Abstract
Streptococcus agalactiae (group B streptococcus [GBS]) is the leading cause of neonatal and maternal sepsis. Penicillin is recommended for intrapartum prophylaxis, but erythromycin or clindamycin is used for penicillin-allergic carriers. Antibiotic resistance (AR) has increased recently and needs to be monitored. We have developed a multiplex PCR-based reverse line blot (mPCR/RLB) hybridization assay to detect, simultaneously, seven genes encoding AR—erm(A/TR), erm(B), mef(A/E), tet(M), tet(O), aphA-3, and aad-6—and two AR-related genes, int-Tn and mreA. We tested 512 GBS isolates from Asia and Australasia and compared mPCR/RLB with antibiotic susceptibility phenotype or single-gene PCR. Phenotypic resistance to tetracycline was identified in 450 (88%) isolates, of which 442 had tet(M) (93%) and/or tet(O) (6%). Of 67 (13%) erythromycin-resistant isolates, 18 were susceptible to clindamycin, i.e., had the M phenotype, encoded by mef(A/E); 39 had constitutive (cMLSB) and 10 inducible clindamycin resistance, and of these, 34 contained erm(B) and 12 erm(A/TR). Of four additional isolates with mef(A/E), three contained erm(B) with cMLSB and one was erythromycin susceptible. Of 61 (12%) clindamycin-resistant isolates, 20 were susceptible to erythromycin and two had intermediate resistance. Based on sequencing, 21 of 22 isolates with mef had mef(E), and 8 of 353 with int-Tn had an atypical sequence. Several AR genes, erm(B), tet(O), aphA-3, aad-6, and mef(A/E), were significantly more common among Asian than Australasian isolates, and there were significant differences in distribution of AR genes between GBS serotypes. Our mPCR/RLB assay is simple, rapid, and suitable for surveillance of antibiotic resistance in GBS.
Streptococcus agalactiae (group B streptococcus [GBS]) is the leading cause of neonatal and maternal infections and an opportunistic pathogen in immunocompromised patients and adults with underlying disease (26). It is usually susceptible to penicillin, which is the antibiotic of choice for treatment or intrapartum antibiotic prophylaxis of GBS infections. Erythromycin (a macrolide) or clindamycin (a lincosamine) are recommended alternatives for intrapartum antibiotic prophylaxis in penicillin-allergic GBS carriers (2). However, an increase in the incidence of resistance to these agents has been observed in the last decade (24, 31), and surveillance is needed.
Tetracycline resistance is common in GBS and usually is due to ribosomal protection encoded by tet(M) or tet(O) or less commonly to an efflux pump encoded by tet(K) or tet(L) (9, 27). tet(M) is often carried by the conjugative transposon Tn916, and its integrase (int-Tn) is commonly present in isolates carrying tet(M). Erythromycin resistance genes (25) are also carried on transposons. They include the erm (B, A/TR, or C) genes, which encode a ribosomal methylase and confer constitutive or inducible cross-resistance to macrolide, lincosamine, and streptogramin B antibiotics (cMLSB or iMLSB phenotypes, respectively), and the mef (A or E) genes, which encode an efflux pump (20) and confer resistance to 14- and 15-membered macrolides, including erythromycin, but not to lincosamines (M phenotype).
The gene mreA was originally reported to encode an alternative efflux pump (6) but has since been shown to be present in all GBS isolates, irrespective of macrolide resistance, and to encode a flavokinase (8). The aminoglycoside resistance genes aphA-3 and aad-6 encode kanamycin phosphorylase and streptomycin adenylylase, respectively (23).
Individual gene-specific and multiplex PCR, with or without microwell probe hybridization, has been used to identify various resistance-related genes in many types of bacteria, including streptococci (14, 29). In this study, we developed a simple and convenient multiplex PCR and reverse line blot (mPCR/RLB) assay to detect nine antibiotic resistance (AR)-related genes in GBS for surveillance of antibiotic resistance and supplementary strain typing and compared it with phenotypic antibiotic resistance profiles and, in a sample of isolates, with individual gene-specific PCRs.
MATERIALS AND METHODS
GBS isolates tested.
A total of 512 GBS isolates were included in the study: 132 from Hong Kong, isolated between 1993 and 2003, of which 52 were from normally sterile sites (mainly blood) and 80 from superficial or nonsterile sites; 118 (mostly invasive) from New Zealand isolated in 2004; 186 from South Korea, isolated between 1993 and 1996 (21), of which 177 were from superficial or nonsterile sites (including 100 from urine) and 11 from sterile sites; and 76 from our own laboratory in Sydney, all from blood cultures, isolated between 2001 and 2004.
DNA extraction.
DNA extraction was performed as described previously (19).
Oligonucleotide design.
For single-gene-specific PCR, we modified a number of previously published primers targeting AR genes associated with resistance to tetracyclines [tet(M), tet(O), and int-Tn], erythromycin [erm(B), erm(A/TR), mef(A/E), and mreA], and aminoglycosides (aphA-3 and aad-6) in gram-positive bacteria (10, 14, 25). To ensure similar amplification efficiencies for each target, these primers were further modified (see Table S1 in the supplemental material) and 5′ biotin labeled (Sigma-Aldrich, Castle Hill, Australia) for use in mPCR. Two specific probes, for each resistance gene, were designed for RLB hybridization and 5′ hexylamine labeled (Sigma-Aldrich, Australia) (see Table S1 in the supplemental material).
In addition, phenotypically erythromycin-resistant isolates, in which none of the three erythromycin resistance genes was detected, were tested by an erm(C)-specific PCR (see Table S1 in the supplemental material) (14).
Individual gene-specific PCR.
All of the 318 isolates from Asia (Hong Kong, 132 isolates, and Korea, 186 isolates) were tested by individual gene-specific PCR. Each individual gene-specific PCR mixture contained the following: 5 μl template DNA, 0.25 μl of forward primer (50 pmol/μl) and reverse primer (50 pmol/μl), 1 μl deoxynucleoside triphosphates (dNTPs) (2.5 mM of each dNTP), 2.5 μl 10× PCR buffer, 0.1 μl Promega DNA Taq polymerase (5 units/μl), and water to 25 μl. PCR was performed as follows: 94°C for 10 s, 65°C for 30 s, 72°C for 30 s, repeated for 35 cycles. A total of 8 μl PCR product was detected by electrophoresis on a 1.5% agarose gel.
Multiplex PCR.
The nine-primer-pair mPCR mixture was prepared as follows: 5 μl template DNA, 0.25 μl of all forward primers (50 pmol/μl) and reverse primers (50 pmol/μl), 1.25 μl dNTPs (2.5 mM of each dNTP), 2.5 μl 10× PCR buffer, 3 μl 25 mM (4.5 mM final) MgCl2, 0.2 μl QIAGEN Hotstart Taq polymerase (5 units/μl), with water added to 25 μl. Multiplex PCR was performed according to instructions from QIAGEN Hotstart Taq polymerase suppliers as follows: 95°C for 15 min one cycle, 94°C for 30 s, 60°C for 30 s, 72°C for 1 min, 35 cycles, 72°C for 10 min 1 cycle, and 22°C hold. A total of 8 μl of PCR product was identified by electrophoresis on a 1.5% agarose gel to confirm the successful amplification. The remaining PCR products (5 μl of PCR product) were used for RLB hybridization.
RLB hybridization.
The RLB hybridization assay was based on the method described previously (32, 34) except that the hybridization temperature was 60°C, and streptavidin-peroxidase conjugate (Roche Diagnostics Co.) was diluted 1:4,000 in 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])-0.5% sodium dodecyl sulfate, and the time of exposure to X-ray film (Hyperfilm; Amersham) was adjusted to 7 min. To optimize hybridization conditions, the probe was tested at several twofold dilutions, starting at a concentration of 2.4 pM.
Phenotypic antibiotic susceptibility testing.
All 512 isolates were tested by disk diffusion for susceptibility to erythromycin, clindamycin, and tetracycline according to the CLSI (formerly NCCLS) method (15). In order to distinguish between the M, iMLSB, and cMLSB phenotypes, for erythromycin-resistant isolates, erythromycin (15 μg) and clindamycin (2 μg) disks were placed approximately 20 mm apart, and the plates were incubated overnight at 35°C in 5% CO2. Flattening of the clindamycin growth inhibition zone, between the two disks, indicated iMLSB resistance; zones of ≤15 mm around both disks indicated cMLSB resistance, and resistance to erythromycin but not to clindamycin indicated the M phenotype.
A small number of isolates, which had apparently inconsistent phenotype and genotypic AR profiles, were tested using the Etest antibiotic gradient strip system (AB Biodisk, Australia; Laboratory Services Pty. Ltd.) according to the manufacturer's instructions, and relevant individual gene specific PCR were repeated, if necessary.
MS identification.
Molecular serotype (MS) identification was performed for all isolates tested as described previously (4, 19)
Sequencing and sequence analysis.
Sequencing was performed as described previously (19). BioManager in ANGIS (Australian National Genomic Information Service; http://biomanager.angis.org.au/) provided all programs used in the study, including Work Bench for sequence file management, BESTFIT for two-sequence comparison, PILEUP and PRETTY in the multiple sequence analysis program group for multiple sequence alignments, and BLASTN for sequence searches.
Statistical analysis.
Statistical analysis was performed using EPI INFO 6 software, and the chi-square test or Fisher's exact test was used where appropriate.
Nucleotide sequence accession number.
A newly identified int-Tn-like sequence (see below) was deposited in GenBank with accession number DQ103589.
RESULTS
Comparison of phenotype with relevant individual PCR results.
Phenotypic tetracycline resistance was identified in 450 of 512 (88%) isolates, of which 413 (92%) had tet(M), 25 (5%) had tet(O), 4 (1%) had both, and 8 (2%) neither. Five isolates with tet(M) had intermediate tetracycline resistance (MIC, 3 to 4 μg/ml), and four were susceptible (MIC, 0.125 to 0.5 μg/ml) based on disk diffusion and Etest.
Phenotypic resistance to erythromycin and clindamycin was identified in 67 (13%) and 61 (12%) isolates, respectively. Erythromycin resistance rates were significantly higher in Hong Kong (31/132; 24%) than in South Korea (20/186; 11% [P = 0.002]), Australia (5/76; 7% [P = 0.002]), and New Zealand (11/118; 9% [P = 0.002]). The corresponding rates of clindamycin resistance were, for Hong Kong, 16/132 (12%); for South Korea, 28/186 (15%); for Australia, 2/76 (3%); and for New Zealand, 15/118 (13%); these rates were significantly higher in Hong Kong (P = 0.02), New Zealand, and Korea (P = 0.004 for both) than in Australia.
Distributions of phenotype and genotype in erythromycin- and/or clindamycin-resistant isolates are shown in Table 1. Three erythromycin-resistant isolates, with the iMLSB phenotype confirmed by retesting and Etest (MIC, >32 μg/ml), contained none of the four erythromycin resistance genes, including erm(C). Three isolates with erm(B) and one with mef(A/E) were susceptible to erythromycin based on disk diffusion and Etest (MIC, 0.19 to 0.25 μg/ml).
TABLE 1.
Relationship of phenotype to genotype in phenotypically erythromycin-resistant (n = 67) and/or clindamycin-resistant (n = 61) GBS isolates
| Phenotypea | No. of strains with genotype
|
Total | ||||
|---|---|---|---|---|---|---|
| erm(B) | erm(A/TR) | mef(A/E) | erm(B) + mef(A/E) | No erythromycin resistance geneb detected | ||
| cMLSBc (eryR; clindR) | 31 | 5 | 3 | 39 | ||
| iMLSBc | 7 | 3 | 10 | |||
| Mc (eryR; clindS) | 18 | 18 | ||||
| eryS; clindS | 1 | 1 | 421 | 423 | ||
| eryS/I; clindR | 2 | 20d | 22 | |||
| Total | 34 | 12 | 19 | 3 | 444 | 512 |
eryS, -R, or -I, erythromycin susceptible, resistant, or intermediate; clindS or -R, clindamycin susceptible or resistant.
Including erm(C).
M, resistant to erythromycin only; cMLSB, constitutively resistant to erythromycin and clindamycin; iMLSB, resistant to erythromycin and inducibly resistant to clindamycin (all of which appeared susceptible to clindamycin on single-disk testing).
Two of these isolates had intermediate resistance to erythromycin.
Of 61 clindamycin-resistant isolates, 9 from Korea (MIC, 8 to 128 μg/ml), 9 from New Zealand (MIC, 1.5 to 32 μg/ml), and 2 from Hong Kong (MIC, 3 μg/ml and 6 μg/ml, respectively) were susceptible to erythromycin (MIC, 0.094 to 0.25 μg/ml), including 2 isolates from Korea containing erm(B); 2 isolates (1 each from Korea and New Zealand) were intermediate to erythromycin (MICs, 0.38 μg/ml and 0.75 μg/ml).
Results of mPCR/RLB and correlation with gene-specific PCR.
The geographic distributions of eight AR-related genes among 512 isolates are shown in Table 2. All isolates contained mreA.
TABLE 2.
Distribution of resistance genes in isolates from four countriesa
| Country (n)b | No. (%) of isolates with AR gene
|
|||||||
|---|---|---|---|---|---|---|---|---|
| tet(M)c | int-Tnd | tet(O) | erm(B)e | erm(A/TR) | mef(A/E)f,g | aphA-3e | aad-6e | |
| Hong Kong (132) | 102 (77) | 81 (61) | 13 (10) | 12 (9) | 3 (2) | 16 (12) | 11 (8) | 12 (9) |
| South Korea (186) | 165 (89) | 157 (84) | 12 (6) | 20 (11) | 0 | 4 (2) | 19 (10) | 22 (12) |
| Australia (76) | 62 (82) | 40 (53) | 1 (1) | 1 (1) | 3 (4) | 1 (1) | 0 | 0 |
| New Zealand (118) | 97 (82) | 67 (57) | 3 (3) | 4 (3) | 6 (5) | 1 (1) | 2 (2) | 2 (2) |
| Total (512) | 426 (83) | 345 (67) | 29 (6) | 37 (7) | 12 (2) | 22 (4) | 32 (6) | 36 (7) |
Boldface areas show data that are significantly different from others in the same column.
n, no. of isolates.
A significantly smaller proportion of Hong Kong isolates than of those from all other countries contained tet(M) (P = 0.03).
A significantly greater proportion of South Korean isolates than of those from all other countries contained int-Tn (P = < 0.0001).
A significantly greater proportion of Asian than of Australasian isolates contained tet(O) (P = 0.01), erm(B) (P = 0.003), aphA-3 (P = 0.0003), and aad-6 (P < 0.0001).
A significantly greater proportion of Hong Kong isolates than of those from all other countries contained mef(A/E) (P < 0.0001).
All but one of the mef genes were mef(E).
Of the 426 isolates that contained tet(M), 340 (80%) also harbored int-Tn and represented 99% of 345 int-Tn positive isolates. Of five isolates containing int-Tn without tet(M), one contained mef(A/E) and the others contained no other AR genes.
Sixty-eight (13%) isolates contained one or more erythromycin resistance genes, including three with both erm(B) and mef(A/E). Thirty-two (47%) isolates contained one (aphA-3, 4; aad-6, 1) or both (27 isolates) aminoglycoside resistance genes. Of 37 isolates that contained erm(B), 21 (57%) contained int-Tn and tet(M), and of these, 16 also had 1 or both aminoglycoside resistance genes. Of 13 isolates with both erm(B) and tet(O), 12 also had aphA-3 and aad-6.
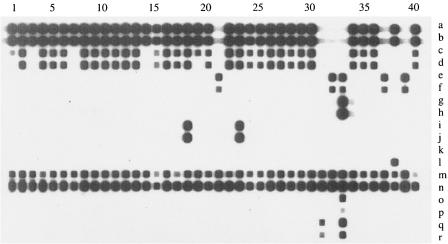
After optimizing the concentration of probes on the membrane, good hybridization signals were detected for all resistance genes by RLB (Fig. 1). For most isolates, there was agreement between mPCR/RLB (two probes per gene target) and individual gene-specific PCR results. The exceptions were 8 of 353 int-Tn PCR-positive and 21 of 22 mef(A/E) PCR-positive isolates, for which mPCR products consistently (tests repeated at least twice) hybridized with only one of two corresponding RLB probes. Sequencing of the amplicons showed that the eight atypical int-Tn-specific PCR products (all from Hong Kong isolates) were identical to each other but different from the GenBank int-Tn sequence (GenBank accession no. U09422) and all existing GenBank sequences. This variant int-Tn sequence was deposited in GenBank (see above). The 21 mef(A/E) amplicons that hybridized with only one probe were identified by sequencing as mef(E). The one that hybridized with both probes was mef(A), from a New Zealand isolate belonging to serotype Ia.
FIG. 1.
Results of multiplex PCR and reverse line blot (RLB) hybridization for 40 consecutive isolates (of 132) from Hong Kong. The 18 labeled probes, from top to bottom are as follows: a, tetmSp; b, tetmAp; c, intSp; d, intAp; e, tetoSp; f, tetoAp; g, ermbSp; h, ermbAp; i, ermatrSp; j, ermatrAp; k, mefSp; l, mefAp; m, mreSp; n, mreAp; o, apha3Sp; p, apha3Ap; q, aad6Sp; r, aad6Ap.
Geographic distribution of resistance genes.
The distributions of eight AR-related genes in four countries are shown in Table 2. The most prevalent resistance-associated genes in all countries were tet(M) and int-Tn. However, int-Tn was significantly more common in South Korea (P < 0.00001) and tet(M) significantly less common in Hong Kong (P = 0.03) than in the other countries combined. Most AR genes, except erm(A/TR) and tet(M), were found significantly more commonly in Asian than in Australasian isolates. For mef(A/E), the rate was significantly higher in Hong Kong than in all of the other countries (P < 0.0001).
A significantly greater proportion of erm(B)-positive isolates from South Korea than from Hong Kong contained int-Tn (15/20 versus 4/12; P = 0.02), usually with tet(M) and aphA-3, indicating that the reported association of erm(B) and tet(M) with int-Tn is stronger in some geographic areas than in others.
Resistance gene distribution among serotypes.
The distributions of eight AR-related genes among nine GBS serotypes are shown in Table 3. The 68 isolates, which contained one or more erythromycin resistance genes, included 59 of 373 (16%) of serotypes III (16%), Ia (12%), V (21%), or II (17%), compared with 9 of 131 (5%) isolates of other serotypes (P < 0.01). The three erythromycin-resistance genes were fairly evenly distributed in serotype V, but there were statistically significant associations between mef(E) and serotype Ia (P = 0.003), erm(A/TR) and serotype II (P = 0.003), and erm(B) and serotype III (P < 0.001).
TABLE 3.
Distribution of AR-related markers among group B streptococcal serotypesa
| MS (nb) | No. (%) of isolates with AR gene
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| tet(M) | int-Tn | tet(O) | erm(B) | erm(A/TR) | mef(A/E) | aphA-3 | aad-6 | None | |
| Ia (92) | 82 (89) | 27 (29)c | 0 | 1 (1) | 0 | 10 (11)d | 1 (1) | 2 (2) | 8 (9) |
| Ib (100) | 85 (85) | 85 (85) | 7 (7) | 7 (7) | 0 | 0 | 7 (7) | 13 (13) | 8 (8) |
| II (30) | 23 (77) | 20 (67) | 5 (17) | 1 | 4 (13)e | 1 (3) | 1 (3) | 1 (3) | 1 (3) |
| III (173) | 147 (85) | 133 (79) | 15 (9) | 22 (13)f | 2 (1) | 5 (3) | 22 (13) | 17 (10) | 11 (6) |
| IV (13) | 9 (69) | 4 (31) | 1 (8) | 0 | 1 (8) | 0 | 0 | 0 | 2 (15) |
| V (78) | 73 (94) | 69 (88) | 1 (1) | 5 (6) | 5 (6) | 6 (8) | 0 | 0 | 3 (4) |
| VI (18) | 2 (11)g | 2 (11) | 0 | 1 (6) | 0 | 0 | 1 (6) | 3 (17) | 13 (72)g |
| VII (3) | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| VIII (3) | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| NTh(2) | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total (512) | 426 (83) | 345 (67) | 29 (6) | 37 (7) | 12 (2) | 22 (4) | 32 (6) | 36 (7) | 49 (10) |
Boldface areas show data that are significantly different from others in the same column.
No. of isolates.
The prevalence of int-Tn in tet(M)-positive isolates was significantly lower in serotype Ia than in other serotypes (P < 0.001).
The prevalence of mef(A/E) in serotype Ia was significantly higher than in other serotypes (P = 0.003).
The prevalence of erm(A/TR) in serotype II was significantly higher than in other serotypes (P = 0.003).
The prevalence of erm(B) in serotype III was significantly higher than in other serotypes (P = 0.0006).
The proportion of isolates without AR markers, especially tet(M), in serotype VI was significantly higher than in other serotypes (P < 0.001).
NT, nontypable by PCR/sequencing, repeated several times.
The majority of isolates of most serotypes contained tet(M) (ranging from 69% to 94%); the exception was serotype VI, of which only 11% contained tet(M) (P < 0.001). Serotype Ia was the only one in which the majority of isolates with tet(M) did not contain int-Tn (66/92 versus 20/334; P < 0.001).
The 49 isolates with none of the 8 AR markers included 13 of 18 (72%) of serotype VI, which was a significantly greater proportion than of all other serotypes (36/494, 7%; P < 0.001).
DISCUSSION
The main aim of this study was to develop and evaluate a multiplex PCR-based RLB method to detect nine AR-related genes, which have been previously identified in GBS (25). The mPCR/RLB method was compared with phenotypic antibiotic susceptibility testing and individual gene-specific PCR to assess its specificity and sensitivity. We selected a total of more than 500 isolates, from four countries, to assess the suitability of the method for epidemiological studies of geographic variation in the distribution of AR-related genes.
To maximize the speed, convenience, and specificity of the mPCR/RLB assay, hybridization targets were generated using a “hotstart” multiplex PCR, which produces short amplicons (355 bp to 553 bp) spanning the oligonucleotide probes on the membrane. Multiplex PCR requires that primers amplify unique DNA regions under the same reaction conditions, whether they are used as single pairs or in combinations of many primers, and individual amplification products must be distinguishable from each other. RLB can simultaneously identify up to 43 mPCR products and, because of its simplicity, speed, and high sensitivity and specificity, can be used to screen large numbers of samples rapidly. It allows cost-effective, rapid, high-throughput and standardized detection of resistance mechanisms in GBS for use in large surveillance studies. The whole procedure takes approximately 11 h, 4 hours for PCR and 7 for RLB; that is, a batch of isolates can be tested in one-and-a-half working days.
The most widely distributed tetracycline resistance determinant in gram-positive bacteria is tet(M), which is carried on the Tn916 conjugative transposon (28). It was the most prevalent AR-associated gene found in this study—in 83% of isolates—which reflects high rates of tetracycline resistance in GBS worldwide (25). By contrast, tet(O) was found in only 6% of isolates; only 2% of tetracycline-resistant isolates had neither tet(M) nor tet(O) but presumably carried one of the other tetracycline resistance genes. Conjugative transposition of Tn916 requires the activity of a transposon-encoded integrase (28) encoded by int-Tn, which was found in 80% of isolates harboring tet(M). The eight isolates from Hong Kong, which hybridized with only one of two int-Tn-specific probes, contained an atypical int-Tn-like sequence. Comparison with the int-Tn sequence in GenBank showed that it carried a mutation in the region corresponding to the probe sequence. RLB was more specific than single-gene-specific PCR in identifying this int-Tn variant but would have been classified as negative according to our criterion that a positive result required hybridization with both probes. This atypical int-Tn-like sequence is a potentially useful molecular marker.
The isolates tested in this study were not collected systematically and represent various proportions of invasive and superficial or colonizing isolates from the four countries represented. However, a number of differences were noted in the geographic distribution of antibiotic resistance and AR-related genes, which deserve further investigation using systematically collected isolates from different countries. Phenotypic resistance to erythromycin was significantly more common in Asian than in Australasian isolates, which correlates with a higher prevalence of erm(B) in Asian than in Australasian isolates and of mef(A/E), especially in Hong Kong. Presumably this reflects differences in antibiotic use between regions.
Like tet(M) and aphA-3, erm(B) in streptococci is often associated with the conjugative Tn916-Tn1514 transposons (9, 13) and therefore with int-Tn (28). However, we found significant geographic differences in the distribution of int-Tn between countries, overall, and among erm(B)-positive isolates. In GBS, erm(B) is usually the predominant erythromycin resistance marker (25), but we found that mef(E) was more common in Hong Kong, where tet(O) was also more common, and tet(M) less common, than elsewhere.
Of the erythromycin-resistant GBS isolates, three had none of the erythromycin resistance genes that we studied, which is consistent with previous reports (1, 16). It is possible that 23S rRNA mutations, which have been recently reported to cause macrolide resistance in S. pneumoniae (11), could be responsible for some erythromycin resistance in GBS. We also found none of the erythromycin resistance genes in 20 clindamycin-resistant, erythromycin-susceptible/intermediate isolates. At least two types of clindamycin resistance have been described for macrolide-sensitive GBS. One is mediated by a lincosamide nucleotidyl transferase, encoded by linB, which was first found in Enterococcus faecium (5) and occurs infrequently in GBS (10, 12). The genetic basis of the other type, described as the lincosamine-streptogramin A phenotype, is unknown. It was recently found in GBS isolates from New Zealand (22), where a relatively high level of clindamycin resistance has been described previously (12, 35). It is therefore of interest that 10 of the 20 clindamycin-resistant, erythromycin-susceptible/intermediate isolates identified in this study were from New Zealand, whereas none were identified among Australian isolates.
The genes mef(A) and mef(E), which were originally discovered in S. pyogenes (7) and S. pneumoniae (30), respectively, and later also in GBS (3), are 90% homologous, and mef-specific PCR usually amplifies both. Restriction enzyme analysis, sequencing, or real-time PCR (18) is needed to distinguish them but is often not done (17). The two probes used in this study, which were designed to identify mef(A), fortuitously allowed us to differentiate mef(A), which bound to both, from mef(E) amplicons, which bound to only one. In the future, we will design two mef(A)-specific and two mef(E)-specific probes to produce more robust results and fulfill our criterion for a positive result. In our study, only 1 of 22 mef-positive isolates (from New Zealand) had mef(A).
As reported by others, erythromycin resistance was most common for serotype V (33), for which all three erythromycin resistance genes were found with similar frequencies. The statistically significant associations between mef(E), erm(B), and erm(A/TR) and serotypes Ia, III, and II, respectively, have not, to our knowledge, been described previously.
In conclusion, we have developed an mPCR/RLB hybridization assay, which can identify nine AR-related genes simultaneously in GBS and will be suitable for large-scale epidemiological studies and molecular “fingerprinting” of GBS in the future. It correlates well, but not completely, with phenotypic susceptibility testing and provides different, complementary information. In particular, it has demonstrated significant differences in the distribution of AR genes between geographic regions and between GBS serotypes and some unusual associations between erythromycin and tetracycline resistance genes and the Tn916 integrase gene, int-Tn, in Hong Kong, which deserve further investigation.
Supplementary Material
Acknowledgments
We thank the following colleagues: Margaret Ip, Department of Microbiology, The Chinese University of Hong Kong, Prince of Wales Hospital, Hong Kong; Julie Morgan, Streptococcus Reference Laboratory, ESR, Wellington, New Zealand; and Kyungwon Lee and Yunsop Chong, Research Institute of Bacterial Resistance, Yonsei University College of Medicine, Seoul, South Korea, for providing isolates; Heather Gidding for statistical analysis; and Damla Power and Edmund Cheuk for performing phenotypic antibiotic susceptibility testing of Australian and Hong Kong isolates, respectively.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Acikgoz, Z. C., E. Almayanlar, S. Gamberzade, and S. Gocer. 2004. Macrolide resistance determinants of invasive and noninvasive group B streptococci in a Turkish hospital. Antimicrob. Agents Chemother. 48:1410-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apgar, B. S., G. Greenberg, and G. Yen. 2005. Prevention of group B streptococcal disease in the newborn. Am. Fam. Physician 71:903-910. [PubMed] [Google Scholar]
- 3.Arpin, C., H. Daube, F. Tessier, and C. Quentin. 1999. Presence of mefA and mefE genes in Streptococcus agalactiae. Antimicrob. Agents Chemother. 43:944-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borchardt, S. M., B. Foxman, D. O. Chaffin, C. E. Rubens, P. A. Tallman, S. D. Manning, C. J. Baker, and C. F. Marrs. 2004. Comparison of DNA dot blot hybridization and Lancefield capillary crecipitin methods for group B streptococcal capsular typing. J. Clin. Microbiol. 42:146-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bozdogan, B., L. Berrezouga, M. S. Kuo, D. A. Yurek, K. A. Farley, B. J. Stockman, and R. Leclercq. 1999. A new resistance gene, linB, conferring resistance to lincosamides by nucleotidylation in Enterococcus faecium HM1025. Antimicrob. Agents Chemother. 43:925-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clancy, J., F. Dib-Hajj, J. W. Petitpas, and W. Yuan. 1997. Cloning and characterization of a novel macrolide efflux gene, mreA, from Streptococcus agalactiae. Antimicrob. Agents Chemother. 41:2719-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clancy, J., J. Petitpas, F. Dib-Hajj, W. Yuan, M. Cronan, A. V. Kamath, J. Bergeron, and J. A. Retsema. 1996. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol. Microbiol. 22:867-879. [DOI] [PubMed] [Google Scholar]
- 8.Clarebout, G., C. Villers, and R. Leclercq. 2001. Macrolide resistance gene mreA of Streptococcus agalactiae encodes a flavokinase. Antimicrob. Agents Chemother. 45:2280-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Culebras, E., I. Rodriguez-Avial, C. Betriu, M. Redondo, and J. J. Picazo. 2002. Macrolide and tetracycline resistance and molecular relationships of clinical strains of Streptococcus agalactiae. Antimicrob. Agents Chemother. 46:1574-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Azavedo, J. C., M. McGavin, C. Duncan, D. E. Low, and A. McGeer. 2001. Prevalence and mechanisms of macrolide resistance in invasive and noninvasive group B streptococcus isolates from Ontario, Canada. Antimicrob. Agents Chemother. 45:3504-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Depardieu, F., and P. Courvalin. 2001. Mutation in 23S rRNA responsible for resistance to 16-membered macrolides and streptogramins in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:319-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desjardins, M., K. L. Delgaty, K. Ramotar, C. Seetaram, and B. Toye. 2004. Prevalence and mechanisms of erythromycin resistance in group A and group B streptococcus: implications for reporting susceptibility results. J. Clin. Microbiol. 42:5620-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doherty, N., K. Trzcinski, P. Pickerill, P. Zawadzki, and C. G. Dowson. 2000. Genetic diversity of the tet(M) gene in tetracycline-resistant clonal lineages of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2979-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrell, D. J., I. Morrissey, S. Bakker, and D. Felmingham. 2001. Detection of macrolide resistance mechanisms in Streptococcus pneumoniae and Streptococcus pyogenes using a multiplex rapid cycle PCR with microwell-format probe hybridization. J. Antimicrob. Chemother. 48:541-544. [DOI] [PubMed] [Google Scholar]
- 15.Fiebelkorn, K. R., S. A. Crawford, M. L. McElmeel, and J. H. Jorgensen. 2003. Practical disk diffusion method for detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci. J. Clin. Microbiol. 41:4740-4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fitoussi, F., C. Loukil, I. Gros, O. Clermont, P. Mariani, S. Bonacorsi, L. Thomas I, D. Deforche, and E. Bingen. 2001. Mechanisms of macrolide resistance in clinical group B streptococci isolated in France. Antimicrob. Agents Chemother. 45:1889-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klaassen, C. H., and J. W. Mouton. 2005. Molecular detection of the macrolide efflux gene: to discriminate or not to discriminate between mef(A) and mef(E). Antimicrob. Agents Chemother. 49:1271-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klomberg, D. M., H. A. de Valk, J. W. Mouton, and C. H. Klaassen. 2005. Rapid and reliable real-time PCR assay for detection of the macrolide efflux gene and subsequent discrimination between its distinct subclasses mef(A) and mef(E). J. Microbiol. Methods 60:269-273. [DOI] [PubMed] [Google Scholar]
- 19.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Serotype identification of group B streptococci by PCR and sequencing. J. Clin. Microbiol. 40:216-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leclercq, R., and P. Courvalin. 2002. Resistance to macrolides and related antibiotics in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:2727-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, K., J. W. Shin, Y. Chong, and H. Mikamo. 2000. Trends in serotypes and antimicrobial susceptibility of group B streptococci isolated in Korea. J. Infect. Chemother. 6:93-97. [DOI] [PubMed] [Google Scholar]
- 22.Malbruny, B., A. M. Werno, T. P. Anderson, D. R. Murdoch, and R. Leclercq. 2004. A new phenotype of resistance to lincosamide and streptogramin A-type antibiotics in Streptococcus agalactiae in New Zealand. J. Antimicrob. Chemother. 54:1040-1044. [DOI] [PubMed] [Google Scholar]
- 23.McKay, G. A., and G. D. Wright. 1996. Catalytic mechanism of enterococcal kanamycin kinase (APH(3′)-IIIa): viscosity, thio, and solvent isotope effects support a Theorell-Chance mechanism. Biochemistry 35:8680-8685. [DOI] [PubMed] [Google Scholar]
- 24.Murdoch, D. R., and L. B. Reller. 2001. Antimicrobial susceptibilities of group B streptococci isolated from patients with invasive disease: 10-year perspective. Antimicrob. Agents Chemother. 45:3623-3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poyart, C., L. Jardy, G. Quesne, P. Berche, and P. Trieu-Cuot. 2003. Genetic basis of antibiotic resistance in Streptococcus agalactiae strains isolated in a French hospital. Antimicrob. Agents Chemother. 47:794-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuchat, A. 1999. Group B streptococcus. Lancet 353:51-56. [DOI] [PubMed] [Google Scholar]
- 27.Speer, B. S., N. B. Shoemaker, and A. A. Salyers. 1992. Bacterial resistance to tetracycline: mechanisms, transfer, and clinical significance. Clin. Microbiol. Rev. 5:387-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Storrs, M. J., C. Poyart-Salmeron, P. Trieu-Cuot, and P. Courvalin. 1991. Conjugative transposition of Tn916 requires the excisive and integrative activities of the transposon-encoded integrase. J. Bacteriol. 173:4347-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tait-Kamradt, A., J. Clancy, M. Cronan, F. Dib-Hajj, L. Wondrack, W. Yuan, and J. Sutcliffe. 1997. mefE is necessary for the erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:2251-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uh, Y., I. H. Jang, G. Y. Hwang, K. J. Yoon, and W. Song. 2001. Emerging erythromycin resistance among group B streptococci in Korea. Eur. J. Clin. Microbiol. Infect. Dis. 20:52-54. [DOI] [PubMed] [Google Scholar]
- 32.van den Brule, A. J., R. Pol, N. Fransen-Daalmeijer, L. M. Schouls, C. J. Meijer, and P. J. Snijders. 2002. GP5+/6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes. J. Clin. Microbiol. 40:779-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Von Both, U., M. Ruess, U. Mueller, K. Fluegge, A. Sander, and R. Berner. 2003. A serotype V clone is predominant among erythromycin-resistant Streptococcus agalactiae isolates in a southwestern region of Germany. J. Clin. Microbiol. 41:2166-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, H., F. Kong, P. Jelfs, G. James, and G. L. Gilbert. 2004. Simultaneous detection and identification of common cell culture contaminant and pathogenic Mollicutes strains by reverse line blot hybridization. Appl. Environ. Microbiol. 70:1483-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Werno, A. M., T. P. Anderson, and D. R. Murdoch. 2003. Antimicrobial susceptibilities of group B streptococci in New Zealand. Antimicrob. Agents Chemother. 47:2710-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.