Abstract
In Staphylococcus aureus strains of human origin, phages which integrate into the chromosomal gene coding for β-hemolysin (hlb) are widely distributed. Most of them encode accessory virulence determinants such as staphylokinase (sak) or enterotoxins. Here, we analyzed the effects of ciprofloxacin and trimethoprim on phage induction and expression of phage-encoded virulence factors by using isolates from patients with cystic fibrosis for which the induction of hlb-converting phages was demonstrated in vivo (C. Goerke, S. Matias y Papenberg, S. Dasbach, K. Dietz, R. Ziebach, B. C. Kahl, and C. Wolz, J. Infect. Dis. 189:724-734, 2004) as well as a φ13 lysogen of phage-cured strain 8325-4. Treatment of lysogens with subinhibitory concentrations of either antibiotic resulted in (i) delysogenization of strains resembling the isolates picked up after chronic lung infection and (ii) replication of phages in the bacterial host in a dose-dependent manner. Ciprofloxacin treatment resulted in enhanced recA transcription, indicating involvement of the SOS response in phage mobilization. Induction of φ13 was linked to elevated expression of the phage-encoded virulence gene sak, chiefly due to the activation of latent phage promoters. In summary, we could show the induction of hlb-converting phages and a subsequent virulence modulation of the host bacterium by ciprofloxacin and trimethoprim.
Staphylococcus aureus asymptomatically colonizes the anterior nares of humans but also causes a wide spectrum of acute and chronic diseases. The ability of S. aureus to cause such a wide variety of diseases is probably due to the fact that it produces a large number of cell-bound and secreted virulence factors (15). Much of the dissimilarity between S. aureus strains is dependent on the presence of mobile genetic elements, such as plasmids, bacteriophages, pathogenicity islands, transposons, and insertions sequences (2, 9, 13). Many virulence factors are encoded on such mobile elements (3, 5, 12, 18, 19, 26). Phages which integrate into the chromosomal gene coding for β-hemolysin (hlb) are widely distributed in S. aureus strains of human origin. Most of them encode accessory virulence determinants such as staphylokinase (sak) and enterotoxins (5, 10). Staphylokinase is a plasminogen activator and has thus been postulated to aid the bacteria in their dissemination from clots and abscesses (1). Additionally, it was shown that S. aureus can resist human defensins by the production of staphylokinase (11).
Many prophages are induced by environmental conditions that lead to DNA damage, including exposure to reactive oxygen species generated by leukocytes or exposure to exogenous agents such as antibiotics (24). We could show that phages are mobilized during chronic infection of the lungs of patients with cystic fibrosis (CF), possibly due to the strong selective pressure exercised on the pathogen by the specific host response and/or by the regular exposure to antibiotics (8). Quinolones (for instance, ciprofloxacin) or trimethoprim are frequently used for the treatment of lung infections in patients with CF. Ciprofloxacin acts as an antibacterial agent by trapping DNA gyrase on DNA and thus blocking the replication fork movement. Trimethoprim prevents the incorporation of thymine into bacterial DNA by inhibiting the dihydrofolate reductase. Blockage of the replication fork by these drugs may trigger the DNA repair system. For Escherichia coli it was shown that damaged bacterial DNA interacts with and activates the multifunctional RecA protein (25). Stimulated RecA promotes the autoproteolysis of the repressor of DNA repair functions, LexA, and also that of the phage repressor CI (20, 25). The decline in active CI levels permits derepression of phage lytic genes and the resumption of lytic growth.
We analyzed the effects of subinhibitory concentrations of ciprofloxacin and trimethoprim on phage induction and the expression of phage-related virulence factors in comparison to those of the classical phage-inducing agent mitomycin C in S. aureus. CF isolates for which induction of hlb-converting phages has been demonstrated in vivo (8) as well as a phage φ13 lysogen of prototypic S. aureus strain 8325-4 were selected for this study. We showed that ciprofloxacin and trimethoprim trigger phage induction and lead to enhanced expression of phage-encoded virulence factors.
MATERIALS AND METHODS
Bacterial strains.
S. aureus isolates from CF patients were described in a previous study (8). Five CF isolates for which induction of an hlb-converting phage was demonstrated in vivo were selected. S. aureus strain 8325-4φ13 is a φ13 lysogen of phage-cured strain 8325-4. Phage φ13 lysate was obtained from strain RN25 (NARSA strain collection) by treatment with mitomycin C and spotted on 8325-4. Colonies located in the center of the plaque were picked and spread on sheep blood agar to screen for negative conversion of Hlb production. Potential lysogens were verified by pulsed-field gel electrophoresis (PFGE) fingerprinting and multiplex PCR. S. aureus phage-sensitive strain R5 was a kind gift from W. van Wamel, Utrecht, The Netherlands, and was used as an indicator strain for spot and plaque assays.
Determination of MICs.
The MIC of ciprofloxacin for the CF isolates was determined by using the Etest, as described by the manufacturer (AB-Biodisk, Solna, Sweden). The MICs of ciprofloxacin (Bayer, Leverkusen, Germany) and trimethoprim (Synopharm, Barsbüttel, Germany) of strain 8325-4 are published at www.narsa.net. MIC determination was additionally performed by the standard growth microdilution method with cation-adjusted Mueller-Hinton broth or Trypticase soy broth (TSB) with a bacterial inoculum of 5 × 105 CFU/ml. In general, Etest and the standard growth microdilution method in Mueller-Hinton broth yielded equivalent results, but incubation in TSB resulted in slightly higher MICs (twofold difference). The MICs used throughout this report refer to those obtained by standard methods in Mueller-Hinton broth.
Screening for restitution of Hlb expression.
Lysogens were grown until exponential phase (optical density at 600 nm [OD600] = 0.8, 2 h) in TSB medium. Different concentrations (one-half the MIC and the MIC) of ciprofloxacin or trimethoprim or medium alone as a control were added, followed by further incubation for 6 h. Serial dilutions of cultures were plated on sheep blood agar. The numbers of CFU/ml and the percentage of Hlb-positive colonies were counted. Although the percentage of Hlb-positive colonies varied between experiments, the relative results were reproducible. Therefore, only the results of representative experiments are shown, and no statistical analysis was performed. Hlb-positive and -negative colonies were picked and stored for PCR and PFGE analysis.
Multiplex PCR.
Multiplex PCR was performed with DNA derived from Hlb-positive and -negative colonies with primers specific for hlb, sak, and the φ13 integrase, as listed in Table 1, by using a multiplex PCR kit (QIAGEN, Hilden, Germany), according to the manufacturer's instructions. The multiplex system was controlled for the exclusion of competition between primers by using single amplifications and mixed templates.
TABLE 1.
Oligonucleotide primers
| Target gene | GenBank accession no. | Primer | Primer sequence | Purpose |
|---|---|---|---|---|
| hlb | X61716 | hlb-2 | AGCTTCAAACTTAAATGTCA | Multiplex PCR |
| hlb-527 | CCGAGTACAGGTGTTTGGTA | |||
| sak | AF424783 | sak-for | GTGCATCAAGTTCATTCGAC | Multiplex PCR, Northern probe |
| sak-rev | TAAGTTGAATCCAGGGTTTT | |||
| φ13 integrase | AF424783 | φ13int-for | GCTTTGAAATCAGCCTGTAG | Multiplex PCR |
| φ13 attP | AF424783 | attPst-for | TCTAGCTTTTGGGGTGTACATTCC | DNA standard |
| attPst-rev | GCTTTGAAATCAGCCTGTAGAG | |||
| attP-for | TTTTATTTTATATGGGGTATTATTGA | Quantitative LightCycler PCR | ||
| attP-rev | GTGTATTCTCATTTGTTAGAAGAAAA | |||
| gyrB | D10489 | gyrB-219 | TTATGGTGCTGGGCAAATACA | DNA standard |
| gyrB-864 | GTACGATTTAATACCGCCCTCATA | |||
| gyrB-297 | TTAGTGTGGGAAATTGTCGATAAT | Quantitative LightCycler PCR | ||
| gyrB-547 | AGTCTTGTGACAATGCGTTTACA | |||
| recA | L25893 | recA-153 | AGATAATGCGCTAGGTGTAGGTGG | Northern probe |
| recA-786 | AGCTACTCTAAATGGTGGTGCCAC |
PFGE analysis.
PFGE was performed after restriction endonuclease digestion of whole chromosomal DNA with SmaI (Roche Biochemicals, Mannheim, Germany), as described in an earlier paper (7).
Spot and plaque assay.
Lysogens were grown until exponential phase (OD600 = 0.8, 2 h) in TSB medium. Different concentrations of mitomycin C (500 ng/ml, 1 μg/ml), ciprofloxacin (one-half the MIC and the MIC), or trimethoprim (one-half the MIC and the MIC) or medium alone as a control were added, followed by further incubation for 6 h. Supernatants were sterilized by using 0.45-μm-pore-diameter membrane filters (Millipore, Schwalbach, Germany). For spot tests, serial dilutions of phage-containing supernatant were prepared in phage buffer (1 mM MgSO4, 4 mM CaCl2, 50 mM Tris pH 7.8, 5.9 g/liter NaCl, 1 g/liter gelatin) and spotted onto strain R5 as an indicator. For plaque assays, serial dilutions of the supernatant were mixed with R5 bacteria in top agar and immediately poured onto TSA plates containing 100 mM CaCl2. The plates were inspected after incubation for 24 h at 32°C.
Quantitative PCR for detection of extrachromosomal phage DNA.
Strain 8325-4φ13 was grown until exponential phase (OD600 = 0.8, 2 h) in TSB medium. Mitomycin C (1 μg/ml), different concentrations of ciprofloxacin (one-quarter the MIC, one-half the MIC, and the MIC) and trimethoprim (one-quarter the MIC, one-half the MIC, and the MIC), or medium alone as the control were added, followed by further incubation for 2 h. For DNA isolation, the bacterial cells were disrupted with glass beads (Sigma) in a high-speed homogenizer (Savant Instrument, Farmingdale, N.Y.) twice for 20 s each time at 6,000 rpm. After the disrupted cells were heated for 2 min at 100°C, 1:10 dilutions of the crude extracts were used for quantitative LightCycler PCR.
Quantification of the attP site of phage φ13 and the S. aureus chromosomal gene gyr was performed by quantitative LightCycler PCR (Roche Biochemicals) with sequence-specific DNA standards for each target DNA. DNA standards were generated by regular PCR by using the primers listed in Table 1. Amplicons were quantified spectrophotometrically after removal of the nucleotides and primers by use of the PCR purification kit (QIAGEN), according to the manufacturer's instructions. Quantitative LightCycler PCR was performed with the LightCycler SYBR green kit (Roche Biochemicals). Master mixtures were prepared by following the manufacturer's instructions and by using the oligonucleotides specific for attP and gyr, as shown in Table 1. The following temperature profile was used for amplification: denaturation for 1 cycle at 95°C for 30 s; 45 cycles at 95°C for 1 s (temperature transition, 20°C/s), 55°C to 50°C (step size, 1°C; step delay, 1 cycle) for 15 s (temperature transition, 20°C/s), and 72°C for 15 s (temperature transition, 2°C/s); and fluorescence acquisition at 72°C in single mode. Melting curve analysis was performed at 45 to 90°C (temperature transition, 0.2°C/s) with stepwise fluorescence acquisition. Sequence-specific standard curves were generated by using 10-fold serial dilutions (104 to 108 copies/μl) of DNA standards. The number of copies of each sample DNA was then determined with the aid of the LightCycler software. At least two separate DNA isolations and two independent PCRs were performed for each of the samples. The specificity of the PCR was verified by ethidium bromide staining on 3% agarose gels and by sequencing of the amplicons (4base lab, Reutlingen, Germany).
Northern analysis.
Strain 8325-4φ13 was grown until exponential phase (OD600 = 0.8, 2 h) in TSB medium. Mitomycin C (300 ng/ml), ciprofloxacin (one-half the MIC), trimethoprim (one-half the MIC), or medium alone as a control was added, followed by further incubation for 2 h. RNA was isolated from the bacterial pellet as described previously (6). Northern blot analysis was done as described previously (6). The primers specific for recA and sak used to generate digoxigenin-labeled probes by PCR-labeling (Roche Biochemicals) are listed in Table 1.
RESULTS AND DISCUSSION
Delysogenization of strains harboring hlb-converting phages by ciprofloxacin and trimethoprim.
Induction of hlb-converting phages could be demonstrated during chronic lung infection in patients with CF (8). These patients regularly receive treatment with ciprofloxacin or trimethoprim. To study whether concentrations in the MIC and sub-MIC ranges of these antibiotics can induce phages, five CF isolates harboring φ13-like phages were selected. Since the genetic background of these isolates is diverse and not well defined, a φ13 lysogen of phage-cured prototypic strain 8325-4 was included in the analysis. Delysogenization of strains was detected by restoration of an Hlb-positive phenotype due to the excision of the phage from the bacterial chromosome.
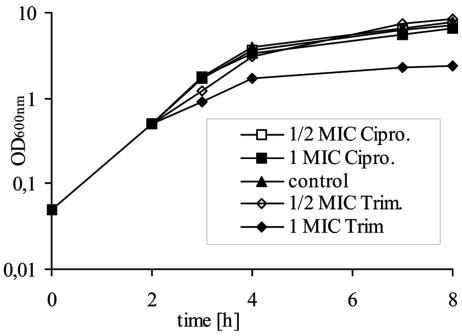
First, the MICs of ciprofloxacin and trimethoprim were determined for strain 8325-4φ13. By using the standard growth microdilution method in Mueller-Hinton broth, an MIC of 0.25 μg/ml each was determined for each of the antibiotics. Growth inhibition was evident only when antibiotics were added at a concentration equivalent to the MIC (Fig. 1). This was accompanied by a decrease in viable cell counts after 8 h of growth to 25% (ciprofloxacin) and 2% (trimethoprim) of the growth of the control without antibiotics. All further investigations of phage induction were performed by using these concentrations near the threshold of growth inhibition (the MIC determined in Mueller-Hinton broth) and below the threshold of growth inhibition (one-half and one-quarter the MIC determined in Mueller-Hinton broth). Antibiotics were added to a growing culture of 8325-4φ13 (2 h after inoculation), and after further incubation for 6 h, screening for restitution of Hlb production on sheep blood agar plates was performed. Hlb-positive single colonies could reliably be detected after treatment with either one-half the MIC or the MIC of ciprofloxacin or trimethoprim (Table 2) but never in the control without antibiotics. However, the actual numbers of Hlb-positive colonies varied between experiments, although the relative results—for instance, the better inducing capacity of ciprofloxacin compared to that of trimethoprim—were reproducible.
FIG. 1.
Growth curve (OD600) of S. aureus strain 8325-4φ13 in TSB (control) and with different concentrations (one-half the MIC and the MIC) of either ciprofloxacin (Cipro.) or trimethoprim (Trim.).
TABLE 2.
Occurrence of Hlb-positive colonies after phage induction in strain 8325-4φ13 and in S. aureus isolates from CF patients harboring φ13-like phages
| Strain | Occurrence of Hlb-positive coloniesa
|
||||
|---|---|---|---|---|---|
| Ciprofloxacin
|
Trimethoprim
|
Controlb | |||
| One-half the MIC | MIC | One-half the MIC | MIC | ||
| 8325-4φ13 | ++ | ++ | + | ++ | − |
| s152 | ++ | ++ | ND | ND | − |
| i102 | + | + | ND | ND | − |
| s87 | + | ++ | ND | ND | − |
| cfs425 | + | + | ND | ND | − |
| s126 | + | + | ND | ND | − |
++, >1% Hlb-positive colonies; +, ≤1% Hlb-positive colonies; −, Hlb-positive colonies not detectable; ND, not determined. Screening for the restitution of Hlb expression was performed on sheep blood agar plates.
Culture without antibiotics.
In further experiments, phage induction by antibiotics was investigated by using the five lysogenic CF isolates. Different concentrations of ciprofloxacin, depending on the corresponding MICs for these strains (strain s152 = 0.75 μg/ml; strain i102 = 0.25 μg/ml; strain s87 = 0.25 μg/ml; strain cfs425 = 0.25 μg/ml; strain s126 = 0.19 μg/ml) were used. For all strains, Hlb-positive colonies were detected after incubation with one-half the MIC or the MIC of ciprofloxacin indicating phage induction (Table 2). However, the frequency of Hlb conversion varied considerably between the strains. For instance, for strain i102 only a very low percentage (<1%) of Hlb-positive colonies were detected, whereas for strain s152, higher percentages of all colonies were Hlb producers. This variation between strains was reproducible and may have been due to the diverse genetic backgrounds of the strains or to differences in the lysogenic phages.
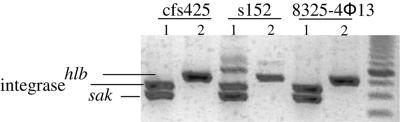
To verify whether colonies became Hlb positive due to phage excision from the bacterial chromosome, a multiplex PCR was used for the detection of intact hlb, φ13 integrase, and the phage-encoded sak. In Fig. 2, phage excision after induction is exemplified in three cases. The original lysogens (lane 1) are positive for the φ13 integrase and sak, whereas the phage-cured derivates (lane 2) show an intact hlb gene. All Hlb-positive colonies analyzed after induction with either ciprofloxacin or trimethoprim showed this pattern.
FIG. 2.
Multiplex PCR for the detection of hlb, φ13 integrase, and the phage-encoded sak. Hlb-negative (lanes 1) and Hlb-positive (lanes 2) colonies of S. aureus isolates from CF patients (strains cfs425 and s152) and from strain 8325-4φ13 were obtained after treatment with either ciprofloxacin, trimethoprim, or mitomycin C. The induction of the phage in colonies with the Hlb-positive phenotype is confirmed by the restitution of the intact hlb gene.
PFGE patterns after phage induction in vitro and in vivo.
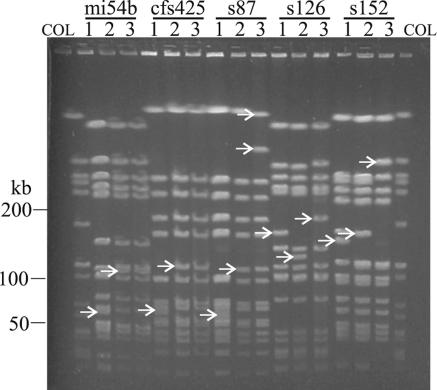
By use of a second approach, lysogens and delysogenized derivatives obtained after phage induction were compared by PFGE analysis. In all cases the excision of the phage from the bacterial genome was evident as fragment shifts when the fingerprints of the delysogenized strains were compared with those of the original lysogens (Fig. 3, lane 2 and lane 1, respectively). The fragment patterns of the phage-cured strains were identical when different agents (mitomycin C, ciprofloxacin, or trimethoprim [data not shown]) were used for phage induction.
FIG. 3.
PFGE patterns after induction of phages in vitro and in vivo. S. aureus isolates from CF patients harboring φ13-like phages (lanes 1) were subjected to treatment with either ciprofloxacin, trimethoprim, or mitomycin C and were screened for delysogenized (Hlb-positive) strains (lane 2). For comparison, the Hlb-positive derivate obtained during chronic lung infection is depicted (lanes 3). COL, molecular weight standard (SmaI-digested whole chromosomal DNA of S. aureus strain COL).
In order to assess whether phage induction alone can account for the observed genome rearrangement during chronic lung infection in patients with CF, the PFGE fingerprints of phage-cured strains obtained in vitro were compared with that of the delysogenized strain obtained during chronic infection (Fig. 3, lanes 2 and 3). The phage-related band shifts were also visible in all in vivo derivatives. In two cases the fragment pattern of the in vitro phage-cured strain was identical to the fingerprint of the delysogen obtained in vivo (strains mi54b and cfs425), thus fully explaining the observed genome rearrangements in these strains during chronic lung infection. Here the fragment shifts added up to the size of the phages. In the three other cases, additional fragment alterations could be distinguished in the fingerprints of the in vivo strains. These additional band shifts may be the result of the integration or the excision of phages not analyzed here or of intrachromosomal rearrangements, such as inversions or duplications mediated by insertion sequence elements (28) or transposons. Another possibility is that phage induction led to comobilization of a pathogenicity island, as was shown for SaPI1 (14, 22). This pathogenicity island is excised and circularized by staphylococcal phages φ13 and 80α. The occurrence of multiple distinct genome rearrangements in the CF strains emphasizes the high degree of selective pressure exerted on the pathogen during chronic infection.
Quantification of phage particles after induction with ciprofloxacin and trimethoprim.
In the next step the amount of liberated phage after antibiotic treatment of lysogens was determined. The phage titers obtained after treatment of 8325-4φ13 with different concentrations of ciprofloxacin, trimethoprim, and mitomycin C were quantified by spot and plaque assays. Incubation with different concentrations of mitomycin and ciprofloxacin resulted in a low titer of liberated phages (Table 3). No plaques were detected when the supernatants of the trimethoprim-treated culture or the control culture without antibiotics were used. In general, the φ13-like phages formed only very small and turbid plaques on the indicator strain. Total lysis of the bacterial culture was never achieved with the inducing agents, even after prolonged incubation (12 h). This failure of host lysis after induction of φ13 may be due to the assay conditions used. It is possible that the phage titers failed to reach the critical threshold for visible lysis of the culture.
TABLE 3.
Phage titers from strain 8325-4φ13 after induction with mitomycin C, ciprofloxacin, and trimethoprim
| Inducer | Phage titer (PFU/ml) at the following antibiotic concna:
|
|
|---|---|---|
| One-half the MIC | MIC | |
| Mitomycin C | 1.3 × 105 | 3 × 104 |
| Ciprofloxacin | 6 × 102 | 9 × 102 |
| Trimethoprim | 0 | 0 |
One-half the and the MIC were as follows: for mitomycin C, 0.5 μg/ml and 1 μg/ml, respectively; for ciprofloxacin, 0.125 μg/ml and 0.25 μg/ml, respectively; for trimethoprim, 0.125 μg/ml and 0.25 μg/ml, respectively.
Not only the hlb-converting phages but also other phages, for instance, phages φ11 and φ12 of strain 8325 or a φETA-like phage of a CF strain (8), could be induced by subinhibitory concentrations of ciprofloxacin and trimethoprim (data not shown). After induction of these phages, the total lysis of the bacterial culture was visible and the lysates obtained formed clear plaques on the indicator strain.
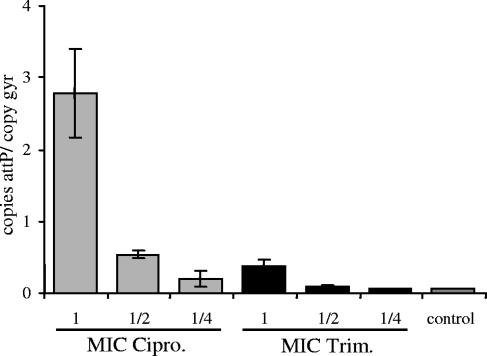
Because of the low phage titers obtained with the φ13 lysogen, we established a method for quantification of extrachromosomal phage DNA by quantitative real-time PCR. After the phage is excised from the bacterial chromosome, the φ13 genome recircularizes and the attP site is restored. Therefore, this site can be used as a marker for phage replication in the host cell (16). For this purpose, primers that span the attP site of φ13 were designed. The specificity of the PCR was verified by melting curve analysis (detection of a single peak), agarose gel electrophoresis (detection of a single band of the expected size), and sequencing of the amplicon (detection of the expected sequence). Quantification was performed with reference to the total amount of bacterial DNA represented by the chromosomal gene gyrase. First, the PCR system was tested with mitomycin C as an inducing agent. A high level of newly synthesized phage DNA was detected after induction for 2 h (12.2 copies of attP/copy of gyr). Using ciprofloxacin and trimethoprim at different concentrations, we were able to detect the dose-dependent formation of extrachromosomal phage DNA (Fig. 4). Overall, the inducing effect of trimethoprim was less pronounced than that of ciprofloxacin. Spontaneous induction of φ13 in the control culture without antibiotic treatment occurs at a low frequency (0.7 copies of attP/copy of gyr). This result is in the same range as the values obtained after spontaneous induction of prophages in lactococci during growth in culture (16).
FIG. 4.
Quantification of newly synthesized phage DNA after induction. Strain 8325-4φ13 was treated with different concentrations (one-quarter the MIC, one-half the MIC, or the MIC) of either ciprofloxacin (Cipro.; gray columns) or trimethoprim (Trim.; black columns) or was left untreated (control). The newly formed attP sites were calculated by quantitative real-time PCR in reference to the total amount of bacterial DNA represented by the chromosomal gene gyrase (number of copies attP/copy of gyr). Values from two separate DNA isolations and two independent PCRs each were used to calculate the mean amount of attP sites (±standard errors of the mean).
When the percentages of Hlb-positive colonies (Fig. 1B) were compared by calculation of the attP sites after ciprofloxacin and trimethoprim treatment of strain 8325-4φ13 (Fig. 4), the correlation seemed to be very poor. This may be explained by the fact that when the Hlb-positive colonies were measured, only the frequency of phage-cured colonies surviving phage induction is determined. In contrast, the attP PCR quantifies the average number of phage particles per bacterium.
Phage induction by activation of the SOS response.
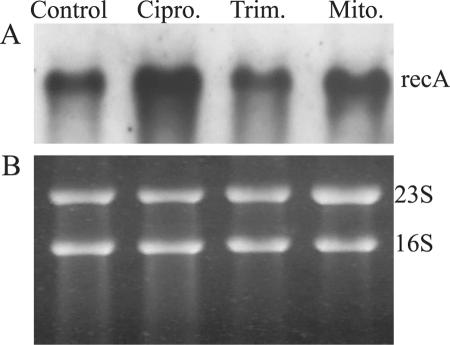
A possible explanation for the observed differences in phage replication between the inducing agents was investigated by analyzing the level of recA transcription after treatment. As part of the SOS response, activated RecA leads to autocleavage of the phage repressor CI and, thereby, to the resumption of the lytic cycle. Expression of recA was analyzed by Northern analysis after treatment of strain 8325-4φ13 with subinhibitory concentrations of ciprofloxacin, trimethoprim, and mitomycin C. The enhanced transcription of recA could be demonstrated after ciprofloxacin and mitomycin C induction, whereas trimethoprim treatment showed no obvious influence (Fig. 5). Thus, the differences in the potential for phage induction between the inducing agents may be explained by the differences in the levels of activated RecA. Trimethoprim treatment resulted in the stimulation of φ13 excision and replication only at a very low level. The influence of ciprofloxacin on the SOS response of S. aureus was also shown by Bisognano et al. (4), who could show activation of recA and, as a consequence, the derepression of the lexA-regulated gene encoding the fibronectin-binding protein FnbB. Transposon insertion in recA resulted in an attenuated virulence in a murine model of bacteremia (17). Recently, Ubeda et al. (22) showed that the antibiotic-induced SOS response promotes the horizontal dissemination of S. aureus pathogenicity islands. These elements are induced to excise and replicate after SOS induction of prophages and are then packaged into phage-like particles and are transferred at a high frequency. They suggested that antibiotics may help to promote the spread of bacterial virulence factors. In summary, the influence of the SOS response on the pathogenicity of S. aureus is probably multifactorial and may depend on the activation of chromosome- or phage-encoded virulence genes.
FIG. 5.
Northern blot analysis for the detection of recA. Strain 8325-4φ13 was treated with one-half the MIC of either ciprofloxacin (Cipro.) or trimethoprim (Trim.) or with 300 ng/ml mitomycin C (Mito.) or was left untreated (Control). (A) The blots were hybridized by using digoxigenin-labeled PCR fragments specific for recA; (B) loading control; the intensities of the 23S and 16S rRNA bands stained with ethidium bromide were equivalent in all the samples before Northern transfer.
Enhanced expression of phage-encoded virulence genes after induction.
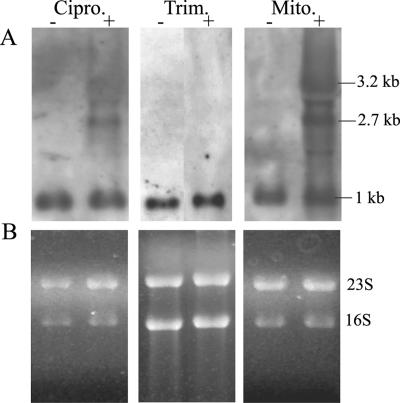
For other bacterial pathogens, it could be shown that phage induction is linked to an enhanced virulence potential (24). Here we analyzed whether phage induction by ciprofloxacin and trimethoprim is linked to virulence modulation in S. aureus. Northern analysis for the detection of sak was performed with strain 8325-4φ13 after treatment with subinhibitory concentrations of mitomycin C, ciprofloxacin, and trimethoprim. Elevated expression of the phage-encoded gene sak was linked to phage excision after induction with mitomycin C and ciprofloxacin (Fig. 6). In addition to the 1-kb monocistronic sak transcript, two additional transcripts (2.7 kb and 3.2 kb) probably originating from phage promoters could be detected after ciprofloxacin treatment. Induction with trimethoprim resulted in only a slight increase in the 1-kb sak transcript.
FIG. 6.
Northern blot analysis for the detection of the phage-encoded sak. Strain 8325-4φ13 was treated with one-half the MIC of either ciprofloxacin (Cipro; lanes +) or trimethoprim (Trim.; lanes +) or with 300 ng/ml mitomycin C (Mito.; lanes +) or was left untreated (lanes −). (A) The blots were hybridized by using digoxigenin-labeled PCR fragments specific for sak; (B) loading control; the intensities of the 23S and 16S rRNA bands stained with ethidium bromide were equivalent in all the samples before Northern transfer.
For Escherichia coli it was observed that the antibiotics commonly used to treat diarrhea induce Shiga toxin-encoding phages (24). For these phages it was demonstrated for the first time that the life cycle of the phage exerts control over virulence factor production: Shiga toxin expression is tightly linked to prophage induction (23, 27). For S. aureus, enhanced expression of the phage-encoded virulence factors was observed upon mitomycin C induction of φSa3ms from strain MSSA476 (21). Induction led to the increased transcription of virulence factors due to an increase in phage copy numbers and to the activation of latent phage promoters upstream of the virulence genes. A 2.7-kb transcript probably originating upstream of the phage holin gene and a 6.8-kb transcript encompassing the virulence factors sea and sak were detected. In our study we could show that induction of the prophage φ13 by ciprofloxacin likewise resulted in the elevated expression of sak, chiefly due to the activation of latent phage promoters. We could also detect the previously described 2.7-kb transcript and a 3.2-kb transcript which probably initiates between open reading frames orf47 and orf49 of phage φ13. The 6.8-kb transcript described by Sumby and Waldor (21) was not detectable here since φ13 does not encode sea, which is encompassed in this transcript in φSa3ms. Trimethoprim resulted in a slightly elevated level of the 1-kb sak transcript; no additional transcripts were detected. This may be because the induction of φ13 by this antibiotic was so weak that the transcripts remained below the detection level.
In summary, for many bacterial species it was shown that prophages are induced by environmental conditions and that induction is linked to enhanced production of phage-encoded virulence genes (24). In the case of E. coli, numerous epidemiological studies have detected an association between an increased severity of infection and treatment with antibiotics. This observation may also hold true for S. aureus, since we could demonstrate the same link between the induction of prophages and subsequent virulence modulation of the bacterium by the antibiotics which are often used for treatment of infection. Suboptimal concentrations of antibiotics are sufficient to exert these effects.
Acknowledgments
We thank W. van Wamel for strain R5 and for very helpful discussions.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (Wo 578/3-3) and the Fortüne Program of the University of Tübingen.
REFERENCES
- 1.Arvidson, S. 1983. Extracellular enzymes from Staphylococcus aureus, p. 745-808. In: C. F. Easmon and C. Adlam (ed.), Staphylococci and staphylococcal infections. Academic Press, London, United Kingdom.
- 2.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 3.Betley, M. J., and J. J. Mekalanos. 1985. Staphylococcal enterotoxin A is encoded by phage. Science 229:185-187. [DOI] [PubMed] [Google Scholar]
- 4.Bisognano, C., W. L. Kelley, T. Estoppey, P. Francois, J. Schrenzel, D. Li, D. P. Lew, D. C. Hooper, A. L. Cheung, and P. Vaudaux. 2004. A recA-lexA-dependent pathway mediates ciprofloxacin-induced fibronectin binding in Staphylococcus aureus. J. Biol. Chem. 279:9064-9071. [DOI] [PubMed] [Google Scholar]
- 5.Coleman, D. C., D. J. Sullivan, R. J. Russell, J. P. Arbuthnott, B. F. Carey, and H. M. Pomeroy. 1989. Staphylococcus aureus bacteriophages mediating the simultaneous lysogenic conversion of beta-lysin, staphylokinase and enterotoxin A: molecular mechanism of triple conversion. J. Gen. Microbiol. 135:1679-1697. [DOI] [PubMed] [Google Scholar]
- 6.Goerke, C., S. Campana, M. G. Bayer, G. Döring, K. Botzenhart, and C. Wolz. 2000. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 68:1304-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goerke, C., K. Kraning, M. Stern, G. Döring, K. Botzenhart, and C. Wolz. 2000. Molecular epidemiology of community-aquired Staphylococcus aureus in families with and without cystic fibrosis patients. J. Infect. Dis. 181:984-989. [DOI] [PubMed] [Google Scholar]
- 8.Goerke, C., S. Matias y Papenberg, S. Dasbach, K. Dietz, R. Ziebach, B. C. Kahl, and C. Wolz. 2004. Increased frequency of genomic alterations in Staphylococcus aureus during chronic infection is in part due to phage mobilization. J. Infect. Dis. 189:724-734. [DOI] [PubMed] [Google Scholar]
- 9.Holden, M. T., E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. Day, M. C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggett, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA 101:9786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iandolo, J. J., V. Worrell, K. H. Groicher, Y. Qian, R. Tian, S. Kenton, A. Dorman, H. Ji, S. Lin, P. Loh, S. Qi, H. Zhu, and B. A. Roe. 2002. Comparative analysis of the genomes of the temperate bacteriophages phi 11, phi 12 and phi 13 of Staphylococcus aureus 8325. Gene 289:109-118. [DOI] [PubMed] [Google Scholar]
- 11.Jin, T., M. Bokarewa, T. Foster, J. Mitchell, J. Higgins, and A. Tarkowski. 2004. Staphylococcus aureus resists human defensins by production of staphylokinase, a novel bacterial evasion mechanism. J. Immunol. 172:1169-1176. [DOI] [PubMed] [Google Scholar]
- 12.Kaneko, J., T. Kimura, S. Narita, T. Tomita, and Y. Kamio. 1998. Complete nucleotide sequence and molecular characterization of the temperate staphylococcal bacteriophage phiPVL carrying Panton-Valentine leukocidin genes. Gene 215:57-67. [DOI] [PubMed] [Google Scholar]
- 13.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 14.Lindsay, J. A., A. Ruzin, H. F. Ross, N. Kurepina, and R. P. Novick. 1998. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol. Microbiol. 29:527-543. [DOI] [PubMed] [Google Scholar]
- 15.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 16.Lunde, M., J. M. Blatny, D. Lillehaug, A. H. Aastveit, and I. F. Nes. 2003. Use of real-time quantitative PCR for the analysis of phiLC3 prophage stability in lactococci. Appl. Environ. Microbiol. 69:41-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mei, J. M., F. Nourbakhsh, C. W. Ford, and D. W. Holden. 1997. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 26:399-407. [DOI] [PubMed] [Google Scholar]
- 18.Narita, S., J. Kaneko, J. Chiba, Y. Piemont, S. Jarraud, J. Etienne, and Y. Kamio. 2001. Phage conversion of Panton-Valentine leukocidin in Staphylococcus aureus: molecular analysis of a PVL-converting phage, phiSLT. Gene 268:195-206. [DOI] [PubMed] [Google Scholar]
- 19.Novick, R. P. 2003. Mobile genetic elements and bacterial toxinoses: the superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid 49:93-105. [DOI] [PubMed] [Google Scholar]
- 20.Ptashne, M. 1992. A genetic switch. Cell Press, Cambridge, Mass.
- 21.Sumby, P., and M. K. Waldor. 2003. Transcription of the toxin genes present within the staphylococcal phage φSa3ms is intimately linked with the phage's life cycle. J. Bacteriol. 185:6841-6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ubeda, C., E. Maiques, E. Knecht, I. Lasa, R. P. Novick, and J. R. Penades. 2005. Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Mol. Microbiol. 56:836-844. [DOI] [PubMed] [Google Scholar]
- 23.Wagner, P. L., J. Livny, M. N. Neely, D. W. Acheson, D. I. Friedman, and M. K. Waldor. 2002. Bacteriophage control of Shiga toxin 1 production and release by Escherichia coli. Mol. Microbiol. 44:957-970. [DOI] [PubMed] [Google Scholar]
- 24.Wagner, P. L., and M. K. Waldor. 2002. Bacteriophage control of bacterial virulence. Infect. Immun. 70:3985-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker, C. W. 1996. The SOS response of Escherichia coli, p. 1400-1416. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. Brooks Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and E. H. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 26.Yamaguchi, T., T. Hayashi, H. Takami, K. Nakasone, M. Ohnishi, K. Nakayama, S. Yamada, H. Komatsuzawa, and M. Sugai. 2000. Phage conversion of exfoliative toxin A production in Staphylococcus aureus. Mol. Microbiol. 38:694-705. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, X., A. D. McDaniel, L. E. Wolf, G. T. Keusch, M. K. Waldor, and D. W. Acheson. 2000. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181:664-670. [DOI] [PubMed] [Google Scholar]
- 28.Ziebuhr, W., K. Dietrich, M. Trautmann, and M. Wilhelm. 2000. Chromosomal rearrangements affecting biofilm production and antibiotic resistance in a Staphylococcus epidermidis strain causing shunt-associated ventriculitis. Int. J. Med. Microbiol. 290:115-120. [DOI] [PubMed] [Google Scholar]