Abstract
Escherichia coli is the most common organism associated with asymptomatic bacteriuria (ABU). In contrast to uropathogenic E. coli (UPEC), which causes symptomatic urinary tract infections (UTI), very little is known about the mechanisms by which these strains colonize the human urinary tract. The prototype ABU E. coli strain 83972 was originally isolated from a girl who had carried it asymptomatically for 3 years. Deliberate colonization of UTI-susceptible individuals with E. coli 83972 has been used successfully as an alternative approach for the treatment of patients who are refractory to conventional therapy. Colonization with strain 83972 appears to prevent infection with UPEC strains in such patients despite the fact that this strain is unable to express the primary adhesins involved in UTI, viz. P and type 1 fimbriae. Here we investigated the growth characteristics of E. coli 83972 in human urine and show that it can outcompete a representative spectrum of UPEC strains for growth in urine. The unique ability of ABU E. coli 83972 to outcompete UPEC in urine was also demonstrated in a murine model of human UTI, confirming the selective advantage over UPEC in vivo. Comparison of global gene expression profiles of E. coli 83972 grown in lab medium and human urine revealed significant differences in expression levels in the two media; significant down-regulation of genes encoding virulence factors such as hemolysin, lipid A, and capsular polysaccharides was observed in cells grown in urine. Clearly, divergent abilities of ABU E. coli and UPEC to exploit human urine as a niche for persistence and survival suggest that these key differences may be exploited for preventative and/or therapeutic approaches.
Urinary tract infection (UTI) is a serious health problem affecting millions of people each year. It is estimated that there are more than 10 million cases in Western Europe alone per year. The recurrence rate is high, and often the infections tend to become chronic with many episodes. UTI usually starts as a bladder infection but often evolves to encompass the kidneys and ultimately can result in renal failure or dissemination to the blood. UTI is the most common infection in patients with a chronic indwelling bladder catheter; bacteriuria is essentially unavoidable in this patient group (9). UTI is classified into disease categories according to the site of infection: cystitis (the bladder), pyelonephritis (the kidney), and bacteriuria (the urine). Colonization of urine in the absence of clinical symptoms is called asymptomatic bacteriuria (ABU). ABU occurs in up to 6% of healthy individuals and 20% of elderly individuals. ABU strains generally do not cause symptoms, and most patients with ABU do not need treatment. Furthermore, colonizing ABU strains may actually help to prevent infection by other more virulent bacteria (8, 14, 44, 45).
Escherichia coli is responsible for more than 80% of all UTIs and causes both ABU and symptomatic UTI (13, 39). These infections are typically caused by a single bacterial clone and are in effect monocultures. The ability of uropathogenic E. coli (UPEC) to cause symptomatic UTI is associated with the expression of a variety of virulence factors, including adhesins (e.g., type 1 and P fimbriae) and toxins (e.g., hemolysin) (20, 33). Bacterial adherence is generally considered to be a pivotal step in the colonization of host tissue surfaces submitted to hydrodynamic flow forces. The human urinary tract is submitted to brutal hydrodynamic shear forces, and adherence to the urinary tract epithelium enables bacteria to resist removal by urine flow. Bacterial adherence not only contributes to colonization, but also to invasion, biofilm formation, and host cell damage. The two primary fimbrial adhesins associated with UPEC strains are type 1 and P fimbriae. Type 1 fimbriae are mainly associated with cystitis and confer binding to α-d-mannosylated proteins, such as uroplakins, which are abundant in the bladder (7, 46). Expression of P fimbriae is primarily linked to pyelonephritic strains. P fimbriae recognize the α-d-galactopyranosyl-(1-4)-β-d-galactopyranoside moiety present in the globoseries of glycolipids located in the human kidney as well as on erythrocytes (21). Both type 1 and P fimbriae trigger host responses that include cytokine production, inflammation, and exfoliation of infected bladder epithelial cells (31, 36, 48).
E. coli strain 83972 is a clinical isolate capable of long-term bladder colonization. The strain was originally isolated from a young Swedish girl with ABU who had carried it for at least 3 years without symptoms (1, 25). It is well adapted for growth in the human urinary tract, where it establishes long-term bacteriuria (1, 14, 47, 49). The strain has been used for prophylactic purposes; as such, it has been used as an alternative treatment in patients with recurrent UTI who are refractory to conventional therapy (8, 14). Here the bladders of patients are deliberately colonized with E. coli 83972 in order to prevent disease-causing strains from colonizing. Deliberate colonization with E. coli 83972 has for example been shown to reduce the frequency of UTI in patients with spinal cord injury and neurogenic bladder (49), and the strain can prevent catheter colonization by bacterial and fungal uropathogens (8, 44, 45). In effect, extensive trials have shown that infection with potentially dangerous UPEC strains often does not take place in such patients as long as the ABU strain stays in the bladder. The mechanism of bladder colonization by E. coli 83972 is not known. Also, the mechanisms underlying its ability to keep other strains away are not known either. Recently, we found that E. coli 83972 is unable to express functional type 1 and P fimbriae (19). The results explained to a large degree why the strain does not cause symptoms in the host. However, important outstanding questions remain, namely, (i) how is the strain capable of efficient bladder colonization and (ii) how does the strain keep pathogenic E. coli from infecting the bladder. In this study we approach these questions.
MATERIALS AND METHODS
Bacterial strains.
The strains used in this study are described in Table 1. E. coli 83972 (OR:K5:H−) was originally isolated from a young Swedish girl (1, 25). E. coli 83972 is a prototype ABU strain and lacks defined O and K surface antigens. It carries adhesin gene clusters homologous to fim, pap, uca, and foc but does not express functional fimbrial adhesins after in vitro culture or when recovered from the urinary tract (1, 15). The E. coli strains 536, CFT073, NU14, and 1177 are all well-characterized uropathogenic strains isolated from patients with severe urinary tract infections (Table 1). Strains 536 and NU14 are naturally resistant to streptomycin. These phenotypes were used to monitor the population composition in growth competition experiments. In the cases of strains CFT073 and 1177, kanamycin-resistant tagged versions were constructed. The CFT073 fim::kan mutant was constructed by using the λRed recombinase gene replacement system. Briefly, the kanamycin gene from plasmid pKD4 was amplified by primers containing 50-nucleotide fim homology extensions (286, 5′-GTCGATTGAGGATTTCGGATATTGATCTTAAGGCAAAGTGGTGTAGGCTGGAGCTGCTTC; 287, 5′-GCTCCTAACGATACCGTGTTATTCGCTGGAATAATCGTACCATATGAATATCCTCCTTAG). This product was digested with DpnI and transformed into CFT073(pKD46), and kanamycin-resistant colonies were selected. The correct double-crossover and recombination event was confirmed by PCR and Southern blotting. The λRed helper plasmid pKD46 was cured by growth at 37°C, and the subsequent strain was designated PK1097. The 1177 fim::kan mutant (PK625) was constructed using the temperature-sensitive suicide plasmid as previously described (37). In short, a kan cassette flanked by DNA segments containing the upstream and downstream regions of the fim gene cluster, respectively, was used for homologous double-crossover recombination. Competition experiments between the kanamycin-resistant strains and their natural/nonresistant counterparts were performed to confirm that addition of the resistance gene had not altered the growth behaviors of the strains.
TABLE 1.
Strains used in this study
| E. coli strain | Relevant characteristics | Reference |
|---|---|---|
| 83972 | ABU isolate (OR:K5:H−) | 1 |
| MG1655 | K-12 reference strain | 3 |
| 536 | UPEC isolate (O6:K15:H31), Smr | 4 |
| CFT073 | UPEC isolate (O6:K2:H1) | 30 |
| PK1097 | CFT073, fim::kan | This study |
| NU14 | UPEC isolate (O18:K1:H7), Smr | 16 |
| 1177 | UPEC isolate (O1:K1:H7) | 28 |
| PK625 | 1177, fim::kan | This study |
Growth conditions for growth characteristics and competition experiments.
For each growth experiment, strains were grown in monocultures or in mixed cultures in LB or human urine collected from at least three healthy men and women volunteers who had no history of UTI or antibiotic use in the prior 2 months. The urine was pooled, filter sterilized, stored at 4°C, and used within the next 1 to 3 days. For measuring growth characteristics, monocultures were grown in 20 ml urine inoculated with an overnight urine culture to an optical density at 600 nm (OD600) of 0.05 and grown in a water bath incubator at 37°C and 130 rpm. The OD600 was measured every 30 min until cells reached stationary phase. Each experiment was performed in triplicate and repeated at least twice.
For competition experiments, mixed cultures were grown in 20 ml urine inoculated 1:1 with freshly grown precultures of E. coli 83972 and either 536, CFT073, NU14, or 1177 to an OD600 of 0.05. Samples from each culture were extracted at the start of the cultivation, diluted, and plated on LB plates with and without antibiotic to confirm the 1:1 ratio between E. coli 83972 and the UPEC strain in question. The cultures were grown at 37°C and 130 rpm, and samples were extracted after 17 h, diluted, and plated on both LB plates and LB plates complemented with the appropriate antibiotic. Each competition experiment was performed in duplicate or triplicate and repeated at least twice. To investigate growth competition in more detail, 83972 and NU14 were inoculated 1:1 to an OD600 of 0.05, in duplicate, and grown as described above, and samples were extracted every hour for measurement of the OD and viable counts.
To monitor the role of the starting ratio of the strains, E. coli 83972 and NU14 cultures were mixed 1:20 in 20 ml urine to an OD600 of 0.05. The starting ratio was confirmed by plating as described above; NU14 constituted 95% (0.95 ± 0.02, mean ± standard deviation) of the cultivation at the starting point. The cultures were grown and analyzed as described above.
In the case of strains CFT073 and 1177, competition experiments between the kanamycin-resistant versions and their natural/nonresistant counterparts were performed as described above. The competitions resulted in equal amounts of each strain.
Growth conditions and stabilization of RNA for microarray experiments.
Human urine was collected from four healthy men and women volunteers who had no history of UTI or antibiotic use in the prior 2 months. The urine was pooled, filter sterilized, stored at 4°C, and used the following day. Overnight cultures of E. coli 83972 were grown in pooled human urine or morpholinepropanesulfonic acid (MOPS) minimal medium supplemented with 0.2% glucose until reaching exponential phase and then used for inoculation of 50 ml urine or MOPS to an OD600 of 0.05. The cultures were grown at 37°C and 130 rpm, and 5-ml samples for isolation of RNA were extracted from three individual cultures at mid-exponential phase (corresponding to an OD600 of approximately 0.4 and 0.5 in urine and MOPS, respectively). Extracted samples were immediately mixed with two volumes of RNAprotect bacteria reagent (QIAGEN AG, Basel, Switzerland), incubated for 5 min at room temperature to stabilize RNA, and centrifuged. The pellets were then stored at −80°C.
RNA isolation and cDNA labeling.
Total RNA was isolated using the RNeasy Mini kit (QIAGEN AG). Eluted RNA samples were treated with DNase I and repurified using RNeasy Mini columns. The quality of the total RNA was examined by agarose gel electrophoresis and by measuring the absorbance at 260 and 280 nm. Purified RNA was precipitated with ethanol and stored at −80°C until further use. Conversion of RNA to cDNA and microarray analysis were performed according to GeneChip Expression Analysis technical manual 701023, rev. 4 (Affymetrix, Inc., Santa Clara, CA). Briefly, 10 μg of RNA was mixed with 750 ng of random hexamer primers (Invitrogen), denatured at 70°C for 10 min, and then allowed to anneal, and cDNA synthesis was performed using SuperScript II reverse transcriptase (Invitrogen). cDNA was purified using a MinElute PCR purification kit (QIAGEN) and fragmented with DNase I. DNase I was inactivated by heating at 98°C for 10 min, and the 3′ terminus of fragmented cDNA was labeled using the GeneChip DNA labeling reagent (Affymetrix).
DNA microarray hybridization.
GeneChip E. coli Genome 2.0 arrays (Affymetrix) were used for hybridization of the labeled cDNA. In total, six samples were hybridized to GeneChip E. coli Genome 2.0 arrays; three chips were hybridized with samples from E. coli 83972 grown in MOPS in triplicate, and three chips were hybridized with cells grown in pooled human urine in three individual flasks. Hybridization, washing, and staining were performed according to GeneChip Expression Analysis technical manual 701023, rev. 4 (Affymetrix), and the microarrays were scanned using the GeneChip Scanner 3000. The GeneChip E. coli genome array contains probe sets to detect transcripts from the K-12 strain of E. coli and three pathogenic strains of E. coli. The GeneChip E. coli Genome 2.0 array includes approximately 10,000 probe sets for all 20,366 genes present in the K-12 (MG1655), CFT073 (uropathogenic), O157:H7-EDL933 (enteropathogenic), and O157:H7-Sakai (enteropathogenic) strains. Due to the high degree of similarity between the E. coli strains, whenever possible a single probe set is tiled to represent the equivalent ortholog in all four strains.
Data analysis.
Array normalization and expression value calculations were performed using the DNA-Chip Analyzer (dChip) 1.3 software program (http://www.dchip.org/) (23). The invariant set normalization method (24) was used to normalize arrays at probe cell level to make them comparable, and the model-based (perfect match/mismatch) method was used for probe selection and computing expression values. These expression levels were attached with standard errors as measurement accuracy, which were subsequently used to compute 90% confidence intervals of changes in a two-group comparison. The three arrays hybridized with samples from E. coli 83972 grown in MOPS were used as the baseline for calculation of changes on the three arrays hybridized with samples from E. coli 83972 grown in urine. In total, 2,633 genes (26%) were filtered as significantly changed in urine compared with MOPS; 1,422 and 850 of these genes were specific for MG1655 and CFT073, respectively.
In dChip, estimation of the percentages of genes identified by chance, the empirical false discovery rate (FDR), can be performed by permutation. Permuting our samples randomly 1,000 times, using the same comparison criteria as when comparing urine- with MOPS-grown samples, resulted in an FDR of 0.3%, or seven false-positive genes.
RT-PCR.
Reverse transcription-PCR (RT-PCR) was performed to confirm DNA microarray gene expression data. Total RNA was isolated exactly as described above and treated with DNase I to remove any traces of DNA. RNA was converted to cDNA using SuperScript II (Invitrogen Life Technologies). cDNA was used directly as template for PCR, and a negative control on the RNA sample (not converted to cDNA) was run in parallel to confirm that all DNA had been removed in the earlier step. The total number of cycles used in PCR ranged from 12 to 30. RT-PCR products were examined by agarose gel electrophoresis. The following primers were used in RT-PCR and PCR: papA, 620 (5′-GTGAAGTTTGATGGGGCGACC) and 621 (5′-CGCAACTGCTGAGAAAGCACC); 16S, 622 (5′-CGGATTGGAGTCTGCAACTCG) and 623 (5′-CACAAAGTGGTAAGCGCCCTC).
Murine model of UTI.
A modification of the murine model of UTI was used for this study (12). Female CBA mice (8 to 10 weeks) were purchased from the Animal Resources Center, Western Australia, and housed in sterile cages with ad libitum access to sterile water. Urine was collected from each mouse 24 h prior to challenge and examined microscopically with a hemocytometer and by culture. Mice with a combination of >5 × 102 CFU of bacteria per ml of urine and >2 × 105 white blood cells in urine were defined as having a preexisting condition and were excluded from the study. Mice (n = 7) were anesthetized by brief inhalation exposure to isoflurane, and the periurethral area was sterilized by swabbing with 10% povidone-iodine solution, which was removed with sterile water. Mice were catheterized using a sterile Teflon catheter (0.28-mm internal diameter, 0.61-mm outer diameter, and 25-mm length; Terumo) by inserting the device directly into the bladder through the urethra. An inoculum of 25 μl, containing 109 CFU of bacteria in phosphate-buffered saline containing 0.1% India ink (for visualization of inocula at autopsy), was instilled directly into the bladder using a 1-ml tuberculin syringe attached to the catheter. The catheter was removed immediately after the challenge, and mice were returned to their cages. Urine was collected from each mouse at 24 h after inoculation for quantitative colony counts.
RESULTS
Growth characteristics of E. coli 83972.
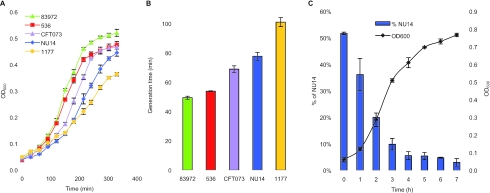
In an attempt to understand the bladder colonization properties of E. coli strain 83972 better, we initially focused on its growth characteristics in human urine. Under normal conditions, an adult human being produces 1 to 2 liters of urine per day, all of which pass through the bladder. Arguably, the ability to grow in human urine must be an important criterion for colonization of the bladder. The strain grows well in human urine in vitro and, depending on the individual batch of urine used, grows exponentially, with doubling times ranging from 45 min to 60 min, and it levels out in stationary phase with an OD600 of 0.5 to 0.9. The E. coli K-12 reference strain MG1655 is unable to grow in urine and cannot be used for comparison. However, in LB medium strain 83972 shows slightly better growth characteristics than the K-12 strain (data not shown). For comparison, we probed the growth characteristics of some well-characterized UPEC strains, viz. E. coli strains 536, CFT073, NU14, and 1177, all isolated from severe cases of urinary tract infections (Table 1). Surprisingly, all four UPEC strains performed significantly poorer than strain 83972 when grown in urine, with doubling times in the exponential phase 10 to 100% longer (two-sample two-tailed t test, P < 0.002), and they entered stationary phase at an OD600 10 to 30% lower than that of 83972 (P < 0.04) (Fig. 1). Accordingly, due to its faster doubling time and higher carrying capacity in urine, it is highly likely that strain 83972 should be able to outcompete these UPEC strains within a relatively modest number of generations.
FIG. 1.
(A) Growth of ABU E. coli 83972 and UPEC strains 536, CFT073, NU14, and 1177 in human urine. The curves are shown as means of triplicates, and error bars indicate standard deviations (σn−1). (B) The doubling time during exponential phase for each strain was calculated and is shown as the mean of triplicates. Bars indicate standard deviations. (C) Competition experiment between 83972 and NU14 mixed 1:1 at the starting point in human urine. Values are means of duplicates, and error bars indicate standard deviations.
E. coli 83972 competition with UPEC strains in human urine.
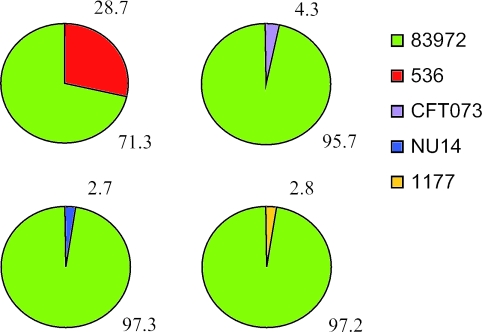
In order to monitor if the superior growth characteristics of strain 83972 were a general phenomenon, we competed it against our panel of well-characterized UPEC strains (Table 1). This was done by incubation of sterile human urine with equal amounts of strain 83972 and the UPEC strain in question and then monitoring the percentage of each strain after overnight cultivation by selective plating. In all cases, strain 83972 outcompeted the UPEC strain; even though the UPEC strain constituted 50% of the population at the start of the cultivation, it ended up constituting only 3% to 4% in the case of CFT073, NU14, and 1177 and 29% in the case of strain 536 (paired two-tailed t test, P < 0.001) (Fig. 2). Figure 1C, displaying the competition between 83972 and NU14 during the first 7 hours, reveals that E. coli 83972 constitutes about 80% of the population already after 2 h. To further investigate the degree of advantage by E. coli 83972, it was competed against NU14 with a starting ratio of 1 to 20; although NU14 constituted 95% of the population at the start of the experiment, it accounted for only 7% of the population after 17 h (paired two-tailed t test, P < 0.01). Taken together the data show that strain 83972 is able to outgrow a representative spectrum of UPEC strains.
FIG. 2.
Results from growth competition experiments (1:1) between E. coli 83972 and UPEC strains after 17 h of growth in human urine. Each value is an average of four individual shake flasks obtained from two separate experiments in different batches of urine. In all four competition experiments, strain 83972 was present in significantly higher numbers than the UPEC isolate (paired two-tailed t test, P < 0.001).
Global gene expression of E. coli 83972 in human urine.
The genomic expression profile of E. coli 83972 grown in pooled, filtered human urine was compared with the strain grown in MOPS-glucose medium. Samples were extracted during mid-exponential growth phase in order to best mimic the situation the cells encounter in the human bladder; in the bladder any colonizing organism will continuously be provided with fresh growth medium (i.e., urine), which most likely will keep the cells in growth phase and prevent them from reaching stationary phase. RNA isolated from exponentially growing cultures (in triplicate) was used for hybridization of microarrays, and the data were analyzed as described in Materials and Methods. For analysis, the GeneChip E. coli Genome 2.0 array was employed. This microarray contains ∼10,000 probe sets for all 20,366 genes present in E. coli strains MG1655, CFT073, EDL933, and O157:H7-Sakai.
The analysis of the microarray data is described in Materials and Methods. Whether a fold change in expression of a gene observed between two different arrays could be considered significant or not was not solely dependent on the magnitude of the change, but also on the absolute signal on the two arrays. A large up-regulation of a gene has to have a signal that is present on the sample array, but not on the baseline array, while in the case of a small up-regulation (low fold change, closer to 1.0), the signal has to be present and high on both the sample and the baseline array, and vice versa for down-regulated genes. Therefore, in our analysis we have only considered an up- or down-regulation to be significant if it meets these criteria, i.e., for a low fold change (1.4 to 1.9) to be considered significant the signal has to be present and high on all three baseline arrays as well as the three sample arrays. Moreover, the comparison criteria were carefully chosen to make sure that the empirical FDR was kept low (0.3%, i.e., seven false-positive genes). The estimation of FDR has become widely accepted as appropriate (10) and, furthermore, it has been argued that FDR is a more natural scale to work on rather than the P value (34).
Overall, 626 genes (10%) were expressed at significantly higher levels in human urine and 1,646 genes (25%) were significantly down-regulated during growth in urine, whereof 383 and 1,039 genes, respectively, were MG1655 transcripts and the remaining genes were CFT073 (uropathogenic) transcripts. Classification of the MG1655 genes into functional groups (41, 42) revealed that a large number of the genes involved in carbohydrate transport and metabolism, energy production and conversion, and inorganic ion transport and metabolism were significantly up-regulated in urine (Table 2). Some of the genes belonging to these groups can also be found among the genes displaying the highest fold changes in urine compared with minimal medium, particularly genes encoding proteins involved in iron transport and metabolism (Table 3).
TABLE 2.
Up- and down-regulated MG1655 genes in each functional group of E. coli 83972 grown in urine versus MOPS
| Functional group | No. (%) of genes up- or down-regulated, urine vs MOPS
|
Total no. of genes | |
|---|---|---|---|
| Up in urine | Down in urine | ||
| Amino acid transport and metabolism | 39 (13) | 81 (27) | 296 |
| Carbohydrate transport and metabolism | 64 (21) | 56 (19) | 298 |
| Cell cycle control, mitosis, and meiosis | 0 (0) | 14 (45) | 31 |
| Cell motility | 1 (1) | 9 (10) | 91 |
| Cell wall/membrane biogenesis | 9 (5) | 78 (39) | 200 |
| Coenzyme transport and metabolism | 4 (4) | 43 (40) | 108 |
| Defense mechanisms | 1 (3) | 12 (34) | 35 |
| Energy production and conversion | 38 (16) | 55 (23) | 240 |
| Function unknown | 28 (11) | 83 (34) | 247 |
| General function prediction only | 22 (8) | 78 (29) | 269 |
| Inorganic ion transport and metabolism | 26 (16) | 39 (24) | 160 |
| Intracellular trafficking and secretion | 2 (6) | 17 (47) | 36 |
| Lipid transport and metabolism | 5 (7) | 22 (31) | 71 |
| Nucleotide transport and metabolism | 10 (13) | 33 (42) | 78 |
| Posttranslational modification, protein turnover, chaperones | 13 (11) | 39 (33) | 119 |
| Replication, recombination, and repair | 3 (2) | 46 (28) | 165 |
| Secondary metabolite biosynthesis, transport, and catabolism | 9 (17) | 9 (17) | 54 |
| Signal transduction mechanisms | 11 (9) | 31 (27) | 116 |
| Transcription | 15 (6) | 54 (23) | 235 |
| Translation | 4 (3) | 49 (31) | 156 |
| Not in COGs | 79 (7) | 191 (18) | 1,065 |
| Total protein-coding genes | 383 (9) | 1039 (26) | 4,070 |
TABLE 3.
Top 25 up-regulated MG1655 and CFT073 genes in E. coli 83972 grown in urine compared with minimal lab medium
| Gene | b/c code | Function | Signal | Fold change |
|---|---|---|---|---|
| c1596 | c1596 | Hypothetical protein | 2,088 | 103.5 |
| iroB | c1254 | Putative glucosyltransferase | 4,953 | 76.4 |
| fepA | c0669 | Ferric enterobactin receptor precursor | 4,538 | 74.8 |
| yeiC | c2701 | Hypothetical sugar kinase yeiC | 4,443 | 55.8 |
| yeiN | b2165 | Hypothetical protein | 1,151 | 55.2 |
| fepA | b0584 | Outer membrane receptor for ferric enterobactin and colicins B and D | 5,347 | 50.8 |
| iucA | c3627 | IucA protein | 2,421 | 48.2 |
| ycdO | b1018 | Hypothetical protein | 3,077 | 46.8 |
| sitA | c1600 | SitA protein | 4,722 | 46.7 |
| chuX | c4315 | Hypothetical protein | 2,400 | 46.0 |
| araD | b0061 | l-Ribulose-5-phosphate 4-epimerase | 2,969 | 45.8 |
| araB | b0063 | l-Ribulokinase | 3,571 | 45.6 |
| iucD | c3624 | lucD protein | 4,015 | 45.2 |
| b1498 | b1498 | Putative sulfatase | 2,883 | 44.5 |
| entC | b0593 | Isochorismate synthase, enterobactin biosynthesis | 2,848 | 44.4 |
| uidB | b1616 | Glucuronide permease | 2,379 | 44.2 |
| b2737 | b2737 | Hypothetical protein | 4,792 | 42.7 |
| sitB | c1599 | SitB protein | 5,082 | 41.3 |
| araE | b2841 | Low-affinity l-arabinose transport system, proton symport protein | 2,921 | 40.0 |
| b2736 | b2736 | Putative dehydrogenase | 4,616 | 38.6 |
| iutA | c3623 | IutA protein | 3,446 | 38.4 |
| ryhB | b4451 | Regulatory RNA-mediating Fur regulon | 4,481 | 38.3 |
| iroN | c1250 | Siderophore receptor IroN | 4,640 | 37.2 |
| srlB | b2704 | PTS family enzyme IIA, glucitol/sorbitol specific | 1,643 | 33.8 |
| c4310 | c4310 | Hypothetical protein | 4,530 | 32.1 |
Iron acquisition genes.
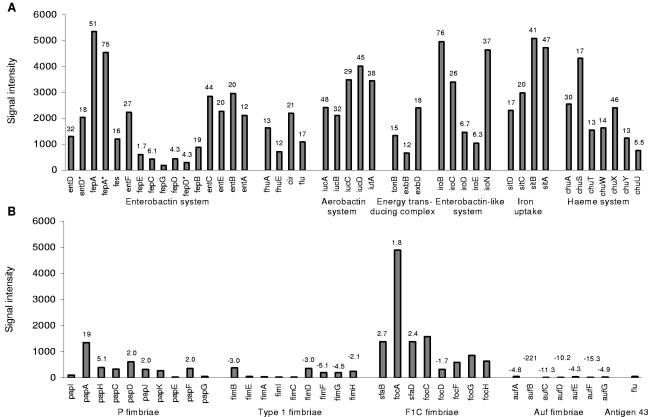
The concentration of soluble forms of iron is very low in the urinary tract and is a growth-limiting factor for bacteria. Consequently, iron acquisition plays an important role in bacterial survival and pathogenicity. In strain 83972 many genes involved in iron uptake and transport were significantly up-regulated in human urine (Fig. 3A). Transcription from genes encoding proteins involved in enterobactin synthesis, entBCDEF, were up-regulated 20- to 44-fold, and the gene encoding the highly specific ferric enterobactin receptor, fepA, revealed one of the highest signals and was up-regulated 51-fold. Genes encoding FhuA, FhuE, and IutA, which transport hydroxamate siderophores, were induced 13-, 12-, and 38-fold, and genes encoding Fiu and Cir, which transport catecholate siderophores, were induced 17- and 21-fold. The transport of iron across the outer membrane is dependent upon the energy-transducing TonB-ExbB-ExbD complex; these genes were up-regulated between 12- and 18-fold in urine.
FIG. 3.
Expression levels of iron acquisition systems (A) and adhesins (B) in E. coli 83972 grown in urine. The numbers indicate fold changes of expression levels for genes significantly changed in urine compared with MOPS. Asterisks indicate CFT073 transcripts in cases where there are two genes with the same name.
Genes encoding proteins responsible for iron uptake without siderophores, sitABCD, were significantly up-regulated (17- to 47-fold); sitB showed the 10th highest signal in urine. It has been demonstrated that the sit cluster is required for full virulence of Salmonella enterica serovar Typhimurium (17). Other virulence-associated genes, iutA and iucABCD, responsible for aerobactin uptake and synthesis, were up-regulated 29- to 48-fold in urine. The iroN gene, encoding a siderophore receptor that contributes to urovirulence (35), was highly up-regulated in urine (37-fold), as were the rest of the genes in the iro cluster (iroBCDE were induced 6.3- to 76-fold). The genes encoding the Chu heme transport system, chuASTWXYU, were up-regulated 5.5- to 46-fold; chuA has been shown to be important for the virulence of a uropathogenic E. coli strain (43). The small regulatory antisense RNA (ryhB) involved in iron homeostasis was up-regulated 38-fold. RyhB down-regulates the mRNA levels for the genes that are known to be positively regulated by Fur (ferric uptake regulator) (29). The fur gene itself showed no change in expression in urine compared with MOPS. Taken together, the data indicate that strain 83972 has numerous genes encoding products that are well suited to cope with an iron-poor environment and that these are highly expressed during growth in urine.
Carbohydrate transport and metabolism.
The ara cluster belonged to one of the most up-regulated in urine (Table 3). The genes araABD, which are responsible for the conversion of arabinose to xylulose-5-phosphate, were up-regulated 32-, 46-, and 46-fold, respectively. The genes encoding arabinose transporters, both the arabinose/proton symporter gene araE and the ABC transporter encoding genes araFGH, were up-regulated (40-, 14-, 17-, and 25-fold, respectively).
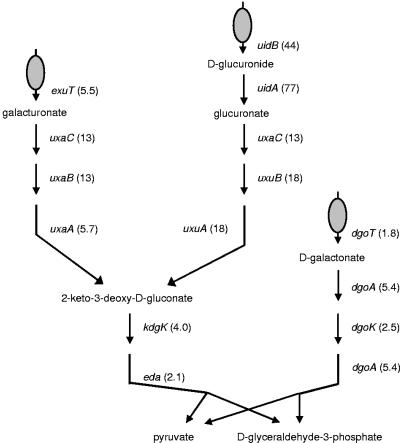
The degradation of sugar acids was highly up-regulated in urine. All the genes involved in the degradation pathways of galacturonate, glucuronate, and galactonate were significantly up-regulated up to 77-fold, and the genes encoding transporters of these compounds were up-regulated up to 44-fold (Fig. 4). The fructuronic acid transporter-encoding gene, gntP, which is divergently transcribed from uxuAB, was up-regulated 4.2-fold. The gene encoding N-acetylneuraminate lyase, nanA, which catalyzes the breakdown of sialic acid, was up-regulated 19-fold. Furthermore, many genes involved in the transport of carbohydrates were up-regulated in urine compared with minimal medium; genes encoding proteins responsible for transport of sorbitol (srlAB), galactose (mglABC), maltose (lamB), xylose (xylFGH), and mannose/fructose (manXYZ) were all significantly up-regulated 2.4- to 34-fold.
FIG. 4.
Transport and degradation pathways of the sugar acids galacturonate, glucuronide, and galactonate. The numbers indicate significant fold changes of expression levels for genes in E. coli 83972 grown in urine compared with the same cells grown in MOPS.
Adhesins.
E. coli 83972 harbors a nonfunctional pap gene cluster, encoding P fimbriae, and a truncated fim gene cluster, encoding type 1 fimbriae (19). Our microarray data revealed that half of the pap genes were significantly up-regulated in urine: papHDJF were up-regulated 2- to 5-fold, and the gene encoding the major subunit, papA, was up-regulated 19-fold (Fig. 3B). While the signals for fimEAIC could not be detected in E. coli 83972 due to a large deletion in the fim cluster, the remaining fim genes, fimBDFGH, displayed low signals and were all down-regulated in urine.
The foc/sfa cluster encoding F1C fimbriae was partly up-regulated in E. coli 83972, with focA, encoding the major subunit, up-regulated 1.8-fold in human urine. Other fimbrial genes significantly changed in urine belonged to the recently characterized auf cluster (6), for which genes were down-regulated 4.3- to 221-fold. Furthermore, the hypothetical fimbrial-like protein precursors yadN and ygiL were down-regulated 4.3- and 115-fold, respectively, and two genes encoding putative adhesins, eaeH and c4424, were down-regulated 12- and 3.8-fold. Expression from the flu gene encoding aggregation factor Ag43 was very low and showed no significant change when grown in urine.
Other virulence factors.
The pore-forming hemolysin (HlyA) is considered an important virulence factor in E. coli extraintestinal infections, such as those of the upper urinary tract. The genes determining the synthesis, activation, and transport of hemolysin, hlyCABD, were all down-regulated in urine (106-, 9.2-, 2.8-, and 90-fold, respectively). This was confirmed by lack of hemolysis on blood agar plates (data not shown). Of the genes encoding proteins involved in biosynthesis of lipopolysaccharides, most were significantly down-regulated in urine, the genes involved in the enterobacterial common antigen biosynthesis pathway (rffACDGHMT, wecBF, and rfe) were all down-regulated 2.0- to 6.3-fold, the genes involved in colonic acid biosynthesis (cpsB, gmd, fcl, and ugd) were down-regulated 4.0- to 26-fold, and the genes involved in the synthesis of lipid A (lpxBHKP, kdsABC, htrB, and kdtA) were down-regulated 1.6- to 6.0-fold. The rcsA gene, encoding a positive regulator of capsular polysaccharide synthesis, was down-regulated 16-fold in urine. The genes kpsEDCS, involved in the transport of polysaccharide to the cell surface, were all down-regulated 2.5- to 6.5-fold. The rfaH gene is another virulence-associated gene which displayed very low signals and significant down-regulation in urine (3.7-fold). RfaH is a global regulator which modifies expression of several virulence factors, and disruption of the rfaH gene in uropathogenic E. coli has been shown to result in a significant decrease in virulence (32).
Taken together the array data show that E. coli 83972 expresses all known iron transport and uptake systems when grown in urine; the strain also showed high expression levels of genes involved in uptake and metabolism of nutrients found in urine, while most of the genes encoding known virulence factors were down-regulated in urine. It is obvious that the strategy to keep and express growth-enhancing genes while shutting off virulence genes that may provoke the host response must be an optimal approach for successful long-term colonization of the urinary tract.
Verification of microarray results.
RT-PCR was performed to verify the transcript levels for an example gene, papA. papA was significantly up-regulated 19-fold in urine and showed very low signals in MOPS. papA could not be detected in the samples from MOPS, not even after 30 cycles of PCR, while papA was detected in all three urine samples, visualized as strong bands on an agarose gel. 16S was used as a normalizing internal standard and was detected with the same intensity in all samples.
The expression levels of the fim genes revealed the sensitivity of the microarrays. Sequencing of the fim cluster of E. coli 83972 revealed that a large part of the cluster is deleted in the strain, i.e., a 4.25-kb deletion between fimB and fimD, resulting in complete absence of fimEAIC, leaving only fimF, fimG, and fimH unaffected (19). The signal from fimEAIC was very low and varied between 7 and 53, with an average of 27 and 35 for MOPS and urine, respectively. The signals of fimBDFGH varied between 136 and 1,209 with an average of 911 and 275 for MOPS and urine, respectively. The signal levels of the different fim genes on the arrays correspond well to the actual presence of fim genes in E. coli 83972.
E. coli 83972 competition with UPEC strain NU14 in mouse bladder.
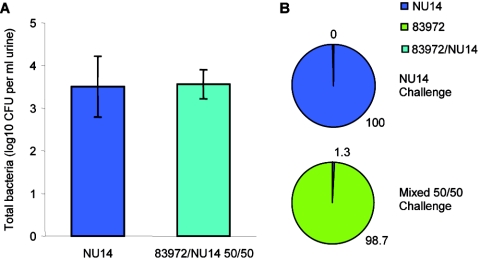
Our growth studies combined with the global gene expression data demonstrate that E. coli 83972 is highly adapted to growth in human urine. Since other studies have demonstrated a prophylactic effect of this strain in the presence of other pathogens, we competed 83972 against UPEC NU14 in the mouse UTI model. Mice were infected with equal numbers of NU14 (alone) or NU14 and 83972 (in combination, 50% mixed infection of each). After 24 h, the total number of bacteria in the urine of mice infected with NU14 or an equal combination of NU14 and 83972 was almost identical (Fig. 5). However, when examining the percentage of each individual strain in the mice receiving the mixed combination of strains, we observed that 83972 was almost totally dominant (98.7% versus 1.3%; paired two-tailed t test, P < 0.001). Thus, 83972 is able to outcompete UPEC in an in vivo UTI infection model in addition to in vitro growth experiments in human urine.
FIG. 5.
Results from mice (n = 7) challenged for 24 h with the UPEC isolate NU14 or E. coli 83972 and NU14 (1:1). (A) Mean total number of CFU per ml of urine ± the standard error of the mean; (B) percentages of NU14 and ABU 83972 calculated from differential colony counts. Total colony counts were not influenced by the presence of 83972. E. coli 83972 was present in significantly higher numbers in urine from mice compared with NU14 (98.7% versus 1.3%; paired two-tailed t test, P < 0.001).
DISCUSSION
Healthy adult humans normally produce 1 to 2 liters of urine per day, which must pass through the bladder, corresponding to average flow rates of 40 to 80 ml per hour. An adult human bladder has a volume of 200 to 400 ml, and micturition causes the release of roughly the same volume of urine; the volume of urine remaining in the bladder following micturition is ∼1 ml (38). The contribution of bladder hydrodynamics on the elimination of bacteria has been recognized for several decades (5, 27), and it has been suggested that without adhesin-assisted attachment to the bladder surface, E. coli would not be able to overcome the losses caused by micturition and, therefore, it would be unable to establish in the urinary tract (2, 40). Implicit in this suggestion is the notion that the growth rate of E. coli in urine is too slow to cope with the losses incurred by micturition. Meanwhile, mathematical modeling suggests that if the growth rate of a strain is high enough it will be able to establish in the bladder in an adhesion-independent manner. Indeed, a theoretical analysis of bacterial growth in the bladder suggested that many urinary tract isolates of E. coli had growth rates that were fully compatible with nonadhesive establishment in the bladder (11). So far, no studies have been published that show that a high growth rate of a UPEC strain can permit colonization of the urinary tract independent of specific adhesion. Recently, Jarboe et al. predicted that pap expression was inversely related to growth rate, indicating that at high growth rates adherence may be unnecessary for persistence, and so a decrease in fimbrial expression at high growth rates potentially conserves cellular resources without decreasing the probability of survival (18). A study on the UPEC isolate NU14 revealed that antibiotic resistance mutations led to significant reduction of biological fitness (defined as decrease in growth rate), providing a possible explanation for the observed negative correlation between fluoroquinolone resistance and bacterial virulence; the study revealed that the loss of fitness could be partially reversed by later mutations (26).
E. coli strain 83972 was recently demonstrated to be unable to express functional versions of type 1 and P fimbriae, the two fimbriae types that are recognized as being the primary adhesins of UTI E. coli strains (19). Furthermore, the strain has never been reported to adhere to any kind of cells originating from the human urinary tract (1, 15, 47). The lack of adhesion could to a large degree account for the inability to cause symptoms in the human host. However, it raises the issue of how the strain is capable of staying in the bladder and how it seems to be able to prevent bladder colonization by pathogenic E. coli. The results presented herein indicate that strain 83972 grows very well in human urine in vitro, with doubling times generally being 45 to 60 min. In some experiments, even shorter doubling times were observed. Indeed, a comparison of the doubling time of the 83972 strain with those of a spectrum of well-characterized UPEC strains showed that it grows faster than any of them. Also, it grows to higher maximum cell densities and exhibits a shorter lag phase than any of the UPEC strains investigated. This enhanced growth capacity in urine was not a unique property of 83972, since other ABU isolates from our laboratory strain collection exhibited similar growth characteristics (data not shown).
When E. coli 83972 was pitted against the UPEC strains in pairwise competition experiments, it outcompeted all of them to a significant degree, i.e., the ABU strain constituted 71 to 97% of the population after overnight growth. Mathematical modeling of the competition experiments, using the observed doubling times (Fig. 1) and assuming exponential growth, resulted in numbers closely resembling the values obtained experimentally (Fig. 2). These results were confirmed in mouse experiments; the 83972 strain was almost totally dominant in the urine of mice 24 h after receiving a mixed challenge consisting of 83972 and the UPEC isolate NU14 (1:1). E. coli 83972 can establish long-term bacteriuria in humans (1, 14, 47, 49) and has been used extensively for treatment of patients with recurrent UTI to avoid the establishment of disease-causing strains (8, 14, 44, 45, 49). Based on the results presented here, we suggest that the ability of E. coli 83972 to establish in the human bladder as well as its prophylactic properties against UPEC strains is first and foremost due to its excellent growth properties in human urine. In theory, strain 83972 could also compete with other strains by killing them. However, we have tested the strain for colicin production and it proved negative in this faculty.
According to the literature, E. coli 83972 had grown in the bladder of a girl with ABU for at least 3 years (1). This represents a substantial number of generations, i.e., more than 30,000, assuming generation times comparable to those observed in the present study. During this period of time the strain must have adapted considerably to this particular environmental niche. The strain lost the ability to express functional type 1 and P fimbriae probably as an evolutionary trade-off with the host defense. This ensured that it did not attract the attention of aggressive host defense mechanisms, such as cytokine production, inflammation, and exfoliation of infected bladder cells. However, in order to avoid being flushed out of the system, it had to adapt to the growth medium, i.e., human urine, and to optimize its growth rate to keep pace with the flow rate in the bladder. Whether this happened during the three-plus years it was carried by the particular girl or before then in other hosts is not possible to conclude. It is, however, interesting that the girl suffered from a voiding problem and was unable to empty her bladder completely, thus leaving a considerable volume of residual urine (1, 25). Arguably, this would provide the 83972 strain with an ideal “training” environment for optimizing its ability to grow in urine.
In a previous study on bacterial growth adaptation to a specific growth medium, it was shown that after 10,000 generations E. coli strains had increased in fitness by ∼50% for growth on glucose medium (22). In line with this notion, strain 83972 must have accrued genetic changes that have favored its fitness for growth in urine. Human urine is a very complex growth medium, and the composition of urine fluctuates daily and varies with the person and diet. It is, however, known that iron availability is a limiting factor. Our array data indicated that strain 83972 has adapted well to growth in this iron-limiting environment, significantly increasing the expression of the majority of all known genes involved in iron uptake and transport when grown in urine compared with minimal medium. Furthermore, the array data revealed high expression levels of genes involved in transportation and degradation pathways of sugar acids and carbohydrates, indicating how E. coli 83972 is able to reach high growth rates in human urine by efficiently utilizing the nutrients available in this growth medium. It should be noted, however, that even though the E. coli arrays employed in this study include transcripts from the uropathogenic strain CFT073, the whole genome of E. coli 83972 has not yet been sequenced and characterized and, therefore, the possibility that strain 83972 carries unique genes enabling a faster growth in urine cannot be ruled out. It remains to be seen whether the mechanisms utilized by E. coli 83972 are superior to those of UPEC strains or whether this strain has evolved additional, yet-undescribed mechanisms for iron sequestration and growth. We are currently examining these possibilities further.
Acknowledgments
We thank Birthe Jul Jørgensen for expert technical assistance and Hugh Connell for helpful discussions regarding the mouse UTI model.
This work was supported by grants from the Danish Medical Research Council (22-03-0462), the Danish Research Agency (2052-03-0013), the Australian National Health and Medical Research Council (301163 and 401714), and the University of Queensland.
Editor: J. B. Bliska
REFERENCES
- 1.Andersson, P., I. Engberg, G. Lidin-Janson, K. Lincoln, R. Hull, S. Hull, and C. Svanborg. 1991. Persistence of Escherichia coli bacteriuria is not determined by bacterial adherence. Infect. Immun. 59:2915-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur, M., C. E. Johnson, R. H. Rubin, R. D. Arbeit, C. Campanelli, C. Kim, S. Steinbach, M. Agarwal, R. Wilkinson, and R. Goldstein. 1989. Molecular epidemiology of adhesin and hemolysin virulence factors among uropathogenic Escherichia coli. Infect. Immun. 57:303-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachmann, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 4.Berger, H., J. Hacker, A. Juarez, C. Hughes, and W. Goebel. 1982. Cloning of the chromosomal determinants encoding hemolysin production and mannose-resistant hemagglutination in Escherichia coli. J. Bacteriol. 152:1241-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boen, J. R., and D. L. Sylwester. 1965. The mathematical relationship among urinary frequency, residual urine, and bacterial growth in bladder infections. Investig. Urol. 15:468-473. [PubMed] [Google Scholar]
- 6.Buckles, E. L., F. K. Bahrani-Mougeot, A. Molina, C. V. Lockatell, D. E. Johnson, C. B. Drachenberg, V. Burland, F. R. Blattner, and M. S. Donnenberg. 2004. Identification and characterization of a novel uropathogenic Escherichia coli-associated fimbrial gene cluster. Infect. Immun. 72:3890-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connell, H., W. Agace, P. Klemm, M. Schembri, S. Marild, and C. Svanborg. 1996. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc. Natl. Acad. Sci. USA 93:9827-9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darouiche, R. O., W. H. Donovan, M. Del Terzo, J. I. Thornby, D. C. Rudy, and R. A. Hull. 2001. Pilot trial of bacterial interference for preventing urinary tract infection. Urology 58:339-344. [DOI] [PubMed] [Google Scholar]
- 9.Foxman, B. 2002. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am. J. Med. 113(Suppl. 1A):5S-13S. [DOI] [PubMed] [Google Scholar]
- 10.Ge, Y., S. Dudoit, and T. P. Speed. 2003. Resampling-based multiple testing for microarray data analysis. Test 12:1-44. [Google Scholar]
- 11.Gordon, D. M., and M. A. Riley. 1992. A theoretical and experimental analysis of bacterial growth in the bladder. Mol. Microbiol. 6:555-562. [DOI] [PubMed] [Google Scholar]
- 12.Hagberg, L., I. Engberg, R. Freter, J. Lam, S. Olling, and C. Svanborg-Eden. 1983. Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect. Immun. 40:273-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hedlund, M., R. D. Duan, A. Nilsson, M. Svensson, D. Karpman, and C. Svanborg. 2001. Fimbriae, transmembrane signaling, and cell activation. J. Infect. Dis. 183(Suppl. 1):S47-S50. [DOI] [PubMed] [Google Scholar]
- 14.Hull, R., D. Rudy, W. Donovan, C. Svanborg, I. Wieser, C. Stewart, and R. Darouiche. 2000. Urinary tract infection prophylaxis using Escherichia coli 83972 in spinal cord injured patients. J. Urol. 163:872-877. [PubMed] [Google Scholar]
- 15.Hull, R. A., D. C. Rudy, W. H. Donovan, I. E. Wieser, C. Stewart, and R. O. Darouiche. 1999. Virulence properties of Escherichia coli 83972, a prototype strain associated with asymptomatic bacteriuria. Infect. Immun. 67:429-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hultgren, S. J., W. R. Schwan, A. J. Schaeffer, and J. L. Duncan. 1986. Regulation of production of type 1 pili among urinary tract isolates of Escherichia coli. Infect. Immun. 54:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janakiraman, A., and J. M. Slauch. 2000. The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol. Microbiol. 35:1146-1155. [DOI] [PubMed] [Google Scholar]
- 18.Jarboe, L. R., D. Beckwith, and J. C. Liao. 2004. Stochastic modeling of the phase-variable pap operon regulation in uropathogenic Escherichia coli. Biotechnol. Bioeng. 88:189-203. [DOI] [PubMed] [Google Scholar]
- 19.Klemm, P., V. Roos, G. C. Ulett, C. Svanborg, and M. A. Schembri. 2005. Molecular characterization of the Escherichia coli asymptomatic bacteriuria strain 83972: the taming of a pathogen. Infect. Immun. 74:781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klemm, P., and M. A. Schembri. 2000. Bacterial adhesins: function and structure. Int. J. Med. Microbiol. 290:27-35. [DOI] [PubMed] [Google Scholar]
- 21.Leffler, H., and C. Svanborg-Eden. 1981. Glycolipid receptors for uropathogenic Escherichia coli on human erythrocytes and uroepithelial cells. Infect. Immun. 34:920-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenski, R. E., J. A. Mongold, P. D. Sniegowski, M. Travisano, F. Vasi, P. J. Gerrish, and T. M. Schmidt. 1998. Evolution of competitive fitness in experimental populations of E. coli: what makes one genotype a better competitor than another? Antonie Leeuwenhoek 73:35-47. [DOI] [PubMed] [Google Scholar]
- 23.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2:Research0032.1-0032.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindberg, U., L. A. Hanson, U. Jodal, G. Lidin-Janson, K. Lincoln, and S. Olling. 1975. Asymptomatic bacteriuria in schoolgirls. II. Differences in Escherichia coli causing asymptomatic bacteriuria. Acta Paediatr. Scand. 64:432-436. [DOI] [PubMed] [Google Scholar]
- 26.Lindgren, P. K., L. L. Marcusson, D. Sandvang, N. Frimodt-Moller, and D. Hughes. 2005. Biological cost of single and multiple norfloxacin resistance mutations in Escherichia coli implicated in urinary tract infections. Antimicrob. Agents Chemother. 49:2343-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mackintosh, I. P., B. J. Hammond, B. W. Watson, and F. O'Grady. 1975. Theory of hydrokinetic clearance of bacteria from the urinary bladder. I. Effect of variations in bacterial growth rate. Investig. Urol. 12:468-472. [PubMed] [Google Scholar]
- 28.Marild, S., U. Jodal, I. Orskov, F. Orskov, and C. Svanborg-Eden. 1989. Special virulence of the Escherichia coli O1:K1:H7 clone in acute pyelonephritis. J. Pediatr. 115:40-45. [DOI] [PubMed] [Google Scholar]
- 29.Massé, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mobley, H. L. T., D. M. Green, A. L. Trifillis, D. E. Johnson, G. R. Chippendale, C. V. Lockatell, B. D. Jones, and J. W. Warren. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58:1281-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulvey, M. A., Y. S. Lopez-Boado, C. L. Wilson, R. Roth, W. C. Parks, J. Heuser, and S. J. Hultgren. 1998. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282:1494-1497. [DOI] [PubMed] [Google Scholar]
- 32.Nagy, G., U. Dobrindt, G. Schneider, A. S. Khan, J. Hacker, and L. Emody. 2002. Loss of regulatory protein RfaH attenuates virulence of uropathogenic Escherichia coli. Infect. Immun. 70:4406-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oelschlaeger, T. A., U. Dobrindt, and J. Hacker. 2002. Virulence factors of uropathogens. Curr. Opin. Urol. 12:33-38. [DOI] [PubMed] [Google Scholar]
- 34.Pawitan, Y., S. Michiels, S. Koscielny, A. Gusnanto, and A. Ploner. 2005. False discovery rate, sensitivity and sample size for microarray studies. Bioinformatics 21:3017-3024. [DOI] [PubMed] [Google Scholar]
- 35.Russo, T. A., C. D. McFadden, U. B. Carlino-MacDonald, J. M. Beanan, T. J. Barnard, and J. R. Johnson. 2002. IroN functions as a siderophore receptor and is a urovirulence factor in an extraintestinal pathogenic isolate of Escherichia coli. Infect. Immun. 70:7156-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samuelsson, P., L. Hang, B. Wullt, H. Irjala, and C. Svanborg. 2004. Toll-like receptor 4 expression and cytokine responses in the human urinary tract mucosa. Infect. Immun. 72:3179-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schembri, M. A., L. Pallesen, H. Connell, D. L. Hasty, and P. Klemm. 1996. Linker insertion analysis of the FimH adhesin of type 1 fimbriae in an Escherichia coli fimH-null background. FEMS Microbiol. Lett. 137:257-263. [DOI] [PubMed] [Google Scholar]
- 38.Shand, D. G., J. C. MacKenzie, W. R. Cattell, and J. Cato. 1968. Estimation of residual urine volume with 131 I-hippuran. Br. J. Urol. 40:196-201. [DOI] [PubMed] [Google Scholar]
- 39.Svanborg, C., and G. Godaly. 1997. Bacterial virulence in urinary tract infection. Infect. Dis. Clin. North Am. 11:513-529. [DOI] [PubMed] [Google Scholar]
- 40.Svanborg-Eden, C., B. Eriksson, L. A. Hanson, U. Jodal, B. Kaijser, G. L. Janson, U. Lindberg, and S. Olling. 1978. Adhesion to normal human uroepithelial cells of Escherichia coli from children with various forms of urinary tract infection. J. Pediatr. 93:398-403. [DOI] [PubMed] [Google Scholar]
- 41.Tatusov, R. L., N. D. Fedorova, J. D. Jackson, A. R. Jacobs, B. Kiryutin, E. V. Koonin, D. M. Krylov, R. Mazumder, S. L. Mekhedov, A. N. Nikolskaya, B. S. Rao, S. Smirnov, A. V. Sverdlov, S. Vasudevan, Y. I. Wolf, J. J. Yin, and D. A. Natale. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 43.Torres, A. G., P. Redford, R. A. Welch, and S. M. Payne. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 69:6179-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trautner, B. W., R. O. Darouiche, R. A. Hull, S. Hull, and J. I. Thornby. 2002. Pre-inoculation of urinary catheters with Escherichia coli 83972 inhibits catheter colonization by Enterococcus faecalis. J. Urol. 167:375-379. [PMC free article] [PubMed] [Google Scholar]
- 45.Trautner, B. W., R. A. Hull, and R. O. Darouiche. 2003. Escherichia coli 83972 inhibits catheter adherence by a broad spectrum of uropathogens. Urology 61:1059-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, X. R., T. T. Sun, and J. J. Medina. 1996. In vitro binding of type 1-fimbriated Escherichia coli to uroplakins Ia and Ib: relation to urinary tract infections. Proc. Natl. Acad. Sci. USA 93:9630-9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wullt, B., G. Bergsten, H. Connell, P. Rollano, N. Gebretsadik, R. Hull, and C. Svanborg. 2000. P fimbriae enhance the early establishment of Escherichia coli in the human urinary tract. Mol. Microbiol. 38:456-464. [DOI] [PubMed] [Google Scholar]
- 48.Wullt, B., G. Bergsten, H. Fischer, G. Godaly, D. Karpman, I. Leijonhufvud, A. C. Lundstedt, P. Samuelsson, M. Samuelsson, M. L. Svensson, and C. Svanborg. 2003. The host response to urinary tract infection. Infect. Dis. Clin. North Am. 17:279-301. [DOI] [PubMed] [Google Scholar]
- 49.Wullt, B., H. Connell, P. Rollano, W. Mansson, S. Colleen, and C. Svanborg. 1998. Urodynamic factors influence the duration of Escherichia coli bacteriuria in deliberately colonized cases. J. Urol. 159:2057-2062. [DOI] [PubMed] [Google Scholar]