, a highly complex process leading to the production of preribosomal particles, which are then released in the nucleoplasm and exported to the cytoplasm as mature ribosomal subunits (1, 29, 36, 48, 50). Recently, new data have suggested that the nucleolus might also play a crucial role in several other key events in cell life. By being the site of transient sequestration and maturation of several factors, the nucleolus might be involved in the control of cell division, aging, and mRNA export (26, 40, 41). In addition, proteins belonging to very distinct species of viruses also have various functions in the nucleolus. These proteins appear to be involved in replication of viral genomes as well as in transcriptional and posttranscriptional regulation of gene expression (11, 12, 32, 44, 52, 55). As a consequence of the various functions of the nucleolus, large amounts of many different components are imported into and exported out of it.
Whereas a clear picture of the general mechanisms governing nucleocytoplasmic transport of proteins is now emerging, how nucleolus components are delivered to their correct site of action for a coordinate activity remains poorly understood (29, 50). Crossing the nuclear pore complex (NPC) requires a signal and is mediated by an energy-dependent process (for a review, see references 39 and 21). Prototypic mono- and bipartite classical nuclear localization signals (cNLS) are those found in simian virus 40 large T antigen and in nucleoplasmin. They are characterized, respectively, by one or two stretches of basic residues (5, 27). Prototypic leucine-rich nuclear export signals (NES) are those found in human immunodeficiency virus type 1 (HIV-1) Rev protein and in cyclic AMP-dependent protein kinase inhibitor (18, 54). They are characterized by a short sequence containing critically spaced leucine. In addition, new transport signals called NS signals (NSS for nucleocytoplasmic shuttling signal) that can direct both nuclear import and nuclear export have been discovered recently (35).
However, many proteins that shuttle between the nucleus and cytoplasm do not display any of these identified transport signals. Several sequences have been defined as nucleolus localization signals (NoLS). However, rather than being NoLS, these signals appear to be nucleolar retention signals (NoRS). Except for a very few proteins (5), these NoRS are generally built with sequences rich in arginine and lysine. NoRS sizes are highly variable, ranging from five amino acids for angiogenin up to several tens or so for nucleolin (9, 38). Therefore, amino acid sequence analysis fails in predicting putative NoRS, especially because they very often overlap NLS.
Infection of human cells with herpes simplex virus type 1 (HSV-1) is followed by important modifications of nucleolus organization (45). At least two HSV-1 proteins involved in posttranscriptional regulation of gene expression, ICP-27 and US11 protein, exhibit localization in nucleoli, the significance of which remains an enigma. Even though the function of US11 protein has not been fully elucidated, it has been demonstrated that this RNA-binding protein is a posttranscriptional regulator of gene expression (7). By its ability to bind target mRNAs and transport them to the cytoplasm, US11 protein displays striking functional, albeit not structural, similarities to HIV-1 Rev and human T-cell leukemia/lymphoma virus type I Rex proteins (12, 16).
However, as for Rev and Rex, intracellular movements of US11 and its localization in nucleoli are critical for its function. Under transient or constitutive expression and in the absence of viral infection, US11 protein shows a localization identical to that observed in infected cells, i.e., in nucleoli and in the cytoplasm, colocalizing with ribosomes (3, 15, 43, 47). These and other results (12, 16) indicate that US11 protein may contain an NoRS and possibly an NES in addition to the expected NLS, the analysis and description of which will be reported elsewhere.
No sequence homology reminiscent of other already-known localization and retention signals has been found within US11 protein. Therefore, we have attempted to delimit and dissect these signals. This was achieved by using a strategy of microinjection of purified glutathione S-transferase (GST)-US11 hybrid proteins into either the nucleus or the cytoplasm, followed by localization of the injected proteins by immunofluorescence. These hybrid proteins were built with either wild-type or modified US11 proteins by amino acid deletion. Using this and another approach combining localization of transiently expressed US11 proteins harboring amino acid substitutions together with tertiary-structure prediction, we have defined the sequence, phosphorylation, and energy requirements for NoRS and NES functions. These results allow us to propose a structure-function relationship of a new kind of bipartite NoRS in US11 protein that also contains an NES, the function of which appears to be regulated by phosphorylation.
MATERIALS AND METHODS
Culture conditions and transfection of HeLa cells.
HeLa cells were grown at 37°C as a monolayer under 5% CO2 in minimal essential medium (MEM) supplemented with 5% heat-inactivated fetal calf serum (FCS). For inhibition of energy-dependent cellular processes, MEM was supplemented with 5 mM HEPES, pH 7.4, and cells were first maintained at room temperature for 10 min and then incubated at 4°C before microinjection for different time courses as indicated. Cells were maintained at 4°C for microinjection.
For actinomycin D treatments, cells were incubated at 37°C for 3 h in MEM containing either 0.05 or 5 μg of actinomycin D per ml. For experiments involving ATP depletion, cells were incubated at 37°C for 2 h in MEM containing either 15 μM 2-deoxyglucose or 12 μM rotenone or a mixture of both compounds at the same concentrations as when used alone. Actinomycin D, 2-deoxyglucose, and rotenone were maintained in the medium for microinjections and after microinjection all along the time course of incubation at 37°C.
Cells were transfected by the calcium phosphate precipitation procedure in the conditions described previously (23). For experiments involving treatment with staurosporine, cells were incubated at 37°C for 15 min in MEM containing 0.2 μM staurosporine before microinjections. Used at this concentration and for a short time, staurosporine was shown to be an efficient inhibitor of several protein kinases, such as protein kinase C (PKC) and mitogen-activated protein (MAP) kinases (2, 20). Staurosporine was maintained in the medium for microinjections and after microinjection all along the 30 min of incubation at 37°C.
Determination of intracellular ATP content.
After treatment of cells with 2-deoxyglucose and/or rotenone, the culture medium containing these compounds was removed and cells were scraped off into ice-cold phosphate-buffered saline (PBS; 130 mM NaCl, 4 mM Na2HPO4 · 7H2O, 1.5 mM KH2PO4). Cells were collected by centrifugation at 500 × g for 15 min at 4°C and lysed in 7% HClO4. The lysate was subjected to centrifugation at 10,000 × g for 10 min at 4°C. The resulting supernatant was neutralized with KOH before quantifying ATP by the UV method using hexokinase and glucose-6-phosphate dehydrogenase.
Construction and preparation of recombinant GST-US11-derived proteins.
We have previously described the construction of five expression vectors used in this study (49). The construction of the other nine vectors was achieved using the same experimental strategy by cloning different parts of the US11 coding sequence (HSV-1 strain KOS) in frame with the GST coding sequence of pGEX-2T (Amersham Pharmacia Biotech). These new expression vectors contain the sequences coding for the entire GST protein. Sequences coding for parts of the US11 protein (KOS strain) are indicated to the right of Fig. 1D.Hybrid proteins containing GST linked to parts of the US11 protein from amino acids 41 to 149, 88 to 149, 1 to 40, and 1 to 83 were previously named GST-US11-m5, GST-US11-m6, GST-US11-m15, and GST-US11-m11, respectively (49).
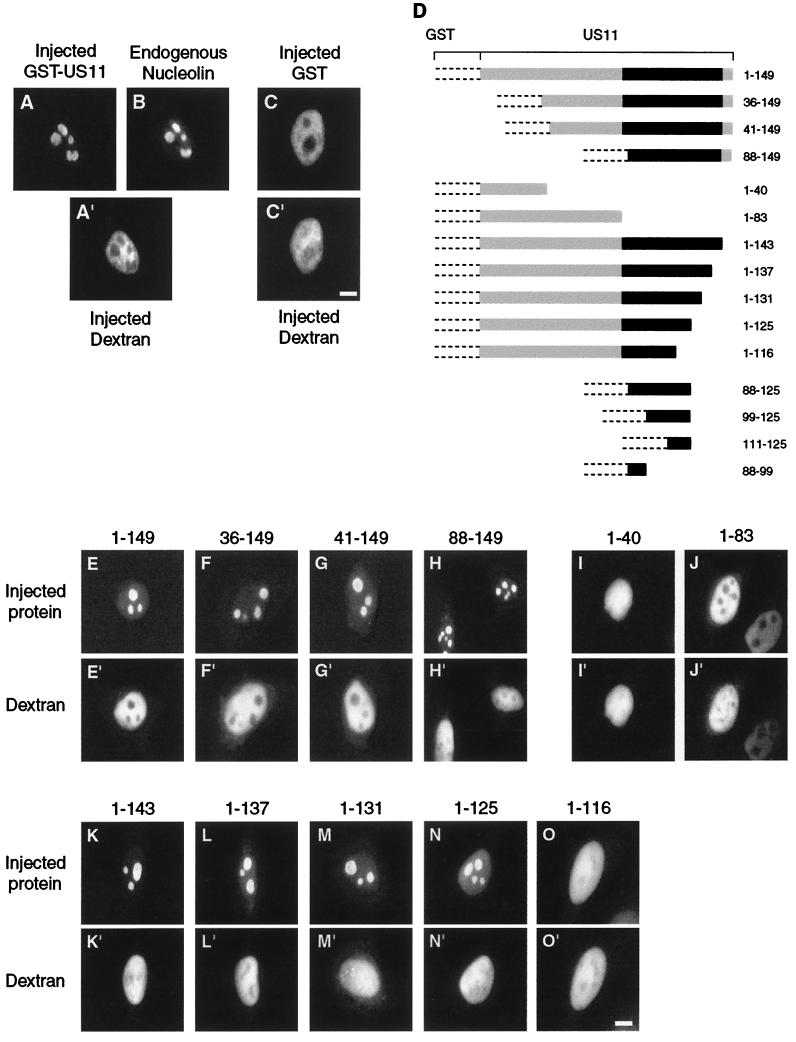
FIG. 1.
Delineation of US11 NoRS. (A to C) Recombinant hybrid proteins GST-US11 and GST were purified, mixed with high-molecular-weight Texas Red-labeled dextran, and injected into the nuclei of HeLa cells. Injected cells were incubated at 37°C for 30 min and fixed. Injected GST-US11 (A), endogenous nucleolin (B), and injected GST (C) were detected by indirect immunofluorescence using a monoclonal mouse anti-GST antibody (A), a polyclonal rabbit antinucleolin antibody (B), and a rabbit polyclonal anti-GST antibody (C), as well as aminomethylcoumarin acetate-labeled (A) and fluorescein isothiocyanate-labeled (B and C) second antibody. The site of injection was monitored in each injected cell by direct observation of fluorescence of Texas Red-labeled dextran under an inverted microscope (A" and C"). (D) Construction of GST-US11 hybrid proteins. Recombinant hybrid proteins were constructed by cloning parts of the US11 coding sequence from the HSV-1 KOS strain in frame with the GST coding sequence. GST protein is represented by dotted lines, and the US11 coding sequence is represented by a plain box, in which the black part represents the 20 repeats of the XPR sequence. GST-US11 contains the full-length US11 coding sequence. The other hybrid proteins contain the same GST but only parts of the US11 protein. Numbers to the right of each construct indicate the positions of amino acids in the US11 protein. (E to O) Intranuclear localization of GST-US11 hybrid proteins. Recombinant hybrid proteins depicted in D were mixed with Texas Red-labeled dextran and injected into the nuclei of HeLa cells. Hybrid proteins were detected by indirect immunofluorescence 30 min after injection using either an anti-US11 antibody (E to H) or an anti-GST antibody (I to O) and fluorescein isothiocyanate-labeled second antibody. Fluorescence of Texas Red-labeled dextran was directly visualized under an inverted microscope (E" to O"). Bars, 10 μm.
Recombinant proteins were produced in Escherichia coli DH5α (Gibco-BRL) and purified from bacterial lysates by affinity chromatography on glutathione-agarose beads (Pharmacia) (49). Solutions of purified proteins were dialyzed against 10 mM Tris-HCl, pH 7.4, concentrated up to 5 mg/ml by filtration (Nanosep; Millipore), and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (28). cNLS-BSA was obtained by coupling the biotinylated peptide GGCYGPKKKRKVGG of simian virus 40 large T antigen to bovine serum albumin (BSA) (8) (Neosystem). The plasmid used for production of GST-M9 was kindly provided by B. Cullen (Department of Genetics and Howard Hughes Medical Institute, Duke University Medical Center, Durham, N.C.) (19). GST-M9 was built by adding the M9 nuclear export signal of hnRNPA1 to GST.
Saturated mutagenesis of the US11 coding sequence.
Base substitutions were introduced into DNA encoding US11 protein by chemical mutagenesis, followed by amplification of randomly modified DNA by PCR (13). For this, the pG-61 vector was first constructed. This plasmid contained the full-length US11 coding sequence under the control of the cytomegalovirus (CMV) immediate-early promoter and was obtained by modification of pCMV-US11 (12). Construction of pG-61 involved destroying the XhoI, ApaI, and KpnI restriction sites within the vector sequence (originally derived from pBC12/CMV [10]) and addition of HindIII and BglII restriction sites at the boundaries of the US11 coding sequence. These modifications facilitated ligation of the mutated fragments of US11 coding sequences within the eukaryotic expression vector. The target pG-61 was dissolved in 250 mM sodium acetate (pH 4.3) containing 1 M sodium nitrite and incubated at 24°C. This resulted in the formation of nitrous acid that induced a deamination of DNA. Aliquots were withdrawn at 10-min intervals and diluted in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.5). The pH was neutralized with sodium acetate, and DNA was precipitated with ethanol.
The DNA of each aliquot was recovered by centrifugation and dissolved in TE buffer for amplification by PCR. Fragments of amplified mutated DNA were excised from the initial US11 coding sequence by endonuclease digestion at the HindIII-XhoI, XhoI-KpnI, and KpnI-BglII restriction sites, giving DNA fragments of 109, 244, and 101 bp, respectively. Each of them was then inserted into the acceptor vector pG-61 in place of the wild-type sequence previously digested by the same enzymes. Three expression libraries were thus generated, each of them containing plasmids harboring point mutations within a defined part of the US11 coding sequence. Several plasmids were purified from each library, and the US11-inserted DNAs were sequenced using the dideoxy chain termination method. From 30 to 60% of the plasmids generated in each library contained codons specifying amino acids different from those of the wild-type US11 protein. Selected modified forms of US11 proteins are reported in Fig. 5. They are designated US11-m plus an arbitrary number.
FIG. 5.
Amino acids involved in NoRS/NES function of US11 protein. (A) Sequences of altered forms of US11 carrying amino acid replacements distributed throughout the domain encompassing the NoRS/NES. The sequence of amino acids 81 to 140 of wild-type US11 protein is reported at the top of the figure. Amino acids are specified by standard single-letter abbreviations. Amino acid sequences from the same region were deduced after sequencing of the 30 different clones of US11 genes and aligned below the wild-type sequence. Amino acids identical to that of the wild-type protein are indicated by dots. Names of the modified forms of US11 are indicated to the right. Nine of the 30 modified US11 proteins exhibited other amino acid replacements or deletions in addition to those shown here, namely, Q for L64 in US11-m45; V for D69 in US11-m65; S for N51 and L for Q57 in US11-m50; E for A55 in US11-m58; S for G41 in US11-m27; L for S73 in US11-m30; M for T144 in US11-m17; and N for I42 and deletion of amino acids 45 to 62 in US11-m33. Seven clones were derived from US11-m14 by a second step of mutagenesis; they are specified by a star after their name. (C to G) Intracellular localization of indicated US11 proteins carrying amino acid substitutions compared to that of wild-type US11 protein (B). Modified US11 proteins were localized by indirect immunofluorescence using an anti-US11 antibody 48 h after transfection. Identical distributions of the different US11 proteins were always obtained whatever the expression level after transfection. Bar, 10 μm.
Microinjection of cells and immunofluorescence analysis.
Solutions used for microinjection were prepared immediately before injection and centrifuged at 10,000 × g for 30 min at 4°C to remove possible particles. Purified proteins were used at a final concentration of 4 mg/ml (except for cNLS-BSA, which was used at 2 mg/ml) and mixed with Texas Red-labeled dextran of 70 kDa (Molecular Probes) at 2 mg/ml as a marker of the injection site. In a typical experiment, 5 × 105 to 1 × 106 copies of protein were injected per cell. When indicated in the text or legends, wheat germ agglutinin (WGA) and/or N-acetylglucosamine (GlcNac) was added to the mixture at 200 μg/ml and 50 μM, respectively. Forty-eight hours before injection, 3 × 105 HeLa cells were plated onto 60-mm-diameter petri dishes (Falcon). Just before injection, the culture medium was replaced and supplemented by 5 mM HEPES, pH 7.4. Cells were injected into either the nucleus or the cytoplasm using a micromanipulator 5171 and an Eppendorf microinjector 5242 installed on an Axiovert 35 M inverted microscope from Zeiss. Microinjections were performed under nitrogen gas pressure using sterile glass capillaries (Femtotips; Eppendorf). About 30 cells were injected in each experiment except when indicated otherwise in the text. Cells were then incubated under the conditions indicated.
For immunofluorescence analyses performed after either transfections or microinjections, cells were fixed with paraformaldehyde and permeabilized with Triton X-100. US11 protein, modified forms of US11 protein, and GST-US11 and other recombinant hybrid GST-US11 proteins were localized with either a rabbit polyclonal anti-US11 antibody (14) or a rabbit polyclonal anti-GST antibody (Eurogentec), unless otherwise specified, and fluorescein isothiocyanate-labeled second antibody (Sigma) or aminomethylcoumarin acetate-labeled second antibody (Jackson Immunoresearch). Rabbit polyclonal antinucleolin antibody was kindly provided by P. Bouvet (Centre National de la Recherche Scientifique, Lyon, France). Fluorescent antibodies and injected Texas Red-labeled dextran were visualized by epifluorescence using an Axiophot microscope from Zeiss. Digital images were captured using a charge-coupled device camera (LH 750 from Lhesa Electronique). Injected recombinant proteins were detected in the cytoplasm and in the nucleus up to 48 h after injection.
32P labeling, immunoprecipitation, and electrophoresis procedures.
HeLa cells (105 cells) were transfected with 15 μg of pCMV-US11 (MP strain), pCMV-US11 KOS, or pCMV-US11m55 (KOS). For in vivo 32P labeling, the medium was replaced by phosphate and serum-free MEM 45 h after transfection. After 1 h, this medium was replaced by MEM supplemented with 5% FCS containing 40 μCi (1.48 MBq) of 32P-labeled H3PO4 (Amersham Pharmacia Biotech) per ml. After 2 h of labeling, cells were lysed in 10 mM Tris-HCl (pH 7.4) buffer containing 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate, 150 mM NaCl, and 1 mM EDTA. The lysate was incubated for 16 h at 4°C with anti-US11 antibody (100-fold dilution). The antigen-antibody complex was captured by protein A coupled to Sepharose CL-4B beads (APC). After washing, proteins were solubilized for separation by electrophoresis in the presence of SDS through a polyacrylamide gel (SDS-PAGE).
Western blot analysis of US11 proteins thus separated was performed as previously described (14), and 32P-labeled proteins were identified by autoradiography after drying the membrane. Phosphorylation of US11 protein was analyzed after separation of acetic acid-extracted proteins by two-dimensional polyacrylamide gel electrophoresis as already described in detail (34). In brief, the first-dimension separation was carried out in a 4% (wt/vol) polyacrylamide gel containing 8 M urea at pH 8.6. The second dimension separation was performed in a 12.5% polyacrylamide gel containing 6 M urea at pH 6.75 in the presence of SDS. Separated proteins were transferred to polyvinylidene difluoride membranes (Immobilon-P from Millipore), and US11 protein was visualized by building a molecular complex with rabbit anti-US11 antibodies (14) and anti-rabbit immunoglobulin conjugated to horseradish peroxidase, the position of which was revealed by fluorography using an ECL kit (Amersham Pharmacia Biotech) together with X-Omat photographic films (Kodak).
Molecular modeling of US11 protein and its modified forms.
Models were generated using the Molecular Simulations (MSI) modules INSIGHTII, BIOPOLYMER, and DISCOVER (version 98), run on a Silicon Graphics O2 workstation. A three-dimensional model of US11 protein was first built by imposing a polyproline II helix structure for the whole sequence. This initial structure was further refined through successive cycles of energy minimization and molecular dynamics. Minimization was performed using the steepest descents for 200 cycles and conjugate gradients for 1,000 cycles, until the maximum derivative was less than 0.1 with a forcing constant of 100 kcal/mol. Dynamics was initialized at 900 K for 250 steps of 1.0 fs. The same protocol was applied to all of the modified forms of US11 protein under study.
RESULTS
NoRS and NES in US11 protein.
Microinjection of purified proteins directly into the nucleus provides a powerful strategy for recognizing localization and retention signals of proteins independently of the signals required for their nuclear import. To validate this strategy, we first verified that a hybrid protein built by adding US11 to the C-terminal end of GST was indeed able to concentrate in nucleoli of noninfected cells. Before injection, GST-US11 protein was purified and mixed with high-molecular-weight Texas Red-labeled dextran, which does not diffuse through an intact nuclear envelope. Injected cells were incubated at 37°C for 30 min, and we localized GST-US11 by immunofluorescence (Fig. 1A) and dextran by direct observation (Fig. 1A"). Thirty minutes after injection, GST-US11 was highly concentrated in discrete structures within the nucleus (Fig. 1A). The same injected cells analyzed by immunofluorescence with an antinucleolin antibody showed colocalization of GST-US11 and nucleolin, demonstrating that these structures were indeed nucleoli (Fig. 1B). As expected for a precise injection into the nucleus, the coinjected dextran was restricted exclusively to the nucleus and was excluded from nucleoli (Fig. 1A"). GST alone, injected into the nucleus, was also excluded from nucleoli (Fig. 1C).
It was next determined whether a single and continuous discrete NoRS could be localized in US11 protein. For this, hybrid proteins that contained parts of US11 linked to GST (Fig. 1D) were localized as described above. Hybrid proteins lacking the N-terminal parts of US11 still displayed a strong concentration in nucleoli, similar to that of the full-length GST-US11 (Fig. 1, compare E with F to H). Accumulation in nucleoli was observed even for a protein lacking the entire N-terminal half of US11 protein (Fig. 1H). This suggested that the NoRS of US11 was localized in its C-terminal half (88 to 149). If this were true, hybrid proteins containing the first 40 and the first 83 amino acids of US11 protein should be excluded from nucleoli after injection into the nucleus. As expected, neither of these two proteins displayed the ability to concentrate in nucleoli (Fig. 1I and 1J).
The C-terminal domain of US11 protein is composed of the XPR tripeptide in 20 repetitions. These 20 XPR repeats are followed by the TARGSV terminal hexapeptide. Therefore, because the C-terminal half of US11 is made of highly repetitive sequences, we determined whether each of these repeats played an equivalent role in allowing the concentration of US11 protein in nucleoli. Hybrid proteins were constructed in which the last six amino acids [GST-US11(1-143)] and then the XPR motifs of US11 protein were progressively and successively removed (Fig. 1D). As above, purified proteins were injected into the nucleus, and their intracellular distribution was visualized by immunofluorescence. Proteins lacking either the last six amino acids (TARGSV) or, in addition, the last two, four, or six XPR motifs retained their ability to concentrate in nucleoli (Fig. 1K, L, M, and N, respectively). Conversely, also removing the three preceding XPR motifs localized between positions 116 and 125 profoundly affected the ability of the hybrid protein to concentrate in nucleoli (Fig. 1O). Therefore, these results suggested strongly that the region between positions 88 and 125 contained the NoRS of US11 protein. However, to prove that the NoRS was indeed localized between amino acids 88 and 125, it was necessary to determine whether this fragment of US11 protein alone was sufficient to direct a foreign protein into nucleoli.
A new hybrid protein containing only amino acids 88 to 125 of US11 was then constructed (Fig. 1D) and injected into the nuclei of HeLa cells. Unexpectedly, the behavior of this hybrid protein was different from that of all the other proteins tested previously. While identical in all cells for all hybrid proteins tested so far, the intracellular distribution of the injected GST-US11(88-125) was heterogeneous from one cell to another, suggesting another role for the 88-125 peptide than that of an NoRS alone. Indeed, when injected into the nuclei of 118 cells, GST-US1(88-125) exhibited two different kinds of intracellular distribution (Fig. 2).
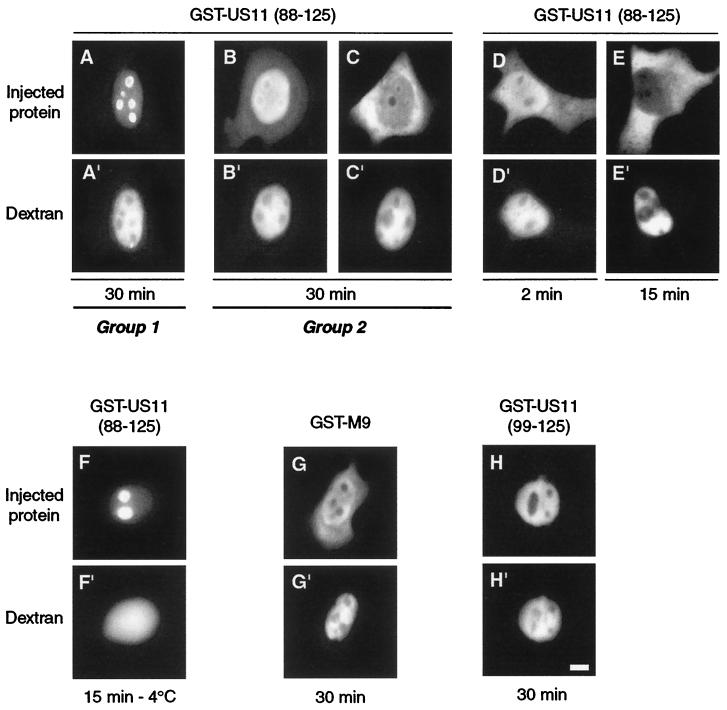
FIG. 2.
Identification of a bifunctional NoRS/NES in US11. Three different recombinant hybrid proteins were injected into the nuclei of HeLa cells, GST-US11(88-125) (A to F), GST-US11(99-125) (H), and GST-M9 (G) as a control for nuclear export (see text). Cells were fixed after either 30 min of incubation at 37°C (A to C and G to H), 2 min at 37°C (D), 15 min at 37°C (E), or 15 min at 4°C (F). Hybrid proteins were detected by indirect immunofluorescence using an anti-GST antibody and fluorescein isothiocyanate-labeled second antibody. The two kinds of localization observed (see text for description) are underlined and labeled group 1 and group 2. Fluorescence of Texas Red-labeled dextran was directly visualized under an inverted microscope (A" to H"). Bar, 10 μm.
In cells belonging to the first group, the hybrid protein was concentrated in nucleoli but was also detected in the nucleoplasm and almost not visible in the cytoplasm (Fig. 2A). In the second group of cells, the hybrid protein was barely detectable in nucleoli. It was concentrated in the region surrounding nucleoli and also abundant in the rest of the nucleoplasm and in the cytoplasm (Fig. 2B and C). Among the 118 injected cells, 45 (38.1%) exhibited a behavior similar to that shown in Fig. 2A, while the 73 (61.9%) remaining behaved like the two samples shown in Fig. 2B and C. These results suggested that the 88-125 US11 peptide was required not only for concentration in nucleoli but also for nuclear export of the hybrid protein.
The appearance of GST-US11(88-125) in the cytoplasm could be observed as soon as 2 min after injection into the nucleus (Fig. 2D). When present, exit of the protein was very efficient after 15 min (compare the fluorescence emitted by the nucleus and the cytoplasm 2 and 15 min after injection in Fig. 2D and E). GST-US11(88-125) was then injected into the nuclei of cells previously incubated at 4°C for 30 min. Injected cells were incubated at 4°C for another 15 min before fixation. In these conditions, the distribution of GST-US11(88-125) in all injected cells was identical to that in cells belonging to group 1 (compare F and A in Fig. 2).
As a control, the behavior of GST-M9 hybrid protein was followed after its injection into the nucleus. As expected, 30 min after injection, GST-M9 was found in the cytoplasm when incubated at 37°C (Fig. 2G) (4). In addition, GST-M9 was retained in the nucleoplasm when incubated at 4°C (not shown). The results presented above strongly suggest the existence of an active NES somewhere between amino acids 88 and 125 of US11 protein.
To delineate more precisely the N-terminal boundary of this signal, three other hybrid proteins were constructed and injected into the nucleus. The first two contained amino acids 99 to 125 and 111 to 125, while the third contained only the 88-99 peptide (Fig. 1D). In all injected cells, all three proteins were found exclusively in the nucleoplasm and were excluded from nucleoli, as shown for GST-US11(99-125) in Fig. 2H.
Taken together, these results indicate that the entire 88-125 peptide contains an NoRS together with an NES. However, they did not allow us to explain why the hybrid proteins were actively transported to the cytoplasm of only two-thirds of the cells when injected into the nucleus (see below).
Mode of accumulation of US11 protein in nucleoli.
To be localized in nucleoli, US11 protein should first be transferred to the nucleus through nuclear pores, then through the nucleoplasm to nucleoli. Crossing the NPC by an active mechanism should allow US11 protein to import GST to the nucleus. This was verified by injecting GST-US11 protein into the cytoplasm and localizing the hybrid protein after incubation at 37°C for 5, 20, 30, and 60 min (Fig. 3).Thirty minutes were necessary to allow GST-US11 to cross through the NPC, followed by its concentration in nucleoli (Fig. 3A to D). However, the hybrid protein was never found entirely accumulated in nucleoli; some of it remained in the cytoplasm.
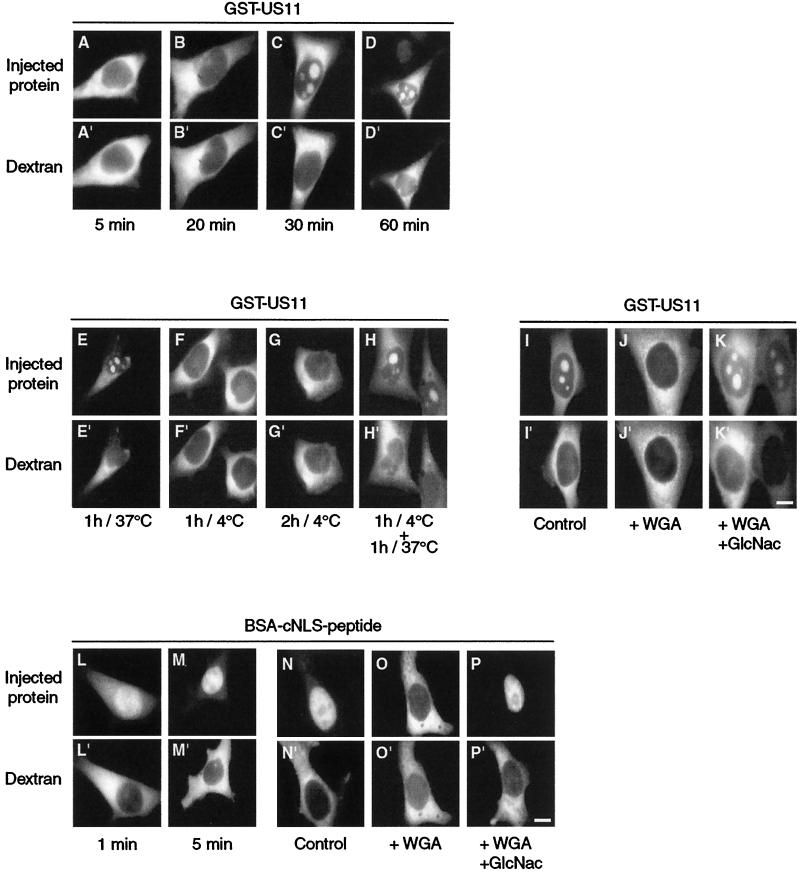
FIG. 3.
Active transport of GST-US11 protein from the cytoplasm to the nucleus. (A to D) Kinetics of nuclear import of GST-US11. Purified GST-US11 protein was mixed with Texas Red-labeled dextran and injected into the cytoplasm of HeLa cells. Cells were fixed after 5, 20, 30, or 60 min of incubation as indicated under each pair of pictures, and GST-US11 was detected by indirect immunofluorescence as described in the legend to Fig. 1. (E to K) Inhibition of nuclear import at 4°C and by WGA. GST-US11 was injected into the cytoplasm, and cells were subsequently incubated at 37 or 4°C for the indicated times (E to G). Injected cells were first incubated at 4°C and then at 37°C for the indicated times (H). GST-US11 was injected either alone (I) or together with WGA (J) or with a mixture of WGA and N-acetylglucosamine (K). (L to P) Nuclear import of BSA-cNLS peptide. As a control for experimental conditions, cNLS conjugated to BSA (see text) was mixed with Texas Red-labeled dextran and injected into the cytoplasm. Kinetics of nuclear import (L and M) and inhibition of nuclear import (N to P) of cNLS-BSA peptide by WGA were analyzed as for that of GST-US11 protein (A to K). BSA-cNLS peptide was detected using fluorescein isothiocyanate-labeled streptavidin. Fluorescence of Texas Red-labeled dextran was directly visualized under an inverted microscope (A" to P"). Bars, 10 μm.
Incubation of the injected cells for more than 18 h did not result in the complete accumulation of GST-US11 in nucleoli (data not shown). Migration of GST-US11 from the cytoplasm to the nucleus and in nucleoli was totally inhibited by incubation of cells at 4°C for 1 or 2 h immediately after injection (Fig. 3E to G). However, a subsequent incubation at 37°C for another hour restored the migration of GST-US11 to the nucleus and its accumulation in nucleoli (Fig. 3H). Conversely, such transport was blocked by coinjection of WGA into the cytoplasm (compare J to I in Fig. 3) but restored when GlcNac was also injected together with WGA and GST-US11 (Fig. 3K). When injected into the cytoplasm, the BSA-cNLS peptide conjugate was transported in less than 1 min to the nucleus (Fig. 3L), where it concentrated almost completely after 5 min (Fig. 3M). This migration was blocked by incubation of cells at 4°C immediately after injection (not shown) and also by coinjection of WGA (compare O to N in Fig. 3). In addition, coinjection of GlcNac together with WGA restored the migration of the BSA-cNLS peptide conjugate to the nucleus (Fig. 3P).
In order to evaluate the time necessary for GST-US11 to cross the NPC, its nucleolar accumulation requiring 30 min after injection into the cytoplasm, as demonstrated above, we determined the time necessary for GST-US11 to concentrate in nucleoli when injected directly into the nucleus. After such an injection, cells were either fixed immediately or incubated at 37°C for short periods (Fig. 4A to D).Less than 1 min was sufficient to provide a strong accumulation of GST-US11 in the nucleoli of all injected cells (Fig. 4A).
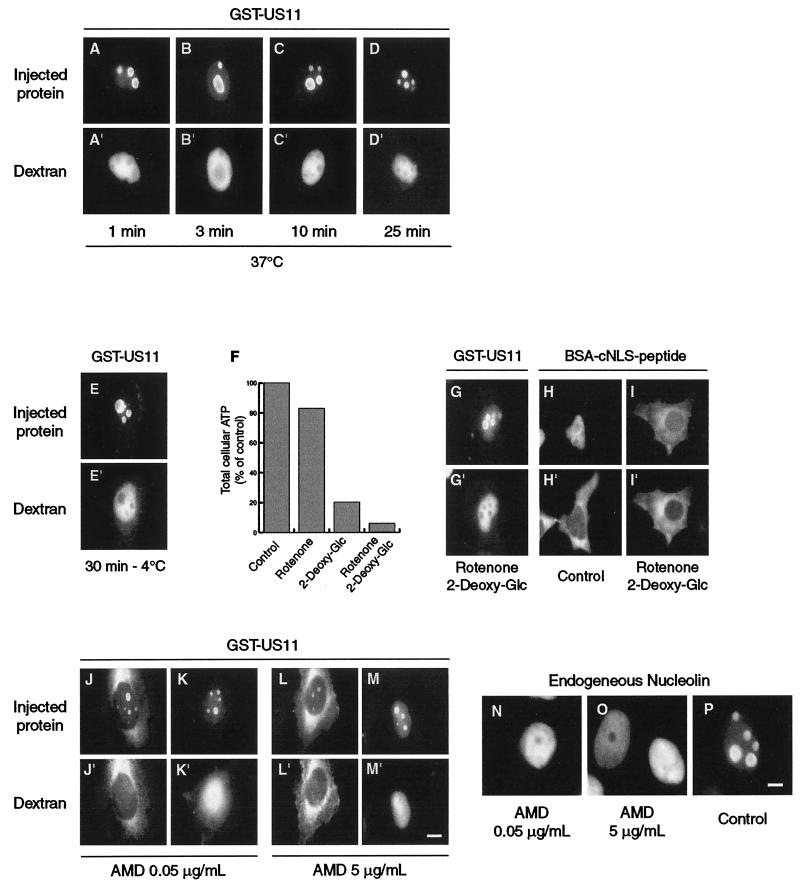
FIG. 4.
Modalities of accumulation in nucleoli of GST-US11 protein. (A to D) Kinetics of accumulation in nucleoli after injection into the nucleus. Purified GST-US11 protein was injected into the nucleus and detected by indirect immunofluorescence after 1, 3, 10, or 25 min of incubation at 37°C. (E and G to I) Accumulation in nucleoli of GST-US11 in the absence of energy. HeLa cells were incubated for 30 min at 4°C before injection of GST-US11 into the nuclei of cells, which were maintained at 4°C for another 30 min before fixation (E). HeLa cells were incubated with either rotenone, 2-deoxyglucose, or both for 2 h before measurement of the total cellular ATP. Results are presented as a percentage of the ATP found in control cells (F). GST-US11 was injected into the nuclei of cells treated with rotenone and 2-deoxyglucose. After 30 min at 37°C in the presence of the two compounds, cells were fixed and GST-US11 was detected by indirect immunofluorescence using an anti-US11 antibody. To control the consequence of ATP depletion for the transport of proteins through the NPC, cNLS-BSA peptide was injected into the cytoplasm of cells treated (I) or not (H) with both rotenone and 2-deoxyglucose for 2 h, and its localization was analyzed 15 min after injection using fluorescein isothiocyanate-labeled streptavidin. (J to M) Accumulation in nucleoli of GST-US11 in the absence of transcription. HeLa cells were treated with either 0.05 or 5 μg of actinomycin D (AMD) per ml for 3 h before they were injected with GST-US11 into either the cytoplasm (J and L) or the nucleus (K and M). GST-US11 was detected by indirect immunofluorescence after either 1 h (J and L) or 30 min (K and M) of incubation at 37°C in the presence of the same concentration of actinomycin D. (A" to M") Fluorescence of Texas Red-labeled dextran was directly visualized under an inverted microscope. (N and O) HeLa cells were treated with actinomycin D for 3 h before fixation. (P) Control untreated HeLa cells. In all cases (N to P), endogenous nucleolin was detected by indirect immunofluorescence using an anti-nucleolin antibody. Bars, 10 μm.
We next determined whether accumulation of GST-US11 in nucleoli was an energy-dependent process. For this, before injection, cells were either incubated at 4°C for 30 min or treated for 2 h with rotenone together with 2-deoxyglucose. Accumulation of GST-US11 in nucleoli was not inhibited at 4°C (Fig. 4E), nor was it inhibited after combined treatment with rotenone and 2-deoxyglucose (Fig. 4G). Such combined treatment induced a severe depletion of the intracellular ATP content (Fig. 4F) (24), thus inhibiting import of the control BSA-cNLS peptide after its injection into the cytoplasm (compare I to H in Fig. 4).
Because accumulation of several proteins in nucleoli is transcription dependent, and because US11 protein possibly binds to rRNA, we determined whether nuclear import and accumulation of GST-US11 in nucleoli were modified by actinomycin D treatment (46). HeLa cells were incubated for 3 h with either 0.05 or 5 μg of actinomycin D per ml in order to inhibit, respectively, the activity of either RNA polymerase I alone or both RNA polymerases I and II. The effectiveness of actinomycin D treatment was assessed by reduction of the size of nucleoli as visualized by phase-contrast light microscopy (data not shown).
When GST-US11 was injected into either the cytoplasm (Fig. 4J and L) or the nucleus (Fig. 4K and M), it still accumulated in the remaining nucleoli, whatever the concentration of actinomycin D and whatever the site of injection. Furthermore, extensive actinomycin D treatment of cells did not disturb the intracellular distribution of US11 protein whatever the means of US11 expression, either transient or constitutive (not shown). As expected, actinomycin D treatment induced a dramatic redistribution of endogenous nucleolin, which appeared to be homogeneously distributed throughout the nucleoplasm, becoming excluded from nucleoli (Fig. 4N to P).
Taken together, these results clearly demonstrated that transport of GST-US11 from the cytoplasm to the nucleus via the NPC was energy dependent. Conversely, its accumulation in nucleoli was very fast and independent of energy as well as of DNA transcription.
Amino acids involved in NoRS and NES function.
Under US11 transient expression, US11 protein was always distributed identically throughout the transfected cells whatever its concentration, i.e., accumulated in all nucleoli, abundant in the cytoplasm, and poorly detectable in the nucleoplasm (Fig. 5B).A similar distribution was observed under US11 constitutive expression (15). In these conditions, US11 protein was distributed in roughly identical amounts between the cytosol and the nucleus, even under cycloheximide treatment, as demonstrated by Western blot analysis (not shown). This suggests that the stability of the protein is identical in both compartments.
To determine which amino acids are critical for NoRS function, we designed a PCR-mediated chemical mutagenesis strategy to saturate the DNA coding for US11 protein with base substitutions. Among the 56 isolated DNA clones exhibiting point mutations all along the US11 coding DNA, 30 were selected for point mutations specifying amino acid substitutions in the above delineated NoRS/NES of US11 protein. The amino acid sequences deduced from these 30 DNA clones are reported in Fig. 5A. All these expression vectors containing mutated DNA were tested by transfection into HeLa cells for their ability to encode modified US11 proteins, the localization of which was determined by immunofluorescence as described above. In addition, the expression of each mutated US11 was verified by Western blot analysis of US11 protein after SDS-PAGE.
Twenty-one of the 30 modified US11 proteins displayed a cellular distribution that was not distinguishable from that of US11 wild-type protein (Fig. 5B). As an example of such similar distribution, that of US11-m53 is shown in Fig. 5C, concentrated in nucleoli, present in the cytoplasm, and barely detectable in the nucleoplasm (compare B with C in Fig. 5). However, intracellular distribution of the remaining nine modified US11 proteins was very different from that of the wild-type protein. US11-m14, US11-m16, and US11-m54 were very abundant in the nucleoplasm, detected in the cytoplasm, and excluded from nucleoli (Fig. 5D). Three other proteins, US11-m15, US11-m56, and US11-m69, were abundant in the nucleoplasm and were also found in nucleoli, although they accumulated in nucleoli apparently less than did wild-type US11 protein. Moreover, none of these three proteins was detected predominantly in the cytoplasm (Fig. 5E). US11-m12 exhibited a nuclear distribution similar to that of US11-m15 but was much more abundant in the cytoplasm (Fig. 5F). Conversely, US11-m55 and US11-m21 accumulated strongly in nucleoli, were apparently absent from the nucleoplasm, and were only poorly detected in the cytoplasm (Fig. 5G).
This fine analysis of the distribution of 30 modified US11 proteins indicated that several amino acid substitutions within the NoRS/NES of US11 appear to be necessary to modify its localization significantly. In general, single point mutations appear to be innocuous, with the remarkable exception of Arg 110 in the middle of the above-delineated NoLS/NES between amino acids 88 and 125, and of Ser 129, close to its C-terminal end but exterior to it.
Phosphorylation of US11 protein at Ser 129.
As demonstrated above, changing Ser 129 to Phe in US11-m55 and US11-m21 led to a strong accumulation of the protein in nucleoli (Fig. 5G). Therefore, we hypothesized that phosphorylation of Ser 129 in US11 wild-type protein could participate in the regulation of its intracellular movements. If this were true, US11-m55 protein should be less phosphorylated than wild-type US11 due to the phosphorylation of the latter on Ser 129.
To verify this point, HeLa cells were transfected with different expression vectors directing the synthesis of either wild-type US11 protein from the macroplaque strain (MP) or wild-type US11 or US11-m55, both from the KOS strain (Fig. 6A).US11 protein originating in the MP strain is 12 amino acids longer than that of the KOS strain. Four XPR repeats account for this additional sequence, which contains an extra serine residue (Fig. 6A) (51). Transfected cells were labeled with 32P, and US11 proteins were immunoprecipitated, separated by SDS-PAGE, and transferred to a nitrocellulose membrane for Western blot analysis using anti-US11 antibodies (Fig. 6B). After immunostaining, the membrane was subjected to autoradiography (Fig. 6C).
FIG. 6.
Identification of the major phosphorylation site of US11 protein and regulation of NoRS/NES function by Ser 129 phosphorylation. (A) Potential phosphorylation sites of US11. Sequences of the XPR repeats in US11 strain MP, US11 strain KOS, and US11-m55 (strain KOS) are indicated in the single-letter code. Potential phosphorylation sites for MAP kinase are indicated by an arrow, and those for casein kinase II and PKC are indicated by a triangle. A gap in the US11 and US11-m55 sequences of the KOS strains points out 12 additional amino acids present only in US11 of the MP strain (see text). (B and C) Immunoprecipitation of in vivo 32P-labeled US11 protein. US11 was immunoprecipitated with anti-US11 antibody in lysates from control HeLa cells (lanes 1 and 5) and from HeLa cells transfected with either CMV-US11 (MP) (lanes 2 and 6), CMV-US11 (KOS) (lanes 3 and 7), or CMV-US11-m55 (lanes 4 and 8) expression vectors. Immunoprecipitated US11 was subjected to SDS-PAGE and transferred to a nitrocellulose membrane for immunodetection with the same anti-US11 antibody (B). After immunostaining, the membrane shown in B was subjected to autoradiography (C). The position of US11 MP is indicated by a black arrow, and that of US11 KOS is indicated by a grey arrow to the left of panel B. (D to F) Analysis of US11 protein phosphorylation by two-dimensional PAGE. Proteins from lysate of HeLa cells previously transfected with the relevant vectors specified above were extracted and lyophilized for separation by two-dimensional PAGE (see text). Proteins were transferred to a nitrocellulose membrane, and US11 was detected with an anti-US11 antibody as described above (B). The positions of the two phosphorylated derivatives of US11 are indicated by a and b, while that of the unphosphorylated protein is labeled US11. Recombinant hybrid proteins GST-US11(88-131) with Ser (G and I) or Phe (H) in position 129 and GST-US11(88-125) (J) were purified, mixed with Texas Red-labeled dextran, and injected into the nuclei of HeLa cells. Injected cells were incubated at 37°C for 30 min in the presence (I and J) or absence (G and H) of 0.2 μM staurosporine and fixed. Hybrid GST-US11s were detected by indirect immunofluorescence using anti-GST antibody (G to J). Fluorescence of Texas Red-labeled dextran was directly visualized under an inverted microscope (G" to J"). Bar, 10 μm.
Both wild-type US11 proteins from the MP and KOS strains exhibited strong phosphorylation (Fig. 6C, lanes 6 and 7). This was not the case for US11-m55 (Fig. 6, lane 8), even though similar amounts of the three proteins were present on the membrane, as assessed by immunostaining (Fig. 6B). When analyzed by Western blot assay after separation by two-dimensional polyacrylamide gel electrophoresis, US11-m55 exhibited only one spot (Fig. 6F), while US11 (MP) and US11 (KOS) exhibited three (Fig. 6D) and two (Fig. 6E) spots, respectively, corresponding to proteins differing in charge. In this electrophoresis system, the phosphorylated derivatives of US11 protein migrated to the left of the main spot, as we have already reported (14, 51). All together, these results indicated that, at least in the absence of viral infection, Ser 129 was phosphorylated in US11 wild-type protein (both KOS and MP strains). The extra Ser 134 was also probably phosphorylated in the US11 protein of the MP strain, as suggested by its two-dimensional PAGE analysis.
Regulation of NoRS/NES activity by Ser 129 phosphorylation.
Because the Phe for Ser 129 substitution made US11 no longer phosphorylable (Fig. 6F), and because US11-m55 and US11-m21 accumulated in nucleoli more efficiently than wild-type US11, we checked for the localization of GST-US11(88-131) with either Ser or Phe in position 129 after microinjection into the nucleus. In 100% of injected cells, GST-US11(88-131)129S was localized in nucleoli, but also in the cytoplasm (Fig. 6G), while most of the GST-US11(88-131)129F was localized almost exclusively in nucleoli (Fig. 6H), 30 min after injection. In these conditions, among the 71 cells injected with GST-US11(88-131)129F, 63 (88.8%) exhibited the behavior shown in Fig. 6H, while the 8 (11.2%) remaining ones behaved like GST-US11(88-125), belonging to group 2 (Fig. 2B and C), i.e., excluded from nucleoli but localized in the nucleoplasm and in the cytoplasm (not shown). These results suggested very strongly that phosphorylation of Ser 129 stimulated the NES activity of GST-US11(88-131).
To verify this point, GST-US11(88-131)129S was injected into the nuclei of cells treated with 0.2 μM staurosporine for 15 min. After another 30 min in the presence of staurosporine, cells were fixed and the protein was localized by immunofluorescence. In these conditions, GST-US11(88-131)129S was localized almost exclusively in nucleoli of all injected cells (Fig. 6I), behaving like GST-US11(88-131)129F, as shown in Fig. 6H. The same treatment with staurosporine did not prevent export of GST-US11(88-125) from the nucleus (Fig. 6J), indicating that 0.2 μM staurosporine was probably sufficient to preclude Ser 129 phosphorylation without inhibiting the nuclear export machinery.
Fine-structure map of the US11 region encompassing the NoRS/NES.
We finally used computer modeling to detect the salient structural characteristics of the NoRS/NES. The thorough analysis of the different forms of US11 obtained by mutagenesis revealed the paramount importance of P112 and R116 in localization of the protein (Fig. 5A). Replacement of these two residues with L and G, respectively, causes a loss of accumulation in nucleoli. We have already pointed out that not all the XPR repeats are functionally equivalent. In the critical RAPRDPRVP (110 to 118) site, two X residues out of three are hydrophobic, namely, A111 and V117. The proline ring and the aliphatic portion of the arginine side chain being also apolar, the link of amino acids in the 111 to 117 sequence permits the formation of a very stable hydrophobic cluster in which A111 is stacked with both the P112 ring and the aliphatic part of the R116 side chain (Fig. 7).Furthermore, P112 is at stacking distance from R113, which is itself stacked with Val 117, while R116 is in stacking interaction with P115. As arginine residues are cornerstones of NoRS (5), we propose that R116 is a major determinant of the US11 NoRS and that its emergence over the numerous other US11 arginine residues results from its particular hydrophobic context.
FIG. 7.
Prediction of the three-dimensional structure of the 88 to 125 domain of US11 protein. An enlarged view of the bipartite NoRS is displayed. The brown color-coded dashed lines show distances compatible with stacking interactions (between 3.6 and 4.9 Å). The NoRS1 and NoRS2 submotifs are represented by the respective 90/95/96 and 111/116/117 residue triads (brown/red/yellow color coding).
Although essential to accumulation in the nucleolus, the 111-117 motif cannot be the sole determinant of the 88-125 region, as shown by the above-described analysis of the behavior of modified US11 proteins. First, two of them, US11-m56 and US11-m69, show an intracellular distribution not too different from that of wild-type US11 protein, even though exhibiting the double P112L and R116G mutation (Fig. 5A). Second, the 99 to 125 domain of US11 protein is not sufficient to specify the NoRS function (Fig. 2H). These observations prompted us to search for another determinant in the 88 to 99 segment.
One obvious candidate was the VPREPRV (90 to 96) motif. It offered the same distribution of hydrophobic residues as the prototypic APRDPRV 111 to 117 and adopted a similar conformation, as evaluated by a root mean square deviation score of 0.4 Å between corresponding carbon α (Cα). The V90I, R92L, and V96A substitutions in the m64, m50, and m58 US11 proteins, respectively, are all innocuous because they preserve hydrophobic side chains at these positions. Therefore, these two homologous motifs, being spatially close to each other, very likely contribute to building a new type of bipartite NoRS, since the distance between R95 and R116 is only 20.6 Å.
One possible sequential description of this motif is NoRS1-(XPR)4-NoRS2, where NoRS1 is the 89-97 peptide and NoRS2 is the 110-118 peptide. They both exhibit the RXhPRXaPRXhP sequence organization, where Xh and Xa stand for hydrophobic and acidic residues, respectively. Finally, this newly defined structural frame was validated by its ability to give a detailed account of the various types of US11 proteins, the behaviors of which are described above (Fig. 5 and data not shown).
DISCUSSION
By microinjecting all or parts of the HSV-1 US11 protein directly into either the nucleus or the cytoplasm, it was possible to study the protein's movements through the NPC and in the nucleus and then to dissect the domains that confer on the protein nucleolar retention and nuclear export ability, which appear to be regulated by phosphorylation. Because US11 protein does not exhibit an obvious well-defined cNLS, and because its small size makes its passive diffusion through the NPC possible, we first demonstrated that US11 protein is transported from the cytoplasm to the nucleus through the NPC by an energy-dependent process.
After injection into the cytoplasm, 30 min was necessary for GST-US11 to concentrate in nucleoli. This time appears longer than that determined previously for import into the nucleus of proteins bearing mono- as well as bipartite cNLS or M9 NLS (21, 22). In order to determine which step in the transport was responsible for this delay, GST-US11 was injected directly into the nucleus, revealing that the concentration in nucleoli required less than 1 min. Therefore, the 30-min delay appears to be mainly the consequence of the time required for US11 protein to cross the NPC, and probably due to as yet undetermined events occurring in the cytoplasm. Since import of the BSA-cNLS peptide conjugate into the nucleus was efficient as early as 1 min after injection into the cytoplasm and complete after 5 min, neither the size of GST-US11 (43.8 kDa) nor our experimental conditions could account for this delay.
Moreover, when injected directly into the nucleus, GST-US11 accumulated in nucleoli even at 4°C as well as after a severe depletion of ATP cellular content. Therefore, migration of GST-US11 inside the nucleus towards nucleoli does not require ATPase or GTPase activity. This is consistent with the absence of temperature influence on migration towards nucleoli of other proteins (42). Our results support the notion of diffusion of proteins inside the nucleus (37). In addition, GST-US11 protein is able to accumulate in nucleoli in experimental conditions leading to inhibition of nuclear import of other cytoplasmic proteins. This demonstrates that cofactors newly imported from the cytoplasm are not required for migration of US11 towards nucleoli. Furthermore, localization of US11 protein in nucleoli is independent of transcription by RNA polymerases I and II, a behavior similar to that of fibrillarin but very different from that of nucleolin and B23 (31, 42). This suggests also that once in the nucleolus, US11 protein interacts specifically with elements of granular and fibrillar components with which it colocalizes, as shown by electron microscopy (3).
By deleting N- and C-terminal parts of US11, we have located the NoRS between amino acids 88 and 125 (the smallest peptide of US11 protein required for retention in nucleoli). Then, by saturated mutagenesis, we showed that critical amino acids for NoRS function were indeed located in this domain. A combination of computer modeling and mutagenesis analysis enabled us to define this domain as a new type of bipartite NoRS that we describe as NoRS1-(XPR)4-NoRS2. NoRS1 and NoRS2 (89-97 and 110-118 peptides, respectively) display a similar structural organization in which a particular hydrophobic cluster ensures a precise orientation of one arginine residue (R95 for NoRS1 and R116 for NoRS2). Moreover, 62% of the injected cells exhibited the 88-125 peptide around nucleoli and dispersed throughout the nucleoplasm and in the cytoplasm. This strongly suggests that the 88 to 125 domain contained a nuclear export activity in addition to the NoRS. Therefore, restricting US11 to the 88-125 peptide allowed us to reveal an NES which was otherwise probably concealed within the full-length GST-US11. A similar result was obtained with the NoRS of MDM2 protein (33).
Because this domain did not display similarities with previously identified nuclear export signals, neither leucine-rich NES nor NSS, we had to find evidence that the 88 to 125 domain of US11 protein contained an active NES. Three arguments allowed us to conclude that this was indeed the case. First, nuclear export of GST-US11(88-125) was very rapid and efficient, since a large amount of the protein was found in the cytoplasm just 2 min after injection into the nucleus. Second, such nuclear export was totally inhibited at 4°C. Third, further deletion of the 88-125 peptide eliminated export of the recombinant proteins, demonstrating that the 88 to 125 domain was the minimum NES. These results demonstrate that the 88 to 125 domain is bifunctional, since it controls both retention within nucleoli and export out of the nucleus.
That retention and localization signals are located in the same domain implies a fine and coordinated regulation of their respective activities. Sequences surrounding the 88 to 125 domain appear to be involved in this regulation. We show that serine 129 is the main phosphorylation site of US11 protein, and changing Ser to Phe results in a stronger accumulation of the protein within nucleoli. This suggests regulation of the NES activity by phosphorylation of this serine residue. In another experiment, extending the 88-125 peptide up to amino acid 131, thus including Ser 129, resulted in export of GST-US11(88-131) protein in 100% of the injected cells, whereas export of GST-US11(88-125) was observed in only 62%. Treatment of cells with staurosporine at low concentration and replacing Ser 129 with Phe in the GST-US11(88-131) protein greatly decreased the ability of the protein to be transported from the nucleus to the cytoplasm. This effect is unlikely to be the consequence of a rapid reimportation of the mutated protein into the nucleus, because more than 20 min are necessary for the appearance of GST-US11 in the nucleus when injected into the cytoplasm. Furthermore, other experiments clearly showed that US11 NLS is excluded from the 88-125 peptide (to be published elsewhere). In addition, phosphorylation of US11 protein in the nucleolus itself may favor its release and subsequent export to the cytoplasm.
The protein kinases responsible for the phosphorylation of US11 protein remain to be identified. PKR could be a good candidate because of its presence within nucleoli and because US11 protein is able to bind it in vitro , thus precluding phosphorylation of eIF-2α by activated PKR (7, 25). Other good candidates are the MAP kinases, since Ser 129 is localized within a very strong potential phosphorylation site for these kinases (53). However, to our knowledge, the presence of MAP kinases in nucleoli has not been demonstrated. From these data, and because the full-length GST-US11 protein is always found concentrated in nucleoli of all injected cells, we propose that US11 may contain a constitutive NoRS and that intracellular distribution of the protein is mainly regulated through the activity of the NES, the unmasking of which might be activated after phosphorylation, thus authorizing subsequent interactions with nuclear export factors.
Some US11 proteins carrying point mutations which are very poorly detected in the cytoplasm do not accumulate in nucleoli. This favors the possibility that transport of US11 protein out of the nucleus requires first crossing the nucleolus. Transport of proteins from nucleoli to the cytoplasm may occur through at least three distinct pathways. The first is transport by the CRM-1/nucleoporin/Ran GTPase system after release from nucleoli into the nucleoplasm. This pathway is used by nucleolar proteins exhibiting a leucine-rich NES, such as Rev (6, 56). The second is direct transport from nucleolus to cytoplasm by a specific class of exportin. Exportin 4 has been shown recently to mediate the nuclear export of eIF-5A, which is probably the first member of this class of exportin (30). The third is direct transport from the nucleolus to the cytoplasm, associated or not with ribosomes, using the ribosome export machinery (1, 17, 36). Because our data show that the nuclear export signal of US11 is different from those already identified, leucine-rich NES and NSS, interaction of the US11 NES with a known export factor or with other still unknown factors remains to be determined.
Acknowledgments
We thank R. Grantham for critical reading of the manuscript and Y. Munari-Silem for generous advice on microinjection experiments.
This work was supported by the Institut National de la Santé et de la Recherche Médicale and by grants from the Association Nationale de Recherches sur le SIDA. F.C. was supported by a fellowship from the Ministère de l'Enseignement Supérieur et de la Recherche.
REFERENCES
- 1.Aitchison, J. D., and M. P. Rout. 2000. The road to ribosomes. Filling potholes in the export pathway. J. Cell Biol. 151:F23-F26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andjelkovic, M., S. M. Maira, P. Cron, P. J. Parker, and B. A. Hemmings. 1999. Domain swapping used to investigate the mechanism of protein kinase B regulation by 3-phosphoinositide-dependent protein kinase 1 and Ser473 kinase. Mol. Cell. Biol. 19:5061-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besse, S., J.-J. Diaz, E. Pichard, K. Kindbeiter, J.-J. Madjar, and F. Puvion-Dutilleul. 1996. In situ hybridization and immunoelectron microscope analyses of the Us11 gene of herpes simplex virus type 1 for transient expression. Chromosoma 104:434-444. [DOI] [PubMed] [Google Scholar]
- 4.Bogerd, H. P., R. E. Benson, R. Truant, A. Herold, M. Phingbodhipakkiya, and B. R. Cullen. 1999. Definition of a consensus transportin-specific nucleocytoplasmic transport signal. J. Biol. Chem. 274:9771-9777. [DOI] [PubMed] [Google Scholar]
- 5.Boulikas, T. 1993. Nuclear localization signals (NLS). Crit. Rev. Eukaryot. Gene Expr. 3:193-227. [PubMed] [Google Scholar]
- 6.Buonomo, S. B., A. Michienzi, F. G. De Angelis, and I. Bozzoni. 1999. The Rev protein is able to transport to the cytoplasm small nucleolar RNAs containing a Rev binding element. RNA 5:993-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassady, K. A., M. Gross, and B. Roizman. 1998. The herpes simplex virus US11 protein effectively compensates for the γ1(34.5) gene if present before activation of protein kinase R by precluding its phosphorylation and that of the alpha subunit of eukaryotic translation initiation factor 2. J. Virol. 72:8620-8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chelsky, D., R. Ralph, and G. Jonak. 1989. Sequence requirements for synthetic peptide-mediated translocation to the nucleus. Mol. Cell. Biol. 9:2487-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Creancier, L., H. Prats, C. Zanibellato, F. Amalric, and B. Bugler. 1993. Determination of the functional domains involved in nucleolar targeting of nucleolin. Mol. Biol. Cell 4:1239-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cullen, B. R. 1986. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell 46:973-982. [DOI] [PubMed] [Google Scholar]
- 11.Dang, C. V., and W. M. Lee. 1989. Nuclear and nucleolar targeting sequences of c-erb-A, c-myb, N-myc, p53, HSP70, and HIV tat proteins. J. Biol. Chem. 264:18019-18023. [PubMed] [Google Scholar]
- 12.Diaz, J.-J., M. Duc Dodon, N. Schaerer-Uthurralt, D. Simonin, K. Kindbeiter, L. Gazzolo, and J.-J. Madjar. 1996. Posttranscriptional transactivation of human retroviral envelope glycoprotein expression by herpes simplex virus Us11 protein. Nature 379:273-277. [DOI] [PubMed] [Google Scholar]
- 13.Diaz, J.-J., and D. J. Roufa. 1992. Fine-structure map of the human ribosomal protein gene RPS14. Mol. Cell. Biol. 12:1680-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diaz, J.-J., D. Simonin, T. Massé, P. Deviller, K. Kindbeiter, L. Denoroy, and J.-J. Madjar. 1993. The herpes simplex virus type 1 Us11 gene product is a phosphorylated protein found to be nonspecifically associated with both ribosomal subunits. J. Gen. Virol. 74:397-406. [DOI] [PubMed] [Google Scholar]
- 15.Diaz-Latoud, C., J.-J. Diaz, N. Fabre-Jonca, K. Kindbeiter, J.-J. Madjar, and A.-P. Arrigo. 1997. Herpes simplex virus Us11 protein enhances recovery of protein synthesis and survival in heat shock treated HeLa cells. Cell Stress Chaperones 2:119-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duc Dodon, M., I. Mikaelian, A. Sergeant, and L. Gazzolo. 2000. The herpes simplex virus 1 Us11 protein cooperates with suboptimal amounts of human immunodeficiency virus type 1 (HIV-1) Rev protein to rescue HIV-1 production. Virology 270:43-53. [DOI] [PubMed] [Google Scholar]
- 17.Dundr, M., and T. Misteli. 2001. Functional architecture in the cell nucleus. Biochem. J. 356:297-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer, U., J. Huber, W. C. Boelens, I. W. Mattaj, and R. Lührmann. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82:475-483. [DOI] [PubMed] [Google Scholar]
- 19.Fridell, R. A., R. Truant, L. Thorne, R. E. Benson, and B. R. Cullen. 1997. Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-beta. J. Cell Sci. 110:1325-1331. [DOI] [PubMed] [Google Scholar]
- 20.Gescher, A. 1998. Analogs of staurosporine: potential anticancer drugs? Gen. Pharmacol. 31:721-728. [DOI] [PubMed] [Google Scholar]
- 21.Gorlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 22.Görlich, D., and I. W. Mattaj. 1996. Nucleocytoplasmic transport. Science 271:1513-1518. [DOI] [PubMed] [Google Scholar]
- 23.Greco, A., D. Simonin, J.-J. Diaz, L. Barjhoux, K. Kindbeiter, J.-J. Madjar, and T. Massé. 1994. The DNA sequence coding for the 5" untranslated region of herpes simplex type 1 ICP22 mRNA mediates high level of gene expression. J. Gen. Virol. 75:1693-1702. [DOI] [PubMed] [Google Scholar]
- 24.Horgan, D. J., and T. P. Singer. 1967. Characteristics of the binding of rotenone in the respiratory chain and the inhibition sites of amytal and piericidin A. Biochem. J. 104:50C-52C. [DOI] [PMC free article] [PubMed]
- 25.Jeffrey, I. W., S. Kadereit, E. F. Meurs, T. Metzger, M. Bachmann, M. Schwemmle, A. G. Hovanessian, and M. J. Clemens. 1995. Nuclear localization of the interferon-inducible protein kinase PKR in human cells and transfected mouse cells. Exp. Cell Res. 218:17-27. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, F. B., R. A. Marciniak, and L. Guarente. 1998. Telomeres, the nucleolus and aging. Curr. Opin. Cell Biol. 10:332-338. [DOI] [PubMed] [Google Scholar]
- 27.Kalderon, D., W. D. Richardson, A. F. Markham, and A. E. Smith. 1984. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311:33-38. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins for the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 29.Lewis, J. D., and D. Tollervey. 2000. Like attracts like: getting RNA processing together in the nucleus. Science 288:1385-1389. [DOI] [PubMed] [Google Scholar]
- 30.Lipowsky, G., F. R. Bischoff, P. Schwarzmaier, R. Kraft, S. Kostka, E. Hartmann, U. Kutay, and D. Gorlich. 2000. Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J. 19:4362-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, H. T., and B. Y. Yung. 1999. In vivo interaction of nucleophosmin/B23 and protein C23 for cell cycle progression in HeLa cells. Cancer Lett. 144:45-54. [DOI] [PubMed] [Google Scholar]
- 32.Liu, J. L., L. F. Lee, Y. Ye, Z. Qian, and H. J. Kung. 1997. Nucleolar and nuclear localization properties of a herpesvirus bZIP oncoprotein, MEQ. J. Virol. 71:3188-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lohrum, M. A., M. Ashcroft, M. H. Kubbutat, and K. H. Vousden. 2000. Identification of a cryptic nucleolar-localization signal in MDM2. Nat. Cell Biol. 2:179-181. [DOI] [PubMed] [Google Scholar]
- 34.Madjar, J.-J., M. Arpin, M. Buisson, and J. P. Reboud. 1979. Spot position of rat liver ribosomal proteins by four different two-dimensional electrophoreses in polyacrylamide gel. Mol. Gen. Genet. 171:121-134. [DOI] [PubMed] [Google Scholar]
- 35.Michael, W. M. 2000. Nucleocytoplasmic shuttling signals: two for the price of one. Trends Cell Biol. 10:46-50. [DOI] [PubMed] [Google Scholar]
- 36.Milkereit, P., O. Gadal, A. Podtelejnikov, S. Trumtel, N. Gas, E. Petfalski, D. Tollervey, M. Mann, E. Hurt, and H. Tschochner. 2001. Maturation and intranuclear transport of preribosomes requires noc proteins. Cell 105:499-509. [DOI] [PubMed] [Google Scholar]
- 37.Misteli, T. 2001. Protein dynamics: implications for nuclear architecture and gene expression. Science 291:843-847. [DOI] [PubMed] [Google Scholar]
- 38.Moroianu, J., and J. F. Riordan. 1994. Identification of the nucleolar targeting signal of human angiogenin. Biochem. Biophys. Res. Commun. 203:1765-1772. [DOI] [PubMed] [Google Scholar]
- 39.Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99:677-690. [DOI] [PubMed] [Google Scholar]
- 40.Olson, M. O., M. Dundr, and A. Szebeni. 2000. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 10:189-196. [DOI] [PubMed] [Google Scholar]
- 41.Pederson, T. 1998. The plurifunctional nucleolus. Nucleic Acids Res. 26:3871-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604-609. [DOI] [PubMed] [Google Scholar]
- 43.Puvion-Dutilleul, F. 1987. Localization of viral-specific 21kDa protein in nucleoli of herpes simplex infected cells. Eur. J. Cell Biol. 43:487-498. [PubMed] [Google Scholar]
- 44.Pyper, J. M., J. E. Clements, and M. C. Zink. 1998. The nucleolus is the site of Borna disease virus RNA transcription and replication. J. Virol. 72:7697-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roizman, B., and A. E. Sears. 1993. Herpes simplex viruses and their replication, p. 11-68. In B. Roizman, R. J. Whitley, and C. Lopez (ed.), The human herpesviruses, 3rd ed. Raven Press, New York, N.Y.
- 46.Roller, R. J., L. L. Monk, D. Stuart, and B. Roizman. 1996. Structure and function in the herpes simplex virus 1 RNA-binding protein U(s)11: mapping of the domain required for ribosomal and nucleolar association and RNA binding in vitro. J. Virol. 70:2842-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roller, R. J., and B. Roizman. 1992. The herpes simplex virus 1 RNA binding protein Us11 is a virion component and associates with ribosomal 60S subunits. J. Virol 66:3624-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rossi, J. J. 1999. Ribozymes in the nucleolus. Science 285:1685.. [DOI] [PubMed] [Google Scholar]
- 49.Schaerer-Uthurralt, N., M. Erard, K. Kindbeiter, J. J. Madjar, and J. J. Diaz. 1998. Distinct domains in herpes simplex virus type 1 US11 protein mediate posttranscriptional transactivation of human T-lymphotropic virus type I envelope glycoprotein gene expression and specific binding to the Rex responsive element. J. Gen. Virol. 79:1593-1602. [DOI] [PubMed] [Google Scholar]
- 50.Scheer, U., and R. Hock. 1999. Structure and function of the nucleolus. Curr. Opin. Cell Biol. 11:385-390. [DOI] [PubMed] [Google Scholar]
- 51.Simonin, D., J.-J. Diaz, K. Kindbeiter, P. Pernas, and J.-J. Madjar. 1995. Phosphorylation of herpes simplex virus type 1 Us11 protein is independent of viral genome expression. Electrophoresis 16:1317-1322. [DOI] [PubMed] [Google Scholar]
- 52.Siomi, H., H. Shida, S. H. Nam, T. Nosaka, M. Maki, and M. Hatanaka. 1988. Sequence requirements for nucleolar localization of human T-cell leukemia virus type I pX protein, which regulates viral RNA processing. Cell 55:197-209. [DOI] [PubMed] [Google Scholar]
- 53.Songyang, Z., K. P. Lu, Y. T. Kwon, L. H. Tsai, O. Filhol, C. Cochet, D. A. Brickey, T. R. Soderling, C. Bartleson, D. J. Graves, A. J. DeMaggio, M. F. Hoekstra, J. Blenis, T. Hunter, and L. C. Cantley. 1996. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol. Cell. Biol. 16:6486-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wen, W., J. L. Meinkoth, R. Y. Tsien, and S. S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82:463-473. [DOI] [PubMed] [Google Scholar]
- 55.Zhai, W., and L. Comai. 1999. A kinase activity associated with simian virus 40 large T antigen phosphorylates upstream binding factor (UBF) and promotes formation of a stable initiation complex between UBF and SL1. Mol. Cell. Biol. 19:2791-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zolotukhin, A. S., and B. K. Felber. 1999. Nucleoporins nup98 and nup214 participate in nuclear export of human immunodeficiency virus type 1 Rev. J. Virol. 73:120-127. [DOI] [PMC free article] [PubMed] [Google Scholar]