Abstract
Mycobacteria are naturally resistant to most common antibiotics and chemotherapeutic agents. The underlying molecular mechanisms are not fully understood. In this paper, we describe a hypersensitive mutant of Mycobacterium smegmatis, MS 2-39, which was isolated by screening for transposon insertion mutants of M. smegmatis mc2155 that exhibit increased sensitivity to rifampin, erythromycin, or novobiocin. The mutant MS 2-39 exhibited increased sensitivity to all three of the above mentioned antibiotics as well as fusidic acid, but its sensitivity to other antibiotics, including isoniazid, ethambutol, streptomycin, chloramphenicol, norfloxacin, tetracycline, and β-lactams, remained unchanged. Uptake experiment with hydrophobic agents and cell wall lipid analysis suggest that the mutant cell wall is normal. The transposon insertion was localized within the asnB gene, which is predicted to encode a glutamine-dependent asparagine synthetase. Transformation of the mutant with wild-type asnB of mc2155 or asnB of Mycobacterium tuberculosis complemented the drug sensitivity phenotype. These results suggest that AsnB plays a role in the natural resistance of mycobacteria.
Mycobacterium tuberculosis, the causative agent of tuberculosis (TB), is responsible for 2 million deaths and 10 million new infections each year. Clinical treatments of TB and other mycobacterial infections are difficult because mycobacteria are naturally resistant to most commonly used antibiotics and chemotherapeutic agents (14, 17). For instance, among the commonly used antibiotics against M. tuberculosis, the only ones found to be effective are rifampin and streptomycin, which are routinely used, in combination with isoniazid, pyrazinamide, and ethambutol, for chemotherapy of TB. Infections caused by nontuberculous mycobacteria such as the Mycobacterium avium complex (MAC) are increasingly common in immunocompromised individuals, which are often more difficult to treat because these organisms are resistant to standard anti-TB drugs including isoniazid and rifampin (14, 17).
The natural resistance of mycobacteria is caused primarily by the impermeability of the mycobacterial cell wall (6, 14, 17). The cell wall forms an asymmetric lipid bilayer with mycolic acids in the inner leaflet and extractable complex lipids in the outer leaflet (17, 20). Mycolic acids are of extraordinary length and are highly saturated; as such, the mycolic acid-containing layer has extremely low fluidity, which forms a strong permeability barrier and contributes to the broad resistance (17, 18, 20). The natural resistance of mycobacteria also involves active efflux processes mediated by various transport systems. Efflux-mediated resistances have been reported for fluoroquinolones (21, 28), tetracyclines (9, 27), isoniazid, ethambutol (7), pyrazinamide (33), erythromycin, and rifamycines (16). These efflux systems, however, often confer only low levels of resistance.
In an attempt to better understand the mechanisms involved in the natural resistance of mycobacteria to antibiotics, we have screened a transposon insertion library of Mycobacterium smegmatis for drug-supersusceptible mutants. Using this strategy, we have previously isolated and characterized a rifampin-hypersensitive mutant, which showed that the presence of a rifampin ADP ribosyltransferase (Arr) in M. smegmatis confers resistance to rifampin (1). In the present work, we describe a different drug-susceptible mutant in which inactivation of asnB, a gene involved in amino acid metabolism, dramatically sensitizes M. smegmatis to multiple antibiotics, including rifampin, erythromycin, novobiocin, and fusidic acid.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Wild-type (WT) Mycobacterium smegmatis strain mc2155 and its mariner insertion mutants were grown in Middlebrook 7H9 broth or Middlebrook 7H11 agar (Difco) supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC; Difco). In preparation for genomic DNA isolation, mycobacteria were grown in 7H9 broth supplemented with 0.1% Tween 80 and 10% albumin-dextrose-catalase (ADC; Difco). Antibiotics (Sigma) were added at the following concentrations: ampicillin, 50 μg/ml; kanamycin, 25 μg/ml for mycobacteria and 50 μg/ml for Escherichia coli; hygromycin, 75 μg/ml for mycobacteria and 150 μg/ml for E. coli.
Generation and screening of M. smegmatis φMycoMar insertion library.
The mariner-based transposon system ΦMycoMarT7 was utilized to generate a transposon insertion mutant library of M. smegmatis mc2155 as described previously (1, 2). Kanamycin-resistant (i.e., transposon-containing) colonies were patched onto Middlebrook 7H11 agar to obtain a library of 7,680 clones (i.e., 80 plates × 96 colonies per plate). To screen this library, clones were replica plated onto Middlebrook 7H11 agar supplemented with either rifampin, erythromycin, or novobiocin, each at one-third the MIC for the WT M. smegmatis mc2155. Clones that failed to grow on the drug-containing plates were deemed hypersensitive. A broth dilution method was used to determine the MIC and confirm the drug hypersensitivity of these clones (19).
Localization of the φMycoMar insertion.
The method used to localize and identify the disrupted gene has been described previously (1, 2). Briefly, total chromosomal DNA of transposon insertion mutant was cleaved with BamHI. Digested DNA was self-ligated with T4 DNA ligase and transformed into competent E. coli DH5α λ pir116 cells. Plasmid DNA was isolated from Kmr E. coli transformants. MycoMar-specific primers were used to determine the DNA sequence of the MycoMar/chromosomal junction. These DNA sequences were compared to the GenBank database and the M. smegmatis mc2155 genome database at the Institute for Genomic Research (http://www.tigr.com/) using the BLASTN algorithm. Nucleotide sequences were also analyzed with NTI Suite software (Informax).
Cloning of asnB gene from M. smegmatis and M. tuberculosis.
The asnB gene of M. smegmatis WT strain mc2155 was amplified by PCR from its genomic DNA using the forward primer 5′-TGCAGCCGCTCAGCAGTAGC-3′ and reverse primer 5′-CCGAGACCCACAAGGAGGAT-3′. The asnB gene of M. tuberculosis H37Rv was amplified by PCR using the forward primer 5′-TACTTGTCCTTGCCCTCTGG-3′ and reverse primer 5′-TCCTAAGGGTGTCGATTTGG-3′. An ordered BAC library of M. tuberculosis H37Rv genome (obtained from S. Cole, Institut Pasteur, Paris, France) was used as the DNA template for cloning. Vector pDrive (QIAGEN) was used for cloning of PCR products. To construct pASNB plasmids, a 2.4-kb EcoRI fragment containing asnB of M. smegmatis or a 3.2-kb StuI fragment containing asnB of M. tuberculosis was cloned into the E. coli-Mycobacterium shuttle vector pNBV1 (11), which contains a hygromycin resistance cassette. The resulting plasmids were named pASNB_MS and pASNB_MTB, respectively. Plasmids pASNB and pNBV1 were transformed into mycobacterial cells by electroporation according to standard protocol. Transformants were selected on Middlebrook 7H11 agar containing hygromycin (75 μg/ml). Drug sensitivities of the transformants were determined by a broth dilution method in Middlebrook 7H9 supplemented with OADC (19).
Uptake assay.
Radiolabeled chemicals were obtained from NEN. Uptake of [14C]erythromycin (specific activity, 55.1 mCi/mmol) and [14C]chenodeoxycholate (specific activity, 48.6 mCi/mmol) by mycobacterial cells was measured as described previously (18, 19). Briefly, [14C]erythromycin or [14C]chenodeoxycholate was added to 1-ml cell suspensions of mc2155 or MS 2-39 in 0.1 M K-phosphate buffer (pH 7.0) at time zero to a final concentration of 10 μM. At various time points thereafter (0, 3, 5, 10, 15, 20, 25, 30 min), 50-μl portions of the suspension were removed, filtered, and washed. The radioactivity retained on the filter was determined by scintillation counting.
Thin-layer chromatrography analysis of cell wall mycolic acids.
Mycolic acids were extracted and analyzed by thin-layer chromatography as previously described (31).
RESULTS
Isolation and characterization of a hypersensitive mutant MS 2-39.
To identify mechanisms that may contribute to the intrinsic drug resistance of mycobacteria, we generated a transposon insertion mutant library of M. smegmatis mc2155 (∼8,000 clones) and screened for mutants that exhibit increased sensitivity to antibiotics. This was achieved by replica plating of each clone onto 7H11 agar plates containing kanamycin (25 μg/ml) and supplemented with either rifampin, erythromycin, or novobiocin, each at one-third the MIC for the WT M. smegmatis mc2155. These plates were incubated at 37°C for 7 days, by which time all colonies on the drug-free (i.e., master) plates fully grew. Colonies on the drug-containing plates were visually inspected to identify mutants that did not grow in the presence of sublethal concentrations of drugs. One mutant, MS 2-39, was unable to grow on plates containing either rifampin, erythromycin, or novobiocin, suggesting that this mutant was hypersensitive to all three drugs.
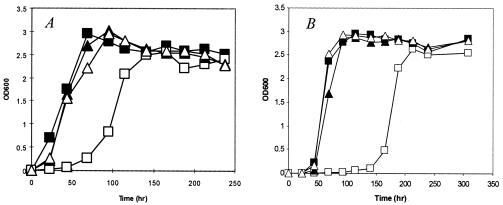
The MS 2-39 mutant was patch purified on 7H11 agar plates, and single colonies were inoculated in 7H9 liquid medium containing kanamycin. The mutant exhibited a growth delay; when grown in 7H9 broth at 37°C, there was an approximately 75-h lagging period at the early stage of growth compared to the WT strain mc2155 (Fig. 1A). However, once it started to grow, the mutant had a similar growth rate as the WT strain and reached the same cell density at stationary phase. Microscopic examination of mutant cells revealed typical rod shape of normal size (not shown).
FIG. 1.
Growth of mutant MS 2-39. Strains of mc2155 (▪), MS 2-39 (□), MS 2-39/pASNB_MS (▴), and MS 2-39/pASNB_MTB (▵) were grown in 7H9 liquid medium at 37°C (A) or 25°C (B). MS 2-39 exhibited a growth delay at both temperatures, which was complemented by plasmid pASNB_MS or pASNB_MTB. The initial inoculate was 106 cells/ml. OD600, optical density at 600 nm.
To determine the influence of temperature on bacterial growth, MS 2-39 and mc2155 were cultured at 25 and 45°C in 7H9 broth. At 25°C, the mutant exhibited a ∼100-h delay at the early growth phase (Fig. 1B). At 45°C, there was no visible growth of the mutant after 10 days, while the WT strain reached stationary phase after only 48 h (data not shown). These results suggest that growth of MS 2-39 is affected at all temperatures but the effect is most pronounced at 45°C.
To confirm the hypersensitivity of the mutant, MICs of MS 2-39 were determined in 7H9 liquid medium at 37°C using the broth dilution method (19). MICs for the WT strain were determined after 4-day incubation, whereas MICs for the mutant were determined after 7-day incubation because the mutant exhibits a 3-day growth delay at 37°C. Prolonged growth (e.g., 10 days) of the mutant did not alter the MIC reading. The result confirmed that MS 2-39 is hypersensitive to erythromycin, rifampin, novobiocin, and fusidic acid (Table 1). MICs of the mutant are between 16- and 64-fold lower than the WT strain for these antibiotics (Table 1). Sensitivity of MS 2-39 to other antibiotics, including isoniazid, ethambutol, streptomycin, chloramphenicol, norfloxacin, nalidixic acid, tetracycline, ampicillin, nafcillin, and cephaloridine, remained unchanged (data not shown).
TABLE 1.
MIC of various antibiotics for M. smegmatis asnB mutant strain MS 2-39
| Strain | MIC (μg/ml)
|
|||
|---|---|---|---|---|
| Rifampicin | Erythromycin | Novobiocin | Fusidic acid | |
| mc2 155 | 64 | 32 | 128 | 128 |
| MS 2-39 | 2 | 0.5 | 4 | 8 |
| MS 2-39/pASNB_MS | 64 | 32 | 128 | 128 |
| MS 2-39/pASNB_MTB | 64 | 32 | 128 | 128 |
Mutant MS 2-39 is defective in AsnB.
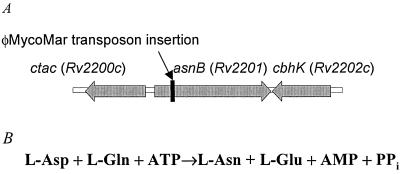
The transposon insertion in MS 2-39 was localized as described in Materials and Methods. Upon comparison, our DNA sequencing data matched the M. smegmatis mc2155 genome data available at The Institute for Genomic Research (www.tigr.org). The transposon had inserted at a TA dinucleotide, 305 bp downstream from the GTG start codon of an open reading frame, which is predicted to encode a 658-amino acid polypeptide (Fig. 2A). A homology search using the BLAST algorithm revealed a significant sequence identity with predicted asparagine synthetase B (AsnB) of various organisms including Mycobacterium avium subsp. paratuberculosis (82% amino acid identity), M. tuberculosis (80%), M. leprae (77%), Rhodococcus erythropolis (72%), Nocardia farcinica (71%), Corynebacterium glutamicum (63%), and Bacillus licheniformis (52%). Based on the sequence homology, we named the gene inactivated in mutant MS 2-39 asnB.
FIG. 2.
(A) The asnB region of M. smegmatis mc2155. The asnB gene and flanking genes are depicted. Block arrows represent open reading frames. Arrow indicates insertion of the transposon to generate the hypersensitive mutant MS 2-39. The genome of M. tuberculosis has the same genetic organization and the corresponding genes are shown (Rv2200c, Rv2201, Rv2202c). (B) Reaction catalyzed by glutamine-dependent asparagine synthetase, AsnB.
AsnB catalyzes the transfer of the γ amino residue of glutamine to the carboxyl residue of aspartate (Fig. 2B) and is a member of the purF family of glutamine-dependent amidotransferases, which includes glucosamine-6-phosphate synthase (GlmS) and glutamine phosphoribosylpyrophosphate amidotransferase (32). Like other AsnB homologs, the deduced sequence of M. smegmatis AsnB is composed of a glutaminase domain (residues 2 to 247) and a synthetase domain (residues 270 to 577). The N-terminal glutaminase domain hydrolyzes glutamine to glutamate and ammonia. It contains a conserved N-terminal second cysteine residue, which is characteristic of all purF enzymes and is essential for glutamine-dependent amidotransferase activity (5, 30). Family members that contain the C-terminal synthetase domain catalyze the conversion of aspartate to asparagine. The three-dimensional architecture of the N-terminal domain of AsnB is similar to that observed for glutamine phosphoribosylpyrophosphate amidotransferase while the molecular motif of the C-terminal domain is reminiscent to that observed for GMP synthetase (15).
Complementation of mutant MS 2-39.
To confirm that the hypersensitive phenotype of MS 2-39 was due to disruption of the asnB gene and not a polar effect caused by the transposon insertion, we cloned the asnB gene from the parental M. smegmatis strain mc2155 and the asnB ortholog of M. tuberculosis into a shuttle vector pNBV1 (11). The resulting plasmids, pASNB_MS and pASNB_MTB, respectively, were transformed into MS 2-39. Recombinant strains of MS 2-39 containing either pASNB_MS or pASNB_MTB exhibited MICs and growth curves that were indistinguishable from the WT strain (Table 1 and Fig. 1A and B), suggesting that the asnB gene of M. smegmatis or M. tuberculosis complemented the mutant phenotypes.
Uptake of erythromycin and chenodeoxycholate.
To determine whether the enhanced drug sensitivity of MS 2-39 is caused by increased cell wall permeability, uptakes of [14C]erythromycin and [14C]chenodeoxycholate by mutant cells were performed and compared with WT cells. Erythromycin was one of the four drugs that showed enhanced activity against mutant MS 2-39. In addition, since all four drugs that showed increased activities against the mutant are hydrophobic molecules, we also measured the uptake of chenodeoxycholate, which is a lipophilic agent that has been widely used for assessing cell wall permeability to hydrophobic agents (18, 19). There is no difference in the rate and levels of uptakes of erythromycin or chenodeoxycholate between mutant and WT cells (data not shown), suggesting that the permeability of the mutant cell wall was not affected. Thin-layer chromatography analysis of mycolic acids, a major cell wall component that plays a critical role on cell wall permeability (18, 19), did not reveal any difference between the WT and the mutant (data not shown).
Mutant MS 2-39 is not an asparagine auxotroph.
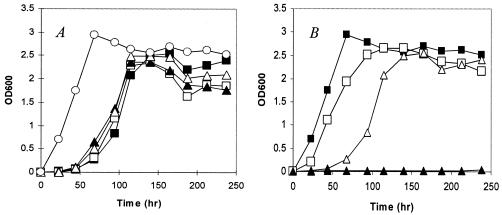
Some bacteria such as E. coli and Klebsiella aerogenes contain two types of asparagine synthetase (13, 25), AsnB and a mechanically distinct asparagine synthetase, AsnA, which utilizes ammonia as an amide donor. AsnB is the only asparagine synthetase found in mycobacterial genomes. To determine whether the asnB mutant is auxotrophic for asparagine, we compared the growth of the mutant in 7H9 supplemented with or without 10 mM asparagine. The result showed that supplementation of asparagine did not facilitate the growth of mutant (Fig. 3A). We also examined the effect of other amino acids including glutamine, glutamate, and aspartate, since the intracellular pools of these amino acids could be affected by an AsnB-mediated reaction (Fig. 2B). Like asparagine, supplementation of glutamine or glutamate had no effect on the growth of mutant MS 2-39 (Fig. 3A). Intriguingly, supplementation of aspartate inhibited the growth of the mutant; there was no viable growth at 37°C after 230 h (Fig. 3B). Supplementation of the same concentration of aspartate to 7H9 caused only a slight growth delay (∼20 h) of the WT strain (Fig. 3B). The mechanism of growth inhibition of aspartate is unknown.
FIG. 3.
Growth of mutant MS 2-39 in 7H9 supplemented with amino acids. (A) MS 2-39 was inoculated (106 cells/ml) in 7H9 supplemented with either asparagine (□), glutamine (▵), or glutamate (▴), each at 10 mM and incubated at 37°C. Growth of MS 2-39 (▪) and mc2155 (○) in 7H9 was shown for comparison. Supplementation of asparagine, glutamine, or glutamate did not facilitate the growth of MS 2-39, which showed a growth delay at the early stage. OD600, optical density at 600 nm. (B) Supplementation of aspartate (10 mM) to 7H9 inhibited the growth of MS 2-39 (▴). Only a slight delay of growth was observed for WT strain mc2155 (□).
DISCUSSION
In this study, we have identified a novel mechanism of natural resistance of mycobacteria. Wild-type M. smegmatis is naturally resistant to rifampin, erythromycin, novobiocin, and fusidic acid. We demonstrated that inactivation of the asnB gene in M. smegmatis confers hypersensitivity to these drugs, indicating that asnB is involved in the natural resistance of M. smegmatis to multiple drugs. To the best of our knowledge, the involvement of an amino acid metabolic enzyme in antibiotic resistance has not been described previously in mycobacteria.
The mutant MS 2-39 was isolated by screening the transposon insertion library for mutants that exhibited increased sensitivity to rifampin, erythromycin, or novobiocin. This mutant turned out to be hypersensitive to all three antibiotics examined, as well as fusidic acid. The transposon insertion was localized within the asnB gene, which is predicted to encode a glutamine-dependent asparagine synthetase. This was somewhat unexpected. However, asnB gene from the WT strain of M. smegmatis and asnB ortholog of M. tuberculosis fully complemented the mutant hypersensitive phenotype, confirming that inactivation of asnB is responsible for the enhanced drug sensitivity of the mutant.
Two families of asparagine synthetase have been reported. One is the AsnA family, whose members are found in prokaryotes such as E. coli and K. aerogenes (13, 25). AsnA is able to use only ammonia as the amino group donor. The other is the AsnB family, whose members are found in both prokaryotes and eukaryotes (12, 29, 30). Members of AsnB are able to use both glutamine and ammonia as the nitrogen donor, but glutamine is preferred. Structure and enzymatic mechanism have been studied extensively for AsnB of E. coli (5, 15). On the basis of sequence analyses and structural studies, AsnB has been shown to belong to a larger family of enzymes referred to as the purF family of glutamine amidotransferases. In all these enzymes, glutamine serves as the preferred source of nitrogen whereby an acceptor molecule is subsequently aminated. A cysteine residue at the N terminus is absolutely required for enzyme activity in the amidotransfer reaction (5, 30).
AsnB of mycobacteria shows a low level of sequence identity to AsnB of E. coli (29% amino acid identity), but it shares significant sequence identity with predicted AsnB of gram-positive bacteria including Rhodococcus, Nocardia, Corynebacterium, and Bacillus. AsnB of gram-positive bacteria has not been studied extensively. Nevertheless, an asnB mutant of Corynebacterium glutamicum was isolated as a lysozyme-sensitive mutant, hence its name ltsA (10). Intriguingly, wild-type asnB (or ltsA) gene of C. glutamicum did not complement asparagine auxotroph of the E. coli asnA asnB double mutant. Based on these observations, it was suggested that AsnB/LtsA of corynebacteria is not an asparagine synthetase; instead, it may catalyze the amidotransfer reaction from glutamine onto some unknown cell surface component(s) (10). AsnB of M. smegmatis shares 63% amino acid identity with AsnB/LtsA of C. glutamicum, suggesting that they may perform a similar function. Like the asnB mutant of C. glutamicum, which is temperature sensitive for growth at 37°C (10), the growth of the M. smegmatis asnB mutant was arrested at a high temperature (45°C). However, unlike the asnB mutant of C. glutamicum, which grows normally at a permissive temperature (30°C) (10), the growth of the M. smegmatis asnB mutant was affected at all temperatures; there was a growth delay at both 25 and 37°C, but once the growth commenced, the actual growth rates were normal. In addition, the asnB mutant of M. smegmatis did not exhibit detectable change on cell size, which was observed for the asnB mutant of C. glutamicum (10), and sensitivity to lysozyme remained unchanged compared to the WT M. smegmatis strain (data not shown).
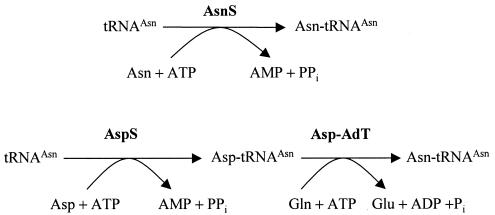
AsnB is the only putative asparagine synthetase found in mycobacterial genomes, and yet the M. smegmatis asnB mutant does not seem to be an asparagine auxotroph. Although the asnB mutant exhibits a delayed growth at 25 and 37°C in 7H9 medium, supplementation of asparagine (10 mM) does not facilitate the mutant growth. The apparent paradox could be explained by the presence of a tRNA-dependent transamidation mechanism (gatCAB) for the conversion of aspartate to asparagine in mycobacteria (Fig. 4) (8, 23). Such a mechanism has been shown to be an essential route to asparagine formation in organisms such as Thermus thermophilus and Deinococcus radiodurans, which lack both asnA and asnB (23, 26).
FIG. 4.
Pathways of Asn-tRNAAsn synthesis. The top reaction is the direct pathway, which synthesizes Asn-tRNAAsn with asparagine either imported or made from aspartate by the AsnA or AsnB enzymes. The bottom reaction is the transamidation pathway, which consists of a nondiscriminating AspS capable of generating the misacylated Asp-tRNAAsn, and a heterotrimeric Asp-tRNAAsn amidotransferase (Asp-AdT, encoded by the gatCAB genes). The genome of M. tuberculosis does not contain asnS, encoding asparaginyl-tRNA synthetase, but does contain the following genes for the transamidation pathway: aspS (Rv2572c), gatCAB (Rv3012c, Rv3011c, Rv3009c).
The mechanism by which AsnB confers antibiotic resistance in mycobacteria is currently unknown. AsnB could catalyze the synthesis of a cell wall component(s) other than asparagine, as suggested for AsnB of C. glutamicum (10). As such, inactivation of asnB disrupts the cell wall structure and confers hypersensitivity to antibiotics. On the other hand, the hypersensitivity of the M. smegmatis asnB mutant is restricted only to several hydrophobic drugs, and sensitivity of the mutant to a panel of other antibiotics including isoniazid, ethambutol, streptomycin, chloramphenicol, norfloxacin, tetracycline, and β-lactams was not affected. This is different from typical cell wall-deficient mutants, which exhibit increased sensitivity to a wide range of antibiotics, for example, a mutant of Pseudomonas aeruginosa in which GlmS (also a member of the purF family of glutamine amidotransferases) was affected (24). In addition, uptake experiments with erythromycin and chenodeoxycholate, as well as analysis of the mycolic acid composition of the mutant cell wall, did not reveal any difference between the WT and the mutant strains.
To our knowledge, there is no published literature on the involvement of ansB in antibiotic resistance. Interestingly, there appears to be an inverse relationship between the susceptibility of leukemia cells to cancer drug therapy and their capacity for intracellular asparagine biosynthesis (3, 4, 22). As a consequence, l-asparaginase, which catalyzes the hydrolysis of asparagine, is widely used in chemotherapeutic protocols for treating acute lymphoblastic leukemia.
In summary, our results indicate that the asnB is involved in the multidrug resistance of mycobacteria. This finding may provide a strategy for designing new drugs or novel combinations of drugs for the treatment of mycobacterial infections including TB.
Acknowledgments
We thank D. Alexander and J. Jones for construction and initial screening of the transposon library.
This work is supported by Canadian Institutes of Health Research (CIHR) grant MOP-15107 and a grant from National Sanitarium Association of Canada (to J.L.).
REFERENCES
- 1.Alexander, D. C., J. R. Jones, and J. Liu. 2003. A rifampin-hypersensitive mutant reveals differences between strains of Mycobacterium smegmatis and presence of a novel transposon, IS1623. Antimicrob. Agents Chemother. 47:3208-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, D. C., J. R. Jones, T. Tan, J. M. Chen, and J. Liu. 2004. PimF, a mannosyltransferase of mycobacteria, is involved in the biosynthesis of phosphatidylinositol mannosides and lipoarabinomannan. J. Biol. Chem. 279:18824-18833. [DOI] [PubMed] [Google Scholar]
- 3.Aslanian, A. M., B. S. Fletcher, and M. S. Kilberg. 2001. Asparagine synthetase expression alone is sufficient to induce l-asparaginase resistance in MOLT-4 human leukaemia cells. Biochem. J. 357:321-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aslanian, A. M., and M. S. Kilberg. 2001. Multiple adaptive mechanisms affect asparagine synthetase substrate availability in asparaginase-resistant MOLT-4 human leukaemia cells. Biochem. J. 358:59-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boehlein, S. K., N. G. Richards, and S. M. Schuster. 1994. Glutamine-dependent nitrogen transfer in Escherichia coli asparagine synthetase B. Searching for the catalytic triad. J. Biol. Chem. 269:7450-7457. [PubMed] [Google Scholar]
- 6.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 64:29-63. [DOI] [PubMed] [Google Scholar]
- 7.Colangeli, R., D. Helb, S. Sridharan, J. Sun, M. Varma-Basil, M. H. Hazbon, R. Harbacheuski, N. J. Megjugorac, W. R. J. Jacobs, A. Holzenburg, J. C. Sacchettini, and D. Alland. 2005. The Mycobacterium tuberculosis iniA gene is essential for activity of an efflux pump that confers drug tolerance to both isoniazid and ethambutol. Mol. Microbiol. 55:1829-1840. [DOI] [PubMed] [Google Scholar]
- 8.Curnow, A. W., D. L. Tumbula, J. T. Pelaschier, B. Min, and D. Soll. 1998. Glutamyl-tRNA(Gln) amidotransferase in Deinococcus radiodurans may be confined to asparagine biosynthesis. Proc. Natl. Acad. Sci. USA 95:12838-12843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Rossi, E., M. C. Blokpoel, R. Cantoni, M. Branzoni, G. Riccardi, D. B. Young, K. A. De Smet, and O. Ciferri. 1998. Molecular cloning and functional analysis of a novel tetracycline resistance determinant, tet(V), from Mycobacterium smegmatis. Antimicrob. Agents Chemother. 42:1931-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirasawa, T., M. Wachi, and K. Nagai. 2000. A mutation in the Corynebacterium glutamicum ltsA gene causes susceptibility to lysozyme, temperature-sensitive growth, and l-glutamate production. J. Bacteriol. 182:2696-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howard, N. S., J. E. Gomez, C. Ko, and W. R. Bishai. 1995. Color selection with a hygromycin-resistance-based Escherichia coli-mycobacterial shuttle vector. Gene 166:181-182. [DOI] [PubMed] [Google Scholar]
- 12.Hughes, C. A., H. S. Beard, and B. F. Matthews. 1997. Molecular cloning and expression of two cDNAs encoding asparagine synthetase in soybean. Plant Mol. Biol. 33:301-311. [DOI] [PubMed] [Google Scholar]
- 13.Humbert, R., and R. D. Simoni. 1980. Genetic and biomedical studies demonstrating a second gene coding for asparagine synthetase in Escherichia coli. J. Bacteriol. 142:212-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarlier, V., and H. Nikaido. 1994. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol. Lett. 123:11-18. [DOI] [PubMed] [Google Scholar]
- 15.Larsen, T. M., S. K. Boehlein, S. M. Schuster, N. G. Richards, J. B. Thoden, H. M. Holden, and I. Rayment. 1999. Three-dimensional structure of Escherichia coli asparagine synthetase B: a short journey from substrate to product. Biochemistry 38:16146-16157. [DOI] [PubMed] [Google Scholar]
- 16.Li, X. Z., L. Zhang, and H. Nikaido. 2004. Efflux pump-mediated intrinsic drug resistance in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 48:2415-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, J., C. E. Barry, and H. Nikaido. 1999. Cell wall: physical structure and permeability, p. 220-239. In C. Ratledge and J. W. Dale (ed.), Mycobacteria: molecular biology and virulence. Blackwell Science, Oxford, United Kindgom.
- 18.Liu, J., C. E. Barry, G. S. Besra, and H. Nikaido. 1996. Mycolic acid structure determines the fluidity of the mycobacterial cell wall. J. Biol. Chem. 271:29545-29551. [DOI] [PubMed] [Google Scholar]
- 19.Liu, J., and H. Nikaido. 1999. A mutant of Mycobacterium smegmatis defective in the biosynthesis of mycolic acids accumulates meromycolates. Proc. Natl. Acad. Sci. USA 96:4011-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, J., E. Y. Rosenberg, and H. Nikaido. 1995. Fluidity of the lipid domain of cell wall from Mycobacterium chelonae. Proc. Natl. Acad. Sci. USA 92:11254-11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, J., H. E. Takiff, and H. Nikaido. 1996. Active efflux of fluoroquinolones in Mycobacterium smegmatis mediated by LfrA, a multidrug efflux pump. J. Bacteriol. 178:3791-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Majlessipour, F., R. Kwock, S. Martin-Aragon, K. I. Weinberg, and V. I. Avramis. 2001. Development of a double-drug-resistant human leukemia model to cytosine arabinoside and L-asparaginase: evaluation of cross-resistance to other treatment modalities. Anticancer Res. 21:11-22. [PubMed] [Google Scholar]
- 23.Min, B., J. T. Pelaschier, D. E. Graham, D. Tumbula-Hansen, and D. Soll. 2002. Transfer RNA-dependent amino acid biosynthesis: an essential route to asparagine formation. Proc. Natl. Acad. Sci. USA 99:2678-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramos-Aires, J., P. Plesiat, L. Kocjancic-Curty, and T. Kohler. 2004. Selection of an antibiotic-hypersusceptible mutant of Pseudomonas aeruginosa: identification of the GlmR transcriptional regulator. Antimicrob. Agents Chemother. 48:843-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reitzer, L. J., and B. Magasanik. 1982. Asparagine synthetases of Klebsiella aerogenes: properties and regulation of synthesis. J. Bacteriol. 151:1299-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruan, B., I. Ahel, A. Ambrogelly, H. D. Becker, S. Bunjun, L. Feng, D. Tumbula-Hansen, M. Ibba, D. Korencic, H. Kobayashi, C. Jacquin-Becker, N. Mejlhede, B. Min, G. Raczniak, J. Rinehart, C. Stathopoulos, T. Li, and D. Soll. 2001. Genomics and the evolution of aminoacyl-tRNA synthesis. Acta Biochim. Pol. 48:313-321. [PubMed] [Google Scholar]
- 27.Silva, P. E., F. Bigi, S. de la Paz, M. I. Romano, C. Martin, A. Cataldi, and J. A. Ainsa. 2001. Characterization of P55, a multidrug efflux pump in Mycobacterium bovis and Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 45:800-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takiff, H. E., M. Cimino, M. C. Musso, T. Weisbrod, R. Martinez, M. B. Delgado, L. Salazar, B. R. Bloom, and W. R. J. Jacobs. 1996. Efflux pump of the proton antiporter family confers low-level fluoroquinolone resistance in Mycobacterium smegmatis. Proc. Natl. Acad. Sci. USA 93:362-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Heeke, G., and S. M. Schuster. 1989. Expression of human asparagine synthetase in Escherichia coli. J. Biol. Chem. 264:5503-5509. [PubMed] [Google Scholar]
- 30.Van Heeke, G., and S. M. Schuster. 1989. The N-terminal cysteine of human asparagine synthetase is essential for glutamine-dependent activity. J. Biol. Chem. 264:19475-19477. [PubMed] [Google Scholar]
- 31.Wang, L., R. A. Slayden, C. E. Barry, and J. Liu. 2000. Cell wall structure of a mutant of Mycobacterium smegmatis defective in the biosynthesis of mycolic acids. J. Biol. Chem. 275:7224-7229. [DOI] [PubMed] [Google Scholar]
- 32.Zalkin, H. 1993. The amidotransferases. Adv. Enzymol. Relat. Areas Mol. Biol. 66:203-309. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, Y., A. Scorpio, H. Nikaido, and Z. Sun. 1999. Role of acid pH and deficient efflux of pyrazinoic acid in unique susceptibility of Mycobacterium tuberculosis to pyrazinamide. J. Bacteriol. 181:2044-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]