Abstract
The Epstein-Barr virus (EBV) has been detected in subsets of breast cancers. In order to elaborate on these observations, we quantified by real-time PCR (Q-PCR) the EBV genome in biopsy specimens of breast cancer tissue as well as in tumor cells isolated by microdissection. Our findings show that EBV genomes can be detected by Q-PCR in about half of tumor specimens, usually in low copy numbers. However, we also found that the viral load is highly variable from tumor to tumor. Moreover, EBV genomes are heterogeneously distributed in morphologically identical tumor cells, with some clusters of isolated tumor cells containing relatively high genome numbers while other tumor cells isolated from the same specimen may be negative for EBV DNA. Using reverse transcription-PCR, we detected EBV gene transcripts: EBNA-1 in almost all of the EBV-positive tumors and RNA of the EBV oncoprotein LMP-1 in a smaller subset of the tissues analyzed. Moreover, BARF-1 RNA was detected in half of the cases studied. Furthermore, we observed that in vitro EBV infection of breast carcinoma cells confers resistance to paclitaxel (taxol) and provokes overexpression of a multidrug resistance gene (MDR1). Consequently, even if a small number of breast cancer cells are EBV infected, the impact of EBV infection on the efficiency of anticancer treatment might be of importance.
The Epstein-Barr virus (EBV), a ubiquitous human herpesvirus, is associated with the development of different epithelial malignancies (28), including nasopharyngeal carcinoma (NPC). It has also been linked with subsets of other types of carcinomas, including gastric carcinoma and lymphoepithelioma-like carcinoma in salivary glands and thymus (32, 44). Several laboratories have reported detection of EBV in a subset of breast tumors (3, 10, 19, 23). However, negative results have also been reported (6, 9, 14, 27). Nevertheless, in most of the studies, a low viral load was detected in breast cancer biopsy specimens but the infected cells were not clearly identified. In the study presented here, we used real-time PCR (Q-PCR) to quantify the copy numbers of the EBV genome in biopsy specimens as well as in microdissected tumor cells. The results show that breast cancer cells harbor the viral genome. However, through microdissection and isolation of pure tumor cells, we now find that even in EBV-positive tumor samples, many tumor cells do not contain EBV genomes and that the breast carcinomas are highly heterogeneous in terms of genome content and distribution. Moreover, using reverse transcription-PCR (RT-PCR), we detected EBNA-1 and BARF-1 transcripts in almost all of the EBV-positive tumors and LMP-1 RNA in 3 of the 15 cases studied. The findings raise the possibility that although EBV is unlikely to have an etiologic role in the genesis of breast cancer, the virus might contribute to tumor progression. Finally, the potential impact of EBV in breast cancer progression was evaluated by estimation of resistance to chemotherapeutic agents on in vitro-infected MDA-MB-231 cells. The results show that EBV confers paclitaxel (taxol) resistance and causes overexpression of a multidrug resistance gene (MDR1). Consequently, if even a small number of breast cancer cells are EBV infected, the impact of EBV infection on the efficiency of anticancer treatment might be of importance.
MATERIALS AND METHODS
Biopsy specimens.
This study includes breast cancer specimens from 37 patients from Institut Gustave Roussy (Villejuif, France) and 95 from the Centre Oscar Lambret (Lille, France). Tumor specimens were obtained with the agreement of the patients and were solely adenocarcinomas. At the time of collection, fat was removed and samples were divided into three parts. The first part was submitted for histological study and histoprognostic grading according to the method of Contesso et al. (8). The second part of the sample was immediately frozen in liquid nitrogen for estrogen receptor (ER) and progesterone receptor (PR) assays (2). The third part was used in molecular biologic assays (30).
The median age of the patients was 58 years (range, 26 to 90 years). Node involvement was detected in 58.3% of the cases. The tumor size was <1 cm in diameter in 2.3% of the cases, >1 and <3 cm in 58.6% of cases, and >3 cm in 39.1% of cases. The tumor type was ductal in 58.6% of the cases, lobular in 11.5% of the cases, and of other types in 29.8% of the cases. The distribution of histoprognostic grading was 8.4% for type I, 41% for type II, and 50.6% for type III (8); 66% of the tumors were ER positive, and 64% were PR positive. Control DNA from two biopsy specimens of nasopharyngeal carcinoma and five pellets of peripheral blood cells was taken from the study described by Grunewald et al. (12).
Laser capture microdissection.
Six frozen breast cancer biopsy specimens collected at Institut Gustave Roussy were taken for microdissection. In each case, one tissue block of breast tumor was cut on a cryostat microtome into 10-μm sections. These were mounted on uncoated slides and fixed immediately in 70% ethanol for 30 s, stained with Mayer's hematoxylin, and then washed in 70%, 90%, and 95% ethanol for 30 s each. Staining was followed by two 60-s dehydration steps in xylene before the samples were air dried. The sections were then microdissected in order to select the tumor cell component. Four samples were dissected with the PixCell II laser capture microdissection (LCM) Arcturus system (Mountain View, California), and two samples were dissected with a Laser Leica LMD. For each tumor block, three or more separate microdissections were taken from different anatomic areas but with no discernible difference in lymphocytic infiltration, cell density, and stromal composition. A minimum of 5,000 cells was captured. The number of cells was evaluated according to the manufacturers' instructions: the number of pulses (Arcturus) or the measurement of the cell surface areas (Leica). Cell populations were estimated to be homogeneous by microscopic visualization. DNA was extracted and subsequently used for Q-PCR.
DNA preparation and Q-PCR.
DNA from pulverized frozen specimens or microdissected cells was extracted with a QIAMP mini kit (QIAGEN, Courtaboeuf, France) according to the manufacturer's instructions. Q-PCR of the EBV thymidine kinase gene (BXLF-1) was performed as described by Brengel-Pesce et al. (4) with omission of uracil DNA glycosylase. Dilutions of DNA extracted from Namalwa cells, containing two integrated copies of EBV genomes per cell (21), were used as the external standard. Samples were measured in duplicate. Amplifications of the EBV genome were performed with the number of cycles ranging from 25 to 38. The calibration curve allowed the establishment of the following equation for the number of EBV genomes: y = −3.320x + 41.50, with an R2 value of 0.99 and an error of 0.079. Genomic DNA was quantified by amplification of the β-globin gene. Standardization was performed with DNA from the LightCycler control kit (Roche Molecular Diagnostic, Meylan, France). The calibration curve allowed the establishment of the following equation: y = −3.312x + 30.06, with an R2 value of 1 and an error of 0.0941.
Preparation of RNA, RT-PCR, and Southern blot hybridization.
Frozen specimens were pulverized, and total RNA was isolated from frozen tumor tissues, using TRIzol reagent (Gibco-BRL, Gaithersburg, MD) according to the manufacturer's instructions. After treatment of 10 μg of extracted RNA with RQ1 RNase-free DNase (Promega Corp., Madison, WI), cDNA was synthesized from 1 μg of treated RNA using Moloney murine leukemia virus reverse transcriptase (Promega Corp., Madison, WI). PCR was carried out with the cDNA samples obtained from 33 ng of total RNA in a final volume of 50 μl, containing 2 μl of first-strand cDNA, 1× PCR buffer, 2.5 mM MgCl2, 200 nM concentrations of each primer (Table 1), and 250 μM deoxynucleoside triphosphates. Reactions were incubated at 95°C for 5 min, and 3 U of Taq DNA polymerase was added to each tube. Forty cycles of cDNA amplification were performed in a DNA thermal cycler (Perkin-Elmer Corp., Foster City, CA) with the following conditions: denaturing at 95°C for 30 s, hybridization at 58°C (55°C for LMP-1 amplification) for 1 min, and elongation at 72°C for 2 min, followed by a final elongation at 72°C for 7 min. Ten microliters of each PCR product was analyzed by electrophoresis in 2% agarose gel and transferred to nylon membranes by Southern blotting (Hybond; Amersham Life Science Inc., Arlington Heights, IL). The blots were prehybridized at 65°C for 2 h with 100 μg/ml of denatured salmon sperm DNA in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5% sodium dodecyl sulfate (SDS), and 5× Denhardt's buffer. Hybridization was performed with an LMP-1 or EBNA-1 cDNA that was labeled with [α-32P]dCTP using the Klenow fragment of DNA polymerase I (106 cpm/ml). Following overnight incubation, membranes were washed twice for 15 min with 2× SSC-0.1% SDS at room temperature, twice with 2× SSC-0.1% SDS at 65°C for 15 min, and twice with 0.1× SSC-0.5% SDS at 65°C for 30 min. The blots were then exposed to X-ray film for imaging.
TABLE 1.
Oligonucleotide sequences
| Target | Primer | Primer sequence (5′→3′) | B95-8 genomic coordinates (nucleotides) |
|---|---|---|---|
| EBNA-1 | E1AS | TTGCAGCCAATGCAACTTGG | 108197-108178 |
| E1S | AGAGAGTAGTCTCAGGGCAT | 67545-67564 | |
| BamH1U | UPUS | GTTCCTCGGTGGCGGGCTTA | 67378-67397 |
| ExonUAS | U172AS | ATGCCCTGAGACTACTCTCT | 67564-67545 |
| LMP-1 | LMP1S | CTGAGGATGGAACACGACCT | 169480-169461 |
| LMP1AS | AATGGAGGGAGAGTCAGTCA | 168099-168108 | |
| BARF-1 | BARF1S | GGCTGTCACCGCTTTCTTGG | 165560-165579 |
| BARF1AS | AGGTGTTGGCACTTCTGTGG | 165762-165743 | |
| BamH1A | BamAS | CGTGGTGAAGCCTCTAACGC | 165883-165902 |
| BamAAS | GGCAAGTGCGTTTATTGCGA | 166180-166161 | |
| BZLF1 | ZS | TTACACCTGACCCATACCAG | 103118-103099 |
| ZAS1 | ACATCTGCTTCAACAGGAGG | 102284-102303 | |
| HPRT | HPRTS | TATGGACAGGACTGAACGTC | |
| HPRTAS | GTTGAGAGATCATCTCCACC |
Cell culture and infection.
MDA-MB-231 cells (a cell line derived from pleural effusions of a breast cancer patient.) (5) were grown in RPMI 1640 (Life Technologies, GIBCO BRL, Gaithersburg, Md.) supplemented with 10% fetal bovine serum and antibiotics at 37°C in a 5% CO2 humidified atmosphere. The Akata cell clone infected with recombinant EBV (rEBV) was maintained in RPMI 1640 containing 10% fetal bovine serum and G418 (700 mg/ml; Invitrogen, The Netherlands). MDA-MB-231 cells were infected by cocultivation with Akata cells producing rEBV following induction by anti-human immunoglobulin G, as previously described (16). Infected clones were selected after cultivation with G418, and four clones were studied: C1D12, C2G6, C3B4, and C4A3.
Evaluation of cell survival.
Three thousand MDA-MB-231 cells attached in a microplate well were subjected to paclitaxel (Sigma). After 3 days, the cell number was estimated by means of the endogenous enzyme hexosaminidase (20).
MDR1 expression.
Evaluation of the expression levels of MDR1 (normalized to the housekeeping β2-microglobulin gene) was performed by real-time quantitative PCR using a LightCycler (Roche Diagnostics, France) as described previously (24). β2-Microglobulin and MDR1 selected sets of primers and fluorescent probe (Eurogentec, Biosense, Italy) are the following: b2m forward, 5′CGC TCC GTG GCC TTA GC 3′; b2m reverse, 5′ GAG TAC GCT GGA TAG CCT CCA 3′; b2m probe, 5′ 6-carboxyfluorescein (FAM) TGC TCG CGC TAC TCT CTC TTT CTG GC 3′ 6-carboxytetramethylrhodamine (TAMRA); MDR1 forward, 5′ TGA TGA CCC TAA AAA CAC CAC TG 3′; MDR1 reverse, 5′ GAA CCT ATA GCC CCT TTA ACT TGA 3′; MDR1 probe, 5′ FAM AGC ATT GAC TAC CAG GCT CGC CAA 3′ TAMRA. Standards for MDR1 and β2-microglobulin were prepared from total normal RNA, amplified by RT-PCR, and cloned using a TOPO II TA cloning kit (Invitrogen, France) following the manufacturer's recommendations.
RESULTS
Viral load in breast cancer tissue specimens.
Q-PCR assays were performed on 95 whole samples of the breast cancer biopsy tissues. In 51 cases, no copies of the EBV genome were amplified or the number was below the threshold of detection. Forty-four samples were considered positive for EBV, since 2 to 6,000 copies were detected in the assay. Table 2 shows the number of copies of the EBV genome detected in each of the 44 EBV-positive tumor samples. The copy number per 1,000 total cells is determined assuming that one cell contains 6 pg of DNA. The number of EBV genomes was very low; we measured in 9 samples less than 0.1 copy per 1,000 total cells, in 18 samples 0.1 to 0.8 copy per 1,000 total cells, and in 14 samples 1 to 19 copies per 1,000 cells. In two samples, an average of 700 copies was detected, corresponding to 303 and 338 copies in 1,000 total cells. In one sample, exhibiting the highest load, 883 copies per 1,000 cells were measured. In two NPC biopsy specimens, 1.8 and 6 million copies of the EBV genome were detected, corresponding to 27 and 25 copies per cell, respectively (Table 2). The copy number of the EBV genome in breast cancer biopsy specimens is thus very low compared to the ones observed in NPC; however, the EBV genomes were detected, and specimens were considered positive compared to the results obtained in five pellets of peripheral blood lymphocytes from healthy donors, in which the level of EBV DNA was below the threshold of detection.
TABLE 2.
Detection of the EBV genome by Q-PCR in biopsied EBV-positive breast cancer tissues
| Cased | DNA (ng)a | Cell equivalentsb | No. of EBV genomesc | Copies of EBV/1,000 cells |
|---|---|---|---|---|
| BK71 | 1,606 | 267,667 | 4 | 0.02 |
| BK66 | 2,386 | 397,667 | 6 | 0.02 |
| BK67 | 2,505 | 417,500 | 8 | 0.02 |
| BK34 | 1,096 | 182,667 | 5 | 0.03 |
| BK88 | 612 | 102,000 | 3 | 0.03 |
| BK69 | 967 | 161,166 | 6 | 0.04 |
| BK72 | 890 | 148,334 | 6 | 0.04 |
| BK61 | 921 | 153,500 | 7 | 0.05 |
| BK62 | 318 | 53,000 | 4 | 0.08 |
| BK47 | 188 | 31,334 | 3 | 0.10 |
| BK56 | 314 | 52,334 | 6 | 0.11 |
| BK70 | 1,651 | 275,166 | 30 | 0.11 |
| BK39 | 181 | 30,166 | 4 | 0.13 |
| BK49 | 291 | 48,500 | 7 | 0.14 |
| BK37 | 193 | 32,166 | 6 | 0.19 |
| BK6 | 379 | 63,166 | 16 | 0.25 |
| BK25 | 134 | 22,334 | 7 | 0.31 |
| BK95 | 84 | 14,000 | 5 | 0.38 |
| BK83 | 48 | 8,000 | 3 | 0.38 |
| BK15 | 141 | 23,500 | 9 | 0.38 |
| BK38 | 31 | 5,166 | 2 | 0.39 |
| BK24 | 94 | 15,667 | 8 | 0.51 |
| BK7 | 248 | 41,334 | 22 | 0.53 |
| BK16 | 76 | 12,667 | 7 | 0.55 |
| BK12 | 17 | 2,833 | 2 | 0.70 |
| BK9 | 73 | 12,166 | 10 | 0.83 |
| BK65 | 43 | 7,166 | 6 | 0.84 |
| BK21 | 36 | 6,000 | 6 | 1 |
| BK51 | 1,277 | 212,833 | 214 | 1 |
| BK68 | 20 | 3,334 | 4 | 1.2 |
| BK13 | 93 | 15,500 | 23 | 1.5 |
| BK22 | 83 | 13,833 | 22 | 1.6 |
| BK30 | 79 | 13,166 | 23 | 1.8 |
| BK27 | 363 | 60,500 | 118 | 2 |
| BK40 | 208 | 34,667 | 93 | 2.7 |
| BK42 | 12 | 2,000 | 7 | 3.5 |
| BK17 | 6 | 1,000 | 4 | 4 |
| BK4 | 163 | 27,166 | 214 | 8 |
| BK8 | 14 | 2,334 | 20 | 9 |
| BK32 | 47 | 7,833 | 93 | 12 |
| BK3 | 54 | 9,000 | 170 | 19 |
| BK2 | 14 | 2,334 | 708 | 303 |
| BK5 | 12 | 2,000 | 676 | 338 |
| BK23 | 47 | 7,833 | 6,918 | 883 |
| NPC1 | 1,315 | 219,166 | 6 × 106 | 27,376 |
| NPC2 | 425 | 70,833 | 1.8 × 106 | 25,400 |
| PBLKu15 | 970 | 161,667 | 0 | 0 |
| PBLKu27 | 6,940 | 1.16 × 106 | 0 | 0 |
| PBLhk35 | 17,350 | 2.9 × 106 | 0 | 0 |
| PBLhk39 | 2,150 | 358,334 | 0 | 0 |
| PBLiKa49 | 6,100 | 106 | 0 | 0 |
Quantified by amplification of the ß-globin gene.
Based on the assumption that one cell contains 6 pg of DNA and that there are two copies of the globin gene per cell.
Expressed as the mean of the results of two assays.
BK corresponds to breast cancer biopsies, NPC to nasopharyngeal carcinoma, and PBL to peripheral blood lymphocytes.
Detection and quantification of the EBV genome in isolated malignant epithelial cells of breast tumor biopsy specimens.
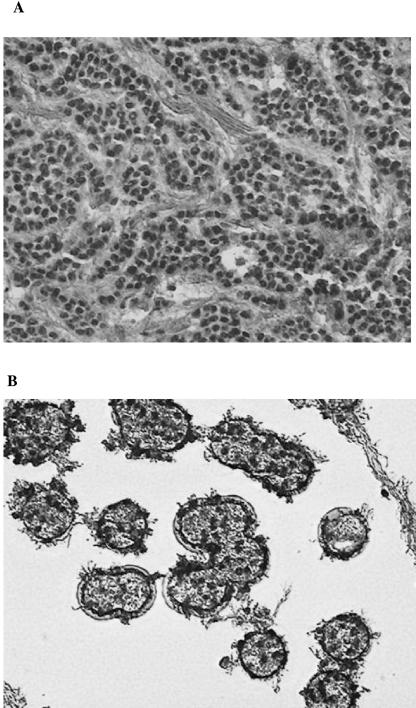
In order to evaluate the relevance of a low number of viral genomes and to identify the EBV-infected cells within the breast cancer biopsy specimens, we determined the EBV DNA load in isolated cancer cell populations. Six breast cancers from Institut Gustave Roussy were analyzed. Four of them were considered EBV positive, since 4, 11, 2, and 5 copies of the EBV genome were detected in a Q-PCR assay in tumor numbers 240, 393, 200, and 275, respectively. Tumor cells were separated from stromal tissue and normal breast cells by laser capture microdissection (LCM). Figure 1A shows a representative target tissue sample before laser dissection, and Fig. 1B shows the malignant epithelial cells captured after LCM. DNA from two to four different areas containing only carcinomatous epithelial cells was isolated, and the viral load was quantified by Q-PCR. No EBV genomes were detected in three different microdissections taken from two of the six tumors (data not shown). The results obtained with the four EBV-positive tumors are summarized in Table 3. In the microdissected specimens, although the tumor cell population appeared to be homogeneous, only one of four clusters of tumor cells taken from tumor 240 contained EBV genomes (100 copies in 1,000 cells). Similarly, in tumor 393, one of the two studied regions was EBV positive (855 copies in 1,000 cells). All studied regions of the other two tumors (200 and 275) were positive, exhibiting viral loads from 15 to 465 and from 49 to 6,333 copies in 1,000 cells, respectively, showing again a high heterogeneity in viral load. The EBV genome was clearly detected in malignant epithelial areas taken from the four EBV-positive biopsy specimens, demonstrating that the virus is harbored by epithelial cells and not accounted for by infiltrating lymphocytes.
FIG. 1.
Laser capture microdissection (LCM) of breast cancer tissue. Frozen tumor sections (10 μm thick) stained with Mayer's hematoxylin and eosin Y were microdissected with the Arcturus PixCell II system to procure homogeneous cell populations. Panel A shows a section before LCM. Panel B shows clusters of the malignant epithelial cells captured by LCM. These are representative sections from the total microdissections summarized in Table 3.
TABLE 3.
Samples in which the EBV genome was detected by Q-PCR in whole tumors and microdissected tumor epithelial cells
| Biopsy no. | DNA (ng)a | No. of cell equivalentsb | No. of EBV genomesc | Copies/ 1,000 cells |
|---|---|---|---|---|
| 240 | 0.03 | 5 | 0 | 0 |
| 0.035 | 5 | 0 | 0 | |
| 0.05 | 8 | 0 | 0 | |
| 0.08 | 13 | 13 | 100 | |
| 393 | 0.015 | 2.5 | 0 | 0 |
| 0.015 | 2.5 | 2 | 855 | |
| 200 | 4 | 667 | 10 | 15 |
| 1.5 | 250 | 20 | 80 | |
| 3.9 | 650 | 54 | 83 | |
| 0.26 | 43 | 20 | 465 | |
| 275 | 0.07 | 12 | 76 | 6,333 |
| 1.2 | 200 | 28 | 140 | |
| 1 | 167 | 12 | 72 | |
| 1.6 | 267 | 13 | 49 |
Quantified by amplification of the β-globin gene.
Based on the assumption that one cell contains 6 pg of DNA and that there are two copies of the globin gene per cell.
Expressed as the mean of the results of two assays.
We compared the average viral load detected in the whole tumor specimen with that determined for different microdissected clusters of tumor cells. As expected from the heterogeneity of viral distribution in the tumor cells, the viral load determined within the entire tumor specimen is much lower than that found within a virus-positive region. We determined that in tumor 393, 0.02 copies of the EBV genome were found per 1,000 total cells, although one microdissected region contained 855 copies per 1,000 cells. This result shows that even if a very low copy number of EBV genomes is detected within a whole biopsy specimen, a high copy number can be present in different parts of the tumor.
Detection of EBV transcripts in breast carcinoma biopsy specimens.
Tumors harboring the highest viral load were chosen for investigation of EBV expression if a sufficient amount of tissue was available. EBNA-1, BARF-1, and LMP-1 RNA expression was studied in whole tumor specimens. We tested for EBNA-1 transcripts by RT-PCR in RNAs from 21 EBV-positive tumors using E1S and E1AS primers (Table 1) and detected EBNA-1 RNA in 17 of the specimens. An amplified band of the expected size (361 bp) hybridized with labeled specific probe in each case. Typical examples are shown in Fig. 2Ba. Tumors 13, 14, 17, 18, 19, and 21 were positive for EBNA-1 cDNA, while 15 and 20 were negative. Amplification of hypoxanthine-guanine phosphoribosyl transferase (HPRT) cDNA was carried out in each case to control for the quality of the cDNAs (36) (data not shown). These results show that an EBV gene is expressed and detected in almost all of the EBV-positive breast tumors.
FIG. 2.
Detection of EBV transcripts by RT-PCR analysis in RNA preparations from breast cancer biopsies. The primer combinations used are shown as arrows in the diagrams presented in panel A. For cDNA detection, the following primers were used: E1AS and E1S were used for EBNA-1 (a), LMP1S and LMP1AS for LMP-1 (b), and BARF1S BARF1AS for BARF-1 (c), and ZS and ZAS1 for BZLF1 (e). For detection of putative DNA contamination, BamAS and BamAAS (c) and U172AS and UPUS (d) were used as controls. Products obtained after RT-PCR are shown in panel B after Southern blotting (Ba, Bb, and Bd) or on agarose gels stained with ethidium bromide (Bc). Amplification of EBNA-1 cDNA is shown in panel Ba. MDA-MB-231 cells were used as the negative control, and EBV-infected MDA-MB-231 cells were used as the positive controls for EBNA-1 cDNA amplification. LMP-1 cDNA amplification is shown in panel Bb. B95-8 and C3B4 (an EBV-infected MDA-MB-231 cell line) served as positive controls. BARF-1 cDNA amplification is shown in panel Bc, and BZLF1 amplification is shown in panel Bd. B95-8 cDNA served as a positive control for BARF-1 and BZLF1, and DNA from B85-8 served as a positive control for DNA amplification in BamH1A and BamH1U.
In order to detect LMP-1 transcripts, we used primers LMP1S and LMP1AS (Table 1 and Fig. 2Ab). Amplification was performed with cDNA obtained from breast carcinoma biopsy specimens. Three of fifteen samples were positive for LMP-1 cDNA amplification, as demonstrated by amplification and specific hybridization of a 377-bp band (Fig. 2Bb).
Expression of the EBV BARF-1 gene was investigated by RT-PCR in 14 different breast cancer biopsy specimens using BARF1S and BARF1AS primers. Figure 2Bc shows that a band of the expected size (202 bp) was amplified from cDNAs of 8 of the 14 biopsy specimens. In order to be sure that the bands obtained correspond to copies of cDNA and not to DNA contamination, PCR of untranscribed regions was performed with combinations of primers lying in the U and A fragments, one primer upstream (UPUS) of the U172 exon and one primer within the U172 exon (U172AS), and also with the combination of one primer (BamAAS) downstream of the BARF-1 sequence and one within the BARF-1 open reading frame (BamAS). No band was obtained with these different cDNA preparations, whereas bands of the expected sizes (185 bp and 317 bp, respectively) were detected after amplification of DNA extracted from B95-8 cells, showing that no genomic DNA was detectable in the cDNA preparations.
BZLF1 expression was assayed in two EBV-negative (986 and BK20) biopsy specimens, eight breast cancer biopsy specimens which harbored the highest viral load for which enough material was available (BK8, BK32, BK3, BK17, BK20, BK9, BK40, and BK23, which contained 9, 12, 19, 4, 0.83, 2.7, and 883 EBV copies/1,000 cells, respectively) (Table 2), and 275 specimens that carried EBV in four of four clusters of tumor cells isolated by LCM (Table 3). RT-PCR followed by Southern blotting was performed. In two tumors, BK3 and BK40, BZLF1 expression was observed after different overexposure of the blot. The pattern of expression is similar to what was seen in NPC (7) and in breast tissues (15).
Paclitaxel resistance of in vitro EBV-infected MDA-MB-231 cells.
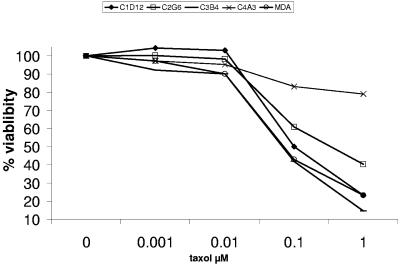
The potential impact of EBV infection on breast carcinoma cells was investigated. The sensitivity of MDA-MB-231 cells, infected or not infected by EBV, to an agent used in chemotherapy was tested. The cells were subjected to treatment of 3 days of different concentrations of paclitaxel (1, 10, and 100 nM and 1 μM). A representative experiment is shown in Fig. 3. When cultured with 100 nM and 1 μM paclitaxel, 40% and 23% of the noninfected cells survived. One of the infected clones, C4A3, was highly resistant to paclitaxel, since 83% of the cells survived when cultured with 100 nM paclitaxel and 79% survived in 1 μM paclitaxel. Another clone, C2G6, exhibited a resistance, since 60% and 40% of the cells survived in 100 nM and 1 μM paclitaxel, respectively. Several potential mechanisms have been proposed to account for the resistance to paclitaxel (for a review, see Orr et al. [29a]); these include overexpression of the multidrug resistance gene (MDR1). We explored the expression of the MRD1 gene in MDA-MB-231 cells as well as in the infected clones. The results are shown in Fig. 4. The copy number of the MDR1 gene transcripts (normalized to the housekeeping β2-microglobulin gene) was sixfold higher in the C2G6 cell line than in the parental noninfected MDA-MB-231 cell line. The levels of MDR1 expression in the other infected clones were not dramatically increased compared to the ones of the noninfected one. However, when cultured in the presence of 100 nM paclitaxel, the level of MDR1 transcripts was 10-fold higher in the C4A3 clone than the level expressed without paclitaxel in the cultured medium. In summary, in the presence of 100 nM paclitaxel, the level of MDR1 transcripts normalized to β2-microglobulin expression is 6- to 10-fold higher in C4A3 and C2G6 EBV-infected clones than in the parental MDA-MB-231 noninfected clone. C4A3 and C2G6 are more resistant to paclitaxel than the two other infected clones, C1D12 and C3B4. The resistance is correlated with overexpression of the MDR1 gene. The mechanism conferring paclitaxel resistance implicated in EBV-infected MDA-MB-231 cells is currently under investigation.
FIG. 3.
Comparative toxicity of paclitaxel to MDA-MB-231 cells and to the different EBV-infected clones. Thirty thousand cells were seeded in 100 μl of culture medium in microtiter plates and incubated for 72 h with various concentrations of paclitaxel. The viability was assessed by means of the endogenous enzyme hexosaminidase. The experiment was performed in triplicate.
FIG. 4.
EBV infection up-regulates MDR1 expression in two different EBV-infected MDA-MB-231 cell clones. Quantitative real-time RT-PCR analysis was performed on RNA extracted from cells exposed during 72 h to 0.1 μM paclitaxel; the transcript copy numbers relative to the reference gene β2-microglobulin (b2m) were determined.
DISCUSSION
Although a part of the literature showed negative results, EBV has been detected in large subsets of infiltrating breast cancer (3, 10, 19, 23). In this study, we detected EBV genomes in various copy numbers in about half of the breast cancer specimens analyzed. We accounted for this variability by microdissection of the specimens, which disclosed that the distribution of EBV genomes in tumor cells was variable within each tumor specimen even though morphologically the tumor cells were indistinguishable. This observation may account for the variable distribution of the EBNA-1 protein detected by immunohistochemistry (3). It is well known that different biological markers (ER and/or PR, epidermal growth factor receptor, and MMP-2) (35, 37, 38) are heterogeneously distributed within breast cancer tissue. Moreover, Glockner et al. determined the amplification status of growth regulatory genes in macroscopically and microscopically separate areas of individual breast cancers and showed marked intratumoral heterogeneity of c-myc and cyclinD1 (11). Intratumoral heterogeneity mirrors subclonal diversity and might affect treatment response. It appears that EBV genomes are also heterogeneously distributed. It would be of interest to identify tissue markers that may correlate with EBV genome content in breast cancer cells that appear otherwise to be morphologically identical. Our results point to heterogeneity in terms of EBV content in populations of breast cancer cells that may be biologically significant.
In different studies, evaluation of EBV in breast cancer has disclosed very low viral loads (10, 27) which have even been defined as EBV negative (31). Murray et al. (27) used 19 sections of paraffin-embedded breast cancer tissues. They measured 2, 12, and 5 samples, respectively, with less than 0.1, 0.1 to 0.9, and 1 to 7 copies of the EBV genome per 1,000 cells. We had similar results but also detected two samples with 303 to 338 copies per 1,000 cells. Moreover, one sample with a load of 883 copies in 1,000 cells was identified among the 95 frozen biopsy tissues. Similarly, one cluster of isolated tumor cells contained 6,333 copies of EBV DNA in 1,000 total cells. As suggested by Huang et al. (15), lytic viral replication might contribute to the detection of the EBV genome in breast cancer, since Zta protein has been immunostained in a small number of cells from breast tumor. We detected BZLF1 transcripts in two of the eight breast cancer biopsy specimens. Moreover, proteins expressed during the productive cycle might be of importance for cell transformation: MMP9 is induced by BZLF1 (43), BRL1 induces expression of FAS (22) and activates S-phase entry through E2F1 induction (33), and BZLF1 interacts with cell cycle progression and is able to form complexes with p53 (45) and p65 (13), which play roles in the control of apoptosis. Moreover, lytic gene expression protects from cell death (17). However, it is also known that EBV reactivation occurs in a few cells in NPC (7) in which infection is predominantly latent, and such a phenomenon does not preclude latent infection.
Since identification of infected cells within a tumor has previously given conflicting results in the literature, evidence of an association of EBV with breast cancer requires clarification. Negative results could arise from several sources: lack of assay sensitivity, storage of samples, use of fixed versus frozen tissues, and heterogeneity of the cancer tissue. False-positive results could arise from nonmonospecific antibody reagents or from EBV-positive infiltrating B lymphocytes, the level of which has been estimated to be 2 to 60 EBV-positive lymphocytes in 106 peripheral blood lymphocytes (25).
In order to test whether the detected EBV genomes were harbored only by lymphocytes, we investigated the viral loads in five pellets of peripheral blood lymphocytes from healthy blood donors (Table 2). As these results were negative, we concluded that most if not all of the EBV DNA detected in the breast cancer biopsy specimens was of epithelial tumor cell origin. We confirmed this conclusion by LCM analysis. Q-PCR performed on epithelial tumor cells purified from EBV-positive tumors by microdissection shows that the isolated tumor cells carry the viral genome and in much higher copy number in several of the microdissected specimens. Six to seven microdissections were also performed in the two biopsied specimens of breast cancer tissue by Fina et al. (10). Their results also showed heterogeneity of the viral load in samples containing only epithelial cancer cells and that infiltrating lymphocytes could not be the only source of the detected EBV.
In contrast, Murray et al. (27) failed to detect EBV genomes in microdissected tissue. However, they did not sample different areas of the same tumor but pooled all of the microdissected cells from each specimen. Thus, dilution of EBV-positive cells by noninfected cells may have resulted. Moreover, they analyzed fixed tissue, which resulted in lower sensitivity.
We show here quantification of EBV DNA in individual microdissections of four different tumors. A large heterogeneity in distribution of viral genomes from one region to another of the same tumor as well as among different tumors was observed. Moreover, the numbers of EBV genomes in some of the microdissected samples are not compatible with contamination with B lymphocytes, since only 1 to 10 per 106 are EBV positive. We also show that even when EBV is detected at a “high” level within one given region, the whole tumor may exhibit a very low viral load overall.
We have detected EBNA-1 staining with two EBNA-1 monoclonal antibodies (1H4, used for frozen sections, and 2B4, used on paraffin sections) of breast cancer tumors (3), and we also detected EBNA-1 transcripts in RNA taken from the same tumor. Xue et al. (41) detected EBV transcripts in samples sorted for the absence of lymphocytes. They detected EBNA-1 and BARF-1 RNA in three of six cases. We show here detection of EBNA-1, BARF-1, and LMP-1, respectively, in 17 of 22, 8 of 14, and 3 of 15 cases. Expression of EBNA-1, BARF-1, and LMP-1 genes is of importance. The EBNA-1 protein is required to replicate and maintain the EBV genome in infected cells, LMP-1 is considered to be the important EBV oncogene (18), and BARF-1 is able to confer transforming properties in cells (29). Both LMP-1 and BARF-1 are expressed in other carcinomas (32), in NPC and in gastric cancers.
In conclusion, it is well known that morphologically indistinguishable breast cancer cells in fact may comprise a heterogeneous population of breast cancer cells Similarly, we have now demonstrated that the distribution of EBV genomes in seemingly identical cells is heterogeneous. Because EBV is detected in only some breast cancer cells, it is unlikely to be a primary etiologic agent.
Herrmann and Niedobitek (14) questioned the implication of EBV in breast cancer, since the virus is detected in only a subset of tumor cells. However, EBV might have a role at an early step in carcinogenesis and then be lost after the development of the tumor. Alternatively, infection with EBV at a late state of tumor development might enhance oncogenic properties, such as invasiveness, angiogenesis, and metastasis. For example, it has been shown that LMP-1 induces matrix metalloproteinase 9 (MMP9), cyclooxygenase-2 (COX2), and vascular endothelial growth factor (VEGF) (26) and induces and causes release of FGF-2 in human epithelial cells (39). Moreover, a correlation between LMP-1 expression and the presence of COX2 in NPC tissue was observed. These findings raise the possibility that EBV might alter the phenotype of a subpopulation of carcinomatous cells so that they become more aggressive in behavior (40). Our results show that EBV infection confers paclitaxel resistance to breast carcinoma cell lines. This resistance might be a consequence of the higher MDR1 expression that is observed in paclitaxel-resistant EBV-infected cells (C4A3 and C2G6). Moreover, MDR is induced by paclitaxel treatment in the C4A3 cells. The manner by which the cells acquired high MDR1 gene expression has not yet been defined. Overexpression of the MDR1 gene product may be intrinsically expressed or acquired following chemotherapeutic drug treatment. Different molecular mechanisms may lead to MDR1 overproduction: increased mRNA stability, increased translational initiation (42), hypomethylation of the promoter (34), and changes in the spatial and temporal pattern of histone modifications (1). How EBV affects those mechanisms is yet to be uncovered. Overexpression of the MRD1 gene upon EBV infection might have an effect on therapeutic treatment of the disease.
Whether EBV might have a role in breast cancer development or progression now needs to be promptly investigated with appropriate cell biologic tools.
Acknowledgments
We are grateful to Joseph Pagano for reading the manuscript and providing suggestions. We thank Nathan LaBorde for careful reading of the manuscript and Fabien Calvo for helpful discussions.
This work was supported by Association de Recherche Contre le Cancer (5512, 4454). Hratch Arbach was supported by grants from Société De Secours Des Amis Des Sciences, Fondation des Treilles, and Association de Recherche Contre le Cancer.
REFERENCES
- 1.Baker, E. K., R. W. Johnstone, J. R. Zalcberg, and A. El-Osta. 8. August 2005. Epigenetic changes to the MDR1 locus in response to chemotherapeutic drugs. Oncogene 10.1038/sj.onc.1208955. [DOI] [PubMed]
- 2.Blankenstein, M. A. 1990. Comparison of ligand binding assay and enzyme immunoassay of oestrogen receptor in human breast cancer cytosols. Experience of the E.O.R.T.C. Receptor Group. Breast Cancer Res. Treat. 17:91-98. [DOI] [PubMed] [Google Scholar]
- 3.Bonnet, M., J. M. Guinebretiere, E. Kremmer, V. Grunewald, E. Benhamou, G. Contesso, and I. Joab. 1999. Detection of Epstein-Barr virus in invasive breast cancers. J. Natl. Cancer Inst. 91:1376-1381. [DOI] [PubMed] [Google Scholar]
- 4.Brengel-Pesce, K., P. Morand, A. Schmuck, M. J. Bourgeat, M. Buisson, G. Bargues, M. Bouzid, and J. M. Seigneurin. 2002. Routine use of real-time quantitative PCR for laboratory diagnosis of Epstein-Barr virus infections. J. Med. Virol. 66:360-369. [DOI] [PubMed] [Google Scholar]
- 5.Cailleau, R., R. Young, M. Olive, and W. J. Reeves, Jr. 1974. Breast tumor cell lines from pleural effusions. J. Natl. Cancer Inst. 53:661-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu, P. G., K. L. Chang, Y. Y. Chen, W. G. Chen, and L. M. Weiss. 2001. No significant association of Epstein-Barr virus infection with invasive breast carcinoma. Am. J. Pathol. 159:571-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cochet, C., D. Martel-Renoir, V. Grunewald, J. Bosq, G. Cochet, G. Schwaab, J. F. Bernaudin, and I. Joab. 1993. Expression of the Epstein-Barr virus immediate early gene, BZLF1, in nasopharyngeal carcinoma tumor cells. Virology 197:358-365. [DOI] [PubMed] [Google Scholar]
- 8.Contesso, G., H. Mouriesse, S. Friedman, J. Genin, D. Sarrazin, and J. Rouesse. 1987. The importance of histologic grade in long-term prognosis of breast cancer: a study of 1,010 patients, uniformly treated at the Institut Gustave-Roussy. J. Clin. Oncol. 5:1378-1386. [DOI] [PubMed] [Google Scholar]
- 9.Deshpande, C. G., S. Badve, N. Kidwai, and R. Longnecker. 2002. Lack of expression of the Epstein-Barr virus (EBV) gene products, EBERs, EBNA1, LMP1, and LMP2A, in breast cancer cells. Lab. Investig. 82:1193-1199. [DOI] [PubMed] [Google Scholar]
- 10.Fina, F., S. Romain, L. Ouafik, J. Palmari, F. Ben Ayed, S. Benharkat, P. Bonnier, F. Spyratos, J. A. Foekens, C. Rose, M. Buisson, H. Gerard, M. O. Reymond, J. M. Seigneurin, and P. M. Martin. 2001. Frequency and genome load of Epstein-Barr virus in 509 breast cancers from different geographical areas. Br. J. Cancer 84:783-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glockner, S., H. Buurman, W. Kleeberger, U. Lehmann, and H. Kreipe. 2002. Marked intratumoral heterogeneity of c-myc and cyclinD1 but not of c-erbB2 amplification in breast cancer. Lab. Investig. 82:1419-1426. [DOI] [PubMed] [Google Scholar]
- 12.Grunewald, V., M. Bonnet, S. Boutin, T. Yip, H. Louzir, M. Levrero, J. M. Seigneurin, M. Raphael, R. Touitou, D. Martel-Renoir, C. Cochet, A. Durandy, P. Andre, W. Lau, Y. Zeng, and I. Joab. 1998. Amino-acid change in the Epstein-Barr-virus ZEBRA protein in undifferentiated nasopharyngeal carcinomas from Europe and North Africa. Int. J. Cancer 75:497-503. [DOI] [PubMed] [Google Scholar]
- 13.Gutsch, D. E., E. A. Holley-Guthrie, Q. Zhang, B. Stein, M. A. Blanar, A. S. Baldwin, and S. C. Kenney. 1994. The bZIP transactivator of Epstein-Barr virus, BZLF1, functionally and physically interacts with the p65 subunit of NF-κB. Mol. Cell. Biol. 14:1939-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrmann, K., and G. Niedobitek. 2003. Lack of evidence for an association of Epstein-Barr virus infection with breast carcinoma. Breast Cancer Res. 5:R13-R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, J., H. Chen, L. Hutt-Fletcher, R. F. Ambinder, and S. D. Hayward. 2003. Lytic viral replication as a contributor to the detection of Epstein-Barr virus in breast cancer. J. Virol. 77:13267-13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imai, S., J. Nishikawa, and K. Takada. 1998. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J. Virol. 72:4371-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inman, G. J., U. K. Binne, G. A. Parker, P. J. Farrell, and M. J. Allday. 2001. Activators of the Epstein-Barr virus lytic program concomitantly induce apoptosis, but lytic gene expression protects from cell death. J. Virol. 75:2400-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kieff, E., and A. B. Rickinson. 2001. Epstein-Barr virus and its replication, p. 2531-2536. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 19.Labrecque, L. G., D. M. Barnes, I. S. Fentiman, and B. E. Griffin. 1995. Epstein-Barr virus in epithelial cell tumors: a breast cancer study. Cancer Res. 55:39-45. [PubMed] [Google Scholar]
- 20.Landegren, U. 1984. Measurement of cell numbers by means of the endogenous enzyme hexosaminidase. Applications to detection of lymphokines and cell surface antigens. J. Immunol. Methods 67:379-388. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence, J. B., C. A. Villnave, and R. H. Singer. 1988. Sensitive, high-resolution chromatin and chromosome mapping in situ: presence and orientation of two closely integrated copies of EBV in a lymphoma line. Cell 52:51-61. [DOI] [PubMed] [Google Scholar]
- 22.Li, Y., J. Webster-Cyriaque, C. C. Tomlinson, M. Yohe, and S. Kenney. 2004. Fatty acid synthase expression is induced by the Epstein-Barr virus immediate-early protein BRLF1 and is required for lytic viral gene expression. J. Virol. 78:4197-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luqmani, A., and S. Shousha. 1995. Presence of Epstein-Barr virus in breast carcinoma. Int. J. Oncol. 6:899-903. [DOI] [PubMed] [Google Scholar]
- 24.Menashi, S., M. Serova, L. Ma, S. Vignot, S. Mourah, and F. Calvo. 2003. Regulation of extracellular matrix metalloproteinase inducer and matrix metalloproteinase expression by amphiregulin in transformed human breast epithelial cells. Cancer Res. 63:7575-7580. [PubMed] [Google Scholar]
- 25.Miyashita, E. M., B. Yang, K. M. Lam, D. H. Crawford, and D. A. Thorley-Lawson. 1995. A novel form of Epstein-Barr virus latency in normal B cells in vivo. Cell 80:593-601. [DOI] [PubMed] [Google Scholar]
- 26.Murono, S., H. Inoue, T. Tanabe, I. Joab, T. Yoshizaki, M. Furukawa, and J. S. Pagano. 2001. Induction of cyclooxygenase-2 by Epstein-Barr virus latent membrane protein 1 is involved in vascular endothelial growth factor production in nasopharyngeal carcinoma cells. Proc. Natl. Acad. Sci. USA 98:6905-6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray, P. G., D. Lissauer, J. Junying, G. Davies, S. Moore, A. Bell, J. Timms, D. Rowlands, C. McConkey, G. M. Reynolds, S. Ghataura, D. England, R. Caroll, and L. S. Young. 2003. Reactivity with a monoclonal antibody to Epstein-Barr virus (EBV) nuclear antigen 1 defines a subset of aggressive breast cancers in the absence of the EBV genome. Cancer Res. 63:2338-2343. [PubMed] [Google Scholar]
- 28.Murray, P. G., G. Niedobitek, E. Kremmer, F. Grasser, G. M. Reynolds, A. Cruchley, D. M. Williams, N. Muller-Lantzsch, and L. S. Young. 1996. In situ detection of the Epstein-Barr virus-encoded nuclear antigen 1 in oral hairy leukoplakia and virus-associated carcinomas. J. Pathol. 178:44-47. [DOI] [PubMed] [Google Scholar]
- 29.Ooka, T. 2001. BARF-1 gene as an EBV encoded oncogene. Epstein-Barr Virus Rep. 8:177-182. [Google Scholar]
- 29a.Orr, G. A., P. Verdier-Pinard, H. McDaid, and S. Band Horwitz. 2003. Mechanisms of Taxol resistance related to microtubules. Oncogene 22:7280-7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pawlowski, V., F. Revillion, M. Hebbar, L. Hornez, and J. P. Peyrat. 2000. Prognostic value of the type I growth factor receptors in a large series of human primary breast cancers quantified with a real-time reverse transcription-polymerase chain reaction assay. Clin. Cancer Res. 6:4217-4225. [PubMed] [Google Scholar]
- 31.Perrigoue, J. G., J. A. den Boon, A. Friedl, M. A. Newton, P. Ahlquist, and B. Sugden. 2005. Lack of association between EBV and breast carcinoma. Cancer Epidemiol. Biomark. Prev. 14:809-814. [DOI] [PubMed] [Google Scholar]
- 32.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2594-2611. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 33.Swenson, J. J., A. E. Mauser, W. K. Kaufmann, and S. C. Kenney. 1999. The Epstein-Barr virus protein BRLF1 activates S phase entry through E2F1 induction. J. Virol. 73:6540-6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tada, Y., M. Wada, K. Kuroiwa, N. Kinugawa, T. Harada, J. Nagayama, M. Nakagawa, S. Naito, and M. Kuwano. 2000. MDR1 gene overexpression and altered degree of methylation at the promoter region in bladder cancer during chemotherapeutic treatment. Clin. Cancer Res. 6:4618-4627. [PubMed] [Google Scholar]
- 35.Talvensaari-Mattila, A., P. Paakko, and T. Turpeenniemi-Hujanen. 2003. Matrix metalloproteinase-2 (MMP-2) is associated with survival in breast carcinoma. Br. J. Cancer 89:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Touitou, R., H. Arbach, C. Cochet, J. Feuillard, A. Martin, M. Raphael, and I. Joab. 2003. Heterogeneous Epstein-Barr virus latent gene expression in AIDS-associated lymphomas and in type I Burkitt's lymphoma cell lines. J. Gen. Virol. 84:949-957. [DOI] [PubMed] [Google Scholar]
- 37.Tsutsui, S., S. Ohno, S. Murakami, A. Kataoka, J. Kinoshita, and Y. Hachitanda. 2002. EGFR, c-erbB2 and p53 protein in the primary lesions and paired metastatic regional lymph nodes in breast cancer. Eur. J. Surg. Oncol. 28:383-387. [DOI] [PubMed] [Google Scholar]
- 38.van Netten, J. P., I. G. Thornton, S. J. Carlyle, M. L. Brigden, P. Coy, N. L. Goodchild, S. Gallagher, and E. J. George. 1987. Multiple microsample analysis of intratumor estrogen receptor distribution in breast cancers by a combined biochemical/immunohistochemical method. Eur. J. Cancer Clin. Oncol. 23:1337-1342. [DOI] [PubMed] [Google Scholar]
- 39.Wakisaka, N., S. Murono, T. Yoshizaki, M. Furukawa, and J. S. Pagano. 2002. Epstein-Barr virus latent membrane protein 1 induces and causes release of fibroblast growth factor-2. Cancer Res. 62:6337-6344. [PubMed] [Google Scholar]
- 40.Wakisaka, N., and J. S. Pagano. 2003. Epstein-Barr virus induces invasion and metastasis factors. Anticancer Res. 23:2133-2138. [PubMed] [Google Scholar]
- 41.Xue, S. A., I. A. Lampert, J. S. Haldane, J. E. Bridger, and B. E. Griffin. 2003. Epstein-Barr virus gene expression in human breast cancer: protagonist or passenger? Br. J. Cancer 89:113-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yague, E., A. L. Armesilla, G. Harrison, J. Elliott, A. Sardini, C. F. Higgins, and S. Raguz. 2003. P-glycoprotein (MDR1) expression in leukemic cells is regulated at two distinct steps, mRNA stabilization and translational initiation. J. Biol. Chem. 278:10344-10352. [DOI] [PubMed] [Google Scholar]
- 43.Yoshizaki, T., H. Sato, S. Murono, J. S. Pagano, and M. Furukawa. 1999. Matrix metalloproteinase 9 is induced by the Epstein-Barr virus BZLF1 transactivator. Clin. Exp. Metastasis 17:431-436. [DOI] [PubMed] [Google Scholar]
- 44.Young, L. S., and P. G. Murray. 2003. Epstein-Barr virus and oncogenesis: from latent genes to tumours. Oncogene 22:5108-5121. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, Q., D. Gutsch, and S. Kenney. 1994. Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr virus latency. Mol. Cell. Biol. 14:1929-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]