Abstract
Functional human telomerase complexes are minimally composed of the human telomerase RNA (hTR) and a catalytic subunit (human telomerase reverse transcriptase [hTERT]) containing reverse transcriptase (RT)-like motifs. The N terminus of TERT proteins is unique to the telomerase family and has been implicated in catalysis, telomerase RNA binding, and telomerase multimerization, and conserved motifs have been identified by alignment of TERT sequences from multiple organisms. We studied hTERT proteins containing N-terminal deletions or substitutions to identify and characterize hTERT domains mediating telomerase catalytic activity, hTR binding, and hTERT multimerization. Using multiple sequence alignment, we identified two vertebrate-conserved TERT N-terminal regions containing vertebrate-specific residues that were required for human telomerase activity. We identified two RNA interaction domains, RID1 and RID2, the latter containing a vertebrate-specific RNA binding motif. Mutations in RID2 reduced the association of hTR with hTERT by 50 to 70%. Inactive mutants defective in RID2-mediated hTR binding failed to complement an inactive hTERT mutant containing an RT motif substitution to reconstitute activity. Our results suggest that functional hTERT complementation requires intact RID2 and RT domains on the same hTERT molecule and is dependent on hTR and the N terminus.
The telomerase enzyme is a ribonucleoprotein that extends the 3" ends of linear eukaryotic chromosomes. It is minimally composed of a protein catalytic subunit, telomerase reverse transcriptase (RT) (TERT; hTERT in humans) and a telomerase RNA (TR; hTR in humans) that contains a short template used for de novo synthesis of telomeric DNA repeats (reviewed in reference 9). Telomerase activity is associated with an increased capacity for cellular proliferation in immortal unicellular eukaryotes and in most immortalized human cancer cells (39). Identifying the mechanisms of telomerase assembly and catalytic function is essential for understanding the role of telomerase in immortalization.
The human telomerase holoenzyme is large (∼1,000 kDa) (43), and mammalian telomerase activity is associated with a number of proteins that may be implicated in ribonucleoprotein assembly, processing, and stability (19, 21, 28, 35, 38). Auxiliary proteins identified in yeast also mediate the access of telomerase to telomeres (15, 22). However, human telomerase that is affinity purified under stringent salt conditions has a molecular mass of 600 kDa, consistent with a minimal complex composed of two hTERTs and two hTRs (45). Recent studies indicate that hTERT proteins functionally (2, 6, 7) and physically (2) multimerize in vivo and in vitro. Functional telomerase multimerization refers to the functional complementation of two distinct, inactive hTERT mutants to reconstitute telomerase activity (6). Physical hTERT multimerization has been demonstrated by the coimmunoprecipitation of rabbit reticulocyte lysate (RRL)-synthesized hTERT proteins with glutathione S-transferase (GST)-hTERT (2). However, the association of hTERT proteins in immunoprecipitates may be indirect and could be mediated by other proteins or RNAs present in RRL. Though yeast and human telomerases contain more than one TR (41, 45), the stoichiometry of TERT molecules in telomerase complexes is unknown.
The TERT component of telomerase is limiting and is required for telomerase function in vivo and in vitro (8, 10, 14, 30, 44). TERT contains motifs (1 and 2 and A to E) that are common to all RTs (reviewed in reference 36). Mutations of most residues conserved between TERTs and members of the broader RT family abolish telomerase activity (for review, see reference 11). The TERT RT domain is essential for catalytic activity but is not sufficient for efficient binding of the Tetrahymena and hTR components (5, 7, 11, 27) nor for complementation of full-length inactive hTERT containing a substitution in the RT domain (6, 7). The N- and C-terminal TERT regions are not conserved among RT family members and therefore may mediate telomerase-specific functions. Most of the C terminus of Tetrahymena and human TERTs is required for in vitro catalytic activity, though this region is not essential for TR binding (5, 7, 27). The C terminus of EST2 (Saccharomyces cerevisiae TERT) influences telomerase processivity (40), and the hTERT C terminus is also implicated in functional multimerization with other hTERT molecules (6).
The TERT N terminus is larger and more highly conserved than the C terminus (40, 46). Most of the N terminus of Tetrahymena, S. cerevisiae, and human TERTs is required to reconstitute wild-type levels of telomerase catalytic activity (2, 5, 7, 11, 16, 27, 34, 44, 46). Portions of the Tetrahymena and human TERT N termini are essential for efficient binding of TR (5, 7, 27), and a newly identified ciliate-specific motif (CP2) in Tetrahymena TERT is one element that defines the enzyme's in vitro 5" RNA template boundary (34). The hTERT N terminus is also implicated in functional multimerization with other hTERT molecules (6, 7), and a recently identified DAT (dissociates activities of telomerase) domain is required for telomere length maintenance but not for in vitro catalytic activity (2).
Six major regions in the N terminus have been identified by sequence alignment of 10 TERT family members (Fig. 1):the nonconserved extreme N terminus (N), motif GQ (also identified as region I, motif T2, or motif N) (16, 31, 34, 46), motif CP (12), a poorly conserved putative linker region between motifs GQ and CP (46), motif QFP (also termed region III) (16, 46), and motif T (also termed region IV) (16, 29, 37). The CP motif, first identified in ciliates and corresponding to region II of EST2 (16), contains residues that are also conserved in nonciliate TERTs (46). An additional ciliate-specific motif, CP2, is located in the linker region of Tetrahymena TERT near the GQ motif boundary (34). The hTERT DAT domain is encoded by the first half of the GQ motif (2). The roles for most of the conserved regions in the TERT N terminus remain to be elucidated.
FIG. 1.
Map of hTERT N terminus, location of N-terminal mutations, and sequence alignment of the extreme N terminus and linker of vertebrate TERTs. (A) A schematic illustration of hTERT and the location of conserved N-terminal subregions previously identified by multiple sequence alignment (46). Alternative nomenclature for N-terminal subregions is described in the introduction. Mutations performed for this study are indicated by black boxes on the linear map of the hTERT N terminus. The DAT domain is a recently identified region (2). (B) Sequence alignment of the extreme N terminus (N) of the vertebrate TERTs demonstrating that this region is conserved among vertebrate TERTs. Alignment of human, M. musculus, M. auratus, and X. laevis TERT sequences (17, 18, 20, 23, 26, 32, 33, 37) was performed using the BLAST program. The symbol “+” indicates nonidentical conserved residues. Residues conserved in all vertebrate TERTs are underlined. (C) Sequence alignment of the vertebrate TERT linker indicating that the vertebrate linker contains conserved subregions, specifically at the C-terminal end. Alignment was performed as described above. Residues conserved in all vertebrate TERTs are underlined. The C-terminal end of the illustrated sequences is continuous with sequences of the CP motif as defined by Xia et al. (46). A highly conserved VSR motif identified in this study is shown in boldface.
The TERT N terminus has been implicated in a number of telomerase-specific functions. However, the domains and mechanisms mediating these functions have not been completely characterized. In this study recombinant human telomerases containing deletions and single amino acid substitutions were expressed in S. cerevisiae and in vitro transcription/translation (RRL) reactions to identify and characterize the regions of the hTERT N terminus involved in in vitro reconstitution of human telomerase activity, hTR binding, and multimerization with other hTERTs.
MATERIALS AND METHODS
Expression constructs.
The construction of the pET28b hTERT, pET28b hTERT D868N, and pET28a GST-hTERT RRL expression plasmids and of the pEGKT GST-hTERT and pEGKT GST-hTERT D868N yeast expression plasmids was previously described (4). pET28a GST-hTERT D868N was constructed by replacing a 1,435-bp fragment from XhoI-digested pET28a GST-hTERT by a similar fragment from XhoI-digested pET28b hTERT D868N. The construction of in vitro transcription and yeast expression plasmids for hTR was previously described (3, 4).
Site-directed mutagenesis, based on the Quik Change Site-Directed Mutagenesis kit from Stratagene, was used to generate pET28b hTERT constructs coding for N-terminal deletions and substitutions. Deletions and substitutions were confirmed by restriction enzyme digest and/or sequencing using a T7 DNA polymerase sequencing kit (USB Corp., Cleveland, Ohio). Yeast pEGKT GST-hTERT expression constructs bearing hTERT N-terminal deletions and substitutions were generated by PCR amplification of pET28b hTERT mutation constructs, using the 5" and 3" primers 5"-TGCTCTAGACCCGCGCGCTCCCCGC-3" and 5"-CCCAAGCTTTCAGTCCAGGATGGTCTTG-3", containing XbaI and HindIII restriction sites, respectively. XbaI-HindIII-digested PCR products were cloned into the pEGKT vector digested with the same enzymes.
Recombinant telomerase production.
The TnT T7-Coupled Reticulocyte Lysate System (Promega) was used to generate hTERT proteins from pET28b hTERT and pET28a GST-hTERT expression constructs, as per the manufacturer's instructions, using 0.8 μCi of [35S]methionine (Perkin-Elmer)/μl. hTR was synthesized from an FspI-linearized phTR+1 plasmid, as previously described (3). Purified hTR was included in the in vitro transcription/translation reactions at a concentration of 80 fmol/μl in the presence of 8 fmol of pET-hTERT plasmid DNA/μl.
Recombinant human telomerase was generated in GST-hTERT- and hTR-expressing yeast as previously described (4), except that yeast was grown in selective medium containing 1% raffinose prior to induction with 4% galactose. Cell pellets were lysed in 3 or 4 volumes of lysis buffer (0.25 mM deoxycholic acid, 10 mM Tris-HCl [pH 8], 1.2 mM MgCl2, 15% glycerol, 0.1 mM EDTA [pH 8], 0.1 mM EGTA [pH 8], 1.5 mM dithiothreitol [DTT], 5 mM β-mercaptoethanol, 1 μM pepstatin A, 1 μM leupeptin, 0.2 mM 4-[2-aminoacyl] benzene sulfonyl fluoride hydrochloride [AEBSF], and 38 U of RNAguard [Amersham Pharmacia]/ml), using a previously described glass bead lysis method (4). Expression of GST-hTERT proteins was confirmed by Western blot analysis using 0.3 μg of an affinity-purified hTERT antibody/ml (described below).
Immunoprecipitations.
Immunoprecipitations were performed with antibodies specific to GST (Amersham Pharmacia) or hTERT. In some experiments, bovine serum albumin and/or Escherichia coli tRNA was added to the immunoprecipitation buffer (10 mM Tris-HCl [pH 7.5], 1 mM EGTA, 1 mM MgCl2, 1% NP-40, 10% glycerol, 150 mM NaCl, 0.2 mM AEBSF, 1 μg of pepstatin/ml, 0.5 μg of leupeptin/ml, and 38 U of RNAguard [Amersham Pharmacia]/ml) as blocking agents (each at 100 ng/ml). Rabbit preimmune serum or GST or hTERT antibodies were prebound to preequilibrated protein A-Sepharose beads (Amersham Pharmacia) by incubation (from 2 h to overnight) at 4°C. RRLs and yeast lysates containing hTERT proteins were precleared with rabbit preimmune serum for 1 h, followed by 2 to 3 h of immunoprecipitation with immobilized GST or hTERT antibodies. Four washes were performed in immunoprecipitation buffer containing 150 mM (washes 1 and 4) or 300 mM (washes 2 and 3) NaCl. Beads suspended in immunoprecipitation buffer were used directly in subsequent assays to detect hTR, hTERT, GST-hTERT, and telomerase activity. hTERT antibody was used at 0.9 to 1.35 μg/ml in immunoprecipitations; efficiency was >25%.
Affinity-purified polyclonal hTERT antibody.
An hTERT peptide-directed polyclonal hTERT antibody was generated in rabbits and affinity-purified using the immunogenic peptide N-SEAEVRQHREARPALLTSRLRFIPKC-C. The peptide used to generate and purify the hTERT antibody was synthesized at the Sheldon Biotechnology Centre of McGill University, and the hTERT antibody was raised and purified by Strategic Biosystems, Ramona, Calif. This peptide is a variant of a peptide previously designed for hTERT antibody generation (20) and falls within motif 1 of the hTERT RT domain. The hTERT antibody detected recombinant GST-hTERT or hTERT expressed in yeast and RRL. The specificity of anti-hTERT was confirmed by Western blot analysis of yeast-expressed GST-hTERT immunoprecipitated by a GST antibody.
Telomerase activity assays and quantification of telomerase activity.
Telomerase activity was detected by a modified, two-step version of the telomeric repeat amplification protocol (TRAP) (3) with minor modifications. Five to 10% of immunoprecipitates, 15 μg of total protein from yeast crude lysates, or 0.5 to 1 μl of RRL was assayed for telomerase activity in a 20-μl final volume during the elongation step of the reaction. Five to 10 μl of this elongation reaction was amplified by PCR. Each 50-μl PCR contained 20 pmol each of TS, NT, and ACX primers and 10−19 moles of the TSNT primer. Amplification of TSNT by TS and NT primers generates an internal control. These primers have been described previously (24). The PCR mix was composed of 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, and 50 μM concentrations of the deoxynucleoside triphosphates, and each reaction contained 2 U of Taq DNA polymerase and 5 μCi of [32P]dGTP (800 Ci/mmol; Perkin-Elmer). PCRs were performed for 25 cycles of 95°C for 30 s, 50°C for 30s, and 72°C for 1 min 30 s. Telomerase activity was quantified from RRL samples expressing equal amounts of hTERT protein. The telomerase elongation product signal generated by individual hTERT mutants was normalized to the PCR internal control signal, and the obtained ratio was expressed as a fraction of the ratio calculated for the elongation products from wild-type telomerase that was always included with each experiment.
hTR binding assay and quantification of RNA binding.
hTERT N-terminal mutants were synthesized and [35S] labeled in RRL in the presence of [32P]-labeled hTR, using a modified version of a method previously developed to detect TR binding to Tetrahymena TERT (11). hTR was synthesized from the FspI-linearized phTR+1 plasmid in the presence of [32P]UTP (800 Ci/mmol). Linearized plasmid (2.5 μg) was combined with T7 RNA polymerase buffer (40 mM Tris-HCl [pH 7.9], 6 mM MgCl2, 2 mM spermidine, and 10 mM DTT), 250 U of T7 RNA polymerase (NEB), and 250 μCi of [32P]UTP (Perkin-Elmer) in a 50-μl reaction mixture containing a 0.5 mM concentration each of ATP, GTP, and CTP. Labeled hTR was diluted to a final concentration of 1 pmol/μl with unlabeled, in vitro-synthesized hTR. The specific activity of diluted, labeled hTR was 106 to 107 cpm/pmol, and 0.5 pmol of hTR was included in a 12.5-μl RRL reaction. hTERT-hTR complexes were immunoprecipitated using hTERT antibody. After washing, 100% of beads were resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (100 mM DTT, 1.67% SDS, 5% glycerol, 0.83% β-mercaptoethanol, and 58 mM Tris HCl [pH 6.8]), and immunoprecipitated hTERT and coprecipitated hTR were resolved on SDS-7.5% PAGE gels and were detected as previously described (11). Bands corresponding to hTR and hTERT were quantified using a Molecular Dynamics Densitometer and ImageQuant software. The amount of hTR in each lane was divided by the amount of protein in the same lane, and this ratio was expressed relative to the same ratio calculated for immunoprecipitated wild-type hTERT and coprecipitated hTR loaded on the same gel. As labeled hTR generated significant levels of nonspecific background in each lane, background signal was subtracted from the signal obtained for hTERT bands, to more accurately quantify the amounts of hTERT present in each lane.
Multimerization assays.
In all mixing experiments, GST-hTERT proteins expressed in yeast or RRL were mixed with hTERT proteins expressed in RRL and were then incubated on ice for 1 h. GST-hTERT/hTERT complexes were immunoprecipitated with a GST antibody (Amersham Pharmacia). Five to 10% of immunoprecipitates were examined for telomerase activity by TRAP, and the remainder was loaded on SDS-7.5% PAGE gels. Immunoprecipitated GST-hTERT proteins and coprecipitated hTERT proteins were detected by Western blot analysis using hTERT antibody and/or by visualization of [35S]-labeled proteins on fixed, dried SDS-PAGE gels exposed to Kodak XAR sensitive film.
For the mixing experiments in which yeast-expressed GST-hTERT mutants were mixed with RRL-synthesized wild type or D868N hTERT (see Fig. 4A and B), 90 μg of total yeast lysate proteins was mixed with 6 μl of RRL. For the mixing experiments in which yeast-expressed D868N GST-hTERT was mixed with hTERT mutants synthesized in RRLs (see Fig. 5A), two quantification steps were performed before mixing. First, 0.5-μl aliquots of RRL-synthesized [35S]-labeled hTERT mutants were subjected to electrophoresis on SDS-7.5% PAGE gels and were quantified relative to wild-type hTERT loaded on the same gel. Second, yeast-generated D868N GST-hTERT was quantified relative to RRL-synthesized wild-type hTERT using Western blot analysis. Yeast-expressed D868N GST-hTERT and RRL-synthesized hTERT were mixed in a 1:1 ratio (∼1 μl of RRL for every 20 μl of yeast lysate). Quantification of input proteins prior to mixing was performed similarly for RRL-based mixing experiments (see Fig. 5B and 6A). Approximately 5 to 10 μl each of RRL-synthesized GST-hTERT and hTERT was mixed in these experiments. Mixes were diluted to 20 μl with yeast lysis buffer prior to incubation on ice.
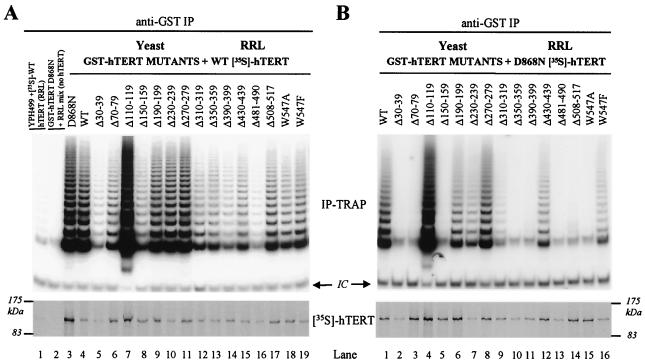
FIG. 4.
Association of GST-hTERT mutants with wild-type and D868N hTERT. Yeast whole-cell extracts were made from S. cerevisiae expressing GST-hTERT fusion proteins (D868N, wild type [WT], and mutants, as indicated) in the presence of hTR. These extracts were mixed with RRL containing [35S]-labeled hTERT proteins synthesized in the presence of hTR. hTERT/GST-hTERT complexes were immunoprecipitated (IP) using a GST antibody and were analyzed for telomerase activity by TRAP (top panels) and for coprecipitation of [35S]-labeled hTERT by SDS-PAGE (bottom panels). IC, internal control. (A) GST-hTERT fusion proteins associate with wild-type hTERT. Extracts prepared from yeast expressing GST-hTERT N-terminal mutants or from the parental YPH499 strain and containing equal amounts of total cellular proteins were mixed with RRL expressing wild-type hTERT. Control reactions were performed to detect nonspecific immunoprecipitation of wild-type hTERT in the absence of GST-hTERT (lane 1) and to determine whether RRL containing only hTR could complement GST-hTERT D868N to reconstitute telomerase activity (lane 2). (B) GST-hTERT fusion proteins physically associate with D868N hTERT, but inactive N-terminal mutants cannot functionally complement the inactive D868N mutant to reconstitute telomerase activity. Extracts prepared from yeast expressing GST-hTERT mutants were mixed with [35S]-labeled D868N hTERT synthesized in RRL and were analyzed as described above.
FIG. 5.
N-terminal mutants are not defective in physical multimerization. Equal amounts of GST-hTERT D868N and hTERT mutants synthesized separately in RRL or yeast in the presence of hTR were mixed and incubated on ice, and telomerase complexes were immunoprecipitated (IP) using a GST antibody. Immunoprecipitated hTERT/GST-hTERT complexes were examined for telomerase activity by TRAP (top panels) and for immunoprecipitation of hTERT and GST-hTERT proteins. WT, wild type. (A) N-terminal mutants interact equally with GST-hTERT D868N expressed in yeast. Equal amounts of yeast-expressed GST-hTERT D868N and RRL-expressed hTERT mutants were mixed and immunoprecipitated. Control reactions were performed to detect nonspecific immunoprecipitation of wild-type hTERT in the absence of GST-hTERT (lane 1) and in the absence of a GST antibody (lane 2). IC, internal control. (Middle panel) Immunoprecipitated GST-hTERT D868N was detected by Western blotting with anti-hTERT. (Bottom panel) Coimmunoprecipitated [35S]-labeled hTERT N-terminal mutants were detected by SDS-PAGE. (B) N-terminal mutants interact equally with GST-hTERT D868N expressed in RRL. Equal amounts of GST-hTERT D868N and hTERT mutants synthesized separately in RRL in the presence of hTR and [35S]methionine were mixed and immunoprecipitated. Control reactions (lanes 3 and 4) were performed as described for panel A. (Bottom panel) Immunoprecipitated GST-hTERT D868N and coprecipitated hTERT mutants were detected by SDS-PAGE. GST-hTERT D868N and wild-type hTERT present in crude RRL prior to immunoprecipitation are shown in lanes 1 and 2, respectively.
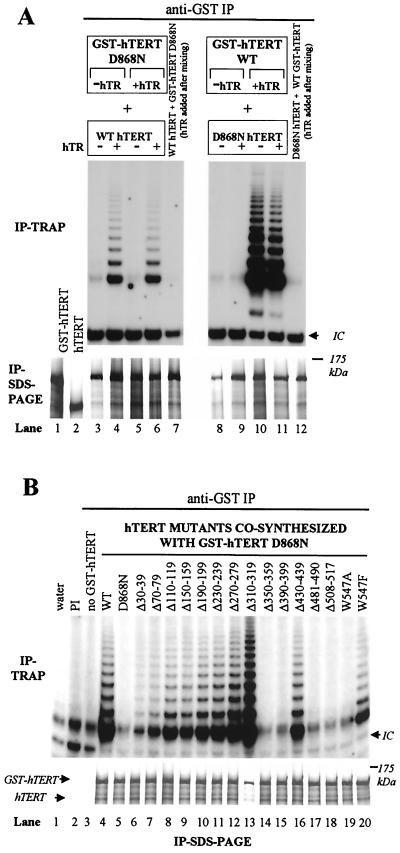
FIG. 6.
Functional but not physical multimerization of hTERT proteins is hTR dependent. (A) Functional and physical interactions of wild-type (WT) and D868N hTERT in the presence or absence of hTR. (Left panels) GST-hTERT D868N and wild-type hTERT proteins were independently synthesized in RRL in the absence or presence of hTR and [35S]methionine and were then mixed in different combinations, as indicated. (Right panels) Wild-type GST-hTERT and D868N hTERT proteins were independently synthesized in RRL in the absence or presence of hTR and were then mixed in different combinations, as indicated. Lanes 7 and 12 show reactions in which GST-hTERT and hTERT proteins were synthesized separately in the absence of hTR and were then mixed with hTR prior to immunoprecipitation (IP). Equal amounts of each protein were mixed, incubated on ice, and immunoprecipitated with a GST antibody. (Top panels) Telomerase activity of the coimmunoprecipitates was measured by TRAP assay. IC, internal control. (Bottom panels) Immunoprecipitated GST-hTERT fusion proteins and coprecipitated hTERT proteins were detected by SDS-PAGE. GST-hTERT D868N and wild-type hTERT present in crude RRL prior to immunoprecipitation are shown in lanes 1 and 2, respectively. (B) N-terminal mutants defective in RID2-mediated hTR interactions cannot functionally complement an inactive RT domain mutant to reconstitute telomerase activity. GST-hTERT D868N and hTERT N-terminal mutants were cosynthesized in RRL in the presence of hTR and [35S]methionine, and telomerase complexes were immunoprecipitated using a GST antibody. Control reactions (lanes 2 and 3) were performed as described for Fig. 5A. (Top panel) Telomerase activity was measured by TRAP assay. (Bottom panel) Immunoprecipitated GST-hTERT D868N and coprecipitated hTERT mutants were detected by SDS-PAGE.
In the experiments where D868N GST-hTERT was cosynthesized with hTERT mutants, equal quantities of plasmids were added to RRLs (20 ng/μl for each plasmid) and synthesis was carried out in the presence of hTR as described above. Immunoprecipitations were performed using 20 μl of each synthesis reaction.
RESULTS
We generated a series of recombinant telomerases containing single amino acid substitutions and 10 amino acid deletions (Fig. 1A) to characterize the roles of the hTERT N terminus in telomerase activity, hTR binding, and multimerization with other hTERT molecules. Deletions were regularly spaced at 40-amino-acid intervals, with the exception of Δ481-490 and Δ508-517, which were designed to overlap with previously characterized EST2 mutations (EST2 residues 303 to 312 and 330 to 339, respectively) (16). Nonconservative and conservative substitutions of the tryptophan at position 547 in the hTERT T motif to alanine (W547A) or phenylalanine (W547F), respectively, were also generated (Fig. 1A). The Tetrahymena TERT expresses a phenylalanine at this location, in contrast to all other TERTs, which contain a tryptophan residue. Deletion of hTERT residues 70 to 79, 150 to 159, 481 to 490, and 508 to 517 removed highly conserved amino acids previously identified by the alignment of TERT sequences from yeasts, ciliates, vertebrates, and plants (Table 1) (46).
TABLE 1.
Telomerase activity, hTERT/hTR association, and functional multimerization of hTERT N terminal mutants
| Mutation | Identity and conservation of deleted residuesa,b | Region of N terminusc | Telomerase activityd
|
hTERT/hTR associatione
|
RIDf | Physical multimerization with GST-hTERT D868Nf | Functional complementation of GST-hTERT D868Ng | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Avg | SD | n | Avg | SD | n | ||||||
| Wild type | 1.00 | 0 | 5 | 1.00 | 0 | 4 | + | + | |||
| D868N | 0 | 0 | 4 | 1.05 | 0.12 | 3 | + | − | |||
| Δ30-39 | RLGPQGWRLV | N | 0.10 | 0.02 | 4 | 0.81 | 0.10 | 3 | RID1 | + | +/− |
| Δ70-79 | SFRQVSCLKE | GQ motif | 0.38 | 0.18 | 5 | 0.87 | 0.13 | 3 | RID1 | + | + |
| Δ110-119 | GPPEAFTTSV | GQ motif (DAT domain) | 0.99 | 0.06 | 3 | 0.83 | 0.11 | 3 | RID1 | + | + |
| Δ150-159 | VHLLARCALF | GQ motif | 0 | 0 | 4 | 0.91 | 0.02 | 3 | RID1 | + | + |
| Δ190-199 | ASGPRRRLGC | GQ linker | 0.92 | 0.16 | 5 | 1.11 | 0.19 | 3 | + | + | |
| Δ230-239 | RSLPLPKRPR | Linker | 0.87 | 0.25 | 4 | 0.90 | 0.25 | 3 | + | + | |
| Δ270-279 | FCVVSPARPA | Linker | 1.16 | 0.18 | 5 | 1.01 | 0.26 | 3 | + | + | |
| Δ310-319 | TSRPPRPWDT | Linker | 0.70 | 0.41 | 4 | 1.07 | 0.07 | 2 | + | + | |
| Δ350-359 | LRPSLTGARR | Linker (VSR motif) | 0.01 | 0.01 | 3 | 0.55 | 0.08 | 2 | RID2 | + | − |
| Δ390-399 | RPLFLELLGN | Linker/CP motif | 0.04 | 0.05 | 4 | 0.52 | 0.11 | 3 | RID2 | + | − |
| Δ430-439 | KPQGSVAAPE | 1.14 | 0.18 | 4 | 0.95 | 0.03 | 3 | + | + | ||
| Δ481-490 | RHNERRFLRN | QFP motif | 0.04 | 0.04 | 3 | 0.41 | 0.1 | 4 | RID2 | + | − |
| Δ508-517 | LTWKMSVRDC | QFP motif | 0.15 | 0.18 | 3 | 0.27 | 0.07 | 4 | RID2 | + | − |
| W547A | T motif | 0.01 | 0.01 | 3 | 0.43 | 0.10 | 4 | RID2 | + | − | |
| W547F | T motif | 1.00 | 0.02 | 3 | 0.77 | 0.13 | 4 | RID2 | + | + | |
Bold face indicates 100% conservation among vertebrate TERTs (Homo sapiens, Mesocricetus auratus, Mus musculus, and Xenopus) (17, 18, 20, 23, 26, 32, 33, 37). Underlined bold face indicates ≥70% conservation among all TERTs (H. sapiens, M. auratus, M. musculus, Xenopus, Arabidopsis, Schizosaccharomyces pombe, Euplotes, Oxytricha, Tetrahymena, S. cerevisiae, and Candida albicans) (46). Underlined lightface indicates >70% conservation of a residue among all TERTs but indicates <100% conservation among vertebrate TERTs.
Alignments were performed with NCBI BLAST and by using the multiple sequence alignment published by Xia et al. (46). See Fig. 1 for sequence alignments of the extreme N-terminal (N) and linker regions of the vertebrate TERTs.
Motifs identified by multiple sequence alignment (46). Additional domains/motifs identified in hTERT are indicated in parentheses.
Telomerase activities of mutant telomerases reconstituted in RRL were expressed relative to the activity of wild-type enzyme.
hTR association with hTERT mutants generated in RRL was expressed relative to hTR association with wild-type hTERT.
The physical association of hTERT mutants with GST-hTERT D868N in vitro was evaluated by SDS-PAGE using (i) immunoprecipitated mixtures of yeast- or RRL-synthesized GST-hTERT D868N and RRL-synthesized hTERT mutants (Fig. 4C and 5A, respectively) and (ii) immunoprecipitated RRL cosynthesis reactions (Fig. 6).
The ability of hTERT mutants to functionally complement inactive GST-hTERT D868N cosynthesized in RRL was evaluated by TRAP assay. The symbol “+” indicates functional complementation to reconstitute telomerase activity. The symbol “−” indicates an inactive mutant that does not complement GST-hTERT D868N. “+/−” indicates weak complementation.
Alignment of TERT sequences from diverse organisms reveals substantial divergence in sequence composition and length in the linker and at the extreme N terminus (46). We aligned four vertebrate TERT sequences (20, 23, 33, 37) using the NCBI BLAST program (1) to determine if any portions of the extreme N-terminal and linker regions were conserved (Fig. 1B and C). The extreme N terminus (hTERT residues 1 to 57) was conserved among vertebrate TERTs (Fig. 1B). The N-terminal two-thirds of the linker were weakly conserved, but the C-terminal end of the linker contained two blocks of highly conserved amino acids (Fig. 1C). One of these conserved linker regions (R381-L396 in hTERT) was continuous with the previously identified CP motif (Fig. 1C). The second conserved block of amino acids in the linker (T355-L366 in hTERT) was flanked on either side by nonconserved residues (Fig. 1C) that are not required for in vitro telomerase activity (2). We named this second conserved linker region the VSR motif (for vertebrate-specific RNA binding motif; see below) (Fig. 1C). Deletion of hTERT residues 30 to 39, 350 to 359, and 390 to 399 removed vertebrate-specific amino acids in the extreme N terminus and linker (Table 1). Deletion of hTERT residues 190 to 199, 230 to 239, 270 to 279, 310 to 319, and 430 to 439 removed sequences that were not conserved among any of the TERTs (Table 1).
Telomerase activity of hTERT N-terminal mutants expressed in S. cerevisiae and in vitro transcription/translation systems.
hTERT N-terminal mutants were expressed in RRL or as GST fusion proteins in S. cerevisiae in the presence of TR (Fig. 2A or B,respectively). The catalytically inactive RT domain point mutant D868N (4) was also synthesized in both systems as a negative control for telomerase activity (Fig. 2A to C, lanes 3, 17, and 3, respectively, top panels). All of the mutant proteins were stably expressed in RRL (Fig. 2A, bottom panel) and were immunoprecipitated using an hTERT antibody (Fig. 2C, bottom panel). However, GST-hTERT mutant protein expression levels varied in yeast. Specifically, GST-hTERTs containing mutations Δ30-39, Δ70-79, Δ481-490, and Δ508-517 were poorly expressed (Fig. 2B, lanes 3, 4, 13, and 14, bottom panel). Three of these mutations partially or fully overlap with mutations associated with EST2 protein instability (16).
FIG. 2.
Reconstitution of telomerase activity in S. cerevisiae and in vitro transcription/translation systems. WT, wild type. (A) Telomerase activity of hTERT N-terminal mutants expressed in RRL. (Top panel) Reconstituted telomerase activity of different hTERT mutants (Fig. 1) was detected using the PCR-based TRAP assay. A PCR amplification control is indicated (IC). Control reactions were performed using RRL containing only hTR (lane 1), no lysate (lane 19; water), or partially purified human 293 cell extracts (lane 20). D868N is a mutation in the RT domain of hTERT that abolishes telomerase activity. (Bottom panel) Expression of hTERT mutants synthesized in RRL in the presence of [35S]methionine was detected by SDS-PAGE. (B) Telomerase activity of GST-hTERT N-terminal mutants expressed in S. cerevisiae. (Top panel) TRAP assays were performed using different GST-hTERT mutants, as indicated. A control reaction was performed using an extract of the parental yeast strain YPH499 not expressing GST-hTERT (lane 1). (Bottom panel) GST-hTERT fusion proteins expressed in S. cerevisiae were detected by Western blotting using a polyclonal hTERT antibody. (C) Immunoprecipitation (IP) of human telomerase from RRLs expressing hTERT N-terminal mutants. Anti-hTERT immunoprecipitates from RRLs expressing different hTERT mutants were analyzed for telomerase activity using TRAP (top panel) and for immunoprecipitation of [35S]-labeled hTERT by SDS-PAGE (bottom panel).
The catalytic activities of reconstituted telomerases were assayed by the TRAP technique, using RRL-expressed enzyme (Fig. 2A, top panel), yeast protein extracts (Fig. 2B, top panel), or immunoprecipitated telomerase from RRL (Fig. 2C, top panel). The relative levels of telomerase activity for mutant enzymes reconstituted in RRL are shown in Fig. 2A. The Δ30-39 mutant, which reconstituted low levels of telomerase activity in crude RRL (Fig. 2A, lane 4), was nearly inactive when expressed in yeast (Fig. 2B, lane 3) or following immunoprecipitation from RRL (Fig. 2C, lane 4). The Δ70-79 mutant, which was active in crude RRL and following immunoprecipitation from RRL (Fig. 2A and C, lane 5), was inactive in yeast (Fig. 2B, lane 4). Overall, the relative telomerase activities of the panel of mutants in yeast and crude or immunoprecipitated RRL were similar (compare Fig. 2A, B, and C). However, the catalytic activity of wild-type and mutant telomerases reconstituted in yeast was generally weaker than the activity observed in crude and immunoprecipitated RRL samples (compare Fig. 2B to 2A and C, respectively). One exception was the GST-hTERT mutant Δ110-119, which reproducibly reconstituted more active telomerase than did wild-type GST-hTERT when expressed in yeast (Fig. 2B, lanes 5 and 2, respectively). This increased activity was not observed when the mutant enzyme was expressed in RRL (Fig. 2A, lane 6). As all hTERTs were expressed at equal levels in RRL, the reconstituted telomerase activities of mutants relative to the wild type were quantified from crude and immunoprecipitated RRL. The average relative telomerase activities of the mutants are indicated in Table 1.
Residues unique to vertebrate TERTs are required for human telomerase activity.
Deletions that removed residues highly conserved among all TERTs (Δ70-79, Δ150-159, Δ481-490, and Δ508-517) severely impaired reconstitution of human telomerase activity (<40% of reconstituted wild-type telomerase activity) (Table 1). The nonconservative W547A substitution abolished telomerase activity, whereas a conservative change to a residue naturally expressed in the Tetrahymena TERT (W547F) had no detectable effect on telomerase activity using the TRAP assay (Table 1). All deletions that removed vertebrate-specific conserved residues, except Δ110-119, which is located within the catalytically nonessential DAT domain of hTERT (2), resulted in inactive or weakly active recombinant enzymes (<10% of reconstituted wild-type telomerase activity) (Table 1, Δ30-39, Δ350-359, and Δ390-399). In contrast, deletion of nonconserved residues (Δ190-199, Δ230-239, Δ270-279, Δ310-319, and Δ430-439) had a negligible effect on telomerase activity (>70% of reconstituted wild-type activity) (Table 1). These results demonstrate a functional role for conserved N-terminal domains previously identified by sequence alignment of TERTs from multiple organisms (46), as all hTERT mutations that altered highly conserved residues impaired in vitro catalytic activity. These and previously published results also suggest that some regions of the TERT N terminus may be implicated in organism-, vertebrate-, or ciliate-specific in vitro catalytic function (34).
Vertebrate-specific conserved regions in hTERT are required for association with hTR.
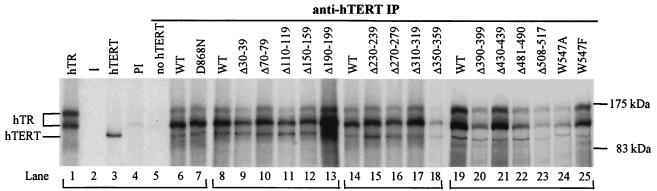
Sequence alignment identified vertebrate-conserved segments of TERT (Fig. 1). These regions may represent domains involved in protein-protein or protein-RNA interactions. Vertebrate TRs share a common structure, including three conserved domains (CR4-CR5, CR7, and Box H/ACA domains) that are not present in the ciliate TRs (13) and which may associate with TERT sequences unique to vertebrates. The association of [32P]-labeled hTR with RRL-synthesized hTERT mutants was examined following immunoprecipitation of hTERT/hTR complexes using an hTERT polyclonal antibody. The hTERT antibody immunoprecipitated all hTERT mutants expressed in RRL (Fig. 2C). Representative results are shown in Fig. 3, and quantification of hTERT-hTR binding from multiple experiments is summarized in Table 1. The catalytically inactive D868N hTERT mutant bound hTR as efficiently as wild-type hTERT (Fig. 3, lanes 7 and 6, respectively; and Table 1). Deletions in the extreme hTERT N terminus and N-terminal two-thirds of the GQ motif, including the DAT domain, resulted in modest (10 to 20%) but reproducible reductions in hTR association with hTERT (Fig. 3 and Table 1, Δ30-39, Δ70-79, Δ110-119, and Δ150-159). We defined the region between amino acids 30 and 159 as hTERT RNA interaction domain 1 (RID1). However, deletion of residues 230 to 239 also resulted in a 10% reduction in hTR association with hTERT, indicating that the RID1 domain might extend to residue 240. Deletion of portions of the hTERT linker that were only weakly conserved among the vertebrate TERTs did not affect hTR binding (>90% of wild-type hTERT-hTR binding) (Fig. 3 and Table 1, Δ190-199, Δ230-239, Δ270-279, and Δ310-319). Deletion of vertebrate-conserved residues in the linker VSR motif and at the CP motif/linker junction resulted in a 50% reduction in hTR binding (Fig. 3 and Table 1, Δ350-359 and Δ390-399, respectively). Mutations in highly conserved residues of the CP, QFP, and T motifs caused a 50 to 70% reduction in hTR association with hTERT (Fig. 3 and Table 1, Δ390-399, Δ481-490, Δ508-517, and W547A), indicating that these regions are likely involved in hTR binding. The conservative substitution W547F resulted in a modest 25% reduction in hTR/hTERT association (Fig. 3 and Table 1) and had no effect on telomerase activity (Table 1). We defined the hTR-interacting region containing the VSR, CP, QFP, and T motifs (residues 350 to 547) as hTERT RID2. However, hTERT containing a deletion in a poorly conserved region between the CP and QFP motifs was catalytically active and bound hTR as efficiently as did wild-type hTERT (Fig. 3 and Table 1, Δ430-439), indicating that RID2 is divided into two noncontiguous regions, defined by residues 350 to 399 and 481 to 547. Mutants demonstrating a 50 to 70% reduction in RID2-mediated hTR binding exhibited a fivefold-or-greater reduction in telomerase activity. Therefore, efficient RID2-mediated hTERT/hTR associations appear to be important for catalytic activity.
FIG. 3.
Association of hTR and hTERT N-terminal mutants. [35S]-labeled hTERT was synthesized in RRL in the presence of [32P]-labeled hTR. hTERT/hTR complexes were immunoprecipitated (IP) using an hTERT polyclonal antibody and were resolved on SDS-7.5% PAGE gels. hTR or hTERT present in crude RRL prior to immunoprecipitation is shown in lanes 1 and 3, respectively. Lane 2 is empty. Control reactions were performed to detect nonspecific immunoprecipitation of wild-type (WT) hTERT/hTR complexes by rabbit preimmune serum (lane 4) and to detect the nonspecific levels of hTR immunoprecipitated in the absence of hTERT (lane 5). Lanes 1 to 7, 8 to 13, 14 to 18, and 19 to 25 represent independent autoradiographs (indicated by brackets below the lane numbers). Levels of hTR coimmunoprecipitated with hTERT mutants are compared to the amounts of coimmunoprecipitated hTR and wild-type hTERT (lanes 6, 8, 14, and 19) loaded on the same gel.
GST-hTERT N-terminal mutants associate with wild-type hTERT.
Previous work suggests that human telomerase functions as a multimer (6, 45). The physical association of hTERT proteins in vitro was demonstrated recently by the coimmunoprecipitation of hTERT with GST-hTERT (2). In an effort to determine the role of the N terminus in multimerization, we investigated the interactions of GST-hTERT N-terminal mutants with wild-type hTERT. We mixed yeast lysates expressing GST-hTERT mutants and hTR with [35S]-labeled wild-type hTERT synthesized in RRL in the presence of hTR. If an interaction occurred between GST-hTERT mutants and wild-type hTERT, immunoprecipitation of inactive GST-hTERT mutants would coprecipitate wild-type hTERT and telomerase activity. Immunoprecipitation of GST protein alone does not coprecipitate hTERT (2; data not shown). Immunoprecipitation of both active and inactive GST-hTERT mutants using a GST antibody coprecipitated [35S]-labeled wild-type hTERT and telomerase activity (Fig. 4A;compare in Fig. 2B and 4A the following: D868N, mutants with deleted residues 30 to 39, 70 to 79, 150 to 159, 350 to 359, 390 to 399, 481 to 490, and 508 to 517; and W547A). Therefore, certain N-terminal regions of hTERT deleted in this study may not be essential for the association of hTERT proteins. Some inactive N-terminal mutants appeared to coprecipitate wild-type hTERT and telomerase activity less efficiently than others (Fig. 4A, lanes 5 and 16). However, these proteins were poorly expressed in yeast compared to most GST-hTERT mutants (Fig. 2B, lanes 3 and 13).
Similar mixing experiments were performed using D868N hTERT (Fig. 4B) to determine if inactive GST-hTERT mutants could functionally complement this catalytically inactive RT domain mutant to reconstitute telomerase activity. Specifically, we mixed yeast lysates expressing GST-hTERT mutants and hTR with [35S]-labeled D868N hTERT synthesized in RRL in the presence of hTR. If functional complementation between two inactive proteins occurred, immunoprecipitation of an inactive GST-hTERT N-terminal mutant would coprecipitate D868N hTERT and telomerase activity. Though all GST-hTERT mutants coprecipitated [35S]-labeled D868N hTERT, inactive GST-hTERT mutants did not functionally complement D868N hTERT to reconstitute telomerase activity (compare Fig. 2B and 4B). Therefore, we concluded that the physical association of hTERT proteins in coimmunoprecipitations is not sufficient for functional complementation of two distinct inactive proteins.
N-terminal mutations do not prevent physical association of hTERT proteins.
Certain GST-hTERT mutants coimmunoprecipitated reduced amounts of [35S]-labeled hTERT and low levels of telomerase activity (Δ30-39 and Δ481-490; Fig. 4A), suggesting that some hTERT N-terminal regions may be implicated in the physical association of hTERT proteins (Fig. 4A). However, some GST-hTERT mutants (including Δ30-39 and Δ481-490) were poorly expressed in yeast (Fig. 2B). Reduced expression levels of certain mutants would affect the amounts of GST-hTERT mutant proteins immunoprecipitated from yeast extracts, resulting in the coprecipitation of less [35S]-labeled hTERT. Thus, experiments similar to those described for Fig. 4 were performed by mixing equal amounts of D868N GST-hTERT expressed in yeast with equal quantities of RRL-synthesized [35S]-labeled hTERT mutants to determine if certain hTERT N-terminal regions were required for efficient physical multimerization. hTR was present in both RRLs and yeast lysates. If an interaction occurred between D868N GST-hTERT and [35S]-labeled hTERT mutants, immunoprecipitation of D868N GST-hTERT would coprecipitate [35S]-labeled hTERT mutant proteins. When equal amounts of GST-hTERT were immunoprecipitated (Fig. 5A,middle panel), similar levels of coprecipitated mutant proteins were detected (Fig. 5A, bottom panel) and inactive N-terminal mutants did not functionally complement the D868N GST-hTERT mutant to reconstitute activity (Fig. 5A, top panel).
Similar experiments were performed using GST-hTERT D868N and N-terminal mutants synthesized in RRL (Fig. 5B) to eliminate the possible effect of yeast lysate components on interactions between hTERT proteins. Again, the levels of coprecipitated hTERT N-terminal mutants were not grossly different (Fig. 5B), and inactive mutants did not complement GST-hTERT D868N to reconstitute telomerase activity. Therefore, we concluded that these N-terminal mutations did not prevent the physical association of hTERT proteins in coimmunoprecipitates.
Physical association of hTERT molecules is hTR independent.
All preceding mixing experiments were performed with hTERT and GST-hTERT proteins synthesized in the presence of hTR and therefore did not address whether hTR is required for the physical and functional association of hTERT proteins. We designed an experiment to determine if physical and functional interactions between hTERT proteins were dependent on the presence of hTR. GST-hTERT, GST-hTERT D868N, hTERT, and D868N hTERT were synthesized separately in RRL in the presence or absence of hTR. GST-hTERT and hTERT proteins were mixed in different combinations, and GST-hTERT/hTERT complexes were immunoprecipitated with a GST antibody (Fig. 6A).Only GST-hTERT/hTERT complexes containing a wild-type hTERT or GST-hTERT synthesized in the presence of hTR were active following immunoprecipitation (Fig. 6A, compare lanes 4, 6, 10, and 11 to lanes 5, 7, 9, and 12). The addition of hTR after GST-hTERT and hTERT proteins were synthesized and mixed did not reconstitute telomerase activity (Fig. 6A, lanes 7 and 12). However, coimmunoprecipitation of hTERT proteins did not require hTR (Fig. 6A, SDS-PAGE, lanes 3 and 8). The independent synthesis of both GST-hTERT and hTERT proteins in the presence of hTR (Fig. 6A, lanes 6 and 11) or the addition of hTR to protein mixtures (Fig. 6A, compare lanes 7 and 12 to lanes 3 and 8) did not increase the efficiency of hTERT coimmunoprecipitation. GST-hTERT proteins seemed to be immunoprecipitated less efficiently in the absence of hTR, and the amounts of coprecipitated hTERT decreased correspondingly (Fig. 6A, lanes 3 and 8). In conclusion, physical association of hTERT proteins was hTR independent, but coimmunoprecipitation of active enzyme required the synthesis of wild-type hTERT molecules in the presence of hTR. These results suggest that the physical association of mixed hTERT proteins in coimmunoprecipitates is not sufficient for functional interactions and that hTR molecules bound to inactive RT mutants cannot be used by another, wild-type hTERT molecule.
N-terminal mutants defective in RID2-mediated hTERT-hTR interactions cannot functionally complement an inactive RT domain mutant.
We found that functional interactions between wild-type and D868N hTERTs required the synthesis of wild-type hTERT proteins in the presence of hTR and that the addition of hTR to hTERT after protein synthesis could not reconstitute telomerase activity (Fig. 6A). Thus, we reasoned that cosynthesis of GST-hTERT and hTERT proteins in the presence of hTR might facilitate the functional complementation between some inactive N-terminal mutants and the inactive GST-hTERT D868N to reconstitute telomerase activity. GST-hTERT D868N and N-terminal mutants were cosynthesized in RRL in the presence of hTR, and immunoprecipitated telomerase complexes were examined for telomerase activity and the coprecipitation of N-terminal mutants (Fig. 6B). All N-terminal mutant proteins, including those defective in RID2-mediated hTR binding, were coimmunoprecipitated equally (Fig. 6B, lanes 6 to 20), supporting our previous conclusions that the physical association of hTERT molecules does not depend on hTR and that the N-terminal mutants that we examined were not defective in physical multimerization. Coexpression of weakly active N-terminal mutants (Δ30-39 and Δ70-79) with GST-hTERT D868N reconstituted increased telomerase activity compared to the activity reconstituted by the Δ30-39 or Δ70-79 mutant alone (compare Fig. 5 to Fig. 6B). The inactive mutant Δ150-159 functionally complemented GST-hTERT D868N to reconstitute telomerase activity (Fig. 6B, lane 9). However, inactive N-terminal mutants Δ350-359, Δ390-399, Δ481-490, Δ508-517, and W547A did not functionally complement GST-hTERT D868N to reconstitute telomerase activity (Fig. 6B). These N-terminal mutants were defective in RID2-mediated hTR binding (Table 1 and Fig. 3). Therefore, we concluded that functional complementation of hTERT molecules was dependent on RID2-mediated hTR interactions and required the presence of intact RT and RID2 domains on the same molecule.
DISCUSSION
Conserved regions of the hTERT N terminus are required for in vitro catalytic activity.
We expressed recombinant human telomerases containing N-terminal mutations in RRL and S. cerevisiae. Our results demonstrate a catalytic role for conserved N-terminal domains previously identified by sequence alignment of TERTs from multiple organisms (46), as all hTERT mutations that altered highly conserved residues impaired in vitro catalytic activity (Table 1; Fig. 7).Mutation of some of these residues in Tetrahymena and yeast TERTs has also been shown to affect telomerase activity (16, 34). Conversely, nonconserved residues were dispensable for in vitro catalytic activity. In addition, alignment of multiple vertebrate TERT sequences revealed vertebrate-specific conserved residues in the extreme N terminus and linker that were essential for human telomerase activity. Though the extreme N terminus and linker are not conserved among all TERTs, they are also implicated in yeast and Tetrahymena telomerase catalytic function (34, 46).
FIG. 7.
Schematic summary of functional domains of the hTERT N terminus. Conserved N-terminal subregions previously identified by alignment of multiple TERT sequences (46) are depicted on a linear map of the hTERT N terminus. Also shown is the recently identified DAT domain (2). In vitro telomerase activity data were derived from this study and reference 2. Detailed mapping of hTERT N-terminal regions implicated in telomerase catalytic activity can be found in reference 2. Data identifying hTR interaction domains (RID1 and RID2) and regions required for trans complementation of the catalytically inactive RT domain mutant GST-hTERT D868N were derived from this study.
hTERT N-terminal RID1.
Mutations in the extreme N terminus and part of the hTERT GQ motif (residues 30 to 159) resulted in modest reductions in association of hTR with hTERT (Table 1 and Fig. 7). N-terminal truncations that delete this region of Tetrahymena and human TERTs appear to cause small reductions in TR binding (7, 27). The corresponding sequences of EST2 are implicated in nonspecific binding of single-stranded nucleic acids (46) and in TR binding (16). Additionally, secondary structure predictions show that the hTERT extreme N terminus and GQ motif are likely to form a continuous, highly structured domain (2); parts of this domain are required for in vivo telomere length maintenance in yeast and human cells (2, 46). Though involved in hTR interactions, our results suggested that RID1 is not the major hTR binding domain of hTERT.
hTERT N-terminal RID2.
In agreement with previous results implicating the CP, QFP, and T motifs of yeast and Tetrahymena TERTs in TR binding (11, 16), mutations in these motifs of hTERT caused severe defects in the association of hTR. In addition, deleting part of the VSR motif in the hTERT linker compromised hTR binding. RID2 (residues 350 to 547) overlaps with the hTERT RNA binding region previously defined by the analysis of hTERT N-terminal truncations (5, 7, 27). Our data further indicated that RID2 contained at least two distinct RNA interaction regions separated by nonconserved sequences between the CP and QFP motifs that are not essential for RNA binding or catalytic activity (Table 1 and Fig. 7). Our results suggested that RID2 is the major hTR binding domain in the hTERT N terminus.
VSR motif.
The VSR motif of hTERT (residues 355 to 366) may be functionally analogous to the ciliate-specific CP2 motif characterized in Tetrahymena TERT (34). Both the VSR and CP2 motifs are required for in vitro catalytic activity and interactions with TR (27, 34). However, the CP2 and VSR motifs are unrelated in sequence and are situated at opposite ends of their respective TERT linkers. The CP2 motif is located near the GQ motif boundary of the Tetrahymena TERT linker. The hTERT linker may contain an additional RNA binding motif near the GQ motif boundary. However, sequence alignments demonstrated that the first half of the linker (near the GQ motif boundary) is poorly conserved among vertebrate TERTs, and our data and others' indicate that this region is not required for human telomerase activity (2) or RNA binding. We hypothesize that differences in the VSR and CP2 motif sequences and in their location with respect to more conserved elements of the RNA binding domain (RID2 in hTERT) may reflect the specific interaction requirements of TRs with different secondary structures. The observation that ciliate- and vertebrate-specific conserved TERT elements are required for interactions with TRs warrants further comparative mapping of the TERT-TR interaction domains from different organisms.
Physical multimerization of hTERT proteins is hTR independent.
Recent studies indicate that (i) the human telomerase complex contains two functional hTR molecules, (ii) separately inactive hTERT fragments can functionally multimerize in vitro and in vivo, and (iii) hTERT molecules physically multimerize in vitro (2, 6, 45). The hTERT N terminus was previously proposed as one of the protein-protein interaction sites in telomerase multimers (6). Interactions between hTERT molecules may be direct or may be mediated by other members of the telomerase complex, such as hTR, or by other proteins, such as molecular chaperones. Though our data demonstrated that hTERT proteins synthesized in yeast and RRL can interact with one another, the physical association of these proteins appeared to be independent of the N terminus. Physical multimerization of hTERT proteins occurred in the absence of hTR, in agreement with the recently published results of Armbruster and colleagues (2), and appeared unaffected by mutations that caused defects in hTR association. Therefore, our data suggest that the physical association of hTERT proteins is not mediated by hTR and may not be mediated by the N terminus.
Functional complementation requires intact RNA binding and RT domains on the same hTERT molecule.
Though hTR and hTR binding were not required for the physical association of hTERT molecules in coimmunoprecipitates, functional hTERT interactions were hTR dependent. Two types of experiments were performed to study the multimerization requirements of human telomerase. First, hTERT proteins were synthesized separately in the presence or absence of hTR and were subsequently mixed. Active enzyme was immunoprecipitated by the inactive GST-hTERT D868N only when hTR was present during the synthesis of wild-type hTERT, suggesting that wild-type hTERT could not functionally interact with hTR bound to inactive GST-hTERT D868N. Our results support the recent observation of Beattie and colleagues that functional complementation requires the association of hTR with only one hTERT subunit (6). Second, hTERT proteins were cosynthesized with GST-hTERT D868N in the presence of the hTR to facilitate the assembly of functional telomerase multimers. Functional complementation that reconstituted telomerase activity occurred between the inactive RT domain mutant and all N-terminal mutants except those with defects in RID2-mediated RNA association (Table 1 and Fig. 7). The results from both types of experiments indicated that functional hTERT multimerization required the presence of intact RID2 and RT domains on the same hTERT molecule.
Role of hTERT N terminus in functional telomerase multimerization.
The human immunodeficiency virus type 1 RT dimerizes in an asymmetric, head-to-tail fashion and contains only one functional catalytic site (25, 42). Beattie and colleagues have proposed an analogous model of hTERT multimerization in which the N terminus of one hTERT molecule acts in trans to allosterically mediate conformational changes in hTR(s) bound to another hTERT containing the RT domain and C terminus (6). Our data indicate that a functional RID2 hTR interaction domain (residues 350 to 547) is also normally required on the same molecule as the RT domain and C terminus. Thus, potential trans-activating portions of the N terminus may be located within the first 350 amino acids of hTERT, a region containing RID1 and the poorly conserved linker. This hypothesis is supported by recent results which demonstrate that the first 300 to 350 amino acids of hTERT can complement a nonoverlapping hTERT protein containing intact RNA binding, RT, and C-terminal domains to reconstitute telomerase activity (6). In addition, our data and others' suggest that the extreme N terminus and GQ motif of TERTs may constitute an independent domain (RID1) that interacts with TR and possibly telomeric DNA (2, 7, 16, 27, 46). These observations suggest a revised model of functional hTERT multimerization in which the extreme N terminus and GQ motif (RID1) modulate the conformation of TRs, perhaps in trans, whereas RID2 mediates RNA binding interactions that functionally link it with the RT and C-terminal domains.
The hTERT N terminus may also be involved in intramolecular N-terminal cis interactions. hTERT truncations lacking part of RID2 and all of the extreme N terminus, GQ motif and linker (6), or RID2 mutants (this study) do not functionally complement an RT-defective mutant. However, hTERT truncations lacking the entire N terminus, including RID2, can complement hTERT mutants defective in the RT domain or C terminus (6). Though it is not clear why complementation is affected differently by partial and complete RID2 disruptions, these latter observations indicate that hTERT RID2 may act in trans with the RT domain and C terminus. This in turn suggests that independent domains of the hTERT N terminus such as RID1 and RID2 may functionally interact in cis. Such an interaction could be mediated by the long, poorly conserved, and catalytically nonessential linker of hTERT.
N terminus-mediated trans interactions between hTERT proteins may occur via direct homomeric protein contacts or may be mediated by hTR, telomeric DNA, or other intervening proteins. None of the hTERT N-terminal mutants tested in our study were defective in physical association with other hTERT proteins. However, these mutations may be too small to disrupt protein associations. Interactions between hTERT proteins also occurred independently of hTR binding. We did not determine whether hTERT interactions might be altered in the presence of telomeric DNA. However, recent work by Armbruster and colleagues indicates that a telomeric oligonucleotide substrate is not essential for the physical association of hTERT proteins (2). These observations can be interpreted in a number of ways. First, the N terminus may not interact with other hTERT molecules in trans. However, some inactive N-terminal mutants functionally complement RT domain mutants containing intact N termini, supporting functional trans interactions 6; this study). Second, physical trans interactions may occur along an extensive surface of the hTERT N terminus and may be identified only by the analysis of larger deletions or substitutions. Third, the primary physical site of hTERT trans interactions may be situated outside the N terminus. Mutations disrupting large subregions of hTERT, especially in the RT and C-terminal domains, will be required to identify and characterize the site(s) and mechanism mediating the physical and functional association of hTERT molecules.
We identified an RID in the hTERT N terminus that was required for the functional association of hTERT proteins, suggesting a role for hTERT-hTR interactions and the N terminus in telomerase multimerization. The potential roles of the RT and C-terminal domains, associated proteins, and telomeric DNA in mediating the molecular mechanism of telomerase multimerization will require further investigation. A better understanding of the mechanisms regulating hTERT multimerization will help to elucidate the role of multimerization in the catalytic function of human telomerase.
Acknowledgments
We thank G. Kukolj, J. Demers, and F. Bachand for critical reading of the manuscript and E. Petroulakis for helpful suggestions related to immunoprecipitations. We thank L. Harrington and C. Counter for communicating data prior to publication.
This work was supported by grant MOP-14026 from the Canadian Institutes of Health Research to C.A.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armbruster, B., S. Banik, C. Guo, A. Smith, and C. Counter. 2001. N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol. Cell. Biol. 21:7775-7786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Autexier, C., R. Pruzan, W. D. Funk, and C. W. Greider. 1996. Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J. 15:5928-5935. [PMC free article] [PubMed] [Google Scholar]
- 4.Bachand, F., and C. Autexier. 1999. Functional reconstitution of human telomerase expressed in Saccharomyces cerevisiae. J. Biol. Chem. 274:38027-38031. [DOI] [PubMed] [Google Scholar]
- 5.Bachand, F., and C. Autexier. 2001. Functional regions of human telomerase reverse transcriptase and human telomerase RNA required for telomerase activity and RNA-protein interactions. Mol. Cell. Biol. 21:1888-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beattie, T., W. Zhou, M. Robinson, and L. Harrington. 2001. Functional multimerization of the human telomerase reverse transcriptase. Mol. Cell. Biol. 21:6151-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beattie, T., W. Zhou, M. Robinson, and L. Harrington. 2000. Polymerization defects within human telomerase are distinct from telomerase RNA and TEP1 binding. Mol. Biol. Cell 11:3329-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beattie, T. L., W. Zhou, M. O. Robinson, and L. Harrington. 1998. Reconstitution of human telomerase activity in vitro. Curr. Biol. 8:177-180. [DOI] [PubMed] [Google Scholar]
- 9.Blackburn, E. 1999. Telomerase, the RNA world, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 10.Bodnar, A. G., M. Ouellette, M. Frolkis, S. E. Holt, C.-P. Chiu, G. B. Morin, C. B. Harley, J. W. Shay, S. Lichtsteiner, and W. E. Wright. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279:349-352. [DOI] [PubMed] [Google Scholar]
- 11.Bryan, T., K. Goodrich, and T. Cech. 2000. Telomerase RNA bound by protein motifs specific to telomerase reverse transcriptase. Mol. Cell 6:493-499. [DOI] [PubMed] [Google Scholar]
- 12.Bryan, T. M., J. M. Sperger, K. B. Chapman, and T. R. Cech. 1998. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc. Natl. Acad. Sci. USA 95:8479-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, J.-L., M. A. Blasco, and C. W. Greider. 2000. Secondary structure of vertebrate telomerase RNA. Cell 100:503-514. [DOI] [PubMed] [Google Scholar]
- 14.Counter, C. M., M. Meyerson, E. N. Eaton, and R. A. Weinberg. 1997. The catalytic subunit of yeast telomerase. Proc. Natl. Acad. Sci. USA 94:9202-9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans, S. K., and V. Lundblad. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286:117-120. [DOI] [PubMed] [Google Scholar]
- 16.Friedman, K. L., and T. R. Cech. 1999. Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev. 13:2863-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenberg, R. A., R. C. Allsopp, L. Chin, G. B. Morin, and R. A. DePinho. 1998. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene 16:1723-1730. [DOI] [PubMed] [Google Scholar]
- 18.Guo, W., M. Okamoto, N. Park, Y. Lee, and N. Park. 2001. Cloning and expression of hamster telomerase catalytic subunit cDNA. Int J. Mol. Med. 8:73-78. [DOI] [PubMed] [Google Scholar]
- 19.Harrington, L., T. McPhail, V. Mar, W. Zhou, R. Oulton, M. B. Bass, I. Arruda, and M. O. Robinson. 1997. A mammalian telomerase-associated protein. Science 275:973-977. [DOI] [PubMed] [Google Scholar]
- 20.Harrington, L., W. Zhou, T. McPhail, R. Oulton, D. S. K. Yeung, V. Mar, M. B. Bass, and M. O. Robinson. 1997. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 11:3109-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holt, S. E., D. L. Aisner, J. Baur, V. M. Tesmer, M. Dy, M. Ouellette, J. B. Trager, G. M. Morin, D. O. Toft, J. W. Shay, W. E. Wright, and M. A. White. 1999. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 13:817-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes, T., S. Evans, R. Weilbaecher, and V. Lundblad. 2000. The Est3 protein is a subunit of yeast telomerase. Curr. Biol. 10:809-812. [DOI] [PubMed] [Google Scholar]
- 23.Kilian, A., D. D. L. Bowtell, H. E. Abud, G. R. Hime, D. J. Venter, P. K. Keese, E. L. Duncan, R. R. Reddel, and R. A. Jefferson. 1997. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum. Mol. Genet. 6:2011-2019. [DOI] [PubMed] [Google Scholar]
- 24.Kim, N. W., and F. Wu. 1997. Advances in quantification and characterization of telomerase activity by the telomeric repeat amplification protocol (TRAP). Nucleic Acids Res. 25:2595-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohlstaedt, L. A., J. Wang, J. M. Friedman, P. A. Rice, and T. A. Steitz. 1992. Crystal structure at 3.5 A resolution of HIV reverse transcriptase complexed with an inhibitor. Science 256:1783-1790. [DOI] [PubMed] [Google Scholar]
- 26.Kuramoto, M., K. Ohsumi, T. Kishimoto, and F. Ishikawa. 2001. Identification and analyses of the Xenopus TERT gene that encodes the catalytic subunit of telomerase. Gene 277:101-110. [DOI] [PubMed] [Google Scholar]
- 27.Lai, C., J. Mitchell, and K. Collins. 2001. RNA binding domain of telomerase reverse transcriptase. Mol. Cell. Biol. 21:990-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le, S., R. Sternglanz, and C. W. Greider. 2000. Identification of two RNA-binding proteins associated with human telomerase RNA. Mol. Biol. Cell 11:999-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lingner, J., T. R. Hughes, A. Shevchenko, M. Mann, V. Lundblad, and T. R. Cech. 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276:561-567. [DOI] [PubMed] [Google Scholar]
- 30.Liu, Y., B. Snow, M. Hande, D. Yeung, N. Erdmann, A. Wakeham, A. Itie, D. Siderovski, P. Lansdorp, M. Robinson, and L. Harrington. 2000. The telomerase reverse transcriptase is limiting and necessary for telomerase function in vivo. Curr. Biol. 10:1459-1462. [DOI] [PubMed] [Google Scholar]
- 31.Malik, H., W. Burke, and T. Eickbush. 2000. Putative telomerase catalytic subunits from Giardia lamblia and Caenorhabditis elegans. Gene 251:101-108. [DOI] [PubMed] [Google Scholar]
- 32.Martin-Rivera, L., E. Herrera, J. P. Albar, and M. A. Blasco. 1998. Expression of mouse telomerase catalytic subunit in embryos and adult tissues. Proc. Natl. Acad. Sci. USA 95:10471-10476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyerson, M., C. M. Counter, E. N. Eaton, L. W. Ellisen, P. Steiner, S. C. Dickinson, L. Ziaugra, R. L. Beijersbergen, M. J. Davidoff, Q. Liu, S. Bacchetti, D. A. Haber, and R. A. Weinberg. 1997. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell 90:785-795. [DOI] [PubMed] [Google Scholar]
- 34.Miller, M., J. Liu, and K. Collins. 2000. Template definition by Tetrahymena telomerase reverse transcriptase. EMBO J. 19:4412-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitchell, J., E. Wood, and K. Collins. 1999. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402:551-555. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura, T., and T. Cech. 1998. Reversing time: origin of telomerase. Cell 92:587-590. [DOI] [PubMed] [Google Scholar]
- 37.Nakamura, T. M., G. B. Morin, K. B. Chapman, S. L. Weinrich, W. H. Andrews, J. Lingner, C. B. Harley, and T. R. Cech. 1997. Telomerase catalytic subunit homologs from fission yeast and human. Science 277:955-959. [DOI] [PubMed] [Google Scholar]
- 38.Nakayama, J., M. Saito, H. Nakamura, A. Matsuura, and F. Ishikawa. 1997. TLP1: a gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell 88:875-884. [DOI] [PubMed] [Google Scholar]
- 39.Oulton, R., and L. Harrington. 2000. Telomeres, telomerase, and cancer: life on the edge of genomic stability. Curr. Opin. Oncol. 12:74-81. [DOI] [PubMed] [Google Scholar]
- 40.Peng, Y., I. Mian, and N. Lue. 2001. Analysis of telomerase processivity: mechanistic similarity to HIV-1 reverse transcriptase and role in telomere maintenance. Mol. Cell 7:1201-1211. [DOI] [PubMed] [Google Scholar]
- 41.Prescott, J., and E. H. Blackburn. 1997. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev. 11:2790-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Restle, T., B. Muller, and R. S. Goody. 1990. Dimerization of human immunodeficiency virus type 1 reverse transcriptase. A target for chemotherapeutic intervention. J. Biol. Chem. 265:8986-8988. [PubMed] [Google Scholar]
- 43.Schnapp, G., H.-P. Rodi, W. J. Rettig, A. Schnapp, and K. Damm. 1998. One-step affinity purification protocol for human telomerase. Nucleic Acids Res. 26:3311-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinrich, S. L., R. Pruzan, L. Ma, M. Ouellette, V. M. Tesmer, S. E. Holt, A. G. Bodnar, S. Lichtsteiner, N. W. Kim, J. B. Trager, R. D. Taylor, R. Carlos, W. H. Andrews, W. E. Wright, J. W. Shay, C. B. Harley, and G. B. Morin. 1997. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 17:498-502. [DOI] [PubMed] [Google Scholar]
- 45.Wenz, C., B. Enenkel, M. Amacker, C. Kelleher, K. Damm, and J. Lingner. 2001. Human telomerase contains two cooperating telomerase RNA molecules. EMBO J. 20:3526-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia, J., Y. Peng, I. Mian, and N. Lue. 2000. Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol. Cell. Biol. 20:5196-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]