Abstract
Molecular modeling studies have identified a putative human immunodeficiency virus (HIV) integrase (IN) inhibitor-binding pocket for l-chicoric acid (l-CA) and other inhibitors of IN (C. A. Sotriffer, H. Ni, and A. McCammon, J. Med. Chem. 43:4109-4117, 2000). By using site-directed mutagenesis of several amino acid residues identified by modeling studies, a common inhibitor-binding pocket on IN was confirmed for l-CA and the diketo acid L-731,988. Specifically, the single mutations E92K, Q148A, K156A, K156R, G140S, and G149S, as well as the double mutations C65S-K156N and H67D-G140A were evaluated for their effects on enzymatic activity and inhibitor susceptibility. Each recombinant IN was attenuated for 3′-end processing and strand transfer activities. Most proteins were also attenuated for disintegration; the IN that contained K156R and C65S-K156N, however, displayed disintegration activity similar to that of IN from HIVNL4-3. All mutant IN proteins demonstrated decreased susceptibility to l-CA, while all mutant proteins except E92K and K156R demonstrated resistance to L-731,988. These data validate the computer modeling data and demonstrate that l-CA and L-731,988 share an overlapping inhibitor-binding pocket that involves amino acids Q148, C65, and H67. The resistance studies confirm that L-731,988 fills one-half of the inhibitor-binding pocket and binds to Q148 but excludes E92, while l-CA fills the entire binding groove and thus interacts with E92. These results provide “wet laboratory” evidence that molecular models of the HIV IN inhibitor-binding pocket can be used for drug discovery.
Drug therapy for the treatment of human immunodeficiency virus (HIV) infection has advanced quickly. Currently, more than 20 inhibitors of HIV type 1 (HIV-1) targeting reverse transcriptase (RT), protease, and, most recently, viral fusion have been licensed by the Food and Drug Administration for clinical use in the United States (23). Unfortunately, current therapies do not eliminate HIV from reservoirs such as the lymph nodes, the testes, and cerebrospinal fluid (15, 49). Furthermore, in part because of the high mutational frequency of RT (35), drug-resistant viruses arise. Indeed, nearly 25% of new infections are with drug-resistant HIV (31, 32, 43, 48, 50); consequently, new therapies and drug targets are a subject of intense research.
The HIV enzyme integrase (IN) is an attractive target for therapy. After viral entry, reverse transcription, and nuclear entry, IN cleaves the terminal two nucleotides from each 3′ end of the viral long terminal repeats (LTRs), located at each 3′ end of the viral cDNA, in a reaction known as 3′-end processing (11, 14). This reaction reveals an absolutely conserved CA dinucleotide that terminates with a free 3′-hydroxyl group. Next, IN catalyzes the strand transfer reaction, a concerted cleavage ligation reaction in which the free 3′-hydroxyl groups undergo nucleophilic attack on the host chromosomal DNA; for HIV, this results in a 5-bp staggered cut in the host DNA (46, 47). Host double-stranded DNA break repair enzymes most likely repair these gaps (10). The stable, integrated provirus is thus permanently incorporated into the host genome and flanked by 5-bp direct repeat sequences.
Integration of the viral cDNA into the host chromosome is essential for productive infection. Indeed, viruses containing mutations that inactivate IN are unable to replicate in culture (6, 12, 27). IN mutations can have pleiotropic effects as well, including effects on reverse transcription, nuclear localization, proteolytic processing, and virion morphology (2, 12, 30, 40, 42). Although host proteins can facilitate integration, IN is the only viral protein required to carry out the integration reaction (9, 22, 39). Moreover, integration can be studied in vitro by using oligonucleotide substrates that mimic the viral LTR ends, IN, and a divalent metal cation (Mg2+ or Mn2+) (7, 39) (Fig. 1). In addition to the 3′-end processing and strand transfer reactions, recombinant IN can also catalyze the reversal of the integration reaction, termed “disintegration.” In disintegration, IN is able to correctly resolve an oligonucleotide resembling viral DNA joined to host DNA into its respective parts (8). While the full-length IN protein is required for the 3′-end processing and strand transfer reactions (45), the catalytic core domain of IN is sufficient for disintegration (5). Whether the disintegration reaction occurs in vivo is unknown, although it is generally believed that it does not.
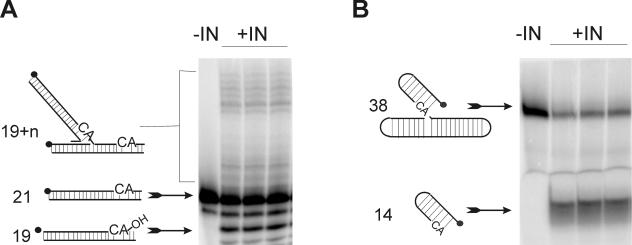
FIG. 1.
Integrase assays. 32P-labeled (dark circles) oligonucleotides homologous to the HIV LTR incubated with recombinant HIV IN can be used to measure the catalytic activities of IN in vitro. In the absence of IN (−IN), no catalysis occurs. In the presence of IN (+IN), products are formed. Products can be separated from substrate by denaturing PAGE. (A) The 3′-end processing and strand transfer reaction. A 21-nucleotide oligomer is processed to the −2 product (19mer). In the same reaction, IN catalyzes the end-joining reaction, resulting in products larger than 21 nucleotides. A preprocessed 19mer can be used to uncouple the 3′-end processing activity from the strand transfer activity. (B) Disintegration assay. A 38-nucleotide substrate representing the partially integrated product can be resolved into its host and viral components. The resulting viral portion is 14 oligonucleotides in length. In both panels the reactions are performed in triplicate. The percent conversion of substrate to products can be quantified via phosphorimager analysis.
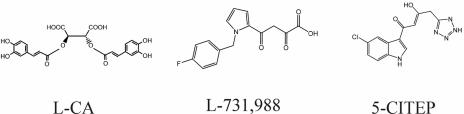
The most potent classes of IN inhibitors with anti-HIV activity remain the dicaffeoylquinic acids (DCQAs), dicaffeoyltartaric acids (DCTAs), and the diketo acids (DKAs) (Fig. 2). The DCQA and DCTA classes of inhibitors were first identified from screens of natural products from Bolivian plant extracts (1). Of these, l-chicoric acid (l-CA), a DCTA, is the most potent (36, 37). l-CA inhibits purified, recombinant IN with 50% inhibitory concentrations (IC50s) of approximately 0.07 μg/ml (145 nM) and 0.15 μg/ml (180 nM) for the 3′-end processing and strand transfer reaction and the disintegration reaction, respectively (36). Furthermore, l-CA inhibits HIV replication in tissue culture with a 50% effective concentration (EC50) of 2 μM (36, 37). Structure-activity relationship studies demonstrate that l-CA analogues can be potent inhibitors of IN and of HIV replication, albeit few are as potent as l-CA itself (25, 29, 34). l-CA resistance maps to a single G140S change (26). When IN containing this single point mutation is introduced into a reference HIV strain, it confers complete resistance to l-CA, as the recombinant virus replicates in the presence of l-CA concentrations as high as 60 μM (26). Recombinant IN containing this mutation is resistant to both l-CA and a DKA, L-731,988 (24).
FIG. 2.
Structures of l-CA, L-731,988, and 5-CITEP.
L-731,988 (Fig. 2) is a potent DKA (19). Similar to l-CA, L-731,988 has an EC50 of 1 to 2 μM in tissue culture (19). In contrast to l-CA, L-731,988 is a selective inhibitor of the strand transfer reaction. The IC50 of L-731,988 for the strand transfer reaction is 0.1 μM, and L-731,988 has little effect on 3′-end processing or disintegration (IC50s = 6 μM and 20 μM, respectively). For L-731,988, several point mutations within IN confer resistance to DKA inhibitors, including T66I (19), S153Y (19), M154I (19), and N155S and N155H (20).
l-CA and DKAs share some structural similarities. Both are small molecules and contain aromatic ring structures, as well as carbonyl groups and at least one free carboxylic acid moiety. l-CA requires one free carboxylic acid for activity (25), while L-731,988 requires both its free carboxylic acid and adjacent carbonyl groups for activity (18). Furthermore, a recent study demonstrated that the DKA resistance mutations T66I, M154I, and S153Y confer resistance to l-CA (28) and, conversely, that IN containing the G140S mutation confers cross-resistance to L-731,988 (24). Thus, it is hypothesized that l-CA and L-731,988 may bind to a common site on IN.
Structural information for IN and IN inhibitors supports this hypothesis. A cocrystal of the catalytic core of IN with the DKA 1-(5-chloroindol-3-yl)-3-(tetrazolyl)-1,3-propanedione enol (5-CITEP) (Fig. 2) has been solved (16). Based on the cocrystal with 5-CITEP, molecular dynamics studies have been completed for several IN inhibitors, including l-CA (4, 36, 41). Figure 3 shows the proposed amino acid contacts between l-CA and IN. These studies of l-CA and the IN core domain have identified amino acids hypothesized to interact with l-CA: D64, C65, T66, H67, E92, D116, D117, Q148, E152, K156, and K159 (36, 41). Based on work by Sotriffer et al., l-CA appears to most completely fill the IN inhibitor-binding groove (41). No computer modeling studies with L-731,988 have been published to date; however, given the structural similarities between 5-CITEP and the DKAs and the similarity of their binding sites to the proposed binding site for l-CA, we hypothesize that DKAs and l-CA bind within this pocket. Indeed, mapping of the resistance mutations T66, S153, M154I, and N155 supports the interpretation that L-731,988, like l-CA and 5-CITEP, interacts near T66, E152, and K156 (28). A similar binding site was identified for some cinnamoyl-based IN inhibitors (4). However, molecular dynamics modeling can be inaccurate; for example, styrylquinolines were hypothesized to bind to the same inhibitor binding pocket, but real-time PCR suggests that RT or an RT-IN interaction may be their site of action (3).
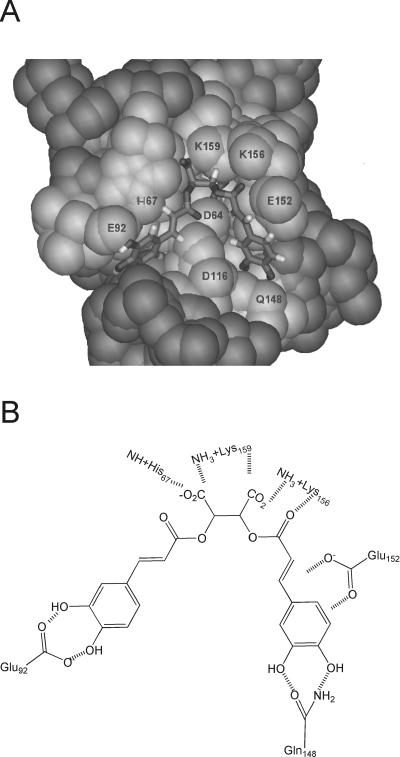
FIG. 3.
Putative l-CA-binding pocket on HIV IN. (A) l-CA modeled into the catalytic core domain of IN. The region in light gray is the area on IN that comprises the putative inhibitor-binding pocket. The indicated amino acids are those proposed to interact with l-CA. This model was provided by T. Charvat and A. R. Chamberlin (published with permission). (B) Schematic of the specific amino acid-inhibitor contacts for l-CA based on studies by Sotriffer et al. (41). Hatched lines, hydrogen bonding and potential salt bridges.
We hypothesized that l-CA and L-731,988 interact with T66, Q148, E152, K156, and K159. In support of this hypothesis, we have already reported that, in addition to conferring resistance to L-731,988, the T66I substitution confers resistance to l-CA (28). To test our hypothesis, mutations to residues within this binding pocket were made. Substitution at E152 cannot be performed, as E152 is a member of the catalytic triad; mutations at this residue lead to inactive protein. We additionally hypothesized that for l-CA, since this molecule is larger and thus should more completely fill an inhibitor-binding pocket, proteins containing substitutions at E92 will be resistant to l-CA but not to L-731,988. In addition to members of the putative binding pocket, mutations at G140 and G149 were studied. Resistance to l-CA was originally mapped to G140S within the IN gene. Previous studies by Greenwald et al. have demonstrated that mutations G140A and G149A lead to attenuation in catalytic activity due to impaired loop mobility (17). Thus, we had proposed that the G140S mutation may adversely affect loop mobility (24). However, molecular modeling has also demonstrated that the G140S mutation decreases the size of the l-CA-binding pocket (unpublished data). The use of a G149S mutation will help us ascertain whether loop mobility or the size of the binding pocket is important for the resistance of the G140S variant. To test our hypotheses, recombinant IN proteins containing point substitutions at the positions described above were evaluated for their enzymatic activities and susceptibilities to both l-CA and L-731,988 to determine the amino acids critical for inhibitor binding.
MATERIALS AND METHODS
Compounds.
l-CA and L-731,988 were synthesized by and are a generous gift from Manfred Reinecke at Texas Christian University, Fort Worth, Tex. Both compounds were >99% pure by nuclear magnetic resonance spectroscopy. Lyophilized L-731,988 was reconstituted to a final concentration of 5 mM in water. Lyophilized l-CA was reconstituted to a final concentration of 2 mM in 20% ethanol.
Oligonucleotides.
All oligonucleotides (Table 1) were synthesized by SigmaGenosys. Mutagenesis oligonucleotides were desalted; oligonucleotides for integration assays were desalted and gel purified prior to use.
TABLE 1.
Oligonucleotide sequences
| Assay and primer | Sequence |
|---|---|
| Integration assays | |
| dBY-1 | 5′-TGCTAGTTCTAGCAGGCCCTTGGGCCGGCGCTTGCGCC-3′ |
| V1 | 5′-ATGTGGAAAATCTCTAGCAGT-3′ |
| V2 | 5′-ACTGCTAGAGATTTTCCACAT-3′ |
| U5V1P | 5′-ATGTGGAAAATCTCTAGCA-3′ |
| Mutagenesisa | |
| C65S+ | 5′-GCCCAGGAATATGGCAGCTAGATAGTACACATTTAGAAGG-3′ |
| C65S− | 5′-CCTTCTAAATGTGTACTATCTAGCTGCCATATTCCTGGGC-3′ |
| H67D+ | 5′-GCCCAGGAATATGGCAGCTAGATTGTACAGATTTAGAAGG-3′ |
| H67D− | 5′-CCTTCTAAATCTGTACAATCTAGCTGCCATATTCCTGGGC-3′ |
| E92K+ | 5′-GCAGAAGTAATTCCAGCAAAGACAGGGCAAGAAACAGC-3′ |
| E92K− | 5′-GCTGTTTCTTGCCCTGTCTTTGCTGGAATTACTTCTGC-3′ |
| G140A+ | 5′-CAAGCAGGAATTTGCCATTCCCTACAATCCCC-3′ |
| G140A− | 5′-GGGGATTGTAGGGAATGGCAAATTCCTGCTTG-3′ |
| Q148A+ | 5′-GCATTCCCTACAATCCCCAAAGTGCAGGAGTAATAG-3′ |
| Q148A− | 5′-CTATTACTCCTGCACTTTGGGGATTGTAGGGAATGC-3′ |
| G149S+ | 5′-GCATTCCCTACAATCCCCAAAGTGGAAGCGTAATAG-3′ |
| G149S− | 5′-CTATTACGCTTCCACTTTGGGGATTGTAGGGAATGC-3′ |
| K156N+ | 5′-GTCAAGGAGTAATAGAATCTATGAAAACAGAATTAAAG-3′ |
| K156N− | 5′-CTTTAATTCTGTTTTCATAGATTCTATTACTCCTTGAC-3′ |
| K156A+ | 5′-GTCAAGGAGTAATAGAATCTATGAATGCAGAATTAAAG-3′ |
| K156A− | 5′-CTTTAATTCTTCATTCATAGATTCTATTACTCCTTGAC-3′ |
| K156R+ | 5′-GTCAAGGAGTAATAGAATCTATGAATAGAGAATTAAAG-3′ |
| K156R− | 5′-CTTTAATTCTCTATTCATAGATTCTATTACTCCTTGAC-3′ |
Underlined nucleotides indicate the substitutions used to introduce each mutation.
Generation of mutant IN.
Each mutation was introduced by site-directed mutagenesis into the reference IN gene from the HIVNL4-3 infectious molecular clone, as described previously (28, 33). Mutant IN genes were sequenced and then cloned into pT7.7, a protein expression vector (28, 33). Recombinant IN was expressed and purified from Escherichia coli, as described previously (25, 28, 33, 34).
Enzymatic activity of mutant IN proteins.
The specific activities of each IN protein in the 3′-end processing and strand transfer and the disintegration assays were determined by using in vitro assays, as described previously (24, 28, 33). For the disintegration reaction, the self-annealing dumbbell substrate dBY-1 was used, whereas the 3′-end processing and strand transfer reactions were performed with the complementary V1 and V2 primers. For direct measure of only the strand transfer reaction, annealing of V2 and the U5V1P oligonucleotides generated a preprocessed LTR substrate. Briefly, for all substrates, increasing concentrations of IN (5 to 450 nM) in triplicate reactions were incubated with 0.1 pmol of oligonucleotide substrate in a 20-μl reaction mixture containing 20 mM HEPES (pH 7.5), 10 mM dithiothreitol, 0.05% Nonidet P-40, 7.5% dimethyl sulfoxide, and 10 mM MnCl2. Following 1 h of incubation at 37°C, the reactions were stopped by addition of EDTA (pH 8.0) to a final concentration of 18 mM and 7 μl of gel loading buffer. The products were separated by 17% denaturing polyacrylamide gel electrophoresis (PAGE) and quantified by phosphorimager analysis, and the specific activities were determined by using SigmaPlot 7.0 linear regression analysis through the linear range of the activity curve.
IC50 determinations.
The susceptibility of IN to inhibition by l-CA and L-731,988 was determined for each mutant IN, as described previously (24, 28, 33). Briefly, 32P-labeled V1-V2, the end-processing and strand transfer substrate, the U5V1P substrate for strand transfer alone, or the dBY-1 substrate for disintegration was incubated in triplicate reactions with recombinant HIV-1 IN and inhibitor at concentrations between 30 nM and 10 μM. The concentration of IN that yielded the maximal activity on the linear portion of the specific activity curve was used. The final volume and composition of the reactions were the same as those for specific activity analyses. The reactions were allowed to proceed for 1 h at 37°C. The reactions were stopped by addition of EDTA to a final concentration of 18 mM. The products were separated from the substrates by denaturing PAGE. The percent conversion of substrate to product was calculated by phosphorimager analysis. The IC50s were computed by using CalcuSyn for Windows software.
RESULTS
Mutant IN proteins are attenuated for catalytic activities.
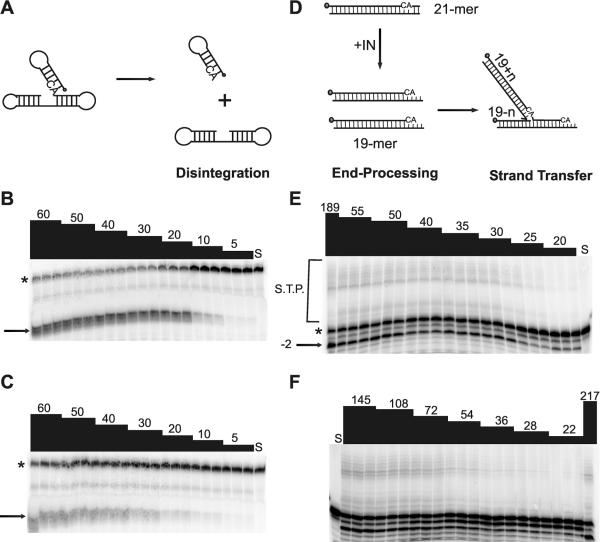
Each mutant IN protein was evaluated for disintegration as well as 3′-end processing and strand transfer activities. Representative enzyme activity gels are shown for K156R and the reference IN for both disintegration (Fig. 4B and C) and 3′-end processing and strand transfer (Fig. 4E and F). Quantification of substrate-to-product conversion through the linear portion of the enzyme activity curve was then carried out to determine the specific activity for each protein. Specific activities (pmol product × pmol IN−1 × h−1) for both reactions for all mutant proteins evaluated are summarized in Table 2. The disintegration assay is useful because it uses a more promiscuous substrate; the core domain can mediate disintegration, while 3′-end processing and strand transfer require the full-length molecule (5, 8). In the disintegration reaction, three classes of attenuation were observed: little to none, modest, and severe. Two proteins, those with the K156R and the C65S-K156N substitutions, demonstrated specific activities similar to that of reference IN. IN proteins containing the G149S, E92K, and Q148A substitutions demonstrated a modest level of attenuation, with specific activities approximately 10 to 20% of that of reference IN. Finally, the K156A and H67D-G140A proteins were severely attenuated for disintegration activity, displaying only 2 to 5% of the reference specific activity (Table 2).
FIG. 4.
Enzymatic activity of representative IN proteins. Increasing amounts of recombinant IN were incubated in the presence of oligonucleotide substrates for 1 h at 37°C in triplicate reactions. The products were separated by denaturing PAGE. (A) dBY-1 disintegration substrate is comprised of viral DNA and target DNA components which, upon addition of IN, are resolved to their respective parts. Gels from disintegration assays are shown for (B) reference IN and (C) IN containing K156R. (D) V1-V2, the 21-mer corresponding to the viral LTR, undergoes 3′-end processing and strand transfer in the presence of IN. (E) Reference IN; (F) IN containing K156R. The numbers at the top of each lane indicate the enzyme concentration (nM); S, substrate control; *, substrate; →, disintegration product; −2→, 3′-end processing product; S.T.P., strand transfer products.
TABLE 2.
Specific activities of mutant IN proteins
| IN | Sp act (pmol product × pmol IN−1 × h−1) for disintegration | r2 for disintegration | 3′-end processing and strand transfer | r2 for 3′-end processing and strand transfer |
|---|---|---|---|---|
| Reference | 0.10 | 0.97 | 0.07 | 0.99 |
| G149S | 0.016 (16)a | 0.99 | 0.005 (7) | 0.99 |
| K156A | 0.005 (5) | 0.98 | 0.005 (7) | 0.98 |
| K156R | 0.083 (83) | 0.97 | 0.03 (43) | 0.98 |
| C65S-K156N | 0.10 (100) | 0.99 | 0.03 (43) | 0.99 |
| E92K | 0.02 (20) | 0.98 | 0.003 (4) | 0.98 |
| Q148A | 0.01 (10) | 0.99 | 0.01 (14) | 0.95 |
| H67D-G140A | 0.002 (2) | 0.98 | 0.002b(3) | 0.99 |
Values in parentheses are the percentage of activity of the reference IN for each mutant.
No strand transfer products were observed.
In contrast to disintegration activity, all mutant IN proteins had decreased 3′-end processing and strand transfer activities compared to that of the reference IN. However, the level of attenuation in the 3′-end processing and strand transfer reaction roughly paralleled that for the disintegration reaction: the K156R and C65S-K156N proteins displayed the least attenuation, while the H67D-G140A protein displayed the most severe attenuation. Indeed, no strand transfer products were observed for IN containing this double mutation; thus, subsequent studies evaluating the effect of H67D-G140A on susceptibility to l-CA and L-731,988 in the strand transfer reaction could not be performed. From Fig. 4 it appears that nonspecific exonuclease activity is increased for the K156R protein, at least at the lowest concentrations of IN tested, as evidenced by more −3 and −4 products.
Mutant IN proteins are resistant to l-CA.
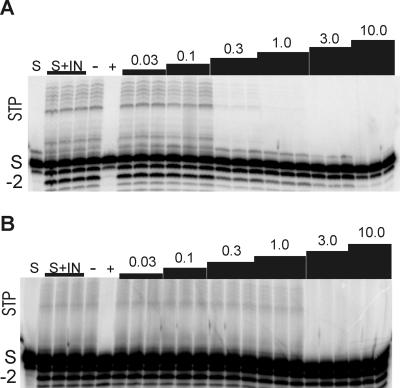
Because l-CA inhibits disintegration, 3′-end processing, and strand transfer, the susceptibility of each protein to l-CA was determined in each of these reactions. For L-731,988, which is a competitive inhibitor of strand transfer (19), susceptibility was evaluated only in the strand transfer reaction. Figure 5 illustrates the results of a representative IC50 analysis in the 3′-end processing and strand transfer reactions for l-CA against reference IN (Fig. 5A) and IN containing the E92K mutation (Fig. 5B). The resistance conferred by the E92K mutation is most evident in the 3′-end processing reaction, where significant −2 product is present, even in the presence of 25 μM l-CA, compared to the amount present in the reaction with reference IN. Resistance in the strand transfer reaction is also apparent, especially with l-CA concentrations of 300 nM and 1 μM. Virtually no strand transfer products were present at these concentrations of l-CA for the reference IN (Fig. 5A), but for IN containing the E92K mutation, the strand transfer reaction was essentially not inhibited in the presence of 1.0 μM l-CA.
FIG. 5.
Inhibitory activity of l-CA against HIVNL4-3 IN and IN containing the E92K mutation. Purified recombinant IN (A) from HIVNL4-3 or (B) containing an E92K mutation was incubated with the V1-V2 3′-end processing substrate for 1 h at 37°C in triplicate reactions in the presence or absence of l-CA. Lanes: S, substrate without IN; S + IN, substrate with IN; −, in the presence of 25 μM L-tartaric acid, a compound that has no activity against HIV IN; +, in the presence of 25 μM l-CA, a concentration that inhibits catalysis by HIV IN. The numbers above the lanes are the concentrations of l-CA (in μM). For the identifiers on the left, STP, strand transfer products; −2, 3′-end processing products; S, input substrate. The reaction products were separated from the substrate by denaturing PAGE. The percent conversion of substrate to products was quantified by phosphorimager analysis and was used to calculate the IC50s presented in Table 3. (B) Darkening to show the strand transfer products, as this protein is significantly attenuated in the strand transfer reaction (Table 2).
Table 3 summarizes data on the susceptibilities of each mutant protein to l-CA and L-731,988. In disintegration, with the exception of the K156R protein, each protein demonstrated resistance to l-CA. The G149S protein demonstrated a modest 4-fold increase in the IC50, while the Q148A and H67D-G140A proteins demonstrated approximately 10-fold resistance in the disintegration reaction. INs containing the E92K or the K156A single mutation and the C65S-K156N double mutation were the most resistant to l-CA in the disintegration reaction, displaying an approximately 20-fold increase in the IC50. In contrast to its susceptibility to l-CA in the disintegration reaction, the K156R protein and all mutant IN proteins demonstrated resistance in both 3′-end processing with the V1-V2 substrate, which mimics the viral LTR recognized by IN, and the strand transfer substrate, U5V1P, which corresponds to a 3′-end processed V1-V2 substrate. Similar to the effect observed in disintegration, the G149S protein displayed a modest threefold increase in the level of resistance. Each of the other mutant IN proteins displayed greater resistance, with the C65S-K156N protein demonstrating a 5-fold increase in resistance to l-CA and the K156A, Q148A, and H67D-G140A proteins and K156R displaying 13- to 20-fold increases in the IC50s. An IC50 could not be determined for IN containing the E92K mutation, as l-CA demonstrated no inhibitory activity against this protein even at 25 μM l-CA (Fig. 5B).
TABLE 3.
IC50s of L-731,988 and l-CA against reference and mutant IN proteins
| IN protein | IC50 (μM)a
|
|||
|---|---|---|---|---|
|
l-CA
|
L-731,988 (ST, U5V1P) | |||
| Disintegration, dBY-1 | 3′-EP, V1-V2 | ST, U5V1P | ||
| Reference | 0.100 (0.03)1 | 0.13 (0.006) | 0.077 (0.02) | 0.48 (0.30) |
| G149S | 0.402 (0.20) | 0.40 (0.09) | 0.55 (0.19) | 5.2 (0.66) |
| K156A | 2.39 (0.46) | 3.6 (1.26) | 0.93 (0.36) | 1.5 (0.49) |
| K156R | 0.074 (0.02) | 2.3 (0.74) | 2.36 (0.48) | 0.20 (0.005) |
| C65S-K156N | 2.89 (1.14) | 0.64 (0.30) | 1.13 (0.25) | 5.0 (0.66) |
| E92K | 2.25 (0.90) | >>25 | 1.02 (0.30) | 0.25 (0.01) |
| Q148A | 0.95 (0.32) | 6.6 (4.65) | 1.60 (0.45) | 10.8 (5.0) |
| H67D-G140A | 1.79 (0.23) | 6.0 (1.23) | ND | ND |
Values in parentheses are 1 standard deviation. 3′-EP, 3′-end processing; ST, strand transfer; ND, not determined since no strand transfer products were observed.
Although the V1-V2 substrate can be used to quantify both 3′-end processing and strand transfer, in this reaction strand transfer is dependent on IN first processing the viral ends. Since all mutant proteins were attenuated for 3′-end processing, the preprocessed U5V1P substrate was used to evaluate the inhibitory effects of l-CA and L-731,988 on strand transfer. In this manner, the strand transfer effect could be measured independently of the effect that each inhibitor and each mutation had on the 3′-end processing reaction. In this reaction, the G149S protein demonstrated only a modest 4- to 6-fold increase in the IC50, while each of the other mutant proteins displayed 10-fold or greater resistance. Thus, taken together, each mutant IN protein demonstrates resistance to l-CA, with the exception of K156 in the disintegration reaction.
The lack of resistance of the K156R protein in the disintegration reaction but not in the 3′-end processing or strand transfer reaction was unexpected, since l-CA inhibits all three reactions. However, the dumbbell substrate is smaller, and the substitution of R for K is conservative, retaining the charge characteristics of the reference protein. Thus, even though R has a bulkier side group, perhaps the smaller dumbbell substrate in combination with the retained side chain charge still allowed l-CA to inhibit and bind in a manner similar to that of reference IN.
E92K and K156R INs are not resistant to L-731,988.
Each mutant IN protein was next evaluated for its susceptibility to L-731,988 by using the preprocessed strand transfer substrate. For some proteins, the results obtained for L-731,988 in the strand transfer reaction differed from those obtained for l-CA. IN containing the G149S substitution demonstrated slightly greater resistance to L-731,988, with an approximately 10-fold increase in the IC50. In contrast, the K156A protein displayed a modest threefold increase in the level of resistance, while neither the K156R nor the E92K protein displayed resistance to L-731,988. INs containing the Q148A single mutation or the C65S-K156N double mutation were the most resistant, demonstrating 10- and 20-fold increases in IC50s, respectively.
Thus, while all mutant IN proteins demonstrated some level of resistance to l-CA, the K156A protein displayed modest resistance and the G149S, C65S-K156N, and Q148A proteins displayed greater resistance. Neither the E92K protein nor the K156R protein displayed statistically significant resistance to L-731,988. Molecular dynamics simulations of 5-CITEP predict these results: l-CA would fill the entire binding groove, including the “left side” of the molecule, which includes E92 (Fig. 3). L-731,988, on the other hand, would fill only the right side; thus, E92 is uninvolved.
DISCUSSION
Several classes of IN inhibitors have been identified, arguably the most promising of which are the DCQA, DCTA, and DKA inhibitors. Goldgur et al. (16) identified amino acids T66, Q148, E152, N155, K156, and K159 as being involved in the binding of 5-CITEP, a DKA, via cocrystallization of the inhibitor with the catalytic core domain of IN. Based on this 5-CITEP cocrystal, computer modeling studies (41) identified additional amino acids, C65, H67, and E92. Previous computer modeling studies had postulated a similar inhibitor-binding pocket (36). In this study, these putative binding pocket amino acids have been mutated to test the validities of the various molecular models.
With the exception of K156R and C65S-K156N in disintegration, all mutant IN proteins demonstrated decreased specific activity for disintegration and 3′-end processing and strand transfer compared to that of the reference IN (Table 2). These data are consistent with, although not identical to, those from previously published reports. For example, in our studies the E92K mutant displayed disintegration specific activity 20% of that of the reference IN and 3′-end processing and strand transfer specific activity approximately 4% of that of the reference IN. Previous reports indicate that substitution of IN at E92 to K had 50 to 100% the disintegration activity and 5 to 20% the 3′-end processing and strand transfer activities of the reference IN at 30 nM (13). In contrast, IN containing substitutions of E92 to A or Q displayed 50 to 100% the disintegration with 3′-end processing and strand transfer activities, comparable to those of the reference IN (13). Previous data indicate that mutation of Q148 to L yields proteins with decreased activities in both reactions for the HIV-2 IN (44). Similarly, our data indicate that Q148A had 10% and 15% the disintegration and 3′-end processing and strand transfer activities compared with those of the reference IN, respectively (Table 2). While our data indicate that the conservative K156R substitution and the C65S-K156N double mutation do not affect disintegration activity, both K156R and K156A are attenuated for 3′-end processing and strand transfer (Fig. 4 and Table 2). Moreover, K156A is attenuated in disintegration. This is consistent with the findings of studies indicating that mutation of K156 to E reduced disintegration activity fourfold, but 3′-end processing and strand transfer activities were undetectable (21). Finally, previous data indicate that the specific activity of IN containing a G140S substitution was attenuated in disintegration, with approximately 20% of the activity compared to that of the reference IN (24). Additional work by Greenwald et al. (17) demonstrated that substitutions of G140A and G149A led to IN proteins with attenuated enzyme activities. Consistent with these studies, substitution of G149S results in disintegration specific activities 16% of that of the reference IN and results in the 3′-end processing and strand transfer specific activities approximately 7% of that of the reference IN (Table 2). Finally, the protein with the double mutation H67D-G140A is severely attenuated in both disintegration and 3′-end processing, with no detectable strand transfer activity (Table 2).
Nevertheless, mutant IN proteins retained enzymatic activity; thus, each protein could be evaluated for susceptibility to l-CA and L-731,988 in order to validate or disprove the proposed binding pocket for these inhibitors. Consistent with the cocrystal of IN and 5-CITEP and modeling of other IN inhibitors, the Q148A protein demonstrated a high, 10- to 20-fold increase in the level of resistance to both l-CA and L-731,988, indicating that Q148 is indeed an important member of the inhibitor-binding pocket for both molecules (Table 3). E92K conferred resistance only to l-CA and not to L-731,988. Furthermore, C65S-K156N conferred resistance in all reactions with both l-CA and L-731,988, indicating that either C65 or K156, or likely, both, are important for binding (Table 3).
The K156A protein, with a single mutation, displayed a modest threefold increase in resistance to L-731,988, but K156R did not confer any resistance (Table 3). These results are in contrast to those for the 5-CITEP crystal, in which K156 was identified as contacting one of the four nitrogen atoms on the tetrazole ring (16). However, recent computer modeling data suggest that 5-CITEP binds closer to K159 than to K156 (38), unlike l-CA. Our results for K156R would indicate that the latter relationship (38) is, indeed, the correct relationship. Unfortunately, we have had no success obtaining recombinant IN containing either the K159D or the K159A mutation to directly assess the roles of K159 in inhibitor binding (data not shown). These data suggest that mutation of K159 leads to gross misfolding of the protein, resulting in either insoluble protein or a six-His tag that is incapable of interacting with the Ni2+ affinity column.
Based on our results for the mutants with the K156A and K156R single mutations, we hypothesize that the double mutation C65S-K156N confers resistance predominantly because the substitution at C65 affects the interaction of L-731,988 with the protein. Furthermore, although mutations at K159 were not studied, we hypothesize that K159, unlike K156, may be more critical for L-731,988 binding. Also consistent with the published cocrystal of the catalytic core of IN with 5-CITEP, E92K did not confer resistance to L-731,988 (Table 3), indicating that E92 is not involved in the binding of this inhibitor to the protein.
The results for l-CA are in agreement with those of published computer modeling studies (36, 41) (Fig. 3). In contrast to their resistance to L-731,988, the E92K and K156A proteins were highly resistant to l-CA, as was the K156R protein, in 3′-end processing and strand transfer reactions (Table 3), demonstrating that these residues are critical for inhibitor binding. Similar to the results for L-731,988, the C65S-K156N protein was resistant (Table 3), demonstrating that these residues are critical for the interaction with l-CA. Previous data have already demonstrated that C65A is resistant to l-CA (51); thus, these data, in combination with the results of previous work, suggest that C65 is critical for the binding of both l-CA and L-731,988.
Similar to the results for L-731,988, the Q148A protein was resistant to l-CA, demonstrating for both molecules a high, 10- to 20-fold increase in IC50s compared to that for the reference IN (Table 3). Thus, the catechol side chain of l-CA and the aromatic ring structure for L-731,988 appear to interact with Q148 to enable inhibitor binding. Finally, H67D-G140A conferred resistance to l-CA (Table 3). These data, in combination with the resistance displayed by the G149S protein and previously published data for the G140S protein (24), suggest that loop mobility affects inhibitor binding. The data obtained for the G149S protein are strikingly parallel to those obtained for the G140S protein (24). Indeed, for both mutant IN proteins, a modest 3- to 5-fold level of resistance was observed with l-CA, while a 10-fold increase in the IC50 was displayed with L-731,988. These data indicate that these residues may confer resistance to both inhibitors by a similar mechanism; since both residues have been implicated in loop mobility, we hypothesize that these residues act by impairing the movement of the disordered loop, thereby distorting the inhibitor binding site. Finally, IN containing the double mutation H67D-G140A is highly resistant to l-CA and much more resistant than the G140S or the G149S protein with a single mutation; thus, H67 appears to interact with l-CA, as predicted by the model.
The data presented herein validate the computer modeling studies and also enhance our understanding of the amino acids critical for inhibitor binding for l-CA and L-731,988. Based on these studies, the Q148 residue is an important member of the binding pocket for both l-CA and L-731,988, probably via interaction of the catechol moiety on l-CA and the aromatic ring on L-731,988 with this residue. This is consistent with resistance and cross-resistance data indicating that L-731,988 and l-CA interact with residues near Q148, specifically, M154 and S153. These data are also consistent with the findings for the cocrystal of 5-CITEP, indicating that the analogous aromatic ring group interacts with Q148. l-CA has two catechol moieties and is essentially twice the size of L-731,988. Thus, the finding that l-CA, but not L-731,988, interacts with E92 is expected and consistent with the cocrystal data and with the results of computer modeling studies for l-CA. In contrast to the cocrystal studies with 5-CITEP and with l-CA, the K156 residue does not appear to play as critical a role in binding of L-731,988, although studies evaluating the requirement for interaction with K159 are ongoing. Finally, in these studies, C65 is also involved with inhibitor binding for both l-CA and L-731,988. Although the cocrystal solved with 5-CITEP does not identify C65 as an amino acid involved in 5-CITEP binding, molecular modeling studies have implicated it (36, 41). The binding of L-731,988 to IN may involve slightly different interactions than those involved with 5-CITEP, the orientation of the carboxylic acid may be more similar to that of the carboxylic acid of l-CA, or perhaps, the binding interactions in solution are slightly different than those in crystals.
Although this work has enabled a further understanding of inhibitor binding to IN, more research should be done to understand the protein-inhibitor interactions involved. Specifically, interactions between inhibitor chemical moieties and specific amino acids on IN have not been confirmed. However, the studies presented herein provide an important foundation which may enable the design of inhibitors with optimal and more potent binding interactions and, thus, better inhibitory activities. Certainly, the data suggest that existing molecular dynamics simulations have given us an adequate model for the rational design of IN inhibitors. The potential of IN as a therapeutic target is also highlighted in this work: mutations within the inhibitor-binding pocket are detrimental to IN activities; therefore, drugs targeted at those amino acids should be potent inhibitors of IN.
Acknowledgments
We are indebted to Manfred G. Reinecke (Texas Christian University) for l-CA and L-731,988. We thank Dustin Gerth and Felicia Hernandez for technical assistance and Trevor Charvat and Richard Chamberlin for providing the computer model. We also acknowledge Hung Fan, Bert Semler, and Roz Sandri-Goldin for thoughtful insights and comments, as well as Joseph Victoria, Bellanira Herrera, and Brenda McDougall for critical review of the manuscript.
This work was supported in part by Public Health Service grants 5RO1-AI41360 (to W.E.R.), 1R21-AI054305 (to W.E.R.), and 5T32-GM08620 (to D.J.L.) and a grant from the Burroughs-Wellcome Fund (grant 99-2609) (to W.E.R.). W.E.R. is a Burroughs-Wellcome Fund Clinical Scientist in Translational Research.
REFERENCES
- 1.Abdel-Malek, S., J. W. Bastien, W. F. Mahler, Q. Jia, M. G. Reinecke, W. E. Robinson, Jr., Y.-H. Shu, and J. Zalles-Asin. 1996. Drug leads from the Kallawaya herbalists of Bolivia. 1. Background, rationale, protocol and anti-HIV activity. J. Ethnopharmacol. 50:157-166. [DOI] [PubMed] [Google Scholar]
- 2.Ansari-Lari, M. A., L. A. Donehower, and R. A. Gibbs. 1995. Analysis of human immunodeficiency virus type 1 integrase mutants. Virology 211:332-335. [DOI] [PubMed] [Google Scholar]
- 3.Bonnenfant, S., C. M. Thomas, C. Vita, F. Subra, E. Deprez, F. Zouhiri, D. Desmaele, J. d'Angelo, J. F. Mouscadet, and H. Leh. 2004. Styrylquinolines, integrase inhibitors acting prior to integration: a new mechanism of action for anti-integrase agents. J. Virol. 78:5728-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buolamwini, J. K., and H. Assefa. 2002. CoMFA and CoMSIA 3D QSAR and docking studies on conformationally-restrained cinnamoyl HIV-1 integrase inhibitors: exploration of a binding mode at the active site. J. Med. Chem. 45:841-852. [DOI] [PubMed] [Google Scholar]
- 5.Bushman, F. D., A. Engelman, I. Palmer, P. Wingfield, and R. Craigie. 1993. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc. Natl. Acad. Sci. USA 90:3428-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannon, P. M., W. Wilson, E. Byles, S. M. Kingsman, and A. J. Kingsman. 1994. Human immunodeficiency virus type 1 integrase: effect on viral replication of mutations at highly conserved residues. J. Virol. 68:4768-4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chow, S. A. 1997. In vitro assays for activities of retroviral integrase. Methods Companion Methods Enzymol. 12:306-317. [DOI] [PubMed] [Google Scholar]
- 8.Chow, S. A., K. A. Vincent, V. Ellison, and P. O. Brown. 1992. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science 255:723-726. [DOI] [PubMed] [Google Scholar]
- 9.Craigie, R., T. Fujiwara, and F. Bushman. 1990. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell 62:829-837. [DOI] [PubMed] [Google Scholar]
- 10.Daniel, R., R. A. Katz, and A. M. Skalka. 1999. A role for DNA-PK in retroviral DNA integration. Science 284:644-647. [DOI] [PubMed] [Google Scholar]
- 11.Ellison, V., H. Abrams, T. Roe, J. Liffson, and P. Brown. 1990. Human immunodeficiency virus integration in a cell-free system. J. Virol. 64:2711-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engelman, A., G. Englund, J. M. Orenstein, M. A. Martin, and R. Craigie. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J. Virol. 69:2729-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelman, A., Y. Liu, H. Chen, M. Farzan, and F. Dyda. 1997. Structure-based mutagenesis of the catalytic domain of human immunodeficiency virus type 1 integrase. J. Virol. 71:3507-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelman, A., K. Mizuuchi, and R. Craigie. 1991. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell 67:1211-1221. [DOI] [PubMed] [Google Scholar]
- 15.Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. Gallant, M. Markowitz, D. D. Ho, D. D. Richman, and R. F. Siliciano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295-1300. [DOI] [PubMed] [Google Scholar]
- 16.Goldgur, Y., R. Craigie, G. H. Cohen, T. Fujiwara, Y. Tomokazu, T. Fujishita, H. Sugimoto, T. Endo, H. Murai, and D. R. Davies. 1999. Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design. Proc. Natl. Acad. Sci. USA 96:13040-13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenwald, J., V. Le, S. L. Butler, F. D. Bushman, and S. Choe. 1999. The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity. Biochemistry 38:8892-8898. [DOI] [PubMed] [Google Scholar]
- 18.Grobler, J. A., K. Stillmock, B. Hu, M. Witmer, P. Felock, A. S. Espeseth, A. Wolfe, M. Egbertson, M. Bourgeois, J. Melamed, J. S. Wai, S. Young, J. Vacca, and D. J. Hazuda. 2002. Diketo acid inhibitor mechanism and HIV-1 integrase: implications for metal binding in the active site of phosphotransferase enzymes. Proc. Natl. Acad. Sci. USA 99:6661-6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hazuda, D. J., P. Pelock, M. Witmer, A. Wolfe, K. Stillmock, J. A. Grobler, A. Espeseth, L. Gabryelski, W. Schleif, C. Blau, and M. D. Miller. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646-650. [DOI] [PubMed] [Google Scholar]
- 20.Hazuda, D. J., S. D. Young, J. P. Guare, N. J. Anthony, R. P. Gomez, J. S. Wai, J. P. Vacca, L. Handt, S. L. Motzel, H. J. Klein, G. Dornadula, R. M. Danovich, M. V. Witmer, K. A. A. Wilson, L. Tussey, W. A. Schleif, L. S. Gabryelski, L. Jin, M. D. Miller, D. R. Casimiro, E. A. Emini, and J. W. Shiver. 2004. Integrase inhibitors and cellular immunity suppress retroviral replication in rhesus macaques. Science 305:528-532. (10.1126/science.1098632.) [DOI] [PubMed] [Google Scholar]
- 21.Jenkins, T. M., D. Esposito, A. Engelman, and R. Craigie. 1997. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 16:6849-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katz, R. A., G. Merkel, J. Kulkosky, J. Leis, and A. M. Skalka. 1990. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell 63:87-95. [DOI] [PubMed] [Google Scholar]
- 23.Kilby, J. M., S. Hopkins, T. M. Venetta, B. DiMassimo, G. A. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 24.King, P. J., D. J. Lee, R. A. Reinke, J. G. Victoria, K. Beale, and W. E. Robinson, Jr. 2003. Human immunodeficiency virus type 1 integrase containing a glycine to serine mutation at position 140 is attenuated for catalysis and resistant to integrase inhibitors. Virology 306:147-161. [DOI] [PubMed] [Google Scholar]
- 25.King, P. J., G. Ma, W. Miao, Q. Jia, B. R. McDougall, M. G. Reinecke, C. Cornell, J. Kuan, T. R. Kim, and W. E. Robinson, Jr. 1999. Structure-activity relationships: analogues of the dicaffeoylquinic and dicaffeoyltartaric acids as potent inhibitors of human immunodeficiency virus type 1 integrase and replication. J. Med. Chem. 42:497-509. [DOI] [PubMed] [Google Scholar]
- 26.King, P. J., and W. E. Robinson, Jr. 1998. Resistance to the anti-human immunodeficiency virus type 1 compound l-chicoric acid results from a single mutation at amino acid 140 of integrase. J. Virol. 72:8420-8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaFemina, R. L., C. L. Schneider, H. L. Robbins, P. L. Callahan, K. LeGrow, E. Roth, W. A. Schleif, and E. A. Emini. 1992. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J. Virol. 66:7414-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, D. J., and W. E. Robinson, Jr. 2004. Human immunodeficiency virus type 1 (HIV-1) integrase: resistance to diketoacid integrase inhibitors impairs HIV-1 replication, integration, and confers cross-resistance to l-chicoric acid. J. Virol. 78:5835-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, Z., N. Neamati, H. Zhao, Y. Kiryu, J. A. Turpin, C. Aberham, K. Strebel, K. Kohn, M. Witvrouw, C. Pannecouque, Z. Debyser, E. DeClercq, W. G. Rice, Y. Pommier, and T. R. Burke, Jr. 1999. Chicoric acid analogues as HIV-1 integrase inhibitors. J. Med. Chem. 42:1401-1414. [DOI] [PubMed] [Google Scholar]
- 30.Lu, R., A. Limon, E. Devroe, P. A. Silver, P. Cherepanov, and A. Engelman. 2004. Class II integrase mutants with changes in putative nuclear localization signals are primarily blocked at a postnuclear entry step of human immunodeficiency virus type 1 replication. J. Virol. 78:12735-12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menéndez-Arias, L. 2002. Targeting HIV: antiretroviral therapy and development of drug resistance. Trends Pharmacol. Sci. 23:381-388. [DOI] [PubMed] [Google Scholar]
- 32.Poveda, E., B. Rodes, J.-L. Labernardiere, J. M. Benito, C. Toro, J. Gonzalez-Lahoz, J.-L. Faudon, F. Clavel, J. Schapiro, and V. Soriano. 2004. Evolution of genotypic and phenotypic resistance to enfuvirtide in HIV-infected patients experiencing prolonged virologic failure. J. Med. Virol. 74:21-28. [DOI] [PubMed] [Google Scholar]
- 33.Reinke, R., N. R. Steffen, and W. E. Robinson, Jr. 2001. Natural selection results in conservation of HIV-1 integrase activity despite sequence variability. AIDS 15:823-830. [DOI] [PubMed] [Google Scholar]
- 34.Reinke, R. A., P. J. King, J. G. Victoria, B. R. McDougall, G. Ma, Y. Mao, M. G. Reinecke, and W. E. Robinson, Jr. 2002. Dicaffeoyltartaric acid analogues inhibit human immunodeficiency virus type 1 (HIV-1) integrase and HIV-1 replication at non-toxic concentrations. J. Med. Chem. 45:3669-3683. [DOI] [PubMed] [Google Scholar]
- 35.Roberts, J. D., K. Bebenek, and T. A. Kunkel. 1988. The accuracy of reverse transcriptase from HIV-1. Science 242:1171-1173. [DOI] [PubMed] [Google Scholar]
- 36.Robinson, W. E., Jr., M. Cordeiro, S. Abdel-Malek, Q. Jia, S. A. Chow, M. G. Reinecke, and W. M. Mitchell. 1996. Dicaffeoylquinic acid inhibitors of human immunodeficiency virus integrase: inhibition of the core catalytic domain of human immunodeficiency virus integrase. Mol. Pharmacol. 50:846-855. [PubMed] [Google Scholar]
- 37.Robinson, W. E., Jr., M. G. Reinecke, S. Abdel-Malek, Q. Jia, and S. A. Chow. 1996. Inhibitors of HIV-1 replication that inhibit HIV integrase. Proc. Natl. Acad. Sci. USA 93:6326-6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schames, J. R., R. H. Henchman, J. S. Siegel, C. A. Sotriffer, H. Ni, and J. A. McCammon. 2004. Discovery of a novel binding trench in HIV integrase. J. Med. Chem. 47:1879-1881. [DOI] [PubMed] [Google Scholar]
- 39.Sherman, P. A., and J. A. Fyfe. 1990. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc. Natl. Acad. Sci. USA 87:5119-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin, C. G., B. Taddeo, W. A. Haseltine, and C. M. Farnet. 1994. Genetic analysis of the human immunodeficiency virus type 1 integrase protein. J. Virol. 68:1633-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sotriffer, C. A., H. Ni, and A. McCammon. 2000. Active site binding modes of HIV-1 integrase inhibitors. J. Med. Chem. 43:4109-4117. [DOI] [PubMed] [Google Scholar]
- 42.Taddeo, B., W. A. Haseltine, and C. M. Farnet. 1994. Integrase mutants of human immunodeficiency virus type 1 with a specific defect in integration. J. Virol. 68:8401-8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.UK Collaborative Group on Monitoring the Transmission of HIV Drug Resistance. 2001. Analysis of prevalence of HIV-1 drug resistance in primary infections in the United Kingdom. Br. Med. J. 322:1087-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Gent, D. C., A. A. Groeneger, and R. H. Plasterk. 1992. Mutational analysis of the integrase protein of human immunodeficiency virus type 2. Proc. Natl. Acad. Sci. USA 89:9598-9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Gent, D. C., C. Vink, A. A. Groeneger, and R. H. Plasterk. 1993. Complementation between HIV integrase proteins mutated in different domains. EMBO J. 12:3261-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vincent, K. A., V. Ellison, S. A. Chow, and P. O. Brown. 1993. Characterization of human immunodeficiency virus type 1 integrase expressed in Escherichia coli and analysis of variants with amino-terminal mutations. J. Virol. 67:425-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vink, C., D. C. van Gent, and R. H. A. Plasterk. 1990. Integration of human immunodeficiency virus types 1 and 2 DNA in vitro by cytoplasmic extracts of Moloney murine leukemia virus-infected mouse NIH 3T3 cells. J. Virol. 64:5219-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wegner, S. A., S. K. Brodine, J. R. Mascola, S. A. Tasker, R. A. Shaffer, M. J. Starkey, A. Barile, G. J. Martin, N. Aronson, W. W. Emmons, K. Stephan, S. Bloor, J. Vingerhoets, K. Hertogs, and B. Larder. 2000. Prevalence of genotypic and phenotypic resistance to anti-retroviral drugs in a cohort of therapy-naive HIV-1 infected US military personnel. AIDS 14:1009-1015. [DOI] [PubMed] [Google Scholar]
- 49.Wong, J. K., M. Hezareh, H. F. Gunthard, D. V. Havlir, C. C. Ignacio, C. A. Spina, and D. R. Richman. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291-1295. [DOI] [PubMed] [Google Scholar]
- 50.Xu, L., A. Pozniak, A. Wildfire, S. A. Stanfield-Oakley, S. M. Mosier, D. Ratcliffe, J. Workman, A. Joall, R. Myers, E. Smit, P. A. Cane, M. L. Greenberg, and D. Pillay. 2005. Emergence and evolution of enfuvirtide resistance following long-term therapy involves heptad repeat 2 mutations within gp41. Antimicrob. Agents Chemother. 49:1113-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu, K., M. L. Cordeiro, J. Atienza, W. E. Robinson, Jr., and S. A. Chow. 1999. Irreversible inhibition of human immunodeficiency virus type 1 integrase by dicaffeoylquinic acids. J. Virol. 73:3309-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]