Abstract
Carbapenem resistance rates in Pseudomonas aeruginosa isolates in Colombia, as in many South American countries, are high for reasons that remain unclear. From our nationwide network, we describe the first detection of the metallo-β-lactamase VIM-2 in clinical isolates of P. aeruginosa from multiple cities within Colombia. Metallo-β-lactamases were not detected in the two centers with the highest imipenem resistance rates. Clonality was noted in five of the eight centers with strains meeting the criteria for molecular typing. The high carbapenem resistance in P. aeruginosa in Colombia may be attributable to a combination of factors, including the presence of metallo-β-lactamases and nosocomial transmission.
Antibiotic-resistant bacteria are a threat to hospital patients worldwide. Among gram-negative bacilli, resistance rates tend to be much higher in Latin America than in the United States and Europe (16). This is true for organisms harboring extended-spectrum β-lactamases (ESBLs), as well as for multiresistant nonfermenters. The reasons for this discrepancy are unclear. Multidrug-resistant Pseudomonas aeruginosa is particularly worrisome because this pathogen can become resistant to all available antibiotics.
During 2002, our research center, the Centro Internacional de Entrenamiento e Investigaciones Medicas (CIDEIM), located in Cali, Colombia, established a network of eight major teaching hospitals from three different cities (Medellín, Bogotá, and Cali) in order to study ESBL prevalence and molecularly characterize ESBLs in Colombia. In 2003, hospitals in three additional cities (Bucaramanga, Pereira, and Barranquilla) entered the network.
High resistance rates were seen in P. aeruginosa in most of the hospitals of our network throughout 2003. At this time, our study was expanded to examine the mechanisms of carbapenem resistance in P. aeruginosa, including identifying and characterizing metallo-β-lactamases (MBLs). We also sought evidence for outbreaks due to inadequate infection control programs by employing molecular typing by pulsed-field gel electrophoresis (PFGE).
MBLs are enzymes that hydrolyze penicillins, cephalosporins, and carbapenems but not aztreonam (1). This phenotype may be helpful in identifying these strains in the laboratory. The two major MBL families, IMP and VIM, show low amino acid homology between the enzymes (approximately 30% to 40%), although their hydrolytic properties are similar (6). Eleven VIM variants have been reported worldwide, mostly in European and Asian countries, occurring in numerous bacterial species. VIM-2, which shares 90% amino acid identity with VIM-1, is the most widespread VIM in terms of both hosts and geography (22).
This report describes the first detection of VIM-2 enzymes from multiple cities within Colombia, noting the current impact of their presence on carbapenem resistance rates within the facilities studied.
MATERIALS AND METHODS
During 2004, CIDEIM conducted a study of nosocomial multidrug-resistant P. aeruginosa with the participation of 10 tertiary-care institutions from the Colombian Nosocomial Resistance Study Group in six different cities. Centers were selected if they provided tertiary care, had microbiologists and infectious disease physicians on site, and agreed to participate.
In order to determine the rates of carbapenem resistance, epidemiological and susceptibility information on all bacteria isolated from patients hospitalized in general wards and intensive care units was received once a month throughout 2004. The information was downloaded from the databases of each microbiology laboratory and sent to CIDEIM by an electronic or magnetic device. The data were transferred to a general database designed by CIDEIM and run on WHONET 5.3 software (18). Simultaneously, all P. aeruginosa isolates from intensive care units and general wards resistant to ceftazidime and imipenem or meropenem were sent to CIDEIM for further analysis. Not all institutions had isolates with this phenotype.
Bacterial identification and antimicrobial susceptibility.
Bacterial identification of the isolates received was confirmed by Vitek (BioMérieux, Lyon, France) with the GNI+ card according to the manufacturer's instructions. Carbapenem and ceftazidime resistance was confirmed by determining the MICs by a twofold standard broth microdilution method in cation-adjusted Mueller-Hinton broth by following the NCCLS M7-A6 guidelines (11). Antimicrobial agents tested included imipenem (Merck Sharp & Dohme, Rahway, NJ), meropenem (AstraZeneca, Alderly Park, United Kingdom), and ceftazidime (Sigma-Aldrich, St. Louis, MO).
Strain typing.
Chromosomal DNA of all strains received by CIDEIM was prepared by following the method described by Gautom (5), with some modifications. In brief, a bacterial cell suspension was made in 100 mM Tris-100 mM EDTA buffer. A plug was prepared from 200 μl of this suspension and 200 μl 2% LPM agarose and then placed in lysis buffer (50 mM Tris, 50 mM EDTA, pH 8.0, 1% N-lauroylsarcosine; 1 mg/ml proteinase K) and incubated at 55°C. The plugs were washed twice with preheated (50°C), sterile, distilled water for 15 min and then washed five times in preheated (50°C) sterile TE (10 mM Tris, pH 8.0, 1 mM EDTA). DNA was digested with XbaI (Promega, Madison, WI) at 37°C for 2 h and run in a 1% agarose gel with Chef Mapper (Bio-Rad, Fremont, CA) at 14°C and 6 V/cm with pulse times of 1 s and 30 s for 19 h. The gels were visualized with ethidium bromide and the results analyzed with Diversity software (Bio-Rad) in order to determine similarities and differences among the bacterial isolates from within each hospital. Band-based dendrograms were produced by using Dice coefficients (4). Indistinguishable (100% related) and closely related (85% to 99% related) pulsotypes were considered clonal.
MBL screening.
One isolate per pulsotype with an imipenem or meropenem MIC of ≥32 μg/ml and a ceftazidime MIC of ≥64 μg/ml was selected for MBL screening. MBL Etest strips (AB Biodisk, Solna, Sweden) were used according to the protocol recommended by the manufacturer (21). MIC ratios (MIC of imipenem alone/MIC of imipenem plus EDTA) of ≥8 were considered positive for MBL production.
PCR amplification and sequencing.
Genomic DNA was prepared by boiling a bacterial culture in 100 μl of distilled water for 10 min. Amplification reactions were carried out with 1 μl of boiled bacterial suspensions, 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 200 μM each deoxynucleoside triphosphate (Invitrogen, Carlsbad, CA), 2.0 mM MgCl2, 25 pmol of each primer, and 1 U of Platinum Taq DNA polymerase (Invitrogen). A 10× Mg-free buffer and 50 mM MgCl2 were supplied by the manufacturer. The PCR program consisted of a bacterial lysis and DNA denaturation step of 10 min at 94°C; 30 cycles with a 1-min denaturation step at 94°C, a 1-min annealing step at 54°C, and a 2.5-min extension at 72°C; and a final 10-min extension step at 72°C.
Primers 5′CS and 3′CS (8), specific for the 5′ and 3′ conserved sequences, respectively, of class 1 integrons and consensus VIM primer sequences (20), were used for blaVIM genes (Table 1). Combinations of these two primer sets were used to amplify the entire blaVIM gene and the surrounding genetic context for sequencing. Products were detected by 1% agarose gel electrophoresis with ethidium bromide staining. PCR products were purified by use of the QIAquick PCR purification kit (QIAGEN, Valencia, CA). Sequencing of both strands was performed by ACGT, Inc. (Wheeling, IL).
TABLE 1.
Oligonucleotides used in PCR amplification
| Primer | Target | Nucleotide sequence (5′ to 3′) | GenBank accession no. | Position | Reference |
|---|---|---|---|---|---|
| 5′-CS | 5′ class 1 integron variable region | GGC ATC CAA GCA GCA AG | M73819 | 1190-1206 | 8 |
| 3′-CS | 3′ class 1 integron variable region | AAG CAG ACT TGA CCT GA | M73819 | 1342-1326 | 8 |
| VIM-F | Conserved region of VIM gene | GTC TAT TTG ACC GCG TC | 20 | ||
| VIM-R | Conserved region of VIM gene | CTA CTC AAC GAC TGA GCG | 20 |
RESULTS
During the study period, great variability was seen in the resistance rates among our hospitals. From P. aeruginosa isolates in general wards, imipenem resistance averaged 13.5% (range, 5.8 to 30.8%), while in intensive care units, the average was 26.6% (range, 2 to 71.4%) (Table 2).
TABLE 2.
Percentages of imipenem resistance and presence of clonality in 10 Colombian hospitals in 2004
| Hospital and department | City |
Pseudomonas aeruginosa
|
|
|---|---|---|---|
| % of isolates resistant (total no.) | Pulsotype(s) (no. of strains)b | ||
| Aa | |||
| Wards | Barranquilla | 8.0 (87) | A1 (4), A2 (3), A3 (unique) |
| ICUc | 9.7 (82) | ||
| B | |||
| Wards | Bucaramanga | 7.4 (27) | ND |
| ICU | 10.5 (19) | ||
| C | |||
| Wards | Bogotá | 15.7 (51) | C1 (2), C2-C9 (unique) |
| ICU | 45.5 (11) | ||
| Da | |||
| Wards | Pereira | 11.4 (35) | D1 (2), D2 (unique), D3-D10 (unique) |
| ICU | 36.7 (30) | ||
| Ea | |||
| Wards | Cali | 5.8 (86) | E1 (2), E2 (unique), E3-E5 (unique) |
| ICU | 2 (49) | ||
| F | |||
| Wards | Medellín | 28 (43) | F1 (5), F2 (4), F3-F16 (unique) |
| ICU | 71.4 (21) | ||
| G | |||
| Wards | Medellín | 6.8 (59) | G1-G6 (unique) |
| ICU | 18.2 (55) | ||
| H | |||
| Wards | Bogotá | 30.8 (107) | H1-H3 (unique) |
| ICU | 40 (30) | ||
| I | |||
| Wards | Bucaramanga | 8 (49) | J1 (unique) |
| ICU | 5.9 (17) | ||
| J | |||
| Wards | Bogotá | 11.8 (34) | ND |
| ICU | 25 (4) | ||
Hospital from which VIM-2-carrying bacteria were isolated.
ND, not determined. Bold indicates a MBL-positive pulsotype.
ICU, intensive care unit.
Out of the 896 imipenem-resistant P. aeruginosa isolates identified within our 10 participating institutions, 68 isolates that met our inclusion criteria of resistance to ceftazidime and imipenem or meropenem were received by CIDEIM in 2004 and subsequently subjected to PFGE analysis. Two institutions, centers B and J, had no isolates with this phenotype. From this collection of 68 isolates, analysis for MBLs was limited to those having an imipenem or meropenem MIC of ≥32 μg/ml and a ceftazidime MIC of ≥64 μg/ml. MIC determinations identified 33 isolates having this phenotype, representing 16 different pulsotypes. One isolate per pulsotype was then screened by MBL Etest for the presence of MBLs. Six were identified as MBL screen positive; two pulsotypes in each participating hospital in the cities of Cali, Pereira, and Barranquilla. All six proceeded to PCR and sequencing analysis. Strain typing had initially been determined within but not between institutions. Pulsotypes D1 from Pereira and E1 from Cali were later recognized as being closely related by PFGE, although neither was related to the pulsotypes from Barranquilla (data not shown). This was an unanticipated observation, and there was no evidence for patient transfer between these two cities that could explain the strain relatedness. Although these isolates from Pereira and Cali would have been designated as clonal, they were analyzed independently for identification of the MBL due to their different geographical origins.
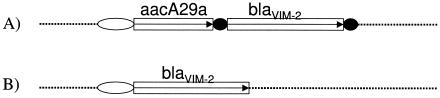
PCR amplification and sequencing detected the presence of blaVIM-2, on a class 1 integron, in each of the six clones of P. aeruginosa. Two different gene cassette organizations were identified in comparing the partially characterized integrons of these isolates (Fig. 1). Pulsotypes D1 and D2 from Pereira and E1 and E2 from Cali exhibited the same integron arrangement, which contained the unusual aminoglycoside-modifying enzyme, aacA29a, as the first gene cassette and VIM-2 as the second cassette. This combination of gene cassettes has only been reported once before in a class 1 integron, In59, from France (12). In pulsotypes A1 and A3 from Barranquilla, however, VIM-2 occurred as the first gene cassette on the integron. We were unable to determine the portions downstream of VIM-2 in these clones as they lacked the 3′ conserved sequence used in the sequencing PCR.
FIG. 1.
Schematic structure (not to scale) of the determined integron arrangement of gene cassettes. (A, Pereira-Cali clones; B, Barranquilla clones). The gene cassettes are boxed. Arrows indicate transcriptional orientation. The open ovals indicate attl recombination sites, and the black circles represent 59-bp elements. Dashed lines represent unknown flanking sequences.
Of the 10 hospitals where imipenem-resistant P. aeruginosa was studied, isolates conforming to our criteria were obtained from 8. Clonality was detected at five of these institutions. In three (centers C, D, and F) of the four hospitals with the highest imipenem resistance rates (the aforementioned and center H), clonality was present (Table 2). In two (centers A and E) of the three centers where MBLs were detected, there were low imipenem resistance rates. In contrast, the center with the highest imipenem resistance rates (center F) had many strains related by PFGE but none were MBL producers. Generally, strain relatedness was observed within, but not between, individual centers, with the exception of the aforementioned Cali-Pereira VIM-2-producing clone.
DISCUSSION
The list of MBL enzymes detected in the Americas continues to grow: IMP-1, IMP-7, IMP-16, VIM-2, VIM-7, VIM-8, and SPM-1 (3, 6, 9, 10, 15, 19). Many of these have been reported exclusively in P. aeruginosa. The first VIM-2 in South America, reported in 2002 by the SENTRY program, occurred in single hospitals in Chile and Venezuela (10). In 2004, a widespread outbreak of multidrug-resistant P. aeruginosa harboring VIM-8 was described in one hospital in Cali, Colombia. Environmental contamination was documented, and disinfection was associated with termination of the outbreak (3).
We report high carbapenem resistance rates among isolates of P. aeruginosa in Colombia. These can be attributed to a combination of factors, including the presence of a carbapenemase, VIM-2, in P. aeruginosa and nosocomial outbreaks, presumably due to inadequate infection control. Of these two factors, the latter appears to have had a greater impact. Carbapenem resistance rates did not correlate with the presence of VIM-2, as two centers harboring VIM-2 producers had low resistance rates while several centers with high resistance rates did not have detectable carbapenemases.
The genes for MBL enzymes, like those for OXA-ESBLs, are often carried as cassettes within integrons. These natural recombination systems assemble series of acquired genes behind a single promoter, with the β-lactamase genes often adjacent to aminoglycoside 6′-N acetyltransferase [aa(6′-1b)] determinants (13), conferring resistance to multiple antibiotic classes. Although MBLs are not very prevalent thus far, they remain a real threat to hospitals and patients because of the risk of dissemination through these mobile genetic elements. Widespread outbreaks have been reported in several countries, including Colombia (2, 3, 6, 7, 14, 17).
Antimicrobial resistance in P. aeruginosa strains may be acquired through either horizontal gene transfer or selection of novel resistant strains. Our documented VIM-2 producers were detected from three different Colombian cities and belonged to five different pulsotypes. This report shows that the same gene cassette arrays, including an unusual aminoglycoside-modifying enzyme, aacA29a, were found in the clones producing VIM-2 from Pereira and Cali. The presence of integrons containing the same organization of cassettes in three different pulsotypes suggests a hypothesis of horizontal gene transfer of the integron. Conversely, the integron organization identified in clones from Barranquilla was unique in comparison, suggesting independent acquisition of blaVIM-2 by the P. aeruginosa isolates from this city.
That clonality was found to exist in five of the eight centers from which strains were typed, independent of VIM-2 production, suggests that standard infection control measures such as hand hygiene, barrier precautions, and perhaps disinfection guidelines for equipment are not being followed. This is a problem in hospitals worldwide.
There are several implications of these data. Given the great variability from center to center, it is essential that local surveillance programs be maintained in each hospital as they can provide an invaluable data source to provide guidance for formulary decision making. In addition, molecular typing has emphasized the need to improve infection control in some centers.
Acknowledgments
The conformation of the network of institutions from the Colombian Nosocomial Resistance Study Group and this study was made possible thanks to the support of Bristol Myers-Squibb, Pfizer, and CIDEIM.
We thank the participating institutions from the Colombian Nosocomial Resistance Study Group, whose members are as follows: CIDEIM, Sandra Reyes and Maria Consuelo Miranda; Cali, Ernesto Martínez, Lena Barrera, Luz Marina Gallardo, and Alba Lucía Bohorquez; Bogotá, Carlos Alquichire, Martha Ruiz, Nancy Obando, Pilar Hurtado, Gladys Ceballos, Andrés Torres, Henry Mendoza, Alba Lucia Sanín, Martha Patricia Meléndez, Sara Maria Quintero, Eugenia Janeth López, Cristina Matiz, Sonia Cuervo, Jorge Cortés, Maria Cristina Paredes, Patricia Arroyo, and Diana Bermudez; Medellín, Carlos Ignacio Gomez, Jaime Lopez, Monica Cuartas, Celina Gomez, Ana Lucia Correa, Julián Betancourth, Esteban Echavarria, Juan David Villa, Ana Cristina Quiroga, Luz Amparo Alvarez, and Claudia Patricia Alvarez; Barranquilla, Rubén Darío Carmargo, Adriana Marín, and Angela Mendoza; Pereira, Carmen Elisa Llano, Myriam Gómez, and Araceli Cano; Bucaramanga, Claudia Bárcenas, Adriana Pinto, and Luis Angel Villar.
REFERENCES
- 1.Bush, K. 1998. Metallo-β-lactamases: a class apart. Clin. Infect. Dis. 27(Suppl. 1):48-53. [DOI] [PubMed] [Google Scholar]
- 2.Cornaglia, G., A. Mazzariol, L. Lauretti, G. M. Rossolini, and R. Fontana. 2000. Hospital outbreak of carbapenem-resistant Pseudomonas aeruginosa producing VIM-1, a novel transferable metallo-β-lactamase. Clin. Infect. Dis. 31:1119-1125. [DOI] [PubMed] [Google Scholar]
- 3.Crespo, M. P., N. Woodford, A. Sinclair, M. E. Kaufmann, J. Turton, J. Glover, J. D. Velez, C. R. Castaneda, M. Recalde, and D. M. Livermore. 2004. Outbreak of carbapenem-resistant Pseudomonas aeruginosa producing VIM-8, a novel metallo-β-lactamase, in a tertiary care center in Cali, Colombia. J. Clin. Microbiol. 42:5094-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dice, L. R. 1945. Measures of the amount of ecological association between species. Ecology 26:297-302. [Google Scholar]
- 5.Gautom, R. K. 1997. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 35:2977-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibb, A. P., C. Tribuddharat, R. A. Moore, T. J. Louie, W. Krulicki, D. M. Livermore, M. F. Palepou, and N. Woodford. 2002. Nosocomial outbreak of carbapenem-resistant Pseudomonas aeruginosa with a new blaIMP allele, blaIMP-7. Antimicrob. Agents Chemother. 46:255-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee, K., J. B. Lim, J. H. Yum, D. Yong, Y. Chong, J. M. Kim, and D. M. Livermore. 2002. blaVIM-2 cassette-containing novel integrons in metallo-β-lactamase-producing Pseudomonas aeruginosa and Pseudomonas putida isolates disseminated in a Korean hospital. Antimicrob. Agents Chemother. 46:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levasque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lolans, K., A. M. Queenan, K. Bush, A. Sahud, and J. P. Quinn. 2005. First nosocomial outbreak of Pseudomonas aeruginosa producing an integron-borne metallo-β-lactamase (VIM-2) in the United States. Antimicrob. Agents Chemother. 49:3538-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendes, R. E., M. Castanheira, P. Garcia, M. Guzman, M. A. Toleman, T. R. Walsh, and R. N. Jones, and the SENTRY Antimicrobial Surveillance Program. 2004. First isolation of blaVIM-2 in Latin America: report from the SENTRY Antimicrobial Surveillance Program. Antimicrob. Agents Chemother. 48:1433-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard M7-A6, 6th edition, vol. 23. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 12.Poirel, L., T. Lambert, S. Turkoglu, E. Ronco, J. Gaillard, and P. Nordmann. 2001. Characterization of class 1 integrons from Pseudomonas aeruginosa that contain the blaVIM-2 carbapenem-hydrolyzing β-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob. Agents Chemother. 45:546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poirel, L., and P. Nordmann. 2002. Acquired carbapenem-hydrolyzing β-lactamases and their genetic support. Curr. Pharm. Biotechnol. 3:117-127. [DOI] [PubMed] [Google Scholar]
- 14.Pournaras, S., M. Maniati, E. Petinaki, L. S. Tzouvelekis, A. Tsakris, N. J. Legakis, and A. N. Maniatis. 2003. Hospital outbreak of multiple clones of Pseudomonas aeruginosa carrying the unrelated metallo-β-lactamase gene variants blaVIM-2 and blaVIM-4. J. Antimicrob. Chemother. 5:1409-1414. [DOI] [PubMed] [Google Scholar]
- 15.Sader, H. S., M. Castanheira, R. E. Mendes, M. Toleman, T. R. Walsh, and R. N. Jones. 2005. Dissemination and diversity of metallo-β-lactamases in Latin America: report from the SENTRY Antimicrobial Surveillance Program. Int. J. Antimicrob. Agents 25:57-61. [DOI] [PubMed] [Google Scholar]
- 16.Sader, H. S., R. N. Jones, A. C. Gales, J. B. Silva, A. C. Pignatari, and the SENTRY Participants Group (Latin America). 2004. SENTRY antimicrobial surveillance program report: Latin American and Brazilian results for 1997 through 2001. Braz. J. Infect. Dis. 8:25-79. [DOI] [PubMed] [Google Scholar]
- 17.Senda, K., Y. Arakawa, K. Nakashima, H. Ito, S. Ichiyama, K. Shimokata, N. Kato, and M. Ohta. 1996. Multifocal outbreaks of metallo-β-lactamase-producing Pseudomonas aeruginosa resistant to broad-spectrum β-lactams, including carbapenems. Antimicrob. Agents Chemother. 40:349-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stelling, J. M., and T. F. O'Brien. 1997. Surveillance of antimicrobial resistance: the WHONET program. Clin. Infect. Dis. 24(Suppl. 1):S157-S168. [DOI] [PubMed] [Google Scholar]
- 19.Toleman, M. A., K. Rolston, R. N. Jones, and T. R. Walsh. 2004. blaVIM-7, an evolutionarily distinct metallo-β-lactamase gene in a Pseudomonas aeruginosa isolate from the United States. Antimicrob. Agents Chemother. 48:329-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toleman, M. A., A. M. Simm, T. A. Murphy, A. C. Gales, D. J. Biedenbach, R. N. Jones, and T. R. Walsh. 2002. Molecular characterization of SPM-1, a novel metallo-β-lactamase isolated in Latin America: report from SENTRY antimicrobial surveillance programme. J. Antimicrob. Chemother. 50:673-679. [DOI] [PubMed] [Google Scholar]
- 21.Walsh, T. R., A. Bolmstrom, A. Qwarnstrom, and A. Gales. 2002. Evaluation of a new Etest for detecting metallo-β-lactamases in routine clinical testing. J. Clin. Microbiol. 40:2755-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh, T. R., M. A. Toleman, L. Poirel, and P. Nordmann. 2005. Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306-325. [DOI] [PMC free article] [PubMed] [Google Scholar]