Abstract
The adaptive and further evolutionary responses of Staphylococcus aureus to selection pressure with the antibiotic rifampin have not been explored in detail. We now present a detailed analysis of these systems. The use of rifampin for the chemotherapy of infections caused by S. aureus has resulted in the selection of mutants with alterations within the β subunit of the target enzyme, RNA polymerase. Using a new collection of strains, we have identified numerous novel mutations in the β subunits of both clinical and in vitro-derived resistant strains and established that additional, undefined mechanisms contribute to expression of rifampin resistance in clinical isolates of S. aureus. The fitness costs associated with rifampin resistance genotypes were found to have a significant influence on their clinical prevalence, with the most common clinical genotype (H481N, S529L) exhibiting no fitness cost in vitro. Intragenic mutations which compensate for the fitness costs associated with rifampin resistance in clinical strains of S. aureus were identified for the first time. Structural explanations for rifampin resistance and the loss of fitness were obtained by molecular modeling of mutated RNA polymerase enzymes.
Resistance to antibiotics arising from point mutations in bacterial genes that encode drug targets is a well-recognized phenomenon (38), and expression of these mechanisms often confers a fitness cost that results from the decreased physiological activity of the altered target (1, 37). Nevertheless, increasing evidence obtained both from laboratory and from epidemiological studies indicates that intragenic compensatory mutations often act to maintain the long-term persistence of resistant bacteria by eliminating or reducing the fitness costs associated with the development of target-based resistance (27). Consequently, bacterial products that are the targets of antibiotic action present interesting systems for the study of structure-function relationships from the perspectives of resistance, fitness, and compensatory evolution (15, 20). Furthermore, fitness costs and compensatory evolution are factors that can influence the prevalence of specific antibiotic resistance genotypes in the clinical setting (5, 19, 20, 30, 43).
We have recently explored the genetic and structural basis of mupirocin resistance and fitness in Staphylococcus aureus and related this to the incidence of mupirocin resistant isoleucyl-tRNA synthetase genotypes arising in the clinic (19, 20). A similar opportunity to examine these paradigms in relation to rifampin resistance now arises.
Mutations that confer resistance to rifampin (Fig. 1) arise in the β subunit (encoded by rpoB) of the target enzyme RNA polymerase (RNAP) and have been mapped to this location in all bacteria examined so far, including Escherichia coli (21), Mycobacterium tuberculosis (5, 36), and S. aureus (32, 43). The fitness burdens and compensatory evolution associated with mutations in rpoB that confer resistance to rifampin have been studied in E. coli (37). However, the structural changes in RNAP responsible for these phenomena have not been explored, and it is not possible to address their impact on the prevalence of the rifampin resistance genotypes found in the clinical setting because rifampin is not used therapeutically for the treatment of infections caused by E. coli (35). However, rifampin is used therapeutically for the chemotherapy of infections caused by S. aureus (35), and preliminary data suggest that the fitness burdens associated with rifampin resistance may influence the prevalence of specific resistance genotypes in clinical isolates of S. aureus (43). Accordingly, we have chosen to examine the molecular and structural bases of rifampin resistance in S. aureus to provide a better understanding of resistance, fitness, and compensatory adaptation in this organism and how these processes relate to the prevalence of rifampin resistance genotypes in clinical strains.
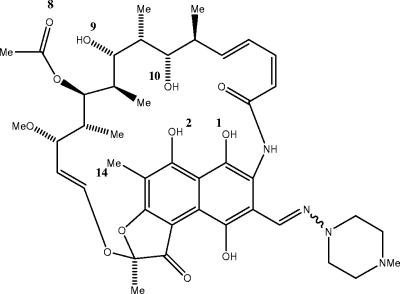
FIG. 1.
Structure and part numbering of rifampin. Me, methyl.
Our work has involved an analysis of clinical S. aureus isolates more extensive than those that have been conducted previously (3, 44). This has enabled a comprehensive examination of the prevalence of rifampin resistance genotypes in clinical isolates and relationships with bacterial fitness, including the effects of mutations on RNAP structure (8) predicted by molecular modeling techniques. The data obtained also support the emerging concept that determination of the fitness of antibiotic-resistant mutants is likely to be a useful predictor for the development of resistance to novel agents in the clinical setting.
MATERIALS AND METHODS
Bacterial strains.
Clinical rifampin-resistant S. aureus strains were acquired from the following sources: BR1 to BR9 were from the Bristol Royal Infirmary (BRI), Bristol, United Kingdom; ST774, ST109, ST3651, and ST1662 were from the Central Public Health Laboratory (PHLS), Colindale, United Kingdom; Red80, Red55, Red4, Red7, G55, G56, G68, G73, 233, 4266, and Y25 were from the Leeds General Infirmary, Leeds, United Kingdom. Strains Mu50 (17), 992 (NJ) (42), and 99-3759 V (18) are rifampin-resistant and vancomycin-intermediate resistant S. aureus kindly provided by the authors who first described them. Strains 560 Australia, 239R, and 472 T/B Taiwan are global methicillin-resistant S. aureus isolates (24). All strains were typed by ribosomal spacer PCR (24).
S. aureus 8325-4 (31) was used as the standard laboratory strain from which resistant mutants were derived and characterized as described previously (32). E. coli XL-10 Gold competent cells (Stratagene, Amsterdam, The Netherlands) and S. aureus RN4220 (12) were used as transformation recipients. Unless otherwise specified, strains were cultured in Luria-Bertani (LB) broth (Fisher, Loughborough, United Kingdom) or on LB agar (Fisher) at 37°C.
Antibiotic susceptibility testing.
MICs were determined by the agar dilution method (7) in Iso-Sensitest agar (Oxoid, Basingstoke, United Kingdom) with inocula in Iso-Sensitest broth (Oxoid) of 106 CFU per spot. After 24 h incubation at 37°C, the MIC was deemed the lowest concentration of antibiotic that prevented visible colonial growth.
Determination of competitive fitness.
The biological fitness of the bacteria was measured by mixed culture competition between resistant mutants and their otherwise isogenic parent strain (strain 8325-4) essentially as described previously (19). Resistant and sensitive strains were coinoculated into LB broth at a ratio of 1:10 and were grown with aeration at 37°C for 24 h. Changes in the relative numbers of strains over this period were established by plating onto selective (0.032 μg rifampin/ml) and nonselective LB agar, and the relative fitness (W) was calculated as follows: W = ln[NR(24)/NR(0)]/ln[NS(24)/NS(0)], where NR(t) and NS(t) represent the population densities of the resistant and sensitive strains, respectively, at time t (0 or 24 h) (9). A minimum of three independent cultures were sampled for each resistant mutant.
DNA manipulation.
Oligonucleotide primers were designed using Oligo 6.0 (MBI, Cascade, Colo.) and were purchased from MWG Biotech (Milton Keynes, United Kingdom). PCR amplifications were performed by using Platinum Pfx (Invitrogen, Paisley, United Kingdom) on genomic DNA templates prepared with the Bacterial Genomic DNA Purification kit (Edge Biosystems, Gaithersburg, Md.) from staphylococcal protoplasts generated by incubation with lysostaphin (100 μg/ml) for 30 min at 37°C. DNA sequencing was performed on an Applied Biosystems 377 DNA sequencer.
Recombinant DNA manipulations were conducted by established procedures (39). Purification of DNA following PCR or restriction was achieved with the MinElute kit or the QIAquick PCR purification kit (both from QIAGEN, Crawley, United Kingdom). Plasmid DNA was purified from E. coli by using the QIAprep Spin Miniprep kit (QIAGEN), in accordance with the manufacturer's instructions. The same kit was used for recovery of plasmid DNA from S. aureus following protoplasting, as described above.
Identification of rifampin resistance mutations in clinical strains of S. aureus.
PCR amplification and sequencing of the rifampin resistance-determining region (RRDR) of S. aureus rpoB was performed with primers F3 and F4 (3). PCR amplification of the entire rpoB gene was performed with primers rpoBF (5′-GTGGATCCATTAGTGTTGCCGTTTTCTTTT [the underlined region represents the engineered restriction site]) and rpoBR (5′-ATTCTGCAGAGTATCTTTTTGCCTGTTTTG), while the rpoA and rpoC genes were amplified with primer pair rpoAI (5′-TAACTGCGATCAGAGACGTTACTCC) and rpoA II (5′-GCTGCATTACGACGAGAAGCTAAAT) and primer pair rpoCI (5′-GACGATGATGTTGTAGAACGCAAAG) and rpoCII (5′-TGTTGTTTGTTAAAGCGTGCAACTT), respectively. The resulting amplicons were sequenced by using both the amplification primers and arbitrarily designed spanning primers.
Examination of the role of putative resistance polymorphisms by overexpression in trans.
PCR amplicons of entire rpoB and rpoC genes (including Shine-Dalgarno sequences) carrying putative resistance polymorphisms were generated with primer pair rpoBF(kpn) (5′-TAGGGTACCGCGGATCACATAATTTTTGAG) and rpoBR(sac) (5′-TAGGAGCTCTTTGCCTGTTTTGTAAATTGC) and primer pair rpoCF(kpn) (5′-AAAGGTACCAGCTAATCTAAATTGAAGGAG) and rpoCR(sac) (5′-TATGAGCTCGTTATTCCGTTACTTCAGTTT), respectively. In addition, the wild-type gene (from strain 8325-4) and known resistant variants with mutations in rpoB recovered from in vitro selections (D471Y and H481N) were amplified by PCR to serve as negative and positive controls, respectively. Amplicons were digested with KpnI and SacI, ligated into tetracycline-regulated expression plasmid pALC2073 (4), and propagated in E. coli before recovery and electroporation (41) into S. aureus RN4220.
Allelic replacement.
For allelic replacement of rifampin resistance genotypes into strain 8325-4, PCR amplicons of the 3′ portion of the rpoB gene from strains carrying alleles of interest were generated with primer rpoU (5′-AGCGGATCCGGATATACAGATGATATTGAC) and primer rpoBR. To generate an amplicon carrying the mutations H481N and L466S, which were not carried by any strain in our collection, primers rpoU (forward) and rpoM (5′-TCCATGAATTGTGATGATTGAGAGCTACCA [reverse primer; the site-directed mutation is shown underlined]) were used to create a mutagenic megaprimer by PCR using DNA from strain R28 (H481N) as the template (Table 1). The agarose gel-purified megaprimer, which carried a mutation coding for amino acid substitution L466S, was used in conjunction with primer rpoBR to generate the final amplicon from R28.
TABLE 1.
Nature of rifampin-resistant mutants of S. aureus 8325-4
| Strain | Rifampin MIC (μg/ml) | Substitution(s) in β subunit | Relative fitness (W) ± SD |
|---|---|---|---|
| 8325-4 | 0.008 | 1 | |
| R31 | 0.125 | D471E | 0.94 ± 0.05 |
| R26 | 0.125 | Q137L | 0.87 ± 0.03 |
| R21 | 0.25 | Q137L | 0.85 ± 0.04 |
| R39 | 0.25 | L466S | 0.96 ± 0.04 |
| R37 | 0.25 | D471E | 0.79 ± 0.05 |
| R22 | 0.25 | Q137L | 0.83 ± 0.05 |
| R28 | 2 | H481N | 0.80 ± 0.01 |
| R50 | 2 | A477V | 0.58 ± 0.03 |
| R61 | 2 | A477V | 0.58 ± 0.03 |
| R53 | 4 | ΔL485 | 0.81 ± 0.03 |
| R34 | 4 | D471Y | 0.84 ± 0.04 |
| R41 | 8 | D471Y | 0.74 ± 0.04 |
| R44 | 16 | I527F | 0.99 ± 0.02 |
| R35 | 16 | S464P | 0.71 ± 0.03 |
| R51 | 128 | L488S | 0.99 ± 0.05 |
| R47 | 256 | P519L | 0.78 ± 0.03 |
| R49 | 512 | D471N/P519L | 0.71 ± 0.01 |
| R38 | 512 | A477D | 0.61 ± 0.03 |
| R24 | 1,024 | S486L | 0.64 ± 0.03 |
| R23 | 1,024 | S486L | 0.75 ± 0.05 |
| R40 | 1,024 | H481D | 0.59 ± 0.02 |
| R15 | 1,024 | H481Y | 0.50 ± 0.02 |
| R19 | 1,024 | H481Y | 0.49 ± 0.03 |
Amplicons were digested with BamHI-PstI, ligated to thermosensitive shuttle vector pCL52.1 (26), and propagated in E. coli. DNA sequencing was performed to ensure that no additional mutations had been introduced into the rpoB fragments during PCR amplification. Following electroporation (41) into S. aureus RN4220, plasmid constructs were recovered and introduced by electroporation into S. aureus 8325-4. Transformants were grown at 30°C with aeration in brain heart infusion (BHI) broth containing 3 μg tetracycline/ml until early log phase and then shifted to the nonpermissive temperature (42°C) and grown for 6 h. Dilutions of this culture were plated onto BHI agar containing 3 μg tetracycline/ml and incubated at 42°C overnight to select integrants. Single colonies were inoculated in BHI broth without antibiotic and incubated at 30°C with aeration for 24 h to allow excision of the vector backbone from the chromosome, and dilutions of the culture were spread onto agar containing 0.032 μg rifampin/ml. Rifampin-resistant colonies that were susceptible to tetracycline represented putative allelic recombinants and were characterized by PCR and DNA sequencing to ensure that (i) the desired mutations had been introduced into rpoB, (ii) that no additional mutations had been introduced into rpoB in the vicinity of the recombination event, and (iii) the overall structure of the rpoB-rpoC region remained otherwise unaffected by these manipulations.
Molecular modeling.
Molecular models of the mutated enzymes were constructed by using the Swiss-PDB viewer V3.7 software (14) in conjunction with the structure file 1I6V.pdb, which contains rifampin cocrystallized within the RNAP structure (8). The lowest-energy conformations of the resulting mutated enzymes were then subjected to full energy minimization by using the MM2 force field within MacroModel (29) on Silicon Graphics O2 or Indy workstations. To simplify the computations, all calculations were performed on a 15-Å spherical portion of the protein centered on rifampin and used an additional force field to simulate a solvent dielectric equivalent to water. For the energy minimizations, a 10-Å spherical region within this substructure centered on the ligand was defined in which all atoms including those from the ligand were allowed to freely move and atoms outside this region were frozen. Additional studies relating to fitness were performed in conjunction with a model of the core protein and nucleic acid RNAP structure previously described by Darst and colleagues (23, 34).
RESULTS AND DISCUSSION
Genetic characterization of rifampin-resistant mutants selected in vitro.
In S. aureus mutations conferring resistance to rifampin are clustered predominantly in a conserved region of the RNAP β subunit spanning amino acids ∼463 to 550 (3, 32, 44), which represents the RRDR. In vitro-derived rifampin-resistant mutants were recovered following plating of saturated cultures of S. aureus 8325-4 onto agar containing rifampin at 4× the MIC (0.032 μg/ml). Mutants were characterized by rifampin susceptibility testing, and their resistance genotypes were established by PCR amplification and DNA sequencing of the RRDR (and, where necessary, the entire rpoB gene) (Table 1). The majority of mutations conferring resistance occurred within the RRDR and predominantly at codons in rpoB previously implicated in rifampin resistance in S. aureus (32, 44). However, several novel resistance mutations that have not previously been described in S. aureus were identified (Table 1), although data exist to link them with rifampin resistance. Mutation P519L (strains R47 and R49) is identical to a rifampin resistance mutation previously identified in E. coli (P564L) (13). Similarly, mutations occurring at the equivalent locus of the L488S substitution (strain R51) have been observed in E. coli (13), although this particular change to serine has not been reported previously. Although D471N is a novel substitution, it occurs at an established rifampin resistance locus (32, 44) and was found in the only strain (R49) carrying two resistance mutations (D471N and P519L) following a single round of rifampin selection (Table 1). Since substitution P519L was also found on its own (strain R47) and is a relatively low-cost mutation, it seems likely that it represents the second mutation event in strain R49, and was selected to rescue what is probably a highly unfit first-step resistance mutation (D471N).
Resistance in strain R53 was achieved by single-codon deletion of L485 (actually, a three-base fragment spanning two codons was lost, but the remaining bases reconstitute one of the disrupted codons). Although rpoB deletions conferring rifampin resistance have been identified in E. coli and M. tuberculosis (16, 21), this is the first rifampin resistance allele resulting from a deletion to be detected in S. aureus.
Fitness of rifampin-resistant mutants selected in vitro.
The biological fitness of the different rifampin resistance genotypes was established by pairwise competition assays (Table 1). The majority of resistance alleles were associated with a reduction in fitness relative to 8325-4. In contrast to previous work that found no correlation between the level of rifampin resistance and the size of the fitness cost, we observed a trend toward higher fitness costs in strains expressing high-level rifampin resistance (Fig. 2). For instance, strains showing a 16- to 32-fold increase in resistance displayed at most a 17% fitness cost, whereas in those strains displaying a 128,000-fold increase in resistance, fitness costs of up to 50% were recorded (Table 1). However, there were exceptions to this trend, since strain R50 (250-fold increase in resistance) displayed a 42% fitness cost and strains R44 and R51, with 2,000- and 16,000-fold increases in resistance, respectively, displayed no significant fitness costs compared with the parental strain 8325-4. The correlation between resistance and fitness is particularly apparent in a graphical form (squared correlation coefficient of 0.74) when the datum points for strains R44, R50, and R51 are not included (Fig. 2).
FIG. 2.
Relative fitness of rifampin-resistant mutants of S. aureus 8325-4 in relation to their susceptibilities to the antibiotic.
In some cases the fitness values that we obtained for particular resistance genotypes were similar to those reported by Wichelhaus et al. (43). However, the fitness costs in our assay were generally greater than those reported previously. For instance, we found that the apparent “no-cost” H481N mutation described by Wichelhaus et al. (43) exhibited a fitness cost of 20% (strain R28; Table 1; Fig. 2). Since the apparent absence of a fitness cost for the H481N mutation has been used to explain its apparent prevalence in rifampin-resistant clinical isolates (43), it was important to establish that the fitness cost that we observed was not due to an additional, unselected mutation(s). Therefore, the H481N mutation was introduced into a clean 8325-4 background by allelic replacement. Competition experiments confirmed the approximate 20% fitness cost (W = 0.84 ± 0.06) for this genotype observed in strain R28 (W = 0.80 ± 0.01) (Table 1). Further differences in fitness costs between the two studies were also noted. Most striking was the situation with mutation H481Y. This exhibited only a very minor fitness cost in the previous study (7%) but caused a ∼50% reduction in fitness in this study (strains R15 and R19; Table 1).
It is not immediately obvious how the differences in fitness costs measured in the two studies have arisen, since the experimental approaches are similar. However, some of the strains in the previous study (43) may have developed second-site compensatory mutations, a phenomenon which can occur relatively quickly under laboratory conditions (20) and which may have led to high fitness values for the reported resistance genotypes. Compensatory mutation may also have occurred in the present study, since in some instances the fitness values for strains with apparently identical resistance genotypes exhibited significant variation (e.g., strains R34 and R41 and strains R23 and R24; Table 1). However, complete sequencing of rpoB from these mutant pairs failed to identify candidate compensatory mutations (data not shown). Therefore, if compensatory mutations occurred in these strains, they must be located at extragenic sites.
Rifampin resistance alleles in rpoB that occur in clinical strains of S. aureus.
Two relatively small studies have already sequenced the rpoB genes from clinical rifampin-resistant S. aureus isolates to identify nucleotide substitutions responsible for resistance (3, 44). We examined an additional collection of isolates (Table 2) to extend the number of clinically occurring rifampin resistance genotypes characterized. Ribosomal spacer PCR typing was used in conjunction with rifampin susceptibility data to ensure that only independent isolates were characterized. This resulted in the identification of 28 independent rifampin-resistant S. aureus isolates, which were subsequently characterized by DNA sequencing of their RRDR regions (Table 2).
TABLE 2.
Resistance to rifampin in clinical isolates of S. aureus and characterization of mutations in the RRDRa
| Strain | Rifampin MIC (μg/ml) | Substitution(s) in β subunit | Reported rifampin MIC (μg/ml) associated with β substitution(s) |
|---|---|---|---|
| 560 Australia | 64 | D471Y | NR |
| A473E | |||
| BR3 | 128 | A477D | 128-512 |
| 239R | 256 | H481N | 128-512 |
| S529L | |||
| 472 T/B | 256 | S463N | NR |
| Taiwan | S464P | ||
| 99-3759V | 256 | H481N | 128-512 |
| S529L | |||
| BR6 | 256 | H481Y | 512-1,024 |
| Red80 | 256 | H481N | 128-512 |
| S529L | |||
| ST774 | 1,024 | D471G | NR |
| R484C | |||
| BR2 | 1,024 | R484H | 128-256 |
| BR8 | 1,024 | H481N | 128-512 |
| S529L | |||
| Red55 | 1,024 | None | |
| ST109 | 1,024 | A477D | 128-512 |
| Red7 | 1,024 | A477D | 128-512 |
| 233 | 1,024 | H481Y | 512-1,024 |
| 4266 | 1,024 | Q468I | NR |
| ST3651 | 1,024 | S486L | 1,024 |
| ST1662 | 1,024 | H481Y | 512-1,024 |
| 992 (NJ) | 1,024 | H481Y | 512-1,024 |
| BR1 | 1,024 | Q468L | 512 |
| BR5 | 1,024 | H481Y | 512-1,024 |
| BR9 | 1,024 | H481N | 1-4 |
| G55 | 1,024 | None | |
| G56 | 1,024 | None | |
| G68 | 1,024 | None | |
| G73 | 1,024 | None | |
| Mu50 | 1,024 | H481Y | 512-1,024 |
| Red4 | 1,024 | D471Y | 4 |
| Y25 | 1,024 | S486L | 1,024 |
On the basis of the results in Table 2, it appears that the nature of rifampin resistance in S. aureus is more complex than previous studies have suggested. For instance, the H481N substitution, detected by Wichelhaus et al. (44) in 27 of 30 (90%) rifampin-resistant S. aureus isolates, either on its own or in combination with other substitutions, was identified in only 5 of the 28 strains (18%) studied here (Table 2). Therefore, this resistance allele may not be as prevalent as was suggested previously. Nevertheless, we found that substitutions at position 481 were common, since 11 of the 28 strains (39%) that we examined had substitutions (either H481N or H481Y) at this position (Table 2).
Several novel resistance genotypes were identified. Although substitutions Q468L (BR1), S464P (472 T/B Taiwan), and S486L (Y25) (Table 2) have previously been described in rifampin-resistant S. aureus isolates generated in vitro (32, 44), they have not been detected in clinical isolates, prior to this study. Additionally, we identified a novel resistance locus in the staphylococcal RRDR (S463N in strain 472 T/B Taiwan; Table 2) for which a direct counterpart has previously been reported only in rifampin-resistant M. tuberculosis (S508T) (28). Four novel amino acid substitutions, Q468I (strain 4266), A473E (strain 560 Australia), and D471G and R484C (both in strain ST774), were also identified (Table 2). These mutations occurred at previously identified resistance loci.
Mutations lying outside the RRDR in clinical rifampin-resistant strains of S. aureus.
Previous studies with clinical strains demonstrated that all staphylococcal rifampin resistance alleles mapped within the RRDR (3, 44). However, we found that five strains (Red55, G55, G56, G68, and G73) (18%) had no mutations in the RRDR (Table 2). Furthermore, three strains (BR2, BR9, and Red4) (11%) likely possess additional mutations outside the RRDR, as they exhibited rifampin resistance levels higher than those conferred simply by the mutations that they possessed in their RRDR alleles (Table 3).
TABLE 3.
Susceptibilities to rifampin and rpoB and rpoC genotypes of various strains of S. aureusa
| Strain | Rifampin MIC (μg/ml) | Substitution(s) in β subunit | MIC (μg/ml) of RN4220 expressing rpoB genotype | Substitution in β′ subunit | MIC (μg/ml) of RN4220 expressing rpoC genotype |
|---|---|---|---|---|---|
| RN4220 | 0.008 | None | 0.008 | None | 0.008 |
| Red4 | 1,024 | D471Y | 4 | G602D | 0.008 |
| BR2 | 1,024 | R484H, Y737F | 256 | None | |
| BR9 | 1,024 | H481N | 1 | None | |
| Red55 | 1,024 | Y737F | 0.008 | None | |
| G68 | 1,024 | Y737F | 0.008 | None | |
| G73 | 1,024 | D320N | 0.008 | None | |
| G55 | 1,024 | None | None | ||
| G56 | 1,024 | None | None |
The strains shown are those from Table 2 in which the mutations have not previously been associated with rifampin resistance or are insufficient to account for the resistance phenotype. Also shown are the levels of rifampin resistance achieved by expression of the rpoB and rpoC genotypes in a rifampin-susceptible heterologous host (S. aureus RN4220).
The entire rpoB genes of these eight strains were sequenced. This failed to identify additional mutations in three of them (Red4, G55, and G56), although strains BR2, Red55, and G68 all carried mutation Y737F and strain G73 carried mutation D320N (Table 3). A review of the reported rifampin resistance genotypes in all bacterial genera examined established that neither of these mutations corresponded with rifampin resistance loci. Hence, it was unclear whether they constituted resistance mutations or were merely natural polymorphisms that exerted no effect on rifampin susceptibility.
In addition, DNA sequencing was performed to detect potential rifampin resistance mutations occurring at loci outside rpoB but within the RNA polymerase complex. The genes that encode the other subunits of the RNA polymerase complex (rpoA, rpoC) were therefore PCR amplified from the eight strains and sequenced in toto. No strain carried coding polymorphisms in rpoA relative to the sequence of S. aureus 8325-4, but strain Red4 carried a mutation encoding substitution G602D in the rpoC gene (Table 3).
Potential roles of the mutations lying outside the RRDR in clinical isolates in expression of rifampin resistance.
We examined the potential roles of the mutations lying outside the RRDR in clinical isolates (Table 3) in the expression of resistance to rifampin. The entire rpoB and rpoC genes and their Shine-Dalgarno regions from the eight relevant strains (Table 3) were cloned and expressed from a tetracycline-regulated promoter in trans in rifampin-susceptible S. aureus strain RN4220.
Expression of wild-type rpoB and rpoC genes in trans had no effect on rifampin susceptibility, while expression of known rifampin resistance rpoB alleles encoding mutations D471Y and H481N conferred the expected levels of rifampin resistance (Table 1; Table 3). Similarly, expression of β (R484H, Y737F) resulted in a level of rifampin resistance (Table 3) consistent with that conferred by the known resistance mutation (R484H) of the doublet, indicating that Y737F does not influence rifampin resistance. Expression of neither β (Y737F) nor β (D320N) conferred resistance to rifampin, and these are therefore probably merely β polymorphisms.
Expression of β′ (G602D) in RN4220 did not result in resistance to rifampin (Table 3). Furthermore, it did not increase the level of rifampin resistance when it was expressed in strains R34 and R41 (data not shown), which carry the D471Y substitution, which is also present in strain Red4 and in conjunction with which the G602D mutation was identified. Consequently, β′ mutation G602D cannot be responsible for the high level of rifampin resistance exhibited by clinical isolate Red4.
Therefore, even though they exhibited high-level rifampin resistance, clinical isolates Red55, G68, G73, G55, and G56 do not carry mutations responsible for rifampin resistance within the core RNAP complex. Furthermore, although strains BR2, BR9, and Red4 do possess mutations in the RRDR of rpoB, these are insufficient to explain the levels of rifampin resistance exhibited by these strains. Possibly, these clinical strains carry mutations that affect the accumulation of rifampin by the cell, e.g., by causing decreased influx or enhanced efflux of the antibiotic.
Clinical prevalence of rifampin resistance alleles in S. aureus and the use of in vitro fitness data to explain the natural occurrence of particular resistance genotypes.
For M. tuberculosis the fitness costs associated with rifampin resistance genotypes reveal a correlation between fitness and the clinical isolation rate of the resistance allele (5). Data for other antibiotics also suggest that resistance mutations which commonly occur in clinical isolates are predominantly those associated with minor or negligible fitness costs in laboratory experiments (1, 6). Therefore, we evaluated the relationship between the in vitro fitness of rifampin resistance alleles and their prevalence in clinical isolates of S. aureus. Although Wichelhaus et al. (43, 44) described 7 rifampin resistance genotypes in clinical S. aureus isolates, this study brings the total to 18 (Fig. 3).
FIG. 3.
Clinical prevalence of rifampin resistance alleles. Data are from this study and previous studies (3, 44).
The four most prevalent genotypes all carried substitutions at position 481 (Fig. 3). Two of these resistance genotypes, including the first and the fourth most frequently detected alleles, were double mutations, i.e., H481N, S529L and H481N, L466S, respectively. Mutants with these double mutations were not represented in our collection of laboratory mutants. Consequently, in vitro fitness data were not available for these genotypes to enable an analysis of in vitro fitness versus clinical prevalence. Attempts were therefore made to select these double mutants in the 8325-4 background by plating strain R28 (H481N; Table 1) onto agar containing rifampin at concentrations above the MIC for this strain, followed by selection and characterization of the mutants. Although a variety of double mutants that have not been detected in the clinic were recovered (e.g., H481N, E478D; H481N, Q468L; and H481N, A487P), the double mutations of specific interest (H481N, S529L and H481N, L466S) were not.
Therefore, the H481N, L466S and H481N, S529L doublets were instead generated in the 8325-4 background by allelic replacement. Competition assays with allelic replacement mutants established that the latter showed essentially complete restoration of fitness (W = 0.99 ± 0.03) compared to that for strains with H481N alone (W = 0.80 ± 0.01; Table 1). Furthermore, this mutation together with H481N in the doublet appears to act cooperatively to increase the level of rifampin resistance from 2 μg/ml to 512 μg/ml. Indeed, this conclusion is supported by molecular modeling analysis (see the subsequent section). Hence, the S529L substitution as a secondary mutation to H481N acts both to increase the level of rifampin resistance and to compensate for the fitness costs of the primary resistance mutation. The high level of rifampin resistance of this genotype, coupled with the absence of a significant fitness cost, provides an explanation for the prevalence of this genotype in the clinic (Fig. 3).
In contrast, double mutation H481N, L466S did result in a fitness cost (W = 0.83 ± 0.04) comparable to that observed with the H481N genotype alone (W = 0.80 ± 0.01) (Table 1). Therefore, it appears that this double mutation has been selected primarily to increase the level of rifampin resistance, especially since L466S and H481N, when each is carried singly, confer only low-level resistance (rifampin MICs, 0.25 and 2 μg/ml, respectively) (Table 1), while the doublet confers high-level resistance (rifampin MIC, 512 μg/ml), presumably by the cooperative actions of the two mutations. These conclusions are supported by molecular modeling analysis (see the subsequent section).
It appears likely that the prevalence of rifampin-resistant genotypes in staphylococcal clinical isolates is influenced primarily by the associated effects on the fitness of the organisms. Thus, the first and the fourth most prevalent mutational combinations found in clinical isolates, i.e., H481N, S529L and H481N, L466S, respectively (Fig. 3), were associated with either no fitness costs (H481N, S529L) or only moderate effects (H481N, L466S) when they were introduced into strain 8325-4. In contrast, the mutations found in rifampin-resistant mutants R50, R61, and R40, which were isolated in the laboratory and which resulted in severe fitness costs (up to 40%) (Table 1), were not represented in clinical isolates (Fig. 3), even though the H481D mutation (strain R40) might be expected to confer a selective advantage following clinical exposure to rifampin due to the high level of resistance to the antibiotic that it mediates. Nevertheless, clinical genotypes that possessed the S486L, D471Y, and A477D mutations (Fig. 3) occurred that were associated with significant fitness costs, ranging from 39% to 26% when they were expressed in strain 8325-4 (Table 1). In clinical isolates these primary rifampin resistance mutations may be associated with compensatory mutations which offset the fitness costs associated with the resistance genotypes. The possibility that compensatory mutations arise to offset fitness costs in clinical isolates possessing rifampin resistance mutations S486L, D471Y, and A477D is considered in the next section.
Several clinical rifampin-resistant isolates possessed alterations in RNAP, including multiple mutations (Fig. 3), that were not represented in our collection of laboratory-derived resistant mutants (Table 1). We do not consider this observation to have any particular significance, as, in our opinion, examination of further laboratory-derived mutants would eventually reveal these mutations.
Do compensatory mutations occur in clinical isolates to offset the fitness costs of mutations conferring rifampin resistance?
The presence of compensatory mutations that alleviate the fitness costs of primary rifampin resistance mutations in RNAP is a well-established phenomenon in laboratory mutants of E. coli (37). We sought evidence for similar mutations in clinical S. aureus strains, focusing in particular on isolates carrying resistance mutations that conferred fitness costs of 20% or greater when they are expressed in laboratory strains.
Compensatory mutations appeared likely to occur in association with the second and the third most prevalent clinical resistance genotypes, i.e., H481Y and H481N, respectively. These alleles, despite their prominence in clinical strains (Fig. 3), are associated with significant fitness costs when they are expressed in strain 8325-4 (W values of about 0.5 and 0.8, respectively) (Table 1). Although our collection of clinical isolates contained the H481N genotype in conjunction with other mutations (Table 2), none contained this mutation alone. Therefore, we were unable to specifically examine the H481N genetic background for compensatory mutations. However, we were able to examine isolates carrying H481Y, which was performed by PCR amplification and sequencing of the entire rpoB genes from strains BR6, ST1662, 992 (NJ), BR5, and 233 and the entire sequence of strain Mu50 (25), by analysis of the database sequence. Although strains BR3 and BR6 carried the Y737F polymorphism (see above), none of the strains examined carried any additional mutations in rpoB. It therefore follows that if any of these strains do carry compensatory mutations, they must be extragenic and may or may not lie within the RNAP complex.
Clinical isolates with genotypes possessing S486L, D471Y, and A477D mutations (Fig. 3) were also represented among the mutants derived from laboratory strain 8325-4 (Table 1). Since these mutations were associated with significant fitness costs in the laboratory strain that ranged from 39% to 26%, their occurrence in clinical isolates may be associated with compensatory mutations.
Sequencing of the entire rpoB genes of clinical strains containing S486L, D471Y, and A477D mutations (Table 2) to detect intragenic compensatory mutations identified a nucleotide substitution that gave rise to a V850A replacement (in addition to primary resistance mutation A477D) in strain Red7. The role of this mutation in compensation was established following introduction of the A477D, V850A genotype into 8325-4 by allelic replacement. The relative fitness of this double mutant was 1.00 ± 0.03, whereas it was 0.61 ± 0.03 for the strain with the A477D mutation alone (Table 1). Although this mutation ameliorated an approximate 40% fitness cost, it had no effect on the level of rifampin resistance in a strain which possessed it (data not shown). Therefore, the V850A mutation can be considered a “true” compensatory mutation.
No evidence for intragenic compensatory mutations was obtained for clinical isolates possessing primary S486L and D471Y rifampin resistance mutations.
Relationship of resistance and fitness to molecular structure: application of molecular modeling.
Although rifampin resistance mutations in core RNAP are believed to cause structural distortions to the rifampin binding pocket in the enzyme (8), the precise effects of these mutations on the binding of rifampin have not been defined. To probe the molecular effects of mutations upon rifampin binding, we used the X-ray crystal structure of the Thermus aquaticus core RNAP in complex with rifampin (8) to construct molecular models of mutants with both single and double RNAP mutations. We then used these models both to quantify the effect of the mutations on the binding energy for rifampin and to establish the precise interactions responsible for any changes in binding energy.
The RNAP mutations to be modeled were selected to give a sample over the broad range of rifampin susceptibility values exhibited by the mutants. The calculated binding energies of rifampin within these complexes are summarized in Table 4. In general, for the mutations selected for this analysis, there appears to be a broad inverse correlation between the calculated rifampin-RNAP binding energies and the measured rifampin MIC exhibited by the mutants. All the mutant enzymes selected were calculated to have binding energies lower than that for the wild-type enzyme (Table 4).
TABLE 4.
Calculated binding energies of rifampin-RNAP complexes in relation to rifampin resistance levels and fitness costs
| Substitution(s) in β subunit of S. aureus 8325-4 | Binding energies of rifampin (ΔERIF; kcal/mol)a | Rifampin MIC (μg/ml) | Relative fitness (W) ± SD |
|---|---|---|---|
| None | 30.16 | 0.008 | 1 |
| H481N | 28.73 | 2 | 0.80 ± 0.01 |
| ΔL485 | 25.32 | 4 | 0.81 ± 0.03 |
| H481N, L466S | 19.39 | 512 | 0.83 ± 0.04 |
| H481N, S529L | 17.56 | 512 | 0.99 ± 0.03 |
| H481Y | 19.87 | 1,024 | 0.50 ± 0.02 |
Obtained from the difference in the sum of the calculated heats of formation of the RNAP (ERNAP) and unbound rifampin, (ERIF-unbound) minus the heat of formation of the rifampin-RNAP complex (Ecomplex), according to the equation ΔERIF = (ERNAP + ERIF-unbound) − Ecomplex.
The detailed interactions involved in the rifampin-RNAP complex from T. aquaticus have been reported previously (8). This work revealed that essentially five amino acid residues make direct H-bonding contact with rifampin, which, using the numbering corresponding to the equivalent residues in the S. aureus enzyme, are F469, Q468, S486, R484, and H481 (Fig. 4). Additional residues lining the rifampin binding pocket, which include Q465, L466, L488, and G489 (data not shown), also appear to make van der Waals contacts with the antibiotic. Detailed examination of the molecular models of the mutant enzymes not only appears to reinforce the importance of these directly contacting residues upon rifampin binding but also throws light on additional allosteric factors that affect the binding efficiency. These aspects are considered in more detail below.
FIG. 4.
Model of rifampin (gold) binding to important residues in the S. aureus wild-type β subunit of RNAP.
H481N.
Histidine 481 (which corresponds to H406 in the T. aquaticus structure) resides on a loop located near to the ansa bridge of rifampin and makes an H-bonding contact to O-10 of rifampin (imidazole N and O-10 separation, 3.6 Å) (Fig. 4). Replacement with asparagine now removes this H bond due to the reduced length of the amide side chain. However, the model (data not shown) predicts that the side chain amide of the asparagine may be able to compensate partially by formation of a weak H bond (amide C—O and O-10 separation of 4.7 Å) to the rifampin O-10. Although there are small movements of various parts of the H481N complex relative to the wild-type structure (data not shown), it appears that the decrease in binding energy of 1.43 kcal/mol (Table 4) results from the weakening of the H bond to O-10 of rifampin and gives rise to a more than 200-fold drop in the activity of the antibiotic against intact bacteria (Table 1).
ΔL485.
Deletion mutant ΔL485 (which corresponds to ΔL410 in the T. aquaticus structure) results in a 500-fold decrease in drug activity against whole cells (Table 1) and is predicted to show a 4.84-kcal/mol decrease in binding energy to rifampin (Table 4).
Examination of the molecular model (Fig. 5i) reveals that L485 is located within a protein loop that makes extensive contacts with the aromatic portion of rifampin. In particular, this residue is flanked by R484 and S486, both of which make H-bond contacts to rifampin O-1 and O-2, respectively (Fig. 4). The L485 deletion results in a “pinching” of this loop, which has the effect of moving these residues closer together and requiring movement of the rifampin molecule, relative to its position in the wild-type complex, in order to maintain the H bond to R484 and S486, respectively (Fig. 4). The net result is that although essentially all of the original H bonding is retained by slight compensatory movements of each of the appropriate amino acid side chains, the hydrophobic interactions involving the C-13 and C-14 methyl groups of rifampin and the alkyl side chain of L488 (L413 in T. aquaticus) are diminished because these methyl groups shift 1.44 Å away from this residue relative to their positions in the wild-type structure. This lessening of the hydrophobic interaction appears to be mainly responsible for the decreased binding of rifampin to this mutant enzyme.
FIG. 5.
Overlay of part of the rifampin-binding region from wild-type RNAP (gray) and (i) mutant ΔL485 (yellow), (ii) mutant H481Y (purple), (iii) mutant H481N, S529L (green), and (iv) mutant H481N, L466S (blue).
H481Y.
The single mutation H481Y (H406Y in T. aquaticus) has a considerable effect on both the calculated binding energy of RNAP for rifampin (which decreases the binding by 10.29 kcal/mol relative to that of the wild-type enzyme) (Table 4) and the MIC (which is increased 128,000-fold from that displayed by the wild type) (Table 1).
Modeling (Fig. 5ii) reveals that due to the increased steric size of the phenolic side chain of tyrosine relative to the steric size of the imidazole ring in histidine, the tyrosine prefers to occupy a conformation which locates the side chain away from the rifampin binding region and therefore does not replace the H bond originally present via the imidazole side chain of H481 (Fig. 4). Additionally, the location of this side chain away from rifampin increases the available space within the binding cavity, resulting in a small movement of the rifampin molecule relative to its location in the wild-type complex. The main effect of this movement again appears to be a decrease in hydrophobic interactions at the C-13 and the C-14 methyl groups, which have moved 0.77 Å away from L488 relative to their positioning in the wild-type enzyme. The alternative conformation, in which the tyrosine side chain protrudes into the rifampin binding cavity, appears to be even more unfavorable and significantly decreases the binding energy even further due to disruption of the H bonding of rifampin with a number of residues within the cavity. It is the combination of the loss of an H bond coupled with a decrease in the hydrophobic interactions within the cavity that appears to be responsible for the significant loss of affinity of rifampin for this mutant enzyme.
H481N, S529L.
In the H481N, S529L double mutation (H406N, S454L in T. aquaticus), the larger size of the alkyl side chain in leucine compared to that of the hydroxymethyl in serine results in steric repulsion between this side chain and the proximal side chain of T480 (R405 in T. aquaticus) (Fig. 5iii). This results in movement of part of the amide backbone containing 480 and 481 and essentially removes the (weak) H bonding between N481 and O-10 of rifampin. Additionally, the model (Fig. 5iii) predicts that the side chain of T480 is pushed toward the C-38 methyl group of rifampin, resulting in a marked tilting of the rifampin molecule, particularly in the region of the phenolic hydroxyl groups. Although the H bonding of the enzyme with these groups is maintained through slight compensatory movements of the corresponding side chains, it is again the marked departure of the C-13 and the C-14 methyl groups from their original positions (by 1.32 Å) and the resulting decrease in hydrophobic interactions that largely result in a diminished affinity of the enzyme for the antibiotic (Table 4).
H481N, L466S.
The substitution of the alkyl group in leucine for the hydroxymethyl in serine has the net effect of decreasing the hydrophobic effect present from L466 on the C-14 methyl group of the antibiotic and also creates more space in the region of the binding cavity near this C-14 methyl group in rifampin. This results from the preference of S466 to be located away from the antibiotic binding cavity due to H bonding with the backbone C—O of F461. This causes a slight movement of the bound rifampin relative to its position in the wild-type complex, with the effect of moving the C-13 and C-14 methyl groups of rifampin away (by 1.34 Å) from L488 (Fig. 5iv).
Effects of rifampin resistance mutations on fitness.
To gain insight into the effects of resistance mutations in RNAP on the fitness of the organism, we used the molecular model of the RNAP-DNA complex (23, 34) to inspect the possible molecular interactions for a small selection of the mutant enzymes with the DNA template.
Major effects on fitness were observed for the rifampin-resistant mutants with the H481Y and A477D mutations (Table 1). In the case of the H481Y mutation, the model (Fig. 6i) reveals that substitution of the imidazole ring in histidine for the phenolic moiety in tyrosine may result in hydrogen bonding between the tyrosyl hydroxyl group and the proximal guanidine unit of R484. As this arginine residue lies at the surface of RNAP and is predicted to be in contact with DNA, this H bonding would move the arginine residue away from its original position, thus weakening the electrostatic attraction to DNA. The results of the modeling studies presented here appear to be consistent with observations on mutations at the equivalent site in the E. coli RNAP (amino acid residue 526), which decrease the stability of the transcription complex (22). The decreased stability of the complex presumably results in a diminished transcriptional efficiency for the rifampin-resistant RNAP, thereby accounting for the fitness cost associated with expression of the H481Y mutation in S. aureus.
FIG. 6.
Molecular models of the S. aureus H481Y (i) and A477D (ii) RNAP mutations based on a model of the T. aquaticus RNAP-DNA complex. Yellow ribbon represents protein backbone of RNAP.
For the A477D mutant, this substitution places a negatively charged carboxylate unit in close proximity to an existing carboxylate from H481, which lies close to the protein-DNA interface (Fig. 6ii). This would increase the negative charge on this surface of the protein and destabilize the enzyme-DNA interaction due to electrostatic repulsion. These conclusions are again consistent with the effects of a mutation at the equivalent position (residue 522) of the E. coli enzyme, which decreases the stability of the transcription complex (22) and which presumably incurs fitness costs on the organism.
Conclusions.
Rifampin is used clinically for the chemotherapy of infections caused by S. aureus, and this has resulted in the selection of mutants with alterations in the RRDR region of RNAP. Although a number of mutations in clinical isolates of S. aureus have already been defined by others (3, 44), we have expanded this analysis by examining a new collection of isolates. This approach led to the identification of several new resistance genotypes in the RRDR of clinical strains. In addition to mutations in the RRDR, our work also suggests that additional mechanisms can contribute to rifampin resistance in clinical isolates. However, we were unable to define these mechanisms. In addition to our work on resistance genotypes in clinical isolates, we also identified a number of novel mutations in the RRDRs of laboratory-derived rifampin-resistant mutants. Consequently, we conclude that the nature of rifampin resistance in S. aureus is more complex than previous studies have suggested.
We also examined, for the first time, the clinical prevalence of rifampin resistance alleles in relation to the fitness costs imposed by these mutations on a laboratory strain. The occurrence of clinical genotypes appears to be strongly influenced by the effects of the resistance mutations on fitness. In some cases clinical genotypes occurred that were associated with significant fitness costs in laboratory strains. We suspect that these resistance mutations in clinical strains could be associated with intragenic or extragenic compensatory mutations to offset fitness costs. Indeed, in a clinical isolate we identified an intragenic compensatory mutation (V850A) responsible for offsetting the fitness cost associated with expression of the rifampin resistance mutation A477D in the RRDR of rpoB.
It is well established that single mutations in β can give rise to high-level rifampin resistance. However, we have shown that a significant proportion of rifampin resistance genotypes from the clinic, including the most common genotype overall (H481N, S529L), involve double mutations. The secondary mutations found in concert with the primary resistance mutations were of three classes: (i) those that serve to increase the level of rifampin resistance (e.g., L466S in the presence of H481N), (ii) those that ameliorate the fitness costs engendered by the primary mutation (e.g., V850A in the presence of A477D), and (iii) those capable of conferring both increased resistance and increased fitness (e.g., S529L in the presence of H481N). Which of these evolutionary routes is chosen will clearly depend on the selection pressures exerted by rifampin and the nature of the primary resistance mutation.
In addition to the clinical implications of the work reported here, we provide a structural understanding of some of the mutations in the RRDR that confer rifampin resistance and their effects upon bacterial fitness. For example, there was a correlation between the binding energies for rifampin-RNAP interactions and the level of resistance to the antibiotic. Furthermore, the severe lack of fitness in two rifampin-resistant mutants could be explained by decreased binding of RNAP to the DNA template, consistent with earlier reports that these mutations affect the stability of the transcription complex.
Reliable models predictive for the likely emergence of bacterial resistance to new antibiotics would be a valuable addition to the preclinical evaluation of novel agents in development (33). The work presented here demonstrates a correlation between the fitness of rifampin-resistant S. aureus mutants determined by laboratory methods and their occurrence in the clinic by use of a larger set of staphylococcal clinical isolates than has been examined previously. The data therefore reinforce the concept that measurement of the fitness of bacterial mutants resistant to antibiotics is an important predictor of their survival in the clinical setting (19, 20, 30). This approach may be of particular value in the evaluation of several new bacterial RNAP inhibitors that are under investigation (2, 10, 11, 40).
Acknowledgments
We thank S. A. Darst (The Rockefeller University) for kindly providing the coordinates for the RNAP-DNA molecular model. The following kindly provided strains or plasmids: A. MacGowan (BRI, United Kingdom), strains BR1 to BR9; C. Y. Lee (University of Kansas), plasmid pCL52.1; and A. L. Cheung (Dartmouth Medical School), plasmid pALC2073. We also thank A. M. Moita for technical assistance.
REFERENCES
- 1.Andersson, D. I., and B. R. Levin. 1999. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 2:489-493. [DOI] [PubMed] [Google Scholar]
- 2.Artsimovitch, I., C. Chu, A. S. Lynch, and R. Landick. 2003. A new class of bacterial RNA polymerase inhibitor affects nucleotide addition. Science 302:650-654. [DOI] [PubMed] [Google Scholar]
- 3.Aubry-Damon, H., C. J. Soussy, and P. Courvalin. 1998. Characterization of mutations in the rpoB gene that confer rifampin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 42:2590-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman, B. T., N. P. Donegan, T. M. Jarry, M. Palma, and A. L. Cheung. 2001. Evaluation of a tetracycline-inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect. Immun. 69:7851-7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Billington, O. J., T. D. McHugh, and S. H. Gillespie. 1999. Physiological cost of rifampin resistance induced in vitro in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 43:1866-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bottger, E. C., B. Springer, M. Pletschette, and P. Sander. 1998. Fitness of antibiotic-resistant microorganisms and compensatory mutations. Nat. Med. 4:1343-1344. [DOI] [PubMed] [Google Scholar]
- 7.British Society for Antimicrobial Chemotherapy. 1991. A guide to sensitivity testing; report of the Working Party on Antibiotic Sensitivity Testing of the British Society for Antimicrobial Chemotherapy. J. Antimicrob. Chemother. 27(Suppl. D):1-50. [PubMed] [Google Scholar]
- 8.Campbell, E. A., N. Korzheva, A. Mustaev, K. Murakami, S. Nair, A. Goldfarb, and S. A. Darst. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901-912. [DOI] [PubMed] [Google Scholar]
- 9.Cohan, F. M., E. C. King, and P. Zawadzki. 1994. Amelioration of the deleterious pleiotropic effects of an adaptive mutation in Bacillus subtilis. Evolution 48:81-95. [DOI] [PubMed] [Google Scholar]
- 10.Darst, S. A. 2004. New inhibitors targeting bacterial RNA polymerase. Trends Biochem. Sci. 29:159-160. [DOI] [PubMed] [Google Scholar]
- 11.Doundoulakis, T., A. X. Xiang, R. Lira, K. A. Agrios, S. E. Webber, W. Sisson, R. M. Aust, A. M. Shah, R. E. Showalter, J. R. Appleman, and K. B. Simonsen. 2004. Myxopyronin B analogs as inhibitors of RNA polymerase, synthesis and biological evaluation. Bioorg. Med. Chem. Lett. 14:5667-5672. [DOI] [PubMed] [Google Scholar]
- 12.Fairweather, N., S. Kennedy, T. J. Foster, M. Kehoe, and G. Dougan. 1983. Expression of a cloned Staphylococcus aureus alpha-hemolysin determinant in Bacillus subtilis and Staphylococcus aureus. Infect. Immun. 41:1112-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garibyan, L., T. Huang, M. Kim, E. Wolff, A. Nguyen, T. Nguyen, A. Diep, K. Hu, A. Iverson, H. Yang, and J. H. Miller. 2003. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair (Amsterdam) 2:593-608. [DOI] [PubMed] [Google Scholar]
- 14.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the swiss-Pdb viewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 15.Hansson, S., R. Singh, A. T. Gudkov, A. Liljas, and D. T. Logan. 2005. Structural insights into fusidic acid resistance and sensitivity in EF-G. J. Mol. Biol. 348:939-949. [DOI] [PubMed] [Google Scholar]
- 16.Herrera, L., S. Jimenez, A. Valverde, M. A. Garcia-Aranda, and J. A. Saez-Nieto. 2003. Molecular analysis of rifampicin-resistant Mycobacterium tuberculosis isolated in Spain (1996-2001). Description of new mutations in the rpoB gene and review of the literature. Int. J. Antimicrob. Agents 21:403-408. [DOI] [PubMed] [Google Scholar]
- 17.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-136. [DOI] [PubMed] [Google Scholar]
- 18.Hood, J., B. Cosgrove, E. Curran, M. Lockhart, B. Thakker, and C. Gemmell. 2000. Vancomycin-intermediate resistant Staphylococcus aureus in Scotland. Abstr., 4th Decennial Int. Conf. Nosocomial Healthcare-Associated Infect.
- 19.Hurdle, J. G., A. J. O'Neill, and I. Chopra. 2004. The isoleucyl-tRNA synthetase mutation V588F conferring mupirocin resistance in glycopeptide-intermediate Staphylococcus aureus is not associated with a significant fitness burden. J. Antimicrob. Chemother. 53:102-104. [DOI] [PubMed] [Google Scholar]
- 20.Hurdle, J. G., A. J. O'Neill, E. Ingham, C. Fishwick, and I. Chopra. 2004. Analysis of mupirocin resistance and fitness in Staphylococcus aureus by molecular genetic and structural modeling techniques. Antimicrob. Agents Chemother. 48:4366-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin, D. J., and C. A. Gross. 1988. Mapping and sequencing of mutations in the Escherichia coli rpob gene that lead to rifampicin resistance. J. Mol. Biol. 202:45-58. [DOI] [PubMed] [Google Scholar]
- 22.Jin, D. J., W. A. Walter, and C. A. Gross. 1988. Characterization of the termination phenotypes of rifampicin-resistant mutants. J. Mol. Biol. 202:245-253. [DOI] [PubMed] [Google Scholar]
- 23.Korzheva, N., A. Mustaev, M. Kozlov, A. Malhotra, V. Nikiforov, A. Goldfarb, and S. A. Darst. 2000. A structural model of transcription elongation. Science 289:619-625. [DOI] [PubMed] [Google Scholar]
- 24.Kumari, D. N. P., V. Keer, P. M. Hawkey, P. Parnell, N. Joseph, J. F. Richardson, and B. Cookson. 1997. Comparison and application of ribosome spacer DNA amplicon polymorphisms and pulsed-field gel electrophoresis for differentiation of methicillin-resistant Staphylococcus aureus strains. J. Clin. Microbiol. 35:881-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 26.Lin, W. S., T. Cunneen, and C. Y. Lee. 1994. Sequence analysis and molecular characterization of genes required for the biosynthesis of type 1 capsular polysaccharide in Staphylococcus aureus. J. Bacteriol. 176:7005-7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maisnier-Patin, S., and D. I. Andersson. 2004. Adaptation to the deleterious effects of antimicrobial drug resistance mutations by compensatory evolution. Res. Microbiol. 155:360-369. [DOI] [PubMed] [Google Scholar]
- 28.Matsiota-Bernard, P., G. Vrioni, and E. Marinis. 1998. Characterization of rpoB mutations in rifampin-resistant clinical Mycobacterium tuberculosis isolates from Greece. J. Clin. Microbiol. 36:20-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohamadi, F., N. G. J. Richards, W. C. Guida, R. Liskamp, M. Lipton, C. Caufield, G. Chang, T. Hendrickson, and W. C. Still. 1990. Macromodel—an integrated software system for modeling organic and bioorganic molecules using molecular mechanics. J. Comput. Chem. 11:440-467. [Google Scholar]
- 30.Nilsson, A. I., O. G. Berg, O. Aspevall, G. Kahlmeter, and D. I. Andersson. 2003. Biological costs and mechanisms of fosfomycin resistance in Escherichia coli. Antimicrob. Agents Chemother. 47:2850-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novick, R. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155-166. [DOI] [PubMed] [Google Scholar]
- 32.O'Neill, A., B. Oliva, C. Storey, A. Hoyle, C. Fishwick, and I. Chopra. 2000. RNA polymerase inhibitors with activity against rifampin-resistant mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 44:3163-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Neill, A. J., and I. Chopra. 2004. Preclinical evaluation of novel antibacterial agents by microbiological and molecular techniques. Expert Opin. Investig. Drugs 13:1045-1063. [DOI] [PubMed] [Google Scholar]
- 34.Opalka, N., M. Chlenov, P. Chacon, W. J. Rice, W. Wriggers, and S. A. Darst. 2003. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell 114:335-345. [DOI] [PubMed] [Google Scholar]
- 35.Parenti, F., and G. Lancini. 1997. Rifamycins, p. 453-459. In F. O'Grady, H. P. Lambert, R. G. Finch, and D. Greenwood (ed.), Antibiotic and chemotherapy, 7th ed. Churchill Livingstone, New York, N.Y.
- 36.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 37.Reynolds, M. G. 2000. Compensatory evolution in rifampin-resistant Escherichia coli. Genetics 156:1471-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell, A. D., and I. Chopra. 1996. Understanding antibacterial action and resistance, 2nd ed. Ellis Horwood, Hertfordshire, United Kingdom.
- 39.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Sarubbi, E., F. Monti, E. Corti, A. Miele, and E. Selva. 2004. Mode of action of the microbial metabolite GE23077, a novel potent and selective inhibitor of bacterial RNA polymerase. Eur. J. Biochem. 271:3146-3154. [DOI] [PubMed] [Google Scholar]
- 41.Schenk, S., and R. A. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133-138. [DOI] [PubMed] [Google Scholar]
- 42.Smith, T. L., M. L. Pearson, K. R. Wilcox, C. Cruz, M. V. Lancaster, B. Robinson-Dunn, F. C. Tenover, M. J. Zervos, J. D. Band, E. White, W. R. Jarvis, et al. 1999. Emergence of vancomycin resistance in Staphylococcus aureus. N. Engl. J. Med. 340:493-501. [DOI] [PubMed] [Google Scholar]
- 43.Wichelhaus, T. A., B. Boddinghaus, S. Besier, V. Schafer, V. Brade, and A. Ludwig. 2002. Biological cost of rifampin resistance from the perspective of Staphylococcus aureus. Antimicrob. Agents Chemother. 46:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wichelhaus, T. A., V. Schafer, V. Brade, and B. Boddinghaus. 1999. Molecular characterization of rpoB mutations conferring cross-resistance to rifamycins on methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 43:2813-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]