Abstract
A critical step toward understanding mitochondrial genetics and its impact on human disease is to identify and characterize the full complement of nucleus-encoded factors required for mitochondrial gene expression and mitochondrial DNA (mtDNA) replication. Two factors required for transcription initiation from a human mitochondrial promoter are h-mtRNA polymerase and the DNA binding transcription factor, h-mtTFA. However, based on studies in model systems, the existence of a second human mitochondrial transcription factor has been postulated. Here we report the isolation of a cDNA encoding h-mtTFB, the human homolog of Saccharomyces cerevisiae mitochondrial transcription factor B (sc-mtTFB) and the first metazoan member of this class of transcription factors to which a gene has been assigned. Recombinant h-mtTFB is capable of binding mtDNA in a non-sequence-specific fashion and activates transcription from the human mitochondrial light-strand promoter in the presence of h-mtTFA in vitro. Remarkably, h-mtTFB and its fungal homologs are related in primary sequence to a superfamily of N6 adenine RNA methyltransferases. This observation, coupled with the ability of recombinant h-mtTFB to bind S-adenosylmethionine in vitro, suggests that a structural, and perhaps functional, relationship exists between this class of transcription factors and this family of RNA modification enzymes and that h-mtTFB may perform dual functions during mitochondrial gene expression.
Human cells maintain essential genetic information in two distinct compartments, the mitochondrion and the nucleus. Mitochondrial DNA (mtDNA) encodes 13 protein components of the mitochondrial oxidative phosphorylation complexes that are essential for aerobic ATP production as well as 22 tRNAs and 2 rRNAs needed for translation of these proteins in the organelle. However, the remainder of mitochondrial proteins (∼1,000) are encoded by nuclear genes and imported into the organelle. This includes regulatory factors required for expression, replication, and maintenance of the mitochondrial genome (38). The realization that mutations in mtDNA or nuclear genes that impact mitochondrial function cause specific diseases (19, 44), and likely contribute to late-onset disorders and aging (43), has heightened interest in understanding human mitochondrial genetics. However, major advancement in this area remains hindered by the fact that even the basic machinery required for these fundamental processes in human cells has not been defined fully.
Transcription of mitochondrial genes is performed by a dedicated mitochondrial RNA (mtRNA) polymerase that is encoded by genes in the nucleus. The first gene encoding an mtRNA polymerase (RPO41) was isolated from the yeast Saccharomyces cerevisiae, which revealed that this class of enzyme is homologous to the RNA polymerases encoded by the bacteriophages T3, T7, and SP6 (24). This breakthrough, coupled with the ability of yeast to survive without mitochondrial respiration, has made this organism an indispensable model system for understanding mitochondrial gene expression and mtDNA maintenance (35). The ability of yeast mtRNA polymerase to initiate transcription from a mitochondrial promoter is dependent on a mitochondrial transcription factor, sc-mtTFB, that is encoded by the MTF1 gene (20, 46). This protein functions in certain respects like a bacterial sigma factor (8, 9, 23); however, amino acid sequence comparisons (7) and mutational analyses (37) do not strongly support the hypothesis that sc-mtTFB is homologous to this class of proteins.
The mitochondrial transcription machinery in humans also consists of a bacteriophage-related RNA polymerase (41) and a transcription factor (h-mtTFA) that, like sc-mtTFB in yeast, is required for high levels of specific transcription initiation (14, 15). However, h-mtTFA (and its yeast homolog sc-mtTFA/Abf2p) is a member of the high-mobility-group box family of DNA binding proteins (27) and bears no sequence or structural resemblance to sc-mtTFB. In addition, sc-mtTFA lacks a C-terminal tail domain present in the human protein and does not exhibit the specific DNA binding capacity or transcriptional activation properties displayed by h-mtTFA (10). These and other differences between yeast and humans have been discussed previously (36), and it remained unclear whether humans possess an sc-mtTFB homolog or if the enhanced function of h-mtTFA in the human system had perhaps bypassed the requirement for this transcription factor. The most convincing evidence to date suggesting that vertebrates do encode an mtTFB homolog came from the characterization of a biochemical activity from Xenopus laevis that displays the predicted properties of an sc-mtTFB-like protein (3, 4). However, direct evidence confirming that this activity is assigned to a Xenopus homolog of sc-mtTFB (e.g., isolation of the gene encoding this activity) has not been reported. Here, we describe the cloning and characterization of human mtTFB (h-mtTFB), the first metazoan homolog of this class of transcription factor to be unequivocally identified.
MATERIALS AND METHODS
Query sequences and Blastp searches.
All Blastp (2) searches were performed against the nonredundant database from the National Center for Biotechnology Information server by using default parameters. The initial query sequence used was the precise open reading frame (ORF) of the S. cerevisiae MTF1 gene encoding sc-mtTFB (GenBank accession no. NP_013955). The results of this search identified a highly significant match (E value, 3e−20) to a putative Schizosaccharomyces pombe homolog of sc-mtTFB (GenBank accession no. CAB65608) and a potentially significant, albeit much lower probability (E value, 0.99), match to a predicted human protein, CGI-75 (GenBank accession no. NP_057104). The precise ORF of the putative S. pombe mtTFB homolog was then used as the query in a subsequent Blastp search. The results of this search revealed a highly significant match (E value, 2e−4) to the human CGI-75 protein, which indicated that the previously identified match between sc-mtTFB and CGI-75 is also likely significant. The precise predicted ORF of CGI-75 was used as the query in subsequent Blastp searches.
Isolation of cDNAs and construction of expression plasmids for CGI-75.
The following primers were used to amplify CGI-75 cDNAs from a human fetal brain and a human B cell library: human B1, 5"-GTGCTTGCCGCGTATCATGG-3", and human B2, 5"-AGTCACATCTGGTCATTGGC-3". A 1.2-kb PCR product that was obtained when each of these libraries was used as a template was ligated into the vector pGEM-T (Promega, Inc.), and the entire nucleotide sequence was determined. Both products were CGI-75 cDNAs that matched the annotated CGI-75 ORF.
The plasmid used for localization studies in HeLa cells was constructed as follows. Using the pGEM-T plasmid containing the 1.2-kb CGI-75 cDNA from B cells as a template, a PCR was performed with the following primers: HBGFP5, 5"-AATTCTCGAGATGGCTGCCTCCGGAAAACTC-3", and HBGFP3, 5"-AATTGGATCCCGGAGTCTGTAATTCTCTGCGTC-3". The resulting PCR product contained the entire CGI-75 ORF minus the stop codon flanked by the XhoI and BamHI restriction sites at the 5" and 3" ends, respectively. This product was digested with XhoI and BamHI and ligated into the plasmid pEGFP-N1 (Clontech Laboratories, Inc.), which was digested with the same enzymes. The resulting plasmid (pN1-HBGFP) contains a fusion allele that encodes the enhanced green fluorescence protein (EGFP) fused to the C terminus of the CGI-75 ORF. This fusion allele was liberated by digesting pN1-HBGFP with BamHI and NotI, and then it was ligated into the plasmid pTRE2 (Clontech Laboratories, Inc.) to generate the plasmid (pTRE2-HBGFP) that is engineered to express the fusion protein in HeLa Tet-On cells.
The plasmid (pGST-CGI75) used to express recombinant CGI-75 in Escherichia coli was constructed in a similar fashion to that described above for the EGFP fusion plasmid, except the PCR was performed with the following primers: 5"-CGI75, 5"-GCGCGGATCCATGGCTGCCTCCGGAAAA-3", and M13 reverse, 5"-GGAAACAGCTATGACCATG-3". The resulting product was digested with BamHI and NotI and ligated into pGEX-4T (Promega, Inc.) to generate a glutathione S-transferase (GST) fusion allele (with the GST tag located at the N terminus of CGI-75) that is under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-regulated bacterial promoter.
Subcellular localization of CGI-75 in HeLa cells by fluorescence microscopy.
HeLa Tet-On cells (Clontech laboratories, Inc.) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with fetal bovine serum (10%) and G418 (100 μg/ml). Approximately 105 cells were seeded onto glass slides and allowed to grow overnight. The following day, the cells were transiently transfected with pTRE2-HBGFP (0.4 μg) by using an Effectene kit (Qiagen, Inc.) as described by the manufacturer. After transfection, the cells were incubated overnight in DMEM supplemented with pyruvate (100 μg/ml), uridine (50 μg/ml), and doxycycline (1 μg/ml). The cells were then washed three times with phosphate-buffered saline and then incubated with 100 nM Mitotracker Red (Molecular Probes, Inc.) in DMEM for 15 min at 37°C. After a 15-min wash at 37°C with DMEM, the cells were fixed in formaldehyde (4%) at room temperature for 30 min. After two washes with phosphate-buffered saline, a coverslip was mounted on the slide using Fluoromount (Sigma Chemical Co., Inc.), and the fluorescence signals from EGFP and Mitotracker Red were observed from the cells by using an Olympus BX60 epifluorescence microscope equipped with a Photometrics Quantix digital camera.
Expression and purification of recombinant CGI-75.
A single colony of E. coli DH5α containing pGST-CGI75 was used to inoculate a 1-liter culture of Luria-Bertani medium (containing 100 μg of ampicillin/ml) that was subsequently grown overnight with shaking at 37°C. After 16 to 20 h of growth, IPTG (0.4 mM) was added and the culture was allowed to grow with shaking for an additional 5 h at room temperature. The cells were collected by centrifugation, and the resulting pellet was resuspended in 50 ml of ice-cold lysis buffer (20 mM Tris · Cl, pH 8.0; 100 mM NaCl; 1 mM EDTA; 0.5% NP-40; 1 mM dithiothreitol [DTT]; 0.5 mM phenylmethylsulfonyl fluoride). The cells were lysed by sonication, and the resulting cell lysate was cleared by centrifugation (10,000 × g, 10 min, 4°C). The cleared supernatant was incubated for 1 h at 4°C with 1 ml of glutathione-Sepharose (Amersham Pharmacia, Inc.) that had been prepared as follows: washed three times with lysis buffer and then resuspended in lysis buffer as a 1:1 (vol/vol) slurry. The protein-bound beads were then washed five times with 1 ml of lysis buffer and resuspended as a 1:1 (vol/vol) slurry in lysis buffer. Thrombin (6 U; Amersham Pharmacia, Inc.) was added to the slurry and allowed to incubate overnight at room temperature with rocking. The slurry was centrifuged (3,000 × g, 1 min, room temperature) to remove the beads, and the supernatant, containing cleaved CGI-75 protein, was concentrated to a final volume of ∼0.5 ml using a Centricon Centriprep-10 column (Amicon, Inc.) as described by the manufacturer. This material was used for most of the experiments. However, where indicated, this sample was subjected to polyacrylamide gel electrophoresis, and the CGI-75 band was excised, eluted, and renatured as described previously for sc-mtTFB (37).
Mitochondrial transcription and gel mobility shift assays.
The light-strand promoter (LSP)-containing DNA template (pBS-LSP) used in the transcription and gel mobility shift assays has been described previously (10). Mitochondrial transcription reactions were performed in a total volume of 25 μl, and all reaction mixtures contained 10 mM Tris · Cl (pH 8.0), 10 mM MgCl2, 1 mM DTT, 0.1 mg of RNase-free bovine serum albumin/ml, 400 μM ATP, 150 μM CTP, 150 μM GTP, 0.2 μM UTP, 5 μl of [α32-P]UTP (specific activity of 3,000 Ci/mmol), 10 μg of BamHI-linearized pBS-LSP/ml, and 5 μl of a transcription-competent mitochondrial extract from HeLa cells that was prepared as described previously (25). Where indicated, 2 μl of recombinant h-mtTFA (10) was added to the reaction mixture as were the indicated amounts of recombinant CGI-75 protein. Transcription reactions were performed for 30 min at 28°C and terminated by the addition of 100 μl of stop buffer containing 0.1% sodium dodecyl sulfate, 0.1 μg of tRNA/ml, 0.2 M NaCl, 1 mM EDTA, and 0.6 μg of proteinase K/μl. This mixture was incubated at 37°C for 30 min, extracted with phenol-chloroform, precipitated with ethanol, and resolved on an 8% polyacrylamide-7 M urea gel.
Gel mobility shift assays were performed using 32P-end-labeled LSP-containing DNA from pBS-LSP as a probe (10). Reactions were performed in a total volume of 25 μl containing 10 mM Tris · Cl (pH 8.0), 10 mM MgCl2, 1 mM DTT, 0.1 mg of bovine serum albumin/ml, 0.05% Triton X-100, 25 ng of radio-labeled DNA probe (final concentration, 2.7 nM), and the indicated amounts of recombinant CGI-75 protein (4 to 12 ng). Where indicated, 30 ng of poly(dI-dC) was also added (final concentration, 1.3 nM). Reaction mixtures were incubated for 30 min at room temperature and resolved by electrophoresis on a 5% native polyacrylamide gel in Tris-borate-EDTA buffer for 4 to 6 h at 4°C. After electrophoresis, gels from the transcription reactions and gel shift assays were dried and subjected to autoradiography.
Solid-phase S-adenosylmethionine binding assays.
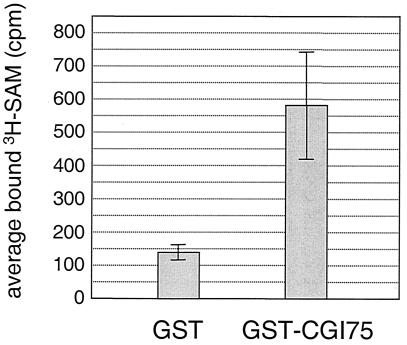
S-Adenosylmethionine binding assays were performed as described (39). Briefly, 10 μl of glutathione-agarose beads were bound with either GST (negative control) or GST::CGI-75 (also called GST::h-mtTFB) as described for the purification of CGI-75 above. Protein-bound beads were then incubated with [3H]S-adenosylmethionine (3H-SAM) (78 Ci/mmol; New England Nuclear, Inc.) in binding buffer (39) for 30 min on ice and then washed five times with binding buffer (1 ml) without 3H-SAM. The beads were transferred to a vial containing 10 ml of scintillation fluid (Ultimagold; Packard, Inc.), and tritium decay was measured in a Wallac 1409 liquid scintillation counter. Three 2-min counts were performed for each sample as well as a background sample (scintillation fluid alone). After subtracting the background counts per minute from each, the average amount of 3H-SAM bound (in background-corrected counts per minute) was determined, and these values were plotted on a graph (see Fig. 7). Western blot analysis was used to assess the relative amount of GST and GST::h-mtTFB fusion protein bound to the beads in each experiment. At least twice as much GST peptide was bound to beads than GST::h-mtTFB fusion protein (data not shown).
FIG. 7.
Recombinant h-mtTFB (CGI-75) binds S-adenosylmethionine. The results of a solid-phase S-adenosylmethionine-binding assay are shown. The amount (in counts per minute) of 3H-SAM bound to beads containing either a GST peptide (GST) or the GST::CGI-75 fusion protein (GST-CGI75) was determined in three experimental trials. The average of these three experiments is plotted on the ordinate, with ± one standard deviation from the mean indicated by brackets.
RESULTS
Identification of a putative human homolog of yeast mitochondrial transcription factor B.
Despite the lack of strong primary sequence conservation between yeast mtTFB homologs (7), we continued to search available databases for homologs of this important mitochondrial regulatory protein using sequence-based strategies. A Blastp search, with the S. cerevisiae amino acid sequence as a query, revealed a strong match to an S. pombe protein that appears to be a fission yeast homolog of sc-mtTFB. The primary sequence of this protein exhibits ∼22% amino acid identity and ∼37% similarity to that of sc-mtTFB (Fig. 1).Using the S. pombe ORF as the query in a subsequent Blastp search, highly significant matches were identified to all four previously identified fungal mtTFB homologs (S. cerevisiae, Saccharomyces douglasii, Saccharomyces kluyveri, and Kluyveromyces lactis) as well as a predicted human protein designated CGI-75. The nucleotide sequence of CGI-75 cDNA in GenBank (accession no. AF151833) is assembled from available human expressed sequence tags by using a comparative gene identification strategy that employed the Caenorhabditis elegans genome sequence as a verification template (21). The primary sequence of CGI-75 exhibits ∼16% identity and ∼26% similarity to the putative S. pombe mtTFB protein. In addition, a triple alignment of the putative S. pombe homolog, CGI-75, and sc-mtTFB revealed a significant degree of amino acid conservation that extended throughout the entire primary sequence, with the amino-terminal two-thirds of these proteins being particularly well conserved (Fig. 1). These observations prompted us to examine whether the CGI-75 protein is the human homolog of sc-mtTFB.
FIG. 1.
Alignment of the human CGI-75 protein with sc-mtTFB and a putative S. pombe sc-mtTFB homolog. Shown is a ClustalW (40) alignment of the amino acid sequence of sc-mtTFB (S. cerevisiae) with putative human (Homo sapiens) and fission yeast (S. pombe) homologs identified in Blastp searches. Amino acids highlighted in gray are identical or chemically similar in all three species. Pairwise comparisons are denoted as follows. Asterisks below the H. sapiens or S. cerevisiae sequence indicate identical amino acid identity between the H. sapiens and S. pombe or S. cerevisiae and S. pombe proteins, respectively. Carets denote chemically similar amino acids in the same manner designated according to the following matrix: S/T, R/K, D/E, N/Q, F/Y/W, I/V/L/M.
The CGI-75 protein is localized to mitochondria in HeLa cells.
Based on the predicted nucleotide sequence of CGI-75, we designed PCR primers and amplified the CGI-75 ORF by using a human fetal brain or B cell cDNA library as a template. An identical full-length cDNA was obtained from each library. To determine the cellular localization of the CGI-75 protein, we fused the ORF from the fetal brain cDNA to the EGFP. When HeLa Tet-On cells transiently transfected with a pTRE-2 vector that expresses the CGI-75::EGFP fusion protein were examined by fluorescence microscopy, a punctuate cytoplasmic fluorescence pattern was observed (Fig. 2).Using a fluorescent dye specific for mitochondria (Mitotracker Red), we confirmed that the GFP fluorescence signals in these cells colocalized with mitochondria, indicating that CGI-75 is a mitochondrial protein.
FIG. 2.
Human CGI-75 is a mitochondrial protein. The mitochondrial localization of a CGI-75::EGFP fusion protein is shown. HeLa Tet-On cells were transiently transfected with a plasmid (pTRE2-HBGFP) that expresses a CGI-75::EGFP fusion protein and stained with the mitochondrion-specific dye Mitotracker Red. Shown are two representative transfected cells analyzed by fluorescence microscopy for EGFP fluorescence in green (labeled GFP), Mitotracker fluorescence in red (labeled MT), and a merge of the two signals (labeled merge), in which a yellow color indicates colocalization. In the panels on the right, the asterisks indicate the signals derived from a cell that was not transfected with the plasmid. As expected, this cell exhibited Mitotracker fluorescence but not EGFP fluorescence.
We next examined the nucleotide sequence of the genomic DNA directly upstream of the CGI-75 gene for transcription factor binding sites using the TFSEARCH program to access the TRANSFAC database (45). Putative binding sites for several nuclear transcription factors were found (Fig. 3).Of particular note were two binding sites for nuclear respiratory factor 2 (NRF-2), a transcription factor known to activate transcription of several nucleus-encoded mitochondrial proteins in mammals, including h-mtTFA (42). Based on these nucleotide sequence and localization data, we hypothesized that the CGI-75 protein is likely a human homolog of mitochondrial transcription factor B, and we embarked on experiments to test this directly.
FIG. 3.
Putative nuclear transcription factor binding sites in the genomic region immediately upstream of the CGI-75 gene. Transcription factor binding sites identified using the TFSEARCH algorithm to access the TRANSFAC database (45) are indicated as follows: SP-1, NFκ-B, and AP-1 are underlined; MZF-1 is printed in bold type; and NRF-2 is highlighted in gray. The sequence shown is the CGI-75 sense DNA strand, and the first two codons of the CGI-75 gene are indicated by lowercase italics.
Recombinant CGI-75 protein binds mtDNA in a nonspecific manner and activates transcription from the human mitochondrial LSP.
We engineered the human CGI-75 protein for regulated expression in E. coli as a fusion protein with GST (GST::CGI-75). This protein was induced by the addition of IPTG to the growth medium, and after breakage of the cells by sonication, it was present in the soluble fraction of crude E. coli lysates. The protein was partially purified from crude lysates by using glutathione-agarose beads and subsequently eluted by cleavage of the GST tag with thrombin, which liberated recombinant CGI-75 protein (data not shown).
Both sc-mtTFB and a biochemical activity designated mtTFB from X. laevis (xl-mtTFB) are reported to bind DNA in a nonspecific manner (4, 31). Using recombinant protein purified as described above, we tested whether CGI-75 has similar activity. We performed a gel mobility shift assay using a radio-labeled double-stranded DNA fragment containing the human mitochondrial LSP. The addition of affinity-purified CGI-75 protein resulted in a shift of the probe to lower mobility, indicating that the protein can bind this DNA fragment (Fig. 4).Addition of nearly stoichiometric amounts of the nonspecific competitor poly(dI-dC) to the reaction strongly inhibited this activity, indicating that the binding event is largely DNA sequence independent. To ensure that this DNA binding activity is due to recombinant CGI-75 protein and not a contaminating activity from the E. coli lysate, we excised the thrombin-cleaved protein from a sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel after following the GST affinity purification protocol and eluted the protein from the gel slice as described for sc-mtTFB (37). Renatured protein purified in this manner displayed the same concentration-dependent DNA binding properties in this assay (data not shown). Similar results were obtained with gel-purified renatured GST-tagged CGI-75 protein (data not shown).
FIG. 4.
Recombinant human CGI-75 protein binds DNA in a non-sequence-specific manner. An autoradiogram from a gel mobility shift assay (see Materials and Methods) using recombinant GST-purified CGI-75 protein and an mtDNA probe containing the mitochondrial LSP is shown. Plus and minus signs indicate the presence and absence of CGI-75 protein and the nonspecific competitive inhibitor poly(dI-dC) in the sample, respectively. Location of the unbound DNA probe is indicated by the arrow, and the position of the retarded CGI-75-bound probe is indicated by the arrowhead. Thirty nanograms of poly(dI-dC) was added where indicated, and the amount of CGI-75 in each reaction is indicated at the top of the figure (numbers indicate nanograms of CGI-75 added).
We next examined the ability of CGI-75 to activate transcription from the mitochondrial LSP in vitro. Mitochondrial lysates from cultured human cells can be separated into two active fractions (14, 15), one containing nonspecific mtRNA polymerase activity as well as other proteins and one containing the transcription factor h-mtTFA. In fact, recombinant h-mtTFA can functionally replace the second fraction, suggesting that it is the only active protein component present with regard to transcription initiation (27). We prepared a mitochondrial extract from HeLa cells that was dependent on recombinant h-mtTFA in order to initiate significant levels of transcription from a linear LSP-containing DNA fragment in vitro. The addition of recombinant h-mtTFA to this mtRNA polymerase fraction resulted in the expected runoff RNA transcript from this template (Fig. 5).However, the addition of recombinant CGI-75 protein to this reaction resulted in significantly enhanced amounts of transcription from this template in a concentration-dependent manner (Fig. 5). As was the case in gel mobility shift assays, gel-purified renatured CGI-75 protein yielded similar results (data not shown). Based on these data, we propose that the CGI-75 protein is the human homolog of mitochondrial transcription factor B (h-mtTFB).
FIG. 5.
Recombinant human CGI-75 protein activates transcription from the human mitochondrial LSP. Mitochondrial transcription was assayed using an in vitro runoff transcription assay that measures specific transcription initiation from the human LSP. The resulting product is a discrete radio-labeled RNA transcript (indicated by an arrowhead) that begins at the LSP and extends to the end of the linear DNA template (10). Protein components present (+) or absent (−) in each reaction are indicated at the top of the figure. All reactions contained 5 μl of an mtTFA-dependent mitochondrial extract containing human mtRNA polymerase (HeLa mt extract). Where indicated, 2 μl of gel-purified and renatured h-mtTFA was added to the reaction mixture. Reaction mixtures contained recombinant human CGI-75 protein in the following amounts: lane 2, 20 ng; lane 4, 20 ng; lane 5, 16 ng; lane 6, 12 ng. The fact that decreasing amounts of CGI-75 were added in the last three reaction mixtures (lanes 4 to 6) is indicated at the bottom of the figure by the decreasing slope of the triangle from left to right.
Human mtTFB (CGI-75) is related to RNA adenine methyltransferases and binds S-adenosylmethionine.
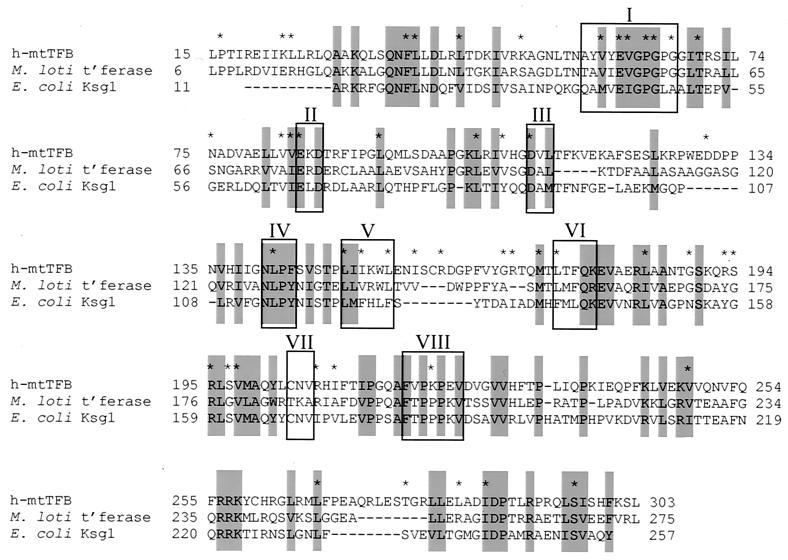
Results of a Blastp database search with the h-mtTFB (i.e., CGI-75) ORF as a query revealed that the two strongest matches were a C. elegans gene product, T03F1.7 (E value, 5e−70), and a Drosophila melanogaster gene product, CG7319 (E value, 2e−52), which are potential candidates for worm and fly homologs of mtTFB, respectively. However, the most significant finding from this database search was the observation that h-mtTFB is closely related to a large number of proteins (>100) that are known, or predicted to be, RNA (adenine-N6,N6)-dimethyltransferase enzymes. Representatives from this extensive list included the homologs of the KsgA rRNA adenine dimethyltransferase enzyme from numerous bacterial species, two homologs of the fungal Dim1p rRNA dimethylase, and predicted fission yeast and human RNA dimethylases (Table 1). Alignment of h-mtTFB with the closest related RNA methyltransferase (from Mesorhizobium loti) and the E. coli KsgA protein revealed a high degree of conservation in the amino-terminal two-thirds of the protein (Fig. 6).Notably, this is also the most conserved region we identified in the triple alignment of human, S. pombe, and S. cerevisiae mtTFB proteins (Fig. 1 and 6). This alignment indicated strong conservation of residues in motifs I, II, III, and VIII of RNA methyltransferases (Fig. 5) that are implicated, based on biochemical and structural data, in binding the S-adenosylmethionine substrate and the adenine ring of the nucleotide to be methylated (1, 5). Although the amino acid sequence conservation with RNA adenine dimethylases is not as pronounced in the S. cerevisiae and S. pombe mtTFB homologs, Blastp searches using the S. pombe ORF did identify a related (29% identity and 46% similarity) RNA adenine dimethylase from Streptomyces venezuelae (GenBank accession no. AF079138) that was also identified in the human Blastp search. The specific region of homology identified here included almost exclusively motifs I, II, and III of the RNA adenine methyltransferase family, which together comprise the S-adenosylmethionine binding pocket.
TABLE 1.
Representative proteins that exhibit a high degree of amino acid similarity to h-mtTFB and are members of an RNA (adenine-N6,N6)-dimethyltransferase superfamily
| Gene product designation | Organism | E value | Accession no. |
|---|---|---|---|
| Dimethyladenosine transferase | M. loti | 1e−44 | NP_108091 |
| Caulobacter crescentus | 7e−36 | AAK23633 | |
| rRNA (adenosine-N6,N6)-dimethylase | Pseudomonas aeruginosa | 2e−30 | H83571 |
| S. pombe | 8e−10 | T43249 | |
| KsgA/dimethyladenosine transferase | Rickettsia prowazekii | 2e−29 | 005952 |
| Bacillus subtilis | 2e−24 | P37468 | |
| E. coli | 1e−22 | P06992 | |
| Dimethyl adenosinetransferase-like protein | Arabadopsis thaliana | 2e−15 | BAB10912 |
| Putative dimethyladenosine transferase | Homo sapiens | 7e−12 | NP_055288 |
| Dimethylase | S. pombe | 2e−10 | T40240 |
| Dim1p/dimethyladenosine transferase | K. lactis | 2e−09 | P78697 |
| S. cerevisiae | 2e−09 | NP_015057 |
FIG. 6.
Human mtTFB (CGI-75) is closely related to RNA adenine methyltransferases. An alignment of the h-mtTFB amino acid sequence with RNA adenine methyltransferase enzymes from M. loti (labeled M. loti t'ferase) and E. coli (E. coli Ksg1) is shown. Amino acids highlighted in gray boxes are identical or chemically similar in all three proteins according to the matrix designated in the legend to Fig. 1. Groups of residues that are boxed are sequence motifs (motifs I to VIII) that are implicated as important structural or catalytic features of RNA methyltransferases (1). Asterisks indicate amino acids that are conserved between sc-mtTFB, h-mtTFB, and the putative S. pombe mtTFB as denoted in Fig. 1.
Given the high degree of similarity of h-mtTFB to the RNA adenine methyltransferase family of proteins, we determined whether this protein can bind S-adenosylmethionine by using a solid-phase binding assay that has been used by others to characterize VP-39 (39), a viral RNA methyltransferase. Glutathione-agarose beads were incubated in crude E. coli lysates containing either GST protein alone (negative control) or an h-mtTFB::GST fusion protein (to allow binding of each of these proteins to the beads), and then the protein-bound beads were incubated with 3H-SAM to assess binding. Similar to that observed with VP-39 (39), an ∼3- to 4-fold-higher increase in bound S-adenosylmethionine was observed with beads containing the GST::h-mtTFB fusion protein than with the GST control (Fig. 7).This difference in binding capacity was observed despite the fact that significantly more GST protein was bound to beads than h-mtTFB fusion protein (data not shown). These data indicate the h-mtTFB can bind S-adenosylmethionine.
DISCUSSION
In the budding yeast S. cerevisiae, mitochondrial transcription factor B (sc-mtTFB) is essential for transcription initiation by mtRNA polymerase and is therefore required for expression and replication of the mitochondrial genome (22, 33, 36). Here we report the isolation of a cDNA encoding h-mtTFB, the first vertebrate homolog of this important mitochondrial regulatory protein to be unequivocally identified. This protein (currently designated CGI-75), which we initially identified based on its amino acid similarity to sc-mtTFB and a putative S. pombe homolog (Fig. 1), is targeted to mitochondria when expressed in HeLa cells (Fig. 2) and displays the documented biochemical properties assigned to other mtTFB homologs (3, 4, 20, 31, 37), namely the ability to bind DNA in a non-sequence-specific manner (Fig. 4) and stimulate transcription from a mitochondrial promoter in vitro (Fig. 5). In addition, analysis of genomic sequences immediately upstream of the CGI-75 gene revealed the presence of two putative binding sites for nuclear respiratory factor 2 (NRF-2) (Fig. 3), a known transcriptional regulator of nucleus-encoded mitochondrial regulatory factors (42). Based on these data, we now refer to CGI-75 as h-mtTFB, according to the proposed nomenclature for mitochondrial transcription proteins (36, 46).
In yeast, sc-mtTFB and mtRNA polymerase are the only two proteins required to initiate transcription from a mitochondrial promoter in vitro (20, 33, 46). Studies in X. laevis, on the other hand, revealed the presence of an mtTFB-like activity that differs from sc-mtTFB in that transcriptional activation by this protein is dependent on Xenopus mtTFA (4). These data, as well as more recent studies of the human transcription system (26, 29), suggest that three proteins are indeed required for optimal transcription initiation from vertebrate mitochondrial promoters. Here, we demonstrate that h-mtTFB is the second mitochondrial transcription factor in humans and, similar to xl-mtTFB, its ability to stimulate transcription in vitro depends on the presence of h-mtTFA in the reaction (Fig. 5). These data confirm the well-documented role for h-mtTFA in mitochondrial transcription initiation (14, 15, 27) and suggest intricate interplay between these two transcription factors and mtRNA polymerase during transcription initiation.
Amino acid comparisons of three yeast mtTFB homologs revealed that this class of protein exhibits a surprisingly low degree of amino acid sequence identity (7). Consistent with this reduced constraint on amino acid sequence for mtTFB function, h-mtTFB does not display a high degree of sequence similarity with its fungal homologs (Fig. 1). However, Blastp searches uncovered an extraordinary relationship between h-mtTFB and a superfamily of S-adenosylmethionine-dependent enzymes that methylates RNA (Table 1), which upon closer inspection is shared by the fungal mtTFB homologs (Fig. 6). The subgroup of this superfamily that is most closely related to h-mtTFB are enzymes that methylate the N6 position of adenine in specific nucleotides in rRNA (1, 5). Certain bacterial enzymes in this class are medically important because they confer differential sensitivity to antibiotics that act by binding to the ribosome (1, 5). The degree of amino acid conservation between h-mtTFB and this group of enzymes is most apparent in those residues that are involved in substrate binding and catalysis in the methyltransferases (Fig. 6). Comparison of the triple alignment of the mtTFB homologs to the alignment of h-mtTFB with the methyltransferases provides additional insight (Fig. 6). For example, while not all of the highly conserved amino acids in the mtTFBs correspond to amino acids conserved in the RNA methyltransferases, many do correspond to conserved residues that are known to be part of important structural or catalytic features of these enzymes (1, 5). In particular, motifs I, II, and III contain amino acids that are critical for binding S-adenosylmethionine (e.g., the conserved glutamic acid and aspartic acid residues in motifs II and III, respectively) (Fig. 6). The fact that h-mtTFB is capable of binding S-adenosylmethionine (Fig. 7) strongly corroborates our amino acid alignment data and supports a main conclusion from this work, that h-mtTFB is structurally, if not also evolutionarily and functionally, related to RNA adenine methyltransferases. To our knowledge this is the first transcription factor documented to display such a relationship with an RNA-modifying enzyme.
While the precise functional significance of the relationship between h-mtTFB and RNA adenine methyltransferases is not known, several possibilities are worthy of discussion. For example, h-mtTFB may be a dual-function protein that serves as a transcription factor and an RNA methyltransferase. It is known that tRNAs and rRNAs are methylated in mammalian mitochondria (11, 12, 17). In fact, the two mitochondrial rRNAs from hamster cells contain several types of methylated nucleotides (12), including one in the 13S subunit that contains an N6 dimethylated adenine, the predicted product of the type of methyltransferases that is most similar to h-mtTFB (Table 1). With regard to a potential function for h-mtTFB in rRNA methylation, it is noteworthy that point mutations in the mitochondrial 12S rRNA in humans are associated with aminoglycoside antibiotic-induced deafness (28). Given that the bacterial homologs of N6 adenine methyltransferases impart sensitivity to antibiotics (1, 5, 18), an intriguing possibility is that h-mtTFB, via an RNA methylation activity, may be one of the nuclear factors that can modify phenotypic expression of this disease (16, 28). A corollary of this dual-function argument is that some RNA methylation events may be coupled to transcription and that h-mtTFB, as part of the transcription machinery, can accomplish this task. It has recently been shown that posttranscriptional events are coupled to transcription in yeast mitochondria via the binding of Nam1p to mtRNA polymerase (32), raising the interesting possibility that certain RNA modification activities are likewise coupled to transcription. In this regard, a report that mitochondrial rRNA processing defects are manifest when human cells are grown in the presence of the methylation inhibitor cycloleucine (30) is compelling because it might suggest that RNA modification, RNA processing, and transcription may all be coupled at some level in mitochondria.
Though it is tempting to speculate, as we have here, that h-mtTFB possesses RNA methyltransferase activity in addition to its transcription factor function, it is important to emphasize an equally likely alternative possibility. That is, that h-mtTFB exhibits structural similarity to RNA methyltransferases but does not possess this enzymatic activity. A salient example in this regard is the recently documented structural similarity between the accessory subunit of mitochondrial DNA polymerase and tRNA synthetase enzymes (13). These structural attributes have been shown to be necessary for function (6), despite the fact that amino acylation activity has not been demonstrated and is perhaps not even expected. Experiments are currently underway to determine the significance of this novel relationship between h-mtTFB and RNA methyltransferases.
The identification of h-mtTFB is an important step forward toward understanding mitochondrial genetics because, in all likelihood, we now have in hand all of the protein components required for transcription initiation in human mitochondria: h-mtRNA polymerase, h-mtTFA, and h-mtTFB. Therefore, the stage is set for future experiments that will allow critical interactions between these factors to be probed at the molecular level and to understand the regulatory properties of these proteins in vivo with regard to mitochondrial gene expression, transcription-dependent mtDNA replication, and human mitochondrial disease.
ADDENDUM
After submission of the manuscript, a study by Schubot et al. (34) came to our attention, which describes the solving of the three-dimensional structure of S. cerevisiae mtTFB. This study revealed that this protein is structurally similar to the ErmC RNA methyltransferase from B. subtilis. These data are entirely consistent with our identification of human mtTFB as a transcription factor that is related to RNA methyltransferases and binds S-adenosylmethionine.
Acknowledgments
This work was supported by a grant from the National Heart, Lung, and Blood Institute of the National Institutes of Health (no. HL-59655) awarded to G.S.S. V.M. is supported by a postdoctoral training grant (no. HL-68459) from the National Institutes of Health.
We thank Alec Hodel and Mary Hodel for insight and technical advice and Matt Rodeheffer for comments on the manuscript.
REFERENCES
- 1.Abad-Zapatero, C., P. Zhong, D. E. Bussiere, K. Stewart, and S. W. Muchmore. 1999. rRNA methyltransferases (ErmC" and ErmAM) and antibiotic resistance, p. 199-205. In X. Cheng and R. M. Blumenthal (ed.), S-Adenosylmethionine-dependent methyltransferases: structure and function. World Scientific Publishing, River Edge, N.J.
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Antoshechkin, I., and D. F. Bogenhagen. 1995. Distinct roles for two purified factors in transcription of Xenopus mitochondrial DNA. Mol. Cell. Biol. 15:7032-7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogenhagen, D. F. 1996. Interaction of mtTFB and mtRNA polymerase at core promoters for transcription of Xenopus laevis mtDNA. J. Biol. Chem. 271:12036-12041. [PubMed] [Google Scholar]
- 5.Bussiere, D. E., S. W. Muchmore, C. G. Dealwis, G. Schluckebier, V. L. Nienaber, R. P. Edalji, K. A. Walter, U. S. Ladror, T. F. Holzman, and C. Abad-Zapatero. 1998. Crystal structure of ErmC", an rRNA methyltransferase which mediates antibiotic resistance in bacteria. Biochemistry 37:7103-7112. [DOI] [PubMed] [Google Scholar]
- 6.Carrodeguas, J. A., and D. F. Bogenhagen. 2000. Protein sequences conserved in prokaryotic aminoacyl-tRNA synthetases are important for the activity of the processivity factor of human mitochondrial DNA polymerase. Nucleic Acids Res. 28:1237-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrodeguas, J. A., S. Yun, G. S. Shadel, D. A. Clayton, and D. F. Bogenhagen. 1996. Functional conservation of yeast mtTFB despite extensive sequence divergence. Gene Expr. 6:219-230. [PMC free article] [PubMed] [Google Scholar]
- 8.Cliften, P. F., J. Y. Park, B. P. Davis, S. H. Jang, and J. A. Jaehning. 1997. Identification of three regions essential for interaction between a sigma-like factor and core RNA polymerase. Genes Dev. 11:2897-2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cliften, P. F., S. H. Jang, and J. A. Jaehning. 2000. Identifying a core RNA polymerase surface critical for interactions with a sigma-like specificity factor. Mol. Cell. Biol. 20:7013-7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dairaghi, D. J., G. S. Shadel, and D. A. Clayton. 1995. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol. 249:11-28. [DOI] [PubMed] [Google Scholar]
- 11.Davenport, L., R. H. Taylor, and D. T. Dubin. 1976. Comparison of human and hamster mitochondrial transfer RNA. Physical properties and methylation status. Biochim. Biophys. Acta 447:285-293. [DOI] [PubMed] [Google Scholar]
- 12.Dubin, D. T. 1974. Methylated nucleotide content of mitochondrial ribosomal RNA from hamster cells. J. Mol. Biol. 84:257-273. [DOI] [PubMed] [Google Scholar]
- 13.Fan, L., P. C. Sanschagrin, L. S. Kaguni, and L. A. Kuhn. 1999. The accessory subunit of mtDNA polymerase shares structural homology with aminoacyl-tRNA synthetases: implications for a dual role as a primer recognition factor and processivity clamp. Proc. Natl. Acad. Sci. USA 96:9527-9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher, R. P., and D. A. Clayton. 1988. Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell. Biol. 8:3496-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher, R. P., J. N. Topper, and D. A. Clayton. 1987. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell 50:247-258. [DOI] [PubMed] [Google Scholar]
- 16.Guan, M. X., N. Fischel-Ghodsian, and G. Attardi. 1996. Biochemical evidence for nuclear gene involvement in phenotype of non-syndromic deafness associated with mitochondrial 12S rRNA mutation. Hum. Mol. Genet. 5:963-971. [DOI] [PubMed] [Google Scholar]
- 17.Helm, M., H. Brule, F. Degoul, C. Cepanec, J. P. Leroux, R. Giege, and C. Florentz. 1998. The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial tRNA. Nucleic Acids Res. 26:1636-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helser, T. L., J. E. Davies, and J. E. Dahlberg. 1972. Mechanism of kasugamycin resistance in Escherichia coli. Nat. New Biol. 235:6-9. [DOI] [PubMed] [Google Scholar]
- 19.Holt, I. J., A. E. Harding, and J. A. Morgan-Hughes. 1988. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature 331:717-719. [DOI] [PubMed] [Google Scholar]
- 20.Jang, S. H., and J. A. Jaehning. 1991. The yeast mitochondrial RNA polymerase specificity factor, MTF1, is similar to bacterial sigma factors. J. Biol. Chem. 266:22671-22677. [PubMed] [Google Scholar]
- 21.Lai, C. H., C. Y. Chou, L. Y. Chang, C. S. Liu, and W. Lin. 2000. Identification of novel human genes evolutionarily conserved in Caenorhabditis elegans by comparative proteomics. Genome Res. 10:703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lisowsky, T., and G. Michaelis. 1988. A nuclear gene essential for mitochondrial replication suppresses a defect of mitochondrial transcription in Saccharomyces cerevisiae. Mol. Gen. Genet. 219:125-128. [DOI] [PubMed] [Google Scholar]
- 23.Mangus, D. A., S. H. Jang, and J. A. Jaehning. 1994. Release of the yeast mitochondrial RNA polymerase specificity factor from transcription complexes. J. Biol. Chem. 269:26568-26574. [PubMed] [Google Scholar]
- 24.Masters, B. S., L. L. Stohl, and D. A. Clayton. 1987. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell 51:89-99. [DOI] [PubMed] [Google Scholar]
- 25.Micol, V., P. Fernandez-Silva, and G. Attardi. 1996. Isolation and assay of mitochondrial transcription termination factor from human cells. Methods Enzymol. 264:158-173. [DOI] [PubMed] [Google Scholar]
- 26.Nam, S. C., and C. Kang. 2001. Expression of cloned cDNA for the human mitochondrial RNA polymerase in Escherichia coli and purification. Protein Expr. Purif. 21:485-491. [DOI] [PubMed] [Google Scholar]
- 27.Parisi, M. A., and D. A. Clayton. 1991. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science 252:965-969. [DOI] [PubMed] [Google Scholar]
- 28.Prezant, T. R., J. V. Agapian, M. C. Bohlman, X. Bu, S. Oztas, W. Q. Qiu, K. S. Arnos, G. A. Cortopassi, L. Jaber, J. I. Rotter, et al. 1993. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat. Genet. 4:289-294. [DOI] [PubMed] [Google Scholar]
- 29.Prieto-Martin, A., J. Montoya, and F. Martinez-Azorin. 2001. A study on the human mitochondrial RNA polymerase activity points to existence of a transcription factor B-like protein. FEBS Lett. 503:51-55. [DOI] [PubMed] [Google Scholar]
- 30.Prince, D. L., R. M. Kotin, and D. T. Dubin. 1986. Evidence that the methylation inhibitor cycloleucine causes accumulation of a discrete ribosomal RNA precursor in hamster mitochondria. Mol. Biol. Rep. 11:51-55. [DOI] [PubMed] [Google Scholar]
- 31.Riemen, G., and G. Michaelis. 1993. A point mutation in the core subunit gene of yeast mitochondrial RNA polymerase is suppressed by a high level of specificity factor MTF1. Mol. Gen. Genet. 237:49-57. [DOI] [PubMed] [Google Scholar]
- 32.Rodeheffer, M. S., B. E. Boone, A. C. Bryan, and G. S. Shadel. 2001. Nam1p, a protein involved in RNA processing and translation, is coupled to transcription through an interaction with yeast mitochondrial RNA polymerase. J. Biol. Chem. 276:8616-8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schinkel, A. H., M. J. Groot Koerkamp, and H. Tabak. 1988. Mitochondrial RNA polymerase of Saccharomyces cerevisiae: composition and mechanism of promoter recognition. EMBO J. 7:3255-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schubot, F. D., C. J. Chen, J. P. Rose, T. A. Dailey, H. A. Dailey, and B. C. Wang. Crystal structure of the transcription factor sc-mtTFB offers insights into mitochondrial transcription. Protein Sci. 10:1980-1988. [DOI] [PMC free article] [PubMed]
- 35.Shadel, G. S. 1999. Yeast as a model for human mtDNA replication. Am. J. Hum. Genet. 65:1230-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shadel, G. S., and D. A. Clayton. 1993. Mitochondrial transcription initiation. Variation and conservation. J. Biol. Chem. 268:16083-16086. [PubMed] [Google Scholar]
- 37.Shadel, G. S., and D. A. Clayton. 1995. A Saccharomyces cerevisiae mitochondrial transcription factor, sc-mtTFB, shares features with sigma factors but is functionally distinct. Mol. Cell. Biol. 15:2101-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shadel, G. S., and D. A. Clayton. 1997. Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem. 66:409-435. [DOI] [PubMed] [Google Scholar]
- 39.Shi, X., P. Yau, T. Jose, and P. D. Gershon. 1996. Methyltransferase-specific domains within VP39, a bi-functional protein that participates in the modification of both mRNA ends. RNA 2:88-101. [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. J. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tiranti, V., A. Savoia, F. Forti, M. F. D'Apolito, M. Centra, M. Rocchi, and M. Zeviani. 1997. Identification of the gene encoding the human mitochondrial RNA polymerase (h-mtRPOL) by cyberscreening of the Expressed Sequence Tags database. Hum. Mol. Genet. 6:615-625. [DOI] [PubMed] [Google Scholar]
- 42.Virbasius, J. V., and R. C. Scarpulla. 1994. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc. Natl. Acad. Sci. USA 91:1309-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wallace, D. C. 1999. Mitochondrial diseases in man and mouse. Science 283:1482-1488. [DOI] [PubMed] [Google Scholar]
- 44.Wallace, D. C., G. Singh, M. T. Lott, J. A. Hodge, T. G. Schurr, A. M. Lezza, L. J. Elsas II, and E. K. Nikoskelainen. 1988. Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science 242:1427-1430. [DOI] [PubMed] [Google Scholar]
- 45.Wingender, E., P. Dietze, H. Karas, and R. Knuppel. 1996. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 24:238-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu, B., and D. A. Clayton. 1992. Assignment of a yeast protein necessary for mitochondrial transcription initiation. Nucleic Acids Res. 20:1053-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]