Abstract
Research interest in microbial biodiversity over the past 25 years has increased markedly as microbiologists have become interested in the significance of biodiversity for ecological processes and as the industrial, medical, and agricultural applications of this diversity have evolved. One major challenge for studies of microbial habitats is how to account for the diversity of extremely large and heterogeneous populations with samples that represent only a very small fraction of these populations. This review presents an analysis of the way in which the field of microbial biodiversity has exploited sampling, experimental design, and the process of hypothesis testing to meet this challenge. This review is based on a systematic analysis of 753 publications randomly sampled from the primary scientific literature from 1975 to 1999 concerning the microbial biodiversity of eight habitats related to water, soil, plants, and food. These publications illustrate a dominant and growing interest in questions concerning the effect of specific environmental factors on microbial biodiversity, the spatial and temporal heterogeneity of this biodiversity, and quantitative measures of population structure for most of the habitats covered here. Nevertheless, our analysis reveals that descriptions of sampling strategies or other information concerning the representativeness of the sample are often missing from publications, that there is very limited use of statistical tests of hypotheses, and that only a very few publications report the results of multiple independent tests of hypotheses. Examples are cited of different approaches and constraints to experimental design and hypothesis testing in studies of microbial biodiversity. To prompt a more rigorous approach to unambiguous evaluation of the impact of microbial biodiversity on ecological processes, we present guidelines for reporting information about experimental design, sampling strategies, and analyses of results in publications concerning microbial biodiversity.
INTRODUCTION
In the mid-1900s, the provocative publications of R. H. MacArthur and G. E. Hutchinson (115, 165, 166) spurred the field of ecology into intense research and debate about the significance of biodiversity. These and other workers claimed that biodiversity is a measure of important ecological processes such as resource partitioning, competition, succession, and community productivity and is also an indicator of community stability. The bulk of this new wave of biodiversity research concerned plant and animal communities. In the 1960s, following in the footsteps of plant and animal ecologists, microbiologists began investigating the impact of biodiversity on the function and structure of microbial communities (97, 257). These questions served to intensify interest in biodiversity, a concept that has been a longstanding foundation of microbiology. Genetic variation among individuals within a population has long been recognized as the starting block for adaptation and evolution among microorganisms as well as among other organisms. Likewise, the consequences of phenotypic variability for the accuracy of disease diagnosis and for establishing taxonomic relationships among microorganisms have been well studied. Following the new research wave of the 1960s, interest in microbial biodiversity has been further bolstered by (i) creation of the Diversitas international research program in 1991 to promote scientific investigations into the origins and conservation of biodiversity and the impact of biodiversity on ecological functions, (ii) the Biodiversity Treaty that issued from the United Nations Conference on Environment and Development in 1992 in Rio de Janeiro, Brazil, and (iii) subsequent initiatives launched by science foundations, scientific societies, and research institutions in a wide range of countries. Microbial biodiversity has also received particular attention in areas where industrial applications are evident—such as for marine, medical, and food biotechnology—and where microbial activity has important implications for Earth's climate and for the bioremediation of polluted sites (46). Nevertheless, in spite of the research devoted to microbial biodiversity and to biodiversity in general, the consequences of biodiversity on the ecological processes cited above are still the object of debate and analysis (263, 264, 290).
If one takes a superficial glance at the study of biodiversity, it seems to be largely a descriptive endeavor: trap, identify, and count. However, as suggested above, the motivation behind these studies arises from specific hypotheses about the nature of biodiversity and its impact on ecological processes. In some studies, the hypotheses are explicit. For example, Hariston et al. (97) sought to test the hypothesis that increasing biodiversity is positively correlated with increasing community stability. Likewise, Kaneko and Atlas (130) focused on the hypothesis that biodiversity is correlated with the density of populations of bacteria in ice, sediment, and water from the Beaufort Sea. In other studies, the hypotheses are implicit, such as for a census of species or groups in a community or for studies of techniques for characterizing biodiversity. The hypotheses in these cases concern the accuracy of the census, the efficiency and biases of the techniques, and, above all, the notion that the observations made are not artifacts.
Tests of such hypotheses are based on demonstrating that one's observations are not due to random error or to factors other than those evoked in the hypothesis. In general, this involves three precautions: (i) experimental design and sampling procedures to reduce the influence of unwanted factors on the resulting observations, (ii) statistical tests to eliminate biases in the judgment of the observer, and (iii) multiple independent tests of a hypothesis to reduce the possibility that random error is the cause of the results observed. The process of hypothesis testing presents some important challenges for the field of microbial biodiversity. The foremost of these is the problem of constituting a sample. How can samples account for the diversity of extremely large and heterogeneous microbial populations when they represent only a very small fraction of these populations or when the methods used have very low resolution? Ecologists studying the diversity of macroscopic organisms have produced a large body of literature devoted to sampling strategies (45, 102, 167, 168, 210). This literature testifies to the important impact that intrinsic properties of a population, such as spatial aggregation, rates of immigration, birth, and death, and the relative frequency of rare species, have on the extent to which the sample represents the population. Unfortunately, microbial ecologists rarely have a priori knowledge about these properties of the populations they study. Furthermore, the work reviewed by Swift (257) concerning the biodiversity of fungi during successional sequences in communities of decomposers provides an early illustration of the wide range of sources of variability that can be encountered on small scales in microbial biodiversity studies. Hence, the initial steps of designing studies of microbial diversity could require considerable reflection and preliminary investigations. A second obvious challenge that one could expect for microbial biodiversity studies resides in multiple, independent tests of hypotheses. This is a challenge because the complexity of identifying microorganisms, and bacteria in particular, can lead to considerable investment in time and labor. As a consequence, the number of strains or samples that can be analyzed is sometimes below the minimal number needed for a single unambiguous test of the hypothesis, and additional tests may be prohibitive.
The field of microbial biodiversity has grown markedly since the Diversitas initiative in 1991 and has resulted in a large body of scientific literature. Through this literature, we have witnessed the development of techniques for characterizing diversity, in particular at the molecular level for both culturable and nonculturable microorganisms (223, 262). Furthermore, this literature has also contributed to a general consensus that the microbial world is much more diverse than we can appreciate at present. However, the abundance of literature published in the field of microbial diversity does not seem proportional to our understanding of the significance of biodiversity for ecological processes in the microbial world or of the ways in which we can manipulate or manage this diversity. The authors of this review wondered about a potential cause of this context. What types of questions—or hypotheses—are being addressed in the field of microbial biodiversity, and how have these hypotheses been tested? Our objective is to elucidate the cause of this context in practical terms: how has the field of microbial biodiversity employed sampling, experimental design, and, ultimately, the process of testing hypotheses over the past 25 years?
To review the approaches used for sampling, experimental design, and hypothesis testing in studies of microbial biodiversity we have, as a first step, traced the growth of research interest in this field for the past 25 years for a wide array of habitats including aquatic systems, soil and rhizosphere systems, mycorrhizae, the phyllosphere, food products, and food-processing factories. These habitats represent the fields of competence of the authors. We established a database of over 2,000 relevant publications and systematically characterized a randomly sampled subset as described in the first part of this review. The second part of the review is dedicated to illustrating the trends in the themes of research concerning the biodiversity of these habitats. This part of the analysis allowed us to identify eight principal themes that have been the main focus of microbial biodiversity studies over the past 25 years. Each theme was then used as a point of reference for analyzing how experimental design and hypothesis testing were treated in each publication. The third part of the review presents our critical analysis of how experimental design and hypothesis testing have been exploited in microbial biodiversity studies for each of the themes identified. We conclude with a series of guidelines for information that should be specified in microbial diversity publications with regard to experimental design and sampling strategies. These guidelines are intended to enhance the contribution of biodiversity studies to elucidating the significance of microbial biodiversity in ecological processes.
METHOD FOR ANALYZING THE SCIENTIFIC LITERATURE
Establishing the Database
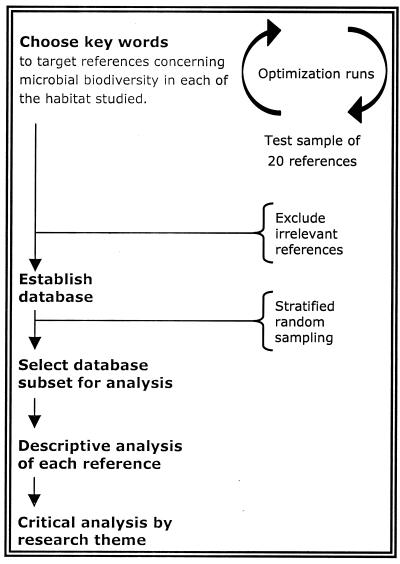
This review is based on studies published in the primary literature concerning eight types of microbial habitats: (i) aquatic systems (continental and marine), (ii) soil, (iii) the rhizosphere, (iv) mycorrhizae, (v) fungus-plant pathosystems, (vi) bacterium-plant systems, (vii) food, and (viii) food-processing factories. For soil and rhizosphere systems, we considered only symbiotic or nonpathogenic bacteria. Plant pathogens were included in the fungus-plant pathosystems and bacterium-plant systems. The latter system also included nonpathogenic bacteria associated with aerial plant parts, i.e., epiphytic bacteria in the phyllosphere. Literature searches were conducted, as illustrated in Fig. 1, using at least one of five bibliographic databases for the years 1975 to 1999: CAB, MedLine, Scifinder, PASCAL, and Food Science and Technology Abstracts. Each database was searched for the scientific fields for which it was relevant: CAB was interrogated for fungus-plant pathosystems, bacterium-plant systems, mycorrhizae, and food systems; MedLine was searched for food and aquatic systems; Scifinder was searched for soil and rhizosphere systems; PASCAL was searched for soil systems; and Food Science and Technology Abstracts was searched for food-processing factory systems. Trial searches were conducted to establish a set of consensus terms and phrases to be used for all habitats. The consensus terms and phrases were diversity, population structure, community structure, variability, dominance, and numerical taxonomy. References in the CAB, MedLine, Scifinder, and PASCAL databases with at least one of these terms in the title, abstract, or key words were retained for further consideration. However, most of these terms and phrases were not appropriate for the Food Science and Technology Abstracts database. This database was searched based on the words diversity, contamination, incidence, prevalence, occurrence, presence, characteristics, and characterization. References were then further selected based on the presence of key words relevant to each habitat. For each habitat, we also established lists of terms and phrases used to exclude the majority of irrelevant references. To exclude the irrelevant references remaining after this step, the database was then screened manually based on titles and contents of abstracts. The references were compiled in EndNote (version 3.0; Niles Software, Inc., Berkeley, Calif.), and duplicates were eliminated. Our analysis of trends is based on the 2,200 references compiled. The database of references may be obtained by contacting the corresponding author.
FIG. 1.
Procedure for establishing and analyzing a database of publications from the primary scientific literature from 1975 to 1999 concerning microbial biodiversity.
Sampling and Descriptive Analysis of Publications
From the database of 2,200 references, 753 publications were randomly selected for analysis. This was accomplished by selecting about 100 of the total publications for each habitat according to random-number tables. Sampling was stratified over the years of publication so that each year was represented in the sample in the same proportion that it was represented in the database. Hence, between 12 and 56% of the publications were analyzed for all habitats except foods and food-processing factories. For these latter fields, all 40 and 39 references, respectively, in the database were analyzed.
For each habitat, the randomly selected publications were characterized according to 17 criteria describing (i) the origin (culture collections, direct environmental samples, etc.) of the microorganisms or microbial nucleic acids studied and the overall approach to collecting samples, (ii) the precise parameters of the reported sampling strategy (use of random sampling protocols; size, weight, and volume of samples taken; and the number of individuals characterized), (iii) the microbiological and molecular biological techniques used to characterize diversity (Table 1), (iv) the calculation of diversity indices and the use of statistics to test the hypotheses evoked concerning microbial diversity, and (v) the use of replicated tests of the principal hypotheses evoked. Furthermore, the principal themes of each study were identified. For the publications analyzed, eight different themes were identified (Table 2). Most publications had only one principal theme, but a few papers had two (27%) or three (3%) principal themes. A descriptive quantitative analysis of the database was conducted to determine the relationships among the different parameters characterizing the publications (cooccurrence of the characteristics described, changes in occurrence with time, etc.). This analysis was based on procedures written in Scilab (http://www-rocq.inria.fr/scilab), a free general-purpose scientific software.
TABLE 1.
Techniques employed for characterizing microbial biodiversity in studies published from 1975 to 1999
| Techniques based on analysis of: | Specific techniques employeda |
|---|---|
| Metabolic properties | API; BIOLOG; presence of certain enzymes; classical phenotypic tests; sensitivity to phages |
| Resistance to antimicrobial compounds | Profiles of resistance to biocides including antibiotics, bacteriocins, pesticides, heavy metals, and polyaromatic hydrocarbons |
| Pathogenicity, pathogenicity-like processes, | Pathogenicity to plants or animals and type of symptoms; capacity for symbiosis; potential |
| and production of toxins | to infect or presence of markers of virulence including cytotoxicity or toxin production |
| Colony and cellular morphology | Morphotype; colony type; cell type |
| Markers of other specific phenotypic properties | Protein profiles; characterization of lipopolysaccharides, antigens, fatty acids, isozymes, and opines |
| Growth on specific media | Growth on selective, semiselective, or differential media |
| Other diverse phenotypic properties | Ice nucleation activity, vegetative compatibility types, mating types, antagonistic activity, etc. |
| Total DNA | RFLP without probes; RFLP with anonymous probes; rep-PCR; AFLP; DAF; microsatellite analysis; PFGE; DNA-DNA hybridization; ASAP; RAPD |
| Targeted DNA sequences | Plasmid profiles; FISH; RFLP with specific probes; hybridization of specific probes; analysis of sequences of ribosomic genes; PCR polymorphisms; PCR-RFLP; ITS polymorphisms; DGGE; ARDRA |
| Other types of genotypic profiles | Genotyping techniques not listed above |
rep-PCR, repetitive-sequence based PCR; AFLP, amplification fragment length polymorphism; DAF, DNA amplification fingerprinting; PFGE, pulsed-field gel electrophoresis; ASAP, arbitrary signature from amplification profiles; RAPD, randomly amplified polymorphic DNA; FISH, fluorescent in situ hybridization; ITS, internal transcribed spacer; DGGE, denaturing gradient gel electrophoresis; ARDRA, amplified R-DNA restriction analysis.
TABLE 2.
Synopsis of the objectives of publications in the primary literature on microbial biodiversity from 1975 to 1999 concerning eight different microbial habitats associated with water, soil, plants, foods, and food-processing factories
| Themea | Objectiveb | Reference(s)c |
|---|---|---|
| Effect: Effect of a defined factor (other than space or time) on microbial diversity | Determine if the following factors or practices influence biodiversity of soil-borne microorganisms: | |
| Application of pesticides | 7, 71, 103, 106, 128, 133, 173, 188, 202, 233 | |
| Chitin amendments | 93 | |
| Compaction | 118 | |
| Crop residues | 194, 236, 283 | |
| Disturbances (pollution, fire, clear cutting, logging) | 28, 52, 60, 64, 91, 123, 127, 138, 181, 225, 250 | |
| Heavy metals | 54, 129, 134, 145, 164, 199, 201, 234, 251, 260, 296 | |
| Introduction of genetically modified microorganisms | 50, 169, 191, 192, 193, 222 | |
| Liming | 1, 8, 59, 69 | |
| Physico-chemical properties of the soil | 135, 211 | |
| Presence and age of specific plant species | 19, 75, 76, 86, 95, 126, 149, 154, 157, 214, 242, 256, 272, 295 | |
| Rotation | 278 | |
| Soil fumigation | 267 | |
| Determine if the following factors influence the biodiversity of microorganisms in aquatic systems: | ||
| Anthropogenic factors (pollutants) | 155 | |
| Natural environmental changes (temperature, salinity, etc.) | 110 | |
| Determine the effect of pesticide application on the variability of pesticide resistance types in a population of plant pathogens | 48, 207 | |
| Determine the effect of characteristics of the host plant population on the pathotype or other phenotypic diversity of plant pathogens | 40, 142, 156, 200 | |
| Dynamics: Spatial or temporal dynamics of the composition or structure of microbial populations or communities | Evaluate: | |
| The effect of location and soil type on the diversity of functional groups of bacteria (oligotrophs, nitrifiers, denitrifiers, cellulose decomposers, and phototrophic, acid-tolerant, or mercury-resistant bacteria) | 111, 144, 203, 204, 247 | |
| Horizontal and vertical variability in biodiversity of aquatic bacteria in water columns | 81, 82 | |
| Regional or within-field variation in composition and structure of plant pathogen populations | 25, 72, 132, 216, 276 | |
| Seasonal variation in diversity of plant-associated or other environmental bacteria | 23, 218, 284 | |
| Changes in composition and structure of plant pathogen populations over time | 40, 151, 180, 293 | |
| Short- and long-term dynamics in planktonic and biofilm bacteria in aquatic systems | 27, 89, 176 | |
| Changes in composition of human pathogen populations in food-processing plants | 66 | |
| Source: Analysis of the structure or composition of microbial populations for the purpose of identifying a source of contamination or infection | Identify the source of human pathogens in food-processing plants | 9, 12, 24, 206, 229, 249 |
| Identify the source of new plant disease outbreaks | 25, 40, 100, 136, 153, 212 | |
| Describe: Evaluation of the composition or structure of a microbial population or community independent of time and specific geographic location | Describe | |
| Pathotypes, races, degrees of aggressiveness, and sensitivity to fungicides in populations of plant-pathogenic fungi | 44, 175, 184, 189, 226 | |
| Vegetative incompatibility groups in populations of mycorrhizal fungi and in plant-pathogenic fungi | 20, 141 | |
| Different phenotypes related to the process of mycorrhization among mycorrhizal fungi | 143, 248, 286 | |
| The variability of rhizobia in nodulating, in fixing nitrogen, and in affecting gene transfer within a population | 31, 78, 84, 148, 185, 187, 219, 237, 255, 268 | |
| Species composition of aquatic bacterial communities based on culture-independent methods | 15, 29, 190, 245 | |
| Relatedness: Phylogenetic or taxonomic analysis of a given group of microorganisms. | Evaluate phylogenetic relationships among uncultured aquatic bacteria | 29, 70, 80, 186, 217, 282 |
| Evaluate the taxonomic variability among plant-pathogenic bacteria and bacteria important in food quality | 16, 18, 55, 63, 73, 101, 291 | |
| Markers: Search for markers potentially useful for diagnosis and identification. | Develop markers: | |
| Of taxonomic identity | 53, 57, 116 | |
| For tracing specific bacteria in ecological studies | 270 | |
| Of pathogenicity and host range of plant-pathogenic microorganisms | 18, 22, 112, 246 | |
| Of other functions | 77, 182, 197, 224 | |
| Methods: Evaluation of methods applicable to the characterization of microbial biodiversity. | Evaluate laboratory methods for characterizing microbial diversity | Studies of methods based on total DNA or targeted DNA sequences are summarized by Theron and Cloete (262) |
| Determine the effect of experimental design and sampling procedures on measures of microbial biodiversity | 6, 30, 74, 122 | |
| New: Search for new species or new taxa. | Determine if an environmental sample contains previously undescribed: | |
| Aquatic bacteria | 232, 235 | |
| Uncultured soil-borne bacteria | 26, 160, 220, 269, 294 | |
| Culturable soil-borne bacteria | 41, 43, 60, 87, 107, 137, 147, 161, 209, 256, 281 |
Each theme is given an acronym (in italics) that is used throughout this paper.
The objectives are listed by theme. The list is not exhaustive.
The publications represent examples which cover the major trends in the 753 publications analyzed in this work.
Critical Analysis
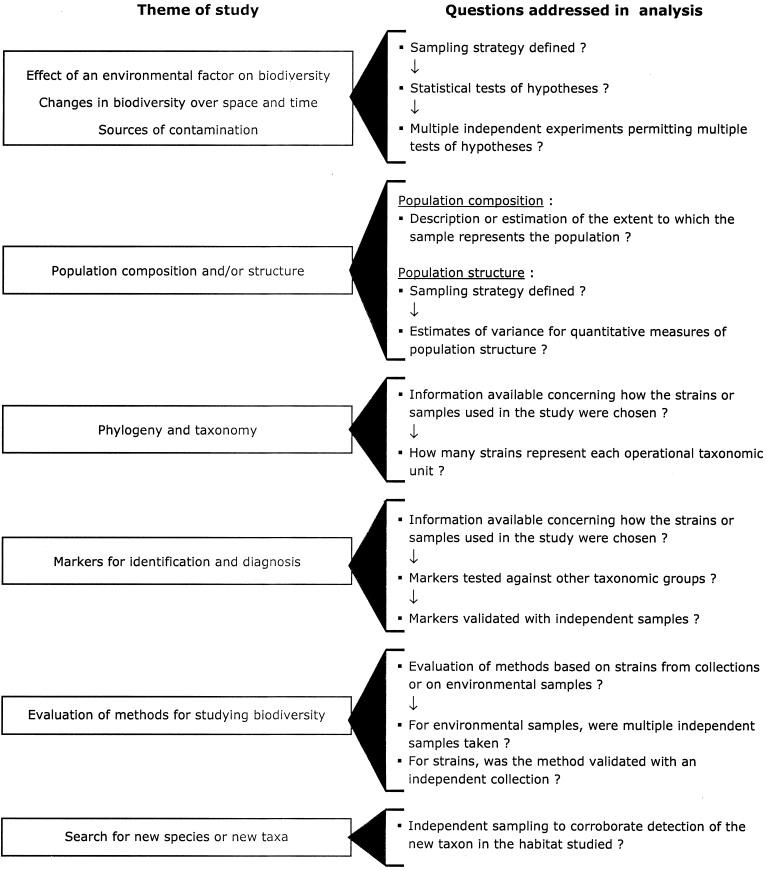
For each of the eight themes identified in our analysis, we constructed a stepwise procedure to evaluate the approaches taken in addressing the principal objectives of the publications. This procedure took into account the overall experimental approach that we would expect for studies under each of the themes. For example, for some of the themes we expected to find specific information in the publication concerning the sampling strategy and statistical tests used as well as descriptions of multiple independent experiments relative to the central hypotheses. The questions we addressed concerning the experimental approaches used for each theme are presented in Fig. 2. Further details concerning these questions are presented below.
FIG. 2.
Procedure for analyzing the role of experimental design and hypothesis testing for eight different themes of research concerning microbial biodiversity. The themes are described in detail in Table 2.
DYNAMICS OF RESEARCH INTEREST IN MICROBIAL BIODIVERSITY SINCE 1975
Dynamics of Publication Rates
For the fields surveyed, the number of publications concerning microbial biodiversity in the primary scientific literature showed a marked increase in the early 1990s (Fig. 3A). This increase was particularly striking for fields concerning plant-pathogenic fungi and aquatic, soil, and rhizosphere systems (including mycorrhizae) (Fig. 3B). For systems implicating food industries, interest in microbial biodiversity did not develop until the 1990s and was prompted by food safety issues and the establishment of hazard analysis protocols (120). The overall increase in the number of publications relevant to microbial biodiversity does not simply reflect the increase in the total number of studies published by the 525 different journals covered in this census. For the 10 journals publishing most frequently in the field of microbial biodiversity (Table 3), the percentage of studies devoted to microbial biodiversity among all the studies published also showed a marked increase in the early 1990s. For example, the publication rate of microbial biodiversity studies increased from only 0.5% of the 589 articles published in 1988 by Applied and Environmental Microbiology to 6% of the 876 studies published by this journal in 1999. Overall, Applied and Environmental Microbiology published the greatest number of studies concerning microbial biodiversity in general (Table 3), as well as for aquatic, soil, and rhizosphere habitats, and was the second most frequent publisher of studies concerning bacterium-plant systems. However, studies concerning the biodiversity of fungus-plant pathosystems were published primarily in Phytopathology and in Mycological Research, those concerning mycorrhizae were published primarily in New Phytologist and Canadian Journal of Botany, and those concerning foods and food industries were published primarily in Journal of Food Protection, Journal of Applied Bacteriology, and International Journal of Food Microbiology.
FIG. 3.
Rate of publication in the primary literature from 1975 to 1999 of studies concerning the biodiversity of eight microbial habitats. (A) Total number of publications per year; (B) number of publications per year concerning fungus-plant pathosystems (◊), the rhizosphere (including mycorrhizae) (▴), microbial habitats in soil (▵), aquatic systems (⧫), bacterium-plant system (---), and the microbiology of food and food-processing factories (▪).
TABLE 3.
Number of publications concerning the biodiversity of microorganisms (bacteria and fungi) in eight different microbial habitats associated with water, soil, plants, foods, and food-processing factories published from 1975 to 1999 by the 10 journals publishing these types of studies most frequently
| Journal | No. of publications |
|---|---|
| Applied and Environmental Microbiology | 263 |
| Phytopathology | 125 |
| Mycological Research (Transactions of the British Mycological Society before 1989) | 89 |
| Soil Biology and Biochemistry | 70 |
| Canadian Journal of Botany | 63 |
| FEMS Microbiology Ecology (FEMS Microbiology Letters before 1985) | 47 |
| Plant Disease (Plant Disease Reporter before 1979) | 45 |
| Plant Pathology | 44 |
| Canadian Journal of Microbiology | 41 |
| Indian Phytopathology | 32 |
Major Research Themes and Objectives
The classification of publications according to themes (Table 2) was an essential step in the elaboration of this review since it permitted us to clearly develop the analysis of approaches used for experimental design and hypothesis testing. We present these themes not only to provide an overview of the types of questions preoccupying the field of microbial biodiversity but also to give insight into how we have classified the studies analyzed here. Each theme has been accorded an acronym to facilitate reference to the theme (Table 2).
Composition and structure of microbial populations and communities.
The description of the composition and structure of microbial populations and communities is an important starting point in studies of microbial biodiversity and sets the stage for fundamental studies concerning how these populations and communities function. Hence, it is not surprising that the basic description of community composition or structure (theme Describe) coupled with the impact of space and time on these parameters (theme Dynamics) was the central theme of over half of the publications analyzed here and was relevant to all habitats surveyed (Table 4). Although descriptive studies (theme Describe) have been dominant in the microbial biodiversity literature considered as a whole, publication of such studies has declined over time whereas reports of spatial and temporal dynamics of microbial biodiversity (theme Dynamics) have become more abundant (Fig. 4).
TABLE 4.
Distribution of publications among the principal themes of studies of microbial biodiversity in the primary literature from 1975 to 1999 concerning eight different microbial habitats associated with water, soil, plants, foods, and food-processing factories
| Themea | % of publications for each habitatb:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Aq | Soil | Rh | Myc | FPP | BP | Food | FI | All systems | |
| Effect | 10 | 52 | 56 | 44 | 15 | 11 | 15 | 3 | 30 |
| Dynamics | 37 | 32 | 25 | 12 | 28 | 20 | 48 | 20 | 26 |
| Source | 0 | 1 | 0 | 1 | 4 | 7 | 10 | 79 | 6 |
| Describe | 18 | 20 | 32 | 24 | 53 | 13 | 15 | 13 | 26 |
| Relatedness | 46 | 8 | 19 | 0 | 7 | 24 | 45 | 0 | 18 |
| Markers | 3 | 4 | 2 | 12 | 4 | 38 | 2 | 0 | 9 |
| Methods | 18 | 26 | 14 | 7 | 7 | 4 | 5 | 13 | 12 |
| New | 5 | 12 | 6 | 0 | 0 | 1 | 0 | 0 | 4 |
| Total no. of publications analyzed | 110 | 116 | 124 | 110 | 109 | 105 | 40 | 39 | 753 |
| Total no. in database | 198 | 413 | 248 | 206 | 867 | 190 | 40 | 39 | 2200 |
The theme corresponding to each acronym is described in Table 2.
For a given habitat, the sum of the percentages of articles published across all themes may be greater than 100% because several principal themes were attributed to some of the articles. The habitats covered in this study were aquatic systems (Aq), soil (Soil), the rhizosphere (Rh), mycorrhizae (Myc), fungus-plant pathosystems (FPP), bacterium-plant systems (BP), food (Food), and food-processing factories (FI).
FIG. 4.
Changes over time in the frequency of publications addressing each of eight different research themes in microbial biodiversity from 1975 to 1999. The themes Effect (⧫), Dynamics (◊), Source (▴), Describe (▵), Relatedness (▪), Markers (□), Methods (+), and New (×) are described in Table 2. Each value represents the percentage of publications addressing a given theme among the total publications for that period. The periods depicted were chosen to represent approximately equal numbers of publications per period.
Microbial populations and communities have been described in terms of a wide range of phenotypic traits, many of which are related to the practical interest in the habitat studied, as listed in Table 2. Numerous studies have also exploited molecular characterization of nucleic acids in order to describe microorganisms; examples of these are cited throughout this review. In aquatic systems in particular, an important set of studies related to the theme Describe has concerned exploratory investigations using culture-independent methods (clone libraries) to determine the species composition of bacterial communities (Table 2).
The impact of time and space on population composition or structure (theme Dynamics) has been investigated to understand the origin and spread of populations of beneficial and of deleterious microorganisms or the probable relationship among organisms from different geographical locations. For systems concerning plant or human pathogens or bacteria involved in food spoilage, these questions have been linked to an interest in disease epidemiology, in the efficiency and durability of practices for control of crop and food losses, and in food safety issues (Table 2). For aquatic systems, studies under this theme have concerned vertical and horizontal spatial variability at a range of scales and short-term as well as seasonal dynamics (Table 2).
Impact of environmental factors on microbial biodiversity.
The effect of environmental factors on microbial diversity (theme Effect) has been a major theme of study for soil and rhizosphere microbial systems and for mycorrhizae (Table 4). Interest in this theme has increased markedly over the past 25 years (Fig. 4). Studies under this theme are apparently motivated by the possibility of modifying the factor of interest via soil or forest management practices, for example. For rhizosphere and soil communities, there has been considerable interest in the relationship between microbial biodiversity and plant health, plant productivity, and the efficiency of ecological processes such as nutrient cycling and phytoremediation (Table 2). For mycorrhizal fungi, interest in their biodiversity arises from the observations that these fungi play a major role in the floristic diversity and structure of plant communities (273).
Studies under this theme relative to soil, rhizosphere, and mycorrhizal systems have addressed the impact of two principal factors on microbial diversity: the properties of the soil and the nature of the plant species. The soil properties studied have been overwhelmingly those related to agricultural and other land management practices and to pollution (Table 2). In studies of aquatic habitats, only a few papers concern the effect of specific factors on the structure of bacterial communities. They are related to the effect of anthropogenic disturbances (mainly pollutants) and to the response of natural communities to natural environmental changes (temperature, salinity, etc.) (Table 2).
Microbial biodiversity as an epidemiological tool.
For all habitats considered together, relatively few publications have clearly linked the concepts of epidemiology and microbial biodiversity (Table 4; Fig. 4). However, studies of microbial biodiversity addressing epidemiological questions (theme Source) have been of interest for habitats concerning human pathogens and occasionally for those containing plant pathogens. Studies under this theme have compared phenotypic and genotypic profiles of strains from different origins as a means of determining possible sources of contamination of foods or sources of outbreaks of plant disease (Table 2).
Markers to facilitate diagnosis and identification.
Characterization of microbial biodiversity as a means of discerning markers useful for diagnosis or microbial identification (theme Markers) has been a major theme of studies concerning plant-associated bacteria (Table 4), where the development of such markers is a matter of particular interest for tracing bacteria in ecological studies and for avoiding the need for time-consuming tests of pathogenicity and host range (Table 2). Markers have also been sought for other types of microorganisms as a means of rapidly identifying phenotypes whose laboratory characterization is laborious, such as the efficiency of mycorrhization, or as a means of facilitating the identification of organisms that are complicated to identify, such as certain mycorrhizal fungi (Table 2).
Methods for studying microbial biodiversity.
Interest in methods for studying microbial biodiversity (theme Methods) has grown steadily since 1975 (Fig. 4). However, among the 94 publications analyzed that evaluated methods applicable to the characterization of microbial biodiversity, 90 concerned the evaluation of laboratory methods for characterizing microorganisms and over half of these focused on evaluating methods for the analysis of total DNA or targeted DNA sequences in a sample. Only four studies focused on the effect of experimental designs or sampling procedures on measures of biodiversity (Table 2). Nevertheless, methodological information was presented in other studies that we analyzed, but to a much more limited extent than in the four publications just mentioned, and it was not the principal objective of the publications.
Phylogenetic and taxonomic studies.
Biodiversity studies motivated by phylogenetic or taxonomic comparisons of a given group of microorganisms (theme Relatedness) have focused primarily on bacteria and reflect the historical tribulations surrounding the definition of bacterial species and the relatedness among individuals of different genotypes. In our database, the frequency of publications under this theme decreased with time (Fig. 4). Since interest in microbial phylogeny probably is not waning, this trend likely reflects the fact that the key words used for our bibliographic searches are employed less and less frequently in publications addressing microbial phylogeny and taxonomy. Nearly half of the biodiversity studies concerning aquatic systems were relevant to the theme Relatedness (Table 4). An important motivation for studies of this habitat is that less than 1% of aquatic picoplankton—the organisms that constitute the fundamental basis for the functioning of these systems—can be cultured. Hence, there is a prevailing interest in describing organisms that were inaccessible prior to the advent of culture-independent techniques. Culture-independent direct retrieval of 16S rDNA was used extensively in the early 1990s to characterize unknown and uncultured species in aquatic systems (Table 2). Most of these studies provide sequences of the isolated 16S rDNA and localize these “phylotypes” in the tree of life. For bacteria that are pathogenic to plants or animals, studies under the theme Relatedness have been motivated by very practical concerns. The taxonomic variability of causal agents of plant and animal diseases has been investigated because it is intimately linked to questions concerning epidemiology (disease origin) and can be useful in subsequent identification of markers for detection (Table 2).
Discovery of new taxa.
The search for new species or taxa (theme New) was a relatively rare theme (Table 4), which has increased slightly in importance over the past few years (Fig. 4). For aquatic systems, studies under this theme generally were derived from phylogenetic or taxonomic studies (theme Relatedness) and focused on the occurrence of new species in a few samples with the aim of finding new DNA sequences (Table 2). For soil and rhizosphere systems, studies under this theme have sought to demonstrate the high degree of bacterial diversity and the presence of nonculturable new species by using direct soil extraction followed by molecular phylogenetic analysis (Table 2). However, the search for new species or taxa in soil and rhizosphere systems has also addressed culturable bacteria (Table 2).
Methods Used for Characterizing Biodiversity
To describe how the different techniques presented in Table 1 were used in biodiversity studies over the last 25 years, we calculated the frequency at which these techniques were reported and their coincidence with the different themes (Table 5). For all publications considered as a whole, and for many of the habitats or themes considered independently, DNA-based characterization techniques—and in particular those based on targeted DNA sequences such as 16S rDNA, specific repeated sequences, or genes for virulence of pathogens—had the most dominant role in biodiversity studies relative to all other types of techniques (Table 5). Furthermore, DNA-based characterization techniques had the greatest growth rate over the past 25 years compared to all other types of techniques for characterizing microbial diversity. The percentage of published articles that reported the use of DNA-based characterization techniques was 9% in the period from 1975 to 1988, 27% from 1989 to 1993, 35% from 1994 to 1995, and 40 to 50% thereafter. Nevertheless, in spite of the important role of DNA-based characterization techniques, tests based on the characterization of phenotypes played a predominant role for four of the habitats. Characterization of microorganisms based on their metabolism, morphology, or ability to grow on selective media were the predominant approaches in studies of soil, mycorrhizae, food, and food-processing factories. The role of phenotypic tests for these systems is related to the frequent use of the BIOLOG system for characterizing soil microorganisms, the ease of characterizing certain mycorrhizal fungi by their morphological features, and the importance of standardized culture methods for food-borne pathogens.
TABLE 5.
Percentage of articles published from 1975 to 1999 reporting the results of studies of microbial biodiversity based on analysis of total or targeted DNA sequences or on characterization of phenotypic properties
| Techniques based on analysis ofb: | % of articles for themea:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| New | Methods | Relatedness | Dynamics | Markers | Describe | Source | Effect | |
| Targeted or total DNA | 54 | 52 | 45 | 36 | 35 | 32 | 24 | 17 |
| Metabolic properties | 17 | 16 | 18 | 18 | 17 | 14 | 26 | 25 |
| Markers of specific phenotypic properties | 7 | 12 | 14 | 12 | 17 | 11 | 14 | 12 |
| Growth on specific media | 7 | 5 | 2 | 7 | 4 | 5 | 23 | 12 |
| Colony and cellular morphology | 2 | 8 | 8 | 8 | 7 | 12 | 4 | 21 |
| Pathogenicity, pathogenicity-like processes, and production of toxins | 5 | 1 | 8 | 10 | 15 | 15 | 6 | 6 |
| Resistance to antimicrobial compounds | 2 | 2 | 5 | 5 | 3 | 4 | 1 | 4 |
| Other diverse phenotypic properties | 5 | 3 | 2 | 4 | 2 | 5 | 1 | 4 |
Although DNA-based characterization techniques had an overall dominant role in studies of microbial biodiversity, their importance was more restricted when studies were classed by theme. In fact, DNA-based techniques had the most dominant role in studies of the phylogenetic and taxonomic relationships among organisms (theme Relatedness) and in the search for new species or taxa (theme New) (Table 5). As indicated above, the evaluation of methods (theme Methods) has also focused principally on characterization of microbial DNA. Conversely, studies concerning the effect of an environmental factor and the description of structure and composition relied more heavily on characterization of microbial phenotypes than on characterization of genotypes. This trend was consistent for all systems except aquatic systems.
These trends led us to wonder if the predominance of phenotypic tests in studies of microbial biodiversity is related to the role of known, specific microbial functions in a habitat. Studies of plant and animal pathogens and of symbiotic microorganisms provided a large enough database to permit us to analyze trends concerning the use of direct tests of pathogenicity and of symbiosis. We based this analysis on 117 publications that concerned the characterization of processes related to pathogenesis or symbiosis: 45 publications for fungus-plant pathosystems, 37 for bacterium-plant systems, 14 for rhizosphere systems, 13 for mycorrhizae, and the remainder for soils, foods, and food-processing factories. The objectives of these studies were, for example, to determine the similarity of populations of a given plant pathogen from different origins or the overall diversity within a specific group of a plant pathogen or food spoilage organism (11, 16, 32, 51, 62, 121, 142, 152, 189, 221). We did not consider studies focused solely on molecular phylogenetics or on the diversity of specific alleles within pathogen or symbiont populations. For the database considered as a whole, pathogenicity or symbiotic host range testing was frequent for studies in all the habitats represented in the 117 publications. However, over time there has been a general tendency for fewer and fewer studies to carry out this type of characterization, as illustrated in Fig. 5. This tendency could possibly be explained by the increasing availability of markers for properties related to pathogenesis (72, 196, 277) or to symbiosis (2, 148, 159, 185). However, we did not find many discussions concerning how markers were good estimates of the pathogenicity or the symbiotic potential of the organisms studied when direct tests of these properties were not used in the studies described in the 117 publications analyzed. This tendency suggests that direct characterization of the pathogenicity or symbiotic potential of microorganisms in biodiversity studies is being progressively abandoned and is not necessarily being replaced by unambiguous markers of these microbial functions. Part of the impetus for reducing the use of pathogenicity tests may reside in the intensification of quarantine and biosecurity constraints in the handling of plant pathogens such as Ralstonia solanacearum and certain species of Cercospora or Fusarium, for example, or for genetically engineered microorganisms. The manipulation of human and animal pathogens in general and the handling of indicator animals used in evaluating pathogenicity are also subject to increasing constraints that may contribute to this trend. For pathogens and symbionts alike, constraints of time and labor may also orient research programs away from approaches requiring these types of phenotypic characterization.
FIG. 5.
Changes over time in the frequency of publications reporting the use of tests of pathogenicity or symbiosis in studies of microbial biodiversity from 1975 to 1999 for systems where these microbial properties are particularly pertinent: fungus-plant pathosystems (▪), bacterium-plant systems (□), mycorrhizae (▴), the rhizosphere (×), and all habitats considered together (⧫).
APPROACHES TO EXPERIMENTAL DESIGN AND HYPOTHESIS TESTING
The objectives of three of the themes described above involve the testing of hypotheses about the effect of the environment, space, or time on microbial biodiversity (themes Effect, Dynamics, and Source). Examples of hypotheses under these themes, stated in a general way, include the following: there is no significant variation in the structure of the population of a given plant pathogen over time or between geographic regions (32, 39), and plant species have a significant effect on the biodiversity of microorganisms in the rhizosphere (19, 95, 171, 205, 252). The test of such hypotheses necessitates well-developed experimental design leading to statistical tests. Because of the analogies that can be made among these three themes, we have grouped them for the analysis presented below. The objectives of the remaining themes focus—to various degrees—on description of the composition or structure of microbial populations or communities. The principal questions we addressed in the analysis of experimental approaches were unique for each of these themes (Fig. 2), and hence, we present each of them independently.
Measuring the Impact of the Environment, Space, and Time
The objectives of more than half of the microbial biodiversity studies published over the past 25 years were to measure the effect of a specific environmental factor on biodiversity, to measure the changes in biodiversity over time and space, or to compare the similarity of a population of pathogens at a given site with that of the population at a suspected source of contamination (themes Effect, Dynamics, and Source) (Table 4). These objectives are analogous to a wide range of seemingly straightforward objectives such as determining the effect of fertilizers on crop yield or the changes in lichen density with increasing distance from a refinery. Hence, we extrapolated the main principles guiding experimental design and hypothesis testing from these latter systems to the analysis of microbial biodiversity in the themes Effect, Dynamics, and Source. As a first step in characterizing the experimental designs reported in studies under these three themes, we noted whether a sampling strategy was clearly described (Fig. 2). Specifically, we determined if either (i) the space and time within which samples were collected were defined or (ii) information was presented concerning how the samples accounted for the variability inherent to the sampling procedure. For these studies, we then noted if statistical tests were used to evaluate the hypotheses evoked concerning biodiversity and if the study reported multiple independent tests of these hypotheses (Fig. 2).
For the themes Effect, Dynamics, and Source considered as a whole, one-third of the 471 publications analyzed did not report a sampling strategy (Table 6). These publications lacked information about the basis for choosing the samples or strains characterized and/or information about the size, weight, volume, or frequency of samples taken. For these studies, there was insufficient information to determine how an independent but comparable sample could be taken. Hence, we could not ascertain the extent to which the characterized organisms represented the population studied or to what extent the results could be compared to those of studies of similar microorganisms. For 6% of these 471 publications, the study was based solely on the use of strains from culture collections. In nearly all studies that employed strains from culture collections, the only information reported concerned their taxonomic identification and the date, location, and environment from which they were isolated. Nevertheless, a few publications indicated how strains from culture collections had been isolated, and we considered this to be part of the description of a sampling strategy. The frequency of studies reporting sampling strategies varied among the habitats. Publications concerning soil and rhizosphere systems, mycorrhizae, fungus-plant pathosystems, and food most frequently reported sampling strategies (Table 6). For all habitats considered together, there was no discernible effect of time on the frequency at which sampling strategies were reported in publications.
TABLE 6.
Number of publications reporting sampling strategies, use of statistical analyses to test hypotheses concerning microbial biodiversity, and replicated tests of hypotheses for the themes Effect, Dynamics, and Source
| Themea | No. of publications analyzed
|
|||
|---|---|---|---|---|
| Totalb | Samplingc | Statisticsd | Replicatione | |
| Effect of a defined factor | ||||
| Habitat | ||||
| Aquatic | 11 | 4 | 0 | 0 |
| Soil | 60 | 51 | 27 | 4 |
| Rhizosphere | 69 | 57 | 29 | 7 |
| Mycorrhizae | 48 | 44 | 35 | 2 |
| Fungal/plant pathosystems | 16 | 14 | 11 | 6 |
| Bacterial/plant systems | 12 | 6 | 4 | 1 |
| Food | 6 | 5 | 2 | 0 |
| Food factories | 1 | 0 | 0 | 0 |
| All systems | 223 | 181 | 108 | 20 |
| Spatial or temporal dynamics | ||||
| Habitat | ||||
| Aquatic | 41 | 10 | 0 | 0 |
| Soil | 37 | 29 | 14 | 2 |
| Rhizosphere | 31 | 24 | 9 | 5 |
| Mycorrhizae | 13 | 12 | 9 | 1 |
| Fungal/plant pathosystems | 30 | 26 | 19 | 5 |
| Bacterial/plant systems | 21 | 5 | 1 | 0 |
| Food | 19 | 12 | 2 | 0 |
| Food factories | 8 | 0 | 2 | 0 |
| All systems | 200 | 118 | 56 | 13 |
| Sources of contamination | ||||
| Habitat | ||||
| Aquatic | 0 | 0 | 0 | 0 |
| Soil | 1 | 0 | 0 | 0 |
| Rhizosphere | 0 | 0 | 0 | 0 |
| Mycorrhizae | 1 | 1 | 0 | 0 |
| Fungal/plant pathosystems | 4 | 3 | 2 | 1 |
| Bacterial/plant systems | 7 | 2 | 0 | 0 |
| Food | 4 | 4 | 0 | 0 |
| Food factories | 31 | 5 | 2 | 0 |
| All systems | 48 | 15 | 4 | 1 |
Themes are described in Table 2.
Total number of publications analyzed for a given habitat within each theme.
Number of publications in which the sampling strategy (the time and space within which samples were collected) was clearly defined.
Number of publications describing sampling strategies and employing statistical tests to evaluate the principal hypotheses concerning microbial biodiversity.
Number of publications reporting replicated or multiple independent tests of the principal hypotheses concerning microbial biodiversity.
Among the 314 publications in the themes Effect, Dynamics, and Source that reported sampling strategies, about 50% also reported values of diversity indices and subsequently employed statistics to test hypotheses concerning biodiversity (Table 6). There were marked differences among the three themes in terms of the frequency with which diversity indices and subsequent statistical tests were employed: 60% of the publications reporting sampling strategies in the theme Effect also employed indices and statistical tests, while 47% of those in the theme Dynamics and 27% of those in the theme Source did so. For the publications in which statistical tests of hypotheses were not employed, many reported experimental designs and data equivalent to those reported in studies employing statistics. For most of these publications, it was not clear why statistical tests were not exploited. A few indicated that they did not have a sufficient number of replications (83). Another (213) clearly stated that statistical tests were considered inappropriate in light of the high variability of mycorrhizal types observed in the large number of samples analyzed. About 8% of the 146 publications that lacked descriptions of sampling strategies under these three themes nevertheless reported statistical evaluations of the hypotheses evoked.
A wide variety of approaches were used for calculating diversity indices and for performing statistical tests of hypotheses. To summarize these approaches, we classified measures of diversity as either primary indices or secondary or composite indices. Primary indices were direct measures of population parameters not requiring any particular calculation or confounding of species (or taxon) richness with relative abundance and identity. In general, these were direct measures of the abundance or relative frequency of phenotypes or genotypes observed in a sample. All other types of indices, such as the Shannon-Wiener diversity index (Hs) (239), Simpson's diversity index (λ) (244), and Nei's index for genetic diversity (IN) (195), were considered to be composite indices. In Table 7 we summarize how primary and composite indices have been coupled with the use of parametric and nonparametric statistical tests.
TABLE 7.
Measures of diversity and statistical tests used to evaluate the effect of environmental factors, space, or time (themes Effect and Dynamics) on microbial biodiversity or to compare microbial populations with the objective of identifying sources of contamination or inoculum (theme Source)
| Habitata | Diversity measuresb
|
Statistical testsc
|
Reference(s) | ||
|---|---|---|---|---|---|
| Primary indices | Secondary indices | Para. | Nonpara. | ||
| Soil | x | x | 17, 21, 37, 54, 69, 108, 117, 128, 198, 271, 283, 292, 296 | ||
| x | x | 99 | |||
| x | x | x | 129 | ||
| x | x | x | 14, 145 | ||
| x | x | 135, 178 | |||
| x | x | 23 | |||
| x | x | x | 52 | ||
| Rh | x | x | 38, 86, 118, 150, 163, 171, 188, 192, 193, 211, 242, 279, 283 | ||
| x | x | 31, 95, 252 | |||
| x | x | x | 35, 285 | ||
| x | x | x | 92, 93, 169 | ||
| x | x | 252 | |||
| x | x | 98 | |||
| Myc | x | x | 3, 4, 5, 58, 68, 123, 131, 146, 183, 172, 214, 225, 227, 231, 240, 241, 258, 275, 280, 289 | ||
| x | x | 36, 250 | |||
| x | x | x | 1, 13, 28, 91, 119, 124, 126, 127, 243, 254 | ||
| x | x | x | 253 | ||
| x | x | x | x | 49, 85, 105 | |
| FPP | x | x | 11, 136 | ||
| x | x | x | 239 | ||
| x | x | 32, 40, 61, 65, 132, 142, 180, 207, 208, 261, 288 | |||
| x | x | x | 39 | ||
| x | x | 24, 25, 32, 40, 142 | |||
| x | x | 94, 179 | |||
| BP | x | x | 79 | ||
| x | x | 122 | |||
| x | x | x | x | 109 | |
| x | x | 276 | |||
| Food | x | x | 67, 125, 170 | ||
| FI | x | x | 34, 140, 229 | ||
| x | x | 249 | |||
The abbreviations used for the different habitats are described in footnote b of Table 4.
Diversity was evaluated in terms of either primary indices of microbial diversity (such as allelic frequencies or number of species) or secondary or composite indices that confound the number of species (or other taxa) and their relative abundance. The measures of diversity indicated here are those that were the basis of the statistical tests reported in the publication. Some publications reported additional indices that were not the basis of subsequent statistical analyses; these are not considered here.
Statistical tests were classified as either parametric (Para.) or nonparametric (Nonpara). The reported parametric tests used to evaluate hypotheses concerning biodiversity included analysis of variance, least-significant-difference test, Tukey's and Newman-Keul's multiple-comparison tests, Duncan's multiple-range test, and t tests. Nonparametric tests included χ2, Fisher's exact test, Spearman's rank correlation, Wilcoxon signed rank test, the Kruskal-Wallis test, and the Mann-Whitney test.
The work of Chen et al. (40) clearly illustrates how the nature of primary and composite diversity indices constrains the choice of parametric and nonparametric statistics for hypothesis testing in microbial biodiversity studies. These workers sought to determine if there were changes in the genetic diversity of populations of the plant pathogenic fungus Mycospharella graminicola during disease epidemics on wheat. Genetic diversity was measured in terms of the frequency of alleles at different restriction fragment length polymorphism (RFLP) loci in isolates of the fungus collected at different times in each of three seasons at one experimental site. For example, to test the difference in genetic diversity between early-season and late-season populations, a contingency χ2 test was employed to compare allelic frequencies at each of eight different RFLP loci determined for each of 444 strains collected in 1990 (about one-third of which were collected early in the season, and the rest were collected late in the season). Likewise, the difference in genetic diversity of populations from three different years was evaluated by the same statistical test by comparing allelic frequencies for each of 10 RFLP loci for the late-season strains collected in 1990 and 58 late-season strains collected in 1991 and 1992. No significant differences were revealed at any of the loci. The authors then calculated various indices of genetic diversity including Nei's index for the different loci at each date. Nei's index is analogous to the Shannon-Wiener index and summarizes the number and relative abundance of different genotypes in a sample without accounting for the identity of the genotypes encountered. Furthermore, the value for such an index calculated from data for a single sample is not associated with a measure of variance. Population variance must be measured from multiple samples from the same population. Alternatively, some authors have exploited methods proposed for estimating the theoretical variance of composite diversity indices via simulation or calculations (25, 276, 277). Hence, for diversity based on Nei's index, Chen et al. (40) used parametric tests (t tests) to evaluate differences in genetic diversity for situations in which they had measures of variance. By using t tests, they compared genetic diversity at different dates or different times in a single year by considering the indices for each of the loci to be replicate measures within a given time. However, the indices for the multiple loci of a single set of strains are not necessarily bona fide independent replicate measures.
Other studies can also serve to illustrate how the types of replication and measures of variability inherent to experimental design influence the coupling of diversity indices to parametric and nonparametric tests, as illustrated in Table 7. Hallmann et al. (93) conducted parametric tests based on composite diversity indices by establishing replicated measures, sensu stricto, of these indices. For chitin-amended and nonamended soils, these authors calculated the number of different bacterial genera and Hill's diversity number (N1) for samples of 35 bacterial strains from each of four replicate plots per treatment. For each index, least-significant-difference tests were used to compare diversity based on the four values measured per treatment. Helm et al. (105) used one-way analysis of variance and the nonparametric Kruskal-Wallis test to evaluate differences in fungal diversity in terms of the infection percentages for different ectomycorrhizal types along a chronosequence of different plant species. Xia et al. (288) employed a nonparametric test to evaluate heterogeneity among primary indices of genetic diversity. To determine variation in diversity of the plant pathogen Magnaporthe grisea among sites within individual fields of rice, these authors determined the DNA fingerprints of 7 to 21 strains collected at each of five sites within two commercial rice fields. Within-field spatial heterogeneity of diversity was determined by a χ2 test based on the frequency of RFLP fingerprint groups in the five locations per field. Although Nei's index of genetic similarity was calculated in this study, it was not employed in statistical tests. The use of nonparametric tests for primary indices was also illustrated by Carraminana et al. (34). These authors used a χ2 test to evaluate the difference in frequency of serotypes among strains of Salmonella collected in poultry slaughterhouses before and after evisceration of carcasses.
Another aspect of experimental design that we noted for the themes Effect, Dynamics, and Source was the reporting of multiple independent tests of hypotheses (Fig 2). Of the 471 publications analyzed in these three themes, about 5% reported multiple independent tests of hypotheses (Table 6). For the 168 that described sampling strategies and employed statistics for testing hypotheses, about 20% reported multiple independent tests of these hypotheses. Nearly all studies under the themes Effect, Dynamics, and Source employed replication to ensure the reliability of the microbial traits characterized in the laboratory. However, most of these studies were based solely on samples from a single location and/or a single date. Hence, multiple independent tests of the principal hypotheses were not possible for these studies.
Examples of studies involving multiple tests of hypotheses include the work of Mahaffee and Kloepper (169), who evaluated the effect of a genetically modified strain of a plant growth-promoting Pseudomonas fluorescens on microbial communities of the cucumber rhizosphere and endorhiza. The microbial diversity of the rhizosphere and endorhiza of inoculated and noninoculated experimental plots did not differ significantly between treatments but was different between 0 and 70 days after planting. Similar results were obtained in each of two field seasons. Jonsson et al. (127) evaluated the effect of wildfires on the ectomycorrhizal fungal communities of Scots pine. To conduct replicated tests of the hypothesis that wildfires have significant effects on mycorrhizal population structure, these authors compared the mycorrhizal populations in burned and adjacent unburned late-successional stands of Scots pine at four different sites in northern Sweden. Asher (11) tested the hypothesis that the aggressiveness of strains of the plant pathogen Gaeumannomyces graminis from fields continuously cultivated with cereal crops is lower than that of strains from fields in which cereals are grown only occasionally. Measures of the aggressiveness of the populations were made from two sites, one representing a short-term (2 or 3 preceding cereal crops) and the other a long-term (12 to 15 preceding cereal crops) cereal sequence. Four tests were done, each with isolates from the two different types of sites. Statistical comparisons among populations from the two types of sites led to the rejection of the hypothesis. Zhan et al. (293) sought to determine the relative contribution of immigration to the genetic structure of Mycosphaerella graminicola populations during the course of an epidemic. They used neutral DNA markers to compare the population structure in wheat plots inoculated with known isolates to that in naturally infected control plots. Field plots were arranged in a randomized complete block design with four replications, and comparisons of allelic frequencies were based on a contingency χ2 test. Significant differences were observed in allelic frequencies in populations from control and inoculated plots, suggesting that immigration was low. These differences were evaluated for numerous different alleles and pairs of alleles for both mid-season and late-season populations but for only one field experiment. Heuer and Smalla (109) compared the diversity of bacteria on leaves of common potato cultivars to that on leaves of genetically modified potato plants expressing the bacteriophage T4 lysozyme. Comparisons were made in a greenhouse experiment as well as in a field trial. Other examples of multiple tests of hypotheses under the themes Effect, Dynamics, and Source are reported by Bever et al. (19), Burdon and Jarosz (32), Chen et al. (39), Garland (75), Handley et al. (95), Hartmann et al. (99), Maloney et al. (171), Marilley et al. (173), Paffetti et al. (205), Safir et al. (227), Sanders (231), and Strain et al. (252).
One of the central purposes of experimental design is to ensure that the measures reported in a publication are repeatable and not due simply to random error. This notion is clearly evoked in the Instructions to Authors of the major journals publishing studies of microbial biodiversity. However, our analysis of the trends in experimental design for publications in the themes Effect, Dynamics, and Source suggests that few publications (about 5%) report repeated measures of the phenomena that are central to the principal objectives of the studies. In other words, publications reporting multiple independent tests of hypotheses were rare. Perhaps verifications of the hypotheses addressed in these publications are, or will be, the subject of subsequent publications. Alternatively, the investment in time and labor for multiple tests of hypotheses may have been prohibitive for some studies. For certain studies, such as those addressing changes in population structure or composition over decades (196) or across a wide range of geographic regions (189), multiple independent tests are virtually impossible or very impractical. However, for other types of studies, we could not identify what led to the publication of results of only a single experiment, a single sampling campaign, or a single set of strains.
The lack of repeated tests of hypotheses and the infrequent use of statistical tests led to some ambiguity in the conclusions presented in many of the publications in the themes Effect, Dynamics, and Source. It is important to note that the conclusions stated in most of the publications analyzed here were cautiously elaborated with regard to interpretation of the results. Hence, we encountered relatively few publications that presented unambiguous conclusions about the effect of specific factors, time, or space on microbial biodiversity. The least ambiguous conclusions were presented in publications reporting sampling strategies and the use of statistical tests. The Discussion sections of many of the publications in these three themes focused on the utility of the techniques employed for microbial characterization rather than on the major ecological theme of the study.
Above, we speculated about why multiple independent tests of hypotheses are not commonly reported in the microbial biodiversity literature. We also wonder why statistical tests are infrequently employed in publications relevant to the themes Effect, Dynamics, and Source. We encountered numerous publications with clearly defined sampling strategies and experimental design for which statistics were not reported but for which they would have been appropriate to test the hypotheses evoked. The reporting of sampling strategies coupled to the use of statistical tests of hypotheses was clearly more typical of studies of certain habitats than of others. In particular, publications concerning soil, the rhizosphere, and fungi in general most frequently reported these aspects of experimental design and hypothesis testing. This tendency has led us to wonder if the scientific literature concerning the microbial biodiversity of a given habitat has a broad impact, in particular on studies of apparently unrelated habitats.
What are the possible barriers to a broad impact of the microbial biodiversity literature among different habitats? One barrier may result from the techniques typically used for isolating and characterizing the microorganisms of a given habitat. These techniques may sharply orient experimental design and may make it difficult to extrapolate experimental designs from one system to another. In particular, for habitats where microorganisms are readily culturable and identifiable, it may be relatively easy to design experiments leading to data sets compatible with the calculation of indices, measures of variability, and statistics. It is interesting that none of the studies analyzed for the themes Effect, Dynamics, and Source concerning aquatic systems employed statistics; most of these studies were based on the characterization of the DNA representing nonculturable organisms. Characterization of microorganisms based on DNA extraction from environmental samples followed by PCR amplification, cloning, and sequencing, as was employed for most of the studies in the themes Effect, Dynamics, and Source for aquatic systems, is very demanding of time and labor. Furthermore, strategies to randomly sample the generated clones so as to represent the relative proportions of organisms in the environment are not obvious. However, this type of characterization has been of great interest because it can lead to identification at the species and strain levels. Recently developed approaches to discriminating isolated nucleotide sequences, such as single-strand conformation polymorphism, terminal RFLP, denaturing gradient gel electrophoresis, and related techniques, may lead to better compatibility of characterization of DNA extracted from the environment with experimental designs leading to statistical tests, as illustrated by the work of van Hannen et al. (274).
Alternatively, other approaches to characterizing noncultured microorganisms have proven to be compatible with statistical analyses. For example, communities of microorganisms have been characterized by direct analysis of the fatty acids in phospholipids or in lipopolysaccharides extracted from environmental samples. The ease of this technique and the obvious relationship between the fatty acid composition in the sample to that in the environment have led to statistical analyses of the resulting fatty acid profiles (37, 69, 117, 211, 242, 292). Nevertheless, this technique is less discriminating and therefore less interesting because the resulting profiles correspond to broad taxonomic groups such as actinomycetes, gram-positive or gram-negative bacteria, or anaerobic bacteria. Nonculturable mycorrhizal fungi, as well as fungi that are obligate plant parasites, have also been the object of studies exploiting statistical analyses (33, 227). Quantitative measures of the biodiversity of these fungi in particular might be readily accessible because they can be identified by morphological characteristics even though they are not culturable. Nevertheless, statistical tests are, in general, infrequently employed even for publications concerning habitats where microorganisms are readily culturable and identifiable. Hence, there is a need for considerable reflection about the barriers to statistical analyses and multiple independent tests of hypotheses in publications concerning microbial biodiversity.
Snapshotting the Composition and Structure of Microbial Populations and Communities
Studies in the theme Describe focused on either describing the composition of microbial populations or evaluating the structure of these populations independent of the effects of time and space. In our analysis, we made a clear distinction between the notions of composition and structure. We defined structural studies as those reporting quantitative measures of the relative abundance of the different groups of organisms characterized and compositional studies as those that either enumerated or identified the different groups detectable in a population or community. Although numerous publications employed “structure” in the title or key words, they were not considered to be studies of structure unless quantitative measures of relative abundances were presented. In the absence of such quantitative measures, publications were considered to represent compositional studies for this analysis. For studies of population structure, our analysis consisted of determining if a sampling strategy was described in the publication and if there was an estimation of the variance associated with the structure of the described population. For studies of population composition in the theme Describe, we noted if the authors attempted to justify or evaluate how well the sample represented the population being studied (Fig. 2).
Studies of microbial populations and communities under the theme Describe have focused primarily on quantification of population structure (Table 8). These studies have quantified population structure by measuring parameters such as (i) the relative abundance of ribotypes or of strains resistant to various antibiotics in rhizosphere bacterial populations to which strains of Pseudomonas fluorescens active as biocontrol agents were introduced (193), (ii) the relative abundance of DNA fingerprint groups of Aspergillus parasiticus from a corn field (177), and (iii) the frequency of alleles within the ribosomal DNA intergenic spacer of Hebeloma cylindrosporium from populations of this haploid ectomycorrhizal basidiomycete (90). Hence, the representativeness of a sample relative to the overall population is a central issue for these studies. Nevertheless, only two-thirds of the 118 studies of population structure that we analyzed reported sampling strategies (Table 8).
TABLE 8.
Characteristics of publications in the theme Describe concerning the evaluation of the composition or structure of a microbial population or community independent of time and location
| Type of publication | No. of publications analyzed for:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Aqe | Soile | Rhe | Myce | FPPe | BPe | Foode | FIe | All systems | |
| Total | 20 | 23 | 40 | 27 | 58 | 14 | 6 | 5 | 193 |
| Population structurea | 9 | 17 | 32 | 20 | 35 | 4 | 1 | 0 | 118 |
| Defined samplingb | 3 | 12 | 32 | 11 | 18 | 3 | 1 | 0 | 80 |
| Population compositionc | 11 | 6 | 8 | 7 | 23 | 10 | 5 | 5 | 75 |
| Justified samplingd | 0 | 0 | 0 | 3 | 5 | 1 | 3 | 0 | 12 |
Number of publications in which the relative frequencies of the different components of the microbial community or population were quantified. Some of these publications also reported qualitative descriptions of the composition of the communities or populations studied.
Number of publications reporting population structure for which the sampling strategy (the time and space within which samples were collected) was clearly defined.
Number of publications that reported only qualitative descriptions (for example, the number of taxa and their identity) of the composition of the communities or populations studied. None of these publications reported quantification of population structure.
Number of publications describing the composition of microbial populations for which the authors justified the sampling strategy exploited.
The habitats covered in this study were aquatic systems (Aq), soil (Soil), the rhizosphere (Rh), mycorrhizae (Myc), fungus-plant pathosystems (FPP), bacterium-plant systems (BP), food (Food), and food-processing factories (FI).
For studies describing the composition of microbial populations, only 16% reported information concerning how the sample was taken or in what way it represented the population (Table 8). Among the publications presenting information concerning the representativeness of the sample, a few included rarefaction curves. Rarefaction curves represent the rate at which new groups (species, genotypes, etc.) are detected as the sample size increases. The asymptote of the curve is considered to be an estimate of the number of different groups in the total population (230, 265). The use of rarefaction curves is standard fare in studies of the biodiversity of macroscopic organisms, and their potential use in microbial diversity studies has been recently well illustrated (114). Among the publications analyzed here, that of Goodman and Trofymow (83) used rarefaction curves to estimate the rate at which new mycorrhizal types were found and also to estimate total richness. Ingleby et al. (119) also used similar curves to compare mycorrhizal diversity (i.e., the cumulative number of different mycorrhizal types) under different plant canopy types. Other examples include the studies by Hantula et al. (96), Ravenschlag et al. (215), and Sakano and Kerkhof (228) for aquatic systems, Clawson and Benson (42), Dunbar et al. (56), and Handley et al. (95) for rhizosphere systems, and Torsvik et al. (266) for soil systems.
The notion of representativeness has a twofold implication for experimental design. First, it is clear that experimental design must take into account the extent to which laboratory tests represent microbial phenotypes and genotypes. This has been of overwhelming interest to microbiologists, as illustrated by the abundance of publications that evaluate assays for phenotypic and genotypic characterizations. Furthermore, the principal types of replication reported in studies of microbial biodiversity pertain to the confirmation of results of such assays. The second implication of representativeness is the extent to which the sample of organisms or their DNA represents a population or a community. For both of these aspects of representativeness, one is compelled to ask if we are studying artifacts and in what way our estimate of the properties of the population or the community is biased. However, it should be kept in mind that the representativeness of a sample vis-à-vis a population is the foundation on which the representativeness of characterization assays rests.
What does “representativeness of a sample” mean? Clearly, for practical and technical reasons, it cannot mean that every taxon in the community is represented in the sample. The growing interest in identifying new taxa and the development of techniques to account for nonculturable microorganisms—major objectives of the Diversitas program and of recent research programs (223)—may be helpful in broadening the scope of biodiversity studies, but they are not intended to address representativeness per se. “Representativeness of a sample” simply means that we can define what the sample represents. The aim of a sampling strategy is to render this definition compatible with the objective of the study. For studies where, for example, the objective is to quantify population structure, the sampling strategy employed should lead to samples in which the proportion of each type or group detected is equivalent to that in the population, within the limits of detection corresponding to the sample size.
Of course, this is a proposition for an ideal world with no sampling biases. Sampling errors and biases are inevitable, and they limit the extent to which approaches such as rarefaction can be used to investigate the representativeness of a sample. Kinkel et al. (139) clearly illustrated how sampling of microbial systems misrepresents the underlying species frequency distributions in the population. Their analysis was based on random sampling of simulated microbial populations having different species abundance distributions (lognormal and negative binomial distributions with different degrees of skewedness) but each having 100 species. Random sampling revealed that depending on sample size, communities with distinctly different species abundance distributions could not be differentiated. Likewise, extrapolations made from identical populations could lead to the conclusion that the populations had distinctly different species abundances. Real-world sampling combines the basic probabilistic errors described by Kinkel et al. with the biological biases of microbiological and molecular biological sampling techniques. Rarefaction and simulated estimates of variance of diversity measures are constrained by the information contained within the sample and do not account for sampling biases unless they have been measured experimentally. Hence, one can understand the importance of replicated sampling and of multiple independent experiments for measures of population structure. Furthermore, it is evident that the results of studies describing the composition of microbial populations are also affected by the way in which the sampling strategy conditions the representativeness of a sample.
Discerning Markers for Diagnosis and Identification
The notion of representativeness is also particularly pertinent to the search for markers. Many of these studies have relied heavily on characterization of strains from culture collections in order to span the geographical and temporal variability of the microorganisms of interest. Hence, we did not consider sampling strategy per se to be a fundamental issue in the overall approach taken for publications in the theme Markers. Rather, for publications in this theme, we noted if the authors explained the basis for choosing the strains or sample from which they identified markers or if there was any discussion about what the strains or samples represented. Furthermore, we also noted the use of strains outside of the taxonomic group for which the markers were identified and the use of independent samples (or collections) to validate the accuracy of the markers (Fig. 2).
The search for markers was dominated by studies of plant-pathogenic and phyllosphere bacteria and of mycorrhizal fungi, although there were examples of such studies for nearly all habitats. For bacteria, 88% of the studies published under this theme sought markers of taxonomic groups, whereas the studies of mycorrhizae were equally divided between the search for taxonomic groups and the search for functional groups. The experimental design and sampling strategies associated with these studies clearly influenced how we interpreted the results. In particular, for nearly all of the studies under the theme Markers, it is difficult to evaluate the potential utility of the markers in terms of the fraction of the naturally occurring population they could detect or identify. This difficulty arises from the fact that none of the 69 studies concerning markers presented information describing how the sample represented the inherent variability of the population and only 2 of these studies tested the validity of the reported markers against a collection or sample of strains independent of that used to develop the markers (158, 251).
To illustrate the impact of representativeness for studies in the theme Markers, we calculated rarefaction curves based on data presented in two studies in which markers of bacterial identity were developed (Fig. 6). In presenting these curves, we keep in mind the limits of rarefaction cited above. These curves suggest that the 19 different RFLP groups described for a collection of 137 strains of Erwinia carotovora (104) represent a value much closer to the asymptotic value than the 28 groups described for the 62 strains of Ralstonia (formerly Pseudomonas) solanacearum (47) (Fig. 6). Hence, in the application of these markers to the identification of strains of these bacteria, there would be a high probability that newly collected strains of R. solanacearum would fall into RFLP groups not described by Cook et al. (47) whereas newly collected strains of E. carotovora would be much more likely to be represented by one of the RFLP groups described by Hélias et al. (104). Such a comparison is based on the assumption that both sets of strains are equally representative of their respective populations. Nevertheless, such estimates could be useful in orienting studies toward the number of strains necessary for characterizing markers with the desired level of accuracy. Ultimately, the validity of such estimates can be confirmed by testing markers against additional samples or collections of strains.
FIG. 6.
Rarefaction curves of the number of different RFLP profiles detected within collections of increasing size for strains of E. carotorovora (wide gray line) and R. solanacearum (narrow black line) based on data presented by Hélias et al. (104) and Cook et al. (47), respectively. Rarefaction curves were calculated by sequentially selecting each of the 137 strains presented by Hélias et al. (104) or the 62 strains presented by Cook et al. (47) in a random order and determining the number of different RFLP profiles obtained as the number of strains increased. The curves represent the mean of five independent randomizations for each data set.
Defining Relatedness among Microorganisms
Many of the publications concerning microbial phylogeny and taxonomy (theme Relatedness), similar to those concerning the theme Markers, were based on the use of strains from culture collections. Nevertheless, there were also numerous examples of studies in this theme involving the isolation of strains via culture methods and of DNA representing nonculturable microorganisms. Hence, in the analysis of publications under the theme Relatedness, we looked for information concerning sampling strategies or other descriptions of the representativeness of the organisms characterized. Furthermore, because of the nature of the numerical analyses employed in these studies, we also noted the number of strains used to represent each operational taxonomic unit described (Fig. 2).
The approach taken for biodiversity studies concerning microbial phylogeny or taxonomy was clearly dependent on the habitat involved. For aquatic systems, nearly 70% of the studies under the theme Relatedness exploited phylogenetic analysis of nucleic acid sequences isolated from diverse aquatic environments. Nevertheless, only one-fourth of the studies of such sequences reported the strategies involved in sampling. For rhizosphere systems, about 40% of the studies under the theme Relatedness were based on strains from culture collections, another 40% were based on strains collected via culture-dependent techniques, and 20% were based on the analysis of nucleic acid sequences isolated from the environment. Only 2 of the 24 studies analyzed under the theme Relatedness for this habitat reported how strains or nucleic acid sequences were sampled. In the case of microorganisms from foods and for plant-pathogenic or epiphytic bacteria, no studies involved the direct isolation of nucleic acid sequences from the environment. About 70% of the studies under this theme for each of these two habitats were based on strains from collections. The few descriptions of sampling strategies reported for these two habitats addressed the isolation of strains via culture-dependent methods expressly for the studies concerned.
For studies under the theme Relatedness based on the analysis of culturable strains, we could not identify a general logic to the number of strains used. These studies were based on analyses of samples ranging from fewer than 10 strains to several hundred strains. Furthermore, although most studies sought to represent a wide range of different groups or types of microorganisms among the strains analyzed, the number of strains chosen to represent each group was highly variable. It is likely that the number of strains employed for each predefined group is limited by the number of such strains that are available, particularly when the strains originate from culture collections. However, we never found a discussion or other justification regarding this subject.
Although it is quite obvious that sample size can have an effect on the results of quantitative studies, we feel that the importance of sample size for the results of descriptive studies, such as those concerning taxonomy, should not be overlooked. As mentioned above, we were intrigued by the unbalanced representation of different taxa in studies under the theme Relatedness. In general, the principal taxa studied were represented by numerous strains and the reference taxa were represented by only a few or single strains. The numerical techniques used for the creation of dendrograms are generally robust and little affected by the number of strains per cluster or taxon. However, if clusters or taxa are represented by unequal numbers of strains, then the variabilities within the populations corresponding to each of these taxa are not estimated with the same precision. In other words, if only a few strains are selected to represent one taxonomic unit and many strains are selected to represent another, then the potential variability within each of these taxa cannot be equally accounted for in numerical analysis. Hence, we feel that there is a need for reflection on how the degree of variability accorded to a taxon affects the estimation of its relatedness to other taxa.
Evaluating Methods for Studying Microbial Biodiversity
As mentioned above, nearly all of the publications addressing the evaluation of methods for studying microbial biodiversity (theme Methods) focused on evaluating laboratory methods for characterizing microorganisms or their DNA. Nevertheless, we could distinguish between methods that were evaluated based on environmental samples and those that were evaluated based on strains from collections. For the former, we noted whether multiple independent samples were employed in the study, and for the latter, we noted whether an independent collection of strains was used to validate the method (Fig. 2). Among the 94 publications analyzed under this theme, 85% were based on environmental samples (aliquots of water or soil, for example) and the remainder were based on strains from collections to evaluate methods of characterization. For the studies employing environmental samples, about 75% exploited multiple samples as the basis for evaluating the method. In the other 25% of studies, the evaluation of the method was based on only a single environmental sample. None of the studies employing strains from laboratories or national collections validated their conclusion with results from a second independent collection of strains. In some cases, independent collections have been combined to determine the genetic diversity within a given species. Tamplin et al. (259) used two laboratory collections (53 environmental and 78 clinical isolates) to determine the genetic variability within Vibrio vulnificus.
Discovering New Species and Other Taxa
Of all of the different themes analyzed here, the discovery of new species or other new taxa of microorganisms (theme New) is particularly dependent on conceptual and technological advances (113, 223). Many of the publications under this theme seek to demonstrate the pertinence of these advances by illustrating how they contribute to the elucidation of new groups of microorganisms. Nevertheless, in the search for new groups of microorganisms, experimental design sets the stage for encountering the new organisms. These discoveries are essential to understanding the richness of the microbial world and may open doors to subsequent studies of new microbial processes and of the impact of these microorganisms on ecosystem functions. Hence, in light of this potential impact, our analysis of experimental design for the theme New consisted of noting if the discovery was corroborated by using additional independent samples from environments that were the same as or similar to those of the original sample.
Of the 28 publications analyzed that addressed the theme New, 6 corroborated the discovery with multiple independent samples. Wright et al. (287) corroborated the discovery of a small-subunit rRNA (16S rRNA) gene lineage (SDAR324) that was initially discovered in the western Sargasso Sea. The corroboration was based on screening clone libraries prepared from the Atlantic and Pacific Oceans by using specific DNA probes. For rhizosphere and soil systems, Khbaya et al. (137) isolated bacteria from 6 sites, Kuske et al. (147) selected 5 independent rhizospheric samples, Wang et al. (281) sampled 11 plants at 2 sites, Coates et al. (43) processed samples from 8 environments, and Lloyd-Jones et al. (161) took samples from 10 different soils to corroborate their discoveries. The other 22 publications reported discoveries based on a single sample.
CONCLUSIONS AND FUTURE DIRECTIONS
The study of biodiversity is motivated by interest in the heterogeneity and variability of organisms. Because of the immense number of microorganisms on Earth and the wide range of habitats they occupy, the potential heterogeneity and variability of microorganisms are probably larger than for any other group of organisms. Microbiologists are well aware of this potential. Nevertheless, the trends illustrated in this review suggest that the field of microbial biodiversity has lost sight of the full meaning of heterogeneity and variability and has not dealt with some of their consequences on experimentation. We have pointed out what we consider to be some of the particular weaknesses of microbial biodiversity publications in this regard: (i) that descriptions of sampling strategies or other information concerning the representativeness of the sample are often missing from publications, (ii) that there is very limited use of nonsubjective tests (statistical tests) of hypotheses, and (iii) that only a very few papers report the results of multiple independent tests of hypotheses. Furthermore, there are very few examples of publications addressing methodological questions about how sampling strategies influence the results of microbial biodiversity studies.
The analysis presented here is based on information contained in publications—the visible face of microbial biodiversity studies. In considering the factors contributing to the criticisms that we have evoked, one must keep in mind all facets of the production of publications. These facets include the conception of studies, experimentation, and elaboration of the publications. The availability of relevant information concerning sources of variability and the appropriate experimental design and statistical tests are likely to have an important influence on how the study is conceived. Furthermore, the costs associated with sampling and laboratory tests are likely to influence how extensively a study is conducted. Finally, in the elaboration of publications, reviewers and journal editors can have important impacts on contents. We need to ask what role they might play in the extent to which information concerning sampling, for example, is presented or in the orientation of Discussion sections.
In light of the vast sources of variability that need to be considered in studies of microbial biodiversity, one is likely to ask if such studies are, in fact, insurmountable tasks. As Kinkel et al. (139) suggest, certain hypotheses concerning microbial biodiversity may be very difficult to test due to problems of sampling inherent to many microbial systems. The task for microbial ecologists is to identify testable hypotheses relative to the sources of variability that can be accounted for in measures of biodiversity. At present, little headway has been made in apprehending the sources of variability in measures of microbial biodiversity. Furthermore, investigations into variability will also need to address the issue of scale. For example, in studies of the microbial biodiversity of soils and waters, there may be remarkable biodiversity at the scale of small aggregates that is not revealed at the scale of large clumps, as illustrated by the work of Grundmann and Normand (88) and of Long and Azam (162). The increasing availability of automation in analysis, particularly for DNA-based characterization, may help in exploring sources of variability.
Obviously, there is no unifying experimental strategy to account for sources of variability that is applicable to all themes and microbial habitats. Neither are there specific recommendations for samples of adequate size or for indices that are the most appropriate and that can be applied across the spectrum of themes and habitats addressed in microbial biodiversity. Biometricians and statisticians have an important role to play in helping microbiologists explore and define the experimental design and sampling strategies that are best adapted to the objectives of their studies. On the other hand, we feel that the presentation of certain basic information in microbial biodiversity publications, as described in Table 9, would contribute greatly to the rigor of these publications and to appreciation of the significance of the results with regard to underlying variability.
TABLE 9.
Guidelines for the content of publications concerning microbial biodiversity
| Information in section of the publication | Information fora:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Effect | Dynamics | Source | Describe | Relatedness | Markers | Methodsb | New | |
| Materials and Methods. Include descriptions of: | ||||||||
| Experimental design indicating how blocks, randomization, and replication were employed | x | x | x | |||||
| The origin of the individualsc characterized (time and place of sampling; size, weight, and/or volume of samples; numbers of individuals characterized per block and per repetition; basis for choosing these individuals)d | x | x | x | x | x | x | x | x |
| Criteria used to determine the number of individuals characterized (per treatment, per block, per taxonomic group, etc., as appropriate) | x | x | x | x | x | x | x | x |
| The quantitative measures of diversity used (indices, proportions, etc.) | x | x | x | xe | ||||
| Statistical criteria used to determine if the differences observed among the biodiversity of the populations or communities studied are significant | x | x | x | |||||
| Criteria for evaluating the representativeness of the sample relative to the population or community (rarefaction, comparison to other independent samples, etc.) | x | x | x | x | ||||
| Results. Report: | ||||||||
| Means and variances of the quantitative measures of diversity used, where appropriate | x | x | x | xe | ||||
| Other measures of the repeatability of results | x | x | x | x | ||||
| Statistical tests based on the criteria described in Materials and Methods | x | x | x | |||||
| Estimates of the representativeness of the sample according to criteria described in Materials and Methods | x | x | x | x | ||||
| Discussion. | ||||||||
| Discussion of the impact of sampling biases on the observations | x | x | x | x | x | x | x | x |
| Discussion of the evidence that the observations are not due to random error | x | x | x | x | x | x | ||
| Unambiguous appraisal of the validity of the principal, explicit hypotheses | x | x | x | |||||
Themes are described in Table 2. The minimal information to present under each theme is indicated.
For the theme Methods, a much wider range of information than indicated here may be appropriate depending on the method studied and on whether specific hypotheses are explicitly stated.
“Individuals” refers to single clonal lines of microorganisms as well as nucleic acids or bulk environmental samples, depending on the techniques used for characterization.
This information is essential for determining the representativeness of a sample and for comparing results to those from other studies. For studies employing strains from culture collections, all these details might not be available. Hence, the publication should contain complementary information about the representativeness of the sample such as results of comparison to other independent samples or collections.
For studies concerning description of population or community structure.
Materials and Methods sections should contain information necessary for evaluating the representativeness of samples. In all cases, this should include specific details about the origins of the microorganisms or nucleic acids characterized and about the criteria used by the authors in determining the number of individuals characterized (Table 9). For studies evoking explicit hypotheses, such as for the themes Effect, Dynamics, and Source, estimates of the variability of quantitative measures of diversity form one approach to evaluating representativeness. For these themes, we do not feel that representativeness needs to be further examined if experimental design and statistical tests are employed as indicated in Table 9. For studies under the themes Describe, Relatedness, Markers, and New, where quantitative measures of diversity and statistical comparisons of populations are not employed, it is important to provide other measures of representativeness (Table 9). Ultimately, these studies can contribute to revealing the biodiversity of total populations or communities. Hence, representativeness is a key issue for these themes. Clearly, almost all authors could state the evident fact that “this study is not exhaustive and could be improved by taking more samples.” However, some daring authors might state that “this study is just a shot in the dark to see what we would find.” Such honest testaments would put the conclusions of the work into perspective and might also lead authors to revise their approach.
The Results sections should report means and variances of quantitative measures of diversity, where appropriate, or other measures of the repeatability of results. Statistical tests should be employed to test the hypotheses explicitly evoked in studies under the themes Effect, Dynamics, and Source or when evoked for other types of studies such as evaluation of the impact of experimental design and sampling strategies on measures of biodiversity (theme Methods). Estimates of the representativeness of samples should be presented for themes Describe, Relatedness, Markers, and New, as described in the respective Materials and Methods sections (Table 9). The body of information presented in the Materials and Methods and the Results sections should provide the foundation for authors to evoke, in the Discussion section, the impact of sampling biases on their observations and to provide sufficient evidence that their observations are not due to random error. Although random error can have an important impact on the results of studies for all the themes we have described, under certain conditions its role could be ignored without greatly influencing the validity of the results (Table 9). For example, a new microbial species is still new even if we do not know the role of random error in its discovery. However, without information about the role of random error, one cannot speculate on the relative frequency of this species in the environment. Likewise, the efficiency or discriminating power of a characterization method can be evaluated without knowing the role of random error when selecting samples used for the evaluation. However, without knowing this role, it would be difficult to extrapolate the properties of the methods to other samples.
Ultimately, the Discussion section should state the significance of the results relative to microbial biodiversity. For the themes Effect, Dynamics, and Source, this should lead to an unambiguous appraisal of the validity of the principal hypotheses used by the authors. Implicit in the guidelines presented in Table 9 is that the objectives and specific hypotheses of studies are clearly stated in publications. The pertinence of experimental design and sampling strategies presented in the Materials and Methods section, of the analyses presented in the Results section and of the focus of the Discussion section fundamentally depends on the objectives.
Application of the guidelines presented in Table 9 when defining protocols and writing and reviewing manuscripts could boost the field of microbial biodiversity into a more active investigation of how experimental design and sampling strategies influence the results of microbial biodiversity studies. These investigations could include the develpoment of models to evaluate the impact of different sources of variability and sample size on the power of statistical tests, thus allowing optimization of the sample size and other parameters of experimental design relative to constraints of labor and cost. The past 25 years of research in microbial biodiversity has contributed greatly to the development of methods for characterizing the multiple facets of the richness of the microbial world. One of the intentions of this review is to prompt research in the upcoming decades toward unambiguous evaluations of the impact of this richness on ecological processes in microbial habitats.
Acknowledgments
We thank the French Network for Microbial Biodiversity and Ecology, funded by the CNRS and coordinated by Jacques Balandreau of the CNRS Laboratory of Soil Microbial Ecology in Lyon, France, for prompting us to establish a working group on sampling strategies in microbial diversity and for subsidizing our meetings. We also thank J. Balandreau for help in establishing the soil ecosystem database, and we thank Philippe Prior, Joël Chadoeuf, and Rachid Senoussi of INRA-Avignon, Jean-Claude Pierrat of INRA-Nancy, and J. Balandreau for critical discussions. For assistance with bibliographic research, we thank Marie-France Cornic and Michelle Fies of INRA-Avignon and Sylvie Cocaud of INRA-Nancy.
REFERENCES
- 1.Agerer, R., A. F. S. Taylor, and R. Treu. 1998. Effects of acid irrigation and liming on the production of fruit bodies by ectomycorrhizal fungi. Plant Soil 199:83-89. [Google Scholar]
- 2.Aguilar, O. M., M. V. Lopez, P. M. Riccillo, R. A. Gonzalez, M. Pagano, D. H. Grasso, A. Puhler, and G. Favelukes. 1998. Prevalence of the Rhizobium etli-like allele in genes coding for 16S rRNA among the indigenous rhizobial populations found associated with wild beans from the southern Andes in Argentina. Appl. Environ. Microbiol. 64:3520-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amaranthus, M. P., D. Page-Dumroese, A. Harvey, E. Cazares, and L. F. Bednar. 1996. Soil compaction and organic matter affect conifer seedling nonmycorrhizal and ectomycorrhizal root tip abundance and diversity. Research paper PNW-RP-494. Pacific Northwest Research Station, USDA Forest Service Portland, Oreg.
- 4.An, Z. Q., J. H. Grove, J. W. Hendrix, D. E. Hershman, and G. T. Henson. 1990. Vertical distribution of endogonaceous mycorrhizal fungi associated with soybean, as affected by soil fumigation. Soil Biol. Biochem. 22:715-719. [Google Scholar]
- 5.An, Z. Q., B. Z. Guo, and J. W. Hendrix. 1993. Mycorrhizal pathogen of tobacco: cropping history and current crop effects on the mycorrhizal fungal community. Crop Prot. 12:527-531. [Google Scholar]
- 6.An, Z. Q., J. W. Hendrix, D. E. Hershman, and G. T. Henson. 1990. Evaluation of the “most probable number” (MPN) and wet-sieving methods for determining soil-borne populations of endogonaceous mycorrhizal fungi. Mycologia 82:576-581. [Google Scholar]
- 7.Annapurna, Y., and P. R. Rao. 1982. Influence of foliar fungicidal sprays on the rhizosphere microflora of Cicer arietinum L. Biol. Bull. India 4:113-116. [Google Scholar]
- 8.Antibus, R. K., and A. E. Linkins III. 1992. Effects of liming a red pine forest floor on mycorrhizal numbers and mycorrhizal and soil acid phosphatase activities. Soil Biol. Biochem. 24:479-487. [Google Scholar]
- 9.Arimi, S. M., E. T. Ryser, T. J. Pritchard, and C. W. Donnelly. 1997. Diversity of Listeria ribotypes recovered from dairy cattle, silage, and dairy processing environments. J. Food Prot. 7:811-816. [DOI] [PubMed] [Google Scholar]
- 10.Artomonova, V. S. 1995. Structural and functional organization of associations of phototrophic microorganisms in virgin soils of Siberia. Eurasian Soil Sci. 27:53-65. [Google Scholar]
- 11.Asher, M. 1980. Variation in pathogenicity and cultural characters in Gaeumannomyces graminis var. tritici. Trans. Br. Mycol. Soc. 75:213-220. [Google Scholar]
- 12.Autio, T., S. Hielm, M. Miettinen, A.-M. Sjöberg, K. Aarnisalo, J. Björkroth, T. Mattila-Sandholm, and H. Korkeala. 1999. Source of Listeria monocytogenes contamination in a cold-smoked rainbow trout processing plant detected by pulsed-field gel electrophoresis typing. Appl. Environ. Microbiol. 65:150-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baar, J. 1996. The ectomycorrhizal flora of primary and secondary stands of Pinus sylvestris in relation to soil conditions and ectomycorrhizal succession. J. Veg. Sci. 7:497-504. [Google Scholar]
- 14.Bardgett, R. D., D. K. Leemans, R. Cook, and P. J. Hobbs. 1997. Seasonality of the soil biota of grazed and ungrazed hill grasslands. Soil Biol. Biochem. 29:1285-1294. [Google Scholar]
- 15.Barns, S. M., R. E. Fundyga, M. W. Jeffries, and N. R. Pace. 1994. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. USA 91:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barret, E. L., R. E. Solanes, J. S. Tang, and N. J. Palleroni. 1986. Pseudomonas fluorescens biovar V: its resolution into distinct component groups and the relationship of these groups to other P. fluorescens biovars, to P. putida, and to psychrotrophic pseudomonads associated with food spoilage. J. Gen. Microbiol. 132:2709-2721. [DOI] [PubMed] [Google Scholar]
- 17.Beese, F., A. Hartmann, T. Beck, R. Rackwitz, and L. Zelles. 1994. Microbial community structure and activity in agricultural soils under different management. J. Plant Nutr. Soil Sci. 157:187-196. [Google Scholar]
- 18.Berthier, Y., V. Verdier, J. L. Guesdon, D. Chevrier, J. B. Denis, G. Decoux, and M. Lemattre. 1993. Characterization of Xanthomonas campestris pathovars by rRNA gene restriction patterns. Appl. Environ. Microbiol. 59:851-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bever, J. D., J. B. Morton, J. Antonovics, and P. A. Schultz. 1996. Host-dependent sporulation and species diversity of arbuscular mycorrhizal fungi in a mown grassland. J. Ecol. 84:71-82. [Google Scholar]
- 20.Bevercombe, G., and A. Rayner. 1984. Population structure of Cryptostroma corticale, the causal fungus of sooty bark disease of sycamore. Plant Pathol. 33:211-217. [Google Scholar]
- 21.Bisset, J., and D. Parkinson. 1980. Long-term effects of fire on the composition and acticity of the soil microflora of a subalpine coniferous forest. Can. J. Bot. 58:1704-1721. [Google Scholar]
- 22.Boccara, M., R. Vedel, D. Lalo, M. H. Lebrun, and J. F. Lafay. 1991. Genetic diversity and host range in strains of Erwinia chrysanthemi. Mol. Plant-Microbe Interact. 4:293-299. [Google Scholar]
- 23.Boehm, M. J., T. Wu, A. G. Stone, B. Kraakman, D. A. Iannotti, G. E. Wilson, L. V. Madden, and H. A. J. Hoitink. 1997. Cross-polarized magic-angle spinning13C nuclear magnetic resonance spectroscopic characterization of soil organic matter relative to culturable bacterial species composition and sustained biological control of Pythium rootrot. Appl. Environ. Microbiol. 63:162-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borchardt, D., H. Welz, and H. Geiger. 1998. Molecular marker analysis of European Setosphaeria turcica populations. Eur. J. Plant Pathol. 104:611-617. [Google Scholar]
- 25.Borchardt, D., H. Welz, and H. Geiger. 1998. Genetic structure of Setosphaeria turcica populations in tropical and temperate climates. Phytopathology 88:322-329. [DOI] [PubMed] [Google Scholar]
- 26.Borneman, J., P. W. Skroch, K. M. O'Sullivan, J. A. Palus, N. G. Rumjanek, J. L. Jansen, J. Nienhuis, and E. W. Triplett. 1996. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl. Environ. Microbiol. 62:1935-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowman, J. P., S. A. McCammon, M. V. Brown, D. S. Nichols, and T. A. McMeekin. 1997. Diversity and association of psychrophilic bacteria in Antarctic sea ice. Appl. Environ. Microbiol. 63:3068-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bradbury, S. M., R. M. Danielson, and S. Visser. 1998. Ectomycorrhizas of regenerating stands of lodgepole pine (Pinus contorta). Can. J. Bot. 76:218-227. [Google Scholar]
- 29.Britschgi, T., and S. Giovannoni. 1991. Phylogenetic analysis of a natural marine bacterioplankton population by rRNA gene cloning and sequencing. Appl. Environ. Microbiol. 57:1707-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brundrett, M. C., L. K. Abbott, and D. A. Jasper. 1999. Glomalean mycorrhizal fungi from tropical Australia. I. Comparison of the effectiveness and specificity of different isolation procedures. Mycorrhiza 8:305-314. [Google Scholar]
- 31.Brunel, B., S. Rome, R. Ziani, and J. C. Cleyet-Marel. 1996. Comparison of nucleotide diversity and symbiotic properties of Rhizobium meliloti populations from annual Medicago species. FEMS Microbiol. Ecol. 19:71-82. [Google Scholar]
- 32.Burdon, J., and A. Jarosz. 1992. Temporal variation in the racial structure of flax rust (Melampsora lini) populations growing on natural stands of wild flax (Linum marginale): local versus metapopulation dynamics. Plant Pathol. 41:165-179. [Google Scholar]
- 33.Caffier, V., U. Brandle, and M. Wolfe. 1999. Genotypic diversity in barley powdery mildew populations in northern France. Plant Pathol. 48:582-587. [Google Scholar]
- 34.Carraminana, J. J., J. Yanguela, D. Blanco, C. Rota, A. I. Agustin, A. Arino, and A. Herrera. 1997. Salmonella incidence and distribution of serotypes throughout processing in a Spanish poultry slaughterhouse. J. Food Prot. 60:1312-1317. [DOI] [PubMed] [Google Scholar]
- 35.Cattelan, A. J., P. G. Hartel, and J. J. Furhmann. 1998. Bacterial composition in the rhizosphere of nodulating and non-nodulating soybean. Soil Sci. Soc. Am. J. 62:1549-1555. [Google Scholar]
- 36.Causin, R., L. Montecchio, and S. M. Accordi. 1996. Probability of ectomycorrhizal infection in a declining stand of common oak. Ann. Sci. For. 53:743-752. [Google Scholar]
- 37.Cavigelli, M. A., G. P. Robertson, and M. J. Klug. 1995. Fatty acid methyl ester (FAME) profiles as measures of soil microbial community structure. Plant Soil 170:99-113. [Google Scholar]
- 38.Cello, F. D., A. Bevivino, L. Chiarini, R. Fani, D. Paffetti, S. Tabacchioni, and C. Dalmastri. 1997. Biodiversity of a Burkholderia cepacia population isolated from the maize rhizosphere at different plant growth stages. Appl. Environ. Microbiol. 63:4485-4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen, D., R. S. Zeigler, H. Leung, and R. J. Nelson. 1995. Population structure of Pyricularia grisea at two screening sites in the Philippines. Phytopathology 85:1011-1020. [Google Scholar]
- 40.Chen, R., J. Boeger, and B. McDonald. 1994. Genetic stability in a population of a plant pathogenic fungus over time. Mol. Ecol. 3:209-218. [Google Scholar]
- 41.Chin, K. J., T. Lukow, S. Stubner, and R. Conrad. 1999. Structure and function of the methanogenic archaeal community in stable cellulose-degrading enrichment cultures at two different temperatures (15 and 30°C). FEMS Microbiol. Ecol. 30:313-326. [DOI] [PubMed] [Google Scholar]
- 42.Clawson, M. L., and D. R. Benson. 1999. Natural diversity of Frankia strains in actinorhizal root nodules from promiscuous hosts in the family Myricaceae. Appl. Environ. Microbiol. 65:4521-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coates, J. D., U. Michaelidou, R. A. Bruce, S. M. O'Connor, J. N. Crespi, and L. A. Achenbach. 1999. Ubiquity and diversity of dissimilatory (per)chlorate-reducing bacteria. Appl. Environ. Microbiol. 65:5234-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coffey, M., L. Klure, and L. Bower. 1984. Variability in sensitivity to metalaxyl of isolates of Phytophthora cinnamomi and Phytophthora citricola. Phytopathology 74:417-422. [Google Scholar]
- 45.Colwell, R. K., and J. A. Coddington. 1994. Estimating terrestrial biodiversity through extrapolation. Philos. Trans. R. Soc. Lond. Ser. B 345:101-118. [DOI] [PubMed] [Google Scholar]
- 46.Colwell, R. R. 1996. Microbial biodiversity and biotechnology, p. 279-287. In M. L. Reaka-Kudla, D. E. Wilson, and E. O. Wilson (ed.), Biodiversity II: understanding and protecting our biological resources. Joseph Henry Press, Washington, D.C.
- 47.Cook, D., E. Barlow, and L. Sequeira. 1989. Genetic diversity of Pseudomonas solanacearum: detection of restriction fragment length polymorphisms with DNA probes that specify virulence and the hypersensitive response. Mol. Plant-Microbe Interact. 2:113-121. [Google Scholar]
- 48.Coskun, H., G. Bateman, and D. Hollomon. 1987. Changes in population structure of carbendazim-resistant eyespot on wheat and barley between spring and summer 1984. Trans. Br. Mycol. Soc. 88:117-119. [Google Scholar]
- 49.Cuenca, G., and E. Meneses. 1996. Diversity patterns of arbuscular mycorrhizal fungi associated with cacao in Venezuela. Plant Soil 183:315-322. [Google Scholar]
- 50.De Leij, F. A. A. M., E. J. Sutton, J. M. Whipps, J. S. Fenlon, and J. M. Lynch. 1995. Impact of field release of genetically modified Pseudomonas fluorescens in indigenous microbial populations of wheat. Appl. Environ. Microbiol. 61:3443-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Denny, T. P. 1988. Phenotypic diversity in Pseudomonas syringae pv. tomato. J. Gen. Microbiol. 134:1939-1948. [DOI] [PubMed] [Google Scholar]
- 52.Derry, A. M., W. J. Staddon, and J. T. Trevors. 1998. Functional diversity and community structure of microorganisms in uncontaminated and creosote-contaminated soils as determined by sole-carbon-source-utilization. World J. Microbiol. Biotechnol. 14:571-578. [Google Scholar]
- 53.Dickinson, T. A., and L. J. Hutchison. 1997. Numerical taxonomic methods, cultural characters, and the systematics of ectomycorrhizal agarics, boletes and gasteromycetes. Mycol. Res. 101:477-492. [Google Scholar]
- 54.Doelman, P., E. Jansen, M. Michels, and M. van Til. 1994. Effects of heavy metals in soil on microbial diversity and activity as shown by the sensitivity-resistance index, an ecologically relevant parameter. Biol. Fertil. Soils 17:177-184. [Google Scholar]
- 55.Doering-Saad, C., P. Kampfer, S. Manulis, G. Kritzman, J. Schneider, J. Zakrzewska-Czerwinska, H. Schrempf, and I. Barash. 1992. Diversity among Streptomyces strains causing potato scab. Appl. Environ. Microbiol. 58:3932-3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dunbar, J., S. Takala, S. M. Barns, J. A. Davis, and C. R. Kuske. 1999. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl. Environ. Microbiol. 65:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eberhardt, U., L. Walter, and I. Kottke. 1999. Molecular and morphological discrimination between Tylospora fibrillosa and Tylospora asterophora mycorrhizae. Can. J. Bot. 77:11-21. [Google Scholar]
- 58.Ellis, J. R., W. Roder, and S. C. Mason. 1992. Grain sorghum-soybean rotation and fertilization influence on vesicular-arbuscular mycorrhizal fungi. Soil Sci. Soc. Am. J. 56:789-794. [Google Scholar]
- 59.El-Tarabily, K. A., G. E. S. J. Hardy, K. Sivasithamparam, and I. D. Kurtboeke. 1996. Microbiological differences between limed and unlimed soils and their relationship with cavity spot disease of carrots (Daucus carota L.) caused by Pythium coloratum in Western Australia. Plant Soil. 183:279-290. [Google Scholar]
- 60.Emerson, D., and J. A. Breznak. 1997. The response of microbial populations from oil-brine contaminated soil to gradients of NaCl and sodium p-toluate in a diffusion gradient chamber. FEMS Microbiol. Ecol. 23:285-300. [Google Scholar]
- 61.Ennos, R. A., and K. W. Swales. 1991. Genetic variability and population structure in the canker pathogen Crumenulopsis sororia. Mycol. Res. 95:521-525. [Google Scholar]
- 62.Ercolani, G. L. 1983. Variability among isolates of Pseudomonas syringae pv. savastanoi from the phylloplane of the olive. J. Gen. Microbiol. 129:901-916 [DOI] [PubMed] [Google Scholar]
- 63.Faucher, E., E. Paradis, C. Goyer, N. C. Hodge, R. Hogue, R. E. Stall, and C. Beaulieu. 1995. Characterization of streptomycetes causing deep-pitted scab of potato in Quebec, Canada. Int. J. Syst. Bacteriol. 45:222-225. [Google Scholar]
- 64.Federle, T. W., R. M. Ventullo, and D. C. White. 1990. Spatial distribution of microbial biomass, activity, community structure, and the biodegradation of linear alkylbenzene sulfonate (LAS) and linear alcohol ethoxylate (LAE) in the subsurface. Microb. Ecol. 20:297-313. [DOI] [PubMed] [Google Scholar]
- 65.Forbes, G., X. Escobar, C. Ayala, J. Revelo, M. Ordonez, B. Fry, K. Doucett, and W. Fry. 1997. Population genetic structure of Phytophthora infestans in Ecuador. Phytopathology 87:375-380. [DOI] [PubMed] [Google Scholar]
- 66.Franco-Abuin, C. M., E. J. Quinto-Fernandez, C. Fente-Sampayo, J. L. Rodriguez-Otero, L. Dominguez-Rodriguez, and A. Cepada-Saez. 1996. Incidence of Listeria species in the environment of a cheese processing plant throughout one year. Arch. Lebensmittelhyg. 47:25-27. [Google Scholar]
- 67.Freitas, A. C., M. J. Sousa, and F. X. Malcata. 1995. Effect of ripening time and the combination of ewe and goat milk on the microflora of Picante cheese. Ital. J. Food Sci. 7:361-377. [Google Scholar]
- 68.Friese, C. F., and R. E. Koske. 1991. The spatial dispersion of spores of vesicular-arbuscular mycorrhizal fungi in a sand dune: microscale patterns associated with the root architecture of American beachgrass. Mycol. Res. 95:952-957. [Google Scholar]
- 69.Frostegard, A., E. Baath, and A. Tunlid. 1993. Shifts in the structure of soil microbial communities in limed forests as revealed by phospholipid fatty acid analysis. Soil Biol. Biochem. 25:723-730. [Google Scholar]
- 70.Fuhrman, J., K. McCallum, and A. Davis. 1993. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans. Appl. Environ. Microbiol. 59:1294-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fulthorpe, R. R., A. N. Rhodes, and J. M. Tiedje. 1996. Pristine soils mineralize 3-chlorobenzoate and 2,4-dichlorophenoxyacetate via different microbial populations. Appl. Environ. Microbiol. 62:1159-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gagnevin, L., J. E. Leach, and O. Pruvost. 1997. Genomic variability of the Xanthomonas pathovar mangiferaeindicae, agent of mango bacterial black spot. Appl. Environ. Microbiol. 63:246-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Garcia, M. C., A. Otero, M. L. Garcia, M. Sierra, and B. Moreno. 1990. Numerical taxonomy of Micrococcaceae isolated from Spanish sheep's milk cheeses. J. Appl. Bacteriol. 68:33-41. [Google Scholar]
- 74.Gardes, M., and T. D. Bruns. 1996. Community structure of ectomycorrhizal fungi in a Pinus muricata forest: above- and below-ground views. Can. J. Bot. 74:1572-1583. [Google Scholar]
- 75.Garland, J. L. 1996. Patterns of potential C source utilization by rhizosphere communities. Soil Biol. Biochem. 28:223-230. [Google Scholar]
- 76.Garland, J. L. 1996. Analytical approaches to the characterization of samples of microbial communities using patterns of potential C source utilization. Soil Biol. Biochem. 28:213-221. [Google Scholar]
- 77.Gay, G., and J. C. Debaud. 1987. Genetic study on indole-3-acetic acid production by ectomycorrhizal Hebeloma species: inter- and intraspecific variability in homo- and dikaryotic mycelia. Appl. Microbiol. Biotechnol. 26:141-146. [Google Scholar]
- 78.Geniaux, E., and N. Amarger. 1993. Diversity and stability of plasmid transfer in isolates from a single field population of Rhizobium leguminosarum bv. viciae. FEMS Microbiol. Ecol. 102:251-260. [Google Scholar]
- 79.Gingell, S. M., R. Campbell, and M. H. Martin. 1976. The effect of zinc, lead and cadmium pollution on the leaf surface microflora. Environ. Pollut. 11:25-37. [Google Scholar]
- 80.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 81.Glockner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gonzalez, J. M., and M. A. Moran. 1997. Numerical dominance of a group of marine bacteria in the alpha-subclass of the class Proteobacteria in coastal seawater. Appl. Environ. Microbiol. 63:4237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goodman, D. M., and J. A. Trofymow. 1998. Comparison of communities of ectomycorrhizal fungi in old-growth and mature stands of Douglas-fir at two sites on southern Vancouver Island. Can. J. For. Res. 28:574-581. [Google Scholar]
- 84.Gordon, D. M., M. Wexler, T. B. Reardon, and P. J. Murphy. 1995. The genetic structure of Rhizobium populations. Soil Biol. Biochem. 27:491-499. [Google Scholar]
- 85.Gould, A. B., and J. W. Hendrix. 1998. Relationship of mycorrhizal activity to time following reclamation of surface mine land in western Kentucky. II. Mycorrhizal fungal communities. Can. J. Bot. 76:204-212. [Google Scholar]
- 86.Grayston, S. J., S. Wang, C. D. Campbell, and A. C. Edwards. 1998. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol. Biochem. 30:369-378. [Google Scholar]
- 87.Grosskopf, R., P. H. Janssen, and W. O. B. Liesack. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grundmann, G. L., and P. Normand. 2000. Microscale diversity of the genus Nitrobacter in soil on the basis of analysis of genes encoding rRNA. Appl. Environ. Microbiol. 66:4543-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gsell, T. C., W. E. Holben, and R. M. Ventullo. 1997. Characterization of the sediment bacterial community in groundwater discharge zones of an alkaline fen: a seasonal study. Appl. Environ. Microbiol. 63:3111-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guidot, A., E. Lumini, J. C. Debaud, and R. Marmeisse. 1999. The nuclear ribosomal DNA intergenic spacer as a target sequence to study intraspecific diversity of the ectomycorrhizal basidiomycete Hebeloma cylindrosporum directly on Pinus root systems. Appl. Environ. Microbiol. 65:903-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hagerman, S. M., M. D. Jones, G. E. Bradfield, M. Gillespie, and D. M. Durall. 1999. Effects of clear-cut logging on the diversity and persistence of ectomycorrhizae at a subalpine forest. Can. J. For. Res. 29:124-134. [Google Scholar]
- 92.Hallmann, J., J. W. Kloepper, and R. Rodriguez-Kabana. 1997. Application of the Scholander pressure bomb to studies on endophytic bacteria of plants. Can. J. Microbiol. 43:411-416. [Google Scholar]
- 93.Hallmann, J., R. Rodriguez-Kabana, and J. W. Kloepper. 1999. Chitin-mediated changes in bacterial communities of the soil, rhizosphere and within roots of cotton in relation to nematode control. Soil Biol. Biochem. 31:551-560. [Google Scholar]
- 94.Hamelin, R. 1996. Genetic diversity between and within cankers of the white pine blister rust. Phytopathology 86:875-879. [Google Scholar]
- 95.Handley, B. A., A. J. Hedges, and J. E. Beringer. 1998. Importance of host plants for detecting the population diversity of Rhizobium leguminosarum biovar viciae in soil. Soil Biol. Biochem. 30:241-249. [Google Scholar]
- 96.Hantula, J., T. T. Koivula, C. Luo, and D. H. Bamford. 1996. Bacterial diversity at surface water in three locations within the Baltic sea as revealed by culture-dependent molecular techniques. J. Basic Microbiol. 36:163-176. [DOI] [PubMed] [Google Scholar]
- 97.Hariston, N. G., J. D. Allan, R. K. Colwell, D. J. Futuyma, J. Howell, M. D. Lubin, J. Mathias, and J. H. Vandermeer. 1968. The relationship between species diversity and stability: an experimental approach with protozoa and bacteria. Ecology 49:1091-1101. [Google Scholar]
- 98.Harrison, S. P., D. G. Jones, and J. P. W. Young. 1989. Rhizobium population genetics: genetic variation within and between populations from diverse locations. J. Gen. Microbiol. 135:1061-1069. [Google Scholar]
- 99.Hartmann, A., J. J. Giraud, and G. Catroux. 1998. Genotypic diversity of Sinorhizobium (formerly Rhizobium) meliloti strains isolated directly from a soil and from nodules of alfalfa (Medicago sativa) grown in the same soil. FEMS Microbiol. Ecol. 25:107-116. [Google Scholar]
- 100.Hartung, J. S., and E. L. Civerolo. 1989. Restriction fragment length polymorphisms distinguish Xanthomonas campestris strains isolated from Florida citrus nurseries from X. c. pv. citri. Phytopathology 79:793-799. [Google Scholar]
- 101.Healy, F. G., and D. H. Lambert. 1991. Relationships among Streptomyces spp. causing potato scab. Int. J. Syst. Bacteriol. 41:479-482. [Google Scholar]
- 102.Heck, K. L., G. V. Belle, and D. Simberloff. 1975. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 56:1459-1461. [Google Scholar]
- 103.Heilmann, B., M. Lebuhn, and F. Beese. 1995. Methods for the investigation of metabolic activities and shifts in the microbial community in a soil treated with a fungicide. Biol. Fertil. Soils 19:186-192. [Google Scholar]
- 104.Hélias, V., A.-C. Le Roux, Y. Bertheau, D. Andrivon, J.-P. Gauthier, and B. Jouan. 1998. Characterization of Erwinia carotovora subspecies and detection of Erwinia carotovora subsp. atroseptica in potato plants, soil and water extracts with PCR-based methods. Eur. J. Plant Pathol. 104:685-699. [Google Scholar]
- 105.Helm, D. J., E. B. Allen, and J. M. Trappe. 1996. Mycorrhizal chronosequence near Exit Glacier, Alaska. Can. J. Bot. 74:1496-1506. [Google Scholar]
- 106.Helmecke, K., B. Hickisch, E. G. Mahn, J. Prasse, and G. Sternkopf. 1977. Contribution to the action of herbicide application on the structure and metabolism of agro-ecosystems. I. Effects on biomass and soil organism properties after several years herbicide application. Hercynia 14:375-398. [Google Scholar]
- 107.Henckel, T., M. Friedrich, and R. Conrad. 1999. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl. Environ. Microbiol. 65:1980-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Herman, R. P. 1997. Shrub invasion and bacterial community pattern in Swedish pasture soil. FEMS Microbiol. Ecol. 24:235-242. [Google Scholar]
- 109.Heuer, H., and K. Smalla. 1999. Bacterial phyllosphere communities of Solanum tuberosum L. and T4-lysozyme-producing transgenic variants. FEMS Microbiol. Ecol. 28:357-371. [Google Scholar]
- 110.Hiraishi, A., T. Umezawa, H. Yamamoto, K. Kato, and Y. Maki. 1999. Changes in quinone profiles of hot spring microbial mats with a thermal gradient. Appl. Environ. Microbiol. 65:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hiroki, M., and M. M. Watanabe. 1996. Microbial community and rate of cellulose decomposition in peat soils in a mire. Soil Sci. Plant Nutr. (Tokyo) 42:893-903. [Google Scholar]
- 112.Hollaway, G. J., M. R. Gillings, and P. C. Fahy. 1997. Use of fatty acid profiles and repetitive element polymerase chain reaction (PCR) to assess the genetic diversity of Pseudomonas syringae pv. pisi and Pseudomonas syringae pv. syringae isolated from field peas in Australia. Aust. Plant Pathol. 26:98-108. [Google Scholar]
- 113.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hutchinson, G. E. 1959. Homage to Santa Rosalia, or why are there so many kinds of animals? Am. Nat. 93:145-159. [Google Scholar]
- 116.Hutchison, L. J. 1990. Studies on the systematics of ectomycorrhizal fungi in axenic culture. III. Patterns of polyphenol oxidase activity. Mycologia 82:424-435. [Google Scholar]
- 117.Ibekwe, A. M., and A. C. Kennedy. 1998. Phospholipid fatty acid profiles and carbon utilization patterns for analysis of microbial community structure under field and greenhouse conditions. FEMS Microbiol. Ecol. 26:151-163. [Google Scholar]
- 118.Ikeda, K., K. Toyota, and M. Kimura. 1997. Effects of soil compaction on the microbial populations of melon and maize rhizoplane. Plant Soil 189:91-96. [Google Scholar]
- 119.Ingleby, K., R. C. Munro, M. Noor, P. A. Mason, and M. J. Clearwater. 1998. Ectomycorrhizal populations and growth of Shorea parvifolia (Dipterocarpaceae) seedlings regenerating under three different forest canopies following logging. For. Ecol. Manage. 111:171-179. [Google Scholar]
- 120.International Commission on Microbiological Specification for Foods. 1988. Application of the hazard analysis critical control point (HACCP) system to ensure microbiological safety and quality. Blackwell Scientific Publications, Oxford, United Kingdom.
- 121.Jindal, J. K., and P. N. Patel. 1984. Variability in xanthomonads of grain legumes. IV. Variations in bacteriological properties of 83 isolates and pathogenic behavior of cultural variants. Phytopathol. Z. 110:63-68. [Google Scholar]
- 122.Johnsen, K., and P. Nielsen. 1999. Diversity of Pseudomonas strains isolated with King's B and Gould's S1 agar determined by repetitive extragenic palindromic-polymerase chain reaction, 16S rDNA sequencing and Fourier transform infrared spectroscopy characterisation. FEMS Microbiol. Lett. 73:155-162. [DOI] [PubMed] [Google Scholar]
- 123.Johnson, C. N. 1995. Interactions between fire, mycophagous mammals, and dispersal of ectomycorrhizal fungi in Eucalyptus forests. Oecologia 104:467-475. [DOI] [PubMed] [Google Scholar]
- 124.Johnson, N. C., D. Tilman, and D. Wedin. 1992. Plant and soil controls on mycorrhizal fungal communities. Ecology 73:2034-2042. [Google Scholar]
- 125.Jones, F. T., and B. E. Langlois. 1977. Microflora of retail fluid milk products. J. Food Prot. 40:693-697. [DOI] [PubMed] [Google Scholar]
- 126.Jones, M. D., D. M. Durall, S. M. K. Harniman, D. C. Classen, and S. W. Simard. 1997. Ectomycorrhizal diversity on Betula papyrifera and Pseudotsuga menziesii seedlings grown in the greenhouse or outplanted in single-species and mixed plots in southern British Columbia. Can. J. For. Res. 27:1872-1889. [Google Scholar]
- 127.Jonsson, L., A. Dahlberg, M. C. Nilsson, O. Zackrisson, and O. Karen. 1999. Ectomycorrhizal fungal communities in late-successional Swedish boreal forests, and their composition following wildfire. Mol. Ecol. 8:205-215. [Google Scholar]
- 128.Ka, J. O., P. Burauel, J. A. Bronson, W. E. Holben, and J. M. Tiedje. 1995. Division S-3-soil biology & biochemistry: DNA probe analysis of microbial community selected in field by long-term 2,4-D application. Soil Sci. Soc. Am. J. 59:1581-1587. [Google Scholar]
- 129.Kandeler, E., C. Kampichler, and O. Horak. 1996. Influence of heavy metals on the functional diversity of soil microbial communities. Biol. Fertil. Soils 23:299-306. [Google Scholar]
- 130.Kaneko, T., and R. Atlas. 1977. Diversity of bacterial populations in the Beaufort Sea. Nature 270:596-599. [Google Scholar]
- 131.Karen, O., and J. E. Nylund. 1997. Effects of ammonium sulphate on the community structure and biomass of ectomycorrhizal fungi in a Norway spruce stand in southwestern Sweden. Can. J. Bot. 75:1628-1642. [Google Scholar]
- 132.Keller, S. M., M. S. Wolfe, J. M. McDermott, and B. A. McDonald. 1997. High genetic similarity among populations of Phaeosphaeria nodorum across wheat cultivars and regions in Switzerland. Phytopathology 87:1134-1139. [DOI] [PubMed] [Google Scholar]
- 133.Kelley, W. D., and R. Rodriguez-Kabana. 1979. Effects of sodium azide and methyl bromide on soil bacterial populations, enzymic activities and other biological variables. Pestic. Sci. 10:207-215. [Google Scholar]
- 134.Kelly, J. J., and R. L. Tate III. 1998. Effects of heavy metal contamination and remediation on soil microbial communities in the vicinity of a zinc smelter. J. Environ. Qual. 27:609-617. [Google Scholar]
- 135.Kennedy, A. C., and K. L. Smith. 1995. Soil microbial diversity and the sustainability of agricultural soils. Plant Soil 170:75-86. [Google Scholar]
- 136.Kerssies, A., A. Bosker-vanZessen, C. Wagemakers, and J. van Kan. 1997. Variation in pathogenicity and DNA polymorphism among Botrytis cinerea isolates sampled inside and outside a glasshouse. Plant Dis. 81:781-786. [DOI] [PubMed] [Google Scholar]
- 137.Khbaya, B., M. Neyra, P. Normand, K. Zerhari, and A. Filali-Maltouf. 1998. Genetic diversity and phylogeny of rhizobia that nodulate Acacia spp. in Morocco assessed by analysis of rRNA genes. Appl. Environ. Microbiol. 64:4912-4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kieliszewska-Rokicka, B., M. Rudawska, and T. Leski. 1997. Ectomycorrhizae of young and mature Scots pine trees in industrial regions in Poland. Environ. Pollut. 98:315-324. [Google Scholar]
- 139.Kinkel, L. L., E. V. Nordheim, and J. H. Andrews. 1992. Microbial community analysis in incompletely or desctructively sampled systems. Microb. Ecol. 24:227-242. [DOI] [PubMed] [Google Scholar]
- 140.Klausner, R. B., and C. W. Donnelly. 1991. Environmental sources of Listeria and Yersinia in Vermont dairy plants. J. Food Prot. 54:607-611. [DOI] [PubMed] [Google Scholar]
- 141.Kohn, L., E. Stasovski, I. Carbone, J. Royer, and J. Anderson. 1991. Mycelial incompatibility and molecular markers identify genetic variability in field populations of Sclerotinia sclerotiorum. Phytopathology 81:480-485. [Google Scholar]
- 142.Kolmer, J. 1992. Effect of sexual recombination in two populations of the wheat leaf rust fungus Puccinia recondita. Can. J. Bot. 70:359-363. [Google Scholar]
- 143.Kope, H. H., and J. A. Fortin. 1991. Genetic variation in antifungal activity by sibling isolates of the ectomycorrhizal fungi Pisolithus arhizus. Soil Biol. Biochem. 23:1047-1051. [Google Scholar]
- 144.Kowalchuk, G. A., J. R. Stephen, W. De Boer, J. I. Prosser, T. M. Embley, and J. W. Woldendorp. 1997. Analysis of ammonia-oxidizing bacteria of the beta-subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl. Environ. Microbiol. 63:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kozdroj, J. 1995. Microbial responses to single or successive soil contamination with Cd or Cu. Soil Biol. Biochem. 27:1459-1465. [Google Scholar]
- 146.Kranabetter, J. M., and T. Wylie. 1998. Ectomycorrhizal community structure across forest openings on naturally regenerated western hemlock seedlings. Can. J. Bot. 76:189-196. [Google Scholar]
- 147.Kuske, C. R., J. D. Busch, D. L. Adorada, J. M. Dunbar, and S. M. Barns. 1999. Phylogeny, ribosomal RNA gene typing and relative abundance of new Pseudomonas species (sensu stricto) isolated from two pinyon-juniper woodland soils of the arid southwest U.S. Syst. Appl. Microbiol. 22:300-311. [DOI] [PubMed] [Google Scholar]
- 148.Laguerre, G., E. Geniaux, S. I. Mazurier, R. R. Casartelli, and N. Amarger. 1993. Conformity and diversity among field isolates of Rhizobium leguminosarum bv. viciae, bv. trifolii, and bv. phaseoli revealed by DNA hybridization using chromosome and plasmid probes. Can. J. Microbiol. 39:412-419. [Google Scholar]
- 149.Laguerre, G., P. Van Berkum, N. Amarger, and D. Prevost. 1997. Genetic diversity of rhizobial symbionts isolated from legume species within the genera Astragalus, Oxytropis, and Onobrychis. Appl. Environ. Microbiol. 63:4748-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Latour, X., L. Philippot, T. Corberand, and P. Lemanceau. 1999. The establishment of an introduced community of fluorescent pseudomonads in the soil and in the rhizosphere is affected by the soil type. FEMS Microbiol. Ecol. 30:163-170. [DOI] [PubMed] [Google Scholar]
- 151.Leach, J. E., M. L. Rhoads, C. M. Vera Cruz, F. F. White, T. W. Mew, and H. Leung 1992. Assessment of genetic diversity and population structure of Xanthomonas oryzae pv. oryzae with a repetitive DNA element. Appl. Environ. Microbiol. 58:2188-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lebreton, L., C. Laurent, and D. Andrivon. 1998. Evolution of Phytophthora infestans populations in the two most important potato production areas of France during 1992-96. Phytopathology 47:427-439. [Google Scholar]
- 153.Lecomte, P., C. Manceau, J. P. Paulin, and M. Keck. 1997. Identification by PCR analysis on plasmid pEA29 of isolates of Erwinia amylovora responsible for an outbreak in Central Europe. Eur. J. Plant Pathol. 103:91-98. [Google Scholar]
- 154.Lemanceau, P., T. Corberand, L. Gardan, X. Latour, G. Laguerre, J.-M. Boeufgras, and C. Alabouvette. 1995. Effect of two plant species, flax (Linum usitatissinum L.) and tomato (Lycopersicon esculentum Mill.), on the diversity of soilborne populations of fluorescent pseudomonads. Appl. Environ. Microbiol. 61:1004-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lemke, M. J., and L. G. Leff. 1999. Bacterial populations in an anthropogenically disturbed stream: comparison of different seasons. Microb. Ecol. 38:234-243. [DOI] [PubMed] [Google Scholar]
- 156.Leung, H., and P. Williams. 1986. Enzyme polymorphism and genetic differentiation among geographic isolates of the rice blast fungus. Phytopathology 76:778-783. [Google Scholar]
- 157.Leung, K., K. Yap, N. Dashti, and P. J. Bottomley. 1994. Serological and ecological characteristics of a nodule-dominant serotype from an indigenous soil population of Rhizobium leguminosarum bv. trifolii. Appl. Environ. Microbiol. 60:408-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Levy, M., J. Romao, M. A. Marchetti, and J. E. Hamer. 1991. DNA fingerprinting with a dispersed repeated sequence resolves pathotype diversity in the rice blast fungus. Plant Cell. 3:95-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lie, T. A., and D. Goktan. 1984. Gene centers, a source for genetic variants in symbiotic nitrogen fixation: the symbiotic response of the cultivated pea to Rhizobium leguminosarum strains from Europe and the Middle East. Plant Soil 82:359-367. [Google Scholar]
- 160.Liesack, W., and E. Stackebrandt. 1992. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J. Bacteriol. 174:5072-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lloyd-Jones, G., A. D. Laurie, D. W. F. Hunter, and R. Fraser. 1999. Analysis of catabolic genes for naphthalene and phenanthrene degradation in contaminated New Zealand soils. FEMS Microbiol. Ecol. 29:69-79. [Google Scholar]
- 162.Long, R. A., and F. Azam. 2001. Microscale patchiness of bacterioplankton assemblage richness in seawater. Aquat. Micbob. Ecol. 26:130-113. [Google Scholar]
- 163.Lottmann, J., and G. Berg. 1999. Antifungal bacteria in the rhizosphere and geocaulosphere of transgenic T4 lysozyme-expressing potato plants. Bull. Pol. Acad. Sci. Biol. Sci. 47:77-84. [Google Scholar]
- 164.Lur'e, N. Y. 1985. Effect of industrial emissions from metallurgy plants on the structure of microbial communities in southern chernozem soils. Khim. Sel'sk. Khoz. 6:52-54. [Google Scholar]
- 165.MacArthur, R. H. 1957. On the relative abundance of bird species. Proc. Natl. Acad. Sci. USA 43:293-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.MacArthur, R. H., and J. W. MacArthur. 1961. On bird species diversity. Ecology 42:594-598. [Google Scholar]
- 167.Magurran, A. E. 1988. Ecological diversity and its measurement. Princeton University Press, Princeton, N.J.
- 168.Magurran, A. E. 1995. Ecological diversity and its measurement, 2nd ed. Princeton University Press, Princeton, N.J.
- 169.Mahaffee, W. F., and J. W. Kloepper. 1997. Bacterial communities of the rhizosphere and endorhiza associated with field-grown cucumber plants inoculated with a plant growth-promoting rhizobacterium or its genetically modified derivative. Can. J. Microbiol. 43:344-353. [DOI] [PubMed] [Google Scholar]
- 170.Majewski, T. 1975. Effect of milking and storing conditions on the psychrotropic and mesophilic microflora of milk. Pol. Arch. Weter. 18:9-15. [Google Scholar]
- 171.Maloney, P. E., A. H. C. van Bruggen, and S. Hu. 1997. Bacterial community structure in relation to the carbon environments in lettuce and tomato rhizospheres and in bulk soil. Microb. Ecol. 34:109-117. [DOI] [PubMed] [Google Scholar]
- 172.Mamoun, M., and J. M. Olivier. 1993. Competition between Tuber melanosporum and other ectomycorrhizal fungi under two irrigation regimes. II. Comparison of soils artificially infested with T. melanosporum and T. brumale. Plant Soil 149:219-225. [Google Scholar]
- 173.Marilley, L., U. A. Hartwig, and M. Aragno. 1999. Influence of an elevated atmospheric CO2 content on soil and rhizosphere bacterial communities beneath Lolium perenne and Trifolium repens under field conditions. Microb. Ecol. 38:39-49. [DOI] [PubMed] [Google Scholar]
- 174.Marks, G. C., and R. Cerra. 1991. Effects of propazine and chlorthal dimethyl on Phytophthora cinnamomi root disease of Pinus radiata seedlings and associated soil microflora. Soil Biol. Biochem. 23:157-164. [Google Scholar]
- 175.Martens, J., and D. Harder. 1983. Incidence and virulence of Puccinia graminis f.sp. avenae in Canada in 1982. Can. J. Plant Pathol. 5:189-193. [Google Scholar]
- 176.Massol-Deya, A., R. Weller, L. Rios-Hernandez, J. Z. Zhou, R. F. Hickey, and J. M. Tiedje. 1997. Succession and convergence of biofilm communities in fixed-film reactors treating aromatic hydrocarbons in groundwater. Appl. Environ. Microbiol. 63:270-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.McAlpin, C. E., D. T. Wicklow, and C. E. Platis. 1998. Genotypic diversity of Aspergillus parasiticus in an Illinois corn field. Plant Dis. 82:1132-1136. [DOI] [PubMed] [Google Scholar]
- 178.McArthur, J. V., D. A. Kovacic, and M. H. Smith. 1988. Genetic diversity in natural populations of a soil bacterium across a landscape gradient. Proc. Natl. Acad. Sci. USA 85:9621-9624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.McCain, J., J. Groth, and A. Roelfs. 1992. Inter- and intrapopulation isozyme variation in collections from sexually reproducing populations of the bean rust fungus, Uromyces appendiculatus. Mycologia 84:329-340. [Google Scholar]
- 180.McDonald, B., and J. Martinez. 1990. DNA restriction fragment length polymorphisms among Mycosphaerella graminicola (anamorph Septoria tritici) isolates collected from a single wheat field. Phytopathology 80:1368-1373. [Google Scholar]
- 181.Meharg, A. A., C. L. Wyatt, I. P. Thompson, M. J. Bailey, R. J. Ellis, and N. Maguire. 1998. Response of soil microbial biomass to 1,2-dichlorobenzene addition in the presence of plant residues. Environ. Toxicol. Chem. 17:1462-1468. [Google Scholar]
- 182.Meysselle, J. P., G. Gay, and J. C. Debaud. 1991. Intraspecific genetic variation of acid phosphatase activity in monokaryotic and dikaryotic populations of the ectomycorrhizal fungus Hebeloma cylindrosporum. Can. J. Bot. 69:808-813. [Google Scholar]
- 183.Miller, S. P., and J. D. Bever. 1999. Distribution of arbuscular mycorrhizal fungi in stands of the wetland grass Panicum hemitomon along a wide hydrologic gradient. Oecologia 119:586-592. [DOI] [PubMed] [Google Scholar]
- 184.Mmbaga, M., J. Steadman, and K. Eskridge. 1996. Virulence patterns of Uromyces appendiculatus from different geographical areas and implications for finding durable resistance to rust of common bean. J. Phytopathol. 144:533-541. [Google Scholar]
- 185.Moawad, H., S. M. S. Badr El-Din, and R. A. Abdel-Aziz. 1998. Improvement of biological nitrogen fixation in Egyptian winter legumes through better management of Rhizobium. Plant Soil 204:95-106. [Google Scholar]
- 186.Moyer, C., F. Dobbs, and D. Karl. 1995. Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl. Environ. Microbiol. 61:1555-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Mpepereki, S., A. G. Wollum, Jr., and F. Makonese. 1996. Diversity in symbiotic specificity of cowpea rhizobia indigenous to Zimbabwean soils. Plant Soil. 186:167-171. [Google Scholar]
- 188.Mudd, P. J., M. P. Greaves, and S. J. L. Wright. 1985. Effects of isoproturon in the rhizosphere of wheat. Weed Res. 25:423-432. [Google Scholar]
- 189.Muller, K., J. McDermott, M. Wolfe, and E. Limpert. 1996. Analysis of diversity in populations of plant pathogens: the barley powdery mildew pathogen across Europe. Eur. J. Plant Pathol. 102:385-395. [Google Scholar]
- 190.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 191.Naseby, D. C., and J. M. Lynch. 1998. Establishment and impact of Pseudomonas fluorescens genetically modified for lactose utilization and kanamycin resistance in the rhizosphere of pea. J. Appl. Microbiol. 84:169-175. [Google Scholar]
- 192.Naseby, D. C., and J. M. Lynch. 1998. Impact of wild-type and genetically modified Pseudomonas fluorescens on soil enzyme activities and microbial population structure in the rhizosphere of pea. Mol. Ecol. 7:617-625. [Google Scholar]
- 193.Natsch, A., C. Keel, N. Hebecker, E. Laasik, and G. Defago. 1998. Impact of Pseudomonas fluorescens strain CHA0 and a derivative with improved biocontrol activity on the culturable resident bacterial community on cucumber roots. FEMS Microbiol. Ecol. 27:365-380. [Google Scholar]
- 194.Naumann, K., and M. Lange-De la Camp. 1976. Effect of plant residues on the parasitic activity of soilborne pathogens and the saprophytic microflora of the soil. I. Model trials with Rhizoctonia solani Kuehn. Zentralbl. Bakteriol. Parasitenkd. Infektionskr. Hyg. Abt. 2 131:378-391. [PubMed] [Google Scholar]
- 195.Nei, M., and F. Tajima. 1981. DNA polymorphisms detectable by restriction endonucleases. Genetics 97:145-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Nelson, R. J., M. R. Baraoidan, C. M. Vera-Cruz, I. V. Yap, J. E. Leach, T. H. Mew, and H. Leung. 1994. Relationship between phylogeny and pathotype for the bacterial blight pathogen of rice. Appl. Environ. Microbiol. 60:3275-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Nguyen, C., W. Yan, F. I. Tacon, and F. Lapeyrie. 1992. Genetic variability of phosphate solubilizing activity by monokaryotic and dikaryotic mycelia of the ectomycorrhizal fungus Laccaria bicolor (Maire) P.D. Orton. Plant Soil 143:193-199. [Google Scholar]
- 198.Niemann, S., S. Keel, A. Puhler, and W. Selbitschka. 1997. Biocontrol strain Pseudomonas fluorescens CHAO and its genetically modified derivative with enhanced biocontrol capability exert comparable effects on the structure of a Sinorhizobium meliloti population in gnotobiotic systems. Biol. Fertil. Soils 25:240-244. [Google Scholar]
- 199.Nordgren, A., T. Kauri, E. Baath, and B. Soederstroem. 1986. Soil microbial activity, mycelial lengths and physiological groups of bacteria in a heavy metal polluted area. Environ. Pollut. Ser. A 41:89-100. [Google Scholar]
- 200.Oard, J. H., and M. D. Simons. 1983. A growth chamber comparison of traits of aggressiveness in sexual and asexual populations of Puccinia coronata. Phytopathology 73:1226-1229. [Google Scholar]
- 201.Obbard, J. P., and K. C. Jones. 1993. The effect of heavy metals on dinitrogen fixation by Rhizobium-white clover in a range of long-term sewage sludge amended and metal-contaminated soils. Environ. Pollut. 79:105-112. [DOI] [PubMed] [Google Scholar]
- 202.Ogawa, M., and Y. Yambe. 1980. Effects of herbicides on the mycorrhizae of Pinus densiflora and soil microorganisms. Ringyo Shikenjo Kenkyu Hokoku 311:93-102. [Google Scholar]
- 203.Okereke, G. U. 1984. Prevalence of nitrous oxide reducing capacity in denitrifiers from a variety of habitats. Plant Soil 81:421-428. [Google Scholar]
- 204.Osborn, A. M., K. D. Bruce, P. Strike, and D. A. Ritchie. 1993. Polymerase chain reaction-restriction fragment length polymorphism analysis shows divergence among mer determinants from gram-negative soil bacteria indistinguishable by DNA-DNA hybridization. Appl. Environ. Microbiol. 59:4024-4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Paffetti, D., C. Scotti, S. Gnocchi, S. Fancelli, and M. Bazzicalupo. 1996. Genetic diversity of an Italian Rhizobium meliloti population from different Medicago sativa varieties. Appl. Environ. Microbiol. 62:2279-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Parish, M. E. 1998. Coliforms, Escherichia coli and Salmonella serovars associated with a citrus-processing facility implicated in a salmonellosis outbreak. J. Food Prot. 3:280-284. [DOI] [PubMed] [Google Scholar]
- 207.Parisi, L., J. Guillaumes, and G. Wuster. 1994. Variability in sensitivity to fenarimol of Venturia inaequalis strains from French orchards. Agronomic 14:387-394. (In French.) [Google Scholar]
- 208.Peever, T. L., and M. G. Milgroom. 1994. Genetic structure of Pyrenophora teres populations determined with random amplified polymorphic DNA markers. Can. J. Bot. 72:915-923. [Google Scholar]
- 209.Pichinoty, F., J. L. Garcia, M. Mandel, C. Job, and M. Durand. 1978. Isolation of bacteria that use nitric oxide as a respiratory electron acceptor under anaerobiosis. C. R. Hebd. Seances Acad. Sci. Ser. D 286:1403-1405. [PubMed] [Google Scholar]
- 210.Pielou, E. C. 1975. Ecological diversity. John Wiley & Sons, Inc., New York, N.Y.
- 211.Priha, O., S. J. Grayston, T. Pennanen, and A. Smolander. 1999. Microbial activities related to C and N cycling and microbial community structure in the rhizospheres of Pinus sylvestris, Picea abies and Betula pendula seedlings in an organic and mineral soil. FEMS Microbiol. Ecol. 30:187-199. [DOI] [PubMed] [Google Scholar]
- 212.Pruvoust, O., A. Couteau, X. Perrier, and J. Luisetti. 1998. Phenotypic diversity of Xanthomonas sp. mangiferaeindicae. J. Appl. Microbiol. 84:115-124. [DOI] [PubMed] [Google Scholar]
- 213.Qian, X. M., I. Kottke, and F. Oberwinkler. 1998. Influence of liming and acidification on the activity of the mycorrhizal communities in a Picea abies (L.) Karst. stand. Plant Soil 199:99-109. [Google Scholar]
- 214.Rao, C. S., G. D. Sharma, and A. K. Shukla. 1997. Distribution of ectomycorrhizal fungi in pure stands of different age groups of Pinus kesiya. Can. J. Microbiol. 43:85-91. [Google Scholar]
- 215.Ravenschlag, K., K. Sahm, J. Pernthaler, and R. Amann. 1999. High bacterial diversity in permanently cold marine sediments. Appl. Environ. Microbiol. 65:3982-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Restrepo, S., and V. Verdier. 1997. Geographical differentiation of the population of Xanthomonas axonopodis pv. manihotis in Colombia. Appl. Environ. Microbiol. 63:4427-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Reysenbach, A. L., G. S. Wickham, and N. R. Pace. 1994. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl. Environ. Microbiol. 60:2113-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Rheims, H., and E. Stackebrandt. 1999. Application of nested polymerase chain reaction for the detection of as yet uncultured organisms of the class Actinobacteria in environmental samples. Environ. Microbiol. 1:137-143. [DOI] [PubMed] [Google Scholar]
- 219.Rice, S. A., J. Bieber, J. Y. Chun, G. Stacey, and B. C. Lampson. 1993. Diversity of retron elements in a population of rhizobia and other gram-negative bacteria. J. Bacteriol. 175:4250-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ritchie, N. J., and D. D. Myrold. 1999. Geographic distribution and genetic diversity of Ceanothus-infective Frankia strains. Appl. Environ. Microbiol. 65:1378-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Roberts, P. D., N. C. Hodge, H. Bouzar, J. B. Jones, R. E. Stall, R. D. Berger, and A. R. Chase. 1998. Relatedness of strains of Xanthomonas fragariae by restriction fragment length polymorphism, DNA-DNA reassociation, and fatty acid analyses. Appl. Environ. Microbiol. 64:3961-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Robleto, E. A., J. Borneman, and E. W. Triplett. 1998. Effects of bacterial antibiotic production on rhizosphere microbial communities from a culture-independent perspective. Appl. Environ. Microbiol. 64:5020-5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Rondon, M. R., P. R. August, A. D. Betterman, S. F. Brady, T. H. Grossman, M. R. Liles, K. A. Loiacono, B. A. Lynch, I. A. MacNeil, C. Minor, C. L. Tiong, M. Gilman, M. S. Osburne, J. Clardy, J. Handelsman, and R. M. Goodman. 2000. Cloning the soil metagenome: a strategy for assessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Rosado, S. C. S., B. R. Kropp, and Y. Piche. 1994. Genetics of ectomycorrhizal symbiosis. II. Fungal variability and heritability of ectomycorrhizal traits. New Phytol. 126:111-117. [Google Scholar]
- 225.Roth, D. R., and T. J. Fahey. 1998. The effects of acid precipitation and ozone on the ectomycorrhizae of red spruce saplings. Water Air Soil Pollut. 103:263-276. [Google Scholar]
- 226.Rowe, R., and M. Beute. 1975. Variability in virulence of Cylindrocladium crotalariae isolates on peanut. Phytopathology 65:422-425. [Google Scholar]
- 227.Safir, G. R., J. O. Siqueira, and T. M. Burton. 1990. Vesicular-arbuscular mycorrhizas in a wastewater-irrigated oldfield ecosystem in Michigan. Plant Soil 121:187-196. [Google Scholar]
- 228.Sakano, Y., and L. Kerkhof. 1998. Assessment of changes in microbial community structure during operation of an ammonia biofilter with molecular tools. Appl. Environ. Microbiol. 64:4877-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Sammarco, M. L., G. Ripabelli, A. Ruberto, G. Ianitto, and G. M. Grasso. 1997. Prevalence of salmonellae, listeriae, and yersiniae in the slaughterhouse environment and on work surfaces, equipment, and workers. J. Food Prot. 60:367-371. [DOI] [PubMed] [Google Scholar]
- 230.Sanders, H. L. 1968. Marine benthic diversity: a comparative study. Am. Nat. 102:243-282. [Google Scholar]
- 231.Sanders, I. R. 1993. Temporal infectivity and specificity of vesicular mycorrhizas in co-existing grassland species. Oecologia 93:349-355. [DOI] [PubMed] [Google Scholar]
- 232.Santegoeds, C. M., S. C. Nold, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis used to monitor the enrichment culture of aerobic chemoorganotrophic bacteria from a hot spring cyanobacterial mat. Appl. Environ. Microbiol. 62:3922-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sato, K., H. Kato, and C. Furusaka. 1987. Comparative study of soil bacterial flora as influenced by the application of a pesticide, pentachlorophenol (PCP). Plant Soil 100:333-343. [Google Scholar]
- 234.Schmieman, E. A., J. N. Petersen, D. R. Yonge, D. L. Johnstone, Y. Bereded-Samuel, W. A. Apel, and C. E. Turick. 1997. Bacterial reduction of chromium. Appl. Biochem. Biotechnol. 63-65:855-864. [DOI] [PubMed] [Google Scholar]
- 235.Schut, F., E.-J. DeVries, J. C. Gottschal, B. R. Robertson, W. Harder, R. A. Prins, and D. K. Button. 1993. Isolation of typical marine bacteria by dilution culture: growth, maintenance and characteristics of isolates under laboratory conditions. Appl. Environ. Microbiol. 59:2150-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Scott, J. S., and G. R. Knudsen. 1999. Soil amendment effects of rape (Brassica napus) residues on pea rhizosphere bacteria. Soil Biol. Biochem. 31:1435-1441. [Google Scholar]
- 237.Segovia, L., D. Pinero, R. Palacios, and E. Martinez-Romero. 1991. Genetic structure of a soil population of nonsymbiotic Rhizobium leguminosarum. Appl. Environ. Microbiol. 57:426-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Semenov, A. M., A. H. C. van Bruggen, and V. V. Zelenev. 1999. Moving waves of bacterial populations and total organic carbon along roots of wheat. Microb. Ecol. 37:116-128. [DOI] [PubMed] [Google Scholar]
- 239.Shannon, C. E., and W. Weaver. 1949. The mathematical theory of communications. University of Illinois Press, Urbana.
- 240.Shaw, P. J. A., J. Dighton, and J. Poskitt. 1993. Studies on the mycorrhizal community infecting trees in the Liphook Forest fumigation experiment. Agric. Ecosyst. Environ. 47:185-191. [Google Scholar]
- 241.Shaw, P. J. A., J. Dighton, J. Poskitt, and A. R. McLeod. 1992. The effect of sulphur dioxide and ozone on the mycorrhizas of Scots pine and Norway spruce in a field fumigation system. Mycol. Res. 96:785-791. [Google Scholar]
- 242.Siciliano, S. D., C. M. Theoret, J. R. De Freitas, P. J. Hucl, and J. J. Germida. 1998. Differences in the microbial communities associated with the roots of different cultivars of canola and wheat. Can. J. Microbiol. 44:844-851. [Google Scholar]
- 243.Simard, S. W., D. A. Perry, J. E. Smith, and R. Molina. 1997. Effects of soil trenching on occurrence of ectomycorrhizas on Pseudotsuga menziesii seedlings grown in mature forests of Betula papyrifera and Pseudotsuga menziesii. New Phytol. 136:327-340. [Google Scholar]
- 244.Simpson, E. H. 1949. Measurement of diversity. Nature (London) 163:688. [Google Scholar]
- 245.Snaidr, J., R. Amann, I. Huber, W. Ludwig, and K. H. Schleifer. 1997. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 63:2884-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Somé, A., and R. Samson. 1996. Isoenzyme diversity in Xanthomonas campestris pv. mangiferaeindicae. Plant Pathol. 45:426-431. [Google Scholar]
- 247.Spirina, E. V., and D. G. Fedorov-Davydov. 1998. Microbiological characterization of cryogenic soils in the Kolymskaya Lowland. Eurasian Soil Sci. 31:1331-1344. [Google Scholar]
- 248.Stahl, P. D., M. Christensen, and S. E. Williams. 1990. Population variation in the mycorrhizal fungus Glomus mosseae: uniform garden experiments. Mycol. Res. 94:1070-1076. [Google Scholar]
- 249.Steele, M., B. McNab, L. Fruhner, S. DeGrandis, D. Woodward, and J. A. Odumeru. 1998. Epidemiological typing of Campylobacter isolates from meat processing plants by pulsed-field gel electrophoresis, fatty acid profile typing, serotyping, and biotyping. Appl. Environ. Microbiol. 64:2346-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Stendell, E. R., T. R. Horton, and T. D. Bruns. 1999. Early effects of prescribed fire on the structure of the ectomycorrhizal fungus community in a Sierra Nevada ponderosa pine forest. Mycol. Res. 103:1353-1359. [Google Scholar]
- 251.Stephen, J. R., Y.-J. Chang, S. J. Macnaughton, G. A. Kowalchuk, K. T. Leung, C. A. Flemming, and D. C. White. 1999. Effect of toxic metals on indigenous soil beta-subgroup Proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl. Environ. Microbiol. 65:95-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Strain, S. R., K. Leung, T. S. Whittam, F. J. de Bruijn, and P. J. Bottomley. 1994. Genetic structure of Rhizobium leguminosarum biovar trifolii and viciae populations found in two Oregon soils under different plant communities. Appl. Environ. Microbiol. 60:2772-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Sturmer, S. L., and M. M. Bellei. 1994. Composition and seasonal variation of spore populations of arbuscular mycorrhizal fungi in dune soils on the island of Santa Catarina, Brazil. Can. J. Bot. 72:359-363. [Google Scholar]
- 254.Stutz, J. C., and J. B. Morton. 1996. Successive pot cultures reveal high species richness of arbuscular endomycorrhizal fungi in arid ecosystems. Can. J. Bot. 74:1883-1889. [Google Scholar]
- 255.Sullivan, J. T., H. N. Patrick, W. L. Lowther, D. B. Scott, and C. W. Ronson. 1995. Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene transfer in the environment. Proc. Natl. Acad. Sci. USA 92:8985-8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Surange, S., A. G. Wollum, II, N. Kumar, and C. S. Nautiyal. 1997. Characterization of Rhizobium from root nodules of leguminous trees growing in alkaline soils. Can. J. Microbiol. 43:891-894. [Google Scholar]
- 257.Swift, M. J. 1974. Species diversity and the structure of microbial communities in terrestrial habitats, p. 185-221 In J. M. Anderson and A. McFadyen (ed.), The role of terrestrial and aquatic organisms in decomposition processes. Blackwell Scientific Publications, Oxford, United Kingdom.
- 258.Sylvia, D. M. 1986. Spatial and temporal distribution of vesicular-arbuscular mycorrhizal fungi associated with Uniola paniculata in Florida foredunes. Mycologia 78:728-735. [Google Scholar]
- 259.Tamplin, M. L., J. K. Jackson, C. Buchrieser, R. L. Murphree, K. M. Portier, V. Gangar, L. G. Miller, and C. W. Kaspar. 1996. Pulse-field gel electrophoresis and ribotype profiles of clinical and environmental Vibio vulnificus isolates. Appl. Environ. Microbiol. 62:3572-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Tang, X. Y., Z. L. Zhang, Y. Cheng, H. F. Wan, and W. M. Zhu. 1997. Effect of Ce accumulation on soil microflora in yellow cinnamon soil. Yingyong Shengtai Xuebao 8:585-588. [Google Scholar]
- 261.Tenzer, I., and C. Gessler. 1997. Subdivision and genetic structure of four populations of Venturia inaequalis in Switzerland. Eur. J. Plant Pathol. 103:565-571. [Google Scholar]
- 262.Theron, J., and T. E. Cloete. 2000. Molecular techniques for determining microbial diversity and community structure in natural environments. Crit. Rev. Microbiol. 26:37-57. [DOI] [PubMed] [Google Scholar]
- 263.Tilman, D., C. L. Lehman, and K. T. Thompson. 1997. Plant diversity and ecosystem productivity: theoretical considerations. Proc. Natl. Acad. Sci. USA 94:1857-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Tilman, D., J. Knops, D. Wedin, P. Reich, M. Ritchie, and E. Siemann. 1997. The influence of functional diversity and composition on ecosystem processes. Science 277:1300-1302. [Google Scholar]
- 265.Tipper, J. C. 1979. Rarefaction and rarefiction—the use and abuse of a method in paleoecology. Paleobiology 5:423-434. [Google Scholar]
- 266.Torsvik, V., K. Salte, R. Sorheim, and J. Goksoyr. 1990. Comparison of phenotypic diversity and DNA heterogeneity in a population of soil bacteria. Appl. Environ. Microbiol. 56:776-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Toyota, K., K. Ritz, S. Kuninaga, and M. Kimura. 1999. Impact of fumigation with Metam sodium upon soil microbial community structure in two Japanese soils. Soil Sci. Plant Nutr. (Tokyo) 45:207-223. [Google Scholar]
- 268.Trinick, M. J. 1980. Growth of Parasponia in agar tube culture and symbiotic effectiveness of isolates from Parasponia spp. New Phytol. 85:37-45. [Google Scholar]
- 269.Ueda, T., Y. Suga, N. Yahiro, and T. Matsuguchi. 1995. Genetic diversity of N2-fixing bacteria associated with rice roots by molecular evolutionary analysis of a nifD library. Can. J. Microbiol. 41:235-240. [DOI] [PubMed] [Google Scholar]
- 270.Ulrich, A., and T. Muller. 1998. Heterogeneity of plant-associated streptococci as characterized by phenotypic features and restriction analysis of PCR-amplified 16S rDNA. J. Appl. Microbiol. 84:293-303. [DOI] [PubMed] [Google Scholar]
- 271.Vahjen, W., J. C. Munch, and C. C. Tebbe. 1995. Carbon source utilization of soil extracted microorganisms as a tool to detect the effects of soil supplemented with genetically engineered and non-engineered Corynebacterium glutamicum and a recombinant peptide at the community level. FEMS Microbiol. Ecol. 18:317-328. [Google Scholar]
- 272.Van Berkum, P., D. Beyene, F. T. Vera, and H. H. Keyser. 1995. Variability among Rhizobium strains originating from nodules of Vicia faba. Appl. Environ. Microbiol. 61:2649-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.van der Heijden, M. J. A., J. N. Klinomoros, M. Ursic, P. Moutoglis, R. Streitwolf-Engel, T. Boller, A. Wiemken, and I. R. Sanders. 1998. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396:69-72. [Google Scholar]
- 274.van Hannen, E. J., G. Zwart, M. P. van Agterveld, H. G. Gons, J. Ebert, and H. J. Laanbroek. 1999. Changes in bacterial and eukaryotic community structure after mass lysis of filamentous cyanobacteria associated with viruses. Appl. Environ. Microbiol. 65:795-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Väre, H., M. Vestberg, and R. Ohtonen. 1997. Shifts in mycorrhiza and microbial activity along an oroarctic altitudinal gradient in Northern Fennoscandia. Arctic Alpine Res. 29:93-104. [Google Scholar]
- 276.Vera Cruz, C. M., E. Y. Ardales, D. Z. Skinner, J. Talag, R. J. Nelson, F. J. Louws, H. Leung, T. W. Mew, and J. E. Leach. 1996. Measurement of haplotypic variation in Xanthomonas oryzae pv. oryzae within a single field by rep-PCR and RFLP analyses. Phytopathology 86:1352-1359. [Google Scholar]
- 277.Verdier, V., S. Restrepo, G. Mosquera, M. C. Duque, A. Gerstl, and R. Laberry. 1998. Genetic and pathogenic variation of Xanthomonas axonopodis pv. manihotis in Venezuela. Plant Pathol. 47:601-608. [Google Scholar]
- 278.Vilich, V. 1997. Assessment of microbial communities by fatty acid analysis. Dev. Plant Pathol. 11:71-74. [Google Scholar]
- 279.Waechter-Kristensen, B., J.-E. Englund, S. Khalil, M. Hultberg, and P. Sundin. 1998. Comparison of sole carbon source profiles monitored at two different wavelengths. J. Microbiol. Methods 34:17-21. [Google Scholar]
- 280.Wallander, H., K. Arnebrant, F. Ostrand, and O. Karen. 1997. Uptake of15N-labelled alanine, ammonium and nitrate in Pinus sylvestris L. ectomycorrhiza growing in forest soil treated with nitrogen, sulphur or lime. Plant Soil 195:329-338. [Google Scholar]
- 281.Wang, E. T., M. A. Rogel, A. Garcia-de los Santos, J. Martinez-Romero, M. A. Cevallos, and E. Martinez-Romero. 1999. Rhizobium etli bv. mimosae, a novel biovar isolated from Mimosa affinis. Int. J. Syst. Bacteriol. 49:1479-1491. [DOI] [PubMed] [Google Scholar]
- 282.Weller, R., M. Bateson, B. Heimbuch, E. Kopczynski, and D. Ward. 1992. Uncultivated cyanobacteria, Chloroflexus-like inhabitants, and spirochete-like inhabitants of a hot spring microbial mat. Appl. Environ. Microbiol. 58:3964-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Weyman-Kaczmarkowa, W., and D. Wojcik-Wojtkowiak. 1991. The dynamics of microflora and the occurrence of phytotoxic substances in different soils with corn residues and inorganic nitrogen. Plant Soil 132:11-20. [Google Scholar]
- 284.Wilson, R. A., B. A. Handley, and J. E. Beringer. 1998. Bacteriocin production and resistance in a field population of Rhizobium leguminosarum biovar viciae. Soil Biol. Biochem. 30:413-417. [Google Scholar]
- 285.Wind, T., and R. Conrad. 1995. Sulfur compounds, potential turnover of sulfate and thiosulfate, and numbers of sulfate-reducing bacteria in planted and unplanted paddy soil. FEMS Microbiol. Ecol. 18:257-266. [Google Scholar]
- 286.Wong, K. K. Y., Y. Piche, D. Montpetit, and B. R. Kropp. 1989. Differences in the colonization of Pinus banksiana roots by sib-monokaryotic and dikaryotic strains of ectomycorrhizal Laccaria bicolor. Can. J. Bot. 67:1717-1726. [Google Scholar]
- 287.Wright, T. D., K. L. Vergin, P. W. Boyd, and S. J. Giovannoni. 1997. A novel delta-subdivision proteobacterial lineage from the lower ocean surface layer. Appl. Environ. Microbiol. 63:1441-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Xia, J., J. Correll, F. Lee, M. Marchetti, and D. Rhoads. 1993. DNA fingerprinting to examine microgeographic variation in the Magnaporthe grisea (Pyricularia grisea) population in two rice fields in Arkansas. Phytopathology 83:1029-1035. [Google Scholar]
- 289.Xiao, G. P., and S. M. Berch. 1996. Diversity and abundance of ericoid mycorrhizal fungi of Gaultheria shallon on forest clearcuts. Can. J. Bot. 74:337-346. [Google Scholar]
- 290.Yachi, S., and M. Loreau. 1999. Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc. Natl. Acad. Aci. USA 96:1463-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Yang, P., P. Rott, L. Vauterin, B. Hoste, P. Baudin, K. Kersters, and J. Swings. 1993. Intraspecific variability of Xanthomonas albilineans. Syst. Appl. Microbiol. 16:420-426. [Google Scholar]
- 292.Zelles, L., R. Rackwitz, Q. Y. Bai, T. Beck, and F. Beese. 1995. Discrimination of microbial diversity by fatty acid profiles of phospholipids and lipopolysaccharides in differently cultivated soils. Plant Soil 170:115-122. [Google Scholar]
- 293.Zhan, J., C. Mundt, and B. McDonald. 1998. Measuring immigration and sexual reproduction in field populations of Mycosphaerella graminicola. Phytopathology 88:1330-1337. [DOI] [PubMed] [Google Scholar]
- 294.Zhou, J. Z., M. E. Davey, J. B. Figueras, E. Rivkina, D. Gilichinsky, and J. M. Tiedje. 1997. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology 143:3913-3919. [DOI] [PubMed] [Google Scholar]
- 295.Zilli, J. E., M. C. P. Neves, and N. G. Rumjanek. 1998. Signalling specificity of rhizobia isolated from nodules of Phaseoleae and Indigofereae tribes. An. Acad. Bras. Cienc. 70:743-750. [Google Scholar]
- 296.Zvyagintsev, D. G., A. V. Kurakov, M. M. Umarov, and Z. Filip. 1997. Microbiological and biochemical indicators of lead pollution in Soddy-podzolic soil. Pochvovedenie. 9:1124-1131. [Google Scholar]