Abstract
The active transport of toxic compounds by membrane-bound efflux proteins is becoming an increasingly frequent mechanism by which cells exhibit resistance to therapeutic drugs. This review examines the regulation of bacterial drug efflux systems, which occurs primarily at the level of transcription. Investigations into these regulatory networks have yielded a substantial volume of information that has either not been forthcoming from or complements that obtained by analysis of the transport proteins themselves. Several local regulatory proteins, including the activator BmrR from Bacillus subtilis and the repressors QacR from Staphylococcus aureus and TetR and EmrR from Escherichia coli, have been shown to mediate increases in the expression of drug efflux genes by directly sensing the presence of the toxic substrates exported by their cognate pump. This ability to bind transporter substrates has permitted detailed structural information to be gathered on protein-antimicrobial agent-ligand interactions. In addition, bacterial multidrug efflux determinants are frequently controlled at a global level and may belong to stress response regulons such as E. coli mar, expression of which is controlled by the MarA and MarR proteins. However, many regulatory systems are ill-adapted for detecting the presence of toxic pump substrates and instead are likely to respond to alternative signals related to unidentified physiological roles of the transporter. Hence, in a number of important pathogens, regulatory mutations that result in drug transporter overexpression and concomitant elevated antimicrobial resistance are often observed.
INTRODUCTION
Drug-resistant microorganisms are a major worldwide health issue, as a number of important human pathogens have now acquired mechanisms that make them largely resistant to all currently available treatment regimens. The action of antimicrobial compounds can be negated at a number of points, including enzymatic inactivation, the employment of alternative metabolic pathways to bypass their activity, sequestration, reduced uptake, and alteration of the target site to render it not susceptible to the effect of otherwise toxic substances (28). A further resistance mechanism that has become increasingly important involves membrane-bound efflux pumps that transport toxic antimicrobial compounds from the cell. ATP-binding cassette (ABC) transporters power this process via the hydrolysis of ATP, whereas secondary transporters utilize the transmembrane electrochemical gradient, typically the proton motive force, to drive drug efflux (161, 173).
The first drug transport proteins to be identified were a family of tetracycline efflux pumps, which provide widespread resistance to tetracycline antibiotics in both gram-negative and gram-positive bacteria (25). More recently, a large number of multidrug resistance (MDR) transport proteins have been found to be involved in the export of a wide range of antimicrobial compounds (161, 173, 185). In contrast to the narrow substrate range of most transporters, including the tetracycline efflux determinants, individual MDR pumps are capable of exporting compounds that have few structural similarities. MDR determinants appear to contribute to the emergence of drug-resistant microorganisms via two mechanisms. First, they can confer low-level protection that facilitates the initial survival of the organism and thus provides it with the opportunity to subsequently acquire one of the high-level specific resistance mechanisms listed above. Alternatively, the MDR transporters themselves can furnish protection against clinically relevant concentrations of many antimicrobial compounds.
In addition to providing many pathogenic bacteria with protection against antibiotics, antiseptics, and disinfectants (161), drug transporters pose a number of additional medical challenges. Host-encoded antimicrobial compounds that are produced to combat infections caused by organisms such as Escherichia coli (143) and Neisseria gonorrhoeae (203) are also exported by some MDR pumps; in Staphylococcus aureus, the presence of an MDR transporter has been correlated with resistance to a cationic antimicrobial peptide (17, 87). Treatment of immunocompromised patients undergoing long-term antifungal therapy is often complicated by development of resistance to antifungal agents by the pathogenic yeast Candida albicans as a result of MDR transporter overexpression (84). Additionally, in human cancer cells, overexpression of an MDR transporter, P-glycoprotein, can provide high-level resistance to antitumor drugs, a finding which has been linked to the failure of chemotherapy (45).
Although these problems have generated a substantial degree of interest in drug transporters, the close association of efflux pumps with cellular membranes has severely hampered efforts to define the exact molecular mechanisms by which these proteins function. Only recently have high-resolution structures for any membrane transport proteins been elucidated, the E. coli proteins MsbA (23) and BtuCD (103), although more limited structural information has also been gained by employing electron microscopy of two-dimensional crystals, e.g., the single-substrate TetA tetracycline exporter has been shown to function as a trimer (240), the MDR transporters EmrE (216) and YvcC (22) form dimers, and P-glycoprotein functions as a monomer (180, 181).
In comparison to the limited achievements in understanding the structure-function relationships of the drug transporters themselves, research into the regulatory pathways that govern the expression of drug transporters has progressed relatively rapidly, in particular for a number of bacterial regulatory proteins. The antimicrobial pumps which are known to be subject to regulatory controls typically belong to either the major facilitator superfamily (MFS) or resistance, nodulation and cell division (RND) superfamily (Table 1), both of which primarily employ the proton motive force to energize drug efflux (161, 173). The requirement for regulatory controls to prevent excessive production of an integral membrane protein that utilizes the proton motive force is demonstrated by the deleterious effect of constitutive expression of the gram-negative tetracycline/H+ antiporters TetA(B) and TetA(C), which, in the absence of tetracycline, place cells at a severe disadvantage when competing with nonconstitutively expressing strains (92, 142). Overproduction of TetA(B) can also be lethal to the cell if the gene is expressed from a strong promoter, resulting in nonspecific cation transport, loss of the membrane H+ potential, and cell death (31, 57).
TABLE 1.
Transcriptional regulators that control expression of genes encoding bacterial drug efflux components
| Organism | Regulatory protein(s) | Regulator familya | Function of regulator | Ligand(s) of regulatory proteinb | Drug efflux gene(s) regulatedc | Reference(s) |
|---|---|---|---|---|---|---|
| RND pump regulators | ||||||
| Acinetobacter baumannii | Orf2-Orf3 | Two-component system? | ? | adeABC* | 111 | |
| Burkholderia pseudomallei | AmrR | TetR | Repressor? | ? | amrAB-oprA | 133 |
| Escherichia coli | AcrR | TetR | Repressor | ? | acrAB* | 107 |
| AcrS | TetR | Repressor? | ? | acrEF* | 156 | |
| BaeR-BaeS | Two-component system | ? | mdtABC | 12, 137 | ||
| EvgA-EvgS | Two-component system | ? | yhiUV | 144 | ||
| MarA/SoxS/Rob | AraC | Global activators | Rob? (MarA/SoxSNA) | acrAB* and tolC | 69, 70, 118, 177 | |
| MarR | MarR | Repressor of marA | DNP, Pg, Sa, Md | acrAB* and tolC, via MarA | 6, 8, 120 | |
| Neisseria gonorrhoeae | MtrA | AraC | Global activator | HAs? | mtrCDE* | 184 |
| MtrR | TetR | Repressor | ? | mtrCDE* and farAB | 106 | |
| Pseudomonas aeruginosa | MexR | MarR | Repressor | ? | mexAB-oprM* | 32, 170 |
| MexT | LysR | Activator | ? | mexEF-oprN* | 81 | |
| MexZ | TetR | Repressor? | ? | mexXY | 4 | |
| NfxB | LacI/GalR | Repressor | ? | mexCD-oprJ* | 169 | |
| Pseudomonas putida | ArpR | TetR | Repressor? | ? | arpABC | 78 |
| Stenotrophomonas maltophilia | SmeR-SmeS | Two-component system? | ? | smeABC | 100 | |
| MFS pump regulators | ||||||
| Bacillus subtilis | BltR | MerR | Activator | ? | blt | 3 |
| BmrR | MerR | Activator | R6G, TPP, Ao, DEC, ABM, ADCP, DDPB | bmr | 2, 227, 244, 245 | |
| Mta | MerR | Global activator | ? | bmr and blt | 13, 43 | |
| Escherichia coli | EmrR | MarR | Repressor | CCCP, DNP, Eb, FCCP, Na, Sa, TCS | emrAB* and mcbABCDEFG | 20, 104, 105, 238 |
| EvgA-EvgS | Two-component system | ? | emrKY | 145 | ||
| TetR | TetR | Repressor | Tc | tetA | 59, 60, 153 | |
| Staphylococcus aureus | ArlR-A1rS | Two-component system | ? | norA*, via 18-kDa protein | 35 | |
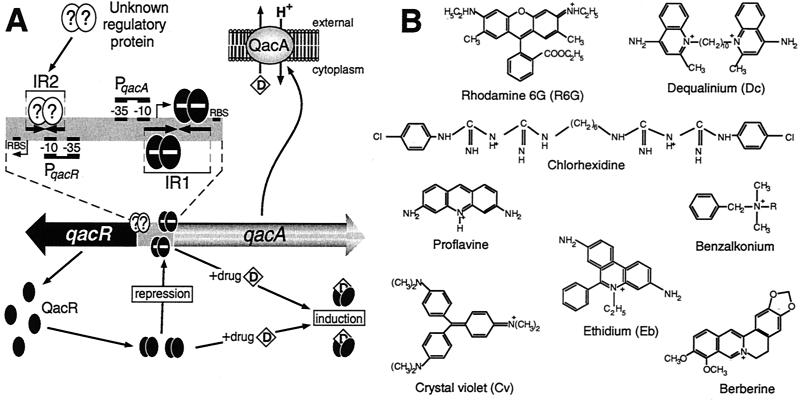
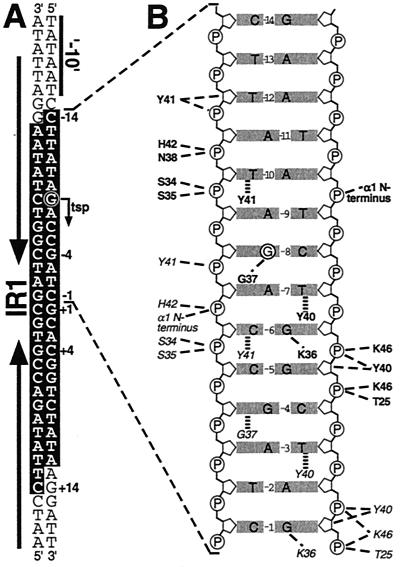
| QacR | TetR | Repressor | Bc, Be, Ch, Cv, Dc, Eb, Mg, Pf, R6G | qacA/qacB | 47, 48, 197, 198 |
Although two-component systems belong to a number of different families, they all consist of a transmembrane sensor of an external signal and a cytoplasmic response protein whose regulatory activities are modulated by reversible phosphorylation. Note that EvgA has been demonstrated to modulate the expression of both an RND type pump and an MFS member, yhiUV and emrKY, respectively.
Ao, astrazon orange; ABM, 5-(1-adamanthylcarboxyethyl)-3-benzyl-4-methylthiazolium; ADCP, 4-amino-3,6-dimethylbenzo[b]cycloheptano[e]pyridinium; Bc, benzalkonium; Be, berberine; CCCP, carbonyl cyanide m-chlorophenylhydrazone; Ch, chlorhexidine; Cv, crystal violet, Dc, dequalinium; DDPB, 5,6-dichloro-1,3-diethyl-2-(phenylaminovinyl)benzoimidazolium; DEC, diethyl-2,4′-cyanine; DNP, 2,4-dinitrophenol; Eb, ethidium bromide; FCCP, carbonyl cyanide p-(trifluoro-methoxy)phenylhydrazone; HAs, hydrophobic agents; Md, menadione; Mg, malachite green; Na, nalidixic acid; Pf, proflavine; Pg, plumbagin; R6G, rhodamine 6G; Sa, salicylate; Tc, tetracycline; TCS, tetrachlorosalicylanilide; TPP, tetraphenylphosphonium; ?, many of these regulatory proteins and two-component transmembrane sensors possess hypothetical ligand-binding domains for which ligands have yet to be identified. NA, not applicable, as the MarA and SoxS proteins do not possess ligand-binding domains.
Drug efflux genes or operons marked with an asterisk (*) have been observed to confer elevated antimicrobial resistance in some clinical isolates due to regulatory mutations that result in overexpression of these determinants.
However, despite these observations, all members of a further family that utilize the proton motive force, the small multidrug resistance family, a subgroup of the drug/metabolite transport superfamily (67), do not appear to be subject to any regulatory controls that can alter the level at which these proteins are synthesized. It is therefore an intriguing question why some proteins that utilize the proton motive force to energize drug efflux are synthesized under strict regulatory controls, yet others appear to be expressed constitutively. This could well reflect currently unknown physiological roles for the unregulated pumps in normal cellular metabolism that require the constant low-level presence of such transporters. The supply of a small but continuous amount of a protein can be easily governed by mechanisms such as low-level production of the relevant mRNA and/or high turnover rates of the mRNA and/or the transport protein, without any need for additional, more complex regulatory controls.
For the bacterial drug efflux genes that are inducible, there are only a few documented cases for which translational controls have been shown to be the primary level at which expression is controlled. For example, translational attenuation has been proposed to modulate the synthesis of the gram-positive tetracycline resistance determinant TetA(K) (206), whereas a second gram-positive tetracycline resistance protein, TetA(L), is regulated by translational reinitiation (209). Experimental evidence suggests that the latter example involves tetracycline-induced stalling of ribosomes during the translation of a short leader peptide. This is likely to facilitate the transfer of ribosomes to the tetA(L) ribosome-binding site, an event which requires the presence of a stem-loop structure that is proposed to correctly orient the tetA(L) ribosome-binding site for ribosome transfer to occur (209).
In contrast to these determinants, expression of the majority of the bacterial drug transporter genes which are known to be subject to regulation is controlled by transcriptional regulatory proteins. Well-characterized proteins with a demonstrated role in controlling the expression of drug efflux genes encompass examples of both repressors and activators of target gene transcription, a process that can occur at either the local or global level. Local regulators of drug transporter genes include the Escherichia coli TetR repressor of tetracycline efflux genes (59) and three regulators of MDR transporter genes, the Bacillus subtilis BmrR activator (2), the Staphylococcus aureus QacR repressor (47), and the E. coli EmrR repressor (105). In some instances, local regulators appear to play only a modulating role, the principal factor controlling transcription instead being global regulatory proteins; e.g., increases in the expression of the E. coli acrAB MDR locus are mediated by the MarA, Rob, and SoxS global activators (7). Two-component regulatory systems are also increasingly being found to be associated with drug efflux genes (Table 1). Sequencing of entire bacterial genomes has identified a large number of additional MDR transporter homologs and their associated regulatory elements, although the functions of the majority of these systems remain to be experimentally investigated (162).
In general, the confirmed regulators of bacterial drug transporter genes belong to one of four regulatory protein families, the AraC, MarR, MerR, and TetR families, despite being from distantly related species, a classification which also shows little correlation with the family of the drug pump whose expression they control (Table 1). However, the assignment of these regulatory proteins to their respective families is based solely on similarities detected within their DNA-binding domains, which typically constitute only one third of each polypeptide. Like the majority of bacterial activators and repressors, the drug transport regulators identified to date all possess α-helix-turn-α-helix (HTH) DNA-binding motifs, which are embedded in larger DNA-binding domains that form a number of different structural environments, such as three-helix bundles and winged helix motifs (155). These serve to create a stable three-dimensional structure that buries the hydrophobic side chains of amino acids in the interior of the DNA-reading head but generally orients the second “recognition” helix of the HTH so that it fits into the major groove of B-DNA. Further detailed information on the structure and function of HTH motifs can be found in a number of excellent reviews (39, 54, 64, 155, 213).
Importantly, for four local regulators of drug efflux determinants, the portions of these proteins not involved in forming the DNA-binding domains have been demonstrated to be capable of directly binding substrates of their cognate pumps, which act as a signal to increase the synthesis of the relevant transport protein(s) in response to the presence of these toxic compounds. This finding has had important ramifications for the field of protein-drug interactions, since, in contrast to the membrane-bound transport proteins, which are notoriously difficult to purify and study in vitro, the soluble cytosolic regulators of drug resistance have provided much more amenable systems for the study of drug recognition and binding. For the TetR (60), QacR (198), and BmrR (245) regulatory proteins in particular, detailed X-ray crystallographic and biochemical data, combined with mutational studies, have been highly successful in providing a wealth of information on the molecular aspects involved in drug binding and the subsequent steps that result in the induction of target gene expression.
By concentrating principally on the better-characterized regulatory pathways, at both the local and global levels, this review is intended to illustrate the extremely varied nature of the transcriptional regulatory controls that act upon the genes encoding bacterial drug efflux systems. Particular emphasis is placed on the contributions that analysis of these regulatory pathways and their attendant proteins have made to the field of drug resistance as a whole. We also discuss the insights that have been gained from evolutionary and medical perspectives.
TRANSPORTER OVEREXPRESSION AND ANTIMICROBIAL RESISTANCE
Almost all of the antimicrobial compounds, both synthetic and natural, which have been employed by humans to combat infectious bacteria are substrates of one or more drug efflux pumps, e.g., the fluoroquinolones; the antibiotics chloramphenicol, tetracycline, β-lactams, and aminoglycosides; and the antiseptics benzalkonium, cetrimide, and chlorhexidine. Tetracycline-specific pumps such as TetA(B) possess regulatory controls that are sensitively attuned for adjusting expression levels in response to the presence of tetracycline. In contrast, the vast majority of bacterial MDR genes need to be expressed at a level substantially greater than that observed in the wild-type organism before significant efflux of clinically relevant antimicrobial compounds occurs. This finding has lent considerable support to the proposal that MDR pumps in general have preexisting physiological roles, such as protection against low levels of toxic hydrophobic molecules encountered in the natural environment of many organisms or the transport of specific metabolites. However, for a limited number of the drug transporters identified to date, such as the tetracycline pumps and perhaps also the plasmid-encoded S. aureus MDR determinant QacA, which exports an impressive array of antiseptics, disinfectants, and related compounds (21), analysis of the pumps and their regulatory controls indicates that the efflux of medically relevant antimicrobial compounds is now the primary function of these systems.
For clinical isolates of the important human pathogens E. coli, Neisseria gonorrhoeae, Pseudomonas aeruginosa, and S. aureus, a large number of regulatory pathway mutations that have resulted in the overexpression of various MDR determinants and a concomitant elevated resistance to antimicrobial compounds have been characterized. For example, it has been well established that low-level multiantibiotic resistance in E. coli can arise from overexpression of the AcrAB-TolC MDR efflux complex as a result of increased production of the MarA global transcriptional activator protein (112, 149, 232). MarA and a number of other E. coli MDR regulators are discussed in greater depth later in this review. Overexpression of MDR transporters has now been identified as a major source of antimicrobial resistance in an alarmingly large number of pathogenic species. The most striking example of this phenomenon is the serious opportunistic pathogen P. aeruginosa, for which the hyperexpression of MDR pumps has been particularly important in the emergence of multiantibiotic-resistant strains.
P. aeruginosa MDR Pumps and Multiantibiotic Resistance
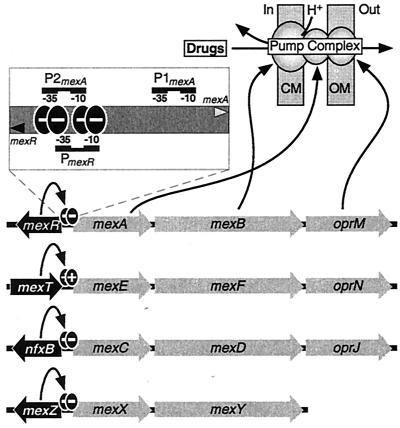
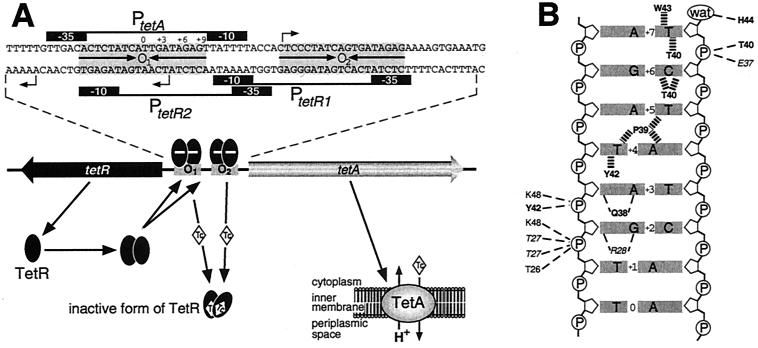
Mutations leading to the overexpression of pump complexes such as MexAB-OprM have now been identified as the predominant cause for acquisition of elevated resistance to structurally unrelated antibiotics by strains of P. aeruginosa (167). The mexAB-oprM operon encodes a tripartite pump complex that comprises the MexB cytoplasmic pump component, the MexA membrane fusion protein, and the OprM outer membrane channel (Fig. 1). Thus, the MexAB-OprM complex can simultaneously transport antibiotics across both the cytoplasmic and outer membranes, which provides a highly effective resistance mechanism when combined with the notoriously low permeability of the P. aeruginosa outer membrane (167). MexAB-OprM together with MexCD-OprJ, MexEF-OprN, and MexXY (which also utilizes the OprM outer membrane channel) form a family of related P. aeruginosa multidrug efflux pump complexes that have an extremely broad substrate range (134, 168). Each of the operons for these four pump complexes has a local transcriptional regulatory protein that is encoded in the immediately adjacent upstream region (Fig. 1). In stark contrast to the high degree of relatedness between the individual components of the pump complexes, the proteins encoded by each of these upstream genes exhibit no homology to each other; instead, they belong to four distinct regulatory protein families (Table 1).
FIG. 1.
Genetic organization of the mexR-mexAB-oprM, mexT-mexEF-oprN, nfxB-mexCD-oprJ, and mexZ-mexXY MDR loci from P. aeruginosa. Each operon contains genes (grey arrows) that encode a drug efflux complex and is regulated by the product of an upstream gene (black arrow) which either represses (−) or activates (+) operon expression, although this is yet to be confirmed for MexZ. The intergenic region separating the mexR and mexA genes is depicted in greater detail, with the positions of the binding sites for the two MexR dimers (black ovals) indicated relative to the −10 and −35 hexamers of the mexR promoter (PmexR), the mexA promoter (P2mexA), and a second potential mexA promoter (P1mexA). A schematic representation of the MexAB-OprM tripartite complex, which can efflux drugs simultaneously across both the cytoplasmic (CM) and outer (OM) membranes, is also shown. MexB, the RND component of the pump complex, transports drugs across the cytoplasmic membrane in exchange for protons (H+).
The regulation of the mexAB-oprM operon is the best-characterized example; the divergently encoded MexR, a member of the MarR family of proteins (Table 1), acts as a repressor of transcription of mexAB-oprM (170). MexR autoregulates expression of its own gene by repressing the mexR promoter (Fig. 1, PmexR) in addition to controlling transcription from the mexA promoter (Fig. 1, P2mexA) (32). A second promoter (Fig. 1, P1mexA) has been proposed to be responsible for the relatively high constitutive level of mexAB-oprM expression, which contributes substantially to the high intrinsic resistance to antibiotics that P. aeruginosa exhibits. However, recent experiments with S1 mapping and reporter gene fusions have suggested that P1mexA is not a functional promoter (189). A large number of mutant strains that contain alterations to the mexR coding region and produce an inactive MexR repressor have been described; all of these mutations result in overproduction of the MexAB-OprM pump complex and enhanced efflux of a broad range of antibiotics, such as β-lactams, fluoroquinolones, tetracycline, and chloramphenicol (98, 186, 208, 246).
It has been proposed that the increased expression from the MexR-controlled promoter (P2mexA) occurring in these strains is responsible for the observed MexAB-OprM hyperexpression (32). In some instances, the inability of the MexR derivatives to repress P2mexA has been confirmed to be due to the production of MexR proteins which are either unstable or compromised in their ability to dimerize or bind DNA (1). However, none of the antibiotics to which these mutants exhibit increased resistance induce expression, suggesting that the MexAB-OprM pump complex has evolved for other physiological roles, which may include the export of secondary metabolites (166) as well as a demonstrated role in the active efflux of an N-(3-oxododecanoyl)-homoserine lactone autoinducer signal associated with quorum sensing (163).
A recent crystal structure for apo-MexR has suggested that the MexR ligand may be an acidic peptide signaling molecule or the C terminus of a protein ligand, either of which could insert between the two winged-helix DNA-binding domains of a MexR dimer and induce a significant reduction in their spacing, thereby rendering the protein incapable of binding DNA (101). Further mutations that alter mexAB-oprM transcription but are located outside of mexR or the intergenic region (171, 208, 246) support the existence of unidentified regulatory proteins that influence the expression of this MDR pump complex, as does the linking of mexAB-oprM expression to the growth phase of cells (33). Interestingly, the MDR systems of P. aeruginosa have also been found to be subject to coordinate regulation, the expression of some systems decreased in response to increased levels of another (99).
Additional multiantibiotic-resistant pseudomonal mutants with an NfxC phenotype display resistance to chloramphenicol, fluoroquinolones, and trimethoprim due to overexpression of the mexEF-oprN MDR efflux operon (81). The locations of the mutations which produce this phenotype are unknown, but the overexpression is dependent on the presence of a functional MexT protein, which is a LysR-type transcriptional activator (Table 1) encoded by an upstream gene transcribed in the same direction as the mexEF-oprN operon (Fig. 1) (82, 123). As for MexR, none of the known substrates of MexEF-OprJ have any effect on the activity of MexT or expression of mexEF-oprJ, although simple overexpression of the MexT activator is sufficient to instigate transcription of this operon (81, 148). However, because most LysR-type regulators become active upon binding a cognate effector molecule, it has been suggested that the NfxC phenotype may result from the overproduction of a MexT effector molecule that normally induces production of the MexEF-OprN pump complex in response to the presence of its physiological substrate(s) (81). An alternative explanation involving mutations in an unidentified suppressor of mexT expression has also been put forward (123).
MexT may possess both repressor and activator functions, as it has been implicated in the downregulation of oprD, a gene which encodes a porin involved in the uptake of the β-lactam imipenem (148). Thus, the resistance to imipenem that nfxC-type mutants exhibit is due to reduced uptake of this antibiotic rather than its efflux by the overproduced MexEF-OprN pump complex. MexEF-OprN has also recently been proposed to influence the intracellular levels of Pseudomonas quinolone signal, a molecule involved in cell-to-cell signaling (83). This has been linked to the surprising finding that nfxC-type strains that overexpress MexEF-OprM, and hence have elevated antibiotic resistance, are actually less virulent due to a decrease in the production of extracellular virulence factors (83).
Overexpression of the mexCD-oprJ operon due to mutations in its divergently transcribed repressor of synthesis, NfxB (Fig. 1), can also confer multiantibiotic resistance on P. aeruginosa (169). NfxB autoregulates its own expression (204) and normally completely represses the mexCD-oprJ operon. Construction of P. aeruginosa strains in which three of the four mex operons are disrupted recently permitted the demonstration of mexCD-oprJ induction by acriflavine, tetraphenylphosphonium (TPP), ethidium bromide, or rhodamine 6G, all of which are substrates of this MDR pump complex (134). Although this result suggests a possible physiological role for MexCD-OprJ in the extrusion of toxic compounds, the mechanism by which the observed induction occurs needs to be identified before more concrete conclusions can be drawn.
Overexpression of the fourth P. aeruginosa multidrug resistance locus, MexXY, has been associated with aminoglycoside resistance (130, 233). In wild-type cells, MexXY also contributes to intrinsic antibiotic resistance by being inducible by the pump substrates tetracycline, erythromycin, and gentamicin, although again the mechanism by which this occurs is unknown (124). mexXY has been proposed to be regulated by MexZ, the product of a divergently transcribed gene (Fig. 1) that encodes a TetR family repressor (4). In addition to the four MDR transporter complexes discussed above, the extremely large P. aeruginosa genome appears to encode a number of additional potential efflux systems which show significant homology to known drug exporters (210).
Despite the recent demonstration that both MexCD-OprJ and MexXY are inducible by some antimicrobials, the majority of the currently available information indicates that the regulatory networks controlling the expression of MDR systems in P. aeruginosa are primarily intended to respond to physiological signals that are not related to the efflux of drugs. In support of this proposal is the identification of a number of Pseudomonas putida pump complexes that exhibit strong homology to MexAB-OprM. Although some of these systems are capable of exporting several structurally dissimilar antibiotics, the predominant physiological role of these efflux complexes appears to be the energy-dependent export of toxic organic solvents, which permits P. putida cells to grow in media containing relatively high concentrations of aromatic hydrocarbons (79, 175, 179). The identification of local and global regulatory proteins, both of which are likely to be involved in controlling the expression of P. putida pumps in response to the presence of toxic organic solvents, provides strong evidence that aromatic hydrocarbon efflux is the authentic function of these systems (30).
In contrast to the P. putida organic solvent pumps, the four P. aeruginosa pump complexes discussed above export an extensive and overlapping array of structurally dissimilar drugs, although none of their local regulatory proteins appear to be involved in responding to the presence of these pump substrates and no alternative induction mechanisms have yet been elucidated. Similarly, ArpABC, another P. putida RND pump which can export structurally unrelated antibiotics, such as carbenicillin, chloramphenicol, erythromycin, and tetracycline, but does not contribute to organic solvent tolerance is also not induced by the addition of antibiotics or solvents (78). However, pseudomonal MDR pump complexes continue to have a substantial impact on antimicrobial resistance due to the many clinical strains which overexpress MDR transporters as a result of regulatory mutations, a trend which is becoming increasingly frequent in other established and emerging human pathogens.
Drug Transporter Expression in Other Pathogens and Antibiotic Producers
Increased synthesis of a chromosomal S. aureus MDR gene which encodes the MFS transporter NorA is particularly important in the emergence of fluoroquinolone-resistant clinical isolates of this species (76, 239, 241). The flqB mutation, a single T-to-G substitution in the 5′ untranslated region upstream of norA, has been shown to increase the half-life of norA mRNA 4.8-fold, resulting in NorA overproduction (37). It has been proposed that the increased mRNA stability is a consequence of changes to its secondary structure, which may affect an RNase III cleavage site involved in the degradation of this transcript (37). A single T-to-A change in the norA promoter region also appears to result in overexpression of NorA and subsequent fluoroquinolone resistance (75). Other mutations that produce increased resistance but lie outside the norA coding and promoter regions are currently uncharacterized (74, 136).
Although the regulation of norA expression is poorly understood, an unidentified 18-kDa protein has been demonstrated to bind to multiple sites in the DNA sequences upstream of the −35 region of the norA promoter (35). The binding of this 18-kDa protein is modified by ArlS, which appears to function as the transmembrane sensor of a two-component regulatory system, ArlR-ArlS. The first component of these systems is typically a transmembrane sensor protein that undergoes autophosphorylation upon binding a signal molecule present in the external environment (158). The phosphate is then transferred to the second component, a cytoplasmic response regulator which can be reversibly phosphorylated at a conserved aspartate residue (158). The activity of the response regulator is thereby altered so that it can then act to either stimulate or repress target gene transcription.
Although disruption of the gene for the ArlS transmembrane sensor resulted in upregulation of NorA expression and also altered the growth phase regulation of this transporter, it is not known if ArlS phosphorylates the 18-kDa protein directly or, alternatively, acts indirectly, e.g., via ArlR altering the transcription of a gene involved in the production of a ligand for the 18-kDa protein (35). The latter scenario could be related to the observation that S. aureus secretes a compound into the external medium that has the effect of lowering norA expression from the late logarithmic growth phase onwards (35). Although ArlR-ArlS also has a role in the regulation of virulence determinants in S. aureus (36), the expression of norA does not appear to be linked to that of known virulence determinants, suggesting that ArlR-ArlS modulates NorA expression in response to other physiological functions (35).
MDR pump overexpression has also made a substantial contribution to the emergence of many other bacterial species as serious human health threats. The intrinsic antibiotic resistance of the opportunistic pathogen Stenotrophomonas maltophilia has been attributed in part to MDR pumps (243), whereas elevated expression of the SmeDEF MDR pump in clinical isolates of this species has been correlated with increased levels of resistance to tetracycline, chloramphenicol, erythromycin, and quinolones (9). Although a second MDR pump homolog identified in this species, SmeABC, is not involved in drug efflux, the SmeC outer membrane component appears to be employed by another, as yet uncharacterized MDR pump which does transport antimicrobial compounds (100). The resistance of a clinical isolate of Acinetobacter baumannii to aminoglycosides and an extensive range of other antimicrobials has been confirmed to be due to an RND-type pump complex encoded by the adeABC gene cluster (111). While the basis for the increased contribution of the adeABC genes to drug efflux is unknown, three divergently transcribed genes which encode a putative two-component regulatory system (Table 1) and a transcriptional terminator-antiterminator have been proposed to be involved in the regulation of this MDR locus (111).
The inherent high-level resistance of the disease-causing bacterium Burkholderia pseudomallei to antimicrobial agents has also been partially attributed to an RND-type pump, AmrAB-OprA (133). Divergently transcribed from amrAB-oprA is the gene for AmrR, a TetR family protein proposed to be a transcriptional repressor of this operon. In the anaerobic pathogen Clostridium perfringens, the most widespread tetracycline resistance determinant is the plasmid-encoded tet(P) operon, comprised of two overlapping genes, which encode the TetA(P) tetracycline efflux pump and TetB(P), a protein likely to be involved in ribosome protection (72). Induction of this operon requires an unidentified, chromosomally encoded regulatory protein (73), whereas a transcriptional attenuation mechanism appears to prevent excessive production of TetA(P) in both the absence and presence of tetracycline (72). Although the mechanisms that lead to MDR overexpression and concomitant elevated antimicrobial resistance have yet to be identified in many of these species, it is immediately obvious that such processes represent one of the primary means by which bacteria subjected to drug exposure can acquire elevated resistance to these compounds.
For several antibiotic-producing bacteria, regulatory proteins have also been identified that control the synthesis of efflux pumps which are employed to provide protection against their own antibiotics. Streptomyces species, in particular, have been the object of considerable attention because these gram-positive bacteria are responsible for the production of the majority of commercially important antibiotics. Uncharacterized regulators designated Pip proteins have been proposed to be involved in the regulation of drug transport genes in a range of Streptomyces species (187, 188). Isolation of the Pip protein from the genetically well-defined Streptomyces coelicolor indicated that it was a TetR family repressor that regulates the expression of the MFS antiporter Ptr, which confers resistance to the antibiotic pristinamycin I (34).
The self-resistance of Streptomyces virginiae to the antibiotic virginiamycin S is provided by the efflux pump VarS, production of which is governed by complex regulatory controls that include the transcriptional repressor BarA, which responds to a γ-butyrolactone autoregulator signal (138). Additionally, VarR, the product of a gene cotranscribed with varS, is a TetR family repressor that regulates varS transcription in a virginiamycin S-dependent manner (138). Another interesting example is the bacitracin peptide antibiotic-producing bacterium Bacillus licheniformis, which utilizes an ABC transporter, BcrABC, rather than a proton motive force-dependent exporter to provide self-resistance (164). An upstream two-component regulatory locus, bacRS, appears to control the expression of the bcrABC genes (139). The doxorubicin- and daunorubicin-producing organism Streptomyces peucetius also uses an ABC transporter, DrrAB, to confer self-resistance to these antibiotics (77). The role of activating expression of the synthesis and resistance operons for doxorubicin and daunorubicin in S. peucetius has been tentatively assigned to DnrI, which itself may be under the control of another positive transcriptional activator from the same gene cluster, DnrN (110).
Both the antibiotic biosynthesis genes and the self-resistance genes in all of these antibiotic-producing organisms appear to be under additional complex regulatory controls that respond to a number of environmental factors, in addition to linking their expression to differentiation events such as sporulation. In stark contrast to the majority of the drug pumps that confer antibiotic resistance to pathogenic bacteria, the expression of the transporters employed by antibiotic-producing organisms to provide self-resistance is intimately linked to the production, and hence the presence, of the relevant pump substrate(s). Understanding the function and regulation of these self-resistance mechanisms has considerable medical relevance, because in addition to aiding the exploitation of these species, their self-resistance mechanisms represent a pool of highly evolved determinants that can potentially be acquired by pathogenic bacteria.
Regulation of N. gonorrhoeae MDR Transporters
The mtrCDE operon of Neisseria gonorrhoeae encodes a multidrug efflux complex that is capable of exporting a range of MDR substrates that includes acriflavine, crystal violet, macrolides, and penicillin. However, the physiological role of this MDR pump is more likely to be reflected by its ability to extrude a range of structurally diverse hydrophobic agents, typically host-derived antimicrobial compounds such as fatty acids, bile salts, gonadol steroids, and antibacterial peptides, many of which coat mucosal sites colonized by N. gonorrhoeae (51). Transcription of mtrCDE can be increased by the addition of Triton X-100, an MtrCDE substrate which has a membrane-acting antimicrobial activity similar to that of the host-derived hydrophobic agents (184). Thus, in addition to being substrates of the efflux pump, hydrophobic agents are likely to act as natural inducers.
Analysis of the completed N. gonorrhoeae genome sequence identified the AraC family activator MtrA (Table 1), which was shown to be required for Triton X-100 enhancement of transcription (184). MtrA, like most AraC proteins (119), has an additional N-terminal domain which is likely to be involved in binding ligands, although it is currently unknown if this putative ligand-binding site plays a role in sensing the presence of toxic hydrophobic agents.
Divergently transcribed from mtrCDE is MtrR (156), a TetR family repressor (Table 1) that binds to a region which encompasses the mtrCDE promoter (106). Although MtrR appears to function solely as a modulator that is not involved in induction of mtrCDE expression (52, 106), mutations in either mtrR, the promoter for this gene, or the MtrR binding site can result in elevated mtrCDE expression and increased levels of resistance to hydrophobic agents (52, 106, 201, 202). Similar mutations have also been shown to produce increased resistance to the antibiotics azithromycin and erythromycin (242). However, disruption of MtrR binding alone does not fully explain the increases in mtrCDE transcription that result from some mutations that lie within the MtrR binding region and/or the mtrR promoter, suggesting the existence of other regulatory mechanisms and demonstrating the impact that cis-acting factors can have on efflux pump regulation (52).
A second N. gonorrhoeae efflux pump, FarAB, which utilizes the same outer membrane protein as the MtrCDE complex, MtrE (203), confers resistance to long-chain fatty acids (91). Surprisingly, although MtrR represses mtrCDE, it appears to enhance farAB expression (91), in addition to being either directly or indirectly responsible for controlling the expression of 14 other genes which may be linked to the establishment and/or maintenance of infections by N. gonorrhoeae (203). Overall, both the substrate range of the pumps and the available information on their regulation suggest that the function and expression of FarAB and MtrCDE are intimately linked to the virulence of N. gonorrhoeae.
The examples discussed to this stage have shown that the MDR systems of pathogenic bacteria are in general ill-adapted for the export of medically relevant drugs because of the requirement for mutations in their regulatory circuits before clinically significant resistance is observed. However, the following closer analysis of MDR regulatory controls from the model gram-negative and gram-positive organisms E. coli and B. subtilis, respectively, will demonstrate that this is not entirely the case. Additionally, the transcriptional repressors of tetracycline-specific pumps and the S. aureus plasmid-encoded QacA MDR determinant respond specifically to substrates of these pumps, and thus these regulatory proteins do not need to be bypassed before significant antimicrobial efflux can occur. These systems serve to illustrate in detail the better-characterized examples of proteins involved in the regulation of drug efflux genes, including examples of global activators and local activators or repressors.
CONTROL OF E. COLI acrAB MDR LOCUS BY GLOBAL ACTIVATORS
The E. coli AcrB RND pump functions as part of a tripartite complex which also consists of the membrane fusion protein AcrA (108) and the outer membrane channel protein TolC (11, 38), which is encoded in a remote part of the chromosome (Fig. 2). This MDR efflux system confers resistance to a diverse range of antimicrobials, such as dyes, detergents, fluoroquinolones, and many other lipophilic antibiotics, e.g., β-lactams, chloramphenicol, erythromycin, and tetracycline (108, 143, 150, 234). Immunoblotting has demonstrated that the AcrA protein was overexpressed in 9 of 10 E. coli clinical isolates which expressed high-level fluoroquinolone resistance (125). This has been attributed primarily to mutations that increase the production of the MarA global regulator, which modulates the expression of many genes, including acrAB and tolC (7, 149, 232). However, a number of insertions and single-amino-acid substitutions, duplications, and deletions that inactivate AcrR, a divergently transcribed local repressor of the acrAB operon (Fig. 2), have also been shown to result in enhanced expression of acrAB and increased fluoroquinolone resistance in clinical E. coli strains (71, 146, 230).
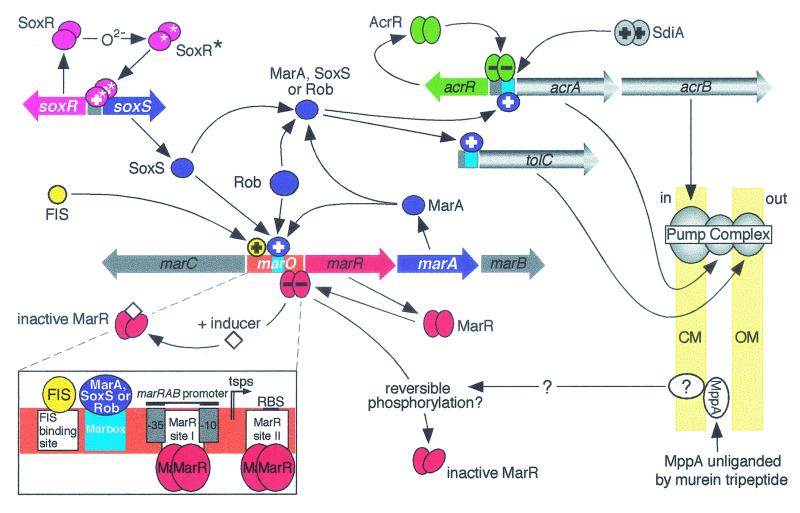
FIG. 2.
Schematic representation of the known regulatory controls governing the expression of the E. coli acrAB and tolC genes. The AcrAB-TolC transport complex extrudes drugs across both the cytoplasmic (CM) and outer (OM) membranes (pale yellow shaded boxes). Excessive production of AcrA and AcrB is prevented (−) by the local dimeric repressor protein AcrR (green), whereas a regulatory protein involved in cell division, SdiA (grey), can increase (+) acrAB expression. However, activation of acrAB and tolC transcription occurs primarily because of the global regulatory proteins MarA, SoxS, and Rob (purple), any one of which can bind to a marbox (cyan) upstream of these genes. The intracellular level of MarA is controlled by MarR (red), a dimeric protein which binds to marO (orange) and represses (−) the expression of its own gene and the two others that constitute the marRAB operon. Binding of inducing compounds (diamonds) such as salicylate by MarR, in addition to the possible phosphorylation of MarR via a putative signal transduction pathway involving the periplasmic binding protein MppA, is proposed to transform MarR into a non-DNA-binding conformation, thereby permitting marRAB transcription to proceed. Hence, MarA protein is produced which can then bind as a monomer to the marO marbox upstream of the marRAB promoter, where it activates (+) transcription of marRAB and enhances the production of MarA. The ensuing highly elevated intracellular levels of MarA can then bind to marboxes adjacent to the promoters of mar regulon genes, such as acrAB and tolC, and activate their transcription. The MarA homologs SoxS and Rob can also bind to the marO marbox and activate marRAB transcription. The positive regulation by all three proteins on marRAB is enhanced by FIS (yellow), an accessory activating protein. SoxS is only produced upon conversion of the SoxR effector protein (magenta) into its active form (SoxR*) by superoxide-generating agents (O2−). Rob, like SoxS, in addition to mediating increases in MarA synthesis, can also directly activate the expression of some genes that belong to the mar regulon, such as acrAB. The regulatory-protein-binding sites within marO are shown in finer detail in the bottom left corner. The positions of the −35 and −10 hexamers of the marRAB promoter, the ribosome-binding site (RBS) and transcription start points (tsps) for the marRAB operon are indicated. See text for other details. (Modified with permission from reference 49.)
AcrR possesses a HTH DNA-binding domain that places it in the TetR family of repressors, together with the multidrug regulators MtrR from N. gonorrhoeae, QacR from S. aureus, and AcrS, a protein which is likely to control the expression of AcrEF (109, 156), an additional MDR transporter from E. coli that is homologous to AcrAB (Table 1). Although AcrR represses both its own and acrAB transcription, it is not involved in the induction of acrAB and acrR expression in response to general stress conditions, such as 4% ethanol, 0.5 M NaCl, and entry into stationary growth phase; instead, these increases in transcription were attributed to an unidentified regulatory protein (107). Thus, it appears that the primary function of AcrR is to modulate acrAB expression, thereby preventing excessive production of the AcrAB pump, whereas MarA and related global regulators are primarily responsible for the actual induction of acrAB and tolC (Fig. 2; described in detail below).
A recent study has demonstrated that AcrAB is also positively regulated by SdiA, a protein which regulates cell division genes in a manner dependent upon quorum sensing (174). The link between AcrAB and quorum sensing, combined with the observation that some signal molecules utilized by bacteria for quorum sensing are similar to known AcrAB substrates, viz., the fluoroquinolones, led to the suggestion that AcrAB may have a physiological role in the export of non-freely diffusible quorum-sensing signals (174). Therefore, increases in AcrAB expression as growth rates slow (107, 176) may be related to increased levels of the quorum-sensing signals produced by E. coli.
MarA Global Activator
Although not involved in the response to the general stress conditions described above, transcriptional activation of acrAB expression is the predominant cause of multidrug resistance in strains that overexpress MarA or the closely related global regulators SoxS and Rob (7). The mar (multiple antibiotic resistance) regulatory locus (Fig. 2) consists of the marRAB operon and the divergently transcribed marC, which encodes a protein of unknown function, as does marB (7). The intracellular levels of the MarA global activator are controlled by the product of the first gene of the marRAB operon, MarR. Both of these proteins bind to marO (7), a region of DNA that separates the two transcriptional units and contains a large number of regulatory protein binding sites, within and around the marRAB promoter (Fig. 2).
MarA, a member of the AraC family of transcriptional activators (Table 1), activates its own transcription and that of a large number of mar regulon genes by binding to 20-bp DNA sequences known as marboxes that are located in the vicinity of the promoters for the target genes. For example, the MarA-binding site within the marO regulatory region is 16 bp upstream of the −35 region of the marRAB promoter (Fig. 2) (118, 177). Importantly, the acrAB promoter is also adjacent to a marbox at which MarA has been demonstrated to bind and activate transcription (7). In addition, overexpression of MarA or its homologs SoxS and Rob has also been demonstrated to result in increased synthesis of the TolC component of the AcrAB-TolC pump complex, which, in combination with the identification of a putative mar/rob/sox-box upstream of the tolC gene, strongly suggests that tolC also belongs to the mar regulon (11).
The transcriptional activation functions of MarA have also been proven to be global in nature by the demonstration that MarA can promote the transcription of genes encoding proteins of diverse functions, both in vivo and in vitro (7, 68). Gene array analysis of a strain constitutively expressing MarA has indicated that more than 60 E. coli genes are differentially regulated by this protein (15), whereas a second study employing an inducible MarA expression system identified an additional 67 MarA-regulated genes (165). However, although the total number of promoters directly activated by MarA has recently been estimated to be less than 40 (122), an even later report has shown that MarA is capable of activating a gene that possess a marbox which diverges substantially from the consensus sequence (14), suggesting that 40 may be an underestimate of the number of mar regulon promoters. One well-documented example of MarA activity is activation of transcription of micF, which produces an antisense RNA that downregulates the expression of ompF, a gene encoding an outer membrane protein that is a site for drug entry (29). Thus, the reduction in the rate of drug influx via OmpF, combined with the increased production of the AcrAB and TolC drug efflux proteins, represents a highly effective mechanism by which MarA can act to coordinate a response to the presence of toxic antimicrobials.
MarA transcriptional activation has several unusual features: it binds DNA as a monomer, and its degenerate 20-bp marbox binding sites are asymmetric, lacking any of the inverted or direct repeats characteristic of bacterial regulatory sequences. Additionally, MarA and the closely related SoxS protein are significantly smaller than other AraC family activators, such as N. gonorrhoeae MtrA, because of the complete lack of a MarA or SoxS ligand-binding domain (119). The crystal structure of MarA in complex with the marbox from the marRAB promoter has been solved (Fig. 3), revealing a protein possessing two separate HTH DNA-binding domains linked by a long α-helix, another highly unusual arrangement for a prokaryotic regulator (177). Since a typical HTH motif is only capable of recognizing 6 bp, in contrast to the requirement of an operator sequence of at least 11 to 12 bp for a DNA-binding protein to recognize sites that do not occur by chance in a bacterial genome (207), most bacterial regulatory proteins acquire two HTH motifs by forming oligomers. However, for MarA, the presence of two HTH motifs in a single polypeptide chain explains how this protein can function as a monomer. In order to facilitate MarA simultaneously contacting two successive major grooves with the recognition helices from its two HTH motifs, the DNA in the MarA-marbox complex is bent by approximately 35° (Fig. 3) (177). Alanine-scanning mutagenesis has confirmed that the N-terminal HTH of MarA, which contacts the more highly conserved portion of the marbox consensus sequence, contributes the majority of the important MarA-DNA interactions (42).
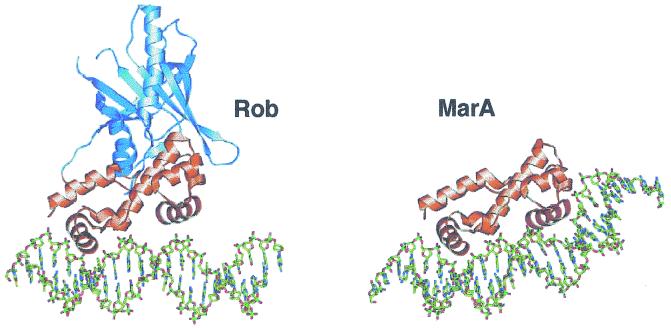
FIG. 3.
Structures of Rob and MarA proteins bound to micF and marRAB marboxes, respectively. The highly homologous DNA-binding domains of Rob and MarA are colored orange, with their respective HTH recognition helices in brown and the additional C-terminal putative ligand-binding domain of Rob in blue. The N-terminal Rob HTH is inserted into the major groove, whereas the C-terminal HTH makes contacts only to the DNA backbone. In contrast, MarA induces a significant bend in the marRAB promoter to facilitate the placement of both HTH motifs in successive major grooves of marRAB marbox DNA. (Reprinted with permission from reference 89; kindly provided by Tom Ellenberger.)
The orientations of marboxes and their distances from the −35 hexamers of mar promoters vary, but both are critical, presumably permitting the efficient formation of MarA-RNA polymerase-DNA ternary complexes (117). Mutagenesis has indicated that distinct solvent-exposed MarA residues are involved in the activation of transcription from each class of promoter, suggesting that MarA employs different mechanisms to promote RNA polymerase activity, depending on the orientation and location of each marbox (42). Utilization of nuclear magnetic resonance techniques to investigate the interaction of MarA with degenerate marboxes in solution revealed that portions of the DNA-bound form of MarA exist in a highly dynamic state (27). This led to the proposal that MarA possesses an inherent flexibility that grants it the ability to undergo significant rearrangements in order to accommodate variations in the DNA sequences of marboxes (27).
More recent evidence suggests that the primary mode of transcriptional activation by MarA first involves the formation of a complex between the activator and RNA polymerase, which can then scan the chromosome for mar regulon promoters more efficiently than RNA polymerase or MarA alone (116). The ability of a MarA-RNA polymerase complex to selectively identify marboxes that are adjacent to promoter sequences was suggested to be the mechanism by which MarA can distinguish real marboxes from the many marbox-like sequences present in the E. coli chromosome (116). A similar mechanism has also been proposed for the activity of the MarA homolog SoxS (see below) (46).
MarR, Antimicrobial-Sensing Repressor of marRAB
The MarR repressor, which is the product of the first gene in the marRAB operon, controls the intracellular levels of MarA and hence plays a crucial role in the MarA-mediated activation of mar regulon promoters (Fig. 2). MarR, like MarA, also binds within marO but at sequences distinct from the marbox, although a degree of competitive binding at marO between the two proteins does exist (118). MarR represses marRAB transcription by binding as a dimer to two distinct regions in marO, site I and site II, which are located downstream from the MarA binding site (Fig. 2) (120). MarR site I is positioned between the −35 and −10 regions of the marRAB promoter, an ideal binding site to prevent access by RNA polymerase, whereas site II does not appear to be required for repression or binding by MarR at site I (Fig. 2) (120).
A crystal structure obtained for MarR in the presence of the compound salicylate indicated that MarR contains a DNA-binding domain belonging to the winged-helix family (8), as was found for another MarR family member, the P. aeruginosa MexR protein (101). The MarR α3 and α4 helices constitute the HTH motif, whereas β-sheets contribute to the formation of the “wings.” The location of the predicted α4 recognition helix, which is likely to make contacts to the DNA major groove, is in good agreement with mutagenesis studies that had previously assigned this region a role in DNA binding (5). In contrast to MexR, for which the proposed induction mechanism was suggested to be facilitated by the inherently high degree of flexibility observed for this protein (101), MarR appears to be a much more rigid protein whose structure is stabilized by a number of salt bridges (8), indicating that these two family members are likely to function by distinct methods.
Overall, MarA activates expression of the mar regulon, including acrAB, tolC, and marRAB, whereas MarR acts to downregulate this response by repressing the synthesis of MarA. Although overexpression of MarA from a plasmid is sufficient to activate the mar regulon genes (40), the addition of the antibiotics tetracycline and chloramphenicol (50), weak aromatic acids, such as salicylate, and a structurally diverse range of other compounds, such as the uncoupling agent carbonyl cyanide m-chlorophenylhydrazone and the redox-cycling compounds menadione and plumbagin (6, 200), have all been shown to cause induction of mar regulon expression. Two independent mechanisms for mar regulon induction have been identified, each of which has been proposed to act by suspending the repressor abilities of MarR. First, salicylate, plumbagin, 2,4-dinitrophenol, and menadione have been demonstrated both to induce the marRAB operon in vivo and also to interfere with the binding of MarR to marO DNA in vitro (6). This suggests that MarR can directly bind a broad range of ligands, the outcome of which is its dissociation from marO, MarA synthesis, and, hence, mar regulon activation (Fig. 2).
Interestingly, in order to produce the aforementioned crystal structure of MarR, high concentrations of the inducer salicylate were found to be necessary. Salicylate was observed to be bound at two sites on the surface of each MarR subunit, in the vicinity of the proposed α4 recognition helix (8). Although these observations were highly suggestive of a potential influence on MarR-DNA interactions, it remains to be seen if binding of salicylate at these sites has any effect on protein conformation or is even physiologically relevant (8).
A second mechanism by which the repressor activities of MarR appear to be interrupted involves the binding of the cell wall component murein tripeptide and its transfer into the cytoplasm by MppA, a periplasmic binding protein (97). An mppA null mutant exhibits phenotypes consistent with derepression of the mar regulon, a process that requires a functional MarA but also alters the expression levels of other genes which are not part of the mar regulon (97). It has been suggested that a low level of murein tripeptide in the periplasm is an indicator of stress, sensed by MppA, which activates a signal transduction pathway that ultimately produces phosphorylated, presumably inactive, MarR (97). However, the suggestion that MarR contains a functional aspartyl phosphorylation-dephosphorylation sequence (97) has been cast in some doubt by the MarR crystal structure (196). Hence, MarR is capable of sensing the presence of deleterious substances and/or environmental conditions by at least one and possibly two independent mechanisms, both of which would cause marRAB to be derepressed and the ensuing activation of the mar regulon, including acrAB.
MarA Homologs SoxS and Rob
SoxS, the effector of the soxRS global superoxide response (sox) regulon, and Rob, which binds the E. coli chromosomal origin of replication, are MarA homologs which, in addition to activating marRAB transcription by binding to marO, have also been shown to directly activate expression from promoters of genes belonging to the mar and sox regulons in vitro and in vivo (Fig. 2) (69, 70, 129, 215). Thus, it is not surprising that the majority of the residues identified in the MarA DNA-bound crystal structure as being important for forming DNA contacts represent amino acids that are conserved in the SoxS and Rob proteins (177). Elevated levels of SoxS and Rob have also been shown to specifically increase the transcription of acrAB (107, 234), which is the most important feature of the mar multidrug resistance phenotype, since even for strains constitutively expressing marA, soxS, or rob, deletion of acrAB results in hypersensitivity to many antimicrobial agents (129, 150, 215).
For SoxS to have a significant effect on acrAB-tolC transcription in wild-type cells, the expression of the soxS gene must first be activated by superoxide-generating agents via the conversion of SoxR, a divergently transcribed local transcriptional activator, into an active form (Fig. 2) (121, 147). Rob, on the other hand, has only a modest effect on the basal level of expression from the marRAB promoter, despite being naturally expressed at a relatively high level (121) and its ability to activate acrAB expression in a mar deletion strain (215). However, unlike SoxS and MarA, Rob contains an additional C-terminal domain, which the crystal structure of Rob complexed with DNA (Fig. 3) has revealed may be involved in the binding of an uncharacterized effector molecule with the potential to alter Rob activity (89). The Rob C-terminal domain has recently been demonstrated to be required for the apparent binding of the compound dipyridyl in vitro and also for the activation of Rob transcriptional stimulation by dipyridyl in vivo, although it is currently uncertain if this occurs via a direct interaction with Rob (182).
An unexpected finding from the Rob-DNA crystal structure (in this case with micF marbox DNA) was that, whereas the N-terminal HTH of Rob was inserted into the DNA major groove, the C-terminal HTH made contacts only to the DNA backbone (Fig. 3) (89). As a result, the Rob-bound micF marbox DNA was not bent, as had been observed for the MarA-bound mar marbox DNA (Fig. 3), although Rob does appear to bend the promoter DNA of some mar regulon genes in solution (70, 119). Because the mar and micF promoters used in the MarA and Rob crystallization studies represent different marbox promoter classes, it is possible that both of these proteins possess the ability to bind DNA with or without their C-terminal HTH inserted into the major groove. Although the role of the Rob C-terminal HTH is still in doubt (119), the use of alternative DNA-binding modes may provide a means by which these proteins can vary the activation mechanisms they employ to suit the different classes of promoters that they bind.
Yet another protein involved in the regulation of acrAB-tolC is FIS, a nucleoid-associated global regulatory protein that modifies transcriptional activity in response to various growth conditions and can also bind to a site within marO just upstream of the marbox (Fig. 2). FIS is proposed to limit the overall level of negative superhelicity and also stabilize the local DNA architecture of certain promoters (225), which, for the marRAB promoter, provides an additional twofold stimulation to MarA-, SoxS-, and Rob-mediated activation of transcription (Fig. 2) (121).
Regulatory systems akin to mar appear likely to be widespread, with evidence suggesting that many bacterial species contain closely related genes (26, 66, 90, 126). For example, in a clinical isolate of Salmonella enterica, increased quinolone resistance has been attributed to a point mutation that produced a constitutively active SoxR protein, presumably leading to elevated expression of sox/mar regulon genes, such as acrAB and tolC (86). Enterobacter aerogenes, an increasingly important cause of respiratory tract infections, also contains a marRAB operon analogous to that of E. coli which has been implicated in antibiotic resistance (24). Altogether, the activation of acrAB and tolC expression by several global regulators in response to a broad range of stress conditions suggests that the primary physiological function of the AcrAB-TolC complex in E. coli is the export of a wide variety of stress-related toxic compounds encountered by this organism in its normal environment, such as a demonstrated role in the efflux of fatty acids and bile salts (108, 143, 221).
Other Regulators of E. coli MDR Pumps
Neither MarR nor MarA has any effect on the expression of another chromosomally encoded E. coli multidrug pump, EmrAB, which, despite being an MFS transporter, also appears to form a tripartite complex with the TolC outer membrane porin in a manner analogous to the function of RND-type pumps (96). Induction of emrAB is controlled by the local repressor EmrR, the product of the first gene of the emrRAB operon. Disruption of the emrR gene via a frameshift mutation has been demonstrated to be responsible for increased expression of EmrAB and elevated resistance to the antibiotic thiolactomycin (105). EmrR is a member of the MarR family of repressors (Table 1) and has been shown to be capable of repressing the marRAB promoter when overexpressed (211). Carbonyl cyanide m-chlorophenylhydrazone, carbonyl cyanide p-(trifluoromethoxy)phenylhydrazone, and 2,4-dinitrophenol, structurally unrelated substrates of the EmrAB pump which induce emrRAB expression (105), have been confirmed to be directly bound by EmrR, one ligand per repressor dimer (20).
EmrR acts by binding to an imperfect inverted repeat centered around the −10 region of the emrRAB promoter, an interaction that can be disrupted in vitro by addition of the ligands carbonyl cyanide m-chlorophenylhydrazone, 2,4-dinitrophenol, tetrachlorosalicylanilide, and nalidixic acid (238). Thus, like MarR, EmrR functions to sense the presence of toxic compounds, but differs in that EmrR exerts direct control over the expression of the EmrAB pump rather than acting indirectly through a global regulator. Interestingly, EmrR was originally described as MprA, the regulator of the plasmid-borne mcb operon, which encodes microcin B17 (104). The addition of compounds that inactivate EmrR repression of emrRAB transcription conversely resulted in EmrR-mediated repression of the main mcb operon promoter. Additionally, EmrR was required for positive regulation of a second mcb promoter that controls expression of microcin export and immunity genes, the products of which have also been implicated in the export of some fluoroquinolone antibiotics (104).
Two-component signal transduction systems involved in modulating the expression of several E. coli drug transporters have also been identified. Overexpression of the EvgA response regulator from the evgSA two-component system produces an elevated level of resistance to MDR substrates, including benzalkonium, crystal violet, deoxycholate, doxorubicin, erythromycin, rhodamine 6G, and sodium dodecyl sulfate (145). These increases have been attributed primarily to increased expression of the yhiUV MDR transporter (144), although the observed increase in resistance to deoxycholate is due in part to enhancement of the expression of another drug transport operon, emrKY, by EvgA (145). It is intriguing that yhiUV and emrKY encode an RND-type and an MFS transporter, respectively, both of which have been shown to require the TolC outer membrane channel (144).
An additional two-component regulatory system, BaeSR, has recently been demonstrated to activate the transcription of yet another E. coli MDR transporter, an RND-type pump complex encoded by the mdtABC operon, which confers resistance to bile salts, novobiocin, and deoxycholate (12, 137). Both mdtB and mdtC appear to encode RND pumps that interact, perhaps as heteromultimers, with the MdtA membrane fusion protein, whereas a fourth gene in the same operon, mdtD, encodes an MFS-type pump that is not required for drug resistance (12, 137). In addition to these E. coli examples, it is interesting that in several other species, such as Acinetobacter baumannii, Stenotrophomonas maltophilia, and S. aureus (Table 1), the expression of a number of MDR transporters also appears to be under the control of two-component regulatory systems which are designed to respond to specific external environmental stimuli (158). Thus, the two-component-regulated MDR transporters, by extension, are likely to perform specific export functions in response to these unknown signals.
A comparison of the nucleotide sequences upstream of known and hypothetical MDR transporters has suggested that the regulation of E. coli MDR efflux genes is likely to be considerably more complicated than the available experimental evidence indicates. Possible regulatory sequences that have been identified include a marbox and an AcrR-binding site upstream of acrEF, which encodes an AcrAB homolog, and an EmrR-binding site upstream of acrAB (178). Sequences upstream of the marRAB and emrRAB operons were also proposed to be bound by a hypothetical regulator (178). In combination with the available information on the local, global, and two-component regulatory systems known to control E. coli drug transport pumps, it would appear that MDR regulatory proteins in this organism are likely to be involved in a complex web of interactions governing the expression of their target genes. Although the individual pumps may have specific physiological roles, E. coli cells may be capable of coordinating the expression of at least some of these MDR transporters in response to multiple threats, making efficient use of their ability to extrude overlapping ranges of substrates.
RELATED LOCAL AND GLOBAL ACTIVATORS CONTROL MDR GENE EXPRESSION IN B. SUBTILIS
The chromosome of the gram-positive bacterium Bacillus subtilis encodes two highly similar MFS MDR transporters, Bmr and Blt. These proteins show 51% amino acid identity and confer very similar levels of resistance to an identical range of toxic substances when overexpressed (3, 141). They are each regulated by the product of an adjacent gene encoding a transcriptional activator belonging to the MerR family, BmrR (2) and BltR (3), respectively (Table 1). Although these activators have similar N-terminal DNA-binding motifs, their C-terminal domains, which are known to be responsible for inducer binding in this family of proteins (113, 212), show little homology to each other or to any other protein sequences currently in the DNA sequence databases, suggesting that distinct ligands are bound by each protein. However, the ligands for BltR have yet to be identified, which, in combination with the inability of known Blt substrates to induce expression of the blt gene, suggests that the multidrug-transporting abilities of Blt are likely to be entirely fortuitous (3).
Interestingly, the second gene in the blt operon, SpAT (Fig. 4A; formerly known as bltD), encodes an enzyme that acetylates polyamines, such as the natural cellular constituent spermidine. Despite this finding, alterations of polyamine levels had no influence on expression of the blt operon (115). Thus, although spermidine is a known substrate of Blt (237), the suggestion that the efflux of polyamines may be the primary role of Blt remains uncertain.
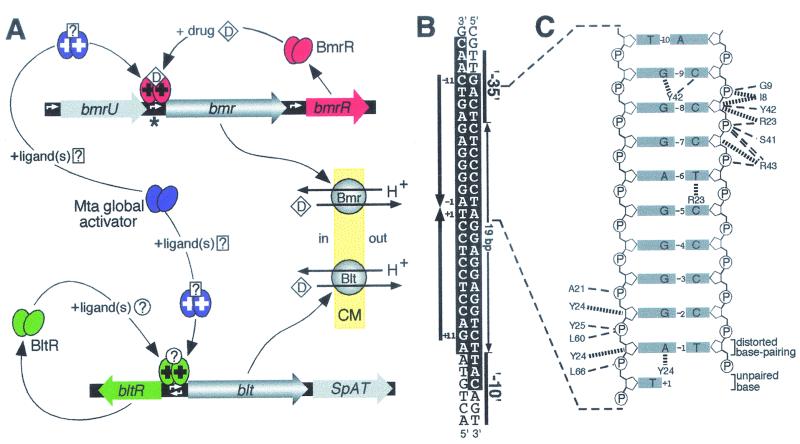
FIG. 4.
(A) Regulation of expression of B. subtilis MDR genes bmr and blt. A BmrR dimer concurrently bound to both bmr promoter DNA and drugs (D) can correctly orient the −10 and −35 hexamers of this promoter to facilitate the binding of RNA polymerase. White arrows indicate the locations of promoters and also the direction in which transcription occurs from these sequences. Because many of the substrates of the Bmr MDR transporter are also ligands of BmrR (red ovals), activation (+) of bmr expression can occur in response to the presence of these deleterious compounds, permitting drug efflux across the cytoplasmic membrane (pale yellow; CM) in exchange for protons (H+) to occur. The global regulatory protein Mta (purple ovals) and the local activator of the blt operon, BltR (green ovals), are likely to act in the same fashion as BmrR, although inducing ligands for these proteins have yet to be identified (?). Mta activates both the bmr and blt genes by binding to the same DNA sequence as the local regulators. The blt operon also encodes SpAT, a polyamine acetyltransferase, whereas Bmr expression can result from transcription initiated at either its own promoter or the promoter of bmrU, an upstream gene of unknown function. (B) The DNA sequence from the region indicated by an asterisk in A, which contains the bmr promoter (Pbmr). The −10 and −35 hexamers of Pbmr and the unusually large 19-bp spacing between these hexanucleotides are indicated, while the large arrows denote the imperfect inverted repeat (labeled −11 to +11) within Pbmr that constitutes the BmrR binding site (2). Bases protected from DNase I digestion by BmrR bound to Pbmr are highlighted in white. (C) DNA contacts made by BmrR to the half-site that encompasses positions −1 to −10 in B. Also shown is the thymine from position +1, which in the BmrR-DNA complex was observed to be no longer base-paired to its partner, whereas the adenine and thymine bases at position −1, although significantly displaced, still formed a distorted base pair. Thin dashed lines indicate hydrogen bonds, and thick broken lines indicate van der Waals interactions between BmrR amino acids and bases (grey boxes) or the phosphates (P) and deoxyribose rings (pentagons) that form the DNA backbone. Note that the sequence depicted in C differs at position −8 from that shown in B because the experimentally determined data best fit a GC base pair at this location rather than the AT that was actually present in the BmrR-DNA complex. (Panel A modified with permission from reference 49; panel C reprinted with permission from reference 245.)
BmrR, Local Transcriptional Activator of bmr Expression
In contrast to Blt, Bmr is expressed under normal growth conditions, and disruption of its gene therefore produces cells that are hypersensitive to MDR substrates. In addition to its own promoter, bmr can also be cotranscribed with bmrU, an upstream gene of unknown function (Fig. 4A), which suggests that Bmr and BmrU may have related but currently unknown physiological roles (115). However, more importantly from a drug resistance perspective, the local transcriptional activator, BmrR, can mediate increases in expression from the promoter immediately upstream of bmr after binding some of the synthetic substrates of the Bmr pump (Fig. 4A), such as astrazon orange, diethyl-2,4′-cyanine, rhodamine 6G, and TPP (2, 113, 114, 227). The drug-bound form of BmrR, in addition to possessing the ability to activate expression from the bmr promoter, also exhibits an improved affinity, in comparison to apo-BmrR, for bmr promoter DNA (2, 196, 227). Thus, it appears likely that the Bmr/BmrR system exists at least partially to provide protection against toxic, lipophilic, cationic substances that B. subtilis may encounter in its natural environment (2).
Like other MerR family members, BmrR binds as a dimer to an imperfect inverted repeat located between the −10 and −35 hexamers of a target promoter that exhibits an unusually large spacing of 19 bp (Fig. 4B) (2). This spatial arrangement places the −10 and −35 regions of the bmr promoter on opposite sides of the DNA helix, a conformation that is incompatible with RNA polymerase binding. In the case of blt, a 1-bp deletion that altered its promoter spacing to 18 bp was sufficient to cause a marked increase in expression (3). For promoters regulated by MerR and another family member, SoxR, 2-bp deletions in their spacer regions in each case produced a promoter that exhibited highly elevated transcription levels independently of their respective activator proteins (58, 157). Binding of the MerR activating ligand is known to have an effect similar to that of the 2-bp deletion, as it allows the protein to initiate the partial untwisting of promoter DNA, which suggested that activation by MerR involves the production of a promoter with a more favorable spacing (212).
Structure of Tripartite BmrR-Drug-Activated DNA Complex
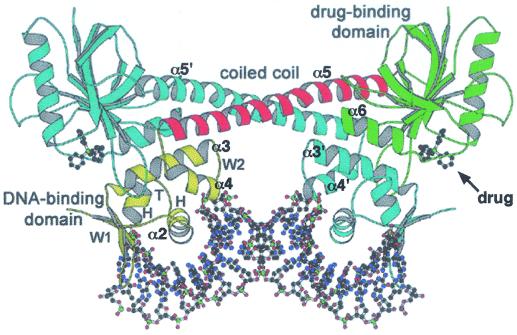
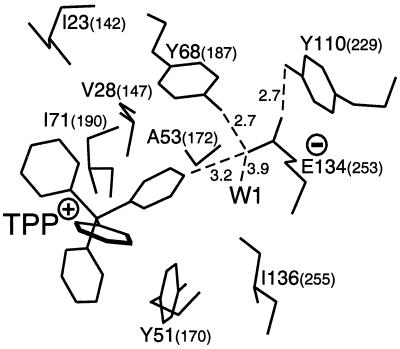
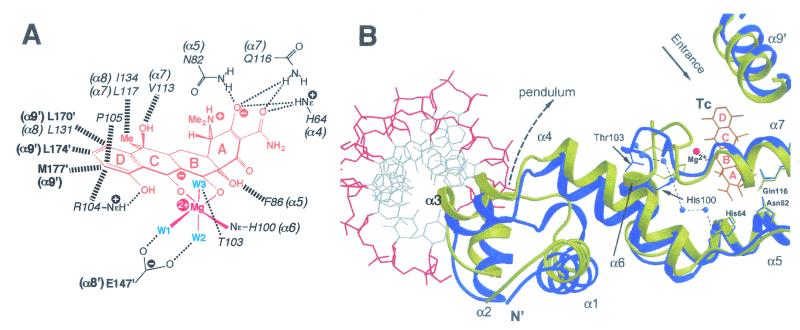
The finer details of the mechanism by which MerR family proteins activate expression from promoters under their control have recently been elucidated by determination of the crystal structure for the tripartite complex of BmrR bound concurrently to DNA and a drug (245). BmrR was found to consist of a DNA-binding domain which is linked to the C-terminal two-thirds of the protein responsible for ligand binding by a long helix, α5 (Fig. 5) (245). The tight packing of the drug-binding domain from each polypeptide against the DNA-binding domain of the second polypeptide in a BmrR dimer, in combination with an antiparallel coiled coil that α5 forms with α5′ from the other subunit, constitutes an extensive dimerization interface (Fig. 5). The BmrR DNA-binding domain belongs to the winged-helix superfamily and consists of an HTH (α1, the α2 recognition helix, and their connecting turn), and two additional wings, W1 and W2 (Fig. 5) (245). The combination of these DNA-binding elements ensures that BmrR makes a large number of contacts with bmr promoter DNA (Fig. 4C), however, at the same time, the promoter remains accessible for RNA polymerase binding, an essential feature for a transcriptional activator protein (245).
FIG. 5.
Structure of a BmrR dimer in the drug- and DNA-bound tripartite complex. One polypeptide chain is colored yellow for the N-terminal winged-helix DNA-binding domain, red for the α5 linker helix, and green for the C-terminal drug-binding domain. The locations of selected helices are indicated for this polypeptide, as are the positions of the α3′, α4′, and α5′ helices in the second (cyan) polypeptide. Also labeled for the first monomer is the HTH (H, α1; T, turn; H, α2 recognition helix) and the two wings, W1 (sheets β2 and β3) and W2 (helices α3 and α4). The DNA and drug (tetraphenylantimonium) molecules are represented as a ball and sticks (carbon, black; nitrogen, blue; oxygen, red; and phosphorus/antimony, green). (Reprinted with permission from reference 245; kindly provided by Richard Brennan.)
A startling revelation from the BmrR-drug-DNA structure was that the bmr promoter had been distorted to reposition the −35 and −10 sequences onto the same face of the DNA, so that both hexamers were now simultaneously available for RNA polymerase binding and subsequent initiation of transcription (245). The distortion of bmr promoter DNA was found to be facilitated by a novel mechanism that disrupted the base-pairing interactions of the 2 bp (AT) at the center of the pseudodyad within the bmr promoter (+1 and −1 bp in Fig. 4B and C). The AT base pair at position +1 was observed to become completely unpaired (Fig. 4C), with the adenine and thymine sliding away from each other, while the AT base pair at position −1, although significantly displaced, was still capable of forming a distorted base-pairing interaction (245). The combined effect produced an operator sequence that was “bunched up” in the middle, shortening the spacing between the −10 and −35 hexamers of the bmr promoter DNA by a distance equivalent to 2 bp (245). Interactions of BmrR residues Tyr24, Tyr25, Lys60, and Lys66 with the DNA backbone serve to stabilize the distorted base pair (Fig. 4C), maintaining bmr promoter DNA in this transcriptionally active configuration. Thus, drug-bound BmrR acts to facilitate the synthesis of Bmr, which can then extrude the deleterious compounds to ensure the continued survival of the cell.
Binding of Antimicrobial Ligands by BmrR
It has been demonstrated that the first 119 residues of BmrR, which comprise the N-terminal DNA-binding region of this protein, can be completely removed and the C-terminal portion expressed as a separate domain, BRC, which retains full dimerization and ligand-binding abilities (113). BRC has been shown to bind one ligand molecule for each BRC and two per BRC dimer (113), consistent with the two drug molecules, one per binding pocket, present in the full-length BmrR-drug-DNA complex (Fig. 5) (245). Prior to the recent work on the full-length protein, earlier X-ray crystallization studies on apo-BRC and BRC bound to the ligand TPP revealed a ligand-binding pocket that was lined with hydrophobic and aromatic amino acids, in addition to the unusual feature of a charged residue deeply buried in the core of the protein, specifically, Glu134 positioned at the base of the binding site (Fig. 6) (244). Normally considered an energetically unfavorable position, the burial of the negatively charged Glu134 residue in the core of unliganded BRC is neutralized by hydrogen bonds to the hydroxyl groups of three BRC tyrosine residues, Tyr33, Tyr68, and Tyr110. Tyr33 is located in the short, flexible α2 helix of BRC (BmrR α6), which is positioned across the entrance to the binding pocket in the absence of a coactivator.
FIG. 6.
Structure of a TPP molecule depicted relative to the location of BRC binding-pocket residues that interact with this ligand. The negatively charged Glu134 BRC residue (the equivalent BmrR amino acid number is shown in parentheses) provides the crucial electrostatic interaction with the delocalized positive charge carried by the TPP phenyl rings. Ile23, Val28, Ala53, Ile71, and Ile136 all form van der Waals contacts with TPP, whereas the aromatic side chains of Tyr51 and Tyr68 stack against TPP rings. Tyr68 also hydrogen bonds to Glu134 to stabilize its negative charge, as does Tyr110 and a water molecule (W1), which replaces the hydrogen bond of Tyr33 (BRC α2) that was broken because of the displacement of BRC α2 by the entry of TPP into the binding site. The stabilizing hydrogen bonds are indicated by broken lines, as is the crucial electrostatic contact, with distances given in angstroms. (Reprinted with permission from reference 244.)
A negative residue (BRC Glu21), located on the surface of the protein adjacent to BRC α2, is thought to initially attract positively charged drugs to the entrance of the binding pocket (244). The BRC α2 appears likely to be highly flexible and fluctuate between a folded and partially unfolded conformation, facilitating entrance of the ligand into the binding site proper. However, the contribution of α2 (BmrR α6) to the specificity of ligand binding appears to be minimal, based on the mutagenesis of several BRC α2 residues, which was observed to have little effect (227). Upon ligand binding, displacement of BRC α2 removes the side chain of Tyr33 from the protein core, thus breaking one of the H bonds stabilizing BRC Glu134. However, a water molecule replaces Tyr33 and forms an H bond with Glu134 (Fig. 6), maintaining the stabilization of the internal BRC negative charge (244). Once the ligand has entered, binding occurs via an electrostatic contact between the negative charge of the carboxylate group of the BRC Glu134 residue at the base of the binding site and the partial positive charge carried by the phenyl rings of TPP (Fig. 6) (244). Van der Waals contacts between TPP and the surrounding hydrophobic amino acids that line the binding pocket strengthen the interaction, in addition to the stacking of TPP rings against aromatic side chains (Fig. 6) (244).
Model building with a second BmrR ligand, rhodamine 6G, demonstrated that the BRC binding pocket is capable of accommodating this compound as well, predicting a much closer electrostatic contact between the carboxylate group of the buried glutamate residue and the positive charge of the amino ethyl group in rhodamine 6G, explaining the 100-fold-greater affinity of BRC for rhodamine 6G than for TPP (244). Site-directed mutagenesis to replace the buried BRC Glu134 with a neutral alanine residue almost completely abolished the binding of five of the six BmrR ligands that were tested, confirming the importance of this electrostatic interaction (227). The binding of the sixth ligand was reduced by less than twofold, suggesting that although Glu134 is important, it is not essential for all BRC ligands (227). Changes to an other six BRC residues that the BRC-TPP structure had indicated were involved in ligand binding had lesser effects, consistent with the weaker nature of their interactions with TPP.
Interestingly, in a number of cases, although an alanine substitution decreased the binding affinities for some compounds, the same change failed to alter the binding of other drugs, whereas still other ligands exhibited an up to threefold increase in their binding affinities for that mutant (227). The inconsistent binding abilities that these mutants demonstrated towards different ligands are thought to reflect the intrinsic nature of the BRC binding site, by which differential interactions of binding pocket residues with various ligands permit the accommodation of structurally diverse drugs (227). The ability of individual compounds to form various interactions with different subsets of the residues lining the binding pocket lends support to the proposal that the ligand-binding sites of MDR proteins are likely to represent a compromise between maximizing their range of ligands and retaining high affinities for the individual compounds.
In addition to Glu253 (BRC Glu134), the structure of the BmrR-drug-DNA tripartite complex revealed that electrostatic interactions also occurred between the positively charged tetraphenylantimonium (a TPP analog) and two other negatively charged BmrR residues, Asp47′ (where the prime indicates the second subunit in a BmrR dimer) and Glu266 (245). This finding helps to explain the unexpected weak effect that the mutation of BRC Glu134 had on the binding of a single compound (227). Asp47′, located in the DNA-binding domain of the second monomer in a BmrR dimer, is of particular interest, because although this residue is absent in BRC, both BmrR and BRC have equal affinities for ligands (114), whereas the residue equivalent to BmrR Glu266 was disordered in the BRC-TPP complex (244). Because of the lack of a structure for the BmrR-DNA complex, it is also uncertain how drug binding is coupled to the transition of BmrR into a state in which it distorts bmr promoter DNA and activates bmr transcription. However, the unwinding of α6 in BmrR (BRC α2) upon drug binding has been proposed to act as the signal for conversion of BmrR into a transcriptional activator (244). The repositioning of α6 could influence DNA binding directly because of alterations in its interactions with α3′ and α4′ from the DNA-binding domain in the second polypeptide of a BmrR dimer (Fig. 5). Alternatively, unwinding of α6 is likely to affect the α5 linker helix (Fig. 5), which could therefore directly transmit a signal from the drug-binding domain to the DNA-binding domain within each polypeptide (245).
Global B. subtilis MDR Gene Regulator Mta
In addition to their local activators, both blt and bmr are known to be subject to global regulation by Mta (Fig. 4), another MerR-like regulator (13). Although full-length Mta did not activate transcription of the blt or bmr gene, expression of MtaN, a truncated version containing only the N-terminal DNA-binding domain, resulted in increased blt and bmr expression. MtaN was found to bind directly to the blt and bmr promoter elements at the same position as the specific BltR and BmrR regulators (Fig. 4) (13). Presumably, deletion of the C-terminal ligand-binding domain of Mta mimics the in vivo effect of a bound inducer molecule. A crystal structure for apo-MtaN revealed a dimeric protein containing a winged HTH DNA-binding domain that is largely structurally comparable to that of BmrR (43). However, differences in the orientation of the α5 dimerization helix and the first wing of the DNA-binding domain suggest that MtaN interacts with DNA in a manner distinct from the transcriptional activation mechanism of BmrR (43).
The ligand-binding domain of Mta, unlike those of BltR and BmrR, shows significant homology to other proteins, most notably to thiostrepton-induced TipA, a global regulator that has been implicated in the control of multidrug transport in Streptomyces lividans (13, 61), although there are currently no clues as to the nature of the compound that binds to Mta and activates its global regulatory functions. Comparable to what has been demonstrated for tipA, mta, in addition to encoding a full-length Mta protein, is also likely to encode an individually expressed C-terminal domain that would sequester the putative Mta inducer in order to limit the positive-feedback loop controlling Mta expression (13).
In summary, despite the assignment of a putative physiological role for Blt, the different expression patterns of blt and bmr during normal growth, and the lack of any common BmrR and BltR ligands, the control of bmr and blt expression by Mta indicates that B. subtilis likely utilizes the very broad substrate specificities of these two MDR pumps as part of a global response to some stress, such as the presence of hydrophobic toxins. In addition, BmrR can independently activate bmr transcription in response to a wide range of toxic compounds. However, the genetic organization of these pumps indicates that they are also both likely to perform unidentified specific physiological roles. From an efficiency viewpoint, it certainly makes sense for cells to employ a single transport protein for multiple functions, provided that this can be achieved without causing any disruption to cellular metabolism.
TetR, EXQUISITELY SENSITIVE REGULATOR OF TETRACYCLINE EFFLUX GENES
The most common form of resistance to tetracycline in gram-negative bacteria is because of a group of related MFS TetA efflux pumps which export tetracycline complexed with a divalent metal cation, normally Mg2+ (25). Unlike the majority of genes for gram-positive tetracycline transporters, the expression of tetA genes in gram-negative organisms is controlled in each case by the product of a divergently transcribed gene, the TetR repressor (59). As for the TetA determinants, the TetR proteins whose genes have been sequenced were classified into eight classes on the basis of amino acid sequence similarity, TetR A to E, G, H, and J, all of which show greater than 40% homology (190). Interestingly, the tetA(Z) and tetR(Z) genes from the gram-positive bacterium Corynebacterium glutamicum also show strong similarity to their gram-negative counterparts (217).
By far the best-characterized TetR protein is the 207-amino-acid Tn10-encoded TetR(B), although a number of detailed structural analyses have also been carried out on the 218-amino-acid TetR(D) determinant encoded on the plasmid RA1, which has 63% amino acid identity with TetR(B) (59). In the absence of tetracycline, TetR binds to the tet operator sequences overlapping the promoter of the tetA gene and represses its transcription (Fig. 7A) (59). Induction occurs on the binding of a tetracycline-Mg2+ complex by TetR, which causes a conformational change in the protein so that it can no longer bind to the tet operator, freeing up the tetA promoter for transcription (60). TetR has a far greater affinity for tetracycline-Mg2+, the intracellular form of the antibiotic, than the drug does for ribosomes, ensuring that expression of tetA occurs well before protein synthesis is inhibited, requiring only nanomolar concentrations of tetracycline-Mg2+ (59).
FIG. 7.
Control of tetA transcription by TetR. (A) The DNA sequence of the Tn10 tet intergenic region containing the tet operators and promoters is shown, with the base pairs that form the O1 and O2 inverted repeats shaded light grey. The tetA promoter PtetA, the two tetR promoters PtetR1 and PtetR2, and their associated transcription start points (right angle arrows) are also indicated. The product of the tetR gene (black ovals) forms dimers and binds to O1 and O2, preventing the expression (−) of both genes. Binding by TetR of the intracellular form of tetracycline, a tetracycline-Mg2+ complex, results in a conformational change so that TetR can no longer bind the tet operators. The ensuing initiation of tetA transcription protects the cell from tetracycline because of subsequent production of the membrane-bound TetA protein, which exports tetracycline-Mg2+ complexes in exchange for protons (H+). See text for other details. (B) DNA contacts formed by operator-bound TetR. The nucleotides from a tet operator half-site, representing the 0 to +7 positions of O1 in A, are depicted as grey boxes attached to the phosphate-ribose backbone. The contacts made by the DNA-reading head of one monomer in a TetR dimer to the bases in that operator half-site are shown as thin dashed lines for hydrogen bonds and thick broken lines for van der Waals interactions. TetR amino acids from the HTH recognition helix (α3) are in boldface type, those from elsewhere in the HTH motif are in italics, and Thr26 and Lys48 are additional residues from outside the HTH which make tet operator DNA contacts. The proline (P39) in the TetR recognition helix transfers binding of the TetR reading head from nucleotides on one strand of the operator half-site to nucleotides on the other strand. A hydrogen bond formed between a water molecule (wat) and His44 in the crystal structure mimics an interaction that would otherwise take place between His44 and the DNA backbone phosphate at position +8 of O1. (Panel B reprinted with permission from reference 153.)
Contribution of tet Promoters and Operators
The oppositely orientated tet genes encoded by Tn10, tetR(B), and tetA(B) represent the system for which regulation has been best characterized. The locations of the tetA(B) promoter, PtetA, and the two divergent tetR(B) promoters, PtetR1 and PtetR2, which partially overlap both the tetA(B) promoter and each other, are illustrated in Fig. 7A. Although the fully induced transcriptional activity of PtetA is 10-fold stronger than that of the combined tetR promoters, it is counterbalanced by a cis-acting sequence in the tetA(B) mRNA that reduces the efficiency of its translation by approximately 10-fold, and therefore, even under induced conditions, excessive synthesis of TetA(B) is prevented (59).
The TetR(B) protein is purified as homodimers that bind noncooperatively to two adjacent inverted repeats known as the tet operators O1 and O2, which overlap the tetA(B) and tetR(B) promoters (Fig. 7A) (128). Thus, TetR(B) binding to DNA can repress tetA(B) transcription and also autoregulate expression of its own gene, an essential aspect of the fine tuning of tetA(B) transcription by this system. Also of importance are the differences between the sequences of O1 and O2 (Fig. 7A), which explains the approximately fourfold-greater affinity that TetR(B) has for O2 (80). Binding of TetR(B) at O1 represses both genes, whereas if only O2 is occupied, tetA(B) continues to be downregulated, but tetR(B) transcription can occur almost completely unhindered from PtetR2 (Fig. 7A) (128). Thus, small decreases in the amount of operator-bound TetR(B) will serve to increase the synthesis of the repressor but not TetA(B), preventing accidental synthesis of the transporter.
Induction of tetA(B) expression can therefore occur only in the presence of tetracycline, as the conversion of operator-bound TetR(B) into the non-DNA-binding, induced TetR-tetracycline-Mg2+ complex is required for both O1 and O2 to be unoccupied by the repressor. The autoregulation of the tetR(B) gene ensures that only the minimum level of the repressor required to fully occupy O1 and O2 will be present in the cell, a situation that, combined with the extreme sensitivity of TetR(B) to the presence of tetracycline, provides for very fast and efficient induction of TetA(B) synthesis in response to the presence of the drug. The concomitant increase in TetR(B) expression ensures that first tetA(B) transcription and then that of tetR(B) can be rapidly brought to a halt once sufficient amounts of the TetA(B) protein have been produced to clear the antibiotic from the cell (59). Thus, by having synthesis of the repressor always occur both prior and subsequent to induction of TetA(B) expression, this regulatory system can maintain strict control of tetA(B) transcription at all times.
Structure of a TetR Dimer
Solving the crystal structure for the inducer-bound form of TetR(D) revealed a protein consisting of 10 α-helices, α1 to α10 (Fig. 8) (60). TetR homodimers contain two polypeptide chains, α1 to α10 from the first monomer and α1′ to α10′ from the second (60). The dimerization of TetR is principally achieved by the antiparallel helices α8 and α10, which interact with the symmetry-related α8′ and α10′ to form a four-helix bundle (Fig. 8) (60). The function of this four-helix bundle has been investigated by site-directed mutagenesis to alter residues in the hydrophobic core of the protein, which produced a TetR(B) derivative with altered dimerization specificity and confirmed the contribution of α8 and α10 to oligomerization (192). In addition to the hydrophobic core of the dimer, solvent-exposed residues on the periphery of the dimerization surface have also been demonstrated to be involved in protein-protein recognition by the four-helix bundle (191, 195).
FIG. 8.
Structure of the TetR dimer-tet operator DNA complex. The α-helices in each polypeptide are colored blue for the DNA-binding domain (α1 to α3), yellow for the rigid α-helices (α5, α8, and α10), and green for those that undergo conformational changes upon induction (α4, α6, α7, and α9). The α3 and α3′ recognition helices fit into successive major grooves of tet operator DNA, which is represented as red for the phosphate-ribose backbone and grey for the bases. A grey line also delineates the curvature of operator DNA induced by TetR binding. (Reprinted with permission from reference 153; kindly provided by Winfried Hinrichs.)
TetR Binding to tet Operator
In each TetR polypeptide chain, the helices α1 to α3 form the DNA-reading head, which is connected to the core of the protein, α5 to α10, through α4 (Fig. 8) (60). Although the HTH motif common to many regulatory proteins comprises α2 and α3, α1 is important for the DNA-binding process because of its stabilizing effect on the HTH structure (153). This confirmed previous studies in which TetR variants that had been constructed with N-terminal deletions of various lengths were found to be incapable of binding DNA (18). Also contributing to DNA binding are the N-terminal residues of α4, which participate in the formation of the hydrophobic center of the DNA-binding domain, further stabilizing it in the operator-bound complex (153).
In the induced TetR-tetracycline-Mg2+ complex, the two HTH DNA recognition helices in the TetR dimer, α3 and α3′, were found to be separated by 39.6 Å, preventing the repressor from binding to successive major grooves in B-form DNA, which are about 34 Å apart (60). This explained the observed inability of TetR to bind the tet operator and the subsequent induction of tetA expression that occurs in response to tetracycline. The recent publication of the crystal structure for a TetR(D)-DNA complex confirmed this, revealing that α3 and α3′ are separated by 36.6 Å when bound to a 15-bp tet operator fragment (−7 to +7 positions of O1; Fig. 7A), compared to the TetR-tetracycline-Mg2+ complex, in which the gap between α3 and α3′ had increased by 3 Å (153).
Each of the HTH motifs in the TetR(D)-DNA complex were found to be bound to the corresponding major groove of operator DNA, whereas no contacts were made with the minor groove (Fig. 8) (153). Except for the central 3 bp, TetR contacted all 15 bp of the operator fragment, either via a series of hydrogen bonds to backbone phosphate groups and the bases themselves or by a lesser number of van der Waals interactions with the bases (Fig. 7B) (153). This proved to be in excellent agreement with data provided by the saturation mutagenesis of one tet operator half-site, which had previously indicated the importance of positions +2 to +7, with bp +1, +8, and +9 making lesser contributions to TetR binding and the central (position 0) base pair being required only for correct spacing of the two half-sites (Fig. 7B) (236).
Although the recognition helix of the TetR HTH is unusually short, the TetR(D)-DNA structure showed that TetR compensates by making a large number of contacts with the DNA (Fig. 7B), involving all recognition helix residues except Leu41, which forms part of the hydrophobic core stabilizing the three-helix bundle (153). Other TetR residues that form DNA contacts are located in the first helix of the HTH, the turn of the HTH, and both of the interhelical loops connecting the HTH to the adjacent helices (Fig. 7B) (153). Many of the TetR residues shown to contact DNA in this structure had been identified previously through mutagenesis (10, 53, 235).
Unlike many protein-DNA interactions, the TetR-DNA interface contains no water molecules, reflecting a high degree of structural complementarity between the DNA-binding domains and the tet operator (153). Contributing to this close fit is Pro39 in the recognition helix of the TetR HTH, a somewhat unusual feature, as proline residues occur infrequently in the α-helices of globular proteins because of their propensity to be helix breakers (229). Pro39 also plays an important role in TetR DNA recognition, as it contributes to the ability of TetR to make contacts to both sides of the tet operator strand by forming van der Waals interactions with each of the nucleotides at the +4 position and a single nucleotide at +5 bp (Fig. 7B) (153). This enables TetR to contact backbone phosphates and nucleotides on one side of the major groove (+2 and +4) before transferring via Pro39 to contact points on the other side of the groove (+4 to +7) (Fig. 7B), an arrangement previously predicted on the basis of methylation protection experiments that utilized mutations in both the tet operator and TetR (56).
TetR-Ligand Interactions
The results of a large number of biochemical, structural, and mutagenesis studies on TetR(B) and TetR(D) are in good agreement, which provides an exceptionally detailed picture of the induction process (55, 60, 135, 193, 194, 205). In addition to identifying amino acids involved in the induction process, random mutagenesis has even produced a TetR mutant that exhibits increased, not decreased, affinity for the tet operator upon inducer binding, which, in combination with the relative nontoxicity of tetracycline to eukaryotic cells, has facilitated its use to tightly regulate gene expression in eukaryotes (16, 44, 62).
The two tetracycline-Mg2+ complexes bound by a TetR homodimer were found to be located in binding tunnels buried in the core of the protein (60). Each of the two binding tunnels accommodates a single tetracycline-Mg2+ complex and is formed by helices contributed by both polypeptide chains, the first by α helices 4 to 8 and 8′ and 9′ and the second by 4′ to 8′ and 8 and 9 (Fig. 8). Although the structure of inducer-bound TetR revealed the specific interactions involved in tetracycline-Mg2+ binding, it failed to identify which of the two similar-sized openings in each tunnel had been used as the entry point to the complex by tetracycline-Mg2+ (60). Solving the structure for tetracycline-free TetR brought to light that the opening adjacent to the C terminus of α9′ (and α9 for the second tunnel) was twice as large as the opening C-terminal to α4 in the inducer-free TetR dimer (151).
Molecular modeling of the protein surface confirmed the opening next to the C terminus of α9 as the site of tetracycline-Mg2+ entry, indicating that the inducer would have a strong preference for attraction to the area surrounding the α9′ and α9 entrances (151). Because of the constricted nature of the binding tunnel, a model for induction could then be proposed in which the tetracycline A ring of the tetracycline-Mg2+ complex (Fig. 9A) is forced to enter through the α9′ opening end-on (151, 152). The subsequent events triggered by the entry of tetracycline-Mg2+ into the binding site are listed below.
FIG. 9.
(A) Diagrammatic illustration of specific interactions between TetR(D) binding-tunnel residues and a tetracycline-Mg2+ complex. Tetracycline is colored orange, with its four rings labeled A to D and the methyl groups shown as Me. The three water molecules coordinated by the Mg2+ atom (red) are represented by W1 to W3 (blue). Residues from the TetR dimer that contribute to the binding of this tetracycline-Mg2+ complex are shown in italics for those from the first polypeptide and in bold type for those from the second polypeptide. The α-helices in which these amino acids are located are also indicated except for the Thr103, Arg104, and Pro105 residues, which form part of the interhelical loop connecting α6 to α7. Thin dotted lines indicate hydrogen bonds, and thick broken lines show hydrophobic interactions. Equivalent residues from the other binding tunnel in a TetR dimer contact a second tetracycline-Mg2+ complex; see text for other details. (B) TetR conformational changes that occur upon binding a tetracycline-Mg2+ complex. The α1 to α8 helices of one monomer and the α9′ helix from the second polypeptide in a dimer are represented in blue for DNA-bound TetR and yellow for the induced tetracycline-Mg2+-bound form. The DNA phosphate-ribose backbone is shown in red, bases in grey, tetracycline (Tc) in orange, the Mg2+ atom in red, and the chain of water molecules that constitute the water zipper in the induced conformation as blue spheres. The pendulum-like motion of α4 upon tetracycline-Mg2+ binding leads to significant displacement of the attached α1-α3 DNA-reading head, so that the α3 recognition helix can no longer contact the major groove of tet operator DNA at the same time as the α3′ recognition helix from the second polypeptide in a TetR dimer. The location of the entrance to the TetR binding tunnel is also indicated, whereas the sliding door motion of α9′ that closes this entrance in the induced form is also apparent. See text for other details. (Panel A reprinted with permission from reference 60; ©1994, American Association for the Advancement of Science. Panel B reprinted with permission from reference 153; kindly provided by Winfried Hinrichs.)
(i) Residues located at the base of the binding site, His64 (α4), Asn82 (α5), and Gln116 (α7), form tight hydrogen bonds to the side chains of the A ring of tetracycline in addition to a hydrophobic contact made by Phe86 (α5) (Fig. 9A). Water molecules displaced by the entry of the inducer are proposed to be released through the tunnel opening C-terminal to α4. (ii) The imidazole group of His100 (α6) binds to Mg2+, which triggers the formation of a hydrogen bond between Thr103 and W3, one of the three Mg2+-coordinated water molecules (Fig. 9A). The resulting 2.5-Å induced movement of Thr103 produces a partial unwinding of α6, which loses its last two residues, Leu101 and Gly102, because of their inclusion in the formation of a type II β-turn with residues His100 and Thr103 (Fig. 9B). Both Gly102 and Thr103 have now undergone a significant displacement from their original positions, an event that triggers the conformational changes associated with induction (Fig. 9B). (iii) Formation of the new β-turn displaces all residues of the adjacent interhelical loop, resulting in reorientation of Arg104 and Pro105 so that they can form part of the hydrophobic pocket surrounding tetracycline ring D (Fig. 9A). Val113, Leu131, Iso134, Leu170′, Leu174′, and Met177′ complete the region of nonpolar van der Waals contacts in this area which serve to guide the tetracycline-Mg2+ complex into its final position (Fig. 9A). Movement of these residues to accommodate tetracycline induces motions in α9′ and the loop connecting α6 and α7, which partially closes the binding tunnel entrance. (iv) By rotating 90°, the carboxylate group of Glu147′ (α8′) can hydrogen bond to Gly102 of the new β-turn and also to the remaining two Mg2+-coordinated water molecules, W1 and W2 (Fig. 9A). (v) Formation of a salt bridge between Asp178′ (α9′) and Arg104 draws α9′ closer and completes the “sliding door” movement of α9′ residues, closing the entrance to the binding pocket. Determination of the interspin distances between nitroxide groups attached to residues near the tetracycline-binding tunnel confirmed the relatively large movement of α9′ that occurs during induction (224).
Conformational Changes in TetR That Transmit the Induction Signal
In the uninduced forms of TetR, the C-terminal residues of α6 form a hydrophobic contact region with the central part of α4, the helix connecting the core of the protein to the DNA-reading head (151, 153). Upon tetracycline-Mg2+ binding, the formation of the new β-turn at the C terminus of α6 creates space at the contact surface with α4, which moves to fill it (Fig. 9B). Because the C terminus of α4 is anchored by the hydrogen bonding of His64 to tetracycline ring A (Fig. 9A and B), the movement of α4 is similar to that of a hinge, serving to reposition the attached α1 to α3 helices in a pendulum-like motion so that they adopt the non-DNA-binding configuration (Fig. 9B). The reorientation of the interhelical turn connecting α6 to α7 permits the formation of a chain of water-mediated hydrogen-bonds (water zipper) that forms along the length of α4 (Fig. 9B), which assists in locking the protein into a non-DNA-binding conformation by linking the DNA-binding domain to the tetracycline-Mg2+ binding site (151).
Of critical importance to the induction process are the hydrophilic interactions which coordinate the Mg2+ atom. Their importance has been validated by solving the structures of TetR-tetracycline-Mg2+ complexes in which Mg2+ has been partially or completely removed by the addition of a divalent metal cation-chelating agent (152). When only tetracycline occupied the binding tunnel, the structure of TetR was almost unchanged from that of inducer-free TetR, confirming the essential role of Mg2+ in initiation of the conformational reorganization associated with induction (152).
The reduced freedom imposed by the interaction of residues from both polypeptides with each tetracycline-Mg2+ complex combined with the hydrogen bond interactions formed by the water zipper serve to freeze TetR into its inducer-bound conformation. It has been suggested that the transition from a free to frozen state has a high entropic potential, which may be partially compensated for by increased motion in some parts of the protein (228). For example, the flexibility of the loop connecting α8′ to α9′ has been shown to increase upon ligand binding, contributing to an overall smaller change in entropy (228). Although this loop is highly variable between the different TetR classes both in length and in composition, deletion of this region produced mutants deficient in tetracycline binding and induction, consistent with a proposed role in the closure of the α9′ sliding door (19). Replacement of the loop with various numbers of alanine residues indicated that the loop has to be a minimum length to permit efficient inducer binding and the associated conformational changes (193). Engineered TetR derivatives that contain cysteine residues in mobile regions of the protein have also been used to confirm, by the formation of ligand-dependent disulfide bonds, that conformational changes in TetR upon tetracycline-Mg2+ binding occur both in vitro (222) and in vivo (223).
A new group of tetracyclines, the glycylcyclines, also demonstrate the highly specific nature of the TetR-ligand interaction. In addition to not being recognized by the TetA transporter, these tetracycline derivatives also cause reduced induction of its synthesis. The glycylcyclines contain a glycyl-amido substituent that causes steric hindrance with the TetR sliding door helix, α9′, thereby preventing the ligand from reaching the binding position within TetR that is necessary to trigger the conformational changes that are required for induction of tetA transcription (154).
QacR, REGULATOR OF S. AUREUS qacA/B MDR GENES
The nearly identical S. aureus qacA and qacB MDR genes encode the first bacterial MDR transporters to be described (218), their discovery closely following that of mammalian P-glycoprotein. Investigation of the S. aureus QacA and QacB MFS pumps has been stimulated by the prevalence of qacA and qacB genes on multiresistance plasmids, such as pSK1, which are commonly isolated from clinical strains of this important human pathogen (88, 160, 219, 220). QacA utilizes the proton motive force to drive the efflux of more than 30 different toxic monovalent or bivalent, cationic, lipophilic compounds which belong to 12 distinct chemical classes (102, 131, 132, 159). Like the majority of compounds exported by MDR pumps, the QacA substrates make ideal antimicrobials because their positive charge attracts them towards the interior of bacterial cells, which are typically negatively charged compared to the exterior environment, while the lipophilic nature of these molecules aids their passage through cellular membranes. In fact, many of the QacA substrates have either a current clinical application, such as the quaternary ammonium antiseptics benzalkonium and cetrimide (199), the antibacterial diamidine compound pentamidine (127), and the biguanidine antiseptic chlorhexidine (41, 65), or they have been used historically as antiseptics, e.g., the dye proflavine.
The qacR gene, which is divergently transcribed from all known qacA/B determinants, encodes a trans-acting transcriptional repressor of the qacA/B genes (47) (Fig. 10A). QacR contains a poorly defined N-terminal HTH DNA-binding motif that places it in the TetR family of regulatory proteins (Table 1) (183). The QacR DNA-binding site, IR1, is a relatively large operator sequence immediately upstream from the qacA promoter (PqacA), consisting of 15 bp in each half-site, separated by a 6-bp spacer sequence. In the absence of QacA substrates, QacR binds IR1 and downregulates qacA transcription. However, the transcription of qacA has been demonstrated to increase in a qacR-dependent manner in response to a structurally diverse range of monovalent and bivalent compounds that represent almost all of the substrate classes exported by the QacA pump (Fig. 10A and B) (47; S. Grkovic, M. H. Brown and R. A. Skurray, unpublished data). A direct interaction of QacR with these inducing compounds was subsequently confirmed by the binding of QacR to IR1 operator DNA that had been disrupted in vitro by the separate addition of eight structurally diverse QacA substrates, including benzalkonium, dequalinium, ethidium bromide, chlorhexidine, and rhodamine 6G (47) Thus, like TetR, QacR acts as a sensor molecule that can facilitate increases in transcription in response to the presence of toxic compounds because the ligand-bound form of this protein is incapable of binding operator DNA (Fig. 10A).
FIG. 10.
(A) Model for regulation of expression of S. aureus qacA MDR gene. QacR (black ovals) represses (−) transcription from the qacA promoter, PqacA, by binding as one dimer per IR1 half-site. Many of the lipophilic cationic drugs (D) exported from the cell by QacA in exchange for protons (H+) are also ligands of QacR. Conformational changes that occur in a QacR dimer upon drug binding result in the ligand-bound form of this protein being incapable of binding IR1, thereby mediating increases in qacA transcription in response to the presence of transporter substrates. The locations of the ribosome-binding site (RBS) and transcription start point (right-angle arrows) are indicated for both the qacA and qacR genes. Preliminary results indicate that an unknown regulatory protein (?) indirectly influences qacA expression by binding IR2, which overlaps the qacR promoter, PqacR. (B) Structurally diverse compounds that are both substrates of the QacA MDR pump and ligands of the QacR regulator. The chemical structures of representative compounds from four distinct chemical families are depicted. Dequalinium and benzalkonium (where R represents a mixture of alkyls, either C12H25, C14H29, or C16H33) are bivalent and monovalent quaternary ammonium compounds, respectively; rhodamine 6G, ethidium bromide, proflavine, and crystal violet are monovalent dyes; chlorhexidine is a bivalent guanidine; and berberine is a monovalent plant alkaloid.
Interestingly, analysis of PqacA-reporter gene fusions (47) indicated that QacR does not repress transcription from PqacA to anywhere near the same extent that the related regulator TetR represses transcription from the tetA promoter (59). Thus, besides inducing expression in response to substrates of the QacA pump, the basal level of qacA expression that is afforded by QacR can mediate protection against compounds which are substrates of the MDR pump but are not strong ligands of the regulatory protein. The demonstration that a simple 2-bp change in the central 6-bp spacer region of IR1 could produce an operator exhibiting greater affinity for QacR (48) supports the proposal that the QacR/IR1 system evolved to provide a substantial basal level of qacA transcription. In comparison, the highly specific nature of the TetA-TetR system does not require any significant level of tetA expression in the absence of tetracycline.
Gene fusions have also permitted demonstration that expression of qacA is induced in response to the plant alkaloid berberine (S. Grkovic, M. H. Brown, and R. A. Skurray, unpublished data), which is an amphipathic, positively charged compound that shows some structural similarities with other QacR ligands (Fig. 10B). In contrast, all previously identified ligands of QacR had been synthetic substances that have been developed only recently. Additional evidence that berberine represents a natural MDR substrate is provided by the demonstration that this plant alkaloid is a substrate of both the plasmid-encoded QacA and the chromosomally encoded NorA S. aureus MDR pumps (63, 94, 95).
Taken together, these results are highly suggestive of a preexisting role for the QacA-QacR system in providing resistance to plant- and other naturally derived, cationic and hydrophobic antimicrobial compounds. Therefore, in order to combat antiseptics and disinfectants prevalent in the hospital environment, S. aureus appears to have recruited a system that was already well adapted for the export of such compounds. The continuing presence of the qacA-qacR locus on multiresistance plasmids associated with clinical S. aureus isolates (88; L. Worton, R. A. Skurray, and N. Firth, unpublished data) provides strong circumstantial evidence that the current function of this system in these strains is now predominantly the export of synthetic antimicrobial compounds. Also supporting this hypothesis is the increased substrate range of QacA compared to QacB (132, 159), which, in combination with the finding that QacB is encoded by the earliest known qacA- or qacB-carrying plasmid (160), suggests that qacA has more recently evolved from qacB in response to antimicrobial agents encountered in the hospital environment.
Although IR2, an inverted repeat that partially overlaps the qacR promoter (Fig. 10A, PqacR), is very similar in sequence to the tet operators (Fig. 7A), overexpression of QacR or TetR in trans had no effect on transcription from PqacR/IR2 in vivo (S. Grkovic, M. H. Brown, and R. A. Skurray, unpublished data), and purified QacR also failed to bind IR2-containing DNA in vitro (47). The failure to define a role for IR2 in the autoregulation of qacR expression is at odds with the results with other divergently transcribed members of the TetR family, which in general autoregulate the expression of their own genes (47). However, mutagenesis of IR2 indicated that this regulatory sequence has an alternative role that involves an as yet undefined chromosomally encoded protein (Fig. 10A) (S. Grkovic, M. H. Brown, and R. A. Skurray, unpublished data).
QacR DNA Binding
QacR, like other TetR family members, self-assembles into dimers. However, QacR-IR1 complexes were found to be equivalent in size to four QacR molecules bound to each IR1-DNA sequence even though IR1 contains only a single palindromic sequence (48). Furthermore, upon addition of QacR ligands, the four DNA-bound protein molecules were observed to dissociate from IR1 DNA as dimers (48). This suggested that QacR does not self-assemble into tetramers, in addition to a pair of QacR dimers appearing to bind IR1 operator DNA without the requirement for any direct contacts between the two DNA-bound dimers (48).
The recent determination of the crystal structure for a QacR-IR1 complex confirmed and extended these findings, permitting the details of an unexpected and highly unusual mode of DNA binding to be elucidated (197). A pair of QacR dimers were found to be bound to opposite sides of a 28-bp DNA fragment so that successive major grooves could be contacted by two HTH motifs, one from each dimer (Fig. 11) (197). A four-helix-bundle dimerization domain was created by the two C-terminal QacR α-helices from each of the subunits in a dimer (Fig. 11; α8 and α9 from the first subunit, and α8′ and α9′ from the second subunit). Other than the three-helix bundle that forms the DNA-binding domain of each monomer (Fig. 11, α1 to α3), the four-helix bundle represented the only region of QacR that exhibited significant structural homology to TetR (197).
FIG. 11.
Structure of the complex formed by a pair of operator-bound QacR dimers. The two QacR dimers bound to a symmetrical version of IR1 operator DNA are depicted as ribbons, whereas the DNA is shown with the phosphate, oxygen, carbon, and nitrogen atoms colored yellow, red, grey, and blue, respectively. The DNA-reading heads from the distal subunit (purple) of dimer 2 and the proximal subunit (green) of dimer 1 contact the first major groove, whereas the second major groove of IR1 DNA is contacted by the DNA-binding domains from the other two subunits, one from each dimer. The individual α-helices (α1 to α9) of the distal subunit from dimer 2 are labeled, as are the N and C termini of that polypeptide. Additionally, the α8′ and α9′ helices from the proximal subunit of dimer 2, which form the four-helix-bundle dimerization domain with α8 and α9, are also indicated. For the distal (purple) polypeptide of dimer 2, an arrow points to the yellow region at the N terminus of α5 that undergoes a coil-to-helix transition upon ligand binding. (Reprinted with permission from reference 197; kindly provided by Maria Schumacher; ©2002, Oxford University Press.)
The combined contacts made by a pair of QacR dimers bound to IR1 DNA were in good agreement with the extended DNase I footprint observed for QacR-IR1 complexes (Fig. 12A) (47). One monomer of a QacR dimer (Fig. 11, green polypeptide of dimer 1) was found to make a large number of base and phosphate contacts to seven out of the eight positions immediately adjacent to the IR1 operator axis of symmetry (Fig. 12A, −1 to −8 bp) (197). This monomer has been designated the proximal (to the center of IR1) subunit to distinguish it from the second, distal dimer 1 subunit (Fig. 11, magenta polypeptide), which binds at a position more remote from the axis of symmetry. The contacts made by the proximal subunit of dimer 1 to 3 bp from the 6-bp IR1 spacer region (Fig. 12B, −1 to −3 bp) predominantly involved the phosphate backbone, which is consistent with the ability of QacR to bind to IR1 sequences in which all 6 of the central 6 bp have been altered (48). Although the base-specific contact made by the proximal subunit to position −3 involved a thymine, the proximal subunit of the second dimer would instead contact an adenine in the corresponding position of the wild-type IR1 sequence (+3 bp; Fig. 12A).
FIG. 12.
DNA contacts formed by QacR to an IR1 half-site. (A) Sequence of the qacA −10 promoter region and the downstream IR1 operator, with the bases that QacR protects from DNase I digestion highlighted (white) against a black background, the qacA transcription start point (tsp) circled, and the location of IR1 indicated by bold arrows that flank the central 6-bp IR1 spacer region (47). (B) The DNA contacts made by a pair of operator-bound QacR dimers to a single IR1 half-site. The bp −1 to −14, which constitute the depicted operator half-site, are shown as a mirror image of A. The QacR residues in bold that contact the DNA in this half-site are from the distal subunit of dimer 2, whereas those in italics are from the proximal subunit of dimer 1 (Fig. 11, purple and green polypeptides, respectively). The contacts made to the bases and DNA backbone in the operator half-site by these amino acids are shown as thin dashed lines for hydrogen bonds and thick broken lines for van der Waals interactions. Also indicated is a DNA contact made by the N terminus of α1. (Panel B reprinted with permission from reference 197.)
The contacts made by the distal subunit from dimer 1 (Fig. 11, magenta polypeptide) involved bases and the phosphate backbone in seven of the positions from +4 to +12 of IR1, i.e., in the half-site opposite that to which the proximal subunit of this dimer binds (Fig. 12) (197). Since the DNA-reading heads from the second QacR dimer have essentially the same interactions with IR1 DNA as the first dimer (197), the contacts illustrated in Fig. 12B for an IR1 half-site depict those formed by the proximal (green) subunit from dimer 1 and the distal (purple) subunit from QacR dimer 2 (Fig. 11). Of note is that the two HTH motifs within each QacR dimer make differential contacts to DNA sequences that lack any obvious twofold symmetry (Fig. 12), a highly unusual interaction for the combination of an inverted repeat sequence and a bacterial regulatory protein that binds to DNA in a dimeric form. However, closer inspection of the distal and proximal subunits illustrated in Fig. 12B, which are from different dimers, reveals that the four base-specific contacts made by each of their DNA-reading heads are very similar and hence constitute a pseudo-direct repeat (Fig. 12B). Thus, the DNA sequence that is bound by the two QacR polypeptides within an individual dimer contains a pseudo-inverted repeat (197).
The QacR-DNA structure indicated that the observed cooperative effect in the binding of a pair of dimers to IR1, which occurs in the absence of any direct dimer-dimer contacts (48), was the results of the successive major grooves of IR1 DNA being contacted by one HTH from each of the two DNA-bound QacR dimers (Fig. 11) (197). It was proposed that the binding of the first dimer requires an induced fit between the protein and IR1 DNA, which produces a significant increase in the width of the major groove and a corresponding undertwisting in QacR-bound DNA, thereby creating the correct conformation for binding of the second dimer (197). The flexibility of the α4 helix, which links the DNA-binding domain to the inducer-binding domain (Fig. 11), has also been suggested to play a role in DNA-binding cooperativity (197). Mutagenesis of the 6-bp IR1 spacer sequence (Fig. 12A) indicated that the proximal subunits of QacR dimers, which make contacts to this region, appear to be highly flexible in accommodating changes to the base composition of these central 6 bp (48). In contrast, the QacR-DNA structure clearly demonstrated that the failure of QacR to bind altered IR1 sequences in which the size of the 6-bp spacer region had been increased or decreased by 2 bp (48) would be predominantly because the change in the spacing of the two IR1 half-sites significantly affected the ability of the four QacR DNA-reading heads to contact successive major groves.
Similar to TetR (59), the abnormally short recognition helix of QacR (α3; residues 36 to 42) is partially compensated for by the large number of residues from this helix that make contacts with operator DNA (Fig. 12B). In fact, the only QacR α3 residue that does not interact with DNA is Leu39, similar to Leu41 of TetR α3. However, whereas TetR employs residues from outside its recognition helix to make contacts to both bases and phosphates (Fig. 7B), the residues that QacR employs from outside α3, which includes the N terminus of α1, contact only phosphates (Fig. 12B). Therefore, in order to ensure that adequate base-specific contacts are made, QacR appears to additionally compensate for its short recognition helix by binding IR1 operator DNA cooperatively as a pair of dimers.
A critical aspect of the interactions between QacR and its operator appears to be the very small side chain of Gly37, which was proposed to facilitate the tight docking of the recognition helix to the DNA, provide room for the second QacR monomer to bind, and also permit the interactions of the Gly37 residues from the various subunits with the conserved guanine at positions −4, −8, +4, and +8 of IR1 (Fig. 12). The contact made to the −8 guanine of IR1 may have extra significance for the repression mechanism, as this base represents the transcription start point of the qacA gene (Fig. 12) (47). The importance of Gly37 and a number of other QacR HTH residues to DNA binding has been recently confirmed by mutagenesis (S. Grkovic, M. H. Brown, and R. A. Skurray, unpublished data).
Versatile QacR Multidrug-Binding Pocket
Structural determinations of QacR-ligand complexes, combined with the data available on ligand binding by BmrR, have produced major advances in our understanding of the molecular basis of multidrug recognition. In the case of QacR, each dimer was found to possess two drug-binding pockets, the first being formed by α4 to α8 from one subunit and α8′ from the other subunit, with the second ligand-binding site involving the reciprocal helices from each of these polypeptides. Although the two drug-binding tunnels possessed by a TetR dimer are also largely formed by the equivalent α-helices, the binding sites of these two related proteins show no significant similarities (198).
QacR-ligand structures have been obtained for a number of different ligands, including rhodamine 6G, ethidium bromide, dequalinium, crystal violet, and berberine. Whereas the monovalent compounds rhodamine 6G (Fig. 13A) and ethidium bromide (Fig. 13B) were found to bind to distinct but overlapping portions of an extended QacR ligand-binding pocket, the flexible linker carbon atoms of the bivalent compound dequalinium enabled the ring systems of this compound to be bound in both of these regions (Fig. 13C) (198). Four negatively charged glutamate residues lining the QacR ligand-binding pocket were observed to be available for forming electrostatic interactions with drugs, viz., Glu90 for neutralization of the positive charge of rhodamine 6G and Glu120 likewise for ethidium bromide, whereas one of the positive charges of dequalinium interacted with Glu57 and Glu58, while the second was neutralized by Glu120 (Fig. 13A to C). The single delocalized positive charge of crystal violet was also found to interact with two glutamates, Glu90 and Glu120 (Fig. 13D) (198). Additionally, the crystal violet structure demonstrated the versatility of the extended QacR ligand-binding pocket, as this less planer compound was bound at an intermediate position between the rhodamine 6G and ethidium bromide binding sites (Fig. 13D). The plant alkaloid berberine was essentially bound in the same manner and to the same portion of the extended QacR binding-pocket as rhodamine 6G (198).
FIG. 13.
(A to D) Binding of structurally diverse compounds in the extended QacR ligand-binding pocket. The key QacR residues and relevant α-helices in the ligand-binding site that interact with the compounds rhodamine 6G (R6G, A), ethidium bromide (Eb, B), dequalinium (Dc, C), and crystal violet (Cv, D) are shown in cyan, whereas the QacR glutamate residue(s) involved in electrostatic interactions with the positive charge(s) of each bound drug is colored red. The carbon, nitrogen, and oxygen atoms of each ligand are colored grey, blue, and red, respectively. (E) The QacR DNA-bound conformation (yellow) has been superimposed on the conformation of the QacR subunit to which a ligand is bound (blue). This illustrates the coil-to-helix transition that extends the N terminus of α5 by a turn and the concomitant shoving effect (blue arrow) of α6 against the DNA-binding domain, which produces a dramatic alteration in the position of this three-helix bundle. The location of rhodamine 6G (red) in the drug-bound structure is also indicated, as are α4 to α9, the HTH recognition helix (Hr), and the N and C termini of the protein. (Reprinted with permission from reference 198; kindly provided by Maria Schumacher.)
Also lining the QacR drug-binding pocket are a large number of aromatic and hydrophobic residues which, similar to the glutamates, present incoming drugs with a broad range of possibilities for the formation of hydrophobic interactions or the stacking of phenyl rings (Fig. 13A to D) (198). In addition, a number of polar residues, such as asparagine, glutamine, serine, and threonine, were observed to be available for interacting with various ligands as hydrogen bond donors or acceptors (Fig. 13A to D) (198). Therefore, the binding of structurally diverse ligands by QacR is facilitated by an incoming drug molecule being presented with a diverse array of potential electrostatic and hydrophobic interactions as well as many possibilities for the stacking of phenyl rings and the formation of hydrogen bonds, so that each ligand can form its own unique subset of interactions to maximize its binding.
A comparison of the ligand-binding domains of the QacR repressor and BmrR activator proteins reveals that although that of QacR is completely helical in nature (198), whereas BmrR is predominantly β-sheet (245), the two proteins have notable similarities, including the aromatic, hydrophobic, and negatively charged residues that line their binding pockets as well as the crucial role played by tyrosines (see below). One important difference between the two proteins is the observation that BmrR does not form any hydrogen bonds to the ligand TPP (Fig. 6). Although it is not known whether BmrR forms hydrogen bonds to ligands other than TPP, it has been suggested that by not employing distance- and orientation-dependent hydrogen bonds to contact ligands, multidrug-binding pockets can enhance their substrate range (115, 140). However, the available QacR-ligand structures clearly demonstrate that an array of residues which are capable of being hydrogen bond donors or acceptors can contribute substantially to multidrug-binding capabilities (198). These variations in the ligand-binding mechanisms used by individual multidrug-binding proteins may reflect the ancestry of each protein, e.g., QacR has a common ancestry with TetR, a protein that employs an extensive network of hydrogen bonds to form highly substrate-specific interactions with tetracycline-Mg2+ complexes (Fig. 9).
The proposal that multidrug transporters and their regulators originally evolved to be a general removal system for hydrophobic compounds may explain the broad ligand range of QacR (140). It has been suggested that because there are only a very few essential intercellular hydrophobic molecules, multidrug-binding proteins do not need to be very specific, as they only need to distinguish between hydrophilic compounds and the vast majority of hydrophobic ones (140). The structural data on the nature of the extended QacR binding pocket, which indicate that this protein is extremely well adapted for binding to a broad range of hydrophobic compounds, is in good agreement with this hypothesis. In contrast, transport machinery involved in the export of hydrophilic molecules requires highly specific interactions to prevent the undesirable efflux of structurally related compounds that are important cellular metabolites.
QacR Induction Mechanism
An intriguing feature of the ligand-binding process observed for QacR was a coil-to-helix transition, which, upon drug binding, resulted in extension of the C terminus of α5 by 4 residues (Fig. 11 and 13E) (198). This coil-to-helix transition was found to eject Tyr92 and Tyr93 from the hydrophobic core of QacR, thereby increasing the volume of the binding pocket available for drug binding; Glu90, previously positioned external to the ligand-binding pocket, was also now repositioned so that it could assist with drug binding (Fig. 13A to D). In the drug-free conformation, Tyr92 and Tyr93 were observed to act as drug “surrogates,” ensuring that the hydrophobic core of QacR was stabilized in the absence of ligands. This situation has many similarities to that of BmrR, in which a flexible α-helix was found to permit the displacement of a stabilizing tyrosine, Tyr152, from the core of the protein upon drug binding.
In the case of QacR, the coil-to-helix transformation also served to switch the dimers into a non-DNA-binding conformation, thereby ensuring induction of qacA transcription. The mechanism by which this occurs involves the lengthening of the α5 C terminus, forcing the attached α6 helix downward (Fig. 13E). Since α6 is anchored to residues 12 to 23 of the DNA-binding domain, the translocation of this helix produces some dramatic changes in the orientation of the DNA-binding domains of both monomers (198). The result is an increase in the HTH center-to-center distance from 37 Å in the DNA-bound form of a QacR dimer to 48 Å in the ligand-bound form (Fig. 13E). Such a drastic change clearly prevents the ligand-bound form of QacR from binding to the successive major grooves of either B-form DNA, which are 34 Å apart, or QacR-bound DNA, which are 37 Å apart (197). This direct “shoving” mechanism observed for QacR is in marked contrast to the induction process of TetR, in which a small increase in the distance between the DNA-reading heads produced by changes transmitted through several helices is locked in place by the formation of a chain of water-mediated hydrogen bonds (Fig. 9B). The QacR induction mechanism appears to be a much more robust process, which may have granted the remainder of the QacR ligand-binding pocket the freedom to maximize the potential for interactions with structurally diverse ligands without compromising the mechanism or the efficiency of the induction switch.
In contrast to TetR, in which a tetracycline-Mg2+ complex is bound in each of the two drug-binding tunnels formed by a TetR dimer (60), the observed QacR-ligand binding stoichiometry was 2:1 despite the presence of two binding pockets in a QacR dimer (198). Closer inspection of QacR-ligand complexes revealed that the coil-to-helix transition that occurs upon ligand binding shifted the position of the adjacent C terminus of that subunit, such that it now blocked the entrance to the second ligand-binding pocket within the same dimer (198). This may give QacR an advantage over BmrR, which binds two drug molecules per dimer (Fig. 5) (245), as the ability of a single ligand bound to a QacR dimer to initiate the induction process may enhance the sensitivity of QacR to drugs, perhaps aiding the induction of expression in response to suboptimally bound ligands. The requirement for two unliganded QacR dimers to bind operator DNA before repression can be established could also have a role in enhancing the sensitivity of this regulator to the presence of inducing compounds.
OVERVIEW
A common theme in the regulation of membrane-bound drug transporter proteins is the need to prevent excessive expression, which is reflected by the fact that these proteins are notoriously difficult to overproduce for purification purposes (231). While it appears that some of the systems that are capable of exporting antimicrobial compounds do not need to be regulated because of a naturally low level of expression, the production of most is subject to some form of transcriptional and/or translational regulation, presumably as a safeguard against the deleterious effects of transporter overproduction. This situation is best exemplified by a number of transporter genes, which, although they are under the control of one or more inducible regulatory proteins, possess additional translational or transcriptional controls to provide added insurance that the fully induced level of expression cannot exceed a certain threshold (59, 72, 107). However, it is equally important to the cell that these transport proteins be available when required to perform their physiological roles. Thus, the expression of many drug transport proteins appears to be inducible, so that, in some instances, upregulation in response to the presence of natural or synthetic antimicrobial molecules has been demonstrated, although the expression of many other proteins is likely to be linked to presently unidentified physiological roles; for example, it has been proposed that the MDR pumps from E. coli and P. aeruginosa are involved in the export of quorum-sensing signals (83, 174).
Perhaps the most striking feature of bacterial drug pump regulation is the incredible diversity in the mechanisms that are employed to control their synthesis: each pump appears to be regulated in its own unique way. Additionally, for the MDR exporters, the majority of these determinants appear to be subject to multiple levels of control, possibly reflecting employment of their diverse substrate capabilities for multiple cellular functions. This is in marked contrast to the transcriptional regulation of the dedicated tetracycline transporters, which need to respond to only one substrate. As it was for TetR, when induction in response to antimicrobials has been clearly demonstrated for an MDR system, the common theme of requiring a protein that is capable of directly or indirectly sensing the presence of pump substrates has also been demonstrated. In comparison, the interaction of tetracycline with the ribosomal machinery involved in translation has provided an alternative method by which translational controls mediate the induction of some tetracycline-specific pumps, particularly in gram-positive organisms.
An important aspect of research into the regulation of bacterial antimicrobial agent export systems has been elucidation of the ligand-binding mechanisms that are employed by some regulatory proteins to sense the presence of pump substrates. Structural analyses of the tetracycline-binding TetR repressor and the QacR and BmrR multidrug-binding regulators have been very successful in providing answers to questions about the mechanisms of protein-drug interactions that have not been forthcoming from the investigation of transport proteins. Of particular interest has been the demonstration of the principles of multidrug binding for the versatile QacR ligand-binding pocket; an extended ligand-binding site that possesses multiple but linked drug-binding pockets (198). These principles are also likely to apply to the MDR transporters themselves, as results from the mutagenesis of specific transporter residues, in addition to biochemical data, suggest that the MDR transporters human P-glycoprotein (214), Saccharomyces cerevisiae Pdr5p (85), Lactococcus lactis LmrP (172, 226), E. coli MdfA (93), and S. aureus QacA (132) are all likely to possess extended substrate-binding sites that are similar in nature to that of QacR.
The detailed pictures of protein-drug interactions for the multidrug-binding regulators as well as the tetracycline-specific system will have important applications in aiding the rational design of new antimicrobial compounds and the development of transporter inhibitors. The latter approach has received considerable attention, as the successful use of compounds that inhibit the function of drug efflux pumps may renew the usefulness of currently ineffective antibiotics (94).
Regardless of whether the primary function of a drug transport system is antimicrobial efflux or the export of such compounds merely occurs fortuitously, these proteins have been recruited by microbial pathogens in a highly successful effort to circumvent the relatively recent widespread use of antimicrobial compounds as therapeutic, prophylactic, and veterinary agents. In particular, the extensive use of antimicrobial compounds in modern hospitals has placed infectious bacteria under immense selective pressures. Investigations into the regulatory pathways controlling the expression of transport proteins capable of drug efflux have provided a fascinating picture of one way in which bacteria have responded to these selective pressures.
The various bacterial drug transport systems can be viewed as being at different stages along the path towards becoming dedicated systems for the provision of resistance to clinically relevant antimicrobial compounds. At one end of the spectrum, there are the acutely sensitive gram-negative TetA/TetR resistance determinants which have evolved specifically to provide the efficient removal of tetracycline. In contrast, at the other end of the scale are many MDR transporters whose broad substrate ranges are being increasingly exploited by microorganisms to cope with the elevated use of antimicrobial compounds. Although some of these MDR transporters are proposed to have had preexisting roles in providing protection against low levels of toxic compounds, many of their regulatory networks instead appear to be generally geared towards other physiological functions, such as the export of specific metabolites. Thus, mutations that disrupt the normal function of local and global regulatory networks, leading to the overexpression of chromosomally encoded MDR transporters in P. aeruginosa, E. coli, and S. aureus, have been required before significant levels of drug efflux occur. This is likely to represent the first step in a process that may culminate in these MDR loci evolving into systems that are specialized for drug efflux.
An interesting example of a drug efflux determinant that would seem to be further along this path, although less well adapted than the tetracycline-specific transport systems, is the plasmid-encoded S. aureus qacA-qacR locus. Evidence supporting this proposal is the apparent recent increase in the substrate range of QacA, the presence of the qacA-qacR locus on multicopy, transmissible, multiresistance-determining plasmids, and the ability of the QacR regulator to respond directly to the presence of antimicrobials, leading to an increase in the transcription of qacA without the need for the regulatory mutations that are characteristic of most other MDR loci involved in drug efflux. Further analysis of drug transporter regulatory networks will reveal whether these systems continue to evolve in order to match the new roles of their target genes, in addition to shedding light on preexisting physiological functions.
The increasing frequency with which bacteria are recruiting transport systems for the purpose of providing antimicrobial resistance makes it essential to gain a fuller understanding of the regulatory controls acting on their expression. This information should prove valuable in developing improved strategies to slow or circumvent the emergence of drug-resistant microorganisms.
Acknowledgments
This work was supported in part by Project Grant 153818 from the National Health and Medical Research Council (Australia) to R.A.S. and M.H.B.
We thank Tom Ellenberger and Winfried Hinrichs for supplying figures and Maria Schumacher and Richard Brennan, with whom we also shared the excitement of collaborating on determining the QacR structures.
REFERENCES
- 1.Adewoye, L., A. Sutherland, R. Srikumar, and K. Poole. 2002. The MexR repressor of the mexAB-oprM multidrug efflux operon in Pseudomonas aeruginosa: characterization of mutations compromising activity. J. Bacteriol. 184:4308-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed, M., C. M. Borsch, S. S. Taylor, N. Vázquez-Laslop, and A. A. Neyfakh. 1994. A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J. Biol. Chem. 269:28506-28513. [PubMed] [Google Scholar]
- 3.Ahmed, M., L. Lyass, P. N. Markham, S. S. Taylor, N. Vázquez-Laslop, and A. A. Neyfakh. 1995. Two highly similar multidrug transporters of Bacillus subtilis whose expression is differentially regulated. J. Bacteriol. 177:3904-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aires, J. R., T. Köhler, H. Nikaido, and P. Plesiat. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alekshun, M. N., Y. S. Kim, and S. B. Levy. 2000. Mutational analysis of MarR, the negative regulator of marRAB expression in Escherichia coli, suggests the presence of two regions required for DNA binding. Mol. Microbiol. 35:1394-1404. [DOI] [PubMed] [Google Scholar]
- 6.Alekshun, M. N., and S. B. Levy. 1999. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J. Bacteriol. 181:4669-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alekshun, M. N., and S. B. Levy. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob. Agents Chemother. 41:2067-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alekshun, M. N., S. B. Levy, T. R. Mealy, B. A. Seaton, and J. F. Head. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat. Struct. Biol. 8:710-714. [DOI] [PubMed] [Google Scholar]
- 9.Alonso, A., and J. L. Martinez. 2001. Expression of multidrug efflux pump SmeDEF by clinical isolates of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45:1879-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschmied, L., R. Baumeister, K. Pfleiderer, and W. Hillen. 1988. A threonine to alanine exchange at position 40 of Tet repressor alters the recognition of the sixth base pair of tet operator from GC to AT. EMBO J. 7:4011-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aono, R., N. Tsukagoshi, and M. Yamamoto. 1998. Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance of Escherichia coli K-12. J. Bacteriol. 180:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baranova, N., and H. Nikaido. 2002. The BaeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J. Bacteriol. 184:4168-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baranova, N. N., A. Danchin, and A. A. Neyfakh. 1999. Mta, a global MerR-type regulator of the Bacillus subtilis multidrug efflux transporters. Mol. Microbiol. 31:1549-1559. [DOI] [PubMed] [Google Scholar]
- 14.Barbosa, T. M., and S. B. Levy. 2002. Activation of the Escherichia coli nfnB gene by MarA through a highly divergent marbox in a class II promoter. Mol. Microbiol. 45:191-202. [DOI] [PubMed] [Google Scholar]
- 15.Barbosa, T. M., and S. B. Levy. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 182:3467-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baron, U., and H. Bujard. 2000. Tet repressor-based system for regulated gene expression in eukaryotic cells: principles and advances. Methods Enzymol. 327:401-421. [DOI] [PubMed] [Google Scholar]
- 17.Bayer, A. S., R. Prasad, J. Chandra, A. Koul, M. Smriti, A. Varama, R. A. Skurray, N. Firth, M. H. Brown, S.-P. Koo, and M. R. Yeaman. 2000. In vitro resistance of Staphylococcus aureus to thrombin-induced platelet microbicidal protein is associated with alterations in cytoplasmic membrane fluidity. Infect. Immun. 68:3548-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berens, C., L. Altschmied, and W. Hillen. 1992. The role of the N terminus in Tet repressor for tet operator binding determined by a mutational analysis. J. Biol. Chem. 267:1945-1952. [PubMed] [Google Scholar]
- 19.Berens, C., D. Schnappinger, and W. Hillen. 1997. The role of the variable region in Tet repressor for inducibility by tetracycline. J. Biol. Chem. 272:6936-6942. [DOI] [PubMed] [Google Scholar]
- 20.Brooun, A., J. J. Tomashek, and K. Lewis. 1999. Purification and ligand binding of EmrR, a regulator of a multidrug transporter. J. Bacteriol. 181:5131-5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown, M. H., and R. A. Skurray. 2001. Staphylococcal multidrug efflux protein QacA. J. Mol. Microbiol. Biotechnol. 3:163-170. [PubMed] [Google Scholar]
- 22.Chami, M., E. Steinfels, C. Orelle, J. M. Jault, A. Di Pietro, J. L. Rigaud, and S. Marco. 2002. Three-dimensional structure by cryo-electron microscopy of YvcC, an homodimeric ATP-binding cassette transporter from Bacillus subtilis. J. Mol. Biol. 315:1075-1085. [DOI] [PubMed] [Google Scholar]
- 23.Chang, G., and C. B. Roth. 2001. Structure of MsbA from E. coli: A homolog of the multidrug resistance ATP binding cassette (ABC) transporters. Science 293:1793-1800. [DOI] [PubMed] [Google Scholar]
- 24.Chollet, R., C. Bollet, J. Chevalier, M. Mallea, J. M. Pages, and A. Davin-Regli. 2002. mar operon involved in multidrug resistance of Enterobacter aerogenes. Antimicrob. Agents Chemother. 46:1093-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen, S. P., W. Yan, and S. B. Levy. 1993. A multidrug resistance regulatory chromosomal locus is widespread among enteric bacteria. J. Infect. Dis. 168:484-488. [DOI] [PubMed] [Google Scholar]
- 27.Dangi, B., P. Pelupessey, R. G. Martin, J. L. Rosner, J. M. Louis, and A. M. Gronenborn. 2001. Structure and dynamics of MarA-DNA complexes: an NMR investigation. J. Mol. Biol. 314:113-127. [DOI] [PubMed] [Google Scholar]
- 28.Davies, J. E. 1997. Origins, acquisition and dissemination of antibiotic resistance determinants, p. 15-35. In D. J. Chadwick and J. Goode (ed.), Antibiotic resistance: origins, evolution, selection and spread. John Wiley and Sons Ltd., Chichester, United Kingdom. [PubMed]
- 29.Delihas, N., and S. Forst. 2001. MicF: an antisense RNA gene involved in response of Escherichia coli to global stress factors. J. Mol. Biol. 313:1-12. [DOI] [PubMed] [Google Scholar]
- 30.Duque, E., A. Segura, G. Mosqueda, and J. L. Ramos. 2001. Global and cognate regulators control the expression of the organic solvent efflux pumps TtgABC and TtgDEF of Pseudomonas putida. Mol. Microbiol. 39:1100-1106. [DOI] [PubMed] [Google Scholar]
- 31.Eckert, B., and C. F. Beck. 1989. Overproduction of transposon Tn10-encoded tetracycline resistance protein results in cell death and loss of membrane potential. J. Bacteriol. 171:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans, E., L. Adewoye, and K. Poole. 2001. MexR repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa: identification of MexR binding sites in the mexA-mexR intergenic region. J. Bacteriol. 183:807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evans, K., and K. Poole. 1999. The MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa is growth-phase regulated. FEMS Microbiol. Lett. 173:35-39. [DOI] [PubMed] [Google Scholar]
- 34.Folcher, M., R. P. Morris, G. Dale, K. Salah-Bey-Hocini, P. H. Viollier, and C. J. Thompson. 2001. A transcriptional regulator of a pristinamycin resistance gene in Streptomyces coelicolor. J. Biol. Chem. 276:1479-1485. [DOI] [PubMed] [Google Scholar]
- 35.Fournier, B., R. Aras, and D. C. Hooper. 2000. Expression of the multidrug resistance transporter NorA from Staphylococcus aureus is modified by a two-component regulatory system. J. Bacteriol. 182:664-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fournier, B., A. Klier, and G. Rapoport. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41:247-261. [DOI] [PubMed] [Google Scholar]
- 37.Fournier, B., Q. C. Truong-Bolduc, X. Zhang, and D. C. Hooper. 2001. A mutation in the 5′ untranslated region increases stability of norA mRNA, encoding a multidrug resistance transporter of Staphylococcus aureus. J. Bacteriol. 183:2367-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fralick, J. A. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178:5803-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gajiwala, K. S., and S. K. Burley. 2000. Winged helix proteins. Curr. Opin. Struct. Biol. 10:110-116. [DOI] [PubMed] [Google Scholar]
- 40.Gambino, L., S. J. Gracheck, and P. F. Miller. 1993. Overexpression of the MarA positive regulator is sufficient to confer multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 175:2888-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gardner, J. F., and M. M. Peel. 1991. Introduction to sterilisation and disinfection. Churchill Livingstone, Melbourne, Australia.
- 42.Gillette, W. K., R. G. Martin, and J. L. Rosner. 2000. Probing the Escherichia coli transcriptional activator MarA with alanine-scanning mutagenesis: residues important for DNA binding and activation. J. Mol. Biol. 299:1245-1255. [DOI] [PubMed] [Google Scholar]
- 43.Godsey, M. H., N. N. Baranova, A. A. Neyfakh, and R. G. Brennan. 2001. Crystal structure of MtaN, a global multidrug transporter gene activator. J. Biol. Chem. 276:47178-47184. [DOI] [PubMed] [Google Scholar]
- 44.Gossen, M., S. Freundlieb, G. Bender, G. Müller, W. Hillen, and H. Bujard. 1995. Transcriptional activation by tetracyclines in mammalian cells. Science 268:1766-1769. [DOI] [PubMed] [Google Scholar]
- 45.Gottesman, M. M., I. Pastan, and S. V. Ambudkar. 1996. P-glycoprotein and multidrug resistance. Curr. Opin. Genet. Dev. 6:610-617. [DOI] [PubMed] [Google Scholar]
- 46.Griffith, K. L., I. M. Shah, T. E. Myers, M. C. O'Neill, and R. E. Wolf, Jr. 2002. Evidence for “pre-recruitment” as a new mechanism of transcription activation in Escherichia coli: the large excess of SoxS binding sites per cell relative to the number of SoxS molecules per cell. Biochem. Biophys. Res. Commun. 291:979-986. [DOI] [PubMed] [Google Scholar]
- 47.Grkovic, S., M. H. Brown, N. J. Roberts, I. T. Paulsen, and R. A. Skurray. 1998. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J. Biol. Chem. 273:18665-18673. [DOI] [PubMed] [Google Scholar]
- 48.Grkovic, S., M. H. Brown, M. A. Schumacher, R. G. Brennan, and R. A. Skurray. 2001. The staphylococcal QacR multidrug regulator binds a correctly spaced operator as a pair of dimers. J. Bacteriol. 183:7102-7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grkovic, S., M. H. Brown, and R. A. Skurray. 2001. Transcriptional regulation of multidrug efflux pumps in bacteria. Seminars Cell. Dev. Biol. 12:225-237. [DOI] [PubMed] [Google Scholar]
- 50.Hächler, H., S. P. Cohen, and S. B. Levy. 1991. marA, a regulated locus which controls expression of chromosomal multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 173:5532-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hagman, K. E., W. Pan, B. G. Spratt, J. T. Balthazar, R. C. Judd, and W. M. Shafer. 1995. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiol. 141:611-622. [DOI] [PubMed] [Google Scholar]
- 52.Hagman, K. E., and W. M. Shafer. 1995. Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae. J. Bacteriol. 177:4162-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hansen, D., and W. Hillen. 1987. Tryptophan in alpha-helix 3 of Tet repressor forms a sequence-specific contact with tet operator in solution. J. Biol. Chem. 262:12269-12274. [PubMed] [Google Scholar]
- 54.Harrison, S. C., and A. K. Aggarwal. 1990. DNA recognition by proteins with the helix-turn-helix motif. Annu. Rev. Biochem. 59:933-969. [DOI] [PubMed] [Google Scholar]
- 55.Hecht, B., G. Müller, and W. Hillen. 1993. Noninducible Tet repressor mutations map from the operator binding motif to the C terminus. J. Bacteriol. 175:1206-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Helbl, V., C. Berens, and W. Hillen. 1995. Proximity probing of Tet repressor to tet operator by dimethylsulfate reveals protected and accessible functions for each recognized base-pair in the major groove. J. Mol. Biol. 245:538-548. [DOI] [PubMed] [Google Scholar]
- 57.Hickman, R. K., L. M. McMurry, and S. B. Levy. 1990. Overproduction and purification of the Tn10-specified inner membrane tetracycline resistance protein Tet with fusions to β-galactosidase. Mol. Microbiol. 4:1241-1251. [DOI] [PubMed] [Google Scholar]
- 58.Hidalgo, E., and B. Demple. 1997. Spacing of promoter elements regulates the basal expression of the soxS gene and converts SoxR from a transcriptional activator into a repressor. EMBO J. 16:1056-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hillen, W., and C. Berens. 1994. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu. Rev. Microbiol. 48:345-369. [DOI] [PubMed] [Google Scholar]
- 60.Hinrichs, W., C. Kisker, M. Düvel, A. Müller, K. Tovar, W. Hillen, and W. Saenger. 1994. Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance. Science 264:418-420. [DOI] [PubMed] [Google Scholar]
- 61.Holmes, D. J., J. L. Caso, and C. J. Thompson. 1993. Autogenous transcriptional activation of a thiostrepton-induced gene in Streptomyces lividans. EMBO J. 12:3183-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoshikawa, Y., K. Amimoto, R. Mizuguchi, and M. Hatakeyama. 1998. Highly controlled heterologous gene expression through combined utilization of the tetracycline-repressible transactivator and the Lac repressor. Anal. Biochem. 261:211-218. [DOI] [PubMed] [Google Scholar]
- 63.Hsieh, P. C., S. A. Siegel, B. Rogers, D. Davis, and K. Lewis. 1998. Bacteria lacking a multidrug pump: a sensitive tool for drug discovery. Proc. Natl. Acad. Sci. USA 95:6602-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huffman, J. L., and R. G. Brennan. 2002. Prokaryotic transcription regulators: more than just the helix-turn-helix motif. Curr. Opin. Struct. Biol. 12:98-106. [DOI] [PubMed] [Google Scholar]
- 65.Hugo, W., and A. Russell. 1992. Evaluation of non-antibiotic antimicrobial agents, p. 258-287. In W. Hugo and A. Russell (ed.), Pharmaceutical microbiology, 5th ed. Blackwell Scientific Publications, Oxford, United Kingdom.
- 66.Ishida, H., H. Fuziwara, Y. Kaibori, T. Horiuchi, K. Sato, and Y. Osada. 1995. Cloning of multidrug resistance gene pqrA from Proteus vulgaris. Antimicrob. Agents Chemother. 39:453-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jack, D. L., N. M. Yang, and M. H. Saier, Jr. 2001. The drug/metabolite transporter superfamily. Eur. J. Biochem. 268:3620-3639. [DOI] [PubMed] [Google Scholar]
- 68.Jair, K.-W., R. G. Martin, J. L. Rosner, N. Fujita, A. Ishihama, and R. E. Wolf, Jr. 1995. Purification and regulatory properties of MarA protein, a transcriptional activator of Escherichia coli multiple antibiotic and superoxide resistance promoters. J. Bacteriol. 177:7100-7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jair, K. W., W. P. Fawcett, N. Fujita, A. Ishihama, and R. E. Wolf, Jr. 1996. Ambidextrous transcriptional activation by SoxS: requirement for the C-terminal domain of the RNA polymerase alpha subunit in a subset of Escherichia coli superoxide-inducible genes. Mol. Microbiol. 19:307-317. [DOI] [PubMed] [Google Scholar]
- 70.Jair, K. W., X. Yu, K. Skarstad, B. Thony, N. Fujita, A. Ishihama, and R. E. Wolf. 1996. Transcriptional activation of promoters of the superoxide and multiple antibiotic resistance regulons by Rob, a binding protein of the Escherichia coli origin of chromosomal replication. J. Bacteriol. 178:2507-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jellen-Ritter, A. S., and W. V. Kern. 2001. Enhanced expression of the multidrug efflux pumps AcrAB and AcrEF associated with insertion element transposition in Escherichia coli mutants selected with a fluoroquinolone. Antimicrob. Agents Chemother. 45:1467-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Johanesen, P. A., D. Lyras, T. L. Bannam, and J. I. Rood. 2001. Transcriptional analysis of the tet(P) operon from Clostridium perfringens. J. Bacteriol. 183:7110-7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Johanesen, P. A., D. Lyras, and J. I. Rood. 2001. Induction of pCW3-encoded tetracycline resistance in Clostridium perfringens involves a host-encoded factor. Plasmid 46:229-232. [DOI] [PubMed] [Google Scholar]
- 74.Kaatz, G. W., and S. M. Seo. 1995. Inducible NorA-mediated multidrug resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 39:2650-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaatz, G. W., S. M. Seo, and T. J. Foster. 1999. Introduction of a norA promoter region mutation into the chromosome of a fluoroquinolone-susceptible strain of Staphylococcus aureus using plasmid integration. Antimicrob. Agents Chemother. 43:2222-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kaatz, G. W., S. M. Seo, and C. A. Ruble. 1993. Efflux-mediated fluoroquinolone resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 37:1086-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaur, P., and J. Russell. 1998. Biochemical coupling between the DrrA and DrrB proteins of the doxorubicin efflux pump of Streptomyces peucetius. J. Biol. Chem. 273:17933-17939. [DOI] [PubMed] [Google Scholar]
- 78.Kieboom, J., and J. de Bont. 2001. Identification and molecular characterization of an efflux system involved in Pseudomonas putida S12 multidrug resistance. Microbiology 147:43-51. [DOI] [PubMed] [Google Scholar]
- 79.Kieboom, J., J. J. Dennis, J. A. M. de Bont, and G. J. Zylstra. 1998. Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J. Biol. Chem. 273:85-91. [DOI] [PubMed] [Google Scholar]
- 80.Kleinschmidt, C., K. Tovar, and W. Hillen. 1991. Computer simulations and experimental studies of gel mobility patterns for weak and strong non-cooperative protein binding to two targets on the same DNA: application to binding of Tet repressor variants to multiple and single tet operator sites. Nucleic Acids Res. 19:1021-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Köhler, T., S. F. Epp, L. K. Curty, and J. C. Pechère. 1999. Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 181:6300-6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Köhler, T., M. Michéa-Hamzehpour, U. Henze, N. Gotoh, L. K. Curty, and J. C. Pechère. 1997. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23:345-354. [DOI] [PubMed] [Google Scholar]
- 83.Köhler, T., C. van Delden, L. K. Curty, M. M. Hamzehpour, and J. C. Pechère. 2001. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 183:5213-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kolaczkowska, A., and A. Goffeau. 1999. Regulation of pleiotropic drug resistance in yeast. Drug Resist. Updates 2:403-414. [DOI] [PubMed] [Google Scholar]
- 85.Kolaczkowski, M., M. van der Rest, A. Cybularz-Kolaczkowska, J. Soumillion, W. N. Konings, and A. Goffeau. 1996. Anticancer drugs, ionophoric peptides, and steroids as substrates of the yeast multidrug transporter Pdr5p. J. Biol. Chem. 271:31543-31548. [DOI] [PubMed] [Google Scholar]
- 86.Koutsolioutsou, A., E. A. Martins, D. G. White, S. B. Levy, and B. Demple. 2001. A soxRS-constitutive mutation contributing to antibiotic resistance in a clinical isolate of Salmonella enterica (serovar Typhimurium). Antimicrob. Agents Chemother. 45:38-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kupferwasser, L. I., R. A. Skurray, M. H. Brown, N. Firth, M. R. Yeaman, and A. S. Bayer. 1999. Plasmid-mediated resistance to thrombin-induced platelet microbicidal protein in staphylococci: role of the qacA locus. Antimicrob. Agents Chemother. 43:2395-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 89.Kwon, H. J., M. H. Bennik, B. Demple, and T. Ellenberger. 2000. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat. Struct. Biol. 7:424-430. [DOI] [PubMed] [Google Scholar]
- 90.Lee, E. H., E. Collatz, I. Podglajen, and L. Gutmann. 1996. A rob-like gene of Enterobacter cloacae affecting porin synthesis and susceptibility to multiple antibiotics. Antimicrob. Agents Chemother. 40:2029-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee, E. H., and W. M. Shafer. 1999. The farAB-encoded efflux pump mediates resistance of gonococci to long-chained antibacterial fatty acids. Mol. Microbiol. 33:839-845. [DOI] [PubMed] [Google Scholar]
- 92.Lee, S. W., and G. Edlin. 1985. Expression of tetracycline resistance in pBR322 derivatives reduces the reproductive fitness of plasmid-containing Escherichia coli. Gene 39:173-180. [DOI] [PubMed] [Google Scholar]
- 93.Lewinson, O., and E. Bibi. 2001. Evidence for simultaneous binding of dissimilar substrates by the Escherichia coli multidrug transporter MdfA. Biochemistry 40:12612-12618. [DOI] [PubMed] [Google Scholar]
- 94.Lewis, K. 2001. In search of natural substrates and inhibitors of MDR pumps. J. Mol. Microbiol. Biotechnol. 3:247-254. [PubMed] [Google Scholar]
- 95.Lewis, K. 1999. Multidrug resistance: versatile drug sensors of bacterial cells. Curr. Biol. 9:R403-R407. [DOI] [PubMed] [Google Scholar]
- 96.Lewis, K. 2000. Translocases: a bacterial tunnel for drugs and proteins. Curr. Biol. 10:R678-R681. [DOI] [PubMed] [Google Scholar]
- 97.Li, H., and J. T. Park. 1999. The periplasmic murein peptide-binding protein MppA is a negative regulator of multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 181:4842-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li, X.-Z., and K. Poole. 1999. Organic solvent-tolerant mutants of Pseudomonas aeruginosa display multiple antibiotic resistance. Can. J. Microbiol. 45:18-22. [DOI] [PubMed] [Google Scholar]
- 99.Li, X. Z., N. Barré, and K. Poole. 2000. Influence of the MexA-MexB-OprM multidrug efflux system on expression of the MexC-MexD-OprJ and MexE-MexF-OprN multidrug efflux systems in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 46:885-893. [DOI] [PubMed] [Google Scholar]
- 100.Li, X. Z., L. Zhang, and K. Poole. 2002. SmeC, an outer membrane multidrug efflux protein of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 46:333-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lim, D. C., K. Poole, and N. Strynadka. 2002. Crystal structure of the MexR repressor of the mexRAB-oprM multi-drug efflux operon of Pseudomonas aeruginosa. J. Biol. Chem. 277:29253-29259. [DOI] [PubMed] [Google Scholar]
- 102.Littlejohn, T. G., I. T. Paulsen, M. T. Gillespie, J. M. Tennent, M. Midgley, I. G. Jones, A. S. Purewal, and R. A. Skurray. 1992. Substrate specificity and energetics of antiseptic and disinfectant resistance in Staphylococcus aureus. FEMS Microbiol. Lett. 95:259-266. [DOI] [PubMed] [Google Scholar]
- 103.Locher, K. P., A. T. Lee, and D. C. Rees. 2002. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science 296:1091-1098. [DOI] [PubMed] [Google Scholar]
- 104.Lomovskaya, O., F. Kawai, and A. Matin. 1996. Differential regulation of the mcb and emr operons of Escherichia coli: role of mcb in multidrug resistance. Antimicrob. Agents Chemother. 40:1050-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lomovskaya, O., K. Lewis, and A. Matin. 1995. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J. Bacteriol. 177:2328-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lucas, C. E., J. T. Balthazar, K. E. Hagman, and W. M. Shafer. 1997. The MtrR repressor binds the DNA sequence between the mtrR and mtrC genes of Neisseria gonorrhoeae. J. Bacteriol. 179:4123-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ma, D., M. Alberti, C. Lynch, H. Nikaido, and J. E. Hearst. 1996. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19:101-112. [DOI] [PubMed] [Google Scholar]
- 108.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 16:45-55. [DOI] [PubMed] [Google Scholar]
- 109.Ma, D., D. N. Cook, J. E. Hearst, and H. Nikaido. 1994. Efflux pumps and drug resistance in gram-negative bacteria. Trends Microbiol. 2:489-493. [DOI] [PubMed] [Google Scholar]
- 110.Madduri, K., and C. R. Hutchinson. 1995. Functional characterization and transcriptional analysis of the dnrR1 locus, which controls daunorubicin biosynthesis in Streptomyces peucetius. J. Bacteriol. 177:1208-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Magnet, S., P. Courvalin, and T. Lambert. 2001. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 45:3375-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Maneewannakul, K., and S. B. Levy. 1996. Identification of mar mutants among quinolone-resistant clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 40:1695-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Markham, P. N., M. Ahmed, and A. A. Neyfakh. 1996. The drug-binding activity of the multidrug-responding transcriptional regulator BmrR resides in its C-terminal domain. J. Bacteriol. 178:1473-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Markham, P. N., J. LoGuidice, and A. A. Neyfakh. 1997. Broad ligand specificity of the transcriptional regulator of the Bacillus subtilis multidrug transporter Bmr. Biochem. Biophys. Res. Commun. 239:269-272. [DOI] [PubMed] [Google Scholar]
- 115.Markham, P. N., and A. A. Neyfakh. 2001. Efflux-mediated drug resistance in gram-positive bacteria. Curr. Opin. Microbiol. 4:509-514. [DOI] [PubMed] [Google Scholar]
- 116.Martin, R. G., W. K. Gillette, N. I. Martin, and J. L. Rosner. 2002. Complex formation between activator and RNA polymerase as the basis for transcriptional activation by MarA and SoxS in Escherichia coli. Mol. Microbiol. 43:355-370. [DOI] [PubMed] [Google Scholar]
- 117.Martin, R. G., W. K. Gillette, S. Rhee, and J. L. Rosner. 1999. Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol. Microbiol. 34:431-441. [DOI] [PubMed] [Google Scholar]
- 118.Martin, R. G., K. W. Jair, R. E. Wolf, and J. L. Rosner. 1996. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J. Bacteriol. 178:2216-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Martin, R. G., and J. L. Rosner. 2001. The AraC transcriptional activators. Curr. Opin. Microbiol. 4:132-137. [DOI] [PubMed] [Google Scholar]
- 120.Martin, R. G., and J. L. Rosner. 1995. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc. Natl. Acad. Sci. USA 92:5456-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Martin, R. G., and J. L. Rosner. 1997. Fis, an accessorial factor for transcriptional activation of the mar (multiple antibiotic resistance) promoter of Escherichia coli in the presence of the activator MarA, SoxS, or Rob. J. Bacteriol. 179:7410-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Martin, R. G., and J. L. Rosner. 2002. Genomics of the marA/soxS/rob regulon of Escherichia coli: identification of directly activated promoters by application of molecular genetics and informatics to microarray data. Mol. Microbiol. 44:1611-1624. [DOI] [PubMed] [Google Scholar]
- 123.Maseda, H., K. Saito, A. Nakajima, and T. Nakae. 2000. Variation of the mexT gene, a regulator of the MexEF-oprN efflux pump expression in wild-type strains of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 192:107-112. [DOI] [PubMed] [Google Scholar]
- 124.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mazzariol, A., Y. Tokue, T. M. Kanegawa, G. Cornaglia, and H. Nikaido. 2000. High-level fluoroquinolone-resistant clinical isolates of Escherichia coli overproduce multidrug efflux protein AcrA. Antimicrob. Agents Chemother. 44:3441-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.McDermott, P. F., D. G. White, I. Podglajen, M. N. Alekshun, and S. B. Levy. 1998. Multidrug resistance following expression of the Escherichia coli marA gene in Mycobacterium smegmatis. J. Bacteriol. 180:2995-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McIvor, R. A., P. Berger, L. L. Pack, A. Rachlis, and C. K. Chan. 1996. An effectiveness community-based clinical trial of Respirgard II and Fisoneb nebulizers for Pneumocystis carinii prophylaxis with aerosol pentamidine in HIV-infected individuals. Toronto Aerosol Pentamidine Study (TAPS) Group. Chest 110:141-146. [DOI] [PubMed] [Google Scholar]
- 128.Meier, I., L. V. Wray, and W. Hillen. 1988. Differential regulation of the Tn10-encoded tetracycline resistance genes tetA and tetR by the tandem tet operators O1 and O2. EMBO J. 7:567-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Miller, P. F., and M. C. Sulavik. 1996. Overlaps and parallels in the regulation of intrinsic multiple-antibiotic resistance in Escherichia coli. Mol. Microbiol. 21:441-448. [DOI] [PubMed] [Google Scholar]
- 130.Mine, T., Y. Morita, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1999. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:415-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mitchell, B. A., M. H. Brown, and R. A. Skurray. 1998. QacA multidrug efflux pump from Staphylococcus aureus: comparative analysis of resistance to diamidines, biguanidines, and guanylhydrazones. Antimicrob. Agents Chemother. 42:475-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mitchell, B. A., I. T. Paulsen, M. H. Brown, and R. A. Skurray. 1999. Bioenergetics of the staphylococcal multidrug export protein QacA: identification of distinct binding sites for monovalent and divalent cations. J. Biol. Chem. 274:3541-3548. [DOI] [PubMed] [Google Scholar]
- 133.Moore, R. A., D. DeShazer, S. Reckseidler, A. Weissman, and D. E. Woods. 1999. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 43:465-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Morita, Y., Y. Komori, T. Mima, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2001. Construction of a series of mutants lacking all of the four major mex operons for multidrug efflux pumps or possessing each one of the operons from Pseudomonas aeruginosa PAO1: MexCD-OprJ is an inducible pump. FEMS Microbiol. Lett. 202:139-143. [DOI] [PubMed] [Google Scholar]
- 135.Muller, G., B. Hecht, V. Helbl, W. Hinrichs, W. Saenger, and W. Hillen. 1995. Characterization of non-inducible Tet repressor mutants suggests conformational changes necessary for induction. Nat. Struct. Biol. 2:693-703. [DOI] [PubMed] [Google Scholar]
- 136.Muñoz-Bellido, J. L., M. Alonzo Manzanares, J. A. Martínez Andrés, M. N. Guttiérrez Zufiaurre, G. Ortiz, M. Segovia Hernández, and J. A. Garcia-Rodríguez. 1999. Efflux pump-mediated quinolone resistance in Staphylococcus aureus strains wild type for gyrA, gyrB, grlA, and norA. Antimicrob. Agents Chemother. 43:354-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nagakubo, S., K. Nishino, T. Hirata, and A. Yamaguchi. 2002. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J. Bacteriol. 184:4161-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Namwat, W., C. K. Lee, H. Kinoshita, Y. Yamada, and T. Nihira. 2001. Identification of the varR gene as a transcriptional regulator of virginiamycin S resistance in Streptomyces virginiae. J. Bacteriol. 183:2025-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Neumüller, A. M., D. Konz, and M. A. Marahiel. 2001. The two-component regulatory system BacRS is associated with bacitracin ‘self-resistance’ of Bacillus licheniformis ATCC 10716. Eur. J. Biochem. 268:3180-3189. [DOI] [PubMed] [Google Scholar]
- 140.Neyfakh, A. A. 2002. Mystery of multidrug transporters: the answer can be simple. Mol. Microbiol. 44:1123-1130. [DOI] [PubMed] [Google Scholar]
- 141.Neyfakh, A. A., V. E. Bidnenko, and L. B. Chen. 1991. Efflux-mediated multidrug resistance in Bacillus subtilis: similarities and dissimilarities with the mammalian system. Proc. Natl. Acad. Sci. USA 88:4781-4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nguyen, T. N., Q. G. Phan, L. P. Duong, K. P. Bertrand, and R. E. Lenski. 1989. Effects of carriage and expression of the Tn10 tetracycline-resistance operon on the fitness of Escherichia coli K12. Mol. Biol. Evol. 6:213-225. [DOI] [PubMed] [Google Scholar]
- 143.Nikaido, H., and H. I. Zgurskaya. 2001. AcrAB and related multidrug efflux pumps of Escherichia coli. J. Mol. Microbiol. Biotechnol. 3:215-218. [PubMed] [Google Scholar]
- 144.Nishino, K., and A. Yamaguchi. 2002. EvgA of the two-component signal transduction system modulates production of the YhiUV multidrug transporter in Escherichia coli. J. Bacteriol. 184:2319-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nishino, K., and A. Yamaguchi. 2001. Overexpression of the response regulator evgA of the two-component signal transduction system modulates multidrug resistance conferred by multidrug resistance transporters. J. Bacteriol. 183:1455-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Notka, F., H. J. Linde, A. Dankesreiter, H. H. Niller, and N. Lehn. 2002. A C-terminal 18 amino acid deletion in MarR in a clinical isolate of Escherichia coli reduces MarR binding properties and increases the MIC of ciprofloxacin. J. Antimicrob. Chemother. 49:41-47. [DOI] [PubMed] [Google Scholar]
- 147.Nunoshiba, T., E. Hidalgo, C. F. Amabile Cuevas, and B. Demple. 1992. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J. Bacteriol. 174:6054-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ochs, M. M., M. P. McCusker, M. Bains, and R. E. Hancock. 1999. Negative regulation of the Pseudomonas aeruginosa outer membrane porin OprD selective for imipenem and basic amino acids. Antimicrob. Agents Chemother. 43:1085-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Oethinger, M., I. Podglajen, W. V. Kern, and S. B. Levy. 1998. Overexpression of the marA or soxS regulatory gene in clinical topoisomerase mutants of Escherichia coli. Antimicrob. Agents Chemother. 42:2089-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Orth, P., F. Cordes, D. Schnappinger, W. Hillen, W. Saenger, and W. Hinrichs. 1998. Conformational changes of the Tet repressor induced by tetracycline trapping. J. Mol. Biol. 279:439-447. [DOI] [PubMed] [Google Scholar]
- 152.Orth, P., W. Saenger, and W. Hinrichs. 1999. Tetracycline-chelated Mg2+ ion initiates helix unwinding in Tet repressor induction. Biochemistry 38:191-198. [DOI] [PubMed] [Google Scholar]
- 153.Orth, P., D. Schnappinger, W. Hillen, W. Saenger, and W. Hinrichs. 2000. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat. Struct. Biol. 7:215-219. [DOI] [PubMed] [Google Scholar]
- 154.Orth, P., D. Schnappinger, P. E. Sum, G. A. Ellestad, W. Hillen, W. Saenger, and W. Hinrichs. 1999. Crystal structure of the Tet repressor in complex with a novel tetracycline, 9-(N,N-dimethylglycylamido)-6-demethyl-6-deoxy-tetracycline. J. Mol. Biol. 285:455-461. [DOI] [PubMed] [Google Scholar]
- 155.Pabo, C. O., and R. T. Sauer. 1992. Transcription factors: structural families and principles of DNA recognition. Annu. Rev. Biochem. 61:1053-1095. [DOI] [PubMed] [Google Scholar]
- 156.Pan, W., and B. G. Spratt. 1994. Regulation of the permeability of the gonococcal cell envelope by the mtr system. Mol. Microbiol. 11:769-775. [DOI] [PubMed] [Google Scholar]
- 157.Parkhill, J., and N. L. Brown. 1990. Site-specific insertion and deletion mutants in the mer promoter-operator region of Tn501; the nineteen-base-pair spacer is essential for normal induction of the promoter by MerR. Nucleic Acids Res. 18:5157-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Parkinson, J. S. 1993. Signal transduction schemes of bacteria. Cell 73:857-871. [DOI] [PubMed] [Google Scholar]
- 159.Paulsen, I. T., M. H. Brown, T. G. Littlejohn, B. A. Mitchell, and R. A. Skurray. 1996. Multidrug resistance proteins QacA and QacB from Staphylococcus aureus: membrane topology and identification of residues involved in substrate specificity. Proc. Natl. Acad. Sci. USA 93:3630-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Paulsen, I. T., M. H. Brown, and R. A. Skurray. 1998. Characterization of the earliest known Staphylococcus aureus plasmids encoding multidrug efflux systems. J. Bacteriol. 180:3477-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Paulsen, I. T., M. H. Brown, and R. A. Skurray. 1996. Proton-dependent multidrug efflux systems. Microbiol. Rev. 60:575-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Paulsen, I. T., J. Chen, K. E. Nelson, and M. H. Saier, Jr. 2001. Comparative genomics of microbial drug efflux systems. J. Mol. Microbiol. Biotechnol. 3:145-150. [PubMed] [Google Scholar]
- 163.Pearson, J. P., C. Van Delden, and B. H. Iglewski. 1999. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Podlesek, Z., A. Comino, B. Herzog-Velikonja, D. Zgurbertok, R. Komel, and M. Grabnar. 1995. Bacillus licheniformis bacitracin-resistance ABC transporter-relationship to mammalian multidrug resistance. Mol. Microbiol. 16:969-976. [DOI] [PubMed] [Google Scholar]
- 165.Pomposiello, P. J., M. H. Bennik, and B. Demple. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Poole, K. 1994. Bacterial multidrug resistance—emphasis on efflux mechanisms and Pseudomonas aeruginosa. J. Antimicrob. Chemother. 34:453-456. [DOI] [PubMed] [Google Scholar]
- 167.Poole, K. 2001. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J. Mol. Microbiol. Biotechnol. 3:255-264. [PubMed] [Google Scholar]
- 168.Poole, K. 2001. Multidrug resistance in gram-negative bacteria. Curr. Opin. Microbiol. 4:500-508. [DOI] [PubMed] [Google Scholar]
- 169.Poole, K., N. Gotoh, H. Tsujimoto, Q. X. Zhao, A. Wada, T. Yamasaki, S. Neshat, J. I. Yamagishi, X. Z. Li, and T. Nishino. 1996. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB multidrug-resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21:713-724. [DOI] [PubMed] [Google Scholar]
- 170.Poole, K., K. Tetro, Q. Zhao, S. Neshat, D. E. Heinrichs, and N. Bianco. 1996. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 40:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pumbwe, L., and L. J. Piddock. 2000. Two efflux systems expressed simultaneously in multidrug-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:2861-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Putman, M., L. A. Koole, H. W. van Veen, and W. N. Konings. 1999. The secondary multidrug transporter LmrP contains multiple drug interaction sites. Biochemistry 38:13900-13905. [DOI] [PubMed] [Google Scholar]
- 173.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Rahmati, S., S. Yang, A. L. Davidson, and E. L. Zechiedrich. 2002. Control of the AcrAB multidrug efflux pump by quorum sensing regulator SdiA. Mol. Microbiol. 43:677-695. [DOI] [PubMed] [Google Scholar]
- 175.Ramos, J. L., E. Duque, P. Godoy, and A. Segura. 1998. Efflux pumps involved in toluene tolerance in Pseudomonas putida DOT-T1E. J. Bacteriol. 180:3323-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Rand, J. D., S. G. Danby, D. L. Greenway, and R. R. England. 2002. Increased expression of the multidrug efflux genes acrAB occurs during slow growth of Escherichia coli. FEMS Microbiol. Lett. 207:91-95. [DOI] [PubMed] [Google Scholar]
- 177.Rhee, S., R. G. Martin, J. L. Rosner, and D. R. Davies. 1998. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc. Natl. Acad. Sci. USA 95:10413-10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Rodionov, D. A., M. S. Gelfand, A. A. Mironov, and A. B. Rakhmaninova. 2001. Comparative approach to analysis of regulation in complete genomes: multidrug resistance systems in gamma-proteobacteria. J. Mol. Microbiol. Biotechnol. 3:319-324. [PubMed] [Google Scholar]
- 179.Rojas, A., E. Duque, G. Mosqueda, G. Golden, A. Hurtado, J. L. Ramos, and A. Segura. 2001. Three efflux pumps are required to provide efficient tolerance to toluene in Pseudomonas putida DOT-T1E. J. Bacteriol. 183:3967-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Rosenberg, M. F., R. Callaghan, R. C. Ford, and C. F. Higgins. 1997. Structure of the multidrug resistance P-glycoprotein to 2.5 nm resolution determined by electron microscopy and image analysis. J. Biol. Chem. 272:10685-10694. [DOI] [PubMed] [Google Scholar]
- 181.Rosenberg, M. F., G. Velarde, R. C. Ford, C. Martin, G. Berridge, I. D. Kerr, R. Callaghan, A. Schmidlin, C. Wooding, K. J. Linton, and C. F. Higgins. 2001. Repacking of the transmembrane domains of P-glycoprotein during the transport ATPase cycle. EMBO J. 20:5615-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Rosner, J. L., B. Dangi, A. M. Gronenborn, and R. G. Martin. 2002. Posttranscriptional activation of the transcriptional activator Rob by dipyridyl in Escherichia coli. J. Bacteriol. 184:1407-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Rouch, D. A., D. S. Cram, D. DiBerardino, T. G. Littlejohn, and R. A. Skurray. 1990. Efflux-mediated antiseptic resistance gene qacA from Staphylococcus aureus: common ancestry with tetracycline- and sugar-transport proteins. Mol. Microbiol. 4:2051-2062. [DOI] [PubMed] [Google Scholar]
- 184.Rouquette, C., J. B. Harmon, and W. M. Shafer. 1999. Induction of the mtrCDE-encoded efflux pump system of Neisseria gonorrhoeae requires MtrA, an AraC-like protein. Mol. Microbiol. 33:651-658. [DOI] [PubMed] [Google Scholar]
- 185.Saier, M. H., Jr., and I. T. Paulsen. 2001. Phylogeny of multidrug transporters. Semin. Cell. Dev. Biol. 12:205-213. [DOI] [PubMed] [Google Scholar]
- 186.Saito, K., H. Yoneyama, and T. Nakae. 1999. nalB-type mutations causing the overexpression of the MexAB-OprM efflux pump are located in the mexR gene of the Pseudomonas aeruginosa chromosome. FEMS Microbiol. Lett. 179:67-72. [DOI] [PubMed] [Google Scholar]
- 187.Salah-Bey, K., V. Blanc, and C. J. Thompson. 1995. Stress-activated expression of a Streptomyces pristinaespiralis multidrug resistance gene (ptr) in various Streptomyces spp. and Escherichia coli. Mol. Microbiol. 17:1001-1012. [DOI] [PubMed] [Google Scholar]
- 188.Salah-Bey, K., and C. J. Thompson. 1995. Unusual regulatory mechanism for a Streptomyces multidrug resistance gene, ptr, involving three homologous protein-binding sites overlapping the promoter region. Mol. Microbiol. 17:1109-1119. [DOI] [PubMed] [Google Scholar]
- 189.Sánchez, P., F. Rojo, and J. L. Martínez. 2002. Transcriptional regulation of mexR, the repressor of Pseudomonas aeruginosa mexAB-oprM multidrug efflux pump. FEMS Microbiol. Lett. 207:63-68. [DOI] [PubMed] [Google Scholar]
- 190.Schnappinger, D., and W. Hillen. 1996. Tetracyclines: antibiotic action, uptake, and resistance mechanisms. Arch. Microbiol. 165:359-369. [DOI] [PubMed] [Google Scholar]
- 191.Schnappinger, D., P. Schubert, C. Berens, K. Pfleiderer, and W. Hillen. 1999. Solvent-exposed residues in the Tet repressor (TetR) four-helix bundle contribute to subunit recognition and dimer stability. J. Biol. Chem. 274:6405-6410. [DOI] [PubMed] [Google Scholar]
- 192.Schnappinger, D., P. Schubert, K. Pfleiderer, and W. Hillen. 1998. Determinants of protein-protein recognition by four helix bundles: changing the dimerization specificity of Tet repressor. EMBO J. 17:535-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Scholz, O., M. Kintrup, M. Reich, and W. Hillen. 2001. Mechanism of Tet repressor induction by tetracyclines: length compensates for sequence in the α8-α9 loop. J. Mol. Biol. 310:979-986. [DOI] [PubMed] [Google Scholar]
- 194.Scholz, O., P. Schubert, M. Kintrup, and W. Hillen. 2000. Tet repressor induction without Mg2+. Biochemistry 39:10914-10920. [DOI] [PubMed] [Google Scholar]
- 195.Schubert, P., D. Schnappinger, K. Pfleiderer, and W. Hillen. 2001. Identification of a stability determinant on the edge of the Tet repressor four-helix bundle dimerization motif. Biochemistry 40:3257-3263. [DOI] [PubMed] [Google Scholar]
- 196.Schumacher, M. A., and R. G. Brennan. 2002. Structural mechanisms of multidrug recognition and regulation by bacterial multidrug transcription factors. Mol. Microbiol. 45:885-893. [DOI] [PubMed] [Google Scholar]
- 197.Schumacher, M. A., M. C. Miller, S. Grkovic, M. H. Brown, R. A. Skurray, and R. G. Brennan. 2002. Structural basis for cooperative DNA binding by two dimers of the multidrug binding protein QacR. EMBO J. 21:1210-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Schumacher, M. A., M. C. Miller, S. Grkovic, M. H. Brown, R. A. Skurray, and R. G. Brennan. 2001. Structural mechanisms of QacR induction and multidrug recognition. Science 294:2158-2163. [DOI] [PubMed] [Google Scholar]
- 199.Scott, E., and S. Gorman. 1992. Chemical disinfectants, antiseptics and preservatives, p. 231-257. In W. Hugo and A. Russell (ed.), Pharmaceutical microbiology. Blackwell Scientific Publications, Oxford, United Kingdom.
- 200.Seoane, A. S., and S. B. Levy. 1995. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J. Bacteriol. 177:3414-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Shafer, W. M., J. T. Balthazar, K. E. Hagman, and S. A. Morse. 1995. Missense mutations that alter the DNA-binding domain of the MtrR protein occur frequently in rectal isolates of Neisseria gonorrhoeae that are resistant to faecal lipids. Microbiology 141:907-911. [DOI] [PubMed] [Google Scholar]
- 202.Shafer, W. M., X.-D. Qu, A. J. Waring, and R. I. Lehrer. 1998. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides because of a member of the resistance/nodulation/division efflux pump family. Proc. Natl. Acad. Sci. USA 95:1829-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Shafer, W. M., W. L. Veal, E. H. Lee, L. Zarantonelli, J. T. Balthazar, and C. Rouquette. 2001. Genetic organization and regulation of antimicrobial efflux systems possessed by Neisseria gonorrhoeae and Neisseria meningitidis. J. Mol. Microbiol. Biotechnol. 3:219-224. [PubMed] [Google Scholar]
- 204.Shiba, T., K. Ishiguro, N. Takemoto, H. Koibuchi, and K. Sugimoto. 1995. Purification and characterization of the Pseudomonas aeruginosa NfxB protein, the negative regulator of the nfxB gene. J. Bacteriol. 177:5872-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Smith, L. D., and K. P. Bertrand. 1988. Mutations in the Tn10 tet repressor that interfere with induction. Location of the tetracycline-binding domain. J. Mol. Biol. 203:949-959. [DOI] [PubMed] [Google Scholar]
- 206.Speer, B. S., N. B. Shoemaker, and A. A. Salyers. 1992. Bacterial resistance to tetracycline: mechanisms, transfer, and clinical significance. Clin. Microbiol. Rev. 5:387-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Spott, S., F. Dong, B. Kisters-Woike, and B. Müller-Hill. 2000. Dimerisation mutants of Lac repressor. II. A single amino acid substitution, D278L, changes the specificity of dimerisation. J. Mol. Biol. 296:673-684. [DOI] [PubMed] [Google Scholar]
- 208.Srikumar, R., C. J. Paul, and K. Poole. 2000. Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 182:1410-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Stasinopoulos, S. J., G. A. Farr, and D. H. Bechhofer. 1998. Bacillus subtilis tetA(L) gene expression: evidence for regulation by translational reinitiation. Mol. Microbiol. 30:923-932. [DOI] [PubMed] [Google Scholar]
- 210.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K.-S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and, M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 211.Sulavik, M. C., L. F. Gambino, and P. F. Miller. 1995. The MarR repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli: prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol. Med. 1:436-446. [PMC free article] [PubMed] [Google Scholar]
- 212.Summers, A. O. 1992. Untwist and shout: a heavy-metal-responsive transcriptional regulator. J. Bacteriol. 174:3097-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Suzuki, M., N. Yagi, and M. Gerstein. 1995. DNA recognition and superstructure formation by helix-turn-helix proteins. Protein Eng. 8:329-338. [DOI] [PubMed] [Google Scholar]
- 214.Tamai, I., and A. R. Safa. 1991. Azidopine noncompetitively interacts with vinblastine and cyclosporin A binding to P-glycoprotein in multidrug resistant cells. J. Biol. Chem. 266:16796-16800. [PubMed] [Google Scholar]
- 215.Tanaka, T., T. Horii, K. Shibayama, K. Sato, S. Ohsuka, Y. Arakawa, K. Yamaki, K. Takagi, and M. Ohta. 1997. RobA-induced multiple antibiotic resistance largely depends on the activation of the AcrAB efflux. Microbiol. Immunol. 41:697-702. [DOI] [PubMed] [Google Scholar]
- 216.Tate, C. G., E. R. Kunji, M. Lebendiker, and S. Schuldiner. 2001. The projection structure of EmrE, a proton-linked multidrug transporter from Escherichia coli, at 7 Å resolution. EMBO J. 20:77-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Tauch, A., A. Puhler, J. Kalinowski, and G. Thierbach. 2000. TetZ, a new tetracycline resistance determinant discovered in gram-positive bacteria, shows high homology to gram-negative regulated efflux systems. Plasmid 44:285-291. [DOI] [PubMed] [Google Scholar]
- 218.Tennent, J. M., B. R. Lyon, M. T. Gillespie, J. W. May, and R. A. Skurray. 1985. Cloning and expression of Staphylococcus aureus plasmid-mediated quaternary ammonium resistance in Escherichia coli. Antimicrob. Agents Chemother. 27:79-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Tennent, J. M., B. R. Lyon, M. Midgley, I. G. Jones, A. S. Purewal, and R. A. Skurray. 1989. Physical and biochemical characterization of the qacA gene encoding antiseptic and disinfectant resistance in Staphylococcus aureus. J. Gen. Microbiol. 135:1-10. [DOI] [PubMed] [Google Scholar]
- 220.Tennent, J. M., J. W. May, and R. A. Skurray. 1984. Multiple antibiotic resistance in Staphylococcus aureus and Staphylococcus epidermidis: plasmids in strains associated with nosocomial infection. Pathology 16:250-255. [DOI] [PubMed] [Google Scholar]
- 221.Thanassi, D. G., L. W. Cheng, and H. Nikaido. 1997. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179:2512-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Tiebel, B., L. M. Aung-Hilbrich, D. Schnappinger, and W. Hillen. 1998. Conformational changes necessary for gene regulation by Tet repressor assayed by reversible disulfide bond formation. EMBO J. 17:5112-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Tiebel, B., K. Garke, and W. Hillen. 2000. Observing conformational and activity changes of Tet repressor in vivo. Nat. Struct. Biol. 7:479-481. [DOI] [PubMed] [Google Scholar]
- 224.Tiebel, B., N. Radzwill, L. M. Aung-Hilbrich, V. Helbl, H. J. Steinhoff, and W. Hillen. 1999. Domain motions accompanying Tet repressor induction defined by changes of interspin distances at selectively labeled sites. J. Mol. Biol. 290:229-240. [DOI] [PubMed] [Google Scholar]
- 225.Travers, A., R. Schneider, and G. Muskhelishvili. 2001. DNA supercoiling and transcription in Escherichia coli: the FIS connection. Biochimie 83:213-217. [DOI] [PubMed] [Google Scholar]
- 226.van Veen, H. W., R. Callaghan, L. Soceneantu, A. Sardini, W. N. Konings, and C. F. Higgins. 1998. A bacterial antibiotic-resistance gene that complements the human multidrug-resistance P-glycoprotein gene. Nature 391:291-295. [DOI] [PubMed] [Google Scholar]
- 227.Vázquez-Laslop, N., P. N. Markham, and A. A. Neyfakh. 1999. Mechanism of ligand recognition by BmrR, the multidrug-responding transcriptional regulator: Mutational analysis of the ligand-binding site. Biochemistry 38:16925-16931. [DOI] [PubMed] [Google Scholar]
- 228.Vergani, B., M. Kintrup, W. Hillen, H. Lami, E. Piemont, E. Bombarda, P. Alberti, S. M. Doglia, and M. Chabbert. 2000. Backbone dynamics of Tet repressor α8-intersection-α9 loop. Biochemistry 39:2759-2768. [DOI] [PubMed] [Google Scholar]
- 229.von Heijne, G. 1991. Proline kinks in transmembrane α-helices. J. Mol. Biol. 218:499-503. [DOI] [PubMed] [Google Scholar]
- 230.Wang, H., J. L. Dzink-Fox, M. Chen, and S. B. Levy. 2001. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob. Agents Chemother. 45:1515-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Ward, A., C. Hoyle, S. Palmer, J. O'Reilly, J. Griffith, M. Pos, S. Morrison, B. Poolman, M. Gwynne, and P. Henderson. 2001. Prokaryote multidrug efflux proteins of the major facilitator superfamily: amplified expression, purification and characterisation. J. Mol. Microbiol. Biotechnol. 3:193-200. [PubMed] [Google Scholar]
- 232.Webber, M. A., and L. J. Piddock. 2001. Absence of mutations in marRAB or soxRS in acrB-overexpressing fluoroquinolone-resistant clinical and veterinary isolates of Escherichia coli. Antimicrob. Agents Chemother. 45:1550-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Westbrock-Wadman, S., D. R. Sherman, M. J. Hickey, S. N. Coulter, Y. Q. Zhu, P. Warrener, L. Y. Nguyen, R. M. Shawar, K. R. Folger, and C. K. Stover. 1999. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 43:2975-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.White, D. G., J. D. Goldman, B. Demple, and S. B. Levy. 1997. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J. Bacteriol. 179:6122-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Wissmann, A., R. Baumeister, G. Muller, B. Hecht, V. Helbl, K. Pfleiderer, and W. Hillen. 1991. Amino acids determining operator binding specificity in the helix-turn-helix motif of Tn10 repressor. EMBO J. 10:4145-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Wissmann, A., I. Meier, and W. Hillen. 1988. Saturation mutagenesis of the Tn10-encoded tet operator O1. Identification of base pairs involved in Tet repressor recognition. J. Mol. Biol. 202:397-406. [DOI] [PubMed] [Google Scholar]
- 237.Woolridge, D. P., N. Vázquez-Laslop, P. N. Markham, M. S. Chevalier, E. W. Gerner, and A. A. Neyfakh. 1997. Efflux of the natural polyamine spermidine facilitated by the Bacillus subtilis multidrug transporter Blt. J. Biol. Chem. 272:8864-8866. [DOI] [PubMed] [Google Scholar]
- 238.Xiong, A., A. Gottman, C. Park, M. Baetens, S. Pandza, and A. Matin. 2000. The EmrR protein represses the Escherichia coli emrRAB multidrug resistance operon by directly binding to its promoter region. Antimicrob. Agents Chemother. 44:2905-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Yamada, H., S. Kurosehamada, Y. Fukuda, J. Mitsuyama, M. Takahata, S. Minami, Y. Watanabe, and H. Narita. 1997. Quinolone susceptibility of norA-disrupted Staphylococcus aureus. Antimicrob. Agents Chemother. 41:2308-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Yin, C. C., M. L. Aldema-Ramos, M. I. Borges-Walmsley, R. W. Taylor, A. R. Walmsley, S. B. Levy, and P. A. Bullough. 2000. The quarternary molecular architecture of TetA, a secondary tetracycline transporter from Escherichia coli. Mol. Microbiol. 38:482-492. [DOI] [PubMed] [Google Scholar]
- 241.Yoshida, H., M. Bogaki, S. Nakamura, K. Ubukata, and M. Konno. 1990. Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J. Bacteriol. 172:6942-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Zarantonelli, L., G. Borthagaray, E. H. Lee, and W. M. Shafer. 1999. Decreased azithromycin susceptibility of Neisseria gonorrhoeae due to mtrR mutations. Antimicrob. Agents Chemother. 43:2468-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Zhang, L., X. Z. Li, and K. Poole. 2001. SmeDEF multidrug efflux pump contributes to intrinsic multidrug resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45:3497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Zheleznova, E. E., P. N. Markham, A. A. Neyfakh, and R. G. Brennan. 1999. Structural basis of multidrug recognition by BmrR, a transcriptional activator of a multidrug transporter. Cell 96:353-362. [DOI] [PubMed] [Google Scholar]
- 245.Zheleznova Heldwein, E. E., and R. G. Brennan. 2001. Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature 409:378-382. [DOI] [PubMed] [Google Scholar]
- 246.Ziha-Zarifi, I., C. Llanes, T. Köhler, J. C. Pechere, and P. Plesiat. 1999. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob. Agents Chemother. 43:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]