Abstract
Nosocomial infections caused by Staphylococcus epidermidis are characterized by biofilm formation on implanted medical devices. Quorum-sensing regulation plays a major role in the biofilm development of many bacterial pathogens. Here, we describe luxS, a quorum-sensing system in staphylococci that has a significant impact on biofilm development and virulence. We constructed an isogenic ΔluxS mutant strain of a biofilm-forming clinical isolate of S. epidermidis and demonstrated that luxS signaling is functional in S. epidermidis. The mutant strain showed increased biofilm formation in vitro and enhanced virulence in a rat model of biofilm-associated infection. Genetic complementation and addition of autoinducer 2-containing culture filtrate restored the wild-type phenotype, demonstrating that luxS repressed biofilm formation through a cell-cell signaling mechanism based on autoinducer 2 secretion. Enhanced production of the biofilm exopolysaccharide polysaccharide intercellular adhesin in the mutant strain is presumably the major cause of the observed phenotype. The agr quorum-sensing system has previously been shown to impact biofilm development and biofilm-associated infection in a way similar to that of luxS, although by regulation of different factors. Our study indicates a general scheme of quorum-sensing regulation of biofilm development in staphylococci, which contrasts with that observed in many other bacterial pathogens.
For a long time, Staphylococcus epidermidis has been regarded as an innocuous commensal bacterium on the skin and mucous membranes of the human body. However, it is now known to be a major nosocomial pathogen, causing infections on implanted medical devices such as central venous catheters (CVCs), urinary catheters, prosthetic heart valves, orthopedic devices, and contact lenses. The investigation of S. epidermidis pathogenesis has become of interest in recent years, as S. epidermidis infections have arisen as a major burden to the public health system. They are often responsible for prolonged hospitalization and may even be fatal (51).
In contrast to Staphylococcus aureus, S. epidermidis produces only a limited amount of toxins and degradative exoenzymes (58). The formation of a biofilm, which is a differentiated structure of surface-attached cells that are embedded in a self-secreted heterogeneous matrix, is thought to be the leading cause for the persistence of S. epidermidis infection. A biofilm constitutes a depository that may continuously release bacteria into the bloodstream. Furthermore, the treatment of biofilm-associated infections is especially difficult due to biofilm resistance to antibiotics and attacks from the human immune system. To date, several genes have been identified that are associated with the different stages of biofilm formation, including those carrying autolysin E (atlE) (20), fibrinogen binding protein (fbe) (41), and the intercellular adhesion operon (ica) (17, 21). However, the exact roles of these genes during biofilm formation are not yet entirely clear (11, 26, 32). Possibly, differential expression of these factors in vitro and in vivo and under different environmental conditions has so far been underestimated.
Quorum sensing (QS) is a bacterial intercommunication system that controls the expression of multiple genes in response to population density (16). QS systems use small signal molecules called autoinducers (AIs). When the AIs accumulate to a threshold concentration, the system is activated and directly or indirectly controls the transcription of target genes. Gram-negative bacteria normally use acylated homoserine lactones (AHLs) as AIs, while gram-positive bacteria use oligopeptide AIs, which act through two-component phosphorelay cascades (36, 44, 53).
In the last decade, a novel quorum-sensing system was discovered in the bioluminescent marine bacterium Vibrio harveyi (3). V. harveyi has three parallel QS systems to induce bioluminescence. One uses AI-1, a typical AHL, as a signal; a second uses CAI-1 (V. cholerae autoinducer 1), a signal-mediating intercommunication in closely related marine bacteria; and finally there is a novel signaling molecule called AI-2 (22). There is ample data to suggest that AI-2s have been designed for interspecies cell-cell communication (53). They are found in both gram-negative and gram-positive bacteria (36, 48, 56). AI-2s are not species specific, as AI-2s produced by other bacteria species can also trigger the bioluminescence of V. harveyi (47). A gene called luxS is required for the synthesis of AI-2 (48). AI-2-like activity and luxS homologues are found in a wide range of bacterial species. Database analysis shows that luxS exists in 35 of the 89 available complete bacterial genomes (National Center for Biotechnology Information) (56). Besides light production in V. harveyi (4) and Vibrio fischeri (28), luxS/AI-2-dependent QS regulates the growth of Escherichia coli (45), Bacillus anthracis (23), and Actinobacillus actinomycetemcomitans (15). It also plays a role in E. coli flagellum motility (45). Furthermore, it modulates the virulence of Streptococcus pyogenes (29), Clostridium perfringens (39), Vibrio vulnificus (25), and Streptococcus pneumoniae (24, 46). Interestingly, AI-2 from the resident microflora in the lungs of cystic fibrosis patients can up-regulate pathogenicity factors in non-AI-2-producing Pseudomonas aeruginosa (12). In some bacteria, it has been reported that luxS/AI-2-dependent QS impacts biofilm formation in various ways. For example, a luxS mutant of Streptococcus gordonii, a major component of dental plaque biofilm, was unable to form a mixed-species biofilm with a luxS-null strain of the periodontal pathogen Porphyromonas gingivalis (34) despite the wild-type strains readily doing so. Furthermore, a luxS mutant of Helicobacter pylori forms biofilm more efficiently than the wild type (9). Finally, a luxS mutant of Streptococcus mutans shows an altered biofilm structure (35).
Staphylococci have one known QS system called agr, for accessory gene regulator, which secretes modified peptides as signals (38, 40). In S. epidermidis, agr represses biofilm formation and influences the structure of a biofilm (49, 50) by regulation of biofilm factors, such as AtlE and δ-toxin (49). luxS homologues are present in the S. aureus and S. epidermidis genomes (54, 58). However, although Winzer et al. have purified the LuxS protein from S. aureus that can synthesize the AI-2 (54), whether S. epidermidis or other staphylococci use an AI-2-type QS system to control gene expression has not been investigated previously.
In this study, we explored the luxS/AI-2-dependent QS system of S. epidermidis by construction and analysis of an allelic replacement mutant of S. epidermidis luxS. We found that the luxS/AI-2-dependent QS system of S. epidermidis is functional. Of note, similarly to the agr system (50), luxS of S. epidermidis limits biofilm formation and virulence in an animal model of device-associated infection. Our data point towards a common scheme of QS control of biofilm formation and biofilm-associated infection in staphylococci.
MATERIALS AND METHODS
Bacterial strains and growth media.
The bacteria and plasmids used are listed in Table 1. E. coli DH5α was grown in Luria-Bertani medium. Plasmid-containing E. coli strains were grown in the same medium but with ampicillin (100 μg/ml) included. S. epidermidis and its derivative strains were grown in TSB (tryptic soy broth, soybean-casein digest medium USP; Oxoid) medium, and when necessary, erythromycin (10 μg/ml), erythromycin (2.5 μg/ml), and chloramphenicol (20 μg/ml) were supplemented. V. harveyi BB170 was kindly provided by B. Bassler (Princeton University) and was grown in autoinducer bioassay (AB) medium at 30°C (48). Media were solidified with 1.5% (wt/vol) agar as needed.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference(s) |
|---|---|---|
| Strains | ||
| S. epidermidis 1457 | luxS wild-type strain, biofilm positive | 33 |
| S. epidermidis 1457 ΔluxS mutant | luxS mutant (LuxS− Ermr) | This study |
| E. coli DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 ΔluxS | 19, 48 |
| V. harveyi BB170 | luxN::Tn5 (sensor 1− sensor 2+), AI-2 reporter strain | 47 |
| Plasmids | ||
| pcDNA2.1 | Cloning vector, ampicillin resistant | Invitrogen |
| pEC4 | A vector containing a 1.45-kb ClaI ermB fragment of Tn551 cloned into the ClaI restriction site of pBluescript KS+ | 6 |
| pBT1 | Shuttle vector, temperature sensitive, ampicillin and chloramphenicol resistant | 6 |
| pQG23 | pcDNA2.1 harboring a 2.5-kb DNA fragment containing the luxS gene | This study |
| pQG24 | pcDNA2.1 harboring an Ermr gene with regions up and down from luxS | This study |
| pQG21 | pBT1 harboring Ermr gene with up- and downstream sequence of luxS gene | This study |
| pQG27 | pBT1 harboring 1.0-kb DNA fragment containing the luxS gene | This study |
DNA manipulation.
Genomic DNA of S. epidermidis 1457 was prepared by a standard protocol for gram-positive bacteria (13). Plasmid DNA from E. coli was extracted using a plasmid purification kit (TianWei Co.) according to the manufacturer's instructions. Plasmid DNA from S. epidermidis and S. aureus was extracted using the same kit except that the cells were incubated for at least 15 min at 37°C in solution P1 with lysostaphin (25 μg/ml; Sigma) before solution P2 was added. Taq DNA polymerase and restriction enzymes were obtained from Takara; and incubation conditions were as recommended by the suppliers. S. epidermidis was transformed by electroporation as described previously (2).
Construction of an S. epidermidis ΔluxS mutant strain.
A 2.5-kb DNA fragment from S. epidermidis 1457 genomic DNA with luxS and 1-kb upstream and 1-kb downstream flanking sequences was PCR amplified using the primers luxS(a)5 and luxS(a)3, introducing BamHI and EcoRI sites, respectively. The amplified PCR products were cloned into pcDNA2.1, yielding pQG23. An erythromycin resistance (Ermr) gene was PCR-amplified by primers erm5 and erm3 from pEC4 (6). The product was ligated to the NdeI and SpeI sites inside the luxS gene on pQG23, yielding pQG24. A BamHI/EcoRI fragment was excised from pQG24, purified, and ligated to the vector pBT1 (6) to create pQG21. Allelic replacement of the native luxS gene with the resulting plasmid in genomic DNA of S. epidermidis 1457 with Ermr was performed as described previously (6). Erythromycin-resistant and chloramphenicol-sensitive colonies were selected. PCR and sequencing were performed to confirm that the desired gene inactivation had occurred by double-crossover recombination.
Construction of pQG27 for complementation.
To complement E. coli DH5α or the ΔluxS mutant strain of S. epidermidis 1457 with a functional copy of luxS, a 1.0-kb fragment containing an intact luxS open reading frame (ORF), including its promoter, was amplified from S. epidermidis 1457 genomic DNA by using primers luxS(b)5 and luxS(b)3. The amplified product was ligated into pBT1, creating pQG27. pQG27 was then introduced into E. coli DH5α or the S. epidermidis 1457 ΔluxS mutant strain by electroporation. Clones exhibiting resistance to ampicillin (100 μg/ml) or chloramphenicol (20 μg/ml) were chosen for further study. The presence of luxS on pQG27 was confirmed by sequencing. To exclude the possible influence from the vector, the empty vector was electroporated into E. coli DH5α or S. epidermidis 1457 as a control strain.
AI-2 assay.
The AI-2 reporter assay was performed according to the method of Surette and Bassler (47) and Surette, Miller, and Bassler (48), which is based on the reporter strain V. harveyi BB170 responding to the presence of exogenous AI-2 by induction of bioluminescence. Cell-free medium (CM) was prepared as follows: tester strains were grown overnight at 37°C and then diluted 1:100 into fresh medium, and 1.5-ml samples were taken every 2 h. Cells were pelleted by centrifugation, and the resulting supernatant was further clarified by filtering through a 0.22-mm-pore-size filter (Millipore). Samples (20 μl) of CM were added into white 96-well microtiter plates. To determine bioluminescence, the V. harveyi BB170 reporter strain, grown for 16 h with aeration (180 rpm) at 30°C in AB medium, was diluted 1:5,000 in fresh AB medium and then 180 μl was added to each sample. The bioassay medium was incubated at 30°C. Luminescence was monitored hourly by Fluoroskan Ascent FL (Labsystems). In every case, conditioned medium from V. harveyi BB170 served as an additional positive control. For all the complementation experiments, strains were grown at 30°C.
Semiquantitative biofilm formation.
The method for quantification of biofilm was performed as described previously (8) and modified as follows. S. epidermidis strains were grown in TSB for 16 h and diluted 1:100 into fresh TSB. The diluted cultures were pipetted into sterile 96-well flat-bottomed tissue culture plates and incubated at 37°C for 24 h. Culture supernatants were gently removed with a pipette, and the wells were washed four times with PBS (phosphate-buffered saline; pH 7.2). The adherent organisms remaining at the bottom of the well were fixed by Bouin fixative. After 1 h, the fixative was removed and wells were washed with PBS. Following that, the adherent bacteria were stained with crystal violet and the excessive stain was washed off gently by slowly running water. After drying, the stained biofilm was read with a MicroELISA autoreader (Bio-Rad Co.) using a wavelength of 570 nm under single-wavelength mode. For all complementation experiments, strains were incubated at 30°C.
SEM.
For SEM (scanning electron microscopy), biofilm was grown in TSB with 0.5% glucose for 24 h at 37°C on the surface of sterile glass slides, which were deposited in advance in each well of 24-well hydroxylapatite disks. The contents of each well then were carefully aspirated with a pipette, rinsed with PBS three times, and then fixed with glutaral fixative solution overnight. Following dehydration through a graded series of ethanol rinses, the slides were coated with gold-palladium. SEM micrographs were taken in the Electron Microscopy Core Facility at Shanghai Medical College, Fudan University.
Quantitative RT-PCR (TaqMan).
Oligonucleotide primers and probes were designed using Primer Express version 2.0 (Applied Biosystems) and were synthesized by Applied Biosystems. The primers (icaC5 and icaC3) and probes (icaCP) used are listed in Table 2. Fluorescent probes were used to continuously monitor formation of PCR products during PCR. Isolated RNA was quantified by spectrophotometry. A total of 100 ng of RNA was used per reaction. Reverse transcription of the RNA template and real-time (RT)-PCR were carried out in a single step using TaqMan one-step RT-PCR autoinducer 2 containing master mix (Applied Biosystems). Reactions were performed in a MicroAmp optical 96-well reaction plate with a model 7700 sequence detector (Applied Biosystems). Reverse transcription was performed at a temperature of 48°C for 30 min. PCRs were performed as described previously (57). Standard curves were determined for each gene by use of purified chromosomal template DNA at concentrations of 0.0005 to 500 ng/ml. Assays were performed in triplicate using 16S rRNA as a control.
TABLE 2.
Oligonucleotides used in this study
| Oligo-nucleotide | DNA sequence (5′→3′)a |
|---|---|
| luxS(a)5 | CGGGATCCATCAAGTTCTTTCCGTGAAG |
| luxS(a)3 | CGGAATTCATAGTGGTTATGATGTAATCG |
| luxS(b)5 | CGGGATCCGCAATATCCGTTTCGATTTC |
| luxS(b)3 | CGGAATTCAGAGAAACATCGAAGGGAAG |
| erm5 | GGAATTGCATATGGATACAAATTCCCCGTAGGC |
| erm3 | GGACTAGTGAAATAGATTTAAAAATTTCGCTG |
| icaC5 | TGCTTACACCAACATATTTGAAGATAATAC |
| icaC3 | GACGCCTATACAAATTCCTAGAATCATT |
| icaCP | TTTCTTGGCGATTTCACT |
Incorporated restriction sites are underlined and boldfaced.
Immuno-dot blot assay.
To quantify polysaccharide intercellular adhesin (PIA) production, equal amounts of S. epidermidis cells were grown to stationary growth phase (24 h) and PIA was extracted by incubation with 0.5 mol/liter EDTA (pH 8.0) for 5 min at 100°C. Two aliquots of the samples were spotted on a nitrocellulose membrane, air-dried, and blocked with 5% skim milk in TBS buffer overnight. PIA was detected using anti-PIA antiserum as described previously (52) by using a scanner and Total Lab version 2003 software (Nonlinear USA, Inc., Durham, NC). Assays were performed in triplicate.
Complementation with conditioned medium.
Cell-free conditioned medium (CM) from S. epidermidis 1457 was prepared as follows. Cultures were grown overnight and then incubated and diluted 1:100 followed by resumption of shaking at 37°C for 4 h for the S. epidermidis wild-type strain and 6 h for the luxS-complemented strain (both achieved exponential phase). Cell-free supernatant was harvested by centrifugation and filtered as described above. CM was added to the culture at the start of incubation of biofilm formation for the ΔluxS mutant in 96-well flat-bottomed tissue culture plates, at 10% (vol/vol) final concentration.
A rat model of intravascular catheter-associated infection.
A rat model of CVC-associated infection was developed according to the method of Mack et al. (43). Thirty healthy male Sprague-Dawley rats (weight, 500 g) (Academia Sinica) were surgically dissected, and Silastic lumen-within-lumen catheters (inside diameter, 0.12 mm) were inserted into their right external jugular veins. The proximal portion of the catheter was tunneled subcutaneously and came out from the midscapular space. A metal column was used to block up the end of the proximal portion of the catheter, preventing contamination from outside. Twenty-four hours after CVC placement, 4 × 105 CFU of S. epidermidis 1457 or its isogenic ΔluxS mutant were inoculated into the catheters. In our study, 15 rats were infected with S. epidermidis 1457 and 15 were infected with its ΔluxS mutant. The catheters were flushed daily with heparin. One week after inoculation, the rats were killed and the bacteria recovered from catheters, blood, hearts, kidneys, and livers were counted.
RESULTS
Identification of luxS from S. epidermidis.
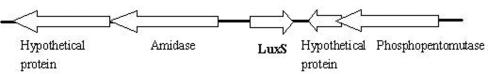
In order to determine whether S. epidermidis has a luxS/AI-2-dependent QS system, we examined the available S. epidermidis genome information (58) and identified a candidate ORF whose predicted protein product was 43% identical and 63% similar to the V. harveyi LuxS protein. S. epidermidis LuxS is very similar to S. aureus LuxS (identity at the amino acid level of 91%; similarity, 98%) and also shows significant similarity (63%) to LuxS of E. coli. The luxS gene of S. epidermidis is an independent ORF (Fig. 1), and its promoter is most likely located upstream of the ORF.
FIG. 1.
Identification of the luxS gene of S. epidermidis. Shown are the luxS gene and its surrounding region according to genetic information (58).
Functional analysis of the AI-2 QS system in S. epidermidis.
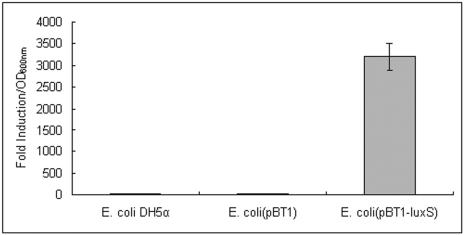
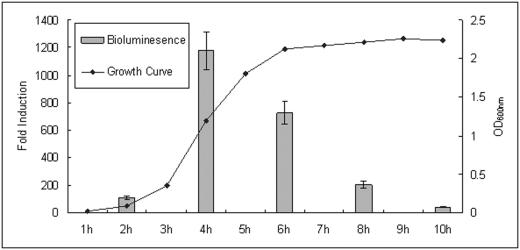
To investigate whether the luxS gene of S. epidermidis is functional and required for AI-2 production, we complemented an E. coli DH5α strain deficient in AI-2 synthesis (47) with a plasmid (pQG27) containing the luxS gene of S. epidermidis under control of its putative native promoter. The presence of the plasmid containing luxS was sufficient to induce light production over 3,000-fold, whereas the control E. coli strains did not produce light (Fig. 2). Furthermore, to determine whether S. epidermidis produces AI-2 and uses AI-2 QS control, we measured AI-2 activity during the growth of the S. epidermidis wild-type strain 1457 using the same reporter assay. AI-2 accumulates during logarithmic phase and is reduced in stationary phase in S. epidermidis (Fig. 3). These analyses demonstrated that S. epidermidis contains a pathway to synthesize and secrete AI-2.
FIG. 2.
Complementation of the AI-2 synthesis deficiency of E. coli DH5α by luxS from S. epidermidis. The capacity to produce AI-2 was examined by an AI-2 reporter assay as described previously (47, 48). V. harveyi BB170 (AI-1 sensor−, AI-2 sensor+) served as a positive control. Cell-free medium was prepared from the strains at logarithmic growth phase, when their light production reached maximum. Luminescence was expressed as fold induction relative to the background values and normalized to cell OD of 1 at 600 nm. Data were obtained from four independent experiments.
FIG. 3.
Detection of AI-2 activity throughout the growth of S. epidermidis 1457. Cells were grown overnight and diluted 1:100 into fresh TSB medium. The absorbance (OD) at 600 nm was then examined hourly by spectrophotometry. Every 2 h, conditioned cell-free supernatants were examined for the capacity to induce light production in V. harveyi BB170. Data were obtained from four independent experiments.
The role of the AI-2 QS system in biofilm formation of S. epidermidis.
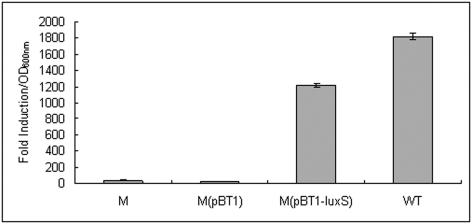
To investigate the role of luxS in the virulence of S. epidermidis, a ΔluxS mutant strain was constructed by an allelic exchange strategy (6). The successful exchange of the luxS gene with the antibiotic resistance cassette was confirmed by PCR and DNA sequencing (data not shown). The mutant and wild-type strains were unaffected in growth rate and extent in a variety of media (data not shown). The ΔluxS mutant of S. epidermidis 1457 produced no detectable AI-2 activity (Fig. 4), and the difference between the wild type and the ΔluxS mutant was most significant after 4 h of growth, at which point both strains entered logarithmic growth phase (optical density, ∼1.2). After complementation of the mutant with plasmid pQG27, AI-2 activity was restored to almost the wild-type level (Fig. 4). These analyses demonstrated that luxS is responsible for AI-2 production in S. epidermidis.
FIG. 4.
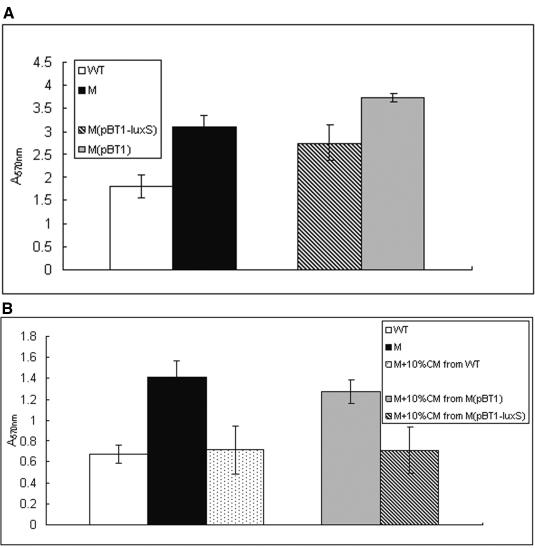
AI-2 activity of S. epidermidis 1457, its isogenic ΔluxS mutant strain and/or the mutant strain with empty vector complemented with luxS. Cell-free medium was prepared from those strains during the log phase. All experiments were performed in triplicate. WT, S. epidermidis 1457 wild-type strain; M, S. epidermidis 1457 isogenic ΔluxS mutant strain; M(pBT1), S. epidermidis ΔluxS mutant strain containing vector pBT1; M(pBT1-luxS), luxS-complemented S. epidermidis mutant strain.
Biofilm formation is considered the main virulence factor of S. epidermidis. To investigate whether luxS influences biofilm formation in S. epidermidis, we first performed semiquantitative biofilm assays using microtiter plates. The ΔluxS mutant of S. epidermidis showed significantly (P < 0.0001, two-tailed t test) increased biomass (1.7 times) compared to that of the wild-type strain (Fig. 5A). Biofilm formation of the complemented strain was reduced compared to the ΔluxS mutant strain (Fig. 5A).
FIG. 5.
Semiquantitative biofilm assay. Each experiment was repeated eight times. A, Biofilm formation of S. epidermidis 1457, its isogenic ΔluxS mutant, the mutant with the empty vector, and the luxS-complemented mutant at 24 h. B, Biofilm formation of S. epidermidis 1457, its isogenic ΔluxS mutant, and the mutants with exogenous AI-2 prepared from wild-type and complemented mutant strains at 24 h using mutant with CM from mutant strain transformed with empty vector as control.
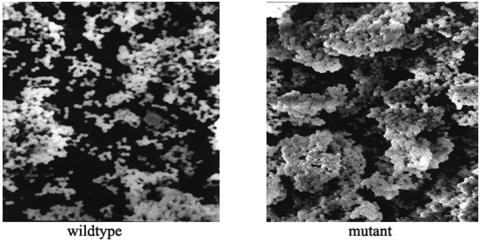
In addition, we used SEM to detect any quantitative or qualitative alteration in the biofilm formation of wild-type and ΔluxS mutant strains. SEM analysis showed that the ΔluxS mutant strain generated a more compact and thicker biofilm than that generated by the wild-type strain (Fig. 6).
FIG. 6.
Scanning electron micrographs of S. epidermidis 1457 and its ΔluxS mutant. Twenty-four-hour biofilms were grown on hydroxyapatite disks that were deposited in 24-well cell culture clusters in TSB with 0.5% (wt/vol) glucose. Images shown are at a magnification of ×3,000.
To examine whether the increased biofilm formation of the S. epidermidis ΔluxS mutant strain was due to a genuine AI-2 signaling event or a mere result of metabolic changes accompanied by the inactivation of the luxS gene, we incubated the ΔluxS mutant strain with exogenous AI-2. To that end, we used CM prepared from exponential-growth-phase cultures of S. epidermidis 1457, the luxS-complemented mutant strain, and the mutant strain with the empty vector as control. CM was mixed with ΔluxS mutant cells grown overnight, which were then incubated for 24 h to test for biofilm formation. The ΔluxS mutant strain showed reduced biofilm formation when 10% CM was added (Fig. 5B) while no growth inhibition was detected (data not shown). These data indicate that the luxS gene of S. epidermidis is involved in repressing biofilm formation through a cell-cell signaling mechanism based on AI-2 secretion.
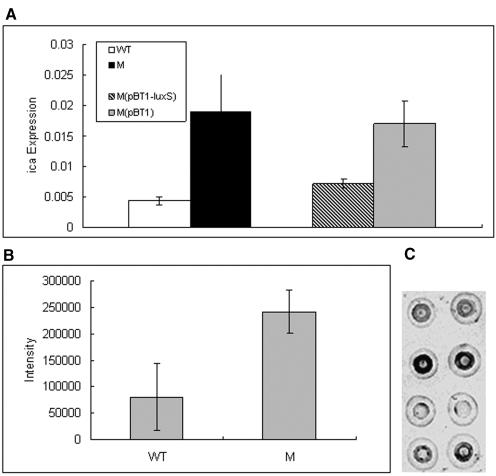
Secretion of the exopolysaccharide PIA is a major factor that determines biofilm formation in S. epidermidis and other bacteria (18, 30, 31). To test whether the altered biofilm formation that we observed in the ΔluxS mutant strain was due to a change in the transcription of the ica operon responsible for PIA production (21), we analyzed transcription of the icaC gene by quantitative real-time PCR. Expression of icaC during mid-exponential growth phase was 4.4 times higher (P = 0.01, two-tailed t test) in the ΔluxS mutant strain than in the wild-type strain (Fig. 7A). The luxS-complemented mutant strain showed an icaC expression level significantly lower than that of the control strain with the empty cloning vector (P = 0.01, two-tailed t test) and comparable to that of the wild-type strain. This indicates that luxS negatively regulates the expression of the ica gene at the transcriptional level. To determine whether luxS regulates the synthesis of PIA, we extracted PIA from stationary-phase cultures of S. epidermidis 1457, the ΔluxS mutant strain, and the complemented and control strains and determined the amount of PIA by use of immuno-dot blot analysis. PIA production was about three times higher in the ΔluxS mutant strain than in the wild-type strain (P = 0.03, two-tailed t test). Thus, luxS/AI-2-dependent QS regulates the expression of PIA. By visual comparison, PIA production was also decreased in the luxS-complemented strain compared to that produced by the mutant strain with the empty vector. Photodigital analysis of this experiment was, however, not possible due to uneven distribution of the reaction on the spotted dots (data not shown).
FIG. 7.
A, TaqMan analysis of ica gene expression. TaqMan analysis was performed using an icaC probe, and cells were grown to mid-exponential phase. The levels of ica mRNA had been normalized to the level of 16S rRNA mRNA. Assays were performed in triplicate. Values given are the means plus or minus standard errors of the means. B, PIA production. PIA samples were isolated from the surface of cells grown to stationary growth phase by boiling with 0.5 mol/liter EDTA. PIA production was determined by immuno-dot blot analysis using anti-PIA antisera and quantified by photodigital analysis. Values are the means plus or minus standard errors of the means from three independent experiments with three measurements (nine dots per strain). C, Representative immuno-dot blot. First row, wild-type strain; second row, isogenic ΔluxS mutant strain; third row, empty vector control strain; fourth row, luxS-complemented strain.
The role of the AI-2 QS system in the virulence of S. epidermidis.
To determine whether luxS has an effect on the pathogenicity of S. epidermidis during biofilm-associated infection, we used a rat intravascular CVC-associated infection model. Twenty-four hours after insertion of Silastic lumen-within-lumen catheters into the necks of the rats, 4 × 105 CFU bacteria were injected into the catheters. Bacteria in blood, the tissue surrounding the catheters, and other organs were counted after sacrificing the rats on day 8 after infection. As shown in Table 3, the ΔluxS mutant had a higher capacity to cause CVC infection (P = 0.006, two-tailed t test) and also showed more significant bacteremia and a greater burden of metastatic disease in liver (P < 0.05, Wilcoxin test). The bacterial counts in other tissues (kidney and heart) did not differ significantly between the two strains, but the ΔluxS mutant strain was found more frequently in tissue samples. Thus, the deletion of luxS in S. epidermidis increased pathogen success during biofilm-associated infection.
TABLE 3.
Metastatic disease caused by S. epidermidis 1457 and its isogenic ΔluxS mutant in the rat CVC-associated infection modelb
| Organ |
S. epidermidis 1457 (n = 15)
|
S. epidermidis 1457 ΔluxS mutant (n = 15)
|
P value | ||||
|---|---|---|---|---|---|---|---|
| No. rats infected/total | Median (range) CFU/g tissue | No. rats infected/total | Median (range) CFU/g tissue | ||||
| Livera | 4/15 | 0 (0-66,000) | 11/15 | 333 (0-2,216,667) | 0.0164 | ||
| Kidneya | 5/15 | 0 (0-308,000) | 9/15 | 2,000 (0-3,673,333) | 0.1002 | ||
| Hearta | 3/15 | 0 (0-66,667) | 6/15 | 0 (0-1,140,000) | 0.2195 | ||
| Blooda | 3/10 | 0 (0-130) | 8/11 | 460 (0-10,980) | 0.0154 | ||
| Tubea | 2,050 (1,750-3,050) | 4,525 (2,050-7,600) | 0.1084 | ||||
| Infection rateb | 27.5 ± 2.8% | 61.5 ± 7.8% | 0.0064 | ||||
The difference in results among liver, kidney, heart, blood, and tube between the two strains was assessed by a Wilcoxon test.
The overall infection rate is defined as recovery of S. epidermidis from the blood, catheter, or organs at the time of sacrifice. The P value for the infection rates of the mutant versus the wild type was 0.0064. The difference of infection rate between the two strains was assessed by a two-tailed t test.
DISCUSSION
Many bacteria use signaling systems to adapt gene expression to environmental changes. For example, QS systems control gene expression in response to cell density. The luxS QS system, which uses AI-2 signal, is a more recently described QS system (7). Importantly, it appears to be the most widespread QS system known so far. However, we lack information about the role of luxS QS control in important bacterial pathogens. Here we describe the luxS/AI-2 QS system in the genus Staphylococcus, which contains several important pathogens such as S. aureus and S. epidermidis.
The luxS system is not always accompanied by endogenous AI-2 activity, suggesting that the AI-2 QS system may not be operative in some bacteria in spite of the existence of luxS. For example, the luxS gene of Borrelia burgdorferi functionally complements a luxS deficiency in E. coli DH5α but AI-2 activity is not detectable in high-density culture supernatants of B. burgdorferi (5). Similar observations have been made in Streptococcus pneumoniae (24). Here we show that the luxS gene of S. epidermidis complements AI-2-deficient E. coli, and AI-2 is endogenously produced by S. epidermidis in a growth-phase-dependent fashion, with a peak observed during exponential growth. In contrast, AI-2 activity was minimal during stationary growth phase. This is consistent with previous reports for E. coli (47), Salmonella typhimurium serovar Typhimurium (47), H. pylori (27), A. actinomycetemcomitans (14), S. mutans (35), and C. perfringens (39).
To date, the V. harveyi AI-2 structure has been determined, but it is possible that the AI-2s synthesized by different bacteria are similar, resulting in a species-nonspecific language. Accordingly, S. epidermidis-conditioned medium containing AI-2 stimulated the luxS gene system in V. harveyi. However, the magnitude of light stimulation in S. epidermidis was lower than that of AI-2 in V. harveyi BB170 (data not shown). This might be due to unique modifications to the basic AI-2 structure or to a lower production level of AI-2 in S. epidermidis under the growth conditions that were used. In fact, interspecies signals secreted by Salmonella serovar Typhimurium have recently been demonstrated to be a distinct form of the AI-2 signal, although they are both derived from the same precursor (37).
Inactivation of the luxS gene of S. epidermidis did not result in noticeable changes in growth patterns compared with those of the wild-type strain in different media. This finding indicates that luxS has no significant effect on basic cellular metabolic processes required for growth of S. epidermidis in vitro. This is in contrast to some other bacteria in which luxS had an effect on growth (23, 29).
Our data showed that biofilm formation, the main virulence mechanism of S. epidermidis, was considerably enhanced in the ΔluxS mutant strain. This repressive effect on biofilm formation by luxS in vitro was also observed in S. mutans (35) and H. pylori (9), while the ΔluxS mutant of Salmonella enterica serovar Typhimurium was unable to develop a complete biofilm (42). Thus, luxS has contrasting effects on biofilm formation in different strains. A repressive effect on biofilm formation in Staphylococcus has also been described for the only other known QS system of Staphylococcus, agr, suggesting a common scheme of QS-dependent repression of the biofilm mode of growth in this genus. In contrast to agr, whose impact on biofilm formation is most likely via regulation of the production of the autolysin AtlE and the detergent-like phenol-soluble modulin peptides and which does not impact PIA production, luxS of S. epidermidis regulates transcription of the ica genes and production of PIA. This impact on the major biofilm exopolysaccharide PIA is most likely the cause for the different degree of biofilm formation of the ΔluxS mutant.
Decreased virulence has been seen in ΔluxS mutants of several pathogenic bacteria. A Serratia marcescens 274 ΔluxS mutant exhibits modestly attenuated virulence in a Caenorhabditis elegans model (10), a Vibrio vulnificus ΔluxS mutant has a significantly delayed time required to kill mice (25), and a Neisseria meningitidis ΔluxS mutant is attenuated for bacteremic infection in an infant rat model (55). In contrast, the S. epidermidis ΔluxS mutant, similar to the agr mutant of S. epidermidis, shows increased virulence in a model of catheter-associated infection. Most likely, the increased virulence may be partly attributed to the increased synthesis of PIA and more-intense biofilm formation.
Absent in humans, the LuxS enzyme is an attractive target for the development of novel therapeutic agents for bacterial infection. Inhibitors of LuxS have been synthesized (1). However, our present data, showing the increased capacity of biofilm formation of the ΔluxS mutant of S. epidermidis in vitro and increased virulence of the mutant in a rat model, suggest that such inhibitors are not suited for the treatment of biofilm-associated S. epidermidis infection.
In conclusion, we show that luxS in S. epidermidis is functional and luxS-dependent gene regulation represses biofilm formation in vitro and pathogen success during biofilm-associated infection. Although by regulating different biofilm factors, the two QS systems of Staphylococcus, agr and luxS, thus have very similar effects on the biofilm mode of growth.
Acknowledgments
We thank Bonnie Bassler for the gift of V. harveyi strain BB170, Friedrich Gotz for providing S. aureus strain RN4220, and Reinhold Bruckner for kindly providing plasmid pBT1. We thank Steven D. Goodman for modification of the manuscript.
This work was supported by the Chinese National Natural Science Foundation Grant (30570087) and 211 Project Grant—Functional Genomics of Important Pathogenic Microorganisms. This work was also supported by the China National 973 Program (2002CB512804).
Editor: F. C. Fang
REFERENCES
- 1.Alfaro, J. F., T. Zhang, D. P. Wynn, E. L. Karschner, and Z. S. Zhou. 2004. Synthesis of LuxS inhibitors targeting bacterial cell-cell communication. Org. Lett. 6:3043-3046. [DOI] [PubMed] [Google Scholar]
- 2.Augustin, J., and F. Gotz. 1990. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol. Lett. 54:203-207. [DOI] [PubMed] [Google Scholar]
- 3.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179:4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 5.Blevins, J. S., A. T. Revel, M. J. Caimano, X. F. Yang, J. A. Richardson, K. E. Hagman, and M. V. Norgard. 2004. The luxS gene is not required for Borrelia burgdorferi tick colonization, transmission to a mammalian host, or induction of disease. Infect. Immun. 72:4864-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruckner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 7.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 8.Christensen, G. D., W. A. Simpson, J. J. Younger, L. M. Baddour, F. F. Barrett, D. M. Melton, and E. H. Beachey. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22:996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole, S. P., J. Harwood, R. Lee, R. She, and D. G. Guiney. 2004. Characterization of monospecies biofilm formation by Helicobacter pylori. J. Bacteriol. 186:3124-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coulthurst, S. J., C. L. Kurz, and G. P. Salmond. 2004. luxS mutants of Serratia defective in autoinducer-2-dependent ‘quorum sensing’ show strain-dependent impacts on virulence and production of carbapenem and prodigiosin. Microbiology 150:1901-1910. [DOI] [PubMed] [Google Scholar]
- 11.Dobinsky, S., K. Kiel, H. Rohde, K. Bartscht, J. K. Knobloch, M. A. Horstkotte, and D. Mack. 2003. Glucose-related dissociation between icaADBC transcription and biofilm expression by Staphylococcus epidermidis: evidence for an additional factor required for polysaccharide intercellular adhesin synthesis. J. Bacteriol. 185:2879-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duan, K., C. Dammel, J. Stein, H. Rabin, and M. G. Surette. 2003. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 50:1477-1491. [DOI] [PubMed] [Google Scholar]
- 13.Flamm, R. K., D. J. Hinrichs, and M. F. Thomashow. 1984. Introduction of pAMβ1 into Listeria monocytogenes by conjugation and homology between native L. monocytogenes plasmids. Infect. Immun. 44:157-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fong, K. P., W. O. Chung, R. J. Lamont, and D. R. Demuth. 2001. Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect. Immun. 69:7625-7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fong, K. P., L. Gao, and D. R. Demuth. 2003. luxS and arcB control aerobic growth of Actinobacillus actinomycetemcomitans under iron limitation. Infect. Immun. 71:298-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerke, C., A. Kraft, R. Sussmuth, O. Schweitzer, and F. Gotz. 1998. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 273:18586-18593. [DOI] [PubMed] [Google Scholar]
- 18.Gotz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 20.Heilmann, C., M. Hussain, G. Peters, and F. Gotz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013-1024. [DOI] [PubMed] [Google Scholar]
- 21.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Gotz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 22.Henke, J. M., and B. L. Bassler. 2004. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J. Bacteriol. 186:6902-6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, M. B., and M. J. Blaser. 2003. Detection of a luxS-signaling molecule in Bacillus anthracis. Infect. Immun. 71:3914-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joyce, E. A., A. Kawale, S. Censini, C. C. Kim, A. Covacci, and S. Falkow. 2004. LuxS is required for persistent pneumococcal carriage and expression of virulence and biosynthesis genes. Infect. Immun. 72:2964-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, S. Y., S. E. Lee, Y. R. Kim, C. M. Kim, P. Y. Ryu, H. E. Choy, S. S. Chung, and J. H. Rhee. 2003. Regulation of Vibrio vulnificus virulence by the LuxS quorum-sensing system. Mol. Microbiol. 48:1647-1664. [DOI] [PubMed] [Google Scholar]
- 26.Kozitskaya, S., S. H. Cho, K. Dietrich, R. Marre, K. Naber, and W. Ziebuhr. 2004. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides. Infect. Immun. 72:1210-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loh, J. T., M. H. Forsyth, and T. L. Cover. 2004. Growth phase regulation of flaA expression in Helicobacter pylori is luxS dependent. Infect. Immun. 72:5506-5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lupp, C., and E. G. Ruby. 2004. Vibrio fischeri LuxS and AinS: comparative study of two signal synthases. J. Bacteriol. 186:3873-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyon, W. R., J. C. Madden, J. C. Levin, J. L. Stein, and M. G. Caparon. 2001. Mutation of luxS affects growth and virulence factor expression in Streptococcus pyogenes. Mol. Microbiol. 42:145-157. [DOI] [PubMed] [Google Scholar]
- 30.Mack, D., W. Fischer, A. Krokotsch, K. Leopold, R. Hartmann, H. Egge, and R. Laufs. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear β-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mack, D., M. Haeder, N. Siemssen, and R. Laufs. 1996. Association of biofilm production of coagulase-negative staphylococci with expression of a specific polysaccharide intercellular adhesin. J. Infect. Dis. 174:881-884. [DOI] [PubMed] [Google Scholar]
- 32.Mack, D., H. Rohde, S. Dobinsky, J. Riedewald, M. Nedelmann, J. K. Knobloch, H. A. Elsner, and H. H. Feucht. 2000. Identification of three essential regulatory gene loci governing expression of Staphylococcus epidermidis polysaccharide intercellular adhesin and biofilm formation. Infect. Immun. 68:3799-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mack, D., N. Siemssen, and R. Laufs. 1992. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 60:2048-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McNab, R., S. K. Ford, A. El-Sabaeny, B. Barbieri, G. S. Cook, and R. J. Lamont. 2003. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J. Bacteriol. 185:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, and W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 37.Miller, S. T., K. B. Xavier, S. R. Campagna, M. E. Taga, M. F. Semmelhack, B. L. Bassler, and F. M. Hughson. 2004. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell 15:677-687. [DOI] [PubMed] [Google Scholar]
- 38.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 39.Ohtani, K., H. Hayashi, and T. Shimizu. 2002. The luxS gene is involved in cell-cell signalling for toxin production in Clostridium perfringens. Mol. Microbiol. 44:171-179. [DOI] [PubMed] [Google Scholar]
- 40.Otto, M., R. Sussmuth, G. Jung, and F. Gotz. 1998. Structure of the pheromone peptide of the Staphylococcus epidermidis agr system. FEBS Lett. 424:89-94. [DOI] [PubMed] [Google Scholar]
- 41.Pei, L., and J. I. Flock. 2001. Functional study of antibodies against a fibrogenin-binding protein in Staphylococcus epidermidis adherence to polyethylene catheters. J. Infect. Dis. 184:52-55. [DOI] [PubMed] [Google Scholar]
- 42.Prouty, A. M., W. H. Schwesinger, and J. S. Gunn. 2002. Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect. Immun. 70:2640-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rupp, M. E., J. S. Ulphani, P. D. Fey, and D. Mack. 1999. Characterization of Staphylococcus epidermidis polysaccharide intercellular adhesin/hemagglutinin in the pathogenesis of intravascular catheter-associated infection in a rat model. Infect. Immun. 67:2656-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider, K. B., T. M. Palmer, and A. D. Grossman. 2002. Characterization of comQ and comX, two genes required for production of ComX pheromone in Bacillus subtilis. J. Bacteriol. 184:410-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stroeher, U. H., A. W. Paton, A. D. Ogunniyi, and J. C. Paton. 2003. Mutation of luxS of Streptococcus pneumoniae affects virulence in a mouse model. Infect. Immun. 71:3206-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA. 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vuong, C., C. Gerke, G. A. Somerville, E. R. Fischer, and M. Otto. 2003. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J. Infect. Dis. 188:706-718. [DOI] [PubMed] [Google Scholar]
- 50.Vuong, C., S. Kocianova, Y. Yao, A. B. Carmody, and M. Otto. 2004. Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J. Infect. Dis. 190:1498-1505. [DOI] [PubMed] [Google Scholar]
- 51.Vuong, C., and M. Otto. 2002. Staphylococcus epidermidis infections. Microbes Infect. 4:481-489. [DOI] [PubMed] [Google Scholar]
- 52.Vuong, C., H. L. Saenz, F. Gotz, and M. Otto. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182:1688-1693. [DOI] [PubMed] [Google Scholar]
- 53.Winans, S. C., and B. L. Bassler. 2002. Mob psychology. J. Bacteriol. 184:873-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winzer, K., K. R. Hardie, N. Burgess, N. Doherty, D. Kirke, M. T. Holden, R. Linforth, K. A. Cornell, A. J. Taylor, P. J. Hill, and P. Williams. 2002. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148:909-922. [DOI] [PubMed] [Google Scholar]
- 55.Winzer, K., Y. H. Sun, A. Green, M. Delory, D. Blackley, K. R. Hardie, T. J. Baldwin, and C. M. Tang. 2002. Role of Neisseria meningitidis luxS in cell-to-cell signaling and bacteremic infection. Infect. Immun. 70:2245-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6:191-197. [DOI] [PubMed] [Google Scholar]
- 57.Yao, Y., D. E. Sturdevant, and M. Otto. 2005. Genomewide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J. Infect. Dis. 191:289-298. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, Y. Q., S. X. Ren, H. L. Li, Y. X. Wang, G. Fu, J. Yang, Z. Q. Qin, Y. G. Miao, W. Y. Wang, R. S. Chen, Y. Shen, Z. Chen, Z. H. Yuan, G. P. Zhao, D. Qu, A. Danchin, and Y. M. Wen. 2003. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 49:1577-1593. [DOI] [PubMed] [Google Scholar]