Abstract
Mesophilic Aeromonas hydrophila strains of serotype O34 typically express smooth lipopolysaccharide (LPS) on their surface. A single mutation in the gene that codes for UDP N-acetylgalactosamine 4-epimerase (gne) confers the O− phenotype (LPS without O-antigen molecules) on a strain in serotypes O18 and O34, but not in serotypes O1 and O2. The gne gene is present in all the mesophilic Aeromonas strains tested. No changes were observed for the LPS core in a gne mutant from A. hydrophila strain AH-3 (serotype O34). O34 antigen LPS contains N-acetylgalactosamine, while no such sugar residue forms part of the LPS core from A. hydrophila AH-3. Some of the pathogenic features of A. hydrophila AH-3 gne mutants are drastically reduced (serum resistance or adhesion to Hep-2 cells), and the gne mutants are less virulent for fish and mice compared to the wild-type strain. Strain AH-3, like other mesophilic Aeromonas strains, possess two kinds of flagella, and the absence of O34 antigen molecules by gne mutation in this strain reduced motility without any effect on the biogenesis of both polar and lateral flagella. The reintroduction of the single wild-type gne gene in the corresponding mutants completely restored the wild-type phenotype (presence of smooth LPS) independently of the O wild-type serotype, restored the virulence of the wild-type strain, and restored motility (either swimming or swarming).
Mesophilic Aeromonas spp. are ubiquitous waterborne bacteria and pathogens of reptiles, amphibians, and fish (4). They can be isolated as part of the fecal flora of a wide variety of other animals, including some used for human consumption, such as pigs, cows, sheep, and poultry. In humans, Aeromonas hydrophila belonging to hybridization groups 1 and 3 (HG1 and HG3), Aeromonas veronii biovar sobria (HG8/HG10), and Aeromonas caviae (HG4) have been associated with gastrointestinal and extraintestinal diseases, such as wound infections of healthy humans, and less commonly with septicemias of immunocompromised patients (24). The pathogenicity of mesophilic aeromonads has been linked to a number of different determinants, such as toxins, proteases, outer membrane proteins (38), lipopolysaccharide (LPS) (35), and flagella (36, 40).
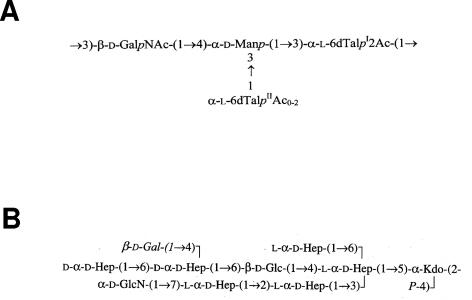
In gram-negative bacteria, lipopolysaccharide is one of the major structural and immunodominant molecules of the outer membrane. It consists of three moieties: the highly conserved and hydrophobic lipid A; the hydrophilic and highly variable O-antigen polysaccharide; and the core oligosaccharide, connecting lipid A and O antigen. The core domain is usually divided into inner and outer core on the basis of sugar composition. O antigen is the most external component of the LPS core, and the LPS core consists of a polymer of oligosaccharide repeating units. Another interesting feature is the high chemical variability shown by the O antigen of the LPS, leading to a similar genetic variation in the genes involved in O-antigen biosynthesis, the so-called wb gene cluster (for a review, see reference 41). The genetics of O-antigen biosynthesis have been studied recently, and it has been shown that the wb gene clusters usually contain genes involved in the biosynthesis of activated sugars, glycosyltransferases, O-antigen polymerases, and O-antigen export (41). Despite the heterogeneity in the structures of O antigens, only three pathways for assembly of O antigens have been recognized (41). The genes involved in core LPS biosynthesis in members of the family Enterobacteriaceae are usually found in the wa (rfa) gene cluster (43), but in some bacteria, they are found in more than one cluster. We fully characterized chemically the O antigen and the core LPS of A. hydrophila strain AH-3 (serotype O34) (Fig. 1) (27, 28), because O34 is one of the most frequently encountered serotypes in mesophilic Aeromonas strains from clinical sources (33).
FIG. 1.
Chemical structures of the O34 antigen LPS (A) and the LPS core (B) from A. hydrophila strain AH-3 (27, 28).
The initial aim of this study was to obtain mutants with altered expression of the O34 antigen LPS. In order to perform this study, we used mini-Tn5 mutagenesis on strain AH-3 (O34) and mutant selection by resistance to bacteriophage PM1; the bacterial surface receptor of PM1 was the O-antigen polysaccharide component of LPS specific for serotype O34 (31).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains used in this study are listed in Table 1. Mesophilic Aeromonas reference strains ATCC 7966 (O1), 987-77 (O2), SL47-79 (O18), and SL37-79 (O34) were also used for serotyping (46).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or phenotypea | Reference or source |
|---|---|---|
| Strains | ||
| Aeromonas hydrophila | ||
| AH-3 | O34, wild type | 38 |
| AH-405 | AH-3, spontaneous Rifr | 38 |
| AH-2767 | AH-3 gne:mini-Tn5Km-1 Rifr Kmr | This study |
| Escherichia coli | ||
| S17-1 | hsdR pro recA, RP4-2 in chromosome, Km::Tn7 (Tc::Mu) | 14 |
| XL1-Blue | endA1 recA1 hsdR17 supE44 thi-1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| DH5α | F−endA hsdR17(rK− mK+)supE44 thi-1 recA1 gyrA96 φ80lacZ | 21 |
| MC1061 | thi thr-1 leu-6 proA2 his-4 argE2 lacY1 galK2 ara-14 xyl-5 supE44 λpir | 45 |
| SM10 | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Kmr λpir | 45 |
| Plasmids | ||
| pRK2073 | Helper plasmid, Spcr | 45 |
| pLA2917 | Cosmid vector, Kmr Tcr | 2 |
| COS-GNE | pLA2917 cosmid with the complete region, Tcr | This study |
| pGEM-T | PCR cloning vector, Ampr | Promega |
| pFS100 | pGP704 suicide plasmid, pir-dependent | 45 |
| pBluescript SK | Apr ori of ColE1 | Stratagene |
| pFS-GNE | pFS100 with an internal fragment of gne, Kmr | This work |
| pACYC184 | Plasmid vector, Cmr Tcr | 20 |
| pACYC-GNE | pACYC184 with the complete gne gene of AH-3, Tcr | This study |
Ampr, ampicillin resistant; Cmr, chloramphenicol resistant; Kmr, kanamycin resistant; Rifr, rifampin resistant; Tcr, tetracycline resistant; Spcr, spectinomycin resistant.
Aeromonas strains were grown on tryptic soy broth (TSB) or tryptic soy agar (TSA) at 30°C, while Escherichia coli strains were grown on Luria-Bertani (LB) Miller broth and LB Miller agar at 37°C. When required, kanamycin (50 μg ml−1), ampicillin (100 μg ml−1), tetracycline (20 μg ml−1), rifampin (100 μg ml−1), or chloramphenicol (20 μg ml−1) was added to the different media. Plasmids used in this study and their characteristics are also shown in Table 1.
Mini-Tn5Km-1 mutagenesis.
Conjugal transfer of transposition element mini-Tn5Km-1 from E. coli S17-1λpirKm-1 (13) to A. hydrophila AH-405 (AH-3 rifampin resistant) was carried out in a conjugal drop incubated for 6 h at 30°C at a 1:5:1 ratio corresponding to S17-1λpirKm-1, AH-405, and HB101 carrying pRK2073 (helper plasmid), respectively. Serial dilutions of the mating mix were plated on TSA supplemented with rifampin and kanamycin in order to select mutants.
General DNA methods.
General DNA manipulations were done essentially as described previously (47). DNA restriction endonucleases, T4 DNA ligase, E. coli DNA polymerase (Klenow fragment), and alkaline phosphatase were used as recommended by the suppliers.
Southern and dot blot hybridizations.
Southern blotting was performed by capillary transfer (47). For dot blot hybridizations, the DNA was denatured by boiling for 5 min, chilled on ice for another 5 min, and spotted onto Hybond N1 (Amersham) nylon membrane. Probe labeling, hybridization, and detection were carried out using the enhanced chemiluminescence labeling and detection system (Amersham) according to the manufacturer's instructions.
DNA sequencing and computer analysis of sequence data.
Double-stranded DNA sequencing was performed by using the dideoxy-chain termination method (48) with the Abi Prism dye terminator cycle sequencing kit (Perkin-Elmer). Oligonucleotides used for genomic DNA amplification experiments and DNA sequencing were purchased from Pharmacia LKB Biotechnology. The DNA sequence was translated in all six frames, and all open reading frames (ORFs) were inspected. Deduced amino acid sequences were compared with those of DNA translated in all six frames from nonredundant GenBank and EMBL databases by using the BLAST (3, 5) network service at the National Center for Biotechnology Information and the European Biotechnology Information Service, respectively. ClustalW was used for multiple-sequence alignments.
DNA amplification and plasmid and mutant construction.
Genomic DNAs from mesophilic Aeromonas strains with different O serotypes were isolated and used as the template in PCR experiments using primers gnefor and gnerev (5′-CAAGAAGAGCGGCAAGAAG-3′ and 5′-ATCAAGGACGGCAGCATAG-3′, respectively) designed to amplify a 1,692-bp band including the complete Aeromonas gne gene. These oligonucleotides were also used to amplify and subclone the gne gene in vector pGEMT (pGEMT-GNE). After EcoRI digestion of plasmid pGEMT-GNE, a single band was obtained and ligated to plasmid pACYC184, yielding plasmid pACYC-GNE where the gne gene is orientated to the promoter. An inner 408-bp DNA fragment of gne was obtained from plasmid pGEMT-GNE by PvuII digestion and subcloned in the pir replication-dependent plasmid pSF100 (45). This plasmid (pSF-GNE) was used to obtain gne deficient mutants from several Aeromonas strains by a single recombination event leading to the generation of two incomplete copies of gne in the chromosomes of these mutants as previously described (45). Plasmid pSF-GNE was isolated, transformed into E. coli SM10 (λpir) (45), and transferred by conjugation from E. coli SM10 to the different Aeromonas strains, which were rifampin-resistant (Rifr) mutants (from our laboratory collection) as previously described (45). Kmr Rifr transconjugants arising from pSF-GNE should contain the mobilized plasmid integrated into the chromosome by homologous recombination between the gne gene and the plasmid, leading to two incomplete copies of the gne gene (defined insertion mutant). Chromosomal DNAs from 10 transconjugants obtained were analyzed by Southern blot hybridization with the appropriate gne DNA probe to obtain defined insertion gne mutants as previously described (38).
Complementation studies.
Complementation analysis of the different gne mutants was performed by conjugal transfer of the wild-type gne gene cloned in pACYC-GNE. Recombinants were selected on LB agar containing tetracycline and rifampin, and LPS was isolated and analyzed in gels.
Cell surface isolation and analyses.
Cell envelopes were prepared by lysis of whole cells in a French press at 16,000 lb/in2. Unbroken cells were removed by centrifugation at 10,000 × g for 10 min, and the envelope fraction was collected by centrifugation at 100,000 × g for 2 h. Cytoplasmic membranes were solubilized twice with sodium N-laurylsarcosinate (16), and the outer membrane fraction was collected as described above. Outer membrane proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by the Laemmli procedure (29). Protein gels were fixed and stained with Coomassie blue. Cultures for analysis of LPS were grown in TSB at 37°C. LPS was purified by the method of Galanos et al. (19), resulting in a 2.3% yield. For screening purposes, LPS was obtained after proteinase K digestion of whole cells (13). LPS samples were separated by SDS-PAGE or SDS-Tricine-PAGE and visualized by silver staining as previously described (13, 22).
Isolation of oligosaccharides.
LPS (20 mg) was hydrolyzed with 1% acetic acid (100°C for 1 h). The resulting precipitate was removed by centrifugation, and the supernatant was analyzed by mass spectrometry (MS). Another sample of LPS was deacylated and purified as previously described (23), obtaining 6 mg of alditol oligosaccharide mixture.
LPS chemical analysis.
For chemical analysis, either purified LPS or core LPS oligosaccharide samples were hydrolyzed with 1 N trifluoroacetic acid for 4 h at 100°C. Alditol acetates and methyl glycoside acetates were analyzed on an Agilent Technologies 5973N mass spectrometer equipped with a 6850A gas chromatograph and an RTX-5 capillary column (Restek; 30 m × 0.25-μm inner diameter; flow rate, 1 ml min−1; He as carrier gas). Acetylated methyl glycoside analysis was performed with the following temperature program: 150 °C for 5 min, 150°C→250 °C at 3 °C min−1, and 250 °C for 10 min. Acetylated methyl ester lipid analysis was performed as follows: 150 °C for 3 min, 150°C→280 °C at 10° min−1, and 280 °C for 15 min. The alditol acetate mixture was analyzed with the following temperature program: 150°C for 5 min, 150°C→300°C at 3°C min−1. For partially methylated alditol acetates, the temperature program was as follows: 90°C for 1 min, 90°C →140°C at 25°C min−1, 140°C→200°C at 5°C min−1, 200°C →280°C at 10°C min−1, and 280°C for 10 min.
Mass spectrometry studies.
Positive- and negative-ion reflectron time-of-flight mass spectra (matrix-assisted laser desorption ionization-time of flight [MALDI-TOF]) were acquired on a Voyager DE-PRO instrument (Applied Biosystems) equipped with a delayed extraction ion source. The ion acceleration voltage was 25 kV, grid voltage was 17 kV, mirror voltage ratio 1.12, and delay time 150 ns. Samples were irradiated at a frequency of 5 Hz by 337-nm photons from a pulsed nitrogen laser. Postsource decay (PSD) was performed using an acceleration voltage of 20 kV. The reflectron voltage was decreased in 10 successive 25% steps. Mass calibration was obtained with a maltooligosaccharide mixture from corn syrup (Sigma). A solution of 2,5-dihydroxybenoic acid in 20% CH3CN in water at a concentration of 25 mg/ml was used as the MALDI matrix. One microliter of matrix solution was deposited on the target followed by loading of 1 μl of the sample. The droplets were allowed to dry at room temperature. Spectra were calibrated and processed under computer control using Applied Biosystems Data Explorer software.
Methylation analysis.
A core oligosaccharide sample (1 mg) obtained from 1% acetic acid hydrolysis were first reduced with NaBH4 and then methylated by the Ciucanu-Kerek procedure (11). Linkage analysis was performed as follows: the methylated sample was carboxymethyl reduced with lithium triethylborohydride (Super-Hydride; Aldrich), mildly hydrolyzed to cleave ketosidic linkage, reduced by means of NaBD4, then totally hydrolyzed, reduced with NaBD4, and finally acetylated as described previously (18).
Cell extract production and enzymatic activity measurements (Gal and GalNAc 4-epimerase assays).
Suspensions of bacteria (25% weight per volume) were washed in 25 mM Tris-HCl buffer (pH 7.5) containing 1 mM MgCl2 and then disrupted in a Branson model 350 sonifier at 0°C. Disrupted bacteria were subjected to high-speed centrifugation (180,000 × g for 2 h) at 5°C to obtain cell extracts. Protein concentrations of extracts were determined by using the Bio-Rad Bradford assay as directed by the manufacturer with bovine serum albumin as the standard.
The assay for UDP-galactose 4-epimerase was performed as previously described (9) by adding 20 to 50 μl of cell extract (approximately 100 to 250 μg of protein) to 1 ml of a solution containing 50 mM Tris-HCl buffer (pH 8.0), 5 mM MgCl2, 1 mM NAD+, and 0.03 U of NAD+-dependent uridine 5′-diphosphoglucose dehydrogenase (Sigma). The assay was begun by addition of 0.5 mM UDP-Gal, and the increase in A340 was followed in a Beckman DU 640 spectrophotometer. Initial rates of NADH formation were determined by using the kinetics program installed in the instrument. A molar extinction (E340) of 6,220 M−1 cm−1 was assumed in all calculations.
The assay for UDP-N-acetylglucosamine 4-epimerase was performed as previously described (12, 52). In this procedure, the conversion of UDP-GalNAc to UDP-GlcNAc is measured after acid hydrolysis by the 3.6-fold increase in the color (A585) of free N-acetylglucosamine over GalNAc in the Morgan-Elson reaction. The reactions were carried out by adding 20 μl of cell extract to a volume containing 0.5 ml of 10 mM glycine, 1 mM MgCl2, 0.1 mM EDTA, and 0.1 mM UDP-GalNAc. Enzyme activity was halted after 5 and 10 min of incubation at 37°C by the addition of 0.8 μl of concentrated HCl. Following hydrolysis and completion of the Morgan-Elson reaction, color development was measured at 585 nm. Control assays with extract alone and with substrate only were run simultaneously. All assays were performed in triplicate. Product formation (i.e., GlcNAc formed by hydrolysis of UDP-GlcNAc) was measured from standard plots prepared by subjecting UDP-GlcNAc, UDP-GalNAc, GlcNAc, and GalNAc (Sigma-Aldrich, St. Louis, Mo.) to the same procedures.
Bacterial survival in human serum.
Bacterial cells (108 CFU) in the logarithmic phase were suspended in phosphate-buffered saline containing 90% serum and incubated at 37°C. Viable counts were made at different times until 3 h by dilution and plating as previously described (32, 34). A pool of nonimmune human sera (NHS) was obtained from healthy volunteers. Control experiments using heat-decomplemented NHS were also performed (32, 34).
ELISAs.
Enzyme-linked immunosorbent assays (ELISAs) using whole cells as antigens were performed as we previously described (6). Specific O34 serum was obtained and purified as we previously described (32).
Adherence assay to HEp-2 cells.
Tissue culture was maintained as described by Thornley et al. (50). The adherence assay of Carrello et al. (10) was performed with slight modifications.
Virulence for fish and mice.
The virulence of the strains grown at 20°C was measured by monitoring their 50% lethal dose (LD50) by the method of Reed and Muench (42).
(i) Fish.
Rainbow trout (12 to 20 g) were maintained in 20-liter static tanks at 17 or 18°C. The fish were injected intraperitoneally with 0.05 ml of the test samples (approximately 109 viable cells). Mortality was recorded for up to 2 weeks; all the deaths occurred within 2 to 8 days.
(ii) Mice.
Albino Swiss female mice (5 to 7 weeks old) were injected intraperitoneally with 0.25 ml of the test samples (approximately 5 × 109 viable cells). Mortality was recorded for up to 1 week; all the deaths occurred within 2 to 5 days.
Motility assays (swarming and swimming).
Freshly grown bacterial colonies were transferred with a sterile toothpick into the center of swarm agar (1% tryptone, 0.5% NaCl, 0.6% agar) or swim agar (1% tryptone, 0.5% NaCl, 0.25% agar). The plates were incubated face up for 16 to 24 h at 30°C, and motility was assessed by examining the migration of bacteria through the agar from the center towards the periphery of the plate. Moreover, swimming motility in liquid media was assessed by light microscopy observations.
Transmission electron microscopy.
Bacterial suspensions were placed on Formvar-coated grids and negative stained with a 2% solution of uranyl acetate, pH 4.1. Preparations were observed on a Hitachi 600 transmission electron microscope.
Nucleotide sequence accession number.
The nucleotide sequence of the A. hydrophila AH-3 gne gene described here has been deposited in GenBank and assigned the following accession number: DQ119103.
RESULTS
After mini-Tn5 mutagenesis of a rifampin-resistant isolate of the A. hydrophila wild-type strain AH-3 (serotype O34), mutants (Kmr) were screened for resistance to bacteriophage PM1 (a bacteriophage specific for O34 antigen LPS [31]). Mutant AH-2767 was found among the 1,500 colonies that were initially screened and showed complete resistance to bacteriophage PM1 compared to the wild type (which is sensitive to this bacteriophage). Strain AH-2767 was sensitive to another Aeromonas bacteriophage (PM2) whose receptor is the lipopolysaccharide core (30), thus indicating that initially, the transposon was affecting some gene(s) involved in O-antigen LPS production. Whole cells of mutant AH-2767 were unable to react in an ELISA with specific antiserum against O34 antigen LPS, and as can be observed in Fig. 2 (lane 2), the LPS of strain AH-2767 was devoid of O34 antigen. Neither whole AH-2767 cells nor its purified LPS were able to react with specific antiserum against O34 antigen LPS in ELISAs. No major differences were found in the growth rates of mutant strain AH-2767 (generation time, 45 min) and the wild-type strain AH-3 (generation time, 40 min). Comparative SDS-PAGE analysis of the outer membrane protein profiles indicates that the mutant and wild-type strains showed essentially the same protein bands (data not shown). Southern blot analysis using a specific probe for the transposon demonstrated that mutant AH-2767 had a single copy of the minitransposon in its genome (data not shown). In order to identify the gene(s) responsible for the observed phenotype, genomic DNA was isolated from mutant AH-2767 and partially digested with EcoRV, and DNA fragments of about 4.0 kb were ligated to vector pBluescript SK. The DNA ligation mixture was transformed into E. coli DH5α, and colonies growing in kanamycin plates were recovered and analyzed again by Southern blotting with the mini-Tn5-specific probe to identify clones containing the transposon and surrounding chromosomal DNA from mutant AH-2767. The nucleotide sequence of the insert of one such recombinant plasmid was determined by using oligonucleotides 5′-AGATCTGATCAAGAGACAG-3′ and 5′-ACTTGTGTATAAGAGTCAG-3′ (from the mini-Tn5Km-1-flanking regions) and oligonucleotides T3 and T7 (from the plasmid vector). Analysis of the sequence allowed the identification of an ORF, interrupted by the mini-Tn5, which encoded a protein with the highest similarity (70% identity) to several nucleotide sugar epimerases from different gram-positive bacteria (Bacillus subtilis or Bacillus cereus) and Shewanella or Haemophilus as gram-negative bacteria.
FIG. 2.
LPS from A. hydrophila AH-3 (wild type) and mutant AH-2767. LPS from A. hydrophila AH-3 (wild type) (lane 1), mutant AH-2767 (gne) alone (lane 2), and AH-2767 with plasmid pACYC184 (lane 3), and complemented with plasmid pACYC-GNE (lane 4) was extracted and analyzed by SDS-PAGE (12%) by the method of Darveau and Hanckok (13) and silver stained (13, 22).
Cloning and sequencing of an A. hydrophila AH-3 genomic region coding for nucleotide sugar epimerase in E. coli K-12 strains.
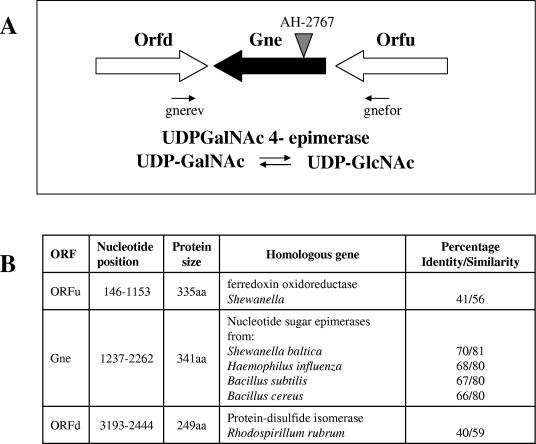
A cosmid-based genomic library of A. hydrophila AH-3 in E. coli DH5α has been constructed as previously described (38). This library was screened by colony blotting using a DNA probe to the previously mentioned ORF of A. hydrophila AH-3. Several positive recombinant clones were identified, of which clone COS-GNE was chosen for further analysis because it was able to completely complement the AH-2767 mutation by rescuing the wild-type strain pattern of bacteriophage sensitivity. The DNA sequence of the ORF (putatively named gne for highest similarity to this nucleotide sugar epimerase), where the mini-Tn5 was inserted, as well as the surrounding region in cosmid COS-GNE, indicates the presence of two ORFs (ORFu and ORFd) flanking gne (Fig. 3). The upstream ORFu transcribed in the same direction of gne was found to be similar (55%) in the BLAST-X to several ferredoxin oxidoreductases from Shewanella, Pseudomonas, and Vibrio. The downstream ORFd transcribed in the opposite direction showed similarity (59%) to several protein-disulfide isomerases from Rhodospirillum rubrum, Rhodobacter, or Mesorhizobium. This genetic organization and the transcriptional directions for these three genes strongly suggest that the mutant AH-2767 phenotype is attributable only to the sugar epimerase insertion mutation, since no polar effects on downstream genes should be expected. This was confirmed, since reintroduction of the single gne gene (plasmid pACYC-GNE) completely rescues the wild-type phenotype in mutant AH-2767, i.e., phage sensitivity and presence of O34 antigen LPS (Fig. 2, lane 4).
FIG. 3.
(A) Genetic organization of the gne region and the role of the encoded product. The positions of the transposon insertion site (AH-2767 mutant) and oligonucleotides (gnefor and gnerev) are indicated. (B) The main homologies of the ORFs and their sizes in nucleotides and amino acid residues (aa) are shown.
Distribution of gne among mesophilic Aeromonas strains.
We studied the presence of gne in different mesophilic Aeromonas strains (n = 50) by PCR fragment DNA amplification using genomic DNAs from these strains and oligonucleotides (gnefor and gnerev [see Materials and Methods]) binding to regions flanking the gne gene (Fig. 3). The mesophilic Aeromonas strains used included environmental and clinical strains. The environmental strains (n = 15) were isolated mainly from water samples (n = 10) and shellfish (n = 5) by using ampicillin dextrin agar or Tergitol agar, while the clinical strains (n = 35) were isolated from blood agar alone or supplemented with ampicillin, depending upon their intestinal (n = 25) or extraintestinal (n = 10) origin. All the strains were identified to the species level by 16S ribosomal RNA gene restriction fragment length polymorphism analysis (15). In all the strains tested, except for two strains, a single 1,692-bp band was amplified. To confirm that gne was indeed present, the nucleotide sequences of the amplified DNA fragments from several Aeromonas strains were determined. The two strains that were negative by PCR were positive in a dot blot using the 1,692-bp band as a DNA probe or in a Southern blot using the internal 408-bp DNA fragment of gne obtained from plasmid pGEMT-GNE by PvuII digestion. Thus, gne coding for a putative sugar nucleotide epimerase was found in all the mesophilic Aeromonas strains tested.
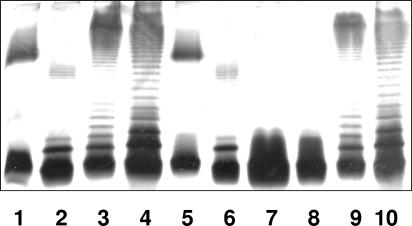
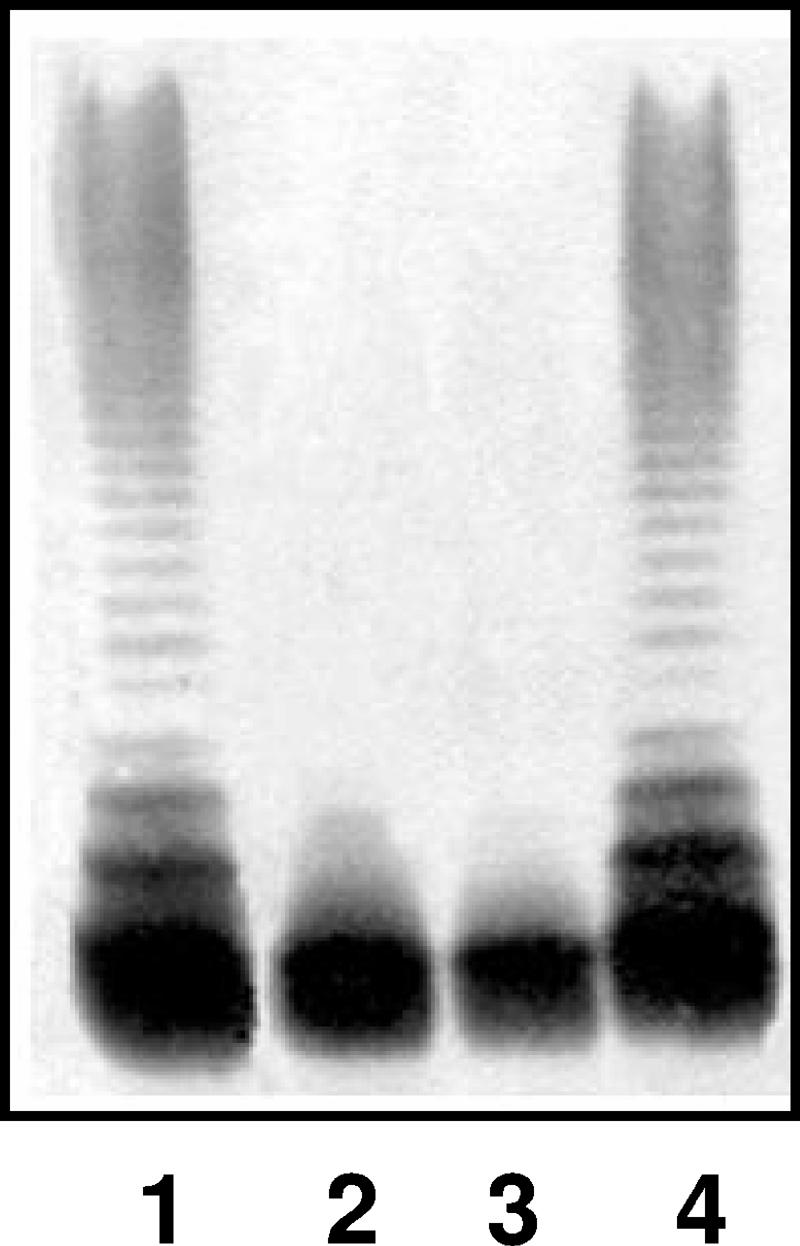
To determine whether the gne gene is also involved in the production of non-O34 antigen LPSs, we constructed gne defined insertion mutants in several mesophilic Aeromonas reference strains belonging to different O serotypes (O1, O2, O18, and O34). LPS from the reference strains that were rifampin resistant and belonged to serotypes O1, O2, O18, and O34 were shown to be O-antigen LPS by SDS-PAGE (Fig. 4, lanes 1, 2, 3, and 4, respectively). All the gne mutants obtained with plasmid pSF-GNE from serotype O18 and O34 strains are in agreement with the AH-2767 mutant phenotype (lack the O-antigen LPS by SDS-PAGE [lanes 7 and 8 of Fig. 4]). All the gne mutants lacking the O-antigen LPS by SDS-PAGE (O18 and O34) can be complemented by the reintroduction of gne in plasmid pACYC-GNE (Fig. 4, lanes 9 and 10). However, genetically confirmed gne defined insertion mutants obtained with plasmid pSF-GNE from serotype O1 and O2 strains exhibited LPS bands (with O-antigen molecules) by SDS-PAGE identical to those of the wild-type strains (Fig. 4, lanes 5 and 6). PCR fragment DNA amplification using genomic DNAs from O1 and O2 strains and gnefor and gnerev oligonucleotides renders a single 1,692-bp band, reconfirmed as gne by DNA sequencing. The gne genes from O1 and O2 strains ligated to plasmid vector pACYC184 as previously done with pACYCGNE were fully able to complement mutant AH-2767 (data not shown).
FIG. 4.
LPS from mesophilic Aeromonas strains. LPS samples from wild-type rifampin-resistant strains of serotypes O1, O2, O18, and O34 (lanes 1 to 4, respectively), gne insertional mutants constructed with pSF-GNE plasmid from strains O1, O2, O18, and O34 (lanes 5 to 8, respectively), and gne mutants from strains O18 and O34 complemented with plasmid pACYC-GNE (lanes 9 and 10, respectively) are shown.
Mutant AH-2767 LPS characterization.
Comparative analysis by SDS-PAGE of LPS obtained from mutant AH-2767 and wild-type strain showed only an absence of O34 antigen in the mutant LPS (Fig. 2, lane 2). Thus, it was hypothesized that mutant AH-2767 LPS would not be truncated at the core level. To test this possibility, LPS obtained from mutant AH-2767 and wild-type strain were subjected to mild acid hydrolysis (1% acetic acid) to cleave the acid-labile ketosidic linkage between 3-deoxy-d-manno-octulosonic acid (Kdo) and lipid A.
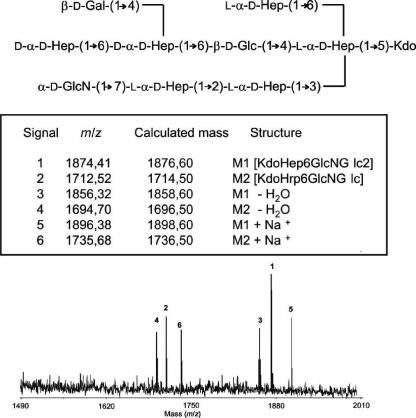
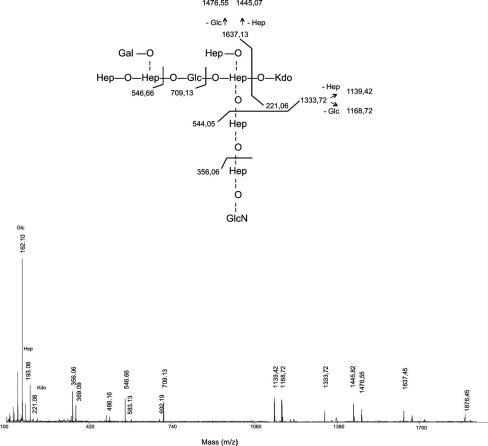
The lipid A fraction was removed by high-speed centrifugation, and the core oligosaccharides were recovered by Sephadex G-50 chromatography. Chemical composition analysis of the core oligosaccharide fraction by gas chromatography-MS of acetylated methyl glycosides revealed the presence of the same residues found in the wild-type core LPS (l-glycero-d-manno-heptose, d-glycero-d-manno-heptose, d-galactose, d-glucose, d-glucosamine, and Kdo), suggesting that in this mutant the core LPS is not truncated (Fig. 2, lane 2). To further prove this point, the oligosaccharide fractions from AH-2767 and its parent strain were analyzed by negative-ion reflectron MALDI-TOF. This experiment revealed the presence of major signals at 1,874.4 and 1,712.5 m/z in both oligosaccharide preparations (Fig. 5 shows data for the AH-2767 mutant). These two signals are in agreement with the previously described structure for the wild-type strain (Kdo-Hep6-GlcN-Glc2 [1,875.6 Da] and Kdo-Hep6-GlcN-Glc [1,713.6 Da]) (25). Signals at 1,856.3 and 1,694.7 m/z (Fig. 5) could correspond to the major oligosaccharide anhydro forms, as previously described for LPS samples hydrolyzed with acetic acid (23). In addition, the major signals were accompanied by the corresponding adducts with sodium ions at m/z 1,896.38 and 1,736.50, respectively. The results of this experiment suggest that the AH-2767 mutant is still able to biosynthesize a wild-type core LPS. This conclusion is supported by determination of the oligosaccharide sequence using the MALDI-PSD technique (Fig. 6). In this spectrum, most fragment ions are attributable to B-type ions, the most relevant signals are shown in the insert at the top of Fig. 6.
FIG. 5.
Negative-ion MALDI-TOF spectrum of acid-released core LPS oligosaccharide from A. hydrophila AH-2767.
FIG. 6.
Positive-ion MALDI-time of flight (TOF)/TOF spectrum of m/z 1,874.40 signal of acid-released core LPS oligosaccharide isolated from A. hydrophila AH-2767. The fragmentation pattern is shown in the insert at the top of the figure.
To further prove that the AH-2767 mutant contains a wild-type LPS core, a methylation analysis experiment of the N-acetylated oligosaccharide alditol mixture obtained from the AH-2767 mutant was performed. In this experiment, the presence of 3,4,6-linked Hep and terminal Hep, Gal, and GlcN confirmed the presence of a wild-type core LPS structure in mutant AH-2767.
These results clearly show that a mutation in the putative nucleotide sugar epimerase produces an LPS with a complete core devoid of the O34 antigen LPS. Since the O34 antigen LPS is known (27), one possible explanation for this defect would be that the putative gne-encoded nucleotide sugar epimerase is involved in the formation of UDP-GalNAc from UDP-GlcNAc. Assuming that UDP-GalNAc is the substrate for GalNAc addition to LPS, a mutation in UDP N-acetylgalactosamine 4-epimerase precluding UDP-GalNAc formation will generate the observed LPS phenotype.
Gal and GalNAc 4-epimerase enzymatic activities.
The genetic analysis as well as the LPS chemical structure of the mutants prompted us to study the enzymatic activities that allow the production of UDP-Gal from UDP-Glc (Gal 4-epimerase) and the production of UDP-GalNAc from UDP- GlcNAc (GalNAc 4-epimerase). A. hydrophila AH-3 (wild-type strain) showed high Gal and GalNAc 4-epimerase activities, grown in glucose or galactose, when measured as described in Materials and Methods (Table 2). However, mutant strain AH-2767 with or without the plasmid vector (pACYC184) showed a complete lack of GalNAc 4-epimerase activity in this assay, which can be rescued by the reintroduction of the single gene using plasmid pACYC-GNE. No significant changes were observed when Gal 4-epimerase activity was measured (Table 2), and the AH-2767 mutant was fully able to grow in minimal medium with galactose as a carbon source. From the results obtained, the named gne gene is in agreement with the UDP-GalNAc 4-epimerase activity of the strains (49).
TABLE 2.
Gal and GalNAc 4-epimerase activities in cell extracts of wild-type A. hydrophila AH-3 and mutant strains
| Strain | Carbon sourcea | Gal 4-epimerase activityb | GalNAc 4-epimerase activityc |
|---|---|---|---|
| AH-3 (wild type) | Glucose | 91.2 ± 1.7 | 24.6 ± 2.2 |
| Galactose | 34.5 ± 1.5 | 12.3 ± 1.6 | |
| AH-405 (AH-3 rifampcin resistant) | Glucose | 90.7 ± 1.9 | 22.4 ± 2.5 |
| Galactose | 33.6 ± 1.4 | 12.0 ± 1.1 | |
| AH-2767 (gne mutant) | Glucose | 89.7 ± 1.8 | <0.03 (NDd) |
| Galactose | 37.1 ± 1.6 | <0.03 (ND) | |
| AH-2767 (pACYC184) | Glucose | 90.0 ± 1.8 | <0.03 (ND) |
| Galactose | 32.6 ± 1.7 | <0.03 (ND) | |
| AH-2767 (pACYC-GNE) | Glucose | 89.5 ± 1.4 | 25.9 ± 1.9 |
| Galactose | 33.4 ± 1.9 | 14.7 ± 2.3 |
Each strain was grown in minimal medium (47) containing glucose or galactose as the carbon source.
Nanomoles of UDP-Gal converted to UDP-Glc min−1 mg protein−1.
Nanomoles of UDP-GalNAc converted to UDP-GlcNAc min−1 mg protein−1.
ND, no detectable activity.
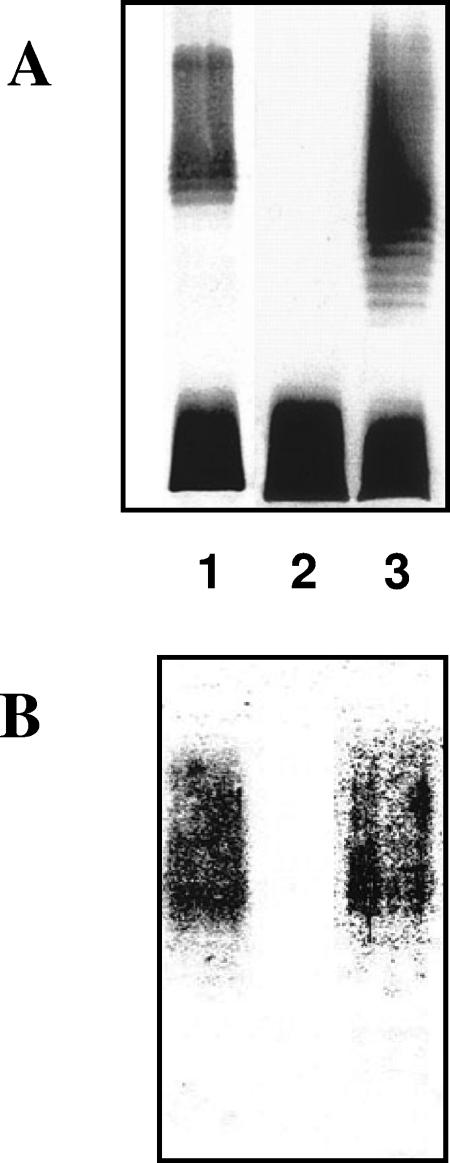
A. hydrophila AH-2767 (gne mutant) was able to produce Klebsiella pneumoniae O5 antigen LPS.
The K. pneumoniae O5 antigen LPS is a homopolymer of mannose (37). When we introduced the COS-KT4 plasmid carrying the complete K. pneumoniae wb05 gene cluster (37) into A. hydrophila AH-2767, we could observe that the strain was able to produce K. pneumoniae O5 antigen LPS as judged by SDS-PAGE and immunoassay (Fig. 7); in addition, this gne mutant was unable to produce Aeromonas O34 antigen LPS.
FIG. 7.
LPS (A) and Western blot (B) using specific antiserum against K. pneumoniae O5 antigen LPS (37) from the following strains: K. pneumoniae serotype O5 wild-type strain (lane 1), A. hydrophila gne mutant AH-2767 (lane 2), and A. hydrophila gne mutant AH-2767 carrying plasmid COS-KT4, which contains the entire K. pneumoniae wb05 gene cluster (lane 3).
Serum killing.
Wild-type serotype O34 strains were resistant to the bactericidal activity of NHS (98% survival after 3 h of incubation with NHS), while the insertion mutant AH-2767 or the same strain with plasmid vector pACYC184 was sensitive (less than 1% survival after 3 h of incubation with NHS). The introduction of the gne gene (pACYC-GNE) renders mutant AH-2767 resistant to the bactericidal activity of NHS (96% survival after 3 h of incubation with NHS), as found for the wild-type strain.
Adherence to HEp-2 cells.
Table 3 shows the adhesion of serotype O34 wild-type strain, gne mutant, and complemented mutant to HEp-2 cells. We previously reported that the O34 antigen LPS facilitates adhesion of these strains to HEp-2 cells (36). The wild-type strains showed a high percentage of adherence to HEp-2 cells, while the gne mutant (with or without the plasmid vector alone) showed a large decrease (60% reduction) in the percent adhesion to HEp-2 cells. The introduction of gne (pACYC-GNE) completely rescues the adherence to Hep-2 cells in the mutant strain. The results obtained in adhesion to Hep-2 cells prompted us to compare the motility (swimming and swarming) of these A. hydrophila strains able to produce a single polar flagellum in liquid media and peritrichous (lateral) flagella in semisolid or solid media (20).
TABLE 3.
Adhesion of different A. hydrophila serotype O34 strains to HEp-2 cells
| Strain (main characteristics) | No. of bacteria/HEp-2 cell (mean ± SD) | % Reduction in adhesiona |
|---|---|---|
| AH-3 (wild type) | 18.3 ± 2.2 | |
| AH-405 (AH-3 rifampin-resistant mutant) | 17.9 ± 2.0 | 2b |
| AH-2767 (gne insertion mutant from AH-405) | 7.2 ± 0.7 | 61 |
| AH-2767(pACYC-GNE) (gne mutant complemented with the single gne gene) | 18.0 ± 2.5 | 1 |
| AH-2767(pACYC184) (gne mutant with plasmid vector alone) | 6.9 ± 1.0 | 62 |
The level of adhesion of strain AH-3 was set at 100%.
This value was significantly different from the value for the wild-type AH-3 strain (P < 0.001 by Student's t test).
Motility assays and electron microscopic examination of gne mutants.
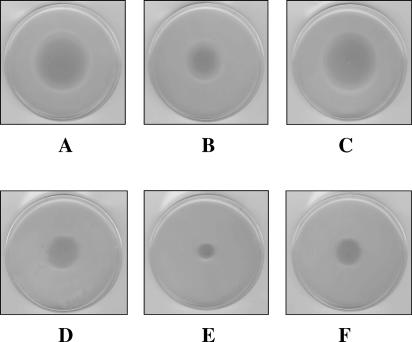
Since there were significant reductions in bacterial swimming and swarming of AH-2767 (gne mutant) in motility plates (Fig. 8B and E) in comparison with the wild type or the complemented strain (Fig. 8A, C, D, and F), the morphology of bacterial cells of A. hydrophila AH-3 (wild type), AH-2767, and AH-2767 complemented with pACYC-GNE were examined by transmission electron microscopy and negative staining. Individual cells of the wild type and complemented mutant produce a single polar flagellum or lateral flagella. Interestingly, the numbers of these surface protein appendages produced by the gne mutants were similar to those produced by the parent strains (data not shown). However, when the motility of the same cells was observed by light microscopy, 40 to 50% of the AH-2767 cells were nonmotile, while the wild type and the complemented strain were fully motile. This situation was similar in the different gne mutants observed that lack the O-antigen LPS, while no differences were observed in gne mutants that retain the O-antigen LPS.
FIG. 8.
Motility phenotypes exhibited in swim (0.25%) and swarm (0.6%) agar by A. hydrophila AH-3 (A and D, respectively), gne mutant AH-2767 (B and E, respectively), and AH-2767 carrying plasmid pACYC-GNE (C and F, respectively).
Virulence studies.
We tested the virulence of the wild-type strain and the corresponding gne insertion mutant (LD50), as shown in Table 4. As can be observed, no differences were found in the mortality of fish and mice infected with the wild-type strain and the rifampin-resistant mutant. However, gne mutant AH-2767 showed a higher LD50 (an increase of 1 to 2 log units) in both fish and mice than the wild-type strain did. Complementation of this insertion mutant with pACYC-GNE (carrying the single gne gene) completely restored the virulence for fish or mice (LD50s similar to those of the wild-type strain [Table 4]), while no changes were observed with the plasmid vector alone. These results suggested that Gne is an important virulence factor for mesophilic Aeromonas serogroup O34 pathogenesis.
TABLE 4.
Virulence for rainbow trout and mice of several A. hydrophila AH-3 (serogroup O34)
| Strain (main characteristic) | LD50a for:
|
|
|---|---|---|
| Rainbow trout | Mice | |
| AH-3 (wild type) | 105.3 | 107.4 |
| AH-405 (AH-3 rifampin-resistant mutant) | 105.4 | 107.6 |
| AH-2767 (gne insertion mutant from AH-405) | 107.2 | 108.6 |
| AH-2767(pACYC-GNE) (gne mutant complemented with the single gne gene) | 105.6 | 107.7 |
| AH-2767(pACYC184) (gne mutant with plasmid vector alone) | 107.3 | 109.0 |
The values are the averages of three independent experiments, and the maximum deviation was always <100.3.
DISCUSSION
The initial isolation of a mutant (AH-2767) resistant to Aeromonas bacteriophage PM1 using the O34 antigen polysaccharide as a receptor led us to the characterization of a gene related to sugar nucleotide epimerases. This gene, named gne (for their similarity to Bacillus subtilis Gne), was clearly located outside the O34 antigen LPS (wb), LPS core (wa), and capsular polysaccharide (wac) gene clusters from the wild-type strain (54, 55). Mutation of the gne gene by either mini-Tn5 or pSF-GNE insertion could not affect the downstream gene by polarity, since both genes are transcribed in opposite directions (Fig. 3). Furthermore, the reintroduction of the single gne wild-type gene by transformation with plasmid pACYC-GNE rescues the wild-type phenotype. The gne was found to be present in all the mesophilic Aeromonas strains tested. Several gne defined insertion mutants showing two different phenotypes were obtained, one like AH-2767 mutant strain lacking the O-antigen LPS and the other one with no changes in LPS compared to the wild-type strain.
Aeromonas Gne (Fig. 9) shares a high level of sequence similarity with nucleotide sugar epimerases and other members of the short-chain dehydrogenase/reductase (SDR) enzyme family of oxidoreductases (25, 39). The SDR family currently includes over 3,000 enzymes from all forms of life. Typically, they carry out oxidation-reduction reactions, usually functioning as dehydrogenases, dehydratases, isomerases, or epimerases. These enzymes share anywhere from 15 to 30% amino acid identity and display two conserved motifs (25, 39) that are present in Aeromonas Gne. The first is a TGXXGXXG motif found in the N terminus that displays a conserved alternating α/β-folding pattern typical of a Rossman fold involved in coenzyme (NAD+) binding (17, 44, 53). The second motif consists of the catalytic triad of Ser, Tyr, and Lys with the YXXXK motif, which has recently been extended to a catalytic tetrad of Asn, Ser, Tyr, and Lys (17). The Ser residue may be replaced by Thr in some members of the SDR family.
FIG. 9.
Alignment of A. hydrophila Gne, its bacterial homologs, and other characterized bacterial epimerases. The A. hydrophila Gne (AH3) was aligned with its homologs in B. subtilis (BS), Haemophilus influenzae (HI), and Y. enterocolitica (YE), as well as the UDP-GlcNAc epimerase WbpP from P. aeruginosa (PA). Identical amino acids are shown on a black background, and similar residues are shown on a gray background. Residues that are involved in nucleotide binding (underlined) and amino acids that have been shown to be important for catalysis (asterisk) are indicated. Amino acids that are underlined twice are thought to be involved in substrate binding. Multiple-sequence alignments were performed with ClustalW available at the ExPASy molecular biology server (au.expasy.org).
It seems clear that the gne gene codes for an UDP-GalNAc 4-epimerase enzyme responsible for the conversion of UDP-GlcNAc to UDP-GalNAc, as can be judged from the results of enzymatic activities assayed. In B. subtilis, in vitro and in vivo interconversion between UDP-Glc and UDP-Gal as well as between their N-acetylated forms is mediated by the same enzyme (49). This situation is thought to be more widespread among prokaryotes than originally believed. Indeed, Yersinia enterocolitica O:8 Gne, in addition to UDP-GlcNAc 4-epimerase has a weak UDP-Glc 4-epimerase activity in vitro (7). Likewise, the Pseudomonas aeruginosa O:6 WbpP protein recognizes acetylated as well as nonacetylated substrates at least in vitro (17). However, Aeromonas Gne seems to be restricted to conversion of UDP-GlcNAc to UDP-GalNAc, and other nucleotide sugar epimerases, such as GalE, would perform conversion between UDP-Glc and UDP-Gal, because Gne seems to be the enzyme that converts UDP-GlcNAc to UDP-GalNAc
The fact that gne mutants in some cases are unable to form O-antigen LPS and in other cases are able to produce the O-antigen LPS may be explained on the basis of the absence or presence of GalNAc sugar residues in the polysaccharide. For instance, GalNAc was present in the chemical composition of the O-antigen LPS in serotype O34 strains (27). It looks like that this is also the case for serotype O18, but its chemical composition is not known. Other strains belonging to different serotypes (O1 and O2) do not appear to have GalNAc in their chemical composition, as judged by the O-antigen production in gne mutants, but their chemical composition is unknown. The gne genes from O1 and O2 strains were fully able to complement mutant AH-2767, so the gne function seems to be the same; they also showed an extremely high identity (but not 100% for DNA) with the gne genes from O34 strains. Two gne copies could not be obtained by PCR in these O1 and O2 strains, which always showed a single DNA band (data not shown). This point is reinforced because when we introduced the K. pneumoniae wb05 genes (which produce an O-antigen LPS with mannose as a unique sugar [37]) from strain AH-3 (serotype O34) into the gne mutant AH-2767, no changes in the O-antigen LPS could be observed (no GalNAc molecules need to be formed for this exogenous O-antigen LPS). In all cases, gne mutants showed a migration of the LPS core similar to that of the wild type. Taking advantage of the fact that the chemical structure of strain AH-3 (Fig. 1) is known (28), we chemically characterized the AH-2767 mutant. Chemical composition, electrospray ionization-MS (Fig. 5), PSD fragmentation (Fig. 6), and permethylation analysis of LPS core obtained from mutant AH-2767 showed that is identical to the LPS core of AH-3. No GalNAc residues are found in the LPS core of strain AH-3. However, a Gal residue is found in the outer core, and no changes could be observed for the LPS core of the AH-2767 mutant. These observations are in agreement with the fact that in A. hydrophila AH-3 has a gene encoding UDP-Gal 4-epimerase activity (GalE) located elsewhere (galE) in the chromosome. Therefore, it seems clear that Aeromonas Gne is essential in the biosyntheses of anionic cell wall polymers where GalNAc is present in the chemical composition of the LPS core, and only a single gne copy could be present in the genomes of Aeromonas serotype O18 and O34 strains.
The gne mutation drastically affected the serum susceptibility of Aeromonas serotype O34 strains, reduced the ability of these strains to adhere to Hep-2 cells, and decreased the virulence in a septicemic model in fish and mice. All the changes observed in the gne mutants in these virulence experiments are rescued by the introduction of the corresponding single wild-type gene, but not by the introduction of the plasmid vector alone. From these results, we can initially conclude that the O34 antigen LPS is essential in Aeromonas serotype O34 pathogenesis. However, some important differences in motility but not in flagellar expression (either polar or lateral flagella [20]) could be observed between mutant strains and the corresponding wild-type or complemented strains. Flagella are much more than just organelles for locomotion; flagella have been shown to be a contributing factor in adhesion, surface colonization, biofilm formation, and invasion (26). Toguchi et al. (51) showed that the lack of O antigen in Salmonella enterica serovar Typhimurium affected the swarming motility of this organism. They suggested that the absence of O antigen affected the surface “wettability” that is required for swarm colony expansion. A report by Bengoechea et al. (8) showed that deficiency in O-antigen biosynthesis in Yersinia enterocolitica O:8 affected virulence and caused down-regulation and up-regulation of the expression of a number of well-characterized virulence genes. On the basis of these findings, these authors suggested that the absence of O antigen in the outer membrane of Y. enterocolitica O:8 might cause cellular or membrane stress that could act as a regulatory signal affecting the expression of a number of virulence-associated genes. Recently, results obtained with P. aeruginosa waaL mutants (lacking the O-antigen LPS) indicated that LPS of P. aeruginosa plays a role in flagellum biogenesis (1). Therefore, apart from the observed defects in flagellum and pilin expression on the surface of P. aeruginosa, the absence of O antigen and less wettability on the waaL mutants provided another explanation for the reduced overall motility in this bacterium. Mutants lacking the O34 antigen LPS in mesophilic Aeromonas strains by gne mutation showed complete flagellar biogenesis, but motility is clearly affected. Due to the fact that the A. hydrophila AH-3 strain had two types of flagella, this is the first time that it has been demonstrated that the absence of O34 antigen LPS affects swimming and swarming motility but is not involved in polar and lateral flagellum biogenesis. Work is in progress to target this enzyme (Gne) for mesophilic Aeromonas serotype O34 experimental therapy.
Acknowledgments
This work was supported by Plan Nacional de I + D and FIS grants (Ministerio de Educación, Ciencia y Deporte and Ministerio de Sanidad of Spain) and from Generalitat de Catalunya. R.C., N.J., and S.V. received predoctoral fellowships from the Universidad de Barcelona and Generalitat de Catalunya.
We also thank Maite Polo for technical assistance.
Editor: D. L. Burns
REFERENCES
- 1.Abeyrathne, P. D., C. Daniels, K. K. H. Poon, M. J. Matewish, and J. S. Lam. 2005. Functional characterization of WaaL, a ligase associated with linking O-antigen polysaccharide to the core of Pseudomonas aeruginosa lipopolysaccharide. J. Bacteriol. 187:3002-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, L. N., and R. S. Hanson. 1985. Construction of broad-host-range cosmid cloning vector: identification of genes necessary for growth of Methylobacterium organophilum on methanol. J. Bacteriol. 161:955-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Austin, B., and C. Adams. 1996. Fish pathogens, p. 197-243. In B. Austin, M. Altwegg, P. J. Gosling, and S. W. Joseph (ed.), The genus Aeromonas. John Wiley and Sons, New York, N.Y.
- 5.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benedí, V. J., B. Ciurana, and J. M. Tomás. 1989. Isolation and characterization of Klebsiella pneumoniae unencapsulated mutants. J. Clin. Microbiol. 27:82-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bengoechea, J. A., E. Pinta, T. Salminen, C. Oertelt, O. Holst, J. Radziejewska-Lebrecht, Z. Piotrowska-Seget, R. Venho, and M. Skurnik. 2002. Functional characterization of Gne (UDP-N-acetylglucosamine-4-epimerase), Wzz (chain length determinant), and Wzy (O-antigen polymerase) of Yersinia enterocolitica serotype O:8. J. Bacteriol. 184:4277-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bengoechea, J. A., H. Najdenski, and M. Skurnik. 2004. Lipopolysaccharide O antigen status of Yersinia enterocolitica O:8 is essential for virulence and absence of O antigen affects the expression of other Yersinia virulence factors. Mol. Microbiol. 52:451-469. [DOI] [PubMed] [Google Scholar]
- 9.Boels, I. C., A. Ramos, M. Kleerebezem, and W. M. de Vos. 2001. Functional analysis of the Lactococcus lactis galU and galE genes and their impact on sugar nucleotide and exopolysaccharide biosynthesis. Appl. Environ. Microbiol. 67:3033-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrello, A., K. A. Silburn, J. R. Budden, and B. J. Chang. 1988. Adhesion of clinical and environmental Aeromonas isolates to HEp-2 cells. J. Med. Microbiol. 26:19-27. [DOI] [PubMed] [Google Scholar]
- 11.Ciucanu, I., and F. Kerek. 1984. A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 131:209-217. [Google Scholar]
- 12.Creuzenet, C., M. Belanger, W. W. Wakarchuk, and J. S. Lam. 2000. Expression, purification, and biochemical characterization of WbpP, a new UDP-GlcNAc C4 epimerase from Pseudomonas aeruginosa serotype O6. J. Biol. Chem. 275:19060-19067. [DOI] [PubMed] [Google Scholar]
- 13.Darveau, R. P., and R. E. Hancock. 1983. Procedure for isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella typhimurium strains. J. Bacteriol. 155:831-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Figueras, M. J., L. Soler, M. R. Chacón, J. Guarro, and A. J. Martínez-Murcia. 2000. Extended method for discrimination of Aeromonas spp. by 16S rDNA RFLP analysis. Int. J. Syst. Evol. Microbiol. 50:2069-2073. [DOI] [PubMed] [Google Scholar]
- 16.Filip, C., G. Fletcher, J. L. Wulff, and C. F. Earhart. 1973. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium lauryl sarcosinate. J. Bacteriol. 115:717-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filling, C., K. D. Berndt, J. Benach, S. Knapp, T. Prozorovski, E. Nordling, R. Ladenstein, H. Jornvall, and U. Oppermann. 2002. Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J. Biol. Chem. 277:25677-25684. [DOI] [PubMed] [Google Scholar]
- 18.Forsberg, L. S., U. Ramadas Bhat, and R. W. Carlson. 2000. Structural characterization of the O-antigenic polysaccharide of the lipopolysaccharide from Rhizobium etli strain CE3. A unique O-acetylated glycan of discrete size, containing 3-O-methyl-6-deoxy-L-talose and 2,3,4-tri-O-methyl-L-fucose. J. Biol. Chem. 275:18851-18863. [DOI] [PubMed] [Google Scholar]
- 19.Galanos, C., O. Lüderitz, and O. Westphal. 1969. A new method for the extraction of R lipopolysaccharides. Eur. J. Biochem. 9:245-249. [DOI] [PubMed] [Google Scholar]
- 20.Gavín, R., A. A. Rabaan, S. Merino, J. M. Tomás, I. Gryllos, and J. G. Shaw. 2002. Lateral flagella of Aeromonas species are essential for epithelial cell adherence and biofilm formation. Mol. Microbiol. 42:383-397. [DOI] [PubMed] [Google Scholar]
- 21.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 22.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izquierdo, L., N. Coderch, N. Piqué, E. Bedini, M. Corsaro, S. Merino, S. Fresno, J. M. Tomas, and M. Regué. 2003. The Klebsiella pneumoniae wabG gene: its role in the biosynthesis of the core lipopolysaccharide and virulence. J. Bacteriol. 185:7213-7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janda, J. M., and S. L. Abbott. 1998. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations, and unanswered questions. Clin. Infect. Dis. 27:332-344. [DOI] [PubMed] [Google Scholar]
- 25.Jornvall, H., B. Persson, M. Krook, S. Atrian, R. Gonzalez-Duarte, J. Jeffery, and D. Ghosh. 1995. Short-chain dehydrogenases/reductases (SDR). Biochemistry 34:6003-6013. [DOI] [PubMed] [Google Scholar]
- 26.Kirov, S. M. 2003. Bacteria that express lateral flagella enable dissection of the multifunctional roles of flagella in pathogenesis. FEMS Microbiol. Lett. 224:151-159. [DOI] [PubMed] [Google Scholar]
- 27.Knirel, Y. A., A. S. Shaskov, S. N. Senchenkova, S. Merino, and J. M. Tomás. 2002. Structure of the O-polysaccharide of Aeromonas hydrophila O:34: a case of random O-acetylation of 6-deoxy-L-talose. Carbohydr. Res. 337:1381-1386. [DOI] [PubMed] [Google Scholar]
- 28.Knirel, Y. A., E. Vinogradov, N. Jimenez, S. Merino, and J. M. Tomás. 2004. Structural studies on the R-type lipopolysaccharide of Aeromonas hydrophila. Carbohydr. Res. 339:787-793. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Merino, S., S. Camprubí, and J. Tomás. 1990. Isolation and characterization of bacteriophage PM2 from Aeromonas hydrophila. FEMS Microbiol. Lett. 68:239-244. [DOI] [PubMed] [Google Scholar]
- 31.Merino, S., S. Camprubí, and J. M. Tomás. 1992. Characterization of an O-antigen bacteriophage from Aeromonas hydrophila. Can. J. Microbiol. 38:235-240. [DOI] [PubMed] [Google Scholar]
- 32.Merino, S., S. Camprubí, and J. M. Tomás. 1992. Effect of growth temperature on outer membrane components and virulence of Aeromonas hydrophila strains of serotype O:34. Infect. Immun. 60:4343-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merino, S., S. Camprubí, and J. M. Tomás. 1993. Incidence of Aeromonas spp. serotypes O:34 and O:11 among clinical isolates. Med. Microbiol. Lett. 2:48-55. [Google Scholar]
- 34.Merino, S., D. Alvarez, S. Hernandez-Alles, and J. M. Tomás. 1994. Effect of the growth temperature on mesophilic Aeromonas spp. serotype O:34 complement-mediated killing. FEMS Microbiol. Lett. 118:163-166. [DOI] [PubMed] [Google Scholar]
- 35.Merino, S., X. Rubires, A. Aguillar, J. F. Guillot, and J. M. Tomas. 1996. The role of the O-antigen lipopolysaccharide on the colonization in vivo of the germfree chicken gut by Aeromonas hydrophila serogroup O:34. Microb. Pathog. 20:325-333. [DOI] [PubMed] [Google Scholar]
- 36.Merino, S., X. Rubirés, A. Aguilar, and J. M. Tomás. 1997. The role of flagella and motility in the adherence and invasion to fish cell lines by Aeromonas hydrophila serogroup O:34 strains. FEMS Microbiol. Lett. 151:213-217. [DOI] [PubMed] [Google Scholar]
- 37.Merino, S., M. Altarriba, L. Izquierdo, M. M. Nogueras, M. Regué, and J. M. Tomás. 2000. Cloning and sequencing of Klebsiella pneumoniae O5 wb gene cluster and its role in pathogenesis. Infect. Immun. 68:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nogueras, M. M., S. Merino, A. Aguilar, V. J. Benedí, and J. M. Tomás. 2000. Cloning, sequencing, and role in serum susceptibility of porin II from mesophilic Aeromonas hydrophila. Infect. Immun. 68:1849-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oppermann, U., C. Filling, M. Hult, N. Shafqat, X. Wu, M. Lindh, J. Shafqat, E. Nordling, Y. Kallberg, B. Persson, and H. Jornvall. 2003. Short chain dehydrogenases/reductases (SDR): the 2002 update. Chem. Biol. Interact. 143-144:247-253. [DOI] [PubMed] [Google Scholar]
- 40.Rabaan, A. A., I. Gryllos, J. M. Tomás, and J. G. Shaw. 2001. Motility and the polar flagellum are required for Aeromonas caviae adherence to Hep-2 cells. Infect. Immun. 69:4257-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raetz, C. R. H., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent end points. Am. J. Hyg. 27:493-497. [Google Scholar]
- 43.Regué, M., N. Climent, N. Abitiu, N. Coderch, S. Merino, L. Izquierdo, M. Altarriba, and J. M. Tomás. 2001. Genetic characterization of the Klebsiella pneumoniae waa gene cluster, involved in core lipopolysaccharide biosynthesis. J. Bacteriol. 183:3564-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rossman, M. G., and P. Argos. 1975. A comparison of the heme binding pocket in globins and cytochrome b5*. J. Biol. Chem. 250:7525-7532. [PubMed] [Google Scholar]
- 45.Rubirés, X., F. Saigí, N. Piqué, N. Climent, S. Merino, S. Albertí, J. M. Tomás, and M. Regué. 1997. A gene (wbbL) from Serratia marcescens N28b (O4) complements the rfb-50 mutation of Escherichia coli K-12 derivatives. J. Bacteriol. 179:7581-7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakazaki, R., and T. Shimada. 1984. O-serogrouping scheme for mesophilic Aeromonas strains. Jpn. J. Med. Sci. Biol. 37:247-255. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y
- 48.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soldo, B., C. Scotti, D. Karamata, and V. Lazarevic. 2003. The Bacillus subtilis Gne (GneA, GalE) protein can catalyse UDP-glucose as well as UDP-N-acetylglucosamine 4-epimerisation. Gene 319:65-69. [DOI] [PubMed] [Google Scholar]
- 50.Thornley, J. P., J. G. Shaw, I. Gryllos, and A. Eley. 1997. Virulence properties of clinically significant Aeromonas species. Rev. Med. Microbiol. 8:61-72. [Google Scholar]
- 51.Toguchi, A., M. Siano, M. Burkart, and R. M. Harshey. 2000. Genetics of swarming motility in Salmonella enterica serovar Typhimurium: critical role for lipopolysaccharide. J. Bacteriol. 182:6308-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, L., S. Huskic, A. Cisterne, D. Rothemund, and P. R. Reeves. 2002. The O-antigen gene cluster of Escherichia coli O55:H7 and identification of a new UDP-GlcNAc C4 epimerase gene. J. Bacteriol. 184:2620-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wierenga, R. K., M. C. De Maeyer, and W. G. J. Hol. 1985. Interaction of pyrophosphate moieties with α-helices in dinucleotide binding proteins. Biochemistry 24:1346-1357. [Google Scholar]
- 54.Zhang, Y. L., E. Arakawa, and K. Y. Leung. 2002. Novel Aeromonas hydrophila PPD134/91 genes involved in O-antigen and capsule biosynthesis. Infect. Immun. 70:2326-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, Y. L., Y. L. Lau, E. Arakawa, and K. Y. Leung. 2003. Detection and genetic analysis of group II capsules in Aeromonas hydrophila. Microbiology 149:1051-1060. [DOI] [PubMed] [Google Scholar]