Abstract
We produced a monoclonal antibody (MAb) (7G10) that has blocking activity against porcine reproductive and respiratory syndrome virus (PRRSV). In this study, we identified the components of the 7G10 MAb-bound complex as cytoskeletal filaments: vimentin, cytokeratin 8, cytokeratin 18, actin, and hair type II basic keratin. Vimentin bound to PRRSV nucleocapsid protein and anti-vimentin antibodies showed PRRSV-blocking activity. Vimentin was expressed on the surface of MARC-145, a PRRSV-susceptible cell line. Simian vimentin rendered BHK-21 and CRFK, nonsusceptible cell lines, susceptible to PRRSV infection. These results suggest that vimentin is part of the PRRSV receptor complex and that it plays an important role in PRRSV binding with the other cytoskeletal filaments that mediate transportation of the virus in the cytosol.
Porcine reproductive and respiratory syndrome (PRRS) is the most important disease causing serious economic losses to the swine industry. Since it was first described in 1987 in the United States as a “mystery swine syndrome,” PRRS has continued to cause severe reproductive failure in sows and respiratory problems in young piglets (8). PRRS virus (PRRSV), the causative agent of PRRS, is a member of the family Arteriviridae, grouped together with the Coronaviridae and Toroviridae in the order Nidovirales (6). Other members of this family are equine arteritis virus, lactate dehydrogenase-elevating virus of mice, and simian hemorrhagic fever virus (33). PRRSV is 40 to 70 nm in diameter and is an enveloped, single-stranded, positive-sense RNA virus. The PRRSV genome, 14.5 kb in length, encodes the viral replicase; the membrane glycoproteins GP2a, GP3, GP4, and GP5; the unglycosylated membrane protein P2b; the matrix protein, M; and the nucleocapsid protein, N.
PRRSV has a highly restricted cell tropism both in vivo and in vitro. PRRSV infects the African green monkey kidney cell line, MA-104 and its derivatives, MARC-145, and CL-2621 in vitro (23). PRRSV preferentially infects the cells of the monocyte/macrophage lineage, especially porcine alveolar macrophages (PAM), in the natural host (13). PRRSV enters host cells through a mechanism of receptor-mediated endocytosis. The entry mechanism is mediated by attachment to one or more cellular receptors and/or coreceptors, which are important determinants of the highly restricted cell tropism (25). To date, only two molecules have been described as putative receptors. One candidate is heparan sulfate (HS). However, addition of soluble HS did not completely block binding and infection of PRRSV, even though it seems to play an important role in the virus infection of PAM (11). The other candidate is sialoadhesin (Sn), which was previously described as a 210-kDa protein (p210) immunoprecipitated by PRRSV blocking monoclonal antibody (MAb) (MAb41D3) on PAM. However, transfection of Sn cDNA did not fully engender PRRSV susceptibility in CRFK cells, even though its antibody (Ab) completely blocked infection (10, 12, 39). Both HS and Sn, which are involved in PRRSV infection of PAM, are not expressed on MARC-145 cells.
In this study, we found that PRRSV binds to vimentin, an intermediate-filament (IF) protein, and anti-vimentin Abs block PRRSV infection of MARC-145 cells. When simian vimentin was delivered to BHK-21 and CRFK cells, they became susceptible to PRRSV. Vimentin is also thought to be involved in the replication and transportation of PRRSV inside the cell by forming a complex with the other components of the intermediate filaments.
MATERIALS AND METHODS
Cell lines and virus.
Cell lines used in this study were derivatives of the African green monkey kidney (MARC-145 and Vero), baby hamster kidney (BHK-21), Crandall Rees feline kidney (CRFK), porcine kidney (PK-15), swine testicle (ST), and Maden-Darby canine kidney (MDCK) cell lines. The cell lines were grown in Eagle's minimum essential medium (Life Technologies, Inc., Gaithersburg, MD) supplemented with 10% fetal bovine serum (FBS) (HyClone, Logan, UT). The ATCC VR 2332 strain of PRRSV was used in the study.
32P-labeled RNA probe preparation.
α-32P-labeled 3′ untranslated region RNA transcript was prepared by in vitro transcription using a T7 RNA synthesis kit, Riboscribe (Epicenter Technologies, Madison, WI), by following the manufacturer's instructions. The probe was purified by Quick Spin columns (Boehringer Mannheim, Indianapolis, IN).
North-Western blotting of RNA-binding proteins.
To screen PRRSV 3′ untranslated region RNA-binding proteins, North-Western blot analysis was performed. Cytoplasmic protein extracts of PRRSV-infected MARC-145 cells were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After the proteins were transferred onto a nitrocellulose membrane (Gelman Sciences, Ann Arbor, MI), the membrane was denatured in 6 M guanidinium hydrochloride for 30 min, followed by sequential renaturation every 10 min with changes of single-binding (SB) buffer (0.05 M NaCl, 10 mM Tris [pH 7.0], 1 mM EDTA, 0.02% [wt/vol] bovine serum albumin, 0.02% [wt/vol] Ficoll, and 0.02% [wt/vol] polyvinyl pyrrolidone). Hybridization was performed in SB buffer containing 32P-labeled RNA probe at 500,000 cpm/ml in the presence of 10 μg/ml of yeast tRNA and 100 μg/ml of denatured sheared salmon sperm DNA overnight. The membrane was washed three times with SB buffer for 30 min and then visualized by autoradiography.
Production and characterization of 7G10 MAb.
The desired protein band on the X-ray film was aligned to a Ponceau S-stained nitrocellulose membrane, and then the band (57 kDa) was excised from the membrane. The protein was eluted from the membrane with 500 μl of elution buffer (1% Triton X-100 in 50 mM Tris-HCl, pH 9.5) per cm2 of the membrane and injected five times at weekly intervals into a BALB/c mouse, along with RIBI adjuvant (RIBI ImmunoChem Research, Inc., Hamilton, MT). Three days before fusion, the mouse was boosted with antigen. The spleen cells were collected and fused with Ag-8 cells. The fused cells in a hypoxanthine-aminopterin-thymidine medium with 20% FBS and 5% hybridoma cloning factor (Fisher Scientific, Pittsburgh, PA) were dispensed into a 96-well tissue culture plate and incubated at 37°C. The MAb supernatants were screened by enzyme-linked immunosorbent assay. Immunolon 1 96-well flat-bottom plates (Dynatech Technologies, Chantilly, VA) were coated with the cytoplasmic proteins of MARC-145 cells, and then each supernatant was used as a primary Ab after being blocked with 0.5% glycine in PBS. The peroxidase-conjugated horse anti-mouse immunoglobulin G (heavy and light chains) (IgG [H+L]) (Vector Laboratories, Inc., Burlingame, CA) was used as a secondary Ab. The presence of 7G10 MAb was detected by the addition of TMB microwell peroxidase substrate (one component) (KPL, Gaithersburg, MD). To confirm the specificity of the 7G10 MAb, Western blot analysis was performed. The cytoplasmic proteins of MARC-145 cells were separated by SDS-PAGE, and 7G10 MAb was used as a primary Ab after it was blocked with 5% skim milk in PBS. The peroxidase-conjugated horse anti-mouse IgG (H+L) was used as a secondary Ab. The band was detected by the addition of TMB membrane peroxidase substrate (one component) (KPL, Gaithersburg, MD).
Purification of 7G10 MAb-bound proteins.
A PRRSV blocking MAb (7G10) was affinity purified using KAPTIV-AE (Tecnogen S. C. P. A., Piana di Monte Verna [CE], Italy), following the manufacturer's instructions. The purified 7G10 MAb was cross-linked to the Affi-Gel active beads (Bio-Rad, Hercules, CA) according to the manufacturer's instructions. The beads (1 ml) were transferred to a column and washed with cold water (3 ml). Approximately 5 mg of 7G10 MAb per ml of beads was mixed and incubated at 4°C overnight. The beads were washed twice with 50 mM sodium phosphate (pH 7.5) and once with 50 mM sodium phosphate (pH 7.5) containing 1 M sodium chloride and blocked by the addition of 100 mM ethanolamine-HCl (pH 7.5) at 4°C overnight, followed by washing with PBS. MARC-145 cytoplasmic proteins were applied to the column and washed with 10 mM sodium phosphate (pH 6.5). The 7G10 MAb-bound proteins were eluted by adding 1 ml of 100 mM glycine (pH 2.5), and the eluted solution was neutralized with 100 μl of 1 M phosphate (pH 8.0).
Plaque reduction assay.
PRRSV was titered by plaque reduction assay in MARC-145 cell cultures that were preincubated with 7G10 MAb. MARC-145 cells were cultured overnight in a six-well tissue culture plate (2.5 × 105 cells/well). The confluent monolayer of cells was incubated with 200 μl of 7G10 MAb. After being washed three times, 100 PFU of PRRSV was added to each well and incubated at 37°C for 1 h. The cells were washed with PBS and overlaid with 1% agarose in minimum essential medium containing 1% FBS. The plate was incubated in an inverted position at 37°C for 24 h. The plaques were counted by staining them with neutral red (0.1% in agarose).
Two-dimensional (2-D) electrophoresis.
Readystrip immobilized pH gradient (IPG) strips (11 cm; pH 3-10NL; Bio-Rad, Hercules, CA) were rehydrated with DeStreak rehydration solution (Amersham Biosciences, Uppsala, Sweden) containing 110 μg of the protein sample at room temperature overnight. Isoelectric focusing was performed in the IPG strips for 10 h for a total of 35,000 V-hours. The IPG strips were incubated in equilibration buffer I containing 6 M urea, 2% (wt/vol) SDS, 0.05 M Tris-HCl (pH 8.8), 20% (vol/vol) glycerol, and 2% (vol/vol) dithiothreitol for 10 min and in equilibration buffer II containing 6 M urea, 2% (wt/vol) SDS, 0.05 M Tris-HCl (pH 8.8), 20% (vol/vol) glycerol, and 2.5% (wt/vol) iodoacetamide. The strips were loaded onto Criterion Precast Gels (10% Tris, 1.0 mm, and IPG + 1 Comb, 11 cm; Bio-Rad, Hercules, CA). SDS-PAGE was performed at 150 V, and then the protein spots were cored.
In-gel tryptic digestion, MALDI-TOF MS, and analysis of peptide sequences.
In-gel tryptic digestion and matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) were performed in the proteomics laboratory at the University of Louisville, Louisville, KY. Protein identification of peptide fragments was performed using Profound and Mascot search engines based on NCBI and/or SWISS-PROT protein databases. All protein identities were in the expected range of size and isoelectric point based on their positions in the gels.
Virus purification.
PRRSV was propagated in MARC-145 cells and purified by sucrose density gradient separation. Polyethylene glycol 8000 (Sigma Chemical Co., St. Louis, MO) at a final concentration of 8% (wt/vol) was added to concentrate the virus. After incubation at 4°C overnight, the precipitated virus was centrifuged at 10,800 × g for 20 min. The virus pellet was resuspended in TNE buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, and 1 mM EDTA). The resuspended virus was layered on 10 to 60% (wt/wt) sucrose gradients and centrifuged at 90,000 × g at 4°C overnight. The purified virus band was collected, and the density (g/cm3) was determined. Centrifugation was performed at 90,000 × g for 1 h to pellet the purified virus, and the pellet was resuspended in TNE buffer.
VOPBA.
To determine that the purified PRRSV could bind to vimentin, a virus overlay protein binding assay (VOPBA) was performed. Vimentin was separated by SDS-PAGE and transferred onto a nitrocellulose membrane. After being blocked in 10% skim milk in PBS at 4°C overnight, the membrane was incubated with 10 μg of the purified PRRSV at 4°C overnight. The membrane was washed with PBS three times and incubated with SR-30 (Rural Technologies, Inc., Brookings, SD), a MAb against PRRSV nucleocapsid protein, or a polyclonal Ab against PRRSV at room temperature for 1 h. The membrane was washed with PBS three times and incubated with the peroxidase-conjugated secondary Abs (horse anti-mouse IgG [H+L] for SR-30; goat anti-porcine IgG [H+L] [ICN Biomedicals, Inc., Aurora, OH] for polyclonal Ab against PRRSV) at room temperature for 1 h. The membrane was washed with PBS three times, and the presence of virus binding to vimentin was detected by the addition of TMB membrane peroxidase substrate (one component).
Total cellular protein isolation.
The total cellular proteins were isolated from MARC-145, BHK-21, CRFK, MDCK, PK-15, ST, or Vero cells. The cells were scraped in the sample preparation buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, and 0.25 M sucrose). One volume of this suspension was placed in a Polyallomer centrifuge tube (Beckman Instruments, Inc., Palo Alto, CA). Four volumes of the extraction buffer {9.6 M urea, 5% (wt/vol) 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS), 25 mM spermine base, and 50 mM dithiothreitol} were added to the tube containing sample preparation buffer and then mixed. Extraction was carried out at room temperature for 1 h. The samples were ultracentrifuged at 250,000 × g at 20°C for 1 h. The protein-containing supernatants were precipitated with 75% acetone containing 10% trichloroacetic acid at 4°C overnight. After ultracentrifugation at 10,000 × g at 4°C for 30 min, the pellets were resuspended in 1 ml of the extraction buffer.
Expression profiles of vimentin in different cell lines by Western blot analysis.
To examine the expression of vimentin in the different cell lines, Western blot analysis was performed. The total cellular proteins from MARC-145, BHK-21, CRFK, MDCK, PK-15, ST, and Vero cells were separated by SDS-PAGE and transferred onto a nitrocellulose membrane. After being blocked in 5% skim milk in 0.05% Tween 20-PBS at 4°C overnight, the membrane was incubated with monoclonal anti-vimentin clone V9 (Sigma-Aldrich Co., St. Louis, MO) at room temperature for 1 h. The membrane was washed with 0.05% Tween 20-PBS three times and incubated with the peroxidase-conjugated horse anti-mouse IgG (H+L) at room temperature for 1 h. The membrane was washed with 0.05% Tween 20-PBS three times, and the presence of vimentin was detected by the addition of TMB membrane peroxidase substrate (one component).
Checkerboard titration assay for measuring blocking activity of anti-vimentin Ab.
To examine the blocking activity of anti-vimentin Ab, a checkerboard titration assay was performed. MARC-145 cells were cultured overnight in a 96-well tissue culture plate (1 × 105 cells/well). The cells were incubated with PRRSV, which were preincubated with either polyclonal rabbit anti-human vimentin Ab (Biomeda Corporation, Foster City, CA) or monoclonal anti-β-galactosidase Ab (Boehringer Mannheim, Indianapolis, IN). The antibodies were prepared as serial twofold dilutions starting with a 1:20 dilution, and the PRRSV preparation was initially diluted 1:10 and in 10-fold dilutions thereafter. The cells were harvested 3 to 4 days postinfection, fixed with cold 80% acetone at 4°C for 10 min, and then incubated at 37°C for 30 min with fluorescein isothiocyanate (FITC)-conjugated SDOW-17 (Rural Technologies, Inc., Brookings, SD), a MAb against PRRSV nucleocapsid protein. After being washed twice in PBS, the cells were examined by fluorescence microscopy.
RT-PCR.
To amplify porcine vimentin cDNA, reverse transcription (RT)-PCR was performed. Total RNA was isolated from ST cells according to the acid guanidinium thiocyanate-phenol-chloroform extraction method (7). RT-PCR was performed using the GeneAmp EZ r Tth RNA PCR Kit (Roche Molecular System, Inc., Branchburg, NJ) with forward (5′-AGAGACGCAGCAGCGCTCCCTT-3′) and reverse (5′-GAAGCAGAACCAAGTTGGTTGG-3′) primers. Reverse transcription was performed at 42°C for 45 min, 95°C for 10 min, and 5°C for 5 min. Standard PCR was done at 95°C for 2 min, 95°C for 30 s, 53°C for 30 s, and 72°C for 90 s for 26 cycles and at 72°C for 30 min. To demonstrate PRRSV RNA production in cells, RT-PCR was performed. RNA was isolated with the RNeasy Mini Kit (QIAGEN, Valencia, CA). RT-PCR was performed using a OneStep RT-PCR kit (QIAGEN, Valencia, CA) with forward (5′-GGTCGCTATTTCACCTGGTA-3′) and reverse (5′-TCAATTCAGGCCTAAAGTTG-3′) primers. The reverse transcription reaction was done at 50°C for 30 min, and the initial PCR was done at 95°C for 15 min. Standard PCR was done at 94°C for 30 s, 55°C for 30 s, and 72°C for 60 s for 30 cycles and at 72°C for 30 min.
Cloning of the RT-PCR product from RNA of ST cells.
The RT-PCR product was purified with a Clontech NucleoTrap gel extraction kit (Clontech Laboratories, Inc., Palo Alto, CA). Purified cDNA was cloned with the pGEM-T Easy vector system I (Promega Corporation, Madison, WI).
Screening of MARC-145 cDNA library for simian vimentin.
The cDNA library from MARC-145 cells was previously constructed in our laboratory in the λ ZAP express vector system (36). The cDNA library was screened by Southern blotting. The cDNA library was combined with Escherichia coli host strain K802, mixed with NZY agarose, and then poured onto NZY agar plates. After being incubated at 37°C for 8 h, the plaques were transferred onto nylon membranes, denatured, neutralized, and fixed at 80°C for 2 h. The porcine vimentin cDNA from ST cells was used as a probe. The cDNA was labeled with [α-32P]CTP by Ready-To-Go DNA-labeling beads (Amersham Biosciences Corp., Piscataway, NJ). The hybridization was performed at 73°C in hybridization solution (5× NaCl-EDTA-Tris-HCl buffer (SET), 5× Denhardt's reagent, 1% SDS, 200 μg/ml denatured salmon sperm DNA, and 200 μg/ml heparin). The nylon membranes were washed for 30 min twice with wash buffer I (2× SET and 5× Denhardt's reagent), for 30 min four times with wash buffer II (2× SET and 0.5% SDS), and for 30 min twice with wash buffer III (0.1× SET and 0.1% SDS). The membranes were exposed to X-ray films, and then the films were developed.
Immunofluorescence microscopy analysis for localization of vimentin.
MARC-145 cells were cultured overnight in a 48-well tissue culture plate (2 × 105 cells/well) and then fixed in cold 80% acetone at 4°C for 10 min. The cells were incubated with FITC-conjugated monoclonal anti-vimentin (V9) Ab (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) at 37°C for 30 min, washed twice with PBS, and then examined by fluorescence microscopy.
Flow cytometric analysis.
MARC-145 cells (5× 105 cells) were washed in staining solution (0.1% bovine serum albumin and 0.1% NaN3 in PBS), blocked in 3% normal goat serum in staining solution on ice for 10 min, and then incubated with polyclonal rabbit anti-vimentin Ab (Biomeda, Foster City, CA) on ice for 30 min. After being washed twice with staining solution, the cells were incubated with FITC-conjugated goat anti-rabbit IgG (H+L) (Bethyl Laboratories, Inc., Montgomery, TX) on ice for 30 min. After being washed twice with staining solution, the cells were resuspended in 1% paraformaldehyde in PBS. Flow cytometric analysis was performed on a FACSCalibur (BD Biosciences, San Jose, CA).
Generation of simian recombinant vimentin.
Simian recombinant vimentin protein was produced in the QIAexpressionist system (QIAGEN, Valencia, CA). The full length of simian vimentin cDNA was digested with KpnI and BamHI, and then ligation was performed with KpnI- and BamHI-digested pQE-30 vector. The ligation-reaction mixture was transformed into competent M15 E. coli. After the expression of recombinant vimentin was induced by IPTG (isopropyl-β-d-thiogalactopyranoside) at a final concentration of 1 mM, recombinant vimentin was purified by Ni-nitrilotriacetic acid columns (QIAGEN, Valencia, CA).
Delivery of recombinant simian vimentin into PRRSV-nonsusceptible cells.
Recombinant simian vimentin was delivered into BHK-21 and CRFK cells by the Chariot protein delivery system (Active Motif, Inc., Carlsbad, CA). BHK-21 and CRFK cells were cultured overnight in a 48-well tissue culture plate (5 × 104 cells/well). Chariot-vimentin complex was formed at room temperature in 30 min. After the medium was removed, the cells were washed twice with PBS. One hundred microliters of Chariot-vimentin complex was added to each well, and then 100 μl of serum-free medium was added to each well. The cells were incubated at 37°C for 1 h, and complete growth medium (300 μl) was added to each well. After incubation at 37°C for 2 h, the cells were infected with PRRSV. At 3 days postinfection, the cells were fixed with cold 80% acetone and stained with FITC-conjugated SDOW-17, a MAb against PRRSV nucleocapsid protein. The cells were examined by fluorescence microscopy for PRRSV.
RESULTS
Production and characterization of PRRSV blocking MAb (7G10) and effect of 7G10 MAb on PRRSV infection.
Production of the 7G10 MAb used, which blocks the infection of MARC-145 cells by PRRSV, was described in Materials and Methods. To study the effect of the 7G10 MAb on PRRSV infection, a plaque reduction assay was performed. The 7G10 MAb reduced the plaque titer by 98%.
Identification of 7G10 MAb-bound host cell protein complex.
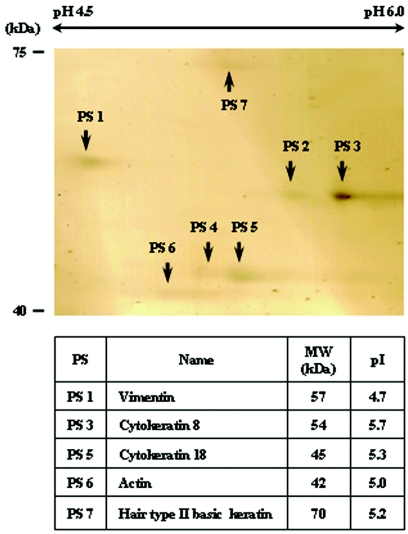
To capture 7G10 MAb-bound host cell proteins, an immunoaffinity column was prepared with 7G10 MAb. The protein complex retained on the 7G10 MAb affinity column was separated by 2-D electrophoresis, cored, and then identified by MALDI-TOF MS. Seven protein spots were obtained from the protein complex by 2-D electrophoresis, and five proteins were identified among the seven proteins (Fig. 1). All proteins were identified as cytoskeletal filaments. Protein spot (PS) 1 was vimentin, PS 3 was cytokeratin 8, PS 5 was keratin 18, PS 6 was actin, and PS 7 was hair type II basic keratin. PS 2 and PS 4 were not identified. Among the complexed proteins, 7G10 MAb bound directly only to vimentin, not the other proteins, in Western blotting (data not shown).
FIG. 1.
Separation of 7G10 MAb-bound proteins by 2-D electrophoresis. A 7G10 MAb-bound host cell protein complex was separated by 2-D elecrophoresis. A Readystrip IPG stip was rehydrated in DeStreak rehydration solution containing the protein complex, and isoelectric focusing was performed. The IPG strip was equilibrated, and the proteins were separated by SDS-PAGE. The protein spots were cored and identified by MALDI-TOF MS (PS 101, vimentin; PS 102, no identification; PS 103, cytokeratin 8; PS 104, no identification; PS 105, keratin 18; PS 106, actin; PS 107, hair type II basic keratin).
Characterization of PRRSV-binding activity of vimentin.
To examine whether vimentin, one of the 7G10 MAb-bound host cell proteins, could interact with PRRSV, VOPBA was performed. PRRSV was purified by polyethylene glycol precipitation, followed by a sucrose density gradient separation. The density of the purified PRRSV was 1.15 g/cm3. As shown in Fig. 2, the purified PRRSV bound to a 57-kDa band of vimentin, which was detected with SR-30, a MAb against PRRSV nucleocapsid protein (lane 1), while no band was observed in the negative control, which was identical except that no virus was added under the same conditions (lane 2). Binding of PRRSV to the 57-kDa band of vimentin was also detected with polyclonal Ab against PRRSV (data not shown).
FIG. 2.
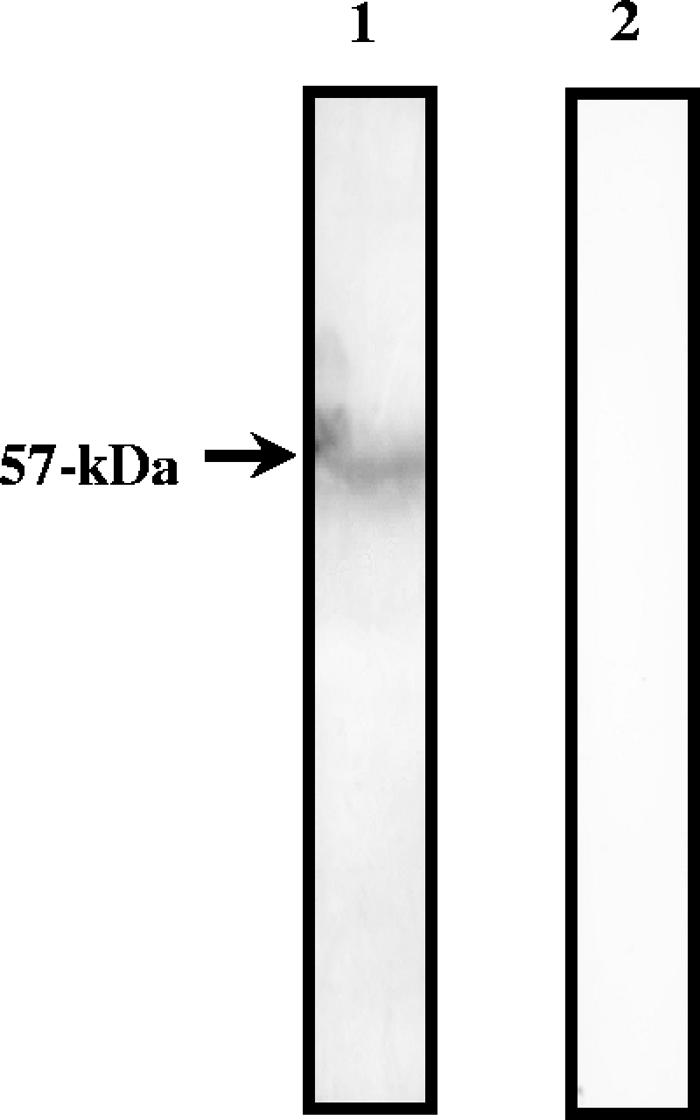
VOPBA of vimentin. Vimentin from bovine lens was separated by SDS-PAGE. The protein was transferred onto a nitrocellulose membrane and incubated with purified PRRSV (lane 1). The protein was incubated with SR-30, a MAb against PRRSV nucleocapsid protein, followed by incubation with the secondary anti-mouse peroxidase-conjugated Ab. The presence of virus binding was detected by the addition of TMB membrane peroxidase substrate (one component). The control reaction was identical, except that no virus was added (lane 2).
Blocking activity of anti-vimentin Ab on PRRSV infection of MARC-145 cells.
To investigate whether anti-vimentin Ab can block PRRSV infection of MARC-145 cells, a checkerboard titration assay was used. As shown in Table 1, PRRSV infection was blocked by polyclonal anti-vimentin Ab in a dose-dependent manner. At the low concentration of the virus (from 10−5 to 10−7 dilution), polyclonal anti-vimentin Ab showed a strong blocking effect, and the effect was dose dependent. At the high concentration of the virus (from 10−1 to 10−4 dilution), the Ab showed a weaker blocking effect. The concentration of 160−1-diluted Ab showed the best blocking effect against PRRSV infection, even at the high concentration of the virus (10−1 or 10−2 dilution). However, an irrelevant Ab, anti-β-galactosidase MAb, did not block PRRSV infection.
TABLE 1.
Checker board titration assay for blocking activity of anti-vimentin Ab
| Virus concn (10-fold dilutions) | Resulta
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ab concn (reciprocal of 2-fold dilutions)
|
No Ab
|
|||||||||||
| 20−1 | 40−1 | 80−1 | 160−1 | 320−1 | 640−1 | 1,280−1 | 2,560−1 | 5,120−1 | ||||
| 10−1 | C | C | C | 1 | 1 | 2 | 3 | 3 | 3 | 3 | 3 | 3 |
| 10−2 | C | C | C | 0 | 1 | 2 | 3 | 3 | 3 | 3 | 3 | 3 |
| 10−3 | C | C | C | 0 | 1 | 2 | 3 | 3 | 3 | 3 | 3 | 3 |
| 10−4 | C | C | C | 0 | 1 | 1 | 2 | 2 | 2 | 3 | 3 | 3 |
| 10−5 | C | C | C | 0 | 1 | 1 | 1 | 1 | 2 | 3 | 3 | 3 |
| 10−6 | C | C | C | 0 | 0 | 1 | 1 | 1 | 1 | 3 | 3 | 3 |
| 10−7 | C | C | C | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 3 | 3 |
| No virus | C | C | C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
MARC-145 cells were cultured with PRRSV and/or anti-vimentin Ab in a 96-well tissue culture plate. Tenfold serial dilutions of the PRRSV preparation were prepared for reaction with serial two-fold dilutions of the polyclonal anti-vimentin Ab. After 3 days of incubation, immunofluorescence microscopy analysis was performed. The cells were fixed with 80% acetone and stained with FITC-conjugated SDOW-17, a Mab against PRRSV nucleocapsid protein. C, cytopathic effect; 0, no fluorescence detected; 1, low-intensity fluorescence; 2, medium-intensity fluorescence; 3, high-intensity fluorescence.
Sequence analysis of the simian cDNA of vimentin from MARC-145 cells.
The full-length cDNA of simian vimentin was screened in a MARC-145 cDNA library (GenBank accession number, DQ190949). As a probe, the partial cDNA of porcine vimentin was used, which was amplified by RT-PCR from ST cells (GenBank accession number, DQ190948). The simian vimentin cDNA sequence has the highest identity (97%) with the human vimentin sequence and also has high identity with the vimentin cDNA sequences of other species. Figure 3 shows the alignment of the simian vimentin amino acid sequence with human, bovine, murine, and porcine vimentin cDNA sequences. The simian vimentin amino acid sequence has the highest identity (99%) with the human vimentin amino acid sequence and also has high identity with the vimentin amino acid sequences of the other species. Nevertheless, the simian vimentin amino acid sequence has a few unique differences. The simian vimentin amino acid sequence has threonine residues at positions 2 (Thr-2) and 179 (Thr-179), while the other vimentin amino acid sequences have a serine residue at position 2 (Ser-2) and an alanine residue at position 179 (Ala-179).
FIG. 3.
Alignment of vimentin amino acid sequences. Simian and porcine vimentin amino acid sequences were generated from the cDNA sequences. The amino acid sequences were aligned with human, murine, and bovine vimentin amino acid sequences. The N-terminal region of the porcine vimentin cDNA sequence is absent because it is a partial sequence. The dashes in the porcine sequence represent deletions. Dots represent similarity of amino acid residues.
Detection of vimentin by Western blot analysis.
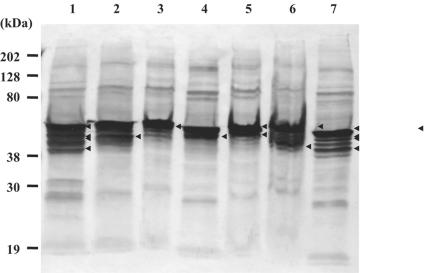
To examine the expression of vimentin in various cell lines, MARC-145, BHK-21, CRFK, MDCK, PK-15, ST, and Vero, Western blot analysis was performed using an anti-vimentin MAb (Fig. 4). In all cell lines, several bands were detected in a broad range, but the major bands were between 40 and 60 kDa. Interestingly, MARC-145 and Vero cells showed very similar patterns of vimentin bands.
FIG. 4.
Expression profiles of vimentin in different cell lines by Western blot analysis. The total cellular proteins were extracted from each cell line and separated by SDS-PAGE (lane 1, MARC-145; lane 2, BHK-21; lane 3, CRFK; lane 4, MDCK; lane 5, PK-15; lane 6, ST; lane 7, Vero). The proteins were transferred onto a nitrocellulose membrane and incubated with monoclonal anti-vimentin Ab, followed by incubation with peroxidase-labeled anti-mouse IgG (H+L). The protein bands were detected by the addition of TMB membrane peroxidase substrate (one component). The major bands are indicated by arrowheads.
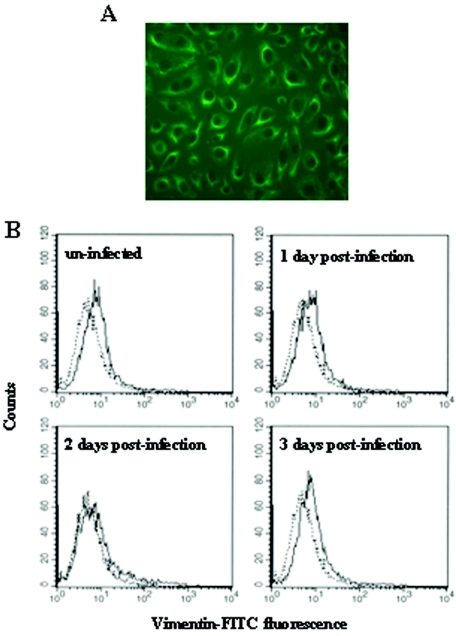
Localization of vimentin in MARC-145 cells.
To investigate the localization of vimentin in MARC-145 cells, immunofluorescence microscopy analysis was performed in permeabilized cells. Vimentin was found in the cytoplasm, especially concentrated around the plasma membrane (Fig. 5A). Next, we addressed whether vimentin is expressed on the cell surface. To examine the cell surface expression of vimentin, flow cytometric analysis was performed on nonpermeabilized MARC-145 cells. As shown in Fig. 5B, vimentin was expressed on the cell surface. We also examined the alteration of the cell surface expression of vimentin by PRRSV infection. The expression of vimentin on the cell surface was changed by PRRSV infection. At 2 days postinfection by PRRSV, the expression level of vimentin was reduced to half compared to uninfected cells. However, the expression level of vimentin was restored to the same level as in uninfected cells at 3 days postinfection.
FIG. 5.
Localization of vimentin in MARC-145 cells. (A) Subcellular localization of vimentin in MARC-145 cells. The cells were permeabilized with 80% acetone and stained with FITC-conjugated monoclonal anti-vimentin (V9) Ab. The cells were examined by fluorescence microscopy and photographed at ×20. (B) Cell surface expression of vimentin on MARC-145 cells at various time points after infection with PRRSV. The nonpermeabilized cells were stained with polyclonal rabbit anti-vimentin Ab, followed by incubation with FITC-conjugated goat anti-rabbit IgG (H+L) (solid lines). An isotype-matched control is represented by the dotted lines.
Delivery of recombinant simian vimentin into nonsusceptible cells.
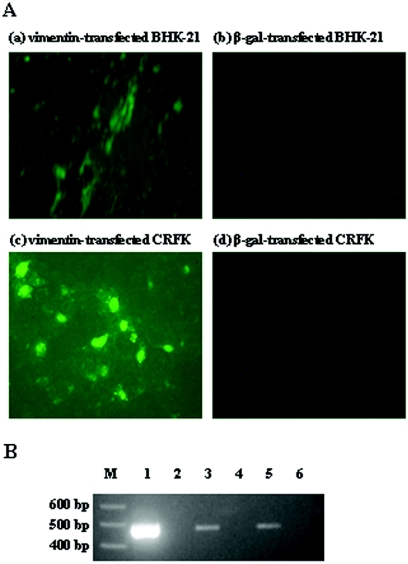
Recombinant protein was made from simian cDNA of vimentin in E. coli and then delivered into PRRSV-nonsusceptible cell lines, BHK-21 and CRFK. The β-galactosidase protein was used as a control in exactly the same way as vimentin. Figure 6A shows immunofluorescence microscopy analysis to detect PRRSV in the simian-vimentin recipient BHK-21 and CRFK cells. Both simian-vimentin recipient BHK-21 (a) and CRFK (c) cells were rendered susceptible to PRRSV, while β-galactosidase recipient BHK-21 (b) and CRFK (d) cells were still nonsusceptible to PRRSV. Figure 6B shows a sensitive RT-PCR assay to demonstrate PRRSV RNA production in the simian-vimentin recipient BHK-21 and CRFK cells. Simian vimentin was delivered into the BHK-21 and CRFK cells, and then the cells were infected with PRRSV. PRRSV RNA was detected in both simian-vimentin recipient BHK-21 (lane 3) and CRFK (lane 5) cells, while PRRSV RNA was not detected in β-galactosidase recipient BHK-21 (lane 4) and CRFK (lane 6) cells.
FIG. 6.
Delivery of simian vimentin into PRRSV-nonsusceptible cell lines. (A) Simian recombinant vimentin or β-galactosidase was delivered into BHK-21 cells (a and b) and CRFK cells (c and d) in the Chariot protein delivery system. Simian vimentin or β-galactosidase recipient cells were infected with PRRSV. The cells were fixed with 80% acetone and stained with FITC-conjugated SDOW-17, a MAb against PRRSV nucleocapsid protein. The cells were examined by fluorescence microscopy and photographed at ×20. (B) A sensitive RT-PCR assay was performed to demonstrate the PRRSV RNA production in simian-vimentin or β-galactosidase recipient cells. M, DNA marker; lane 1, PRRSV-infected MARC-145; lane 2, PRRSV-uninfected MARC-145; lane 3, PRRSV-infected, vimentin recipient BHK-21; lane 4, PRRSV-infected, β-galactosidase recipient BHK-21; lane 5, PRRSV-infected, vimentin recipient CRFK; lane 6, PRRSV-infected, β-galactosidase recipient CRFK.
DISCUSSION
PRRSV is endemic in most swine-producing areas of the world, except a few countries in Europe (4) and Oceania (19). To decrease economic losses from PRRSV, effectively controlling the virus is one of the most important tasks. In many viruses, the viral receptors have been one of the targets for control of viral infection. PRRSV infects well-differentiated cells of the monocyte/macrophage lineage and cells derived from African green monkey kidney cells by receptor-mediated endocytosis (13, 23, 25). So far, two putative receptors have been reported on primary macrophages: HS and Sn. Both HS and Sn are expressed only on PAM, not MARC-145 cells, which are widely used for virus isolation in vitro (10, 11, 12, 39). In this study, we identified the components of PRRSV blocking MAb (7G10)-bound complex. The complex was composed of vimentin and known vimentin-bound proteins; cytokeratin 8, cytokeratin 18, actin, and hair type II basic keratin. We also demonstrated that PRRSV bound to vimentin.
Vimentin is the most abundant component of IFs, which comprise cytoskeleton networks with microfilaments and microtubules. IFs are divided into five different types: types I, II, III, IV, and V. Vimentin is included in type III IFs with desmin, glial fibrillary acidic protein, and peripherin (16). Vimentin is widely expressed in various cell types, such as monocytes, macrophages, multinuclear giant cells, white blood cells, astrocytes, mesenchymal cells, muscle cells, and some neurons (3). Previous studies have reported that vimentin is localized in the cytoplasm, the perinuclear region, the Golgi apparatus, and the endoplamic reticulum (3, 17, 29). It has also been demonstrated that vimentin is expressed on the cell surface and even secreted into the extracellular environment by activated macrophages (29). In this study, the cellular distribution of vimentin in MARC-145 cells was examined. Vimentin was localized on the cell surface, as well as in the cytoplasm, of MARC-145 cells as previously reported in other cells, such as monocytes and macrophages (29). Based on our findings, we propose that PRRSV binds to vimentin expressed on the surfaces of MARC-145 cells. Interestingly, the expression level of vimentin on the cell surface was modulated by PRRSV infection. Modulation of vimentin on the cell surface after PRRSV infection may occur due to binding of the PRRSV with the receptor, leading to opsonization and endocytosis of PRRSV. After 3 days postinfection, the expression level of vimentin returned to the preinfection level due to recovery of the cells. Delivery of simian vimentin into PRRSV-nonsusceptible cell lines, BHK-21 and CRFK, also supports the notion that vimentin might be a receptor of PRRSV. Both simian-vimentin recipient BHK-21 and CRFK cells were rendered susceptible to PRRSV infection. Both BHK-21 and CRFK cells normally lack PRRSV-binding activity (24).
Vimentin plays an important role in stabilizing the architecture of the cytoplasm in various cells. Additionally, vimentin and the other cytoskeletal filaments play important roles in virus entry and/or infection for many viruses, such as human immunodeficiency virus type 1 (22, 28, 37), Junin virus (9), human cytomegalovirus (1), vaccinia virus (34), Theiler's murine encephalomyelitis virus (31), Epstein-Barr virus (35), adenovirus type 2 (2), human T-cell leukemia virus type I (27), influenza A virus (40), frog virus 3 (30), African swine fever virus (5), human respiratory syncytial virus (18), hemagglutinating virus of Japan (20), bluetongue virus (14), and herpes simplex virus type 1 (32). Especially in human immunodeficiency virus type 1 infection of differentiated macrophages and activated T lymphocytes, vimentin binds to the third hypervariable region (the V3 loop) of the viral envelope protein gp120 and subsequently induces the entry of the virus and translocation of the viral DNA (37).
Multifunctional proteins are often regulated by posttranslational modifications. Vimentin is one of the most prominent phosphoproteins in various cells and is phosphorylated by a variety of protein kinases (15, 38). The phosphorylation of vimentin may affect the properties of vimentin. For example, a previous study reported that vimentin was secreted into the extracellular space through the Golgi apparatus by phosphorylation, and the secreted vimentin might be related to the response to pathogens (29). In this study, vimentins from different cell lines were shown to migrate differently due to posttranslational modifications, such as phosphorylation. Interestingly, two PRRSV-susceptible cell lines, MARC-145 and Vero, show similar patterns of vimentin expression. Thus, phosphorylation of vimentin may be related to the restricted cell tropism of PRRSV. Also, a threonine residue which is present at position 2 (Thr-2) in the amino acid sequence of simian vimentin may determine the use of vimentin as a host range determinant for the simian cell line. The hydroxyl group of threonine is buried and offers less flexibility in contact with PRRSV. However, the hydroxyl group of serine (Ser-2) of vimentin in the other species is more exposed and makes general contact in a less specific manner. Thus, simian vimentin can function as a specific viral receptor, while vimentins in the other species lack a receptor function. The fact that very few amino acids are critical in virus-receptor interaction has been reported in several viruses. It has been recently reported that the difference of 4 amino acids in the receptor-binding domain of the human epidemic strain of severe acute respiratory syndrome coronavirus (SARS-CoV) causes more than a 1,000-fold difference in binding affinity to its receptor, human angiotensin-converting enzyme 2 (ACE2). Especially, the change of an amino acid residue at position 487 (serine to threonine) of the spike proteins between civet and human virus strains is strongly related to the adaptation of SARS-CoV to humans (21). Also, in its receptor, highly conserved among mammals, few amino acids are critical in virus-receptor binding activity. Human ACE2 has a lysine at position 353 (Lys-353) that is critical for the interaction with Thr-487 in the receptor binding domain of SARS-CoV. However, rat and mouse ACE2, which inefficiently support infection by SARS-CoV, have a histidine at position 353 (His-353). Replacement of His-353 with lysine in mouse ACE2 allows high-level infection of murine cells (26).
In the present study, it was shown that PRRSV bound to vimentin, and PRRSV infection of MARC-145 cells was blocked by anti-vimentin Abs. In MARC-145 cells, vimentin was expressed on the cell surface, as well as in the cytoplasm, and associated with the other cytoskeletal filaments in the cytoplasm. Here, we report that vimentin may be a putative receptor of PRRSV and may play an important role with the other cytoskeletal filaments in PRRSV infection. To our knowledge, this is the first demonstration of the interaction of PRRSV with cytoskeletal filaments, such as vimentin.
Acknowledgments
This project was supported by USDA Health Funds (NC 229 Project).
We thank Teresa Yeary for excellent editorial assistance.
REFERENCES
- 1.Arcangeletti, M. C., F. Pinardi, M. C. Medici, E. Pilotti, F. De Conto, F. Ferraglia, M. P. Landini, C. Chezzi, and G. Dettori. 2000. Cytoskeleton involvement during human cytomegalovirus replicative cycle in human embryo fibroblasts. New Microbiol. 23:241-256. [PubMed] [Google Scholar]
- 2.Belin, M. T., and P. Boulanger. 1987. Processing of vimentin occurs during the early stages of adenovirus infection. J. Virol. 61:2559-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cain, H., B. Kraus, R. Krauspe, M. Osborn, and K. Weber. 1983. Vimentin filaments in peritoneal macrophages at various stages of differentiation and with altered function. Virchows Arch. B 42:65-81. [DOI] [PubMed] [Google Scholar]
- 4.Canon, N., L. Audige, H. Denac, M. Hofmann, and C. Griot. 1998. Evidence of freedom from porcine reproductive and respiratory syndrome virus infection in Switzerland. Vet. Rec. 142:142-143. [DOI] [PubMed] [Google Scholar]
- 5.Carvalho, Z. G., A. P. De Matos, and C. Rodrigues-Pousada. 1988. Association of African swine fever virus with the cytoskeleton. Virus Res. 11:175-192. [DOI] [PubMed] [Google Scholar]
- 6.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 7.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 8.Collins, J. E., D. A. Benfield, W. T. Christianson, L. Harris, J. C. Hennings, D. P. Shaw, S. M. Goyal, S. McCullough, R. B. Morrison, H. S. Joo, D. Gorcyca, and D. Chladek. 1992. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J. Vet. Diagn. Investig. 4:117-126. [DOI] [PubMed] [Google Scholar]
- 9.Cordo, S. M., and N. A. Candurra. 2003. Intermediate filament integrity is required for Junin virus replication. Virus Res. 97:47-55. [DOI] [PubMed] [Google Scholar]
- 10.Delputte, P. L., and H. J. Nauwynck. 2004. Porcine arterivirus infection of alveolar macrophages is mediated by sialic acid on the virus. J. Virol. 78:8094-8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delputte, P. L., N. Vanderheijden, H. J. Nauwynck, and M. B. Pensaert. 2002. Involvement of the matrix protein in attachment of porcine reproductive and respiratory syndrome virus to a heparinlike receptor on porcine alveolar macrophages. J. Virol. 76:4312-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duan, X., H. J. Nauwynck, H. W. Favoreel, and M. B. Pensaert. 1998. Porcine reproductive and respiratory syndrome virus infection of alveolar macrophages can be blocked by monoclonal antibodies against cell surface antigens. Adv. Exp. Med. Biol. 440:81-88. [DOI] [PubMed] [Google Scholar]
- 13.Duan, X., H. J. Nauwynck, and M. B. Pensaert. 1997. Effects of origin and state of differentiation and activation of monocytes/macrophages on their susceptibility to porcine reproductive and respiratory syndrome virus (PRRSV). Arch. Virol. 142:2483-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eaton, B. T., A. D. Hyatt, and J. R. White. 1987. Association of bluetongue virus with the cytoskeleton. Virology 157:107-116. [DOI] [PubMed] [Google Scholar]
- 15.Evans, R. M. 1989. Phosphorylation of vimentin in mitotically selected cells. In vitro cyclic AMP-independent kinase and calcium-stimulated phosphatase activities. J. Cell Biol. 108:67-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs, E., and K. Weber. 1994. Intermediate filaments: structure, dynamics, function, and disease. Annu. Rev. Biochem. 63:345-382. [DOI] [PubMed] [Google Scholar]
- 17.Gao, Y., and E. Sztul. 2001. A novel interaction of the Golgi complex with the vimentin intermediate filament cytoskeleton. J. Cell Biol. 152:877-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Barreno, B., J. L. Jorcano, T. Aukenbauer, C. Lopez-Galindez, and J. A. Melero. 1988. Participation of cytoskeletal intermediate filaments in the infectious cycle of human respiratory syncytial virus (RSV). Virus Res. 9:307-321. [DOI] [PubMed] [Google Scholar]
- 19.Garner, M. G., L. J. Gleeson, P. K. Holyoake, R. M. Cannon, and W. J. Doughty. 1997. A national serological survey to verify Australia's freedom from porcine reproductive and respiratory syndrome. Aust. Vet. J. 75:596-600. [DOI] [PubMed] [Google Scholar]
- 20.Hirayama, E., M. Ama, K. Takahashi, A. Hiraki, and J. Kim. 2002. Changes in fused cells induced by hvj (Sendai virus): redistribution of cytoplasmic organelles and cytoskeletal reorganization. Cell Biol. Int. 26:347-353. [DOI] [PubMed] [Google Scholar]
- 21.Holmes, K. V. 2005. Adaptation of SARS coronavirus to human. Science 309:1822-1823. [DOI] [PubMed] [Google Scholar]
- 22.Khakhulina, T. V., G. K. Vorkunova, E. E. Manukhina, and A. G. Bukrinskaia. 1999. Vimentin intermediate filaments are involved in replication of human immunodeficiency virus type I. Dokl. Akad. Nauk. 368:706-708. [PubMed] [Google Scholar]
- 23.Kim, H. S., J. Kwang, I. J. Yoon, H. S. Joo, and M. L. Frey. 1993. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch. Virol. 133:477-483. [DOI] [PubMed] [Google Scholar]
- 24.Kreutz, L. C. 1998. Cellular membrane factors are the major determinants of porcine reproductive and respiratory syndrome virus tropism. Virus Res. 53:121-128. [DOI] [PubMed] [Google Scholar]
- 25.Kreutz, L. C., and M. R. Ackermann. 1996. Porcine reproductive and respiratory syndrome virus enters cells through a low pH-dependent endocytic pathway. Virus Res. 42:137-147. [DOI] [PubMed] [Google Scholar]
- 26.Li, F., W. Li, M. Farzan, and S. C. Harrison. 2005. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309:1864-1868. [DOI] [PubMed] [Google Scholar]
- 27.Lilienbaum, A., M. Duc Dodon, C. Alexandre, L. Gazzolo, and D. Paulin. 1990. Effect of human T-cell leukemia virus type I tax protein on activation of the human vimentin gene. J. Virol. 64:256-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, B., R. Dai, C. J. Tian, L. Dawson, R. Gorelick, and X. F. Yu. 1999. Interaction of the human immunodeficiency virus type 1 nucleocapsid with actin. J. Virol. 73:2901-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mor-Vaknin, N., A. Punturieri, K. Sitwala, and D. M. Markovitz. 2003. Vimentin is secreted by activated macrophages. Nat. Cell Biol. 5:59-63. [DOI] [PubMed] [Google Scholar]
- 30.Murti, K. G., R. Goorha, and M. W. Klymkowsky. 1988. A functional role for intermediate filaments in the formation of frog virus 3 assembly sites. Virology 162:264-269. [DOI] [PubMed] [Google Scholar]
- 31.Nedellec, P., P. Vicart, C. Laurent-Winter, C. Martinat, M. C. Prevost, and M. Brahic. 1998. Interaction of Theiler's virus with intermediate filaments of infected cells. J. Virol. 72:9553-9560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Norrild, B., V. P. Lehto, and I. Virtanen. 1986. Organization of cytoskeleton elements during herpes simplex virus type 1 infection of human fibroblasts: an immunofluorescence study. J. Gen. Virol. 67:97-105. [DOI] [PubMed] [Google Scholar]
- 33.Plagemann, P. G., and V. Moennig. 1992. Lactate dehydrogenase-elevating virus, equine arteritis virus, and simian hemorrhagic fever virus: a new group of positive-strand RNA viruses. Adv. Virus Res. 41:99-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Risco, C., J. R. Rodriguez, C. Lopez-Iglesias, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 2002. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J. Virol. 76:1839-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sairenji, T., Q. V. Nguyen, B. Woda, and R. E. Humphreys. 1987. Immune response to intermediate filament-associated, Epstein-Barr virus-induced early antigen. J. Immunol. 138:2645-2652. [PubMed] [Google Scholar]
- 36.Shanmukhappa, K., and S. Kapil. 2001. Cloning and identification of MARC-145 cell proteins binding to 3′ UTR and partial nucleoprotein gene of porcine reproductive and respiratory syndrome virus. Adv. Exp. Med. Biol. 494:641-646. [DOI] [PubMed] [Google Scholar]
- 37.Thomas, E. K., R. J. Connelly, S. Pennathur, L. Dubrovsky, O. K. Haffar, and M. I. Bukrinsky. 1996. Anti-idiotypic antibody to the V3 domain of gp120 binds to vimentin: a possible role of intermediate filaments in the early steps of HIV-1 infection cycle. Viral Immunol. 9:73-87. [DOI] [PubMed] [Google Scholar]
- 38.Turowski, P., T. Myles, B. A. Hemmings, A. Fernandez, and N. J. Lamb. 1999. Vimentin dephosphorylation by protein phosphatase 2A is modulated by the targeting subunit B55. Mol. Biol. Cell 10:1997-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vanderheijden, N., P. L. Delputte, H. W. Favoreel, J. Vandekerckhove, J. Van Damme, P. A. van Woensel, and H. J. Nauwynck. 2003. Involvement of sialoadhesin in entry of porcine reproductive and respiratory syndrome virus into porcine alveolar macrophages. J. Virol. 77:8207-8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wheeler, J. G., L. S. Winkler, M. Seeds, D. Bass, and J. S. Abramson. 1990. Influenza A virus alters structural and biochemical functions of the neutrophil cytoskeleton. J. Leukoc. Biol. 47:332-343. [DOI] [PubMed] [Google Scholar]