Abstract
Streptococcus pneumoniae M22 is a multidrug-resistant mutant selected after exposure of capsulated wild-type S. pneumoniae NCTC 7465 (strain M4) to ciprofloxacin. DNA microarray analysis comparing the gene expression profiles of strain M22 with those of strain M4 showed that strain M22 constitutively expressed 22 genes at levels higher than those observed in strain M4 under all conditions studied. These included the genes encoding the enzymes involved in branched-chain amino acid biosynthesis and two genes (patA and patB) with sequences suggestive of ABC transporter proteins. Expression of the patA and patB genes was induced by ciprofloxacin in both strains, but in strain M4 it only reached the levels observed in strain M22 after long incubation with high concentrations of ciprofloxacin. The altered expression profile observed with strain M22 suggested that the mutation or mutations acquired during resistance selection bring the cell into a state in which the expression of critical genes is preemptively altered to correct for the potential effects of ciprofloxacin on gene expression in the parent strain.
Streptococcus pneumoniae is an important cause of respiratory illnesses, including pneumonia, as well as meningitis and otitis (3). Clinical failures due to fluoroquinolone-resistant organisms have occurred when either ciprofloxacin or ofloxacin was used to treat respiratory tract infections (4, 27, 28). Epidemiological studies have identified resistance in S. pneumoniae with mutated gyrase and topoisomerase IV genes (10, 34, 35), but efflux may also cause fluoroquinolone resistance (1, 6-8, 13, 14, 19, 30, 36, 37, 46). Markham (31) reported that the efflux inhibitor reserpine prevents development of resistance to ciprofloxacin in S. pneumoniae. Gill et al. (19) described a putative efflux pump, PmrA, that mediates low-level resistance to norfloxacin, ethidium bromide, and acriflavine, and recent studies have suggested that efflux pumps besides PmrA may export fluoroquinolones from S. pneumoniae (8, 39).
S. pneumoniae M22 is a multidrug-resistant mutant selected with ciprofloxacin during a study of mutational resistance development (38). The mutation frequency of 6.9 × 10−8 and subsequent stable resistance without antibiotic pressure suggested a single point mutation (38). Strain M22 was more resistant than strain M4 to several fluoroquinolones, acriflavine, ethidium bromide, doxorubicin, tetracycline, erythromycin, and cetrimide. Characterization of strain M22 suggested that it had a fluoroquinolone efflux system since accumulation of ciprofloxacin, gatifloxacin, and ofloxacin was significantly less than in strain M4 (38). We describe here the results of a genome-wide analysis of transcriptional responses of strains M22 and M4 to ciprofloxacin that was designed to characterize the effects of the mutation in strain M22. DNA microarrays have been used to investigate antimicrobial resistance and the mechanism of action of antibiotics (9, 25). A basic tenet of gene expression analysis is that bacteria will respond to externally imposed toxic stress by inducing the expression of defense mechanisms that can combat the effects of the imposed stress. Antibiotics and other toxic chemicals are known to induce energy-dependent efflux systems; for example, salicylic acid, bile salts, and methyl viologen induce expression of the AcrAB-TolC broad-spectrum proton-coupled efflux system of Escherichia coli (33, 41, 42), and Van Dyk et al. (44) demonstrated that aromatic carboxylic acids induce the expression of a specific efflux system, AaeAB, in E. coli. Thus, if efflux systems play a role in fluoroquinolone resistance in S. pneumoniae, one expects that they will be induced by exposure to these agents. The two genome sequences of S. pneumoniae now available (23, 43) contain a number of potential efflux systems that could contribute to fluoroquinolone resistance. As expected from previous analysis of the transcriptome of Haemophilus influenzae after exposure to ciprofloxacin (20), the expression of many genes was altered by exposure to ciprofloxacin. Surprisingly, pmrA was not one of these and it appears that resistance in strain M22 involves proteins belonging to the ABC transporter family.
MATERIALS AND METHODS
Bacteria and growth conditions.
S. pneumoniae NCTC 7465 (M4) and M22 (38), obtained from the University of Birmingham, were maintained at −80°C on Protect beads (Protect Bacterial Preservers; TSC Ltd., Heywood, United Kingdom) without antibiotic and grown overnight in Todd-Hewitt broth at 37°C in 5% CO2 to provide inocula for expression experiments.
Microarray analysis.
Sense (ROEZ06s) and antisense (ROEZ06a) arrays custom fabricated by Affymetrix (Santa Clara, Calif.) to cover the genomes of both S. pneumoniae and H. influenzae were used. Probe selection, open reading frame coverage, and array design for ROEZ06s and ROEZ06a were described by Hakenbeck et al. (21) and de Saizieu et al. (16). The array area covering S. pneumoniae has over 130,000 oligonucleotide probes that are complementary to the S. pneumoniae KNR.7/87 genome (16) sequence published as TIGR4 (45). A total of 1,968 putative genes, predicted by GeneMark software, and 323 intergenic regions longer than 200 bp from S. pneumoniae are represented. Each gene is represented by at least 20 probe pairs (for short genes) and in general by 25 probe pairs. The probe pairs (25-residue oligonucleotides) comprise a perfect-match (PM) probe and a mismatch (MM) probe that differs by a single base change at the central position. The designation antisense or sense refers to the target nucleic acid; i.e., the oligonucleotide probes on microarray have, respectively, the sequence of the coding strand and the sequence complementary to the coding strand. For experiments with ROEZ06s, bacteria were grown in Todd-Hewitt medium and chromosomal DNA was prepared with the QIAGEN Genomic DNA Purification Kit. DNA was fragmented and labeled as described by Hakenbeck et al. (21). For experiments with ROEZ06a, bacteria were grown on at least two separate occasions to an optical density at 600 nm of 0.3 in Todd-Hewitt medium and the cells harvested by centrifugation and frozen in liquid nitrogen. The effect of ciprofloxacin on gene expression was examined by harvesting cultures after 10, 40, or 60 min of exposure to 2, 12, or 80 μg of ciprofloxacin/ml. Antibiotic-free cultures were analyzed in parallel. RNA extraction and cDNA labeling were performed as described by de Saizieu et al. (16). Fragmented biotin-labeled cDNA was hybridized to the chips and stained as previously described (16), with minor modifications. The hybridization mixtures contained 5 μg of biotin-labeled cDNA, and TOP-BLOCK (Juro) was used instead of acetylated bovine serum albumin (Sigma) at 1.5 g/liter in the hybridization solutions and 2 g/liter in the staining solution. The microarrays were scanned at 570 nm, 3-μm resolution, with an Affymetrix gene chip scanner and analyzed as previously described (29). The reproducibility and validation of the genomic hybridization on the microarray were assessed as previously described (15). The signal for each gene was calculated as the average intensity difference (AID) represented by Σ(intensity PM − intensity MM)/number of probe pairs. All experiments were performed twice and the AID values averaged. The intensity ratios were defined as the AID under the conditions where the gene was expressed at the highest level divided by the AID under the condition where the gene was expressed at the lowest level.
PCR and DNA sequencing of topoisomerase genes.
The quinolone resistance-determining regions (QRDRs) of gyrA (nucleotides [nt] 137 to 408), gyrB (nt 1096 to 1553), parC (nt 104 to 465), and parE (nt 981 to 1334) were amplified by PCR performed on whole-cell lysates. The primers were designed with Primer software (Cambridge Scientific) from the DNA sequences available in the EMBL database (GenBank accession numbers: parC and parE, X95717; gyrA, X95718; gyrB, Z67740). The DNA sequences of all amplimers were determined by MWG Biotech.
Preparation of total RNA and real-time PCR.
Total RNA was isolated with the TRizol Max bacterial RNA isolation kit (Invitrogen, Carlsbad, CA) and subsequently treated with DNase I (Ambion, Austin, TX). A one-step real-time hot-start reverse transcription (RT)-PCR assay was performed with a QuantiTect SYBR Green RT-PCR kit (QIAGEN, Basel, Switzerland) and 50 to 150 ng of total RNA on an ABI Prism 7000 Sequence Detection System (PE Applied Biosystems, Foster City, CA). Relative quantitation of mRNA transcription was done by the relative standard curve method (K. J. Livak, ABI Prism 7700 Sequence Detection System, User Bulletin 2, PE Applied Biosystems, 1997). The following gene-specific primers were designed with Primer Express software (PE Applied Biosystems): SP2073, AAATGTGACGCTGGCTCTCA (forward) and GCTGGAGGTTGGTGTATTTGC (reverse); SP2075, CCTTCTTGAGCGCATCAATG (forward) and CTGTAAACTTAGCAAATTGCTCTTTTTC (reverse); SP0446, ACCTTTCCGTGCAACAGTAGTAGA (forward) and GGCGAATGACTCGCAATAGG (reverse); SP0450, CCTTTAAAATTCGTGGTGCCTATT (forward) and TCCCTGCGCATGATTTCC (reverse); SP1202, GCCGTGTAAATGGTCAGATGGT (forward) and ACGCATTAACTCCTCATGGTCAT (reverse); SP1219, AGGAGATGAAGGCAAGTTTTATCG (forward) and AATGCGACGGTG AACAGGTT (reverse); 16srRNA, TGGAGCATGTGGTTTAATTCGA (forward) and CACCTCTGTCCCGAAGGAAA (reverse).
RESULTS
Lack of mutation in topoisomerase genes from M4 and M22.
Sequencing of the QRDRs of the parC, parE, gyrA, and gyrB genes in strains M4 and M22 revealed that gyrB in both strains contained two silent mutations in the codons for Val57 and Asp159. The QRDR sequences of the other three genes were identical in both strains.
Global analysis.
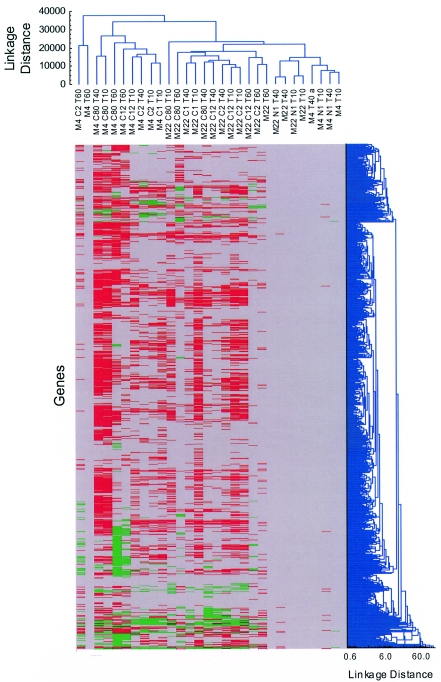
Of the 1,968 putative genes represented on the chip, 1,312 with an AID in strains M4 and M22 of >50 were selected for further analysis. A boundary condition of change factor (CHF) of ±1.6 was chosen for a significant effect because >98% of the variance between repeated samples is encompassed within the limits −1.6 < CHF < 1.6 (data not shown). Hierarchical cluster analysis with STATISTICA (Statsoft Inc.) revealed several characteristic response patterns (Fig. 1). The principal components of the variance between samples were incubation time, constitutive differential expression between M4 and M22 fixed by the mutation, and exposure to ciprofloxacin. Incubation time appeared to be the major component of clustering of samples derived from strain M22, whereas fluoroquinolone exposure was more important for clustering of samples derived from strain M4. For an overview of the number of genes whose expression is affected by the different parameters, see Fig. 3.
FIG. 1.
Global analysis of the transcriptomes of S. pneumoniae M4 and M22 (FQr). The horizontal dendrogram to the right of the colored block shows the clustering of genes according to the CHFs exhibited under different conditions. The vertical dendrogram at the top of the block shows the clustering of conditions according to the similarity between absolute levels of gene expression (AID) under each condition. The dendrograms were constructed with weighted pair group averages and “Manhattan city block” distances by using STATISTICA. Expression changes induced by ciprofloxacin are indicated in the colored block, with up-regulated genes (CHF, >1.6) shown in green, down-regulated genes (CHF, <−1.6) shown in red, and “unaffected” genes (−1.6 ≤ CHF ≤ 1.6) shown in gray. The controls not exposed to ciprofloxacin are, by definition, unaffected. Conditions are indicated by strain (M4, M22), quinolone antibiotic (N &= norfloxacin, C = ciprofloxacin; values indicate concentrations in milligrams per liter), and incubation time (T; values indicate times in minutes).
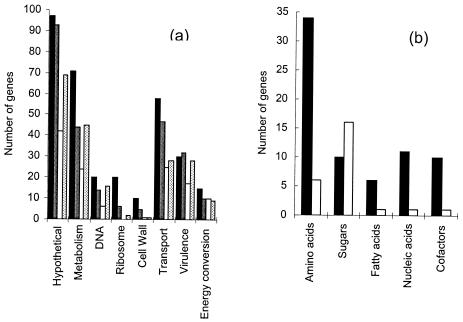
FIG. 3.
Functions of genes showing differential expression between strains and after exposure to ciprofloxacin. (a) Histogram showing the relative frequencies of genes with different functions that showed significantly altered expression profiles. The columns show constitutive overexpression in strain M22 (black), constitutive repression in strain M22 (white), induction by ciprofloxacin at its MIC in strain M4 (dark gray), and repression by ciprofloxacin at its MIC in strain M4 (light gray). The genes are grouped into categories according to assignment of function in the TIGR database (45). “Hypothetical” includes genes for which there is no specific function prediction. “Metabolism” includes genes assigned to metabolic pathways. “DNA” includes genes assigned to a role either in synthesis of DNA from nucleotides or in repair or the maintenance topology. “Ribosome” includes structural genes and genes for ancillary proteins such as elongation factors. “Cell wall” includes genes assigned to a role in cell wall precursor biosynthesis and polymerization. “Transport” includes genes assigned a role in uptake or efflux of small molecules and ions. “Virulence” includes genes associated with signal transduction, growth control, expression of virulence factors, and competence (45). “Energy conversion” includes genes associated with ATP synthesis, maintenance of proton motive force, and redox reactions. (b) Histogram showing the relative proportions of genes involved in the metabolism and biosynthesis of amino acids, sugars, fatty acids, and nucleic acid precursors that were significantly overexpressed (black) or repressed (white) in strain M22 compared to strain M4.
Effects of incubation time.
Clustering according to absolute expression levels (vertical dendrogram above the colored block, Fig. 1) revealed a significant difference between the expression profiles at 10 min and 40 min on the one hand and 60 min on the other. In the absence of antibiotic, both strains were still in exponential growth phase at the 10- and 40-min time points. At the 60-min time point, the growth of both strains had slowed down and they were entering stationary phase. The change from 40 min to 60 min was greater for the M4 samples than for the M22 samples, with more than 50% of the genes exhibiting a CHF of <−1.6 or >1.6 as the cells moved from exponential growth (10- and 40-min time points) into stationary phase (60-min time point).
Constitutive differences in gene expression between M4 and M22.
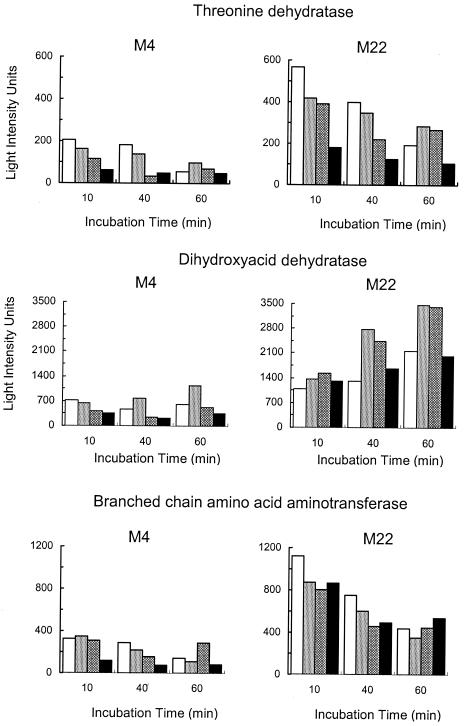
The two genomes were isogenic on chip ROEZ06s, with all genes in strain M4 being found in strain M22 (data not shown). Clustering genes according to CHF (horizontal dendrogram right of the colored block, Fig. 1) revealed distinct clusters with common regulation patterns. After 10 min of incubation, 40 (3%) of the genes showed a CHF of >1.6. After 40 min of incubation the number rose to 73 genes (6%) and after 60 min it rose to 193 (15%) genes. Altogether, 249 genes showed higher expression in strain M22 than in strain M4 at one or more time points (Fig. 2) and 22 were always overexpressed in strain M22 (Table 1). Many of the up-regulated genes encode proteins that are involved in amino acid biosynthesis and in transport (Fig. 3). The entire pathway for isoleucine and valine biosynthesis is represented, with the expression of the genes encoding the subunits of acetolactate synthase being as much as sevenfold higher in strain M22 (Fig. 4). The genes encoding the subunits of the GlnPQ glutamine transporter were expressed at an up to 24-fold higher level in strain M22. Three genes encoding putative transporters were consistently expressed at higher levels in strain M22. One (SP0159) is a homologue of a transporter for Mn(II), Mn(III), and Fe(II) of the NRAMP family (43). The other two encode a homologous pair of proteins that are juxtaposed in the genome and are each homologous to components of ABC-type efflux systems involved in antibiotic resistance (32). The two genes, here designated patA (TIGR4 gene number SP2075) and patB (TIGR4 gene number SP2073), were expressed at three- to fivefold higher levels in strain M22 (Fig. 5). The pmrA gene encoding a putative proton-coupled symporter implicated in norfloxacin resistance (19) was not among those genes that had constitutive differential expression. Only one regulatory protein was strongly overexpressed in strain M22; this was the Rgg protein that is involved in coordinating virulence factor synthesis and metabolism in streptococci (11).
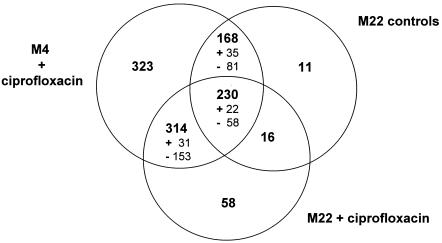
FIG. 2.
Summary of genes showing differential expression between strains and after exposure to ciprofloxacin. The Venn diagram shows the numbers of genes differentially expressed under the different conditions used in this study. The set “M4 plus ciprofloxacin versus M4 controls” contains all genes that are differentially expressed, with a CHF greater than 1.6 or less than −1.6, between strain M4 in the presence of ciprofloxacin, at any one concentration and time, and the corresponding unexposed control. The set “M22 versus M4 controls” contains all genes that are differentially expressed with 1.6 < CHF < −1.6, between strains M4 and M22 at any time, in the absence of ciprofloxacin. The set “M22 plus ciprofloxacin versus M22 controls” contains all genes that are differentially expressed with 1.6 < CHF < −1.6, between strain M22 in the presence of ciprofloxacin, at any one concentration and time, and the corresponding unexposed control. The values in smaller font indicate the numbers of genes consistently induced (+) or repressed (−) under all conditions in that set.
TABLE 1.
Genes constitutively expressed at higher levels in strain M22 under all conditions
| TIGR4 IDa | Description | CHF, M22/M4
|
||
|---|---|---|---|---|
| T10b | T40 | T60 | ||
| SP0141 | Transcriptional regulator (mutR homologue) | 3.8 | 2.8 | 1.8 |
| SP0159 | Mn(II) and Fe(II) symporter of NRAMP family | 4.3 | 2.7 | 1.8 |
| SP0445 | Acetolactate synthase large subunit | 3.3 | 3.8 | 6.8 |
| SP0446 | Acetolactate synthase small subunit | 4.8 | 4.9 | 7.3 |
| SP0447 | Ketol-acid reductoisomerase | 2.4 | 3.7 | 4.3 |
| SP0448 | DNA helicase II | 2.9 | 4.3 | 2.9 |
| SP0449 | Hypothetical protein | 4.0 | 3.7 | 2.2 |
| SP0450 | Threonine dehydratase | 2.0 | 1.9 | 2.5 |
| SP0757 | Cell division ABC transporter, permease protein FtsX | 2.7 | 2.3 | 6.8 |
| SP0789 | Predicted transcriptional regulator | 3.2 | 3.1 | 7.8 |
| SP0790 | Hypothetical transmembrane protein | 2.9 | 2.6 | 5.4 |
| SP0823 | Glutamine ABC transporter, permease protein (Glnp) | 3.6 | 2.8 | 26.4 |
| SP0824 | Glutamine ABC transporter, ATP-binding protein (Glnq) | 2.5 | 1.8 | 24.5 |
| SP0856 | Branched-chain amino acid aminotransferase | 1.7 | 2.4 | 2.0 |
| SP1394 | Glutamine-binding periplasmic protein precursor | 2.8 | 2.6 | 3.6 |
| SP1429 | Peptidase of U32 family | 1.7 | 1.7 | 2.4 |
| SP1460 | Amino acid ABC transporter, ATP-binding protein YckI | 2.2 | 2.8 | 2.1 |
| SP1461 | Amino acid ABC transporter permease | 2.6 | 2.4 | 2.3 |
| SP1624 | 1-Acyl-sn-glycerol-3-phosphate acyltransferase | 2.6 | 6.4 | 2.3 |
| SP2073 | ABC transporter homolog z | 2.6 | 2.5 | 2.0 |
| SP2075 | Hypothetical ABC transporter, ATP-binding protein | 3.2 | 4.4 | 4.0 |
| SP2126 | Dihydroxy acid dehydratase | 1.6 | 2.4 | 2.6 |
ID, identification.
Time of exposure to ciprofloxacin in minutes.
FIG. 4.
Differential expression of genes from the branched-chain amino acid biosynthetic pathway. The histograms show the AIDs for the genes encoding threonine deaminase, dihydroxy acid dehydratase, and the branched-chain amino acid aminotransferase. The genes for the two subunits of acetolactate synthase had an expression profile similar to that of threonine deaminase. Expression levels are indicated as follows: unexposed controls (white), samples exposed to 2 μg/ml ciprofloxacin (light gray), samples exposed to 12 μg/ml ciprofloxacin (dark gray), and samples exposed to 80 μg/ml ciprofloxacin (black).
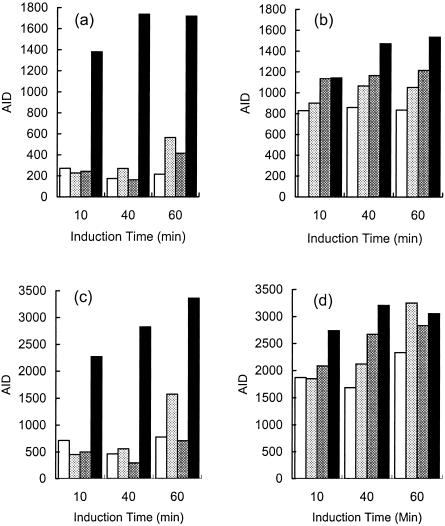
FIG. 5.
Differential expression of genes encoding putative transport proteins PatA and PatB. The histograms show the AIDs for the genes encoding PatA in M4 (a) and M22 (b) and PatB in M4 (c) and M22 (d). Expression levels are indicated as follows: unexposed controls, white; samples exposed to 2 μg/ml ciprofloxacin, light gray; samples exposed to 12 μg/ml ciprofloxacin, dark gray; samples exposed to 80 μg/ml ciprofloxacin, black.
Changes in gene expression between parent and mutant strains induced by exposure to ciprofloxacin and norfloxacin.
The MICs of ciprofloxacin and norfloxacin for strain M4 observed after overnight incubation were 2 μg/ml and 4 μg/ml, respectively (32). Ciprofloxacin caused a rapid cessation of growth of strain M4; no effect was evident after 10 min of incubation, but by 40 min growth was reduced by 34% and 41% for 12 and 80 μg/ml ciprofloxacin, respectively, and after 60 min strain M4 had stopped growing. The fluoroquinolones only affected strain M22 (MICs of ciprofloxacin and norfloxacin, 12 and 32 μg/ml, respectively) at high concentrations (12 and 80 μg/ml) and after a long incubation (60 min); strain M22 only stopped growing after 200 min of incubation with 80 μg/ml ciprofloxacin. The fluoroquinolones both had a much stronger effect on gene expression in strain M4 than in strain M22, and almost twice as many genes were down-regulated as were up-regulated (Fig. 2 and 3). Many of the genes affected in strain M4 were reported to respond to fluoroquinolones in H. influenzae (20). In general, the changes in gene expression observed were time and concentration dependent in strain M4 but not in strain M22. Norfloxacin had much less of an effect on growth (16% inhibition of strain M4 at 4 μg/ml after 60 min and no inhibition of strain M22 at 32 μg/ml after 60 min) and induced fewer changes in gene expression under every condition investigated (Fig. 1). We therefore concentrated on ciprofloxacin in the rest of the analysis. At 2 μg/ml ciprofloxacin, the expression of <1% of the genes in strain M4 was altered. After exposure to 80 μg/ml ciprofloxacin, the expression of 15% of the genes was altered and the CHFs were larger (Fig. 2). Fewer genes were affected in strain M22 under any condition (Fig. 2 and 3). This indicates that many of the gene expression changes stem from responses common to both strains and therefore not involved in the resistance of strain M22. Among the genes showing clear differences in regulation between M4 and M22 are putative transporters, DNA topoisomerases, and a number of genes that participate in DNA repair.
Expression of genes encoding putative transporters.
Expression of PatA and PatB in strain M4 was induced by ciprofloxacin at 80 μg/ml at all times and by lower concentrations after 60 min (Fig. 5). In contrast, modest induction (CHFs, 1.3 to 1.9) from the already high level in strain M22 was both time and concentration dependent. Expression reached similar levels in both strains after induction at 80 μg/ml for 60 min. The expression changes induced by ciprofloxacin under selected conditions were confirmed by RT-PCR (Table 2). Both genes were also induced by norfloxacin at its MICs for the respective strains (CHF of 1.7 for strain M4 at 4 μg/ml, CHF of 1.4 for strain M22 at 32 μg/ml). Six other transporter genes had higher expression in strain M22: SP0287, a putative member of the xanthine/uracil permease family; SP0786, an ATP-binding protein; SP1282, a homologue of MsrA which confers resistance to 14-membered ring macrolides and type B streptogramins in Staphylococcus epidermidis (40); SP1587, a homologue of an oxalate/formate antiporter; SP1861, an ABC transporter homologous to osmoprotectant transporters; and SP2169, a homologue of ABC transporters involved in Zn(II) uptake. The responses of these genes to fluoroquinolones will be discussed in more detail elsewhere (32). Expression of pmrA by either strain was not affected by ciprofloxacin or norfloxacin (−1.6 ≤ CHF ≤ 1.6 under all conditions).
TABLE 2.
Changes in expression of selected genes after exposure to ciprofloxacin monitored by RT-PCR
| TIGR4: IDa | Description | Ciprofloxacin exposure
|
Mean n-fold change in expression level ± SEM (CHF)
|
||||
|---|---|---|---|---|---|---|---|
| Time (min) | Concn (μg/ml) | M4
|
M22
|
||||
| RT-PCR | Chip | RT-PCR | Chip | ||||
| SP2073 | PatB | 10 | 80 | 7.1 ± 0.31 (6.1) | (4.1) | 1.09 ± 0.65 (0.1) | (0.5) |
| SP2075 | PatA | 10 | 80 | 4.6 ± 0.18 (3.6) | (2.2) | 1.35 ± 0.25 (0.4) | (0.4) |
| SP0450 | Threonine dehydratase | 10 | 80 | 0.26 ± 0.19 (−2.9) | (−2.3) | 0.93 ± 0.40 (−0.1) | (−2.1) |
| SP1202 | RecN | 40 | 2 | 1.37 ± 0.43 (0.37) | (−1.3) | 1.7 ± 0.49 (0.7) | (−0.5) |
| SP1219 | GyrA | 40 | 2 | 0.95 ± 0.93 (−0.053) | (−0.5) | 1.05 ± 0.27 (0.05) | (−0.1) |
ID, identification.
Expression of topoisomerase genes.
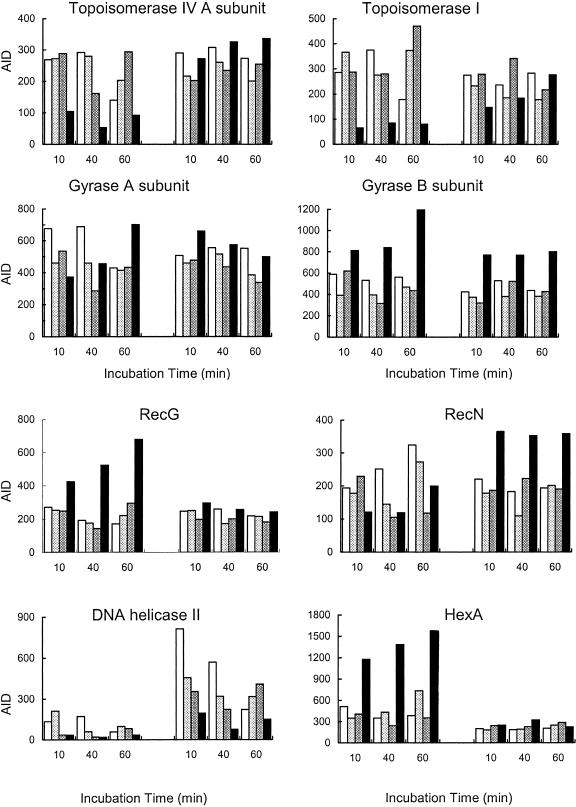
The basal expression levels of the genes encoding topoisomerase I (topA), topoisomerase IV subunits A and B (parC and parE, respectively), and gyrase subunits A and B (gyrA and gyrB, respectively) were similar in both strains. Expression of topA, parC, and parE was repressed in strain M4 at 80 μg/ml ciprofloxacin and not at all in strain M22. Expression of gyrA was not strongly affected in either strain under any condition (Fig. 6 and Table 2), and gyrB was significantly induced by 80 μg/ml ciprofloxacin in strain M4 but not strain M22 (Fig. 6).
FIG. 6.
Differential expression of genes involvement in maintenance of DNA topology and integrity. The histograms show the AIDs for the genes encoding topoisomerase IV subunit A (the expression profile of subunit B was very similar), topoisomerase I, gyrase subunits A and B, the RecG protein, the RecN protein, DNA helicase II, and the HexA protein. Expression levels are indicated as follows: unexposed controls, white; samples exposed to 2 μg/ml ciprofloxacin, light gray; samples exposed to 12 μg/ml ciprofloxacin, dark gray; samples exposed to 80 μg/ml ciprofloxacin, black.
Expression of genes involved in DNA repair and replication.
Some genes were repressed in strain M22 irrespective of the condition (Fig. 6). These included dnaG (replication primase), recA (break repair and recombination), and hexA (mismatch repair). Expression of primase was affected by ciprofloxacin in both strains (data not shown), whereas ciprofloxacin had much less of an effect on the expression of recA and, especially, hexA (Fig. 6) in strain M22 than it did in strain M4. Other genes had similar levels of expression in both strains, except after exposure to the higher concentrations, especially 80 μg/ml ciprofloxacin, which had a strong effect on expression levels in strain M4 but not in strain M22. These genes included recF and recG, both induced in strain M4 (Fig. 6); polA (DNA polymerase I) and lig (DNA ligase), both repressed in strain M4 (data not shown); and recN, which was repressed in strain M4 but induced in strain M22 (Fig. 6). The gene encoding DNA helicase II was expressed at a much higher level in strain M22, but the expression level responded to the ciprofloxacin concentration in the same way as in strain M4 (Fig. 6). At high concentrations, the residual expression level in strain M22 was similar to the expression level in strain M4 in the absence of the agent.
DISCUSSION
Quinolones form a quinolone-topoisomerase-DNA cleavable complex that can be converted into a double-stranded break but which also interferes with the replication fork, thereby inhibiting DNA synthesis and cell growth (18, 22, 26). The physiological consequences of fluoroquinolone action observable in the microarray analysis of strain M4 were an increase in the expression of the genes involved in the SOS pathway. These included recA, recF, recG, uvrD, mut, and ruvA. Increased expression of DNA primase, the chromosomal replication initiator dnaA, and single-stranded DNA-binding proteins may be seen as compensating for inhibition of replication. The expression of hexA and hexB, which are part of a mismatch repair system analogous to the MutSL system in E. coli (24), was induced by ciprofloxacin in strain M4, but both genes were constitutively repressed and ciprofloxacin insensitive in strain M22. Spontaneous mutator strains are defective in mismatch repair pathways, often because the mutS gene is inactivated (17), which also tends to increase a hyperrecombination phenotype (40). Abrogation of fidelity mechanisms such as proofreading and mismatch correction increases the frequency of mutations, some of which might lead to resistance. Blasquez et al. (5) reported that there is a relationship between the molecular mechanisms of hypermutability and acquisition of resistance. The constitutive repression of the hexA and hexB genes in strain M22 suggests that mutations are less likely to be corrected, increasing the possibility for the cell to acquire mutations leading to resistance.
Resistance in strain M22 was not associated with mutation or altered regulation of the target protein gyrase or topoisomerase IV (12, 30). These genes were only affected by exposure to high concentrations of ciprofloxacin in strain M4 and are clearly not involved in the short-term reaction to growth inhibition in either organism.
The constitutive high-level expression of the two efflux transporter homologues PatA and PatB in strain M22 strongly suggests a role for these two proteins in its efflux resistance phenotype. The role of PatA and PatB in fluoroquinolone transport will be investigated elsewhere (32). Expression of PmrA, which has been implicated in fluoroquinolone resistance (19), was not affected by the fluoroquinolones used in this study. It has been noted before that expression of PmrA in clinical isolates does not correlate with a phenotype suggestive of a fluoroquinolone efflux mutant (39).
The response of strain M4 to ciprofloxacin is a complicated one involving a network of genes implicated in the transport of nutrients and waste products, sensing environmental stresses, replication, transcription, and DNA repair. The mutant M22 has acquired a regulatory pattern that anticipates many of these induced changes. The regulation of this network of genes is still under investigation, but the frequency of mutation observed during selection of strain M22 suggests a point mutation (38), while the transcriptome analysis suggests that the mutation affects a global regulator. Such a mutation could affect numerous unlinked genes and be analogous to a marR(O) mutation in E. coli, where increased expression of MarA alters the expression of over 60 unlinked genes, including some involved in antibiotic efflux (2).
The net effect of the mutation in strain M22 is overexpression of the PatAB putative efflux system, which could lower cytoplasmic concentrations of fluoroquinolones; overexpression of nutrient uptake systems and metabolic pathways, which could increase fitness; and repression of repair mechanisms, which could allow a hypermutator phenotype. These three predictions are being examined in more detail; the role of PatA/PatB will be discussed elsewhere (32). The three factors add up not only to an organism that has an established resistance but also to a “superbug” that is equipped to face further challenges and to evolve to meet them.
.
Acknowledgments
We thank Antoine de Saizieu for sharing microarray expertise and both Antoine de Saizieu and Martin Stieger for helpful discussions.
REFERENCES
- 1.Baranova, N. N., and A. A. Neyfakh. 1997. Apparent involvement of a multidrug transporter in the fluoroquinolone resistance of Streptococcus pneumoniae. Antimicrob. Chemother. 41:1396-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbosa, T. M., and S. B. Levy. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 182:3467-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartlett, J. G., and L. M. Grundy. 1995. Community acquired pneumonia. N. Engl. J. Med. 333:1618-1624. [DOI] [PubMed] [Google Scholar]
- 4.Bernard, L., J.-C. Ngyen Van, and J. L. Mainardi. 1995. In vivo selection of Streptococcus pneumoniae resistant to quinolones including sparfloxacin. Clin. Microbiol. Infect. 1:60-61. [DOI] [PubMed] [Google Scholar]
- 5.Blasquez, J., O. A.Gomez, and J. M. Gomez. 2002. Mutation and evolution of antibiotic resistance: antibiotics as promoters of antibiotic resistance? Curr. Drug Targets 3:345-349. [DOI] [PubMed] [Google Scholar]
- 6.Brenwald, N. P., M. J. Gill, and R. Wise. 1997. The effect of reserpine, an inhibitor of multi-drug efflux pumps, on the in vitro susceptibilities of fluoroquinolone-resistant strains of Streptococcus pneumoniae to norfloxacin. J. Antimicrob. Chemother. 40:458-460. [DOI] [PubMed] [Google Scholar]
- 7.Brenwald, N. P., M. J. Gill, and R. Wise. 1998. Prevalence of a putative efflux mechanism among fluoroquinolone-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2032-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brenwald, N. P., P. Appelbaum, T. Davies, and M. J. Gill. 2003. Evidence for efflux pumps other than PmrA associated with fluoroquinolone resistance in Streptococcus pneumoniae. Clin. Microbiol. Infect. 9:140-143. [DOI] [PubMed] [Google Scholar]
- 9.Call, D. R., M. K. Bakko, M. J. Krug, and M. C. Roberts. 2003. Identifying antimicrobial resistance genes with DNA microarrays. Antimicrob. Agents Chemother. 47:3290-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canton, R., M. Morosini, M. C. Enright, and I. Morrissey. 2003. Worldwide incidence, molecular epidemiology and mutations implicated in fluoroquinolone-resistant Streptococcus pneumoniae: data from the PROTEKT surveillance programme. J. Antimicrob. Chemother. 52:944-952. [DOI] [PubMed] [Google Scholar]
- 11.Chaussee, M. S., G. L. Sylva, D. E. Sturdevant, L. M. Smoot, M. R. Graham, R. O. Watson, and J. M. Musser. 2002. Rgg influences the expression of multiple regulatory loci to coregulate virulence factor expression in Streptococcus pyogenes. Infect. Immun. 70:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi, H., H.-J. Lee, and Y. Lee. 1998. A mutation in QRDR in the ParC subunit of topoisomerase IV was responsible for fluoroquinolone resistance in clinical isolates of Streptococcus pneumoniae. Yonsei Med. J. 39:541-545. [DOI] [PubMed] [Google Scholar]
- 13.Daporta, M. T., J. L. Munoz Bellido, G. Y. Guirao, M. S. Hernandez, and J. A. Garcia-Rodriguez. 2004. In vitro activity of older and newer fluoroquinolones against efflux-mediated high-level ciprofloxacin-resistant Streptococcus pneumoniae. Int. J. Antimicrob. Agents 24:185-187. [DOI] [PubMed] [Google Scholar]
- 14.Davies, T. A., G. A. Pankuch, B. E. Dewasses, M. R. Jacobs, and P. C. Appelbaum. 1999. In vitro development of resistance to five quinolones and amoxicillin-clavulanate in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 43:1177-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Saizieu, A., U. Certa, J. Warrington, C. Gray, W. Keck, and J. Mous. 1998. Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nat. Biotechnol. 16:45-48. [DOI] [PubMed] [Google Scholar]
- 16.de Saizieu, A., C. Gardès, N. Flint, C. Wagner, M. Kamber, T. J. Mitchell, W. Keck, K. E. Amrein, and R. Lange. 2000. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J. Bacteriol. 182:4696-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Visser, G. M. 2002. The fate of microbial mutators. Microbiology 148:1247-1252. [DOI] [PubMed] [Google Scholar]
- 18.Fournier, B., X. Shako, T. Lu, K. Delia, and D. C. Hooper. 2000. Selective targeting of topoisomerase IV and DNA gyrase in Staphylococcus aureus: different patterns of quinolone-induced inhibition of DNA synthesis. Antimicrob. Agents Chemother. 44:2160-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gill, M. J., N. P. Brenwald, and R. Wise. 1999. Identification of an efflux pump gene, pmrA, associated with fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 43:187-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gmuender, H., K. Kuratli, K. Padova, C. P. Gray, W. Keck, and S. Evers. 2001. Gene expression changes triggered by exposure of Haemophilus influenzae to novobiocin or ciprofloxacin: combined transcription and translation analysis. Genome Res. 11:28-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hakenbeck, R., N. Balmelle, B. Weber, C. Gardès, W. Keck, and A. de Saizieu. 2001. Mosaic genes and mosaic chromosomes: Intra- and interspecies genomic variation of Streptococcus pneumoniae. Infect. Immun. 69:2477-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heddle, J. G., F. M. Barnard, L. M. Wentzell, and A. Maxwell. 2000. The interaction of drugs with DNA gyrase: a model for the molecular basis of quinolone action. Nucleosides Nucleotides Nucleic Acids 19:1249-1264. [DOI] [PubMed] [Google Scholar]
- 23.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. De Hoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. Le Blanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. Mc Ahren, M. Mc Henney, K. Mc Leaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Humbert, O., M. Prudhomme, R. Hakenbeck, C. G. Dowson, and J. P. Claverys. 1995. Homologous recombination and mismatch repair during transformation in Streptococcus pneumoniae: saturation of the Hex mismatch repair system. Proc. Natl. Acad. Sci. USA 92:9052-9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutter, B., C. Schaab, S. Albrecht, M. Borgmann, N. A. Brunner, C. Freiberg, K. Ziegelbauer, C. O. Rock, I. Ivanov, and H. Loferer. 2004. Prediction of mechanisms of action of antibacterial compounds by gene expression profiling. Antimicrob. Agents Chemother. 48:2838-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khodursky, A. B., and N. R. Cozzarelli. 1998. The mechanism of inhibition of topoisomerase IV by quinolone antibacterials. J. Biol. Chem. 273:42:27668-27677. [DOI] [PubMed] [Google Scholar]
- 27.Korner, R. J., D. S. Reeves, and A. P. MacGowen. 1994. Dangers of oral fluoroquinolone treatment in community acquired upper respiratory tract infections. BMJ 308:191-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, B. L., A. M. Padula, R. C. Kimbrough, S. R. Jones, R. E. Chaisson, J. Mills, and M. A. Sandy. 1991. Infection complications with respiratory pathogens despite ciprofloxacin therapy. N. Engl. J. Med. 325:520. [DOI] [PubMed] [Google Scholar]
- 29.Lockhart, D. J., H. Dong, M. C. Byrne, M. T. Follettie, M. V. Gallo, M. S. Chee, M. Mittmann, C. Wang, M. Kobayashi, H. Horton, and E. L. Brown. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14:1675-1680. [DOI] [PubMed] [Google Scholar]
- 30.Madaras-Kelly, K. J., C. Daniels, M. Hegbloom, and M. Thompson. 2002. Pharmacodynamic characterization of efflux and topoisomerase IV-mediated fluoroquinolone resistance in Streptococcus pneumoniae. J. Antimicrob. Chemother. 50:211-218. [DOI] [PubMed] [Google Scholar]
- 31.Markham, P. N. 1999. Inhibition of the emergence of ciprofloxacin resistance in Streptococcus pneumoniae by the multidrug efflux inhibitor reserpine. Antimicrob. Agents Chemother. 43:988-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marrer, E., K. Schad, A. T. Satoh, M. G. P. Page, M. M. Johnson, and L. J. V. Piddock. Submitted for publication.
- 33.Martin, R. G., and J. L. Rosner. 1995. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc. Natl. Acad. Sci. USA 92:5456-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montanari, M. P., E. Tili, I. Cochetti, M. Mingoia, A. Manzin, and P. E. Varaldo. 2004. Molecular characterization of clinical Streptococcus pneumoniae isolates with reduced susceptibility to fluoroquinolones emerging in Italy. Microb. Drug Res. 10:209-217. [DOI] [PubMed] [Google Scholar]
- 35.Perichon, B., J. Tankovic, and P. Courvalin. 1997. Characterization of a mutation in the parE gene that confers fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:1166-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piddock, L. J. V., Y.-F. Jin, and M. J. Everett. 1997. Non-gyrA-mediated ciprofloxacin resistance in laboratory mutants of Streptococcus pneumoniae. J. Antimicrob. Chemother. 39:609-615. [DOI] [PubMed] [Google Scholar]
- 37.Piddock, L. J. V., M. M. Johnson, V. Ricci, and S. L. Hill. 1998. Activities of new fluoroquinolones against fluoroquinolone-resistant pathogens of the lower respiratory tract. Antimicrob. Agents Chemother. 42:2956-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piddock, L. J. V., and M. M. Johnson. 2002. Accumulation of ten fluoroquinolones by wild-type and efflux mutant Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:813-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piddock, L. J. V., M. M. Johnson, S. Simjee, and L. Pumbe. 2002. Expression of the efflux pump gene pmrA in fluoroquinolone-resistant and -susceptible clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:808-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rayssiguier, C., D. S. Thaler, and M. Radman. 1989. The barrier of recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature 342:396-401. [DOI] [PubMed] [Google Scholar]
- 41.Reynolds, E., J. I. Ross, and J. H. Cove. 2003. Msr(A) and related macrolide/streptogramin resistance determinants: incomplete transporters? Int. J. Antimicrob. Agents 22:228-236. [DOI] [PubMed] [Google Scholar]
- 42.Rosenberg, E. Y., D. Bertenthal, M. L. Nilles, K. P. Bertrand, and H. Nikkaido. 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 48:1609-1619. [DOI] [PubMed] [Google Scholar]
- 43.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 44.Van Dyk, T., L. J. Templeton, K. A. Cantera, P. L. Sharpe, and F. S. Sariaslani. 2004. Characterization of the Escherichia coli AaeAB efflux pump: a metabolic relief valve? J. Bacteriol. 186:7196-7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner, C., A. de Saizeau, H.-J. Schönfeld, M. Kamber, R. Lange, C. J. Thompson, and M. G. P. Page. 2002. Genetic analysis and functional characterization of the Streptococcus pneumoniae vic operon. Infect. Immun. 70:6121-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeller, V., C. Janoir, M.-D. Kitzis, L. Gutmann, and N. J. Moreau. 1997. Active efflux as a mechanism of resistance to ciprofloxacin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:1973-1978. [DOI] [PMC free article] [PubMed] [Google Scholar]