Abstract
Lamivudine was the first approved inhibitor of hepatitis B virus (HBV) reverse transcriptase (RT). Lamivudine resistance develops in 53% to 76% of patients after 3 years of treatment. We extensively characterized the dynamics of HBV quasispecies variant populations in four HBV-infected patients who developed lamivudine resistance. Virological breakthrough was preceded by 2 to 4 months by the emergence of quasispecies variants bearing amino acid substitutions at RT position 204, i.e., within the YMDD catalytic motif (rtM204V/I). Three patients had a gradual switch from a YMDD variant population at baseline to a 100% lamivudine-resistant variant population, whereas the remaining patient had a fluctuating pattern of resistance variant dynamics. Careful analysis of amino acid substitutions located outside domain C of HBV RT, including those known to partially restore replication capacities in vitro, showed that the in vivo replication of HBV variants is driven by multiple forces, including intrinsic replicative advantages conferred by mutations accumulating outside domain C and the changing environment in which these variants replicate. Our findings also suggest that individual treatment optimization will require sensitive methods capable of detecting the emergence of viral resistance before the relevant variants acquire optimal replicative capacities.
Hepatitis B virus (HBV) infection is a major public health problem, with approximately 350 million individuals chronically infected worldwide (19). Chronic HBV carriers are exposed to a risk of complications, such as chronic hepatitis, cirrhosis, and hepatocellular carcinoma, of which HBV is currently the most frequent cause (13). Up to one million people die every year from complications of HBV infection (19).
HBV infection is characterized by high levels of virus production and turnover (28, 39), whereas the HBV reverse transcriptase (RT), like the human immunodeficiency virus (HIV) RT, is an error-prone enzyme lacking 3′-5′-exonuclease proofreading capacity (3, 14). As a result, HBV, like other viruses with error-prone polymerases, such as HIV, hepatitis C virus, and poliovirus, has a quasispecies distribution in infected individuals (14). This means that HBV circulates as a complex mixture of genetically distinct but closely related variants that are in equilibrium at a given time point of infection in a given replicative environment. The quasispecies distribution of HBV implies that any newly generated mutation conferring a selective advantage to the virus in a given replicative environment will allow the corresponding viral population to overtake the other variants, following a classical Darwinian evolutionary process (10).
Treatment of chronic hepatitis B is aimed at driving viral replication to the lowest possible level, and thereby to halt the progression of liver disease and prevent the onset of complications. However, HBV infection cannot be fully eradicated, because of covalently closed circular proviral DNA persistence in host cells. The first HBV RT inhibitor to be approved for the treatment of chronic hepatitis B was lamivudine (dideoxy-2′,3′-thiacytidine) (8, 9, 17, 22, 23). Its principal target is the YMDD catalytic motif of HBV RT, located in domain C of the polymerase molecule (5). Lamivudine exerts its anti-polymerase/RT activity by inhibiting the elongation of the HBV DNA minus strand through competition with the natural polymerase substrate dCTP and by acting as a chain terminator through its incorporation in the nascent DNA strand (37, 41). Lamivudine therapy results in a 3 to 4 log decline in plasma HBV DNA after a few weeks and also reduces disease activity and improves liver histological status (8, 9, 17, 22, 23). Long-term lamivudine administration frequently elicits viral resistance characterized by a reincrease of viral replication in an adherent patient. The incidence of lamivudine resistance is 14% to 32% after 1 year of treatment, 38% after 2 years, and 53% to 76% after 3 years (18). The principal mutations associated with lamivudine resistance are located in domain C of the YMDD motif. They include rtM204V (YVDD sequence), rtM204I (YIDD) (1), and the more recently identified rtM204S (YSDD) (2, 27). Lamivudine-resistant mutants with amino acid substitutions in the YMDD motif appear to replicate less efficiently than the wild-type virus in vitro. However, additional substitutions that are often coselected with the resistance substitutions at RT position 204 of domain C, such as rtL180M and rtV173L, which are located in the B domain, can compensate for this loss of replication efficiency in vitro (1, 6, 12, 36).
A proper understanding of the mechanisms underlying viral resistance to a specific inhibitor implies the need to unravel the quasispecies variant dynamics at work during the emergence and amplification of resistance, together with the role of the relevant amino acid substitutions in viral replication in vivo. In contrast to HIV and hepatitis C virus (4, 31, 32), little such information on resistance to specific RT inhibitors is available for HBV.
MATERIALS AND METHODS
Patients.
We studied four male patients, aged 39 to 52 years, who had chronic HBeAg-positive HBV infection (genotype A was present in two cases, genotype C was present in one case, and genotype E was present in one case) and who had developed viral resistance to lamivudine. Resistance was characterized by an initial decline in HBV DNA levels during lamivudine therapy followed by a reincrease despite full adherence to ongoing lamivudine therapy. All four patients were HIV seronegative and immunocompetent. Patients A, B, and C had not responded to previous treatment with standard alpha interferon, while patient D had not been treated before receiving lamivudine. All the patients received lamivudine monotherapy at 100 mg/day. Serum samples were taken at various time points before and during lamivudine therapy to monitor the virological response to lamivudine and HBV RT quasispecies dynamics (12 samples each for patients A and C, 18 samples each for patients B and D).
Methods.
Aspartate aminotransferase and alanine aminotransferase levels were determined before treatment and for each available sample taken during lamivudine therapy. HBV DNA levels were measured at each time point by means of a branched DNA-based assay (Versant HBV DNA 1.0 assay; Bayer Corporation, Tarrytown, N.Y.) with a lower detection limit of 125,000 IU/ml (5.1 log IU/ml). Serum samples testing negative in the branched DNA assay (HBV DNA level, <100,000 IU/ml or 5.0 log IU/ml) were retested with an in-house quantitative real-time PCR assay based on Lightcycler technology (Roche Molecular Systems, Pleasanton, California) with a lower detection limit of 50 IU/ml.
HBV RT quasispecies were characterized for all available samples. For this, DNA was extracted from 200 μl of serum and a 630-bp fragment encompassing domains A to E of the HBV RT was amplified with primers POL4b and PORV2 as previously described (35). When HBV DNA level was below 100,000 IU/ml, a nested PCR technique was used to amplify a 132-bp region spanning the YMDD catalytic motif of domain C. The first round consisted of the 630-bp amplification. The second round used upstream primer CO1 (5′CTGGGCTTCCGCAAAATACCT3′, nucleotide positions 619 to 639) and downstream primer CO2 (5′ ATAATGGGACTCACGATGCTG-3′, nucleotide positions 772 to 792). Purified PCR products were cloned, recombinant plasmid DNA was transformed into Escherichia coli competent cells, and transformants were grown on ampicillin plates. Cloned DNA was reamplified by PCR for sequencing. Approximately 30 clones per sample were sequenced. HBV RT clone nucleotide and amino acid sequences were aligned by using the CLUSTAL W program, version 1.5 (38). The PHYLIP program, version 3.572, was used to construct phylogenetic trees by means of the neighbor-joining method (11, 33) with a sequence matrix determined by the two-parameter method of Kimura. Phylogenetic trees were constructed for each patient.
Nucleotide sequence accession numbers.
Sequence data from this article have been deposited in GenBank under accession numbers AM073272 to AM073968.
RESULTS
Biochemical and virological outcomes.
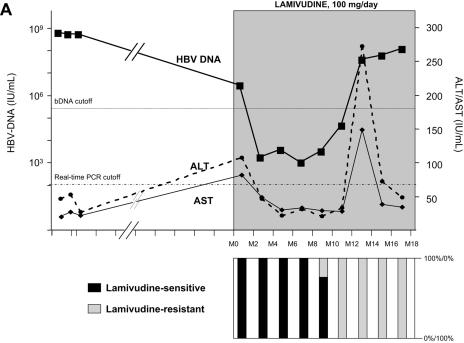
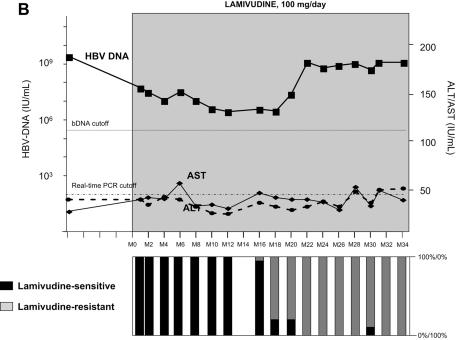
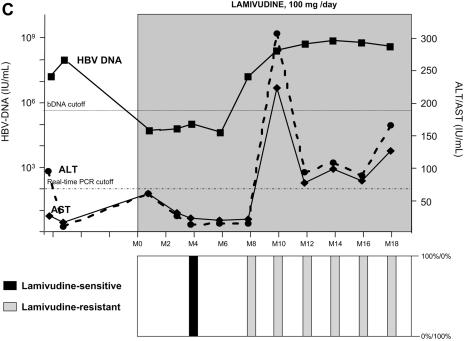
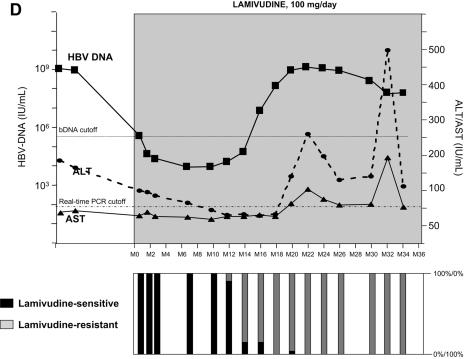
As shown in Fig. 1, lamivudine administration induced a sharp decline in HBV DNA in all four patients, ranging from −2.5 to −6.0 log IU of HBV DNA/ml. All patients, having started with high viral loads, maintained a relatively high residual level of HBV replication during lamivudine administration (Table 1). A breakthrough characterized by a significant increase in HBV DNA levels by more than 1.0 log above the nadir was observed in all four patients after several months of stable, reduced HBV DNA loads. HBV DNA levels returned to baseline within a few months in all four patients despite continued lamivudine therapy. Virological breakthrough was followed in three patients by biochemical breakthrough (alanine aminotransferase and aspartate aminotransferase peaks); this occurred when the HBV DNA level had reached baseline level in patients A and C and several months later in patient D. No biochemical breakthrough occurred in patient B (Fig. 1).
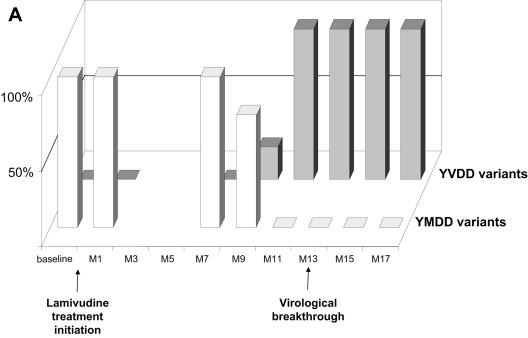
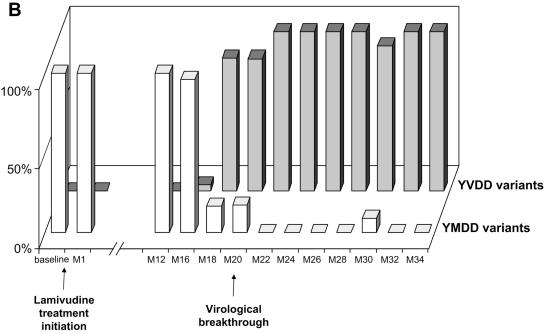
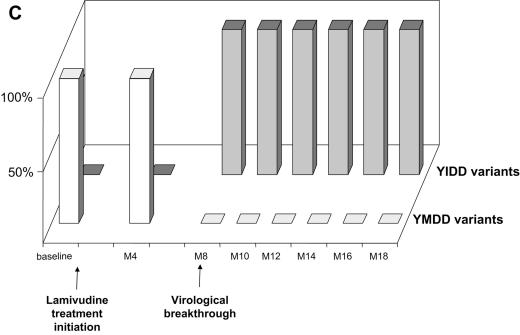
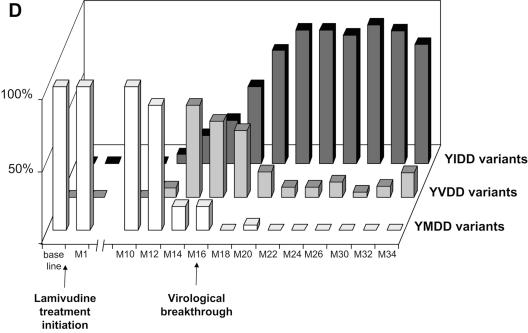
FIG. 1.
Dynamics of serum HBV DNA levels, serum aminotransferase activities, and lamivudine-sensitive and -resistant HBV viral populations before and during lamivudine therapy in patients A to D (panels A, B, C, and D, respectively). For each patient, the dynamics of HBV DNA and aminotransferase levels are shown at the top, while the dynamics of HBV variants are shown at the bottom as respective proportions of lamivudine-sensitive and lamivudine-resistant variant populations within the viral quasispecies at each time point. The lamivudine treatment period is shown in gray. The patients continued to receive lamivudine after this study. Abbreviations: bDNA, branched DNA; ALT, alanine aminotransferase; AST, aspartate aminotransferase; M, month.
TABLE 1.
Dynamics of HBV populations with mutations located outside the YMDD motif according to the amino acid at position 204 in the four patients during lamivudine therapya
| Patient | Time point | Amt of HBV DNA (log IU/ml) | YMDD | M204V | M204V + L180M | M204V + V173L | M204V + V207I | M204V + L180M + V173L | M204V + L180M + V207I | M204V + V173L + V207I | M204V + L180M + V173L + V207I | M204I | M204I + L180M | M204I + V173L | M204I + V207I | M204I + L180M + V173L | M204I + L180M + V207I | M204I + V173L + V207I | M204I + L180M + V173L + V207I |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | Start | 8.6 | 100.0% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| M1 | 6.4 | 100.0% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| M3 | 2.1 | 100.0% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| M5 | 3.5 | 100.0% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| M7 | 2.9 | 100.0% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| M9 | 3.4 | 78.5% | - | 21.5% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| M11 | 3.6 | - | - | 96.9% | - | - | 3.1% | - | - | - | - | - | - | - | - | - | - | - | |
| M13 | 7.5 | - | - | 96.8% | - | - | 3.2% | - | - | - | - | - | - | - | - | - | - | - | |
| M15 | 7.7 | - | - | 91.9% | - | - | 8.1% | - | - | - | - | - | - | - | - | - | - | - | |
| M17 | 8.0 | - | - | 93.8% | - | - | 6.2% | - | - | - | - | - | - | - | - | - | - | - | |
| B | Start | 9.4 | 100.0% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| M1 | 7.7 | 100.0% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| M2 | 7.4 | 100.0% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| M4 | 7.0 | 100.0% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| M6 | 7.5 | 100.0% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| M8 | 7.0 | 100.0% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| M10 | 6.6 | 100.0% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| M12 | 6.4 | 100.0% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| M16 | 6.5 | 96.2% | - | 3.8% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| M18 | 6.4 | 16.6% | 19.8% | 9.9% | - | - | 53.7% | - | - | - | - | - | - | - | - | - | - | - | |
| M20 | 7.3 | 17.0% | - | 20.6% | - | - | 62.4% | - | - | - | - | - | - | - | - | - | - | - | |
| M22 | 9.0 | - | - | 10.0% | - | - | 90.0% | - | - | - | - | - | - | - | - | - | - | - | |
| M24 | 8.7 | - | - | 13.6% | - | - | 86.4% | - | - | - | - | - | - | - | - | - | - | - | |
| M26 | 8.9 | - | - | 8.3% | - | - | 91.7% | - | - | - | - | - | - | - | - | - | - | - | |
| M28 | 9.0 | - | 3.6% | 14.4% | - | - | 82.0% | - | - | - | - | - | - | - | - | - | - | - | |
| M30 | 8.6 | 9.0% | 2.2% | 4.4% | - | - | 84.4% | - | - | - | - | - | - | - | - | - | - | - | |
| M32 | 9.1 | - | - | 3.6% | - | - | 96.4% | - | - | - | - | - | - | - | - | - | - | - | |
| M34 | 9.0 | - | - | 8.8% | - | - | 91.2% | - | - | - | - | - | - | - | - | - | - | - | |
| C | Start | 8.0 | 100.0% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| M4 | 5.0 | 100.0% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| M8 | 7.2 | - | - | - | - | - | - | - | - | - | 100.0% | - | - | - | - | - | - | - | |
| M10 | 8.4 | - | - | - | - | - | - | - | - | - | 96.9% | 3.1% | - | - | - | - | - | - | |
| M12 | 8.7 | - | - | - | - | - | - | - | - | - | 67.9% | 32.1% | - | - | - | - | - | - | |
| M14 | 8.9 | - | - | - | - | - | - | - | - | - | 49.9% | 50.1% | - | - | - | - | - | - | |
| M16 | 8.8 | - | - | - | - | - | - | - | - | - | 40.1% | 59.9% | - | - | - | - | - | - | |
| M18 | 8.6 | - | - | - | - | - | - | - | - | - | 31.3% | 68.7% | - | - | - | - | - | - | |
| D | Start | 8.8 | 100.0% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| M1 | 5.6 | 100.0% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| M2 | 4.6 | 100.0% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| M3 | 4.4 | 100.0% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| M7 | 4.0 | 100.0% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| M10 | 4.0 | 100.0% | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| M12 | 4.0 | 87.2% | - | 6.4% | - | - | - | - | - | - | 3.2% | - | - | 3.2% | - | - | - | - | |
| M14 | 4.7 | 16.7% | 11.1% | 52.8% | - | - | - | - | - | - | 8.3% | - | - | 11.1% | - | - | - | - | |
| M16 | 6.8 | 16.9% | 6.7% | 46.0% | - | - | - | - | - | - | 6.8% | - | - | 23.6% | - | - | - | - | |
| M18 | 8.1 | - | 6.8% | 39.7% | - | - | - | - | - | - | 36.6% | - | - | 16.9% | - | - | - | - | |
| M20 | 8.9 | 3.6% | - | 14.2% | - | - | - | - | - | - | 53.7% | - | 7.2% | 14.1% | - | - | 3.6% | - | |
| M22 | 9.0 | - | - | 6.8% | - | - | - | - | - | - | 69.2% | - | 10.2% | 13.8% | - | - | - | - | |
| M24 | 8.9 | - | - | 6.8% | - | - | - | - | - | - | 48.1% | 3.5% | 20.8% | 17.3% | 3.5% | - | - | - | |
| M26 | 8.8 | - | - | 10.4% | - | - | - | - | - | - | 31.0% | - | 20.6% | 38.0% | - | - | - | - | |
| M30 | 8.3 | - | - | 3.7% | - | - | - | - | - | - | 11.1% | 7.4% | 25.9% | 44.5% | - | 7.4% | - | - | |
| M32 | 7.7 | - | - | 3.7% | - | - | - | - | - | - | 11.1% | - | 26.0% | 44.4% | - | - | 7.4% | 3.7% | |
| M34 | 7.7 | - | - | 3.7% | 6.8% | - | 6.8% | - | - | - | 7.0% | - | 20.8% | 41.5% | 6.8% | - | - | 6.8% |
Each possible combination of the L180M, V173L, or V207I mutation with either the M204V or M205I mutation has been taken into consideration. The proportion of each variant population is shown as a percentage of the circulating quasispecies at each time point. The HBV DNA load at each time point is given in log international units/milliliter. M, month of therapy. -, no such population was found within the quasispecies at the indicated time point.
Quasispecies distribution of HBV RT.
A total of 1,710 sequences were generated and analyzed, allowing us to fully characterize nearly full-length HBV RT quasispecies at each time point before and during lamivudine therapy in the four patients. The risk of resampling errors, which may result from the analysis of quasispecies from a commonly sampled source that contains a small number of viral genomes (generally fewer than 100 viral copies/ml) (16, 24), was minimal in our study, in which HBV DNA levels remained high before as well as during lamivudine therapy (Table 1). Before treatment, the lamivudine-sensitive (YMDD) catalytic motif was fully conserved in all quasispecies variants of the four patients. No minor quasispecies populations were found to bear amino acid substitutions known to confer lamivudine resistance. Note that this did not mean that such variants were absent but that their prevalence in the quasispecies populations was very low. Sporadic amino acid substitutions were found in minor variants at various positions outside the YMDD motif.
Dynamics of HBV variants during lamivudine therapy: amino acid substitutions within the YMDD motif.
In all four patients, virological breakthrough was preceded by 2 to 4 months by the detection of quasispecies variants bearing amino acid substitutions within the YMDD motif that are known to confer lamivudine resistance (Fig. 1A to 1D, bottom panels). Gradually, these resistant variants, which initially represented a minority of the quasispecies, fully replaced the wild-type YMDD variants (Fig. 2A to 2D). This switch occurred in parallel to the increase in HBV DNA load. The absolute amount of resistant HBV variants increased exponentially such that they represented 100% of the viral quasispecies by the time the HBV DNA load had returned to baseline. In the case of patient B (Fig. 1B and 2B), HBV DNA load was low and stable at month 16, when 3.8% of circulating variants already bore the rtM204V substitution. Two months later, 83.4% of the variants bore the rtM204V substitution, but the HBV DNA load remained low. In contrast, at month 20, when the proportion of resistant variants was still high, HBV DNA load had increased by approximately 1 log, reflecting the significant increase in the absolute amount of circulating resistant variants. At month 22, the HBV DNA load had returned to baseline, and 100% of the variants bore the rtM204V substitution. The analytical sensitivity of the techniques used here allowed us to observe the last steps of the exponential expansion of preexisting resistant HBV variants in a context in which the replication of wild-type (sensitive) HBV variants was continually inhibited by lamivudine.
FIG. 2.
Dynamics of HBV variant populations before and during lamivudine therapy in patients A to D (panels A, B, C, and D, respectively). This figure illustrates the respective losses and gains of in vivo fitness by the different HBV populations over time, relative to treatment outset and virological breakthroughs. M, month.
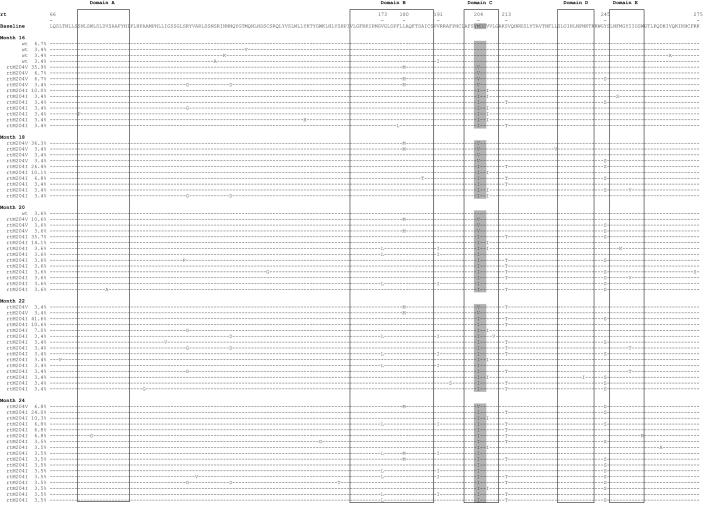
The dynamics of the RT quasispecies during lamivudine therapy (Fig. 2A to 2D) showed two distinct patterns. The first pattern, which was observed in patients A, B, and C, involved a simple, gradual switch from a 100% YMDD variant population at baseline and during the first months of lamivudine therapy to a 100% lamivudine-resistant variant population, including rtM204V (YVDD) in patients A and B and rtM204I (YIDD) in patient C (Fig. 1 and Table 1). In contrast, patient D had a much more complex, fluctuating pattern of resistance variant dynamics, in which the sensitive YMDD variant was gradually replaced by a resistant YVDD variant, which started to be detected 4 months before virological breakthrough (Fig. 2D). As the population of this resistant variant increased, another population of resistant variants bearing the YIDD motif also appeared (Fig 3). After month 14, the YVDD population started to decline and was gradually replaced by the YIDD population, which had apparently gained in replication capacity. A steady state was reached after a few months, with the YIDD and YVDD variants representing approximately 90% and 10% of circulating variants, respectively (Fig. 2D).
FIG. 3.
Distribution of HBV RT quasispecies at different time points, showing successive switches in viral populations between months 16 and 24 in patient D. At each time point, the variant amino acid sequences are shown, in order of decreasing frequency, within each population of catalytic motif variants (sensitive YMDD variants and resistant variants rtM204V and rtM204I), with their relative proportions among the quasispecies (percentages to the left of each sequence). The functional domains of the HBV RT are highlighted, and the catalytic motif YMDD is shaded. The reference sequence is the most frequent variant sequence at baseline. wt, wild type.
Dynamics of HBV variants during lamivudine therapy: amino acid substitutions outside the YMDD motif.
The principal substitutions outside the YMDD motif were rtV173L and rtL180M (located outside the C domain), which have been reported to partially restore the in vitro replication capacity of variants with substitutions in the YMDD motif (1, 6, 12, 36), and rtV207I, found within the C domain in patient D. These three substitutions were always associated with rtM204V or rtM204I, except for one minor variant detected at a single time point in patients B and D. The dynamics of the different HBV populations during lamivudine therapy are summarized in Table 1. (i) In patient A, the wild-type YMDD population was rapidly replaced by selected viruses bearing both the rtM204V and rtL180M substitutions. The latter appeared to confer a replication advantage in this patient's replicative environment. The rtV173L substitution did not appear to provide a significant replication advantage in this patient, as variants bearing all three substitutions, although detected early during virological breakthrough, remained present as a minor population until the end of follow-up. (ii) In contrast, in patient B, the HBV population bearing triple substitutions (rtM204V, rtL180M, and rtV173L) rapidly overtook HBV populations with single (rtM204V) or dual (rtM204V and rtL180M) substitutions, suggesting optimal replication capacities in patient B's specific replicative environment. (iii) In patient C, we saw a gradual switch from a situation where most variants bore only the rtM204I substitution to one where most variants bore both the rtM204I and rtL180M substitutions. This switch was not associated with a major gain in replication, as the HBV DNA load had already increased sharply when the single mutant was still predominant (Fig. 2C). (iv) The HBV quasispecies dynamics were far more complex in patient D (Fig. 2D, Fig. 3, and Table 1). Variants with both the rtM204V and rtL180M substitutions quickly overtook the YVDD population. No additional substitutions were selected in this mutated population, whose in vivo replication capacity gradually declined relative to the YIDD population. The gain in replication capacity of the YIDD population was apparently associated with the selection of a population bearing substitutions rtM204I and rtV173L and a population bearing substitutions rtM204I and rtV207I. These populations coexisted, representing the majority of circulating quasispecies variants, but variants bearing both mutations outside domain C were very rare.
In summary, the rtL180M substitution conferred a substantial replicative advantage to lamivudine-resistant variants in patient A but conferred only a moderate advantage in patient C. In patient D, the same substitution conferred a replicative advantage to YVDD variants, but this advantage was lost as YIDD populations gradually took over. The rtV173L substitution apparently had no effect in patient A, but it provided significant added replication capacity to YVDD variants in patient B and to YIDD variants in patient D, in whom the additional rtV207I substitution also contributed to takeover by the corresponding variants. Additional substitutions, such as rtS213T and rtY245S, were found outside known functional domains of HBV RT. Whether or not they influenced in vivo replication is unclear.
DISCUSSION
Our findings show that extensive analysis of HBV quasispecies dynamics aids understanding of the fine mechanisms of resistance to antiviral drugs. Experience with highly active antiretroviral therapy in HIV-infected patients shows that resistance to HIV RT inhibitors is acquired gradually, through the selection of preexisting resistant variants and gradual accumulation of new amino acid substitutions that confer stepwise increases in the level of drug resistance (reviewed in reference 4). Schematically, partial resistance conferred by a substitution in a preexisting viral population impairs drug efficacy sufficiently to restore a level of replication compatible with the accumulation of further mutations. The latter may restore the in vivo replication capacity of the resistant virus, allowing viral replication to return to pretreatment levels. There are some exceptions to this rule, however, such as the M184V substitution in the HIV RT, which immediately confers full resistance to lamivudine together with a significant fitness hit (34), and single mutations in the hydrophobic pocket of the HIV RT, which also confer strong resistance to nonnucleoside RT inhibitors (15). HBV resistance to lamivudine appears to be more closely related to the latter mechanism, for the following reasons: (i) HBV lamivudine resistance is characterized by a rapid, one-step reincrease in viral replication to baseline levels (18), as confirmed here; and (ii) full resistance can be conferred by a single substitution (V or I) at RT position 204, in the YMDD motif; this results in a more than 1,000-fold increase in resistance in vitro (1), and it led to a return to baseline replication levels in our patients B and C.
Our results suggest that rtM204V and rtM204I are frequent preexisting polymorphisms in HBV-infected patients and that they appear randomly in viral populations which have a replication disadvantage relative to YMDD variants in the absence of lamivudine. Their selection during lamivudine therapy appears to depend principally on their presence at the outset of treatment, as only rtM204V variants were selected in our patients A and B, while only rtM204I variants were selected in patient C. In patient D, who harbored both variants prior to treatment, the two were selected together, and their dynamics fluctuated as additional mutations accumulated. A steady state was reached when YIDD variants overtook YVDD variants, even though both variants were still present at the end of follow-up. It is theoretically possible, however, that YMDD variants could arise after the initiation of therapy, given the relatively high levels of residual replication during lamivudine administration (Table 1).
This study provides new information on the influence of amino acid substitutions located outside the C domain of HBV RT in vivo. Two substitutions, rtL180M and rtV173L, are frequently associated with the C domain during the onset of lamivudine resistance. Both partially restore the replication capacities of C-domain HBV mutants in vitro (1, 6, 12, 36). In our study, the in vivo replicative advantages conferred by these substitutions varied substantially from one patient to another and also from one time point to another in a given patient. Thus, the in vivo replication of HBV variants appears to be driven by multiple forces, including the following: (i) intrinsic replicative advantages potentially conferred to lamivudine-resistant variants by mutations accumulating outside domain C, such as rtL180M and rtV173L, and (ii) the fluctuating environment in which these variants replicate during lamivudine administration to a given patient, potentially favoring one variant population over another, regardless of their intrinsic replication capacity. Direct host protein-viral protein interactions and the host immune response probably play a key role in determining relative HBV variant replication capacities in vivo. As a result, the influence of specific resistance mutations on the development and establishment of HBV resistance to antiviral drugs in vivo cannot be inferred solely from in vitro replication experiments and cannot be fully predicted by structural analysis in silico.
Our results also have major implications for virological follow-up and diagnosis of resistance in the clinical setting. Currently, the most sensitive method for detecting early selection of resistant variants is to generate large numbers of clones at regular intervals during therapy. By analyzing approximately 30 clones per time point and carrying out a highly sensitive quantitative HBV DNA assay based on real-time PCR technology, we started to detect resistant variants between 2 and 4 months before virological breakthrough. However, this approach is too cumbersome to be used in clinical practice. Reverse hybridization with the line probe assay, which can detect variant populations representing approximately 10% of the HBV quasispecies, is usable in practice and was recently reported to reveal the emergence of resistance variants an average of 2 weeks before the HBV DNA load started to increase (26, 30). The time of virological breakthrough is too late for diagnosis of HBV resistance, because most of the quasispecies variants are already both resistant and highly fit. This is a particularly important issue, because lamivudine resistance is associated with gradual progression of liver disease (7, 20, 21, 23, 25, 29, 40).
REFERENCES
- 1.Allen, M. I., M. Deslauriers, C. W. Andrews, G. A. Tipples, K. A. Walters, D. L. Tyrrell, N. Brown, and L. D. Condreay. 1998. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Hepatology 27:1670-1677. [DOI] [PubMed] [Google Scholar]
- 2.Bozdayi, A. M., O. Uzunalimoglu, A. R. Turkyilmaz, N. Aslan, O. Sezgin, T. Sahin, G. Bozdayi, K. Cinar, S. B. Pai, R. Pai, H. Bozkaya, S. Karayalcin, C. Yurdaydin, and R. F. Schinazi. 2003. YSDD: a novel mutation in HBV DNA polymerase confers clinical resistance to lamivudine. J. Viral Hepat. 10:256-265. [DOI] [PubMed] [Google Scholar]
- 3.Cane, P. A., D. Mutimer, D. Ratcliffe, P. Cook, G. Beards, E. Elias, and D. Pillay. 1999. Analysis of hepatitis B virus quasispecies changes during emergence and reversion of lamivudine resistance in liver transplantation. Antivir. Ther. 4:7-14. [PubMed] [Google Scholar]
- 4.Clavel, F., and A. J. Hance. 2004. HIV drug resistance. N. Engl. J. Med. 350:1023-1035. [DOI] [PubMed] [Google Scholar]
- 5.Das, K., X. Xiong, H. Yang, C. E. Westland, C. S. Gibbs, S. G. Sarafianos, and E. Arnold. 2001. Molecular modeling and biochemical characterization reveal the mechanism of hepatitis B virus polymerase resistance to lamivudine (3TC) and emtricitabine (FTC). J. Virol. 75:4771-4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delaney, W. E., IV, H. Yang, C. E. Westland, K. Das, E. Arnold, C. S. Gibbs, M. D. Miller, and S. Xiong. 2003. The hepatitis B virus polymerase mutation rtV173L is selected during lamivudine therapy and enhances viral replication in vitro. J. Virol. 77:11833-11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dienstag, J. L., R. D. Goldin, E. J. Heathcote, H. W. Hann, M. Woessner, S. L. Stephenson, S. Gardner, D. F. Gray, and E. R. Schiff. 2003. Histological outcome during long-term lamivudine therapy. Gastroenterology 124:105-117. [DOI] [PubMed] [Google Scholar]
- 8.Dienstag, J. L., R. P. Perrillo, E. R. Schiff, M. Bartholomew, C. Vicary, and M. Rubin. 1995. A preliminary trial of lamivudine for chronic hepatitis B infection. N. Engl. J. Med. 333:1657-1661. [DOI] [PubMed] [Google Scholar]
- 9.Dienstag, J. L., E. R. Schiff, T. L. Wright, R. P. Perrillo, H. W. Hann, Z. Goodman, L. Crowther, L. D. Condreay, M. Woessner, M. Rubin, and N. A. Brown. 1999. Lamivudine as initial treatment for chronic hepatitis B in the United States. N. Engl. J. Med. 341:1256-1263. [DOI] [PubMed] [Google Scholar]
- 10.Duarte, E. A., I. S. Novella, S. C. Weaver, E. Domingo, S. Wain-Hobson, D. K. Clarke, A. Moya, S. F. Elena, J. C. de la Torre, and J. J. Holland. 1994. RNA virus quasispecies: significance for viral disease and epidemiology. Infect. Agents Dis. 3:201-214. [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 12.Fu, L., and Y. C. Cheng. 1998. Role of additional mutations outside the YMDD motif of hepatitis B virus polymerase in L(-)SddC (3TC) resistance. Biochem. Pharmacol. 55:1567-1572. [DOI] [PubMed] [Google Scholar]
- 13.Ganem, D., and A. M. Prince. 2004. Hepatitis B virus infection—natural history and clinical consequences. N. Engl. J. Med. 350:1118-1129. [DOI] [PubMed] [Google Scholar]
- 14.Gunther, S., L. Fischer, I. Pult, M. Sterneck, and H. Will. 1999. Naturally occurring variants of hepatitis B virus. Adv. Virus Res. 52:25-137. [DOI] [PubMed] [Google Scholar]
- 15.Hanna, G. J., V. A. Johnson, D. R. Kuritzkes, D. D. Richman, A. J. Brown, A. V. Savara, J. D. Hazelwood, and R. T. D'Aquila. 2000. Patterns of resistance mutations selected by treatment of human immunodeficiency virus type 1 infection with zidovudine, didanosine, and nevirapine. J. Infect. Dis. 181:904-911. [DOI] [PubMed] [Google Scholar]
- 16.Henderson, G. J., N. G. Hoffman, L. H. Ping, S. A. Fiscus, I. F. Hoffman, K. M. Kitrinos, T. Banda, F. E. Martinson, P. N. Kazembe, D. A. Chilongozi, M. S. Cohen, and R. Swanstrom. 2004. HIV-1 populations in blood and breast milk are similar. Virology 330:295-303. [DOI] [PubMed] [Google Scholar]
- 17.Lai, C. L., R. N. Chien, N. W. Leung, T. T. Chang, R. Guan, D. I. Tai, K. Y. Ng, P. C. Wu, J. C. Dent, J. Barber, S. L. Stephenson, and D. F. Gray. 1998. A one-year trial of lamivudine for chronic hepatitis B. N. Engl. J. Med. 339:61-68. [DOI] [PubMed] [Google Scholar]
- 18.Lai, C. L., J. Dienstag, E. Schiff, N. W. Leung, M. Atkins, C. Hunt, N. Brown, M. Woessner, R. Boehme, and L. Condreay. 2003. Prevalence and clinical correlates of YMDD variants during lamivudine therapy for patients with chronic hepatitis B. Clin. Infect. Dis. 36:687-696. [DOI] [PubMed] [Google Scholar]
- 19.Lee, W. M. 1997. Hepatitis B virus infection. N. Engl. J. Med. 337:1733-1745. [DOI] [PubMed] [Google Scholar]
- 20.Leung, N. W., C. L. Lai, T. T. Chang, R. Guan, C. M. Lee, K. Y. Ng, S. G. Lim, P. C. Wu, J. C. Dent, S. Edmundson, L. D. Condreay, and R. N. Chien. 2001. Extended lamivudine treatment in patients with chronic hepatitis B enhances hepatitis B e antigen seroconversion rates: results after 3 years of therapy. Hepatology 33:1527-1532. [DOI] [PubMed] [Google Scholar]
- 21.Liaw, Y. F., R. N. Chien, C. T. Yeh, S. L. Tsai, and C. M. Chu. 1999. Acute exacerbation and hepatitis B virus clearance after emergence of YMDD motif mutation during lamivudine therapy. Hepatology 30:567-572. [DOI] [PubMed] [Google Scholar]
- 22.Liaw, Y. F., N. W. Leung, T. T. Chang, R. Guan, D. I. Tai, K. Y. Ng, R. N. Chien, J. Dent, L. Roman, S. Edmundson, and C. L. Lai. 2000. Effects of extended lamivudine therapy in Asian patients with chronic hepatitis B. Gastroenterology 119:172-180. [DOI] [PubMed] [Google Scholar]
- 23.Liaw, Y. F., J. J. Sung, W. C. Chow, G. Farrell, C. Z. Lee, H. Yuen, T. Tanwandee, Q. M. Tao, K. Shue, O. N. Keene, J. S. Dixon, D. F. Gray, and J. Sabbat. 2004. Lamivudine for patients with chronic hepatitis B and advanced liver disease. N. Engl. J. Med. 351:1521-1531. [DOI] [PubMed] [Google Scholar]
- 24.Liu, S. L., A. G. Rodrigo, R. Shankarappa, G. H. Learn, L. Hsu, O. Davidov, L. P. Zhao, and J. I. Mullins. 1996. HIV quasispecies and resampling. Science 273:415-416. [DOI] [PubMed] [Google Scholar]
- 25.Lok, A. S., C. L. Lai, N. Leung, G. B. Yao, Z. Y. Cui, E. R. Schiff, J. L. Dienstag, E. J. Heathcote, N. R. Little, D. A. Griffiths, S. D. Gardner, and M. Castiglia. 2003. Long-term safety of lamivudine treatment in patients with chronic hepatitis B. Gastroenterology 125:1714-1722. [DOI] [PubMed] [Google Scholar]
- 26.Lok, A. S., F. Zoulim, S. Locarnini, A. Mangia, G. Niro, H. Decraemer, G. Maertens, F. Hulstaert, K. De Vreese, and E. Sablon. 2002. Monitoring drug resistance in chronic hepatitis B virus (HBV)-infected patients during lamivudine therapy: evaluation of performance of INNO-LiPA HBV DR assay. J. Clin. Microbiol. 40:3729-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niesters, H. G., R. A. De Man, S. D. Pas, E. Fries, and A. D. Osterhaus. 2002. Identification of a new variant in the YMDD motif of the hepatitis B virus polymerase gene selected during lamivudine therapy. J. Med. Microbiol. 51:695-699. [DOI] [PubMed] [Google Scholar]
- 28.Nowak, M. A., S. Bonhoeffer, A. M. Hill, R. Boehme, H. C. Thomas, and H. McDade. 1996. Viral dynamics in hepatitis B virus infection. Proc. Natl. Acad. Sci. USA 93:4398-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papatheodoridis, G. V., E. Dimou, A. Laras, V. Papadimitropoulos, and S. J. Hadziyannis. 2002. Course of virologic breakthroughs under long-term lamivudine in HBeAg-negative precore mutant HBV liver disease. Hepatology 36:219-226. [DOI] [PubMed] [Google Scholar]
- 30.Pas, S. D., R. A. de Man, E. Fries, A. D. Osterhaus, and H. G. Niesters. 2002. The dynamics of mutations in the YMDD motif of the hepatitis B virus polymerase gene during and after lamivudine treatment as determined by reverse hybridisation. J. Clin. Virol. 25:63-71. [DOI] [PubMed] [Google Scholar]
- 31.Pawlotsky, J. M. 2003. Hepatitis C virus genetic variability: pathogenic and clinical implications. Clin. Liver Dis. 7:45-66. [DOI] [PubMed] [Google Scholar]
- 32.Pawlotsky, J. M. 2003. Mechanisms of antiviral treatment efficacy and failure in chronic hepatitis C. Antivir. Res. 59:1-11. [DOI] [PubMed] [Google Scholar]
- 33.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 34.Schuurman, R., M. Nijhuis, R. van Leeuwen, P. Schipper, D. de Jong, P. Collis, S. A. Danner, J. Mulder, C. Loveday, and C. Christopherson. 1995. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC). J. Infect. Dis. 171:1411-1419. [DOI] [PubMed] [Google Scholar]
- 35.Seigneres, B., C. Pichoud, S. S. Ahmed, O. Hantz, C. Trepo, and F. Zoulim. 2000. Evolution of hepatitis B virus polymerase gene sequence during famciclovir therapy for chronic hepatitis B. J. Infect. Dis. 181:1221-1233. [DOI] [PubMed] [Google Scholar]
- 36.Seigneres, B., C. Pichoud, P. Martin, P. Furman, C. Trepo, and F. Zoulim. 2002. Inhibitory activity of dioxolane purine analogs on wild-type and lamivudine-resistant mutants of hepadnaviruses. Hepatology 36:710-722. [DOI] [PubMed] [Google Scholar]
- 37.Severini, A., X. Y. Liu, J. S. Wilson, and D. L. Tyrrell. 1995. Mechanism of inhibition of duck hepatitis B virus polymerase by (−)-β-l-2′,3′-dideoxy-3′-thiacytidine. Antimicrob. Agents Chemother. 39:1430-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whalley, S. A., J. M. Murray, D. Brown, G. J. Webster, V. C. Emery, G. M. Dusheiko, and A. S. Perelson. 2001. Kinetics of acute hepatitis B virus infection in humans. J. Exp. Med. 193:847-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuen, M. F., T. Kato, M. Mizokami, A. O. Chan, J. C. Yuen, H. J. Yuan, D. K. Wong, S. M. Sum, I. O. Ng, S. T. Fan, and C. L. Lai. 2003. Clinical outcome and virologic profiles of severe hepatitis B exacerbation due to YMDD mutations. J. Hepatol. 39:850-855. [DOI] [PubMed] [Google Scholar]
- 41.Zoulim, F., E. Dannaoui, C. Borel, O. Hantz, T. S. Lin, S. H. Liu, C. Trepo, and Y. C. Cheng. 1996. 2′,3′-Dideoxy-β-l-5-fluorocytidine inhibits duck hepatitis B virus reverse transcription and suppresses viral DNA synthesis in hepatocytes, both in vitro and in vivo. Antimicrob. Agents Chemother. 40:448-453. [DOI] [PMC free article] [PubMed] [Google Scholar]