Abstract
Enteropathogenic Escherichia coli (EPEC) is an important cause of infant diarrhea in developing countries and is useful for general investigations of the bacterial infection process. However, the study of the molecular pathogenesis of EPEC has been hampered by the lack of genetically tractable, convenient animal models. We have therefore developed the use of the nematode Caenorhabditis elegans as a small animal model of infection for this diarrheal pathogen. We found that nematodes died faster on nematode growth medium in the presence of EPEC pathogens than in the presence of the laboratory control strain MG1655. Increased numbers of pathogens in the gut, determined by standard plate count assays and fluorescence microscopy using green fluorescent protein-expressing bacteria, correlated with killing. Deletion of the gene encoding the global regulator Ler severely reduced the ability of EPEC to colonize the nematode gut and could be complemented by providing the ler gene on a multicopy plasmid in trans. Neither the type III secretion system nor the type IV bundle-forming pilus was required for colonization. Combined, the similarities and distinct differences between EPEC infection of nematodes and that of humans offer a unique opportunity to study several stages of the infection process, namely, attachment, colonization, and persistence, in a genetically tractable, inexpensive, and convenient in vivo system.
Our laboratory investigates the molecular pathogenesis of enteropathogenic Escherichia coli (EPEC), a leading cause of infant diarrhea in developing countries. Like the case for other bacterial pathogens, disease caused by EPEC is multifactorial (41). EPEC causes attaching and effacing (AE) intestinal lesions, which are characterized by disruption of the epithelial cytoskeleton and the formation of pedestals that protrude out from the host cell surface to cup the bacterium (40). The formation of AE lesions is mediated by a type III secretion system (TTSS) (37), a syringe-like structure that connects the host cell cytoplasm to the bacterial cytoplasm. The EPEC TTSS is encoded by the locus of enterocyte effacement (LEE) pathogenicity island, and expression of the TTSS is controlled by the LEE-encoded regulator (Ler) (8, 15, 38, 47, 51, 57). Proteins injected into the host cell by the TTSS, including Tir, EspF, EspG, EspH, Map, and NleA, subvert host cell signaling events, leading to diarrhea (for recent reviews, see references 9, 26, and 31). Tight attachment occurs when Tir, localized to the host cell membrane, is bound to the EPEC outer membrane protein intimin (3, 32, 35).
Early events in the infection process, namely, attachment and colonization, are less understood, and the bacterial adhesin responsible for initial attachment to the host epithelium has not been clearly established (43). The type IV, bundle-forming pilus (BFP) was hypothesized to be the initial adhesin when it was first described (22). However, AE lesions can form in the absence of the BFP (28), leading investigators to posit that the BFP is not the initial adhesin. The BFP mediates microcolony formation on host epithelial cells and is encoded on the E. coli attachment factor (EAF) plasmid along with the PerABC regulatory proteins (25, 50, 55). Other candidates possibly involved in initial attachment include the EspA filament of the TTSS (10), the intimin protein, which has also been shown to bind to β1 integrins (17), flagella (23), and the large, 385-kDa protein LifA, and lymphostatin, which has been associated with adhesion by enterohemorrhagic E. coli strain O157:H7 (42).
In order to extend our knowledge of bacterial pathogenesis, understand its effects on host cells, study colonization and innate immunity processes, and formulate effective vaccine and chemical therapies, the scientific community must have animal models of infection. Animal models are currently available for a number of bacterial pathogens, such as the infant mouse model for Salmonella enterica or Vibrio cholerae. The mouse model of infection has also been used for the LEE-containing AE pathogen Citrobacter rodentium (36). C. rodentium does not cause diarrhea in mice, but rather a disease known as transmissible murine colonic hyperplasia, inducing colitis and crypt cell proliferation. All models can be problematic because they often do not truly represent disease in humans. It is the use of a diverse group of animal models that offers the best hope of fully understanding bacterial molecular pathogenesis, and thus there is a great need for the development of simple animal models of infection for bacterial pathogens (16).
Recent reports suggested that the soil nematode Caenorhabditis elegans can be used as an infection model for diverse groups of bacterial pathogens, including those of both animals and plants (11, 53). Bacterial pathogens kill nematodes by at least two distinct mechanisms, a slowly killing (slow-kill) phenotype associated with infection and a fast-kill phenotype associated with diffusible toxins (2). The ability to use C. elegans as a small animal model for EPEC infection not only would significantly contribute to our understanding of EPEC pathogenesis but would also enhance our understanding of the mechanisms of infection of many related gram-negative pathogens. C. elegans is a well-studied organism which is used as a model for genetics, development, and cell biology in higher organisms, and the genome of the nematode has been sequenced, thus making it an attractive, convenient small animal with which to model EPEC infection.
MATERIALS AND METHODS
Bacterial and nematode strains and growth media.
The bacterial and nematode strains used for this study are listed in Table 1. The clinical isolates TB135A and TB156A were obtained from children with diarrhea living in the Seattle area (6); TB135A (Aps) and TB156A (Apr) have the genotype eae+ stx EAF− and thus are atypical EPEC because they lack the EAF plasmid, are both positive for localized adherence and fatty acid synthase, and caused chronic diarrhea. Spontaneous rifampin-resistant mutants of the above strains were isolated to limit contamination and prevent growth of the feeding strain OP50 in colonization assays. We obtained C. elegans strain DH26 fer-15(b26)II, which is sterile at 25°C, from the Caenorhabditis Genetic Center to ensure a constant number of nematodes during the assays due to their inability to reproduce when incubated at 26°C. Nematodes were propagated on pregrown lawns of the E. coli food strain OP50 at 15°C prior to synchronization for the assays described below.
TABLE 1.
Bacterial and nematode strains and plasmids used for this study
| Strain or plasmid | Genotype or description | Reference |
|---|---|---|
| E. coli strains | ||
| MG1655 | F− λ− | 5 |
| OP50 | Uracil auxotrophy | 7 |
| E2348/69 | Prototypical EPEC serotype O127:H6 | 34 |
| JPN15 | EAF plasmid-cured E2348/69 | 30 |
| CVD452 | E2348/69 ΔescN::aphA3 Kmr | 29 |
| SE796 | E2348/69 Δler::aphA3 Kmr | 15 |
| B171-8 | Wild-type EPEC serotype O111:NM, contains EAF plasmid | 22 |
| B171-8ΔA | B171-8 deleted for bfpA Cmr | 4 |
| TB135A | Atypical EPEC serotype O128:H2, eae+ EAF− | 6 |
| TB156A | Atypical EPEC serotype O55:H7, eae+ EAF− | 6 |
| C. elegans strain | ||
| DH26 | fer-15(b26)II; sterile at 25°C | 46 |
| Plasmids | ||
| pGEN91 | ori15A gfpuv bla Apr | 19 |
| pKH91 | ori15A gfpuv bla Aprtet Tcr | This work |
Bacterial strains were grown and colonization assays were performed on nematode growth medium (NGM) agar (3 g NaCl, 2.5 g peptone, and 17 g agar to 1 liter in H2O; after autoclaving, add 1 ml 1 M CaCl2, 1 ml 1 M MgSO4, 1 ml 2-mg/ml uracil, 1 ml 5-mg/ml cholesterol in ethanol, and 25 ml 1 M KPO4) supplemented with the following antibiotics where appropriate: rifampin at 100 μg/ml, ampicillin at 100 μg/ml, and tetracycline at 15 μg/ml. The NGM agar was supplemented with uracil because E. coli strain OP50 is a uracil auxotroph.
Plasmids.
The plasmids used for this study are listed in Table 1. Plasmid pKH91 was constructed by excising the tetracycline resistance gene from plasmid pBR322, using EcoRI and AvaI, and overhangs were filled in with the Klenow fragment of DNA polymerase I to create blunt ends. The DNA fragment was then blunt end ligated into the pGEN91 plasmid, previously digested with EcoRI, and 5′ overhang ends were filled in using the Klenow fragment. The correct plasmid construct was confirmed by selecting for pKH91-transformed E. coli on tetracycline-containing LB agar and observing fluorescence.
Kill and standard plate count assays.
Prior to the colonization assays, nematodes were age synchronized by a bleaching procedure. Nematodes/embryos grown on E. coli strain OP50 at 15°C were harvested by washing the seeded NGM agar plate with M9 buffer (3 g KH2PO4, 6 g Na2HPO4, 5 g NaCl, 1 ml 1 M MgSO4 in 1 liter H2O), were placed into a microcentrifuge tube, and then were washed three times with 1 ml M9 buffer after spinning for 10 seconds at 12,000 rpm. Nematodes/embryos were resuspended in 100 μl M9 buffer and bleach treated by adding 350 μl 280 mM KOH and 50 μl bleach. Nematodes/embryos were agitated gently and mixed intermittently for 10 min. After a 10-second spin at 12,000 rpm, the supernatant was discarded, and embryos were washed twice more in 1 ml M9 buffer as described above. After a final spin, the embryos and dead nematodes were resuspended in 50 μl M9 buffer; the suspension was placed on NGM agar plates with the food strain OP50, without antibiotic selection, and incubated at 26°C.
After 3 to 4 days at 26°C, L4 nematodes were removed from feeding by use of a platinum wire and placed on rifampin-containing NGM agar plates with pregrown EPEC and control strains that were incubated at 37°C overnight. Prior to seeding of C. elegans, NGM agar plates were shifted to 26°C, the temperature where they remained for the duration of the assay. For the kill assay, 12 nematodes were fed on the test strains, and nematode viability was monitored over a 12-day period. Nematodes were considered dead when they did not respond to gentle prodding with a platinum wire. The times until 50% of animals were dead (LT50s) were subjected to statistical analyses using the paired t test in StatView, version 5.0.1 (SAS Institute, Inc., Cary, NC).
For the standard plate count assay, nematodes were fed on EPEC and control strains for 24 h at 26°C. Individual nematodes were then chilled in M9 buffer for 24 h at 4°C to loosen bacteria adherent to the cuticle, washed three times in M9 buffer, treated with 100 μg/ml gentamicin at 37°C for 1 hour to kill exterior bacteria, again washed three times with M9 buffer, treated with 50% chloroform-saturated M9 buffer for 10 minutes, washed three times in M9 buffer containing 1% saponin and 1% Triton X-100, pulverized for 1 minute using a sterile plastic pestle, and finally plated on LB agar containing rifampin. Extreme washing measures were taken because viable bacteria are difficult to remove from the nematode cuticle and the pharynx and because EPEC strain-to-strain susceptibilities to chemical agents can vary greatly (data not shown). The thorough treatment of infected nematodes ensured that only viable bacteria within the nematode gut were quantified in our assays. Plate count data were analyzed by negative binomial regression (see Results), using Stata, version 7.0 (Stata Corporation, College Station, TX).
Fluorescence microscopy.
Synchronized L4 nematodes were subjected to infection by EPEC and control strains containing the green fluorescent protein (GFP)-producing plasmid pGEN91 or pKH91 on NGM supplemented with ampicillin and rifampin or tetracycline and rifampin, respectively, at 26°C as described above. Twenty-four hours after infection, nematodes were removed using a platinum wire and placed in 500 μl of M9 buffer. For the persistence assay, nematodes were removed from GFP-labeled strains, washed in M9 buffer, and then placed on nonselective medium containing overnight cultures of the feeding strain OP50. Immediately prior to microscopy, 500 μl of a saturated solution of chloroform in M9 buffer was added to the nematodes, and they were incubated at room temperature for 10 min to kill and remove any bacteria adherent to the exterior. Nematodes were washed three times in 1 ml M9 buffer and then three times with 100 μl of M9 buffer. Nematodes were chilled, transferred to 1-day-old 1% agarose pads on glass microscope slides to control the rate of desiccation, and visualized using an Olympus BX60 microscope fitted with an Optronics Microfire digital camera (Optronics, Goleta, CA).
RESULTS
EPEC kills nematodes faster than laboratory control strains.
To initiate studies to determine whether C. elegans is an appropriate model system for EPEC infection, we performed simple kill assays on NGM agar at 26°C. Nematode death was monitored in the presence of EPEC and control bacteria over a period of ∼12 days. The mean LT50 for nematodes grown on the laboratory control strain MG1655 was 247 h, whereas the mean for strain E2348/69 was 161 h (Table 2). By a paired t test, the LT50 for nematodes in the presence of strain E2348/69 was significantly less than that for control strain MG1655 (P = 0.03). EPEC strains JPN15, CVD452, and B171-8 killed nematodes with mean LT50s of 150, 149, and 185 h, respectively, which were statistically significantly less than the mean LT50 for MG1655 (Table 2). (Strain JPN15 is an E2348/69 derivative lacking the BFP-encoding EAF plasmid; CVD45 is isogenic to E2348/69, with a deletion of the gene encoding the TTSS ATPase EscN; and strain B171-8 is a wild-type EPEC strain of serotype O111:NM containing the EAF plasmid.) The mean LT50 values for strains B171-8ΔA (deleted for the gene encoding the major subunit of the type IV pilus), TB135A, and TB156A (recent clinical isolates from infants living in the Seattle area) were 181, 161, and 157, respectively. These values were similar to the mean LT50 for E2348/69 but did not reach statistical significance compared to the LT50 for MG1655 (Table 2). Importantly, the LT50 values for all strains tested, except the control strain MG1655, were statistically the same as the LT50 value for the prototypical pathogen E2348/69 (Table 2). One strain included in this group was strain SE796, with a deletion of ler, which did not kill nematodes significantly slower than the prototypical E2348/69 strain (P = 0.3), even though we observed a defect in colonization for this strain (see below). We therefore concluded that EPEC strains, particularly E2348/69, consistently kill nematodes faster than the control strain MG1655, but the simple kill assay was not sensitive enough to investigate EPEC virulence in the C. elegans infection model.
TABLE 2.
Statistical analyses of C. elegans kill assaysa
| Strain | LT50b | P valuec (MG1655) | P valued (E2348/69) |
|---|---|---|---|
| MG1655 | 247 ± 16 | 0.03* | |
| E2348/69 | 161 ± 34 | 0.03* | |
| JPN15 | 150 ± 15 | 0.03* | 0.6 |
| CVD452 | 149 ± 26 | 0.006* | 0.7 |
| SE796 | 175 ± 40 | 0.06 | 0.3 |
| B171-8 | 185 ± 9 | 0.02* | 0.5 |
| B171-8ΔA | 181 ± 33 | 0.1 | 0.7 |
| TB135A | 161 ± 46 | 0.07 | 0.95 |
| TB156A | 157 ± 36 | 0.1 | 0.9 |
Nematodes were subjected to infection with EPEC and control bacteria on NGM agar at 26°C, and survival was monitored as described in Materials and Methods.
Mean LT50 values from at least three replicate assays using 12 nematodes each are presented in mean hours ± 1 SD.
Obtained by paired t test comparing LT50 values with those obtained for control strain MG1655. Asterisks indicate P values of <0.05.
Obtained by paired t test comparing LT50 values with those obtained for prototypical EPEC strain E2348/69. Asterisks indicate P values of <0.05.
EPEC bacteria displayed a slow-kill phenotype, similar to the slow-kill phenotype of Salmonella enterica serovars in C. elegans (1, 33), killing nematodes over several days as opposed to a few hours in the case of killing by diffusible toxins. Neither heat-killed EPEC bacteria nor EPEC bacteria grown on 0.2-μm filters and then removed prior to placing nematodes on the NGM agar plates killed nematodes faster than combining C. elegans with the control strain MG1655 (data not shown). Thus, we concluded that EPEC bacteria killed nematodes by infection as opposed to production of either a heat-labile or heat-stable diffusible toxin.
EPEC bacteria are found within the nematode gut in larger numbers than the laboratory control strain MG1655.
Because EPEC bacteria killed nematodes by infection faster than the control strain MG1655, we predicted that these pathogens would be found in greater numbers within the nematode gut than strain MG1655 cells. Age-synchronized L4 nematodes were placed on rifampin-containing NGM agar with pregrown lawns of rifampin-resistant bacteria as described for the kill assay.
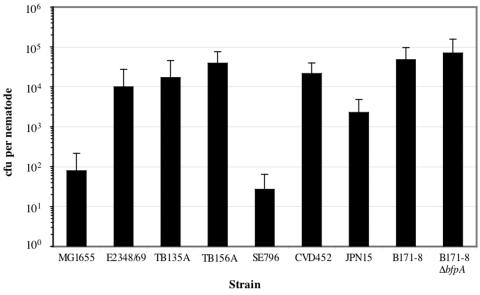
Twenty-four hours after initial infection, ∼1 × 104 CFU of strain E2348/69 were found within the nematode gut, whereas we observed ∼1 × 102 CFU per nematode of control strain MG1655, a difference of 2 orders of magnitude (Fig. 1). Because these data did not fit a Poisson model due to overdispersion, we analyzed CFU-per-nematode results by negative binomial regression (58). The CFU value per nematode for strain E2348/69 was greater than that observed for the control strain MG1655, with statistical significance (P < 0.001). Similarly, the CFU per nematode for the clinical isolates TB135A and TB156A were 2 × 104 and 4 × 104, respectively, and these values were statistically greater than those for strain MG1655 (P < 0.001).
FIG. 1.
EPEC pathogens colonize the nematode gut in greater numbers than the laboratory control strain. Bleach-synchronized L4 nematodes were subjected to infection by EPEC strains and the MG1655 control on NGM supplemented with rifampin. After 24 h of infection, three nematodes were harvested, washed to remove exterior bacteria (see Materials and Methods), pulverized, and plated on selective LB agar. Average CFU per nematode exposed to the indicated rifampin-resistant bacterial strains for 24 h are presented. Values represent the means of three replicate assays, and error bars indicate 1 standard deviation (SD).
The global regulator Ler is necessary for colonization of the nematode gut.
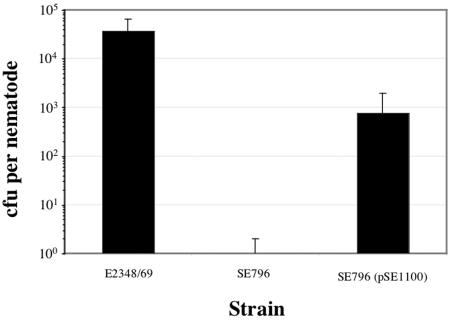
We determined whether virulence factors known to play a role in the infection of humans contributed to the ability of EPEC pathogens to colonize the nematode gut. To this end, we tested strain SE796, deleted for the global regulator ler, which activates expression of the LEE required for the AE phenotype (15), in a standard plate count assay. Regulatory proteins of both gram-positive and gram-negative bacteria appear to play important roles in C. elegans pathogenicity models (12, 20, 54). Deletion of the gene encoding Ler severely reduced the ability of EPEC to colonize, from ∼1 × 104 to 30 CFU per nematode (P < 0.001) (Fig. 1). Providing the ler gene on a multicopy plasmid restored the ability of EPEC to colonize, though not completely to wild-type levels (∼1 × 103 CFU per nematode) (Fig. 2).
FIG. 2.
Ler expressed from a multicopy plasmid complemented the noncolonizing Δler strain SE796. Nematodes were infected with the indicated bacterial strains as described in the legend to Fig. 1. Average CFU per nematode exposed to the indicated rifampin-resistant bacteria for 24 h are presented. Values represent the means of three replicate assays, and error bars indicate 1 SD.
We found that the TTSS was not necessary for colonization of the nematode gut because strain CVD452, deleted for the gene encoding EscN, the ATPase necessary for secretion in EPEC and related bacteria (14, 21, 62), colonized nematode guts in numbers similar to those for strain E2348/69 (∼2 × 104 CFU per nematode) (Fig. 1). We also determined whether BFP played a role in colonization. Strain JPN15, lacking the E. coli attachment factor (EAF) plasmid and thus the bfp operon, exhibited a partial defect in its ability to colonize the nematode gut, with a reduction of 5- to 20-fold compared to E2348/69 and the recent clinical isolates TB135A and TB156A, but the difference between CFU per nematode for strains JPN15 and E2348/69 did not reach statistical significance (P = 0.091) (Fig. 1). To ask specifically whether BFP played a role in colonization, we tested EPEC strain B171-8 and an isogenic derivative deleted for the gene encoding the type IV pilus subunit, bfpA (4), in our assay. We found that the deletion of bfpA did not affect the B171-8 strain's ability to colonize the nematode gut and that the CFU recovered were similar to those observed for E2348/69 (∼2 × 104 to 8 × 104 CFU per nematode) (Fig. 1). We thus concluded that the global regulator Ler is necessary for colonization of the nematode gut but that neither the TTSS nor the type IV BFP is necessary for this phenotype.
EPEC bacteria persist within the nematode gut.
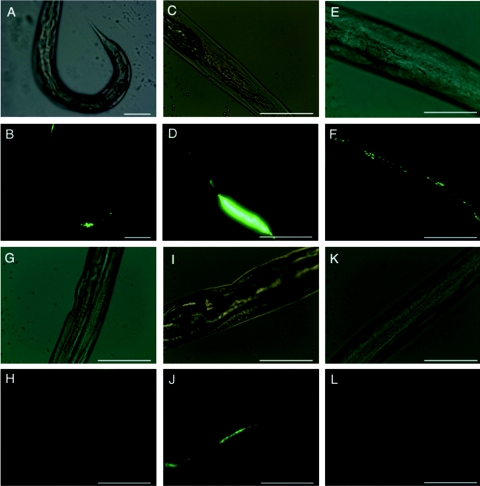
We transformed EPEC pathogens and control strains with GFP-encoding plasmids to monitor colonization by fluorescence microscopy. Consistent with the values obtained by standard plate count assays, we observed greater numbers of fluorescent bacteria in nematodes exposed to strain E2348/69 and the clinical isolate TB135A than in nematodes exposed to the control strain MG1655 (Fig. 3B, D, and L, respectively). We often observed colonization differentially near the anus (Fig. 3B and 4B, F, H, J, and L); similar observations have been made for uropathogenic strains of E. coli tested in the C. elegans infection model (Creg Darby, personal communication). Strain SE796, deleted for ler, showed reduced colonization compared to the parent strain E2348/69 by fluorescence microscopy (Fig. 3H), and introduction of a plasmid encoding a minimal DNA fragment expressing Ler, pSE1100, in trans restored visible GFP-expressing bacteria to levels comparable to those for strain E2348/69 within the nematode gut (Fig. 3J). Thus, the fluorescence microscopy images showing decreased colonization by the ler deletion strain SE796 and subsequent complementation were consistent with the results observed with standard plate count assays (Fig. 1 and 2).
FIG. 3.
Fluorescence microscopy images of EPEC pathogens within the nematode gut. After synchronization, L4 nematodes were subjected to infection by EPEC and control strains containing GFP-producing plasmids on NGM agar supplemented with appropriate antibiotics. After 24 h, bacterial strains E2348/69(pGEN91) (A, B), TB135A(pGEN91) (clinical isolate) (C, D), JPN15(pGEN91) (E, F), SE796(pGEN91) (G, H), SE796(pSE1100) (pKH91) (I, J), and MG1655(pGEN91) (K, L) were viewed by fluorescence and light microscopy. Light microscopy images (A, C, E, G, I, and K) appear adjacent to corresponding fluorescence microscopy images (B, D, F, H, J, and L). Representative images are presented. Magnification, ×200 (A, B) or ×400 (C to L). Bars, 100 μm.
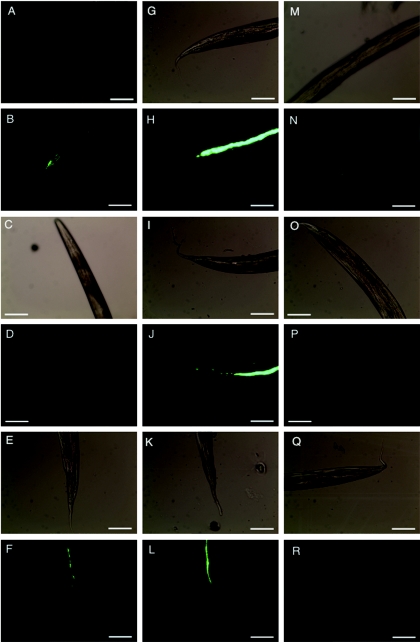
FIG.4.
EPEC persistence within the C. elegans gut. Synchronized L4 nematodes were subjected to infection with GFP-labeled, pGEN91-containing strain E2348/69 (A to F), TB135A (G to L), or MG1655 (M to R) on NGM agar supplemented with the appropriate antibiotics. At 24 hours postinfection, nematodes were rinsed gently with M9 buffer and then transferred to NGM agar containing lawns of the feeding strain OP50 without antibiotic selection. Nematodes were prepared for light and fluorescence microscopy as described in Materials and Methods 0 (A, B, G, H, M, N), 24 (C, D, I, J, O, P), and 48 (E, F, K, L, Q, R) hours after transfer to coculture with strain OP50. Light microscopy images (A, C, E, G, I, K, M O, and Q) appear adjacent to corresponding fluorescence microscopy images (B, D, F, H, J, L, N, P, and R). Representative images are presented. Magnification, ×200. Bars, 100 μm.
To test whether EPEC bacteria persist within the nematode gut, we transferred age-synchronized nematodes to separate agar plates containing strain E2348/69, clinical isolate TB135A, and strain MG1655 labeled with GFP. After 24 h of exposure, nematodes were gently washed with M9 buffer and transferred to agar plates containing the unlabeled food strain OP50. Fluorescence microscopy images were captured 0, 24, and 48 h after the shift to the unlabeled food strain and are presented in Fig. 4. Forty-eight hours after the shift, GFP-labeled E2348/69 and TB135A bacteria were visible within the nematode gut (Fig. 4F and L). At the 24-hour time point, we observed minimal colonization by strain E2348/68 (Fig. 4D), and not surprisingly, more robust colonization by the recent clinical isolate TB135A (Fig. 4J). The GFP-labeled control strain MG1655 was not visible within C. elegans at either the 24- or 48-hour time point (Fig. 4P and R). We therefore concluded that EPEC bacteria could persist within the nematode gut for at least 48 h after initial infection.
Microcolony formation.
EPEC strain E2348/69 formed microcolonies, mimicking the three-dimensional structures observed on human epithelial cells in culture (Fig. 3B and 4B) (48, 59). Clinical isolate TB135A formed large aggregates inside the nematode gut, but we also observed adherent microcolonies of this strain (Fig. 3D and 4J). Although TB135A is characterized as a bfp mutant, it does form microcolonies on cultured human epithelial cells (6). In contrast to strain E2348/69 and isolate TB135A, we observed more diffuse adherence throughout the gut by strain JPN15 lacking the BFP (Fig. 3F). Strains B171-8 and B171-8ΔA, deleted for bfpA, also tended to colonize throughout the nematode, but there were no apparent differences in CFU per nematode or colonization phenotype for these strains (Fig. 1 and data not shown). We hypothesized that like the case on human tissue, the type IV BFP of EPEC may be involved in microcolony formation in nematodes but that additional EPEC factors might also contribute to this phenotype, depending on the EPEC strain, host, and environmental conditions.
DISCUSSION
The goal of this study was to determine whether the nematode C. elegans could be used as a small animal model of EPEC infection, and we found several parallels between EPEC pathogenesis in humans and that in nematodes. Firstly, EPEC disease in humans is caused by colonization and by definition is not toxin mediated. EPEC killed nematodes by colonization over several days and not by a fast-kill phenotype generally associated with a diffusible toxin(s) (2). Secondly, we found that EPEC formed microcolonies in the gut of the nematode similar to those observed on human epithelial cells in culture.
Our most significant finding was that the global regulator Ler was necessary for colonization of the nematode gut. Deletion of the gene encoding Ler in EPEC causes a severe reduction in the bacterium's ability to cause AE lesions on cultured epithelial cells (15). A clinical isolate of the related pathogen E. coli serotype O157:H7 with a point mutation in the gene encoding Ler did not form AE lesions on HEp-2 cells in culture, signifying the importance of Ler in human disease for the related O157:H7 pathogen (44). Thus, a third parallel between EPEC disease in humans and colonization of the nematode gut is that both processes require the global regulator Ler.
Ler expressed from a plasmid only partially complemented the attenuated colonization of the Δler strain SE796 (Fig. 2). An explanation for this result might be that the ler gene carried on plasmid pSE1100 is controlled by its native promoter and is repressed at temperatures lower than 37°C (57). Because our colonization assays were performed at 26°C, it is likely that the lack of complete complementation was due to decreased Ler expression. PerC increases the expression of Ler (8, 38, 45), and thus we were surprised to find that the EAF plasmid-deficient, PerC-lacking strain JPN15 was not significantly deficient in the ability to colonize the nematode gut. It is likely that signals other than temperature and proteins other than PerC are important for EPEC virulence gene expression inside the nematode and within the human intestine. Quorum-sensing signals (51), integration host factor (18), Fis (24), BipA (27), and GadX (49) are known to control Ler expression in vivo either directly or indirectly, and a combination of environmental cues and regulatory proteins likely controls EPEC pathogenesis in the nematode.
Ler is deemed a global regulator because it controls the expression of the LEE-encoded TTSS (8, 18, 38), the enterotoxin EspC (15), located within a second pathogenicity island in EPEC (39, 52), and a non-BFP fimbrial adhesin (15). Although Ler controls the expression of the TTSS necessary for AE lesion formation (15) and thus disease in humans, EPEC strain CVD452, which is deficient in TTSS formation, colonized nematodes in numbers similar to those of the parent strain E2348/69 (Fig. 1). By immunoblot analysis, we found that the EspA protein, which forms the molecular syringe of the TTSS, was expressed inside the guts of nematodes (data not shown). The lack of a role of the TTSS in colonization of the nematode gut therefore could not be explained by a lack of EspA expression. These results were consistent with recent studies using C. elegans to model Pseudomonas aeruginosa pathogenesis, where the TTSS is expressed in the nematode intestine but is not necessary for colonization or virulence in the nematode model (60). Although the LEE-encoded intimin protein and EAF-located BFP have been established as factors necessary for full virulence in humans (4, 13), additional uncharacterized virulence factors important for EPEC pathogenesis undoubtedly exist.
Like the case for EPEC disease in humans, colonization of the nematode gut is most likely multifactorial, and we hypothesize that a Ler-regulated factor or set of factors is responsible for this phenotype. We tested the mutant strain CVD206, which is defective in the production of intimin, also regulated by Ler (38, 47), in the colonization assay and found a modest decrease in colonization, of approximately threefold, compared to the parent strain E2348/69 (data not shown). Interestingly, intimin does not require the TTSS to be localized to the outer membrane (61). C. elegans shares many protein homologues with humans, including β1 integrins, to which intimin is known to bind, and thus it is possible that intimin binds to β1 integrins within the nematode gut, but this protein cannot be solely responsible for the phenotype observed when ler is deleted, i.e., a 100-fold reduction in CFU per nematode. The EPEC genome sequence was recently completed (http://www.sanger.ac.uk/Projects/Escherichia_Shigella/), elucidating at least 9 regions with similarity to potential fimbrial adhesins and 10 regions encoding putative nonfimbrial adhesins (56). A combination of the intimin protein, fimbriae, EspA, LifA, flagella, and/or other Ler-regulated factors could contribute to colonization of the C. elegans gut and the human intestine.
Here we describe an EPEC infection model using the nematode C. elegans. In a recent report, Anyanful et al. (2a) described paralysis and killing of C. elegans by EPEC grown on Fluorocult ECD agar, whereby killing occurs not by direct contact with the nematodes but rather by a diffusible toxin. In contrast, killing of nematodes in our NGM agar-based system occurs from infection, not from a diffusible toxin, more closely mimicking EPEC disease in humans. Because there is a need to understand more completely the initial events involved in EPEC infection (31), we envision this simple, convenient in vivo system to be useful for studying previously uncharacterized EPEC attachment factors, colonization, and persistence.
Acknowledgments
We thank Iruka Okeke for critically reading the manuscript, Jim Galen for generously providing the plasmid pGEN91, Jim Kaper, Phil Tarr, and Alexy Merz for bacterial strains, and the Caenorhabditis Genetics Center for providing the nematode strain DH26. We also thank Greg Hermann for his expertise on manipulating C. elegans strains. We acknowledge Steve Black for lending time and technical expertise on fluorescence microscopy, Albyn Jones and Marc Schneiberg for their expertise in statistical analyses, and Derek Galligan and Lindsay Swanson for completing informative preliminary experiments.
This work was supported by DARPA grant DAAD19-03-1-0055 awarded to J.L.M. and was funded in part by a Hughes Undergraduate Research Program award to A.M.S.B., resulting from an HHMI grant awarded to the Biology Department of Reed College, and a Corporate Activities student travel grant awarded to A.M.S.B. for attending the ASM National Meeting, 5 to 9 June 2005, Atlanta, Ga.
Editor: J. B. Bliska
REFERENCES
- 1.Aballay, A., P. Yorgey, and F. M. Ausubel. 2000. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr. Biol. 10:1539-1542. [DOI] [PubMed] [Google Scholar]
- 2.Alegado, R. A., M. C. Campbell, W. C. Chen, S. S. Slutz, and M. W. Tan. 2003. Characterization of mediators of microbial virulence and innate immunity using the Caenorhabditis elegans host-pathogen model. Cell Microbiol. 5:435-444. [DOI] [PubMed] [Google Scholar]
- 2a.Anyanful, A., J. M. Dolan-Livengood, T. Lewis, S. Sheth, M. N. Dezalia, M. A. Sherman, L. V. Kalman, G. M. Benian, and D. Kalman. 2005. Paralysis and killing of Caenorhabditis elegans by enteropathogenic Escherichia coli requires the bacterial tryptophanase gene. Mol. Microbiol. 57:988-1007. [DOI] [PubMed] [Google Scholar]
- 3.Batchelor, M., S. Prasannan, S. Daniell, S. Reece, I. Connerton, G. Bloomberg, G. Dougan, G. Frankel, and S. Matthews. 2000. Structural basis for recognition of the translocated intimin receptor (Tir) by intimin from enteropathogenic Escherichia coli. EMBO J. 19:2452-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieber, D., S. W. Ramer, C. Y. Wu, W. J. Murray, T. Tobe, R. Fernandez, and G. K. Schoolnik. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114-2118. [DOI] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Bokete, T. N., T. S. Whittam, R. A. Wilson, C. R. Clausen, C. M. O'Callahan, S. L. Moseley, T. R. Fritsche, and P. I. Tarr. 1997. Genetic and phenotypic analysis of Escherichia coli with enteropathogenic characteristics isolated from Seattle children. J. Infect. Dis. 175:1382-1389. [DOI] [PubMed] [Google Scholar]
- 7.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bustamante, V. H., F. J. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39:664-678. [DOI] [PubMed] [Google Scholar]
- 9.Clarke, S. C., R. D. Haigh, P. P. Freestone, and P. H. Williams. 2003. Virulence of enteropathogenic Escherichia coli, a global pathogen. Clin. Microbiol. Rev. 16:365-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cleary, J., L. C. Lai, R. K. Shaw, A. Straatman-Iwanowska, M. S. Donnenberg, G. Frankel, and S. Knutton. 2004. Enteropathogenic Escherichia coli (EPEC) adhesion to intestinal epithelial cells: role of bundle-forming pili (BFP), EspA filaments and intimin. Microbiology 150:527-538. [DOI] [PubMed] [Google Scholar]
- 11.Couillault, C., and J. J. Ewbank. 2002. Diverse bacteria are pathogens of Caenorhabditis elegans. Infect. Immun. 70:4705-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darby, C., J. W. Hsu, N. Ghori, and S. Falkow. 2002. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417:243-244. [DOI] [PubMed] [Google Scholar]
- 13.Donnenberg, M. S., C. O. Tacket, S. P. James, G. Losonsky, J. P. Nataro, S. S. Wasserman, J. B. Kaper, and M. M. Levine. 1993. Role of the eaeA gene in experimental enteropathogenic Escherichia coli infection. J. Clin. Investig. 92:1412-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichelberg, K., C. C. Ginocchio, and J. E. Galan. 1994. Molecular and functional characterization of the Salmonella typhimurium invasion genes invB and invC: homology of InvC to the F0F1 ATPase family of proteins. J. Bacteriol. 176:4501-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott, S. J., V. Sperandio, J. A. Girón, S. Shin, J. L. Mellies, L. Wainwright, S. W. Hutcheson, T. K. McDaniel, and J. B. Kaper. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finlay, B. B. 1999. Bacterial disease in diverse hosts. Cell 96:315-318. [DOI] [PubMed] [Google Scholar]
- 17.Frankel, G., O. Lider, R. Hershkoviz, A. P. Mould, S. G. Kachalsky, D. C. Candy, L. Cahalon, M. J. Humphries, and G. Dougan. 1996. The cell-binding domain of intimin from enteropathogenic Escherichia coli binds to beta1 integrins. J. Biol. Chem. 271:20359-20364. [DOI] [PubMed] [Google Scholar]
- 18.Friedberg, D., T. Umanski, Y. Fang, and I. Rosenshine. 1999. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol. Microbiol. 34:941-952. [DOI] [PubMed] [Google Scholar]
- 19.Galen, J. E., J. Nair, J. Y. Wang, S. S. Wasserman, M. K. Tanner, M. B. Sztein, and M. M. Levine. 1999. Optimization of plasmid maintenance in the attenuated live vector vaccine strain Salmonella typhi CVD 908-htrA. Infect. Immun. 67:6424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garsin, D. A., C. D. Sifri, E. Mylonakis, X. Qin, K. V. Singh, B. E. Murray, S. B. Calderwood, and F. M. Ausubel. 2001. A simple model host for identifying gram-positive virulence factors. Proc. Natl. Acad. Sci. USA 98:10892-10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gauthier, A., J. L. Puente, and B. B. Finlay. 2003. Secretin of the enteropathogenic Escherichia coli type III secretion system requires components of the type III apparatus for assembly and localization. Infect. Immun. 71:3310-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Girón, J. A., A. S. Ho, and G. K. Schoolnik. 1991. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254:710-713. [DOI] [PubMed] [Google Scholar]
- 23.Girón, J. A., A. G. Torres, E. Freer, and J. B. Kaper. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44:361-379. [DOI] [PubMed] [Google Scholar]
- 24.Goldberg, M. D., M. Johnson, J. C. Hinton, and P. H. Williams. 2001. Role of the nucleoid-associated protein Fis in the regulation of virulence properties of enteropathogenic Escherichia coli. Mol. Microbiol. 41:549-559. [DOI] [PubMed] [Google Scholar]
- 25.Gómez-Duarte, O. G., and J. B. Kaper. 1995. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect. Immun. 63:1767-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goosney, D. L., R. DeVinney, R. A. Pfuetzner, E. A. Frey, N. C. Strynadka, and B. B. Finlay. 2000. Enteropathogenic E. coli translocated intimin receptor, Tir, interacts directly with alpha-actinin. Curr. Biol. 10:735-738. [DOI] [PubMed] [Google Scholar]
- 27.Grant, A. J., M. Farris, P. Alefounder, P. H. Williams, M. J. Woodward, and C. D. O'Connor. 2003. Co-ordination of pathogenicity island expression by the BipA GTPase in enteropathogenic Escherichia coli (EPEC). Mol. Microbiol. 48:507-521. [DOI] [PubMed] [Google Scholar]
- 28.Hicks, S., G. Frankel, J. B. Kaper, G. Dougan, and A. D. Phillips. 1998. Role of intimin and bundle-forming pili in enteropathogenic Escherichia coli adhesion to pediatric intestinal tissue in vitro. Infect. Immun. 66:1570-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jarvis, K. G., J. A. Girón, A. E. Jerse, T. K. McDaniel, M. S. Donnenberg, and J. B. Kaper. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. USA 92:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 32.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 33.Labrousse, A., S. Chauvet, C. Couillault, C. L. Kurz, and J. J. Ewbank. 2000. Caenorhabditis elegans is a model host for Salmonella typhimurium. Curr. Biol. 10:1543-1545. [DOI] [PubMed] [Google Scholar]
- 34.Levine, M. M. 1987. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic, and enteroadherent. J. Infect. Dis. 155:377-389. [DOI] [PubMed] [Google Scholar]
- 35.Luo, Y., E. A. Frey, R. A. Pfuetzner, A. L. Creagh, D. G. Knoechel, C. A. Haynes, B. B. Finlay, and N. C. Strynadka. 2000. Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex. Nature 405:1073-1077. [DOI] [PubMed] [Google Scholar]
- 36.Luperchio, S. A., J. V. Newman, C. A. Dangler, M. D. Schrenzel, D. J. Brenner, A. G. Steigerwalt, and D. B. Schauer. 2000. Citrobacter rodentium, the causative agent of transmissible murine colonic hyperplasia, exhibits clonality: synonymy of C. rodentium and mouse-pathogenic Escherichia coli. J. Clin. Microbiol. 38:4343-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23:399-407. [DOI] [PubMed] [Google Scholar]
- 38.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 39.Mellies, J. L., F. Navarro-Garcia, I. Okeke, J. Frederickson, J. P. Nataro, and J. B. Kaper. 2001. espC pathogenicity island of enteropathogenic Escherichia coli encodes an enterotoxin. Infect. Immun. 69:315-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moon, H. W., S. C. Whipp, R. A. Argenzio, M. M. Levine, and R. A. Giannella. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicholls, L., T. H. Grant, and R. M. Robins-Browne. 2000. Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol. Microbiol. 35:275-288. [DOI] [PubMed] [Google Scholar]
- 43.Nougayrede, J. P., P. J. Fernandes, and M. S. Donnenberg. 2003. Adhesion of enteropathogenic Escherichia coli to host cells. Cell. Microbiol. 5:359-372. [DOI] [PubMed] [Google Scholar]
- 44.Ogierman, M. A., A. W. Paton, and J. C. Paton. 2000. Up-regulation of both intimin and eae-independent adherence of Shiga toxigenic Escherichia coli O157 by ler and phenotypic impact of a naturally occurring ler mutation. Infect. Immun. 68:5344-5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Porter, M. E., P. Mitchell, A. Free, D. G. Smith, and D. L. Gally. 2005. The LEE1 promoters from both enteropathogenic and enterohemorrhagic Escherichia coli can be activated by PerC-like proteins from either organism. J. Bacteriol. 187:458-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts, T. M., and S. Ward. 1982. Membrane flow during nematode spermiogenesis. J. Cell Biol. 92:113-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanchez-SanMartin, C., V. H. Bustamante, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of the orf19 gene and the tir-cesT-eae operon of enteropathogenic Escherichia coli. J. Bacteriol. 183:2823-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scaletsky, I. C., M. L. Silva, and L. R. Trabulsi. 1984. Distinctive patterns of adherence of enteropathogenic Escherichia coli to HeLa cells. Infect. Immun. 45:534-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shin, S., M. P. Castanie-Cornet, J. W. Foster, J. A. Crawford, C. Brinkley, and J. B. Kaper. 2001. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol. Microbiol. 41:1133-1150. [DOI] [PubMed] [Google Scholar]
- 50.Sohel, I., J. L. Puente, S. W. Ramer, D. Bieber, C. Y. Wu, and G. K. Schoolnik. 1996. Enteropathogenic Escherichia coli: identification of a gene cluster coding for bundle-forming pilus morphogenesis. J. Bacteriol. 178:2613-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stein, M., B. Kenny, M. A. Stein, and B. B. Finlay. 1996. Characterization of EspC, a 110-kilodalton protein secreted by enteropathogenic Escherichia coli which is homologous to members of the immunoglobulin A protease-like family of secreted proteins. J. Bacteriol. 178:6546-6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan, M. W., S. Mahajan-Miklos, and F. M. Ausubel. 1999. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. USA 96:715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan, M. W., L. G. Rahme, J. A. Sternberg, R. G. Tompkins, and F. M. Ausubel. 1999. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 96:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tobe, T., G. K. Schoolnik, I. Sohel, V. H. Bustamante, and J. L. Puente. 1996. Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 21:963-975. [DOI] [PubMed] [Google Scholar]
- 56.Torres, A. G., X. Zhou, and J. B. Kaper. 2005. Adherence of diarrheagenic Escherichia coli strains to epithelial cells. Infect. Immun. 73:18-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Umanski, T., I. Rosenshine, and D. Friedberg. 2002. Thermoregulated expression of virulence genes in enteropathogenic Escherichia coli. Microbiology 148:2735-2744. [DOI] [PubMed] [Google Scholar]
- 58.Venables, W. N., and B. D. Ripley. 2002. Modern applied statistics with S, 4th ed., p. 206-210. Springer, New York, N.Y.
- 59.Vuopio-Varkila, J., and G. K. Schoolnik. 1991. Localized adherence by enteropathogenic Escherichia coli is an inducible phenotype associated with the expression of new outer membrane proteins. J. Exp. Med. 174:1167-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wareham, D. W., A. Papakonstantinopoulou, and M. A. Curtis. 2005. The Pseudomonas aeruginosa PA14 type III secretion system is expressed but not essential to virulence in the Caenorhabditis elegans-P. aeruginosa pathogenicity model. FEMS Microbiol. Lett. 242:209-216. [DOI] [PubMed] [Google Scholar]
- 61.Wentzel, A., A. Christmann, T. Adams, and H. Kolmar. 2001. Display of passenger proteins on the surface of Escherichia coli K-12 by the enterohemorrhagic E. coli intimin EaeA. J. Bacteriol. 183:7273-7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woestyn, S., A. Allaoui, P. Wattiau, and G. R. Cornelis. 1994. YscN, the putative energizer of the Yersinia Yop secretion machinery. J. Bacteriol. 176:1561-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]