Abstract
Several Borrelia burgdorferi outer surface proteins have been identified over the past decade that are up-regulated by temperature- and/or mammalian host-specific signals as this spirochete is transmitted from ticks to mammals. Given the potential role(s) that these differentially up-regulated proteins may play in B. burgdorferi transmission and Lyme disease pathogenesis, much attention has recently been placed on identifying additional borrelial outer surface proteins. To identify uncharacterized B. burgdorferi outer surface proteins, we previously performed a comprehensive gene expression profiling analysis of temperature-shifted and mammalian host-adapted B. burgdorferi. The combined microarray analyses revealed that many genes encoding known and putative outer surface proteins are down-regulated in mammalian host-adapted B. burgdorferi. At the same time, however, several different genes encoding putative outer surface proteins were found to be up-regulated during the transmission and infection process. Among the putative outer surface proteins identified, biochemical and surface localization analyses confirmed that seven (Bb0405, Bb0689, BbA36, BbA64, BbA66, BbA69, and BbI42) are localized to the surface of B. burgdorferi. Furthermore, enzyme-linked immunosorbent assay analysis using serum from tick-infested baboons indicated that all seven outer surface proteins identified are immunogenic and that antibodies are generated against all seven during a natural infection. Specific antibodies generated against all seven of these surface proteins were found to be bactericidal against B. burgdorferi, indicating that these newly identified outer surface proteins are prime candidates for analysis as second-generation Lyme disease vaccinogens.
Lyme disease, caused by the pathogenic spirochete Borrelia burgdorferi, is a debilitating multisystem disease that can chronically affect patients for decades (58, 61). The disease is typically transmitted to humans by the bite of infected Ixodes ticks and is currently the most common arthropod-borne infection in the United States (8, 49, 60). B. burgdorferi is maintained in nature through a complex enzootic cycle involving the horizontal transmission of spirochetes between ticks and mammalian hosts (39). As a consequence of its unique life cycle, this organism must adapt to very distinct environmental niches as it migrates from the tick to the mammalian host. Given the extracellular lifestyle of this pathogen, the outer surface of this organism is the interface between B. burgdorferi and its tick and mammalian hosts during infection. Therefore, to better examine Lyme disease pathogenesis and identify possible vaccine candidates, many investigations have focused on identifying new B. burgdorferi outer surface proteins (Osps). Additionally, since it is now well recognized that many surface proteins, such as OspA, expressed by B. burgdorferi are down-regulated or completely turned off during tick transmission and mammalian infection (1, 25), the identification of surface proteins that are expressed during infection has become a priority.
Since the elucidation of the B. burgdorferi genome by Fraser and colleagues (23), numerous genes encoding putative outer surface proteins have been identified using computer-based algorithms. The putative surface proteins identified all share an N-terminal signal peptide, which is needed to direct protein export through the B. burgdorferi cytoplasmic membrane (26, 51). Unfortunately, while many putative surface proteins have been described, few have been empirically verified to be surface exposed and expressed during both tick transmission and mammalian infection. However, using the combined genome sequence information in conjunction with the transcriptional profiling studies previously performed in our laboratory (6, 48), we were able to identify a subset of putative Osps that are expressed during tick transmission and mammalian infection.
A majority of the genes encoding putative Osps that were identified in the microarray studies were down-regulated by temperature and mammalian host factors (6, 48). However, at least 10 putative Osps were identified that were up-regulated by these environmental cues and were subsequently selected for further study. Here we show, using a combination of Triton X-114 phase partitioning and cellular localization experiments, that 7 of the 10 candidates were determined to be bona fide Osps that are surface exposed in B. burgdorferi. Consistent with the cellular localization analyses, specific antibodies generated against all seven Osps were bactericidal towards B. burgdorferi. The combined studies have identified seven previously unrecognized Osps from B. burgdorferi that can now be further examined for their role(s) in Lyme disease pathogenesis and for their ability to be used as novel vaccinogens.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
B. burgdorferi strain B31 MI was cultivated in BSK-II medium supplemented with 6% rabbit serum (4). For temperature shift experiments, spirochetes were first cultivated in BSK-II medium supplemented with 6% rabbit serum at 23°C to mid-logarithmic phase (5 × 107 per ml) before being seeded at a concentration of 1,000 spirochetes per ml into medium prewarmed to 37°C. To examine protein expression profiles from mammalian host-adapted spirochetes, organisms were cultivated in dialysis membrane chambers implanted into the peritoneal cavities of rats as previously described (1, 28) All cloning experiments and purification of recombinant proteins were performed using Escherichia coli DH5α as the host strain and tryptone-yeast broth or agar medium supplemented with the appropriate antibiotic.
Hydrophilicity analysis and identification of signal peptides.
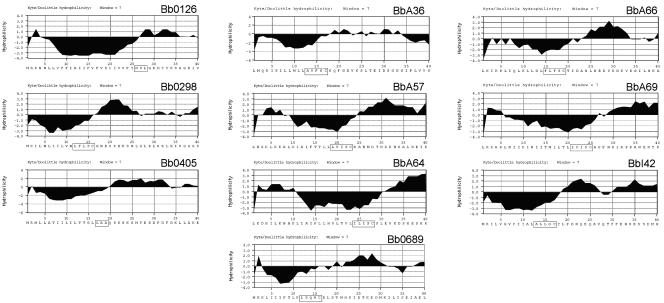
DNA sequences were downloaded from The Institute for Genomic Research website (http://www.tigr.org/tigr-scripts/CMR2/GenomePage3.spl?database = gbb), and hydrophilicity plots were generated using MacVector version 6.5.3 sequence analysis software (Oxford Molecular Ltd., Madison, WI) according to the method of Kyte and Doolittle (37) using a window size of 7. To identify signal peptide export sequences, the first 40 N-terminal amino acids of each protein were subjected to the SignalP 3.0 (5) and LipoP 1.0 (34) algorithms to identify putative signal peptidase I and II processing sites, respectively (see Fig. 1).
FIG. 1.
Hydrophilicity analysis of candidate B. burgdorferi surface proteins. Hydrophilicity plots of the first 40 amino acids for each protein were generated using the algorithm of Kyte and Doolittle with a window size of 7. All 10 proteins contain putative signal peptides at their N terminus. Potential signal peptidase I (Bb0126 and Bb0405) and signal peptidase II cleavage motifs are indicated as boxed regions.
Generation of recombinant proteins.
Gene-specific primers used to amplify the 10 B. burgdorferi open reading frames for generation of fusion proteins are listed in Table 1. Amplicons corresponding to the region encoding the mature portion of each protein (i.e., lacking the signal peptide) were ligated into the pGEX-4T-3 vector (Pharmacia Biotech Inc., Piscataway, NJ) or the TOPO-TA pBAD/thio vector (Invitrogen, Carlsbad, CA) for overexpression in E. coli DH5α. All clones generated were verified not to contain PCR errors by DNA sequence analysis. Recombinant glutathione S-transferase (GST) fusion proteins were purified and cleaved free of the GST moiety by use of procedures previously described (2, 3). For histidine-tagged fusion protein purification, transformed E. coli cultures were grown to an optical density at 600 nm (OD600) of 0.8 in tryptone-yeast broth containing 100 μg of ampicillin per ml before being induced with 0.2% l-arabinose (Sigma Chemical Co., St. Louis, MO) overnight. Cells were subsequently harvested by centrifugation at 8,000 × g for 20 min and lysed by sonication in a buffer containing 50 mM Tris-HCl, 50 mM Tris-Base (pH 7.9) (Sigma Chemical Co., St. Louis, MO), 12.5 mM β-mercaptoethanol before affinity column purification was performed according to the manufacturer's instructions (QIAGEN, Valencia, CA). Purified protein was eluted from the column in 20 mM Tris-HCl, 20 mM Tris-Base (pH 7.9), 100 mM KCl, 12.5 mM β-mercaptoethanol, 10% glycerol, 200 mM imidazole. All GST and His-tagged recombinant proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and examined by silver staining (45) to verify purity.
TABLE 1.
Oligonucleotides used for the generation of fusion proteins
| Gene | Oligonucleotidea
|
|
|---|---|---|
| Forward | Reverse | |
| Bb0126 | GCGGGATCCTCTTTAGGCAAGGATTATGTAAA | GCGCTCGAGTTAATTTTGCTTAAGTTCTAAAAT |
| Bb0298 | GCGGGATCCGGCAATGAATCTAAAGAAAAATCA | GCGCTCGAGTTACTTCAAATTACTCATGAT |
| Bb0405 | GCGGGATCCCAATCCAAAAGCAAAAGTATGACT | TATATATATTTTTATAAAGCCTGT |
| Bb0689 | GCGGGATCCCATAAAATAGATACAAAAGAAGAT | ATTCTTATATTTTCTTTTTCC |
| BbA36 | GCGGGATCCAAGCAGTTTGGCGATGTTAAA | GCGCTCGAGTTAAACATTTCCATAATTTTTCAA |
| BbA57 | GCGGGATCCAATGCAAATATGGATACAAACGAT | GCGCTCGAGTTATTGATAATTTTTTTCTACCAATAA |
| BbA64 | GCGGGATCCGAAGTCAAAGACAGCAATGAAAGC | CTGAATTGGAGCAAGAATATT |
| BbA66 | GCGGGATCCAATCTAAACGAAGATTATAAAAAC | CATTATACTAATGTATGCTTCAAG |
| BbA69 | GCGGGATCCCCTTTTAACAAAATCAATCCCAAG | GCGCTCGAGTTAATAAAAGGCAGATTGTAAAGAATC |
| BbI42 | GCGGGATCCTTGCCTGATAATCAGGAACAA | GCGCTCGAGTTATGTAGGTAAAATAGGAAC |
Nucleotides in bold indicate restriction sites used for cloning.
Generation of monospecific, polyclonal antibodies.
Thirty micrograms of each recombinant protein, GST, thioredoxin, or OspA was added to 200 μl of phosphate-buffered saline (PBS; pH 7.4) and vigorously mixed with 200 μl of Freund's complete adjuvant (Difco Laboratories, Detroit, MI) before intraperitoneal injection into Sprague-Dawley rats (Harlan, Indianapolis, IN). At 2 and 4 weeks following the primary immunization, intraperitoneal booster immunizations were performed with 30 μg of each protein in Freund's incomplete adjuvant (Difco Laboratories). All rats were exsanguinated 2 weeks after the second booster immunization to collect sera. Antibody specificity was determined by immunoblot analysis of both B. burgdorferi B31 whole-cell lysates and/or the purified recombinant protein. B. burgdorferi B31 FlaB protein was purified (14), and polyclonal antibodies were generated in rabbits as described previously (52).
SDS-PAGE and immunoblot analysis.
Whole-cell lysates of B. burgdorferi B31 were boiled for 10 min in final sample buffer (62.5 μM Tris-HCl [pH 6.8], 10% [vol/vol] glycerol, 5% [vol/vol] β-mercaptoethanol, 5% SDS, 0.001% bromophenol blue) before electrophoresis through a 2.4% stacking and 12.5% separating gel. Gels were transferred electrophoretically to nitrocellulose (Schleicher and Schuell, Keene, N.H.) for immunoblot analysis. Immunoblots were blocked for 45 min in PBS (pH 7.4) containing 0.2% Tween 20 (PBS-T). To analyze specific reactivity of immunized rat sera to whole-cell lysates and the various recombinant proteins, transferred proteins were incubated with a 1:1,000 dilution of the primary rat serum for 45 min followed by three washes with PBS-T. Subsequently, 1:1,000 dilutions of horseradish peroxidase (HRP)-conjugated goat anti-rat antibody or anti-rabbit antibody (Zymed, San Francisco, CA) were incubated with the membranes for 45 min. Membranes were washed for 30 min with PBS before being developed with the chromogenic substrate 4-chloro-1-naphthol. For enhanced chemiluminescence immunoblotting, membranes were initially blocked for 45 min in PBS-T followed by blocking overnight in PBS-T-10% fetal calf serum (Difco Laboratories, Detroit, MI). Membranes were then washed three times in PBS-T for 10 min before being incubated for 45 min with a 1:2,000 dilution of the primary rat antibodies in Blotto HD (2% IGEPAL, 0.2% SDS, 28 mM Tris-HCl, 22 mM Tris-Base, 1.5 mM calcium chloride, and 80 mM sodium chloride). After incubation with primary antibodies, the membranes were washed three times in Blotto HD-0.2% bovine serum albumin and a 1:5,000 dilution of HRP-conjugated goat anti-rat or anti-rabbit antibody in Blotto HD was added and allowed to incubate with the membranes for 45 min. Membranes were then washed three times for 10 min with PBS (pH 7.4) before being developed with the enhanced chemiluminescence plus reagent provided by the manufacturer (Amersham Pharmacia Biotech, Piscataway, NJ).
Triton X-114 phase partitioning.
To examine the amphiphilic characteristics of the candidate Osps, 1 × 109 spirochetes were sonicated on ice in 800 μl PBS four times for 20 s. Two hundred microliters of 10% Triton X-114 (Sigma Chemical Co.) was then added to the sonicated samples before being rocked overnight at 4°C. Following incubation at 4°C, phase-partitioned lysates were centrifuged at 13,000 × g for 15 min at 4°C to remove insoluble cell debris. The resulting supernatant was removed and placed at 37°C to allow for phase separation. The resulting aqueous and detergent-enriched phases were then each washed five times as previously described (7). A total of 10 volumes of ice-cold acetone was then added to both the detergent and aqueous phases followed by centrifugation at 13,000 × g for 15 min to precipitate proteins. Protein pellets were resuspended in PBS (pH 7.4), and equivalent amounts of cell lysates from each phase (∼1 × 108 organisms) were subjected to SDS-PAGE before transfer to nitrocellulose for immunoblot analysis as described above.
PK surface localization.
To remove adherent serum proteins from spirochetes, 2 × 108 organisms were gently washed three times in 1 ml of PBS (pH 7.4); spirochetes were gently collected between after wash by centrifugation at 4,000 × g for 4 min. Subsequently, the washed spirochetes were resuspended in 1 ml of PBS and split into two equal 500-μl volumes. One aliquot received 200 μg of proteinase K (PK) (Sigma Chemical Co.), while the other aliquot received an equal volume of PBS without PK. Both aliquots were incubated for 1 h at room temperature before 10 μl of phenylmethylsulfonyl fluoride (Sigma Chemical Co.) was added to stop PK activity. Spirochete suspensions were subsequently pelleted by centrifugation at 10,000 × g for 10 min and resuspended in PBS for immunoblot analysis.
Bactericidal assay.
Antibodies generated against FlaB, GST, thioredoxin, OspA, Bb0405, Bb0689, BbA66, BbA64, BbA36, BbA69, and BbI42 were used for B. burgdorferi bactericidal assays. B. burgdorferi organisms (4 × 106) were seeded into wells of a 96-well flat-bottom microtiter plate containing either 2 units of normal guinea pig complement alone (Sigma Chemical Co.) or normal guinea pig complement with a 1:10 dilution of the antibody preparations. Plates were sealed and incubated at 34°C for 72 h. Aliquots of each well were examined by dark-field microscopy to quantitate cell survival and percent inhibition (44). All assays were performed in triplicate. Statistical significance was determined using the unpaired, two-tailed Student's t test (15).
Enzyme-linked immunosorbent assay (ELISA).
One hundred nanograms of each recombinant protein was diluted in 50 μl of PBS (pH 7.4) prior to plating in triplicate in 96-well Maxisorp Nunc-Immuno plates (Nalge Nunc International, Naperville, Ill.). Samples were allowed to coat overnight at 4°C. For end point titers, serum samples taken from tick-infested baboons 0, 42, 90, 180, and 360 days post tick infestation were initially diluted 1:100 and then serially diluted twofold to a final dilution of 1:25,600. Secondary antibodies were HRP-conjugated sheep anti-human immunoglobulin, isotypes G (gamma chain) and M (mu chain) specific (The Binding Site, San Diego, CA), which were previously demonstrated to react with baboon antibodies (27). Antibody end point titers were determined as the highest dilution for which the mean OD405 was 3 standard deviations above the mean OD405 of the same dilution of serum added to wells coated with blocking reagent alone.
RESULTS
Selection of candidate surface proteins.
To identify B. burgdorferi outer surface proteins (Osps), we first identified all B. burgdorferi genes that were up-regulated by temperature- or mammalian host-specific factors in our prior microarray analyses (6, 48). We focused our efforts on cataloguing up-regulated genes, since these would be the most likely candidates to be expressed during a natural infection. Of the 310 different up-regulated genes identified in the prior studies, 51 were observed to encode putative N-terminal signal peptides, indicating that the genes encoding these proteins are most likely (i) exported to the periplasmic compartment, (ii) exported to the outer membrane, or (iii) secreted from the bacterial cell into the extracellular environment. Among the 51 up-regulated genes identified, 10 that had not previously been characterized were chosen for further analysis. As shown in Fig. 1, hydrophilicity plots of the 10 candidate Osps chosen for further study revealed a common hydrophobic region at the N terminus approximately 15 to 30 amino acids in length, which is typical of bacterial N-terminal signal peptides. Additionally, no other membrane-spanning hydrophobic regions were identified within any of the proteins analyzed, indicating that all 10 lacked transmembrane-spanning domains that could anchor them to the borrelial inner membrane. A closer examination of the putative leader peptides revealed that eight (Bb0298, Bb0689, BbA36, BbA57, BbA64, BbA66, BbA69, and BbI42) contained typical signal peptidase II cleavage and processing motifs, suggesting that they are lipoproteins and are lipid modified during export. Bb0126 and Bb0405 were the only two candidate surface proteins that appeared to contain signal peptides processed by signal peptidase I (Fig. 1).
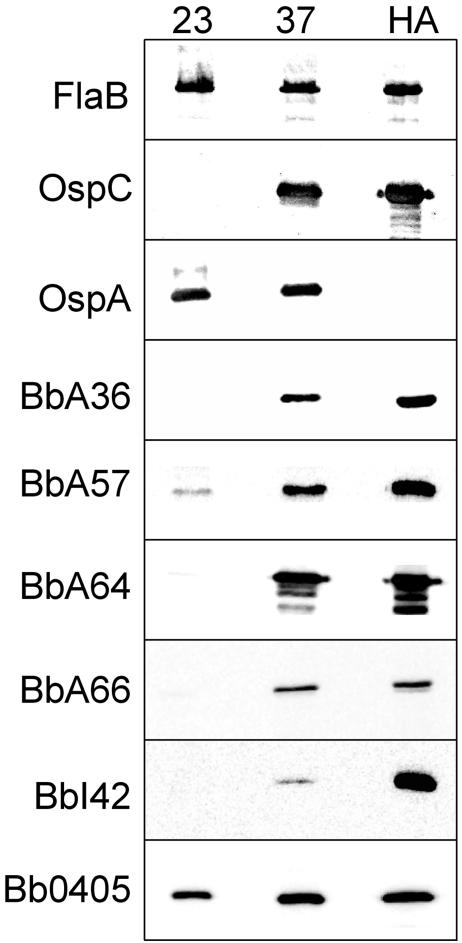
To confirm that the expression patterns of these proteins corresponded to the prior transcriptional profiles determined by microarray analyses, we also performed chemiluminescent immunoblot analyses for 6 of the 10 candidate outer surface proteins identified. As shown in Fig. 2, all six proteins analyzed (BbA36, BbA57, BbA64, BbA66, BbI42, and Bb0405) were up-regulated by temperature shift and by cultivation within a mammalian host. These data were consistent with the transcriptional regulation previously observed in microarray analyses (6, 48). As a control and to confirm that spirochetes responded appropriately to temperature shift and mammalian host adaptation, the constitutively expressed FlaB protein was not observed to be differentially expressed by temperature- or by mammalian host-adapted spirochetes. By contrast, OspC was dramatically up-regulated by temperature and in mammalian host-adapted spirochetes whereas OspA was dramatically down-regulated in mammalian host-adapted organisms, as expected (1, 12, 29, 57).
FIG. 2.
Candidate B. burgdorferi surface proteins are up-regulated by temperature-shifted and/or mammalian host-adapted spirochetes. Whole-cell lysates (1 × 107) of B. burgdorferi B31 cultivated at 23°C, temperature shifted from 23 to 37°C, or cultivated in a mammalian host environment (HA) were subjected to SDS-PAGE and transferred to nitrocellulose. The membranes were blotted with 1:1,000 dilutions of the respective rat polyclonal antibodies before being visualized by enhanced chemiluminescence. Antibodies utilized are indicated at the left of each panel.
To examine the overall conservation of the 10 proteins chosen for further analysis, we also compared the overall sequence relationship of these B. burgdorferi strain B31 proteins with their respective orthologs from B. burgdorferi strains N40 and 297 (Sherwood R. Casjens, John J. Dunn, Benjamin J. Luft, Claire M. Fraser, Wei-Gang Qiu, and Steven E. Schutzer, unpublished data) as well as from the B. garinii PBi strain. This comparison was performed since one of the important parameters for generating a second-generation Lyme disease vaccine revolves around the overall conservation of potential vaccine candidates among strains and genospecies of spirochetes. As shown in Table 2, among the proteins for which sequences are currently available, all were highly conserved between all B. burgdorferi strains (greater than 96% identity) while the proteins from the B. garinii genospecies were less well conserved (ranging from 23 to 95% identity).
TABLE 2.
Percent identity between B. burgdorferi B31 candidate outer membrane proteins and proteins identified in B. garinii strain PBi and B. burgdorferi strains N40 and 297
| B31 gene designation | % Identity with:
|
||
|---|---|---|---|
| B. burgdorferi N40 | B. burgdorferi 297 | B. garinii Pbi (open reading frame) | |
| Bb0126 | 99 | NAb | 93 (Bg0407) |
| Bb0298 | 100 | NAb | 95 (Bg0302) |
| Bb0405 | 100 | 100 | 94 (Bg0407) |
| Bb0689 | 98 | NAb | 85 (Bg0713) |
| BbA36 | 99 | 97 | 64 (BgA30) |
| BbA57 | 99 | 98 | 67 (BgA58) |
| BbA64 | 99 | 99 | 63 (BgA63) |
| BbA66 | 99 | 99 | 50 (BgA65) |
| BbA69 | 99 | 99 | 23 (BgA63) |
| BbI42 | —a | 96 | 72 (BgP310) |
The plasmids from strain N40 have been partially sequenced but are not yet fully completed. Some plasmid telomere regions are still unsequenced in the N40 strain. Therefore, a strain B31 ortholog of the I42 gene, which is located on the telomere of 1p28-4 in strain B31, may be encoded by strain N40 but was not identified in the strain N40 sequence that is currently available (Dr. Sherwood Casjens, personal communication).
NA, not available.
Characterization of candidate Osps using Triton X-114 phase partitioning and proteinase K surface localization assays.
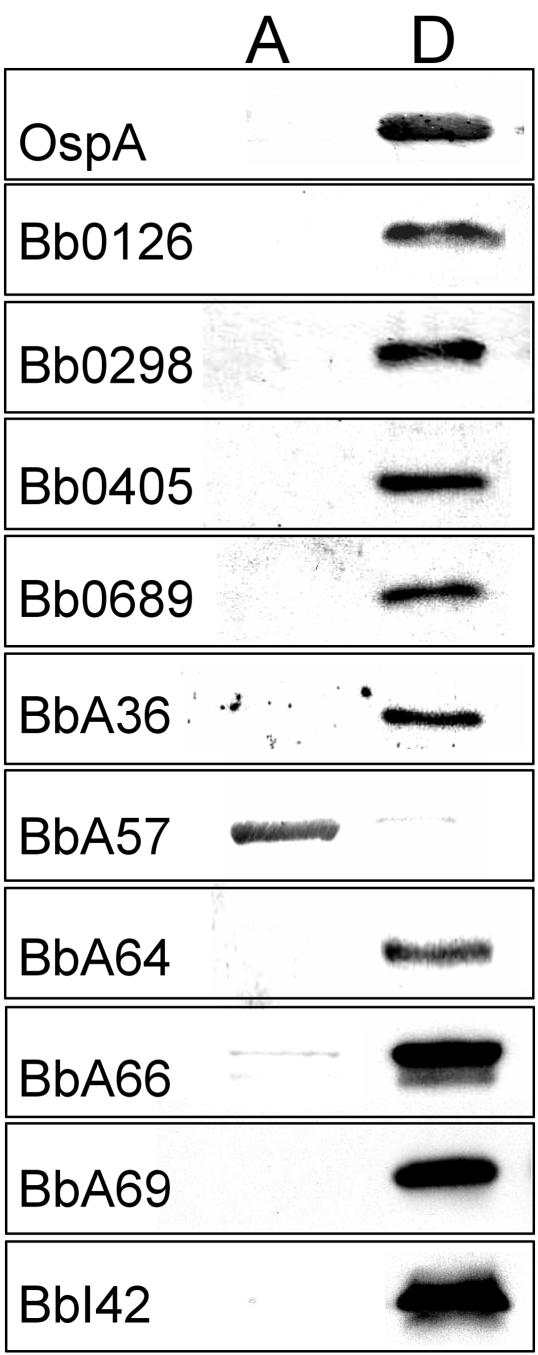
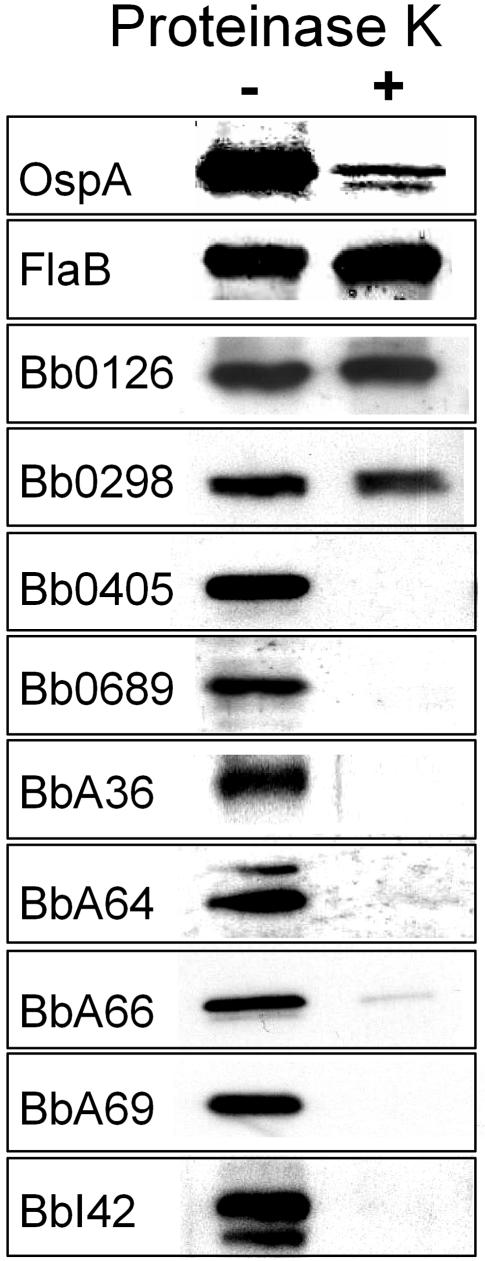
Triton X-114 phase partitioning can quickly and easily assess whether an unknown protein has the amphiphilic properties expected of a bona fide outer membrane protein (7). At the same time, however, amphiphilic lipid-modified proteins also will partition into the detergent phase after Triton X-114 phase partitioning (3). Therefore, in addition to Triton X-114 phase partitioning, cellular localization of lipid-modified proteins must be confirmed by other means, because it is well recognized that many lipoproteins are not located on the surface of B. burgdorferi; rather, they are located in the periplasmic space, where they are anchored to the outer leaflet of the inner membrane or the inner leaflet of the outer membrane (26). Therefore, we chose to confirm that all 10 candidate outer membrane proteins are both amphiphilic (i.e., partition into the Triton X-114 detergent phase) and proteinase K accessible (i.e., are located on the surface) before further studies were undertaken. As shown in Fig. 3, Triton X-114 phase-partitioning experiments on whole-cell lysates of B. burgdorferi B31 revealed that the native proteins for all but BbA57 selectively partitioned into the detergent phase. As a control, antibodies directed against the amphiphilic OspA lipoprotein were also included. As expected, OspA was found to selectively partition into the detergent phase. Among the candidate outer surface proteins that were found to partition into the detergent phase, seven also were found to be sensitive to PK digestion (Fig. 4). Intact, viable spirochetes incubated in the absence of PK were used as a control for these experiments. Specific antibodies directed against the periplasmic protein FlaB also were used as a control to ensure that there was no apparent degradation of this periplasmic protein, which verified that the organisms analyzed in these experiments were not disrupted and that their outer membranes were intact. Additionally, the outer membrane OspA lipoprotein was used as a positive control to show successful PK digestion of a known outer surface protein. Although a small amount of OspA was found to be undegraded by the PK treatment, this was expected since it has been shown that a proportion of OspA is found in the periplasmic compartment in viable spirochetes (14). Combined, the phase-partitioning and PK accessibility experiments strongly suggested that Bb0405, Bb0689, BbA36, BbA64, BbA66, BbA69, and BbI42 are surface-exposed B. burgdorferi proteins. Among the newly identified surface proteins, only Bb0405 would be predicted to contain membrane-spanning domains, while the other six are likely lipid modified and anchored to the borrelial surface by N-terminal fatty acids.
FIG. 3.
Triton X-114 phase partitioning of candidate surface proteins. Whole-cell lysates of B. burgdorferi B31 were subjected to Triton X-114 phase partitioning prior to SDS-PAGE and transfer to nitrocellulose for immunoblot analysis using specific antibodies. Candidate surface proteins were found to be either soluble and selectively partition into the aqueous (A) phase or were amphiphilic and partitioned into the detergent (D) phase.
FIG. 4.
Proteinase K surface localization of candidate surface proteins. Viable spirochetes (1 × 108 bacteria) were incubated with (+) or without (−) proteinase K before being subjected to SDS-PAGE and transfer to nitrocellulose for immunoblot analysis using specific antibodies. Proteins found to be sensitive to proteinase K treatment are located on the surface of B. burgdorferi. OspA and FlaB were utilized as internal controls for proteinase K sensitivity and resistance, respectively.
Specific antibodies directed against borrelial surface proteins are bactericidal.
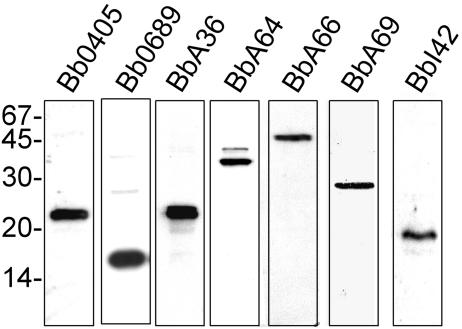
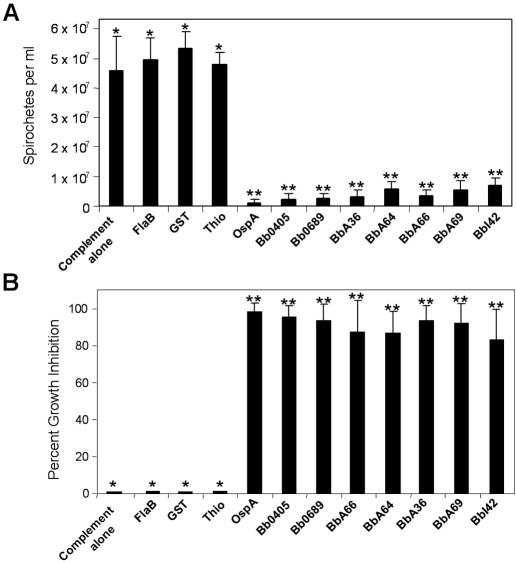
To verify that the antibodies generated against Bb0405, Bb0689, BbA36, BbA64, BbA66, BbA69, and BbI42 were specific, we next performed immunoblot analyses with whole-cell lysates from B. burgdorferi strain B31. As shown in Fig. 5, one major reactive band of the correct size was observed for all seven proteins by immunoblot analysis. To help verify that all seven proteins were on the surface of B. burgdorferi, we next determined whether the specific antibodies generated against the recombinant proteins could kill live spirochetes. Additionally, these experiments also were aimed at elucidating the relative protection that could potentially be provided by specific antibodies directed against these surface proteins and determine whether any were viable Lyme disease vaccine candidates. To assess the bactericidal activity of antibodies specific for the newly identified surface proteins, spirochetes were seeded into 96-well plates containing either guinea pig complement alone or guinea pig complement plus specific antibody (55). Antibodies specific for thioredoxin, GST, and FlaB were also included as negative controls for all experiments. Additionally, OspA antibodies were used as a positive control to demonstrate that antibodies against a known surface protein could kill B. burgdorferi as previously shown (55). Spirochetes were incubated with 1:10 dilutions of the various antibody preparations for 72 h before enumeration of live spirochetes using dark-field microscopy. Antibodies against all seven surface proteins exhibited significant bactericidal activity and inhibited growth (P < 0.01) compared to the negative control antibody sample results (Fig. 6). Antibodies directed against Bb0405, Bb0689, BbA36, and BbA66 had the greatest impact on growth, limiting the bacterial cell growth to less than 5% (i.e., greater than 95% killing), while antibodies against BbI42 had the least effect on bacterial cell growth, with 86% of spirochetes being killed. As expected, antibodies against OspA resulted in almost 99% killing at a 1:10 dilution of antibody whereas antibodies specific for thioredoxin, GST, and FlaB had no effect on growth. Although there was some variation in the overall killing ability of the antibodies directed against the new outer surface proteins identified, there was no statistical difference in killing activity between these antibodies and the antibodies specific for OspA (P < 0.01).
FIG. 5.
Polyclonal antibodies generated against all seven surface proteins recognize a single, dominant protein in B. burgdorferi whole-cell lysates. Whole-cell lysates (1 × 107) of temperature-shifted B. burgdorferi B31 were subjected to SDS-PAGE and transferred to nitrocellulose. The membranes were blotted with 1:1,000 dilutions of the respective rat polyclonal antibodies before being visualized by enhanced chemiluminescence. Antibodies utilized are indicated at the top of each panel, and molecular mass markers, in kilodaltons, are shown at left.
FIG. 6.
Antibodies generated against all seven surface proteins are borreliacidal. Antibodies generated against recombinant forms of all seven surface proteins were used to examine their bactericidal activity. Spirochetes (4 × 106) were seeded into wells of a 96-well flat-bottom microtiter plate containing either 2 units of guinea pig complement alone per well or 2 units of guinea pig complement with a 1:10 dilution of antibody specific for each surface protein. Plates were sealed and incubated at 34°C for 72 h. After incubation, aliquots of each well were examined by dark-field microscopy to quantitate spirochete viability. Graphs show spirochete viability (A) and percent growth inhibition (B). All experiments were performed in triplicate; samples with one asterisk were found to be significantly different from samples with two asterisks (P < 0.01).
Antibodies specific for Bb0405, Bb0689, BbA36, BbA64, BbA66, BbA69, and BbI42 are generated and maintained during infection.
To determine whether the seven identified surface proteins generated a specific antibody response during primate infection, serum from nonhuman primates infected with B. burgdorferi by natural tick infestation was examined (Table 3). Serum from baboons taken 0, 42, 90, 180, and 365 days post tick infestation were examined by ELISA, which confirmed that antibodies were generated against all seven surface proteins during a natural infection and that antibodies were maintained for at least 1 year postinfection. This observation suggests that all seven proteins are immunogenic and that B. burgdorferi actively expressed all seven proteins at some point during nonhuman primate infection. Within 42 days post tick infestation, antibody titers of between 1:200 and 1:12,800 were observed for all seven proteins and by 1 year post tick infestation the antibody titers were still observed to be between 1:200 and 1:3,200. The least-immunogenic proteins among the seven analyzed corresponded to BbA69 and BbI42, where antibody titers were never observed to be greater than 1:400. OspA, OspC, and FlaB antibody titers also were examined in these studies as controls. As expected, tick-infested animals never generated a humoral response against OspA that was greater than 1:100 (the lowest dilution analyzed), which is consistent with minimal expression of this protein during infection (46). On the other hand, a robust antibody response was observed to OspC and FlaB, which are both known to be expressed during transmission and infection (27-29, 57).
TABLE 3.
Reciprocal antibody endpoint titers of baboon serum against candidate surface proteinsa
| Protein designation | Titer at indicated day post tick infestation
|
||||
|---|---|---|---|---|---|
| 0 | 42 | 90 | 180 | 365 | |
| BbA15 (OspA) | <100 | <100 | <100 | <100 | <100 |
| BbB19 (OspC) | <100 | 800 | 12,800 | 6,400 | 3,200 |
| Bb0147 (FlaB) | <100 | 6,400 | 3,200 | 3,200 | 3,200 |
| Bb0405 | <100 | 1,600 | 1,600 | 1,600 | 1,600 |
| Bb0689 | <100 | 6,400 | 3,200 | 1,600 | 1,600 |
| BbA36 | <100 | 12,800 | 12,800 | 3,200 | 3,200 |
| BbA64 | <100 | 6,400 | 3,200 | 1,600 | 800 |
| BbA66 | <100 | 3,200 | 3,200 | 1,600 | 400 |
| BbA69 | <100 | 200 | 400 | 200 | 200 |
| BbI42 | <100 | 200 | 200 | 400 | 200 |
A total of 100 ng of recombinant protein for each candidate surface protein was subjected to ELISA using serum from tick-infested baboons at time points 0, 42, 90, 180, and 365 days post tick infestation.
DISCUSSION
The unique enzootic life cycle of B. burgdorferi places several challenges on this organism as it migrates between its tick and mammalian host environments (19, 42, 43). While there is little immune pressure on spirochetes in the tick environment, to survive in the mammalian host, B. burgdorferi must evade both the innate immune response and the adaptive immune response during infection (13, 58). Given the extracellular lifestyle of this pathogen, the humoral arm of the immune system has been noted as the most effective mechanism for clearing B. burgdorferi from infected mammals (13, 31-33, 35, 36). Therefore, prior studies have focused on identifying outer surface proteins that are differentially expressed and also targets of the humoral immune response. These prior studies not only led to the identification of vaccine candidates but also helped to characterize novel virulence determinants that are located on the surface of B. burgdorferi (2, 9, 16, 18, 22, 28, 38, 42, 53). The major goal of the studies outlined here was to combine prior functional genomic analyses with cellular and biochemical studies to identify genes that are up-regulated during transmission and infection that encode previously unrecognized B. burgdorferi surface proteins.
Among the seven new surface proteins identified, six were observed to contain typical signal peptidase II processing and modification sites, indicating that they are anchored to the borrelial surface by their lipid moieties. Currently, only the sec-dependent pathway has been identified in B. burgdorferi for the export of proteins. However, this secretion system still has not been fully characterized and some of the secretion components found in other bacteria either appear to be absent in B. burgdorferi or have not yet been identified (23, 26). It currently is believed that all proteins destined for the borrelial surface contain a typical N-terminal signal peptide and are exported by the sec-dependent pathway. Lipoproteins that are destined for translocation across the inner membrane can be identified based on a well-conserved signal peptidase II cleavage and modification site terminated by a cysteine residue that becomes lipid modified during transport (56). During export, glycerol modification of the terminal cysteine in the mature protein and subsequent lipid modification results in two fatty acids being covalently attached to the glycerol; a third fatty acid is then added directly to the N-terminal cysteine during modification within the periplasm. The three hydrophobic fatty acid moieties then anchor otherwise hydrophilic proteins to hydrophobic membrane bilayers. To identify lipid-modified proteins, phase partitioning using Triton X-114 has been shown to be a useful technique because it can selectively separate molecules on the basis of their amphipathic characteristics (7). This unique property of Triton X-114 was exploited to examine the solubility properties of the native proteins identified. As shown in Fig. 3, all candidate lipoproteins partitioned into the detergent phase as expected. This finding strongly suggests that the otherwise hydrophilic Bb0689, BbA36, BbA64, BbA66, BbA69, and BbI42 proteins are lipid modified. The native form of Bb0405 was found to selectively partition into the detergent phase, which is consistent with this protein containing membrane-spanning domains that traverse the outer membrane.
The PK experiments also indicated that Bb0405, the integral outer membrane protein identified, also is surface exposed. This observation is important, since only one other integral outer membrane protein has been identified thus far in B. burgdorferi. The other integral outer membrane protein, designated p66, that has been characterized has been shown to be an integrin binding protein (10, 17). Based on the observation that p66 binds beta (3)-chain integrins (10), which is thought to help B. burgdorferi bind platelets and megakaryocytes in the mammalian host, it is tempting to speculate that Bb0405 also plays an important role in host-pathogen interactions during infection. Furthermore, the observation that bb0405 is up-regulated by temperature and stays up-regulated during mammalian infection (6, 48) suggests that Bb0405 is important during both tick transmission and the early dissemination stage of Lyme disease. This conjecture could be tested by deleting or mutating the bb0405 gene in a virulent strain of B. burgdorferi and determining whether this alters borrelial transmission or dissemination within the mammalian host. Experiments of this nature are now feasible given recent advances that have made it possible to genetically manipulate at least some virulent strains of B. burgdorferi (20, 21, 30, 40).
Of the seven surface proteins identified in this study, one was recently characterized for its expression by B. burgdorferi in mice and for its ability to protect mice from experimental Lyme disease infection. Consistent with our prior microarray data and the data shown here, these recent studies showed that bbA36 is expressed during mammalian infection for at least 4 months (41) and that antibodies directed against BbA36 could passively protect immunodeficient mice from infection (62). However, when bbA36 was specifically inactivated in a virulent B. burgdorferi strain in a recent study by Norgard and coworkers (54), no differential phenotype was observed for this mutant in either the mouse or tick environment. While the function of BbA36 on the surface of B. burgdorferi is still unknown, the combined studies provide empirical evidence that our strategy for identifying viable second-generation Lyme disease vaccinogens is promising.
A major outcome of these studies is the finding that antibodies generated against all seven of the newly identified surface proteins could kill B. burgdorferi. Therefore, all seven can be considered candidates for further vaccine studies in animals. However, this observation also raises a paradox since B. burgdorferi can persist in infected animals even in the presence of specific antibodies against all seven of these proteins. One possible explanation for this phenomenon is that B. burgdorferi colonizes immune-privileged niches during early infection (24, 50) or localizes to tissues with a low penetration of antibody and/or immune effector cells, which would allow organisms to persist in the presence of a specific and robust immune response (11, 24, 47, 50, 59). Therefore, as long as B. burgdorferi can colonize immune-restricted tissue niches before an immune response develops, they are protected and can chronically persist in the mammalian host. Examining the protective nature of antibodies generated in animals prior to infection will allow us to assess this important issue. Preliminary studies have supported this notion. For example, when immunocompetent C3H/HeJ mice were actively immunized with Bb0689 and BbA36, both antigens were able to protect mice from subsequent challenge with 1 × 105 B. burgdorferi bacteria by syringe inoculation (C. S. Brooks and D. R. Akins, unpublished observations). These studies will be expanded in future experiments to include all seven surface proteins identified using animals infected by tick infestation.
In conclusion, we identified seven B. burgdorferi surface proteins by focusing our efforts on a subset of genes previously identified as up-regulated during transmission and infection by use of microarray analyses (6, 48). Among the candidate surface proteins identified in the microarray analyses, a subset of 10 was found to be up-regulated and putatively located on the surface of B. burgdorferi. PK accessibility experiments verified that 7 of the initial 10 proteins are bona fide outer surface proteins in B. burgdorferi. Given the paucity of outer surface proteins that have been identified in B. burgdorferi over the last decade, the results of this study are an important advance in the field and lay the foundation for future work aimed at defining the roles of these proteins in B. burgdorferi physiology and host-pathogen interactions. Tick-infested baboons were observed to generate specific humoral responses to all seven of the surface proteins during the course of infection. Additionally, baboons maintained circulating antibodies against all seven proteins for at least 1 year postinfection while bactericidal assays revealed that all seven surface proteins are targets for killing B. burgdorferi in vitro. The combined data suggest that these newly identified surface proteins are (i) actively expressed during a natural tick-derived infection, (ii) immunogenic during infection, (iii) exposed targets for bactericidal antibody, and (iv) candidate vaccine molecules for Lyme disease. Furthermore, since these molecules are surface exposed and up-regulated during infection, they are likely integral to the parasitic strategy utilized by B. burgdorferi and may be important virulence determinants. With the recent advances in borrelial mutagenesis techniques, future studies will include selectively targeting these genes and disrupting their function to better understand their role(s) in borrelial virulence and Lyme disease pathogenesis.
Acknowledgments
This work was supported in part by grants AI059373 from the National Institutes of Health (NIAID) and 0555550Z from the American Heart Association to D.R.A. C.S.B. was supported in part by Molecular Pathogenesis Training Grant AI07364 from NIAID.
We acknowledge the Borrelia sequencing group of Sherwood R. Casjens, John J. Dunn, Benjamin J. Luft, Claire M. Fraser, Wei-Gang Qiu, and Steven E. Schutzer, working under grants from the Lyme Disease Association and National Institutes of Health (AI37256 and AI49003), for access to unpublished sequence information.
Editor: J. D. Clements
REFERENCES
- 1.Akins, D. R., K. W. Bourell, M. J. Caimano, M. V. Norgard, and J. D. Radolf. 1998. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J. Clin. Investig. 101:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akins, D. R., S. F. Porcella, T. G. Popova, D. Shevchenko, S. I. Baker, M. Li, M. V. Norgard, and J. D. Radolf. 1995. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homolog. Mol. Microbiol. 18:507-520. [DOI] [PubMed] [Google Scholar]
- 3.Akins, D. R., B. K. Purcell, M. Mitra, M. V. Norgard, and J. D. Radolf. 1993. Lipid modification of the 17-kilodalton membrane immunogen of Treponema pallidum determines macrophage activation as well as amphiphilicity. Infect. Immun. 61:1202-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbour, A. G. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521-525. [PMC free article] [PubMed] [Google Scholar]
- 5.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 6.Brooks, C. S., P. S. Hefty, S. E. Jolliff, and D. R. Akins. 2003. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect. Immun. 71:3371-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brusca, J. S., and J. D. Radolf. 1994. Isolation of integral membrane proteins by phase partitioning with Triton X-114. Methods Enzymol. 228:182-193. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control. 2004. Lyme disease-United States, 2001-2002. Morbid. Mortal. Wkly. Rep. 53:365-369. [PubMed] [Google Scholar]
- 9.Champion, C. I., D. R. Blanco, J. T. Skare, D. A. Haake, M. Giladi, D. Foley, J. N. Miller, and M. A. Lovett. 1994. A 9.0-kilobase-pair circular plasmid of Borrelia burgdorferi encodes an exported protein: evidence for expression only during infection. Infect. Immun. 62:2653-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coburn, J., W. Chege, L. Magoun, S. C. Bodary, and J. M. Leong. 1999. Characterization of a candidate Borrelia burgdorferi beta(3)-chain integrin ligand identified using a phage display library. Mol. Microbiol. 34:926-940. [DOI] [PubMed] [Google Scholar]
- 11.Coburn, J., M. Medrano, and C. Cugini. 2002. Borrelia burgdorferi and its tropisms for adhesion molecules in the joint. Curr. Opin. Rheumatol. 14:394-398. [DOI] [PubMed] [Google Scholar]
- 12.Coleman, J. L., J. A. Gebbia, J. Piesman, J. L. Degen, T. H. Bugge, and J. L. Benach. 1997. Plasminogen is required for efficient dissemination of B. burgdorferi in ticks and for enhancement of spirochetemia in mice. Cell 89:1111-1119. [DOI] [PubMed] [Google Scholar]
- 13.Connolly, S. E., and J. L. Benach. 2005. The versatile roles of antibodies in Borrelia infections. Nat. Rev. Microbiol. 3:411-420. [DOI] [PubMed] [Google Scholar]
- 14.Cox, D. L., D. R. Akins, K. W. Bourell, P. Lahdenne, M. V. Norgard, and J. D. Radolf. 1996. Limited surface exposure of Borrelia burgdorferi outer surface lipoproteins. Proc. Natl. Acad. Sci. USA 93:7973-7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daniel, W. W. 1999. Biostatistics: a foundation for analysis in the health sciences. John Wiley and Sons, Inc., New York, N.Y.
- 16.Das, S., S. W. Barthold, S. S. Giles, R. R. Montgomery, S. R. Telford, and E. Fikrig. 1997. Temporal pattern of Borrelia burgdorferi p21 expression in ticks and mammalian host. J. Clin. Investig. 99:987-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Defoe, G., and J. Coburn. 2001. Delineation of Borrelia burgdorferi p66 sequences required for integrin αIIbβ3 recognition. Infect. Immun. 69:3455-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Silva, A. M., and E. Fikrig. 1997. Arthropod- and host-specific gene expression by Borrelia burgdorferi. J. Clin. Investig. 99:377-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Silva, A. M., E. Fikrig, E. Hodzie, F. S. Kantor, S. R. Telford III, and S. W. Barthold. 1998. Immune evasion by tickborne and host-adapted Borrelia burgdorferi. J. Infect. Dis. 177:395-400. [DOI] [PubMed] [Google Scholar]
- 20.Eggers, C. H., M. J. Caimano, M. L. Clawson, W. G. Miller, D. S. Samuels, and J. D. Radolf. 2002. Identification of loci critical for replication and compatibility of a Borrelia burgdorferi cp32 plasmid and use of a cp32-based shuttle vector for the expression of fluorescent reporters in the Lyme disease spirochaete. Mol. Microbiol. 43:281-295. [DOI] [PubMed] [Google Scholar]
- 21.Elias, A. F., P. E. Stewart, D. Grimm, M. J. Caimano, C. H. Eggers, K. Tilly, J. L. Bono, D. R. Akins, J. D. Radolf, T. G. Schwan, and P. Rosa. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect. Immun. 70:2139-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fikrig, E., S. W. Barthold, W. Sun, W. Feng, S. R. Telford, and R. A. Flavell. 1997. Borrelia burgdorferi p35 and p37 proteins, expressed in vivo, elicit protective immunity. Immunity 6:531-539. [DOI] [PubMed] [Google Scholar]
- 23.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J.-F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidman, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 24.Georgilis, K., M. Peacocke, and M. S. Klempner. 1992. Fibroblasts protect the Lyme disease spirochete, Borrelia burgdorferi, from ceftriaxone in vitro. J. Infect. Dis. 166:440-444. [DOI] [PubMed] [Google Scholar]
- 25.Grimm, D., K. Tilly, R. Byram, P. E. Stewart, J. G. Krum, D. M. Bueschel, T. G. Schwan, P. F. Policastro, A. F. Elias, and P. A. Rosa. 2004. Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc. Natl. Acad. Sci. USA 101:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haake, D. A. 2000. Spirochaetal lipoproteins and pathogenesis. Microbiology 146:1491-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hefty, P. S., C. S. Brooks, A. M. Jett, G. L. White, S. K. Wikel, R. C. Kennedy, and D. R. Akins. 2002. OspE-related, OspF-related, and Elp lipoproteins are immunogenic in baboons experimentally infected with Borrelia burgdorferi and in human Lyme disease patients. J. Clin. Microbiol. 40:4256-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hefty, P. S., S. E. Jolliff, M. J. Caimano, S. K. Wikel, and D. R. Akins. 2002. Changes in temporal and spatial patterns of outer surface lipoprotein expression generate population heterogeneity and antigenic diversity in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 70:3468-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hefty, P. S., S. E. Jolliff, M. J. Caimano, S. K. Wikel, J. D. Radolf, and D. R. Akins. 2001. Regulation of OspE-related, OspF-related, and Elp lipoproteins of Borrelia burgdorferi strain 297 by mammalian host-specific signals. Infect. Immun. 69:3618-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hubner, A., X. Yang, D. M. Nolen, T. G. Popova, F. C. Cabello, and M. V. Norgard. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc. Natl. Acad. Sci. USA 98:12724-12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson, R. C., C. Kodner, and M. Russell. 1986. Active immunization of hamsters against experimental infection with Borrelia burgdorferi. Infect. Immun. 54:897-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson, R. C., C. Kodner, and M. Russell. 1986. Passive immunization of hamsters against experimental infection with the Lyme disease spirochete. Infect. Immun. 53:713-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson, R. C., C. L. Kodner, and M. E. Russell. 1986. Vaccination of hamsters against experimental infection with Borrelia burgdorferi. Zentralbl. Bakteriol. Mikrobiol. Hyg. 263:45-48. [DOI] [PubMed] [Google Scholar]
- 34.Juncker, A. S., H. Willenbrock, G. von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kochi, S. K., and R. C. Johnson. 1988. Role of immunoglobulin G in killing of Borrelia burgdorferi by the classical complement pathway. Infect. Immun. 56:314-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kochi, S. K., R. C. Johnson, and A. P. Dalmasso. 1991. Complement-mediated killing of the Lyme disease spirochete Borrelia burgdorferi. Role of antibody in formation of an effective membrane attack complex. J. Immunol. 146:3964-3970. [PubMed] [Google Scholar]
- 37.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 38.Lahdenne, P., S. F. Porcella, K. E. Hagman, D. R. Akins, T. G. Popova, D. L. Cox, J. D. Radolf, and M. V. Norgard. 1997. Molecular characterization of a 6.6-kilodalton Borrelia burgdorferi outer membrane-associated lipoprotein (lp6.6) which appears to be downregulated during mammalian infection. Infect. Immun. 65:412-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lane, R. S., J. Piesman, and W. Burgdorfer. 1991. Lyme borreliosis: relation of its causative agent to its vectors and hosts in North America and Europe. Annu. Rev. Entomol. 36:587-609. [DOI] [PubMed] [Google Scholar]
- 40.Lawrenz, M. B., H. Kawabata, J. E. Purser, and S. J. Norris. 2002. Decreased electroporation efficiency in Borrelia burgdorferi containing linear plasmids lp25 and lp56: impact on transformation of infectious B. burgdorferi. Infect. Immun. 70:4798-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lerder, S., C. Brenner, T. Stehle, L. Gern, R. Wallich, and M. M. Simon. 2005. Quantitative analysis of Borrelia burgdorferi gene expression in naturally (tick) infected mouse strains. Med. Microbiol. Immunol. 194:81-90. [DOI] [PubMed] [Google Scholar]
- 42.Liang, F. T., F. K. Nelson, and E. Fikrig. 2002. DNA microarray assessment of putative Borrelia burgdorferi lipoprotein genes. Infect. Immun. 70:3300-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang, F. T., F. K. Nelson, and E. Fikrig. 2002. Molecular adaptation of Borrelia burgdorferi in the murine host. J. Exp. Med. 196:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma, J., and R. T. Coughlin. 1993. A simple, colorimetric microtiter assay for borreliacidal activity of antisera. J. Microbiol. Methods 145:145-153. [Google Scholar]
- 45.Morrissey, J. H. 1981. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal. Biochem. 117:307-310. [DOI] [PubMed] [Google Scholar]
- 46.Narasimhan, S., M. J. Caimano, F. T. Liang, F. Santiago, M. Laskowski, M. T. Philipp, A. R. Pachner, J. D. Radolf, and E. Fikrig. 2003. Borrelia burgdorferi transcriptome in the central nervous system of non-human primates. Proc. Natl. Acad. Sci. USA 100:15953-15958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nocton, J. J. and A. C. Steere. 1995. Lyme disease. Adv. Intern. Med. 40:69-117. [PubMed] [Google Scholar]
- 48.Ojaimi, C., C. Brooks, S. Casjens, P. Rosa, A. Elias, A. Barbour, A. Jasinskas, J. Benach, L. Katona, J. Radolf, M. Caimano, J. Skare, K. Swingle, D. Akins, and I. Schwartz. 2003. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun. 71:1689-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orloski, K. A., E. B. Hayes, G. L. Campbell, and D. T. Dennis. 2000. Surveillance for Lyme disease—United States, 1992-1998. MMWR CDC Surveill. Summ. 49:1-9. [PubMed] [Google Scholar]
- 50.Pachner, A. R., J. Basta, E. Delaney, and D. Hulinska. 1995. Localization of Borrelia burgdorferi in murine Lyme borreliosis by electron microscopy. Am. J. Trop. Med. Hyg. 52:128-133. [DOI] [PubMed] [Google Scholar]
- 51.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Radolf, J. D., K. W. Bourell, D. R. Akins, J. S. Brusca, and M. V. Norgard. 1994. Analysis of Borrelia burgdorferi membrane architecture by freeze-fracture electron microscopy. J. Bacteriol. 176:21-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramamoorthy, R., L. Povinelli, and M. T. Philipp. 1996. Molecular characterization, genomic arrangement, and expression of bmpD, a new member of the bmp class of genes encoding membrane proteins of Borrelia burgdorferi. Infect. Immun. 64:1259-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Revel, A. T., J. S. Blevins, C. Almazan, L. Neil, K. M. Kocan, J. de la Fuente, K. E. Hagman, and M. V. Norgard. 2005. bptA (bbe16) is essential for the persistence of the Lyme disease spirochete, Borrelia burgdorferi, in its natural tick vector. Proc. Natl. Acad. Sci. USA 102:6972-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sadziene, A., P. A. Thompson, and A. G. Barbour. 1993. In vitro inhibition of Borrelia burgdorferi by antibodies. J. Infect. Dis. 167:165-172. [DOI] [PubMed] [Google Scholar]
- 56.Sankaran, K., S. D. Gupta, and H. C. Wu. 1995. Modification of bacterial lipoproteins. Methods Enzymol. 250:683-697. [DOI] [PubMed] [Google Scholar]
- 57.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 92:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sigal, L. H. 1997. Lyme disease: a review of aspects of its immunology and immunopathogenesis. Annu. Rev. Immunol. 15:63-92. [DOI] [PubMed] [Google Scholar]
- 59.Steere, A. C. 1989. Lyme disease. N. Engl. J. Med. 321:586-597. [DOI] [PubMed] [Google Scholar]
- 60.Steere, A. C. 1994. Lyme disease: a growing threat to urban populations. Proc. Natl. Acad. Sci. USA 91:2378-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steere, A. C. 1995. Borrelia burgdorferi (Lyme disease, Lyme borreliosis), p. 2143-2155. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases. Churchill Livingstone, New York, N.Y.
- 62.Wallich, R., O. Jahraus, T. Stehle, T. T. Tran, C. Brenner, H. Hofmann, L. Gern, and M. M. Simon. 2003. Artificial-infection protocols allow immunodetection of novel Borrelia burgdorferi antigens suitable as vaccine candidates against Lyme disease. Eur. J. Immunol. 33:708-719. [DOI] [PubMed] [Google Scholar]