Abstract
TOR (target of rapamycin) is a phosphatidylinositol kinase-related protein kinase that controls cell growth in response to nutrients. Rapamycin is an immunosuppressive and anticancer drug that acts by inhibiting TOR. The modes of action of TOR and rapamycin are remarkably conserved from S. cerevisiae to humans. The current understanding of TOR and rapamycin is derived largely from studies with S. cerevisiae. In this review, we discuss the contributions made by S. cerevisiae to understanding rapamycin action and TOR function.
INTRODUCTION
The immunosuppressive and anticancer drug rapamycin acts by binding the highly conserved immunophilin FKBP12 (FK506-binding protein of 12 kDa, encoded by the FPR1 gene in Saccharomyces cerevisiae), and the FKBP12-rapamycin complex then binds and inhibits the kinase TOR (target of rapamycin, encoded by the homologous TOR1 and TOR2 genes in S. cerevisiae) (63, 64, 135) (Fig. 1). This mechanism of action is conserved from yeasts to humans. Indeed, a number of important discoveries that contributed to elucidating rapamycin's mode of action, including the discovery of TOR, were made in S. cerevisiae. Since its discovery, TOR has been widely investigated and has been recognized as a central controller of cell growth in eukaryotes (52, 130). S. cerevisiae also played a significant part in elucidating the cellular role of TOR and in defining TOR signaling pathways. In this review, we discuss the contributions made by S. cerevisiae to understanding rapamycin action and TOR function.
FIG. 1.
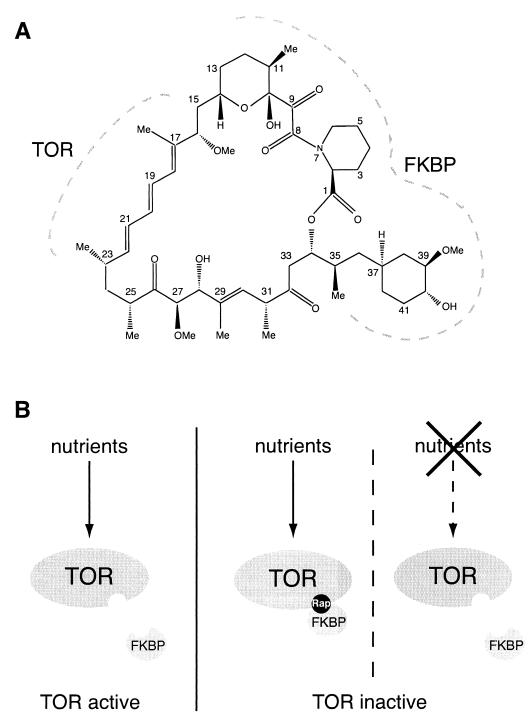
Rapamycin-FKBP complex binds and inhibits TOR. (A) Chemical structure of rapamycin. The TOR- and FKBP-interacting regions of rapamycin (30) are indicated by dashed lines. (B) TOR is active in the presence of nutrients and inactive upon nutrient limitation or FKBP-rapamycin (Rap) binding.
RAPAMYCIN ACTION
Rapamycin is a potent antibiotic produced by a strain of Streptomyces hygroscopicus isolated from a soil sample collected in Rapa-Nui (Easter Island) (156). The pharmaceutical potential of rapamycin was originally discovered in a screen for novel antifungal agents. Much later, rapamycin was found to exhibit immunosuppressive activity due to its capacity to block the growth and proliferation of T cells (136, 142). More recently, rapamycin has been found to display anticancer properties (57, 70, 73).
Rapamycin binds 12-kDa FK506-binding protein (FKBP12) with high affinity. FKBP12 was first identified in vitro as a receptor of FK506, an immunosuppressant structurally related to rapamycin, and was later shown to be a cytoplasmic peptidylprolyl rotamase (62, 135). The observations that S. cerevisiae mutants lacking FKBP12 are viable and resistant to rapamycin toxicity and that rapamycin analogs still bind and inhibit FKBP12 rotamase activity but do not immunosuppress indicated that FKBP12 is not the target through which rapamycin blocks growth (14, 63, 64, 90, 135, 164). Rather, an FKBP12-rapamycin complex is the toxic agent that then acts on another target to inhibit cell growth.
The findings that dominant mutations in either TOR1 (TOR1-1, Ser1972Arg) or TOR2 (TOR2-1, Ser1975Ile) confer complete resistance to the growth-inhibitory properties of rapamycin and that such mutations prevent the binding of FKBP12-rapamycin to TOR demonstrated that TOR is the relevant target through which rapamycin blocks cell growth (19, 63, 101, 145, 171). Finally, the observation that loss of TOR function in S. cerevisiae mimics rapamycin treatment indicated that FKBP12-rapamycin inhibits TOR function (92). More details are given below concerning the mode of action of rapamycin and the evolutionary conservation of this mode of action.
TOR IS CONSERVED IN LOWER AND HIGHER EUKARYOTES
After the original identification of TOR in the yeast Saccharomyces cerevisiae, TOR was identified in fungi, mammals, flies, worms, and plants, suggesting that TOR is conserved in all eukaryotic life forms. In the opportunistic fungal pathogen Cryptococcus neoformans, a TOR1 homologue was cloned by degenerate PCR, and a TOR2 homologue was detected by sequencing expressed sequence tags (36). Moreover, a C. neoformans FKBP12 homologue was cloned based on its ability to interact with the FKBP12-rapamycin binding domain in TOR1 (36). These findings revealed that the antifungal activity of rapamycin in C. neoformans is mediated via conserved complexes involving FKBP12 and TOR homologues. More recently, a TOR1 homologue has also been identified and characterized in the fungal pathogen Candida albicans (37), and TOR1 and TOR2 homologues have been identified and characterized in the fission yeast Schizosaccharomyces pombe (161, 162).
Unlike most lower eukaryotes, which contain two TOR genes, higher eukaryotes appear to possess only one TOR gene. The first TOR identified in a higher eukaryote was mammalian TOR (mTOR; also known as FRAP, RAFT, and RAPT). mTOR was discovered based on its ability to interact in vitro with the FKBP12-rapamycin complex (18, 127, 128) or by a two-hybrid screen (29). The subsequent demonstration that an mTOR variant constructed to contain a mutation (Ser2035Ile) analogous to the previously identified yeast TOR2-1 mutation (Ser1975Ile) confers rapamycin resistance in mammalian cells indicated that mTOR is the in vivo target of FKBP12-rapamycin and that rapamycin action is conserved from yeasts to mammals (19). More recently, the isolation of Drosophila melanogaster mutants and the sequencing of the Caenorhabditis elegans and Arabidopsis thaliana genomes allowed the cloning and characterization of TOR genes (dTOR, CeTOR, and AtTOR, respectively) from these organisms (106, 114, 169; J. Avruch and F. Mueller, personal communication for CeTOR).
The S. cerevisiae TOR1 and TOR2 genes encode two large (approximately 280 kDa) and highly homologous (67% identical) TOR1 and TOR2 proteins. TOR1 and TOR2, as suggested by their similarity, are functionally redundant. However, TOR2 has an additional function that TOR1 is unable to perform (see below). In S. pombe, the two TOR proteins Tor1 and Tor2 are 52% identical to each other and 42 to 48% identical to the S. cerevisiae TOR1 and TOR2 proteins (161). mTOR, dTOR, CeTOR, and AtTOR show approximately 38%, 37%, 28%, and 36% identity with the S. cerevisiae TOR proteins, respectively (114, 169) (Table 1).
TABLE 1.
TOR is evolutionarily conserveda
| TOR protein | % Identity
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| ScTOR1 | ScTOR2 | Sptor1 | Sptor2 | CnTOR1 | CeTOR | AtTOR | dTOR | mTOR | |
| ScTOR1 | — | 67 | 42 | 47 | 39 | 28 | 36 | 37 | 38 |
| ScTOR2 | — | 43 | 48 | 40 | 28 | 38 | 38 | 40 | |
| SpTOR1 | — | 52 | 42 | 28 | 38 | 40 | 42 | ||
| SpTOR2 | — | 44 | 29 | 42 | 42 | 44 | |||
| CnTOR1 | — | 26 | 35 | 38 | 39 | ||||
| CeTOR | — | 28 | 32 | 35 | |||||
| AtTOR | — | 38 | 40 | ||||||
| dTOR | — | 53 | |||||||
| mTOR | — | ||||||||
The table shows the identity (expressed as a percentage) between the TOR proteins so far identified in eukaryotes. ScTOR1, Saccharomyces cerevisiae TOR1; ScTOR2, Saccharomyces cerevisiae TOR2; SpTOR1, Schizosaccharomyces pombe TOR1; SpTOR2, Schizosaccharomyces pombe TOR2; CnTOR1, Cryptococcus neoformans TOR1; CeTOR, Caenorhabditis elegans TOR; AtTOR, Arabidopsis thaliana TOR; dTOR, Drosophila TOR; mTOR, mammalian TOR (human in this case).
DOMAIN STRUCTURE OF TOR
A feature common to all TOR proteins is a conserved C-terminal region with strong homology to the related catalytic domains of phosphatidylinositol 3-kinase (PI3K) and phosphatidylinositol 4-kinase (Fig. 2). The presence of this characteristic phosphatidylinositol kinase (PIK) homology domain has defined a TOR-related family of kinases termed the PIK-related kinases. In addition to TOR, this family includes the mammalian ATM (ataxia telangiectasia mutated), ATR (ataxia telangiectasia related), DNA-dependent protein kinase, the Drosophila mei-41, and the yeast MEC1, RAD53, and TEL1 proteins (for a recent review, see reference 2). All these proteins are involved in diverse cellular functions, such as control of cell growth, regulation of cell cycle progression, a DNA damage checkpoint, recombination, and maintenance of telomere length. Despite significant homology to lipid kinases, no lipid kinase activity has been demonstrated for any of the PIK-related kinases. However, yeast and mammalian TORs have been reported to exhibit Ser/Thr protein kinase activity (see below). Genetic studies in S. cerevisiae have revealed that the integrity of the TOR kinase domain is essential for TOR function (3, 67, 92, 171).
FIG. 2.
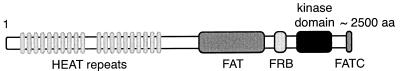
Structure of the TOR kinases. Functional domains conserved in TOR proteins are depicted, including the N-terminal HEAT repeats, the central FAT domain, and the C-terminal FKBP-rapamycin binding (FRB), kinase, and FATC domains. aa, amino acids.
N-terminal to the kinase domain, the TORs contain a potential regulatory region of approximately 100 amino acids, the FKBP12-rapamycin binding (FRB) domain (Fig. 2). The location of the FRB domain in TOR was initially identified in S. cerevisiae by TOR1 and TOR2 point mutations that prevent FKBP12-rapamycin binding and confer dominant rapamycin resistance (22, 67). The FRB domain, the minimal protein domain capable of binding FKBP12-rapamycin, was then defined by deletion analysis of the region in mTOR corresponding to the region in S. cerevisiae TOR containing the rapamycin resistance-conferring mutations (28). Analysis of the original rapamycin resistance-conferring TOR alleles revealed that a Ser residue (Ser1972 in TOR1 and Ser1975 in TOR2) is important for the interaction of TOR with the FKBP12-rapamycin complex (22, 67, 145). The yeast Ser1972/1975 is conserved in mTOR (18, 127). Subsequent genetic studies identified two additional residues (Trp2042 and Phe2049 in TOR2) also conserved in mTOR that play a critical role in FKBP12-rapamycin binding to the yeast TOR proteins (101).
The crystal structure of FKBP12-rapamycin bound to the FRB domain of mTOR has been determined (30). This structure revealed that FKBP12 and the FRB domain of mTOR interact primarily via rapamycin. Rapamycin simultaneously occupies a hydrophobic binding pocket in FKBP12 and a hydrophobic pocket in the FRB domain and thus “glues” FKBP12 and mTOR together (30). The residues that form the rapamycin-binding pocket of mTOR are conserved in the S. cerevisiae TOR1 and TOR2 proteins, and thus all three proteins are likely to contain a hydrophobic pocket with similar architecture (30). The protein-protein contacts between FKBP12-rapamycin and mTOR, although a minor contribution to the overall interaction between FKBP12-rapamycin and mTOR, may explain why rapamycin by itself cannot interact with TOR. The mechanism by which FKBP12-rapamycin inhibits TOR function is unknown. The FKBP12-rapamycin complex may inhibit TOR kinase activity directly or, for example, may block access to substrates or partner proteins (46, 117). mTOR kinase activity has been shown to be inhibited by the FKBP12-rapamycin complex in vitro (19, 20, 137), but this inhibition remains controversial (21, 117).
In addition to the catalytic and FRB domains, TOR proteins also contain up to 20 tandemly repeated HEAT motifs at their N termini (5) (Fig. 2). The term HEAT motif is derived from the four proteins where this domain was originally identified: huntingtin, elongation factor 3, the A subunit of type 2A protein phosphatase (PP2A), and TOR. Each HEAT repeat consists of an antiparallel α-helical motif of approximately 40 amino acids (56, 68) and is thought to mediate protein-protein interactions. Recently, it has been proposed that HEAT repeats anchor S. cerevisiae TOR2 to the plasma membrane, possibly by mediating an interaction with a membrane-associated protein (93). The N-terminal part of mTOR has been shown to interact with gephyrin, a protein involved in the postsynaptic clustering of glycine receptors in spinal cord neurons (126).
Additional domains found in TOR and in other members of the PIK-related kinase family are the FAT and FATC domains (Fig. 2). The FAT domain, spanning approximately 500 amino acids N-terminal to the FRB and catalytic domains in TOR, is found only in members of the PIK-related kinase family (3, 17). Although the function of the FAT domain remains to be elucidated, it has been proposed that this domain could serve as a scaffold or as a protein-protein interaction domain, similar to the HEAT repeats (17). Finally, the FATC domain, a 35-amino-acid sequence in the extreme C terminus, occurs only in combination with the FAT domain and may be important for catalytic activity of PIK-related kinases (17, 87).
TOR SIGNALING IN YEASTS AND HIGHER EUKARYOTES
Organization of the Actin Cytoskeleton
S. cerevisiae TOR2 has two essential signaling functions. One function is shared with TOR1 and is required for activation of translation initiation and early G1 progression in response to nutrients (see below) (7, 44). The second essential function is unique to TOR2 and mediates the cell cycle-dependent polarization of the actin cytoskeleton (132, 134). Polarization of the actin cytoskeleton is essential for targeting secretion to the bud site and thus for establishing and maintaining cell polarity (133). The shared function of TOR2 is sensitive to rapamycin, whereas the unique TOR2 function is not (134, 171). The molecular basis for the selective inhibition of only one of the two TOR2 functions by rapamycin is unknown.
TOR2 signaling to the actin cytoskeleton is mediated by activation of the small GTPase RHO1 via the exchange factor ROM2 (132, 133). RHO1, in turn, signals to the actin cytoskeleton via its direct effector protein kinase C1 (PKC1) and a PKC1-activated mitogen-activated protein (MAP) kinase cascade (65, 66). Although the PKC1-controlled MAP kinase cascade maintains cell integrity by activating the transcription of genes required for cell wall synthesis (74, 170), it remains to be determined whether control of the actin cytoskeleton by this MAP kinase cascade also occurs at the transcriptional level or in a more direct fashion. To date, it is not known if the rapamycin-insensitive, TOR2-unique function is conserved in other organisms. Rapamycin-insensitive TOR signaling has not been detected in mammalian cells, possibly because studies on TOR signaling in mammalian cells have relied exclusively on rapamycin to inhibit mTOR.
Translation Initiation
Initiation of protein synthesis in eukaryotes is a highly regulated process mediated by several polypeptide initiation factors (for a review, see Hershey and Merrick [69]). The heterotrimeric initiation factor 4F (eIF4F) mediates translation initiation by facilitating ribosome binding to the 5′ cap (m7GpppN, where m is a methyl group and N is any nucleotide) structure of the mRNA. In S. cerevisiae, the eIF4F complex is composed of the cap-binding subunit eIF4E (encoded by CDC33), eIF4G (TIF4631 and TIF4632, encoding two similar proteins termed eIF4G1 and eIF4G2, respectively), and eIF4A (encoded by TIF1 and TIF2).
The loss of TOR function in yeast cells results in an early and severe inhibition of translation initiation (7). It is as a consequence of this translation defect that TOR-inhibited cells arrest in the G1 phase of the cell cycle. The mechanism by which TOR1 and TOR2 activate translation initiation is uncertain, although the most plausible hypothesis is that the TOR pathway positively controls translation initiation through activation of eIF4E (7). Several observations suggest that TOR may control translation at the level of eIF4E. First, cdc33 and tor mutants display remarkably similar phenotypes (7, 39). Second, the EAP1 protein has recently been identified in S. cerevisiae based on its capacity to interact with eIF4E (33). EAP1 blocks cap-dependent translation via competition with eIF4G, and disruption of the EAP1 gene confers partial resistance to rapamycin, suggesting a role for EAP1 similar to the one described for mammalian eIF4E-binding protein 1 (4E-BP1; see below).
Translational inhibition upon TOR inactivation may involve degradation of the initiation factor eIF4G because degradation of eIF4G protein has been reported in rapamycin-treated cells (12). However, it is unknown whether the degradation of eIF4G is a primary cause of translation inhibition or a secondary effect of the translational downregulation caused by TOR inhibition.
Several lines of evidence indicate that the G1 cell cycle arrest observed in rapamycin-treated cells is, at least in part, a consequence of the inhibited translation of CLN3 (7, 44), a cyclin involved in G1 progression (111, 154). The abundance of CLN3 depends on its relative rates of synthesis and degradation (166) and therefore, regulation of CLN3 synthesis or stability is a critical step in controlling progression through the cell cycle (7, 39, 48, 119). The finding that the G1 arrest in response to TOR inactivation is suppressed by cap-independent expression of CLN3 supports a model in which TOR stimulates cap-dependent translation initiation, including translation of CLN3 and other G1 cyclins, to drive cells through G1 and into S phase (7).
The eIF4F complex is highly conserved. Formation of the mammalian eIF4F complex is regulated by the 4E-BP family of translational repressors (11, 100, 108, 120). The 4E-BPs compete with the eIF-4G proteins for binding to eIF4E, and binding of 4E-BPs to eIF4E is regulated by the phosphorylation state of 4E-BPs (124). Low phosphorylation of 4E-BP promotes the formation of a 4E-BP-eIF4E complex, whereas high phosphorylation of 4E-BP inhibits this interaction. mTOR, in conjunction with the PI3K signaling pathway, modulates 4E-BP phosphorylation. mTOR immunoprecipitates phosphorylate 4E-BP1 in vitro (21, 50), although it remains to be determined whether mTOR phosphorylates all or some of the phosphorylated sites in 4E-BP1 (51). mTOR and PI3K signaling may also contribute to control translation initiation in response to amino acids or insulin by regulating the phosphorylation state of eIF-4GI (123).
The 40S ribosomal phosphoprotein S6 has been proposed to affect translation initiation of a group of mRNAs possessing a 5′-terminal oligopyrimidine tract (5′ TOP) in mammals and Drosophila melanogaster (reviewed by Meyuhas and Hornstein [107]). Most 5′ TOP mRNAs encode components of the translational apparatus, such as ribosomal proteins, elongation factors, and the poly(A)-binding protein. mTOR, in the presence of amino acids, promotes phosphorylation of S6 through activation of p70 S6 kinase (p70S6k). Phosphorylation of S6, in turn, results in the upregulation of translation initiation. mTOR phosphorylates p70S6k in vitro, suggesting that mTOR may act on this protein directly (21, 78). p70S6k phosphorylation is also controlled by the PI3K signaling pathway in response to growth factors such as insulin (152). Interestingly, the mTOR and PI3K inputs on p70S6k can be separated (43, 104). Thus, mTOR and PI3K also control translation initiation by regulating p70S6k activity.
Early on, mTOR was known to control translation only via p70S6k and S6 phosphorylation (31, 47, 94, 122). However, the observation that phosphorylation of the yeast equivalent of S6 (S10) is not important for growth led to the model in S. cerevisiae that TOR controls translation via eIF-4E and cap-dependent translation (7, 82). This model from S. cerevisiae, in turn, led to the finding that mTOR controls translation via eIF-4E (and 4E-BP) in addition to p70S6k (10). 4E-BP phosphorylation was originally thought to be controlled by MAP kinase independently of mTOR (100, 115) rather than by mTOR independently of MAP kinase (10, 157).
Ribosome Biogenesis
The production of ribosomes is an energetically very costly process that requires the action of all three RNA polymerases and involves more than 100 gene products (160). Thus, to couple the rate of protein synthesis to the energetic and metabolic demands of the cell, the abundance of the components of the translation machinery must be finely regulated according to growth conditions and nutrient availability. In S. cerevisiae, TOR signaling regulates ribosome biogenesis at both the transcriptional and translational levels and plays an important role in coupling nutrient availability to the transcription of genes involved in the formation of ribosomes. Inhibition of the TOR pathway by rapamycin treatment or nutrient starvation leads to a downregulation of transcription of ribosomal protein mRNAs by polymerase II (24, 60, 121) as well as transcription of rRNA and tRNA by polymerase I and polymerase III (121, 168). TOR also controls processing of at least the 35S precursor rRNA (121).
All the above findings point to TOR signaling as an essential pathway in the control of ribosome biogenesis. However, the mechanism by which TOR controls the synthesis of ribosomes remains largely unknown. TAP42, an essential phosphoprotein that interacts with the catalytic subunits of protein phosphatase 2A (PP2A) and PP2A-related phosphatases, is under control of the TOR pathway (see below) (44, 81). The finding that a mutation in TAP42 inhibits polyribosome formation suggests that TAP42/protein phosphatase functions upstream of translational initiation (44). Furthermore, a role for TAP42/protein phosphatase in regulating ribosomal protein and rRNA gene expression has also been proposed (121).
In mammalian cells, mTOR controls the synthesis of rRNA in a process that involves activation of p70S6k (96, 103). mTOR also regulates the abundance of ribosomal proteins and other components of the translation machinery, such as the poly(A)-binding protein, by promoting translation of 5′ TOP mRNAs (80) (see above). Thus, like yeast TOR, mTOR controls ribosome biogenesis at both the transcriptional and translational levels.
Control of Phosphatases by TOR
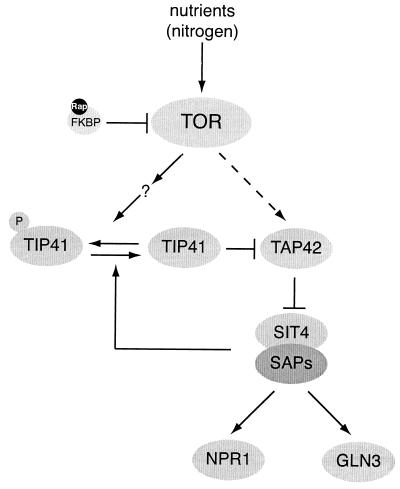
The regulation of phosphatase activity constitutes a particularly important branch in TOR signaling (130). Genetic screens in S. cerevisiae have identified TAP42 (a phosphatase-associated protein of 42 kDa), the type 2A phosphatase (PP2A) catalytic subunits PPH21 and PPH22, and the type 2A-related phosphatase SIT4 as components of the TOR signaling pathway (44). TAP42 is an essential, conserved protein that independently associates with SIT4 and with the catalytic subunits of PP2A in response to nutrient availability and TOR activity. In the presence of a good nitrogen source such as ammonium or glutamine, TOR keeps SIT4 inactive by promoting the binding of TAP42 to SIT4 (Fig. 3). Upon nitrogen starvation or rapamycin treatment (conditions that inactivate TOR), SIT4 is released from TAP42 and thereby activated (8, 44). As described below, activated SIT4 dephosphorylates target proteins such as NPR1 and GLN3. Interestingly, TAP42-associated SIT4 may not be inactive for substrates other than NPR1 and GLN3 (38).
FIG. 3.
TOR controls phosphatases in S. cerevisiae. Under good nutrient (nitrogen) conditions, TOR inhibits the phosphatase SIT4 by promoting the association of SIT4 with TAP42. Two different models have been proposed for the mechanism by which TOR controls the SIT4-TAP42 complex. Jiang and Broach (81) proposed that TOR controls the interaction between SIT4 and TAP42 by phosphorylating TAP42 directly (indicated by a dashed arrow). Jacinto et al. (79) suggested that the association of SIT4 and TAP42 is controlled primarily by the TAP42 interactor TIP41. TOR may phosphorylate and inactivate TIP41 by an unknown mechanism. Dephosphorylated TIP41 positively regulates SIT4 by binding and inhibiting TAP42. The association of TIP41 with TAP42 enhances SIT4 phosphatase activity, allowing free SIT4 subunits to associate with SAPs and activate target phosphoproteins such as NPR1 and GLN3. Dephosphorylation and activation of TIP41 are mediated by SIT4, indicating that TIP41 is part of a feedback loop that amplifies SIT4 activity. Arrows indicate activation; bars indicate inhibition. Adapted from Jacinto et al. (79). Rap, rapamycin.
TOR may control the association of TAP42 with the PP2A catalytic subunits and with SIT4 by phosphorylating TAP42 directly (81). Phosphorylated TAP42 effectively competes with the PP2A regulatory subunits CDC55 and TPD3 for binding to the PP2A catalytic subunits, whereas dephosphorylated TAP42 does not compete for binding (81). However, although rapamycin treatment causes dephosphorylation of TAP42 in vivo (81), it is unlikely that the phosphorylation state of TAP42 plays a major role in SIT4 regulation (79). Upon rapamycin treatment, dephosphorylation of TAP42 occurs much more slowly than dissociation of the SIT4-TAP42 complex (44) and also much more slowly than SIT4 appears to be activated (8, 131). Thus, another protein(s) may participate in the regulation of SIT4. Recently, a conserved TAP42-interacting phosphoprotein, TIP41, was identified in S. cerevisiae (79). TIP41 positively regulates SIT4 by binding to and inhibiting TAP42 (79). The binding of TIP41 to TAP42 is stimulated by rapamycin treatment via SIT4-dependent dephosphorylation of TIP41, suggesting that TIP41 is part of a feedback loop that rapidly amplifies SIT4 phosphatase activity under TOR-inactivating conditions (79) (Fig. 3). Whether TOR phosphorylates TIP41 directly (or indirectly) remains to be determined.
TAP42 is conserved in mammals and plants (61, 109), suggesting that a regulation of PP2A activity similar to that found in S. cerevisiae may exist in higher eukaryotes. Indeed, the murine α4 phosphoprotein, the mammalian homologue of TAP42, binds directly to the catalytic subunits of PP2A (77, 109), PP4, and PP6 (27, 110). However, there is substantial controversy regarding the rapamycin sensitivity of the α4-phosphatase interaction (27, 109). Although it is unclear which component(s) of the α4-phosphatase complex is sensitive to rapamycin, inactivation of mTOR by rapamycin causes rapid dephosphorylation of the ribosomal S6 kinase p70s6k by PP2A (41, 118), suggesting that TOR also negatively controls phosphatase activity in mammals.
Regulation of Amino Acid Permeases
Yeast cells can utilize a wide variety of compounds as nitrogen or carbon sources, including sugars, amino acids, and peptides. This versatility is determined by the ability of cells to transport these nutrients across the plasma membrane, followed either by their direct utilization or by conversion to metabolites required by the cell. Amino acids are essential for cell growth because they constitute the building blocks for protein synthesis. However, in S. cerevisiae, some amino acids (such as glutamine, glutamate, and asparagine) are also important because they serve as nitrogen sources. Therefore, amino acid permeases play an important role in cell growth and viability.
Based on their function and regulation, yeast amino acid permeases can be divided into two classes (144). Permeases of one class, including the general amino acid permease GAP1, are regulated in response to the available nitrogen source. In the presence of a good nitrogen source, such as ammonium or glutamine, the uptake activity of these permeases is low, whereas in medium containing a poor nitrogen source, such as proline or urea, transport activity is strongly induced. The second class of amino acid permeases consists of transporters that are specific for single amino acids or a small set of structurally related amino acids. The histidine permease HIP1 and the tryptophan permease TAT2 belong to this group of specific amino acid permeases.
Studies in S. cerevisiae have revealed that the TOR pathway plays a prominent role in regulation of amino acid permease activity. Inhibition of TOR function by rapamycin or nitrogen starvation induces ubiquitination and degradation of TAT2 and, as a consequence, leads to a decrease in tryptophan import (9, 131). Starvation-induced downregulation of amino acid permeases also applies to HIP1 and possibly to all specific amino acid permeases (9). In contrast to TAT2 and HIP1, rapamycin treatment causes a significant increase in GAP1 protein (9, 131). Thus, TOR proteins appear to regulate inversely the high-specificity permeases, such as TAT2 and HIP1, and the broad-specificity permease GAP1 in response to nutrient availability.
Upregulation of GAP1 upon nitrogen starvation is mediated by the Ser/Thr nitrogen permease reactivator kinase NPR1 (40, 54, 55, 155). In the presence of a poor nitrogen source, NPR1 promotes GAP1 function (54, 55), probably by phosphorylating and protecting GAP1 from degradation (40, 146). In agreement with the opposite regulation of GAP1 and TAT2, NPR1 has been proposed to function as a negative regulator of TAT2. Indeed, tryptophan import decreases upon NPR1 overexpression (131). How does the cell modulate NPR1 to inversely regulate GAP1 and TAT2 in response to the nitrogen source? NPR1 is a phosphoprotein whose phosphorylation state is controlled by the TOR signaling pathway in response to the nitrogen source (131). In response to a good nitrogen source, TOR keeps NPR1 phosphorylated and in an inactive form that is unable to protect GAP1 from ubiquitination. Under poor nitrogen conditions, NPR1 becomes dephosphorylated and activated in a SIT4- and TIP41-dependent manner (79). Activation of NPR1 leads to GAP1 protection and to TAT2 ubiquitination and degradation (131). It is unknown whether NPR1 directly phosphorylates GAP1 or TAT2.
At present, it is not known whether TOR plays a role in regulating the traffic of nutrient permeases through the membrane in higher eukaryotes. However, in a recent report, it was proposed that the mTOR pathway may be involved in a process that rapidly mobilizes the glucose transporter GLUT4 to a highly insulin-responsive compartment upon insulin stimulation (16). mTOR has also been proposed to play an important role in the stimulation of another glucose transporter, GLUT1, by insulin, although this effect appears to be at the level of translation (148).
Autophagy
In response to nitrogen or carbon limitation, yeast cells undergo a catabolic membrane-trafficking process known as autophagy (for recent reviews, see references 1 and 89). During this process, a large number of cytoplasmic components are nonselectively enclosed within a double-membrane structure (autophagosome) and delivered to the vacuole for degradation. Although autophagy was first described in developing kidney cells (32), the most important advances in the understanding of autophagy were obtained much later with the discovery of autophagy in S. cerevisiae and the isolation of the first autophagy mutants (149, 153).
Inactivation of TOR function by rapamycin induces autophagy even in rich nutrient conditions, indicating that TOR inhibits autophagy (113). The mechanism by which TOR inhibits autophagy is being elucidated by Ohsumi and coworkers. Kamada et al. (85) reported that the protein kinase APG1 is essential for autophagy and plays a pivotal role in the control of autophagy by TOR. APG1 associates with APG13 and APG17 to form the APG1 protein complex (105). The role of TOR in the regulation of autophagy is to maintain APG13 in a phosphorylated form with low affinity for APG1 and thereby to inhibit APG1 activity (85) (Fig. 4). Inactivation of TOR by rapamycin treatment or nutrient starvation causes rapid dephosphorylation of APG13, which increases the affinity of this protein for APG1 and enhances APG1 kinase activity (85). How TOR promotes APG13 phosphorylation is currently unknown. The finding that mutations in the TOR-controlled protein TAP42 have no effect on either APG1 activity or autophagy induction suggests that the APG1-APG13 interplay comprises a novel TOR signaling pathway regulating autophagy (85).
FIG. 4.
TOR inhibits autophagy in S. cerevisiae. Under good nutrient conditions, TOR inhibits autophagy by promoting phosphorylation (P) of APG13 and thereby preventing the formation of an APG1-APG13 complex, which is essential for the induction of autophagy. Inactivation of TOR by rapamycin (Rap) treatment or nutrient deprivation results in rapid dephosphorylation of APG13. Dephosphorylated APG13 associates with APG1, and the active APG13-APG1 complex induces autophagy. Arrows indicate activation; bars indicate inhibition.
Autophagy also occurs in animal cells that are serum starved or challenged with specific hormonal stimuli (for recent reviews, see references 1 and 147). A number of yeast autophagy genes, such as APG1, APG5, APG7, APG12, and AUT7, have homologues in mammals (83, 129, 150), but only some of these homologues have been demonstrated to participate in autophagy in mammalian cells. This is the case for the mammalian gene beclin-1, a homologue of yeast APG6 that promotes autophagy in autophagy-defective S. cerevisiae (97). Interestingly, the mTOR signaling pathway may play a role in the regulation of autophagy, as rapamycin addition to mammalian cells in culture induces autophagy even in nutrient-rich medium (15, 141). Inhibition of autophagy by mTOR may involve the p70S6k signaling branch (15, 141).
Transcriptional Control of Nutrient Metabolism
Control of gene expression at the level of transcription represents an important branch of TOR signaling. Genome-wide expression analysis of yeast cells treated with rapamycin or of cells shifted from a rich to a poor nitrogen or carbon source revealed a prominent role of TOR in the coordination of transcription of nutrient-regulated genes (24, 60, 91, 140). Rapamycin rapidly and strongly modulates the expression of several hundred genes involved in various metabolic pathways, including nitrogen metabolism, the glycolytic pathway, and the tricarboxylic acid cycle. The most striking set of genes affected by rapamycin treatment, however, are those involved in the uptake and assimilation of different nitrogen sources. Specifically, rapamycin causes a decrease in the expression of genes participating in the uptake and metabolism of preferred nitrogen sources (glutamine and ammonium) and a pronounced increase in the expression of genes involved in the uptake and use of poor nitrogen sources (urea and proline). Thus, rapamycin induces a nitrogen starvation response.
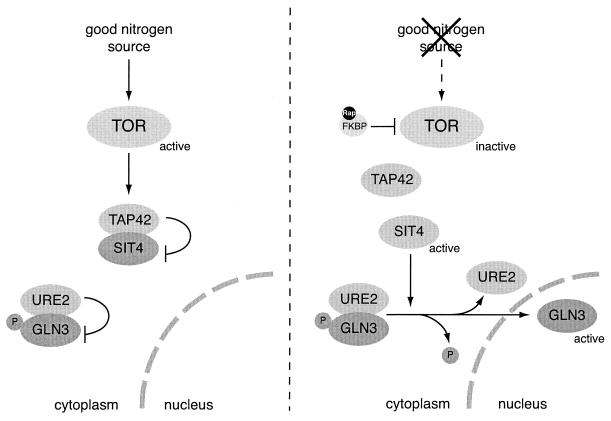
TOR controls the expression of nutrient-regulated genes by sequestering several nutrient-responsive transcription factors in the cytoplasm (8). The expression of most of the nitrogen-responsive genes is regulated by the GATA transcription factors GLN3 and GAT1 and their cytoplasmic repressor URE2 (102). Under good nitrogen conditions, GLN3 is retained in the cytoplasm by URE2. The binding of GLN3 to URE2 requires TOR-dependent phosphorylation of GLN3 by a mechanism that involves TAP42-mediated inhibition of the phosphatase SIT4 (8) (Fig. 5). Rapamycin treatment or nitrogen starvation causes GLN3 to become dephosphorylated and to dissociate from URE2. GLN3 then translocates into the nucleus to activate its target genes (8) (Fig. 5). GLN3 importin and exportin have recently been identified as SRP1 and CRM1, respectively (25). URE2 is also phosphorylated in a rapamycin-sensitive manner (24, 60), but the regulation of URE2 phosphorylation is TAP42 independent. TOR also prevents the access of GAT1 to the nucleus (8), but, although GAT1 inhibition might be mediated by URE2, a direct interaction between GAT1 and URE2 has not been shown. Several lines of evidences suggest that GLN3 and GAT1 are regulated in response to different stimuli (35, 95).
FIG. 5.
TOR prevents nuclear accumulation of the nitrogen-regulated transcription activator GLN3 via TAP42-mediated inhibition of the phosphatase SIT4. Under good nitrogen conditions, GLN3 is phosphorylated and retained in the cytoplasm by URE2. Upon nitrogen starvation or rapamycin treatment, SIT4 is released from TAP42 and activated. Activated SIT4 dephosphorylates the GATA transcription factor GLN3. Dephosphorylated GLN3 dissociates from URE2 and translocates into the nucleus, where it activates transcription of target genes. Arrows indicate activation; bars indicate inhibition.
Inactivation of TOR by rapamycin also activates the partially redundant Zn2+ finger transcription factors MSN2 and MSN4 (8), both of which respond to different types of cellular stress, including carbon source limitation (53, 143, 151). TOR may inhibit MSNs (MSN2 and MSN4) in response to nutrients by promoting the association of MSNs with the abundant 14-3-3 proteins BMH1 and/or BMH2 (8). In agreement with this model, BMH1 and BMH2 have been shown to positively regulate rapamycin-sensitive signaling (13). Glucose withdrawal or rapamycin treatment causes a release of MSN from BMH2. Contrary to what was observed for GLN3, this process is independent of TAP42 and SIT4 (8), suggesting that another, unknown TOR pathway regulates MSN.
TOR also controls the heterodimeric bHLH/Zip transcription factor composed of RTG1 and RTG3 (91). RTG1 and RTG3 were originally identified as genes required under conditions in which mitochondrial respiratory function is impaired (98, 99). RTG1 and RTG3 regulate the expression of tricarboxylic acid and glyoxylate cycle genes that participate in the synthesis of intermediates, primarily α-ketoglutarate, required for de novo synthesis of some amino acids, such as glutamine and glutamate. Similar to GLN3 regulation, inhibition of TOR by rapamycin or nitrogen starvation results in both rapid nuclear accumulation of RTG1 and RTG3 and induction of their target genes (91). RTG2, a positive regulator of RTG1 and RTG3, is essential for the TOR- and nitrogen-inhibited nuclear accumulation of RTG1 and RTG3 (91).
How TOR maintains RTG1 and RTG3 in the cytoplasm is not well understood. It has been reported that RTG3 is phosphorylated upon rapamycin-mediated inactivation of TOR (91). Whether RTG3 phosphorylation is influenced by TAP42 and/or SIT4 (PP2A) remains to be determined. Recently, it was reported that MKS1, a phosphoprotein controlled by TOR, is a negative regulator of RTG1 and RTG3, suggesting a high degree of complexity in regulation of the RTG branch of TOR signaling (45, 138).
mTOR signaling also controls transcription in mammalian cells. Recently, it was reported that mTOR phosphorylates the transcriptional activator STAT3 (167). Activation of STAT3 occurs in response to the neuropoietic cytokine ciliary neurotrophic factor and requires phosphorylation on tyrosine and serine residues (163). While members of the Jun-associated kinase/Tyk family of tyrosine kinases mediate phosphorylation of STAT3 on Tyr705, mTOR appears to phosphorylate STAT3 on Ser727 directly.
The TOR signaling pathway in yeast and mammalian cells controls gene expression via mRNA stability in addition to mRNA synthesis. Inhibiting TOR signaling, through either nutrient limitation or rapamycin treatment, causes the accelerated turnover of a subset of mRNAs (4, 6). Moreover, the inhibition of TOR appears to destabilize mRNAs by multiple mechanisms (4).
TOR RESPONDS TO NUTRIENTS
TOR inactivation by rapamycin treatment results in a nutrient starvation response, suggesting that TOR responds to nutrient availability (see above) (7). In S. cerevisiae, TOR signaling has been proposed to respond to nitrogen and possibly carbon sources (8, 35, 95, 131, 140). However, the specific nitrogen or carbon metabolites that act upstream of TOR are unknown. Recent evidence argues that TOR signaling, or at least a specific subset of the TOR pathways, responds to the amino acid glutamine, suggesting that glutamine is a particularly important indicator of nutrient status (35). Indeed, glutamine is a preferred nitrogen source and controls carbon metabolism via the tricarboxylic acid cycle (35, 102). Glutamine depletion activates the TOR-controlled transcription factors GLN3, RTG1, and RTG3. However, other TOR readouts, such as localization of MSN2/4 and GAT1 and downregulation of ribosomal protein mRNAs, are not activated under the same glutamine starvation conditions, suggesting that TOR must also respond to other yet-to-be identified nutrients (35). Schreiber and coworkers have proposed that TOR may act as a multichannel processor that can elicit different responses to distinct nutrient signals (95, 140).
Glutamine may play a particularly important role in TOR signaling in both yeast and mammalian cells. TOR in S. cerevisiae (35), as described above, and in mammalian cells (75) appears to respond to glutamine. Furthermore, a recent transcriptional profiling of mammalian cells revealed that rapamycin treatment mimics glutamine or leucine starvation more than glucose starvation (116). Interestingly, a decrease in blood glutamine levels causes immunosuppression in humans and mice similar to that caused by rapamycin treatment (23, 84).
In mammalian cells, amino acid deprivation causes rapid dephosphorylation and activation of the eIF-4E binding protein (4E-BP) and dephosphorylation and inhibition of p70S6k (59, 158, 165). Both of these processes are mediated by mTOR, indicating that amino acids signal to 4E-BP and p70S6k via mTOR. It has been proposed that the aminoacylation state of tRNA may be responsible for the regulation of p70S6k phosphorylation (76). In yeast cells, the GCN2 kinase senses intracellular amino acid availability through a tRNA-binding domain that exhibits similarity to histidyl-tRNA synthetase. Upon amino acid limitation, levels of uncharged tRNA increase significantly and activate the kinase domain of GCN2 kinase, leading to activation of GCN4, a transcriptional activator of several genes involved in amino acid biosynthesis (reviewed by Hinnebusch [71, 72]). Although the GCN2 pathway is conserved in mammals (88) and despite the similarity of the mechanism of amino acid sensing proposed for GCN2 and mTOR, a connection between these two signaling pathways has never been demonstrated. Dennis et al. (42) argue that amino acid pools rather than the amount of aminoacylated tRNA are important for mTOR signaling. Moreover, sensing of the intracellular level of glutamine in S. cerevisiae seems to be independent of the aminoacylated state of glutaminyl-tRNA (our unpublished data). Thus, how TOR may sense amino acid pools is unknown.
In mammals, both TOR and PI3K signaling are required for regulation of common downstream effectors such as 4E-BP and p70S6k (52, 130). Although the nature of the link between mTOR and the PI3K signaling pathway is controversial, most evidence suggests that mTOR activity is not regulated by PI3K. Whereas some studies have presented evidence that protein kinase B (PKB), a downstream effector of PI3K, stimulates phosphorylation and activity of mTOR in response to insulin and that mTOR is a direct substrate of PKB (112, 137, 139), other studies have failed to detect significant alteration of mTOR kinase activity in response to amino acids or insulin (58, 59). Furthermore, mTOR mutants bearing Ala substitutions at two PKB-dependent phosphorylation sites (Ser2448 and Thr2446) indicate that PKB-dependent phosphorylation of mTOR is not essential for mTOR function (139). Finally, cells deficient in the PI3K effector PDK1 fail to activate PKB yet display normal mTOR activity, indicating that PKB activity is not necessary for mTOR activity (159). Thus, mTOR and PI3K, although connected by common targets, may respond separately to amino acids and growth factors, respectively.
Recently, two different, novel mechanisms have been proposed for regulation of mTOR. Dennis et al. (42) have shown that the mTOR pathway is influenced by the intracellular concentration of ATP. Although the mechanism by which ATP acts on TOR is unknown, it appears to be different from that used by amino acids, as both ATP and amino acids are required for mTOR signaling (42). Fang et al. (46) reported that phosphatidic acid, which accumulates in mammalian cells upon mitogenic stimulation, is required for activation of mTOR downstream effectors. Phosphatidic acid interacts directly with the FRB domain in mTOR, and this rapamycin-sensitive interaction correlates with the ability of mTOR to activate downstream effectors (46). The finding that phosphatidic acid has no effect on mTOR kinase activity (and the effect of rapamycin or insulin on mTOR kinase activity is controversial, as discussed above) suggests that the inhibitory effect of rapamycin on mTOR may derive from its competition with phosphatidic acid for binding to the FRB, independent of an effect on intrinsic mTOR kinase activity (46). In other words, displacement of phosphatidic acid by rapamycin may affect the interaction of mTOR with a substrate rather than intrinsic mTOR kinase activity.
TOR AND STRESS
TOR promotes cell growth in response to nutrient availability (125, 130). Several lines of evidence suggest that TOR also plays a role in cell growth under stress conditions other than nutrient limitation. First, TOR controls the transcription factors MSN2 and MSN4 (8), which activate expression of genes in response to several different environmental stress conditions, including heat shock and H2O2 treatment (26, 49). As described above, TOR controls cellular localization of MSN2 and MSN4 by preventing their access to the nucleus (8). The specific environmental conditions in response to which TOR controls MSN remains to be determined. Second, S. pombe cells lacking TOR1 are sensitive to osmotic stress, oxidative stress, high external pH, and high or low temperature (86, 161). Third, tor1 mutants of S. cerevisiae are sensitive to high concentrations of salt, suggesting that TOR1 is necessary for the proper cellular response to saline stress (34). Although the role of TOR in salt stress signaling is at present unknown, a connection between the TOR-controlled transcription factors GLN3 and GAT1 and the lithium and sodium extrusion pump ENA1, which is essential for survival under saline stress conditions, has been shown (34). GLN3 and GAT1 mediate induction of ENA1 transcription upon rapamycin treatment and, like TOR1, are required for growth under salt stress (34).
Why does TOR respond to environmental stress? One plausible explanation is that TOR, as a central controller of cell growth, may respond to several different types of stress to ensure that growth occurs only when overall conditions are favorable.
CONCLUSION
TOR appears to activate a number of different effector pathways in response to a wide variety of stimuli, including the nitrogen source, amino acids, ATP, phosphatidic acid, and stress. The mechanisms by which TOR responds to and integrates different inputs remain to be determined. However, the answers to these open questions will most likely come, at least in part, from S. cerevisiae.
Acknowledgments
We thank Tobias Schmelzle, Robbie Loewith, and Estela Jacinto for helpful discussion.
We acknowledge the Federation of European Biochemical Societies (J.L.C.) and the Swiss National Science Foundation and the Canton of Basel (M.N.H.) for support.
REFERENCES
- 1.Abeliovich, H., and D. J. Klionsky. 2001. Autophagy in yeast: mechanistic insights and physiological function. Microbiol. Mol. Biol. Rev. 65:463-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. [DOI] [PubMed] [Google Scholar]
- 3.Alarcon, C. M., J. Heitman, and M. E. Cardenas. 1999. Protein kinase activity and identification of a toxic effector domain of the target of rapamycin TOR proteins in yeast. Mol. Biol. Cell 10:2531-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albig, A. R., and C. J. Decker. 2001. The target of rapamycin signaling pathway regulates mRNA turnover in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 12:3428-3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrade, M. A., and P. Bork. 1995. HEAT repeats in the Huntington's disease protein. Nat. Genet. 11:115-116. [DOI] [PubMed] [Google Scholar]
- 6.Banholzer, R., A. P. Nair, H. H. Hirsch, X. F. Ming, and C. Moroni. 1997. Rapamycin destabilizes interleukin-3 mRNA in autocrine tumor cells by a mechanism requiring an intact 3′ untranslated region. Mol. Cell. Biol. 17:3254-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbet, N. C., U. Schneider, S. B. Helliwell, I. Stansfield, M. F. Tuite, and M. N. Hall. 1996. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7:25-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beck, T., and M. N. Hall. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402:689-692. [DOI] [PubMed] [Google Scholar]
- 9.Beck, T., A. Schmidt, and M. N. Hall. 1999. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J. Cell Biol. 146:1227-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beretta, L., A. C. Gingras, Y. V. Svitkin, M. N. Hall, and N. Sonenberg. 1996. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 15:658-664. [PMC free article] [PubMed] [Google Scholar]
- 11.Bernal, A., and D. A. Kimbrell. 2000. Drosophila Thor participates in host immune defense and connects a translational regulator with innate immunity. Proc. Natl. Acad. Sci. USA 97:6019-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berset, C., H. Trachsel, and M. Altmann. 1998. The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95:4264-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertram, P. G., C. Zeng, J. Thorson, A. S. Shaw, and X. F. Zheng. 1998. The 14-3-3 proteins positively regulate rapamycin-sensitive signaling. Curr. Biol. 8:1259-1267. [DOI] [PubMed] [Google Scholar]
- 14.Bierer, B. E., P. K. Somers, T. J. Wandless, S. J. Burakoff, and S. L. Schreiber. 1990. Probing immunosuppressant action with a nonnatural immunophilin ligand. Science 250:556-559. [DOI] [PubMed] [Google Scholar]
- 15.Blommaart, E. F., J. J. Luiken, P. J. Blommaart, G. M. van Woerkom, and A. J. Meijer. 1995. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J. Biol. Chem. 270:2320-2326. [DOI] [PubMed] [Google Scholar]
- 16.Bogan, J. S., A. E. McKee, and H. F. Lodish. 2001. Insulin-responsive compartments containing GLUT4 in 3T3-L1 and CHO cells: regulation by amino acid concentrations. Mol. Cell. Biol. 21:4785-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bosotti, R., A. Isacchi, and E. L. Sonnhammer. 2000. FAT: a novel domain in PIK-related kinases. Trends Biochem. Sci. 25:225-227. [DOI] [PubMed] [Google Scholar]
- 18.Brown, E. J., M. W. Albers, T. B. Shin, K. Ichikawa, C. T. Keith, W. S. Lane, and S. L. Schreiber. 1994. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 369:756-758. [DOI] [PubMed] [Google Scholar]
- 19.Brown, E. J., P. A. Beal, C. T. Keith, J. Chen, T. B. Shin, and S. L. Schreiber. 1995. Control of p70 s6 kinase by kinase activity of FRAP in vivo. Nature 377:441-446. [DOI] [PubMed] [Google Scholar]
- 20.Brunn, G. J., C. C. Hudson, A. Sekulic, J. M. Williams, H. Hosoi, P. J. Houghton, J. C. Lawrence, Jr., and R. T. Abraham. 1997. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 277:99-101. [DOI] [PubMed] [Google Scholar]
- 21.Burnett, P. E., R. K. Barrow, N. A. Cohen, S. H. Snyder, and D. M. Sabatini. 1998. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. USA 95:1432-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cafferkey, R., P. R. Young, M. M. McLaughlin, D. J. Bergsma, Y. Koltin, G. M. Sathe, L. Faucette, W. K. Eng, R. K. Johnson, and G. P. Livi. 1993. Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol. Cell. Biol. 13:6012-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calder, P. C., and P. Yaqoob. 1999. Glutamine and the immune system. Amino Acids 17:227-241. [DOI] [PubMed] [Google Scholar]
- 24.Cardenas, M. E., N. S. Cutler, M. C. Lorenz, C. J. Di Como, and J. Heitman. 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13:3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carvalho, J., P. G. Bertram, S. R. Wente, and X. F. Zheng. 2001. Phosphorylation regulates the interaction between Gln3p and the nuclear import factor Srp1p. J. Biol. Chem. 276:25359-25365. [DOI] [PubMed] [Google Scholar]
- 26.Causton, H. C., B. Ren, S. S. Koh, C. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander, and R. A. Young. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12:323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen, J., R. T. Peterson, and S. L. Schreiber. 1998. Alpha 4 associates with protein phosphatases 2A, 4, and 6. Biochem. Biophys. Res. Commun. 247:827-832. [DOI] [PubMed] [Google Scholar]
- 28.Chen, J., X. F. Zheng, E. J. Brown, and S. L. Schreiber. 1995. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc. Natl. Acad. Sci. USA 92:4947-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiu, M. I., H. Katz, and V. Berlin. 1994. RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc. Natl. Acad. Sci. USA 91:12574-12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi, J., J. Chen, S. L. Schreiber, and J. Clardy. 1996. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science 273:239-242. [DOI] [PubMed] [Google Scholar]
- 31.Chung, J., C. J. Kuo, G. R. Crabtree, and J. Blenis. 1992. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell 69:1227-1236. [DOI] [PubMed] [Google Scholar]
- 32.Clark, S. L. 1957. Cellular differentiation in the kidneys of newborn mice studied with the electron microscope. J. Biophys. Biochem. Cytol. 3:349-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cosentino, G. P., T. Schmelzle, A. Haghighat, S. B. Helliwell, M. N. Hall, and N. Sonenberg. 2000. Eap1p, a novel eukaryotic translation initiation factor 4E-associated protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:4604-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crespo, J. L., K. Daicho, T. Ushimaru, and M. N. Hall. 2001. The GATA transcription factors GLN3 and GAT1 link TOR to salt stress in Saccharomyces cerevisiae. J. Biol. Chem. 276:34441-34444. [DOI] [PubMed] [Google Scholar]
- 35.Crespo, J. L., T. Powers, B. Fowler, and M. N. Hall. 2002. The TOR-controlled transcription activators GLN3, RTG1 and RTG3 are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. USA 99:6784-6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cruz, M. C., L. M. Cavallo, J. M. Gorlach, G. Cox, J. R. Perfect, M. E. Cardenas, and J. Heitman. 1999. Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in Cryptococcus neoformans. Mol. Cell. Biol. 19:4101-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cruz, M. C., A. L. Goldstein, J. Blankenship, M. Del Poeta, J. R. Perfect, J. H. McCusker, Y. L. Bennani, M. E. Cardenas, and J. Heitman. 2001. Rapamycin and less immunosuppressive analogs are toxic to Candida albicans and Cryptococcus neoformans via FKBP12-dependent inhibition of TOR. Antimicrob. Agents Chemother. 45:3162-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cutler, N. S., X. Pan, J. Heitman, and M. E. Cardenas. 2001. The TOR signal transduction cascade controls cellular differentiation in response to nutrients. Mol. Biol. Cell 12:4103-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Danaie, P., M. Altmann, M. N. Hall, H. Trachsel, and S. B. Helliwell. 1999. CLN3 expression is sufficient to restore G1-to-S-phase progression in Saccharomyces cerevisiae mutants defective in translation initiation factor eIF4E. Biochem. J. 340:135-141. [PMC free article] [PubMed] [Google Scholar]
- 40.De Craene, J. O., O. Soetens, and B. Andre. 2001. The Npr1 kinase controls biosynthetic and endocytic sorting of the yeast Gap1 permease. J. Biol. Chem. 276:43939-43948. [DOI] [PubMed] [Google Scholar]
- 41.Dennis, P. B., S. Fumagalli, and G. Thomas. 1999. Target of rapamycin (TOR): balancing the opposing forces of protein synthesis and degradation. Curr. Opin. Genet. Dev. 9:49-54. [DOI] [PubMed] [Google Scholar]
- 42.Dennis, P. B., A. Jaeschke, M. Saitoh, B. Fowler, S. C. Kozma, and G. Thomas. 2001. Mammalian TOR: a homeostatic ATP sensor. Science 294:1102-1105. [DOI] [PubMed] [Google Scholar]
- 43.Dennis, P. B., N. Pullen, S. C. Kozma, and G. Thomas. 1996. The principal rapamycin-sensitive p70s6k phosphorylation sites, T-229 and T-389, are differentially regulated by rapamycin-insensitive kinase kinases. Mol. Cell. Biol. 16:6242-6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Como, C. J., and K. T. Arndt. 1996. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 10:1904-1916. [DOI] [PubMed] [Google Scholar]
- 45.Dilova, I., C. Y. Chen, and T. Powers. 2002. Mks1 in concert with TOR signaling negatively regulates RTG target gene expression in S. cerevisiae. Curr. Biol. 12:389-395. [DOI] [PubMed] [Google Scholar]
- 46.Fang, Y., M. Vilella-Bach, R. Bachmann, A. Flanigan, and J. Chen. 2001. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science 294:1942-1945. [DOI] [PubMed] [Google Scholar]
- 47.Ferrari, S., R. B. Pearson, M. Siegmann, S. C. Kozma, and G. Thomas. 1993. The immunosuppressant rapamycin induces inactivation of p70s6k through dephosphorylation of a novel set of sites. J. Biol. Chem. 268:16091-16094. [PubMed] [Google Scholar]
- 48.Gallego, C., E. Gari, N. Colomina, E. Herrero, and M. Aldea. 1997. The Cln3 cyclin is downregulated by translational repression and degradation during the G1 arrest caused by nitrogen deprivation in budding yeast. EMBO J. 16:7196-7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gasch, A. P., M. Huang, S. Metzner, D. Botstein, S. J. Elledge, and P. O. Brown. 2001. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol. Biol. Cell 12:2987-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gingras, A. C., S. P. Gygi, B. Raught, R. D. Polakiewicz, R. T. Abraham, M. F. Hoekstra, R. Aebersold, and N. Sonenberg. 1999. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13:1422-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gingras, A. C., B. Raught, S. P. Gygi, A. Niedzwiecka, M. Miron, S. K. Burley, R. D. Polakiewicz, A. Wyslouch-Cieszynska, R. Aebersold, and N. Sonenberg. 2001. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 15:2852-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gingras, A. C., B. Raught, and N. Sonenberg. 2001. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 15:807-826. [DOI] [PubMed] [Google Scholar]
- 53.Gorner, W., E. Durchschlag, M. T. Martinez-Pastor, F. Estruch, G. Ammerer, B. Hamilton, H. Ruis, and C. Schuller. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12:586-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grenson, M. 1983. Inactivation-reactivation process and repression of permease formation regulate several ammonia-sensitive permeases in the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 133:135-139. [DOI] [PubMed] [Google Scholar]
- 55.Grenson, M. 1983. Study of the positive control of the general amino-acid permease and other ammonia-sensitive uptake systems by the product of the NPR1 gene in the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 133:141-144. [DOI] [PubMed] [Google Scholar]
- 56.Groves, M. R., N. Hanlon, P. Turowski, B. A. Hemmings, and D. Barford. 1999. The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell 96:99-110. [DOI] [PubMed] [Google Scholar]
- 57.Guba, M., P. von Breitenbuch, M. Steinbauer, G. Koehl, S. Flegel, M. Hornung, C. J. Bruns, C. Zuelke, S. Farkas, M. Anthuber, K. W. Jauch, and E. K. Geissler. 2002. Rapamycin inhibits primary and metastatic tumor growth by antiangiogenesis: involvement of vascular endothelial growth factor. Nat. Med. 8:128-135. [DOI] [PubMed] [Google Scholar]
- 58.Hara, K., K. Yonezawa, M. T. Kozlowski, T. Sugimoto, K. Andrabi, Q. P. Weng, M. Kasuga, I. Nishimoto, and J. Avruch. 1997. Regulation of eIF-4E BP1 phosphorylation by mTOR. J. Biol. Chem. 272:26457-26463. [DOI] [PubMed] [Google Scholar]
- 59.Hara, K., K. Yonezawa, Q. P. Weng, M. T. Kozlowski, C. Belham, and J. Avruch. 1998. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 273:14484-14494. [DOI] [PubMed] [Google Scholar]
- 60.Hardwick, J. S., F. G. Kuruvilla, J. K. Tong, A. F. Shamji, and S. L. Schreiber. 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 96:14866-14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harris, D. M., T. L. Myrick, and S. J. Rundle. 1999. The Arabidopsis homolog of yeast TAP42 and mammalian alpha4 binds to the catalytic subunit of protein phosphatase 2A and is induced by chilling. Plant Physiol. 121:609-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heitman, J., N. R. Movva, and M. N. Hall. 1992. Proline isomerases at the crossroads of protein folding, signal transduction, and immunosuppression. New Biol. 4:448-460. [PubMed] [Google Scholar]
- 63.Heitman, J., N. R. Movva, and M. N. Hall. 1991. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253:905-909. [DOI] [PubMed] [Google Scholar]
- 64.Heitman, J., N. R. Movva, P. C. Hiestand, and M. N. Hall. 1991. FK 506-binding protein proline rotamase is a target for the immunosuppressive agent FK 506 in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 88:1948-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Helliwell, S. B., I. Howald, N. Barbet, and M. N. Hall. 1998. TOR2 is part of two related signaling pathways coordinating cell growth in Saccharomyces cerevisiae. Genetics 148:99-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Helliwell, S. B., A. Schmidt, Y. Ohya, and M. N. Hall. 1998. The Rho1 effector Pkc1, but not Bni1, mediates signalling from Tor2 to the actin cytoskeleton. Curr. Biol. 8:1211-1214. [DOI] [PubMed] [Google Scholar]
- 67.Helliwell, S. B., P. Wagner, J. Kunz, M. Deuter-Reinhard, R. Henriquez, and M. N. Hall. 1994. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol. Biol. Cell 5:105-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hemmings, B. A., C. Adams-Pearson, F. Maurer, P. Muller, J. Goris, W. Merlevede, J. Hofsteenge, and S. R. Stone. 1990. alpha- and beta-forms of the 65-kDa subunit of protein phosphatase 2A have a similar 39 amino acid repeating structure. Biochemistry 29:3166-3173. [DOI] [PubMed] [Google Scholar]
- 69.Hershey, J. W. B., and W. C. Merrick. 2000. The pathway and mechanism of initiation of protein synthesis, p. 33-88. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 70.Hidalgo, M., and E. K. Rowinsky. 2000. The rapamycin-sensitive signal transduction pathway as a target for cancer therapy. Oncogene 19:6680-6686. [DOI] [PubMed] [Google Scholar]
- 71.Hinnebusch, A. G. 2000. Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes, p. 185-243. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 72.Hinnebusch, A. G. 1997. Translational regulation of yeast GCN4. A window on factors that control initiator-tRNA binding to the ribosome. J. Biol. Chem. 272:21661-21664. [DOI] [PubMed] [Google Scholar]
- 73.Hosoi, H., M. B. Dilling, L. N. Liu, M. K. Danks, T. Shikata, A. Sekulic, R. T. Abraham, J. C. Lawrence, Jr., and P. J. Houghton. 1998. Studies on the mechanism of resistance to rapamycin in human cancer cells. Mol. Pharmacol. 54:815-824. [DOI] [PubMed] [Google Scholar]
- 74.Igual, J. C., A. L. Johnson, and L. H. Johnston. 1996. Coordinated regulation of gene expression by the cell cycle transcription factor Swi4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 15:5001-5013. [PMC free article] [PubMed] [Google Scholar]
- 75.Iiboshi, Y., P. J. Papst, S. P. Hunger, and N. Terada. 1999. l-Asparaginase inhibits the rapamycin-targeted signaling pathway. Biochem. Biophys. Res. Commun. 260:534-539. [DOI] [PubMed] [Google Scholar]
- 76.Iiboshi, Y., P. J. Papst, H. Kawasome, H. Hosoi, R. T. Abraham, P. J. Houghton, and N. Terada. 1999. Amino acid-dependent control of p70(s6k). Involvement of tRNA aminoacylation in the regulation. J. Biol. Chem. 274:1092-1099. [DOI] [PubMed] [Google Scholar]
- 77.Inui, S., H. Sanjo, K. Maeda, H. Yamamoto, E. Miyamoto, and N. Sakaguchi. 1998. Ig receptor binding protein 1 (alpha4) is associated with a rapamycin-sensitive signal transduction in lymphocytes through direct binding to the catalytic subunit of protein phosphatase 2A. Blood 92:539-546. [PubMed] [Google Scholar]
- 78.Isotani, S., K. Hara, C. Tokunaga, H. Inoue, J. Avruch, and K. Yonezawa. 1999. Immunopurified mammalian target of rapamycin phosphorylates and activates p70 S6 kinase alpha in vitro. J. Biol. Chem. 274:34493-34498. [DOI] [PubMed] [Google Scholar]
- 79.Jacinto, E., B. Guo, K. T. Arndt, T. Schmelzle, and M. N. Hall. 2001. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol. Cell 8:1017-1026. [DOI] [PubMed] [Google Scholar]
- 80.Jefferies, H. B., S. Fumagalli, P. B. Dennis, C. Reinhard, R. B. Pearson, and G. Thomas. 1997. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J. 16:3693-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jiang, Y., and J. R. Broach. 1999. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 18:2782-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johnson, S. P., and J. R. Warner. 1987. Phosphorylation of the Saccharomyces cerevisiae equivalent of ribosomal protein S6 has no detectable effect on growth. Mol. Cell. Biol. 7:1338-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kabeya, Y., N. Mizushima, T. Ueno, A. Yamamoto, T. Kirisako, T. Noda, E. Kominami, Y. Ohsumi, and T. Yoshimori. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720-5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kafkewitz, D., and A. Bendich. 1983. Enzyme-induced asparagine and glutamine depletion and immune system function. Am. J. Clin. Nutr. 37:1025-1030. [DOI] [PubMed] [Google Scholar]
- 85.Kamada, Y., T. Funakoshi, T. Shintani, K. Nagano, M. Ohsumi, and Y. Ohsumi. 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150:1507-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kawai, M., A. Nakashima, M. Ueno, T. Ushimaru, K. Aiba, H. Doi, and M. Uritani. 2001. Fission yeast tor1 functions in response to various stresses including nitrogen starvation, high osmolarity, and high temperature. Curr. Genet. 39:166-174. [DOI] [PubMed] [Google Scholar]
- 87.Keith, C. T., and S. L. Schreiber. 1995. PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science 270:50-51. [DOI] [PubMed] [Google Scholar]
- 88.Kilberg, M. S., R. G. Hutson, and R. O. Laine. 1994. Amino acid-regulated gene expression in eukaryotic cells. FASEB J. 8:13-19. [DOI] [PubMed] [Google Scholar]
- 89.Klionsky, D. J., and Y. Ohsumi. 1999. Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:1-32. [DOI] [PubMed] [Google Scholar]
- 90.Koltin, Y., L. Faucette, D. J. Bergsma, M. A. Levy, R. Cafferkey, P. L. Koser, R. K. Johnson, and G. P. Livi. 1991. Rapamycin sensitivity in Saccharomyces cerevisiae is mediated by a peptidyl-prolyl cis-trans isomerase related to human FK506-binding protein. Mol. Cell. Biol. 11:1718-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Komeili, A., K. P. Wedaman, E. K. O'Shea, and T. Powers. 2000. Mechanism of metabolic control. Target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J. Cell Biol. 151:863-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kunz, J., R. Henriquez, U. Schneider, M. Deuter-Reinhard, N. R. Movva, and M. N. Hall. 1993. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 73:585-596. [DOI] [PubMed] [Google Scholar]
- 93.Kunz, J., U. Schneider, I. Howald, A. Schmidt, and M. N. Hall. 2000. HEAT repeats mediate plasma membrane localization of Tor2p in yeast. J. Biol. Chem. 275:37011-37020. [DOI] [PubMed] [Google Scholar]
- 94.Kuo, C. J., J. Chung, D. F. Fiorentino, W. M. Flanagan, J. Blenis, and G. R. Crabtree. 1992. Rapamycin selectively inhibits interleukin-2 activation of p70 S6 kinase. Nature 358:70-73. [DOI] [PubMed] [Google Scholar]
- 95.Kuruvilla, F. G., A. F. Shamji, and S. L. Schreiber. 2001. Carbon- and nitrogen-quality signaling to translation are mediated by distinct GATA-type transcription factors. Proc. Natl. Acad. Sci. USA 98:7283-7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leicht, M., A. Simm, G. Bertsch, and J. Hoppe. 1996. Okadaic acid induces cellular hypertrophy in AKR-2B fibroblasts: involvement of the p70S6 kinase in the onset of protein and rRNA synthesis. Cell Growth Differ. 7:1199-1209. [PubMed] [Google Scholar]
- 97.Liang, X. H., S. Jackson, M. Seaman, K. Brown, B. Kempkes, H. Hibshoosh, and B. Levine. 1999. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402:672-676. [DOI] [PubMed] [Google Scholar]
- 98.Liao, X., and R. A. Butow. 1993. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell 72:61-71. [DOI] [PubMed] [Google Scholar]
- 99.Liao, X. S., W. C. Small, P. A. Srere, and R. A. Butow. 1991. Intramitochondrial functions regulate nonmitochondrial citrate synthase (CIT2) expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:38-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lin, T. A., X. Kong, T. A. Haystead, A. Pause, G. Belsham, N. Sonenberg, and J. C. Lawrence, Jr. 1994. PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science 266:653-656. [DOI] [PubMed] [Google Scholar]
- 101.Lorenz, M. C., and J. Heitman. 1995. TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J. Biol. Chem. 270:27531-27537. [DOI] [PubMed] [Google Scholar]
- 102.Magasanik, B. 1992. Regulation of nitrogen utilization, p. 283-317. In E. W. Jones, J. R. Pringle, and J. R. Broach (ed.), The molecular and cellular biology of the yeast Saccharomyces: gene expression, vol. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 103.Mahajan, P. B. 1994. Modulation of transcription of rRNA genes by rapamycin. Int. J. Immunopharmacol. 16:711-721. [DOI] [PubMed] [Google Scholar]
- 104.Mahalingam, M., and D. J. Templeton. 1996. Constitutive activation of S6 kinase by deletion of amino-terminal autoinhibitory and rapamycin sensitivity domains. Mol. Cell. Biol. 16:405-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Matsuura, A., M. Tsukada, Y. Wada, and Y. Ohsumi. 1997. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192:245-250. [DOI] [PubMed] [Google Scholar]
- 106.Menand, B., T. Desnos, L. Nussaume, F. Berger, D. Bouchez, C. Meyer, and C. Robaglia. 2002. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. USA 99:6422-6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Meyuhas, O., and E. Hornstein. 2000. Translational control of TOP mRNAs, p. 671-693. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 108.Miron, M., J. Verdu, P. E. Lachance, M. J. Birnbaum, P. F. Lasko, and N. Sonenberg. 2001. The translational inhibitor 4E-BP is an effector of PI(3)K/Akt signalling and cell growth in Drosophila. Nat. Cell Biol. 3:596-601. [DOI] [PubMed] [Google Scholar]
- 109.Murata, K., J. Wu, and D. L. Brautigan. 1997. B cell receptor-associated protein alpha4 displays rapamycin-sensitive binding directly to the catalytic subunit of protein phosphatase 2A. Proc. Natl. Acad. Sci. USA 94:10624-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nanahoshi, M., Y. Tsujishita, C. Tokunaga, S. Inui, N. Sakaguchi, K. Hara, and K. Yonezawa. 1999. Alpha4 protein as a common regulator of type 2A-related serine/threonine protein phosphatases. FEBS Lett. 446:108-112. [DOI] [PubMed] [Google Scholar]
- 111.Nasmyth, K. 1993. Control of the yeast cell cycle by the Cdc28 protein kinase. Curr. Opin. Cell Biol. 5:166-179. [DOI] [PubMed] [Google Scholar]
- 112.Nave, B. T., M. Ouwens, D. J. Withers, D. R. Alessi, and P. R. Shepherd. 1999. Mammalian target of rapamycin is a direct target for protein kinase B: identification of a convergence point for opposing effects of insulin and amino-acid deficiency on protein translation. Biochem. J. 344:427-431. [PMC free article] [PubMed] [Google Scholar]
- 113.Noda, T., and Y. Ohsumi. 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273:3963-3966. [DOI] [PubMed] [Google Scholar]
- 114.Oldham, S., J. Montagne, T. Radimerski, G. Thomas, and E. Hafen. 2000. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev. 14:2689-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pause, A., G. J. Belsham, A. C. Gingras, O. Donze, T. A. Lin, J. C. Lawrence, Jr., and N. Sonenberg. 1994. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371:762-767. [DOI] [PubMed] [Google Scholar]
- 116.Peng, T., T. R. Golub, and D. M. Sabatini. 2002. The immunosuppressant rapamycin mimics a starvation-like signal distinct from amino Acid and glucose deprivation. Mol. Cell. Biol. 22:5575-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Peterson, R. T., P. A. Beal, M. J. Comb, and S. L. Schreiber. 2000. FKBP12-rapamycin-associated protein (FRAP) autophosphorylates at serine 2481 under translationally repressive conditions. J. Biol. Chem. 275:7416-7423. [DOI] [PubMed] [Google Scholar]
- 118.Peterson, R. T., B. N. Desai, J. S. Hardwick, and S. L. Schreiber. 1999. Protein phosphatase 2A interacts with the 70-kDa S6 kinase and is activated by inhibition of FKBP12-rapamycinassociated protein. Proc. Natl. Acad. Sci. USA 96:4438-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Polymenis, M., and E. V. Schmidt. 1997. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev. 11:2522-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Poulin, F., A. C. Gingras, H. Olsen, S. Chevalier, and N. Sonenberg. 1998. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J. Biol. Chem. 273:14002-14007. [DOI] [PubMed] [Google Scholar]
- 121.Powers, T., and P. Walter. 1999. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 10:987-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Price, D. J., J. R. Grove, V. Calvo, J. Avruch, and B. E. Bierer. 1992. Rapamycin-induced inhibition of the 70-kilodalton S6 protein kinase. Science 257:973-977. [DOI] [PubMed] [Google Scholar]
- 123.Raught, B., A. C. Gingras, S. P. Gygi, H. Imataka, S. Morino, A. Gradi, R. Aebersold, and N. Sonenberg. 2000. Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J. 19:434-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Raught, B., A. C. Gingras, and N. Sonenberg. 2000. Regulation of ribosomal recruitment in eukaryotes, p. 245-293. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 125.Rohde, J., J. Heitman, and M. E. Cardenas. 2001. The TOR kinases link nutrient sensing to cell growth. J. Biol. Chem. 276:9583-9586. [DOI] [PubMed] [Google Scholar]
- 126.Sabatini, D. M., R. K. Barrow, S. Blackshaw, P. E. Burnett, M. M. Lai, M. E. Field, B. A. Bahr, J. Kirsch, H. Betz, and S. H. Snyder. 1999. Interaction of RAFT1 with gephyrin required for rapamycin-sensitive signaling. Science 284:1161-1164. [DOI] [PubMed] [Google Scholar]
- 127.Sabatini, D. M., H. Erdjument-Bromage, M. Lui, P. Tempst, and S. H. Snyder. 1994. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 78:35-43. [DOI] [PubMed] [Google Scholar]
- 128.Sabers, C. J., M. M. Martin, G. J. Brunn, J. M. Williams, F. J. Dumont, G. Wiederrecht, and R. T. Abraham. 1995. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J. Biol. Chem. 270:815-822. [DOI] [PubMed] [Google Scholar]
- 129.Sagiv, Y., A. Legesse-Miller, A. Porat, and Z. Elazar. 2000. GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. EMBO J. 19:1494-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schmelzle, T., and M. N. Hall. 2000. TOR, a central controller of cell growth. Cell 103:253-262. [DOI] [PubMed] [Google Scholar]
- 131.Schmidt, A., T. Beck, A. Koller, J. Kunz, and M. N. Hall. 1998. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 17:6924-6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schmidt, A., M. Bickle, T. Beck, and M. N. Hall. 1997. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell 88:531-542. [DOI] [PubMed] [Google Scholar]
- 133.Schmidt, A., and M. N. Hall. 1998. Signaling to the actin cytoskeleton. Annu. Rev. Cell Dev. Biol. 14:305-338. [DOI] [PubMed] [Google Scholar]
- 134.Schmidt, A., J. Kunz, and M. N. Hall. 1996. TOR2 is required for organization of the actin cytoskeleton in yeast. Proc. Natl. Acad. Sci. USA 93:13780-13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Schreiber, S. L. 1991. Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science 251:283-287. [DOI] [PubMed] [Google Scholar]
- 136.Schreiber, S. L. 1992. Immunophilin-sensitive protein phosphatase action in cell signaling pathways. Cell 70:365-368. [DOI] [PubMed] [Google Scholar]
- 137.Scott, P. H., G. J. Brunn, A. D. Kohn, R. A. Roth, and J. C. Lawrence, Jr. 1998. Evidence of insulin-stimulated phosphorylation and activation of the mammalian target of rapamycin mediated by a protein kinase B signaling pathway. Proc. Natl. Acad. Sci. USA 95:7772-7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sekito, T., Z. Liu, J. Thornton, and R. A. Butow. 2002. RTG-dependent mitochondria-to-nucleus signaling is regulated by MKS1 and is linked to formation of yeast prion [URE3]. Mol. Biol. Cell 13:795-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sekulic, A., C. C. Hudson, J. L. Homme, P. Yin, D. M. Otterness, L. M. Karnitz, and R. T. Abraham. 2000. A direct linkage between the phosphoinositide 3-kinase-AKT signaling pathway and the mammalian target of rapamycin in mitogen-stimulated and transformed cells. Cancer Res. 60:3504-3513. [PubMed] [Google Scholar]
- 140.Shamji, A. F., F. G. Kuruvilla, and S. L. Schreiber. 2000. Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr. Biol. 10:1574-1581. [DOI] [PubMed] [Google Scholar]
- 141.Shigemitsu, K., Y. Tsujishita, K. Hara, M. Nanahoshi, J. Avruch, and K. Yonezawa. 1999. Regulation of translational effectors by amino acid and mammalian target of rapamycin signaling pathways. Possible involvement of autophagy in cultured hepatoma cells. J. Biol. Chem. 274:1058-1065. [DOI] [PubMed] [Google Scholar]
- 142.Sigal, N. H., and F. J. Dumont. 1992. Cyclosporin A, FK-506, and rapamycin: pharmacologic probes of lymphocyte signal transduction. Annu. Rev. Immunol. 10:519-560. [DOI] [PubMed] [Google Scholar]
- 143.Smith, A., M. P. Ward, and S. Garrett. 1998. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 17:3556-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sophianopoulou, V., and G. Diallinas. 1995. Amino acid transporters of lower eukaryotes: regulation, structure and topogenesis. FEMS Microbiol. Rev. 16:53-75. [DOI] [PubMed] [Google Scholar]
- 145.Stan, R., M. M. McLaughlin, R. Cafferkey, R. K. Johnson, M. Rosenberg, and G. P. Livi. 1994. Interaction between FKBP12-rapamycin and TOR involves a conserved serine residue. J. Biol. Chem. 269:32027-32030. [PubMed] [Google Scholar]
- 146.Stanbrough, M., D. W. Rowen, and B. Magasanik. 1995. Role of the GATA factors Gln3p and Nil1p of Saccharomyces cerevisiae in the expression of nitrogen-regulated genes. Proc. Natl. Acad. Sci. USA 92:9450-9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Stromhaug, P. E., and D. J. Klionsky. 2001. Approaching the molecular mechanism of autophagy. Traffic 2:524-531. [DOI] [PubMed] [Google Scholar]
- 148.Taha, C., Z. Liu, J. Jin, H. Al-Hasani, N. Sonenberg, and A. Klip. 1999. Opposite translational control of GLUT1 and GLUT4 glucose transporter mRNAs in response to insulin. Role of mammalian target of rapamycin, protein kinase b, and phosphatidylinositol 3-kinase in GLUT1 mRNA translation. J. Biol. Chem. 274:33085-33091. [DOI] [PubMed] [Google Scholar]
- 149.Takeshige, K., M. Baba, S. Tsuboi, T. Noda, and Y. Ohsumi. 1992. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119:301-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tanida, I., E. Tanida-Miyake, T. Ueno, and E. Kominami. 2001. The human homolog of Saccharomyces cerevisiae Apg7p is a Protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J. Biol. Chem. 276:1701-1706. [DOI] [PubMed] [Google Scholar]
- 151.Thevelein, J. M., and J. H. de Winde. 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33:904-918. [DOI] [PubMed] [Google Scholar]
- 152.Thomas, G., and M. N. Hall. 1997. TOR signalling and control of cell growth. Curr. Opin. Cell Biol. 9:782-787. [DOI] [PubMed] [Google Scholar]
- 153.Tsukada, M., and Y. Ohsumi. 1993. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333:169-174. [DOI] [PubMed] [Google Scholar]
- 154.Tyers, M., G. Tokiwa, and B. Futcher. 1993. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 12:1955-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Vandenbol, M., J. C. Jauniaux, S. Vissers, and M. Grenson. 1987. Isolation of the NPR1 gene responsible for the reactivation of ammonia-sensitive amino-acid permeases in Saccharomyces cerevisiae. RNA analysis and gene dosage effects. Eur. J. Biochem. 164:607-612. [DOI] [PubMed] [Google Scholar]
- 156.Vezina, C., A. Kudelski, and S. N. Sehgal. 1975. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J. Antibiot. (Tokyo) 28:721-726. [DOI] [PubMed] [Google Scholar]
- 157.von Manteuffel, S. R., A. C. Gingras, X. F. Ming, N. Sonenberg, and G. Thomas. 1996. 4E-BP1 phosphorylation is mediated by the FRAP-p70s6k pathway and is independent of mitogen-activated protein kinase. Proc. Natl. Acad. Sci. USA 93:4076-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang, X., L. E. Campbell, C. M. Miller, and C. G. Proud. 1998. Amino acid availability regulates p70 S6 kinase and multiple translation factors. Biochem. J. 334:261-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang, X., W. Li, M. Williams, N. Terada, D. R. Alessi, and C. G. Proud. 2001. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 20:4370-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437-440. [DOI] [PubMed] [Google Scholar]
- 161.Weisman, R., and M. Choder. 2001. The fission yeast TOR homolog, tor1+, is required for the response to starvation and other stresses via a conserved serine. J. Biol. Chem. 276:7027-7032. [DOI] [PubMed] [Google Scholar]
- 162.Weisman, R., S. Finkelstein, and M. Choder. 2001. Rapamycin blocks sexual development in fission yeast through inhibition of the cellular function of an FKBP12 homolog. J. Biol. Chem. 276:24736-24742. [DOI] [PubMed] [Google Scholar]
- 163.Wen, Z., Z. Zhong, and J. E. Darnell, Jr. 1995. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82:241-250. [DOI] [PubMed] [Google Scholar]
- 164.Wiederrecht, G., L. Brizuela, K. Elliston, N. H. Sigal, and J. J. Siekierka. 1991. FKB1 encodes a nonessential FK 506-binding protein in Saccharomyces cerevisiae and contains regions suggesting homology to the cyclophilins. Proc. Natl. Acad. Sci. USA 88:1029-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Xu, G., G. Kwon, C. A. Marshall, T. A. Lin, J. C. Lawrence, Jr., and M. L. McDaniel. 1998. Branched-chain amino acids are essential in the regulation of PHAS-I and p70 S6 kinase by pancreatic beta-cells. A possible role in protein translation and mitogenic signaling. J. Biol. Chem. 273:28178-28184. [DOI] [PubMed] [Google Scholar]
- 166.Yaglom, J., M. H. Linskens, S. Sadis, D. M. Rubin, B. Futcher, and D. Finley. 1995. p34Cdc28-mediated control of Cln3 cyclin degradation. Mol. Cell. Biol. 15:731-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yokogami, K., S. Wakisaka, J. Avruch, and S. A. Reeves. 2000. Serine phosphorylation and maximal activation of STAT3 during ciliary neurotrophic factor signaling is mediated by the rapamycin target mTOR. Curr. Biol. 10:47-50. [DOI] [PubMed] [Google Scholar]
- 168.Zaragoza, D., A. Ghavidel, J. Heitman, and M. C. Schultz. 1998. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol. Cell. Biol. 18:4463-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang, H., J. P. Stallock, J. C. Ng, C. Reinhard, and T. P. Neufeld. 2000. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 14:2712-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhao, C., U. S. Jung, P. Garrett-Engele, T. Roe, M. S. Cyert, and D. E. Levin. 1998. Temperature-induced expression of yeast FKS2 is under the dual control of protein kinase C and calcineurin. Mol. Cell. Biol. 18:1013-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zheng, X. F., D. Florentino, J. Chen, G. R. Crabtree, and S. L. Schreiber. 1995. TOR kinase domains are required for two distinct functions, only one of which is inhibited by rapamycin. Cell 82:121-130. [DOI] [PubMed] [Google Scholar]