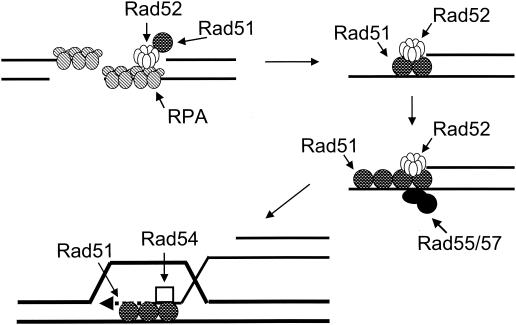
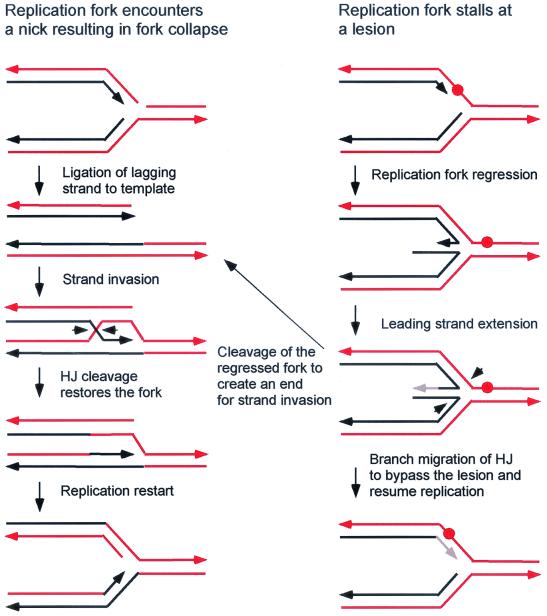
Abstract
The process of homologous recombination is a major DNA repair pathway that operates on DNA double-strand breaks, and possibly other kinds of DNA lesions, to promote error-free repair. Central to the process of homologous recombination are the RAD52 group genes (RAD50, RAD51, RAD52, RAD54, RDH54/TID1, RAD55, RAD57, RAD59, MRE11, and XRS2), most of which were identified by their requirement for the repair of ionizing-radiation-induced DNA damage in Saccharomyces cerevisiae. The Rad52 group proteins are highly conserved among eukaryotes, and Rad51, Mre11, and Rad50 are also conserved in prokaryotes and archaea. Recent studies showing defects in homologous recombination and double-strand break repair in several human cancer-prone syndromes have emphasized the importance of this repair pathway in maintaining genome integrity. Although sensitivity to ionizing radiation is a universal feature of rad52 group mutants, the mutants show considerable heterogeneity in different assays for recombinational repair of double-strand breaks and spontaneous mitotic recombination. Herein, I provide an overview of recent biochemical and structural analyses of the Rad52 group proteins and discuss how this information can be incorporated into genetic studies of recombination.
INTRODUCTION
Recombination refers to the exchange or transfer of information between DNA molecules and is found in all organisms that have been studied in detail. Homologous recombination involves the exchange of DNA between sequences of perfect or near perfect homology over several hundreds of base pairs. In contrast, nonhomologous recombination occurs between sequences with little or no sequence homology. The process of homologous recombination plays essential roles in the mitotic and meiotic cell cycles of most eukaryotic organisms. In meiosis, the primary function of recombination is to establish a physical connection between homologous chromosomes to ensure their correct disjunction at the first meiotic division. In addition, meiotic recombination contributes to diversity by creating new linkage arrangements between genes, or parts of genes. It is now widely recognized that the primary function of homologous recombination in mitotic cells is to repair double-strand breaks (DSBs) that form as a result of replication fork collapse, from processing of spontaneous damage, and from exposure to DNA-damaging agents. Recombination is also required to repair the DSBs that initiate programmed rearrangements, such as mating-type switching in Saccharomyces cerevisiae.
Most of the genes in the RAD52 epistasis group (RAD50, RAD51, RAD52, RAD54, RAD55, RAD57, RAD59, RDH54 [TID1], MRE11 [RAD58], and XRS2) were identified by their requirement in the repair of ionizing-radiation (IR)-induced DNA damage. Mutations in these genes lead to defects in meiotic and/or mitotic recombination, providing evidence for a link between DSB repair (DSBR) and homologous recombination. Homologues of the RAD52 group of genes have been identified in many other eukaryotes, and in some cases in prokaryotes and archaea, indicating high conservation of the recombinational repair pathway. The recent discovery that several human cancer-prone syndromes, for example, Nijmegen breakage syndrome (NBS) and ataxia-telangiectasia-like disorder (A-TLD), are caused by defects in DSBR has highlighted the importance of this repair pathway in maintaining genome integrity and cancer avoidance (47, 358, 411).
The goal of this review is to provide an update of the role of the RAD52 group genes in spontaneous and DSB-induced homologous recombination. Other aspects of DSBR, cell cycle checkpoints, and meiosis are beyond the scope of this review, and the reader is referred to several excellent reviews on these topics (212, 271, 432). Most of the studies described in this review were performed with S. cerevisiae and with proteins corresponding to the S. cerevisiae gene products. When studies with other organisms are described, the gene or protein is prefixed by the abbreviated species name. The first part of the review describes models for DSBR and physical and genetic assays used to monitor repair and recombination. In the second part, the biochemical functions of the Rad52 group proteins and phenotypes of the rad52 group mutants in various recombination assays are described.
CURRENT VIEW OF THE MECHANISMS OF HOMOLOGOUS RECOMBINATION
Much of our understanding of the mechanisms of recombination is based on organisms, such as S. cerevisiae, in which all of the products of an individual meiosis can be recovered for analysis in the form of asci containing four haploid spores. Two types of recombination events have been identified based on the segregation of heterozygous markers during meiosis: crossing over and gene conversion. A crossover between linked heterozygous markers results in new linkage arrangements for two spore products, but the markers are still recovered in Mendelian ratios. Gene conversion represents the nonreciprocal transfer of information between two homologous sequences to duplicate one of the alleles, with the corresponding loss of the other, resulting in a non-Mendelian segregation. Studies with yeast and other fungi have shown that gene conversion events are frequently associated with the exchange of flanking markers (145). This association is thought to be due to alternate resolution of a Holliday junction containing intermediate to generate crossover or noncrossover products (135). Several models have been proposed to explain the molecular mechanisms of recombination, and of these the DSBR and synthesis-dependent strand-annealing (SDSA) models are most consistent with the available genetic data (135, 236, 257, 381).
Double-Strand Break Repair Model
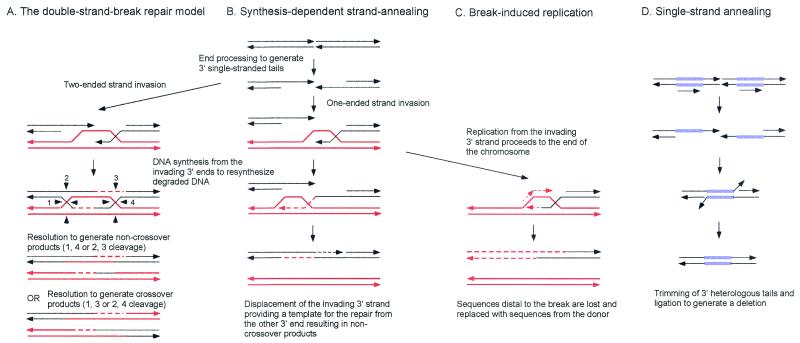
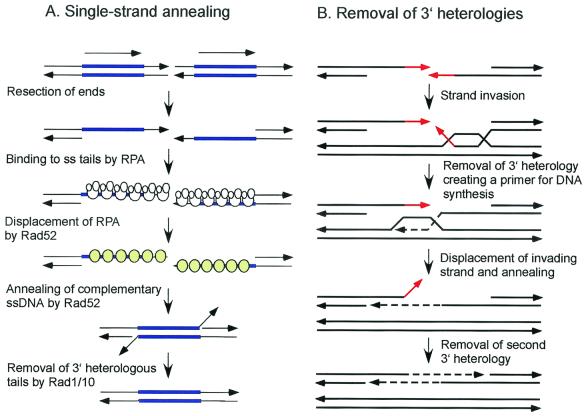
The observation that IR stimulates recombination suggested that recombination is initiated by DSBs. Studies by Orr-Weaver et al. demonstrated that a nonreplicating plasmid containing a DSB made within a region of the plasmid with homology to genomic sequences recombined with high efficiency following transformation of yeast (269). The plasmid DNA integrated at the homologous locus and was flanked by a duplication of the region of yeast DNA carried on the plasmid. When plasmid DNA was digested with restriction endonucleases to delete an internal fragment from the yeast DNA, it was still found to transform yeast at high frequency. The resulting transformants were indistinguishable in structure from those obtained using uncut DNA, indicating that repair of the gapped region had occurred during integration. This repair of a double-strand gap is equivalent to gene conversion without mismatch repair. Using linearized replicating plasmids, Orr-Weaver and Szostak reported the recovery of approximately equal numbers of integrated and non-integrated plasmids and concluded that gene conversion by DSBR can occur with or without crossing over (268). These observations formed the basis for the DSBR model of recombination (381) (Fig. 1A). In this model, the ends of the break are resected to form 3′ single-stranded tails that are active in strand invasion with a homologous duplex. Following strand invasion, the 3′ end is extended by DNA synthesis. The D-loop formed by strand invasion is able to pair with the other side of the DSB, and the 3′ end of the noninvading strand is also extended by DNA synthesis, forming a double-Holliday-junction (dHJ) intermediate. Random resolution of the two Holliday junctions is expected to yield equal numbers of crossover and noncrossover products.
FIG. 1.
Models for the repair of DSBs. (A) In the DSBR model, the ends are processed to yield 3′ single-stranded tails. The 3′ ends invade the homologous duplex, priming DNA synthesis. After ligation, a dHJ intermediate is formed, which can subsequently be resolved by endonucleolytic cleavage of the two Holliday junctions to generate crossover or noncrossover products. (B) In the SDSA model, the ends are processed to yield 3′ single-stranded tails, one of which invades the homologous duplex, priming DNA synthesis. The displacement loop (D-loop) formed by strand invasion could be extended by DNA synthesis or could migrate with the newly synthesized DNA. After displacement from the donor duplex, the nascent strand pairs with the other 3′ single-stranded tail and DNA synthesis completes repair. (C) The initial steps in the BIR model are the same as in the SDSA model, but DNA synthesis from the invading strand continues to the end of the DNA molecule. (D) In the SSA model, a DSB made between direct repeats is subject to resection to generate 3′ single-stranded tails. When complementary sequences are revealed due to extensive resection, the single-stranded DNA anneals, resulting in deletion of one of the repeats and the intervening DNA. The 3′ tails are endonucleolytically removed, and the nicks are ligated. The 3′ ends are indicated by arrowheads.
Further support of the DSBR model as a general model for recombination came from the observation of transient DSBs at meiotic recombination hot spots (46, 369). The ends of meiotic DSBs are processed to yield 3′ single-stranded tails of about 600 nucleotides (370). DSBs made by the HO endonuclease in mitotic cells are also resected to generate long 3′ single-stranded tails (419). The length of the single-stranded tails at the ARG4 locus correlates well with the length of gene conversion tracts within the ARG4 gene, suggesting that most conversion events arise from repair of heteroduplex DNA and that four-strand branch migration is limited (370). Branched DNA molecules with the topology expected from dHJ intermediates have been detected by two-dimensional agarose gel electrophoresis of meiotic DNA, providing further support for the model (61, 327, 328).
Synthesis-Dependent Strand-Annealing Model
Subsequent studies of plasmid gap repair in yeast and Ustilago maydis and chromosomal DSBR following P element excision in Drosophila melanogaster have shown a lower association of crossing over with gene conversion (20, 96, 257, 291). Similarly, mating-type switching in S. cerevisiae, which is a gene conversion event initiated by a site-specific DSB at the MAT locus made by the HO endonuclease (183, 360), is rarely associated with crossing over. To account for the low levels of associated crossing over observed for some DSB-induced gene conversion events, the SDSA/migrating D-loop model was proposed (96, 257, 360) (Fig. 1B). In this model, strand invasion occurs as envisioned in the DSBR model, but after extensive DNA synthesis primed from the invading strand, the elongated invading strand is displaced and pairs with the other side of the break. DNA synthesis can then be primed from the noninvading 3′ end to repair the DSB or gap. An alternative scenario for gap repair involves coupling of lagging-strand DNA synthesis to leading-strand synthesis from the invading strand (137). Further evidence in support of the SDSA model comes from the observation that the donor sequences are generally unchanged during DSB-induced gene conversion (273).
Recent studies of meiotic recombination intermediates have identified a second class of branched molecules, termed single-end invasion intermediates (SEI) (144). These could be precursors to dHJ intermediates or could be processed directly into noncrossover products as predicted by the SDSA model. The relatively long-lived dHJ intermediates appear to be precursors for crossover products. The observation that formation of crossover, but not noncrossover, products is blocked by an ndt80 mutation is consistent with the suggestion that these two classes of recombinants are derived from different intermediates (11). Allers and Lichten propose that noncrossovers are generated by SDSA and crossovers are generated from the resolution of dHJ intermediates that are formed as envisioned by the DSBR model (11).
Other Mechanisms for Homology-Dependent Double-Strand Break Repair
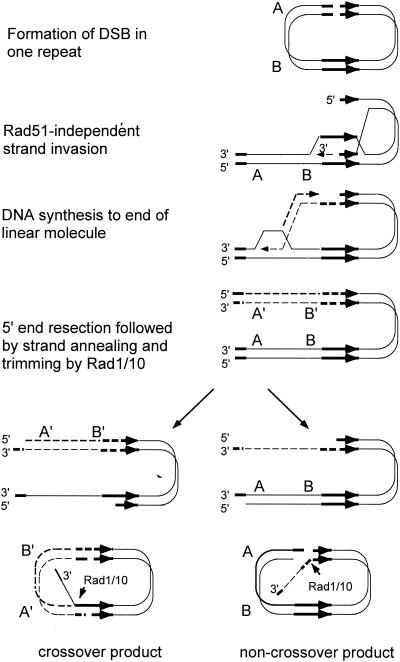
When a DSB is induced at the MAT locus on one chromosome III homologue of diploid cells, the break is generally repaired by gene conversion with about 20% associated crossing over (217). In certain genetic backgrounds, gene conversion is eliminated and repair occurs by invasion of the donor duplex by the broken chromosome followed by replication to the end of the donor chromosome (217) (Fig. 1C). This process is known as break-induced replication (BIR). This nonreciprocal process is likely to be important for telomere maintenance in the absence of telomerase (190, 388). Homology-dependent duplication of an entire chromosome arm to the end of a transformed linearized plasmid has also been detected in yeast and is presumed to occur by BIR (248). Another pathway of homology-dependent repair, single-strand annealing (SSA), is restricted to DSBs that occur between direct repeats (Fig. 1D). These events have been detected in yeast and animal cells by using artificial direct repeats and could conceivably be important for repair in genomes of higher eukaryotes that contain many repeated sequences (101, 202, 224). After formation of the DSB, the ends are resected to produce 3′ single-stranded tails, which can anneal when resection is sufficient to reveal complementary single-stranded regions. Single-stranded tails are removed by nucleases, and the resulting gaps and nicks are filled in by DNA repair synthesis and ligation. This process is accompanied by deletion of one of the repeats and the DNA between the direct repeats and is therefore considered to be mutagenic.
GENETIC AND PHYSICAL ASSAYS FOR RECOMBINATION
Although sensitivity to IR is a universal feature of rad52 group mutants, the mutants show considerable heterogeneity in different assays for recombinational repair of DSBs and spontaneous mitotic recombination. A brief description of the commonly used recombination assays and the products that are recovered from wild-type cells follows. Later sections discuss in detail the phenotype of the rad52 group mutants in these assays.
Spontaneous and Induced Heteroallelic Recombination
The term “spontaneous recombination” refers to events that occur during normal growth rather than following treatment of cells with DNA-damaging agents such as UV light or IR. These events most probably result from recombination initiated by spontaneous lesions that arise from replication fork arrest or collapse, intermediates in base excision repair, aberrant repair, or transcription. Strand breaks that occur in the context of the replication fork are most likely to be repaired using the sister chromatid, an event that is normally genetically silent. Spontaneous recombination can be detected between chromosome homologues in diploid cells or between artificial duplications in haploids or diploids. In most cases, the two recombining sequences contain mutant alleles (heteroalleles) of a selectable gene to allow selection for recombination events during growth of a culture. Heteroallelic recombination in diploids generally occurs by gene conversion with low (10 to 20%) associated exchange of flanking markers. Understanding the mechanisms of spontaneous recombination is difficult because these events occur at low frequency (around 10−6/cell/generation) and are usually selected. Although both products of a G1 event should be retained, the products of an event initiated during the S or G2 phase of the cell cycle will segregate to different daughter cells 50% of the time and only one of these will be recovered by selection (Fig. 2). The frequency of heteroallelic recombination can be increased by up to 1,000-fold when cells are treated with UV light or IR; these events are referred to as induced recombination. Insertion of the recognition sequence for a rare-cutting endonuclease, such as HO, within one of the repeats results in very high levels of recombination when HO endonuclease is induced and allows the analysis of unselected events. Analysis of HO-induced gene conversion has shown that the level of associated crossing over is similar to that for spontaneous events (217, 261).
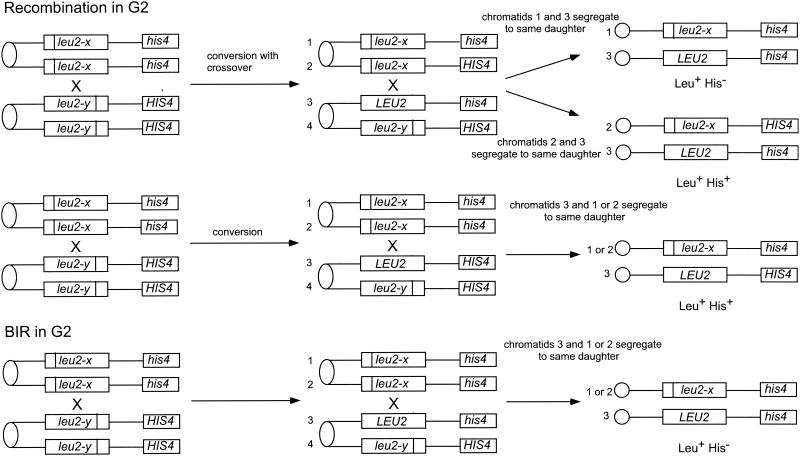
FIG. 2.
Mitotic recombination in diploids. A diploid containing leu2 heteroalleles and heterozygous for HIS4 is shown. Gene conversion associated with crossing over gives rise to Leu+ His+ or Leu+ His− recombinants, whereas gene conversion events maintain heterozygosity at HIS4. A BIR event cannot be distinguished from a G2 crossover unless the reciprocal product is recovered.
Haploid strains containing duplicated genes have been used extensively to study mechanisms and genetic control of mitotic recombination (177). Chromosomal direct repeats can be generated by targeting the integration of a plasmid containing a yeast gene to the homologous chromosomal location to form a nontandem duplication (Fig. 3). Recombination between the repeats can occur by conversion, maintaining the structure of the duplication, or by a deletion event that removes one repeat and the intervening DNA (176, 319, 391). Deletion events between direct repeats can be directly selected if the intervening DNA contains a marker with counterselection, for example URA3, or if the repeats are truncated genes that, by deletion, restore a functional copy of the gene. Conversion events between repeats can occur by an intrachromatid or sister chromatid interaction. Gene conversion between misaligned sister chromatids can generate a triplication on one sister while retaining the direct repeat on the other chromatid. Triplications can also occur by unequal crossing over between sister chromatids, but in this case the reciprocal product is equivalent to a deletion event. Because spontaneous recombination events occur at low frequency, events are usually selected and therefore the reciprocal product is not recovered. Intrachromatid gene conversion associated with crossing over should generate a deletion on the chromosome and an extrachromosomal circular product. It is clear that deletions are produced at high frequency between direct repeats, but the reciprocal product expected from a conservative crossover event is produced in only about 7% of deletions (316). This suggests that intrachromatid gene conversion is rarely associated with crossing over or that most gene conversion events occur between misaligned sisters rather than by intramolecular interactions.
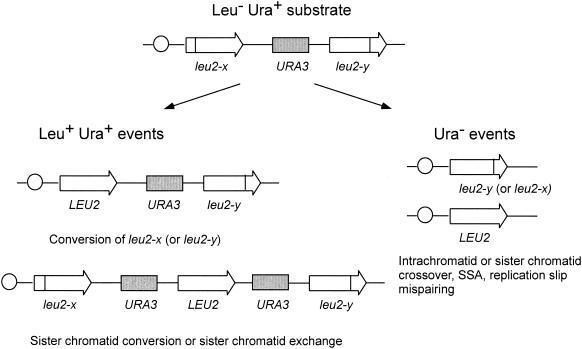
FIG. 3.
Use of a direct-repeat substrate to measure mitotic recombination. The substrate contains leu2 heteroalleles separated by a copy of the URA3 gene. Leu+ Ura+ recombinants can be generated by intrachromatid or sister chromatid conversion or by unequal sister chromatid exchange. Ura− recombination events can occur by an intrachromatid or unequal sister chromatid crossover, by SSA, or by replication slip mispairing.
HO-induced recombination between repeats can occur at high enough frequency to be studied nonselectively in order to recover both products of a recombination event and to monitor the kinetics of DSBR in synchronized populations of cells (260, 312). When the HO-cut site is within one of the repeats, the same spectrum of recombination events found for spontaneous events is recovered, including conversions, deletions, and triplications. The high frequency of triplication or deletion events recovered in one study suggests that sister chromatid interactions are at least as frequent as intrachromatid events (416). Spontaneous deletion events between direct repeats are assumed to occur by SSA, mispairing during replication, or replication slippage rather than by a reciprocal exchange between the repeats. A study of HO-induced recombination involving direct repeats of lacZ contained on a CEN plasmid supports the idea that SSA is a major pathway for DSB-induced deletion formation between direct repeats (101). However, in another study, evidence for DSB-induced reciprocal recombination between direct repeats was presented. An ARS1 element and TRP1 marker were introduced between direct repeats so that an intrachromatid reciprocal exchange could be detected as unstable Trp+ colonies (300). No stable Trp− recombinants were recovered following HO induction, indicating that SSA was not playing a significant role in repair. Of the recombinants, 6% had the unstable Trp+ phenotype indicative of a reciprocal exchange. The reasons for these conflicting results are unclear but could include the length of the repeats, the distance between the repeats, and the location (chromosomal versus plasmid). Deletions are the primary products recovered when the HO-cut site is within unique sequences between the repeats and appear to result from SSA (347, 362) (Fig. 1D).
One concern about the use of direct repeats to study recombination is that deletions can occur by a variety of mechanisms (177). Furthermore, direct repeat recombination still occurs at high frequency in most of the rad52 group mutants (199, 233). In efforts to avoid SSA and replication-associated events, a number of studies have used repeats in inverted orientation (Fig. 4). These generally contain additional sequences between the repeats because perfect palindromes are unstable (118, 313). In most cases the repeats consist of heteroalleles or truncated genes, and in one study inversions were selected by increased expression of a reporter gene present between the repeats (4, 5, 421). To facilitate the detection of recombination events by a colony color assay, Rattray and Symington generated an inverted-repeat substrate from ade2 heteroalleles (299). ade2 mutants accumulate a red pigment, resulting in the formation of red colonies; recombination events that restore a functional ADE2 gene result in white sectors within the red colony. Recombination events within the ade2 reporter can occur by gene conversion or by inversion, and these events occur with equal frequency in wild-type strains (299). Although inversions between inverted repeats could conceivably occur by an intrachromatid crossover, recent studies suggest other mechanisms. Chen and Jinks-Robertson (56) presented evidence in support of long conversion tracts between misaligned sister chromatids to generate inversions (Fig. 4B), as originally suggested by Rothstein et al. (308). Another model suggests that BIR followed by SSA could generate inversions between inverted repeats (166).
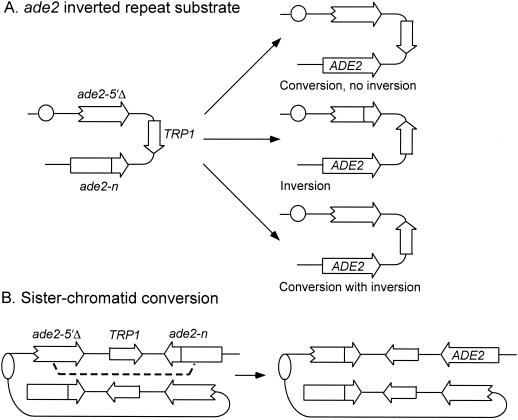
FIG. 4.
Inverted-repeat substrates. (A) The ade2 heteroalleles can recombine to Ade+ by conversion or inversion (apparent crossover) of the intervening DNA. (B) A long-tract conversion event between sister chromatids can result in an Ade+ product with the intervening DNA inverted.
Repeats present on nonhomologous chromosomes (ectopic), usually as heteroalleles, have also been used to study recombination (156). If the repeats have the same orientation with respect to the centromeres, reciprocal crossovers can occur to form viable products. The association of crossing over with gene conversion is about 20%, similar to that observed for recombination between homologous chromosomes (156). The advantage of using an ectopic recombination assay is that simple SSA events cannot give rise to recombinants; the disadvantage is the low frequency at which ectopic recombination occurs, compared with direct or inverted repeats.
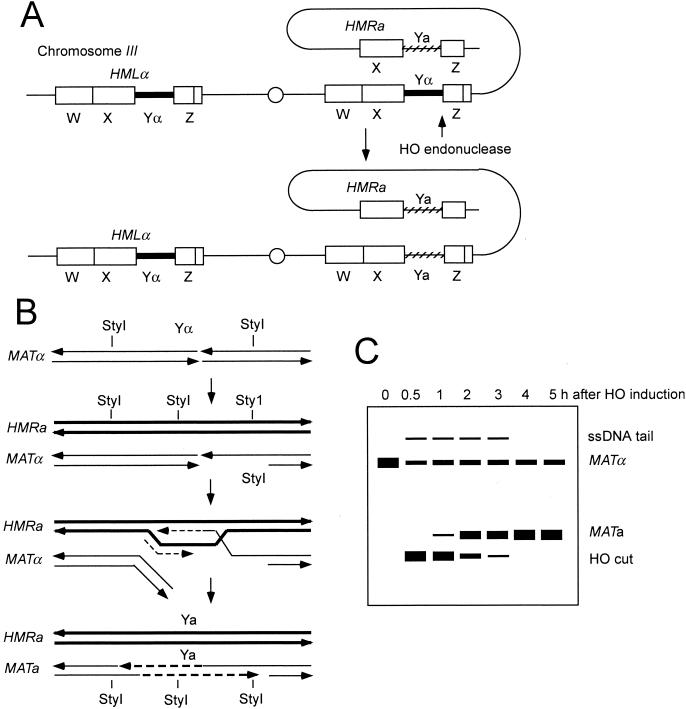
Mating-Type Switching
The budding yeast mating-type switching system provides a useful tool to study intermediates in DSBR and the genetic control of this process (124) (Fig. 5). The HO gene is normally expressed in late G1, but expression can be regulated using a GAL1-HO cassette so that HO endonuclease can be synchronously induced in a population of cells by addition of galactose to the growth medium (132). After the formation of a DSB at the MAT locus by the HO endonuclease, the ends are processed to form 3′ single-stranded tails (419). The 3′ single-stranded tail CEN distal to the cut site invades the donor cassette to initiate DNA synthesis by using components of both leading- and lagging-strand synthesis (137). The initial strand invasion and extension step can be monitored by PCR, and the formation of completed products can be detected by using novel restriction fragments (419). Branched DNA molecules have not been reported as intermediates during mating-type switching, suggesting that these intermediates are less stable than intermediates in meiotic recombination. The ability of mutant strains to switch mating type can also be determined by monitoring the survival of cells following HO expression and analyzing the survivors for mating phenotype (221, 414).
FIG. 5.
Mating-type switching. (A) Cartoon of chromosome III showing the silent cassettes, HMLα and HMRa and the expressed MATα locus. HO endonuclease cleaves between the Y and Z sequences, and repair using the HMRa donor results in switching to MATa. Regions of homology flanking Yα and Ya are indicated by W, X, and Z. (B) After HO cleavage, the 5′ end on the distal side of the break is resected and the resulting 3′ single-stranded tail invades the HMRa locus. DNA synthesis is primed from the invading strand, duplicating Ya sequences. Lagging-strand synthesis initiated from the D-loop results in synthesis of the other strand. The mechanism for removal of the Yα strands is currently unknown. (C) Schematic representation of a Southern blot showing the kinetics of mating-type switching. Switching to Ya results in transfer of a novel StyI site and therefore can be monitored by Southern blot analysis of DNA extracted after HO induction and digested with StyI. Resection of the 5′ strands beyond the distal StyI site results in resistance to digestion to StyI because the site is within ssDNA.
As described above, the HO cut site can be inserted at other locations to create an initiation site for recombination. The advantage of this system over inducing events by irradiation is that the precise location of the initiating lesion is known. Furthermore, the high efficiency of cutting by HO endonuclease allows physical monitoring of recombination, as described for mating-type switching, and recovery of all the products of a recombination event. The Haber laboratory has used the HO system extensively to characterize the mechanisms of direct- and inverted-repeat recombination, as well as allelic recombination in diploids (271). The high efficiency of cleavage by HO endonuclease is currently being exploited to identify proteins that associate with DSBs in vivo by the chromatin immunoprecipitation method (91).
To study DSB-induced recombination in mammalian cells, several groups have made use of the rare-cutting HO and I-SceI endonucleases. I-SceI is a site-specific endonuclease responsible for intron mobility in mitochondria of yeast and initiates a gene conversion event that resembles HO-induced gene conversion (292). Adenoviruses expressing the HO gene or containing the HO cut site can be used to infect human cells in culture (262). Coinfection with the two viruses results in efficient cutting of viral genomes containing the cut site, and these are substrates for end-joining repair (262). Direct-repeat recombination substrates containing a cleavage site for I-SceI have been stably transfected into several mammalian cell lines (157, 310). The I-SceI endonuclease was expressed by transient transfection using the constitutive cytomegalovirus promoter. Although the efficiency of cleavage was lower than that found for I-SceI or HO endonucleases in yeast, the frequency of homologous recombination was induced more than 100-fold in the presence of I-SceI.
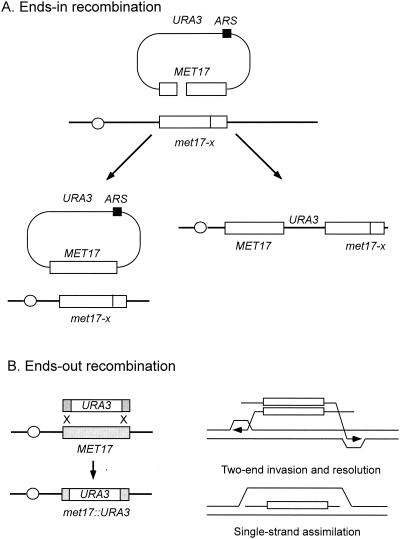
Ends-In and Ends-Out Recombination
The early studies by Orr-Weaver et al. demonstrated efficient repair of DSBs and gaps in plasmids by homologous recombination during yeast transformation (268, 269). The repair of breaks and gaps on plasmids is referred to as “ends-in” recombination. Subsequent studies showed that homologous linear DNA fragments could be used to replace chromosomal sequences during transformation (309). This method of gene replacement or gene targeting is routinely used to knock out genes. Since the orientation of the two ends is opposite to that for plasmid gap repair, gene replacement is sometimes referred to as “ends-out” recombination. The major difference between ends-in and ends-out events is that during ends-in recombination one invading end primes DNA synthesis toward the other end of the broken plasmid, much the same as in a chromosomal DSB, whereas replication is primed away from the fragment during ends-out recombination (Fig. 6). There is also evidence that ends-out recombination can occur by assimilation of a single strand from the linear transforming fragment into the genome to form a displacement loop (D-loop) (194). Subsequent repair of the D-loop restores the original marker or incorporates information from the transforming fragment into the chromosomal locus.
FIG. 6.
Gene targeting. (A) When yeast cells are transformed with a replicating plasmid (ARS plasmid) containing a double-strand break or gap within a region of the plasmid with homology to the yeast genome, the break or gap is repaired by gene conversion from the chromosomal locus. The plasmid remains episomal if gene conversion without an associated crossover occurs but is integrated into the genome if conversion is associated with crossing over. If the plasmid contains a CEN sequence, only conversion events can give rise to viable transformants. If the plasmid contains no origin of replication (ARS), only integration events are observed. (B) Ends-out gene targeting refers to replacement of chromosomal sequences with sequences present on a linear DNA fragment introduced into cells by transformation. Gene targeting is thought to occur by invasion of the two ends into the chromosomal locus followed by resolution of the resulting Holliday junctions. Extensive DNA synthesis could be primed from the invading 3′ ends prior to Holliday junction resolution, or resolution could occur by replication to the end of the chromosome. Gene targeting could also result from integration of a single strand of the targeting fragment followed by trimming the D-loop and mismatch repair.
Genetic and Physical Assays for Meiotic Recombination
Analysis of the segregation of heterozygous markers during meiosis is used to measure recombination in sporulation-proficient strains. Gene conversion is detected as a departure from the normal 2:2 segregation of heterozygous markers and crossing over by new linkage arrangements between linked heterozygous markers. However, this method is not very useful for strains that fail to sporulate or that give rise to inviable spores. Diploid cells induced to undergo sporulation can return to vegetative growth when plated on rich medium (87, 334). The time at which cells commit to high levels of recombination precedes the time when they commit to completion of the sporulation program; therefore, when diploids are plated on rich medium after several hours of growth in sporulation medium, they experience high levels of recombination but retain the diploid state. The frequency of heteroallelic recombination increases 100- to 1,000-fold when cells initiate meiotic recombination and are returned to vegetative growth. Because the return-to-growth protocol does not rely on the formation of viable spores, it can be used to measure the induction of meiotic recombination in sporulation-defective mutants.
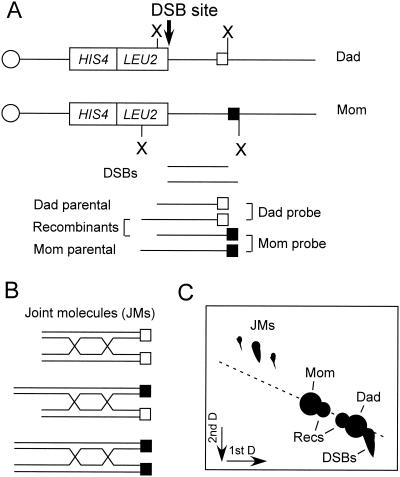
The development of physical assays to monitor the kinetics of formation and processing of DNA intermediates has been invaluable for understanding the mechanisms of recombination (Fig. 7). Also, physical methods can be used to detect recombination intermediates in strains that fail to produce viable spores. In wild-type cells, DSBs are formed about 1 h after DNA synthesis at recombination initiation sites (defined by genetic methods) (46, 369). The ends are processed to form 3′ single-stranded tails of about 600 nucleotides that are the substrates for strand invasion to form heteroduplex DNA (370). Heteroduplex DNA can be detected when the mismatches formed are poorly recognized by the mismatch repair system and therefore persist in DNA (198, 255). Restriction site polymorphisms flanking an initiation site can be used to detect branched intermediates by two-dimensional gel electrophoresis and the formation of crossover products (36, 46, 61, 328). Two types of branched-DNA intermediates have been detected during meiosis: SEIs and dHJ intermediates (11, 144). Kinetic studies suggest that SEIs arise before dHJ intermediates and could be precursors to them or could be processed independently to form noncrossover products (11, 144). The formation of crossover products detected by novel restriction fragments occurs almost simultaneously with the meiosis I division, suggesting a tight coupling between resolution of recombination intermediates, exit from pachytene, and chromosome segregation.
FIG. 7.
Physical assays for meiotic recombination. (A) The hot spot created by insertion of the LEU2 gene at HIS4 in the SK1 strain background is shown. The end of the LEU2 gene contains a site for meiosis-specific DSBs flanked by restriction sites that can be used to distinguish between DSB fragments, and the two parental and two recombinant fragments. The white and black boxes indicate insertions of heterologous sequences used to distinguish the parental molecules by using strand-specific probes. (B) Three types of joint molecules are expected: Mom-Mom intersister joint molecules, Dad-Dad intersister joint molecules, and Mom-Dad interhomologue joint molecules. Size and hybridization to strand specific probes distinguish the three types of joint molecules. (C) Cartoon of a neutral-neutral two-dimensional gel showing separation of joint molecules from the parental and recombinant fragments. The interhomologue joint molecules are more abundant than the intersister joint molecules in wild-type cells. (Adapted from Fig. 1 of reference 329 with permission)
GENETIC AND BIOCHEMICAL PROPERTIES OF THE RAD52 GROUP GENES AND PROTEINS
The RAD52 group genes can be broadly grouped into the MRE11, RAD50, XRS2 (NBS1) subgroup and the RAD51, RAD52, RAD54, RAD55, RAD57, RAD59, RDH54/TID1 subgroup. MRE11, RAD50, and XRS2 are implicated in the formation and processing of DSBs during meiotic recombination and also function in the end-joining pathway of repair, telomere maintenance, in DNA replication-associated repair, and in the DNA damage checkpoint in mitotic cells. The RAD51 subgroup appears to function only in homologous recombination and is described separately.
Mre11-Rad50-Xrs2 (Nbs1) Complex
Yeast strains with null mutations of MRE11, RAD50, or XRS2 have very similar phenotypes. All of the mutants show poor vegetative growth, high sensitivity to IR, and defects in meiosis (7, 110, 149). The three proteins interact in the two-hybrid system, and coimmunoprecipitation studies have confirmed that they form a stable complex (160, 407). Although all three proteins can be coimmunoprecipitated from wild-type cells with antibodies directed against any one of the components, Rad50 and Xrs2 fail to interact in the absence of Mre11, suggesting a central role for Mre11 in complex formation (407). The Mre11 and Rad50 proteins are conserved in all kingdoms of life, whereas Xrs2 appears to be weakly conserved and is present only in eukaryotes. The human gene, HsMRE11, was fortuitously recovered in a two-hybrid screen for DNA ligase I-interacting proteins (282). Using antibodies against HsMre11, Petrini and coworkers identified a large protein complex containing HsRad50 and a 95-kDa protein (78). The 95-kDa protein was subsequently shown to be mutated in humans with the rare autosomal recessive trait NBS (47, 411). Consequently, p95 is referred to as Nbs1 or Nibrin. Nbs1 is considered functionally analagous to Xrs2. Hypomorphic alleles of MRE11 are found in persons with a different human chromosomal instability syndrome, A-TLD (358). At the cellular level, ataxia-telangiectasia (A-T), A-TLD, and NBS are very similar and are characterized by sensitivity to IR, radioresistant-DNA synthesis, and chromosome instability (primarily translocations) (280).
Although mre11, rad50, and xrs2 null mutants of yeast are viable, MRE11, RAD50, and NBS1 are all essential for the viability of vertebrate cell lines and for mouse early embryonic development (215, 426, 428, 431). An mre11 conditional chicken cell line was generated by deleting both copies of MRE11 in the DT40 cell line, in the presence of a transgene expressing MRE11 from a tetracycline-repressible promoter (428). Down regulation of the gene resulted in rapid depletion of Mre11 concomitant with increased radiosensitivity, increased levels of spontaneous chromosome breaks, arrest in the G2 phase of the cell cycle, and eventual cell death. Similarly, antibody depletion of Mre11 from Xenopus extracts resulted in aberrant DNA synthesis accompanied by the formation of DSBs (67). These studies suggest an essential role of the Mre11 complex in the repair of lesions generated during S phase in vertebrates.
Localization of the MRN complex to DSBs in vivo.
Immunofluorescence has been used to localize the Mre11-Rad50-Nbs1 (MRN) complex in response to DNA damage in vertebrate cells. In unirradiated cells, Mre11 and Rad50 are uniformly distributed throughout the nucleus, but after exposure to IR, both proteins form discrete nuclear foci (226). Foci that are microscopically visible are thought to represent sites of repair. Discrete foci were not detected after UV irradiation, suggesting that Mre11 and Rad50 associate specifically with DSBs. Formation of Mre11 and Rad50 IR-induced foci was eliminated in cells from NBS and A-TLD patients, consistent with a defect in the DNA damage response (47, 358). In a highly innovative study, soft X rays were used to form discrete areas of DNA damage within the nuclei of fibroblasts and areas of repair localized by incorporation of bromodeoxyuridine (BrdU) (assayed using an anti-bromodeoxyuridine monoclonal antibody) by terminal deoxynucleotidyltransferase (258). This study suggests that DSBs are held in a relatively fixed position during the early stages of DNA repair. At 30 min after irradiation, Mre11 colocalized with BrdU, suggesting that the MRN complex migrates to the sites of DSBs within the nucleus. Discrete areas of DNA damage have also been generated using a laser scissor method (279). The phosphorylated form of H2AX (γ-H2AX), a nonessential member of the histone H2A family (48), associated with the sites of damage within 30 min, and HsRad50 was found to colocalize extensively with γ-H2AX. Treatment of cells with IR also resulted in the formation of γ-H2AX foci, but in this case Rad50 did not colocalize until 6 to 8 h after irradiation. Preirradiation exposure of cells to the fungal metabolite wortmannin, which inhibits phosphatidylinositol 3-kinases, prevented the formation of γ-H2AX and Rad50 foci, suggesting that γ-H2AX is required to recruit Rad50 to sites of damage (279).
Mre11 is associated with chromatin in replicating Xenopus extracts and appears to be important either for prevention of replication-induced breaks or for their repair (67). Immunofluorescence studies of detergent extracted cells also show association of Mre11 with sites of replication based on colocalization of Mre11 with proliferating-cell nuclear antigen during S phase (225). The Mre11 complex could play an important role during replication or could be associated with the fork in preparation for damage signaling or repair.
Structural and biochemical studies.
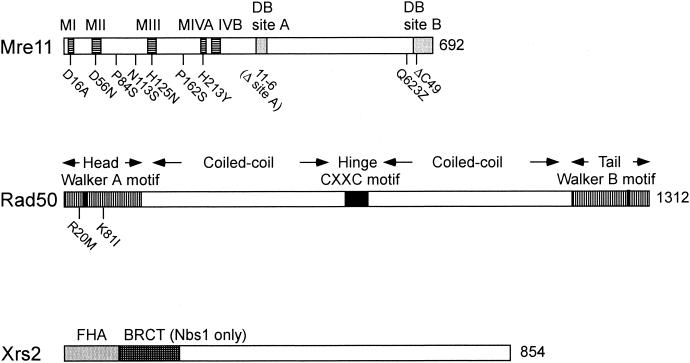
The 83-kDa Mre11 protein is highly conserved among eukaryotes, and the N-terminal region has several sequence motifs shared by a large family of phosphodiesterases, including the Escherichia coli SbcD and bacteriophage T4 gp46 nucleases and protein phosphatases (333) (Fig. 8). Yeast and human Mre11 proteins have single-stranded DNA (ssDNA) endonuclease and weak 3′-to-5′ exonuclease activities (108, 242, 277, 397, 407). Like SbcD (62, 63), the Mre11 nuclease activities are dependent on manganese as a cofactor. An allele of MRE11, mre11-58 (rad58), originally thought to represent a new gene in the RAD52 epistasis group, has a point mutation within the fourth phosphodiesterase motif (H213Y), and the encoded protein is devoid of nuclease activity in vitro (400, 407). Several other mutations have been made at conserved residues within the other phosphodiesterase motifs (mre11-D16A, mre11-D56N, and mre11-H125N), and all abolish the endonuclease and 3′-to-5′ exonuclease activities in vitro (108, 241). The aspartic acid residues at positions 16 and 56 are directly involved in coordination of two Mn2+ ions at the active site, and His125 stabilizes the transition state intermediate (139). Two-hybrid and size exclusion chromatography analyses indicate that Mre11 forms a dimer (108, 160). Consistent with these findings, diploid strains expressing an N-terminal mre11 mutation (mre11S) and a C-terminal insertion mutation showed intragenic complementation for meiosis and methyl methanesulfonate (MMS) resistance (256).
FIG. 8.
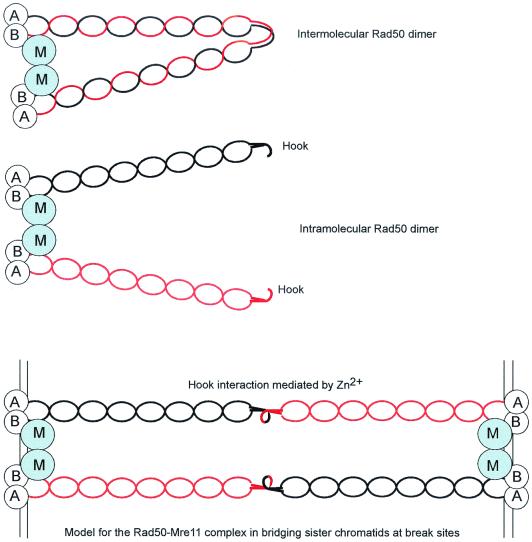
Schematic representation of Mre11, Rad50, and Xrs2 (Nbs1). The phosphodiesterase motifs of Mre11 are labeled MI through MIV, and DNA binding sites are labeled DB site A and B. The Mre11-D16A, Mre11-D56N, Mre11-H125N, Mre11-H213Y, and Mre11-6 mutants are nuclease defective in vitro. Residue Pro84 is mutated in the mre11-S allele, and Pro162 is mutated in the mre11-1 temperature-sensitive allele. The mre11-N113S and mre11-Q623Z alleles correspond to the mutations in the A-TLD patients. Rad50 contains two coiled-coil domains separating the Walker A and B motifs for NTP binding and hydrolysis. The hook domain, containing the conserved CXXC motif, is located between the two coiled-coil domains. The positions of the rad50S alleles, rad50-R20M and rad50-K81I, are shown.
Rad50, like Mre11, is conserved in all kingdoms of life. In bacteria, the Rad50 homologue, SbcC, forms a complex with SbcD, the homologue of Mre11. The SbcCD complex has ATP-dependent 3′-to-5′ exonuclease and ATP-independent single-stranded endonuclease activity (62, 63). The 150-kDa Rad50 protein is related to the SMC proteins, which have the Walker A and B motifs characteristic of nucleotide triphosphate (NTP)-binding proteins separated by a long coiled-coil region (9) (Fig. 8). The SMC proteins, including the Rad50 subgroup, have a conserved hinge region within the coiled region. The hinge region of the Rad50 subfamily is distinct from that of other SMC proteins and contains a conserved Cys-X-X-Cys motif (140). Electron microscopy studies of Rad50 suggest a dimeric structure to bring together the Walker A and B motifs, forming two catalytic sites. The dimer could result from two protomers in an antiparallel configuration or by hinge-mediated interactions between two intramolecularly coiled protomers (12, 139) (Fig. 9). Atomic force microscopy of the human Rad50-Mre11 complex identified two highly flexible intramolecular coiled coils emanating from the globular Rad50-Mre11 DNA binding domain (73). The intramolecular coiled coil places the Walker A and B motifs of one Rad50 monomer together juxtaposed to Mre11. The size of the globular domain is consistent with a dimer of Mre11 and a dimer of the Rad50 ATPase domain. Dimerization of Rad50 is mediated by the CXXC motif within the hinge region (140). Structural analysis of the hinge region revealed a hook structure at the apex of the coiled coil, creating half of a composite metal binding site. Two hook regions coordinate a single Zn2+ ion with the two coiled-coil regions extending in almost opposite directions. This arrangement could potentially bridge sister chromatids because the distance between the head domains of the Mre112Rad502 complex is approximately equal to the distance between sister chromatids (1,200 Å) (140). Another class of SMC proteins, called cohesins, is known to mediate sister chromatid cohesion (128). The Mre11-Rad50 complex could function specifically in sister chromatid interactions during DSBR, as suggested by genetic studies. Electron microscopy studies have provided direct evidence for end binding by the human and yeast Mre11-Rad50 complex and bridging of different DNA molecules (53, 73).
FIG. 9.
Models for the Rad50-Mre11 complex. A dimer of Rad50 could form by antiparallel intermolecular interaction to position the Walker A motif from one monomer next to the Walker B motif of the other. A dimer of Mre11 binds adjacent to the head-tail region of Rad50. Recent results are more consistent with an intramolecular interaction between the coiled-coil domains of Rad50, with the dimer held together by a dimer of Mre11. Interactions between the hinge domains could connect two dimers of Rad50 and two dimers of Mre11. The length of the Rad50 tetramer is consistent with the distance between sister chromatids in eukaryotes. The Mre11 and Rad50 head-tail domains are envisioned to interact with DNA.
Structural studies of the Pyrococcus furiosis Rad50 catalytic domain (Rad50cd) indicate a structure similar to the ATP binding cassette transporter family of ATPases (142). Binding of the Rad50cd to the ATP analog AMP-PNP induces a structural change to align the Walker A and B motifs. The ATP-bound form of the Rad50cd binds DNA more tightly, suggesting that ATP regulates DNA binding by conformational switching. Mutation of the conserved lysine residue within the Walker A box confers a null phenotype in yeast, indicating the importance of ATP binding and/or hydrolysis to function (8). The DNA binding activity of yeast Rad50 is stimulated by ATP, but no ATPase activity has been observed for the purified protein (301).
Rad50 stimulates the nuclease activities of yeast and human Mre11. Unlike the EcSbcC-SbcD complex and the PfMre11-Rad50 complex, the complex of HsMre11 with HsRad50, or HsMre11, HsRad50, and Nbs1 has no demonstrated ATP-dependent exonuclease activity (63, 141). ATP stimulates a weak DNA unwinding activity of the HsMre11-Rad50-Nbs1 complex, and hairpin cleavage has also been observed for the complex (278). However, the yeast Mre11-Rad50 complex cleaves hairpin structures in the absence of Xrs2 (396).
In mammals, Nbs1 (p95) appears to be the functional homologue of Xrs2 in that it is tightly associated with Mre11 and Rad50, but sequence similarity to Xrs2 is limited to the N-terminal 115 amino acids (47, 411). Xrs2 and Nbs1 have no obvious sequence motifs indicative of function. Nbs1 contains two domains in the N-terminal region that are found in DNA damage-responsive cell cycle checkpoint proteins (95). A forkhead-associated (FHA) domain, which is thought to be important for interactions between phosphorylated proteins, is found at the N terminus of Nbs1. The BRCT (breast cancer carboxy-terminal) domain, which is adjacent to the FHA domain in the N-terminal region of Nbs1, is found in a variety of proteins that participate in DNA damage-responsive cell cycle checkpoints. To date, the only biochemical activity assigned to Nbs1 is stimulation of the unwinding and hairpin cleavage activities of HsMre11 and HsRad50 (278).
Meiotic phenotype of mre11, rad50, and xrs2 mutants.
Diploid strains homozygous for mre11, rad50, or xrs2 fail to form meiosis-specific DSBs and thus are unable to initiate meiotic recombination (8, 160). This results in the formation of aneuploid (inviable) spores in some yeast strain backgrounds (e.g., SK1) or a complete block of the sporulation pathway in others (e.g., W303). The spo13 mutation causes cells to bypass the first meiotic division, and thus spo13 strains no longer require recombination for segregation of homologous chromosomes. spo13 suppresses the sporulation and spore viability defects of mutant strains unable to initiate DSB formation, including mre11, rad50, and xrs2 (7, 149, 222). Because of the failure to induce meiosis-specific DSBs, mre11, rad50, and xrs2 null mutants show no induction of meiotic heteroallelic recombination in the return-to-growth protocol or recombinants among viable dyads derived from spo13 meiosis (7, 149, 222). Several separation-of-function alleles of RAD50 have been identified that are proficient for DNA repair in mitotic cells but sporulation defective (8). The sporulation defect conferred by rad50S alleles cannot be suppressed by spo13 but can be suppressed by spo13 plus spo11 (defective for initiation of recombination). Meiotic DSBs are still formed in rad50S mutants, but the ends are not processed to generate 3′ single-stranded tails and are stable rather than transient (8, 46, 370).
The role of MRE11, RAD50, and XRS2 in DSB formation is unclear. MRE11 is required for a meiosis-specific alteration of chromatin structure at recombination hot spots, and the chromatin alteration and DSB formation functions of Mre11 are eliminated by a deletion removing the C-terminal 49 residues (108, 266). This region of Mre11 contains one of the two DNA binding sites of Mre11 defined by in vitro studies (407). At least 10 genes are required for DSB formation; one of these, SPO11, is known to play a direct role since it encodes the catalytic subunit of the DSB-forming complex (170). Spo11 is an atypical type II topoisomerase that catalyzes DSB formation by a transesterification mechanism. MRE11 and RAD50 also appear to function after the formation of meiosis-specific DSBs as evidenced by several separation-of-function alleles that are still proficient for DSB formation but are defective in DSB processing. In rad50S mutants, Spo11 remains covalently associated with the 5′ ends at break sites (170). Several alleles of MRE11 have been identified that confer a similar phenotype to rad50S in meiosis (mre11S, mre11-58, mre11-6, mre11-D16A, and mre11-H125N) (108, 241, 256, 400, 407). Although covalent attachment of Spo11 at break sites has not been demonstrated for all of these mutants, it is assumed to occur because they all result in the accumulation of unprocessed breaks during meiosis. mre11S was isolated in a screen for mutants that showed RAD50-dependent spore inviability (256). From the same genetic screen, the SAE2/COM1 gene was identified (295), as well as from a screen for SPO11-dependent spore inviability (235). Null mutations in SAE2/COM1 confer a phenotype very similar to that of rad50S and mre11S mutants; accumulation of unprocessed DSBs in meiosis.
Two models have been proposed for the removal of Spo11 from break sites. First, Spo11 could be removed while still covalently attached to the 5′ strand by the endonucleolytic activity of the Mre11-Rad50-Xrs2 (MRX) (Sae2?) complex. Second, Mre11, Rad50, and Sae2 could be required for the reversal of the Spo11 transesterification reaction. The first model predicts that resection of the 5′ strand is intrinsic to removal of Spo11, whereas the second model predicts that resection of the 5′ strand occurs after Spo11 removal. Support for the first model comes from the observation that strains containing nuclease-defective alleles of MRE11 (mre11-D16A, mre11-H125N and mre11-58) have unprocessed DSBs.
Role of the MRX complex in mitotic recombination.
The complex phenotype of mre11, rad50, and xrs2 mutants is even more apparent in vegetative cells. Although it is generally assumed that yeast cells repair IR-induced DNA damage by homologous recombination, mre11, rad50, and xrs2 mutants show little or no defect in spontaneous mitotic recombination. Spontaneous heteroallelic recombination is elevated about 10-fold in mre11, rad50, and xrs2 diploids and still shows some induction by DNA-damaging agents, but not to the extent observed in wild-type cells (7, 149, 314). Because diploids in the G2 stage of the cell cycle preferentially repair lesions from a sister chromatid instead of a homologue, it has been suggested that MRE11, RAD50, and XRS2 are specifically involved in sister chromatid recombination (8, 42, 149). In support of this hypothesis, mre11 diploids are more resistant to irradiation than are haploids; G1 diploids show the same sensitivity to IR as do wild-type strains, and mutant G2 haploids and diploids both show high IR sensitivity (42). In a genetic assay designed specifically to detect sister chromatid recombination (94), mre11 mutants showed a slight decrease in the rate of recombination but not to the extent expected if MRE11 was essential for this process (42). rad52 mutants show a 50-fold reduction in sister chromatid recombination in the same assay system. The hyperrecombination phenotype exhibited by mre11, rad50, and xrs2 mutants for heteroallelic recombination in diploids could be due to channeling lesions from the normal sister chromatid repair pathway into interactions between homologues (8, 149).
Spontaneous deletion between direct repeats occurs at wild-type frequency in rad50 mutants (119, 233). However, the recombination products were not studied in sufficient detail to determine whether there were defects in sister chromatid events using this system. Using the ade2 inverted-repeat assay (Fig. 4), a threefold reduction in spontaneous recombination was reported for rad50 and xrs2 mutants, with the same distribution of events as found in the wild-type strain (298). Ectopic recombination between ura3 heteroalleles on chromosomes II and V was unaffected by a rad50 mutation in haploids but showed a threefold increase in rad50 homozygous diploids (357). As suggested above for heteroallelic recombination, the hyperrecombination phenotype could be due to channeling lesions from sisters to interchromosomal interactions.
Studies of HO-induced recombination have also revealed only modest defects in these mutants (151, 400). The most striking finding that emerged from studies of mating-type switching is the decreased extent of processing of the 5′ strand at the HO-cut site in the null mutants (151, 362, 400). This result, in combination with the defect in processing of meiosis-specific breaks in certain mre11 and rad50 mutants, led to the suggestion that the MRX complex resects ends to produce 3′ single-stranded tails. However, even in mre11, rad50, or xrs2 null mutants, processing does occur, and while there is a delay in mating-type switching, most cells are able to complete the process with fairly high efficiency. HO-induced SSA between chromosomal direct repeats is also delayed by 1 to 2 h in rad50 mutants, but cells are able to repair the break with only a twofold decrease in viability (362).
Role of the Mre11 nuclease in processing DSBs in mitotic cells.
The observation that HO-induced DSBs are processed more slowly in mre11, rad50, and xrs2 null mutants suggests that the MRX complex is directly involved in end processing or regulates the activity of a nuclease. Because the exonuclease activity of Mre11 is of the opposite polarity to that expected for resection of DSBs, the endonuclease activity is thought to be targeted to the 5′ strand by an as yet unknown mechanism. The role of the Mre11 nuclease in resection has been investigated by generating nuclease-defective alleles of MRE11 for analysis of end processing in vivo. The phenotype of the mre11-58 (rad58) strain is due to mutation of a conserved residue in phosphodiesterase motif IV (H213Y) (400). The Mre11-58 mutant protein is proficient for DNA binding but lacks exonuclease and endonuclease activities in vitro and fails to interact with Rad50 and Xrs2 when immunoprecipitated from yeast extracts (407). The phenotype of the mre11-58 strain is very similar to that of the mre11 null mutant: high sensitivity to MMS, delayed kinetics of mating-type switching, and elevated rates of mitotic heteroallelic recombination (400). The Mre11-D56N and Mre11-H125N mutant proteins also lack nuclease activity (241), and Mre11-H125N retains interaction with Xrs2 and Rad50 (S. Moreau and L. S. Symington, unpublished data). These two mutations confer much less severe mitotic phenotypes than does the mre11Δ allele. Strains containing either the mre11-D56N or mre11-H125N allele show intermediate sensitivity to IR and normal levels of spontaneous mitotic recombination and plasmid gap repair, and the kinetics of mating-type switching are indistinguishable from those in the wild type (241, 379). These observations suggest that the Mre11 nuclease may not be involved in the resection of mitotic DSBs or is redundant with another nuclease. The more severe phenotype conferred by the mre11-58 allele could be a consequence of defective complex formation rather than the nuclease defect. Bressan et al. (43) generated mutations within all four phosphodiesterase motifs. The mre11-2 and mre11-4 alleles conferred phenotypes indistinguishable from those conferred by the null mutation for radiation sensitivity and spontaneous mitotic recombination. These two proteins both contain nonconservative substitutions of two amino acids. Both mutants failed to interact with Rad50 in the yeast two-hybrid system, suggesting that the severity of the mitotic DNA repair defect could be due to lack of the MRX complex rather than just loss of the Mre11 nuclease. The mre11-11 and mre11-3 alleles, containing substitutions within motifs I and III, respectively, conferred less severe phenotypes for sensitivity to IR than did the mre11Δ mutation, and both retained interaction with Rad50. Thus, the severity of the phenotype conferred by these alleles is directly correlated with the ability of the mutant proteins to form complexes. The Mre11-D16A protein lacks exo- and endonuclease activities but retains DNA binding; interaction with Rad50 and Xrs2 has not been determined (108). The mre11-D16A mutant shows intermediate sensitivity to MMS, intermediate-length telomeres, and normal rates of spontaneous mitotic recombination, but has not been tested for processing of HO-induced DSBs in vivo (108).
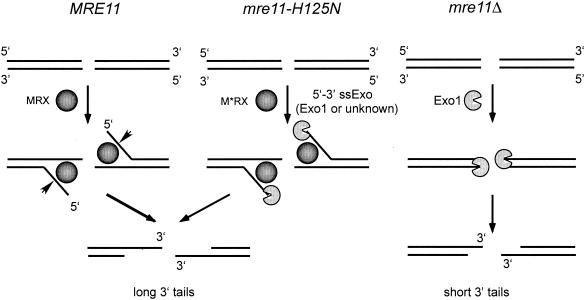
Because mre11-H125N strains are unable to process meiosis-specific DSBs but are proficient at repair of HO-induced breaks, we have suggested that the complex unwinds ends and that nucleases redundant with Mre11 can process free 5′ ends but not ends bound by Spo11 (241). A redundant endonuclease would be expected to substitute for Mre11 in both mitotic and meiotic cells, suggesting that it is a 5′-to-3′ exonuclease that processes ends in the absence of Mre11 in mitotic cells. Alternatively, the redundant activity could be an endonuclease that either is not expressed during meiosis or is excluded from the meiotic DSB-processing complex. The EXO1 gene, which encodes a 5′-to-3′ exonuclease with a twofold preference for double-stranded over ssDNA (98), was found to suppress the mitotic DNA repair defect of mre11 strains when present at high copy number (195, 242, 399). This suppression was also observed for rad50 and xrs2 mutants, suggesting that EXO1 in more than one copy can bypass the requirement for the MRX complex in DNA repair. Furthermore, exo1 mre11 double mutants have a severe growth defect (only about 30% plating efficiency), higher sensitivity to IR and MMS, and a longer delay in mating-type switching compared with mre11 single mutants (242, 399). The exo1 null mutation alone confers no significant sensitivity to IR or mating-type switching defects, and the exo1 mre11-H125N strain has normal kinetics of mating-type switching (98, 242). This contrasts with the severe defect observed for the exo1 mre11Δ double mutant. We envision inefficient processing of duplex ends by Exo1 in the absence of the MRX complex, but the efficiency of processing is greater when EXO1 is present on a high-copy-number plasmid. In mre11-H125N cells there must be a single-strand-specific nuclease activity that can substitute for either the Mre11 or Exo1 nuclease activity but is unable to process duplex ends (Fig. 10).
FIG. 10.
Models for DSB processing by the MRX complex and Exo1. In wild-type cells, ends are processed by unwinding of ends and endonucleolytic cleavage of the 5′ strand by the Mre11 nuclease. Unwinding could be mediated by the weak unwinding activity of the Mre11 complex or by association with a DNA helicase. In the absence of the MRX complex, Exo1 inefficiently processes the ends. The M*RX complex in cells expressing the mre11-H125N allele is still able to unwind ends, and other nucleases remove the 5′ single-stranded tails.
An alternative hypothesis to explain the normal processing of HO-induced breaks in the mre11-H125N strain is that the Mre11 nuclease plays no role in resection and the role of the MRX complex is to recruit the resection nuclease to break sites or to clean up ends for the resection nuclease. The mre11-H125N diploid is unable to process DSBs with Spo11 covalently bound to the 5′ ends and does show sensitivity to high doses of IR. IR causes base and sugar damage in addition to strand breaks, and termini frequently have phosphate or phosphoglycolate groups. One attractive model is for the endonuclease activity of Mre11 to remove damaged nucleotides or protein-DNA covalent adducts (such as Spo11) from ends to provide the substrate for the resection nuclease. Further support for this model comes from recent studies suggesting that Mre11 removes the terminal protein of adenonvirus during infection (359). Adenoviruses use a protein-priming mechanism for replication, resulting in linear duplexes with a protein covalently bound to the 5′ ends. Viruses in which the E4 region is deleted fail to package viruses due to concatemerization of viral genomes (413). Concatemerization of E4-deleted viral genomes was found to require DNA-PKcs, ligase IV, Mre11, and Nbs1, suggesting that concatemers are formed by the end-joining pathway (359). Concatemer formation was restored to the ATLD3 cell line by transfection with wild-type Hs MRE11 but not with the Hs mre11-3 nuclease-defective allele. Infection of cells with wild-type virus results in depletion of Mre11, and this is dependent on the E4 region. Together, these results suggest that one function of E4 is to promote the degradation of Mre11, thus preventing cleavage of the terminal protein from 5′ ends and subsequent concatemerization by end joining.
Recent studies suggest the Mre11 nuclease is important for processing unusual DNA structures, such as hairpins. Insertion of a 323-bp quasipalindrome derived from human Alu elements into the yeast LYS2 gene stimulates the rate of ectopic recombination (with a different lys2 allele) 1,000-fold. This stimulation is dependent on MRE11, RAD50, and XRS2 (207). Interestingly, mre11-D56N, mre11-H125N, rad50S, and sae2Δ mutations also completely eliminate the hyperrecombination induced by the palindrome. The palindrome appears to be extruded to form a cruciform structure, which is cleaved at the base by an unknown activity to form two hairpin ends. These hairpins fail to be resolved in the mutant strains and are replicated, resulting in the formation of an inverted duplication. The failure of the mre11-D56N, mre11-H125N, rad50S, and sae2Δ mutants to resolve the hairpin intermediate directly implicates the nuclease activity of Mre11, the Rad50 function compromised by the K81I mutation, and Sae2 in cleaving these structures in vivo. rad32 (MRE11)- and rad50-dependent stimulation of mitotic recombination by a 160-bp palindrome is also observed in Schizosaccharomyces pombe (93).
An allele of SAE2 was isolated in a screen for mutants that aberrantly process DSBs within an inverted repeat (297). In the sae2 mutant, DSB-stimulated events occurred normally 46% of the time but the other 54% of the events were characterized by a duplication of part of the inverted repeat. These events were hypothesized to occur by break-induced replication through the inverted repeat followed by an end-joining event to form a palindrome structure. These aberrant events were not detected in the wild-type strain, suggesting that they do not occur, or that the palindrome is not stably maintained, in wild-type cells. The mre11-H125N and rad50S mutations conferred the same phenotype as did the sae2 mutation in this assay, again suggesting that the Mre11 nuclease and Sae2 resolve palindromes. The results of these two studies are consistent with the observation that palindromes are stabilized in sbcC and sbcD mutants of E. coli (49, 115).
The RAD27 gene encodes a flap endonuclease that removes RNA primers from Okazaki fragments during DNA synthesis. rad27 mutants are viable but depend on homologous recombination functions (RAD50, RAD51, RAD52, RAD54, RAD55, RAD57, RAD59, MRE11, and XRS2) for viability (378, 393). The sae2Δ, mre11S, mre11-H125N, and rad50S mutations all cause death in a rad27 strain (72, 241). The Mre11 nuclease could be partially redundant with the Rad27 nuclease or could be required to process aberrant DNA structures that accumulate in rad27 mutants. An alternative explanation is that the large number of lesions generated in a rad27 mutant overloads the homologous recombination system so that mutants with subtle DNA repair defects are unable to repair all of them. The similarity in the phenotypes of mre11S, mre11-H125N, rad50S, and sae2Δ mutants suggests that the nuclease activity of Mre11 is absent in these mutants. This has clearly been shown for the Mre11-H125N protein, but the Mre11S protein retains endo- and exonuclease activities (E. A. Morgan and L. S. Symington, unpublished data). This suggests the possibility that the Mre11 nuclease is active in vivo only in the presence of Sae2 and when Rad50 is fully functional. An attractive model is for Sae2 to interact with the MRX complex to activate the Mre11 nuclease and for the Sae2-MRX complex to be disrupted by rad50S and mre11S mutations.
Role of the MRX complex in nonhomologous end joining.
The end-joining pathway of repair requires Ku70 and Ku80, encoded by the YKU70 (HDF1) and YKU80 (HDF2) genes, respectively, a specialized DNA ligase encoded by the DNL4 gene, and a ligase stimulatory factor, Lif1 (XRCC4 in mammals) (271). In mammals, the Ku heterodimer associates with a kinase (DNA-PKcs) to form the DNA-dependent protein kinase (DNA-PK), but to date a similar kinase has not been identified in yeast (83, 120). An additional factor, Lif2/Nej1, which interacts with Lif1 and is regulated by mating type, is required for end joining in yeast (104, 171, 267, 408). Defects in any of the components of this pathway, with the exception of MRE11, RAD50, and XRS2, do not cause IR sensitivity but do increase the IR sensitivity of a rad52 strain in stationary phase. This suggests that the homologous pathway is the primary means of repairing IR-induced damage and that end joining can be used as a backup pathway.
Several assays have been used to measure end joining in yeast. The transformation efficiency of autonomously replicating plasmid DNA that has been linearized with a restriction enzyme to produce cohesive ends is used to measure precise rejoining (38). In wild-type cells, ligation occurs with high fidelity, with no loss or gain of nucleotides at the junction most of the time (38). A similar assay to monitor the repair of chromosomal breaks measures cell survival in response to induction of EcoRI endonuclease in a strain containing a GAL1-regulated EcoRI gene (196). Imprecise end joining can be assayed by survival in response to HO endonuclease induction of a strain that cannot repair the break at the MAT locus by homologous recombination (either by deletion of the donor cassettes or by deletion of RAD52) (240). In all three assays, mre11, rad50, xrs2, and yku70 strains have similar phenotypes and appear to be epistatic (37, 240). Although mre11 and yku70 mutations cause a similar reduction in the efficiency of joining cohesive ends of plasmids, the types of products recovered are different. Repaired plasmids recovered from yku70 and yku80 strains have large deletions flanking the break site and rejoin through short sequence homologies, whereas plasmids recovered from mre11, rad50, and xrs2 strains show mainly faithful repair (37, 38). In the HO assay, faithful repair restores the HO-cut site, which can then be recut by HO. Survivors of continuous HO expression have small deletions or insertions at the HO cut site, which prevent further cutting by HO. The frequency of survivors is reduced in mre11, rad50, and xrs2 mutants, but the survivors have large deletions (240). Although it has been suggested that the Mre11 nuclease could function in processing ends for the end-joining pathway, the characterization of junctions produced in mre11 mutants in yeast is inconsistent with this hypothesis. Furthermore, the mre11-H125N strain is proficient for end joining and EXO1 present on a high-copy-number plasmid is unable to suppress the end-joining defect of mre11 strains (195, 241, 242). S. pombe rad32 (mre11) and rad50 mutants show no defect in a plasmid recircularization assay, raising the issue of how general the S. cerevisiae findings are (223, 422).
The purified MRX complex stimulates intermolecular DNA joining by the Dnl4-Lif1 complex (53). Atomic force microscopy analysis revealed juxtaposition of DNA ends by the MRX complex to form linear concatemers, suggesting that the MRX complex can align DNA molecules for ligation. Interaction between the MRX complex and Dnl4-Lif1 appears to be mediated by Xrs2, suggesting that Xrs2 might function to recruit Dnl4 to ends held together by Mre11 and Rad50. Intermolecular end joining promoted by the human DNLIV-XRCC4 complex and DNA-PK is also stimulated by the addition of the MRN complex (143).
Role of the MRX complex in telomere maintenance.
Strains with a mutation of MRE11, RAD50, or XRS2 have short, but stable telomeres (37, 174). The length of the telomeric repeat tracts in these strains is similar to that of strains with a mutation of TEL1, the yeast homologue of the human ATM gene (121, 216, 249). The tel1 mutation is epistatic to mre11, rad50, and xrs2 for telomere length, and, like tel1, a rad50 mutation confers senescence to mec1 strains (305, 306). Thus, Tel1 and the MRX complex work in the same pathway to maintain normal telomere clength. One attractive hypothesis for the role of the MRX complex in telomere maintenance is to produce a substrate for telomerase, perhaps by generation of a single-stranded tail. However, the mre11-D56N and mre11-H125N strains, defective for the Mre11 nuclease, have normal length telomeres (241; Morgan and Symington, unpublished). Furthermore, the mre11-D56N and mre11-H125N mutations do not cause senescence in a mec1 strain (401). As suggested for processing of HO-induced breaks, there may be other nucleases that can substitute for the Mre11 nuclease as long as the complex is present. Interestingly, overexpression of EXO1 is unable to suppress the short-telomere defect of mre11, rad50, or xrs2 strains, suggesting that the nuclease activity is not important for the function of MRX at telomeres (50, 242, 399). The length of the single-stranded tail at native telomeres in mre11, rad50, and xrs2 strains is sufficient for binding of the TG1-3 DNA binding protein, Cdc13, in asynchronous cells (401). Targeting of the telomerase catalytic subunit (Est2) to telomeres by using a Cdc13-Est2 chimera suppresses the telomere length and senescence phenotype of mre11 and mre11 mec1 strains, respectively, suggesting that one function of the MRX complex at telomeres is to recruit telomerase (401).
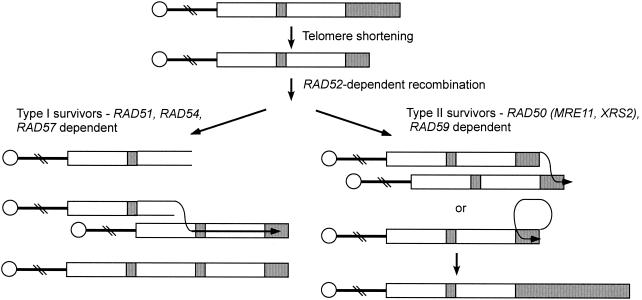
In the absence of telomerase, telomeres become progressively shorter, which leads to cellular senescence (214). Suppressors of this growth defect arise at high frequency due to RAD52-dependent recombination to restore telomere length either by amplification of the subtelomeric Y′ elements (type I survivors) or by formation of very long telomeric tracts (type II survivors) (213, 389) (Fig. 11). The type I survivors have multiple copies of Y′ elements but still have very short TG1-3 tracts. Both types of survivors are generated in a tlc1 strain (deficient for the RNA component of telomerase). Generation of type I survivors requires RAD51, RAD54, RAD55, and RAD57 (55, 190, 388). The type I survivors eventually convert to the type II survivor pattern after several hundred divisions due to the improved growth of type II survivors compared with type I survivors. In rad50 mutants, type I survivors are predominantly recovered, indicating a role for the MRX complex in the generation of long telomeric tracts in the absence of telomerase (55, 388). The type II survivors are thought to arise by inter- or intramolecular recombination between TG1-3 tracts (Fig. 11). The requirement for RAD50 for this process suggests that Rad50 could be important for pairing between short homologies.
FIG. 11.
Telomere maintenance by recombination in the absence of telomerase. In the absence of telomerase, telomeres become progressively shorter and are maintained by RAD52-dependent recombination. Two types of recombination events give rise to survivors: RAD51-, RAD54-, and RAD57-dependent recombination between Y′ sequences, and RAD50- and RAD59-dependent recombination between T G1-3 tracts. Y′ elements are marked by open boxes, and tracts of TG1-3 are indicated by shaded boxes.
Role of the MRX complex in suppression of gross chromosome rearrangements.
Chen et al. described a genetic assay to detect gross chromosome rearrangements (GCR) by measuring the simultaneous loss of two linked subtelomeric markers in haploid yeast strains (52). In wild-type cells such events are rare (3 × 10−10/cell/generation) and are due primarily to loss of the markers followed by telomere addition. The rate of GCRs is increased by 600-fold in mre11, rad50, and xrs2 strains. One-third of the events represent telomere additions after loss of the markers, indicating that de novo telomere formation can still occur in mre11, rad50, and xrs2 mutants. The other 70% of the events recovered from mre11 mutants are chromosome translocations, most of which have no significant homology at the breakpoints (52). The increased rate of GCRs observed in the mre11, rad50, and xrs2 mutants could be due to increased accumulation of lesions and/or channeling of lesions from the normal nonmutagenic repair pathway to mutagenic repair. mec1 mutants, defective for the S-phase and damage checkpoints, show a 200-fold increase in GCRs, and these are due primarily to telomere addition (254). The tel1 mutation by itself confers no increase in the rate of chromosome rearrangements in wild-type or mre11 strains, but the tel1 mec1 and mre11 mec1 double mutants show a synergistic increase (≈10,000-fold) in GCR events (254). The high rates of GCRs observed in the mec1 tel1 and mec1 mre11 double mutants are most probably due to the combination of checkpoint, telomere addition, and DNA repair defects and are consistent with Tel1 and MRX functioning in the same pathway.
Defects in MRE11 and NBS1 are causes of human chromosomal instability syndromes.
The human chromosomal instability syndrome A-T is caused by mutation of the ATM gene, which encodes a protein kinase homologous to the yeast Tel1 and Mec1 proteins (335). Cells established from A-T patients show sensitivity to IR, chromosomal instability, and radioresistant-DNA synthesis (the failure to suppress replication initiation in the presence of DNA DSBs). The cellular features of A-T are shared by two related chromosomal instability syndromes, NBS and A-TLD (335, 358). Although the clinical presentations of these syndromes are different, individuals with A-T and NBS show growth retardation, immunodeficiency, and cancer predisposition. Of individuals with NBS, 90% are homozygous for the 657del5 allele, a truncating mutation of NBS1 that causes premature termination at codon 219 (411). This was originally assumed to be a null mutation. However, recent studies provide evidence for a 70-kDa Nbs1 protein that is produced by internal translation initiation within the NBS1 mRNA (227). The 70-kDa Nbs1 protein is present at reduced levels but retains association with Mre11 and Rad50. Thus, the most common NBS1 allele encodes a partially functional protein, consistent with recent studies showing cell death for a NBS1 null mutation (227, 431). The N-terminal truncation of Nbs1, which removes the FHA domain, results in defects in checkpoint signaling and localization of the complex but retains the essential function of Nbs1 (47, 227).
Two mutations within the MRE11 gene have been found for the two families with A-TLD-affected members (358). In one family, the mutation is a stop codon at position 633, producing a truncated protein. The truncated form of Mre11 produced by these patients is less abundant, and the levels of Rad50 and Nbs1 are also reduced. The mutation in the other family is a change of Asn117 to Ser. Both of these mutations have been generated in the Sc MRE11 gene. As expected based on analysis of other yeast mre11 mutations, the truncating mutation confers a meiotic defect but no effect on survival to IR whereas the missense allele confers a slight increase in sensitivity to IR and reduced spore viability. Interestingly, the mre11-N113S strain has short telomeres and this defect is partially complemented by the mre11-58 allele, suggesting intragenic complementation between N-terminal alleles (191). The integrity of the intra-S phase checkpoint has not been determined for the yeast mre11-Q623Z or mre11-N113S strains.
Although no human cancer-prone syndrome has been assigned to a defect in Hs RAD50, a rad50 hypomorphic mutant mouse has been generated (26). The rad50-K22M/rad50-K22M mutation corresponding to one of the rad50S alleles identified by Alani et al. (8) supports the viability of mouse embryonic stem cells. The rad50-K22M allele conferred no obvious sensitivity to clastogens and no apparent growth defect, and the intra-S-phase checkpoint was unaffected. However, the rad50-K22M allele caused a profound defect at the organismal level. Mice homozygous for this allele showed decreased birth weight, and most died with complete bone marrow depletion due to progressive hematopoietic stem cell failure. Rare long-term survivors were highly predisposed to malignancy, and cell lines derived from these tumors showed chromosome aberrations. Surprisingly, both male and female mice were fertile and there were no obvious defects in ovarian or testicular development.
Function of the MRX complex in the DNA damage checkpoint.
DNA damage and stalled replication forks cause temporary arrest of the cell cycle to allow time for DNA repair to occur before progression through DNA synthesis or mitosis (415). Defects in the signaling pathway result in sensitivity to DNA-damaging agents and replication inhibitors, such as hydroxyurea (HU), and genome instability. The failure of cells from NBS and A-TLD patients to arrest DNA synthesis in response to IR was the first evidence suggesting a role for the MRN complex in the DNA damage checkpoint (281). ATM phosphorylates the Nbs1 protein in response to IR, and mutation of the Nbs1 residue that is phosphorylated results in loss of the S-phase checkpoint (114, 201). However, Nbs1 is still phosphorylated in ATLD3 cells, which are checkpoint defective, suggesting that Nbs1 phosphorylation is not sufficient for checkpoint activation.
Recent studies with yeast also implicate the MRX complex in the DNA damage response (68, 122, 192, 406). The presence of a single, unrepairable DSB induces a long G2/M arrest, but cells adapt to the damage and resume the cell cycle (192, 315). mre11 mutants arrest normally in response to the DSB but adapt faster than do wild-type cells (192). Conditions that result in increased amounts of ssDNA (from processing the break) correlate with inability to adapt, leading to the suggestion that ssDNA or a ssDNA-protein complex is recognized as the damage signal. Strains containing the nuclease-defective mre11-3 allele show normal adaptation, consistent with studies by Moreau et al. showing normal resection of HO-induced breaks in the mre11-H125N strain (191, 241). The MRX complex is required for phosphorylation of the Rad53 kinase in response to IR but not to UV irradiation in G1-phase cells, and G1-arrested mre11 mutants fail to delay DNA synthesis in response to IR (68, 122). mre11, rad50, and xrs2 mutants are hypersensitive to HU and show no delay in DNA synthesis in the presence of HU or bleomycin, indicating a defect in the intra-S checkpoint (68). One attractive model is for the MRX complex to recognize unusual structures that arise during replication and to signal through Rad53 to arrest S phase. D'Amours and Jackson suggested that the nuclease activity of Mre11 is required for the intra-S checkpoint, but the mre11-58 and mre11-P162S strains utilized in their study are defective for MRX complex formation (50, 407). In contrast, the mre11-H125N strain is not hypersensitive to HU and is proficient for complex formation (S. Moreau, B. Krogh, and L. S. Symington, unpublished data). The Mre11 and Xrs2 proteins are also phosphorylated in response to IR, and this phosphorylation is dependent on Tel1 (68, 406). The physiological relevance of the Tel1-mediated phosphorylation is unknown. Studies in other systems, however, are not consistent with the MRX complex functioning in the S-phase checkpoint. S. pombe rad50 mutants delay DNA synthesis in the presence of HU and MMS, suggesting that the replication checkpoint is still functional (129). Similarly, initiation of DNA replication is still delayed in the presence of DNA damage in Xenopus cell extracts following depletion of Mre11 (67).
RAD51, RAD52, RAD54, RDH54/TID1, RAD55, RAD57, RAD59, and RFA1 Subgroup.
Genetic studies place the RAD51, RAD52, RAD54, RAD55, RAD57, RAD59, RDH54, and RFA1 genes in the homologous recombination pathway. Within this group, the RAD51, RAD52, RAD54, RAD55, and RAD57 genes are essential for conservative DSB repair, resulting in gene conversion (and associated crossing over), and RAD52 and RAD59 have additional functions in the nonconservative BIR and SSA pathways. The requirement for replication protein A (RPA) has been more difficult to address in vivo because of the essential function of this complex in DNA replication. However, several non-null alleles of RFA1 exhibit recombination and repair deficiencies. RDH54/TID1 is discussed with RAD54 (see below) because it encodes a protein with homology to Rad54 and shows redundancy with RAD54 in some assays. In this section, the biochemical activities of each of the proteins are described, followed by characterization of mutant phenotypes and implications for the mechanism of recombination.
Rad51.
Sc RAD51 encodes a 43-kDa protein with 30% identity to bacterial RecA proteins; the highest homology is with the catalytic domain of RecA, encompassing the Walker A and B motifs for nucleotide binding and/or hydrolysis (1, 22, 338). In E. coli, recA is the most important gene for homologous recombination and the purified RecA protein promotes homologous pairing and strand exchange in vitro (184). Rad51 is conserved in all eukaryotes for which sequence information is available, and the mouse and human proteins are 59% identical to ScRad51 (246, 337). Alignment of the RecA and Rad51 amino acid sequences reveals an N-terminal extension of 100 amino acids in the eukaryotic Rad51 proteins that is absent from RecA (337). This region is well conserved among eukaryotic Rad51 proteins, suggesting that it is important, and recent studies implicate the N-terminal domain in DNA binding and protein-protein interactions (6, 185).
Yeast rad51 null mutants are viable but show high sensitivity to IR and meiotic inviability. Surprisingly, deletion of RAD51 in vertebrates results in cell inviability and early embryonic death in mice (200, 404). Trophoblast-like cells derived from mouse rad51−/− blastocysts are sensitive to IR, suggesting that the normal function of MmRad51 is to repair DSBs. A RAD51 conditional cell line has been made by deleting both copies of RAD51 in DT40 chicken cells in the presence of Hs RAD51 regulated by a tetracycline-repressible promoter (352). Down regulation of the gene, resulting in depletion of HsRad51, is concomitant with a G2/M-phase arrest, accumulation of cytologically visible chromosomal breaks, and eventual cell death. These data suggest that the essential role of RAD51 in vertebrates is to repair breaks generated during DNA replication. In support of this hypothesis, Hs RAD51 is expressed primarily during S phase and forms foci in proliferating cells (387). The RAD51 transcript is also highly induced during meiosis and following treatment of cells with DNA-damaging agents such as MMS (22, 30, 338). Rad51 foci are detected during meiosis coincident with the timing of meiotic recombination and colocalize with the meiosis-specific RecA homologue, Dmc1 (31). The formation of Rad51 foci is dependent on Spo11 and can be induced in spo11 mutants by treatment of meiotic cells with IR (113). Meiosis-induced Rad51 foci are not formed in rad52, rad55, or rad57 mutants, consistent with biochemical studies implicating these factors in assembly of the Rad51 presynaptic filament (113). Radiation-induced Rad51 foci form at reduced levels in rad55 and rad52 mutants but are absent from rad52 rad55 double mutants, suggesting some redundancy for presynaptic complex formation between these factors in mitotic cells (112a).
(i) Biochemical studies.
Purified Rad51 forms right-handed helical filaments on double-stranded DNA with structural similarity to those formed by RecA (265). Formation of filaments on ssDNA is stimulated in the presence of the heterotrimeric DNA binding protein RPA, although short filaments of HsRad51 have been observed on tailed duplex molecules in the absence of RPA (234, 374). Rad51 binds with higher affinity to DNA duplexes with single-stranded tails than to duplex or single-stranded oligonucleotides (232). Although Rad51 is expected to bind to any DNA sequence independent of base composition, preferred binding sites have been identified. Rad51, RecA, and the Sulfolobus solfataricus RadA protein all bind to GT-rich sequences with higher affinity than to random sequences (331, 394, 395). Binding of HsRad51 to DNA can occur in the absence of ATP, but ATP is required for DNA binding by ScRad51 (76). Binding to ssDNA activates the Rad51 ATPase, but the kcat for the ATPase is about 30-fold lower than found for the RecA protein (371). Mutation of the conserved lysine residue within the Walker A motif to alanine (Rad51-K191A) abolishes DNA binding and ATPase activities of ScRad51 (375). When the same lysine residue is replaced by arginine (Rad51-K191R), the protein retains ATP-dependent DNA binding, but no significant hydrolysis of ATP (375). The ATPase activity of Rad51 is higher when poly(dT) is used as a substrate compared with M13 DNA, and RPA stimulates the ATPase activity of Rad51 only when M13 is used as a cofactor, not poly(dT) (368). E. coli single-strand binding protein can substitute for RPA in stimulation of the Rad51 ATPase, indicating that Rad51 and RPA are unlikely to interact. These observations suggest that the role of RPA in presynapsis is to remove secondary structures from ssDNA to allow the formation of a continuous Rad51 nucleoprotein filament.
Once assembled, the Rad51 nucleoprotein filament is capable of interacting with a second DNA molecule, either ssDNA or dsDNA, to initiate strand exchange. Synapsis entails alignment of the nucleoprotein filament with homologous sequences within the second molecule. Although the details of this step of the reaction have not been well characterized for Rad51, studies with RecA protein suggest that the homology search is rapid and involves random collisions of the two molecules. Both RecA and HsRad51 form coaggregates of ssDNA and duplex DNA that are independent of sequence homology during the homology search (23, 398). Once homology is found, strand exchange occurs between the two aligned molecules if one of the partners has a free end to allow stable intertwining of the complementary strands.
Strand exchange can be measured in vitro using a variety of DNA substrates (Fig. 12). The three-strand exchange reaction using circular ssDNA and homologous linear duplex has been used extensively to characterize Rad51-promoted strand exchange. The reaction requires ATP but is also supported by nonhydrolyzable analogs of ATP (375). Furthermore, the Rad51-K191R mutant protein catalyzes extensive strand exchange when present at high concentration. The polarity of strand exchange is 5′ to 3′ with respect to the complementary strand of the DNA duplex, opposite to the polarity observed for RecA but the same as for the U. maydis Rec2 protein (27, 181, 374). The strand exchange activity of Rad51 is greatly stimulated by the addition of RPA to the reaction mixture if RPA is added after Rad51 has already nucleated onto the initiating ssDNA substrate (373). However, addition of RPA simultaneously with or prior to Rad51, to mimic the likely in vivo situation, results in a severe reduction in strand exchange products. The inhibition of Rad51-mediated strand exchange imposed by RPA can be overcome by the addition of Rad52 or the Rad55-Rad57 heterodimer to the reaction mixture (28, 259, 339, 372, 373). Both Rad52 and Rad55-Rad57 are thought to mediate the assembly of the Rad51 presynaptic filament, although most biochemical studies have focussed on the effects of these proteins in the strand exchange assay.
FIG. 12.
Substrates used for in vitro strand exchange assays. (A) Pairing of a circular single-stranded molecule with linear duplex results in the formation of a nicked circular duplex product and a displaced linear single strand. (B) Invasion of a linear single-stranded molecule into supercoiled circular DNA results in the formation of a joint molecule with a D-loop. (C) Strand exchange between an unlabeled single-stranded oligonucleotide and labeled double-stranded oligonucleotides is detected by displacement of a labeled single-stranded oligonucleotide.
Oligonucleotide substrates have been used in fluorometric assays to distinguish between the pairing and strand displacement phases of strand exchange (123). When the G+C content of the oligonucleotides is very low (16%), HsRad51 promotes efficient pairing and strand exchange, but when the G+C content is increased to 40%, pairing still occurs but strand displacement does not (123). These results suggest that the initial homology recognition step requires partial base pairing between the incoming and outgoing strands prior to the full base-pairing interactions necessary for strand exchange. The use of single-stranded tailed duplex oligonucleotides improves the yield of strand exchange products; however, the effects of G+C content have not been determined with these substrates (232).
The D-loop assay, which requires the invasion of intact duplex DNA by ssDNA, appears to most closely model the expected situation in vivo (Fig. 12) (136). Rad51 inefficiently promotes D-loop formation between single-stranded oligonucleotides and homologous supercoiled DNA (232). HsRad51 promotes efficient D-loop formation between large linear single-stranded molecules or duplex molecules with single-stranded tails of either 5′ or 3′ polarity and supercoiled plasmid DNA (234). This reaction does not require stimulatory factors, whereas the equivalent reaction catalyzed by ScRad51 is inefficient in the absence of RPA and Rad54 (283). The reason for this difference is unclear.
(ii) Behavior of rad51 mutants.
Comparison of the sequences of RecA and Rad51 shows that several residues are invariant, notably in the regions assigned to ATP binding or hydrolysis (1, 22, 338). Site-directed mutation of the conserved lysine present in the Walker A-site for NTP binding to alanine results in a protein that is unable to bind DNA or promote strand exchange, while the Rad51-K191R protein is proficient for DNA binding and strand exchange (375). When the rad51-K191A allele is present in a rad51Δ mutant on a low-copy-number vector or overexpressed, the phenotype is indistinguishable from that due to a rad51 null mutation, but this allele confers a semidominant phenotype when expressed in wild-type cells (79, 338, 375). In vivo, the rad51-K191R allele suppresses the MMS sensitivity of a rad51Δ strain when overexpressed, but when present on a low-copy-number plasmid it shows only partial activity (338, 375). Constitutive high-level expression of the Hs rad51-K133R allele supports growth and radiation resistance of chicken DT40 rad51−/− cells but not gene targeting, leading to the suggestion that ATP hydrolysis is not important for the essential function of Rad51 but is required for some homologous recombination functions (247). Expression of the Hs rad51-K133R allele in wild-type mouse embryonic stem cells causes increased sensitivity to IR and cross-linking agents and reduced efficiency of homologous recombination (355). These dominant negative effects support the hypothesis that ATP hydrolysis is important for Rad51 function in vivo.
In contrast, when the rad51-K191R allele is expressed as the only chromosomal allele of RAD51 in budding yeast, the phenotype conferred is quite similar to that conferred by the null allele with respect to radiation sensitivity, mating-type switching and spontaneous mitotic recombination (244). However, diploids homozygous for the rad51-K191R allele show normal levels of sporulation and high spore viability and are much more resistant to IR than is the rad51-K191R haploid. The suppression of the DNA repair defect conferred by the rad51-K191R allele in diploids is due to heterozygosity at the MAT locus. One attractive hypothesis to explain these results is that the rad51-K191R mutant protein has a defect in recycling and repair can occur when there is sufficient mutant protein in the cell. Based on this idea, it would be expected that other factors that promote recycling of Rad51 should be able to suppress the rad51-K191R phenotype when overexpressed and might be regulated by mating type and/or during meiosis. Alternatively, the Rad51-K191R nucleoprotein filament might be less stable than the filament formed from wild-type Rad51 and suppression by MATa/α could occur by stabilization of the mutant protein filament by an unknown factor. RAD54 present at high copy number also suppresses the radiation sensitivity of the rad51-K191R strain (244). The partial suppression conferred by RAD54 is consistent with either of the proposed models because Rad54 could function to stabilize the Rad51-DNA interaction by binding to the Rad51 nucleoprotein filament or could function to displace Rad51 from DNA by translocation activity (231). The HsRad51K133R protein forms more stable D-loops between a 90-mer oligonucleotide and supercoiled plasmid DNA than does wild-type HsRad51; this is attributed to decreased turnover of Rad51 in the absence of ATP hydrolysis (346).
Chanet and coworkers isolated a number of rad51 alleles that suppress the MMS sensitivity of srs2 diploids (1, 51). SRS2 encodes a DNA helicase that is suggested to limit heteroduplex extension and/or reverse abnormal recombination intermediates (51, 307). Mutations in SRS2 suppress the UV sensitivity of rad6 and rad18 mutants, and this suppression is thought to occur by channeling lesions from error-prone repair into the recombinational repair pathway (1, 189, 321). The srs2 diploids show elevated rates of spontaneous and induced mitotic recombination and sensitivity to both ionizing and UV irradiation. The UV sensitivity of srs2 diploids is suppressed by homozygous mutations in RAD51, RAD52, RAD55, and RAD57. The rad51 alleles isolated as suppressors of the MMS sensitivity of srs2 diploids were all semidominant, requiring one functional allele of RAD51 for suppression. When present in haploids or as hemizygous diploids, all of the alleles were as defective in DNA repair and recombination as was a rad51 null allele. Thus, the screen required decreased recombination to suppress lethal recombination events in the srs2 cells but, at the same time, required residual recombination activity to survive the lethal effects of MMS. Sequencing of the 26 rad51 mutations recovered revealed substitutions of 18 different amino acids, most of which are conserved in the RecA family of proteins. Ten of the mutations correspond to regions of RecA directly involved in ATP binding, ATP hydrolysis, or ATP-induced allosteric changes; six are in putative monomer-monomer interaction domains. Presumably, the mutant monomers can be assembled with wild-type monomers to yield a partially functional filament.
Fortin and Symington isolated a novel class of RAD51 gain-of-function alleles that partially suppress the requirement for RAD55 and RAD57 in DNA repair (102). Five of the six mutations map to the region of Rad51 that, on the basis of modeling with RecA, corresponds to one of the DNA binding sites. The other mutation is in the N terminus of Rad51, in a domain implicated in protein-protein interactions or DNA binding (6, 185). These rad51 alleles were unable to suppress the DNA repair defect conferred by a partially functional rad52 allele, demonstrating genetic separation of the mediator functions. The Rad51-I345T mutant protein showed increased binding to ssDNA and dsDNA and was proficient in displacement of RPA from ssDNA, suggesting that the normal function of Rad55-Rad57 is to promote and stabilize Rad51-ssDNA complexes.
(iii) Rad51-interacting proteins.
The yeast two-hybrid system has been used to identify Rad51-interacting proteins and to map domains of Rad51 involved in protein-protein interactions. Rad51 self-interacts and interacts with Rad52, Rad54, Rdh54/Tid1, and Rad55 (57, 79, 82, 130, 155, 159, 237). Rad51 self-association, as well as Rad51-Rad52 interaction, is mediated via a domain in the amino terminus of Rad51 (79). However, single-amino-acid substitutions within Rad51 that disrupt the interaction with Rad52 map to the C terminus of Rad51 (185). Some of the mutations that disrupt the Rad51-Rad52 interaction also disrupt the interaction between Rad51 and Rad54, suggesting that interactions between these proteins and Rad51 are likely to be dynamic. Mutations within the N terminus of Rad51 that reduce Rad51 homotypic interactions also disrupt the interaction with Rad55 (185). The rad51 mutations that disrupt interaction with Rad52 and Rad54 confer sensitivity to MMS, confirming the importance of these interactions (185).
Most of the interactions determined by the two-hybrid system have been verified using biochemical methods (57, 117, 283, 285, 338). However, the interaction between Rad51 and Rad55 is extremely weak in vitro (373), and the interaction between MmRad51 and MmRad54 by coimmunoprecipitation is detected only following exposure to IR (384). HsRad51 associates directly with two important tumor suppressor proteins, p53 and Brca2 (54, 361, 424). Colocalization of HsRad51 and Brca1 has been detected on meiotic chromosomes, but this is probably due to the interaction between Brca1 and Brca2 rather than a direct interaction between HsRad51 and Brca1 (330). Rad51 interacts with the BRC repeats of Brca2, which are important for resistance to MMS (54), indicating the functional significance of this interaction. Furthermore, cell lines expressing hypomorphic alleles of BRCA2 exhibit chromosome instability, radiation sensitivity, and defective homologous recombination (252, 332, 412, 430). HsRad51 also interacts directly with BLM, a member of the RecQ family of DNA helicases (425). The BLM gene is mutated in individuals with Bloom's syndrome, a cancer predisposition syndrome.
To analyze the dynamics of Rad51, Rad52, and Rad54 in living cells, Essers et al. introduced green fluorescent protein (GFP)-tagged versions of all three genes into CHO cells (89). On treatment with IR, the proteins assembled into discrete nuclear foci. Rad51, Rad52, and Rad54 colocalized, as demonstrated previously by immunofluorescence of fixed cells (89, 384). The mobility of these proteins was determined in irradiated cells by using fluorescence redistribution after photobleaching (FRAP). While Rad51 was stably associated with damage-induced structures, Rad52 and Rad54 rapidly and reversibly interacted with these structures. These results suggest that interactions between Rad51, Rad52, and Rad54 are dynamic and argue against a stable complex of all three proteins (89). In yeast, most of the Rad52 present in extracts is associated with Rad51, suggesting that this subcomplex is quite stable (372).
Rad51 paralogs. (i) Fungal Rad51 paralogs.
The RAD55 and RAD57 genes of S. cerevisiae encode proteins with sequence similarity to RecA and Rad51 and are considered to be Rad51 paralogs (167, 210). Null mutations of either RAD55 or RAD57 cause cold sensitivity for DNA repair (130, 159, 211). At 23°C, rad57 mutants show the same sensitivity to γ-irradiation as rad51 mutants, and rad51 rad57 double mutants have the same sensitivity as the single mutants, whereas rad57 mutants are 10- to 100-fold more resistant than rad51 or the rad51 rad57 double mutant at 30°C (102). Cold sensitivity is usually indicative of proteins that act as components, or stabilizers, of protein complexes. Consistent with this idea, Rad55 and Rad57 form a stable heterodimer and Rad55 also interacts with Rad51 (130, 159, 373). Unlike Rad51, neither Rad55 nor Rad57 exhibits self-interaction in the two-hybrid system, but the Rad55-Rad57 heterodimer has the anticipated molecular weight as determined by molecular sieve chromatography (130, 159, 373). RAD51 present on a high-copy-number plasmid partially suppressed the radiation sensitivity of rad55 and rad57 mutants, and further suppression occurred when RAD52 was also present in more than one copy (130, 159). These studies are suggestive of an accessory role for Rad55 and Rad57 in DNA repair, consistent with results of in vitro studies showing a stimulation of the Rad51-promoted strand exchange reaction by the Rad55-Rad57 heterodimer. Meiosis-specific Rad51 foci are not observed in rad55 or rad57 mutants, supporting the idea that Rad55 and Rad57 act in the formation or stabilization of the Rad51 nucleoprotein filament. Mutation of the invariant lysine residue of the Walker A box of Rad57 confers no defect in DNA repair or sporulation, but mutation of the corresponding residue in Rad55 does cause sensitivity to IR and prevents sporulation (159). The Rad55 Walker A-box mutants have not been tested for mediator function either in vitro or in vivo; therefore, it is not clear whether ATP hydrolysis is important for this function of Rad55.
Diploids homozygous for rad55 or rad57 mutations are more resistant to IR than are haploids (159, 211, 314). This suppression is due to mating-type heterozygosity and can be recapitulated in haploids by coexpression of MATa and MATα alleles (by a sir mutation or introduction of MAT alleles on plasmids) (211). Because overexpression of RAD51 also suppresses the radiation sensitivity of rad55 and rad57 mutants, it seemed possible that RAD51 might be regulated by mating-type heterozygosity. However, there is no evidence from microarray analyses for increased expression of RAD51 in diploids, and Western blot analysis indicates that there does not appear to be an increase in the Rad51 protein level in haploids expressing both MAT alleles (244). RAD54 present at high copy number also partially suppresses the radiation sensitivity of rad57 mutants, but, like Rad51, Rad54 protein levels do not appear to be affected by MAT heterozygosity (58, 244).
Rad55 is phosphorylated in response to DNA-damaging agents. This phosphorylation is dependent on Mec1 and partially dependent on Rad53 but not on other checkpoint functions (21). rad55 mutants show normal responses to DNA damage, indicating that Rad55 is not required for the damage checkpoint. Interestingly, mec1 mutants are defective in both spontaneous and MMS-induced heteroallelic recombination (21). This reduction is greater than reported for rad55 mutants, suggesting that there might be RAD55-dependent and independent pathways for recombination, both of which require MEC1 (211, 314).
The S. pombe Rad51 paralogs are encoded by the rhp55 and rhp57 genes (172, 403). Like their counterparts in S. cerevisiae, deletion mutations of rhp55 and rhp57 confer higher sensitivity to DNA-damaging agents at low temperatures than at 30°C and their DNA repair defects can be suppressed by rhp51 present on a high-copy-number plasmid. Replacement of the conserved lysine residue of the Walker A motif by alanine of either Rhp55 or Rhp57 confers a modest DNA repair defect, but a strain containing the Lys-to-Ala substitutions in both genes is as sensitive to DNA-damaging agents as are the deletion mutants (402). This result suggests that the heterodimer may require only one active ATP binding site.
The fungus U. maydis encodes two Rad51-like proteins. One is orthologous to Rad51, and the other, encoded by the REC2 gene, is much more divergent, is more than twice the size of Rad51, and has no clear structural equivalent in databases (97, 311). Unlike the rad55 and rad57 mutants of S. cerevisiae, the DNA repair defect of rec2 strains is not cold sensitive, is not suppressed by Um RAD51 on a multicopy plasmid, and is not suppressed in the diploid state (97, 182). The rec2-1 mutant was the first eukaryotic recombination-defective mutant identified and shows decreased radiation-induced recombination, defective plasmid gap repair, reduced levels of gene targeting, and meiotic death (97, 134). A proteolytic form of Rec2 purified by conventional methods from U. maydis extracts and referred to as the rec1 protein was shown to promote homologous pairing and strand exchange in vitro (180). The full-length product of the recombinant REC2 gene catalyzes similar reactions in vitro and has the same polarity and preference for tailed duplex molecules as shown for Rad51 (27, 232). The pairing and strand exchange activities of Rec2 require ATP but are not dependent on ATP hydrolysis. Consistent with this observation, replacement of the conserved lysine residue of the Walker A site by alanine results in a null mutant phenotype whereas the rec2-K257R allele confers no apparent DNA repair defect (311).
(ii) Characterization of vertebrate Rad51 paralogs.
In addition to Rad51, vertebrates appear to encode five Rad51 paralogs, Xrcc2, Xrcc3, Rad51B/Rad51L1, Rad51C/Rad51L2, and Rad51D/Rad51L3 (392). These proteins have 20 to 30% sequence identity to Rad51 and to each other. The XRCC2 and XRCC3 genes were identified by complementation of the mitomycin C sensitivity of the irs1 and irs1SF hamster cell lines (161, 204). The irs1 and irs1SF cell lines show high sensitivity to DNA-cross-linking agents and are about twofold more sensitive to IR than are the parental cell lines (161). Rad51B, Rad51C, and Rad51D were identified through database analysis or by PCR amplification on the basis of their similarity to Rad51 (10, 81, 290). Multiple protein alignments suggest that Xrcc2 is most similar to yeast Rad55 and Rad51D and that Xrcc3 is closest to yeast Rad57. The irs3 and CL-V4B Chinese hamster cell lines have recently been shown to have mutations in the RAD51C gene, and the mitomycin C sensitivity of these cell lines was complemented by expression of the Hs RAD51C gene (106, 116). Two- and three-hybrid studies have shown interactions among the Rad51 paralogs but no evidence for self-associations (324). Rad51 interacts with Xrcc3 and weakly with Rad51C; the Rad51-Rad51C interaction is improved in the presence of Xrcc3. Rad51C interacts with Xrcc3, Rad51B, and Rad51D; and Rad51D interacts with Xrcc2. Many of these interactions have been confirmed by coimmunoprecipitation of the Rad51 paralogs from HeLa cells or following expression of recombinant proteins in E. coli or baculovirus-infected insect cells (40, 188, 229, 324). Recent studies have identified two discrete complexes of Rad51 paralogs, one containing Rad51C and Xrcc3 and the other containing Rad51B, Rad51C, Rad51D, and Xrcc2 (205, 230, 420). Although Rad51 was found to interact with Xrcc3 in the two-hybrid system, recent biochemical studies have failed to detect this interaction (205, 230, 420).
The Rad51D protein has been purified following expression in E. coli and exhibits preferential binding to ssDNA over dsDNA and a weak DNA-stimulated ATPase activity (40). The complex of Rad51C and Xrcc3 has also been purified (188, 229). The two proteins form a heterodimer with 1:1 stoichiometry, similar to that observed for Rad55-Rad57 and Rad51D-Xrcc2 (40, 188, 229, 373). The Rad51C-Xrcc3 complex binds to ssDNA but only weakly to dsDNA and aggregates ssDNA to form large networks (188, 229). The DNA binding and aggregation activities of the complex are independent of ATP (229). The Rad51C-Xrcc3 complex also promotes ATP-independent D-loop formation and strand exchange between short oligonucleotides (188). The Rad51B-Rad51C complex was purified from insect cells and shown to bind to ssDNA with higher affinity than dsDNA. Although DNA binding is ATP independent, the complex exhibits a weak ssDNA-stimulated ATPase activity. The Rad51B-Rad51C complex has mediator activity in the HsRad51-promoted strand exchange reaction. HsRad51 can efficiently make joint molecules between circular ssDNA and homologous linear duplex DNA in the absence of RPA (24), but the formation of complete strand exchange products requires RPA (344). As described for the yeast system, when RPA is added to the reaction mixture at the same time as Rad51, the formation of products in inhibited. The addition of Rad51B-Rad51C to the reaction mixture partially alleviates the RPA inhibition (345). The Rad51B, Rad51C, Rad51D, and Xrcc2 proteins form a stoichiometric complex that has similar DNA binding and ATPase activities to those of the constituent subcomplexes (230). The Rad51B-Rad51C-Rad51D-Xrcc2 complex has the interesting property of specific binding to nicked duplex DNA (230), which could be relevant to the function of these proteins in repair.
Knockout mutants of each of the Rad51 paralog-encoding genes have been generated in the DT40 chicken cell line (382, 383). All of the cell lines are viable but show higher spontaneous cell death, a large increase in spontaneous chromosomal aberrations (chromosome and chromatid breaks), and increased sensitivity to IR and cross-linking agents compared with the parental cell line (382, 383). The sensitivity of each of the mutant cell lines to cross-linking agents can be suppressed by overexpression of Hs RAD51. This result illustrates the similarity in phenotype between the chicken and yeast rad51 paralog mutants. The number of cells containing γ-radiation-induced Rad51 foci and the number of foci produced are reduced in the mutant cell lines, again consistent with a mediator function for the Rad51 paralogs (382, 383). The hamster cell lines deficient for Rad51C show a reduction in spontaneous and mitomycin C-induced sister chromatid exchanges, in addition to sensitivity to DNA cross-linking agents and chromosome aberrations (106, 116). Interestingly, a reduced level of sister chromatid cohesion was observed in the CL-V4B cell line, which could contribute to chromosome instability (116). Although Rad51C is a component of both paralog complexes, the rad51c−/− DT40 cell line exhibits less severe defects in gene targeting and formation of spontaneous chromosome aberrations than do the other Rad51 paralog-defective cell lines (383). This could potentially be due to redundancy between Rad51C and Rad52 (107). Chicken DT40 cell lines deficient in Xrcc2, Rad51B, and Rad51D are viable; however, homozygous mutant mice undergo embryonic death (71, 289, 342). Xrcc2−/− embryos are recovered from early embryonic stages (before day 8.5), with embryonic death occurring from midgestation. Embryos that survive to late stages of development exhibit developmental abnormalities and neurological defects (71). The large increase in the number of apoptotic cells in neural tissues from xrcc2−/− mice is similar to that observed for ligase 4 and Xrcc4-deficient mice (103, 112).
The role of the Rad51 paralogs in homologous recombination has been assessed using recombination reporters containing two nonfunctional copies of the neomycin phosphotransferase (neo) gene or the GFP gene (41, 158, 287). These reporters contain an insertion of an I-SceI-cut site to inactivate one copy of the gene, and the other copy has inactivating 5′ and/or 3′ truncations. In the presence of I-SceI, a DSB is made within the reporter and can be repaired by homologous recombination or end joining. In the irs1 (xrcc2−) and irs1SF (xrcc3−) cell lines, 100- and 25-fold decreases, respectively, in the efficiency of repair by homologous recombination were observed (158, 287). Analysis of the recombinants derived from the irs1SF (xrcc3−) cell line revealed altered produce spectra compared with the XRCC3-complemented cell line (41). These alterations included increased gene conversion tract lengths, discontinuities of the tracts, and frequent rearrangements. Presumably, recombination still initiates in the cell line but occurs with lower fidelity. This could be due to Rad51-independent events, such as the BIR events seen in yeast that would be expected to be associated with longer tracts, or could be due to a late role of Xrcc3 in stabilizing Rad51-promoted recombination intermediates.
(iii) Dmc1.
DMC1 is not considered to be a member of the RAD52 epistasis group because mutants are resistant to IR, but DMC1 is essential for the repair of DSBs during meiotic recombination. DMC1 was identified in a screen for meiosis-specific prophase-induced genes that, when disrupted, resulted in meiotic prophase arrest (33). Mutation of DMC1 leads to the accumulation of DSBs in meiosis and a reduction in the level of reciprocal recombinants, as measured by a physical assay. DMC1 encodes a 334-amino-acid polypeptide with significant similarity to RecA and Rad51. Dmc1 is 45% identical to Rad51 along its entire length, and biochemical studies indicate a higher functional conservation than the other Rad51 paralogs to RecA-Rad51. Both Dmc1 and Rad51 form foci during meiosis, and the foci indicate significant colocalization of the two proteins (31). RAD51 and DMC1 are both required for high levels of meiotic recombination and play overlapping but nonidentical roles in meiosis (336). Homologues of Dmc1 have been identified in a number of eukaryotes, including mouse and human, and dmc1−/− knockout mice show the expected sterile phenotype (127, 288, 429).
The purified HsDmc1 protein binds to both ssDNA and dsDNA DNA, has a weak ATPase activity, and catalyzes strand exchange between oligonucleotides or from linear DNA to a single-stranded circular molecule (197, 228). The yeast Dmc1 protein also promotes annealing of complementary ssDNA (138). Unlike Rad51, Dmc1 does not appear to form helical filaments on DNA and instead forms octameric rings with DNA in the central channel (228, 276).
Rad52.
Deletion of RAD52 in budding yeast results in severe defects in homology-dependent DSBR and meiosis. rad52 mutants are defective in BIR and SSA in addition to the RAD51-dependent gene conversion pathway; consequently, they show the most severe recombination defects of all the rad52 group mutants. However, chicken and human cell lines lacking RAD52 are viable, and, in contrast to RAD51, the deletion of RAD52 does not cause embryonic death in mice (303, 427). Furthermore, mutant cell lines show no increase in sensitivity to DNA-damaging agents and the efficiency of gene targeting is only marginally reduced. S. pombe encodes two Rad52-like proteins, Rad22 and Rti1 (376, 409). The rad22 mutant has more severe defects in mitotic recombination and repair of IR-induced damage than does the rti1 mutant, but the double mutant is even more defective than the rad22 single mutant (409). This suggests some redundancy between these genes in fission yeast and raises the possibility that another Rad52-like protein exists in vertebrates or that vertebrates have another function that is redundant with Rad52. Evidence for the latter comes from recent experiments showing inviability of a rad52 xrcc3 DT40 chicken cell line even though the single mutants are able to proliferate (107, 353). The death of the double-mutant cell line is overcome by overexpression of RAD51, consistent with defects in two mediator functions. This result suggests that the Rad52 and Xrcc3 proteins are redundant or that they function in two different homologous recombination pathways that contribute to survival.
RAD52 of budding yeast is expressed throughout the cell cycle and is induced 2- to 3-fold by DNA-damaging agents and about 10-fold during meiosis (59, 60). Rad52 is nuclear and forms discrete foci in response to IR and during S phase of unirradiated cells (203). Surprisingly, only one or two foci are observed using a Rad52-GFP fusion, independent of the dose of IR, suggesting that damaged DNA is sequestered at one or two sites within the nucleus, perhaps at replication factories. Rad52 foci are also observed during meiosis, are dependent on SPO11, and show extensive colocalization with RPA (113, 203).
(i) Structural and biochemical studies.
The published Sc RAD52 open reading frame encodes a protein of 504 amino acids (3). The actual length is likely to be shorter because there are five putative start codons near the N terminus of the RAD52 gene and mutation of either of the first two methionine residues confers no obvious DNA repair defect (251). Furthermore, the homologues of Rad52 identified in S. pombe and vertebrates lack the 34-amino-acid N-terminal extension present in ScRad52, suggesting that the start site is likely to be the third putative initiating codon (251). There is less sequence identity between ScRad52 and other family members than for other Rad52 epistasis group proteins, and most of the homology is restricted to the N-terminal 200 residues. The C terminus, which includes the Rad51 interaction domain, is less highly conserved. RAD51 present on a high-copy-number plasmid suppresses the MMS sensitivity of strains expressing a deletion of Rad52 C terminus, suggesting that the primary function of this domain is to recruit Rad51 (237).
Both yeast and human Rad52 are multimeric and form ring structures as visualized by electron microscopy (296, 340, 356). Using both conventional and scanning transmission electron microscopy, the HsRad52 protein was observed to form heptameric rings with a strong pinwheel appearance and a central channel (356). It is generally accepted that DNA is bound within the central channel of hexameric helicases, but to date there is no evidence that DNA lies within the central channel of the Rad52 heptamer. Rad52 appears to have two modes of self-association. Assembly of monomers into rings requires sequences in the conserved N-terminal domain of Rad52, whereas the formation of higher-order multimers is mediated by the C terminus (296). The N-terminal 192 residues of Rad52 form rings with an average mass of 277 kDa, consistent with an average of 10 subunits, while the full-length protein forms a ring with an average mass of 298 kDa, consistent with the heptameric ring form (296). Formation of multimers is consistent with genetic studies showing intragenic complementation between N- and C-terminal mutations of rad52 (39). The crystal structure of the N-terminal domain (residues 1 to 212) of the HsRad52 protein has been determined and reveals an 11-member ring (163). The overall structure resembles a mushroom, consisting of a stem containing highly conserved hydrophobic residues and a domed cap. A positively charged groove was identified under the domed cap, and mutational analysis was consistent with this region comprising a DNA binding surface (163). One of the residues identified as important for DNA binding, Arg55, is equivalent to Arg70 of ScRad52, previously shown to be important for Rad52 function in vivo (16). Surprisingly, several of the residues identified as being important for DNA binding do not appear to be important in vivo in the context of the full-length ScRad52 (251).
The purified Rad52 protein binds preferentially to ssDNA and promotes annealing of complementary ssDNA (250). Rad52-promoted annealing of long molecules is stimulated by RPA, whereas Rad52 efficiently anneals oligonucleotides in the absence of RPA (250, 340, 367). The probable role of RPA in strand annealing is to remove secondary structures from ssDNA to allow annealing by Rad52, but the stimulation of annealing also requires a species-specific interaction between Rad52 and RPA. RPA alone inhibits the annealing of oligonucleotides; this inhibition is overcome by Rad52 (367). HsRad52 binds directly to HsRPA, and a similar interaction is thought to occur in yeast based on two-hybrid and genetic studies (99, 131). Because Rad52 interacts with both Rad51 and RPA, the contemporary models hold that Rad52 replaces RPA from ssDNA with Rad51 or that Rad52 provides a seeding site within the RPA-bound ssDNA for subsequent cooperative binding by Rad51 (351). Rad52 forms a complex with RPA-coated ssDNA, but does not displace RPA (366). Rad51 can displace RPA from ssDNA following interaction with Rad52 bound to RPA-coated DNA (366).
Studies with the HsRad52 protein have shown preferential binding to the ends of ssDNA of tailed duplex molecules (275). The terminal nucleotide is protected, and the region of ssDNA bound shows highly regular sensitivity to hydroxyl radicals. The periodicity of the hydroxyl radical sensitivity of DNA within Rad52-DNA complexes is thought to be due to wrapping of ssDNA on the outside of the Rad52 ring (275, 356). Genetic studies with yeast suggest a RAD52-dependent, RAD51-independent pathway for strand invasion, and recent studies show the formation of D-loops by HsRad52 (162). This activity, as well as DNA binding, is retained in a truncated form of the protein comprising the first 237 residues (162). The formation of D-loops by Rad52 probably occurs by annealing between the incoming ssDNA and transiently ssDNA present in the supercoiled plasmid.
(ii) Protein interactions.
As described above, Rad52 self-associates to form a ring structure. It also interacts with Rad51 via the C-terminal domain of Rad52, and deletion of residues 409 to 412 eliminates Rad51 binding (186, 237). The Rad51 interaction domain of Rad52 is necessary for overcoming the RPA inhibition to strand exchange in vitro, consistent with the model that the mediator function of Rad52 requires interaction between Rad52 and Rad51 (186, 339). Although ScRad52 shows genetic and two-hybrid interaction with the large subunit of RPA (Rfa1) (99, 131), studies with the human proteins suggest a direct interaction between the 34-kDa subunit of RPA and Rad52 (274). The central domain of HsRad52 is required for interaction with RPA (131, 274). Rad52 also interacts with Rad59, raising the possibility of formation of a heteromeric Rad52-Rad59 ring (70).
(iii) Behavior of rad52 mutants.
Some point mutations in the C-terminal region of RAD52 and deletions that encode only the first 210, 252, or 327 residues of Rad52 can be partially suppressed by RAD51 present on a high-copy-number plasmid (15, 39, 237, 323). However, rad52 null alleles, and most inactivating point mutations within the N-terminal region of RAD52, cannot be suppressed by RAD51 at high copy number. No extragenic or high-copy-number suppressors of the rad52Δ allele have been identified. These observations suggest that the N-terminal region of Rad52 comprises a core domain with discrete functions in DSB repair that are independent of Rad51 (15). The first two alleles of RAD52 identified, rad52-1 (A90V) and rad52-2 (P64L), both cause high sensitivity to IR and meiotic death, but rad52-2 strains show increased levels of spontaneous heteroallelic recombination compared with RAD52 strains. This suggests that spontaneous lesions might have a less stringent requirement for RAD52 than do radiation-induced lesions. A non-null allele of RAD52, rad52-R70K, was identified in a screen for mutants defective in RAD51-independent recombination of inverted repeats (16). The rad52-R70K strain showed only a 4-fold reduction in inverted-repeat recombination, compared with a 3,000-fold decrease observed for the rad52 null strain and a 1,300-fold decrease found for the rad51 rad52-R70K strain. The rad52-R70K mutation conferred partial sensitivity to γ-irradiation and was synergistic with a rad59 null mutation for inverted-repeat recombination, SSA, mating-type switching, and sporulation, suggesting that some weak alleles of rad52 are highly dependent on RAD59 function.
In a systematic analysis, positively charged, aromatic, and hydrophobic residues within the N-terminal region of Rad52 were replaced by alanine and tested for radiation resistance and spontaneous mitotic recombination (251). From this analysis, mutations that conferred a phenotype similar to that for a null mutation were identified, along with several classes of separation-of-function mutations. One class, defined by a single allele, conferred intermediate sensitivity to IR but was completely defective for heteroallelic recombination. Another class, represented by several alleles, showed intermediate sensitivity to IR but wild-type or higher rates of heteroallelic recombination. The last class was partially defective for heteroallelic recombination but showed only mild sensitivity to IR. To date, there is no information on the biochemical activities of these Rad52 mutants.
Rad59.
The RAD59 gene was identified in a screen for mutants defective for RAD51-independent spontaneous mitotic recombination between inverted repeats (17). The rad59 mutation was shown to cause modest defects in several mitotic recombination assays and moderate sensitivity to IR (16). In the chromosomal inverted-repeat recombination assay, rad52 was epistatic to rad51 and rad59 and rad59 was synergistic with rad51. The level of recombination in a rad51 rad59 double mutant is higher than in a rad52 mutant, indicating either that there is an additional pathway or that Rad52 is able to carry out some recombination functions by itself. RAD59 is important for SSA between chromosomal direct repeats, and the requirement for RAD59 increases as the repeat length decreases (16, 363). This suggests that Rad52 becomes more dependent on Rad59 when the repeats to be annealed are short or when short homologies are embedded within extensive regions of heterology.
The RAD59 gene encodes a 238-residue protein with significant homology to the N-terminal half of Rad52 (17). The RAD52 gene, when present on a CEN-based plasmid (one to three copies/cell), partially suppressed the radiation sensitivity of the rad59 strain, suggesting that Rad52 and Rad59 have overlapping functions and/or that they interact. Interaction between Rad52 and Rad59 was demonstrated using the two-hybrid system and by coimmunoprecipitation of the two proteins from yeast extracts (70). A complex of Rad51, Rad52, and Rad59 can be immunoprecipitated from yeast extracts, but Rad51 and Rad59 fail to interact in the absence of Rad52 (A. P. Davis and L. S. Symington, unpublished data). In addition, Rad52 and Rad59 share some biochemical activities. Rad59 binds to DNA, with a preference for ssDNA, and anneals complementary ssDNA (70, 284). Annealing of long molecules is not stimulated by RPA, but Rad59 can overcome the inhibitory effect of RPA on annealing of oligonucleotides. Despite the similarity of their biochemical activities, RAD59 cannot substitute for RAD52 in vivo, even in RAD51-independent recombination events such as SSA (70).
Rad54 and Rdh54/Tid1 (Rad54B).
Rad54 and the Rad54 homologue Rdh54 (Tid1) have sequence motifs characteristic of DNA helicases and are members of the SNF2/SW12 family of DNA-dependent ATPases (84). As with the other RAD52 group genes, RAD54 is not essential for viability in yeast but is required for resistance to IR. Rdh54 was found by database analysis and independently identified in a two-hybrid screen as Tid1, a Dmc1-interacting protein (82, 179, 341). Diploid yeast cells disrupted for RDH54/TID1 show slight sensitivity to high levels of MMS, but haploid mutants show no significant sensitivity to DNA-damaging agents (179). The rad54 rdh54 haploid strains have similar growth rates and MMS sensitivities to rad54 haploids, but homozygous rad54 rdh54 diploids grow slowly and are more sensitive to MMS than are rad54 diploids. Interestingly, the growth defect of the diploid double mutant can be suppressed by mutation of RAD51, suggesting that the poor growth is due to inappropriate recombination (179). A rad54 mutation causes death in srs2 strains; this is suppressed by mutation of RAD51, RAD52, RAD55, or RAD57 or by elimination of the checkpoint functions, RAD17, RAD24, or MEC3 (178, 270, 323). Although srs2 rdh54 haploids are viable, the homozygous diploids are not; again, this is suppressed by mutation or checkpoint or homologous recombination functions (178, 179). The suppression of the srs2 rad54 or srs2 rdh54 lethality by rad51, rad52, rad55, and rad57 suggests that Rad54 and Rdh54 function later in recombination than Rad51, Rad52, Rad55, and Rad57. The intermediates that accumulate in rad54 srs2 strains presumably trigger the damage checkpoint, leading to cell cycle arrest.
Homologues of Rad54 have been identified in vertebrates by PCR amplification on the basis of their similarity to ScRad54 and SpRad54 (165). The mouse and human Rad54 proteins have 48% identity with ScRad54. A second Rad54 homologue, called Rad54B, has been identified in humans and is thought to be more closely related to ScRdh54/Tid1 than to ScRad54 (385, 386). Mouse and chicken rad54−/− cells are sensitive to IR, MMS, and mitomycin C, but not to UV light, and they show reduced frequency of gene targeting (29, 88). Human rad54B−/− cell lines are defective for gene targeting but do not show sensitivity to DNA-damaging agents (238). In contrast to RAD51, the absence of RAD54 is compatible with normal mouse development. B- and T-cell development and immunoglobulin class switch recombination all occur normally in rad54−/− mice, indicating that these recombination events are independent of homologous recombination; however, immunoglobulin light-chain gene conversion is reduced in chicken bursal cells (29, 88). Although Mm rad54−/− embryonic cells are sensitive to IR, adult rad54−/− mice show high survival following IR (90). Survival of adult mice following IR is dependent on DNA PKcs, suggesting that the relative contribution of homologous recombination and end joining changes during development of the animal (90).
(i) Biochemical characterization.
Both Rad54 and Rdh54/Tid1 proteins show dsDNA-dependent ATPase activity and promote a conformational change of closed-circular duplex due to the creation of positive and negative writhe (231, 286, 377, 384, 410). However, neither protein shows helicase activity in the standard strand displacement assay. ATP-dependent translocation of Rad54 along duplex DNA generates both negative and positive supercoiled domains and is stimulated by Rad51-ssDNA filaments (231, 304, 410). DNA remodeled by Rad54 becomes more sensitive to the ssDNA-specific nuclease, P1, indicating transient strand separation (410). Rad54 and Rdh54/Tid1 both stimulate Rad51-promoted D-loop formation in the presence of either oligonucleotides or long ssDNA and supercoiled plasmid DNA (283, 285). Presumably, the change in writhe by Rad54 facilitates the invasion of duplex DNA by the Rad51 nucleoprotein filament by creating transiently unwound DNA. Rad54 also stimulates Rad51-promoted strand exchange between circular ssDNA and linear duplex DNA (350). The stimulation is greatest at the initial stage of synapsis, and the rate of heteroduplex extension is increased three- to fourfold by Rad54. All of the biochemical activities of Rad54 and Rdh54, except binding to dsDNA, are dependent on ATP hydrolysis and are eliminated by mutation of the invariant lysine residue within the Walker A motif to either arginine or alanine (284, 285, 350). However, rad54-K341A and rad54-K191R homozygous diploids retain high levels of interchromosomal, but not intrachromosomal, gene conversion (286).
Replication protein A.
RPA was discovered by the requirement for an ssDNA binding protein in the simian virus 40 in vitro replication assay (423). Yeast RPA was found to substitute for human RPA in stabilization of unwound DNA by T antigen, providing an assay for purification (45). The RPA complex consists of three subunits of 70, 34, and 14 kDa, encoded by the RFA1, RFA2, and RFA3 genes, respectively. All three subunits of the heterotrimeric complex are essential for viability in yeast, confirming the requirement for RPA in DNA replication (44, 133). Several alleles of RFA1 have been identified in screens for altered rates of recombination. Firmenich et al. recovered the rfa1-44 allele in a screen for mutants defective in DSB-induced mitotic recombination (99). This allele of RFA1 failed to complement certain rad52 alleles, and the DNA repair defects of the rfa1-44 strain were suppressed by RAD52 expressed from a high-copy-number plasmid. These results suggest that RPA and Rad52 are likely to interact, a prediction subsequently verified by experiments with the two-hybrid system (131). The temperature-sensitive rfa1-M2 allele confers sensitivity to UV and displays an increased spontaneous mutation rate and decreased rates of direct repeat recombination (209).
Another allele of RFA1, rfa1-D228Y, was identified as a suppressor of the spontaneous direct-repeat recombination defect of a rad1 rad52 strain (348). The rfa1-228Y strain by itself conferred a hyperrecombination phenotype for deletions between direct repeats that was independent of RAD52 and partially dependent on RAD1. Physical studies showed an increased level of deletion product by SSA in the rad52 rfa1-D228Y strain and the disappearance of long single-stranded intermediates that are characteristic of rad52 strains (347). By cell fractionation, there appeared to be less of the RPA complex from the rfa1-D228Y strain compared with a wild-type strain (348). These results suggest that Rad52 is required to displace RPA from ssDNA to promote strand annealing; in the absence of Rad52, RPA is inhibitory to spontaneous annealing or to another factor that is unable to displace RPA. Rad59 does not appear to be this factor because the rad59 rad52 rfa1-D228Y strain exhibits equivalent levels of SSA to those for the rad52 rfa1-D228Y strain (70).
Random mutagenesis of the RFA1 gene resulted in the identification of 19 recessive alleles of RFA1 that confer sensitivity to UV and MMS (405). Of these, the rfa1-t11 (rfa1-K45E) mutation was characterized in detail and shown to be defective in mating-type switching and HO-induced SSA. The rfa1-t11 and rfa1-t48 homozygous diploids also showed greatly reduced meiotic recombination and spore viability (354). These studies confirm an important role for RPA in homologous recombination, presumably to remove secondary structure from ssDNA in order to allow more efficient binding by Rad51.
Role of the Rad51 subgroup in meiotic recombination.
Diploids homozygous for rad51, rad52, rad55, or rad57 mutations either fail to sporulate or give rise to inviable spores (111, 294, 338). The sporulation defect is not rescued by spo13, but the sporulation and spore viability of rad52 diploids are restored by rad50 and spo13 mutations (219, 222). This suggests that the defect is subsequent to DSB formation and can be bypassed by preventing the initiation of recombination. Suppression of the spore inviability of spo13 rad52 strains has been used to identify genes required the initiation of meiotic recombination (220). Induction of meiotic recombination in sporulation-defective strains can be measured by using a return-to-mitotic-growth protocol. By this assay, rad51, rad52, rad57, and rfa1 mutants show a reduction in the level of recombinants, but significant induction over mitotic levels is observed (111, 338, 354). In rad52 mutants, recombinants appear to be unstable and the yield is reduced if the cells are held in rich growth medium before being plated onto selective medium (302). By physical analysis, rad51, rad52, rad55, rad57, and rfa1 mutants form normal levels of DSBs, but the ends are hyperresected due to the defect in strand invasion (264, 338, 354). Levels of crossover products, monitored using restriction fragment length polymorphisms flanking the initiating DSB, are reduced about fivefold in both rad51 and rad52 mutants (36, 264, 338). Mutation of the gene encoding the meiosis-specific RecA homologue, DMC1, results in a similar defect to that observed in rad51 mutants; however, the double mutant shows a greater reduction in the level of crossover products (33, 336). In a rad51 rad55 rad57 triple mutant, levels of interhomologue dHJ intermediates are greatly reduced indicating an important role for the Rad51 pathway in the generation of these intermediates (329).
Unlike other members of this subgroup, rad54 and rdh54 homozygous diploids exhibit only about a twofold decrease in sporulation efficiency and spore viability compared with wild-type strains (179, 325, 341). However, the rad54 rdh54 double mutant is severely defective for sporulation and spore viability, indicating functional redundancy between these proteins in meiosis. The results of physical and genetic assays to measure meiotic recombination are also consistent with redundancy between Rad54 and Rdh54. Rdh54 was identified in a two-hybrid screen for Dmc1-interacting proteins, and the phenotype of the mutants is consistent with a role in interhomologue recombination (82). Although RAD54 does not appear to play an important role in meiotic recombination, the high viability of dmc1 strains when returned to vegetative growth medium during meiosis is dependent on RAD54 (14). Furthermore, the pachytene arrest of dmc1 mutants can be suppressed by RAD54 on a high-copy-number plasmid (32). The interpretation of these results is that RAD54 is required for sister chromatid interactions; hence the important role in mitotic but not meiotic recombination. In dmc1 mutants, cells retain high viability when returned to vegetative growth because sister interactions can still occur; however, in the absence of RAD54, meiosis-induced DSBs cannot be repaired from the sister. High-copy-number expression of RAD54 is envisioned to direct some meiosis-specific DSBs from the interhomologue to the intersister pathway.
RAD51-dependent mitotic recombination.
The observation that rad51, rad54, rad55, and rad57 mutants show high sensitivity to IR indicates an important role for these genes in mitotic DSBR. However, in several assays for spontaneous and DSB-induced recombination, rad51, rad54, rad55, and rad57 mutants show high levels of recombination. RAD52 is required in the same assays, leading to the suggestion of RAD51-dependent and -independent pathways for mitotic recombination (the RAD51 pathway includes RAD54, RAD55, and RAD57), both of which are dependent on RAD52 (199, 233, 299, 364). RAD51, RAD52, RAD54, and RDH54 are required for high levels of spontaneous heteroallelic recombination in diploids, which is thought to occur primarily by gene conversion, and there is no induction of recombination by DNA-damaging agents in the corresponding mutants (179, 314). Diploids homozygous for rad55 or rad57 show wild-type levels of spontaneous heteroallelic recombination but show a reduction in UV-induced recombination at 23°C (211, 314). Because the DNA repair defect of rad55 mutants is suppressed by mating-type heterozygosity, the natural state of diploids, it is possible that the recombination defect in rad55 diploids is suppressed (211). Consistent with this hypothesis, ectopic gene conversion between heteroalleles in haploids is slightly reduced in rad55 and rad57 mutants (105, 199). However, rad51 and rad54 mutants show a much greater decrease in the rate of ectopic heteroallelic gene conversion in haploids (10- to 30-fold), and in rad52 mutants the rate is reduced more than 50-fold (105, 199). Thus, the requirement for RAD55 and RAD57 in spontaneous mitotic gene conversion is less stringent than for RAD51, RAD52, and RAD54. HO-induced gene conversion between allelic MAT sequences in diploids is dependent on RAD51, RAD52, and RAD54 but not on RDH54 (217, 343). As observed for radiation sensitivity, rad55 and rad57 diploids are more defective in HO-induced gene conversion at 23 than at 30°C, and the recombination defect at 23°C is comparable to that in rad51 mutants (343). Diploids homozygous for rad59 are not defective for HO-induced gene conversion between allelic MAT sequences, and they show elevated rates of spontaneous heteroallelic recombination (17, 343). In contrast, ectopic gene conversion in haploids is slightly reduced by mutation of RAD59 (105, 152). Because the radiation sensitivity of rad59 mutants is suppressed by mating-type heterozygosity, the recombination proficiency of rad59 diploids could be due to suppression of a mild defect by MATa/α (Y. Bai and L. Symington, unpublished data).
Very few studies have addressed the role of the RAD52 group genes in spontaneous mitotic crossing over. Because half of the crossover-associated conversion events are masked by chromatid segregation, it is difficult to distinguish between true reciprocal recombination and BIR (Fig. 2). Furthermore, mitotic crossover events that are selected by homozygosis of a recessive drug resistance marker are indistinguishable from BIR events. One assay system that avoids some of these problems is ectopic recombination in haploids. If the repeats are arranged in the same orientation relative to the centromeres of the two chromosomes, then viable reciprocal recombination products can be recovered. When essential genes are distal to the repeats, BIR events cannot give rise to viable recombinants. The ectopic repeats can be manipulated so that only crossover or only conversion products can be recovered as recombinants (105). Using such a system, Freedman and Jinks-Robertson showed a 10- to 20-fold decrease in crossovers in rad51, rad52, and rad54 mutants, a 3-fold decrease in crossovers in rad55, rad57, and rad59 mutants, and a 3-fold elevation in crossovers in rad50 mutants (105). These results confirm that true reciprocal exchanges occur during mitotic recombination and require RAD51, RAD52, and RAD54 for their formation. The mitotic crossovers that accompany rare gene conversions in rad52 diploids are aberrant and usually associated with the loss of one of the two chromosomes that participated in the recombination event (125).
HO-induced recombination occurs at a high enough frequency to monitor unselected events and therefore to determine whether crossovers are reciprocal. Using a chromosome VII disome, Galgoczy and Toczyski showed that repair of an HO-induced break was accompanied by reciprocal exchange in 27% of recombinants in wild-type strains, but fewer than 1% of the reciprocal products were recovered from rad51 and rad52 mutants (109).
Mating-type switching in S. cerevisiae occurs by a DSB-induced gene conversion event that is rarely associated with crossing over. Early studies using diploids heterozygous for HO and RAD52 demonstrated that RAD52 is essential for mating-type switching (221, 414). RAD51 is also essential, but some switched products are recovered from rad54, rad55, and rad57 mutants (1, 102, 159, 326). Physical monitoring of DNA during mating-type switching has shown more extensive resection of sequences distal to the HO break site in rad51, rad52, rad54, rad55, and rad57 mutants, and no strand invasion products are detected by PCR or Southern blot analysis (364, 419). The disparity between the physical monitoring and genetic assays for mating-type switching in rad54, rad55, and rad57 mutants may be due to the decreased sensitivity of physical assays compared with the genetic assay.
Gene conversion between heteroalleles oriented as a direct repeat in haploid strains is reduced 100-fold by mutation of RAD52 and 20- to 90-fold by mutation of RAD51 or RAD54 (153, 176, 286, 338). The rate of gene conversion between direct repeats has not been determined for rad55, rad57, and rad59 mutants. RAD52 is also required for mitotic recombination between the cluster of delta sequences oriented as direct and inverted repeats around the SUP4 locus (308). Spontaneous gene conversion between heteroalleles oriented as an inverted repeat is reduced more than 5,000-fold in rad52 mutants (299). However, the rate of gene conversion (unassociated with inversion) is reduced only 19-fold in rad51 mutants, and inversion events occur at 50% of the wild-type rate (299). The rate of gene conversion between inverted repeats is reduced about 60-fold in rad54 and rad57 mutants, but the rad51 rad54 and rad51 rad57 double mutants show the same rate as the rad51 single mutant (298). This epistasis could reflect channeling of recombinogenic lesions into an alternate pathway that is used less effectively when Rad51 is present and the mediator proteins are absent, perhaps because of unproductive association of Rad51 with ssDNA. When Rad51 is also removed, the alternate pathway functions more efficiently. As described above, RAD59 was identified as a component of the alternate pathway because recombination of inverted repeats was reduced more than 1,000-fold in the rad51 rad59 double mutant (17).
RAD51-independent mitotic recombination.
When HO is used to create a DSB in one of the two MAT alleles in wild-type diploids (in strains with HMR and HML deleted to prevent intramolecular recombination), repair occurs by gene conversion (217). As described above, gene conversion is eliminated in rad51 mutants but repair of the broken chromosome can still occur by a mechanism that results in homozygosis of all markers distal to the DSB. This mechanism is known as BIR and is thought to occur by one-ended strand invasion followed by duplication of the sequences from the break site to the telomere. The process is inefficient in rad51 mutants and appears to be preceded by cell cycle arrest and checkpoint adaptation (109, 217). BIR is observed in rad51, rad54, rad55, and rad57 mutants but not in rad52 mutants (343). BIR in rad51 diploids requires RAD50 (and presumably MRE11 and XRS2), as well as RAD59 and RDH54. Interestingly, RDH54 is required for checkpoint adaptation; therefore, the requirement in rad51-independent BIR could be due to the failure of cells to adapt (193). Because BIR in rad51 mutants involves the degradation or about 30 kb of DNA from the HO-cut site to a preferred site for BIR, it is possible that the requirement for RAD50 is in DNA degradation rather than in the strand invasion step (218). The genetic requirements for BIR and telomere maintenance in the absence of telomerase are quite similar, leading to the suggestion that BIR plays an important role in recombination between telomeres. Telomerase-deficient survivors that arise by recombination between the subtelomeric repeated Y′ elements require the RAD51 pathway genes, suggesting that this pathway does promote BIR, but when two ends are present, gene conversion is favored over BIR (388).
Ends-in and ends-out gene targeting involves the introduction of naked DNA into cells by transformation followed by homologous recombination with the chromosomal locus. Both types of events are dependent on RAD52 (20, 269, 322). RAD51 and RAD57 are required for ends-in targeting but show only a 3- to 10-fold reduction in ends-out targeting (rad54 and rad55 mutants were not tested) (20, 322). Several models have been proposed for ends-out gene targeting; presumably, they are all dependent on RAD52, but only one of the proposed pathways may require RAD51 function (194).
Analysis of recombination between repeated sequences has caused the greatest confusion with regard to the requirement for the RAD52 group genes in homologous recombination. As described above, recombination between sequences oriented as direct repeats can occur by gene conversion, maintaining the structure of the duplication, or by deletion, removing one repeat and the intervening sequences. Some direct-repeat substrates are designed only to select for deletion events, whereas those employing heteroalleles can be used to measure gene conversion and/or deletion events. Most studies have focused on spontaneous deletion events between direct repeats, and these events are actually elevated in rad51, rad54, rad55, and rad57 mutants (199, 233). The higher rate of deletions in RAD51 pathway mutants is presumably due to channeling lesions from the favored gene conversion pathway to mutagenic pathways such as SSA, sister strand mispairing, and replication slippage (177). Studies in which HO endonuclease is used to create a DSB between repeats have shown that SSA occurs with high efficiency and with normal kinetics in rad51, rad54, rad55, and rad57 mutants (150). These results are consistent with the hypothesis that spontaneous DSBs cannot be repaired by gene conversion in RAD51 pathway mutants but can be efficiently repaired by SSA when direct repeats are available.
In contrast to rad51, rad54, rad55, and rad57 mutants, rad52 mutants show a 2- to 10-fold reduction in the rate of spontaneous deletions between direct repeats (176, 319, 390). DSB-induced deletion formation between plasmid-borne direct repeats is reduced about eightfold in rad52 mutants; however, deletions are not detected by physical methods between chromosomal ura3 or leu2 repeats in rad52 strains (101, 347, 362). This difference could be due to plasmid versus chromosomal sequences or to the location of the HO-cut site within one of the repeats in the plasmid study and between them in the chromosomal studies. These results suggest that RAD52 is important for SSA, consistent with biochemical experiments demonstrating annealing of complementary ssDNA by Rad52 (250).
Studies of spontaneous direct-repeat recombination led to the discovery of a role for the nucleotide excision repair gene, RAD1, in this process. The effect of a rad1 mutation by itself is quite variable in different direct-repeat assays, but when combined with rad52, rad1 causes a synergistic decrease in the rate of direct-repeat recombination (176, 319, 391). Rad1 interacts with Rad10, and, as anticipated, rad10 mutants have similar defects to rad1 mutants in direct-repeat recombination (320). Although the rad1 mutation confers a defect in deletion formation, there is no effect on spontaneous gene conversion (319). Studies of HO-induced recombination have demonstrated a role for Rad1 in SSA (148), and, consistent with the prediction from the genetic studies, the Rad1/10 heterodimer is a structure-specific nuclease that cleaves 3′ tails from branched intermediates (Fig. 13) (19, 100).
FIG. 13.
Role of the Rad1-Rad10 nuclease in recombination. (A) In the SSA reaction, resected 3′ single-stranded tails are first coated with RPA; the RPA is then displaced by Rad52 to promote annealing of the complementary single strands. After annealing, 3′ heterologous tails are removed by the Rad1-Rad10 endonuclease, which cleaves at the junction between dsDNA and ssDNA. Gaps are filled by repair synthesis and ligated. (B) If the invading 3′ end is heterologous to the donor, then the 3′ end cannot be used to template DNA synthesis. The branched structure is cleaved by Rad1-Rad10, creating a primer for DNA synthesis. The second 3′ heterologous tail is also removed by Rad1-Rad10 for the second round of DNA synthesis predicted by the SDSA model (shown) or DSBR model.
Inverted repeats have been used to study recombination with the expectation that viable recombinants cannot be generated by nonconservative pathways such as SSA (80, 299). Although recombination between inverted repeats is highly dependent on RAD52, the rate of recombination in the other RAD52 group mutants is reduced by only 4- to 30-fold (4, 17, 80, 299, 421). Recombinants can be generated by gene conversion, inversion of the intervening DNA (apparent crossover), or gene conversion associated with inversion when the inverted-repeat substrate consists of heteroalleles. In rad51 mutants, most of the recombinants recovered are inversions, suggesting that RAD51 is more important for gene conversion than for crossing over (299). Using a plasmid containing inverted copies of the lacZ gene, one containing an HO-cut site, Sugawara et al. reported no effect of the rad51 mutation on the efficiency of HO-induced recombination (364). However, RAD51 was required for repair of an HO-induced break within plasmid-borne inverted repeats when the donor was in heterochromatin. This interesting result suggests that the RAD51 pathway can efficiently use donor DNA assembled into heterochromatin but that the RAD51-independent pathway, presumably mediated by Rad52, cannot. Analysis of the products generated from the lacZ inverted repeat plasmid in rad51 strains revealed a higher proportion of inversions (crossovers), consistent with the study of chromosomal inverted repeats (166). This observation suggests that the mechanism of repair in rad51 mutants is different from that in wild-type strains.
Although RAD51 is not required for DSB-induced recombination of plasmid-borne inverted repeats that are transcribed, rad51 mutants are severely deficient in the repair of gapped plasmids introduced into cells by transformation from either plasmid or chromosomal donor sequences (20). RAD51 is also required for the repair of an HO-induced DSB present on a plasmid from a chromosomal donor (86). These results suggests that plasmid context is not the determinant of RAD51-independent recombination. Instead, the results are more consistent with intermolecular recombination events requiring RAD51 function and intramolecular recombination events being largely independent of RAD51. HO-induced recombination between inverted repeats, like spontaneous events, requires RAD52 (364). Because BIR and SSA can both occur in the absence of RAD51 but are dependent on RAD52, we have suggested that recombination between inverted repeats could occur by a combination of these two processes (Fig. 14) (20, 166). The BIR-SSA model proposes that one end at the break site invades the homologous repeat and primes DNA synthesis to the end of the molecule. The small repeats generated by BIR could then be substrates for SSA, generating either inversion (crossover), or noninversion (conversion) products. One of the attractive features of this model is that apparent crossover products can be generated by a mechanism that does not involve the resolution of a dHJ intermediate. Experimental support for this model is derived from the observations that rad1 rad51 double mutants show a lower efficiency of repair of inverted-repeat plasmids and that more inversion products are recovered from rad51 strains (166). The RAD51-independent pathway for HO-induced inverted-repeat recombination is still efficient even when the homologies are short (30- bp) and requires RAD50 and RAD59 (147). Although this mechanism might be functionally active in rad51 mutants, there is no evidence for these events in wild-type cells.
FIG. 14.
The BIR/SSA model for repair of DSBs within inverted repeat plasmids. After formation of the DSB, ends are processed and one end invades the other repeat (drawn here as an intramolecular event, but it could also occur intermolecularly). If DNA synthesis extends to the end of the molecule, small repeats will be present at both ends of the linear molecule. Following resection of the ends, the repeats at either end of the molecule can pair with an internal repeat by SSA, generating either noncrossover or apparent crossover products.
Because RAD52 is the only known gene required for BIR and other RAD51-independent recombination events, it is assumed that the annealing activity of Rad52 can be used for strand invasion (217, 299). Presumably some donor sequences are transiently single stranded and can be annealed to ssDNA generated from a resected DSB by Rad52. Once this annealing step has occurred, the 3′ end could be extended by DNA synthesis to the end of the molecule. For plasmids, DNA synthesis would need to occur for only a few kilobases to complete replication of the molecule whereas synthesis might have to occur over several hundred kilobases to repair a chromosome arm.
In summary, the RAD51 pathway is clearly important for gene conversion unassociated with crossing over and for true reciprocal crossover recombination. These events probably occur as envisioned in the DSBR and SDSA models (Fig. 1), with the Rad51 protein playing a central role in homologous pairing and strand invasion. The mediator proteins, Rad52, Rad55, and Rad57, function with Rad51 to promote the assembly of the Rad51 nucleoprotein filament on ssDNA (Fig. 15). Rad54 is required during synapsis to promote unwinding of the donor duplex in order to allow pairing between the incoming ssDNA and the complementary strand of the donor duplex. Rad54 could play additional roles in heteroduplex extension and removal of Rad51 from the newly formed joint molecule to allow access of the 3′ end to the DNA replication apparatus (349, 350). In the absence of RAD51, RAD54, RAD55, and RAD57, some types of recombinational repair can still occur but are dependent on the context of the recombining sequences. SSA is extremely efficient in the absence of the RAD51, RAD54, RAD55, and RAD57 but is a viable option only if direct repeats are available flanking the break site. RAD52 is clearly important for SSA, and RAD59 is required when the direct repeats are short (362, 363). RAD51-independent BIR may be restricted because certain sequences are required for strand invasion, and unscheduled replication through the centromere has not been documented (218, 248). Thus, RAD51-independent BIR may be an option only for cells that have undergone prolonged cell cycle arrest and adaptation to damage and that have a BIR facilitator sequence centromere proximal to the break site.
FIG. 15.
Model for strand invasion by Rad51 and the mediator proteins, Rad52, Rad54, and Rad55-Rad57. Single-stranded tails produced at break sites are coated by RPA. Rad52 interacts with RPA targeting Rad51 to the ssDNA. Rad52 is thought to displace RPA and promote the binding of Rad51 to ssDNA. This initial interaction between Rad51 and ssDNA is thought to be stabilized by Rad55-Rad57 to allow cooperative binding by Rad51. Rad54 interacts with the Rad51 nucleoprotein filament and promotes the unwinding of duplex DNA for pairing between the donor DNA and the incoming single strand.
RAD51 is essential for S-phase progression in chicken cells and is proposed to function in restoration of collapsed replication forks (Fig. 16). Repair of collapsed forks involves the invasion of one broken end into an intact duplex. This configuration is similar to the BIR reaction modeled by Malkova et al. (217), except that the donor during replication restart is a sister chromatid instead of a homologue. Because BIR can occur in the absence of RAD51 in yeast, why is RAD51 essential for viability in vertebrates? There are several possible explanations for this conundrum. First, RAD51-independent BIR is inefficient in yeast, raising the possibility that this pathway would be unable to repair multiple replication-induced breaks in more complex genomes. Second, the genetic requirements for BIR between sister chromatids might be different from the requirement for BIR between homologues. Studies of telomere maintenance have demonstrated RAD51-dependent recombination between subtelomeric repeats, and this most probably occurs by BIR. It seems possible that RAD51 could be required for BIR under most circumstances and that rare BIR events could occur in the absence of RAD51, but these would probably be inefficient and would only rarely contribute to cell survival.
FIG. 16.
Role of recombination in restoring collapsed or regressed replication forks. The replication fork collapses if it encounters a nick on the template strand. After ligation of the lagging strand to the template, the broken arm invades, forming a D-loop. Replication can then be primed from the invading strand. Resolution of the Holliday junction (HJ) restores the replication fork. A replication-blocking lesion stalls replication, and the fork regresses by pairing of the nascent strands. If the lagging strand has progressed ahead of the leading strand, the single-strand extension on the lagging strand can serve as a primer for synthesis of the leading strand. If the Holliday junction is cleaved, an end is made for strand invasion, as shown in the left panel. Alternatively, branch migration of the regressed fork can restore the replication fork.
HONORARY MEMBERS OF THE RECOMBINATIONAL REPAIR GROUP
Rad1/Rad10
The Rad1/10 heterodimer is a structure-specific nuclease that cleaves at the 5′ side of UV-induced photoproducts and bulky lesions during nucleotide excision repair (19, 74). As described above, the Rad1/10 nuclease is thought to remove 3′ flaps during SSA (Fig. 13). In addition, it is required to remove heterologies from the 3′ ends of DSB breaks to allow initiation of DNA synthesis during gene conversion. This activity was first discovered when a substrate containing an insertion of a 117-bp HO-cut site within one copy of the lacZ gene was used to monitor the efficiency of HO-induced gene conversion (100). rad1 mutants were found to be defective for gene conversion using this substrate, but there was no defect in repair when the donor sequence contained a 117-bp insertion at the homologous site that differed from the recipient allele by a nucleotide change preventing cleaving by HO. Thus, the requirement for RAD1 and RAD10 in homologous recombination is specific for events that require removal of heterologies, either during strand invasion or during SSA (Fig. 13). The mismatch repair proteins Msh2 and Msh3 are also required for SSA and for removal of heterologies of more than 30 nucleotides from DSBs during gene conversion (272, 317, 365). Mutation of MSH2 or MSH3 is epistatic to rad1 in these repair processes. These results have led to the suggestion that branched intermediates are stabilized by Msh2/Msh3 binding in preparation for cleavage by Rad1/Rad10 (365).
In mammalian cells, XPF and ERCC1 encode the Rad1 and Rad10 homologues, respectively (74). Although ERCC1 is not required for spontaneous recombination between repeated sequences (318), ercc1−/− cell lines do exhibit defects in ends-inand ends-out gene targeting (Fig. 6) (2, 263). ERCC1 is required for ends-in gene targeting when the ends of the transformed DNA have long heterologies; this is consistent with studies from yeast (2). Surprisingly, ercc1−/− cell lines were found to be completely defective for ends-out gene targeting, even when the ends of the targeting fragment were homologous to the target sequence (263). Because gene-targeting constructs generally have an insertion of a selectable marker creating a large heterology, the defect in targeting could be due to the central heterology. Repair of large-loop heteroduplexes during meiotic recombination requires RAD1 and RAD10 in addition to MSH2 (169, 173). Studies with yeast have shown a reduction in ends-out gene targeting in rad1 mutants but of only 3- to 10-fold (319, 379). The reason for the disparity between yeast and vertebrates is unclear but could involve alternate mechanisms for targeting in yeast, which are unavailable in vertebrate cells.
Holliday Junction Resolution Activities
Molecular and biochemical studies of the Rad52 group proteins have shown that most are required at early steps during recombinational repair. The absence of a yeast X-ray-sensitive mutant defective for Holliday junction resolution is surprising and suggests that (i) redundant activities exist in eukaryotes, (ii) it is an essential activity, or (iii) Holliday junction resolution is not obligatory for recombinational repair in yeast. For example, the repair of DSBs by the SDSA model is predicted to occur without Holliday junction resolution (Fig. 1). However, some mitotic recombination events do result in reciprocal exchange, and integration of linearized plasmids into the genome is thought to occur by resolution of Holliday junctions as predicted by the DSBR model (Fig. 1). The identification of branched-DNA molecules with the expected topology for Holliday junctions during meiotic recombination is consistent with the hypothesis that these structures are normal intermediates during meiotic recombination in yeast (61, 327, 328). Branched-DNA molecules have been detected during mitotic S phase within the tandemly repeated rDNA locus, and formation of these intermediates is dependent on RAD52, suggesting that they correspond to recombination intermediates (433). Postreplicative, DNA replication-dependent X-shaped molecules have also been detected between sister chromatids in Physarum polycephalum (25). The X-forms are readily detected, indicating that they form at high frequency; this suggests that resolution of Holliday junctions could in fact be essential for chromosome segregation in eukaryotes.
In E. coli, the RuvC nuclease promotes the resolution of Holliday junctions and works in concert with the RuvA/RuvB branch migration complex (417). The ruv genes are required for resistance of E. coli to UV, X rays, and mitomycin C and for plasmid and conjugal recombination. No homologues of RuvC or the bacteriophage-encoded resolvases have been identified among sequenced eukaryotic genomes, except for the mitochondrial activity, Cce1 (13). Biochemical approaches have identified three Holliday junction-resolving activities from fractionated extracts of mitotic yeast cells (154, 380, 418). Of these, Cce1 (Mgt1) is mitochondrial and is important for the segregation of mitochondrial genomes (92, 175, 208, 380). The identity of the other two yeast resolvase activities is still unknown. Holliday junction resolvases have also been identified from fractionated extracts of mammalian cells (65, 85). The resolvase identified by Constantinou et al. (65) was found to cofractionate with an ATP-dependent branch migration activity during several chromatographic steps. These activities were shown to be nuclear and independent of the BLM and WRN helicases, which had previously been shown to promote ATP-dependent branch migration of Holliday junctions (65, 66, 168). The mammalian activity promotes a concerted branch migration/resolution reaction similar to that catalyzed by RuvABC (65).
Recent studies have identified the Mus81-Mms4 (Eme1) heterodimer as a putative Holliday junction resolvase (126). Mus81 and Mms4 were identified in two-hybrid screens by interaction with Rad54, S. pombe Cdc1 (the orthologue of S. cerevisiae Rad53), and the meiosis-specific checkpoint kinase, Mek1 (35, 75, 146). The MMS4 gene was originally identified by the sensitivity of mutants to MMS and rediscovered in a screen for mutations that cause synthetic lethality with sgs1, along with mus81 (253, 293). SGS1 encodes a helicase of the RecQ family and is homologous to the human BLM and WRN helicases, which play important roles in genome stability and life span (111a, 239). Mus81 has sequence similarity to the nuclease domain of Rad1, and Mms4 has limited similarity to Rad10, suggesting that they are components of a structure-specific nuclease (146, 253). Eme1 was identified by interaction with SpMus81 and is the presumptive orthologue of Mms4 (34). The Mus81-Mms4/Eme1 heterodimer cleaves a variety of branched molecules, including simple Y structures, duplex Y structures and X forms, with a strong preference for duplex Y structures (34, 164). The preferred substrate for Mus81-Mms4/Eme1 is structurally related to a stalled replication fork, leading to the hypothesis that Sgs1 and Mus81-Mms4 are redundant functions for processing stalled replication forks (Fig. 16) (77, 164). Alternatively, unusual structures may accumulate in sgs1 mutants requiring cleavage by Mus81-Mms4.
MUS81 and MMS4/EME1 are also required for meiotic DNA metabolism (34, 75, 146, 164, 253). In S. pombe, mus81 and eme1 diploids produce only 1% viable spores and the high inviability is partially suppressed by the rec12 mutation, which prevents initiation of meiotic recombination (34). Most striking is the observation that expression of the E. coli Holliday junction resolvase, RusA, in mus81 diploids restores spore viability to 35%. While these data provide a compelling argument for Mus81-Eme1 being the elusive Holliday junction resolvase, studies with S. cerevisiae are not fully compatible with this hypothesis. First, mus81 mutants are resistant to IR (146). Although one could argue that IR-induced damage is repaired by mechanisms that do not require Holliday junction resolution, mus81 mutants also do not show any defect in integration of plasmids (B. Llorente and L. S. Symington, unpublished data). Second, mms4 and mus81 diploids show quite high spore viability in some strain backgrounds and crossover products are readily detected by both genetic and physical analyses (75). Third, Holliday junctions cleaved by Mus81-Mms4/Eme1 cannot be ligated, indicating that the cleavage sites are not symmetrical (64). The bacterial and bacteriophage-encoded resolvases cleave Holliday junctions symmetrically to generate products that can be ligated. This result has led to the suggestion that Mus81-Mms4/Eme1 cleaves branched structures that form during SDSA, instead of the dHJ intermediates that are destined to become crossovers (75).
The discrepancy between the S. pombe and S. cerevisiae data suggests that budding yeast could have another activity for resolving Holliday junctions and that this activity might be absent or less effective during S. pombe meiosis. Support for the hypothesis of two resolvase activities comes from recent studies by Constantinou et al. (64). Fractionation of HeLa cell extracts revealed two discrete Holliday junction resolvase activities, one corresponding to Mus81 and the other corresponding to the previously described resolvase that cofractionates with a branch migration activity, referred to as resolvase A (64, 65). In addition to their distinct chromatographic properties, resolvase A activity was not depleted by anti-Mus81 polyclonal antibodies and the substrate specificities of the two activities were different (64). Resolvase A shows high specificity for Holliday junctions, whereas Mus81 cleaves 3′ flap and Y-shaped molecules more efficiently than Holliday junctions (64, 77, 164). Thus, genetic and biochemical data support the hypothesis that Mus81 cleaves stalled replication forks and 3′ flaps that form during meiotic SDSA. Resolvase A has the hallmarks of a true resolvase, and we eagerly await the identification of the gene encoding this activity.
CONCLUSIONS AND FUTURE DIRECTIONS
In summary, significant advances have been made in our understanding of the structure and biochemical functions of the Mre11-Rad50 complex, but the function of the complex in various aspects of DNA metabolism remains elusive. Recent studies support the hypothesis that the primary function of the Mre11-Rad50 complex is structural and serves to bridge sister chromatids and/or DNA ends (53, 73, 140). The nuclease activity of Mre11 is required to remove covalent adducts from DNA ends and to process unusual DNA structures, but it may not play a significant role in resection of ends to produce long 3′ single-stranded tails (108, 207, 241, 359, 407). Xrs2/Nbs1 is restricted to eukaryotes and may function to recruit other factors for specialized functions within the cell, such as the Dn14-Lif1 complex for end joining (53). The RAD51 pathway is clearly important for gene conversion unassociated with crossing over and for true reciprocal-crossover recombination. These events probably occur as envisioned in the DSBR or SDSA models, with the Rad51 protein playing a central role in homologous pairing and strand invasion with the assistance of the mediator proteins. In the absence of RAD51, RAD54, RAD55, and RAD57, some types of recombinational repair can still occur but are dependent on the context of the recombining sequences. SSA is only a viable option if direct repeats are available flanking the break site, and RAD51-independent BIR may be restricted because certain sequences are required for strand invasion. RAD52 is the most important recombination gene in S. cerevisiae, and this is presumably due to the important role of Rad52 as a mediator for Rad51 and in strand annealing for RAD51-independent recombination. While many advances have been made in this field, we still know little about the nucleases that function in early and late stages of recombination, the roles of the Rad51 accessory proteins, and coordination of homologous recombination with other cellular processes. Understanding the intimate relationship between damage sensing, replication, and homologous recombination and the ways in which these processes function in tumor suppression is likely to keep the field busy for many years to come.
Acknowledgments
I am grateful to members of my laboratory, past and present, and W. K. Holloman for stimulating discussions and critical reading of the manuscript.
Research performed in my laboratory that is cited in this review was supported by grants from the National Institutes for Health (GM41784 and GM54099).
REFERENCES
- 1.Aboussekhra, A., R. Chanet, A. Adjiri, and F. Fabre. 1992. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol. Cell. Biol. 12:3224-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adair, G. M., R. L. Rolig, D. Moore-Faver, M. Zabelshansky, J. H. Wilson, and R. S. Nairn. 2000. Role of ERCC1 in removal of long non-homologous tails during targeted homologous recombination. EMBO J. 19:5552-5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adzuma, K., T. Ogawa, and H. Ogawa. 1984. Primary structure of the RAD52 gene in Saccharomyces cerevisiae. Mol. Cell. Biol. 4:2735-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aguilera, A. 1995. Genetic evidence for different RAD52-dependent intrachromosomal recombination pathways in Saccharomyces cerevisiae. Curr. Genet. 27:298-305. [DOI] [PubMed] [Google Scholar]
- 5.Aguilera, A., and H. L. Klein. 1989. Yeast intrachromosomal recombination: long gene conversion tracts are preferentially associated with reciprocal exchange and require the RAD1 and RAD3 gene products. Genetics 123:683-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aihara, H., Y. Ito, H. Kurumizaka, S. Yokoyama, and T. Shibata. 1999. The N-terminal domain of the human Rad51 protein binds DNA: structure and a DNA binding surface as revealed by NMR. J. Mol. Biol. 290:495-504. [DOI] [PubMed] [Google Scholar]
- 7.Ajimura, M., S. H. Leem, and H. Ogawa. 1993. Identification of new genes required for meiotic recombination in Saccharomyces cerevisiae. Genetics 133:51-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alani, E., R. Padmore, and N. Kleckner. 1990. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61:419-436. [DOI] [PubMed] [Google Scholar]
- 9.Alani, E., S. Subbiah, and N. Kleckner. 1989. The yeast RAD50 gene encodes a predicted 153-kD protein containing a purine nucleotide-binding domain and two large heptad-repeat regions. Genetics 122:47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albala, J. S., M. P. Thelen, C. Prange, W. Fan, M. Christensen, L. H. Thompson, and G. G. Lennon. 1997. Identification of a novel human RAD51 homolog, RAD51B. Genomics 46:476-479. [DOI] [PubMed] [Google Scholar]
- 11.Allers, T., and M. Lichten. 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106:47-57. [DOI] [PubMed] [Google Scholar]
- 12.Anderson, D. E., K. M. Trujillo, P. Sung, and H. P. Erickson. 2001. Structure of the Rad50-Mre11 DNA repair complex from Saccharomyces cerevisiae by electron microscopy. J. Biol. Chem. 276:37027-37033. [DOI] [PubMed] [Google Scholar]
- 13.Aravind, L., K. S. Makarova, and E. V. Koonin. 2000. Survey and Summary. Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 28:3417-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arbel, A., D. Zenvirth, and G. Simchen. 1999. Sister chromatid-based DNA repair is mediated by RAD54, not by DMC1 or TID1. EMBO J. 18:2648-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asleson, E. N., R. J. Okagaki, and D. M. Livingston. 1999. A core activity associated with the N terminus of the yeast RAD52 protein is revealed by RAD51 overexpression suppression of C-terminal rad52 truncation alleles. Genetics 153:681-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai, Y., A. P. Davis, and L. S. Symington. 1999. A novel allele of RAD52 that causes severe DNA repair and recombination deficiencies only in the absence of RAD51 or RAD59. Genetics 153:1117-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai, Y., and L. S. Symington. 1996. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev 10:2025-2037. [DOI] [PubMed] [Google Scholar]
- 18.Reference deleted.
- 19.Bardwell, A. J., L. Bardwell, A. E. Tomkinson, and E. C. Friedberg. 1994. Specific cleavage of model recombination and repair intermediates by the yeast Rad1-Rad10 DNA endonuclease. Science 265:2082-2085. [DOI] [PubMed] [Google Scholar]
- 20.Bartsch, S., L. E. Kang, and L. S. Symington. 2000. RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates. Mol. Cell. Biol. 20:1194-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bashkirov, V. I., J. S. King, E. V. Bashkirova, J. Schmuckli-Maurer, and W. D. Heyer. 2000. DNA repair protein Rad55 is a terminal substrate of the DNA damage checkpoints. Mol. Cell. Biol. 20:4393-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basile, G., M. Aker, and R. K. Mortimer. 1992. Nucleotide sequence and transcriptional regulation of the yeast recombinational repair gene RAD51. Mol. Cell. Biol. 12:3235-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baumann, P., F. E. Benson, and S. C. West. 1996. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell 87:757-766. [DOI] [PubMed] [Google Scholar]
- 24.Baumann, P., and S. C. West. 1999. Heteroduplex formation by human Rad51 protein: effects of DNA end- structure, hRP-A and hRad52. J. Mol. Biol. 291:363-374. [DOI] [PubMed] [Google Scholar]
- 25.Benard, M., C. Maric, and G. Pierron. 2001. DNA replication-dependent formation of joint DNA molecules in Physarum polycephalum. Mol. Cell. 7:971-980. [DOI] [PubMed] [Google Scholar]
- 26.Bender, C. F., M. L. Sikes, R. Sullivan, L. E. Huye, M. M. Le Beau, D. B. Roth, O. K. Mirzoeva, E. M. Oltz, and J. H. Petrini. 2002. Cancer predisposition and hematopoietic failure in Rad50(S/S) mice. Genes Dev. 16:2237-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennett, R. L., and W. K. Holloman. 2001. A RecA homologue in Ustilago maydis that is distinct and evolutionarily distant from Rad51 actively promotes DNA pairing reactions in the absence of auxiliary factors. Biochemistry 40:2942-2953. [DOI] [PubMed] [Google Scholar]
- 28.Benson, F. E., P. Baumann, and S. C. West. 1998. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature 391:401-404. [DOI] [PubMed] [Google Scholar]
- 29.Bezzubova, O., A. Silbergleit, Y. Yamaguchi-Iwai, S. Takeda, and J. M. Buerstedde. 1997. Reduced X-ray resistance and homologous recombination frequencies in a RAD54−/− mutant of the chicken DT40 cell line. Cell 89:185-193. [DOI] [PubMed] [Google Scholar]
- 30.Bezzubova, O. Y., H. Schmidt, K. Ostermann, W. D. Heyer, and J. M. Buerstedde. 1993. Identification of a chicken RAD52 homologue suggests conservation of the RAD52 recombination pathway throughout the evolution of higher eukaryotes. Nucleic Acids Res. 21:5945-5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bishop, D. K. 1994. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 79:1081-1092. [DOI] [PubMed] [Google Scholar]
- 32.Bishop, D. K., Y. Nikolski, J. Oshiro, J. Chon, M. Shinohara, and X. Chen. 1999. High copy number suppression of the meiotic arrest caused by a dmc1 mutation: REC114 imposes an early recombination block and RAD54 promotes a DMC1-independent DSB repair pathway. Genes Cells 4:425-444. [DOI] [PubMed] [Google Scholar]
- 33.Bishop, D. K., D. Park, L. Xu, and N. Kleckner. 1992. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69:439-456. [DOI] [PubMed] [Google Scholar]
- 34.Boddy, M. N., P. H. Gaillard, W. H. McDonald, P. Shanahan, J. R. Yates, 3rd, and P. Russell. 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107:537-548. [DOI] [PubMed] [Google Scholar]
- 35.Boddy, M. N., A. Lopez-Girona, P. Shanahan, H. Interthal, W. D. Heyer, and P. Russell. 2000. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 20:8758-8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borts, R. H., M. Lichten, and J. E. Haber. 1986. Analysis of meiosis-defective mutations in yeast by physical monitoring of recombination. Genetics 113:551-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boulton, S. J., and S. P. Jackson. 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17:1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boulton, S. J., and S. P. Jackson. 1996. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 15:5093-5103. [PMC free article] [PubMed] [Google Scholar]
- 39.Boundy-Mills, K. L., and D. M. Livingston. 1993. A Saccharomyces cerevisiae RAD52 allele expressing a C-terminal truncation protein: activities and intragenic complementation of missense mutations. Genetics 133:39-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braybrooke, J. P., K. G. Spink, J. Thacker, and I. D. Hickson. 2000. The RAD51 family member, RAD51L3, is a DNA-stimulated ATPase that forms a complex with XRCC2. J. Biol. Chem. 275:29100-29106. [DOI] [PubMed] [Google Scholar]
- 41.Brenneman, M. A., B. M. Wagener, C. A. Miller, C. Allen, and J. A. Nickoloff. 2002. XRCC3 controls the fidelity of homologous recombination: role for XRCC3 in late stages of recombination. Mol. Cell 10:387-395. [DOI] [PubMed] [Google Scholar]
- 42.Bressan, D. A., B. K. Baxter, and J. H. Petrini. 1999. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7681-7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bressan, D. A., H. A. Olivares, B. E. Nelms, and J. H. Petrini. 1998. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics 150:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brill, S. J., and B. Stillman. 1991. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 5:1589-1600. [DOI] [PubMed] [Google Scholar]
- 45.Brill, S. J., and B. Stillman. 1989. Yeast replication factor-A functions in the unwinding of the SV40 origin of DNA replication. Nature 342:92-95. [DOI] [PubMed] [Google Scholar]
- 46.Cao, L., E. Alani, and N. Kleckner. 1990. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61:1089-1101. [DOI] [PubMed] [Google Scholar]
- 47.Carney, J. P., R. S. Maser, H. Olivares, E. M. Davis, M. Le Beau, J. R. Yates III, L. Hays, W. F. Morgan, and J. H. Petrini. 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93:477-486. [DOI] [PubMed] [Google Scholar]
- 48.Celeste, A., S. Petersen, P. J. Romanienko, O. Fernandez-Capetillo, H. T. Chen, O. A. Sedelnikova, B. Reina-San-Martin, V. Coppola, E. Meffre, M. J. Difilippantonio, C. Redon, D. R. Pilch, A. Olaru, M. Eckhaus, R. D. Camerini-Otero, L. Tessarollo, F. Livak, K. Manova, W. M. Bonner, M. C. Nussenzweig, and A. Nussenzweig. 2002. Genomic instability in mice lacking histone H2AX. Science 296:922-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chalker, A. F., D. R. Leach, and R. G. Lloyd. 1988. Escherichia coli sbcC mutants permit stable propagation of DNA replicons containing a long palindrome. Gene 71:201-205. [DOI] [PubMed] [Google Scholar]
- 50.Chamankhah, M., T. Fontanie, and W. Xiao. 2000. The Saccharomyces cerevisiae mre11(ts) allele confers a separation of DNA repair and telomere maintenance functions. Genetics 155:569-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chanet, R., M. Heude, A. Adjiri, L. Maloisel, and F. Fabre. 1996. Semidominant mutations in the yeast Rad51 protein and their relationships with the Srs2 helicase. Mol. Cell. Biol. 16:4782-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen, C., and R. D. Kolodner. 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 23:81-85. [DOI] [PubMed] [Google Scholar]
- 53.Chen, L., K. Trujillo, W. Ramos, P. Sung, and A. E. Tomkinson. 2001. Promotion of Dn14-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8:1105-1115. [DOI] [PubMed] [Google Scholar]
- 54.Chen, P. L., C. F. Chen, Y. Chen, J. Xiao, Z. D. Sharp, and W. H. Lee. 1998. The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc. Natl. Acad. Sci. USA 95:5287-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen, Q., A. Ijpma, and C. W. Greider. 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 21:1819-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen, W., and S. Jinks-Robertson. 1998. Mismatch repair proteins regulate heteroduplex formation during mitotic recombination in yeast. Mol. Cell. Biol. 18:6525-6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clever, B., H. Interthal, J. Schmuckli-Maurer, J. King, M. Sigrist, and W. D. Heyer. 1997. Recombinational repair in yeast: functional interactions between Rad51 and Rad54 proteins. EMBO J. 16:2535-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clever, B., J. Schmuckli-Maurer, M. Sigrist, B. J. Glassner, and W. D. Heyer. 1999. Specific negative effects resulting from elevated levels of the recombinational repair protein Rad54p in Saccharomyces cerevisiae. Yeast 15:721-740. [DOI] [PubMed] [Google Scholar]
- 59.Cole, G. M., D. Schild, S. T. Lovett, and R. K. Mortimer. 1987. Regulation of RAD54- and RAD52-lacZ gene fusions in Saccharomyces cerevisiae in response to DNA damage. Mol. Cell. Biol. 7:1078-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cole, G. M., D. Schild, and R. K. Mortimer. 1989. Two DNA repair and recombination genes in Saccharomyces cerevisiae, RAD52 and RAD54, are induced during meiosis. Mol. Cell. Biol. 9:3101-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Collins, I., and C. S. Newlon. 1994. Meiosis-specific formation of joint DNA molecules containing sequences from homologous chromosomes. Cell. 76:65-75. [DOI] [PubMed] [Google Scholar]
- 62.Connelly, J. C., E. S. de Leau, E. A. Okely, and D. R. Leach. 1997. Overexpression, purification, and characterization of the SbcCD protein from Escherichia coli. J. Biol. Chem. 272:19819-19826. [DOI] [PubMed] [Google Scholar]
- 63.Connelly, J. C., and D. R. Leach. 1996. The sbcC and sbcD genes of Escherichia coli encode a nuclease involved in palindrome inviability and genetic recombination. Genes Cells 1:285-291. [DOI] [PubMed] [Google Scholar]
- 64.Constantinou, A., X.-B. Chen, C. H. McGowan, and S. C. West. 2002. Holliday junction resolvases in human cells: two activities with distinct substrate specificities. EMBO J. 21:5577-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Constantinou, A., A. A. Davies, and S. C. West. 2001. Branch migration and Holliday junction resolution catalyzed by activities from mammalian cells. Cell 104:259-268. [DOI] [PubMed] [Google Scholar]
- 66.Constantinou, A., M. Tarsounas, J. K. Karow, R. M. Brosh, V. A. Bohr, I. D. Hickson, and S. C. West. 2000. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 1:80-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Costanzo, V., K. Robertson, M. Bibikova, E. Kim, D. Grieco, M. Gottesman, D. Carroll, and J. Gautier. 2001. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol. Cell 8:137-147. [DOI] [PubMed] [Google Scholar]
- 68.D'Amours, D., and S. P. Jackson. 2001. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 15:2238-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reference deleted.
- 70.Davis, A. P., and L. S. Symington. 2001. The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single-strand annealing. Genetics 159:515-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deans, B., C. S. Griffin, M. Maconochie, and J. Thacker. 2000. Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J. 19:6675-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Debrauwere, H., S. Loeillet, W. Lin, J. Lopes, and A. Nicolas. 2001. Links between replication and recombination in Saccharomyces cerevisiae: a hypersensitive requirement for homologous recombination in the absence of Rad27 activity. Proc. Natl. Acad. Sci. USA 98:8263-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Jager, M., J. van Noort, D. C. van Gent, C. Dekker, R. Kanaar, and C. Wyman. 2001. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell 8:1129-1135. [DOI] [PubMed] [Google Scholar]
- 74.de Laat, W. L., N. G. Jaspers, and J. H. Hoeijmakers. 1999. Molecular mechanism of nucleotide excision repair. Genes Dev. 13:768-785. [DOI] [PubMed] [Google Scholar]
- 75.de los Santos, T., J. Loidl, B. Larkin, and N. M. Hollingsworth. 2001. A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics 159:1511-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.De Zutter, J. K., and K. L. Knight. 1999. The hRad51 and RecA proteins show significant differences in cooperative binding to single-stranded DNA. J. Mol. Biol. 293:769-780. [DOI] [PubMed] [Google Scholar]
- 77.Doe, C. L., J. S. Ahn, J. Dixon, and M. C. Whitby. 2002. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 277:32753-32759. [DOI] [PubMed] [Google Scholar]
- 78.Dolganov, G. M., R. S. Maser, A. Novikov, L. Tosto, S. Chong, D. A. Bressan, and J. H. Petrini. 1996. Human Rad50 is physically associated with human Mre11: identification of a conserved multiprotein complex implicated in recombinational DNA repair. Mol. Cell. Biol. 16:4832-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Donovan, J. W., G. T. Milne, and D. T. Weaver. 1994. Homotypic and heterotypic protein associations control Rad51 function in double-strand break repair. Genes Dev. 8:2552-2562. [DOI] [PubMed] [Google Scholar]
- 80.Dornfeld, K. J., and D. M. Livingston. 1992. Plasmid recombination in a rad52 mutant of Saccharomyces cerevisiae. Genetics 131:261-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dosanjh, M. K., D. W. Collins, W. Fan, G. G. Lennon, J. S. Albala, Z. Shen, and D. Schild. 1998. Isolation and characterization of RAD51C, a new human member of the RAD 51 family of related genes. Nucleic Acids Res. 26:1179-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dresser, M. E., D. J. Ewing, M. N. Conrad, A. M. Dominguez, R. Barstead, H. Jiang, and T. Kodadek. 1997. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics 147:533-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dvir, A., S. R. Peterson, M. W. Knuth, H. Lu, and W. S. Dynan. 1992. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc. Natl. Acad. Sci. USA 89:11920-11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eisen, J. A., K. S. Sweder, and P. C. Hanawalt. 1995. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 23:2715-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Elborough, K. M., and S. C. West. 1990. Resolution of synthetic Holliday junctions in DNA by an endonuclease activity from calf thymus. EMBO J. 9:2931-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Elias-Arnanz, M., A. A. Firmenich, and P. Berg. 1996. Saccharomyces cerevisiae mutants defective in plasmid-chromosome recombination. Mol. Gen. Genet. 252:530-538. [DOI] [PubMed] [Google Scholar]
- 87.Esposito, R. E., and M. S. Esposito. 1974. Genetic recombination and commitment to meiosis in Saccharomyces. Proc. Natl. Acad. Sci. USA 71:3172-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Essers, J., R. W. Hendriks, S. M. Swagemakers, C. Troelstra, J. de Wit, D. Bootsma, J. H. Hoeijmakers, and R. Kanaar. 1997. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell 89:195-204. [DOI] [PubMed] [Google Scholar]
- 89.Essers, J., A. B. Houtsmuller, L. van Veelen, C. Paulusma, A. L. Nigg, A. Pastink, W. Vermeulen, J. H. Hoeijmakers, and R. Kanaar. 2002. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 21:2030-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Essers, J., H. van Steeg, J. de Wit, S. M. Swagemakers, M. Vermeij, J. H. Hoeijmakers, and R. Kanaar. 2000. Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J. 19:1703-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Evans, E., N. Sugawara, J. E. Haber, and E. Alani. 2000. The Saccharomyces cerevisiae Msh2 mismatch repair protein localizes to recombination intermediates in vivo. Mol. Cell 5:789-799. [DOI] [PubMed] [Google Scholar]
- 92.Ezekiel, U. R., and H. P. Zassenhaus. 1993. Localization of a cruciform cutting endonuclease to yeast mitochondria. Mol. Gen. Genet. 240:414-418. [DOI] [PubMed] [Google Scholar]
- 93.Farah, J. A., E. Hartsuiker, K. Mizuno, K. Ohta, and G. R. Smith. 2002. A 160-bp palindrome is a Rad50.Rad32-dependent mitotic recombination hotspot in Schizosaccharomyces pombe. Genetics 161:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fasullo, M. T., and R. W. Davis. 1987. Recombinational substrates designed to study recombination between unique and repetitive sequences in vivo. Proc. Natl. Acad. Sci. USA 84:6215-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Featherstone, C., and S. P. Jackson. 1998. DNA repair: the Nijmegen breakage syndrome protein. Curr. Biol. 8:R622-R625. [DOI] [PubMed] [Google Scholar]
- 96.Ferguson, D. O., and W. K. Holloman. 1996. Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc. Natl. Acad. Sci. USA 93:5419-5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ferguson, D. O., M. C. Rice, M. H. Rendi, H. Kotani, E. B. Kmiec, and W. K. Holloman. 1997. Interaction between Ustilago maydis REC2 and RAD51 genes in DNA repair and mitotic recombination. Genetics 145:243-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fiorentini, P., K. N. Huang, D. X. Tishkoff, R. D. Kolodner, and L. S. Symington. 1997. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol. Cell. Biol. 17:2764-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Firmenich, A. A., M. Elias-Arnanz, and P. Berg. 1995. A novel allele of Saccharomyces cerevisiae RFA1 that is deficient in recombination and repair and suppressible by RAD52. Mol. Cell. Biol. 15:1620-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fishman-Lobell, J., and J. E. Haber. 1992. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science 258:480-484. [DOI] [PubMed] [Google Scholar]
- 101.Fishman-Lobell, J., N. Rudin, and J. E. Haber. 1992. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol. Cell. Biol. 12:1292-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fortin, G. S., and L. S. Symington. 2002. Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing stability of Rad51/DNA complexes. EMBO J. 21:3160-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Frank, K. M., J. M. Sekiguchi, K. J. Seidl, W. Swat, G. A. Rathbun, H. L. Cheng, L. Davidson, L. Kangaloo, and F. W. Alt. 1998. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature 396:173-177. [DOI] [PubMed] [Google Scholar]
- 104.Frank-Vaillant, M., and S. Marcand. 2001. NHEJ regulation by mating type is exercised through a novel protein, Lif2p, essential to the ligase IV pathway. Genes Dev. 15:3005-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Freedman, J. A., and S. Jinks-Robertson. 2002. Genetic requirements for spontaneous and transcription-stimulated mitotic recombination in Saccharomyces cerevisiae. Genetics 162:15-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.French, C. A., J. Y. Masson, C. S. Griffin, P. O'Regan, S. C. West, and J. Thacker. 2002. Role of mammalian RAD51L2 (RAD51C) in recombination and genetic stability. J. Biol. Chem. 277:19322-19330. [DOI] [PubMed] [Google Scholar]
- 107.Fujimori, A., S. Tachiiri, E. Sonoda, L. H. Thompson, P. K. Dhar, M. Hiraoka, S. Takeda, Y. Zhang, M. Reth, and M. Takata. 2001. Rad52 partially substitutes for the Rad51 paralog XRCC3 in maintaining chromosomal integrity in vertebrate cells. EMBO J. 20:5513-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Furuse, M., Y. Nagase, H. Tsubouchi, K. Murakami-Murofushi, T. Shibata, and K. Ohta. 1998. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 17:6412-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Galgoczy, D. J., and D. P. Toczyski. 2001. Checkpoint adaptation precedes spontaneous and damage-induced genomic instability in yeast. Mol. Cell. Biol. 21:1710-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Game, J. C., and R. K. Mortimer. 1974. A genetic study of x-ray sensitive mutants in yeast. Mutat. Res. 24:281-292. [DOI] [PubMed] [Google Scholar]
- 111.Game, J. C., T. J. Zamb, R. J. Braun, M. Resnick, and R. M. Roth. 1980. The role of the radiation (rad) genes in meiotic recombination in yeast. Genetics 94:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111a.Gangloff, S., J. P. McDonald, C. Bendixen, L. Arthur, and R. Rothstein. 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14:8391-8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gao, Y., Y. Sun, K. M. Frank, P. Dikkes, Y. Fujiwara, K. J. Seidl, J. M. Sekiguchi, G. A. Rathbun, W. Swat, J. Wang, R. T. Bronson, B. A. Malynn, M. Bryans, C. Zhu, J. Chaudhuri, L. Davidson, R. Ferrini, T. Stamato, S. H. Orkin, M. E. Greenberg, and F. W. Alt. 1998. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell 95:891-902. [DOI] [PubMed] [Google Scholar]
- 112a.Gasior, S. L., H. Olivares, U. Ear, D. M. Hari, R. Weichselbaum, and D. K. Bishop. 2001. Assembly of RecA-like recombinases: distinct roles for mediator proteins in mitosis and meiosis. Proc. Natl. Acad. Sci. USA 98:8411-8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gasior, S. L., A. K. Wong, Y. Kora, A. Shinohara, and D. K. Bishop. 1998. Rad52 associates with RPA and functions with Rad55 and Rad57 to assemble meiotic recombination complexes. Genes Dev. 12:2208-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gatei, M., D. Young, K. M. Cerosaletti, A. Desai-Mehta, K. Spring, S. Kozlov, M. F. Lavin, R. A. Gatti, P. Concannon, and K. Khanna. 2000. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat. Genet. 25:115-119. [DOI] [PubMed] [Google Scholar]
- 115.Gibson, F. P., D. R. Leach, and R. G. Lloyd. 1992. Identification of sbcD mutations as cosuppressors of recBC that allow propagation of DNA palindromes in Escherichia coli K-12. J. Bacteriol. 174:1222-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Godthelp, B. C., W. W. Wiegant, A. van Duijn-Goedhart, O. D. Scharer, P. P. van Buul, R. Kanaar, and M. Z. Zdzienicka. 2002. Mammalian Rad51C contributes to DNA cross-link resistance, sister chromatid cohesion and genomic stability. Nucleic Acids Res. 30:2172-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Golub, E. I., O. V. Kovalenko, R. C. Gupta, D. C. Ward, and C. M. Radding. 1997. Interaction of human recombination proteins Rad51 and Rad54. Nucleic Acids Res. 25:4106-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gordenin, D. A., K. S. Lobachev, N. P. Degtyareva, A. L. Malkova, E. Perkins, and M. A. Resnick. 1993. Inverted DNA repeats: a source of eukaryotic genomic instability. Mol. Cell. Biol. 13:5315-5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gottlieb, S., J. Wagstaff, and R. E. Esposito. 1989. Evidence for two pathways of meiotic intrachromosomal recombination in yeast. Proc. Natl. Acad. Sci. USA 86:7072-7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gottlieb, T. M., and S. P. Jackson. 1993. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell 72:131-142. [DOI] [PubMed] [Google Scholar]
- 121.Greenwell, P. W., S. L. Kronmal, S. E. Porter, J. Gassenhuber, B. Obermaier, and T. D. Petes. 1995. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell 82:823-829. [DOI] [PubMed] [Google Scholar]
- 122.Grenon, M., C. Gilbert, and N. F. Lowndes. 2001. Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nat. Cell Biol. 3:844-847. [DOI] [PubMed] [Google Scholar]
- 123.Gupta, R. C., E. Folta-Stogniew, S. O'Malley, M. Takahashi, and C. M. Radding. 1999. Rapid exchange of A:T base pairs is essential for recognition of DNA homology by human Rad51 recombination protein. Mol. Cell 4:705-714. [DOI] [PubMed] [Google Scholar]
- 124.Haber, J. E. 1995. In vivo biochemistry: physical monitoring of recombination induced by site-specific endonucleases. Bioessays 17:609-620. [DOI] [PubMed] [Google Scholar]
- 125.Haber, J. E., and M. Hearn. 1985. Rad52-independent mitotic gene conversion in Saccharomyces cerevisiae frequently results in chromosomal loss. Genetics 111:7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Haber, J. E., and W. D. Heyer. 2001. The fuss about Mus81. Cell 107:551-554. [DOI] [PubMed] [Google Scholar]
- 127.Habu, T., T. Taki, A. West, Y. Nishimune, and T. Morita. 1996. The mouse and human homologs of DMC1, the yeast meiosis-specific homologous recombination gene, have a common unique form of exon-skipped transcript in meiosis. Nucleic Acids Res. 24:470-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Haering, C. H., J. Lowe, A. Hochwagen, and K. Nasmyth. 2002. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9:773-788. [DOI] [PubMed] [Google Scholar]
- 129.Hartsuiker, E., E. Vaessen, A. M. Carr, and J. Kohli. 2001. Fission yeast Rad50 stimulates sister chromatid recombination and links cohesion with repair. EMBO J. 20:6660-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hays, S. L., A. A. Firmenich, and P. Berg. 1995. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc. Natl. Acad. Sci. USA 92:6925-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hays, S. L., A. A. Firmenich, P. Massey, R. Banerjee, and P. Berg. 1998. Studies of the interaction between Rad52 protein and the yeast single-stranded DNA binding protein RPA. Mol. Cell. Biol. 18:4400-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Herskowitz, I., and R. E. Jensen. 1991. Putting the HO gene to work: practical uses for mating-type switching. Methods Enzymol. 194:132-146. [DOI] [PubMed] [Google Scholar]
- 133.Heyer, W. D., M. R. Rao, L. F. Erdile, T. J. Kelly, and R. D. Kolodner. 1990. An essential Saccharomyces cerevisiae single-stranded DNA binding protein is homologous to the large subunit of human RP-A. EMBO J. 9:2321-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Holliday, R. 1967. Altered recombination frequencies in radiation sensitive strains of Ustilago. Mutat. Res. 4:275-288. [DOI] [PubMed] [Google Scholar]
- 135.Holliday, R. 1964. A mechanism for gene conversion in fungi. Genet. Res. 5:282-304. [DOI] [PubMed] [Google Scholar]
- 136.Holloman, W. K., R. Wiegand, C. Hoessli, and C. M. Radding. 1975. Uptake of homologous single-stranded fragments by superhelical DNA: a possible mechanism for initiation of genetic recombination. Proc. Natl. Acad. Sci. USA 72:2394-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Holmes, A. M., and J. E. Haber. 1999. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell 96:415-424. [DOI] [PubMed] [Google Scholar]
- 138.Hong, E. L., A. Shinohara, and D. K. Bishop. 2001. Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single- strand DNA (ssDNA) and assimilation of ssDNA into homologous super- coiled duplex DNA. J. Biol. Chem. 276:41906-41912. [DOI] [PubMed] [Google Scholar]
- 139.Hopfner, K., A. Karcher, L. Craig, T. T. Woo, J. P. Carney, and J. A. Tainer. 2001. Structural biochemistry and interaction architecture of the DNA double- strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105:473-485. [DOI] [PubMed] [Google Scholar]
- 140.Hopfner, K. P., L. Craig, G. Moncalian, R. A. Zinkel, T. Usui, B. A. Owen, A. Karcher, B. Henderson, J. L. Bodmer, C. T. McMurray, J. P. Carney, J. H. Petrini, and J. A. Tainer. 2002. The Rad50 zinc-hook is a structure joining Mre 11 complexes in DNA recombination and repair. Nature 418:562-566. [DOI] [PubMed] [Google Scholar]
- 141.Hopfner, K. P., A. Karcher, D. Shin, C. Fairley, J. A. Tainer, and J. P. Carney. 2000. Mre11 and Rad50 from Pyrococcus furiosus: cloning and biochemical characterization reveal an evolutionarily conserved multiprotein machine. J. Bacteriol. 182:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hopfner, K. P., A. Karcher, D. S. Shin, L. Craig, L. M. Arthur, J. P. Carney, and J. A. Tainer. 2000. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101:789-800. [DOI] [PubMed] [Google Scholar]
- 143.Huang, J., and W. S. Dynan. 2002. Reconstitution of the mammalian DNA double-strand break end-joining reaction reveals a requirement for an Mre11/Rad50/NBS1-containing fraction. Nucleic Acids Res. 30:667-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hunter, N., and N. Kleckner. 2001. The single-end invasion: an asymmetric intermediate at the double- strand break to double-Holliday junction transition of meiotic recombination. Cell 106:59-70. [DOI] [PubMed] [Google Scholar]
- 145.Hurst, D. D., S. Fogel, and R. K. Mortimer. 1972. Conversion-associated recombination in yeast. Proc. Natl. Acad. Sci. USA 69:101-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Interthal, H., and W. D. Heyer. 2000. MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet. 263:812-827. [DOI] [PubMed] [Google Scholar]
- 147.Ira, G., and J. E. Haber. 2002. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 22:6384-6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ivanov, E. L., and J. E. Haber. 1995. RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:2245-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ivanov, E. L., V. G. Korolev, and F. Fabre. 1992. XRS2, a DNA repair gene of Saccharomyces cerevisiae, is needed for meiotic recombination. Genetics 132:651-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ivanov, E. L., N. Sugawara, J. Fishman-Lobell, and J. E. Haber. 1996. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics 142:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ivanov, E. L., N. Sugawara, C. I. White, F. Fabre, and J. E. Haber. 1994. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:3414-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jablonovich, Z., B. Liefshitz, R. Steinlauf, and M. Kupiec. 1999. Characterization of the role played by the RAD59 gene of Saccharomyces cerevisiae in ectopic recombination. Curr. Genet. 36:13-20. [DOI] [PubMed] [Google Scholar]
- 153.Jackson, J. A., and G. R. Fink. 1981. Gene conversion between duplicated genetic elements in yeast. Nature 292:306-311. [DOI] [PubMed] [Google Scholar]
- 154.Jensch, F., H. Kosak, N. C. Seeman, and B. Kemper. 1989. Cruciform cutting endonucleases from Saccharomyces cerevisiae and phage T4 show conserved reactions with branched DNAs. EMBO J. 8:4325-4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jiang, H., Y. Xie, P. Houston, K. Stemke-Hale, U. H. Mortensen, R. Rothstein, and T. Kodadek. 1996. Direct association between the yeast Rad51 and Rad54 recombination proteins. J. Biol. Chem. 271:33181-33186. [DOI] [PubMed] [Google Scholar]
- 156.Jinks-Robertson, S., and T. D. Petes. 1986. Chromosomal translocations generated by high-frequency meiotic recombination between repeated yeast genes. Genetics 114:731-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Johnson, R. D., and M. Jasin. 2001. Double-strand-break-induced homologous recombination in mammalian cells. Biochem. Soc. Trans. 29:196-201. [DOI] [PubMed] [Google Scholar]
- 158.Johnson, R. D., N. Liu, and M. Jasin. 1999. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature 401:397-399. [DOI] [PubMed] [Google Scholar]
- 159.Johnson, R. D., and L. S. Symington. 1995. Functional differences and interactions among the putative RecA homologs Rad51, Rad55, and Rad57. Mol. Cell. Biol. 15:4843-4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Johzuka, K., and H. Ogawa. 1995. Interaction of Mre11 and Rad50: two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics 139:1521-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jones, N. J., R. Cox, and J. Thacker. 1987. Isolation and cross-sensitivity of X-ray-sensitive mutants of V79-4 hamster cells. Mutat. Res. 183:279-286. [DOI] [PubMed] [Google Scholar]
- 162.Kagawa, W., H. Kurumizaka, S. Ikawa, S. Yokoyama, and T. Shibata. 2001. Homologous pairing promoted by the human Rad52 protein. J. Biol. Chem. 276:35201-35208. [DOI] [PubMed] [Google Scholar]
- 163.Kagawa, W., H. Kurumizaka, R. Ishitani, S. Fukai, O. Nureki, T. Shibata, and S. Yokoyama. 2002. Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. Mol. Cell. 10:359-371. [DOI] [PubMed] [Google Scholar]
- 164.Kaliraman, V., J. R. Mullen, W. M. Fricke, S. A. Bastin-Shanower, and S. J. Brill. 2001. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 15:2730-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kanaar, R., C. Troelstra, S. M. Swagemakers, J. Essers, B. Smit, J. H. Franssen, A. Pastink, O. Y. Bezzubova, J. M. Buerstedde, B. Clever, W. D. Heyer, and J. H. Hoeijmakers. 1996. Human and mouse homologs of the Saccharomyces cerevisiae RAD54 DNA repair gene: evidence for functional conservation. Curr. Biol. 6:828-838. [DOI] [PubMed] [Google Scholar]
- 166.Kang, L. E., and L. S. Symington. 2000. Aberrant double-strand break repair in rad51 mutants of Saccharomyces cerevisiae. Mol. Cell. Biol. 20:9162-9172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kans, J. A., and R. K. Mortimer. 1991. Nucleotide sequence of the RAD57 gene of Saccharomyces cerevisiae. Gene 105:139-140. [DOI] [PubMed] [Google Scholar]
- 168.Karow, J. K., A. Constantinou, J. L. Li, S. C. West, and I. D. Hickson. 2000. The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl. Acad. Sci. USA 97:6504-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kearney, H. M., D. T. Kirkpatrick, J. L. Gerton, and T. D. Petes. 2001. Meiotic recombination involving heterozygous large insertions in Saccharomyces cerevisiae: formation and repair of large, unpaired DNA loops. Genetics 158:1457-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Keeney, S., C. N. Giroux, and N. Kleckner. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88:375-384. [DOI] [PubMed] [Google Scholar]
- 171.Kegel, A., J. O. Sjostrand, and S. U. Astrom. 2001. Nej1p, a cell type-specific regulator of nonhomologous end joining in yeast. Curr. Biol. 11:1611-1617. [DOI] [PubMed] [Google Scholar]
- 172.Khasanov, F. K., G. V. Savchenko, E. V. Bashkirova, V. G. Korolev, W. D. Heyer, and V. I. Bashkirov. 1999. A new recombinational DNA repair gene from Schizosaccharomyces pombe with homology to Escherichia coli RecA. Genetics 152:1557-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kirkpatrick, D. T., and T. D. Petes. 1997. Repair of DNA loops involves DNA-mismatch and nucleotide-excision repair proteins. Nature 387:929-931. [DOI] [PubMed] [Google Scholar]
- 174.Kironmai, K. M., and K. Muniyappa. 1997. Alteration of telomeric sequences and senescence caused by mutations in RAD50 of Saccharomyces cerevisiae. Genes Cells 2:443-455. [DOI] [PubMed] [Google Scholar]
- 175.Kleff, S., B. Kemper, and R. Sternglanz. 1992. Identification and characterization of yeast mutants and the gene for a cruciform cutting endonuclease. EMBO J. 11:699-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Klein, H. L. 1988. Different types of recombination events are controlled by the RAD1 and RAD52 genes of Saccharomyces cerevisiae. Genetics 120:367-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Klein, H. L. 1995. Genetic control of intrachromosomal recombination. Bioessays 17:147-159. [DOI] [PubMed] [Google Scholar]
- 178.Klein, H. L. 2001. Mutations in recombinational repair and in checkpoint control genes suppress the lethal combination of srs2 delta with other DNA repair genes in Saccharomyces cerevisiae. Genetics 157:557-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Klein, H. L. 1997. RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics 147:1533-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kmiec, E., and W. K. Holloman. 1982. Homologous pairing of DNA molecules promoted by a protein from Ustilago. Cell 29:367-374. [DOI] [PubMed] [Google Scholar]
- 181.Kmiec, E. B., and W. K. Holloman. 1983. Heteroduplex formation and polarity during strand transfer promoted by Ustilago rec 1 protein. Cell 33:857-864. [DOI] [PubMed] [Google Scholar]
- 182.Kojic, M., C. W. Thompson, and W. K. Holloman. 2001. Disruptions of the Ustilago maydis REC2 gene identify a protein domain important in directing recombinational repair of DNA. Mol. Microbiol. 40:1415-1426. [DOI] [PubMed] [Google Scholar]
- 183.Kostriken, R., J. N. Strathern, A. J. Klar, J. B. Hicks, and F. Heffron. 1983. A site-specific endonuclease essential for mating-type switching in Saccharomyces cerevisiae. Cell 35:167-174. [DOI] [PubMed] [Google Scholar]
- 184.Kowalczykowski, S. C., D. A. Dixon, A. K. Eggleston, S. D. Lauder, and W. M. Rehrauer. 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58:401-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Krejci, L., J. Damborsky, B. Thomsen, M. Duno, and C. Bendixen. 2001. Molecular dissection of interactions between Rad51 and members of the recombination-repair group. Mol. Cell. Biol. 21:966-976. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 186.Krejci, L., B. Song, W. Bussen, R. Rothstein, U. H. Mortensen, and P. Sung. 2002.. Interaction with Rad51 is indispensable for recombination mediator function of Rad52. J. Biol. Chem. 277:40132-40141. [DOI] [PubMed]
- 187.Reference deleted.
- 188.Kurumizaka, H., S. Ikawa, M. Nakada, K. Eda, W. Kagawa, M. Takata, S. Takeda, S. Yokoyama, and T. Shibata. 2001. Homologous-pairing activity of the human DNA-repair proteins Xrcc3.Rad51C. Proc. Natl. Acad. Sci. USA 98:5538-5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lawrence, C. W., and R. B. Christensen. 1979. Metabolic suppressors of trimethoprim and ultraviolet light sensitivities of Saccharomyces cerevisiae rad6 mutants. J. Bacteriol 139:866-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Le, S., J. K. Moore, J. E. Haber, and C. W. Greider. 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152:143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lee, S.-E., D. A. Bressan, J. H. Petrini, and J. E. Haber. 2002. Complementation between N-terminal Saccharomyces cerevisiae mre11 alleles in DNA repair and telomere length maintenance. DNA Repair 1:27-40. [DOI] [PubMed] [Google Scholar]
- 192.Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. D. Kolodner, and J. E. Haber. 1998. Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94:399-409. [DOI] [PubMed] [Google Scholar]
- 193.Lee, S. E., A. Pellicioli, A. Malkova, M. Foiani, and J. E. Haber. 2001. The Saccharomyces recombination protein Tid1p is required for adaptation from G2/M arrest induced by a double-strand break. Curr. Biol. 11:1053-1057. [DOI] [PubMed] [Google Scholar]
- 194.Leung, W., A. Malkova, and J. E. Haber. 1997. Gene targeting by linear duplex DNA frequently occurs by assimilation of a single strand that is subject to preferential mismatch correction. Proc. Natl. Acad. Sci. USA 94:6851-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lewis, L. K., G. Karthikeyan, J. W. Westmoreland, and M. A. Resnick. 2002. Differential suppression of DNA repair deficiencies of yeast rad50, mre11 and xrs2 mutants by EXO1 and TLC1 (the RNA component of telomerase). Genetics 160:49-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lewis, L. K., J. M. Kirchner, and M. A. Resnick. 1998. Requirement for end-joining and checkpoint functions, but not RAD52-mediated recombination, after EcoRI endonuclease cleavage of Saccharomyces cerevisiae DNA. Mol. Cell. Biol. 18:1891-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Li, Z., E. I. Golub, R. Gupta, and C. M. Radding. 1997. Recombination activities of HsDmc1 protein, the meiotic human homolog of RecA protein. Proc. Natl. Acad. Sci. USA 94:11221-11226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lichten, M., C. Goyon, N. P. Schultes, D. Treco, J. W. Szostak, J. E. Haber, and A. Nicolas. 1990. Detection of heteroduplex DNA molecules among the products of Saccharomyces cerevisiae meiosis. Proc. Natl. Acad. Sci. USA 87:7653-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Liefshitz, B., A. Parket, R. Maya, and M. Kupiec. 1995. The role of DNA repair genes in recombination between repeated sequences in yeast. Genetics 140:1199-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lim, D. S., and P. Hasty. 1996. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol. 16:7133-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lim, D. S., S. T. Kim, B. Xu, R. S. Maser, J. Lin, J. H. Petrini, and M. B. Kastan. 2000. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 404:613-617. [DOI] [PubMed] [Google Scholar]
- 202.Lin, F. L., K. Sperle, and N. Sternberg. 1984. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol. Cell. Biol. 4:1020-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lisby, M., R. Rothstein, and U. H. Mortensen. 2001. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. USA 98:8276-8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Liu, N., J. E. Lamerdin, R. S. Tebbs, D. Schild, J. D. Tucker, M. R. Shen, K. W. Brookman, M. J. Siciliano, C. A. Walter, W. Fan, L. S. Narayana, Z. Q. Zhou, A. W. Adamson, K. J. Sorensen, D. J. Chen, N. J. Jones, and L. H. Thompson. 1998. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell 1:783-793. [DOI] [PubMed] [Google Scholar]
- 205.Liu, N., D. Schild, M. P. Thelen, and L. H. Thompson. 2002. Involvement of Rad51C in two distinct protein complexes of Rad51 paralogs in human cells. Nucleic Acids Res. 30:1009-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Reference deleted.
- 207.Lobachev, K. S., D. A. Gordenin, and M. A. Resnick. 2002. The Mre11 complex is required for repair of hairpin-capped double- strand breaks and prevention of chromosome rearrangements. Cell 108:183-193. [DOI] [PubMed] [Google Scholar]
- 208.Lockshon, D., S. G. Zweifel, L. L. Freeman-Cook, H. E. Lorimer, B. J. Brewer, and W. L. Fangman. 1995. A role for recombination junctions in the segregation of mitochondrial DNA in yeast. Cell 81:947-955. [DOI] [PubMed] [Google Scholar]
- 209.Longhese, M. P., P. Plevani, and G. Lucchini. 1994. Replication factor A is required in vivo for DNA replication, repair, and recombination. Mol. Cell. Biol. 14:7884-7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Lovett, S. T. 1994. Sequence of the RAD55 gene of Saccharomyces cerevisiae: similarity of RAD55 to prokaryotic RecA and other RecA-like proteins. Gene 142:103-106. [DOI] [PubMed] [Google Scholar]
- 211.Lovett, S. T., and R. K. Mortimer. 1987. Characterization of null mutants of the RAD55 gene of Saccharomyces cerevisiae: effects of temperature, osmotic strength and mating type. Genetics 116:547-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lowndes, N. F., and J. R. Murguia. 2000. Sensing and responding to DNA damage. Curr. Opin. Genet. Dev. 10:17-25. [DOI] [PubMed] [Google Scholar]
- 213.Lundblad, V., and E. H. Blackburn. 1993. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell 73:347-360. [DOI] [PubMed] [Google Scholar]
- 214.Lundblad, V., and J. W. Szostak. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57:633-643. [DOI] [PubMed] [Google Scholar]
- 215.Luo, G., M. S. Yao, C. F. Bender, M. Mills, A. R. Bladl, A. Bradley, and J. H. Petrini. 1999. Disruption of m Rad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc. Natl. Acad. Sci. USA 96:7376-7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Lustig, A. J., and T. D. Petes. 1986. Identification of yeast mutants with altered telomere structure. Proc. Natl. Acad. Sci. USA 83:1398-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Malkova, A., E. L. Ivanov, and J. E. Haber. 1996. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. USA 93:7131-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Malkova, A., L. Signon, C. B. Schaefer, M. L. Naylor, J. F. Theis, C. S. Newlon, and J. E. Haber. 2001. RAD51-independent break-induced replication to repair a broken chromosome depends on a distant enhancer site. Genes Dev. 15:1055-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Malone, R. E. 1983. Multiple mutant analysis of recombination in yeast. Mol. Gen. Genet. 189:405-412. [DOI] [PubMed] [Google Scholar]
- 220.Malone, R. E., S. Bullard, M. Hermiston, R. Rieger, M. Cool, and A. Galbraith. 1991. Isolation of mutants defective in early steps of meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics 128:79-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Malone, R. E., and R. E. Esposito. 1980. The RAD52 gene is required for homothallic interconversion of mating types and spontaneous mitotic recombination in yeast. Proc. Natl. Acad. Sci. USA 77:503-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Malone, R. E., and R. E. Esposito. 1981. Recombinationless meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1:891-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Manolis, K. G., E. R. Nimmo, E. Hartsuiker, A. M. Carr, P. A. Jeggo, and R. C. Allshire. 2001. Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J. 20:210-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Maryon, E., and D. Carroll. 1991. Characterization of recombination intermediates from DNA injected into Xenopus laevis oocytes: evidence for a nonconservative mechanism of homologous recombination. Mol. Cell. Biol. 11:3278-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Maser, R. S., O. K. Mirzoeva, J. Wells, H. Olivares, B. R. Williams, R. A. Zinkel, P. J. Farnham, and J. H. Petrini. 2001. Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol. Cell. Biol. 21:6006-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Maser, R. S., K. J. Monsen, B. E. Nelms, and J. H. Petrini. 1997. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell. Biol. 17:6087-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Maser, R. S., R. Zinkel, and J. H. Petrini. 2001. An alternative mode of translation permits production of a variant NBS1 protein from the common Nijmegen breakage syndrome allele. Nat. Genet. 27:417-421. [DOI] [PubMed] [Google Scholar]
- 228.Masson, J. Y., A. A. Davies, N. Hajibagheri, E. Van Dyck, F. E. Benson, A. Z. Stasiak, A. Stasiak, and S. C. West. 1999. The meiosis-specific recombinase hDmc1 forms ring structures and interacts with hRad51. EMBO J. 18:6552-6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Masson, J. Y., A. Z. Stasiak, A. Stasiak, F. E. Benson, and S. C. West. 2001. Complex formation by the human RAD51C and XRCC3 recombination repair proteins. Proc. Natl. Acad. Sci. USA 98:8440-8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Masson, J. Y., M. C. Tarsounas, A. Z. Stasiak, A. Stasiak, R. Shah, M. J. McIlwraith, F. E. Benson, and S. C. West. 2001. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 15:3296-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Mazin, A. V., C. J. Bornarth, J. A. Solinger, W. D. Heyer, and S. C. Kowalczykowski. 2000. Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol. Cell 6:583-592. [DOI] [PubMed] [Google Scholar]
- 232.Mazin, A. V., E. Zaitseva, P. Sung, and S. C. Kowalczykowski. 2000. Tailed duplex DNA is the preferred substrate for Rad51 protein-mediated homologous pairing. EMBO J. 19:1148-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.McDonald, J. P., and R. Rothstein. 1994. Unrepaired heteroduplex DNA in Saccharomyces cerevisiae is decreased in RAD1 RAD52-independent recombination. Genetics 137:393-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.McIlwraith, M. J., E. Van Dyck, J. Y. Masson, A. Z. Stasiak, A. Stasiak, and S. C. West. 2000. Reconstitution of the strand invasion step of double-strand break repair using human Rad51 Rad52 and RPA proteins. J. Mol. Biol. 304:151-164. [DOI] [PubMed] [Google Scholar]
- 235.McKee, A. H., and N. Kleckner. 1997. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics 146:797-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Meselson, M. S., and C. M. Radding. 1975. A general model for genetic recombination. Proc. Natl. Acad. Sci. USA 72:358-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Milne, G. T., and D. T. Weaver. 1993. Dominant negative alleles of RAD52 reveal a DNA repair/recombination complex including Rad51 and Rad52. Genes Dev. 7:1755-1765. [DOI] [PubMed] [Google Scholar]
- 238.Miyagawa, K., T. Tsuruga, A. Kinomura, K. Usui, M. Katsura, S. Tashiro, H. Mishima, and K. Tanaka. 2002. A role for RAD54B in homologous recombination in human cells. EMBO J. 21:175-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Mohaghegh, P., and I. D. Hickson. 2001. DNA helicase deficiencies associated with cancer predisposition and premature ageing disorders. Hum. Mol. Genet. 10:741-746. [DOI] [PubMed] [Google Scholar]
- 240.Moore, J. K., and J. E. Haber. 1996. Cell Cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2164-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Moreau, S., J. R. Ferguson, and L. S. Symington. 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19:556-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Moreau, S., E. A. Morgan, and L. S. Symington. 2001. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics 159:1423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Reference deleted.
- 244.Morgan, E. A., N. Shah, and L. S. Symington. 2002. The requirement for ATP hydrolysis by Saccharomyces cerevisiae Rad51 is bypassed by mating-type heterozygosity or RAD54 in high copy. Mol. Cell. Biol. 22:6336-6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Reference deleted.
- 246.Morita, T., Y. Yoshimura, A. Yamamoto, K. Murata, M. Mori, H. Yamamoto, and A. Matsushiro. 1993. A mouse homolog of the Escherichia coli recA and Saccharomyces cerevisiae RAD51 genes. Proc. Natl. Acad. Sci. USA 90:6577-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Morrison, C., A. Shinohara, E. Sonoda, Y. Yamaguchi-Iwai, M. Takata, R. R. Weichselbaum, and S. Takeda. 1999. The essential functions of human Rad51 are independent of ATP hydrolysis. Mol. Cell. Biol. 19:6891-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Morrow, D. M., C. Connelly, and P. Hieter. 1997. “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics 147:371-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Morrow, D. M., D. A. Tagle, Y. Shiloh, F. S. Collins, and P. Hieter. 1995. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell 82:831-840. [DOI] [PubMed] [Google Scholar]
- 250.Mortensen, U. H., C. Bendixen, I. Sunjevaric, and R. Rothstein. 1996. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl. Acad. Sci. USA 93:10729-10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Mortensen, U. H., N. Erdeniz, Q. Feng, and R. Rothstein. 2002. A molecular genetic dissection of the evolutionary conserved N terminus of yeast Rad52. Genetics 161:549-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Moynahan, M. E., A. J. Pierce, and M. Jasin. 2001. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 7:263-272. [DOI] [PubMed] [Google Scholar]
- 253.Mullen, J. R., V. Kaliraman, S. S. Ibrahim, and S. J. Brill. 2001. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157:103-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Myung, K., A. Datta, and R. D. Kolodner. 2001. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104:397-408. [DOI] [PubMed] [Google Scholar]
- 255.Nag, D. K., and T. D. Petes. 1993. Physical detection of heteroduplexes during meiotic recombination in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 13:2324-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Nairz, K., and F. Klein. 1997. mre11S—a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev. 11:2272-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Nassif, N., J. Penney, S. Pal, W. R. Engels, and G. B. Gloor. 1994. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 14:1613-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Nelms, B. E., R. S. Maser, J. F. Mackay, M. G. Lagally, and J. H. Petrini. 1998. In situ visualization of DNA double-strand break repair in human fibroblasts. Science 280:590-592. [DOI] [PubMed] [Google Scholar]
- 259.New, J. H., T. Sugiyama, E. Zaitseva, and S. C. Kowalczykowski. 1998. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391:407-410. [DOI] [PubMed] [Google Scholar]
- 260.Nickoloff, J. A., J. D. Singer, M. F. Hoekstra, and F. Heffron. 1989. Double-strand breaks stimulate alternative mechanisms of recombination repair. J. Mol. Biol. 207:527-541. [DOI] [PubMed] [Google Scholar]
- 261.Nickoloff, J. A., D. B. Sweetser, J. A. Clikeman, G. J. Khalsa, and S. L. Wheeler. 1999. Multiple heterologies increase mitotic double-strand break-induced allelic gene conversion tract lengths in yeast. Genetics 153:665-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Nicolas, A. L., P. L. Munz, E. Falck-Pedersen, and C. S. Young. 2000. Creation and repair of specific DNA double-strand breaks in vivo following infection with adenovirus vectors expressing Saccharomyces cerevisiae HO endonuclease. Virology 266:211-224. [DOI] [PubMed] [Google Scholar]
- 263.Niedernhofer, L. J., J. Essers, G. Weeda, B. Beverloo, J. de Wit, M. Muijtjens, H. Odijk, J. H. Hoeijmakers, and R. Kanaar. 2001. The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J. 20:6540-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Ogawa, T., A. Shinohara, A. Nabetani, T. Ikeya, X. Yu, E. H. Egelman, and H. Ogawa. 1993. RecA-like recombination proteins in eukaryotes: functions and structures of RAD51 genes. Cold. Spring. Harbor Symp. Quant. Biol. 58:567-576. [DOI] [PubMed] [Google Scholar]
- 265.Ogawa, T., X. Yu, A. Shinohara, and E. H. Egelman. 1993. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science 259:1896-1899. [DOI] [PubMed] [Google Scholar]
- 266.Ohta, K., A. Nicolas, M. Furuse, A. Nabetani, H. Ogawa, and T. Shibata. 1998. Mutations in the MRE11, RAD50, XRS2, and MRE2 genes alter chromatin configuration at meiotic DNA double-stranded break sites in premeiotic and meiotic cells. Proc. Natl. Acad. Sci. USA 95:646-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Ooi, S. L., D. D. Shoemaker, and J. D. Boeke. 2001. A DNA microarray-based genetic screen for nonhomologous end-joining mutants in Saccharomyces cerevisiae. Science 294:2552-2556. [DOI] [PubMed] [Google Scholar]
- 268.Orr-Weaver, T. L., and J. W. Szostak. 1983. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc. Natl. Acad. Sci. USA 80:4417-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Orr-Weaver, T. L., J. W. Szostak, and R. J. Rothstein. 1981. Yeast transformation: a model system for the study of recombination. Proc. Natl. Acad. Sci. USA 78:6354-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Palladino, F., and H. L. Klein. 1992. Analysis of mitotic and meiotic defects in Saccharomyces cerevisiae SRS2 DNA helicase mutants. Genetics 132:23-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Paques, F., and J. E. Haber. 1997. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6765-6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Paques, F., W. Y. Leung, and J. E. Haber. 1998. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol. Cell. Biol. 18:2045-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Park, M. S., D. L. Ludwig, E. Stigger, and S. H. Lee. 1996. Physical interaction between human RAD52 and RPA is required for homologous recombination in mammalian cells. J. Biol. Chem. 271:18996-19000. [DOI] [PubMed] [Google Scholar]
- 275.Parsons, C. A., P. Baumann, E. Van Dyck, and S. C. West. 2000. Precise binding of single-stranded DNA termini by human RAD52 protein. EMBO J. 19:4175-4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Passy, S. I., X. Yu, Z. Li, C. M. Radding, J. Y. Masson, S. C. West, and E. H. Egelman. 1999. Human Dmc1 protein binds DNA as an octameric ring. Proc. Natl. Acad. Sci. USA 96:10684-10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Paull, T. T., and M. Gellert. 1998. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol. Cell 1:969-979. [DOI] [PubMed] [Google Scholar]
- 278.Paull, T. T., and M. Gellert. 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 13:1276-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Paull, T. T., E. P. Rogakou, V. Yamazaki, C. U. Kirchgessner, M. Gellert, and W. M. Bonner. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886-895. [DOI] [PubMed] [Google Scholar]
- 280.Petrini, J. H. 1999. The mammalian Mre11-Rad50-Nbs1 protein complex: integration of functions in the cellular DNA-damage response. Am. J. Hum. Genet. 64:1264-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Petrini, J. H. 2000. The Mre11 complex and ATM: collaborating to navigate S phase. Curr. Opin. Cell Biol. 12:293-296. [DOI] [PubMed] [Google Scholar]
- 282.Petrini, J. H., M. E. Walsh, C. DiMare, X. N. Chen, J. R. Korenberg, and D. T. Weaver. 1995. Isolation and characterization of the human MRE11 homologue. Genomics 29:80-86. [DOI] [PubMed] [Google Scholar]
- 283.Petukhova, G., S. Stratton, and P. Sung. 1998. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature 393:91-94. [DOI] [PubMed] [Google Scholar]
- 284.Petukhova, G., S. A. Stratton, and P. Sung. 1999. Single strand DNA binding and annealing activities in the yeast recombination factor Rad59. J. Biol. Chem. 274:33839-33842. [DOI] [PubMed] [Google Scholar]
- 285.Petukhova, G., P. Sung, and H. Klein. 2000. Promotion of Rad51-dependent D-loop formation by yeast recombination factor Rdh54/Tid1. Genes Dev. 14:2206-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Petukhova, G., S. Van Komen, S. Vergano, H. Klein, and P. Sung. 1999. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J. Biol. Chem. 274:29453-29462. [DOI] [PubMed] [Google Scholar]
- 287.Pierce, A. J., R. D. Johnson, L. H. Thompson, and M. Jasin. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13:2633-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Pittman, D. L., J. Cobb, K. J. Schimenti, L. A. Wilson, D. M. Cooper, E. Brignull, M. A. Handel, and J. C. Schimenti. 1998. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell 1:697-705. [DOI] [PubMed] [Google Scholar]
- 289.Pittman, D. L., and J. C. Schimenti. 2000. Midgestation lethality in mice deficient for the RecA-related gene, Rad51D/Rad51L3. Genesis 26:167-173. [DOI] [PubMed] [Google Scholar]
- 290.Pittman, D. L., L. R. Weinberg, and J. C. Schimenti. 1998. Identification, characterization, and genetic mapping of Rad51D, a new mouse and human RAD51/RecA-related gene. Genomics 49:103-111. [DOI] [PubMed] [Google Scholar]
- 291.Plessis, A., and B. Dujon. 1993. Multiple tandem integrations of transforming DNA sequences in yeast chromosomes suggest a mechanism for integrative transformation by homologous recombination. Gene 134:41-50. [DOI] [PubMed] [Google Scholar]
- 292.Plessis, A., A. Perrin, J. E. Haber, and B. Dujon. 1992. Site-specific recombination determined by I-SceI, a mitochondrial group I intron-encoded endonuclease expressed in the yeast nucleus. Genetics 130:451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Prakash, L., and S. Prakash. 1977. Isolation and characterization of MMS-sensitive mutants of Saccharomyces cerevisiae. Genetics 86:33-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Prakash, S., L. Prakash, W. Burke, and B. Montelone. 1980. Effects of the RAD52 gene on recombination in Saccharomyces cerevisiae. Genetics 94:31-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Prinz, S., A. Amon, and F. Klein. 1997. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics 146:781-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Ranatunga, W., D. Jackson, J. A. Lloyd, A. L. Forget, K. L. Knight, and G. E. Borgstahl. 2001. Human RAD52 exhibits two models of self-association. J. Biol. Chem. 276:15876-15880. [DOI] [PubMed] [Google Scholar]
- 297.Rattray, A. J., C. B. McGill, B. K. Shafer, and J. N. Strathern. 2001. Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae. A role for sae2/com1. Genetics 158:109-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Rattray, A. J., and L. S. Symington. 1995. Multiple pathways for homologous recombination in Saccharomyces cerevisiae. Genetics 139:45-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Rattray, A. J., and L. S. Symington. 1994. Use of a chromosomal inverted repeat to demonstrate that the RAD51 and RAD52 genes of Saccharomyces cerevisiae have different roles in mitotic recombination. Genetics 138:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Ray, A., I. Siddiqi, A. L. Kolodkin, and F. W. Stahl. 1988. Intra-chromosomal gene conversion induced by a DNA double-strand break in Saccharomyces cerevisiae. J. Mol. Biol. 201:247-260. [DOI] [PubMed] [Google Scholar]
- 301.Raymond, W. E., and N. Kleckner. 1993. RAD50 protein of S. cerevisiae exhibits ATP-dependent DNA binding. Nucleic Acids Res. 21:3851-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Resnick, M. A., J. Nitiss, C. Edwards, and R. E. Malone. 1986. Meiosis can induce recombination in rad52 mutants of Saccharomyces cerevisiae. Genetics 113:531-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Rijkers, T., J. Van Den Ouweland, B. Morolli, A. G. Rolink, W. M. Baarends, P. P. Van Sloun, P. H. Lohman, and A. Pastink. 1998. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol. 18:6423-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Ristic, D., C. Wyman, C. Paulusma, and R. Kanaar. 2001. The architecture of the human Rad54-DNA complex provides evidence for protein translocation along DNA. Proc. Natl. Acad. Sci. USA 98:8454-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Ritchie, K. B., J. C. Mallory, and T. D. Petes. 1999. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6065-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Ritchie, K. B., and T. D. Petes. 2000. The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics 155:475-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Rong, L., and H. L. Klein. 1993. Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 268:1252-1259. [PubMed] [Google Scholar]
- 308.Rothstein, R., C. Helms, and N. Rosenberg. 1987. Concerted deletions and inversions are caused by mitotic recombination between delta sequences in Saccharomyces cerevisiae. Mol. Cell. Biol. 7:1198-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Rothstein, R. J. 1983. One-step gene disruption in yeast. Methods Enzymol. 101:202-211. [DOI] [PubMed] [Google Scholar]
- 310.Rouet, P., F. Smith, and M. Jasin. 1994. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl. Acad. Sci. USA 91:6064-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Rubin, B. P., D. O. Ferguson, and W. K. Holloman. 1994. Structure of REC2, a recombinational repair gene of Ustilago maydis, and its function in homologous recombination between plasmid and chromosomal sequences. Mol. Cell. Biol. 14:6287-6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Rudin, N., E. Sugarman, and J. E. Haber. 1989. Genetic and physical analysis of double-strand break repair and recombination in Saccharomyces cerevisiae. Genetics 122:519-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Ruskin, B., and G. R. Fink. 1993. Mutations in POL1 increase the mitotic instability of tandem inverted repeats in Saccharomyces cerevisiae. Genetics 134:43-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Saeki, T., I. Machida, and S. Nakai. 1980. Genetic control of diploid recovery after gamma-irradiation in the yeast Saccharomyces cerevisiae. Mutat. Res. 73:251-265. [DOI] [PubMed] [Google Scholar]
- 315.Sandell, L. L., and V. A. Zakian. 1993. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell 75:729-739. [DOI] [PubMed] [Google Scholar]
- 316.Santos-Rosa, H., and A. Aguilera. 1994. Increase in incidence of chromosome instability and non-conservative recombination between repeats in Saccharomyces cerevisiae hpr1 delta strains. Mol. Gen. Genet. 245:224-236. [DOI] [PubMed] [Google Scholar]
- 317.Saparbaev, M., L. Prakash, and S. Prakash. 1996. Requirement of mismatch repair genes MSH2 and MSH3 in the RAD1-RAD10 pathway of mitotic recombination in Saccharomyces cerevisiae. Genetics 142:727-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Sargent, R. G., R. L. Rolig, A. E. Kilburn, G. M. Adair, J. H. Wilson, and R. S. Nairn. 1997. Recombination-dependent deletion formation in mammalian cells deficient in the nucleotide excision repair gene ERCC1. Proc. Natl. Acad. Sci. USA 94:13122-13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Schiestl, R. H., and S. Prakash. 1988. RAD1, an excision repair gene of Saccharomyces cerevisiae, is also involved in recombination. Mol. Cell. Biol. 8:3619-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Schiestl, R. H., and S. Prakash. 1990. RAD10, an excision repair gene of Saccharomyces cerevisiae, is involved in the RAD1 pathway of mitotic recombination. Mol. Cell. Biol. 10:2485-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Schiestl, R. H., S. Prakash, and L. Prakash. 1990. The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics 124:817-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Schiestl, R. H., J. Zhu, and T. D. Petes. 1994. Effect of mutations in genes affecting homologous recombination on restriction enzyme-mediated and illegitimate recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:4493-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Schild, D. 1995. Suppression of a new allele of the yeast RAD52 gene by overexpression of RAD51, mutations in srs2 and ccr4, or mating-type heterozygosity. Genetics 140:115-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Schild, D., Y. Lio, D. W. Collins, T. Tsomondo, and D. J. Chen. 2000. Evidence for simultaneous protein interactions between human Rad51 paralogs. J. Biol. Chem. 275:16443-16449. [DOI] [PubMed] [Google Scholar]
- 325.Schmuckli-Maurer, J., and W. D. Heyer. 2000. Meiotic recombination in RAD54 mutants of Saccharomyces cerevisiae. Chromosoma 109:86-93. [DOI] [PubMed] [Google Scholar]
- 326.Schmuckli-Maurer, J., and W. D. Heyer. 1999. The Saccharomyces cerevisiae RAD54 gene is important but not essential for natural homothallic mating-type switching. Mol. Gen. Genet. 260:551-558. [DOI] [PubMed] [Google Scholar]
- 327.Schwacha, A., and N. Kleckner. 1995. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83:783-791. [DOI] [PubMed] [Google Scholar]
- 328.Schwacha, A., and N. Kleckner. 1994. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell 76:51-63. [DOI] [PubMed] [Google Scholar]
- 329.Schwacha, A., and N. Kleckner. 1997. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90:1123-1135. [DOI] [PubMed] [Google Scholar]
- 330.Scully, R., J. Chen, A. Plug, Y. Xiao, D. Weaver, J. Feunteun, T. Ashley, and D. M. Livingston. 1997. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88:265-275. [DOI] [PubMed] [Google Scholar]
- 331.Seitz, E. M., and S. C. Kowalczykowski. 2000. The DNA binding and pairing preferences of the archaeal RadA protein demonstrate a universal characteristic of DNA strand exchange proteins. Mol. Microbiol. 37:555-560. [DOI] [PubMed] [Google Scholar]
- 332.Sharan, S. K., M. Morimatsu, U. Albrecht, D. S. Lim, E. Regel, C. Dinh, A. Sands, G. Eichele, P. Hasty, and A. Bradley. 1997. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386:804-810. [DOI] [PubMed] [Google Scholar]
- 333.Sharples, G. J., and D. R. Leach. 1995. Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol. Microbiol. 17:1215-1217. [DOI] [PubMed] [Google Scholar]
- 334.Sherman, F., and H. Roman. 1963. Evidence for two types of allelic recombination in yeast. Genetics 48:255-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Shiloh, Y. 1997. Ataxia-telangiectasia and the Nijmegen breakage syndrome: related disorders but genes apart. Annu. Rev. Genet. 31:635-662. [DOI] [PubMed] [Google Scholar]
- 336.Shinohara, A., S. Gasior, T. Ogawa, N. Kleckner, and D. K. Bishop. 1997. Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes Cells 2:615-629. [DOI] [PubMed] [Google Scholar]
- 337.Shinohara, A., H. Ogawa, Y. Matsuda, N. Ushio, K. Ikeo, and T. Ogawa. 1993. Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA. Nat. Genet. 4:239-243. [DOI] [PubMed] [Google Scholar]
- 338.Shinohara, A., H. Ogawa, and T. Ogawa. 1992. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69:457-470. [DOI] [PubMed] [Google Scholar]
- 339.Shinohara, A., and T. Ogawa. 1998. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature 391:404-407. [DOI] [PubMed] [Google Scholar]
- 340.Shinohara, A., M. Shinohara, T. Ohta, S. Matsuda, and T. Ogawa. 1998. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells 3:145-156. [DOI] [PubMed] [Google Scholar]
- 341.Shinohara, M., E. Shita-Yamaguchi, J. M. Buerstedde, H. Shinagawa, H. Ogawa, and A. Shinohara. 1997. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics 147:1545-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Shu, Z., S. Smith, L. Wang, M. C. Rice, and E. B. Kmiec. 1999. Disruption of mu REC2/RAD51L1 in mice results in early embryonic lethality which can Be partially rescued in a p53(−/−) background. Mol. Cell. Biol. 19:8686-8693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Signon, L., A. Malkova, M. L. Naylor, H. Klein, and J. E. Haber. 2001. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol. Cell. Biol. 21:2048-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Sigurdsson, S., K. Trujillo, B. Song, S. Stratton, and P. Sung. 2001. Basis for avid homologous DNA strand exchange by human Rad51 and RPA. J. Biol. Chem. 276:8798-8806. [DOI] [PubMed] [Google Scholar]
- 345.Sigurdsson, S., S. Van Komen, W. Bussen, D. Schild, J. S. Albala, and P. Sung. 2001. Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev. 15:3308-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Sigurdsson, S., S. Van Komen, G. Petukhova, and P. Sung. Homologous DNA pairing by human recombination factors Rad51 and Rad54. J. Biol. Chem. in press. [DOI] [PubMed]
- 347.Smith, J., and R. Rothstein. 1999. An allele of RFA1 suppresses RAD52-dependent double-strand break repair in Saccharomyces cerevisiae. Genetics 151:447-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Smith, J., and R. Rothstein. 1995. A mutation in the gene encoding the Saccharomyces cerevisiae single-stranded DNA-binding protein Rfa1 stimulates a RAD52-independent pathway for direct-repeat recombination. Mol. Cell. Biol. 15:1632-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Solinger, J. A., and W. D. Heyer. 2001. Rad54 protein stimulates the postsynaptic phase of Rad51 protein-mediated DNA strand exchange. Proc. Natl. Acad. Sci. USA 98:8447-8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Solinger, J. A., G. Lutz, T. Sugiyama, S. C. Kowalczykowski, and W. D. Heyer. 2001. Rad54 protein stimulates heteroduplex DNA formation in the synaptic phase of DNA strand exchange via specific interactions with the presynaptic Rad51 nucleoprotein filament. J. Mol. Biol. 307:1207-1221. [DOI] [PubMed] [Google Scholar]
- 351.Song, B., and P. Sung. 2000. Functional interactions among yeast Rad51 recombinase, Rad52 mediator, and replication protein A in DNA strand exchange. J. Biol. Chem. 275:15895-15904. [DOI] [PubMed] [Google Scholar]
- 352.Sonoda, E., M. S. Sasaki, J. M. Buerstedde, O. Bezzubova, A. Shinohara, H. Ogawa, M. Takata, Y. Yamaguchi-Iwai, and S. Takeda. 1998. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 17:598-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Sonoda, E., M. Takata, Y. M. Yamashita, C. Morrison, and S. Takeda. 2001. Homologous DNA recombination in vertebrate cells. Proc. Natl. Acad. Sci. USA 98:8388-8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Soustelle, C., M. Vedel, R. Kolodner, and A. Nicolas. 2002. Replication protein A is required for meiotic recombination in Saccharomyces cerevisiae. Genetics 161:535-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Stark, J. M., P. Hu, A. J. Pierce, M. E. Moynahan, N. Ellis, and M. Jasin. 2002. ATP hydrolysis by mammalian RAD51 has a key role during homology-directed DNA repair. J. Biol. Chem. 277:20185-20194. [DOI] [PubMed] [Google Scholar]
- 356.Stasiak, A. Z., E. Larquet, A. Stasiak, S. Muller, A. Engel, E. Van Dyck, S. C. West, and E. H. Egelman. 2000. The human Rad52 protein exists as a heptameric ring. Curr. Biol. 10:337-340. [DOI] [PubMed] [Google Scholar]
- 357.Steele, D. F., M. E. Morris, and S. Jinks-Robertson. 1991. Allelic and ectopic interactions in recombination-defective yeast strains. Genetics 127:53-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Stewart, G. S., R. S. Maser, T. Stankovic, D. A. Bressan, M. I. Kaplan, N. G. Jaspers, A. Raams, P. J. Byrd, J. H. Petrini, and A. M. Taylor. 1999. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99:577-587. [DOI] [PubMed] [Google Scholar]
- 359.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11 Rad50 NBS1 DNA repair complex. Nature 418:348-352. [DOI] [PubMed] [Google Scholar]
- 360.Strathern, J. N., A. J. Klar, J. B. Hicks, J. A. Abraham, J. M. Ivy, K. A. Nasmyth, and C. McGill. 1982. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell 31:183-192. [DOI] [PubMed] [Google Scholar]
- 361.Sturzbecher, H. W., B. Donzelmann, W. Henning, U. Knippschild, and S. Buchhop. 1996. p53 is linked directly to homologous recombination processes via RAD51/RecA protein interaction. EMBO J. 15:1992-2002. [PMC free article] [PubMed] [Google Scholar]
- 362.Sugawara, N., and J. E. Haber. 1992. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol. Cell. Biol. 12:563-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Sugawara, N., G. Ira, and J. E. Haber. 2000. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 20:5300-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Sugawara, N., E. L. Ivanov, J. Fishman-Lobell, B. L. Ray, X. Wu, and J. E. Haber. 1995. DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature 373:84-86. [DOI] [PubMed] [Google Scholar]
- 365.Sugawara, N., F. Paques, M. Colaiacovo, and J. E. Haber. 1997. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc. Natl. Acad. Sci. USA 94:9214-9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Sugiyama, T., and S. C. Kowalczykowski. 2002. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 277:31663-31672. [DOI] [PubMed] [Google Scholar]
- 367.Sugiyama, T., J. H. New, and S. C. Kowalczykowski. 1998. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl. Acad. Sci. USA 95:6049-6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Sugiyama, T., E. M. Zaitseva, and S. C. Kowalczykowski. 1997. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 272:7940-7945. [DOI] [PubMed] [Google Scholar]
- 369.Sun, H., D. Treco, N. P. Schultes, and J. W. Szostak. 1989. Double-strand breaks at an initiation site for meiotic gene conversion. Nature 338:87-90. [DOI] [PubMed] [Google Scholar]
- 370.Sun, H., D. Treco, and J. W. Szostak. 1991. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell 64:1155-1161. [DOI] [PubMed] [Google Scholar]
- 371.Sung, P. 1994. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265:1241-1243. [DOI] [PubMed] [Google Scholar]
- 372.Sung, P. 1997. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 272:28194-28197. [DOI] [PubMed] [Google Scholar]
- 373.Sung, P. 1997. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 11:1111-1121. [DOI] [PubMed] [Google Scholar]
- 374.Sung, P., and D. L. Robberson. 1995. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell 82:453-461. [DOI] [PubMed] [Google Scholar]
- 375.Sung, P., and S. A. Stratton. 1996. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem. 271:27983-27986. [DOI] [PubMed] [Google Scholar]
- 376.Suto, K., A. Nagata, H. Murakami, and H. Okayama. 1999. A double-strand break repair component is essential for S phase completion in fission yeast cell cycling. Mol. Biol. Cell 10:3331-3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Swagemakers, S. M., J. Essers, J. de Wit, J. H. Hoeijmakers, and R. Kanaar. 1998. The human RAD54 recombinational DNA repair protein is a double-stranded DNA-dependent ATPase. J. Biol. Chem. 273:28292-28297. [DOI] [PubMed] [Google Scholar]
- 378.Symington, L. S. 1998. Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res. 26:5589-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Symington, L. S., L. E. Kang, and S. Moreau. 2000. Alteration of gene conversion tract length and associated crossing over during plasmid gap repair in nuclease-deficient strains of Saccharomyces cerevisiae. Nucleic Acids Res. 28:4649-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Symington, L. S., and R. Kolodner. 1985. Partial purification of an enzyme from Saccharomyces cerevisiae that cleaves Holliday junctions. Proc. Natl. Acad. Sci. USA 82:7247-7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Szostak, J. W., T. L. Orr-Weaver, R. J. Rothstein, and F. W. Stahl. 1983. The double-strand-break repair model for recombination. Cell 33:25-35. [DOI] [PubMed] [Google Scholar]
- 382.Takata, M., M. S. Sasaki, E. Sonoda, T. Fukushima, C. Morrison, J. S. Albala, S. M. Swagemakers, R. Kanaar, L. H. Thompson, and S. Takeda. 2000. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol. Cell. Biol. 20:6476-6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Takata, M., M. S. Sasaki, S. Tachiiri, T. Fukushima, E. Sonoda, D. Schild, L. H. Thompson, and S. Takeda. 2001. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol. 21:2858-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Tan, T. L., J. Essers, E. Citterio, S. M. Swagemakers, J. de Wit, F. E. Benson, J. H. Hoeijmakers, and R. Kanaar. 1999. Mouse Rad54 affects DNA conformation and DNA-damage-induced Rad51 foci formation. Curr. Biol. 9:325-328. [DOI] [PubMed] [Google Scholar]
- 385.Tanaka, K., T. Hiramoto, T. Fukuda, and K. Miyagawa. 2000. A novel human rad54 homologue, Rad54B, associates with Rad51. J. Biol. Chem. 275:26316-26321. [DOI] [PubMed] [Google Scholar]
- 386.Tanaka, K., W. Kagawa, T. Kinebuchi, H. Kurumizaka, and K. Miyagawa. 2002. Human Rad54B is a double-stranded DNA-dependent ATPase and has biochemical properties different from its structural homolog in yeast, Tid1/Rdh54. Nucleic Acids Res. 30:1346-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Tashiro, S., N. Kotomura, A. Shinohara, K. Tanaka, K. Ueda, and N. Kamada. 1996. S phase specific formation of the human Rad51 protein nuclear foci in lymphocytes. Oncogene 12:2165-2170. [PubMed] [Google Scholar]
- 388.Teng, S. C., J. Chang, B. McCowan, and V. A. Zakian. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell 6:947-952. [DOI] [PubMed] [Google Scholar]
- 389.Teng, S. C., and V. A. Zakian. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:8083-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 391.Thomas, B. J., and R. Rothstein. 1989. The genetic control of direct-repeat recombination in Saccharomyces: the effect of rad52 and rad1 on mitotic recombination at GAL10, a transcriptionally regulated gene. Genetics 123:725-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Thompson, L. H., and D. Schild. 2001. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res. 477:131-153. [DOI] [PubMed] [Google Scholar]
- 393.Tishkoff, D. X., N. Filosi, G. M. Gaida, and R. D. Kolodner. 1997. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88:253-263. [DOI] [PubMed] [Google Scholar]
- 394.Tracy, R. B., J. K. Baumohl, and S. C. Kowalczykowski. 1997. The preference for GT-rich DNA by the yeast Rad51 protein defines a set of universal pairing sequences. Genes Dev. 11:3423-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Tracy, R. B., and S. C. Kowalczykowski. 1996. In vitro selection of preferred DNA pairing sequences by the Escherichia coli RecA protein. Genes Dev. 10:1890-1903. [DOI] [PubMed] [Google Scholar]
- 396.Trujillo, K. M., and P. Sung. 2001. DNA Structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50-Mre11 complex. J. Biol. Chem. 276:35458-35464. [DOI] [PubMed] [Google Scholar]
- 397.Trujillo, K. M., S. S. Yuan, E. Y. Lee, and P. Sung. 1998. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 273:21447-21450. [DOI] [PubMed] [Google Scholar]
- 398.Tsang, S. S., S. A. Chow, and C. M. Radding. 1985. Networks of DNA and RecA protein are intermediates in homologous pairing. Biochemistry 24:3226-3232. [DOI] [PubMed] [Google Scholar]
- 399.Tsubouchi, H., and H. Ogawa. 2000. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol. Biol. Cell 11:2221-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Tsubouchi, H., and H. Ogawa. 1998. A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol. Cell. Biol. 18:260-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Tsukamoto, Y., A. K. Taggart, and V. A. Zakian. 2001. The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 11:1328-1335. [DOI] [PubMed] [Google Scholar]
- 402.Tsutsui, Y., F. K. Khasanov, H. Shinagawa, H. Iwasaki, and V. I. Bashkirov. 2001. Multiple interactions among the components of the recombinational DNA repair system in Schizosaccharomyces pombe. Genetics 159:91-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Tsutsui, Y., T. Morishita, H. Iwasaki, H. Toh, and H. Shinagawa. 2000. A recombination repair gene of Schizosaccharomyces pombe, rhp57, is a functional homolog of the Saccharomyces cerevisiae RAD57 gene and is phylogenetically related to the human XRCC3 gene. Genetics 154:1451-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Tsuzuki, T., Y. Fujii, K. Sakumi, Y. Tominaga, K. Nakao, M. Sekiguchi, A. Matsushiro, Y. Yoshimura, and Morita T. 1996. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl. Acad. Sci. USA 93:6236-6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Umezu, K., N. Sugawara, C. Chen, J. E. Haber, and R. D. Kolodner. 1998. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics 148:989-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Usui, T., H. Ogawa, and J. H. Petrini. 2001. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7:1255-1266. [DOI] [PubMed] [Google Scholar]
- 407.Usui, T., T. Ohta, H. Oshiumi, J. Tomizawa, H. Ogawa, and T. Ogawa. 1998. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95:705-716. [DOI] [PubMed] [Google Scholar]
- 408.Valencia, M., M. Bentele, M. B. Vaze, G. Herrmann, E. Kraus, S. E. Lee, P. Schar, and J. E. Haber. 2001. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature 414:666-669. [DOI] [PubMed] [Google Scholar]
- 409.van den Bosch, M., K. Vreeken, J. B. Zonneveld, J. A. Brandsma, M. Lombaerts, J. M. Murray, P. H. Lohman, and A. Pastink. 2001. Characterization of RAD52 homologs in the fission yeast Schizosaccharomyces pombe. Mutat. Res. 461:311-323. [DOI] [PubMed] [Google Scholar]
- 410.Van Komen, S., G. Petukhova, S. Sigurdsson, S. Stratton, and P. Sung. 2000. Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol. Cell 6:563-572. [DOI] [PubMed] [Google Scholar]
- 411.Varon, R., C. Vissinga, M. Platzer, K. M. Cerosaletti, K. H. Chrzanowska, K. Saar, G. Beckmann, E. Seemanova, P. R. Cooper, N. J. Nowak, M. Stumm, C. M. Weemaes, R. A. Gatti, R. K. Wilson, M. Digweed, A. Rosenthal, K. Sperling, P. Concannon, and A. Reis. 1998. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell 93:467-476. [DOI] [PubMed] [Google Scholar]
- 412.Venkitaraman, A. R. 2001. Chromosome stability, DNA recombination and the BRCA2 tumour suppressor. Curr. Opin. Cell Biol. 13:338-343. [DOI] [PubMed] [Google Scholar]
- 413.Weiden, M. D., and H. S. Ginsberg. 1994. Deletion of the E4 region of the genome produces adenovirus DNA concatemers. Proc. Natl. Acad. Sci. USA 91:153-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Weiffenbach, B., and J. E. Haber. 1981. Homothallic mating type switching generates lethal chromosome breaks in rad52 strains of Saccharomyces cerevisiae. Mol. Cell. Biol. 1:522-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Weinert, T. A., G. L. Kiser, and L. H. Hartwell. 1994. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8:652-665. [DOI] [PubMed] [Google Scholar]
- 416.Weng, Y. S., and J. A. Nickoloff. 1998. Evidence for independent mismatch repair processing on opposite sides of a double-strand break in Saccharomyces cerevisiae. Genetics 148:59-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.West, S. C. 1997. Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet. 31:213-244. [DOI] [PubMed] [Google Scholar]
- 418.West, S. C., and A. Korner. 1985. Cleavage of cruciform DNA structures by an activity from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 82:6445-6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.White, C. I., and J. E. Haber. 1990. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 9:663-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Wiese, C., D. W. Collins, J. S. Albala, L. H. Thompson, A. Kronenberg, and D. Schild. 2002. Interactions involving the Rad51 paralogs Rad51C and XRCC3 in human cells. Nucleic Acids Res. 30:1001-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Willis, K. K., and H. L. Klein. 1987. Intrachromosomal recombination in Saccharomyces cerevisiae: reciprocal exchange in an inverted repeat and associated gene conversion. Genetics 117:633-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Wilson, S., N. Warr, D. L. Taylor, and F. Z. Watts. 1999. The role of Schizosaccharomyces pombe Rad32, the Mre11 homologue, and other DNA damage response proteins in non-homologous end joining and telomere length maintenance. Nucleic Acids Res. 27:2655-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Wold, M. S., D. H. Weinberg, D. M. Virshup, J. J. Li, and T. J. Kelly. 1989. Identification of cellular proteins required for simian virus 40 DNA replication. J. Biol. Chem. 264:2801-2809. [PubMed] [Google Scholar]
- 424.Wong, A. K., R. Pero, P. A. Ormonde, S. V. Tavtigian, and P. L. Bartel. 1997. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J. Biol. Chem. 272:31941-31944. [DOI] [PubMed] [Google Scholar]
- 425.Wu, L., S. L. Davies, N. C. Levitt, and I. D. Hickson. 2001. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J. Biol. Chem. 276:19375-19381. [DOI] [PubMed] [Google Scholar]
- 426.Xiao, Y., and D. T. Weaver. 1997. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 25:2985-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Yamaguchi-Iwai, Y., E. Sonoda, J. M. Buerstedde, O. Bezzubova, C. Morrison, M. Takata, A. Shinohara, and S. Takeda. 1998. Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol. Cell. Biol. 18:6430-6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Yamaguchi-Iwai, Y., E. Sonoda, M. S. Sasaki, C. Morrison, T. Haraguchi, Y. Hiraoka, Y. M. Yamashita, T. Yagi, M. Takata, C. Price, N. Kakazu, and S. Takeda. 1999. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 18:6619-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Yoshida, K., G. Kondoh, Y. Matsuda, T. Habu, Y. Nishimune, and T. Morita. 1998. The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol. Cell 1:707-718. [DOI] [PubMed] [Google Scholar]
- 430.Yu, V. P., M. Koehler, C. Steinlein, M. Schmid, L. A. Hanakahi, A. J. van Gool, S. C. West, and A. R. Venkitaraman. 2000. Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 14:1400-1406. [PMC free article] [PubMed] [Google Scholar]
- 431.Zhu, J., S. Petersen, L. Tessarollo, and A. Nussenzweig. 2001. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr. Biol. 11:105-109. [DOI] [PubMed] [Google Scholar]
- 432.Zickler, D., and N. Kleckner. 1999. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33:603-754. [DOI] [PubMed] [Google Scholar]
- 433.Zou, H., and R. Rothstein. 1997. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell 90:87-96. [DOI] [PubMed] [Google Scholar]