Abstract
The secreted protein hedgehog (Hh) plays a critical role in the developmental patterning of multiple tissues. In Drosophila melanogaster, a cytosolic multiprotein signaling complex appears necessary for Hh signaling. Genes that encode components of this Hh signaling complex (HSC) were originally identified and characterized based on their genetic interactions with hh, as well as with each other. It is only in recent years that the mechanistic functions of these components have begun to be unraveled. Here, we have investigated the relationship between two components of the HSC, the serine/threonine protein kinase Fused (Fu) and the kinesin-related protein Costal2 (Cos2). We have reconstituted a Fu/Cos2 complex in vitro and shown that Fu is able to directly associate with Cos2, forming a complex whose molecular size is similar to a previously described complex found in Drosophila cell extracts. We have also determined that the carboxyl-terminal domain of Fu is necessary and sufficient for the direct binding of Fu to Cos2. To validate the physiological relevance of this interaction, we overexpressed the carboxyl-terminal domain of Fu in wild-type flies. These flies exhibit a phenotype similar to that seen in fu mutants and consistent with an hh loss-of-function phenotype. We conclude that the carboxyl-terminal domain of Fu can function in a dominant negative manner, by preventing endogenous Fu from binding to Cos2. Thus, we provide the first evidence that Hh signaling can be compromised by targeting the HSC for disruption.
The secreted protein hedgehog (Hh) exerts potent tissue patterning activity during the development of a diverse array of organisms (15). During Drosophila melanogaster development Hh is responsible for the correct patterning of a variety of embryonic and adult tissues (5, 11, 26, 30, 35, 54). hh mutations are embryonically lethal, exhibiting a strong segment polarity phenotype (35). hh is also expressed within the posterior (P) compartment of imaginal discs that go on to form the various adult structures (26, 53). Hh then acts on adjacent anterior (A) compartment cells to specify cell fates in a concentration-dependent manner (26, 39, 54). The various concentrations of Hh are translated into different cell fates through a series of poorly understood molecular events that go through, at least in part, components of a large intracellular Hh signaling complex (HSC) (16, 19, 33, 43, 51, 52).
The HSC includes the kinesin-related protein Cos2 (43, 47), the Ser/Thr protein kinase Fu (41, 57), the pioneer protein Suppressor of fused [Su(fu)] (38), and the transcription factor Cubitus interruptus (Ci) (1, 13, 37). Ci exists in at least four different forms: a repressor form (Ci75) (4), a cytosolic default form (Ci155) (4, 32), an active full-length form (actCi155), and an activated labile form (Ci*) (1, 9, 12, 22, 36, 59, 60, 64). It has been hypothesized that A cells interpret their position within the Hh concentration gradient by regulating the conversion between the various forms of Ci, in turn repressing or activating various Ci-dependent target genes (3, 45). This processing of Ci would be regulated by the other components of the HSC. In the absence of Hh, Cos2, Fu, and Ci155 are enriched on microtubules, where Ci processing to Ci75 has been proposed to occur (4, 43). Consistent with this, a disruption in Ci155-to-Ci75 processing is observed in cos2 or fu mutants (2, 29, 47, 61, 63). Upon Hh activation, the complex no longer enriches on microtubules and Ci155 processing to Ci75 is blocked, resulting in an increased cytosolic concentration of Ci155 (4, 9, 43, 62). This allows actCi155 to accumulate in the nucleus (9, 61, 63, 64), without the other members of the HSC (29, 50). Anterior cells that abut the A/P border are exposed to the highest concentration of Hh. These cells cease to produce Ci75 and convert their Ci155 to Ci*, the most active form of Ci (4, 36). In cells that lack Cos2 or Fu, the conversion of Ci155 to Ci* is also lost (2, 28, 36, 61, 63, 64).
fu mutations are embryonically lethal, displaying a segment-polarity defect consistent with fu being a positive regulator of Hh signaling (16, 35). This embryonic lethality can be rescued by maternal copies of fu mRNA, allowing the mutant embryos to develop into adults. fu adults display a variety of phenotypes, including a fusion of longitudinal veins 3 and 4 (LV3 and LV4) (14, 31). Additionally, the wings of fu flies have a posterior extension of the double row of marginal bristles, into the intervein region of LV3 and LV4. This posterior extension of double-row bristles is indicative of a loss of hh-dependent anterior en expression (20, 46, 55).
Two major classes of fu alleles have been characterized, class I and II, based on their genetic interactions with Su(fu) (42, 56). Class I alleles encode mutations that appear to affect the protein kinase domain, while class II alleles encode frame shift mutations that truncate the carboxyl-terminal domain of Fu (42, 56). Fu isolated from class I mutant flies can associate with Cos2, whereas Fu isolated from class II mutant flies cannot (43). Thus, the kinase activity of Fu is not required for its interaction with Cos2, but the carboxyl-terminal domain of Fu is required. These results suggest that the carboxyl-terminal domain of Fu plays a structural role in maintaining HSC integrity. However, it was also recently shown that the carboxyl-terminal domain of a human homolog of Fused (hFU) is an activator of the human Hh pathway (34). Murone et al. showed that when hFU is transfected into cells it can activate a reporter gene construct containing Gli (the vertebrate homolog of Ci) DNA binding consensus sites. hFU also acted synergistically with GLI2 to activate this reporter gene. hFU did not seem to require an intact kinase domain for function, as constructs expressing the carboxyl-terminal domain of hFU appear sufficient to activate the various assays performed. In light of these results, we decided to investigate, in vitro and in vivo, the role that the carboxyl-terminal domain of Drosophila Fu plays in Hh signal transduction.
Here, we show that Fu associates directly with Cos2 to form a high-affinity complex. The carboxyl-terminal domain of Fu is both necessary and sufficient for this high-affinity association with Cos2. Furthermore, transgenic flies that overexpress the carboxyl-terminal domain of Fu exhibit a phenotype similar to that of known fu mutants, consistent with disruption of the Hh pathway. We conclude that the carboxyl-terminal domain of Fu acts in a dominant negative manner to disrupt Hh signaling by preventing the kinase domain of endogenous Fu from accessing its substrates.
MATERIALS AND METHODS
Generation of Fu mutant constructs.
See Fig. 1for schematic details. The kinase-inactive Fu mutant, G13V, was made by replacing glycine 13 with a valine residue, using a PCR-based site-directed mutagenesis kit (Quikchange, Stratagene). ΔFu is a carboxyl-terminal truncated form of Fu containing the first 305 amino acids (aa) (kinase domain) of Fu. ΔFu was subcloned into pBacPak8 by use of a 5" SmaI site on the vector and an internal NruI site in Fu. Fu-tail (aa 421 to 805 of Fu) was subcloned into pFastBacHTb, in frame with a His6 tag, by use of HindIII flanking sites. Fu-tail was also subcloned into pAc5.1V5-His in frame with a 5"-Flag epitope and out of frame with the 3" tandem V5-His6 epitope by use of HindIII flanking sites. A slightly larger carboxyl-terminal domain of Fu (aa 270 to 805), which was made by amplifying a portion of the fu D6 cDNA by PCR, was used to generate transgenic flies (57). A His6-tagged version was subcloned into the pUAST vector (7). More-detailed subcloning procedures are available upon request.
Generation and characterization of Cos MAb 5D6.
Eight-week-old female BALB/c mice (B&K Universal) were immunized with decreasing doses of antigen (glutathione S-transferase [GST]-heptad repeat region of Cos2, kindly provided by M. Scott [47]) over a period of approximately 4 months by standard techniques. Mice having serum antibody titers of approximately 1:400,000 to 1:800,000 were selected for hybridoma development. Three days prior to fusion, mice were administered a prefusion intraperitoneal injection of antigen (5 μg) in phosphate-buffered saline (PBS), and then isolated splenocytes were fused to murine myeloma FO (ATCC no. CRL 1646; American Type Culture Collection). Selected hybridomas were cloned by limiting dilution, and the immunoglobulin subtype was determined for each resulting monoclonal antibody (MAb). Subtypes of selected MAbs were determined with IsoStrip Antibody Subtyping Dipsticks (Boehringer-Mannheim). Antibody 5D6 was determined to be immunoglobulin G1(κ) [IgG1(κ)].
Spinner flasks containing BD Cell MAb Medium (Becton Dickinson Microbiology Systems) plus 10% fetal bovine serum, 2% l-glutamine, and penicillin-streptomycin were inoculated with hybridoma cell lines at 2 × 105 cells/ml. Cultures were incubated at 37°C in 5% CO2 for approximately 2 weeks. Hybridoma cells were removed from the culture medium by centrifugation, and MAbs were purified by protein G-Sepharose (Amersham Pharmacia Biotech) affinity chromatography, followed by elution with a glycine buffer, pH 2.5. The pH of the antibodies was adjusted to 7.0 and then dialyzed against PBS.
Cell culture and transfection.
Sf21 cells were cultured in Grace's insect medium (Life Technologies, Inc.) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Drosophila S2 cells were cultured in Schneider's Drosophila medium (Life Technologies, Inc.) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Transfection of S2 cells was carried out with Lipofectin according to the manufacturer's instructions (Life Technologies, Inc.).
Preparation of baculovirus.
Baculoviruses were produced and titered according to the manufacturer's instructions (Life Technologies, Inc.). Infections were carried out with Sf21 cells at a total multiplicity of infection of 4 to 6. Wild-type (wt) baculovirus was used to normalize the coinfections done in this study. The infected Sf21 cells were allowed to incubate postinfection for 44 to 48 h.
Cellular lysates.
Sf21 or S2 cells were washed twice with PBS at 4°C and then lysed in Nonidet P-40 (NP-40) buffer (150 mM NaCl, 50 mM HEPES [pH 7.6], 1 mM dithiothreitol, 1 mM EDTA, 1% NP-40, and 1:250 protease inhibitor cocktail [PIC]). PIC contains 1 mM benzamidine, 1 mg of aprotinin per ml, 1 mg of leupeptin per ml, and 1 mg of pepstatin per ml in 100% ethanol. The various cellular lysates were centrifuged at 16,000 × g for 20 min at 4°C. The supernatants were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then analyzed by immunoblotting or subjected to immunoprecipitation analysis.
For gel filtration analysis, the infected Sf21 cells were washed twice with PBS at 4°C and then lysed in a hypertonic lysis buffer (325 mM NaCl, 50 mM Tris-HCl [pH 7.6], 1 mM dithiothreitol, 1 mM EDTA, 5% glycerol, 0.0005% NP-40, and 1:250 PIC) using a glass Dounce homogenizer (Kontes). The cellular lysates were centrifuged at 16,000 × g for 20 min at 4°C. The resulting supernatants were then centrifuged at 100,000 × g for 30 min prior to gel filtration analysis.
Immunoprecipitation of cytosolic lysates.
The baculovirus-infected Sf21 lysates were normalized according to Cos2 expression, to correct for any variation in protein expression seen in multiple infections. Two micrograms of mouse Flag M2 (Sigma) or 5D6 MAbs per sample was used for each immunoprecipitation. Mouse IgG1 was used as the isotype-matched control for both MAbs. The cellular lysates were immunoprecipitated as previously described (43). The immunoprecipitates were washed five times with 1.25 ml of NP-40 lysis buffer, followed by the addition of 2× Laemmli protein gel loading buffer. The resulting samples were separated by SDS-PAGE and immunoblotted for the appropriate proteins.
Size exclusion chromatography.
For gel filtration analysis, baculovirus-infected Sf21 lysates were fractionated by size with a Superose 6 gel filtration column by fast-performance liquid chromatography (Amersham-Pharmacia), equilibrated with 650 mM NaCl, 50 mM Tris-HCl (pH 7.6), 1 mM EDTA, 10% glycerol, and 0.001% NP-40.
Purification of a Fu/Cos2 complex.
One billion Sf21 cells were coinfected with Cos2 and flag-tagged G13V baculovirus and then lysed 48 h later in hypertonic lysis buffer. The resulting supernatants were made isotonic. One milliliter of anti-Flag (M2) affinity resin (Sigma) was added to the supernatant and shaken for 4 h. The supernatant and resin suspension was applied to a column and washed extensively with ≈100 ml of isotonic buffer (150 mM NaCl, 50 mM Tris-HCl [pH 7.6], 1 mM dithiothreitol, 1 mM EDTA, 5% glycerol, and 1:250 PIC). Wash fractions were monitored for UV absorbance at 280 nm on a Spectronic Genesys 5 UV/VIS spectrophotometer until the readings were near background levels. Bound proteins were then eluted with 0.2 mg of Flag peptide per ml in the same buffer. Fractions were collected, resolved by SDS-PAGE, and then immunoblotted with Fu or Cos2 antibodies, or Coomassie blue stained. Fraction 8 contained the peak of Fu and Cos and was injected onto a Superose 6 gel filtration column and fractionated as above.
Construction of Saccharomyces cerevisiae vectors and yeast two-hybrid analysis
The directed yeast two-hybrid screen was performed essentially as previously described (24). Briefly, Cos2 and Fu-tail cDNA was subcloned into both pGAD-C1 (GAL4 activation domain) and pGBDU-C1 (GAL4 DNA binding domain) vectors and transformed into the yeast strain PJ69-4A. Each cDNA was also subcloned in its antisense orientation, as a negative control. Yeasts that were able to grow in the histidine and adenine screening plates were interpreted as true and positive clones. A more detailed description of our yeast two-hybrid screening procedure is available upon request.
Drosophila strains and generation of transgenic flies.
Drosophila strains were maintained as previously described (58). Transgenic flies were made according to the method of Rubin and Spradling (44) with yw flies as hosts and maintained at 18°C. dpp-GAL4 (48, 49) and ptc-GAL4 (21) were kind gifts of the Kornberg laboratory. ap-GAL4 (8) was a kind gift from C. Micchelli. Crosses to detect adult wing phenotype were conducted at 29°C. For wing preparations, whole flies were placed in isopropanol overnight. The wings were then dissected and placed into 100% ethanol. The wings were subsequently washed twice in ethanol, mounted in a 6:5 lactic acid-to-ethanol solution, and imaged using an Axiophot microscope with an Axiocam CCD camera and software.
Immunofluorescence of Drosophila wing imaginal discs.
Wing imaginal discs of wandering third instar larvae were dissected. The discs were fixed in PBS-4% formaldehyde (Ted Pella)-0.2% Triton X-100 for 10 min and then washed three times with PBT (PBS plus 0.1% Tween 20). The discs were then incubated with primary antibody in PBT plus 5% donkey serum (Jackson Labs) for 1 h as previously described (4). Anti-Fused antiserum (43) was used at 1:1,000, and MAb 2A1 (anti-Ci) was used at 1:2 (4). Texas Red-conjugated antirabbit (Jackson Labs) and fluorescein isothiocyanate (FITC)-conjugated antirat (Jackson Labs) antibodies were both used at 1:400. Imaging was conducted on a Leica TCS NT confocal microscope (Leica America). Images were processed with Adobe Photoshop.
RESULTS
Fu and Cos2 associate with each other quantitatively.
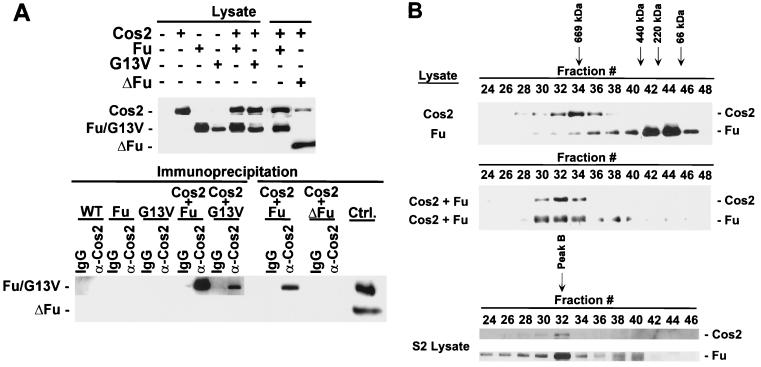
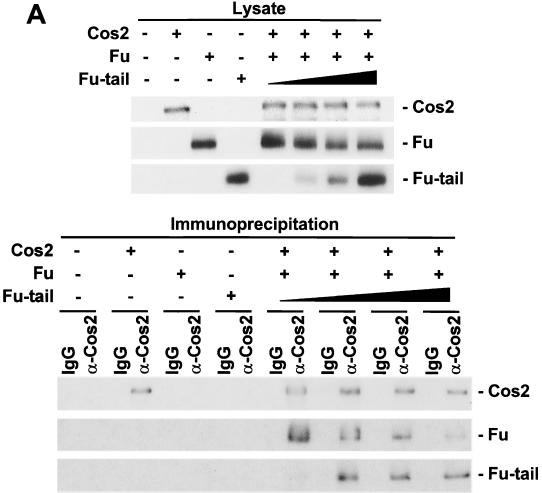
We have previously shown that while Cos2 can associate with kinase-inactive Fu mutants isolated from class I fu flies, it is unable to associate with carboxyl-terminal truncated forms of Fu isolated from class II fu flies. (43). There are at least two models that might explain these observations: (i) the carboxyl-terminal domain is necessary for regulating an event, or other protein, which allows Fu to bind to Cos2; or (ii) the carboxyl-terminal domain is both sufficient and necessary to bind Fu to Cos2. To test these models, we decided to reconstitute these interactions. Therefore, we made recombinant baculoviruses that express a kinase-inactive Fu (G13V) or a carboxyl-terminal truncation of Fu containing just the kinase domain (ΔFu) (Fig. 1). In flies, G13V is a class I fu allele (56), whereas ΔFu is similar to class II fu alleles that encode a mutant protein lacking the carboxyl-terminal domain. G13V, ΔFu, and full-length Fu were expressed in Sf21 cells individually, or coexpressed with a Cos2 baculovirus. Using a Cos2 MAb (5D6; see Materials and Methods), we were able to coimmunoprecipitate both Fu and G13V with Cos2 from the appropriate coinfected lysates (Fig. 2A). Under similar conditions we were unable to coimmunoprecipitate ΔFu or GST, expressed by a GST baculovirus (data not shown), with Cos2. Thus, using a baculoviral system, we are able to recapitulate the interactions previously observed in fly imaginal disc lysates between Fu and Cos2; Fu does not require a functional kinase domain to associate with Cos2 but does require the carboxyl-terminal domain.
FIG. 1.
Various Fu mutants. (A) Schematic diagram of wt, Class I (Fu62) and Class II (FuRX16) Fu gene products. Class I fu alleles encode mutations within the kinase domain that are thought to inactivate Fu kinase activity. Class II fu alleles encode frameshift mutations that effectively truncate the carboxyl-terminal domain (extra catalytic domain). Fu62 has three amino acids deleted (positions 139 to 141) within the kinase domain as indicated by the symbol ^. Hatch marks indicate the 84 extra amino acids coded as a result of a frameshift mutation in the FuRX16 mutant. (B) Schematic diagram illustrating the various Fu gene mutants used in this study. G13V contains a mutation that replaces a conserved Gly with Val at position 13; such a mutation behaves like a class I allele. The asterisk within the G13V schematic denotes the mutated amino acid. ΔFu contains only the kinase domain of Fu and is similar to various class II mutants. Fu-tail contains only the carboxyl-terminal domain of Fu, aa 421 to 805.
FIG. 2.
Cos2 binds Fu quantitatively. (A) Fu and a kinase-inactive mutant of Fu (G13V) associate with Cos2, but ΔFu does not. Immunoblots of various lysates from Sf21 cells infected with baculovirus expressing Cos2, Fu, G13V, ΔFu, or various combinations are shown (top panel). The various lysates were immunoprecipitated with the Cos2 5D6 MAb or an isotype-matched IgG control. The resulting immunoprecipitates were resolved by SDS-PAGE and then immunoblotted for the various Fu mutants (bottom panel). GST served as a negative control in this experiment (data not shown). The immunoblots were probed with 5D6 to verify that Cos2 was immunoprecipitated (data not shown). Lysates containing Fu or ΔFu were used as immunoblotting positive controls in this experiment (Ctrl.) (B) Gel filtration analysis of lysates from Sf21 cells expressing Cos2, Fu, or both. Cos2, when expressed alone, elutes at fraction 34. Fu, expressed alone, elutes at fraction 44 with a molecular size consistent with it being a monomer or dimer (top panel). In lysates from cells coinfected with Fu and Cos2, both proteins comigrate at fraction 32 (middle panel). Various protein standards were used to calibrate the Superose 6 gel filtration column, i.e., thyroglobulin (669 kDa), ferritin (440 kDa), catalase (220 kDa), and bovine serum albumin (66 kDa), which eluted at fractions 34, 41, 43, and 45, respectively. A Drosophila S2 cellular lysate fractionated on a Superose 6 column illustrates peak B, where endogenous Fu and Cos2 comigrate (bottom panel). Peak A is a complex that contains Cos2, Fu, and likely other proteins in S2 cells. However, peak A is an unstable complex, whose appearance in S2 cell lysates fractionated by gel filtration, while reproducible, remains variable. The calculated exclusion volume for the column elutes at fraction 22 and is reported to be 4 × 107 daltons for globular proteins.
To establish that the Fu/Cos2 association was direct, we assessed whether the interactions were quantitative under conditions where the two proteins are greatly overexpressed. Lysates from Sf21 cells infected with Fu or Cos2 baculovirus individually, or coinfected, were subjected to size exclusion chromatography (Fig. 2B). Upon size exclusion chromatography of Drosophila cellular lysates, Fu is found in three distinct peaks (A, B, and C) (43). Peak A, which contains Fu, Cos2, and Ci, migrates as a large megadalton-sized protein complex. Peak B, which contains the vast majority of Fu and Cos2, migrates as an approximately 700-kDa complex. Peak C is approximately 100 to 200 kDa in size. Fu, from Fu-infected cells, elutes at fraction 44 with a molecular size (100 to 200 kDa) consistent with it being monomeric or dimeric, and similar to that previously described for peak C. Cos2, from Cos2-infected cells, elutes at fraction 34, comigrating with a 669-kDa protein standard. The anomalous migration of Cos2 (calculated molecular mass, ≈130 kDa) through the gel filtration column is consistent either with it being multimeric, binding some abundant Sf21 cellular proteins or, more likely (see below), with Cos2 migrating through the gel filtration column with a rod-like structure. Such uncharacteristic behavior, upon gel filtration, has been described for other kinesin-related proteins. When extracts from Fu- and Cos2-coinfected cells are analyzed by gel filtration, the peaks of Fu and Cos2 immunoreactivity comigrate at fraction 32. The peak of Fu shifts quantitatively, from 100 to 200 kDa to greater than 669 kDa (from fractions 44 to 32), whereas the peak of Cos2 now elutes two fractions larger than when expressed alone (from fractions 34 to 32). Fu and Cos2 associate with high affinity, as these associations occur in a 650 mM NaCl buffer. These two proteins also appear sufficient to form a complex that migrates at a molecular size similar to that of Fu peak B, from S2 cell extracts (Fig. 2B, bottom panel). These results support the idea that the interaction between Fu and Cos2 is direct and suggest that the previously described peak B is composed primarily of Cos2 and Fu.
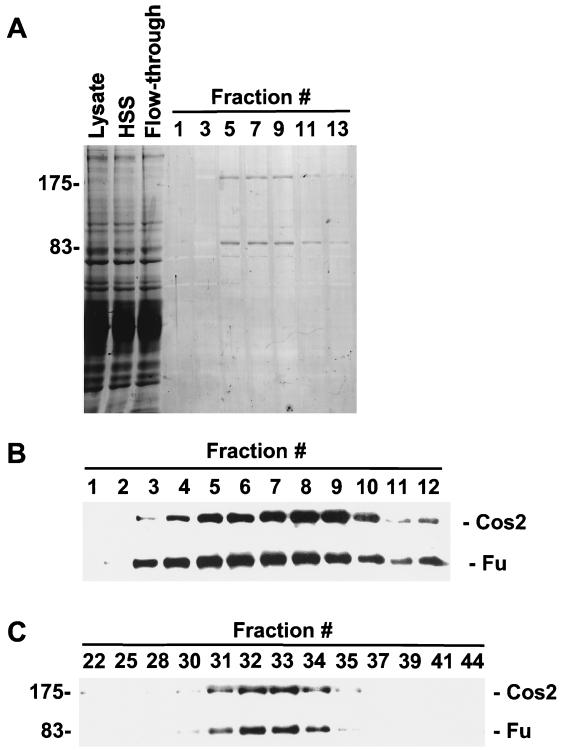
To verify that Fu and Cos2 associate directly, given their large apparent molecular size upon gel filtration analysis, we purified the Fu/Cos2 complex. Extracts of Sf21 cells, expressing Cos2 and Flag-tagged Fu, were purified over an anti-Flag MAb affinity column and eluted with excess Flag peptide. Fractions from this column were resolved by SDS-PAGE and then either Coomassie blue stained or immunoblotted with antibodies to Fu or Cos2 (Fig. 3A and B). After Coomassie blue staining, two major proteins that migrated at the apparent molecular weights of Fu and Cos2 and comigrated with Fu and Cos2 immunoreactivity were detected. A fraction containing the peak of this purified material (fraction 8) was also analyzed by gel filtration chromatography, to determine if these two proteins alone were sufficient to migrate at the molecular size of peak B. We found that highly purified Fu and Cos2 still comigrate on a sizing column at fraction 32, suggesting that Fu and Cos2 are sufficient to form the bulk of peak B found in extracts of Drosophila cells (Fig. 3C, compare to Fig. 2B, bottom panel).
FIG. 3.
Purification of a recombinant Fu/Cos2 complex. Lysates of Sf21 cells infected with Cos2 and Flag G13V were purified over an anti-flag MAb (M2) affinity column and washed extensively. Bound protein was eluted with excess Flag peptide. The various fractions were resolved by SDS-PAGE and then Coomassie blue stained (A) or immunoblotted with Fu or Cos2 antibodies (B). (A) Coomassie blue-stained SDS-PAGE showing purification of a Cos2 Fu complex. Starting fractions containing the infected Sf21 lysate (Lysate), the high speed supernatant (HSS), the flowthrough fraction from the Flag M2 affinity resin (Flow-through), and the fractions collected after addition of excess Flag peptide are shown (lanes 1 to 13, every other fraction). Two major proteins that correspond to the apparent molecular weights of Fu and Cos2 are seen in lanes 5 to 9. (B) Immunoblot analysis of the Cos2 Fu purification. Fu and Cos2 immunoreactivity comigrated with the two major Coomassie blue-stained proteins, at molecular weights of 83,000 and 175,000. (C) Fraction 8 off the M2 affinity resin was loaded onto a Superose 6 gel filtration column to determine quantitative association between purified Cos2 and G13V. Both purified proteins comigrated at fraction 32, verifying that Fu and Cos2 are sufficient to form the bulk of peak B seen in S2 lysates.
The carboxyl-terminal domain of Fu is necessary and sufficient to bind Cos2.
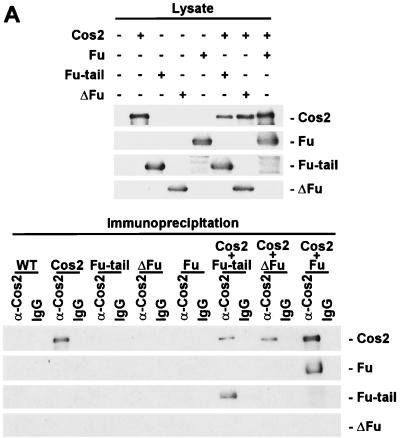
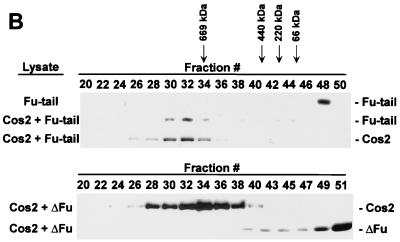
We hypothesized that the region of Fu that binds Cos2 is located in the carboxyl-terminal domain, given the inability of Fu mutants lacking this domain to bind Cos2 (43). To test this hypothesis we constructed a recombinant baculovirus expressing only the carboxyl-terminal domain of Fu (Fu-tail). We then coexpressed Fu-tail with or without Cos2 in Sf21 cells, using ΔFu coinfected with Cos2 as a negative control for Cos2 binding (Fig. 4A, top panel). When Cos2 was immunoprecipitated from extracts of these various infected cells, Fu-tail coimmunoprecipitated with Cos2 whereas ΔFu did not (Fig. 4A, bottom panel). To verify that the interaction between Fu-tail and Cos2 is quantitative, we subjected the various cellular lysates to gel filtration analysis. Fu-tail, when expressed alone, elutes at a molecular size (fraction 48) consistent with its calculated molecular weight. When Fu-tail is coexpressed with Cos2, its peak shifts to fraction 32, where it comigrates with Cos2 (Fig. 4B, top panel). Under these conditions, Cos2 elutes in fraction 32 instead of fraction 34 (see Fig. 2B, top panel). When lysates from ΔFu- and Cos2-coinfected cells are fractionated, ΔFu elutes at a size inconsistent with it being associated with Cos2.
FIG. 4.
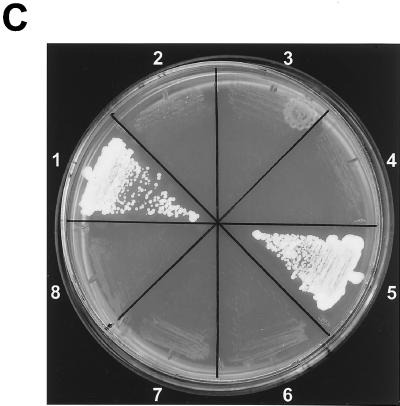
The carboxyl-terminal domain of Fu tethers Fu to Cos2. (A) Fu-tail associates with Cos2. An immunoblot of Cos2, Fu, Fu-tail, and ΔFu from the various Sf21 lysates is shown (top panel). Fu-tail coimmunoprecipitates with Cos2 from lysates that contain Cos2 and Fu-tail (bottom panel). A lysate containing Cos2 and Fu served as a positive control, while one containing Cos2 and ΔFu served as a negative control for binding in this experiment. A lysate containing Fu was used as an immunoblotting control in the bottom panel (data not shown). (B) Fu-tail quantitatively associates with Cos2. Gel filtration analysis of Sf21 lysates containing Cos2 coexpressed with either Fu-tail (top panel) or ΔFu (bottom panel). Fu-tail elutes at fraction 48 when expressed on its own in Sf21 cells. Cos2 and Fu-tail, from the coinfected Sf21 lysate, comigrate at fraction 32, indicating that Fu-tail can quantitatively associate with Cos2. In a Cos2 and ΔFu coinfection, Cos2 elutes at fraction 34 while ΔFu elutes at fraction 51, verifying that these proteins do not interact significantly, at least not with high affinity. (C) A directed yeast two-hybrid assay demonstrating that Fu-tail directly associates with Cos2. Yeast organisms transformed with various vectors were plated on minimal media without leucine, uracil, or adenine. Yeast transformed with the GAL4 activation domain/Fu-tail and the GAL4 DNA binding domain/Cos2 vectors were able to grow (sector 1). As negative controls, Fu-tail (sectors 2 and 4) and Cos2 (sectors 3 and 4) were also subcloned in their antisense orientation. The reciprocal experiment, in which Fu-tail was subcloned as a fusion protein with the GAL4 DNA binding domain and Cos2 was subcloned as a fusion protein with the GAL4 activation domain (sector 5), was also performed. As before, both Fu-tail (sectors 7 and 8) and Cos2 (sectors 6 and 8) were also subcloned in their antisense orientation.
Although we highly purified the Fu/Cos2 complex (Fig. 3C), it is still possible that there were numerous copies of both proteins present in this complex, bound together by a cross-bridging protein. Such a putative cross-bridging protein would have to be provided by the Sf21 cells and be present as a minor contaminant in our highly purified Fu/Cos2 complex. To bolster our argument that Fu-tail interacts directly with Cos2, we conducted a directed yeast two-hybrid assay. We expressed either Fu-tail or Cos2 as a GAL4 DNA binding domain fusion protein (bait) and as a GAL4 activation domain fusion protein (prey) (Fig. 4C). We observed that Fu-tail and Cos2 are able to associate sufficiently to permit the growth of yeast under the stringent conditions of our yeast two-hybrid system (Fig. 4C, sectors 1 and 5). This association was independent of whether Cos2 or Fu-tail served as the bait fusion protein or if both Cos2 and Fu-tail were subcloned in their antisense orientation. Thus, Fu-tail is necessary and sufficient to recapitulate the high affinity quantitative association seen with Cos2 and full-length Fu.
The carboxyl-terminal domain of Fu can compete with endogenous Fu for Cos2 binding.
We hypothesized that overexpression of Fu-tail in vivo might allow it to act as a dominant inhibitor of the Hh pathway, by associating with Cos2 to compromise the interaction between Cos2 and endogenous full-length Fu. To test this hypothesis we coexpressed Cos2 and Fu with increasing amounts of Fu-tail in Sf21 cells (Fig. 5A, top panel). These lysates were then immunoprecipitated using the 5D6 antibody and subjected to immunoblot analysis. Cos2 and full-length Fu are able to associate tightly in the absence of Fu-tail (Fig. 5A, bottom panel). As Fu-tail expression is increased, the association between Cos2 and Fu is concomitantly reduced. At the highest levels of Fu-tail expression, Cos2 associated predominantly with Fu-tail, instead of full-length Fu, in a manner consistent with our hypothesis.
FIG. 5.
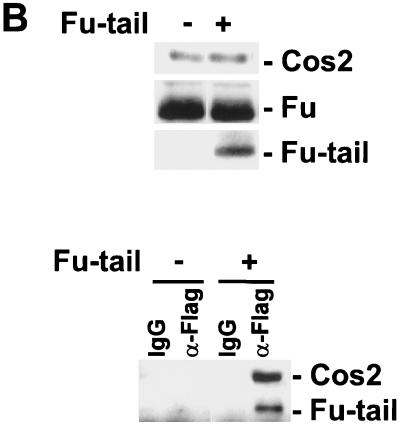
Fu-tail can compete with full-length Fu for association with Cos2 in vitro and in vivo. (A) Fu-tail disrupts a full-length Fu/Cos2 association. Immunoblots of lysates containing Cos2, Fu, and increasing amounts of Fu-tail are shown (top panel). These lysates were then immunoprecipitated with the 5D6 MAb, resolved by SDS-PAGE, and immunoblotted for Cos2, Fu, and Fu-tail. As shown, the full-length Fu/Cos2 association is disrupted with an increasing expression of Fu-tail (bottom panel). (B) Fu-tail can compete with endogenous Fu for association with Cos2 in S2 cells. S2 cells were transfected with a vector control or a vector expressing Flag Fu-tail. Forty-eight hours posttransfection the various transfected S2 cells were lysed in an NP-40 lysis buffer, resolved by SDS-PAGE, and then immunoblotted with Cos2 or Fu antibodies (top panel) or immunoprecipitated with either an anti-flag MAb or an isotype-matched IgG control (bottom panel).
To verify that Fu-tail could compete with full-length Fu, for Cos2, under conditions where more physiologically relevant levels of Fu and Cos2 are present, we expressed Flag-tagged Fu-tail in S2 cells. Lysates from transfected cells were separated by SDS-PAGE and imunoblotted to verify the normalized expression of Fu-tail and Cos2 (Fig. 5B, top panel). The lysates were then immunoprecipitated with an anti-Flag MAb and subjected to immunoblot analysis. Cos2 specifically coimmunoprecipitated with Fu-tail. These results suggest that Fu-tail can specifically associate with endogenous Cos2 (Fig. 5B, bottom panel). This association does not appear to be Hh sensitive (data not shown). Furthermore, our results suggest that Fu-tail can disrupt an association between Cos2 and full-length Fu, since the majority of Cos2 is normally complexed with Fu (Fig. 2B) (43). Thus, Fu-tail can compete with endogenous Fu for Cos2.
Fu-tail can disrupt the Hh signaling pathway.
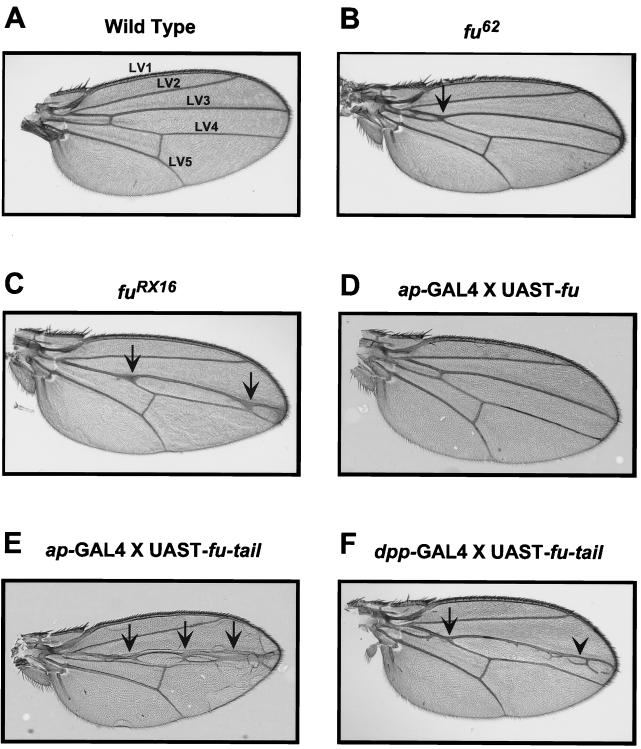
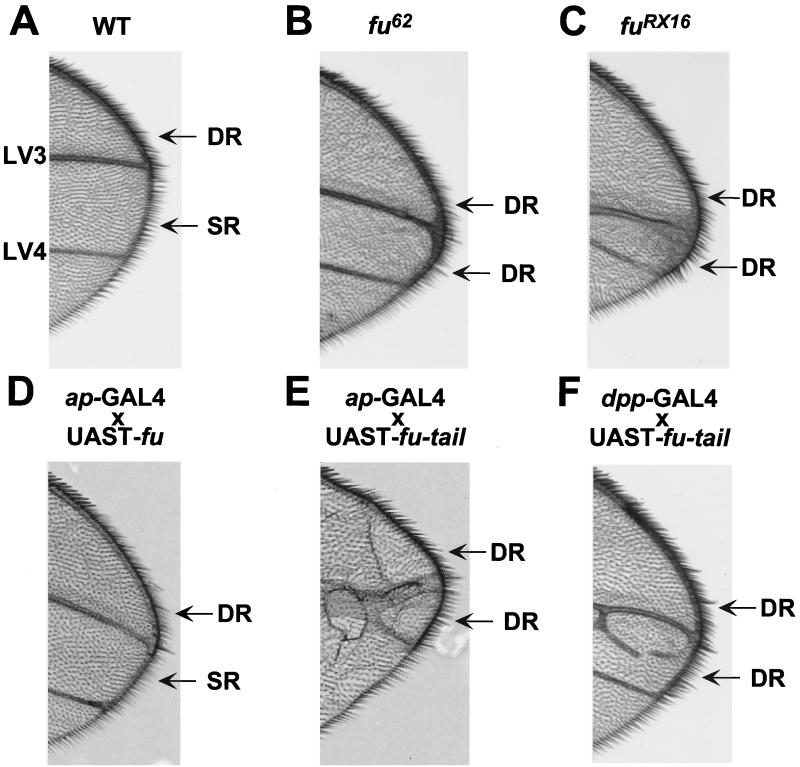
To define the role of the carboxyl-terminal domain of Fu in Hh signaling in vivo, we generated transgenic flies expressing fu-tail or wt fu. A fu mutant adult fly, which displays a classic fu LV3-LV4 fusion wing phenotype, can be rescued by expressing full-length UAST-fu under the control of either the dpp-GAL4 or the ptc-GAL4 driver. These results suggested that these domains of expression delineate where Fu is required in the wing imaginal disc (2). Therefore, we expressed UAST-fu-tail, or UAST-fu, driven by dpp-GAL4 or ptc-GAL4 in otherwise wt flies, to assess whether Fu-tail could act as a dominant inhibitor of the Hh pathway in vivo. Additionally, we expressed fu-tail using the apterous promoter because this promoter, unlike dpp-GAL4 or ptc-GAL4, is not Hh sensitive. Regardless of the promoter used to drive expression, when fu-tail is expressed in wing imaginal discs, we observe a fusion of LV3 and LV4, a phenotype identical to fu loss-of-function mutations (Fig. 6 and data not shown) (16, 31). This phenotype is indicative of a loss of tissue at the A/P border of the wing imaginal disc and consistent with decreased Hh signaling (14). Moreover, overexpression of Fu-tail also results in the posterior extension of the wing margin double-row bristles (Fig. 7E and F). In fu mutants, the double row of bristles extends as far as LV4, consistent with a loss of anterior en expression (Fig. 7B and C, compare to Fig. 7A) (6, 46). Therefore, expression of Fu-tail disrupts anterior en expression, as determined by the loss of the single-row bristles and subsequent expansion of the double-row bristles. These findings are distinct from those seen for flies overexpressing full-length Fu, which do not exhibit any apparent wing phenotype (Fig. 6 and 7; data not shown) (2), even though wt fu is overexpressed to the same degree as fu-tail (Fig. 8).
FIG. 6.
Fu-tail expression in vivo results in a loss-of-function phenotype. Overexpression of Fu-tail in wing imaginal discs yields a phenotype similar to that seen with fu loss-of-function mutations. (A) A wt Drosophila wing blade. There is continuous separation between longitudinal wing veins 3 and 4 (LV3 and LV4). (B) A wing from a fu62 class I homozygous adult fly (Fig. 1). Proximal fusions between LV3 and LV4 (arrow) as well as narrowing of the distance between these veins distally can be seen. (C) A wing from a fuRX16 class II homozygous adult fly. Fusion between LV3 and LV4 can be seen as well as narrowing of this region (arrows). (D) A wing from an ap-GAL4 × UAST-fu fly. The wing blade appears normal and has no fusion of LV3 and LV4. (E) A wing from an ap-GAL4 × UAST-fu-tail fly. The wing has severe proximal and distal fusions of LV3 and LV4 (arrows). (F) A wing from a dpp-GAL4 × UAST-fu-tail fly. The wing has proximal fusions of LV3 and LV4 (arrow) as well as some distal ectopic veins (arrowhead).
FIG. 7.
Fu-tail disrupts anterior en expression. (A) The wing margins, from wt flies, have a double row (DR) of bristles from LV2 to LV3, and a single row (SR) of fine bristles in the intervein region between LV3 and LV4. The SR bristles are indicative of Hh-dependent anterior en expression (see text). In class I (B) and class II (C) fu alleles, DR bristles are found to extend beyond vein 3 to vein 4. (D) This extension of DR bristles is not observed when wt Fu is overexpressed using ap-GAL4, but it is clearly seen when Fu-tail is overexpressed with either ap-GAL4 (E) or dpp-GAL4 (F) drivers.
FIG. 8.
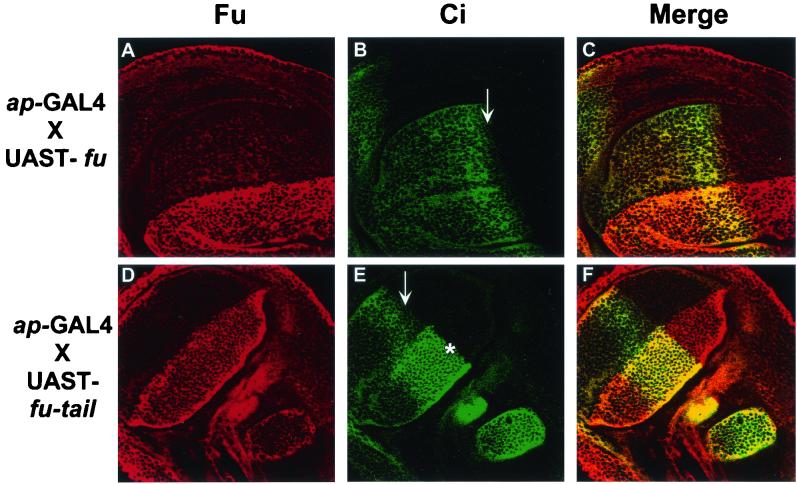
Fu-tail expression in vivo affects Ci155 processing. Staining for Fu, Ci, or both (merge) in third-instar wing imaginal discs isolated from transgenic Drosophila expressing UAST-fu-tail or UAST-fu driven by ap-GAL4. (A and D) Fu immunofluorescence, with an antiserum that recognizes both endogenous Fu and expressed Fu-tail. (B and E) Ci immunofluorescence with the 2A1 MAb that detects full-length Ci. (C and F) Composite images for top and bottom rows, respectively. Overexpression of Fu (A) does not appear to affect the staining of Ci (B) in the anterodorsal compartment of the wing pouch. Flies overexpressing Fu are phenotypically wt. The accumulation of full-length Ci at the A/P border can clearly be seen. In addition, Ci155 levels decrease slightly at a region immediately adjacent to the A/P border (arrows in panels B and E). This region contains the most active form of Ci, which has the ability to upregulate anterior en expression. Fu-tail is expressed across the entire dorsal compartment of the imaginal disc as indicated by the increased Fu staining (D). In the dorsal compartment, where Fu-tail is expressed, Ci155 levels do not decrease in the region immediately adjacent to the A/P border (∗) in discs expressing Fu-tail (E), in contrast to the reduced staining of Ci in the ventral region of the same disc (arrow in E), where Fu-tail is not expressed.
Wild-type discs have increased Ci155 levels in a tight band close, but not adjacent, to the A/P border, reflecting this area's increased exposure to Hh (2, 4, 32, 36, 61, 63). However, Ci155 levels noticeably decrease in the region directly adjacent to the A/P border, as previously described. These cells contains Ci*, which is responsible for anterior en transcription late in wing disk development (6, 36). To verify that overexpression of Fu-tail had resulted in changes in Ci processing to its various forms, we performed indirect immunofluorescence for Ci in wing imaginal discs expressing either wt fu or fu-tail. Ci staining in discs overexpressing full-length Fu is not significantly different from that observed for wt (Fig. 8B and data not shown). However, in disc regions where Fu-tail is overexpressed, Ci155 levels are not reduced at the A/P border, compared to either wt discs or more ventral regions of the A/P border (Fig. 8E). These results suggest that Fu-tail may block the ability of Ci to be processed into its most active labile form, Ci*. We find that in anterior regions distant from the A/P border, Ci155 does not accumulate, despite the overexpression of Fu-tail (Fig. 8D and E). These results suggest that overexpression of Fu-tail may not be sufficient to disrupt the proteolytic processing of Ci155 to Ci75. Thus, overexpression of the carboxyl-terminal domain of Fu disrupts the HSC in a specific manner that perturbs formation of Ci* but not Ci75.
DISCUSSION
In this study we show that Fu associates directly with Cos2 to form a high-affinity complex, which appears to comigrate with the peak of an endogenous Drosophila Fu/Cos2 complex (Peak B). We also show that the carboxyl-terminal domain of Fu is both necessary and sufficient to form this high-affinity association with Cos2. Finally, we test the physiological relevance of this association by demonstrating that overexpression of Fu-tail, in wing imaginal discs, disrupts Hh signaling. These results provide evidence that a targeted disruption of the Fu-Cos2 interaction effectively prevents Hh signaling, demonstrating the physiological importance of the HSC. We propose that Fu-tail is acting in a dominant negative manner in these discs, disrupting the interaction between Cos2 and endogenous Fu, for the following reasons: (i) Fu-tail is sufficient to bind to Cos2, (ii) Fu-tail is able to compete with endogenous wt Fu for binding to Cos2, (iii) overexpression of Fu-tail in a wt background results in phenotypes similar to loss of function fu mutants, and (iv) overexpression of wt Fu yields no phenotype.
Earlier experiments showed that class II fu alleles encode carboxyl-terminal truncations of Fu that do not bind Cos2, while class I alleles expressed point mutations or in-frame deletions in the kinase domain that do bind Cos2 (43). Those experiments, however, did not determine if the carboxyl-terminal domain was sufficient for binding. Based on the results presented here, we suggest that the main function of the carboxyl-terminal domain of Fu is to target the kinase domain to Cos2. Cos2 could then act as a substrate of Fu or could in turn present another substrate to Fu. This model would explain the similar phenotypes that result from either class I or class II mutations, at least in the presence of Su(fu) (40, 43). Class I mutant Fu would be unable to phosphorylate its substrate, and class II mutant Fu would be unable to locate its substrate. A similar model, based on experiments that used a recombinant fu construct lacking a kinase domain to decrease the severity of a fu class II/Su(fu)−/− phenotype, has been previously suggested (58). Alternatively, Fu-tail may also have some other function in vivo that results in a fu phenotype.
It has been proposed, based on the distribution of the various forms of Ci in either fu or cos2 mutants (2, 3, 28, 36, 47, 61, 63), that both Fu and Cos2 regulate Ci processing to either Ci75 or Ci*. However, our results indicate that while the expression of Fu-tail in wing imaginal discs blocks the conversion of Ci155 to Ci*, it does not appear to block the processing of Ci155 to Ci75. Thus, our Ci immunostaining pattern is similar to that described for fu class I mutant discs, which display an expanded stripe of Ci155 accumulation close to the A/P border (2, 56, 64), whereas fu class II mutant imaginal discs appear to accumulate Ci155 throughout most of, if not the entire, anterior compartment. We propose the following model to account for our results. When Fu-tail is overexpressed in wing imaginal discs, it displaces endogenous Fu from binding to Cos2. If the kinase domain of Fu is not bound to Cos2, Ci conversion to Ci* is blocked. Absence of Ci* results in a loss of anterior en expression and in the reduction or lack of ptc upregulation (2, 36, 64). The increased region of Ci155 stabilization, observed in wing imaginal discs expressing Fu-tail, would result as a secondary consequence of Hh being able to diffuse farther into the anterior regions of the imaginal disc, as previously suggested (2). However, overexpression of Fu-tail did not appear to affect Ci conversion to Ci75 at more anterior regions of the wing imaginal discs, even though wt Fu would also be separated from Cos2 in this part of the disc. These results suggest that the carboxyl-terminal domain of Fu has two functions. In the absence of Hh, the carboxyl-terminal domain of Fu acts to stabilize the HSC, in part with Cos2, and efficiently process Ci to Ci75, as has been recently suggested (27). At high levels of Hh signaling, the carboxyl-terminal domain of Fu is required to target its kinase domain to Cos2. Cos2 could then act as a substrate of Fu or could in turn present another substrate to Fu.
We propose, based on our data and those of others (2, 34, 43, 56), that Fu-tail plays a structural role in maintaining a functional HSC. Given the role that components of the human Hh pathway play in a variety of cancers (10, 18, 23, 25, 65), functional inhibitors of this pathway will be therapeutically beneficial. Therefore, it would be of interest to determine whether disrupting a human FU/COS2 complex with a human FU-tail would inhibit the human Hh signaling pathway. However, while we present a model whereby Fu-tail acts as a dominant inhibitor of Hh signaling, it has been previously suggested that hFU-tail can activate the mammalian Hh pathway (34). This study also showed that hFU and the mammalian Ci homolog GLI2 synergize to activate a Gli-reporter assay, an assay that appeared not to require a functional kinase domain. In contrast, during the revision of the manuscript for this report, another group has shown that the kinase activity of Drosophila Fu is required for Ci-dependent transcriptional activity (17). The reasons for these apparent discrepancies between hFu and Drosophila Fu remain unknown.
Acknowledgments
We thank the members of the Robbins' laboratory, A. Capobianco and members of his laboratory, M. Wathelet, and Y. Sanchez (University of Cincinnati) for helpful discussions. We thank the members of the Sanchez laboratory for their help in our yeast two-hybrid work. We thank P. Aza-Blanc and T. Kornberg (University of California, San Francisco) for assistance with confocal microscopy and K. Hill (University of California, San Francisco) for assistance in generating the transgenic flies. We also thank J. M. Bishop (University of California, San Francisco), in whose laboratory part of this work was done.
This work was supported by National Institutes of Health grant CA82628-01 (to D.J.R.) and NCI National Institutes of Health training grant 5T32 ES07250 (to M.A.). D.J.R. is a recipient of a Burroughs Wellcome Fund Career Award in the Biomedical Sciences.
M.A. and K.E.N. contributed equally to this paper.
REFERENCES
- 1.Alexandre, C., A. Jacinto, and P. W. Ingham. 1996. Transcriptional activation of hedgehog target genes in Drosophila is mediated directly by the cubitus interruptus protein, a member of the GLI family of zinc finger DNA-binding proteins. Genes Dev. 10:2003-2013. [DOI] [PubMed] [Google Scholar]
- 2.Alves, G., B. Limbourg-Bouchon, H. Tricoire, J. Brissard-Zahraoui, C. Lamour-Isnard, and D. Busson. 1998. Modulation of Hedgehog target gene expression by the Fused serine-threonine kinase in wing imaginal discs. Mech. Dev. 78:17-31. [DOI] [PubMed] [Google Scholar]
- 3.Aza-Blanc, P., and T. B. Kornberg. 1999. Ci: a complex transducer of the hedgehog signal. Trends Genet. 15:458-462. [DOI] [PubMed] [Google Scholar]
- 4.Aza-Blanc, P., F. A. Ramirez-Weber, M. P. Laget, C. Schwartz, and T. B. Kornberg. 1997. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell 89:1043-1053. [DOI] [PubMed] [Google Scholar]
- 5.Basler, K., and G. Struhl. 1994. Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature 368:208-214. [DOI] [PubMed] [Google Scholar]
- 6.Blair, S. S. 1992. Engrailed expression in the anterior lineage compartment of the developing wing blade of Drosophila. Development 115:21-33. [DOI] [PubMed] [Google Scholar]
- 7.Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 8.Calleja, M., E. Moreno, S. Pelaz, and G. Morata. 1996. Visualization of gene expression in living adult Drosophila. Science 274:252-255. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C. H., D. P. von Kessler, W. Park, B. Wang, Y. Ma, and P. A. Beachy. 1999. Nuclear trafficking of Cubitus interruptus in the transcriptional regulation of Hedgehog target gene expression. Cell 98:305-316. [DOI] [PubMed] [Google Scholar]
- 10.Chidambaram, A., A. M. Goldstein, M. R. Gailani, B. Gerrard, S. J. Bale, J. J. DiGiovanna, A. E. Bale, and M. Dean. 1996. Mutations in the human homologue of the Drosophila patched gene in Caucasian and African-American nevoid basal cell carcinoma syndrome patients. Cancer Res. 56:4599-4601. [PubMed] [Google Scholar]
- 11.DiNardo, S., E. Sher, J. Heemskerk-Jongens, J. A. Kassis, and P. H. O'Farrell. 1988. Two-tiered regulation of spatially patterned engrailed gene expression during Drosophila embryogenesis. Nature 332:604-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dominguez, M., M. Brunner, E. Hafen, and K. Basler. 1996. Sending and receiving the hedgehog signal: control by the Drosophila Gli protein Cubitus interruptus. Science 272:1621-1625. [DOI] [PubMed] [Google Scholar]
- 13.Eaton, S., and T. B. Kornberg. 1990. Repression of ci-D in posterior compartments of Drosophila by engrailed. Genes Dev. 4:1068-1077. [DOI] [PubMed] [Google Scholar]
- 14.Fausto-Sterling, A. 1978. Pattern formation in the wing veins of the fused mutant (Drosophila melanogaster). Dev. Biol. 63:358-369. [DOI] [PubMed] [Google Scholar]
- 15.Fietz, M. J., J. P. Concordet, R. Barbosa, R. Johnson, S. Krauss, A. P. McMahon, C. Tabin, and P. W. Ingham. 1994. The hedgehog gene family in Drosophila and vertebrate development. Dev. Suppl. 1994:43-51. [PubMed] [Google Scholar]
- 16.Forbes, A. J., Y. Nakano, A. M. Taylor, and P. W. Ingham. 1993. Genetic analysis of hedgehog signalling in the Drosophila embryo. Dev. Suppl. 1993:115-124. [PubMed] [Google Scholar]
- 17.Fukumoto, T., R. Watanabe-Fukunaga, K. Fujisawa, S. Nagata, and R. Fukunaga. 2001. The fused protein kinase regulates hedgehog-stimulated transcriptional activation in drosophila schneider 2 cells. J. Biol. Chem. 276:38441-38448. [DOI] [PubMed] [Google Scholar]
- 18.Gailani, M. R., M. Stahle-Backdahl, D. J. Leffell, M. Glynn, P. G. Zaphiropoulos, C. Pressman, A. B. Unden, M. Dean, D. E. Brash, A. E. Bale, and R. Toftgard. 1996. The role of the human homologue of Drosophila patched in sporadic basal cell carcinomas. Nat. Genet. 14:78-81. [DOI] [PubMed] [Google Scholar]
- 19.Heemskerk, J., and S. DiNardo. 1994. Drosophila hedgehog acts as a morphogen in cellular patterning. Cell 76:449-460. [DOI] [PubMed] [Google Scholar]
- 20.Hidalgo, A. 1994. Three distinct roles for the engrailed gene in Drosophila wing development. Curr. Biol. 4:1087-1098. [DOI] [PubMed] [Google Scholar]
- 21.Hinz, U., B. Giebel, and J. A. Campos-Ortega. 1994. The basic-helix-loop-helix domain of Drosophila lethal of scute protein is sufficient for proneural function and activates neurogenic genes. Cell 76:77-87. [DOI] [PubMed] [Google Scholar]
- 22.Hui, C. C., D. Slusarski, K. A. Platt, R. Holmgren, and A. L. Joyner. 1994. Expression of three mouse homologs of the Drosophila segment polarity gene cubitus interruptus, Gli, Gli-2, and Gli-3, in ectoderm- and mesoderm-derived tissues suggests multiple roles during postimplantation development. Dev. Biol. 162:402-413. [DOI] [PubMed] [Google Scholar]
- 23.Ingham, P. W. 1998. The patched gene in development and cancer. Curr. Opin. Genet. Dev. 8:88-94. [DOI] [PubMed] [Google Scholar]
- 24.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson, R. L., A. L. Rothman, J. Xie, L. V. Goodrich, J. W. Bare, J. M. Bonifas, A. G. Quinn, R. M. Myers, D. R. Cox, E. H. Epstein, Jr., and M. P. Scott. 1996. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science 272:1668-1671. [DOI] [PubMed] [Google Scholar]
- 26.Lee, J. J., D. P. von Kessler, S. Parks, and P. A. Beachy. 1992. Secretion and localized transcription suggest a role in positional signaling for products of the segmentation gene hedgehog. Cell 71:33-50. [DOI] [PubMed] [Google Scholar]
- 27.Lefers, M. A., Q. T. Wang, and R. A. Holmgren. 2001. Genetic dissection of the Drosophila Cubitus interruptus signaling complex. Dev. Biol. 236:411-420. [DOI] [PubMed] [Google Scholar]
- 28.Methot, N., and K. Basler. 1999. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell 96:819-831. [DOI] [PubMed] [Google Scholar]
- 29.Methot, N., and K. Basler. 2000. Suppressor of fused opposes hedgehog signal transduction by impeding nuclear accumulation of the activator form of Cubitus interruptus. Development 127:4001-4010. [DOI] [PubMed] [Google Scholar]
- 30.Mohler, J. 1988. Requirements for hedgehog, a segmental polarity gene, in patterning larval and adult cuticle of Drosophila. Genetics 120:1061-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan, T. H., and C. B. Bridges. 1916. Sex-linked inheritance in Drosophila. Carnegie Inst. Washington Publ. 237:55-58. [Google Scholar]
- 32.Motzny, C. K., and R. Holmgren. 1995. The Drosophila cubitus interruptus protein and its role in the wingless and hedgehog signal transduction pathways. Mech. Dev. 52:137-150. [DOI] [PubMed] [Google Scholar]
- 33.Mullor, J. L., and I. Guerrero. 2000. A gain-of-function mutant of patched dissects different responses to the hedgehog gradient. Dev. Biol. 228:211-224. [DOI] [PubMed] [Google Scholar]
- 34.Murone, M., S. M. Luoh, D. Stone, W. Li, A. Gurney, M. Armanini, C. Grey, A. Rosenthal, and F. J. de Sauvage. 2000. Gli regulation by the opposing activities of fused and suppressor of fused. Nat. Cell Biol. 2:310-312. [DOI] [PubMed] [Google Scholar]
- 35.Nusslein-Volhard, C., and E. Wieschaus. 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287:795-801. [DOI] [PubMed] [Google Scholar]
- 36.Ohlmeyer, J. T., and D. Kalderon. 1998. Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature 396:749-753. [DOI] [PubMed] [Google Scholar]
- 37.Orenic, T. V., D. C. Slusarski, K. L. Kroll, and R. A. Holmgren. 1990. Cloning and characterization of the segment polarity gene cubitus interruptus Dominant of Drosophila. Genes Dev. 4:1053-1067. [DOI] [PubMed] [Google Scholar]
- 38.Pham, A., P. Therond, G. Alves, F. B. Tournier, D. Busson, C. Lamour-Isnard, B. L. Bouchon, T. Preat, and H. Tricoire. 1995. The Suppressor of fused gene encodes a novel PEST protein involved in Drosophila segment polarity establishment. Genetics 140:587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Porter, J. A., D. P. von Kessler, S. C. Ekker, K. E. Young, J. J. Lee, K. Moses, and P. A. Beachy. 1995. The product of hedgehog autoproteolytic cleavage active in local and long-range signalling. Nature 374:363-366. [DOI] [PubMed] [Google Scholar]
- 40.Preat, T. 1992. Characterization of Suppressor of fused, a complete suppressor of the fused segment polarity gene of Drosophila melanogaster. Genetics 132:725-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Preat, T., P. Therond, C. Lamour-Isnard, B. Limbourg-Bouchon, H. Tricoire, I. Erk, M. C. Mariol, and D. Busson. 1990. A putative serine/threonine protein kinase encoded by the segment-polarity fused gene of Drosophila. Nature 347:87-89. [DOI] [PubMed] [Google Scholar]
- 42.Preat, T., P. Therond, B. Limbourg-Bouchon, A. Pham, H. Tricoire, D. Busson, and C. Lamour-Isnard. 1993. Segmental polarity in Drosophila melanogaster: genetic dissection of fused in a Suppressor of fused background reveals interaction with costal-2. Genetics 135:1047-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robbins, D. J., K. E. Nybakken, R. Kobayashi, J. C. Sisson, J. M. Bishop, and P. P. Therond. 1997. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell 90:225-234. [DOI] [PubMed] [Google Scholar]
- 44.Rubin, G. M., and A. C. Spradling. 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218:348-353. [DOI] [PubMed] [Google Scholar]
- 45.Ruiz i Altaba, A. 1997. Catching a Gli-mpse of Hedgehog. Cell 90:193-196. [DOI] [PubMed] [Google Scholar]
- 46.Sanchez-Herrero, E., J. P. Couso, J. Capdevila, and I. Guerrero. 1996. The fu gene discriminates between pathways to control dpp expression in Drosophila imaginal discs. Mech. Dev. 55:159-170. [DOI] [PubMed] [Google Scholar]
- 47.Sisson, J. C., K. S. Ho, K. Suyama, and M. P. Scott. 1997. Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell 90:235-245. [DOI] [PubMed] [Google Scholar]
- 48.Staehling-Hampton, K., and F. M. Hoffmann. 1994. Ectopic decapentaplegic in the Drosophila midgut alters the expression of five homeotic genes, dpp, and wingless, causing specific morphological defects. Dev. Biol. 164:502-512. [DOI] [PubMed] [Google Scholar]
- 49.Staehling-Hampton, K., F. M. Hoffmann, M. K. Baylies, E. Rushton, and M. Bate. 1994. dpp induces mesodermal gene expression in Drosophila. Nature 372:783-786. [DOI] [PubMed] [Google Scholar]
- 50.Stegman, M. A., J. E. Vallance, G. Elangovan, J. Sosinski, Y. Cheng, and D. J. Robbins. 2000. Identification of a tetrameric hedgehog signaling complex. J. Biol. Chem. 275:21809-21812. [DOI] [PubMed] [Google Scholar]
- 51.Strigini, M., and S. M. Cohen. 1999. Formation of morphogen gradients in the Drosophila wing. Semin. Cell Dev. Biol. 10:335-344. [DOI] [PubMed] [Google Scholar]
- 52.Strigini, M., and S. M. Cohen. 1997. A Hedgehog activity gradient contributes to AP axial patterning of the Drosophila wing. Development 124:4697-4705. [DOI] [PubMed] [Google Scholar]
- 53.Tabata, T., S. Eaton, and T. B. Kornberg. 1992. The Drosophila hedgehog gene is expressed specifically in posterior compartment cells and is a target of engrailed regulation. Genes Dev. 6:2635-2645. [DOI] [PubMed] [Google Scholar]
- 54.Tabata, T., and T. B. Kornberg. 1994. Hedgehog is a signaling protein with a key role in patterning Drosophila imaginal discs. Cell 76:89-102. [DOI] [PubMed] [Google Scholar]
- 55.Tabata, T., C. Schwartz, E. Gustavson, Z. Ali, and T. B. Kornberg. 1995. Creating a Drosophila wing de novo, the role of engrailed, and the compartment border hypothesis. Development 121:3359-3369. [DOI] [PubMed] [Google Scholar]
- 56.Therond, P., G. Alves, B. Limbourg-Bouchon, H. Tricoire, E. Guillemet, J. Brissard-Zahraoui, C. Lamour-Isnard, and D. Busson. 1996. Functional domains of fused, a serine-threonine kinase required for signaling in Drosophila. Genetics 142:1181-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Therond, P., D. Busson, E. Guillemet, B. Limbourg-Bouchon, T. Preat, R. Terracol, H. Tricoire, and C. Lamour-Isnard. 1993. Molecular organisation and expression pattern of the segment polarity gene fused of Drosophila melanogaster. Mech. Dev. 44:65-80. [DOI] [PubMed] [Google Scholar]
- 58.Therond, P. P., J. D. Knight, T. B. Kornberg, and J. M. Bishop. 1996. Phosphorylation of the fused protein kinase in response to signaling from hedgehog. Proc. Natl. Acad. Sci. USA 93:4224-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Von Ohlen, T., D. Lessing, R. Nusse, and J. E. Hooper. 1997. Hedgehog signaling regulates transcription through cubitus interruptus, a sequence-specific DNA binding protein. Proc. Natl. Acad. Sci. USA 94:2404-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walterhouse, D., M. Ahmed, D. Slusarski, J. Kalamaras, D. Boucher, R. Holmgren, and P. Iannaccone. 1993. gli, a zinc finger transcription factor and oncogene, is expressed during normal mouse development. Dev. Dyn. 196:91-102. [DOI] [PubMed] [Google Scholar]
- 61.Wang, G., K. Amanai, B. Wang, and J. Jiang. 2000. Interactions with Costal2 and suppressor of fused regulate nuclear translocation and activity of cubitus interruptus. Genes Dev. 14:2893-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, G., B. Wang, and J. Jiang. 1999. Protein kinase A antagonizes Hedgehog signaling by regulating both the activator and repressor forms of Cubitus interruptus. Genes Dev. 13:2828-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, Q. T., and R. A. Holmgren. 2000. Nuclear import of cubitus interruptus is regulated by hedgehog via a mechanism distinct from Ci stabilization and Ci activation. Development 127:3131-3139. [DOI] [PubMed] [Google Scholar]
- 64.Wang, Q. T., and R. A. Holmgren. 1999. The subcellular localization and activity of Drosophila cubitus interruptus are regulated at multiple levels. Development 126:5097-5106. [DOI] [PubMed] [Google Scholar]
- 65.Xie, J., R. L. Johnson, X. Zhang, J. W. Bare, F. M. Waldman, P. H. Cogen, A. G. Menon, R. S. Warren, L. C. Chen, M. P. Scott, and E. H. Epstein, Jr. 1997. Mutations of the PATCHED gene in several types of sporadic extracutaneous tumors. Cancer Res. 57:2369-2372. [PubMed] [Google Scholar]