Abstract
The ecological impact of antibiotic resistance in the absence of selective pressure has been poorly studied. We assessed the carriage of tetracycline resistance genes, persistence in the microbiota, fecal population counts and virulence factor genes in 309 commensal, intestinal Escherichia coli strains obtained from 128 Swedish infants followed during the first year of life with regular quantitative fecal cultures. No infant was given tetracycline, but 25% received other antibiotics. Tetracycline resistance was identified in 12% of strains, all of which carried either tet(A) (49%) or tet(B) (51%) genes. Resistance to other antibiotics occurred in 50% of tet(A)-positive strains, 42% of tet(B)-positive strains and 13% of tetracycline-sensitive strains. However, colonization with tetracycline-resistant strains was unrelated to treatment with antibiotics. Strains that were tet(B)- or tet(A)-positive carried the genes for P fimbriae and aerobactin, respectively, more often than susceptible strains. Tetracycline-resistant and -susceptible strains were equally likely to persist among the intestinal microbiota for ≥3 weeks and had similar population numbers. However, when a resistant strain and a susceptible strain colonized a child simultaneously, the resistant variety showed lower counts (P = 0.03). In cases of long-term colonization by initially tetracycline-resistant E. coli strains, loss of tet genes occurred in 3 of 13 cases with variable effects on population counts. The results indicate that there is limited pressure against the carriage of tet genes in the infantile gut microbiota even in the absence of antibiotics. Resistant strains may possess colonization factors that balance the cost of producing resistance elements.
Treatment of humans and animals by antibiotics affects not only the targeted pathogenic bacteria, but also the complex commensal microbial communities that inhabit the skin and mucosal membranes. The emergence of resistance among commensal bacteria is a serious side effect of antibiotic usage in human and veterinary medicine because the commensals may, at a later stage, cause extraintestinal infections (15), spread to other hosts (27), or transfer genetic resistance elements to other members of the microbiota (18). Despite this, the dynamics of antibiotic resistance in commensals has not been studied to any great extent. For example, it is not known whether antibiotic resistance influences the capacity of a given strain to persist in the normal microbiota, or whether resistance genes are kept or lost during colonization. In theory, resistance elements which do not confer any advantage should be eliminated from the gene pool. However, the persistence of resistant clones in the absence of selective pressure from antibiotics has been observed (31).
Escherichia coli is the most common facultative bacterium in the intestinal microbiota of humans and many animals. Most people have one or a few E. coli strains in the large intestine at any point in time. Some strains, termed resident, may persist for many months in the microbiota of the individual, while so-called transient strains have low colonizing capacities and disappear within some weeks (32). E. coli strains colonizing the large intestine vary in their carriage of genes encoding adhesins, iron-trapping compounds and hemolysin (23). These are virulence factors known to enable bacteria to cause extraintestinal infections, such as urinary tract infection and septicemia (36, 37). Some of these factors, especially P fimbriae, also contribute to persistence of E. coli in the colon (23).
Tetracycline resistance is an attractive model for studying the ecology of antibiotic resistance. Tetracycline exerts its antibacterial action by binding to the bacterial ribosome and inhibiting protein synthesis (30). It is one of the earliest broad-spectrum antimicrobials, and since it was developed in the 1940s, it has been produced at the largest scale of all antibiotics (28). Not only has it been used in human and veterinary medicine, but it has also been used as a growth promoter in animal husbandry (34). Tetracycline is never given to children, due to its effect on growing bones and teeth (29). Tetracycline is not used to treat E. coli infections in humans, but resistance to tetracycline is still common in E. coli (4, 7), which suggests that resistance has been selected by a bystander effect on commensal E. coli, during treatment of other pathogens in humans or animals. Bacterial resistance to tetracycline is most commonly mediated by energy-dependent pumping of tetracycline out of the bacterial cell. The tet(A), -(B), -(C), -(D), -(E), -(Y), and -(I) genes in gram-negative bacteria encode such efflux systems (6).
In the present study, the occurrence of phenotypic tetracycline resistance and the carriage of tet resistance genes in intestinal E. coli strains obtained from Swedish infants, followed longitudinally over their first year of life with regular stool sampling, is investigated. The strains were characterized with respect to the time of persistence in the individual infant, fecal population counts and carriage of virulence factor genes. The stability of tetracycline resistance gene carriage and the effect of resistance on persistence in the bowel microbiota in a human population not exposed to tetracycline could, thereby, be assessed. Furthermore, the relation between virulence and antibiotic resistance gene carriage could be studied.
MATERIALS AND METHODS
Isolation, culture and identification of intestinal E. coli.
Commensal Escherichia coli strains from 128 Swedish infants born in the period from 1998 to 2001 at the Sahlgrenska University Hospital were analyzed. The infants were part of the ALLERGYFLORA birth-cohort, in which the compositions of their microbiota were analyzed by quantitative cultivation of serial fecal specimens and correlated to later development of allergies (1). In brief, a rectal swab was obtained from each child at 3 days of age and cultured semiquantitatively for facultative bacteria. Fecal samples were obtained at 1, 2, 4, and 8 weeks and at 6, 12 and, in some cases, 18 months and 3 years of age and cultured quantitatively for all major groups of facultative and anaerobic bacteria (1). The parents registered feeding patterns, illnesses and medications in a diary. Almost all infants were breast-fed until four months of age. When solid foods were introduced, 75% of the infants still received some breast milk until at least 6 months of age.
Twenty-five percent of the infants received antibiotics at least once during their first year of life, namely, penicillin V or G (n = 22), amoxicillin/ampicillin (n = 13), trimethoprim (n = 4), erythromycin (n = 4), cefuroxime/ceftibuten (n = 2), nitrofurantoin (n = 1), tobramycin (n = 1), and ceftazidime (n = 1). No child received tetracycline or related compounds.
For the isolation of E. coli, serially diluted fecal samples were cultivated on Drigalski agar, which is selective for Enterobacteriaceae. One to five different colony types, which were judged to differ in size, shape, color, or mucoid appearance, were isolated from each sample. Each colony type was enumerated separately, speciated using API 20E identification strips (bioMerieux, Marcy-l'Etoile, France), and assigned a strain identity by random amplified polymorphic DNA (RAPD) analysis (24, 25). Several isolates were defined as belonging to the same strain if their RAPD profiles showed the same major bands and differed in no more than two to three minor bands (5). If several isolates in a sample were found to belong to the same strain by this definition, their population counts were added together. Thus, one or several different strains could be present in a single sample, each of which had an individual population count. The time of persistence of each strain was determined by its appearance in multiple samples. On average, each child carried 2.2 E. coli strains during the first year of life, of which 81% persisted for ≥3 weeks in the microbiota. The population counts of individual strains varied from 103.5 to 1012.52, the level of detection being 102.52 (300 colonies/g of feces).
PFGE.
In selected cases, the strain identities determined by RAPD were confirmed by pulsed-field gel electrophoresis (PFGE) analysis, as described elsewhere (35). The PFGE types were interpreted according to the method of Tenover et al. (33). Isolates whose PFGE patterns differed by ≤3 bands were defined as belonging to the same strain (33).
Carriage of virulence genes.
At least one isolate of each strain was assessed, using multiplex PCRs, for the following virulence factor genes: type 1 fimbriae (fimA), P fimbriae (papC), S fimbriae (sfaDE), aerobactin (iutA), and hemolysin (hlyA), as previously described (24). Previous analyses showed that, among 15 strains analyzed, all isolates of the same strain showed identical virulence gene patterns (own unpublished results).
Phenotypic resistance to tetracycline and other antibiotics.
At least one isolate of each E. coli strain was screened for resistance to tetracycline, using the agar disk diffusion method as outlined by the Swedish Reference Group for Antibiotics (SRGA) (32a). Strains with clearance zones of <19 mm in diameter were further analyzed for tetracycline MICs by the Etest, as outlined by SRGA (32a).
Phenotypic resistance to the following antibiotics was also screened using disk-diffusion (AB Biodisk): ampicillin, amdinocillin, norfloxacin (10 μg), cefadroxil, ceftazidime, chloramphenicol, gentamicin, tobramycin (30 μg), nitrofurantoin (100 μg), and trimethoprim (5 μg), according to methodology specified by SRGA (32a).
Multiplex PCR detection of tet genes.
All isolates of tetracycline-resistant E. coli strains were assessed for carriage of the tetracycline resistance genes tet(A), -(B), -(C), -(D), and -(E) by multiplex PCR, using previously published primers (21). The method was optimized using the following reference strains with known tet genes: E. coli K-12 NC 50078-02 [tet(A)], E. coli K-12 NC 50019 [tet(B)], E. coli K-12 NC 50270-01 [tet(C)], E. coli K-12 J53-1 RA1 NC 50073-02 [tet(D)], and E. coli HB 101 pSL 1456 NC 50273-01 [tet(E)] (all from the Public Health Laboratory Service, London, United Kingdom). The following procedure was used: a small amount of biomass from a bacterial colony was added to a sterile thin-walled reaction tube (PerkinElmer, Foster City, CA) containing HotStarTaq Master Mix (QIAGEN, Spånga, Sweden), primers, MgCl2, and distilled water. The first multiplex PCR included primers for tet(B) and tet(C) (0.25 μM each) and 1.5 mM MgCl2. The second PCR included primers for tet(A) and tet(E) (1.0 μM each) and tet(D) (3 μM) and 2 mM MgCl2. The tubes were sealed with a drop of mineral oil and heated to 95°C for 15 min in a thermocycler (model 480; PerkinElmer Cetus) to activate the HotStarTaq DNA polymerase. After 5 min DNA template denaturation at 94°C, 25 PCR cycles followed, with DNA denaturation at 94°C for 1 min, primer annealing at 60°C for 1 min, and primer extension at 72°C for 1.5 min.
PCR products were separated electrophoretically in a 2% agarose gel, visualized by staining with 0.5 μg/ml ethidium bromide, and examined in UV light.
Statistics.
Frequencies were compared using Fisher's exact test, MIC values were compared using an unpaired t test, and population counts were compared using a paired t test. When analyzing the occurrences of carriage of virulence factors in different bacterial groups, only significances at P values of <0.01 and <0.001 were considered, in order to reduce the risk of random significances with multiple comparisons.
RESULTS
Resistance to tetracycline and association of tet gene to other antibiotic resistance.
Thirty-seven (12%) of the commensal E. coli strains were phenotypically resistant to tetracycline. Tetracycline-resistant strains were analyzed with respect to the carriage of tet(A) through tet(E) genes and MICs. All tetracycline-resistant strains carried at least one of the tet genes examined. tet(B) was the most commonly observed, found in 51% (19 of 37) of resistant strains, followed by tet(A) (49%, 18 of 37). tet(C) was found in one strain, in combination with tet(A). Neither tet(D) nor tet(E) was detected.
The MICs of tetracycline-resistant strains ranged from 32 to 256 mg/liter (median, 128 mg/liter), with significantly higher values for strains carrying tet(B) than for those carrying tet(A) (median, 256 versus 96 mg/liter; P < 0.0001).
All strains were also tested for their susceptibilities to ampicillin, cefadroxil, ceftazidime, amdinocillin, chloramphenicol, gentamicin, tobramycin, nitrofurantoin, norfloxacin, and trimethoprim. Tetracycline resistance occurred in combination with resistance to ampicillin (n = 12), trimethoprim (n = 11), chloramphenicol (n = 2), and cefadroxil (n = 1). Strains carrying tet(A) or tet(B) were more often resistant to other antibiotics than were tetracycline-susceptible strains (50% and 42% versus 13%, P = 0.0003 and P = 0.002, respectively).
Resistance to tetracycline and carriage of virulence genes.
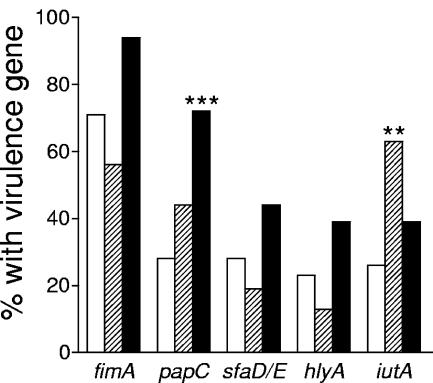
Tetracycline-resistant and -susceptible strains were screened for the carriage of a range of virulence factor genes shown to be of importance for extra-intestinal infections, such as P, type 1 and S fimbriae, aerobactin and hemolysin (36, 37). As shown in Fig. 1, strains carrying tet(B) had the P-fimbrial gene papC significantly more often than susceptible strains (72% versus 28%, P = 0.0002). Strains carrying tet(A) also had the aerobactin gene iutA more often than susceptible strains (63% versus 26%, P = 0.003). The genes fimA, sfaDE, and hlyA all tended to be more common in tet(B)-positive strains than in other strains, but the differences were not significant. Thus, in general, the tet(B)-positive strains appeared to be more virulent than tet(A)-positive and susceptible strains (Fig. 1).
FIG. 1.
Frequencies of various virulence factor genes in E. coli strains carrying tet(A) or tet(B) and in strains susceptible to tetracycline. The white bars denote tetracycline-susceptible strains, striped bars tet(A)-positive strains, and black bars tet(B)-positive strains. **, P < 0.01, and ***, P < 0.001, compared to susceptible strains by Fisher's exact test.
Persistence and population counts of tetracycline-resistant and -susceptible strains.
Since the production of resistance elements is energy consuming, resistant strains in the complex intestinal microbiota may be disadvantaged in competition with strains without resistance markers. In order to investigate this, resistant and susceptible isolates were examined with respect to whether they are equally likely to be resident in the microbiota, defined as being present for at least 3 weeks in an infant. Strains appearing only once in samples, spaced >1 month, were excluded, since their colonizing period could not be determined adequately. Among all strains, 109 could be defined as resident and 25 as transient. The proportion of strains that colonized for ≥3 weeks was 64% (7 of 11) among tet(A)-positive strains, 86% (6 of 7) among tet(B)-positive strains and 83% (96 of 116) among susceptible strains. Thus, most tetracycline-resistant strains were capable of long-term persistence in the infantile microbiota.
Another measure of colonizing success is the capacity to form populations of high bacterial numbers. When comparing population levels of different strains, one must take into account that the population levels of facultative bacteria like E. coli decrease with time, in parallel with the acquisition of an increasingly complex microbiota (2). Accordingly, the average strain population counts decreased from 108.7 CFU/g feces at the age of one week to 107.7 CFU/g feces at 1 year (P = 0.0025). In cross-sectional analysis, no significant differences were observed in average population counts at any time point between tet(A)-positive, tet(B)-positive and tet susceptible strains, (e.g., 108.42, 108.34, and 108.31, respectively, at 6 months of age).
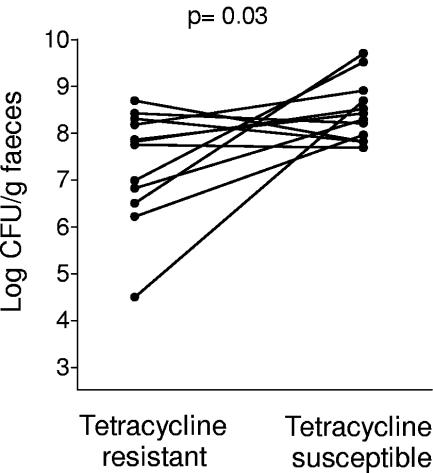
Even if resistant strains expand to high population numbers, they may be at a disadvantage when competing with susceptible strains of the same species. To examine this, the population counts of resistant and susceptible E. coli strains that colonized simultaneously the same child were compared. Eleven infants carried one tetracycline-resistant strain and one tetracycline-susceptible strain at the same time point. The population counts of resistant and susceptible E. coli in these infants were compared by paired t test. Tetracycline-resistant E. coli strains showed significantly lower population counts than tetracycline-susceptible strains in the same stool samples (P = 0.03) (Fig. 2).
FIG. 2.
Fecal population counts (log 10 values) for tetracycline-resistant strains and tetracycline-susceptible strains colonizing simultaneously in an infant. Eleven infants were colonized with one tetracycline-resistant and one tetracycline-susceptible strain simultaneously at a time point. The P value was calculated by paired t test.
Colonization by tetracycline-resistant strains in relation to treatment with antibiotics.
No infant was treated with tetracycline, but tetracycline resistance was not uncommon among intestinal E. coli, and resistant strains appeared to persist well in the microbiota. The relationship between antibiotic treatments of the individual infants and the appearance of tetracycline-resistant strains in the intestinal microbiota of the children was assessed in order to examine whether cross-selection by other antibiotics contributed to colonization by tetracycline-resistant E. coli.
Twenty-five percent of the infants received antibiotics during their first year of life, mostly penicillin or amoxicillin (see Materials and Methods for details). Only 6 of the 28 tetracycline-resistant strains (21%) isolated from the cohort were derived from infants treated with antibiotics, which corresponded closely with the fact that 25% of the infants had received antibiotics during the period of study. Furthermore, three of these six tetracycline-resistant strains were found in children before they were treated with antibiotic. The remaining three resistant strains isolated from children treated with antibiotics appeared at 12 months of age in infants treated between 6 and 12 months with penicillin V or amoxicillin. Two of these strains were mono-resistant to tetracycline, while one was resistant to tetracycline and trimethoprim. Thus, in no case was a tetracycline-resistant strain established after treatment with antibiotics to which it was resistant.
Stability of tetracycline resistance.
Tetracycline resistance is often carried on mobile genetic elements, such as plasmids or transposons. In theory, the loss of such elements might occur during colonization in the microbiota. In the cohort examined for this study, 13 infants were found to harbor one tetracycline-resistant E. coli strain each in their microbiota for at least 3 weeks. All isolates of these 13 strains were analyzed for phenotypic and genotypic resistance to tetracycline. Ten of the strains kept their resistance genes during the entire colonization period. Three strains (colonizing children no. 91, 120, and 123) lost their resistance genes during intestinal colonization. All isolates of these strains were also analyzed by PFGE and their virulence gene profiles were determined, to confirm the strain identity that had been determined by RAPD analysis.
Table 1 shows the details of the three strains that changed resistance patterns during colonization. Strain 91A was present in child 91 at 3 days of age and persisted in the microbiota until 6 months of age (Table 1, isolate identification numbers [IDs] 1 through 5). In the 2-month sample, an isolate (isolate ID 4) lacking the aerobactin gene, iutA, was recovered from the stool specimen. This isolate belonged to strain 91A, according to RAPD profile analysis, and also had the identical PFGE pattern as another isolate from the same stool sample which had the iutA gene (Fig. 3, lanes 3 and 4). The variety of strain 91A that had lost the aerobactin gene was present at population counts 2 log units higher than the variety that carried iutA (Table 1, isolate IDs 3 and 4). At 6 months of age, the isolate of strain 91A recovered from the stools lacked not only the aerobactin gene but also tet(B) and was susceptible to tetracycline (Table 1). No change in the PFGE pattern was seen (Fig. 3, lanes 2 and 5). Loss of the tet(B) gene was associated with minimal changes in population levels between 2 and 6 months of age. After 6 months, strain 91A was lost.
TABLE 1.
E. coli strains which changed tetracycline resistance pattern during persistence in the colonic microbiotaa
| Patient and time of sampling | Strain ID | Isolate ID | Virulence gene(s) | Tetracycline zone diam (mm) | tet gene | Population count (log CFU/g feces) |
|---|---|---|---|---|---|---|
| Child 91 | ||||||
| 3 days | 91A | 1 | fimA, iutA | <6 | tet(B) | NDb |
| 4 wk | 91A | 2 | fimA, iutA | <6 | tet(B) | 8.5 |
| 2 mo | 91A | 3 | fimA, iutA | <6 | tet(B) | 5.8 |
| 2 mo | 91A | 4 | fimA | <6 | tet(B) | 7.6 |
| 6 mo | 91A | 5 | fimA | 27 | 7.8 | |
| Child 120 | ||||||
| 1 wk | 120A | 6 | fimA | <6 | tet(A) | 9.2 |
| 4 wk | 120A | 7 | fimA | <6 | tet(A) | 7.2 |
| 2 mo | 120A | 8 | fimA | 27 | 9.9 | |
| 2 mo | 120A | 9 | fimA | <6 | tet(A) | 6.5 |
| 6 mo | 120A | 10 | fimA | 28 | 8.0 | |
| Child 123 | ||||||
| 1 wk | 123A | 11 | fimA, kfiC | <6 | tet(A) | 8.2 |
| 2 mo | 123A | 12 | fimA, kfiC | <6 | tet(A) | 9.4 |
| 2 mo | 123A | 13 | fimA, kfiC | 26 | 7.3 | |
| 6 mo | 123A | 14 | fimA, kfiC | <6 | tet(A) | 8.5 |
All isolates presented in the table were analyzed for their virulence gene profile. Strain identities were determined by RAPD and confirmed by PFGE (Fig. 3). Phenotypic tetracycline resistance was analyzed by disk diffusion, and the carriage of tet genes was analyzed by multiplex PCR. The population counts were determined by counting colonies of spread plates from serial dilutions of stool specimens.
ND, not determined. The 3-day sample consisted of a rectal swab, and the strain's fecal population could not be quantified.
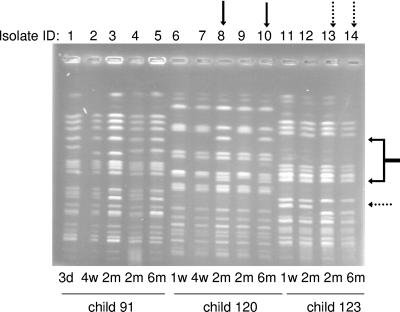
FIG. 3.
Strain genomic DNA profiles obtained with PFGE. The figure shows the results for isolates of three tetracycline-resistant E. coli strains obtained from infants 91, 120, and 123, respectively. The solid arrows show changed positions of two bands in the strain from infant 120, and the dotted arrows show the changed position of one band in the strain from infant 123. Lanes are numbered in correspondence with Table 1. d, days; w, week(s); m, months.
Child no. 120 was colonized with strain 120A from the ages of 1 week to 6 months (Table 1, isolate IDs 6 to 10). The strain was initially phenotypically tetracycline resistant and carried the tet(A) gene. At two months of age, two morphologically distinct isolates of the strain were recovered from the stool culture, one of which carried tet(A) and was phenotypically tetracycline resistant, while the other was susceptible and lacked tet genes (Table 1, isolates IDs 8 and 9). The susceptible variety of the strain (Fig. 3, lane 8) showed different positions of two PFGE bands (indicated by black arrows). According to the definition applied for PFGE profiles, these two variants belonged to the same strain (differing in <3 bands). The tetracycline-susceptible variant had a population level more than 3 log units greater than the resistant variant (Table 1, isolates IDs 8 and 9). In the 6-month stool sample, only the susceptible variety was detected (Fig. 3, lane 10).
In child no. 123, a tetracycline-resistant, tet(A)-positive strain, 123A, was established in the microbiota when the child was 1 week old and persisted until 6 months of age (Table 1, isolate IDs 11 through 14). In the 2-month sample, an isolate of the strain showed a susceptible phenotype and lacked tet genes. This variety had a population size of 107.3 CFU/g, compared to 109.4 CFU/g for the resistant variety (Table 1, isolate IDs 13 and 12, respectively). In the 6-month sample, the resistant variety was still present, but the susceptible variety was not found. The PFGE pattern of the susceptible isolate (Fig. 3, lane 13), as well as the resistant isolate from the 6-month sample (lane 14), had a difference in the position of one band compared with the original strain. By definition, they still belonged to the same strain.
Taken together, 10 of 13 strains kept their tetracycline resistance genes during colonization of the infantile bowel. In the remaining three cases, loss of the tet gene resulted in retained population counts of the strain in one case, higher fecal population counts in one case, and lower population counts in one case. None of the three infants harboring these strains received any antibiotics.
DISCUSSION
In this study, commensal E. coli isolated from Swedish infants was assessed for phenotypic and genotypic tetracycline resistance and virulence gene profiles. The infants were followed with regular sampling of the colonic microbiota over the first year of life, which enabled the determination of population counts and persistence in the microbiota of each individual strain. As it was desirable to find as many different strains as possible in individual stool samples, picking of different colonies, even if they differed very slightly in morphology, was carried out in a liberal fashion. This enabled the isolatation of quantitatively subdominant strains.
No child was given tetracycline or related compounds. Nevertheless, 12% of the E. coli strains were resistant to tetracycline. Resistance to tetracycline was second only to resistance to ampicillin in the strain collection (N. Karami, E. Lindberg, F. Nowrouzian, A. E. Wold, and I. Adlerberth, unpublished data). As a comparison, 9% of E. coli strains isolated from the rectal flora of healthy Swedish school children from 1971 to 1974 were tetracycline resistant (20). The use of tetracycline as a growth promoter for livestock was banned during the 1980s in Sweden and its use in veterinary medicine has also declined (http://www.regeringen.se). However, tetracycline is still used frequently in human clinical practice, the consumption in Sweden being slightly above three defined daily doses per 1,000 inhabitants per day (http://en.strama.se), second only to the usage of the β-lactams. The fact that tetracycline resistance in E. coli has not been reduced in the last 20 years might suggest that the frequent use of tetracycline in human medicine balances the reduced usage of tetracycline in farming animals in selection for tetracycline-resistant strains among commensal E. coli.
All phenotypically resistant isolates carried either tet(A) or tet(B), suggesting that other tet genes should be of minor importance. Accordingly, tet(A) and tet(B), both of which encode efflux mechanisms, have been identified previously as the most common tetracycline resistance genes in E. coli of both human and animal origin (7, 16).
The tet genes often occur on mobile genetic elements, such as plasmids, transposons and integrons (19). Plasmids harboring tetracycline resistance may also carry other antibiotic resistance and virulence factor genes (9, 26). A selective advantage for any gene carried on the plasmid may enhance the persistence of plasmid-bearing strains. This phenomenon, termed cross-selection, is thought to have contributed significantly to the increase in multiple drug resistance in bacteria over the last 40 years (6, 22). tet(A)- and tet(B)-positive isolates were often observed to be resistant to other antibiotics as well. However, cross-selection, by treatment with antibiotics other than tetracycline, did not explain the successful persistence of tetracycline-resistant strains in these Swedish infants. Children who had received antibiotics (mostly β-lactams) were colonized by multiresistant strains no more often than other children. In fact, colonization by a tetracycline-resistant strain was never preceded by treatment with antibiotics to which the strain was resistant.
Synthesis of genetic resistance elements costs the bacterium energy and antibiotic-resistant isolates should, theoretically, be at a disadvantage in the absence of antibiotics (8, 17). Interestingly, when a tetracycline-resistant strain and a tetracycline-susceptible strain were simultaneously present in an infant's stool sample, the resistant variety had significantly lower population counts. However, in cross-sectional analysis of the average counts of all resistant and susceptible strains found in infants of a certain age, no difference between resistant and susceptible strains was observed. This observation suggests that resistant strains can attain high population levels in the absence of direct competition from other members of the same species. In the bowel microbiota, different species occupy different ecological niches and mainly compete with members of the same bacterial group (10). The Swedish infant cohort examined in this study had, on average, only 1.5 E. coli strains in their microbiota during the first 6 months of life, which can be compared with 8.5 in Pakistani infants followed for an equal period of time (3). The limited competition with other E. coli strains may permit tetracycline-resistant strains to expand in the microbiota.
The loss of tet resistance genes was observed during colonization of the infant bowel in 3 cases, while the resistant phenotype was unaltered through colonization for as long as a year in 10 cases. In one case out of three, the susceptible mutant attained higher population levels than previously observed for the resistant variety of the strain. In the second case, the susceptible mutant disappeared from the microbiota, while the previously resistant phenotype persisted, and in the third case, population counts were retained after loss of the tet gene. Furthermore, tetracycline-resistant strains were, on average, equally as capable as susceptible strains of persisting in the microbiota. Taken together, these findings indicate a limited pressure against the carriage of tetracycline resistance gene elements in the microbiota of young Swedish infants.
An explanation for the colonizing success of resistant strains could be that they possess inherent traits that endow them with colonizing ability. tet(B)-positive strains were more often P fimbriated and tet(A)-positive strains more often had the aerobactin gene than susceptible strains. P fimbriae and aerobactin are associated not only with enhanced capacity to cause extraintestinal infection but also with persistence of E. coli in the intestinal microbiota (13, 14, 23). Because P-fimbriated strains are adapted to the human host and tend to persist in the human bowel, they may be more likely to become exposed to antibiotics used in clinical medicine than strains lacking such colonization traits. Unnecessary use of antibiotics in humans may therefore be more hazardous than misuse of antibiotics in animal husbandry, because the former targets strains with capacities to persist in the human microbiota and also to cause clinical disease in humans.
Traditionally, the risk of inducing resistance in the bacterial species to which the antibiotic treatment is directed has been considered, while potential effects on commensal bacteria have been of less concern. However, a commensal strain may later cause symptomatic infection in the same or another host or transfer its resistance genes to other bacteria. The fate of antibiotic-resistant commensal strains, therefore, deserves more attention. To our knowledge, this study is the first to examine the effects of antibiotic resistance on the colonizing capacity of individual strains in the human commensal microbiota. Although the findings of this study may not be generalized to other host groups and resistance phenotypes, it is clear that the ecological effects of antibiotic resistance in commensal bacteria are complex and deserve further study.
Acknowledgments
We thank U. Jodal, P. Larsson, T. Åhren, and E. R. B. Moore for critical reading of the manuscript. We thank Flemming Scheutz and Lykke Fisher for generously providing reference E. coli strains for development of the tet gene PCR analysis.
The study was supported by grants from the Swedish Medical Research Council (no. K98-06X-12612-01A), the Swedish Strategic Program for the Rational Use of Antimicrobial Agents and Surveillance of Resistance, the Capio Research Foundation, the Magnus Bergvall Foundation, the Wilhelm and Martina Lundgren foundation, and the Mayflower Foundation.
REFERENCES
- 1.Adlerberth, I., E. Lindberg, N. Åberg, B. Hesselmar, R. Saalman, I. L. Strannegård, and, A. E. Wold. Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel—an effect of hygienic life-style? Ped. Res., in press. [DOI] [PubMed]
- 2.Adlerberth, I., L. A. Hanson, and A. E. Wold. 1999. The ontogeny of the intestinal flora, p. 279-292. In I. Sanderson and W. A. Walker (ed.), Development of the gastrointestinal tract. B. C. Decker, Hamilton, Ontario, Canada.
- 3.Adlerberth, I., F. Jalil, B. Carlsson, L. Mellander, L. A. Hanson, P. Larsson, K. Khalil, and A. E. Wold. 1998. High turnover rate of Escherichia coli strains in the intestinal flora of infants in Pakistan. Epidemiol. Infect. 121:587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calva, J. J., J. Sifuentes-Osornio, and C. Ceron. 1996. Antimicrobial resistance in fecal flora: longitudinal community-based surveillance of children from urban Mexico. Antimicrob. Agents Chemother. 40:1699-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cave, H., E. Bingen, J. Elion, and E. Denamur. 1994. Differentiation of Escherichia coli strains using randomly amplified polymorphic DNA analysis. Res. Microbiol. 145:141-150. [DOI] [PubMed] [Google Scholar]
- 6.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dominguez, E., M. Zarazaga, Y. Saenz, L. Brinas, and C. Torres. 2002. Mechanisms of antibiotic resistance in Escherichia coli isolates obtained from healthy children in Spain. Microb. Drug Resist. 8:321-327. [DOI] [PubMed] [Google Scholar]
- 8.Godwin, D., and J. H. Slater. 1979. The influence of the growth environment on the stability of a drug resistance plasmid in Escherichia coli K12. J. Gen. Microbiol. 111:201-210. [DOI] [PubMed] [Google Scholar]
- 9.Gophna, U., A. Parket, J. Hacker, and E. Z. Ron. 2003. A novel ColV plasmid encoding type IV pili. Microbiology 149:177-184. [DOI] [PubMed] [Google Scholar]
- 10.Herias, M. V., T. Midtvedt, L. A. Hanson, and A. E. Wold. 1995. Role of Escherichia coli P fimbriae in intestinal colonization in gnotobiotic rats. Infect. Immun. 63:4781-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reference deleted.
- 12.Reference deleted.
- 13.Johnson, J. R. 1991. Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 4:80-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korhonen, T. K., R. Virkola, B. Westurlund, H. Holthofer, and J. Parkkinen. 1990. Tissue tropism of Escherichia coli adhesins in human extraintestinal infections. Curr. Top. Microbiol. Immunol. 151:115-127. [DOI] [PubMed] [Google Scholar]
- 15.Kucheria, R., P. Dasgupta, S. H. Sacks, M. S. Khan, and N. S. Sheerin. 2005. Urinary tract infections: new insights into a common problem. Postgrad. Med. J. 81:83-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lanz, R., P. Kuhnert, and P. Boerlin. 2003. Antimicrobial resistance and resistance gene determinants in clinical Escherichia coli from different animal species in Switzerland. Vet. Microbiol. 91:73-84. [DOI] [PubMed] [Google Scholar]
- 17.Lee, S. W., and G. Edlin. 1985. Expression of tetracycline resistance in pBR322 derivatives reduces the reproductive fitness of plasmid-containing Escherichia coli. Gene 39:173-180. [DOI] [PubMed] [Google Scholar]
- 18.Levy, J. 2000. The effects of antibiotic use on gastrointestinal function. Am. J. Gastroenterol. 95:S8-S10. [DOI] [PubMed] [Google Scholar]
- 19.Levy, S. B. 1989. Evolution and spread of tetracycline resistance determinants. J. Antimicrob. Chemother. 24:1-3. [DOI] [PubMed] [Google Scholar]
- 20.Lidin-Janson, G., E. Falsen, U. Jodal, B. Kaijser, and K. Lincoln. 1977. Characteristics of antibiotic-resistant Escherichia coli in the rectum of healthy school-children. J. Med. Microbiol. 10:299-308. [DOI] [PubMed] [Google Scholar]
- 21.Ng, L. K., I. Martin, M. Alfa, and M. Mulvey. 2001. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 15:209-215. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen, H. U., A. M. Hammerum, K. Ekelund, D. Bang, L. V. Pallesen, and N. Frimodt-Moller. 2004. Tetracycline and macrolide co-resistance in Streptococcus pyogenes: co-selection as a reason for increase in macrolide-resistant S. pyogenes? Microb. Drug Resist. 10:231-238. [DOI] [PubMed] [Google Scholar]
- 23.Nowrouzian, F., I. Adlerberth, and A. E. Wold. 2001. P fimbriae, capsule and aerobactin characterize colonic resident Escherichia coli. Epidemiol. Infect. 126:11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nowrouzian, F., B. Hesselmar, R. Saalman, I. L. Strannegard, N. Aberg, A. E. Wold, and I. Adlerberth. 2003. Escherichia coli in infants' intestinal microflora: colonization rate, strain turnover, and virulence gene carriage. Pediatr. Res. 54:8-14. [DOI] [PubMed] [Google Scholar]
- 25.Nowrouzian, F., A. E. Wold, and I. Adlerberth. 2001. Computer-based analysis of RAPD (random amplified polymorphic DNA) fingerprints for typing of intestinal Escherichia coli. Mol. Biol. Today 2:5-10. [Google Scholar]
- 26.Olasz, F., P. Z. Fekete, G. Blum-Oehler, Z. Boldogkoi, and B. Nagy. 2005. Characterization of an F18(+) enterotoxigenic Escherichia coli strain from post weaning diarrhoea of swine, and of its conjugative virulence plasmid pTC. FEMS Microbiol. Lett. 244:281-289. [DOI] [PubMed] [Google Scholar]
- 27.Orskov, I., and F. Orskov. 1985. Escherichia coli in extra-intestinal infections. J. Hyg. (London) 95:551-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts, M. C. 1996. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol. Rev. 19:1-24. [DOI] [PubMed] [Google Scholar]
- 29.Sanchez, A. R., R. S. Rogers III, and P. J. Sheridan. 2004. Tetracycline and other tetracycline-derivative staining of the teeth and oral cavity. Int. J. Dermatol. 43:709-715. [DOI] [PubMed] [Google Scholar]
- 30.Schnappinger, D., and W. Hillen. 1996. Tetracyclines: antibiotic action, uptake, and resistance mechanisms. Arch. Microbiol. 165:359-369. [DOI] [PubMed] [Google Scholar]
- 31.Schrag, S. J., V. Perrot, and B. R. Levin. 1997. Adaptation to the fitness costs of antibiotic resistance in Escherichia coli. Proc. R. Soc. Lond. Ser. B 264:1287-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sears, H. J., H. James, R. Saioum, I. Brownlee, and L. F. Lamereaux. 1956. Persistence of individual strains of Escherichia coli in man and dog under varying conditions. J. Bacteriol. 71:370-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Swedish Reference Group for Antibiotics. 28 April 2004, revision date. www.srga.org.
- 33.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van den Bogaard, A. E., and E. E. Stobberingh. 2000. Epidemiology of resistance to antibiotics. Links between animals and humans. Int. J. Antimicrob. Agents 14:327-335. [DOI] [PubMed] [Google Scholar]
- 35.Welinder-Olsson, C., E. Kjellin, M. Badenfors, and B. Kaijser. 2000. Improved microbiological techniques using the polymerase chain reaction and pulsed-field gel electrophoresis for diagnosis and follow-up of enterohaemorrhagic Escherichia coli infection. Eur. J. Clin. Microbiol. Infect. Dis. 19:843-851. [DOI] [PubMed] [Google Scholar]
- 36.Wilson, H. D., and H. F. Eichenwald. 1974. Sepsis neonatorum. Pediatr. Clin. N. Am. 21:571-582. [DOI] [PubMed] [Google Scholar]
- 37.Winberg, J., H. J. Andersen, T. Bergstrom, B. Jacobsson, H. Larson, and K. Lincoln. 1974. Epidemiology of symptomatic urinary tract infection in childhood. Acta Paediatr. Scand. Suppl. 252:1-20. [DOI] [PubMed] [Google Scholar]