Abstract
Insulin receptor substrate 1 (IRS-1) plays an important role in the insulin signaling cascade. In vitro and in vivo studies from many investigators have suggested that lowering of IRS-1 cellular levels may be a mechanism of disordered insulin action (so-called insulin resistance). We previously reported that the protein levels of IRS-1 were selectively regulated by a proteasome degradation pathway in CHO/IR/IRS-1 cells and 3T3-L1 adipocytes during prolonged insulin exposure, whereas IRS-2 was unaffected. We have now studied the signaling events that are involved in activation of the IRS-1 proteasome degradation pathway. Additionally, we have addressed structural elements in IRS-1 versus IRS-2 that are required for its specific proteasome degradation. Using ts20 cells, which express a temperature-sensitive mutant of ubiquitin-activating enzyme E1, ubiquitination of IRS-1 was shown to be a prerequisite for insulin-induced IRS-1 proteasome degradation. Using IRS-1/IRS-2 chimeric proteins, the N-terminal region of IRS-1 including the PH and PTB domains was identified as essential for targeting IRS-1 to the ubiquitin-proteasome degradation pathway. Activation of phosphatidylinositol 3-kinase is necessary but not sufficient for activating and sustaining the IRS-1 ubiquitin-proteasome degradation pathway. In contrast, activation of mTOR is not required for IRS-1 degradation in CHO/IR cells. Thus, our data provide insight into the molecular mechanism of insulin-induced activation of the IRS-1 ubiquitin-proteasome degradation pathway.
Insulin receptor substrate (IRS) proteins are key molecules in the insulin signaling cascade (79, 80). Upon insulin stimulation, IRS proteins are tyrosyl phosphorylated by insulin receptor tyrosine kinase, forming a signaling complex with many SH2 domain-containing proteins, including phosphatidylinositol (PI) 3-kinase, Grb-2, SHP-2, Fyn, and Nck, and initiating multiple insulin intracellular signals (70, 77, 79, 80). Decreased cellular levels of IRS-1 and IRS-2 have been shown to be associated with insulin resistance in animal and human subjects (1, 3, 6, 26, 29, 50, 53, 55, 73, 84). Furthermore, genetic ablation of either IRS-1 or IRS-2 in mice showed impaired insulin action (3, 6, 73, 83, 84), indicating that appropriate levels of IRS proteins are crucial for maintaining normal insulin-regulated metabolism.
The cellular levels of IRS-1 are influenced by many factors, including growth factors and cytokines (35, 47, 49, 56, 65, 85). Studies using cell lines or isolated primary cells chronically exposed to insulin (a condition known to induce impaired insulin signaling) have revealed that the reduced level of IRS-1 protein is due to enhanced degradation (35, 49, 59, 61, 62, 68). This is consistent with the fact that chronic insulin treatment can induce insulin resistance in healthy human subjects and rats (14, 25, 32, 39), suggesting that one mechanism of insulin resistance is a reduction of the IRS-1 protein level in the major insulin-sensitive tissues.
Early studies proposed that the degradation was mediated by calpain, a calcium-dependent protease (61, 62). However, recent studies from several laboratories, including ours, have shown that proteasome-mediated degradation is the likely cause of IRS-1 degradation (23, 35, 48, 68, 72). In contrast, IRS-2 was resistant to proteasome degradation in the same cell background (23, 48, 68, 72).
Interestingly, PI 3-kinase activation was consistently shown to be required in both calpain-mediated and proteasome-mediated IRS-1 degradation in cultured cells (23, 35, 48, 63, 68, 72). PI 3-kinase is a dual kinase that possesses lipid and protein kinase activities which are inhibited by LY294002 and wortmannin (15, 75). The lipid kinase activity of PI 3-kinase is well documented in its ability to phosphorylate phosphoinositol at the D-3 position, producing PI 3,4-disphosphate (PI3,4P2) or PI3,4,5P3, which function to activate downstream signaling molecules, including PDK-1 and Akt (7). Akt is a serine kinase that is activated by PDK-1 in a PI3,4P2-dependent manner (66, 67). When activated, Akt phosphorylates various substrates, including GSK-3, Bad, Raf, and mTOR (2, 11-13, 52, 86). mTOR has been shown to phosphorylate IRS-1 and implicated in IRS-1 degradation (23, 48, 72). However, the exact molecular requirement for activation of the IRS-1 proteasome degradation pathway remains controversial.
The proteasome degradation system is normally composed of two distinct and successive steps, ubiquitin conjugation and proteasome degradation (10). Conjugation of ubiquitin to the substrate proceeds via a three-step mechanism (10, 33). Ubiquitin is first activated by a single ubiquitin-activating enzyme, E1. Following activation, one of several E2 enzymes (ubiquitin-conjugating proteins) transfers ubiquitin from E1 to a member of the ubiquitin-protein ligase family, E3, to which the substrate protein is specifically bound. E3 catalyzes the last step in the conjugation process, covalent attachment of ubiquitin to the substrate (10, 33). Ubiquitin-tagged proteins are then recognized by the 26S proteasome and degraded. Control of protein degradation by the proteasome degradation pathway depends mostly on the ubiquitination of targeted proteins and often on modifications such as serine phosphorylation (10). Understanding the molecular mechanism of IRS-1 proteasome degradation during prolonged insulin exposure promises to shed light on the molecular mechanism of insulin resistance.
The current study investigated the signaling pathway that leads to the activation of ubiquitination and proteasome degradation of IRS-1 and also the structural elements in IRS-1 that are important for its degradation. We have confirmed that ubiquitination is a prerequisite for insulin-induced degradation of IRS-1 and now show that the N-terminal region of IRS-1 is required for ubiquitin-proteasome degradation. Furthermore, we have excluded mTOR in IRS-1 degradation in CHO cells. Instead, our results show that tyrosyl phosphorylation of IRS-1 and PI 3-kinase activation are both required for the activation of the IRS-1 ubiquitin-proteasome degradation pathway. However, activation of PI 3-kinase alone is insufficient.
MATERIALS AND METHODS
Cell culture.
CHO cells and CHO cells overexpressing wild-type human insulin receptors (CHO/IR) or mutant insulin receptors (CHO/IRA960 and CHO/IRF960) or both insulin receptor and rat IRS-1 (CHO/IR/IRS-1) were grown in F-12 medium supplemented with 10% fetal bovine serum (68). They were cultured at 37°C in a humidified atmosphere composed of 95% air and 5% CO2.
CHO/IR cells overexpressing mutant IRS-1with 18 potential tyrosine residues replaced by phenylalanines (CHO/IR/IRS-1F18) and chimeras (IRS-1/IRS-2) were obtained by transfection of CHO/IR cells with pCMV/His/IRS-1F18 or pCMV/His chimeras (IRS-1/IRS-2) by the calcium phosphate precipitation method and selected by resistance to 10 mM histidinol (Sigma) (69) for 3 weeks before the experiment.
CHO/IR/IRS-1 cells overexpressing hemagglutinin (HA)-tagged mouse Akt (CHO/IR/IRS-1/HA-Akt) were obtained by cotransfection of CHO/IR/IRS-1 cells with pCMV/HA-Akt (10 μg) and pEBVHis A (1 μg; Invitrogen) by the calcium phosphate precipitation method and selected by resistance to hygromycin B (400 μg/ml; Sigma) (69). Stable CHO/IR/IRS-1/HA-Akt cell lines were cloned and further selected by immunoblotting with anti-HA tag (αHA) antibody (F-7; Santa Cruz).
E36 and ts20 cells (kind gifts of Simon Wing from McGill University) were grown in Dulbecco's modified Eagle's medium (DMEM) with high glucose (DMEM/H) supplemented with 10% fetal bovine serum (37). E36 and ts20 cells overexpressing human insulin receptors (E36/IR and ts20/IR, respectively) were obtained by cotransfection of E36 and ts20 cells with pSG5/hIR (10 μg) and pEBVHis A (1 μg) by the calcium phosphate precipitation method and selected by resistance to hygromycin B (200 μg/ml; Sigma) (69). Stable E36/IR and ts20/IR cell lines were cloned and further selected by immunoblotting with an antibody to the β subunit of the insulin receptor (αIRβ) (C-19; Santa Cruz).
Prolonged insulin exposure.
Subconfluent cells were made quiescent in DMEM/H for at least 4 h and then incubated with or without insulin (100 nM or as indicated) at 37°C for various times (68). In some cases, inhibitors were added at indicated times before or after the addition of insulin. At the end of the prolonged insulin exposure, cells were stimulated again with or without insulin for 2 min before being lysed.
Immunoprecipitation and immunoblotting analysis.
For immunoprecipitation, cells were washed once with ice-cold buffer A (20 mM Tris-HCl [pH 7.5], 137 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 100 μM Na3VO4) and lysed in 1 ml of lysis buffer (buffer A containing 1 mM phenylmethylsulfonyl fluoride [PMSF], 100 μM Na3VO4, 10% glycerol, and 1% NP-40 [Calbiochem] plus 50 μg of aprotinin and 50 μg of leupeptin per ml). Lysates were centrifuged at 13,000 × g for 15 min, and the supernatants were incubated with αIRS-1 (VT01 immune serum used at 1:100), αIRS-2 (VT11 immune serum used at 1:100), αIRβ (C-19 at 2 μg/ml; Santa Cruz), antiphosphotyrosine (αPY; PY-20 at 2 μg/ml; Santa Cruz), αHA (F-7 at 2 μg/ml; Santa Cruz), or anti-p70S6K (2 μg/ml; Cell Signaling) for 1 h at 4°C. Immune complexes were captured with protein A-agarose (Gibco-BRL), washed three times with the lysis buffer, and denatured in Laemmli sample buffer containing 0.1 M dithiothreitol (DTT) (68).
For whole-cell lysate immunoblotting analysis, cells were lysed directly in Laemmli sample buffer containing 0.1 M DTT and sonicated. Proteins in immune complexes or in whole-cell lysates were further denatured by boiling for 5 min, separated by sodium dodecyl sulfate-7.5% or 6% polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to nitrocellulose membranes. Membranes were blocked overnight at 4°C in TBS (20 mM Tris-HCl [pH 8.0], 0.15 M NaCl) containing 1% milk and 1% bovine serum albumin (BSA), then incubated for 1 h at room temperature in TBST (TBS containing 0.05% Tween 20) containing the following antibodies: αIRS-1 (VT 01) and αIRS-2 (VT 11), both used at a 1:400 dilution, and αHA (F-7), αIRβ (C-19), αphospho-Akt (Ser 473), and αp70S6K according to the recommendations of the manufacturers. The membranes were washed three times with TBST and probed with horseradish peroxidase (HRP)-conjugated protein A or protein G (Santa Cruz) at 1:3,000 for 30 min.
For the detection of tyrosyl phosphorylation, the membranes were directly probed with HRP-conjugated PY-20 or PY-99 (Santa Cruz) at 1:3,000 for 30 min. The membranes were washed three times with TBST and once with TBS and then developed with SuperSignal substrate (Pierce) for 5 min. Bands were detected by exposing the membranes to Kodak BioMax MR film (68). In some cases, Western blots were scanned and quantified using NIH Image version 1.62.
PI 3-kinase activity assay.
PI 3-kinase activity was determined by incorporation of [32P]phosphate into phosphatidylinositol using immune complexes. Immune complexes obtained by αIRS-1 or αPY immunoprecipitation were washed successively in phosphate-buffered saline (PBS) containing 1% NP-40 and 100 μM Na3VO4 (three times), 10 mM Tris-HCl (pH 7.5) containing 500 mM LiCl and 100 μM Na3VO4 (three times), and 100 mM Tris-HCl (pH 7.5) containing 100 mM NaCl, 1 mM EDTA, and 100 μM Na3VO4 (two times). The washed immune complexes were resuspended in 50 μl of 10 mM Tris-HCl (pH 7.5) containing 100 mM NaCl and 1 mM EDTA and combined with 10 μl of 100 mM MgCl2, and 10 μl of 2-μg/μl phosphatidylinositol (Avanti) which had been sonicated in 10 mM Tris-HCl (pH 7.5) containing 1 mM EGTA. The assay was initiated by addition of 5 μl of [γ-32P]ATP solution (0.88 mM ATP containing 30 μCi of [γ-32P]ATP and 20 mM MgCl2). The reaction proceeded for 10 min at room temperature and was terminated by addition of 20 μl of 8 N HCl and 160 μl of CHCl3-methanol (1:1). The samples were vortexed and centrifuged. The lower organic phase was removed and applied to silica gel thin-layer chromatography plates (Merck). The plates were developed in CHCl3-CH3OH-H2O-NH4OH (60:47:11.3:2), dried, and visualized by autoradiography (69).
Construction of His6-tagged IRS-1 and IRS-2.
A His6 tag was inserted into the C terminus of IRS-1 and IRS-2 by a PCR-mediated reaction (19). The PCR product for IRS-1 was obtained using 5"-CGC GGA TCC TTT AAG CAC ACC CAG CGC-3" as the 5"-end primer and 5"-CCA TGA CGT CTA GTG ATG GTG GTG ATG GTG TTG ACG GTC CTC TGG TTG-3" as the 3"-end primer, which contained sequence for His6, and pBluescript/IRS-1 as a template. The PCR product for IRS-2 was obtained using 5"-CGC GGA TCC GCT AAG GTC ATC CGT GCA G-3" as the 5"-end primer, 5"-CAT GGC CAA GCT TCA GTG ATG GTG GTG ATG GTG CTC TTT CAC GAC TGT GGC-3" as the 3"-end primer, which contained sequence for His6, and pBluscript/IRS-2 as a template.
The IRS-1 PCR product and pBluescript/IRS-1 were digested with BamHI and AatII, and the PCR fragment was inserted in place of the wild-type sequence, resulting in pBluescript/IRS-1CH. The IRS-2 PCR product and pBluescript/IRS-2 were digested with AatII and HindIII, and the PCR fragment was inserted in place of the wild-type sequence, resulting in pBluescript/IRS-2CH. The presence of the His6 tag in the right reading frame in IRS-1CH and IRS-2CH was confirmed by sequencing the recombinant molecules on a 373 XL DNA sequencer in the Vermont Cancer Center DNA Analysis Facility at the University of Vermont. IRS-1CH and IRS-2CH were isolated from pBluescript by digestion with SacI and HindIII and subcloned into pCMV/His expression vector between the SacI and HindIII sites.
Construction of chimeric IRS-1/IRS-2.
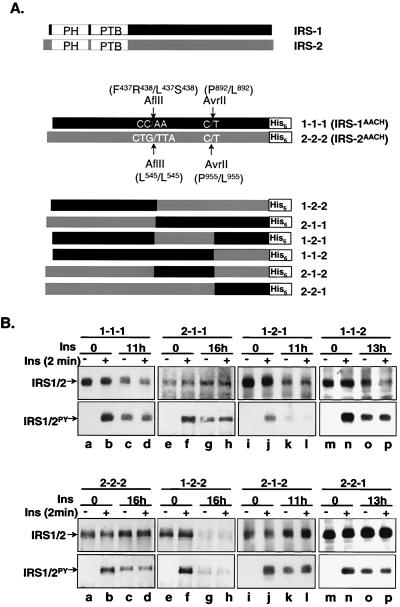
AflII and AvrII sites were introduced into IRS-1CH and IRS-2CH cDNAs at similar positions by PCR-mediated oligonucleotide-directed mutagenesis (19). To create AflII sites, the mutagenic primers for IRS-1 were 5"-CGA AGT TCC TTA AGC AGT GTC ACC-3" and 5"-GGT GAC ACT GCT TAA GGA ACT TCG-3", and for IRS-2, they were 5"-GGA TAG ACC CTT AAG CCA CTG TGG-3" and 5"-CAG TGG CTT AAG GGT CTA TCC ATG-3". To create AvrII sites, the mutagenic primers for IRS-1 were 5"-GCC CAA AAG CCT AGG AGA ATA TG-3" and 5"-CAT ATT CTC CTA GGC TTT TGG GC-3", and for IRS-2, they were 5"-TTC AGG AAC CCT AGG CAC CAG CAG-3" and 5"-TGC TGG TGC CTA GGG TTC CTG AAC-3".
pBluescript/IRS-1CH, pBluescript/IRS-2CH, and mutant PCR products were digested with appropriate restriction enzymes, and mutant PCR fragments were inserted in place of the wild-type sequences to create AflII and AvrII mutants of IRS-1CH (pBluescript/IRS-1AACH) and IRS-2CH (pBluescript/IRS-2AACH). The presence of the desired mutations was confirmed by sequencing the recombinant DNA on a 373 XL DNA sequencer in the Vermont Cancer Center DNA Analysis Facility at the University of Vermont. Both IRS-1AACH and IRS-2AACH mutants were digested with AflII, AvrII, and/or HindIII, and fragments were isolated and religated in all combinations to create six chimeras with each fragment in its original position (see Fig. 8A). The chimeras were verified by restriction analysis. Chimeras were isolated from pBluescript by digestion with SacI and HindIII and subcloned into the pCMV/His expression vector between the SacI and HindIII sites.
FIG. 8.
Insulin-induced degradation of IRS-1/IRS-2 chimeric proteins. (A) Construction of IRS-1/IRS-2 chimeras. A His6 tag was first inserted into the C terminus of IRS-1 and IRS-2, resulting in IRS-1CH, and IRS-2CH. AflII and AvrII restriction sites were then introduced into the IRS-1CH and IRS-2CH cDNAs at similar positions by PCR-based site-directed mutagenesis, resulting in IRS-1AACH and IRS-2 AACH. IRS-1AACH and IRS-2 AACH were digested with AflII and/or AvrII, and fragments were isolated and religased in all combinations to create six chimeras with each fragment similar to its original position. Each chimera was given a code, indicated on the right. (B) CHO/IR/chimera cells were exposed to 100 nM insulin or nothing for the indicated times and rechallenged with 100 nM insulin for 2 min before being lysed in Laemmli sample buffer containing 0.1 M DTT. Proteins were separated by SDS-7.5% PAGE and transferred to nitrocellulose membranes. The protein levels of the chimeras were detected by immunoblotting analysis with α-His tag antibody and tyrosyl phosphorylation with αPY. The results are representative of at least two experiments.
RESULTS
Insulin receptor is required for proteasome degradation of IRS-1.
Degradation of endogenous IRS-1 and IRS-2 was examined in CHO cells (expressing only 3,000 hamster insulin receptors/cell) and CHO cells overexpressing 100,000 human insulin receptors/cell (CHO/IR) (81). Cells were exposed to insulin for 12 h to see the chronic effect and rechallenged for 2 min to see the acute effect of insulin on tyrosyl phosphorylation and protein levels of IRS-1.
Acute insulin stimulation elicited low levels of tyrosyl phosphorylation on IRS-1 and IRS-2 in CHO cells, and overexpressing the insulin receptor in CHO cells significantly increased tyrosyl phosphorylation of IRS-1 and IRS-2 (Fig. 1A and B,lower panels, lanes a, b, g, and h). Moderate degradation of IRS-1 was detected in CHO cells after prolonged insulin exposure (Fig. 1A, upper panel, lanes a to d); however, overexpressing the insulin receptor greatly accelerated IRS-1 degradation, which was completely blocked by either MG132 (a potent proteasome inhibitor) (Fig. 1A, upper panel, lanes g to l) or epoxomicin (the most specific proteasome inhibitor) (38) (Fig. 1A, upper panel, lanes m to r). In contrast, IRS-2 in the same cell background failed to show significant degradation during prolonged insulin exposure (Fig. 1B, upper panel, lanes a to r).
FIG. 1.
Degradation of IRS-1 and IRS-2 in CHO, CHO/IR, and CHO/IR/IRS-1 cells. (A and B) CHO and CHO/IR cells were untreated or treated with insulin (Ins, 100 nM) for 12 h in the presence or absence of 25 μM MG132 or 10 μM epoxomicin and rechallenged or not with insulin (100 nM) for 2 min before being lysed. Endogenous IRS-1 and IRS-2 were immunoprecipitated (IP) with αIRS-1 (A) or αIRS-2 (B). (C) CHO/IR/IRS-1 cells were incubated with or without actinomycin (Act., 0.5 μM) or cycloheximide B (Cyc., 0.5 mM) for 30 min before being treated with 100 nM insulin for 8 h or left untreated. Some cells were rechallenged with insulin for 2 min before being lysed in Laemmli sample buffer containing 0.1 M DTT. αIRS-1 and αIRS-2 immune complexes and lysates were separated by SDS-7.5% PAGE and transferred to nitrocellulose membranes. Tyrosine phosphorylation (IRS-1PY or IRS-2PY) and protein levels of IRS-1 and IRS-2 were detected by immunoblotting (IB) analysis using αPY, αIRS-1, and αIRS-2, respectively. These results are representative of at least two experiments.
Preincubation of cells with actinomycin (a transcription inhibitor) or cycloheximide (a protein translation inhibitor) had no effect on insulin-induced degradation of IRS-1 (Fig. 1C), suggesting that the degradation of IRS-1 was not a secondary effect of RNA or protein synthesis. Together, these data suggest that although the insulin receptor mediates the tyrosyl phosphorylation of both IRS-1 and IRS-2, it activates an existing proteasome degradation pathway leading specifically to IRS-1 degradation.
Ubiquitin conjugation to IRS-1 is a prerequisite for its degradation.
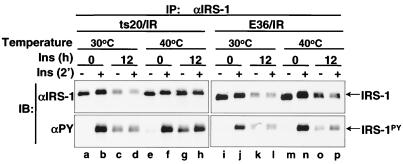
We and others have previously shown, by utilizing an antiubiquitin antibody, that IRS-1 is ubiquitinated (35, 68, 85). However, ubiquitination of a protein does not necessarily lead to its degradation. To address whether ubiquitin conjugation to IRS-1 is required for its degradation, we examined insulin-induced degradation of IRS-1 in ts20 cells, which are derived from E36 cells and are temperature sensitive for the ubiquitin-activating enzyme E1 (34, 37). In ts20 cells, the E1 enzyme is active at the permissive temperature of 30°C, whereas at the nonpermissive temperature of 40°C, E1 is inactivated, and substrate proteins accumulate because they cannot be ubiquitinated.
Like CHO cells, both E36 and ts20 cells showed moderate IRS-1 degradation during prolonged insulin exposure due to the low endogenous insulin receptor expression (data not shown). However, when the human insulin receptors were overexpressed in E36 cells (E36/IR), insulin-induced degradation of IRS-1 occurred at both 30 and 40°C (Fig. 2,lanes i to p). In contrast, degradation of IRS-1 occurred at 30°C but not at 40°C in ts20 cells overexpressing insulin receptor (ts20/IR) (Fig. 2, lanes a to h), suggesting that E1 is absolutely required for IRS-1 degradation. This result confirms that ubiquitin conjugation to IRS-1 is necessary for its degradation during prolonged insulin exposure.
FIG. 2.
Insulin-induced IRS-1 degradation requires the activity of ubiquitin-activating enzyme E1. E36 and ts20 cells overexpressing the human insulin receptor (E36/IR and ts20/IR, respectively) were exposed to nothing or 100 nM insulin for 12 h at 30 or 40°C. Cells were unstimulated or stimulated with insulin for 2 min before being lysed. IRS-1 was immunoprecipitated from lysates with αIRS-1, separated by SDS-7.5% PAGE, and transferred to nitrocellulose membranes. Tyrosine phosphorylation of IRS-1 (IRS-1PY) and protein levels of IRS-1 were detected by immunoblotting analysis with αPY and αIRS-1, respectively. These results are representative of at least two experiments.
Tyrosyl phosphorylation of IRS-1 is required for the activation of the IRS-1 ubiquitin-proteasome degradation pathway but not for degradation per se.
Mutation at tyrosine960 of the insulin receptor β-subunit significantly decreases its ability to phosphorylate IRS-1 and impairs insulin action without any effect on its autophosphorylation and kinase activity (27, 81). CHO cells overexpressing wild-type insulin receptor (CHO/IR) or the tyrosine960 to alanine mutant (CHO/IRA960) were used to test if tyrosyl phosphorylation of IRS-1 is required for activation of the insulin-induced IRS-1 degradation signal.
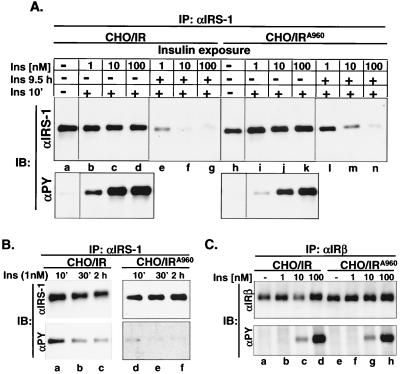
As expected, acute insulin-stimulated tyrosine phosphorylation of endogenous IRS-1 was significantly lower in CHO/IRA960 cells than in CHO/IR cells (Fig. 3A,lower panel, lanes a to d versus h to k). A 10-fold higher concentration of insulin was required for CHO/IRA960 cells to achieve the same level of tyrosyl phosphorylation of IRS-1 as in CHO/IR cells (Fig. 3A, lower panel, lanes b and c versus j and k). Consistent with the defect of the IRA960 mutant in tyrosyl phosphorylation of IRS-1, insulin-induced PI 3-kinase activation was dramatically reduced in CHO/IRA960 cells (Fig. 4B,lanes g and h versus e and f).
FIG. 3.
Insulin-induced proteasome degradation of IRS-1 in CHO cells overexpressing the IRA960 mutant. (A) CHO/IR and CHO/IRA960 cells were exposed to various concentrations of insulin for 9.5 h or 10 min, and IRS-1 was immunoprecipitated (IP) from cell lysates with αIRS-1. (B) CHO/IR and CHO/IRA960 cells were exposed to 1 nM insulin for up to 2 h, and IRS-1 was immunoprecipitated from cell lysates with αIRS-1. (C) CHO/IR and CHO/IRA960 cells were exposed to various concentrations of insulin for 10 min, and insulin receptors were immunoprecipitated from cell lysates with αIRβ. Immunoprecipitated IRS-1 or insulin receptor were separated by SDS-7.5% PAGE and transferred to nitrocellulose membranes. Protein levels of IRS-1 and insulin receptor and their tyrosine phosphorylation were detected by immunoblotting analysis using αIRS-1, αIRβ, and αPY, respectively.
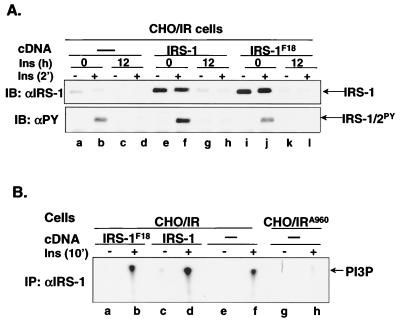
FIG. 4.
Insulin-induced degradation of the IRS-1F18 mutant in CHO/IR cells. (A) CHO/IR, CHO/IR/IRS-1, and CHO/IR/IRS-1F18 cells were exposed to nothing or 100 nM insulin for 12 h. Cells were unstimulated or stimulated with insulin for 2 min before being lysed in Laemmli sample buffer containing 0.1 M DTT. Proteins were separated on SDS-7.5% PAGE and transferred to nitrocellulose membranes, which were then immunoblotted with αIRS-1 or αPY. The results are representative of six experiments. Three Western blots were scanned and quantified using NIH Image ver. 1.62. Results for αIRS-1: 538 ± 141, 260 ± 119, 4 ± 3, 8 ± 8, 5,956 ± 648, 5,063 ± 724, 489 ± 58, 264 ± 109, 6,704 ± 216, 6,441 ± 382, 212 ± 106, and 180 ± 95. Results for αPY: 120 ± 16, 3,123 ± 304, 123 ± 65, 137 ± 69, 185 ± 85, 6126 ± 275, 372 ± 57, 353 ± 55, 181 ± 58, 2,972 ± 215, 165 ± 54, and 165 ± 76. (B) CHO/IR/IRS-1F18, CHO/IR/IRS-1, CHO/IR, and CHO/IRA960 cells were unstimulated or stimulated with 100 nM insulin for 10 min. IRS-1 was immunoprecipitated from cell lysates by αIRS-1, and PI 3-kinase activity was measured in IRS-1 immune complexes. The results are representative of three experiments.
Prolonged exposure of cells to various concentrations of insulin revealed a significant reduction in insulin-induced degradation of IRS-1 in CHO/IRA960 cells compared with CHO/IR cells (Fig. 3A, upper panel, lanes l to n versus e to g). The reduced degradation of IRS-1 in CHO/IRA960 cells correlated very well with its reduced insulin-induced tyrosyl phosphorylation, since a 10-fold-higher concentration of insulin was required for CHO/IRA960 cells to achieve the same degree of IRS-1 degradation as in CHO/IR cells (Fig. 3A, upper panel, lanes m and n versus e and f). This was not due to the different levels of the expressed insulin receptor, since both cell lines expressed approximately the same level of human insulin receptors and showed the same autophosphorylation in response to various insulin concentrations (Fig. 3C). The decreased tyrosyl phosphorylation of IRS-1 in CHO/IRA960 could not be overcome by prolonged insulin exposure, since tyrosyl phosphorylation of IRS-1 did not increase for the 2 h of insulin exposure in either cell line (Fig. 3B). Identical results were obtained with CHO cells overexpressing the tyrosine960 to phenylalanine mutant of insulin receptor (data not shown). Thus, tyrosyl phosphorylation of IRS-1 is required for IRS-1 degradation.
It is possible that tyrosyl phosphorylation of IRS-1 is required for activation of the ubiquitin-proteasome degradation pathway or IRS-1 degradation per se. To address this question, IRS-1 degradation was investigated in CHO/IR cells overexpressing an IRS-1F18 mutant in which 18 major tyrosine phosphorylation sites have been replaced by phenylalanines (41). Although IRS-1 protein levels (IRS-1F18 mutant plus endogenous IRS-1) in CHO/IR/IRS-1F18 cells were much higher than in CHO/IR cells and comparable to those in CHO/IR/IRS-1 cells (Fig. 4A, upper panel, lanes a to l), insulin-induced tyrosyl phosphorylation of IRS-1/IRS-2- and IRS-1-associated PI 3-kinase activity in CHO/IR/IRS-1F18 cells was comparable to that in CHO/IR cells (Fig. 4A, lower panel, lanes i and j versus a and b, and Fig. 4B, lanes a to f) and significantly lower than that in CHO/IR/IRS-1 cells (Fig. 4A, lower panel, lanes i and j versus e and f, and Fig. 4B, lanes a to f). This suggests that endogenous IRS-1 but not IRS-1F18 mutant is tyrosyl phosphorylated and activates PI 3-kinase in response to insulin in CHO/IR/IRS-1F18 cells.
Surprisingly, the IRS-1F18 mutant was also targeted for ubiquitin-proteasome degradation during prolonged insulin exposure (Fig. 4A, upper panel, lanes i to l versus a to h) although it was not tyrosyl phosphorylated. Thus, endogenous IRS-1 mediated the insulin-induced activation of the IRS-1 ubiquitin-proteasome degradation pathway, resulting in degradation of IRS-1F18.
Insulin-induced IRS-1 degradation pathway is tightly associated with PI 3-kinase activity.
To further establish the signaling pathway that mediates the ubiquitin-proteasome degradation of IRS-1, various inhibitors were tested for their ability to prevent the insulin-induced degradation of IRS-1. These inhibitors included H7, a broad serine/threonine kinase inhibitor; bisindolylmaleimide I (BIM), a highly selective protein kinase C (PKC) inhibitor for PKCα, -βI, -βII, -γ, -δ, and -ɛ isozymes; rapamycin, a highly selective inhibitor of mTOR; Ro-31-8220, a specific inhibitor for atypical PKCs, including ξ and λ; LY294002, a specific inhibitor of PI 3-kinase; and PD169316 and SB203580, specific inhibitors for p38 mitogen-activated protein (MAP) kinase (data not shown).
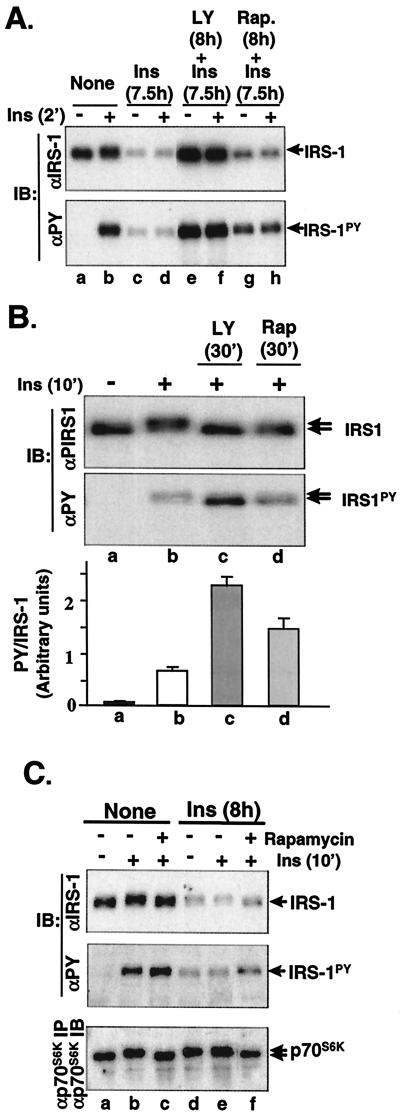
Among all the tested inhibitors, LY294002 was the only compound that completely blocked insulin-induced degradation of IRS-1 (Fig. 5A,upper panel, lanes e and f versus a to d), suggesting that PI 3-kinase activity is necessary for the activation of the IRS-1 ubiquitin-proteasome degradation pathway. Rapamycin is a specific inhibitor of mTOR, a downstream serine kinase of PI 3-kinase, and has been shown to inhibit serine phosphorylation of IRS-1 (serine636/639) and p70S6K. As with LY294002, preincubation of cells with rapamycin also abolished insulin-induced IRS-1 mobility shift on SDS-PAGE (an indication of the inhibition of serine/threonine phosphorylation of IRS-1) (Fig. 5B, upper panel, lane d versus b, and 5C, upper panel, lanes c and f versus b and e), slightly increased insulin-stimulated tyrosyl phosphorylation of IRS-1 (Fig. 5B, middle panel, lane d versus b, and lower panel, and Fig. 5C, middle panel, lanes c and f versus b and e), and inhibited insulin-induced mTOR serine kinase activity (judged by an abolished mobility shift of p70S6K [8, 57]) (Fig. 5C, lower panel, lanes c and f versus b and e). However, it had little effect on the insulin-induced degradation of IRS-1 in CHO/IR/IRS-1 cells (Fig. 5A, lanes g and h versus c and d, and 5C, upper panel, lanes d to f versus a to c). These results indicate that PI 3-kinase but not mTOR plays a major role in the activation of the IRS-1 ubiquitin-proteasome degradation pathway.
FIG. 5.
Effect of inhibitors on the insulin-induced degradation of IRS-1. (A) CHO/IR/IRS-1 cells were untreated or treated with LY294002 (LY, 50 μM) or rapamycin (Rap., 200 nM) for 30 min before being exposed to insulin (Ins, 100 nM) for 7.5 h. Cells were rechallenged with insulin for 2 min or not, lysed in Laemmli sample buffer containing 0.1 M DTT, separated by SDS-7.5% PAGE, and transferred to nitrocellulose membranes. IRS-1 and tyrosyl-phosphorylated IRS-1 were detected by immunoblotting (IB) analysis using αIRS-1 and αPY, respectively. (B) CHO/IR/IRS-1 cells were untreated or treated with LY294002 (50 μM) or rapamycin (200 nM) for 30 min, followed by 100 nM insulin for 10 min before being lysed. Proteins were separated by SDS-6% PAGE, and transferred to nitrocellulose membranes. Tyrosine phosphorylation (IRS-1PY) and levels of IRS-1 protein were detected by immunoblotting analysis using αPY and αIRS-1, respectively. Mobility shifts of IRS-1 are indicated by arrows. In the lower panel, Western blots from three separate experiments were scanned and quantified by NIH image ver. 1.62. The data are expressed as a density ratio of phospho-IRS-1 to IRS-1 protein. The standard deviation is indicated. (C) CHO/IR/IRS-1 cells were untreated or treated with 200 nM rapamycin for 30 min, followed by exposure to nothing or 100 nM insulin for 8 h. Cells were rechallenged with 100 nM insulin for 10 min or not before being lysed. IRS-1 and phosphorylated IRS-1 were immunoblotted with αIRS-1 and αPY, respectively (upper and middle panels), and p70 S6K was immunoprecipitated with αp70S6K and immunoblotted with αp70S6K (lower panel).
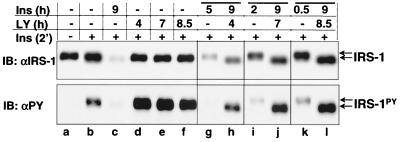
Since preincubation of cells with LY294002 completely inhibited PI 3-kinase activity and the insulin-induced IRS-1 degradation signal, we addressed whether IRS-1 degradation would be sustained if LY294002 was added once the IRS-1 ubiquitin-proteasome degradation pathway had been activated. LY294002 alone had no effect on the IRS-1 protein level except that it potentiated the insulin-induced tyrosyl phosphorylation of IRS-1 (Fig. 6,lanes a to f). CHO/IR/IRS-1 cells that were exposed to insulin for 9 h showed a significant degradation of IRS-1 (Fig. 6, upper panel, lane c versus a and b). In contrast, when LY294002 was added after an initial 30 min of insulin stimulation, insulin-induced degradation of IRS-1 was completely arrested for the rest of the 8.5-h exposure to both insulin and LY294002, as if the cells had only been treated with insulin for 30 min instead of 9 h (Fig. 6, upper panel, lane l versus k and c). Similar results were obtained when LY294002 was added after an initial insulin exposure of 2 or 5 h (Fig. 6, upper panel, lanes h and j versus g, i, and c).
FIG. 6.
Effect of LY294002 on IRS-1 degradation in pre-insulin-activated cells. CHO/IR/IRS-1 cells were exposed to nothing (lanes a and b) or 100 nM insulin for 9 h (lanes c, h, j, and l); to some cells, LY294002 (50 μM) was added after 30 min (lane l), 2 h (lane j), or 5 h (lane h) of insulin exposure. Control cells were exposed to LY294002 alone for 4, 7, or 8.5 h (lanes d to f) or to insulin alone for 30 min (lane k), 2 h (lane i), or 5 h (lane g). At the end of the incubation, cells were stimulated with 100 nM insulin for 2 min (lanes b to l) before being lysed in Laemmli sample buffer containing 0.1 M DTT. Proteins were separated by SDS-6% PAGE and transferred to nitrocellulose membranes. Protein levels and tyrosyl phosphorylation of IRS-1 were detected by immunoblotting analysis with αIRS-1 and αPY, respectively. The results are representative of two experiments.
The levels of IRS-1 protein were determined not by the overall time of insulin exposure (9 h), but rather by the time of insulin exposure in the absence of LY294002 (0.5, 2, and 5 h). This result suggested that an LY294002-sensitive molecule, most likely PI 3-kinase, is required to maintain the activation of the IRS-1 ubiquitin-proteasome degradation pathway.
Activation of PI 3-kinase is not sufficient for IRS-1 degradation.
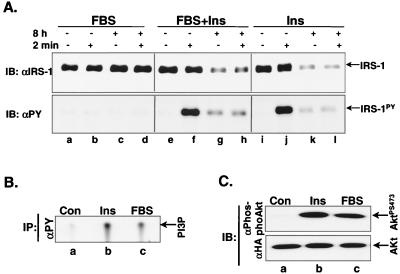
Since PI 3-kinase appears to be critical for IRS-1 proteasome degradation, we wanted to determine whether activation of PI 3-kinase is sufficient for the degradation of IRS-1 without inducing tyrosyl phosphorylation of IRS-1. CHO/IR/IRS-1/HA-Akt cells were exposed to 10% fetal bovine serum or insulin, and the activity of PI 3-kinase and Akt and the tyrosyl phosphorylation of IRS-1 were examined. Both PI 3-kinase and Akt were activated by 10% fetal bovine serum to a similar extent as by 100 nM insulin (Fig. 7B and C)during acute stimulation. In contrast, compared with insulin, 10% fetal bovine serum failed to induce significant tyrosyl phosphorylation of IRS-1 during acute stimulation (Fig. 7A, lower panel, lanes a and b versus lanes i and j) and failed to induce IRS-1 degradation after prolonged exposure (Fig. 7A, upper panel, lanes a to d versus i to l). This is not due to an inhibitory effect of fetal bovine serum on the IRS-1 degradation pathway, since IRS-1 degradation was comparable when cells were chronically exposed to both fetal bovine serum and insulin (Fig. 7A, upper panel, lanes e to h versus i to l). This suggests that activation of PI 3-kinase alone is not sufficient to activate the IRS-1 ubiquitin-proteasome degradation pathway.
FIG. 7.
Effect of 10% fetal bovine serum on activation of PI 3-kinase, Akt, and IRS-1 degradation. (A) CHO/IR/IRS-1/HA-Akt cells were stimulated with nothing, 10% fetal bovine serum, 100 nM insulin, or 10% fetal bovine serum plus 100 nM insulin for 8 h and rechallenged with nothing, 10% fetal bovine serum, 100 nM insulin, or 10% fetal bovine serum plus 100 nM insulin for 2 min before being lysed. Protein levels and tyrosyl phosphorylation of IRS-1 were measured by immunoblotting analysis with αIRS-1 and αPY, respectively. (B and C) CHO/IR/IRS-1/HA-Akt cells were stimulated with nothing, 10% fetal bovine serum, or 100 nM insulin for 10 min and lysed in lysate buffer. PI 3-kinase activity was measured in αPY immune complexes (B). Phosphorylation of Akt at serine473 and protein levels of Akt were measured in the lysates by immunoblotting analysis with αphosphoAkt and αHA, respectively (C). Results are representative of at least three experiments.
N-terminal region of IRS-1 is responsible for its ubiquitin-proteasome degradation.
The insulin-induced ubiquitin-proteasome pathway degrades IRS-1 but not IRS-2 in the same cell background (Fig. 1A and 1B), suggesting that a unique structural element in IRS-1 is required for this specific process (68). To identify this structural element, His6-tagged chimeras were constructed in which various regions of IRS-1 and IRS-2 were exchanged (Fig. 8A). They were expressed in CHO/IR cells, and insulin-induced degradation of the chimeras was examined.
Although insulin acutely stimulated tyrosyl phosphorylation of both IRS-1AACH and IRS-2 AACH, IRS-1AACH but not IRS-2 AACH showed evident degradation during prolonged insulin exposure (Fig. 8B, 1-1-1 and 2-2-2), suggesting that introducing internal mutations (AlfII and AvrII) and His6 tag at the C terminus did not alter the ability of IRS-1 to be specifically degraded by the ubiquitin-proteasome pathway during prolonged insulin exposure. All chimeras remained competent for insulin-induced tyrosyl phosphorylation during acute insulin stimulation (Fig. 8B, second and fourth panels). All three chimeras which contained the N-terminal region of IRS-1 were degraded during prolonged insulin exposure (Fig. 8B, 1-2-1, 1-1-2, and 1-2-2), whereas the chimeras containing the N-terminal region of IRS-2 were not affected (Fig. 8A). Thus, the N-terminal region of IRS-1 contains a structural element that is crucial for the specificity of ubiquitination and proteasome degradation of IRS-1.
DISCUSSION
Although insulin is generally considered an anabolic hormone that supports protein synthesis and inhibits general protein degradation (21, 30, 72), evidence has accumulated over the past 5 years suggesting that prolonged exposure of cells to insulin promotes IRS-1 degradation (23, 35, 48, 68, 72, 85). We previously identified the proteasome degradation pathway as a major mechanism that specifically regulates the cellular level of IRS-1 but not IRS-2 during prolonged insulin exposure in a cell culture system (68). In the current study, we have demonstrated, for the first time, that ubiquitin conjugation of IRS-1 is a prerequisite for insulin-induced IRS-1 proteasome degradation. The N-terminal region of IRS-1 contains the structural element that specifically targets IRS-1 to the ubiquitin-proteasome degradation pathway. Both tyrosyl phosphorylation of IRS-1 and PI 3-kinase activation are needed to activate the IRS-1 ubiquitin-proteasome degradation pathway. We have provided evidence against mTOR's playing a significant role in the process of IRS-1 proteasome degradation in CHO cells. Together, these results provide insight into the mechanism of activation of the IRS-1 ubiquitin-proteasome degradation pathway during prolonged insulin exposure, which may underlie the molecular mechanism of insulin resistance.
Proteasome degradation is an important cellular system that not only functions to eliminate malfunctioning or misfolded proteins but also acts as a regulatory mechanism to precisely control the levels of cellular proteins and thus their biological functions in a timely manner (9, 10, 33, 36, 45, 46). Proteasome degradation is a highly controlled and specific process and depends largely on ubiquitination of the targeted protein (10). In addition to the three-step ubiquitination reaction involving E1, E2, and E3 ligases, recent studies have shown that ubiquitination of targeted proteins is regulated by a signaling pathway which activates a specific E3 ligase (10, 33).
Although we and others have previously shown by immunoblotting with antiubiquitin antibodies, or a similar technique, that IRS-1 is indeed ubiquitinated (35, 68, 85), the requirement for ubiquitination of IRS-1 in terms of proteasome degradation had not been established prior to this study. ts20 cells were derived from chemically mutagenized E36 cells and shown to contain a temperature-sensitive mutation in the ubiquitin-activating enzyme E1 (22, 34). ts20 cells which overexpress the human insulin receptor (ts20/IR) were used to test the requirement for ubiquitination in the insulin-induced degradation of IRS-1. Inactivating E1 in ts20/IR cells by raising the temperature to 40°C completely blocked the insulin-induced degradation of IRS-1, clearly demonstrating that insulin-induced degradation of IRS-1 is a ubiquitin-dependent process. This result further suggests that prolonged insulin exposure activates a signaling pathway that mediates the ubiquitination and proteasome degradation of IRS-1 (IRS-1 ubiquitin-proteasome degradation pathway).
The insulin receptor is absolutely required for the insulin-induced degradation of IRS-1, suggesting that the activation of the IRS-1ubiquitin degradation pathway is mediated by insulin receptor signaling. Binding of insulin to its receptor activates the intrinsic protein tyrosyl kinase, which subsequently phosphorylates its intracellular substrates on tyrosine residues, including IRS proteins (77, 78). The NPXY960 motif at the juxtamembrane region of the insulin receptor has been shown to interact with the PTB domain of IRS-1 when it is phosphorylated (20, 24, 27, 43, 81), and mutation at tyrosine960 significantly decreases its ability to phosphorylate IRS-1 (27, 81). Tyrosyl phosphorylation of IRS proteins is an important step to activate insulin intracellular signaling (77, 78).
Here we showed that tyrosyl phosphorylation of IRS-1 is also a critical step for the insulin-induced degradation of IRS-1. First, cells (CHO, E36, and ts20 cells) with low levels of insulin receptors displayed only a modest tyrosyl phosphorylation of IRS-1 in response to acute insulin stimulation and a modest degradation of endogenous IRS-1 during prolonged insulin exposure. However, overexpression of insulin receptor in these cells dramatically increased insulin-induced tyrosyl phosphorylation of endogenous IRS-1 and concomitantly increased insulin-induced ubiquitin-proteasome-dependent degradation of IRS-1. Second, a good correlation between decreased tyrosyl phosphorylation of IRS-1 and decreased insulin-induced degradation of IRS-1 in cells overexpressing IRA960 or IRF960 was observed at all insulin concentrations tested (from 1 to 100 nM) compared with the cells overexpressing wild-type insulin receptors. Finally, fetal bovine serum failed to induce tyrosyl phosphorylation of IRS-1 and degradation of IRS-1, although it activated PI 3-kinase and Akt. Taken together, these data strongly suggest that tyrosyl phosphorylation of IRS-1 is an initial step in the activation of the IRS-1 ubiquitin-proteasome degradation pathway.
An IRS-1 mutant in which 18 potential tyrosyl phosphorylation sites were replaced by phenylalanines (IRS-1F18) is unable to undergo tyrosyl phosphorylation or mediate activation of PI 3-kinase and p70S6K during insulin stimulation (41). To determine if it can be degraded by the ubiquitin-proteasome system, IRS-1F18 was overexpressed in CHO/IR cells, in which the IRS-1 ubiquitin-proteasome degradation pathway is activated by endogenous IRS-1 during prolonged insulin exposure. The insulin-stimulated tyrosyl phosphorylation of the endogenous IRS proteins and PI 3-kinase activity in CHO/IR/IRS-1F18 cells was comparable to that in CHO/IR cells suggesting that IRS-1F18 does not function as a dominant negative molecule to block endogenous IRS-1 signaling. Therefore, it is reasonable to assume that in CHO/IR/IRS-1F18 cells, the IRS-1 ubiquitin-proteasome degradation pathway is activated normally through the endogenous IRS-1 during prolonged insulin exposure. Interestingly, IRS-1F18 was degraded at the same rate as wild-type IRS-1 in CHO/IR cells during prolonged insulin exposure, suggesting that unphosphorylated IRS-1 can be targeted for ubiquitin-proteasome degradation.
Tyrosyl phosphorylation of IRS-1 promotes the interaction of IRS-1 and SH2 domain-containing proteins, including PI 3-kinase, Grb2, Nck, Fyn, and SHP-2, leading to the activation of signaling pathways mediated by these molecules (77, 79). To identify the downstream molecules that mediate the activation of the IRS-1 ubiquitin-proteasome degradation pathway, we used a wide range of inhibitors for serine kinases, including PKCs, MAP kinases, mTOR, and PI 3-kinase. Among the inhibitors tested, LY294002 and wortmannin effectively blocked the insulin-induced degradation of IRS-1, suggesting that PI 3-kinase activity is essential for the IRS-1 ubiquitin-proteasome degradation pathway.
The insulin-induced activation of PI 3-kinase is mediated by tyrosyl-phosphorylated IRS-1 (4, 5,40, 54), consistent with the requirement of tyrosyl phosphorylation of IRS-1 for activation of the IRS-1 ubiquitin-proteasome degradation pathway. More interestingly, LY294002 was able to completely block IRS-1 degradation even after the IRS-1 ubiquitin-proteasome degradation pathway had been activated for up to 5 h. This suggests that the activity of PI 3-kinase is essential not only to activate but also to maintain the IRS-1 ubiquitin-proteasome degradation pathway.
The essential role of PI 3-kinase in IRS-1 degradation raises the important question of whether the activation of PI 3-kinase is sufficient for IRS-1 degradation. Fetal bovine serum contains various growth factors which can effectively activate PI 3-kinase and downstream signaling molecules, including Akt and mTOR. Our data clearly demonstrate that fetal bovine serum failed to induce IRS-1 degradation, suggesting that PI 3-kinase activation alone is not sufficient for IRS-1 degradation. The fact that fetal bovine serum failed to induced tyrosyl phosphorylation of IRS-1 further supports the notion that activation of PI 3-kinase via interaction with tyrosyl-phosphorylated IRS-1 is essential for the IRS-1 ubiquitin-proteasome degradation pathway.
The location of the activated PI 3-kinase may be important, since platelet-derived growth factor (PDGF)-activated-PI 3-kinase associates mainly with the PDGF receptor at the plasma membrane, whereas the majority of insulin-stimulated IRS-1-associated PI 3-kinase activity is in the low-density microsomes (42, 51, 82). Overexpressing a constitutively active form of PI 3-kinase, p110CAAX, in the adenovirus system has been shown to induce IRS-1 degradation in the absence of insulin in 3T3-L1 adipocytes (16, 17, 23, 72), suggesting that PI 3-kinase activation is sufficient for IRS-1 degradation. The reason for this contradiction is not clear. It is possible that overexpression of a large quantity of a constitutively active form of PI 3-kinase may overcome the need for tyrosyl phosphorylation of IRS-1. Another possibility is that p110CAAX is a membrane-targeted construct and may target PI 3-kinase activity to a compartment to which wild-type PI 3-kinase is never translocated without binding tyrosyl-phosphorylated IRS-1.
Rapamycin, a specific inhibitor of the mammalian target of rapamycin (mTOR) and a downstream molecule of PI 3-kinase (58), has been shown to block the insulin-induced degradation of IRS-1 (23, 48) and tumor necrosis factor alpha-induced serine636/serine639 phosphorylation in IRS-1 in 3T3-L1 adipocytes (44). Our data agree with these studies in that rapamycin did prevent serine phosphorylation of IRS-1 manifested by prevention of the IRS-1 mobility shift on SDS-PAGE, and increased insulin-induced tyrosyl phosphorylation of IRS-1 to some degree, and the activation of p70S6K in CHO/IR cells. However, rapamycin failed to block the insulin-induced degradation of IRS-1, suggesting that a rapamycin-sensitive molecule (most likely mTOR) is not a major player in the activation of the IRS-1 ubiquitin-proteasome degradation pathway in CHO/IR cells. The reason for these different findings remains unknown. Insulin-induced degradation of IRS-1 in 3T3-L1 adipocytes can be blocked by a calcium chelator or specific inhibitors for calpain, indicating a Ca2+-dependent and calpain-mediated process (62, 63). Furthermore, IRS-1 can be degraded by calpain in vitro (61). One possible explanation is that in 3T3-L1 adipocytes, in addition to the proteasome degradation pathway (a Ca2+-independent process), IRS-1 degradation may also be controlled by a Ca2+-dependent calpain degradation pathway which may be sensitive to rapamycin.
Serine phosphorylation has been shown to be an important regulatory mechanism in the ubiquitin-proteasome degradation pathway. A growing number of E3 ligases have been identified as SCF (Skp/Cullin/F-box protein) complexes which couple the protein kinase signaling pathway to the control of protein abundance (18, 28, 60, 76). F-box proteins in SCF complexes serve as receptors that recruit serine-phosphorylated substrates to the SCF ubiquitin-ligase complexes, a mechanism that controls not only the substrate specificity but also the timing (60). For example, the IκB-ubiquitin ligase complex only ubiquitinates IκBα when it is phosphorylated (64), whereas β-catenin ubiquitination is preceded by phosphorylation via the glycogen synthase kinase 3β/Axin kinase complex, which coexists in the ubiquitin ligase complex (31).
Studies using in vivo 32P metabolic labeling of CHO/IR/IRS-1 have shown that insulin induces serine/threonine phosphorylation of IRS-1 (69), which is prevented by preincubation of cells with wortmannin (unpublished data). Inactivation of PI 3-kinase by LY294002 before or after insulin stimulation not only blocked the degradation of IRS-1, but also prevented the mobility shift of IRS-1 on SDS-PAGE (an indication of serine phosphorylation of IRS-1) and increased the insulin-induced tyrosyl phosphorylation of IRS-1, suggesting that PI 3-kinase-dependent serine phosphorylation of IRS-1 might be involved in the regulation of IRS-1 ubiquitin-proteasome degradation. Although direct evidence is lacking, it is tempting to speculate that the IRS-1 ubiquitin ligase complex may associate with an LY294002-sensitive serine kinase which phosphorylates IRS-1, triggering ubiquitination. Since PI 3-kinase has been reported to phosphorylate IRS-1 in αIRS-1 immune complexes in a wortmannin-sensitive manner (74), it is possible that PI 3-kinase is the serine kinase. Further work needs to be done to test this idea.
The IRS-1 ubiquitin-proteasome degradation pathway is specific for IRS-1 and is completely ineffective for IRS-2 in both CHO/IR cells and 3T3-L1 adipocytes (48, 68, 72). This suggests that a unique structural element present in IRS-1 but absent in IRS-2 is important for IRS-1 degradation. The results from IRS chimeras confirmed that this structural element does exist. IRS-1 with its N-terminal region replaced by that of IRS-2 lost the ability to be degraded, whereas IRS-2 with its N-terminal region replaced by that of IRS-1 gained the ability to be degraded, suggesting that the structural element for IRS-1 degradation resides within the N-terminal region of IRS-1, where the PH, PTB, and IH3 domains (71) are located. Interestingly, this region of IRS-1 shows the highest homology with IRS-2 (71).
Exactly how the N-terminal region of IRS-1 plays a role in controlling the ubiquitin-proteasome degradation of IRS-1 is not known. It may contain ubiquitination sites, phosphorylation sites for recognition by the IRS-1 ubiquitin ligase complex, or a domain that interacts with the ubiquitin ligase. Further investigation should answer these key questions.
Acknowledgments
We thank S. Wing at McGill University for kindly providing us with E36 and ts20 cells and helpful discussions, M. White at the Joslin Diabetes Center for the IRS-1F18 construct, and J. Backer at Albert Einstein College of Medicine for the CHO/IRA960 and CHO/IRF960 cell lines used in this study. We thank J. Leahy for his constructive suggestions and comments on the manuscript.
This work was supported by an ADA research grant (X.J.S.) and NIH grant AI41426-02 (X.J.S.).
REFERENCES
- 1.Anai, M., M. Funaki, T. Ogihara, J. Terasaki, K. Inukai, H. Katagiri, Y. Fukushima, Y. Yazaki, M. Kikuchi, Y. Oka, and T. Asano. 1998. Altered expression levels and impaired steps in the pathway to phosphatidylinositol 3-kinase activation via insulin receptor substrates 1 and 2 in Zucker fatty rats. Diabetes 47:13-23. [DOI] [PubMed] [Google Scholar]
- 2.Aoki, M., E. Blazek, and P. K. Vogt. 2001. A role of the kinase mTOR in cellular transformation induced by the oncoproteins P3k and Akt. Proc. Natl. Acad. Sci. USA 98:136-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Araki, E., M. A. Lipes, M. E. Patti, J. C. Bruning, B. Haag, 3rd, R. S. Johnson, and C. R. Kahn. 1994. Alternative pathway of insulin signalling in mice with targeted disruption of the IRS-1 gene. Nature 372:186-190. [DOI] [PubMed] [Google Scholar]
- 4.Backer, J. M., M. G. Myers, Jr., S. E. Shoelson, D. J. Chin, X. J. Sun, M. Miralpeix, P. Hu, B. Margolis, E. Y. Skolnik, J. Schlessinger, and M. F. White. 1992. Phosphatidylinositol 3"-kinase is activated by association with IRS-1 during insulin stimulation. EMBO J. 11:3469-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Backer, J. M., M. G. Myers, Jr., X. J. Sun, D. J. Chin, S. E. Shoelson, M. Miralpeix, and M. F. White. 1993. Association of IRS-1 with the insulin receptor and the phosphatidylinositol 3"-kinase. Formation of binary and ternary signaling complexes in intact cells. J. Biol. Chem. 268:8204-8212. [PubMed] [Google Scholar]
- 6.Bruning, J. C., J. Winnay, S. Bonner-Weir, S. I. Taylor, D. Accili, and C. R. Kahn. 1997. Development of a novel polygenic model of NIDDM in mice heterozygous for IR and IRS-1 null alleles. Cell 88:561-572. [DOI] [PubMed] [Google Scholar]
- 7.Burgering, B. M., and P. J. Coffer. 1995. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature 376:599-602. [DOI] [PubMed] [Google Scholar]
- 8.Chung, J., T. C. Grammer, K. P. Lemon, A. Kazlauskas, and J. Blenis. 1994. PDGF- and insulin-dependent pp70S6k activation mediated by phosphatidylinositol-3-OH kinase. Nature 370:71-75. [DOI] [PubMed] [Google Scholar]
- 9.Ciechanover, A. 1994. The ubiquitin-proteasome proteolytic pathway. Cell 79:13-21. [DOI] [PubMed] [Google Scholar]
- 10.Ciechanover, A. 1998. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 17:7151-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cross, D. A. E., D. R. Alessi, P. Cohen, M. Andjelkovich, and B. A. Hemmings. 1995. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378:785-789. [DOI] [PubMed] [Google Scholar]
- 12.Datta, S. R., H. Dudek, X. Tao, S. Masters, H. Fu, Y. Gotoh, and M. E. Greenberg. 1997. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 91:231-241. [DOI] [PubMed] [Google Scholar]
- 13.del Peso, L., M. Gonzalez-Garcia, C. Page, R. Herrera, and G. Nunez. 1997. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science 278:687-689. [DOI] [PubMed] [Google Scholar]
- 14.Del Prato, S., F. Leonetti, D. C. Simonson, P. Sheehan, M. Matsuda, and R. A. DeFronzo. 1994. Effect of sustained physiologic hyperinsulinaemia and hyperglycaemia on insulin secretion and insulin sensitivity in man. Diabetologia 37:1025-1035. [DOI] [PubMed] [Google Scholar]
- 15.Dhand, R., I. Hiles, G. Panayotou, S. Roche, M. J. Fry, I. Gout, N. F. Totty, O. Truong, P. Vicendo, K. Yonezawa, M. Kasuga, S. A. Courtneidge, and M. D. Waterfield. 1994. PI-3-kinase is a dual specificity enzyme -- autoregulation by an intrinsic protein-serine kinase activity. EMBO J. 13:522-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egawa, K., N. Nakashima, P. M. Sharma, H. Maegawa, Y. Nagai, A. Kashiwagi, R. Kikkawa, and J. M. Olefsky. 2000. Persistent activation of phosphatidylinositol 3-kinase causes insulin resistance due to accelerated insulin-induced insulin receptor substrate-1 degradation in 3T3-L1 adipocytes. Endocrinology 141:1930-1935. [DOI] [PubMed] [Google Scholar]
- 17.Egawa, K., P. M. Sharma, N. Nakashima, Y. Huang, E. Huver, G. R. Boss, and J. M. Olefsky. 1999. Membrane-targeted phosphatidylinositol 3-kinase mimics insulin actions and induces a state of cellular insulin resistance. J. Biol. Chem. 274:14306-14314. [DOI] [PubMed] [Google Scholar]
- 18.Feldman, R. M., C. C. Correll, K. B. Kaplan, and R. J. Deshaies. 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91:221-230. [DOI] [PubMed] [Google Scholar]
- 19.Gilliland, G., S. Perrin, and H. F. Bunn. 1990. Competitive PCR for quantitation of mRNA, p. 60-75. In M. A. Innis, D. H. Geland, J. J. Sninsky, and T. J. White (ed.), PCR protocols, a guide to methods and applications. Academic Press, Inc., San Diego, Calif.
- 20.Gustafson, T. A., W. He, A. Craparo, C. D. Schaub, and T. J. O'Neill. 1995. Phosphotyrosine-dependent interaction of Shc and IRS-1 with the NPEY motif of the insulin receptor via a novel non-SH2 domain. Mol. Cell. Biol. 15:2500-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamel, F. G., R. G. Bennett, K. S. Harmon, and W. C. Duckworth. 1997. Insulin inhibition of proteasome activity in intact cells. Biochem. Biophys. Res. Commun. 234:671-674. [DOI] [PubMed] [Google Scholar]
- 22.Handley-Gearhart, P. M., J. S. Trausch-Azar, A. Ciechanover, and A. L. Schwartz. 1994. Rescue of the complex temperature-sensitive phenotype of Chinese hamster ovary E36ts20 cells by expression of the human ubiquitin-activating enzyme cDNA. Biochem. J. 304:1015-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haruta, T., T. Uno, J. Kawahara, A. Takano, K. Egawa, P. M. Sharma, J. M. Olefsky, and M. Kobayashi. 2001. A rapamycin-sensitive pathway down-regulates insulin signaling via phosphorylation and proteasomal degradation of insulin receptor substrate-1. Mol. Endocrinol. 14:783-794. [DOI] [PubMed] [Google Scholar]
- 24.He, W., T. J. O'Neill, and T. A. Gustafson. 1995. Distinct modes of interaction of SHC and insulin receptor substrate-1 with the insulin receptor NPEY region via non-SH2 domains. J. Biol. Chem. 270:23258-23262. [DOI] [PubMed] [Google Scholar]
- 25.Iozzo, P., T. Pratipanawatr, H. Pijl, C. Vogt, V. Kumar, R. Pipek, M. Matsuda, L. J. Mandarino, K. J. Cusi, and R. A. DeFronzo. 2001. Physiological hyperinsulinemia impairs insulin-stimulated glycogen synthase activity and glycogen synthesis. Am. J. Physiol. Endocrinol. Metab. 280:E712-E719. [DOI] [PubMed] [Google Scholar]
- 26.Jiang, Z. Y., Y. W. Lin, A. Clemont, E. P. Feener, K. D. Hein, M. Igarashi, T. Yamauchi, M. F. White, and G. L. King. 1999. Characterization of selective resistance to insulin signaling in the vasculature of obese Zucker (fa/fa) rats. J. Clin. Investig. 104:447-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaburagi, Y., K. Momomura, R. Yamamoto-Honda, K. Tobe, Y. Tamori, H. Sakura, Y. Akanuma, Y. Yazaki, and T. Kadowaki. 1993. Site-directed mutatgenesis of the juxtamembrane domain of the human insulin receptor. J. Biol. Chem. 268:16610-16622. [PubMed] [Google Scholar]
- 28.Kaplan, K. B., A. A. Hyman, and P. K. Sorger. 1997. Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell 91:491-500. [DOI] [PubMed] [Google Scholar]
- 29.Kerouz, N. J., D. Horsch, S. Pons, and C. R. Kahn. 1997. Differential regulation of insulin receptor substrates-1 and -2 (IRS-1 and IRS-2) and phosphatidylinositol 3-kinase isoforms in liver and muscle of the obese diabetic (ob/ob) mouse. J. Clin. Investig. 100:3164-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kettelhut, I. C., S. S. Wing, and A. L. Goldberg. 1988. Endocrine regulation of protein breakdown in skeletal muscle. Diabetes Metab. Rev. 4:751-772. [DOI] [PubMed] [Google Scholar]
- 31.Kitagawa, M., S. Hatakeyama, M. Shirane, M. Matsumoto, N. Ishida, K. Hattori, I. Nakamichi, A. Kikuchi, and K. Nakayama. 1999. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of beta-catenin. EMBO J. 18:2401-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koopmans, S. J., L. Ohman, J. R. Haywood, L. J. Mandarino, and R. A. DeFronzo. 1997. Seven days of euglycemic hyperinsulinemia induces insulin resistance for glucose metabolism but not hypertension, elevated catecholamine levels, or increased sodium retention in conscious normal rats. Diabetes 46:1572-1578. [DOI] [PubMed] [Google Scholar]
- 33.Kornitzer, D., and A. Ciechanover. 2001. Modes of regulation of ubiquitin-mediated protein degradation. J. Cell Physiol. 182:1-11. [DOI] [PubMed] [Google Scholar]
- 34.Kulka, R. G., B. Raboy, R. Schuster, H. A. Parag, G. Diamond, A. Ciechanover, and M. Marcus. 1988. A Chinese hamster cell cycle mutant arrested at G2 phase has a temperature-sensitive ubiquitin-activating enzyme, E1. J. Biol. Chem. 263:15726-15731. [PubMed] [Google Scholar]
- 35.Lee, A. V., J. L. Gooch, S. Oesterreich, R. L. Guler, and D. Yee. 2000. Insulin-like growth factor I-induced degradation of insulin receptor substrate 1 is mediated by the 26S proteasome and blocked by phosphatidylinositol 3"-kinase inhibition. Mol. Cell. Biol. 20:1489-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maniatis, T. 1999. A ubiquitin ligase complex essential for the NF-kappaB, Wnt/Wingless, and Hedgehog signaling pathways. Genes Dev. 13:505-510. [DOI] [PubMed] [Google Scholar]
- 37.Meerovitch, K., S. Wing, and D. Goltzman. 1998. Proparathyroid hormone-related protein is associated with the chaperone protein BiP and undergoes proteasome-mediated degradation. J. Biol. Chem. 273:21025-21030. [DOI] [PubMed] [Google Scholar]
- 38.Meng, L., R. Mohan, B. H. Kwok, M. Elofsson, N. Sin, and C. M. Crews. 1999. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc. Natl. Acad. Sci. USA 96:10403-10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miles, P. D., S. Li, M. Hart, O. Romeo, J. Cheng, A. Cohen, K. Raafat, A. R. Moossa, and J. M. Olefsky. 1998. Mechanisms of insulin resistance in experimental hyperinsulinemic dogs. J. Clin. Investig. 101:202-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myers, M. G., Jr., J. M. Backer, X. J. Sun, S. E. Shoelson, P. Hu, J. Schlessinger, M. Yoakim, B. Schaffhausen, and M. F. White. 1992. IRS-1 activates phosphatidylinositol 3"-kinase by associating with src homology 2 domains of p85. Proc. Natl. Acad. Sci. USA 89:10350-10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Myers, M. G., Jr., Y. Zhang, G. A. Aldaz, T. Grammer, E. M. Glasheen, L. Yenush, L. M. Wang, X. J. Sun, J. Blenis, J. H. Pierce, and M. F. White. 1996. YMXM motifs and signaling by an insulin receptor substrate 1 molecule without tyrosine phosphorylation sites. Mol. Cell. Biol. 16:4147-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nave, B. T., R. J. Haigh, A. C. Hayward, K. Siddle, and P. R. Shepherd. 1996. Compartment-specific regulation of phosphoinositide 3-kinase by platelet-derived growth factor and insulin in 3T3-L1 adipocytes. Biochem. J. 318:55-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Neill, T. J., A. Craparo, and T. A. Gustafson. 1994. Characterization of an interaction between insulin receptor substrate-1 and the insulin receptor by using the two-hybrid system. Mol. Cell. Biol. 14:6433-6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ozes, O. N., H. Akca, L. D. Mayo, J. A. Gustin, T. Maehama, J. E. Dixon, and D. B. Donner. 2001. A phosphatidylinositol 3-kinase/Akt/mTOR pathway mediates and PTEN antagonizes tumor necrosis factor inhibition of insulin signaling through insulin receptor substrate-1. Proc. Natl. Acad. Sci. USA 98:4640-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pagano, M. 1997. Cell cycle regulation by the ubiquitin pathway. FASEB J. 11:1067-1075. [DOI] [PubMed] [Google Scholar]
- 46.Palombella, V. J., O. J. Rando, A. L. Goldberg, and T. Maniatis. 1994. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell 78:773-785. [DOI] [PubMed] [Google Scholar]
- 47.Patti, M. E., A. Virkamaki, E. J. Landaker, C. R. Kahn, and H. Yki-Jarvinen. 1999. Activation of the hexosamine pathway by glucosamine in vivo induces insulin resistance of early postreceptor insulin signaling events in skeletal muscle. Diabetes 48:1562-1571. [DOI] [PubMed] [Google Scholar]
- 48.Pederson, T. M., D. L. Kramer, and C. M. Rondinone. 2001. Serine/threonine phosphorylation of IRS-1 triggers its degradation: possible regulation by tyrosine phosphorylation. Diabetes 50:24-31. [DOI] [PubMed] [Google Scholar]
- 49.Rice, K. M., M. A. Turnbow, and C. W. Garner. 1993. Insulin stimulates the degradation of IRS-1 in 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 190:961-967. [DOI] [PubMed] [Google Scholar]
- 50.Ricort, J. M., J. F. Tanti, E. Van Obberghen, and Y. Le Marchand-Brustel. 1995. Alterations in insulin signalling pathway induced by prolonged insulin treatment of 3T3-L1 adipocytes. Diabetologia 38:1148-1156. [DOI] [PubMed] [Google Scholar]
- 51.Ricort, J. M., J. F. Tanti, E. Van Obberghen, and Y. Le Marchand-Brustel. 1996. Different effects of insulin and platelet-derived growth factor on phosphatidylinositol 3-kinase at the subcellular level in 3T3-L1 adipocytes. A possible explanation for their specific effects on glucose transport. Eur. J. Biochem. 239:17-22. [DOI] [PubMed] [Google Scholar]
- 52.Rommel, C., B. A. Clarke, S. Zimmermann, L. Nunez, R. Rossman, K. Reid, K. Moelling, G. D. Yancopoulos, and D. J. Glass. 1999. Differentiation stage-specific inhibition of the Raf-MEK-ERK pathway by Akt. Science 286:1738-1741. [DOI] [PubMed] [Google Scholar]
- 53.Rondinone, C. M., L. M. Wang, P. Lonnroth, C. Wesslau, J. H. Pierce, and U. Smith. 1997. Insulin receptor substrate (IRS) 1 is reduced and IRS-2 is the main docking protein for phosphatidylinositol 3-kinase in adipocytes from subjects with non-insulin-dependent diabetes mellitus. Proc. Natl. Acad. Sci. USA 94:4171-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rordorf-Nikolic, T., D. J. Van Horn, D. Chen, M. F. White, and J. M. Backer. 1995. Regulation of phosphatidylinositol 3-kinase by tyrosyl phosphoproteins. Full activation requires occupancy of both SH2 domains in the 85 kDa regulatory subunit. J. Biol. Chem. 270:3662-3666. [DOI] [PubMed] [Google Scholar]
- 55.Saad, M. J., E. Araki, M. Miralpeix, P. L. Rothenberg, M. F. White, and C. R. Kahn. 1992. Regulation of insulin receptor substrate-1 in liver and muscle of animal models of insulin resistance. J. Clin. Investig. 90:1839-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saad, M. J., F. Folli, and C. R. Kahn. 1995. Insulin and dexamethasone regulate insulin receptors, insulin receptor substrate-1, and phosphatidylinositol 3-kinase in Fao hepatoma cells. Endocrinology 136:1579-1588. [DOI] [PubMed] [Google Scholar]
- 57.Sabatini, D. M., H. Erdjument-Bromage, M. Lui, P. Tempst, and S. H. Snyder. 1994. RAFT1: A mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 78:35-43. [DOI] [PubMed] [Google Scholar]
- 58.Schmelzle, T., and M. N. Hall. 2001. TOR, a central controller of cell growth. Cell 103:253-262. [DOI] [PubMed] [Google Scholar]
- 59.Singh, T. J. 1993. Insulin receptor serine kinase activation by casein kinase 2 and a membrane tyrosine kinase. Mol. Cell. Biochem. 121:167-174. [DOI] [PubMed] [Google Scholar]
- 60.Skowyra, D., K. L. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91:209-219. [DOI] [PubMed] [Google Scholar]
- 61.Smith, L. K., M. Bradshaw, D. E. Croall, and C. W. Garner. 1993. The insulin receptor substrate (IRS-1) is a PEST protein that is susceptible to calpain degradation in vitro. Biochem. Biophys. Res. Commun. 196:767-772. [DOI] [PubMed] [Google Scholar]
- 62.Smith, L. K., K. M. Rice, and C. W. Garner. 1996. The insulin-induced down-regulation of IRS-1 in 3T3-L1 adipocytes is mediated by a calcium-dependent thiol protease. Mol. Cell. Endocrinol. 122:81-92. [DOI] [PubMed] [Google Scholar]
- 63.Smith, L. K., C. J. Vlahos, K. K. Reddy, J. R. Falck, and C. W. Garner. 1995. Wortmannin and LY294002 inhibit the insulin-induced down-regulation of IRS-1 in 3T3-L1 adipocytes. Mol. Cell. Endocrinol. 113:73-81. [DOI] [PubMed] [Google Scholar]
- 64.Spencer, E., J. Jiang, and Z. J. Chen. 1999. Signal-induced ubiquitination of IkappaBalpha by the F-box protein Slimb/beta-TrCP. Genes Dev. 13:284-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stephens, J. M., J. Lee, and P. F. Pilch. 1997. Tumor necrosis factor-alpha-induced insulin resistance in 3T3-L1 adipocytes is accompanied by a loss of insulin receptor substrate-1 and GLUT4 expression without a loss of insulin receptor-mediated signal transduction. J. Biol. Chem. 272:971-976. [DOI] [PubMed] [Google Scholar]
- 66.Stephens, L., K. Anderson, D. Stokoe, H. Erdjument-Bromage, G. F. Painter, A. B. Holmes, P. R. J. Gaffney, C. B. Reese, F. McCormick, P. Tempst, J. Coadwell, P. T. Hawkins, and P. R. Gaffney. 1998. Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B. Science 279:710-714. [DOI] [PubMed] [Google Scholar]
- 67.Stokoe, D., L. R. Stephens, T. Copeland, P. R. Gaffney, C. B. Reese, G. F. Painter, A. B. Holmes, F. McCormick, and P. T. Hawkins. 1997. Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science 277:567-570. [DOI] [PubMed] [Google Scholar]
- 68.Sun, X. J., J. L. Goldberg, L. Y. Qiao, and J. J. Mitchell. 1999. Insulin-induced insulin receptor substrate-1 degradation is mediated by the proteasome degradation pathway. Diabetes 48:1359-1364. [DOI] [PubMed] [Google Scholar]
- 69.Sun, X. J., M. Miralpeix, M. G. Myers, Jr., E. M. Glasheen, J. M. Backer, C. R. Kahn, and M. F. White. 1992. The expression and function of IRS-1 in insulin signal transmission. J. Biol. Chem. 267:22662-22672. [PubMed] [Google Scholar]
- 70.Sun, X. J., S. Pons, L.-M. Wang, Y. Zhang, L. Yenush, D. Burks, M. G. Myers, Jr., E. M. Glasheen, N. G. Copeland, N. A. Jenkins, J. H. Pierce, and M. F. White. 1997. The IRS-2 gene on murine chromosome 8 encodes a unique signaling adapter for insulin and cytokine action. Mol. Endocrinol. 11:251-262. [DOI] [PubMed] [Google Scholar]
- 71.Sun, X. J., L. M. Wang, Y. Zhang, L. Yenush, M. G. Myers, Jr., E. Glasheen, W. S. Lane, J. H. Pierce, and M. F. White. 1995. Role of IRS-2 in insulin and cytokine signalling. Nature 377:173-177. [DOI] [PubMed] [Google Scholar]
- 72.Takano, A., I. Usui, T. Haruta, J. Kawahara, T. Uno, M. Iwata, and M. Kobayashi. 2001. Mammalian target of rapamycin pathway regulates insulin signaling via subcellular redistribution of insulin receptor substrate 1 and integrates nutritional signals and metabolic signals of insulin. Mol. Cell. Biol. 21:5050-5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tamemoto, H., T. Kadowaki, K. Tobe, T. Yagi, H. Sakura, T. Hayakawa, Y. Terauchi, K. Ueki, Y. Kaburagi, S. Satoh, H. Sekihara, S. Yoshioka, H. Horikoshi, Y. Furuta, Y. Ikawa, M. Kasuga, Y. Yazaki, and S. Aizawa. 1994. Insulin resistance and growth retardation in mice lacking insulin receptor substrate-1. Nature 372:182-186. [DOI] [PubMed] [Google Scholar]
- 74.Tanti, J. F., T. Gremeaux, E. Van Obberghen, and Y. Le Marchand-Brustel. 1994. Insulin receptor substrate 1 is phosphorylated by the serine kinase activity of phosphatidylinositol 3-kinase. Biochem. J. 304:17-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vanhaesebroeck, B., K. Higashi, C. Raven, M. Welham, S. Anderson, P. Brennan, S. G. Ward, and M. D. Waterfield. 1999. Autophosphorylation of p110delta phosphoinositide 3-kinase: a new paradigm for the regulation of lipid kinases in vitro and in vivo. EMBO J. 18:1292-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Verma, R., R. S. Annan, M. J. Huddleston, S. A. Carr, G. Reynard, and R. J. Deshaies. 1997. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 278:455-460. [DOI] [PubMed] [Google Scholar]
- 77.Virkamaki, A., K. Ueki, and C. R. Kahn. 1999. Protein-protein interaction in insulin signaling and the molecular mechanisms of insulin resistance. J. Clin. Investig. 103:931-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.White, M. F. 1996. The IRS-signalling system in insulin and cytokine action. Phil. Trans. R. Soc. Lond. Ser. B Biol. Sci. 351:181-189. [DOI] [PubMed] [Google Scholar]
- 79.White, M. F. 1997. The insulin signalling system and the IRS proteins. Diabetologia 40(Suppl. 2):S2-S17. [DOI] [PubMed] [Google Scholar]
- 80.White, M. F. 1998. The IRS-signaling system: a network of docking proteins that mediate insulin and cytokine action. Recent Prog. Hormone Res. 53:119-138. [PubMed] [Google Scholar]
- 81.White, M. F., J. N. Livingston, J. M. Backer, V. Lauris, T. J. Dull, A. Ullrich, and C. R. Kahn. 1988. Mutation of the insulin receptor at tyrosine 960 inhibits signal transmission but does not affect its tyrosine kinase activity. Cell 54:641-649. [DOI] [PubMed] [Google Scholar]
- 82.Wiese, R. J., C. C. Mastick, D. F. Lazar, and A. R. Saltiel. 1995. Activation of mitogen-activated protein kinase and phosphatidylinositol 3"-kinase is not sufficient for the hormonal stimulation of glucose uptake, lipogenesis, or glycogen synthesis in 3T3-L1 adipocytes. J. Biol. Chem. 270:3442-3446. [DOI] [PubMed] [Google Scholar]
- 83.Withers, D. J., D. J. Burks, H. H. Towery, S. L. Altamuro, C. L. Flint, and M. F. White. 1999. Irs-2 coordinates Igf-1 receptor-mediated beta-cell development and peripheral insulin signalling. Nat. Genet. 23:32-40. [DOI] [PubMed] [Google Scholar]
- 84.Withers, D. J., J. S. Gutierrez, H. Towery, D. J. Burks, J. M. Ren, S. Previs, Y. Zhang, D. Bernal, S. Pons, G. I. Shulman, S. Bonner-Weir, and M. F. White. 1998. Disruption of IRS-2 causes type 2 diabetes in mice. Nature 391:900-904. [DOI] [PubMed] [Google Scholar]
- 85.Zhang, H., H. Hoff, and C. Sell. 2000. Insulin-like growth factor 1-mediated degradation of insulin receptor substrate-1 is inhibited by epidermal growth factor (EGF) in prostate epithelial cells. J. Biol. Chem. 275:22558-22562. [DOI] [PubMed] [Google Scholar]
- 86.Zimmermann, S., and K. Moelling. 1999. Phosphorylation and regulation of Raf by Akt (protein kinase B). Science 286:1741-1744. [DOI] [PubMed] [Google Scholar]