Abstract
Recent research has provided evidence that interference with bacterial cell-to-cell signaling is a promising strategy for the development of novel antimicrobial agents. Here we report on the computer-aided design of novel compounds that specifically inhibit an N-acyl-homoserine lactone-dependent communication system that is widespread among members of the genus Burkholderia. This genus comprises more than 30 species, many of which are important pathogens of animals and humans. Over the past few years, several Burkholderia species, most notably Burkholderia cenocepacia, have emerged as important opportunistic pathogens causing severe pulmonary deterioration in persons with cystic fibrosis. As efficient treatment of Burkholderia infections is hampered by the inherent resistance of the organisms to a large range of antibiotics, novel strategies for battling these pathogens need to be developed. Here we show that compounds targeting the B. cenocepacia signaling system efficiently inhibit the expression of virulence factors and attenuate the pathogenicity of the organism.
The gram-negative bacterium Burkholderia cepacia was first described as a phytopathogen associated with a soft rot of onion bulbs (2). Taxonomic studies of the past few years have shown that B. cepacia-like organisms comprise a group of closely related strains, collectively referred to as the B. cepacia complex. Strains of the B. cepacia complex are ubiquitously distributed in nature and have been isolated from soil, water, plants, and industrial settings. Over the past few years, B. cepacia complex strains have also emerged as opportunistic pathogens of humans (3, 5, 16, 26). These strains can cause life-threatening lung infections in patients requiring mechanical ventilation and in individuals with chronic granulomatous disease or cystic fibrosis (CF). The clinical outcome of infection with B. cepacia complex in CF patients is varied and unpredictable, ranging from asymptomatic carriage to a fulminant and fatal pneumonia, the so-called cepacia syndrome (10). Although all nine B. cepacia genomovars (Gv.) described to date have been isolated from CF patients, Burkholderia multivorans (Gv. II) and Burkholderia cenocepacia (Gv. III) are most commonly found in clinical samples (18). A major problem with Burkholderia infections is the intrinsic resistance of the organism to various antibiotics and biocides, impeding effective medical treatment.
B. cenocepacia utilizes an N-acyl homoserine lactone (AHL)-dependent quorum-sensing (QS) system to express certain genes in a cell density-dependent manner. This regulatory system consists of the AHL synthase CepI, which directs the synthesis of the signal molecule N-octanoylhomoserine lactone (C8-HSL) and the transcriptional regulator CepR (4, 14). When the concentration of C8-HSL reaches a critical threshold concentration, the signal molecule binds to the cognate receptor protein CepR, which in turn leads to the induction or repression of target genes. The cep system was shown to positively regulate the expression of extracellular proteases and chitinases, swarming motility, and biofilm formation (14, 9) and to repress the synthesis of the siderophore ornibactin (13).
B. cenocepacia mutants with a defective cep quorum-sensing system were demonstrated to be attenuated in a Caenorhabditis elegans pathogenesis model (12) as well as in a short-term intranasal colonization mouse model and a chronic agar bead infection model in rats (24). Hence, this regulatory system represents highly attractive targets for therapeutic intervention of Burkholderia infections.
Previous work identified synthetic derivatives of halogenated furanones, which are produced by the marine red alga Delisea pulchra, that were demonstrated to effectively antagonize AHL-mediated quorum sensing in the opportunistic pathogen Pseudomonas aeruginosa (6, 7). In a mouse pulmonary infection model, one of these compounds promoted the clearance of P. aeruginosa by the mouse immune response and thus dramatically reduced the severity of the infection (7). Furthermore, by altering moieties of the homoserine lactone molecules by employing combinatory chemistry in a trial-and-error approach, several compounds with AHL antagonistic activity that were capable of inhibiting the expression of QS-regulated phenotypes in P. aeruginosa were identified (11, 22, 23). To date, however, no QS inhibitor for Burkholderia has been described. In fact, furanone compounds were proven to be ineffective due to the rapid inactivation of the molecules by the bacteria (unpublished results). Here we report on the rational design of a novel anti-infective agent targeting the Burkholderia cep QS system.
MATERIALS AND METHODS
Computer-aided design of quorum-sensing antagonists.
Virtual screening was performed with 4SCan, which combines a molecular alignment tool with an iterative database screening and prioritization procedure (21). Initially, the 4SCan algorithm aligns a relatively small number of randomly chosen molecules (typically 5,000) from an in-house database, which contains 1.1 million commercially available compounds. 4SCan searches for molecules that are most similar to the reference molecule in terms of shape and possible intermolecular interactions. The alignment algorithm was implemented on a 770 Intel central processing unit cluster. This allowed the screening of the entire virtual database within 2 hours, resulting in a ranking list that arranges the molecules according to their predicted biological activity. Virtual compounds were treated as flexible at all acyclic single bonds, while the structure of the reference molecule was kept rigid. We used the energy-minimized structures (20) (CORINA) of N-(3-oxo-dodecanoyl)-l-homoserine lactone (3-oxo-C12-HSL) and C8-HSL as reference templates in this study.
Compounds from the initial screening were tested in various bioassays. On the basis of these results, more-focused virtual combinatorial libraries were generated by a descriptor-based learning algorithm. Iterative cycles of alignment, screening, and testing were used to improve the activities of the compounds. Additionally, on the basis of the biological activities and molecular field properties (steric and electrostatic) around the aligned molecules, several comparative molecular field analysis models were developed during the screening cycles by using the TRIPOS (St. Louis, MO) software SYBYL and the alignments of 4SCan.
Purchase/synthesis of compounds 1, 2, and 3.
Compound 1 was purchased from Maybridge Chemical Company (United Kingdom). For the synthesis of compound 3, a solution of 3-(tert-butyl)-1-methylpyrazole-5-carbonyl chloride (1.2 eq) in dimethylformamide was added to a solution of 5-chloro-2-thiophenecarboxylic acid hydrazide (1 eq) in dimethylformamide and (1.2 eq) triethylamine at 0°C. The reaction mixture was stirred for 1 h at room temperature and then concentrated in vacuo. The resulting solid was purified by preparative thin-layer chromatography (column, 20 by 20 cm; Silica Gel 60 F254; Merck) (CH2Cl2-methanol, 100:1). For the synthesis of compound 2, a solution of 3-chloro-thiophene-2-carbonyl chloride (1.2 eq) in dimethylformamide was added to a solution of hydrazine monohydrate (1 eq) in dimethylformamide and (1.2 eq) triethylamine at 0°C. The reaction mixture was stirred for 5 h at room temperature and then concentrated in vacuo. The resulting mixture was purified by preparative thin-layer chromatography (column, 20 by 20 cm; Silica Gel 60 F254; deactivated with NH3; Merck) (petrol ether-ethyl acetate, 1:1). The identities and purity of the synthesized compounds were determined by liquid chromatography-positive-electron spray ionization and liquid chromatography-negative-electron spray ionization mass spectrometry and nuclear magnetic resonance analysis.
High-throughput screening of QS blockers by the aid of GFP-based AHL biosensors.
Quorum-sensing antagonistic activities of compounds were tested with the aid of two AHL biosensors: Pseudomonas putida F117(pKR-C12) and P. putida F177(pAS-C8) (25). The AHL monitor plasmid pKR-C12 contains a PlasB-gfp(ASV) translational fusion together with the lasR gene placed under the control of Plac. This sensor strain is most sensitive for 3-oxo-C12-HSL. Plasmid pAS-C8 was constructed from components of the cep system of B. cenocepacia H111 and contains a PcepI-gfp(ASV) translational fusion together with the cepR gene transcribed from the Plac promoter of the broad-host-range plasmid pBBR1MCS-5. This sensor plasmid responds most efficiently to C8-HSL and, with a lower efficiency, to related AHL molecules. The sensor strains were grown overnight in Luria-Bertani (LB) medium (1) at 30°C, diluted fourfold in fresh medium, and grown for another hour. Following the addition of appropriate AHLs (50 nM for 3-oxo-C12-HSL for pKR-C12 and 50 nM C8-HSL for pAS-C8), 100-μl aliquots of cultures were pipetted into the wells of microtiter plates (FluoroNunc; Polysorp). Compounds were dissolved in dimethyl sulfoxide (100 mM) and added to the wells at final concentrations ranging from 8 to 500 μM. The microtiter plates were incubated at 30°C for 4 h before green fluorescent protein (GFP) fluorescence was measured with a Lambda Fluoro 320 Plus reader (Bio-Tek Instruments) equipped with filters with an excitation wavelength of 485 nm and emission wavelength of 530 nm. Inhibitor-mediated reduction of the fluorescent signal was correlated to the value obtained when the compounds were not added.
Effects of compound 3 on expression of QS-regulated functions in B. cenocepacia H111.
Proteolytic activity was determined by applying 50-μl samples of sterile filtered supernatants of cultures grown in the absence or presence of a 1 mM concentration of compound 3 for 24 h onto skim milk agar plates. Protease activity is indicated by clearing zones. Biofilm formation and swarming motility were assayed in the presence or absence of a 0.5 mM concentration of compound 3 as described previously (9). Cellular levels of AidA were determined by the aid of anti-AidA antibodies (8). For nematode killing, strains were grown in the absence or presence of a 0.5 mM concentration of compound 3 before appropriate aliquots were plated on 6.0-cm-diameter agar plates containing either no compound 3 or a 0.5 mM concentration (12). Following 24 h of incubation at 37°C, the plates were allowed to cool to 23°C before adult worms were inoculated. Plates were then incubated at 23°C and scored for live worms after 24 h.
Growth conditions.
Bacterial strains were routinely cultivated in LB medium. The growth of liquid cultures was monitored spectrophotometrically by an Ultrospec Plus spectrophotometer (Pharmacia) by measurement of optical density at 600 nm (OD600).
Two-dimensional gel electrophoresis.
The B. cenocepacia wild-type strain H111 was grown in the absence or presence of a 1 mM concentration of compound 3 in LB medium to an OD600 of 2.0. The cepI mutant B. cenocepacia H111-I was included as a control. Sample preparation of intracellular proteins, two-dimensional gel electrophoresis, and comparison of protein patterns were performed as described previously (19).
RESULTS AND DISCUSSION
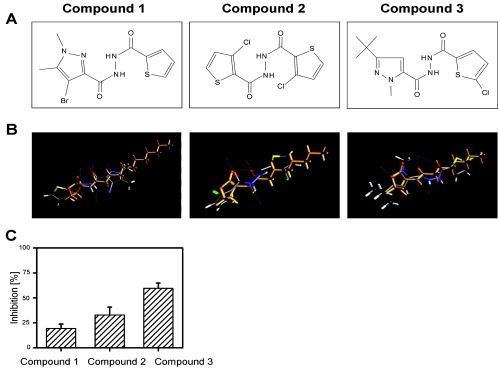
A database of commercially available compounds was screened in silico with 4SCan on the basis of molecular alignment scores for molecules that are structurally similar to 3-oxo-C12-HSL, one of the two main quorum-sensing molecules produced by P. aeruginosa (17). The hundred compounds with the highest alignment scores from this virtual screening were tested for antagonizing quorum sensing with the aid of two different AHL biosensors, which, depending on the components used for construction, respond to different types of AHL molecules (25). P. putida pKR-C12 is most sensitive for 3-oxo-C12-HSL, while P. putida pAS-C8 is most sensitive for C8-HSL, a signal molecule that is utilized by most Burkholderia species (4, 15, 28). Antagonistic activity was assessed by adding decreasing concentrations of the substances to cultures of the different biosensors in the presence of appropriate AHLs. In the case of AHL-inhibitory activity, a reduction in GFP expression was anticipated. Eleven percent of the tested compounds showed inhibitory activity in at least one of the two bioassays without affecting the growth of the biosensor. The active compounds were totally unrelated to AHLs or furanones and could be grouped on the basis of their central moieties into distinct chemical classes. Surprisingly, even though the primary screening was based on 3-oxo-C12-HSL, several compounds showed no antagonistic activity in the 3-oxo-C12-HSL-specific bioassay but inhibited the C8-HSL-specific biosensor. Furthermore, 8% of the compounds belonging to different chemical classes activated one or both of the biosensors and thus acted as AHL agonists (data not shown). The hydrazide derivative compound 1 (Fig. 1A), which most effectively inhibited the C8-HSL-based biosensor, was chosen for the further design of a QS blocker specific for the Burkholderia cep system. To this end, we developed a computer model that combines molecular alignment with comparative molecular field analysis. This model, which was now based on the structure of C8-HSL instead of 3-oxo-C12-HSL, was permanently refined as new experimental data became available. Combinatorial libraries of lead substances were synthesized and tested for AHL antagonistic activity using the biosensor P. putida (pAS-C8), and this information served as an additional input parameter for refining the model. After several iterative rounds of molecule design and the testing of over 400 substances, we obtained compound 3, which so far represents the endpoint of the optimization procedure. The evolution of this molecule from the initial substance, compound 1, via the intermediate compound 2 to the final compound 3 is shown in Fig. 1. Interestingly, exogenous addition of C8-HSL reversed the inhibitory effect of compound 3, suggesting a competitive inhibition mechanism (data not shown).
FIG. 1.
Evolution of the QS blocker compound 3. Compound 1, which was originally identified as a structural homologue of 3-oxo-C12-HSL, provided the basis for the design of QS antagonists that specifically block the Burkholderia cep system. The activity of the compound was successively improved by iterative rounds of rational molecule design and activity testing. (A) Structures of compounds 1, 2, and 3. (B) Alignment of compound 1 with 3-oxo-C12-HSL and of compounds 2 and 3 with C8-HSL. (C) Antagonistic activities of the compounds in the P. putida F117(pAS-C8)-based bioassay.
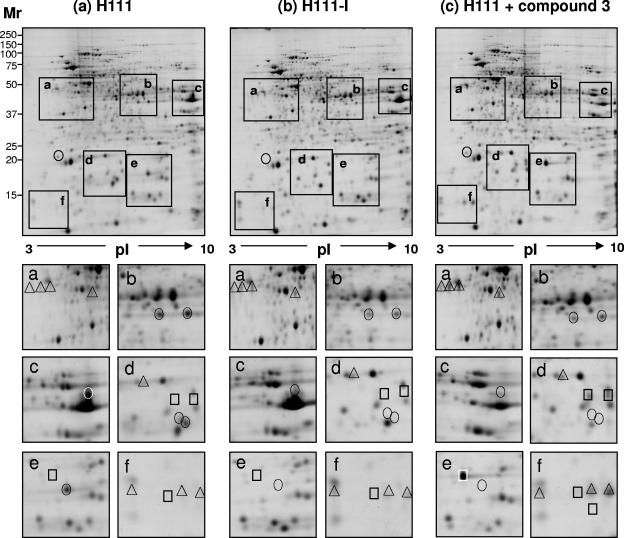
To assess the specificity of compound 3, we compared the protein patterns of the B. cenocepacia wild-type strain H111 (4) grown in the absence or presence of a 1 mM concentration of compound 3 with those of the B. cenocepacia mutant H111-I (9), which is defective in C8-HSL production due to inactivation of cepI (Fig. 2). In the presence of compound 3, the expression of only 1 out of 15 QS-regulated protein spots was not inhibited and only few changes (4 out of 532 protein spots) in the expression profile of H111 that were not related to quorum sensing were observed, indicating that compound 3 specifically inhibits the signaling system of B. cenocepacia with high specificity.
FIG. 2.
Comparative two-dimensional gel electrophoresis of intracellular proteins of the B. cenocepacia wild-type strain H111, grown in the absence (A) or presence (C) of a 1 mM concentration of compound 3, and the cepI mutant H111-I (B). The proteins were separated on Immobiline Dry strips with nonlinear pH gradients from 3 to 10 (Amersham Bioscience, Uppsala, Sweden), followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 13% polyacrylamide gels. Gels were stained with Coomassie brilliant blue G-250. Regions of interest are boxed (a to f) and shown below in better detail. Circles indicate positively QS-regulated spots, triangles indicate negatively QS-regulated spots, and squares indicate changes in the protein profile not related to quorum sensing.
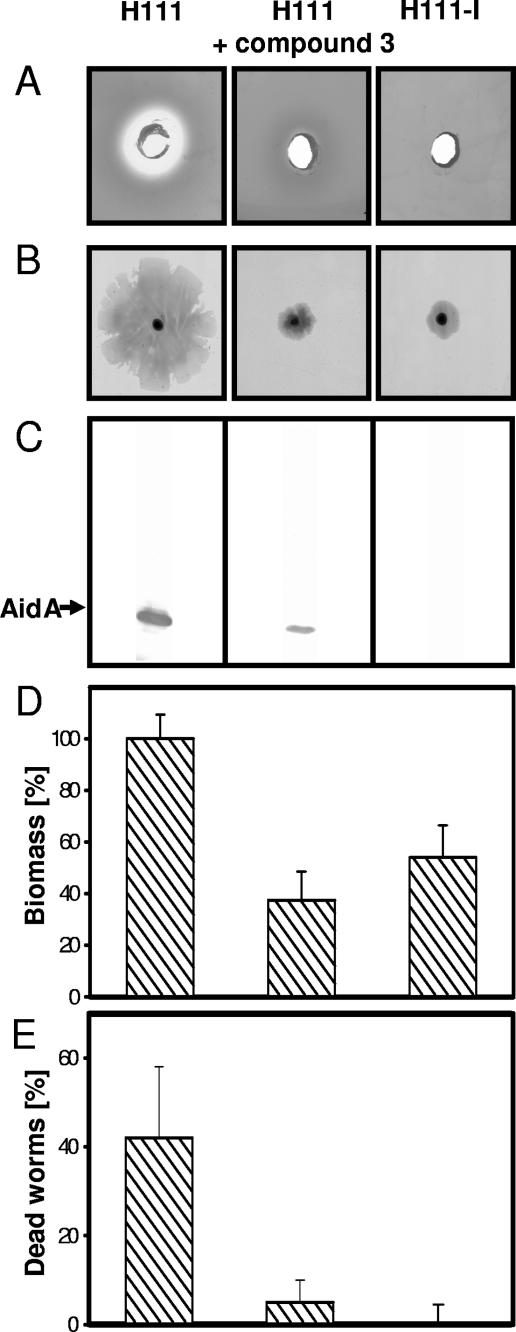
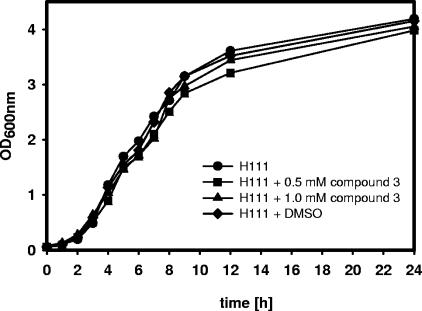
We next tested the ability of compound 3 to interfere with the expression of known QS-regulated functions in B. cenocepacia H111 (9, 14, 19). Expression of extracellular proteolytic activity (Fig. 3A), swarming motility (a specialized form of surface translocation) (Fig. 3B), and biofilm formation (Fig. 3D) were strongly inhibited in the presence of compound 3 and were virtually indistinguishable from the those of the AHL-negative mutant H111-I. Likewise, the expression of AidA, a QS-regulated virulence factor produced by many Burkholderia species (8), was greatly reduced, albeit not entirely to the level of mutant H111-I (Fig. 3C). Finally, we tested whether compound 3 affects the virulence of B. cenocepacia in a C. elegans pathogenesis model (12). Figure 3E shows that the rate of nematode killing was markedly reduced in the presence of a 0.5 mM concentration of compound 3 relative to that of the control and was only slightly higher than that of mutant H111-I. Importantly, the compound affected neither bacterial growth (Fig. 4) nor the developmental cycle or life span of the nematode.
FIG. 3.
Compound 3 inhibits QS-regulated functions in B. cenocepacia H111. Various phenotypes of B. cenocepacia H111 grown in the absence or presence of compound 3 were determined. The QS mutant H111-I was included as a control. (A) Extracellular proteolytic activity; (B) swarming motility; (C) expression of the virulence factor AidA; (D) biofilm formation; (E) killing of the nematode C. elegans.
FIG. 4.
Compound 3 does not affect the growth of B. cenocepacia H111. Cultures were grown in the presence (0.5 or 1.0 mM) or absence of compound 3 dissolved in dimethyl sulfoxide (DMSO).
In conclusion, employing rational drug design, we have developed novel agents that antagonize the cep QS system, which is present in many members of the genus Burkholderia. These substances are totally unrelated to AHL signal molecules or furanone compounds that were previously shown to interfere with quorum sensing (6, 7, 11). Interestingly, further cycles of modeling and testing did not substantially improve the potency of the compounds but with a high frequency gave rise to compounds exhibiting agonistic rather than antagonistic activity, suggesting that the structures of these compounds are very similar to that of C8-HSL (unpublished results). As compound 3 is capable of efficiently blocking the expression of pathogenic traits, it may have potential as a therapeutic option for treating B. cenocepacia infections, which due to the intrinsic resistance of the organisms are extremely difficult to eradicate. Given that Burkholderia mallei and Burkholderia pseudomallei utilize homologous signaling systems to control pathogenicity (27, 28), compound 3 may also prove useful for tackling glanders and melioidosis.
Acknowledgments
This work was supported by grants from the BMBF and the Deutsche Forschungsgemeinschaft.
The technical assistance of Dorothea Begert, Heike Endress, Stefan Kirchner, Heidi Mommerskamp, and Tanja Wieber is greatly acknowledged.
REFERENCES
- 1.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burkholder, W. H. 1950. Sour skin, a bacterial rot of onion bulbs. Phytopathology 40:115-117. [Google Scholar]
- 3.Coenye, T., and P. Vandamme. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5:719-729. [DOI] [PubMed] [Google Scholar]
- 4.Gotschlich, A., B. Huber, O. Geisenberger, A. Tögl, A. Steidle, K. Riedel, et al. 2001. Synthesis of multiple N-acylhomoserine lactones is wide-spread among the members of the Burkholderia cepacia complex. Syst. Appl. Microbiol. 24:1-14. [DOI] [PubMed] [Google Scholar]
- 5.Govan, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hentzer, M., K. Riedel, T. B. Rasmussen, A. Heydorn, J. B. Andersen, M. R. Parsek, et al. 2002. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148:87-102. [DOI] [PubMed] [Google Scholar]
- 7.Hentzer, M., H. Wu, J. B. Andersen, K. Riedel, T. B. Rasmussen, N. Bagge, et al. 2003. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22:3803-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huber, B., F. Feldmann, M. Köthe, P. Vandamme, J. Wopperer, K. Riedel, et al. 2004. Identification of a novel virulence factor in Burkholderia cenocepacia H111 required for efficient slow killing of Caenorhabditis elegans. Infect. Immun. 72:7220-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huber, B., K. Riedel, M. Hentzer, A. Heydorn, A. Gotschlich, M. Givskov, et al. 2001. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147:2517-2528. [DOI] [PubMed] [Google Scholar]
- 10.Isles, A., I. Maclusky, M. Corey, R. Gold, C. Prober, P. Fleming, et al. 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr. 104:206-210. [DOI] [PubMed] [Google Scholar]
- 11.Kline, T., J. Bowman, B. H. Iglewski, T. de Kievit, Y. Kakai, and L. Passador. 1999. Novel synthetic analogs of the Pseudomonas autoinducer. Bioorg. Med. Chem. Lett. 9:3447-3452. [DOI] [PubMed] [Google Scholar]
- 12.Köthe, M., M. Antl, B. Huber, K. Stoecker, D. Ebrecht, I. Steinmetz, et al. 2003. Killing of Caenorhabditis elegans by Burkholderia cepacia is controlled by the cep quorum-sensing system. Cell. Microbiol. 5:343-351. [DOI] [PubMed] [Google Scholar]
- 13.Lewenza, S., B. Conway, E. P. Greenberg, and P. A. Sokol. 1999. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181:748-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewenza, S., and P. A. Sokol. 2001. Regulation of ornibactin biosynthesis and N-acyl-l-homoserine lactone production by CepR in Burkholderia cepacia. J. Bacteriol. 183:2212-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lutter, E., S. Lewenza, J. J. Dennis, M. B. Visser, and P. A. Sokol. 2001. Distribution of quorum-sensing genes in the Burkholderia cepacia complex. Infect. Immun. 69:4661-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahenthiralingam, E., A. Baldwin, and P. Vandamme. 2002. Burkholderia cepacia complex infection in patients with cystic fibrosis. J. Med. Microbiol. 51:533-538. [DOI] [PubMed] [Google Scholar]
- 17.Pearson, J. P., K. M. Gray, L. Passador, K. D. Tucker, A. Eberhard, B. H. Iglewski, et al. 1994. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 91:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reik, R., T. Spilker, and J. J. LiPuma. 2005. Distribution of Burkholderia cepacia complex species among isolates recovered from persons with or without cystic fibrosis. J. Clin. Microbiol. 43:2926-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riedel, K., C. Arevalo-Ferro, G. Reil, A. Görg, F. Lottspeich, and L. Eberl. 2003. Analysis of the quorum-sensing regulon of the opportunistic pathogen Burkholderia cepacia H111 by proteomics. Electrophoresis 24:740-750. [DOI] [PubMed] [Google Scholar]
- 20.Sadowski, J., J. Gasteiger, and G. Klebe. 1994. Comparison of automatic three-dimensional model builders using 639 X-ray structures. J. Chem. Infect. Comput. Sci. 34:1000-1008. [Google Scholar]
- 21.Seifert, M. H. J., K. Wolf, and D. Vitt. 2003. Virtual high-throughput in silico screening. Biosilico 1:143-149. [Google Scholar]
- 22.Smith, K. M., Y. Bu, and H. Suga. 2003. Induction and inhibition of Pseudomonas aeruginosa quorum sensing by synthetic autoinducer analogs. Chem. Biol. 10:81-89. [DOI] [PubMed] [Google Scholar]
- 23.Smith, K. M., Y. Bu, and H. Suga. 2003. Library screening for synthetic agonists and antagonists of a Pseudomonas aeruginosa autoinducer. Chem. Biol. 10:563-571. [DOI] [PubMed] [Google Scholar]
- 24.Sokol, P. A., U. Sajjan, M. B. Visser, S. Gingues, J. Forstner, and C. Kooi. 2003. The CepIR quorum-sensing system contributes to the virulence of Burkholderia cenocepacia respiratory infections. Microbiology 149:3649-3658. [DOI] [PubMed] [Google Scholar]
- 25.Steidle, A., K. Sigl, R. Schuhegger, A. Ihring, M. Schmid, S. Gantner, et al. 2001. Visualization of N-acylhomoserine lactone-mediated cell-cell communication between bacteria colonizing the tomato rhizosphere. Appl. Environ. Microbiol. 67:5761-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomich, M., A. Griffith, C. A. Herfst, J. L. Burns, and C. D. Mohr. 2003. Attenuated virulence of a Burkholderia cepacia type III secretion mutant in a murine model of infection. Infect. Immun. 71:1405-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ulrich, R. L., D. Deshazer, H. B. Hines, and J. A. Jeddeloh. 2004. Quorum sensing: a transcriptional regulatory system involved in the pathogenicity of Burkholderia mallei. Infect. Immun. 72:6589-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valade, E., F. M. Thibault, Y. P. Gauthier, M. Palencia, M. Popoff, and D. R. Vidal. 2004. The PmlI-PmlR quorum-sensing system in Burkholderia pseudomallei plays a key role in virulence and modulates production of the MprA protease. J. Bacteriol. 186:2288-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]