Abstract
The ability of many retroviruses to cause disease can be correlated to their cytopathic effect (CPE) in tissue culture characterized by an acute period of cell death and viral DNA accumulation. Here, we show that mutants of a subgroup B avian retrovirus (Alpharetrovirus) cause a very dramatic CPE in certain susceptible avian cells that is coincident with elevated levels of apoptosis, as measured by nuclear morphology, and persistent viral DNA accumulation. These mutants also have a broadly extended host range that includes rodent, cat, dog, monkey, and human cells (31). Previously, we have shown that the mutants exhibit diminished resistance to superinfection. The results presented here have important implications for the process of evolution of retroviruses to use distinct cellular receptors.
Avian retroviruses cause a variety of diseases in vivo. Subgroup B and D avian (Alpharetrovirus) retroviruses and reticuloendotheliosis virus cause a wasting syndrome in chickens characterized by anemia (36), stunted growth, and atrophy of lymphoid organs leading to immunosuppression. Other alpharetroviruses have been shown to cause obesity, metabolic defects, myocarditis, osteopetrosis, and immunosuppression, as well as a variety of malignancies (30).
Subgroup B, D, and F alpharetroviruses cause plaques in cultured cells overlaid with agar, while subgroup A, C, and E viruses cannot (12, 14, 20, 23). While most studies defining these subgroups as cytopathic have relied on plaque assays, some reports have described a cytopathic effect (CPE) exhibited by cells grown in normal liquid culture. Cells infected with reticuloendotheliosis virus and some isolates of subgroup B, D, and F alpharetroviruses exhibit a generalized CPE characterized by a transient decrease in cell number in the first few days postinfection, followed by normal growth. The CPE correlates well with a transient accumulation of integrated and unintegrated viral DNA in cells infected with these viruses (21, 38, 42, 43). An early study of subgroup B alpharetrovirus-induced CPE documented cells that exhibited rounding up, detachment, and DNA laddering (43), features now known to be characteristic of cells undergoing apoptosis (19).
Viral DNA accumulation can be correlated with CPE and apoptosis in other groups of retroviruses as well. Variants of feline leukemia virus that cause feline AIDS show accumulation of both unintegrated and integrated viral DNA in vivo (25, 33). Similar mutants cause CPE in cultured cells resulting in a 1,000-fold decrease in cell number, and this effect probably maps to the SU subunit of the envelope (Env) glycoprotein (26). Similarly, T cells infected with human immunodeficiency virus (HIV) IIIB accumulate up to 80 copies of viral DNA per cell, while cells treated with neutralizing antibody after infection only accumulate 4 copies (32), again implying a role for SU in the process (8, 9). A T-cell line infected with HIV-1 accumulated viral DNA and exhibited CPE, but treatment with zidovudine to inhibit reinfection abrogated the CPE without having a major effect on HIV production by infected cells (29). Mink cell focus-forming murine leukemia virus has been shown to cause accumulation of unintegrated viral DNA as cells undergo CPE involving apoptosis (44). Recent reports describe the role of mutations in variable regions of murine leukemia virus in expanding host range and increasing syncytium formation and cytopathogenicity (17, 18).
Receptors for subgroup B, D, and E alpharetroviruses have been cloned and shown to belong to the tumor necrosis factor (TNF) receptor superfamily (1, 2, 7). These receptors are encoded by various alleles of the gene tv-b in chickens and homologous genes in other avian species. These proteins contain extracellular cysteine-rich domains and cytoplasmic death domains typical of death receptors. The subgroup B receptor can mediate apoptosis in cells stimulated by subgroup B SU, demonstrating that the death domain is functional and that virus binding can trigger a proapoptotic signal (11). Further, incubation of cells susceptible to alpharetrovirus subgroup B-mediated killing with nonsusceptible cells expressing subgroup B env led to cell death characterized by apoptosis (11). These results have led to the hypothesis that ALV-B kills cells as a result of signals mediated by Env-receptor interaction and that infected cells can kill uninfected cells in a “bystander killing” model (11). In contrast, another report has recently shown that alpharetrovirus cytopathic effects can be triggered in the absence of death receptor signaling (22). Further, a noncytopathic subgroup E SU can send proapoptotic signals in the presence of cycloheximide and subgroup E receptor (6). All tv-b-related receptors cause apoptosis when overexpressed, presumably by clustering of death domains in a manner similar to normal signaling resulting from cross-linking of death receptors by natural ligands (6). These findings suggest that receptor interaction is involved in the CPE documented for subgroup B alpharetroviruses. The ability of subgroup E receptor to send proapoptotic signals despite its inability to cause a CPE underscores that receptor interaction is not the whole story. Other factors, such as superinfection and accumulation of viral DNA, must be involved in making the CPE subgroup specific.
Recently, CPE resulting from alpharetrovirus infection has been shown to occur in the absence of death receptors (22). This effect can be caused by alpharetroviruses of subgroups A, B, and E and has been documented under conditions of high receptor expression and with viruses that grow to very high titers. In these experiments, elevated levels of viral DNA are strongly correlated with the observed CPE. This study demonstrates that CPE can result from high levels of infection and DNA accumulation alone but indicate that under normal replicative conditions, the receptor probably plays a role in concert with viral DNA to send coordinated signals leading to apoptosis and death (22).
Previously, we have described two groups of variants that expand host range. The first of these variant viruses is NTRE4, a recombinant between Prague Rous sarcoma virus B (PrB), and Rous-associated virus 0, with a subgroup B/E chimeric envelope that has the combined host range of these subgroups. NTRE4 reciprocally interferes with superinfection by viruses from both of its parental subgroups, indicating that it is capable of using both a subgroup B and E receptor (13, 41). The second virus, PrB LT154/155SI, expands host range to avian cells lacking tv-b, as well as to mammalian cells (37). We have shown that the L154S mutation is sufficient to expand host range, while the T155I mutant has the same host range as wild-type PrB. Infection of cells with L154S and LT154/155SI induces strong interference to subgroup B virus, but interference is diminished in the reciprocal situation (31). This result implies that host range extension mutants retain the ability to use the subgroup B receptor but can exploit an alternate means of entry.
In this work, we characterize CPE caused by host range extension mutants of subgroup B alpharetrovirus. We measure viral replication, cell viability, nuclear morphology, and viral DNA accumulation in a single course of infection for the host range extension mutants compared to classic cytopathic and noncytopathic alpharetroviruses. We found that the host range extension mutants cause a dramatic CPE in DF1 cells, an immortalized line of chicken embryo fibroblasts (CEFs), but not in a CEF cell strain. The level of CPE correlated with elevated levels of viral DNA accumulation and apoptosis, as measured by nuclear morphology. Finally, we show that a small fraction of infected DF1 cells survives the initial infection but becomes sensitive to induction of apoptosis by cycloheximide treatment, as do cells chronically infected with wild-type (WT) subgroup B and host range extension mutant viruses.
MATERIALS AND METHODS
Cells and viruses.
All cells were grown in modified Richter's minimal essential medium (Tufts Formulation, Irvine Scientific) supplemented with 10% fetal calf serum (Sigma). C/E CEFs were prepared from fertilized eggs from Lansing line 0 chickens (USDA Poultry Station, East Lansing, MI). QT6 cells (QT6/BD) are a continuous cell line derived from a fibrosarcoma induced by methylcholanthrene in Japanese quail (24). Q24 cells (a generous gift from Jürgen Brojatsch) are QT6 cells that have been stably transfected with a vector expressing the tv-bs3 gene, encoding susceptibility to subgroup B and D viruses (31). DF1 cells are a continuously dividing C/E line derived from a spontaneous transformant of Lansing line 0 CEFs (3, 15, 34). Viruses were harvested from chronically infected Q24 cells as previously described (31).
Infections.
Cells were infected with equal reverse transcriptase (RT) units of virus for 1 h at 37°C in the presence of 1.5-μg/ml polybrene (Aldrich). No polybrene was used for RCASBP(A) infections (40). From the measurement of viral production over time (Fig. 1), it is clear that a minority of cells was infected in the initial round, given that it took 6 to 8 days (CEF) or 8 to 12 days (DF1) for the cultures to become completely infected and for viral production to plateau. Cells were passaged every 3 to 4 days, when mock-infected cultures were confluent. At each passage, cells were counted and replated at a standard concentration (1 × 106 or 2 × 106 cells/100-mm dish). At each passage and every 1 to 2 days between passages, adherent and nonadherent cells were pooled and processed for Hoechst staining and DNA isolation, and medium was sampled for RT assays. Material for all assays was obtained from the same set of infected cells over a single time course experiment.
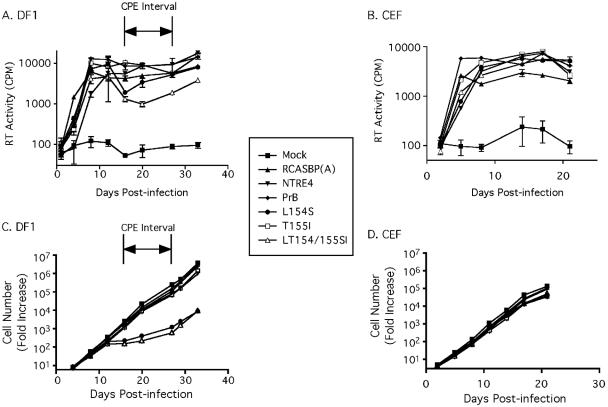
FIG. 1.
Viral replication and cell growth. Cells were infected and passaged at equal density every 3 to 4 days. RT activity was measured and plotted to give viral growth curves for the DF1 cell line (A) and a CEF cell strain (B). The average of three viable cell counts was used to determine the relative increase in cell number for the DF1 cells (C) and CEF cells (D). Results are the average of three replicates (± standard error of the mean [SEM]) from a single experiment, representative of three independent experiments. The interval during which the strongest CPE was evident for DF1 cells infected with L154S and LT154/155SI is indicated (A and C).
RT assay.
Virus-containing cell supernatant was harvested and frozen in 10-μl aliquots. All samples to be compared to each other were assayed on the same day. A total of 50 μl of reverse transcriptase assay buffer, consisting of 50 mM Tris (pH 8.3), 1% β-mercaptoethanol, 10 mM MgCl2, 6.5 mM NaCl, 100 μM ATP, 0.2% NP-40, 0.91-U/ml poly(A)/oligo(dT) (Sigma), and 2-μl/ml [α-32P]dTTP (3,000 Ci/mmol) was added to each 10-μl sample of virus and incubated at 37°C for 1 h. The samples were then transferred to DEAE filters and washed under vacuum with 2× SSC (0.3 M NaCl and 0.03 M Na citrate; 1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Incorporated radioactivity was determined by scintillation counting.
Total cellular DNA isolation.
Adherent and nonadherent cells were pooled, washed in phosphate-buffered saline (PBS), resuspended in lysis buffer (10 mM Tris [pH 7.5]-1 mM EDTA [TE], 100 mM NaCl, 1-mg/ml pronase, 1% sodium dodecyl sulfate), and incubated at 37°C for 1 to 4 days. The samples were extracted with phenol:chloroform:isoamyl alcohol (25:24:1) and then chloroform:isoamyl alcohol (24:1) and ethanol precipitated. The pellets were dissolved and treated with RNase (TE with 100-μg/ml RNase A and 0.1% sodium dodecyl sulfate), extracted, precipitated as described above, and dissolved in TE. DNA concentration was assayed, adjusted to a standard concentration, and assayed again to determine that all concentrations were equal.
Southern blotting and hybridization.
A total of 5 μg of genomic DNA was subjected to electrophoresis through 0.8% agarose gels at 20 V for 20 to 30 h at 4°C. Linearized PrB plasmid DNA standards were run on each gel for quantitation. The gels were soaked in 0.25 N HCl, in denaturation solution (1.5 M NaCl, 0.5 N NaOH), twice in neutralization buffer (1 M Tris [pH 7.5], 1.5 M NaCl), and finally in 10× SSC. The DNA was transferred to charged nylon membranes (Schleicher & Schuell) and UV cross-linked using a Stratalinker (Stratagene). The 1.2-kb SacII gag fragment from PrB plasmid DNA was labeled by random priming using the PrimeIt RmT kit (Stratagene) according to the manufacturer's instructions. The probe was purified on a Sephadex G50 spin column (Roche) and 2 × 106 counts/min/ml were used for hybridization overnight at 42°C in 50% formamide. The blots were washed with a final stringency of 0.1× SSC at 65°C/15 min and exposed to phosphorimager screens. Using ImageQuant 2 software (Molecular Dynamics), intensities of integrated and unintegrated species of viral DNA were obtained and compared to a standard curve of viral DNA loaded onto every gel.
Hoechst staining.
Adherent and nonadherent cells were pooled and fixed in 0.5% glutaraldehyde-1× PBS for 5 min and then stained in 1-μg/ml Hoechst dye-1× PBS for 3 min. The cells were then washed twice in 1× PBS and mounted on slides to be scored by fluorescence microscopy. For each sample, the percentage of morphologically apoptotic nuclei was scored by averaging three counts of at least 200 cells. The results were graphed using Prism 3 (Graphpad). To represent the total amount of apoptosis that had occurred in each sample over the course of the experiment, the integral of each curve was calculated from time zero until time x and the cumulative area under the curve was plotted as a function of time using Prism 3 (Graphpad).
Cycloheximide treatment.
Chronically infected DF1 cells were plated at 2 × 106 cells/100-mm dish and allowed to attach overnight. Cycloheximide (10 μg/ml) was added to half of the dishes, and adherent and nonadherent cells were harvested at 24-h intervals, pooled, and processed for Hoechst staining as described above.
RESULTS
Growth and rebound of mutant viruses.
To compare the growth of virus on DF1, a cell line derived from CEF, with that on a related cell strain, we infected CEF and DF1 cells with the wild-type, recombinant, and mutant viruses and used RT assays to determine virus production over time, allowing us to measure and compare their rate of spread. With the data shown in Fig. 1A and B, we demonstrate the growth curves of NTRE4, PrB, L154S, T155I, and LT154/155SI, as well as that of a noncytopathic subgroup A control [RCASBP(A)]. In both cell types, the viruses infected a minority of cells in the first round of infection and then spread throughout the cell cultures, reaching a peak level at 6 to 12 days postinfection. In DF1 cells, L154S and LT154/155SI showed reduced RT activity 16 to 27 days postinfection, immediately following the peak level of virus production. This reduction in RT concentration occurred despite normalization of viable cell concentrations among all infected cultures each time they were passaged.
The reduction in virus concentration in DF1 cultures infected with L154S and LT154/155SI coincided with a steep decrease in the growth rate of the infected cells (Fig. 1C and D). In DF1 cells infected with L154S and LT154/155SI, viable cells increased only about fivefold during the CPE interval. The DF1 cells that were left uninfected or infected with other viruses, by contrast, grew by an average of 300 fold during this time. DF1 cells infected with the host range extension point mutants had largely recovered by 27 days postinfection, doubling during the 2 days following this time point, identical to cells infected with the other viruses. CEF cells, on the other hand, showed no major difference in viability among any of the infected cultures and the uninfected control, even up to 2 weeks after virus production had plateaued in the cultures. It is noteworthy that we did not observe any CPE in CEF or DF1 cells infected with wild-type PrB. It is important to bear in mind that recent studies of CPE with wild-type subgroup B viruses have been conducted with RCAS vectors, which have been engineered to replicate to very high levels. The studies presented here use a viral isolate (PrB), which replicates to much lower levels and only has very mild CPE in infected cells, requiring agar overlays and neutral red staining for reproducible CPE measurement (14, 20, 23). Therefore, the CPE that we observed with host-range-extended mutant PrB viruses in infected cultures represents a dramatic increase in cytopathogenicity, allowing clear direct observation of CPE in a manner similar to, and exceeding in magnitude, that which has been observed with engineered RCAS-based viruses.
Next, we documented visual characteristics of cells infected with the panel of viruses throughout the time course study. Figure 2 shows the cell cultures at the onset of the CPE interval for DF1 and for an analogous time point for CEF. It was clear that cultures of DF1 cells infected with L154S and LT154/155SI were much more sparsely populated and a higher proportion of the cells were rounded up than in the other cultures. DF1 cells infected with the other viruses and all of the CEF cultures were similar to mock-infected cultures. Notably, no syncytia were observed during the course of these experiments, indicating that syncytium induction could not account for the decrease in cell viability observed.
FIG. 2.
Micrographs of infected cells. Cells were examined using phase-contrast microscopy and photographed with a charge-coupled device camera. Typical fields are shown for cells infected with each virus for DF1 (16 days) and CEF (14 days) cells. Black bars, 100 μm.
Apoptosis in infected cultures.
Brojatsch et al. (7) reported that cells infected with subgroup B virus undergo apoptosis under some circumstances. Given that DF1 cells infected with the extended-host-range mutant viruses exhibited many characteristics of apoptosis, including rounding up, shriveling, and detachment from the plate, we decided to examine the levels of apoptosis in our system. At regular intervals, adherent and nonadherent cells were collected, pooled, and scored for nuclear morphology consistent with apoptosis by being stained with Hoechst 33342 dye (Fig. 3). L154S- and LT154/155SI-infected DF1 cells showed elevated levels of apoptosis, particularly during the time period during which the CPE was most visible. Uninfected DF1 cells (Fig. 3A) had a background level of around 1.5% apoptotic nuclei at any given time, ranging from 0.5% to 2.5% positive nuclei over the course of the assay. L154S-infected cells exhibited elevated levels of apoptosis throughout the entire time course, peaking at 8%. LT154/155SI-infected cells underwent apoptosis at background levels except during the CPE period, where they peaked at 5%. This level of apoptosis was four- to sixfold over background. CEFs (Fig. 3B) did not exhibit any differences in apoptosis levels among infected cells and uninfected controls, consistent with the similar rate of viable cell growth in all cultures (Fig. 1D). The background level of apoptosis in these cells was higher than in DF1, averaging 2.5% in both mock and infected cells, with values ranging from 1% to 4% apoptosis.
FIG. 3.
Apoptosis in infected cultures. Adherent and nonadherent cells were pooled and stained with Hoechst 33342 dye. Apoptotic nuclei were scored as a percentage of total nuclei by fluorescence microscopy. (A) DF1 cell line. The interval with the strongest CPE is indicated. (B) CEF cell strain. Results are the average of three counts (± SEM) from a single experiment, representative of two independent experiments. The area under the curve was determined to indicate cumulative apoptosis levels (arbitrary units) in infected cells and plotted versus time for DF1 (C) and CEF (D) cells.
To more clearly demonstrate the differences in apoptosis among DF1 cells infected with L154S and LT154/155SI versus the other viruses, we integrated (determined the area under the curve) the apoptosis plots around the time period flanking maximal CPE (Fig. 3C), yielding a cumulative sum of apoptosis over time. Cumulative apoptosis in uninfected cells and cells infected with RCASBP(A), NTRE4, PrB, and T155I increased linearly with time at very similar rates, reflecting the low constant level of apoptosis observed with these cultures. Infection with L154S and LT154/155SI, on the other hand, led to a clear increase in slope preceding the onset of the CPE interval and leveling off just prior to the cells returning to normal growth. The correlation between apoptosis and the CPE interval clearly showed that the events were related. A similar analysis of infected CEF cells showed no differences among infected and uninfected cultures (Fig. 3D).
Accumulation of viral DNA.
Viral DNA accumulation is a hallmark of CPE caused by alpharetroviruses and other retroviruses (18, 21, 25, 29, 42-44). In situations in which CPE can be observed for subgroup B viruses, viral DNA accumulation accompanies this phase and resolves as the effect goes away. It is believed that this accumulation of DNA results from reduced superinfection resistance early after infection, since incubation of infected cells with neutralizing antibodies blocks both accumulation of viral DNA and the cytopathic effect (42, 44). To determine whether DNA accumulation in our mutants correlated with CPE, we examined the DNA forms present in cells infected with our panel of viruses. We harvested total cellular DNA from pooled adherent and nonadherent cells throughout the course of infection and analyzed it by gel electrophoresis without restriction digestion, followed by Southern blotting and hybridization with a virus-specific probe (Fig. 4A). This assay was used to determine the levels of integrated and unintegrated viral DNA present over time. Figure 4B to E shows quantitation of these results normalized to a loading control present on every gel.
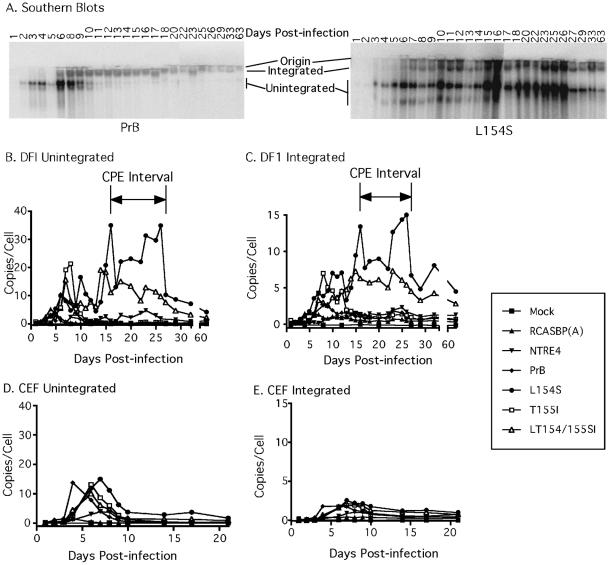
FIG. 4.
Accumulation of viral DNA. Southern blots of total cellular DNA were hybridized to a labeled gag probe. Data from DF1 cells infected with PrB and L154S are shown (A), and these as well as the others were quantitated with a phosphorimager and plotted (B to E). Integrated high-molecular-weight (C and E) and unintegrated (B and D) DNA forms were measured for DF1 (B and C) and CEF (D and E) cells, and the DNA copy number was obtained by comparison to a standard curve present on every gel. Results are presented from a single experiment, representative of two independent experiments.
PrB and RCASBP (A) (Fig. 4B and D) gave results similar to those reported previously (43), with an initial peak of unintegrated DNA 5 to 10 days after infection, followed by a decrease of unintegrated DNA to below the level of detection. T155I-infected cells had a profile similar to that of cells infected with PrB. With our cell viability and apoptosis assays, we were unable to observe any cytopathic effects caused by PrB or T155I under these conditions (Fig. 1 and 3). This result is not surprising, given the limited cytopathogenicity under normal culturing conditions exhibited by the wild-type isolate used in this study.
DF1 cells infected with NTRE4, L154S, and LT154/155SI, by contrast, had persistent unintegrated DNA that peaked 15 to 26 days after infection, correlating perfectly with the CPE interval (16 to 27 days postinfection), and persisted at low levels for >2 months. Levels of integrated DNA corresponded to those of unintegrated DNA, leveling off at about 0.5 to 1 copy per cell for the noncytopathic infections and 5- to 10-fold more for DF1 cells infected with L154S viruses. The reduction of the number of copies of integrated DNA in the latter cells was most likely due to overgrowth of a small fraction of cells that survived the infection. Although persistent unintegrated viral DNA was also present in NTRE4-infected cells during this time, its level was less than in cells infected with the other mutants, correlating with the lack of CPE.
CEF cells infected with the host range extension mutants had a pattern of accumulation of viral DNA similar to that of DF1, with a virtually identical initial peak at 5 to 10 days. However, although unintegrated viral DNA persisted in CEF, it did so at a level 5- to 15-fold lower than in DF1 cells, similar to conditions in which we did not see CPE in DF1 cells.
As has been reported previously, the transient accumulation of unintegrated DNA was higher in subgroup B viruses than in the noncytopathic RCASBP(A) (42, 43). Interestingly, this transient first wave of viral DNA accumulation was similar in the two cell types and was not associated with elevated levels of apoptosis. In DF1 cells infected with L154S and LT154/155SI, there were several apparent waves of both unintegrated and integrated viral DNA accumulation. The first wave corresponded to that seen with WT PrB, NTRE4, and T155I, 5 to 10 days postinfection. Subsequent waves could be observed at 14 to 16, 18 to 22, and 22 to 27 days postinfection (Fig. 4B and C). The dramatic CPE observed for the L154S and LT154/155SI viruses correlated well with very high levels of viral DNA present in the second and subsequent waves. This comparison suggests that accumulation of viral DNA must persist over a longer period of time and at a higher level for the dramatic CPE caused by the host range extension mutants to take place.
Apoptosis in cycloheximide-treated chronically infected cells.
We next wanted to determine the ability of viral Env proteins to trigger apoptosis. We hypothesized that receptor signaling acts in concert with DNA damage signals to lead to apoptosis in cells infected with the host range extension mutant viruses. If this hypothesis is correct, one would predict that the mutant Env proteins in chronically infected cells would be competent to trigger apoptosis, but that because these cells lack the high levels of viral DNA observed earlier in the course of infection, no apoptosis occurs. Cells chronically infected with a subgroup B alpharetrovirus (J. Brojatsch, personal communication) or cells treated with a soluble form of SU (7, 11) undergo apoptosis in the presence of cycloheximide, while cells infected with noncytopathic viruses do not. To determine whether the host range extension mutant Env proteins could trigger apoptosis, chronically infected DF1 cells were treated with cycloheximide 60 days postinfection.
Cells that were not treated with cycloheximide showed the normal background levels of apoptosis typical for DF1 cells (0.5 to 2%) when harvested after 24, 48, and 72 h of growth (Fig. 5). For some viruses, cycloheximide treatment led to elevated levels of apoptosis at all three time points, with an increase in apoptosis over time. Cells infected with L154S showed the highest levels of apoptosis by the end of the 72 h, with LT154/155SI- and PrB-infected cells showing slightly lower levels. T155I-infected cells showed only very low apoptosis levels throughout the experiment, correlating with a lower level of functional envelope protein on such cells (31). NTRE4 induced lower levels of apoptosis than PrB, correlating with its decreased ability to use Tv-bs3 as a receptor (31). Finally, mock-infected cells and cells infected with the noncytopathic RCASBP(A) did not show any increase in apoptosis levels upon cycloheximide treatment.
FIG. 5.
Induction of apoptosis by cycloheximide in chronically infected DF1 cells. Chronically infected cells (60 days postinfection) were treated with cycloheximide (10 μg/ml), harvested at indicated times, and scored for apoptosis as described in the legend to Fig. 3. Results are the average of three replicates (± SEM) from a single experiment, representative of three independent experiments.
DISCUSSION
Host range point mutant viruses cause a dramatic CPE in DF1 but not CEF cells.
In previous studies, we identified an alpharetrovirus mutant with expanded host range attributable to a single base change (L154S) in the hr1 region of env (31, 37). During the course of measuring the replication of this mutant on various cell types, we observed a dramatic CPE in DF1 cells, a cell line derived from CEF cells. This CPE affected a larger fraction of cells in the culture than that caused by other cytopathic retroviruses, leading to a >50-fold decrease in viable cells 16 to 27 days after infection, compared to a typical 2-fold reduction in viability resulting from infection of CEF with subgroup B, D, and F alpharetroviruses (42). The reduction in viability was associated with a substantial accumulation of both unintegrated and integrated viral DNA. The levels of apoptosis, as measured by nuclear morphology, in DF1 cells infected with the host range mutants increased 15 days after infection, immediately before the reduction in cell viability was observed (16 days after infection), suggesting that induction of apoptosis was responsible for this loss of cell viability. Similarly, levels of apoptosis decreased 23 to 27 days postinfection, immediately preceding the return to normal growth rate of these cultures around 27 days postinfection. Following this period, the cultures resumed normal growth and virus production as the fraction of cells that survived and contained reduced amounts of viral DNA expanded. We favor a model in which cells that are resistant to superinfection and killing are selected over a model in which less-cytopathic viral variants develop, because we have performed bulk sequencing of the SU regions of the viruses at the beginning and end of the time course and found no sequence difference (data not shown).
The dramatic killing effect was specific for DF1 cells infected with the extended-host-range mutants and was not observed in CEF cultures derived from the same line of chickens or in an established quail cell line (our unpublished results). This observation suggests that some difference in replicative behavior between these two cell culture systems was responsible for the difference observed in their cytopathogenicity. Such a difference most likely affects the proclivity of the cells to accumulate viral DNA over multiple waves of infection but could also affect the sensitivity of the cell to apoptotic signals related to interaction of the virus with its receptor, Tvb. One possibility that could account for greater superinfection of DF1 cells would be higher levels of receptor expression. However, studies of SU binding to cells demonstrated that CEFs have about twice as many binding sites as DF1 cells for all of the PrB-derived SU proteins described here, arguing against a role for expression level of Tvb or some other CPE variant receptor in enhancing superinfection of DF1 cells (data not shown). Genetic lesions resulting from, and possibly related to, isolation of DF1 as a permanent cell line may underlie this difference. In support of the latter hypothesis, DR5, the putative human homologue of Tvb, has been shown to induce apoptosis in tumor cells but not in normal cells (27). As an immortal cell line, DF1 may have some characteristics in common with tumor cells and distinct from CEF. It is important to remember, however, that PrB and the T155I mutant also use Tvb as a receptor but do not cause observable CPE in these cells under the conditions of the experiments presented. We conclude that DF1 cells serve as a sensitive system in which to test the cytopathic nature of alpharetrovirus variants, amplifying their cytopathogenicity relative to that observed in primary cells. Clearly, experiments with DF1 cells demonstrate the ability of these viruses to exhibit greatly enhanced cytopathic behavior. Further studies with chickens are required to validate any increased pathogenic potential these viruses may exhibit in vivo. Previously, another group observed higher susceptibility to virus-mediated killing for DF-1 cells than for CEFs; however, no direct comparison was presented (15).
Interestingly, we did not observe CPE induced in either cell type by wild-type PrB, a virus that has been characterized as cytopathic. The reason for this discrepancy is likely that reports that found CPE caused by subgroup B viruses have largely relied on plaque assays (12, 14, 15, 20, 23). While providing a useful quantitative measure of cytopathogenicity, plaque assays likely contribute to the cytopathic effect that they are designed to measure by promoting high local concentrations of virus and overconfluency of cells. Further, inclusion of stains, such as neutral red, that make the plaques easier to observe may contribute to the observed CPE. A few reports have described CPE in cell culture, but these have employed different subgroup B viral strains that may have higher replicative capacity (11, 42, 43).
Relationship of viral DNA to CPE and apoptosis.
A hallmark of retrovirus-induced CPE is the accumulation of large amounts of viral DNA (25, 26, 42, 44). The dramatic CPE caused by the host range extension mutants was accompanied by persistently high levels of both integrated and unintegrated viral DNA species. The profiles of both forms of viral DNA were quite similar to each other and to the apoptosis profile. The major difference between these DNA forms was the level observed within infected cells. Interestingly, DF1 cells that grew out and survived as chronically infected cells in which no CPE was apparent continued to contain elevated levels of viral DNA compared to that in WT PrB, but much lower levels of DNA than we observed during the peak of CPE. This lower level of viral DNA in the surviving cells suggests that viral DNA must be present above a certain threshold to be associated with apoptosis and cell killing. The lower amounts of viral DNA in the surviving cells are likely due to a reduced susceptibility to superinfection.
Mechanism of apoptosis induction.
Tvb, the receptor used by subgroup B alpharetroviruses, is a member of a family of proteins involved in proapoptotic signaling in response to ligands like TNF and TNF-related apoptosis-inducing ligand (7, 28). Consistent with this relationship, it has been observed that cells can be driven into apoptosis by infection with subgroup B virus or interaction of subgroup B Env proteins with cellular receptors containing death domains (6, 11). Recent evidence has been presented indicating that infected cells can kill uninfected cells in a “bystander killing” mechanism (11). However, the killing observed with host range extension mutant alpharetroviruses far exceeds that observed with WT subgoup B viruses, consistent with the idea that other triggers may contribute to this effect. We observed induction of apoptosis in cycloheximide-treated DF1 cells infected with NTRE4, PrB, and the point mutants. This result is consistent with the ability of SU from viruses that interact with Tvb to cause apoptosis in those cells (6, 7, 11), indicating that the viruses are capable of triggering apoptosis via an Env-receptor interaction. DNA repair mechanisms are capable of sending signals leading to cell growth arrest or apoptosis, depending upon the degree of damage (4, 10, 35, 39). These signals may play a role in the apopotosis that we observe, either by shifting the balance of the Tvb-mediated signal toward apoptosis or by acting independently of Tvb and triggering apoptosis on their own. Consistent with this idea, cells that overexpress receptors for cytopathic and noncytopathic alpharetroviruses devoid of signaling capacity can still lead to apoptosis and death when high levels of superinfection are achieved (22).
The data presented here suggest a model where relative insensitivity to superinfection resistance by the host range mutants leads to massive superinfection, causing a buildup of viral DNA forms that alone or in collaboration with Tvb triggers apoptosis and cell death. Other possibilities such as more potent receptor triggering by the mutants cannot be excluded, nor can some unknown cause that leads to all of these phenomena, creating a strong correlation among them. However, given our study of CEFs, a very similar system where we do not observe CPE, the correlation between the growth characteristics of the viruses and CPE is quite striking. In CEFs, interference with superinfection is apparently stronger, with the accumulation of much less viral DNA that persists for a shorter time, no detectable increase in apoptosis, and no CPE, despite the presence of the same receptors in CEF and DF1 cells.
Host range extension and viral evolution.
Among all viruses known, alpharetroviruses and gammaretroviruses are unique in their ability to adapt to the use of different cell surface proteins as primary receptors. Clearly, these viruses have responded to selective pressures to be able to replicate in a variety of hosts. The selective pressure for host range variation is most likely due to both innate superinfection resistance due to expression of endogenous proviruses (5) and genetic polymorphism in receptors (16). Evolution of alpharetroviruses to recognize novel receptors has been accompanied by extensive modification of hr1 and hr2 sequences in env and must therefore have proceeded sequentially through a series of intermediates. To better understand this process, we have manipulated selective pressures in the laboratory to isolate the expanded host range viruses used in this study (37). Initially, it would seem that such viruses would have a significant selective advantage over their more restricted parents, and it is surprising that such viruses have not been isolated from nature, particularly since they differ from the wild type by only one or two amino acids. The results presented here, however, suggest that although such viruses may be intermediate forms in evolution, enhanced cytopathogenicity may be detrimental to their survival in the long term. Therefore, it is likely that these mutants rapidly undergo additional mutations to attenuate their cytopathogenicity and focus in on a discreet subset of available receptors in their changing environment.
REFERENCES
- 1.Adkins, H. B., J. Brojatsch, J. Naughton, M. M. Rolls, J. M. Pesola, and J. A. Young. 1997. Identification of a cellular receptor for subgroup E avian leukosis virus. Proc. Natl. Acad. Sci. USA 94:11617-11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adkins, H. B., J. Brojatsch, and J. A. Young. 2000. Identification and characterization of a shared TNFR-related receptor for subgroup B, D, and E avian leukosis viruses reveal cysteine residues required specifically for subgroup E viral entry. J. Virol. 74:3572-3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aschoff, J. M., D. Foster, and J. M. Coffin. 1999. Point mutations in the avian sarcoma/leukosis virus 3′ untranslated region result in a packaging defect. J. Virol. 73:7421-7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein, C., H. Bernstein, C. M. Payne, and H. Garewal. 2002. DNA repair/pro-apoptotic dual-role proteins in five major DNA repair pathways: fail-safe protection against carcinogenesis. Mutat. Res. 511:145-178. [DOI] [PubMed] [Google Scholar]
- 5.Boeke, J. D., and J. P. Stoye. 1997. Retrotransposons, endogenous retroviruses, and the evolution of retroelements, p. 343-435. In J. M. Coffin, S. H. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 6.Brojatsch, J., J. Naughton, H. B. Adkins, and J. A. Young. 2000. TVB receptors for cytopathic and noncytopathic subgroups of avian leukosis viruses are functional death receptors. J. Virol. 74:11490-11494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brojatsch, J., J. Naughton, M. M. Rolls, K. Zingler, and J. A. Young. 1996. CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell 87:845-855. [DOI] [PubMed] [Google Scholar]
- 8.Cheng-Mayer, C., D. Seto, M. Tateno, and J. A. Levy. 1988. Biologic features of HIV-1 that correlate with virulence in the host. Science 240:80-82. [DOI] [PubMed] [Google Scholar]
- 9.Cheng-Mayer, C., T. Shioda, and J. A. Levy. 1991. Host range, replicative, and cytopathic properties of human immunodeficiency virus type 1 are determined by very few amino acid changes in tat and gp120. J. Virol. 65:6931-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel, R., R. A. Katz, and A. M. Skalka. 1999. A role for DNA-PK in retroviral DNA integration. Science 284:644-647. [DOI] [PubMed] [Google Scholar]
- 11.Diaz-Griffero, F., S. A. Hoschander, and J. Brojatsch. 2003. Bystander killing during avian leukosis virus subgroup B infection requires TVBS3 signaling. J. Virol. 77:12552-12561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorner, A. J., and J. M. Coffin. 1986. Determinants for receptor interaction and cell killing on the avian retrovirus glycoprotein gp85. Cell 45:365-374. [DOI] [PubMed] [Google Scholar]
- 13.Dorner, A. J., J. P. Stoye, and J. M. Coffin. 1985. Molecular basis of host range variation in avian retroviruses. J. Virol. 53:32-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graf, T. 1972. A plaque assay for avian RNA tumor viruses. Virology 50:567-578. [DOI] [PubMed] [Google Scholar]
- 15.Himly, M., D. N. Foster, I. Bottoli, J. S. Iacovoni, and P. K. Vogt. 1998. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 248:295-304. [DOI] [PubMed] [Google Scholar]
- 16.Hunter, E. 1997. Viral entry and receptors, p. 71-119. In J. M. Coffin, S. H. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 17.Jung, Y. T., and C. A. Kozak. 2003. Generation of novel syncytium-inducing and host range variants of ecotropic moloney murine leukemia virus in Mus spicilegus. J. Virol. 77:5065-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung, Y. T., T. Wu, and C. A. Kozak. 2004. Novel host range and cytopathic variant of ecotropic Friend murine leukemia virus. J. Virol. 78:12189-12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasof, G., K. Degenhardt, D. Perez, A. Thomas, and E. White. 1999. Overview: a matter of life and death, p. 1-28. In M. Lavin and D. Watters (ed.), Signalling pathways in apoptosis. Harwood Academic, Amsterdam, The Netherlands.
- 20.Kawai, S., and H. Hanafusa. 1972. Plaque assay for some strains of avian leukosis virus. Virology 48:126-135. [DOI] [PubMed] [Google Scholar]
- 21.Keshet, E., and H. M. Temin. 1979. Cell killing by spleen necrosis virus is correlated with a transient accumulation of spleen necrosis virus DNA. J. Virol. 31:376-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klucking, S., A. S. Collins, and J. A. Young. 2005. Avian sarcoma and leukosis virus cytopathic effect in the absence of TVB death domain signaling. J. Virol. 79:8243-8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moscovici, C., D. Chi, L. Gazzolo, and M. G. Moscovici. 1976. A study of plaque formation with avian RNA tumor viruses. Virology 73:181-189. [DOI] [PubMed] [Google Scholar]
- 24.Moscovici, C., M. G. Moscovici, H. Jimenez, M. M. Lai, M. J. Hayman, and P. K. Vogt. 1977. Continuous tissue culture cell lines derived from chemically induced tumors of Japanese quail. Cell 11:95-103. [DOI] [PubMed] [Google Scholar]
- 25.Mullins, J. I., C. S. Chen, and E. A. Hoover. 1986. Disease-specific and tissue-specific production of unintegrated feline leukaemia virus variant DNA in feline AIDS. Nature 319:333-336. [DOI] [PubMed] [Google Scholar]
- 26.Overbaugh, J., E. A. Hoover, J. I. Mullins, D. P. Burns, L. Rudensey, S. L. Quackenbush, V. Stallard, and P. R. Donahue. 1992. Structure and pathogenicity of individual variants within an immunodeficiency disease-inducing isolate of FeLV. Virology 188:558-569. [DOI] [PubMed] [Google Scholar]
- 27.Ozawa, F., H. Friess, J. Kleeff, Z. W. Xu, A. Zimmermann, M. S. Sheikh, and M. W. Buchler. 2001. Effects and expression of TRAIL and its apoptosis-promoting receptors in human pancreatic cancer. Cancer Lett. 163:71-81. [DOI] [PubMed] [Google Scholar]
- 28.Pan, G., J. Ni, Y. F. Wei, G. Yu, R. Gentz, and V. M. Dixit. 1997. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science 277:815-818. [DOI] [PubMed] [Google Scholar]
- 29.Pauza, C. D., J. E. Galindo, and D. D. Richman. 1990. Reinfection results in accumulation of unintegrated viral DNA in cytopathic and persistent human immunodeficiency virus type 1 infection of CEM cells. J. Exp. Med. 172:1035-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Payne, L. N. 1992. Biology of avian retroviruses, p. 299-404. In J. A. Levy (ed.), The retroviridae, vol. 1. Plenum Press, New York, N.Y. [Google Scholar]
- 31.Rainey, G. J., A. Natonson, L. F. Maxfield, and J. M. Coffin. 2003. Mechanisms of avian retroviral host range extension. J. Virol. 77:6709-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson, H. L., and D. M. Zinkus. 1990. Accumulation of human immunodeficiency virus type 1 DNA in T cells: results of multiple infection events. J. Virol. 64:4836-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohn, J. L., M. S. Moser, S. R. Gwynn, D. N. Baldwin, and J. Overbaugh. 1998. In vivo evolution of a novel, syncytium-inducing and cytopathic feline leukemia virus variant. J. Virol. 72:2686-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaefer-Klein, J., I. Givol, E. V. Barsov, J. M. Whitcomb, M. VanBrocklin, D. N. Foster, M. J. Federspiel, and S. H. Hughes. 1998. The EV-O-derived cell line DF-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors. Virology 248:305-311. [DOI] [PubMed] [Google Scholar]
- 35.Skalka, A. M., and R. A. Katz. 2005. Retroviral DNA integration and the DNA damage response. Cell Death Differ. 12(Suppl. 1):971-978. [DOI] [PubMed] [Google Scholar]
- 36.Smith, R. E., and E. V. Schmidt. 1982. Induction of anemia by avian leukosis viruses of five subgroups. Virology 117:516-518. [DOI] [PubMed] [Google Scholar]
- 37.Taplitz, R. A., and J. M. Coffin. 1997. Selection of an avian retrovirus mutant with extended receptor usage. J. Virol. 71:7814-7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Temin, H. M., and V. K. Kassner. 1974. Replication of reticuloendotheliosis viruses in cell culture: acute infection. J. Virol. 13:291-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomson, B. J. 2001. Viruses and apoptosis. Int. J. Exp. Pathol. 82:65-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toyoshima, K., and P. K. Vogt. 1969. Enhancement and inhibition of avian sarcoma viruses by polycations and polyanions. Virology 38:414-426. [DOI] [PubMed] [Google Scholar]
- 41.Tsichlis, P. N., K. F. Conklin, and J. M. Coffin. 1980. Mutant and recombinant avian retroviruses with extended host range. Proc. Natl. Acad. Sci. USA 77:536-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weller, S. K., A. E. Joy, and H. M. Temin. 1980. Correlation between cell killing and massive second-round superinfection by members of some subgroups of avian leukosis virus. J. Virol. 33:494-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weller, S. K., and H. M. Temin. 1981. Cell killing by avian leukosis viruses. J. Virol. 39:713-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshimura, F. K., T. Wang, and S. Nanua. 2001. Mink cell focus-forming murine leukemia virus killing of mink cells involves apoptosis and superinfection. J. Virol. 75:6007-6015. [DOI] [PMC free article] [PubMed] [Google Scholar]