Abstract
Papillomaviruses are internalized via clathrin-dependent endocytosis. However, the mechanism by which viral genomes pass endosomal membranes has not been elucidated. In this report we show that the minor capsid protein L2 is required for egress of viral genomes from endosomes but not for initial uptake and uncoating and that a 23-amino-acid peptide at the C terminus of L2 is necessary for this function. Pseudogenomes encapsidated by L1 and L2 lacking this peptide accumulated in vesicular compartments similar to that observed with L1-only viral particles, and these mutant pseudoviruses were noninfectious. This L2 peptide displayed strong membrane-disrupting activity, induced cytolysis of bacteria and eukaryotic cells in a pH-dependent manner, and permeabilized cells after exogenous addition. Fusions between green fluorescent protein and the L2 peptide integrated into cellular membranes like the wild type but not like C-terminal mutants of L2. Our data indicate that the L2 C terminus facilitates escape of viral genomes from the endocytic compartment and that this feature is conserved among papillomaviruses. Furthermore, the characteristic of this peptide differs from the classical virus-encoded membrane-penetrating peptides.
After endocytic uptake, access of most ligands to the cytosol is limited and tightly regulated. Therefore, a large variety of transmembrane channels have evolved to mediate the passage of hydrophilic substances across the limiting cellular membrane. The mechanism and control of these processes have been intensely studied. Likewise, viruses have necessarily developed mechanisms for membrane penetration, which are essential for the establishment of a productive infection. The access of enveloped viruses to the cytosol requires the fusion of the viral envelope with a target cell membrane (13). This fusion event may occur at the cell surface or after internalization of the virus. Nonenveloped viruses either lyse the limiting vesicular membrane (26) or generate a pore through it to allow escape of the viral genome into the cytosol (20). There are several descriptions of the mechanisms that nonenveloped viruses have evolved for endosome escape, notably for poliovirus, reovirus, and adenovirus. However, pore formation through the endosomal membrane by a nonenveloped virus has been definitively demonstrated only for reoviruses and polioviruses. In the case of reoviruses, acid-dependent proteolytic disassembly within endosomes results in the production of infectious subvirion particles. These activated particles can induce membrane channels that allow delivery of a partially uncoated particle directly into the cytoplasm (8, 43-45). Essentially nothing is known about these mechanisms for other nonenveloped viruses, including Papillomaviridae.
Papillomaviruses are nonenveloped viruses with icosahedral symmetry that induce a variety of benign tumors of the skin and mucosa. Some types of human papillomaviruses (HPV) are also associated with cervical carcinoma and other tumors of anogenital epithelia. The replication of papillomavirus (PV) is strictly dependent on the terminal differentiation of the stratified squamous epithelium or mucosa. The PV particles are composed of 360 copies of the major capsid protein, L1, organized into pentameric capsomers (1, 19, 27), and a less well defined number of the minor capsid protein, L2 (2, 34, 46).
The biology of papillomaviruses has been difficult to study, since they could not be propagated in vitro. However, the advent of technologies for the in vitro generation of infectious papillomavirus capsids has significantly increased our knowledge of many aspects of the papillomavirus life cycle (reviewed in reference 38). Pseudovirions carrying a marker plasmid instead of the viral genome have been especially useful in the study of the infectious entry pathway (6, 31, 42, 47). It was shown that PV binding to heparan sulfate proteoglycans on the cell surface is an essential step for infection (17, 22). After a delay of several hours, in which the state of binding changes, the virions are taken up via a clathrin-dependent pathway, which requires intact actin filaments. Both acidification of endosomes and intact microtubules have been demonstrated to be essential for efficient papillomavirus infection (11, 37).
The minor capsid protein, L2, does not contribute to the initial binding of papillomaviruses to the cell surface. DNA-free virus-like particles (VLPs) consisting of L1 alone or L1 and L2 are equally effective in competing with virions for cell binding and infection (29, 32). In addition, L1-only particles are internalized with kinetics comparable to L1- and L2-containing particles, suggesting that L2 contributes little to the early steps of internalization (36). L2 appears to be dispensable for DNA encapsidation for some PV types in in vitro packaging systems, including HPV type 16 (HPV16) and HPV33 (7, 47) but seems to contribute somewhat to encapsidation in raft culture systems (21). Despite this, L2 is essential for PV infection, since pseudovirions containing the L1 protein only display a strongly reduced infectivity (6, 23, 47). Recently, it was demonstrated that the L2 protein accompanies the viral genome to specific nuclear domains to establish infection (10), suggesting that it is important for later steps in the infection process.
We now show that the L2 protein contains a membrane-penetrating activity, which is absolutely essential for efficient papillomavirus infection. This activity was mapped to the C terminus of L2. Deletion or single point mutations that affect this membrane disturbance completely abrogate the L2-induced enhancement of pseudovirus infectivity and prevent endosomal escape of the viral genome. To our knowledge, this is the first report that identifies a specific amino acid sequence in one of the capsid proteins of small DNA tumor viruses responsible for passing the endosomal membrane, a prerequisite for establishing infection and, consequently, the induction of virus-induced tumors.
MATERIALS AND METHODS
Cell lines, antibodies, and peptides.
All cell lines were grown in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal calf serum and antibiotics at 37°C. Human 293TT cells were grown in the same medium supplemented with 1% GlutaMAX and 1% nonessential amino acids (Invitrogen). The osteosarcoma cell line HuTK−143B was obtained from Bernard Moss (28). HP3 cells and C127 cells were propagated as previously described (10, 11). L1- and L2-specific mouse monoclonal antibodies have previously been described (33, 48, 49). Green fluorescent protein (GFP)- and Calnexin-specific antibodies were purchased from Dianova, and Hsc70-specific antibody was purchased from Stressgen. A CD138/syndecan-1-specific antibody (clone B = B4) was obtained from Chemicon, Ltd. The 33L2-445/467-specific rabbit polyclonal antiserum was obtained from BioScience, Göttingen, Germany. The hemagglutinin (HA) tag-reactive ascites was purchased from Babco (Richmond, CA). Promyelocytic leukemia protein was detected with a rabbit polyclonal antiserum (Chemicon). The rat anti-lamp-2 antibody ABL-93 was a kind gift from Jonathan Yewdell (NIAID, NIH). Synthetic peptides were purchased from BioScience (crude preparations) and from GENOSPHERE Biotechnologies (highly purified preparations).
ATP bioluminescence assay.
ATP levels in cultures of HeLa cells after incubation with peptides were measured by luciferase-driven bioluminescence. HeLa cells were grown in 96-well plates (4 × 104/well) and treated with different concentrations of peptides. Peptide solutions were prepared in Hank's balanced salt solution (HBSS) (pH 7.2; Invitrogen) or in McIlvaine's phosphate buffer (pH 6.0). After the indicated period of time at 37°C, cells were suspended in 150 μl lysis buffer (1% Triton X-100), diluted 1:2 in H2O, and analyzed in a luminometer using the ATP bioluminescence assay kit CLS II (Roche). Results are expressed in bioluminescence relative light units, and controls without peptides were set at 100% ATP.
Immunofluorescence.
HeLa cells were grown on coverslips and incubated with peptide solutions (100 μg/ml in HBSS) at pH 7.2 for 5 h. Cells were then fixed with methanol-0.02 M EGTA (−20°C) for at least 20 min, washed twice with phosphate-buffered saline (PBS), and blocked in 5% goat serum dissolved in PBS. Coverslips were incubated for 1 h at 37°C with the indicated antibodies. After washing with PBS, coverslips were again blocked for 30 min with 5% goat serum and subsequently incubated at 37°C with Cy3-conjugated Affinipure goat anti-rabbit immunoglobulin G (IgG) and Cy2-conjugated Affinipure goat anti-mouse IgG (Jackson Immunochemicals) for 1 h. Coverslips were washed with PBS and mounted onto slides using Fluroprep mounting medium (bioMérieux). Pictures were taken using a Zeiss Axiovert 200 M microscope and a Zeiss Axiocam digital camera. Axiovision software 3.0 was used for merging pictures.
The method for microscopic detection of uncoated PV pseudovirus has been previously described (10). For colocalization of uncoated L2-HA and lamp-2 the staining was completed in four stages. The initial incubation with mouse anti-HA and subsequent detection with fluorescein isothiocyanate-conjugated goat anti-mouse IgG were completed prior to incubation with the rat anti-lamp-2 monoclonal antibody and detection with Texas Red-conjugated goat anti-rat antibody. Control experiments confirmed the correct antigen recognition.
Flow cytometry.
To test the effect of L2 peptides on plasma membranes, we measured the influx of propidium iodide into HeLa cells. Cells were grown in six-well plates (1 × 106/well) and incubated for 5 h at 37°C with increasing concentrations of peptides dissolved as described above in HBSS. Cells were washed with PBS and harvested with PBS containing 2.5 mM EDTA. Cell pellets were washed three times and finally resuspended in 900 μl of PBS-2% fetal calf serum. Prior to flow cytometric analysis, cells were supplemented with 100 μl propidium iodide to yield a final concentration of 0.5 μg/ml. Cells were then analyzed in a FACSCalibur flow cytometer (Becton Dickinson) and evaluated using the CellQuest software.
Generation of GFP2-peptide fusions.
GFP2-peptide fusions were generated by PCR using pEGFP-C1 (Clontech) as a template and oligonucleotide 5′-ATTGAATTCATGGTGAGCAAGGGC GAG-3′ (ON-GFP-5′) as the forward primer. The following oligonucleotides were used as reverse primers: 5′-TTTGGATCCTCATGCAGATGCACGCTTGCGCCTTGTAGATCGTTTGTGTCTCATCTTGTACAGCTCGTCCATGCC-3′ (ON-GFP-33L2-1/14-3′), 5′-TTTGGATCCTCAAAAATATGGAAAACGTTTACGCCTGCGACGTAAAATAAAATAACTCTTGTACAGCTCGTCCATGCC-3′ (ON-GFP-33L2-445/459-3′), 5′-TTTGGATCCTCAACGTTTACGCCTGCGACGTAA-3′ (ON-GFP-33L2-449/456), 5′-TTTGGATCCTCAGGCCGCCACTCGGACATCTGTAAAAAAATATGGAAAACGTTTACGCCTGCGACGTAAAATAATAACTCTTGTACAGCTCGTCCATGCC-3′ (ON-GFP-33L2-445/467-3′). Upstream of the ATG and downstream of the stop codon, recognition sites for EcoRI and BamHI have been added (highlighted in boldface type). Fragments were cut with EcoRI and BamHI and cloned into correspondingly cut pEGFP-C1 to obtain pEGFP2-33L2-1/14, pEGFP2-33L2-445/459, and pEGFP2-33L2-445/467. Generation of the GFP2-33L1-485/499 (GFP2-33L1-NLS) construct was previously described (15).
The bovine papillomavirus type 1 (BPV-1) L2 C-terminal deletion 1/460-HA construct was modified from the HA-tagged BPV-1 L2 that has been previously described (10). An oligomer primer corresponding to the coding strand was synthesized. This primer (GGGCAGCGGCGTGCGCAGAAGCCTGATCCCT) is upstream of a BstEII site. The primer on the noncoding strand (GGCGGCCGCTCTAGATTAAGCGTAATCTGGAACATCGTATGGGTACAGCAGGGAGGGATGCAGGGTGTAGTTGCTGC) bridges the deleted region and includes an XbaI site downstream of this deletion. The PCR product was gel purified, digested with BstEII and XbaI, and ligated into the original construct, likewise digested to remove the corresponding wild-type sequence. The correct construct was confirmed by DNA sequencing of both strands encompassing the entire region that was subjected to PCR.
Mutagenesis and generation of recombinant vaccinia viruses.
C-terminal deletion mutants of HPV33 L2 were constructed by PCR using pCMV33L2 as the template and oligonucleotide 5′-GGTGAATTCCATGAGACACAAACGATCTAC-3′ (ON-33L2-1-5′) as the forward primer. The following oligonucleotides were used as reverse primers (recognition sites for EcoRI and BamHI are highlighted in boldface type): 5′-AAAGGATCCCTATGGGCTAGATGTGGGAA-3′ (ON-33L2-420S-3′), 5′-AAAGGATCCCTATAAAATAAAATAACTAGG-3′ (ON-33L2-449S-3′), 5′-GCGGGATCCCTAACGTTTACGCCTGCGACG-3′ (ON-33L2-456S-3′). The resulting fragments were cloned into pCR2.1topo (Invitrogen). Fragments were excised using EcoRI and BamHI and cloned into the vaccinia virus transfer vector pTM1 to obtain pTM33L2-1/420, -1/449, and -1/456. Numbers indicate 33L2 amino acids still present in the construct. Replacement of arginine 452 and 453 with aspartate was carried out by overlap extension PCR using pTM-33L2 as a template and two pairs of primers, 5′-ACAGAATTCATGCCTGCTTTTTTAACATCG-3′ (ON-33L2-M240-5′) and 5′-TGTTAGCAGCCGGATCGTC-3′ (ON-pTM-3′) together with 5′-TGGAAAACGTTTATCGTCGCGACGTAAAATAAAATAACTAGG-3′ (ON-33L2-DD452-3′) and 5′-CCTAGTTATTTTATTTTACGTCGCGACGATAAACGTTTTCCA-3′ (ON-33L2-DD452-5′), respectively. The resulting fragment was cut with StuI and BamHI and cloned into pTM-33L2 to obtain pTM-33L2-DD452. Corresponding recombinant vaccinia viruses were generated by cotransfection of these plasmids with wild-type vaccinia virus DNA following published procedures (47). Construction of vac33L1 and vac33L2 has been described (47). The helper virus VTF7-3, recombinant for the T7 RNA polymerase, was a generous gift from Bernard Moss (28). pUF-16L2-1/464 and −1/454 were obtained by amplifying 16L2 from pUF3AAV#893-hum16L2 (24) using oligonucleotide 5′-CGGAATTGTACCCGCGGC-3′ as a forward primer and 5′-TTTAAGCTTTCAGTAGGGCAGCCTCTTCCTCC or 5′-TTTAAGCTTTCACAGCATGTAGTAGCTGGGGTG-3′ as a reverse primer. The resulting fragment was cloned into pUF3AAV using the NotI and HindIII restriction sites (shown in boldface type). pUF16L2-EE458 was obtained by PCR using the megaprimer approach. In a first amplification step, oligonucleotides 5′-CCCAGCTACATGCTGAGGAAGGAGGAGAAGAGGCTGCCCTACTTC-3′ (16L2-EE458-5′) and 5′-CAAAAAGCTTTCAAGCGTAATCTGGAACATCGTATGGGTAGGCGGCCAGGC-3′ (16L2-HA-S-3′) and template pUF3AAV#893-hum16L2 were used to generate a C-terminal 16L2 fragment harboring the point mutation and a C-terminal HA tag. This fragment was isolated after agarose gel electrophoresis and subsequently used as megaprimer together with 5′-CGGAATTGTACCCGCGGC-3′ (16L2-1-5′) and template pUF3AAV#893-hum16L2 to amplify the complete L2 gene. The resulting fragment was cloned into pUF3AAV using the NotI and HindIII restriction sites.
Fractionation of cellular membranes.
The fractionation of cellular membranes was carried out by following the protocol of Gaynor et al. (16) with some modifications. COS7 cells (2 × 107 cells in 15-cm2 wells) were transfected via electroporation with the indicated constructs encoding the GFP2-peptide fusion proteins. At 24 h after transfection, cells were harvested by centrifugation for 5 min at 300 × g and subsequently suspended in 1 ml of lysis buffer (10 mM Tris, pH 7.5, 0.25 M sucrose, 1 mM EDTA, 5 μg/ml aprotinin, and 5 μg/ml leupeptin). The suspension was incubated for 30 min on ice and then transferred to a prechilled Dounce homogenizer. Cells were disrupted by 25 strokes with a tight-fitting pestle. Nuclei were pelleted for 5 min at 800 × g at 4°C (p800). Supernatants (s800) were then spun for 10 min at 13,000 × g and 4°C. The resulting pellet, containing membranes (p13000 membranes), was resuspended in 200 μl of lysis buffer. The supernatant (s13000) contains soluble cytosolic proteins. Membranes (p13000) were loaded onto the top of a 1.2 M/1.5 M sucrose step gradient and centrifuged for 1 h at 85,000 × g and 4°C. Finally, six fractions at the interphase of the sucrose steps were collected as described in the protocol. Proteins were precipitated by chloroform-methanol. Precipitates were subsequently analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. Proteins were stained using the indicated primary antibodies and horseradish peroxidase-coupled secondary antibody (Jackson Immunoresearch Products). The signal was visualized by enhanced chemiluminescence (Amersham Pharmacia).
Alkaline treatment of cellular membranes.
Treatment of isolated membranes with carbonate at pH 11.5 allows distinction between associated and integral membrane proteins. COS7 cells were transfected as described above, and HuTK− cells (1 × 107 cells in 10-cm2 wells) were infected with the indicated recombinant vaccinia viruses. At 24 h after transfection and 16 h after infection, cells were harvested and treated as described above. Membranes (p13000) were suspended in 2 ml of lysis buffer, added to 1.7 ml of 10% sucrose, and then centrifuged for 45 min at 200,000 × g and 4°C. Pellets were resuspended in 1 ml of 100 mM Na2CO3 (pH 11.5), homogenized via passage through a 25-gauge needle and incubated for 1 h on ice. The suspension was supplemented with 2.7 ml of 100 mM Na2CO3 (pH 11.5) and spun for 30 min at 140,000 × g and 4°C. Supernatants were neutralized with acetic acid (7.5 μl/ml), and membrane associated proteins were precipitated with trichloroacetic acid and resuspended in 100 μl of lysis buffer. Pellets containing integral membrane proteins were resuspended in 100 μl of lysis buffer supplemented with 1% Nonidet P-40. Pellet fractions, nuclei (p800), and the cytosol fractions (s13000) were analyzed by immunoblotting.
Infection of cells with recombinant vaccinia viruses.
Confluent HuTK−143B cells were split 1:6 and grown for 24 h at 37°C. Cells were washed once with PBS (pH 7.3) and subsequently infected with recombinant vaccinia viruses diluted in serum-free DMEM at a multiplicity of infection of 0.1 for each virus. After incubation for 1 h at room temperature, virus-containing medium was replaced by supplemented DMEM. After the indicated period of time at 37°C, membranes were isolated and treated with alkaline buffer as described.
Preparation of VLPs and pseudovirions.
HPV33 pseudovirions were generated in vivo as described previously (17, 47). HPV16 pseudovirions were prepared following the protocol of Buck et al. (6). VLPs were extracted from HuTK−143B cells infected with indicated recombinant vaccinia viruses for 48 h as described. BrdU-labeled HPV16 pseudovirions were prepared as described previously (10).
Sucrose gradient analysis.
Incorporation of L2 into HPV33 pseudovirions and VLPs was determined by sucrose gradient analysis, as published recently (2).
Plasmid extraction from pseudovirions and transformation of Escherichia coli.
Plasmid extraction from pseudovirions and transformation of E. coli were performed as described (35).
Pseudoinfection assay.
Cells were grown in 24-well plates (5 × 104 cells/well) and infected with 5 μl of the pseudovirus preparation in a total volume of 300 μl of serum-free DMEM. After 1 h at 4°C, the pseudovirions were replaced by 1 ml of supplemented culture medium. Subsequently, cells were grown for 72 h at 37°C before infectious events were determined by counting cells with nuclear green fluorescence.
RESULTS
Block of endosome escape in the absence of L2.
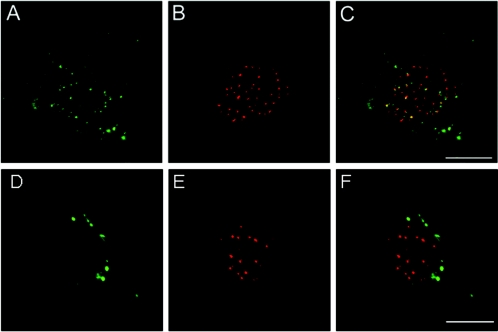
To evaluate whether the presence of the PV L2 capsid protein is necessary for uncoating of the viral particle and the subsequent trafficking of the genome into the nucleus, we took advantage of the ability of HPV16 L1-only pseudovirus to package DNA (6). It was recently demonstrated that L2 and the genome colocalize at the intranuclear domains, ND10, but the fate of the genome in the absence of L2 was not examined (10). Uncoating was evaluated by the acquisition of accessibility of BrdU-labeled pseudogenome to antibody as previously described (10). In this assay, anti-BrdU binding is only detectable after the initiation of particle disassembly, which occurs intracellularly. The detection of the BrdU-labeled pseudogenome after addition of the L1-only capsids to cells clearly demonstrated that the minor capsid protein is dispensable for the initial uncoating process (Fig. 1). Since the timely detection of BrdU-labeled pseudogenome was similar for L1-only and L1-L2 pseudovirions (data not shown), L2 does also not affect the kinetics of uptake. However, the distribution of the genome of the L1-only pseudovirions and the L1-L2 pseudovirions observed at late times after binding was strikingly different. As previously reported, by 24 h postentry, the genome delivered by L1-L2 capsids is partially localized to intranuclear domains (10) (Fig. 1), which is in line with previous observations of a slow uptake of papillomaviruses (9, 36). In contrast, the genome in the pseudovirions lacking L2 appeared to be retained in a vesicular compartment. These results suggest that the L2 protein mediates egress of the viral genome from the endocytic compartment.
FIG. 1.
L2 is not required for viral uncoating but is necessary for egress from the endocytic pathway. HPV16 particles composed of both L1 and L2 capsid proteins (panels A to C) or particles composed of only the L1 protein (panels D to F) were prepared with encapsidated BrdU-labeled DNA. Particles were added to HP3 cells for 24 h. Disassembled virus is detected with anti-BrdU antibodies (panels A and D). Entry of the viral DNA into the nucleus is monitored by colocalization of the genome with PML (shown in panels B and E), a marker of ND10. The merges are shown in panels C and F.
Role of the C terminus of L2 in egress from endosomes.
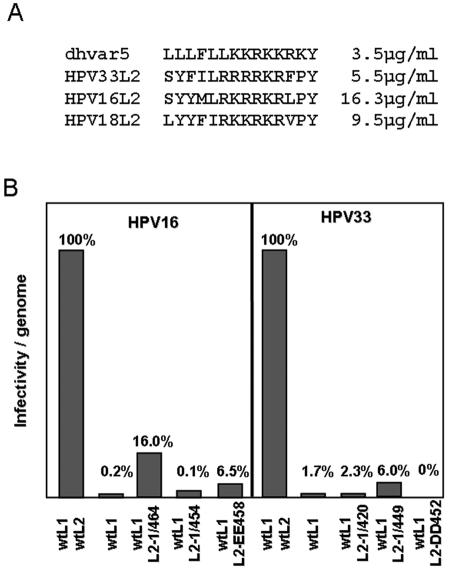
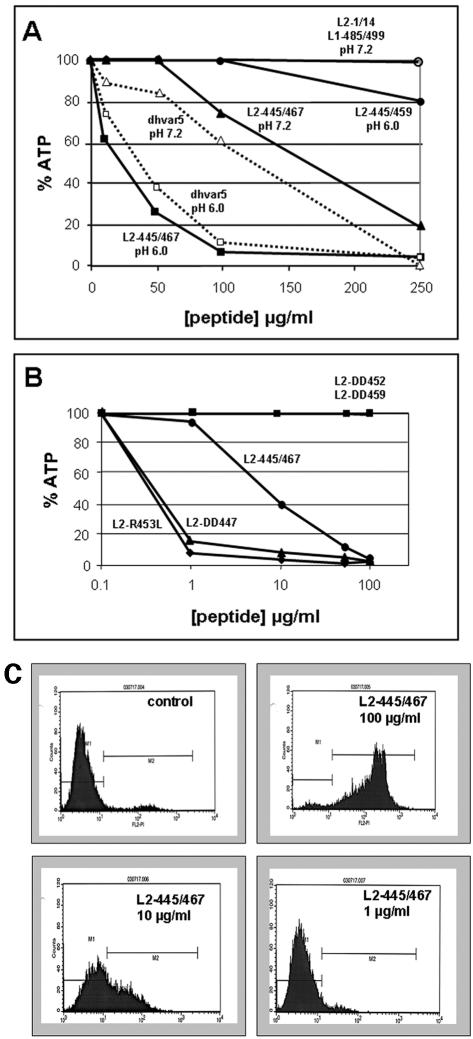
One possible mechanism for endosome escape is via a destabilization of the limiting membrane. In a search for possible membrane-disturbing domains within the PV L2 protein, we observed similarity of a carboxyl-terminal L2 peptide with the synthetic peptide dhvar5 (LLLFLLKKRKKRKY) (Fig. 2A), which was recently shown to destabilize and depolarize membranes of bacteria and fungi, resulting in strong microbicidal and fungicidal activities (12). Peptide dhvar5 is a derivative of histatin 5, a human antimicrobial and antifungal peptide found in human saliva. Characteristic features of dhvar5 are a stretch of hydrophobic amino acids adjacent to a cluster of basic amino acids. The sequence similarity of L2 peptides to dhvar5 was strongest for HPV39, -68, and -70, but the general characteristic of the peptide is conserved among all papillomaviruses, even though the stretch of hydrophobic amino acids is interrupted in some papillomavirus types (data not shown). We evaluated the L2-derived peptides of HPV16, -18, and -33 L2 for microbicidal and fungicidal activities. We measured 50% lethal concentrations (LC50) of 16.3 μg/ml, 9.5 μg/ml, and 5.5 μg/ml, respectively, compared to 3.5 μg/ml for dhvar5 (Fig. 2A). In addition, survival of Escherichia coli was strongly affected by these peptides (data not shown), further demonstrating their microbicidal and fungicidal potency.
FIG. 2.
The L2 C terminus is required for efficient papillomavirus infection. (A) Comparison of dhvar5 and selected HPV L2 peptides. Numbers to the right indicate LC50s using Candida albicans. (B) The infectivity of HPV16 and HPV33 pseudovirions harboring the indicated capsid proteins was quantified. Values shown were normalized to the amount of encapsidated marker plasmid DNA to correct for variation in pseudovirus yields.
To investigate the importance of this region for infection, we generated HPV16 capsids consisting of L1-, L1/L2-, and L1/mutant L2. The C-terminally truncated L2 mutants used comprise deletion mutants 16L2-1/464 and −1/454, harboring truncations of 8 and 18 amino acids, respectively, as well as the point mutation 16L2-EE458, which has arginine residues at positions 458 and 459 replaced by glutamate. Neither DNA encapsidation nor L2 incorporation in mutant pseudoviruses was negatively affected (data not shown). As previously reported, the pseudovirions composed of L1 alone were at least 50 times less infectious than those containing both capsid proteins (Fig. 2B) (6, 47). Interestingly, the deletion of the C-terminal membrane-active peptide (16L2-1/454) completely abrogated this L2-induced enhancement of infectivity. Destruction of the basic cluster of amino acids by replacement of arginines with glutamate (16L2-EE458) as well as the deletion of a second cluster of hydrophobic amino acids located just downstream of the basic cluster (16L1-1/464) strongly reduced infectivity of mutant pseudovirions, too. These defects are not due to impairment of uptake of pseudoviruses, as we observed no differences in the internalization of pseudoviruses composed of L1 alone, L1/wild-type (wt) L2, and L1/mutant L2 (36; data not shown).
Conserved function of the L2 C terminus.
To monitor the importance of this region of L2 for infection by other papillomavirus types, we generated HPV33 pseudoviruses harboring mutant 33L2 proteins carrying truncations at the C terminus (33L2-1/449 and 33L2-1/420) or point mutations at positions 452 and 453 in the basic cluster (33L2-DD452). Again, infectivity of the mutant HPV33 pseudoviruses was reduced to the level of L1-only pseudovirions for the deletion and point mutations (Fig. 2B), even though DNA encapsidation and L2 incorporation were not negatively affected (data not shown). These data suggest that this region of L2 plays a vital role in the infectious process. As shown here for HPV33 and HPV16, this is a conserved feature of papillomaviruses. Additionally, it was previously shown that deletion of the C-terminal basic cluster of BPV-1 L2 (BPVL2-1/460) also abrogated infection (30). Pseudovirions produced with a C-terminally truncated L2 protein were shown to assemble with L1 with normal stoichiometry and encapsidate wild-type levels of DNA but were noninfectious due to an undetermined deficit.
We recently showed that Hsc70 transiently associates with the C terminus of L2 during virion assembly (14). Therefore, we examined whether this interaction was abrogated by the L2 mutation, as this could conceivably result in a misfolded particle. However, we found that Hsc70 accompanied 33L2-DD452 to the nucleus and cosedimented with VLPs in a manner indistinguishable from wt 33L2, suggesting that Hsc70 also interacts with this mutant (data not shown). As previously reported by us, Hsc70 is not associated with C-terminally truncated L2 protein (14).
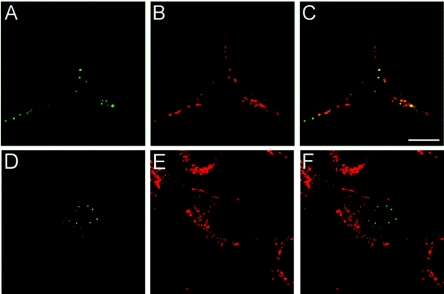
To confirm that the C-terminally truncated L2 mutant pseudovirions are noninfectious due to a defect in endosome escape, we examined the localization and trafficking of BPV particles that harbored a C-terminally truncated L2 (Fig. 3). Unsurprisingly, uncoating occurred normally with a clear appearance of genome and L2 in endocytic vesicles. However, similar to that seen for the viral genome packaged in the L1-only particles, the truncated L2 protein was retained in an endosomal compartment. Therefore, it seems likely that this region of L2 is responsible for mediating the passage of the genome and itself across the vesicular membrane. Double staining experiments with an anti-lamp-2 antibody indicated that this staining pattern represented retention of L2 in late endosomal and/or lysosomal vesicles (Fig. 3). We also analyzed the trafficking properties of the HPV16 pseudovirions assembled with the truncated L2. The microscopic analysis confirmed that pseudovirus made with these L2 proteins was also defective at exit from endocytic vesicles. Additionally, the two amino acid changes within this region of 16L2 generated pseudovirions with identical phenotypes (data not shown).
FIG. 3.
The C-terminal truncation of BPV L2 prevents exit from the endocytic compartment. C127 cells were allowed to internalize BPV-1 L1+L2-1/460-HA (A to C) or BPV-1 L1-wtL2-HA (D to F) pseudovirions for 24 h. The HA-tagged L2 protein was detected with an anti-HA monoclonal antibody (panels A and D). The staining was colocalized with lamp-2 as a marker for the late endosomal and lysosomal vesicles (panels B and E). The merged images are shown in panels C and F.
Cytotoxicity of the C-terminally truncated L2 peptide.
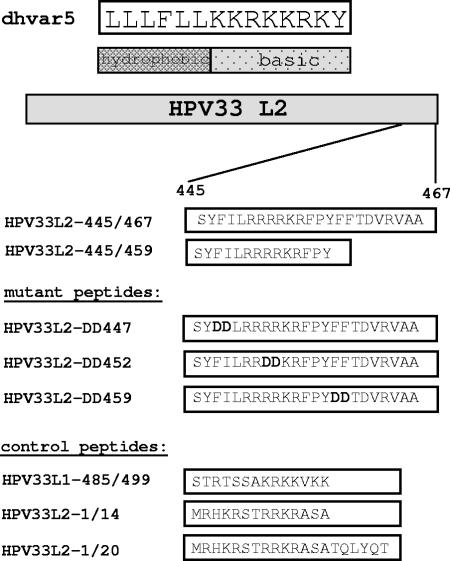
To further strengthen our results, we performed a biochemical characterization of the dhvar5 analogue of HPV33, 33L2-445/459. Since an additional stretch of predominantly hydrophobic amino acids is present further downstream from this peptide, we also analyzed a C-terminally elongated peptide 23 amino acids in length (33L2-445/467) (Fig. 4). We first determined if these peptides exhibit cytotoxic activity against mammalian cells. HeLa cells were incubated with peptides at neutral pH, and cytolysis was determined by measurement of intracellular ATP levels (Fig. 5A). Peptide 33L2-445/467 induced a weak but reproducible reduction of ATP with an LC50 of approximately 170 μg/ml, whereas control peptides did not significantly reduce intracellular ATP levels. Interestingly, this activity was greatly enhanced when the incubations were performed at pH 6.0 (LC50 = 40 μg/ml) (Fig. 5A). The use of highly purified peptide preparations (>95%) instead of the initially used cruder preparation (>70%) increased the activity of the peptide to an LC50 of 8 μg/ml (Fig. 5B). To our surprise, 33L2-445/459 did not significantly reduce ATP levels in this assay, in contrast to dhvar5, which also displayed a pH dependence (Fig. 5A). These data suggested that the N-terminal hydrophobic stretch of amino acids and the cluster of basic amino acids of the HPV33 L2 are not sufficient. This was confirmed by the analysis of mutant peptides with acidic substitutions in the N-terminal and C-terminal hydrophobic stretch as well as in the basic cluster (Fig. 4). Whereas the exchange in the N-terminal hydrophobic region even increased the activity of 33L2-445/467, as shown for 33L2-DD447, changes in the basic cluster and the C-terminal stretch of hydrophobic amino acids (33L2-DD452 and 33L2-DD459) completely prevented the reduction of intracellular ATP levels (Fig. 5B). These data indicate that membrane destabilization requires both the basic cluster and the C-terminal hydrophobic stretch of amino acids.
FIG. 4.
Listing of peptides used throughout the study. Mutations introduced into the HPV33 L2-445/467 peptide are highlighted in boldface type.
FIG. 5.
Cytotoxicity of peptides. (A) HeLa cells were incubated with the indicated concentrations of crude preparations of peptides (70% purity) for 15 h at the indicated pH. Intracellular ATP levels were determined using a luciferase-based assay. (B) Highly purified peptides (>95% purity) were added to HeLa cells for 15 h at pH 7.2 at the indicated concentrations, and ATP levels were determined. ATP levels from mock-treated cells were set at 100%. (C) Propidium iodide was added to HeLa cells treated with the indicated concentrations of 33L2-445/467 for 5 h at pH 7.2. The influx of propidium iodide into cells was determined by fluorescence-activated cell sorter analysis.
Permeabilization of cells.
To test whether cytotoxicity is due to permeabilization of cells for hydrophilic substances, we measured the influx of propidium iodide into HeLa cells treated with increasing concentrations of various L2 peptides. As shown in Fig. 5C and Table 1, more than 90% of cells subjected to 100 μg/ml of peptide 33L2-445/467 became permeable for propidium iodide as measured by flow cytometric analysis. Slightly fewer than 40% of the cells treated with 10 μg/ml of this peptide accumulated propidium iodide, whereas incubation of cells with 1 μg/ml did not increase their permeability. Again, the shorter dhvar5 analogue (peptide 33L2-445/459), as well as the control peptides, did not affect the uptake of propidium iodide into HeLa cells even at concentrations of 100 μg/ml (Table 1). Substitution of amino acids at positions 452/453 or 459/460 but not at positions 447/448 in the N-terminal cluster of hydrophobic amino acids strongly reduced the membrane permeabilizing activity of 33L2-445/467 at the 10-μg/ml concentration.
TABLE 1.
Permeability of HeLa cells to propidium iodide after peptide treatment
| Peptide | % of cells penetrable (mean ± SD) at peptide concn (μg/ml) of:
|
||
|---|---|---|---|
| 100 | 10 | 1 | |
| Control | 6.1 | ||
| 33L2-445/467 | 92.2 ± 2.5 | 37.1 ± 3.3 | 4.8 ± 0.5 |
| 33L2-445/459 | 7.0 ± 0.7 | 3.0 ± 0.0 | 3.1 ± 0.2 |
| 33L2-1/14 | 3.9 ± 0.9 | 3.1 ± 0.2 | 3.9 ± 0.3 |
| 33L2-1/20 | 4.0 ± 0.1 | 5.0 ± 0.3 | 3.9 ± 0.4 |
| 33L2-DD447 | 83.8 ± 11.2 | 41.0 ± 5.6 | 3.1 ± 0.5 |
| 33L2-DD452 | 70.7 ± 2.7 | 5.9 ± 2.6 | 4.0 ± 0.1 |
| 33L2-DD459 | 52.8 ± 0.5 | 12.1 ± 5.8 | 3.2 ± 0.3 |
| 33L1-485/499 | 7.0 ± 0.8 | 4.9 ± 1.1 | 6.1 ± 0.0 |
Membrane integration of GFP-L2 fusion proteins.
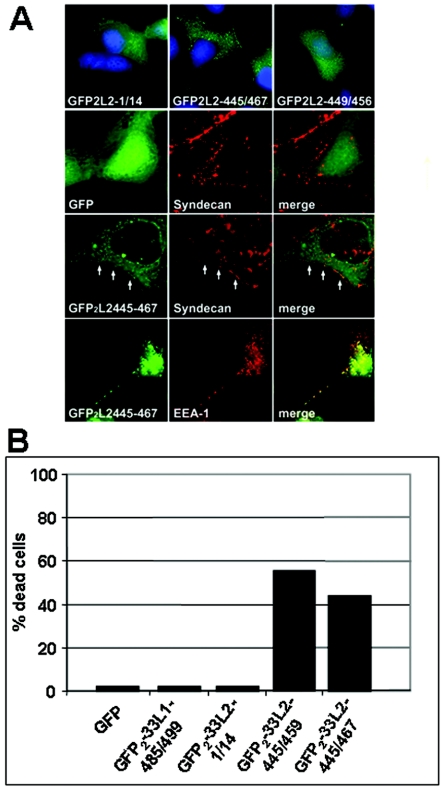
To analyze the intracellular affinity of L2 peptides for cellular membranes, we generated fusion proteins with dimeric GFP under the control of the cytomegalovirus promoter for expression in mammalian cell lines. We analyzed the subcellular distribution of these fusion proteins after transfection of HuTK−143B cells (Fig. 6A). We found that GFP2-33L2-445/467 partially colocalized with syndecan-1 and EEA-1, markers of the plasma membrane and early endosomes, respectively, whereas the control transfected proteins, either GFP alone or GFP fused to an N-terminal L2 peptide (GFP2-33L2-1/14), displayed a diffuse intracellular staining pattern. The shorter L2 peptide (33L2-445/459) was also found to preferentially localize to cellular membranes (data not shown). Fusion protein 33L2-449/456, which misses the N- and C-terminal hydrophobic clusters of amino acids, was not associated with membranes, again pointing to the importance of these hydrophobic regions for membrane association. During this analysis we reproducibly observed that a high percentage of cells expressing GFP2-33L2-445/459 or -445/467 but not of cells expressing GFP or GFP fused to control peptides died within 24 h (Fig. 6B). These data again demonstrate the ability of these C-terminally truncated L2 peptides to perturb cellular membranes and mediate cytolysis.
FIG. 6.
Membrane association of GFP2-33L2-445/467. (A) COS7 cells were transfected with pEGFP-C1 (GFP), pGFP2-33L2-1/14 (GFP2L2-1/14), -33L2-449/456 (GFP2L2-449/456), or -33L2-445/467 (GFP2L2-445/467). GFP fluorescence was determined at 24 h posttransfection (upper panel). Cells displayed in the three lower panels were additionally stained for syndecan-1 or EEA-1. Colocalization of syndecan-1 with GFP2-33L2-445/467 is indicated by arrows. (B) The percentage of necrotic cells transfected with the indicated plasmids was determined.
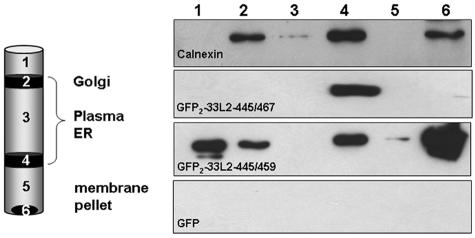
Membrane association was confirmed biochemically by subcellular fractionation analyses. Cellular membranes were isolated from COS-7 cells that expressed the L2-derived GFP fusion proteins. After fractionation through sucrose step gradient centrifugation, proteins were analyzed by GFP-specific immunoblotting, as shown in Fig. 7. Calnexin served as a marker for integral membrane proteins and was predominantly found in fractions 2, 4, and 6, which would be expected to be enriched for membranes. Both GFP2-33L2-445/467 and -445/459 (Fig. 7) but not GFP and GFP fused to control peptides (data not shown) copurified with cellular membranes. Whereas GFP2-33L2-445/467 was reproducibly associated with both the plasma and ER membranes, the distribution of GFP2-33L2-445/459 was more variable. Since GFP2-33L2-445/459 was also repeatedly found in the first fraction containing soluble proteins, we postulate that a significant fraction of it is attached to membranes rather weakly.
FIG. 7.
GFP-L2 fusion proteins copurify with membranes. Membranes were isolated from COS7 cells transfected with the indicated GFP constructs and subjected to a sucrose step gradient centrifugation. Fractions were analyzed by Western blotting using calnexin- or GFP-specific antibodies. C-terminally truncated L2 peptides induce membrane association of GFP fusions.
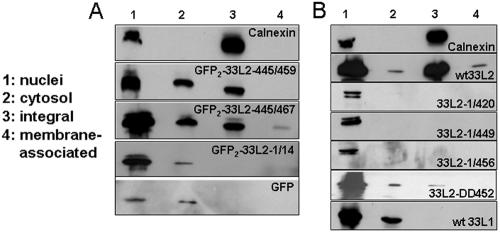
To distinguish between fusion proteins integrated into rather than associated with membranes, we treated isolated membranes with carbonate buffer at pH 11.5 to remove membrane-associated proteins. We used calnexin again as a marker for the integral membrane protein fraction. All fusions were expressed and could be detected in nuclear fractions and in fractions containing the soluble cytosolic proteins. In contrast to the GFP control proteins, dimeric GFP fused to C-terminally truncated L2 peptides was again found in the membrane fractions. The majority of GFP2-33L2-445/467 and -445/459 could not be extracted from membranes by alkaline treatment, suggesting they are integrated into cellular membranes (Fig. 8A).
FIG. 8.
Wild-type but not C-terminally truncated L2 are integrated into membranes. (A) Isolated membranes from transfected COS7 cells were treated with alkaline buffer, pH 11.5, prior to Western blot analysis. C-terminally truncated L2 peptides induce integration of GFP fusion protein. (B) HuTK−143B cells were infected with vaccinia virus recombinant for the indicated HPV33 capsid genes. Membranes were isolated, alkaline treated, and analyzed by Western blotting using calnexin-, L1-, or L2-specific antibodies.
To exclude that our observations are artifactual due to the utilization of peptides, we analyzed the membrane association of full-length and C-terminally truncated L2 protein using the same approach (Fig. 8B). These proteins were expressed in HuTK−143B cells using recombinant vaccinia viruses, and membranes were isolated and subjected to treatment with carbonate buffer, pH 11.5. A substantial fraction of the full-length 33L2 protein, but not of the C-terminally truncated mutant proteins, was detected as an integral membrane protein (Fig. 8C). As an additional control, we show that the 33L1 protein was also not detectable in the membrane fraction. The amount of mutant L2 protein with acidic substitutions at positions 452 and 453 (33L2-DD452) that copurified with membranes was strongly reduced compared to wt 33L2 protein, confirming the observations made with the peptides (Table 1).
DISCUSSION
We have presented several lines of evidence that the papillomavirus minor capsid protein mediates the endosomal escape of the viral genome. A C-terminal 23-amino-acid peptide from the L2 protein was identified that is essential for transit of the viral DNA across the endosomal membrane during papillomavirus infection. This peptide is also highly cytotoxic to bacteria, fungi, and mammalian cells after exogenous application due to membrane depolarization and permeabilization and induces the integration of GFP fusion proteins and full-length L2 protein into cellular membranes when expressed in mammalian cells, eventually resulting in cell death. Since the full-length L2 protein but not C-terminally mutated L2 nor L1 protein integrated into cellular membranes, an artificial effect of the observations with peptides and fusion proteins can be excluded. These critical functions render this peptide sequence indispensable for PV infection. The activity of this peptide is highest at modestly acidic pH, which corresponds well with the observation that acidification of endosomes is an essential step in papillomavirus infection (11, 37) and that viral uncoating occurs within the milieu of the endocytic compartment (10).
The characteristic features of the C-terminally truncated L2 peptide are distinct in comparison with other membrane-affecting peptides. The latter are mostly amphipathic in nature, i.e., have alternating basic and hydrophobic amino acids and often form a pore or channel through the membrane bilayer. In contrast, the L2 peptide is composed of a cluster of basic amino acids adjacent to at least one cluster of hydrophobic amino acids. Both regions are required for full activity of the peptide. At present, the mechanism of membrane permeabilization is unclear. A likely possibility is that the positive charge of the basic amino acid cluster mediates tight association with negatively charged lipids of membranes and that subsequent insertion of the hydrophobic cluster(s) into membranes induces a torsional stress, which results in membrane disruption. The peptide concentrations required for cytotoxicity suggest that it probably functions via the carpet or detergent-like mechanism rather than pore formation resulting in the collapse of the target membrane (41). Future experiments utilizing artificial membranes and purified L2 and/or L2 peptides should allow identification of the lipids required for the peptide's activity and determination of the size of the rupture or pore that is subsequently formed. This, in turn, would allow insight into the precise mechanism of peptide action. Using liposomes, it was recently shown for dhvar5 that negatively charged phospholipids are required for the peptide to translocate across and permeabilize membranes (12).
Introduction of mutations within the peptide, which abrogate the membrane perturbation, have pronounced effects on the infectivity of pseudovirions. Mutations within this region that do not interfere with membrane destabilization also do not show any defect in infectivity (data not shown). The observed effects are not at the level of virion assembly, since mutant L2 incorporation and DNA encapsidation were not negatively affected. To exclude that mutations within the 23-amino-acid peptide prevent Hsc70/L2 interaction, thus interfering with a functional conformation of the virion-incorporated L2, we also determined that the transient complex formation of Hsc70 and L2 was unaffected by the mutations. We previously identified a role of the constitutively expressed chaperone Hsc70 in L2 nuclear translocation and subsequent L2 incorporation into papillomavirus particles and found that it exerts its function through interaction with the C terminus of L2 (14). The observation that this interaction is unaffected suggests that the observed defect in the infectivity of pseudovirions is not due to an effect on viral morphogenesis.
We also excluded that the introduced mutations interfere with pseudovirion adsorption and internalization into target cells. Capsids were taken up with equal efficiency as wt pseudovirions. This is in agreement with previous reports that L2 does not contribute to the initial steps of papillomavirus infection (29, 32). The C terminus of L2 is probably located within the assembled capsid (25), and a recent report confirms that this region is only accessible to antibody binding relatively late in infection, after virion disassembly has commenced (10). Since accessibility of L2 coincides with the detection of encapsidated genomes, uncoating is likely to be a prerequisite for the membrane-disrupting function of L2. Whereas a fraction of wt L2 escapes intracellular vesicles together with the viral genome, resulting in nuclear translocation, genomes encapsidated by wt L1 alone or wt L1 and C-terminally truncated L2 mutants are not able to pass this cellular membrane. Since the HA tag on the truncated L2 can be detected after infection with mutant pseudovirions, the introduction of the truncation does not interfere with uncoating. In this instance, the L2 colocalizes with lamp-2 at late time points. Furthermore, viral DNA is retained within vesicles after the addition of HPV16 L1-only pseudovirions. Taken together, these data are strongly suggestive that the defect is at the level of endosome escape. Even though colocalization of L2 and viral DNA with lamp-2 is observed after infection with truncated L2 pseudovirions, it is not clear if the viral genome and L2 escape from the late endosomes or if it reflects the compartment where the mutant L2 and DNA accumulate after failing to leave the endocytic system.
Our recent observations that Hsc70 accompanies L2 to the nucleus by binding to its C terminus (14), in addition to the data presented here, point to Hsc70 as a key regulator of L2 function. Hsc70 binding may mask L2's membrane-destabilizing activity during the productive phase of the life cycle, preventing premature integration of L2 into membranes and thus allowing virus assembly. Since Hsc70 is not incorporated into virions, it cannot interfere with the endosomolytic activity of L2 once the virus is partially disassembled.
Although long recognized as essential for infection, the study of the passage across the limiting membrane by nonenveloped viruses or their components has been difficult, and as a result, little insight has been acquired for most of these viruses. The best-studied virus in this respect is poliovirus, whose interaction with its cellular receptor PVR/CD155 induces a defined, temperature-sensitive conformational change in two of the viral capsid proteins, VP1 and VP4. Hydrophobic amino acids exposed by this structural change insert into the host cell membrane and form a pore, which can be used by ions and the viral RNA to pass through the membrane (3, 43, 45). Reports are still conflicting if this process occurs at the plasma membrane or from within endocytic vesicles after internalization. Due to structural and sequence homology, similar processes are believed to occur in other members of the Picornaviridae (4, 5). In addition, adenoviruses have long been known to induce endosomolysis in the target cell. A role of the viral capsid protein, penton base, and the virus-encoded 23-kDa adenovirus cysteine protease in this process has been suggested (18, 39, 40). In addition, the adenovirus uptake receptor αvβ5 integrin is required for efficient endosomal release of adenoviruses (50, 51). Recently, it was demonstrated that an amphipathic peptide within the capsid protein VI mediates membrane disruption (52). This capsid protein is initially incorporated into capsids as a preprotein which requires cleavage by the 23-kDa cysteine protease for activation, explaining the important role of the protease in endosomal escape mechanisms.
In contrast to the amphipathic or exclusively hydrophobic nature of virus-encoded membrane-disrupting activities in adenoviruses and polioviruses, respectively, the herein identified L2 peptide of papillomaviruses contains adjacent hydrophobic and basic clusters of amino acids. It may, therefore, represent a new class of virus-encoded proteins mediating membrane destabilization to facilitate egress from endosomes. This report, in addition to its contribution to the understanding of the papillomavirus life cycle, may also help to elucidate the mechanisms of membrane passage of other nonenveloped DNA viruses.
Acknowledgments
We are grateful to Katrin Anne Becker for initial experiments, Rolf E. Streeck for continuous support, and Sucharit Bhakdi and Iwan Valev for establishment of the ATP assays.
This work was supported by grants from the Deutsche Forschungsgemeinschaft to M.S. (SFB490/B5).
REFERENCES
- 1.Baker, T. S., W. W. Newcomb, N. H. Olson, L. M. Cowsert, C. Olson, and J. C. Brown. 1991. Structures of bovine and human papillomaviruses. Analysis by cryoelectron microscopy and three-dimensional image reconstruction. Biophys. J. 60:1445-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, K. A., C. Sapp, L. Florin, G. G. Maul, and M. Sapp. 2004. Nuclear localization but not PML is required for incorporation of papillomavirus minor capsid protein L2 into virus-like particles. J. Virol. 78:1121-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belnap, D. M., D. J. Filman, B. L. Trus, N. Cheng, F. P. Booy, J. F. Conway, S. Curry, C. N. Hiremath, S. K. Tsang, A. C. Steven, and J. M. Hogle. 2000. Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. J. Virol. 74:1342-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brabec, M., G. Baravalle, D. Blaas, and R. Fuchs. 2003. Conformational changes, plasma membrane penetration, and infection by human rhinovirus type 2: role of receptors and low pH. J. Virol. 77:5370-5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brabec, M., D. Schober, E. Wagner, N. Bayer, R. F. Murphy, D. Blaas, R. Fuchs, and G. Baravalle. 2005. Opening of size-selective pores in endosomes during human rhinovirus serotype 2 in vivo uncoating monitored by single-organelle flow analysis [DOI] [PMC free article] [PubMed]
- 6.Buck, C. B., D. V. Pastrana, D. R. Lowy, and J. T. Schiller. 2004. Efficient intracellular assembly of papillomaviral vectors. J. Virol. 78:751-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buck, C. B., C. D. Thompson, Y. Y. S. Pang, D. R. Lowy, and J. T. Schiller. 2005. Maturation of papillomavirus capsids. J. Virol. 79:2839-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandran, K., D. L. Farsetta, and M. L. Nibert. 2002. Strategy for nonenveloped virus entry: a hydrophobic conformer of the reovirus membrane penetration protein micro 1 mediates membrane disruption. J. Virol. 76:9920-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen, N. D., N. M. Cladel, and C. A. Reed. 1995. Postattachment neutralization of papillomaviruses by monoclonal and polyclonal antibodies. Virology 207:136-142. [DOI] [PubMed] [Google Scholar]
- 10.Day, P. M., C. C. Baker, D. R. Lowy, and J. T. Schiller. 2004. Establishment of papillomavirus infection is enhanced by promyelocytic leukemia protein (PML) expression. Proc. Natl. Acad. Sci. USA 101:14252-14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day, P. M., D. R. Lowy, and J. T. Schiller. 2003. Papillomaviruses infect cells via a clathrin-dependent pathway. Virology 307:1-11. [DOI] [PubMed] [Google Scholar]
- 12.Den Hertog, A. L., H. W. Wong Fong Sang, R. Kraayenhof, J. G. Bolscher, W. Van't Hof, E. C. Veerman, and A. V. Nieuw Amerongen. 2004. Interactions of histatin 5 and histatin 5-derived peptides with liposome membranes: surface effects, translocation and permeabilization. Biochem. J. 379:665-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Earp, L. J., S. E. Delos, H. E. Park, and J. M. White. 2005. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 285:25-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Florin, L., K. A. Becker, C. Sapp, C. Lambert, H. Sirma, M. Müller, R. E. Streeck, and M. Sapp. 2004. Nuclear translocation of papillomavirus minor capsid protein L2 requires Hsc70. J. Virol. 78:5546-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Florin, L., C. Sapp, R. E. Streeck, and M. Sapp. 2002. Assembly and translocation of papillomavirus capsid proteins. J. Virol. 76:10009-10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaynor, E. C., S. te Heesen, T. R. Graham, M. Aebi, and S. D. Emr. 1994. Signal-mediated retrieval of a membrane protein from the Golgi to the ER in yeast. J. Cell Biol. 127:653-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giroglou, T., L. Florin, F. Schäfer, R. E. Streeck, and M. Sapp. 2001. Human papillomavirus infection requires cell surface heparan sulfate. J. Virol. 75:1565-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greber, U. F., P. Webster, J. Weber, and A. Helenius. 1996. The role of the adenovirus protease on virus entry into cells. EMBO J. 15:1766-1777. [PMC free article] [PubMed] [Google Scholar]
- 19.Hagensee, M. E., N. H. Olson, T. S. Baker, and D. A. Galloway. 1994. Three-dimensional structure of vaccinia virus-produced human papillomavirus type 1 capsids. J. Virol. 68:4503-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hogle, J. M. 2002. Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu. Rev. Microbiol. 56:677-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmgren, S. C., N. A. Patterson, M. A. Ozbun, and P. F. Lambert. 2005. The minor capsid protein L2 contributes to two steps in the human papillomavirus type 31 life cycle. J. Virol. 79:3938-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joyce, J. G., J.-S. Tung, C. T. Przysiecki, J. C. Cook, E. D. Lehman, J. A. Sands, K. U. Jansen, and P. M. Keller. 1999. The L1 major capsid protein of human papillomavirus type 11 recombinant virus-like particles interacts with heparin and cell-surface glycosaminoglycans on human keratinocytes. J. Biol. Chem. 274:5810-5822. [DOI] [PubMed] [Google Scholar]
- 23.Kawana, K., H. Yoshikawa, Y. Taketani, K. Yoshiike, and T. Kanda. 1998. In vitro construction of pseudovirions of human papillomavirus type 16: incorporation of plasmid DNA into reassembled L1/L2 capsids. J. Virol. 72:10298-10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leder, C., J. A. Kleinschmidt, C. Wiethe, and M. Müller. 2001. Enhancement of capsid gene expression: preparing the human papillomavirus type 16 major structural gene L1 for DNA vaccination purposes. J. Virol. 75:9201-9209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, W. J., L. Gissmann, X. Y. Sun, A. Kanjanahaluethai, M. Muller, J. Doorbar, and J. Zhou. 1997. Sequence close to the N-terminus of L2 protein is displayed on the surface of bovine papillomavirus type 1 virions. Virology 227:474-483. [DOI] [PubMed] [Google Scholar]
- 26.Meier, O., and U. F. Greber. 2004. Adenovirus endocytosis. J. Gene Med. 6(Suppl. 1):S152-S163. [DOI] [PubMed] [Google Scholar]
- 27.Modis, Y., B. L. Trus, and S. C. Harrison. 2002. Atomic model of the papillomavirus capsid. EMBO J. 21:4754-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moss, B., O. Elroy-Stein, T. Mizukami, W. A. Alexander, and T. R. Fuerst. 1990. New mammalian expression vectors. Nature 348:91-92. [DOI] [PubMed] [Google Scholar]
- 29.Müller, M., L. Gissmann, R. J. Cristiano, X. Y. Sun, I. H. Frazer, A. B. Jenson, A. Alonso, H. Zentgraf, and J. Zhou. 1995. Papillomavirus capsid binding and uptake by cells from different tissues and species. J. Virol. 69:948-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roden, R. B., P. M. Day, B. K. Bronzo, W. H. Yutzy IV, Y. Yang, D. R. Lowy, and J. T. Schiller. 2001. Positively charged termini of the L2 minor capsid protein are necessary for papillomavirus infection. J. Virol. 75:10493-10497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roden, R. B., H. L. Greenstone, R. Kirnbauer, F. P. Booy, J. Jessie, D. R. Lowy, and J. T. Schiller. 1996. In vitro generation and type-specific neutralization of a human papillomavirus type 16 virion pseudotype. J. Virol. 70:5875-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roden, R. B., R. Kirnbauer, A. B. Jenson, D. R. Lowy, and J. T. Schiller. 1994. Interaction of papillomaviruses with the cell surface. J. Virol. 68:7260-7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sapp, M., U. Kraus, C. Volpers, P. J. Snijders, J. M. Walboomers, and R. E. Streeck. 1994. Analysis of type-restricted and cross-reactive epitopes on virus-like particles of human papillomavirus type 33 and in infected tissues using monoclonal antibodies to the major capsid protein. J. Gen. Virol. 75:3375-3383. [DOI] [PubMed] [Google Scholar]
- 34.Sapp, M., C. Volpers, M. Müller, and R. E. Streeck. 1995. Organization of the major and minor capsid proteins in human papillomavirus type 33 virus-like particles. J. Gen. Virol. 76:2407-2412. [DOI] [PubMed] [Google Scholar]
- 35.Schäfer, F., L. Florin, and M. Sapp. 2002. DNA binding of L1 is required for human papillomavirus morphogenesis in vivo. Virology 295:172-181. [DOI] [PubMed] [Google Scholar]
- 36.Selinka, H. C., T. Giroglou, T. Nowak, N. D. Christensen, and M. Sapp. 2003. Further evidence that papillomavirus particles exist in two distinct conformations. J. Virol. 77:12961-12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selinka, H. C., T. Giroglou, and M. Sapp. 2002. Analysis of the infectious entry pathway of human papillomavirus type 33 pseudovirions. Virology 299:279-287. [DOI] [PubMed] [Google Scholar]
- 38.Selinka, H. C., and M. Sapp. 2003. Papillomavirus/cell-interactions initiating the infectious entry pathway. Papillomavirus Rep. 14:259-265. [Google Scholar]
- 39.Seth, P., D. Fitzgerald, H. Ginsberg, M. Willingham, and I. Pastan. 1984. Evidence that the penton base of adenovirus is involved in potentiation of toxicity of Pseudomonas exotoxin conjugated to epidermal growth factor. Mol. Cell. Biol. 4:1528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seth, P., I. Pastan, and M. C. Willingham. 1985. Adenovirus-dependent increase in cell membrane permeability. J. Biol. Chem. 260:9598-9602. [PubMed] [Google Scholar]
- 41.Shai, Y. 2002. Mode of action of membrane active antimicrobial peptides. Biopolymers 66:236-248. [DOI] [PubMed] [Google Scholar]
- 42.Stauffer, Y., K. Raj, K. Masternak, and P. Beard. 1998. Infectious human papillomavirus type 18 pseudovirions. J. Mol. Biol. 283:529-536. [DOI] [PubMed] [Google Scholar]
- 43.Tosteson, M. T., and M. Chow. 1997. Characterization of the ion channels formed by poliovirus in planar lipid membranes. J. Virol. 71:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tosteson, M. T., M. L. Nibert, and B. N. Fields. 1993. Ion channels induced in lipid bilayers by subvirion particles of the nonenveloped mammalian reoviruses. Proc. Natl. Acad. Sci. USA 90:10549-10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tosteson, M. T., H. Wang, A. Naumov, and M. Chow. 2004. Poliovirus binding to its receptor in lipid bilayers results in particle-specific, temperature-sensitive channels. J. Gen. Virol. 85:1581-1589. [DOI] [PubMed] [Google Scholar]
- 46.Trus, B. L., R. B. Roden, H. L. Greenstone, M. Vrhel, J. T. Schiller, and F. P. Booy. 1997. Novel structural features of bovine papillomavirus capsid revealed by a three-dimensional reconstruction to 9 Å resolution. Nat. Struct. Biol. 4:413-420. [DOI] [PubMed] [Google Scholar]
- 47.Unckell, F., R. E. Streeck, and M. Sapp. 1997. Generation and neutralization of pseudovirions of human papillomavirus type 33. J. Virol. 71:2934-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Volpers, C., M. Sapp, C. A. Komly, P. Richalet-Secordel, and R. E. Streeck. 1993. Development of type-specific and cross-reactive serological probes for the minor capsid protein of human papillomavirus type 33. J. Virol. 67:1927-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Volpers, C., M. Sapp, P. J. Snijders, J. M. Walboomers, and R. E. Streeck. 1995. Conformational and linear epitopes on virus-like particles of human papillomavirus type 33 identified by monoclonal antibodies to the minor capsid protein L2. J. Gen. Virol. 76:2661-2667. [DOI] [PubMed] [Google Scholar]
- 50.Wang, K., T. Guan, D. A. Cheresh, and G. R. Nemerow. 2000. Regulation of adenovirus membrane penetration by the cytoplasmic tail of integrin beta5. J. Virol. 74:2731-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wickham, T. J., E. J. Filardo, D. A. Cheresh, and G. R. Nemerow. 1994. Integrin alpha v beta 5 selectively promotes adenovirus mediated cell membrane permeabilization. J. Cell Biol. 127:257-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiethoff, C. M., H. Wodrich, L. Gerace, and G. R. Nemerow. 2005. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J. Virol. 79:1992-2000. [DOI] [PMC free article] [PubMed]