Abstract
We have analyzed changes to proviral Env gp120 sequences and the development of neutralizing antibodies (NAbs) during 1 year of simian/human immunodeficiency virus SHIV-89.6P infection in 11 Macaca nemestrina macaques. Seven macaques had significant env divergence from that of the inoculum, and macaques with greater divergence had higher titers of homologous NAbs. Substitutions in sequons encoding potential N-linked glycosylation sites (PNGs) were among the first to be established, although overall the total number of sequons did not increase significantly. The majority (19 of 23) of PNGs present in the inoculum were conserved in the sequences from all macaques. Statistically significant variations in PNGs occurred in multiple macaques within constrained regions we term “hot spots,” resulting in the selection of sequences more similar to the B consensus. These included additions on V1, the N-terminal side of V4, and the outer region of C2. Complex mutational patterns resulted in convergent PNG shifts in V2 and V5. Charge changes in Env V1V2, resulting in a net acidic charge, and a proline addition in V5 occurred in several macaques. Molecular modeling of the 89.6P sequence showed that the conserved glycans lie on the silent face of Env and that many are proximal to disulfide bonds, while PNG additions and shifts are proximal to the CD4 binding site. Nonsynonymous-to-synonymous substitution ratios suggest that these changes result from selective pressure. This longitudinal and cross-sectional study of mutations in human immunodeficiency virus (HIV) env in the SHIV background provides evidence that there are more constraints on the configuration of the glycan shield than were previously appreciated.
The high degree of variability that is tolerated by the human immunodeficiency virus type 1 (HIV-1) envelope protein (Env), coupled with the extensive use of carbohydrates as a means of immune escape, has contributed to the formidable challenge of developing an effective vaccine against HIV. HIV Env is one of the most heavily glycosylated proteins currently known, with carbohydrates comprising >50% of its molecular weight (26). It has been appreciated for some years that Env glycosylation can constrain T-cell epitope (6) as well as neutralizing antibody (NAb) (11, 32, 41, 62) recognition while at the same time contributing to the overall integrity and conformation of the glycoprotein (38, 63). Recently, Wei et al. (61) proposed a model wherein the carbohydrate moieties of HIV comprise a glycan shield that creates a fluid canopy around the conserved neutralizing epitopes of the protein domain beneath. This shield is proposed to evolve in response to pressure from NAbs, creating a continuously changing landscape on the viral Env. The functionality of the Env protein and space constraints appear to limit the number of potential N-linked glycosylation (PNG) sites, or sequons, resulting in the maintenance of 18 to 33 N-linked glycans and redistribution of their placement (66).
In recent years, insight has been gained from structural analyses of the bound (as a ligand) and unbound gp120 core (12, 36) as well as from several studies looking at the dynamics of the glycan shield in both simian immunodeficiency virus (SIV) (11) and HIV (15, 17, 61). Such studies have documented that the acquisition or rearrangement of carbohydrates can allow for neutralization escape mutants (8, 13, 52), while many of the individual glycans play important roles in protecting the CD4 binding site (CD4bs) and gp41 ectodomain from neutralizing antibodies (42).
Neutralizing antibodies are known to influence the evolution of HIV within a host and to play an important role in disease progression. NAb titers correlate with a lack of disease progression in some HIV-infected long-term nonprogressors (10), and viral persistence may depend on the selection of NAb escape variants (16, 61). In some patients, the NAb response can show evidence of affinity maturation and recognition of common determinants conserved among distinct isolates (55). However, this maturation is a multiyear process and is not universal, occurring in a minority of patients (43). Thus, an understanding of the mechanism(s) governing this response would be valuable for vaccine design.
A major question that remains unanswered involves the boundaries of variation that exist as the virus evolves in individual hosts and the relationship between these changes and the adaptive immune response. This has been difficult to address for HIV-1 infection of humans due to the distinct viruses present in each individual. This question can be addressed in part by comparing the sequences from different individuals over time during disease progression and when there are alterations in Env functionality, such as its coreceptor usage. Such a comparison was performed using published sequences found in the Los Alamos National Laboratory (LANL) HIV database. An analysis of 49 published full-length HIV-1 gp160 sequences in the LANL database revealed that approximately half of these glycans are in conserved regions (66). For those that are not conserved, there appear to be small windows, or “hot spots,” where the glycans can vary in relative position and number as coreceptor switching occurs and immune responses develop. However, this type of analysis cannot directly link Env mutations to the development of antibody or cytotoxic T lymphocyte (CTL) responses.
Another approach to addressing this question is to determine whether key changes are similar in different individuals infected with the same virus. Using molecularly cloned or low-diversity virus isolates to infect nonhuman primates, it is possible to monitor the fate of viral sequences over months or years postinfection in multiple individuals with greater or lesser degrees of control of viremia. These types of studies have been valuable for understanding viral mutations leading to immune escape from cytotoxic T cells (1). Much of the data available from the analysis of mutations in Env in nonhuman primates have utilized SIV, where changes to PNGs can directly affect both resistance to neutralization by sera from infected macaques and the pathogenicity of the virus itself (11, 31). SIV is closely related to HIV-2 and differs significantly in Env from HIV-1; until recently, SIV and HIV-1 were not known to share any neutralization determinants. A recent report demonstrated that CD4-induced antibodies cross-neutralize primate lentiviruses in the presence of soluble CD4 (20). Despite this shared epitope, it is likely that few specific escape mutations in SIV Env will be directly informative regarding HIV-1 in humans. In contrast, chimeric simian/human immunodeficiency viruses (SHIVs) bearing HIV-1 env genes have the potential for direct comparison with HIV-1 env genes in their native context.
For this study, we performed an analysis of 11 macaques that were infected with SHIV-89.6P (49) for approximately 1 year. The Env proteins in seven of these macaques underwent significant divergence from that in the inoculum, while four had minimal divergence. SHIV-89.6P was constructed with the HIV-89.6P (a subtype B virus) env gene (50). Subtype B viruses, unlike A and C viruses, do not seem to accumulate PNGs over time from infection, while A and C viruses tend to have reduced numbers of PNGs in early infection and gradually acquire more (14, 22). Despite this difference among subtypes, the population of subtype B viruses in the HIV database tends to have comparable numbers of PNG sites and levels of diversity to those in the other subtypes. In this study, using a subtype B env gene in an SIV background (SHIV-89.6P), there was no inherent tendency for the total number of PNG sites to gradually increase, although Env proteins from macaques with divergent viruses did acquire significantly more PNGs than those from macaques with minimally divergent viruses. We observed a remarkable degree of conservation in patterns of change in Env PNGs and alterations in V2 and V5 in all seven macaques with divergent viruses, despite variations in overall amino acid sequences that were unique to each macaque. The majority of these changes made the SHIV-89.6P virus more similar to the typical patterns seen for HIV-1 clade B. The data from this study suggest that the glycan shield and certain other key regions of HIV-1 Env may be under considerable constraints during viral divergence.
MATERIALS AND METHODS
SHIV-89.6p challenge stock.
A SHIV-89.6P bulk culture was kindly provided by Norm Letvin. The virus was passaged twice through Macaca nemestrina peripheral blood mononuclear cells (PBMCs), titrated in vivo, and then used to challenge M. nemestrina intrarectally with 50 50% macaque infectious doses (24). Sequencing of proviral gp120 from the PBMCs used to grow the virus revealed that the consensus sequence was identical to the published sequence KB9 (accession no. U89134 and data not shown). While there was some minor variation in the inoculum used to infect macaques (Fig. 1), all but two PNGs were identical in all inoculum variants. The presence of residues N139C and N397 was heterogeneous, as they were found in 88% and 38% of the inoculum variants, respectively (see Table 2). SHIV-89.6P gp120 carries 23 PNGs, which is below the median for HIV-1 Env.
FIG. 1.
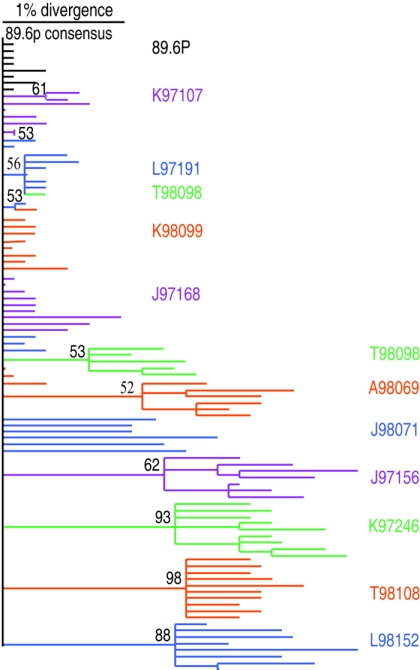
Phylogenetic analysis of individual HIV-1 envelope gp120 proviral clones from macaques infected with SHIV-89.6P at 32 weeks postinfection. The maximum likelihood tree is rooted on the consensus of the inoculum. Sequences from each macaque are clustered together and uniformly colored. Branches were collapsed if the bootstrap value was below 50; bootstrap values over 50 are labeled on the tree.
TABLE 2.
Convergent PNG changes in gp120 at 32 wpia
| Sample | (N)b | Prevalence (%) of indicated PNG
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V1V2
|
C2
|
C3
|
V4
|
V5
|
||||||||||
| 136 | 139 | 139C | 141 | 186 | 188 | 234 | 276 | 362 | 386 | 397 | 460 | 462/3 | ||
| Inoculumc | 8 | 100 | 100 | 88 | 0 | 100 | 0 | 100 | 0 | 100 | 0 | 38 | 100 | 0 |
| K97107 | 8 | 100 | 100 | 100 | 25 | 75 | 25 | 75 | 0 | 100 | 0 | 0 | 100 | 0 |
| K98099 | 9 | 100 | 100 | 100 | 0 | 100 | 0 | 100 | 0 | 100 | 0 | 0 | 100 | 0 |
| L97191 | 9 | 100 | 100 | 33 | 0 | 100 | 0 | 100 | 0 | 100 | 0 | 0 | 100 | 0 |
| J97168 | 9 | 100 | 100 | 56 | 0 | 89 | 0 | 100 | 0 | 100 | 0 | 33 | 100 | 0 |
| A98069 | 9 | 100 | 100 | 89 | 56 | 33 | 0 | 100 | 0 | 100 | 67 | 22 | 100 | 0 |
| T98098 | 9 | 100 | 100 | 78 | 55 | 44 | 56 | 100 | 33 | 78 | 56 | 0 | 44 | 0 |
| J97156 | 7 | 100 | 100 | 71 | 43 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 0 | 86 |
| K97246 | 9 | 100 | 100 | 100 | 78 | 0 | 100 | 100 | 100 | 89 | 100 | 100 | 0 | 0 |
| L98152 | 8 | 0 | 100 | 88 | 13 | 0 | 100 | 0 | 25 | 100 | 0 | 100 | 25 | 75 |
| T98108 | 10 | 100 | 100 | 80 | 70 | 0 | 100 | 100 | 100 | 40 | 100 | 30 | 0 | 100 |
| J98071 | 6 | 100 | 33 | 67 | 83 | 100 | 0 | 100 | 100 | 100 | 17 | 100 | 0 | 67 |
Percent prevalence of each PNG within the quasispecies. Sites that differ significantly from the inoculum are shown in bold (Fisher's exact P value, <0.05).
Ten proviral clones were sequenced from each animal. Duplicate or missense sequences were removed from analysis. N, number of unique clones incorporated for analysis from each animal.
Proviral clones from the PBMCs used to grow the viral stock of SHIV-89.6P were analyzed as a measure of heterogeneity in the inoculum.
PBMC extraction, PCR, cloning, and sequencing.
All extractions were performed in a separate PCR containment hood using unidirectional workflow, separate storage of templates, primers, and other reagents, and other cleaning precautions to avoid sample cross-contamination. DNAs were extracted using QIAGEN QiaAmp DNA mini kits and QiaAmp DNA Blood mini kits according to the manufacturer's instructions. Nested PCR was performed on 200 to 500 ng genomic DNA in order to amplify proviral gp120. To avoid amplification bias of the template, five replicates were performed with each sample. Control amplifications with no template were included in each experiment to monitor for carryover contamination. First-round PCR was performed with the primers 3′half start (GCATGCTGTAGAGCAAGAAATGGAGC) and 1548 (GCTCCCAAGAACCCAAGGAAC), using the following cycling conditions: denaturation at 94°C for 3 min and 35 cycles of 94°C for 15 s, 55°C for 3 min, and 68°C for 3 min, with a 5-s increase in each cycle, followed by a final extension at 68°C for 7 min.
Nested PCR was performed on a 1:25 dilution of first-round PCR product, using the primers 3′gp120 89.6 (CGATTCATCTTTTTTCTCTTTGCACTGT) and EO (TAGAGCCCTGGAAGCATCCAGGAAGTCAGCCTA). The following cycling conditions were used: denaturation at 94°C for 2 min and 25 cycles of 94°C for 15 s, 64°C for 3 min, and 68°C for 3 min, with a 5-s increase in each cycle, followed by a final extension at 68°C for 7 min. PCR products were purified using a QIAGEN QIAquick PCR purification kit according to the manufacturer's instructions. For each sample, the purified nested PCR products were pooled and ligated into the 2.1 TOPO-TA vector (Invitrogen, Carlsbad, CA), using 4 μl of PCR product and 1 μl of vector. Two microliters of ligation mix was transformed into TOP10F′ chemically competent cells (Invitrogen). Transformed cells were selected with ampicillin and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Ten clones were obtained from each sample, and DNAs were prepared using a QIAGEN miniprep kit according to the manufacturer's instructions. Inserts were confirmed by digestion with EcoRI. Cloned V1-C5 was sequenced using the primer 319 (CATGGTAGAACAGATGCATGAGG) and the TOPO-TA cloning primers M13F and M13R. Sequencing was performed with Big Dye chemistry, version 3, on an Applied Biosystems 3100 genetic analyzer. Sequences giving missense predicted proteins were removed from analysis, as were duplicate sequences, resulting in an average of nine clones analyzed per animal per time point. The proviral env gene of the inoculation virus was amplified from the PBMC stock from which it was prepared in accordance with the same methods described above. A consensus was created from the eight sequenced clones obtained. The consensus sequence was identical to the published sequence of SHIV-89.6p KB9 (accession no. U89134).
Sequence analysis.
Sequences were assembled using Lasergene DNAstar Seqman and Editseq (2) and aligned with ClustalX (58). Nucleotide alignments were manually edited using BioEdit 5.0.9 (27). A maximum likelihood tree of the nucleotide sequences was constructed with PAUP*4.0 (57). Maximum likelihood bootstrapping was performed with 100 replicates, using the HKY85 model with a subtree-pruning-regrafting-branch-swapping algorithm. The starting tree was obtained by neighbor joining, and the starting branch lengths were obtained using the Rogers-Swofford approximation method. Branches with a bootstrap value of <50 were collapsed, and the tree was rooted on the consensus of 89.6P KB9 (accession no. U89134). For an analysis of diversity, nucleotide alignments were analyzed in MEGA3.0 (34), using the Kimura two-parameter model with pairwise deletions (transition-to-transversion ratio of 2). For an analysis of divergence, a consensus sequence from eight clones of the inoculating strain was obtained using the LANL Seqpublish program (http://www.hiv.lanl.gov/content/hiv-db/SeqPublish/seqpublish.html). The consensus of the inoculation virus was compared to each clone sequence, and divergence was measured with MEGA3.0, using the Kimura two-parameter model with pairwise deletions (transition-to-transversion ratio of 2). The average divergence at each time point for each animal is reported. All sequences were compared to the published sequence to produce the percent difference from the published sequence. All of the clones from the inoculum were then compared to the clones from each animal in order to test whether or not the inoculum was significantly different from the animal's quasispecies by using a Mann-Whitney test (http://eatworms.swmed.edu/∼leon/stats/utest.html). In order to compute selective pressure, the ratio of synonymous (dS) to nonsynonymous (dN) rates of change was calculated with the program PAML (65), using maximum likelihood analyses. Calculations utilized the tree topology shown in Fig. 1. A dN/dS ratio of 1 denotes neutral selection, ratios of >1 denote positive selection, and ratios of <1 denote purifying selection. N-linked glycosylation sites were tracked and tallied using the LANL N-Glycosite program (http://www.hiv.lanl.gov/content/hiv-db/GLYCOSITE/glycosite.html). The frequency of use of each PNG was determined by manually creating a matrix of PNG locations in Excel (Microsoft, Redmond, WA). The nucleotide sequences were translated to amino acids using JavaScript DNA Translator 1.1 (https://deathstar.bcc.washington.edu/public/dnatranslater/index.html), aligned with ClustalX, and manually edited with BioEdit 5.0.9. The reference sequence HIV-HXB2 (accession no. K03455) was included in the alignment in order to standardize the numbering position of each amino acid residue, as recommended by Korber et al. (33). Briefly, each aligned residue was numbered according to the corresponding position in HXB2, and insertions were given the number of the residue prior to the insertion, with a letter to denote each inserted residue. Each clone was then scored for the presence or absence of the canonical PNG motif (Asn-X-Ser/Thr, where X represents any residue other than proline). The presence or absence of a given PNG motif was then compared to the consensus of the inoculum and reported as a percentage of use for a given PNG site. The prevalence of a PNG motif within an animal's quasispecies was compared to the prevalence of the same PNG motif within the inoculum, using Fisher's exact t test (http://www.unc.edu/∼preacher/fisher/fisher.htm). P values of <0.05 are reported as significant.
Three-dimensional mapping of PNG sites.
The PNG sites were mapped on a model structure of gp120. The core structure corresponded to the X-ray structure of YU2 (35) (pdb code 1RZK). For completeness and for the visualization of all PNG sites, model structures for the V1, V2, and V3 loops corresponding to YU2 were added to the core structure. Added loops were minimized while restraining the rest of the atoms of gp120 with a large force constant that was gradually relaxed to 500 kcal/mol-Å2 for backbone atoms of the crystal structure. Molecular dynamic simulations were carried out with flexible loops on a rigid core in a vacuum at high temperature. The resulting structure was solvated with 16,307 water molecules and minimized with restrained core backbone atoms. Next, all-atom molecular dynamic simulations were carried out, with periodic boundary conditions of 300 K for 2 ns. In the simulations, only water molecules and V1/V2/V3 loops were allowed to equilibrate on a rigid core. This provided an efficient sampling of loops on the solvated core structure. The above calculations were carried out using a modified version of AMBER software, and simulation settings and conditions conformed to the program defaults (46). We used these modeled loops on the CD4-bound structure of gp120 to guide us in modeling the relative locations of the N-glycosylation sites. Three-dimensional images were made using VMD software.
Construction of homologous pseudovirus.
In order to test for homologous neutralizing antibodies, a single round of infection with a SHIV-89.6P pseudovirus was utilized. The viral backbone plasmid Q23ΔEnv was kindly provided by Julie Overbaugh (40). The 89.6P Env-encoding plasmid was constructed in pEMC*. The proviral 89.6P gp160 env gene was first cloned into the 2.1 TOPO-TA vector as described above. The first-round PCR primers were EO (TAGAGCCCTGGAAGCATCCAGGAAGTCAGCCTA) and EO1 (TCCAGTCCCCCCTTTTCTTTTAAAAA), and the cycling conditions used were as follows: denaturation at 95°C for 5 min and 35 cycles of 94°C for 40 s, 61°C for 30 s, and 72°C for 3 min 15 s, with a final elongation step of 72°C for 10 min. The second-round primers were 89.6p 3′ gp41 ClaI (GGCGGCGGCATCGATTCACAAGAGAGTGAGCTCAAGC) and 89.6p 5′ gp120 NheI (GCGGCGGCGGCTAGCACAGAAAAATTGTGGGTCACAG), and the cycling conditions used were as follows: 92°C for 5 min, 40°C for 1 min, 68°C for 5 min, 92°C for 1 min, 45°C for 1 min, 68°C for 5 min, 2 cycles (92°C for 1 min, 50°C for 1 min, and 68°C for 5 min), 2 cycles (92°C for 1 min, 55°C for 1 min, and 68°C for 5 min), 25 cycles (92°C for 1 min, 60°C for 1 min, and 68°C for 5 min), and 68°C for 10 min. The second-round primers inserted an in-frame NheI restriction site 3′ of the env leader sequence and a ClaI restriction site at the 3′ end of gp41. Transformants were generated as described above. The purified plasmid was digested with NheI (Roche, Indianapolis, IN) and ClaI (Roche, Indianapolis, IN), and the gp160 fragment was ligated into pEMC* with T4 DNA ligase (NEB, Beverly, MA). Total plasmid DNA (5.5 ng) was transformed into 50 μl of Max Efficiency DH10B competent cells (Invitrogen, Carlsbad, CA) per the manufacturer's instructions and grown at 30°C for 24 h. The plasmid was purified with a QIAGEN miniprep kit as described above and sequenced for verification of the insert. 293T cells were transfected with Fugene 6 (Roche, Indianapolis, IN) per the manufacturer's instructions. Cells were plated to 50% confluence in a T75 flask, and 4 μg total DNA with a 2:1 backbone-to-Env ratio was prepared in a mixture of 12 μl Fugene and 188 μl Dulbecco's modified Eagle's medium. Virus was harvested 48 h later, spun at 2,000 rpm for 10 min, and stored at −70°C until use. The pseudovirus was titrated on Tzm-bl cells, and 200 50% tissue culture infective doses were added to each neutralization assay.
Neutralization assay.
Serum from each animal was tested at 0, 12, 20, 32, and 40 weeks postinfection. Sera were heat inactivated at 56°C for 1 h and spun at 14,000 rpm for 5 min to remove coagulants. The Tzm-bl neutralization assay was performed in 96-well plates (60). Briefly, 200 50% tissue culture infective doses of virus were added to serial dilutions of sera in the presence of 7.5 μg/ml DEAE-dextran and incubated in a total volume of 150 μl medium (Dulbecco's modified Eagle's medium, 10% fetal calf serum, 1% l-glutamine, 1% penicillin-streptomycin) for 1 h at 37°C. Each well received 100 μl of Tzm-bl cells resuspended in medium at 1 × 105 cells/ml. Forty-eight hours later, cells were lysed for 2 min directly in the neutralization plate, using 100 μl of Bright-Glo luciferase assay substrate (Promega, Madison, WI), and immediately analyzed for luciferase activity on a luminometer. The reciprocal dilution of serum necessary to achieve 50% neutralization is reported. Note that the collection of late sera ranged from week 39 to week 53 but is referred to as week 40 for ease of reporting. Neutralization values greater than three times the prebleed background level are considered significant. The lowest dilution of prebleed sera tested (1:100) never reached 50% neutralization; therefore, values above 300 were considered significant. Any sample that did not neutralize at a dilution of 1:100 is reported as 50. All values were calculated with respect to virus only [(value for virus only minus cells only) minus (value for serum minus cells only)] divided by (value for virus minus cells only).
Nucleotide sequence accession numbers.
All sequences included in this study have been deposited in GenBank (accession numbers DQ105094 to DQ105195).
RESULTS
SHIV-89.6P-infected Macaca nemestrina.
The goal of this study was to examine viral envelope divergence, glycosylation site variation, and homologous neutralizing antibodies in a group of macaques that were challenged with the same virus stock. We performed a vaccine challenge study with M. nemestrina (23), using a stock of SHIV-89.6P grown in M. nemestrina peripheral blood mononuclear cells and titrated in vivo in this species. The macaques were challenged by the intrarectal route, and all were infected, as determined by the presence of viral plasma RNA. These animals were monitored for approximately 1 year. The vaccines used in the study included a recombinant vaccinia virus (expressing SHIV-89.6 Gag-Pol and Env), DNA expression vectors (expressing the entire SHIV-89.6 genome and Env-89.6P), and whole inactivated SHIV-89.6 treated with AT-2. Statistically significant differences in virus control and CD4+ T-cell decline were observed in the 12 animals that received DNA plus vaccinia virus vaccines, and improved outcomes were correlated with both cellular and humoral immunity, including the early development of NAbs. In contrast, macaques that received DNA priming and either DNA or protein boosting neither controlled viremia nor maintained their CD4+ T-cell counts. Eleven macaques from this study were chosen as representative of each vaccine group; seven of the animals were not protected from CD4+ T-cell decline and had high viral load set points (>104 RNA copies/ml), and four of the animals were protected from CD4+ T-cell decline and had lower viral load set points (<104 RNA copies/ml) (Table 1).
TABLE 1.
Characteristics of SHIV-89.6P-infected Macaca nemestrina
| Group | Animal no. | Vaccine group | Viral load (RNA copies/ml plasma at 32 wpi) | CD4+ T cells (cells/μl at 32 wpi) | Ratio of dN/dSa | % Divergence at wk 32b | % Diversity at wk 32 | Avg no. of PNGsc |
|---|---|---|---|---|---|---|---|---|
| Minimally divergent | K97107 | Control | 2.8 × 102 | 1,155 | 0.3 | 0.3 | 0.6 | 22.0 |
| K98099 | DNA-vaccinia virus | 1.2 × 103 | 601 | 0.2 | 0.3* | 0.5 | 21.9 | |
| L97191 | DNA-vaccinia virus | 2.5 × 105 | 73 | 0.5 | 0.3* | 0.5 | 21.3 | |
| J97168 | Vaccinia virus-DNA | 5 × 101 | 1,372 | 0.3 | 0.4 | 0.7 | 21.7 | |
| Divergent | T98098 | DNA-protein | 1.6 × 105 | 40 | 0.8 | 0.9* | 1.2 | 20.2d |
| A98069 | DNA-DNA | 1.2 × 104 | 141 | 1.3 | 1.2* | 1.6 | 22.2 | |
| J98071 | Control | 1.4 × 105 | 29 | 1.8 | 1.4** | 1.3 | 23.7 | |
| J97156 | DNA-protein | 5.6 × 104 | 56 | 2.2 | 1.8** | 1.6 | 24.9 | |
| K97246 | DNA-DNA | 1.6 × 105 | 11 | 2.4 | 1.9 | 1.5 | 24.1 | |
| T98108 | DNA-protein | 5.9 × 104 | 5 | 2.0 | 1.9** | 1.0 | 24.3 | |
| L98152 | Vaccinia virus-DNA | 5 × 101 | 1,820 | 2.9 | 1.9** | 1.4 | 21.1 |
Average ratio within the proviral clones of nonsynonymous change to synonymous change, calculated using PAML codeml.
Average percent divergence of each animal's cloned quasispecies away from the inoculum. *, Mann-Whitney P value of <0.05; **, P value of <0.001.
Average number of PNGs per env region from V1 to gp41. Note that N88 is not included in this region and therefore not represented in the average but is likely well conserved.
T98098 sequence reads did not cover the N130 and N136 motifs, which were well conserved in the other macaques. Therefore, the average is likely a low estimate.
Envelope divergence rates of SHIV-89.6P infection at 32 weeks postinfection are similar to those for HIV-1 infection in humans.
A median of 9 (range, 6 to 10) unique gp120 sequences free of missense mutations were obtained from the proviral DNAs in each animal. A maximum likelihood phylogenetic tree was constructed from the gp120 clones obtained at 32 weeks postinfection (wpi). The published SHIV-89.6P sequence (accession no. U89134) and eight clones obtained from the preparation of the viral inoculum were also included for comparison (Fig. 1).
Further analysis of the aligned env nucleotide sequences allowed for quantification of the horizontal distance depicted on the phylogenetic tree. Divergence is defined as viral evolution away from the original sequence, while the quasispecies or sequence variation present within an animal at a given time defines diversity. As expected, there was only slight heterogeneity (0.3%) within the inoculum. Sequences obtained from 4 of the 11 animals showed little divergence (mean, 0.35%; range, 0.3% to 0.4%) and little diversity beyond those of the inoculum (mean, 0.58%; range, 0.5% to 0.7%). These animals included K97107, K98099, L97191, and J97168 and are referred to as “minimally divergent” from here on. In contrast, the sequences obtained from each of the remaining seven animals showed marked divergence (mean, 1.5%; range, 0.9% to 1.9%) and diversification (mean, 1.4%; range, 1.1% to 1.6%). These animals included T98098, A98069, J98071, J97156, K97246, T98108, and L98152 and are referred to as “divergent” from here on (Table 1). The average divergence of all 11 animals at 32 wpi was 1.1%, giving an average rate of 1.8% divergence in gp120 per year. This is comparable to the 1 to 2% per year rate of divergence reported for HIV-1-infected patients (5, 54).
Several clones within the four minimally divergent quasispecies and the inoculum had a limited number of unique changes from the consensus sequence of SHIV-89.6P. These are represented as individual short terminal branches in the phylogenetic analysis (Fig. 1). In contrast, the seven macaques with divergent sequences each formed unique branches. This supports the conjecture that the clustered sequences were derived from the same macaque and that each quasispecies is unique to an animal. None of the sequences obtained from the inoculum cluster matched with any of the seven divergent quasispecies, suggesting that the divergence does not represent the common outgrowth of a minor preexisting sequence.
The rate of nonsynonymous to synonymous change was calculated for each clone, and the average for each animal is reported in Table 1. All four minimally divergent animals had dN/dS ratios of <1, while six of the seven divergent animals had evidence of selective pressure, with dN/dS ratios of >1.
Relationship between viral load and Env divergence.
Six of the seven macaques with high viral set points (>104 RNA copies/ml) developed quasispecies that diverged from the inoculum (Table 1). The one exception was animal L97191, which had the highest viral load set point but whose viral population was only 0.3% divergent from the inoculum at 32 wpi. In contrast, only one of the four animals with low viral set points (<104 RNA copies/ml) developed quasispecies that diverged from the inoculum. The control animals and those that received ineffective vaccines (DNA-DNA or DNA-protein) tended to be the animals with higher viral loads and greater divergence of their quasispecies. This trend did not reach significance in a two-tailed t test (P value = 0.09).
Total number of predicted glycans increases in macaques with divergent Env proteins.
The published 89.6P KB9 sequence (accession no. U89134) has 23 total PNGs in gp120. This is less than the median number of 26 PNGs seen in gp120 of HIV-1 clade B viruses (66). Therefore, it was of interest to analyze if the total number of PNGs in 89.6P increased over time to become more similar to the median for HIV-1 clade B viruses. Sequences from each animal were obtained from V1 through the end of gp120 and did not span a known PNG in C1 (N88). Therefore, the sequenced region in this study included 22 PNGs present in the consensus of the inoculum (and, accordingly, 25 in HIV-1 clade B). Env sequences from macaque T98098 did not span the complete V1 region and were therefore not considered for this analysis. When sequences from all remaining macaques were included in the analysis, we found moderate variation in the total number of PNGs per Env but no significant trend overall to suggest that the Env of 89.6P becomes more heavily glycosylated by 32 wpi. However, there is a difference in total PNGs when the sequences are grouped according to divergence levels. Of the six divergent macaques analyzed, five had sequences with a net gain of PNGs (A98069, J98071, J97156, K97246, and T98108), while only one demonstrated a net loss (L98152). In contrast, three of four minimally divergent macaques demonstrated a net loss of PNGs (K98099, L97191, and J97168), while one (K97107) had sequences that remained the same as the inoculum (Table 1). As Env diverges in these animals, the number of potential N-linked glycosylation sites increases; Env proteins from minimally divergent macaques show an average of 21.7 PNGs, while those from divergent macaques show an average of 23.4 PNGs (unpaired two-tailed t test with unequal variances; P < 0.001).
Many predicted glycans are highly conserved in gp120 even as Env diverges.
The SHIV-89.6P consensus contained 22 PNGs from V1 through the end of gp120 (Fig. 2A). Nineteen were present at the same locations in over 90% of all sequences analyzed as a group (Fig. 2B, blue lines). Seven of the highly conserved PNGs were located within the variable loops, including four in V1 (N130, N136, N139, and N156), one in V3 (N301), and two in V4 (N392 and N406) (Fig. 2B). The remaining 12 PNG motifs that are highly conserved (N197, N230, N234, N241, N262, N289, N295, N332, N339, N356, N362, and N448) lie within the conserved regions, or core, of gp120 (Fig. 2B). When the quasispecies of each macaque were analyzed individually, 11 of the 19 PNGs described above were found to be well conserved in each of the macaques tested (N156, N197, N230, N241, N289, N295, N301, N392, N406, N332, and N339).
FIG. 2.
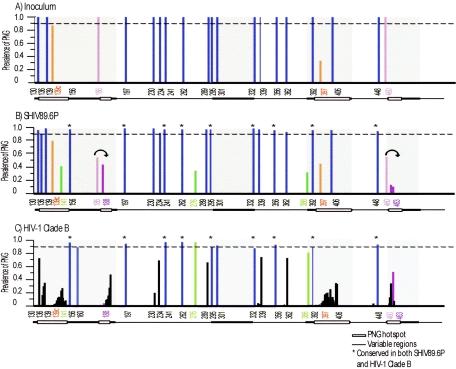
Graphical representation of potential N-linked glycosylation sites in SHIV-89.6P Env and HIV-1 clade B Env. N-Glycosite was used to quantify the locations and prevalence of PNGs within the SHIV-89.6P inoculum (A), pooled SHIV-89.6P variants from all macaques (B), and an HIV-1 clade B reference alignment (C). The amino acid number of each PNG is labeled on the x axis. The prevalence of each PNG is graphed as a fraction of the total on the y axis. Variable regions are denoted as a bar at the bottom of each figure; shaded boxes within the variable regions represent hot spots of PNG variation. PNGs that are conserved (>90% prevalent) are shown in blue, PNGs that are heterologous within the inoculum are shown in orange, convergent PNG additions are shown in green, and convergent PNG shifts are shown in purple. The PNGs that are conserved in both SHIV-89.6P and HIV-1 clade B are denoted with asterisks in panels B and C.
Many of the same PNGs were also conserved when a cross-sectional analysis of published HIV-1 clade B env sequences was analyzed in the same manner. The SHIV-89.6P sequences obtained in this study were compared to an alignment of 176 HIV-1 clade B sequences representing only one sequence per person (http://www.hiv.lanl.gov/content/hiv-db/ALIGN_CURRENT /ALIGN_INDEX.html). These sequences were aligned with the reference strain HXB2 (accession no. K03455) and analyzed for the presence or absence of PNGs. Of the 19 highly conserved PNGs described for the SHIV-89.6P data set, 10 are also conserved in HIV-1 clade B (N156, N197, N241, N262, N295, N301, N332, N356, N392, and N148). These are shown with asterisks in Fig. 2B and C. This result supports the hypothesis that lentiviral envelopes maintain a core set of glycans that are important in producing a functional protein, regardless of the HIV or SHIV background.
Convergent PNG changes occur in the quasispecies of diverged Envs from independent macaques.
An analysis of the published HIV-1 env sequences revealed that the majority of PNG changes fall within limited windows, or hot spots, of the HIV envelope. These hot spots include the N-terminal side of V1, a small region on the C-terminal side of V2, most of V4, and a small region of V5. These hot spots, first noted by Zhang et al. (66), are highlighted at the bottom of Fig. 2C. It is interesting that although all of these hot spots fall within variable regions, there are many PNGs within the variable loops that remain relatively conserved (Fig. 2C).
For each macaque's quasispecies, an average of nine gp120 clones were analyzed for changes in PNGs. The sequences were aligned with that of the reference strain HXB2 (accession no. K03455), and each clone was scored for the presence or absence of a given PNG. For each macaque and the inoculum, the prevalence of each PNG site is reported as a percentage. Each PNG location that underwent a significant level of change from the inoculum (Fisher's exact t test; P < 0.05) is reported in Table 2. Within the SHIV-89.6P data set, the variation in PNGs within independent macaques was shown to occur in remarkably similar ways. We found a total of six convergent PNG changes that occurred independently in multiple macaques, including four additions (N141 [8/11 animals], N276 [6/11 animals], N386 [6/11 animals], and N397 [6/11 animals, although this site was a mixture in the inoculum]) and two shifts (N186 to N188 [6/11 animals] and N460 to N462/3 [4/11 animals]). Five of these six PNG changes in the SHIV-89.6P model fall within the hot spots defined in the HIV-1 sequences (Fig. 2B). The addition of PNGs at all four of the convergent sites was predominantly due to simple G-to-A transitions common in HIV infection, although some sequences did acquire a new PNG by a relatively rare transversion event. These nucleotide changes are described in Table 3. For each PNG addition described, the prevalence of the site within each macaque quasispecies is shown in Table 2, while the location of the site and how it relates to HIV-1 clade B are shown in Fig. 2B and C. PNG N141 in V1 was significantly added (Fisher's exact t test; P < 0.05) in five of the seven divergent animals and increased as a minor species in three additional macaques. This site lies within a hot spot of HIV-1 clade B, and PNG additions within this region are common. PNG N276 in the outer region of C2 was significantly added (Fisher's exact test; P < 0.05) in four of the seven divergent animals and increased in two additional macaques. This site has been defined as part of the CD4bs (36) and is well conserved in HIV-1 clade B. PNG N386 on the N-terminal side of V4 was significantly added (Fisher's exact t test; P < 0.05) in five of the seven divergent animals, while trending towards an increase in one additional macaque. This site is part of the 2G12 epitope (53) and is commonly present in HIV-1 clade B. PNG N397 on the N-terminal side of V4 was present in 38% of the 89.6P inoculum clones used for this study. The prevalence of this site significantly increased (Fisher's exact test; P < 0.05) in four of the seven divergent animals. This site lies within a hot spot of HIV-1 clade B, and PNG additions within this region are common. It is interesting that sequences from several macaques also showed a decreased prevalence of this site compared to the inoculum. The observed mixture in the inoculum suggests that stochastic events at transmission rather than convergent selection may have impacted the observed pattern of occurrence for this particular PNG.
TABLE 3.
Mutations resulting in recurrent additions of PNG sites
| gp120 region and virus | Amino acid and nucleotides at PNGa | Type of mutation | Macaque no. | ||
|---|---|---|---|---|---|
| V1 | 141 | 142 | 143 | ||
| 89.6P | S | S | S | ||
| AGT | AGC | AGC | |||
| Variant | N | S | S | Transition | K97107, J97156, K97246, A98069, J98071, T98098, T98108, L98152 |
| AAT | AGC | AGC | |||
| C2 | 276 | 277 | 278 | ||
| 89.6P | D | F | T | ||
| GAT | TTC | ACA | |||
| Variant | N | F | T | Transition | J97156, K97246, J98071, T98098, T98108, L98152 |
| AAT | TTC | ACA | |||
| V4 | 386 | 387 | 388 | ||
| 89.6P | N | T | A | ||
| AAT | ACA | GCA | |||
| Variant 1 | N | T | T | Transition | J97156, K97246, A98069, J98071, T98098, T98108 |
| AAT | ACA | ACA | |||
| Variant 2 | N | T | S | Transversion | K97246 |
| AAT | ACA | TCA | |||
Underlined amino acids and nucleotides indicate mutations.
A PNG shift is defined here as the removal of one PNG with the concurrent addition of a new PNG within two to three residues. A shift occurred in V2, from N186 to N188, in five of the seven divergent animals. This shift was also detected in one minimally divergent animal (K97107). There is evidence that several mutation strategies were used to achieve this end (Table 4), and it occurred in 100% of the clones in four of the animals. This shift lies within the small region of V2 defined as a hot spot within HIV-1 clade B (Fig. 2C). Interestingly, the residues within the V2 hot spot of HIV-1 clade B are skewed towards the C-terminal end of the hot spot (Fig. 2C). The shift of N186 to N188 in SHIV-89.6P moves the PNG toward the C terminus (Fig. 2B). A second PNG shift occurred in V5, from N460 to N462/3, in four of the seven divergent macaques (Table 2). This shift lies within a hot spot of HIV-1 clade B, and PNG additions within this region are common (Fig. 2C). This V5 shift, like that in V2, involves a complex variety of mutational strategies (Table 4).
TABLE 4.
Mutations resulting in recurrent shifting PNG sites
| gp120 region and virus | Amino acid and nucleotides at PNGa | Type of mutation | Macaque no. | ||||||
|---|---|---|---|---|---|---|---|---|---|
| V2 | 186 | 187 | 187a | 188 | 189 | 190 | |||
| 89.6P | N | T | S | N | T | ||||
| AAT | ACT | AGT | AAT | ACT | |||||
| Variant 1 | N | N | N | T | Deletion, transition | K97107 | |||
| AAT | AAT | AAT | ACT | ||||||
| Variant 2 | N | T | N | N | T | Transition | J97168, A98069, | ||
| AAT | ACT | AAT | AAT | ACT | T98098 | ||||
| Variant 3 | N | T | N | N | N | T | Insertion, transition | A98069 | |
| AAT | ACT | AAT | AAT | AAT | ACT | ||||
| V5 | 460 | 461 | 462 | 462a | 463 | 464 | 465 | ||
| 89.6P | N | S | T | E | T | E | T | ||
| AAT | AGT | ACT | GAG | ACT | GAG | ACT | |||
| Variant 1 | I | S | A | E | N | E | T | Two transversions, transition | J97156 |
| ATT | AGT | GCT | GAG | AAT | GAG | ACT | |||
| Variant 2 | P | E | N | E | T | Two transversions | J97156 | ||
| CCT | GAG | AAT | GAG | ACT | |||||
| Variant 3 | N | S | A | E | N | E | T | Transition, transversion | J98071 |
| AAT | AGT | GCT | GAG | AAT | GAG | ACT | |||
| Variant 4 | N | I | A | E | N | E | T | Transition, transversion | J98071 |
| AAT | ATT | GCT | GAG | AAT | GAG | ACT | |||
| Variant 5 | N | S | N | K | T | D | T | Transition, transversion | L98152 |
| AAT | AGT | AAT | AAG | ACT | GAG | ACT | |||
| Variant 6 | N | S | N | K | T | D | P | Two transversions, transition | L98152 |
| AAT | AGT | AAT | AAG | ACT | GAG | CCT | |||
Underlined amino acids and nucleotides indicate mutations.
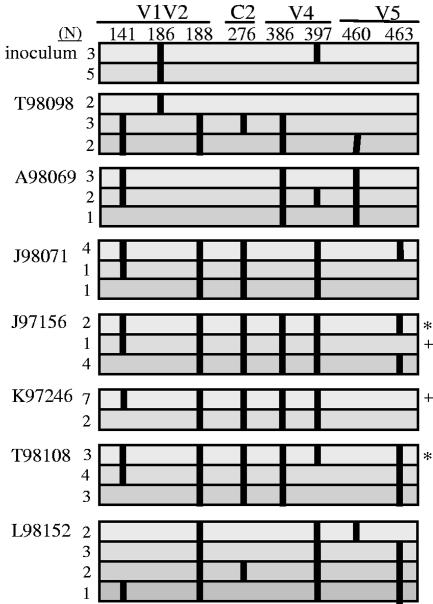
Although the six PNG changes described above were common to multiple animals, identical combinations of these changes occurred in only a few macaques (J97156 shares one type with K97246 and one type with T98108) (Fig. 3), all of which had different amino acid sequences surrounding the sequons (see Fig. 5; data not shown). These data (Fig. 3) further support the concept that these changes represent convergent evolution rather than the outgrowth of a minor variant, with the exception of N397.
FIG. 3.
PNG combinations within the Env quasispecies members of each macaque at 32 weeks postinfection. Each horizontal bar corresponds to a combination of PNG changes that exists within the quasispecies of each animal. The number of clones with a given combination is shown to the left of the bars. The position of each PNG is shown along the top. If the PNG is present, it is shown as a vertical line. Identical combinations found within different animals are denoted with symbols on the right.
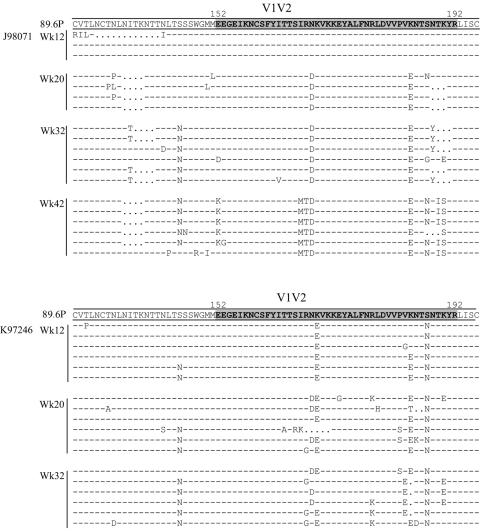
FIG. 5.
Accumulation of Env sequence changes over time in two macaques. The variable regions of representative longitudinal sequences from two macaques with significant divergence are shown. The consensus sequence of Env in the SHIV-89.6P inoculum is shown along the top. A charged 40-residue region from positions 152 to 192 is highlighted in V1/V2.
Some PNG changes are unique to individual macaque quasispecies.
In addition to the changes described above, there were significant unique PNG changes that occurred within individual macaques. Although these do not represent convergent PNG changes, they are of interest as they represent plausible changes to the glycan shield of 89.6P. These changes are shown in Table 2. The majority of unique changes happen within the described V1 hot spot (Fig. 2C). PNG 136 on the N-terminal side of V1, though well conserved in the overall data, was removed in one divergent animal (L98152). PNG 139 on the N-terminal side of V1, also well conserved in the overall data, was removed in one divergent animal (J98071). PNG 139C is present in the consensus of the inoculum but is missing in one of eight clones, putting its prevalence within the inoculum at 88%. This PNG is carried by the sequon NLT and is removed by either changing L to P (NPT) or deleting LT. This PNG was removed to various degrees in many of the macaques, but this change only reached significance in the minimally divergent macaque L97191. The convergent change in V2 described above includes a shift, wherein N186 is removed and a new site at N188 is added. There was one divergent macaque (A98069) with significant removal of N186 without the addition of a new site at N188. This trend also occurred in J98071 and K97107, though neither reached significance. PNG 234 in C2 and PNG 362 in C3 are both well conserved in the overall data, but each was removed in one divergent animal (L98152 and T98108, respectively). Both of these sites are approximately 75% prevalent in HIV-1 clade B.
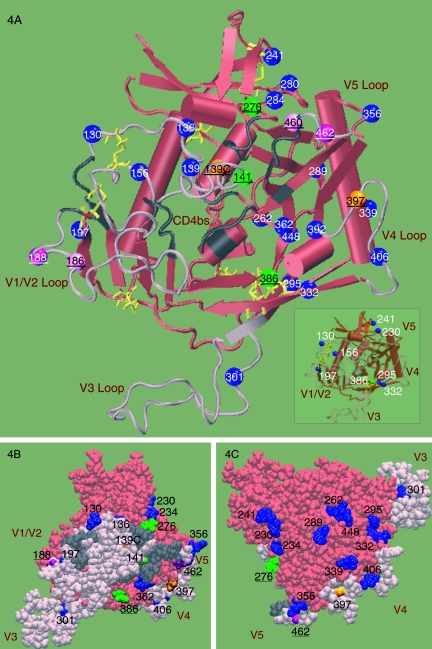
Mapping of PNG residues onto the gp120 structure.
PNGs were mapped onto a gp120 structure that was created by modeling the variable loop sequences of HIV-1 YU2 onto the core crystal structure of CD4-bound YU2 gp120 (35, 36). This three-dimensional mapping facilitates interpretation of the PNG patterns in the SHIV 89.6P Env proteins that we observed within the context of the modeled protein structure (Fig. 4A). All six convergent PNG changes described above map to the neutralizing face of gp120 and are proximal to the CD4bs (Fig. 4B). In contrast, the PNGs on the silent face are predominantly conserved (Fig. 4C, blue residues). The only PNG within the V3 loop (N301) is highly conserved in both 89.6P and HIV-1 clade B; it is in close proximity to the chemokine receptor binding site and can influence coreceptor usage (44).
FIG. 4.
Three-dimensional structural illustration of locations of N-glycosylation sites. Locations of PNGs from the SHIV-89.6P clones were mapped onto a model of gp120 based on the X-ray structure of the CD4-bound YU2 gp120 core (35) (PDB code 1RZK). The V1, V2, and V3 loops (pink) of YU2 are modeled onto the core (red) for completeness and identification of N-glycosylation sites. In the gp120 orientation shown, the viral and target membranes would be located above and below the protein, respectively. The yellow lines show the disulfide bridges. The N-glycosylation sites that are stable (blue), added (green), shifted (purple), and heterologous within the inoculum (orange) are shown as balls and labeled with the corresponding amino acid numbers. The predicted CD4bs is shown in gray; residues were included based on the contacts identified within 5 Å of CD4 in the CD4-bound YU2 crystal structure (35). (A) Cartoon diagram showing the locations of all PNGs in 89.6P. The inset highlights a subset of PNGs proximal to the disulfide bridges. (B) Space-filling model of the inner face. The predicted CD4bs (gray) is partially occluded by the modeled V1V2 loop (pink). All variant PNGs (underlined) lie on the inner face of Env. (C) Space-filling model of the outer face. The PNGs on the outer domain of gp120 are predominantly conserved.
Seven conserved PNG sites are located proximal to conserved disulfide bonds (Fig. 4A, inset, blue residues). Three of these sites are at the base of V1V2 (N130, N197, and N156), and two of these sites are at the base of V3 (N295 and N332). The convergent addition of N386, a site highly prevalent in HIV-1 clade B, contributes an additional PNG adjacent to the disulfide bond at the base of V4 (Fig. 4B, green residue). PNGs at N230 and N241 frame the disulfide bond between the β5 and β7 regions in the core. We observed that some of the PNGs that lie far apart in the gp120 amino acid sequence are in fact spatially close in the three-dimensional model (Fig. 4). An analysis of the locations of Asn residues of PNGs in the YU2-based model of Env gp120 found two sets of three (triads of) PNGs that are within 12 Å (C-alpha distance) of each other. The first of these includes N362, N386, and N392 on the neutralizing or inner domain of gp120, and the second includes N262, N295, and N448 on the silent face. With the exception of N386, all of these spatially proximal PNGs are highly conserved in these variants, and N386 is a site that is prevalent in HIV-1 clade B and is added to SHIV-89.6P in multiple macaques. The PNGs N392, from the first group, and N295, from the second group, form part of the monoclonal antibody (MAb) 2G12 epitope (59).
Longitudinal changes in PNGs occur at different rates.
The viral quasispecies from a subset of four macaques were analyzed longitudinally, comprising two animals with rapid PNG changes (K97246 and J97156) and two animals with delayed PNG changes (J98071 and L98152); sequences derived from one animal in each group are shown as representatives in Fig. 5. The addition of N386 is an example of how some PNG changes occur more rapidly in some macaques than in others; this addition began to occur in clones from macaque K97246 by 12 wpi, while it was still only seen in one clone from macaque J98071 at 32 wpi. All four macaques underwent a steady accumulation of consistent changes over time, yet all of the changes within an animal did not occur together temporally. This is demonstrated with the quasispecies of macaque K97246, which underwent a complete PNG shift in V2 from N186 to N188 by 12 wpi, while the addition of N276 in C2 did not occur until 32 wpi.
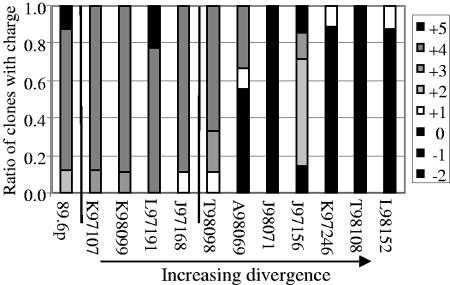
Changes in charge and potential changes in structure are conserved in multiple macaques.
Although the majority of common sequence changes occur at PNG sites, we observed two additional types of changes in the hypervariable regions. There is a 40-residue stretch of V1V2 that has a strong net positive charge in the consensus sequence of the SHIV-89.6P virus used to challenge the macaques. In the cloned inoculum, 88% of the sequences have a net charge of +4 or +5. The four animals with viral sequences that did not diverge from the inoculum shared this charge profile (91% of sequences had a net charge of +4 or +5). In all seven macaques with divergent quasispecies, this region of V1V2 became more acidic by 32 wpi, with only 17% of the clones having a net charge of +4 or +5 (Fig. 6). Also, a proline was added to V5 by 32 wpi in five of the seven macaques with divergent quasispecies (mean, 66%; range, 38% to 100%). This mutation occurred at either residue 462 or 465. This region of V5 is flanked by residues that are predicted to be involved in the CD4bs (36).
FIG. 6.
Net charge of the V2 region becomes more acidic at week 32 in animals with diversified quasispecies. A 40-residue region from positions 152 to 192 had a median charge of +4 in the inoculum. The four macaques with minimal divergence (K97107, K98099, L97191, and J97168) maintained this charge. The seven animals with viruses that diverged from the inoculum (A98069, T98098, J97156, K97246, L98152, T98108, and J98071) had mutations in this region that resulted in a more acidic (more negative) net charge. The vertical line divides animals that do not have any significant PNG changes from those that do. Macaque numbers on the x axis are arrayed in order of increasing levels of divergence.
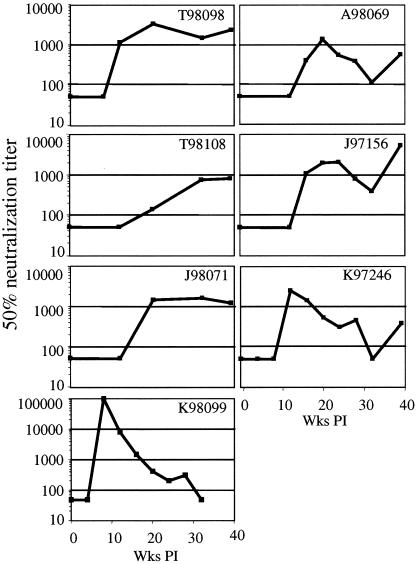
Macaques with divergent Env quasispecies have higher levels of homologous neutralizing antibodies.
Although none of the macaques in this study had detectable NAbs at the time of challenge, those from protected vaccine groups receiving vaccinia virus-DNA or DNA-vaccinia virus were positive in gamma interferon-specific enzyme-linked immunospot assays for one or more of the peptide pools for Gag and Env on the day of challenge and showed accelerated development of NAbs in the first 4 weeks postchallenge (23). In addition to the lack of NAbs, macaques in the other vaccine groups and the control group had no T cells expressing gamma interferon against Gag, Env, Pol, or accessory peptide pools on the day of challenge, and these macaques were immunologically indistinguishable postchallenge. For the 11 animals examined here, we extended the analysis of NAbs through 40 weeks postchallenge, utilizing a luciferase-based assay with a pseudovirus bearing the inoculum Env protein (Fig. 7). Three of the four minimally divergent macaques never developed detectable levels of homologous neutralizing antibodies within the 1-year span of this study (K97107, L97191, and J97168). Macaque K98099 showed no divergence from the inoculum yet rapidly developed homologous NAbs by 12 wpi, with the highest titer of any in this study. This response also declined rapidly, showing a weak response by 20 wpi and an undetectable response by 32 wpi. As is characteristic of the minimally divergent animals, this animal had a low viral load set point, likely as a result of the DNA-vaccinia virus vaccine that it received (23); the early development of NAbs may also be attributable to priming by the vaccine.
FIG. 7.
Development of homologous NAbs as a function of time postinfection. Sera from the SHIV-89.6P-infected macaques were tested against pseudotyped SHIV-89.6P in a Tzm-bl cell assay. Reciprocal titers for 50% neutralization are shown. The y axis for animal K98099 is different (higher values) from the rest. Each sample was run in triplicate, and most data points represent the geometric means of three independent assays. Macaques K97107, L97191, J97168, and L98152 had no detectable NAbs at any time point tested (not shown).
Of the macaques with more divergent quasispecies, six of seven developed homologous NAbs (Fig. 7). By 12 wpi, sera from two macaques (K97246 and T98098) achieved appreciable levels of homologous NAbs. An additional three animals developed homologous NAbs by 20wpi (A98069, J97156, and J98071). The remaining animal, T98108, began to develop a low-titer homologous NAb response by week 32. All six macaques maintained a homologous response through the latest time point tested. Three of the macaques (J98071, T98108, and T98098) maintained steady titers of homologous NAbs. J98071 is a control animal that developed a homologous response at 20 wpi that was maintained through 45 wpi. Macaque T98098 received a DNA-protein vaccine and developed a homologous response by 12 wpi that was maintained through 45 wpi. T98108 also received a DNA-protein vaccine but did not develop a detectable homologous response until 20 wpi. The other three animals (A98069, J97156, and K97246) developed strong homologous NAbs at 20, 20, and 12 wpi, respectively, but showed declined titers at 32 wpi. The titers of homologous NAbs then continued to increase in all three macaques until week 45. The one divergent animal that developed weak homologous NAbs that declined after week 4 (L98152) is also unusual in that it is the only divergent animal with a low viral load.
DISCUSSION
This longitudinal and cross-sectional study of mutations in HIV env in the SHIV background provides evidence that there are more constraints on the configuration of the glycan shield than was previously appreciated. The majority of PNGs are relatively invariant, as 19 of the 23 PNGs present in the inoculum were well conserved in Envs from all macaques studied and 11 were rarely altered in any individual. With one exception, these PNGs are maintained in the human clade B population, shown here in Fig. 2 and presented in a cross-sectional analysis of published HIV Env sequences from multiple subtypes (66). As the virus diversified in independent macaques, there was an increase in the total number of PNGs per Env, and the virus converged upon several additions and shifts of PNG locations within regions that we term “hot spots.” Our modeling data show that the hot spots primarily correspond to residues within the variable loops that are proximal to the CD4bs. This divergence was often associated with higher viral loads and high-titer homologous NAbs and may represent a complex interplay between antigen availability, viral fitness and escape, and the development of neutralizing antibodies.
The remarkable level of glycan conservation seen in this study could result from several different selective pressures. The monomeric form of gp120 consists of an inner domain, an outer domain, and a bridging sheet that connects the two (36). In the trimeric form, the outer domain of gp120 is theoretically the region most accessible to neutralizing antibodies, yet few antibodies are known to recognize this heavily glycosylated “silent face” (63, 64). Nevertheless, removal of carbohydrates from this region renders the virus more susceptible to neutralization (42). Mapping the conserved PNGs identified in this study onto the HIV-1 gp120 core demonstrated that most of the 19 highly conserved sites are located on the outer domain of gp120. The removal of the conserved glycans on the silent face may be selected against since exposure of the epitopes beneath may readily elicit neutralizing antibodies. Many of the conserved PNG sites also lie within close proximity to the disulfide bonds that define the variable loops. These strategically placed PNG sites may define the conformational space of loops and influence their flexibility and orientation. Alternatively, these sites may be conserved due to cotranslational folding requirements; for example, it has been proposed that N-glycans either protect the cysteine residue pair or recruit the necessary cellular machinery to the correct location for the formation of proper disulfide bonds in influenza virus hemagglutinin (18). In support of this, N-linked glycosylation plays a role in ensuring the proper folding of newly synthesized gp120 in the endoplasmic reticulum (ER) (39), and carbohydrates are known to prevent aggregation, allow indirect interactions through the resident lectin chaperons calnexin and calreticulin of the ER (37), and provide spatial constraints that facilitate folding (18). Thus, the conserved PNGs near cysteines in HIV-1 Env may guide the formation of correct disulfide bonds during cotranslational folding and direct the loops to functionally relevant spatial regions in folded gp120.
SHIV-89.6P contains a clade B envelope that is less glycosylated than those of the average HIV-1 clade B viruses in the LANL database. Surprisingly, we observed four additions and two shifts of PNGs that resulted in more “typical” representations of the clade B glycosylation profiles. Two of the PNG additions that occur in SHIV-89.6P are at locations outside the hot spots that are lacking in the inoculum yet highly conserved in HIV-1 clade B (N276 and N386). The other two additions (N141 and N397) occur within hot spots identified on HIV-1 clade B. Two PNG shifts (N186 to N188 and N460 to N462) occur within HIV-1 clade B hot spots, in each case shifting the PNG N-terminally to a region within the hot spot with a higher prevalence of PNGs. Although the four PNG additions resulted from simple G-to-A transitions, the recurrent shifts in V2 and V5 were much more mutationally complex, involving transversions and multiple base substitutions, and were realized in multiple ways in different macaques. When considering these six PNG changes, it is noteworthy that although multiple changes occurred within the same clone, the same combination of changes rarely occurred in clones from different macaques, and the temporal development of the changes was not consistent. In the two incidents where the same combination of PNG changes was found in two macaques, these PNGs occurred in different sequence backgrounds. This suggests that the sequon changes were arrived at in an independent manner and did not arise from the outgrowth of a minor variant from the inoculum. There is a scattering of unique PNG changes that occur in only one animal or at low levels; these may reflect positions that are not vital to but are generally favored for the function of the virus. Other studies have documented that specific PNGs may be less important than the density of PNGs in a given region; the unique PNG changes seen in this model may lend further evidence to this phenomenon (45, 48).
It has been shown that the relative positions and number of PNG sites can vary without sacrificing infectivity (28, 45); however, these sites do have functional roles. Several of the convergent PNG changes occur within hot spots proximal to the CD4bs, a conformational determinant of Env that is conserved among the primate lentivirus family. At least some of the broadly neutralizing activity in human serum has been shown to map to this region (55), and human MAbs such as immunoglobulin G1b12 have confirmed that some antibodies targeted to this region can neutralize divergent isolates (9). An interesting study of HIV resistance to plant lectins hints at an interpretation for the PNG shift seen in V2. Plant lectins have specificity for high-mannose oligomers and thus can inhibit the entry of HIV into its target cell (3). Intriguingly, under selective pressure by high-mannose-specific lectins, the PNG at site 186 of HIV-1 (IIIb) shifted two residues downstream to position 188 (4), comparable to the shift we see here. It is possible that the shifting PNG site at 186/188 is involved in the entry of HIV into target cells. It is unlikely, however, that a unique functional role can be assigned to each PNG change seen in this study. The PNGs may collectively perform a function, or alternatively, a single PNG may participate in many functions. For example, the addition of a PNG at N386 was seen both in the SHIV-89.6P Envs in this study and in clade B viruses (Fig. 2C). N386 is proximal to the coreceptor binding site as well as a disulfide bond, mutations at N386 influence antibody accessibility to the CD4 binding site (32), and changes at N386 can impact mutational pressure at the tip of the V3 loop (30). A mass spectroscopic approach that assigned 25 of 26 consensus glycosylation sites of HIV-1 SF2 gp120 found that high-mannose carbohydrates and complex carbohydrates each cluster together, with little overlap between the two forms (67). An examination of the fully glycosylated SIV gp120 crystal structure also shows clustering of high-mannose carbohydrates (12). In that structure, two of the three clusters arise from a group of three PNGs where the Asn residues are within 12 Å of each other (C-alpha distance). The mapping of PNGs in 89.6P Env gp120 also revealed two groups of three PNGs that are structurally close to each other, within a 12-Å distance according to this model. The close proximity of these Asn residues does not necessarily imply that they form a glycan cluster, but it is interesting to speculate whether they might interact. Interestingly, these groups appear on opposite faces of gp120, and a residue from each group is also part of the epitope of the MAb 2G12.
Since the coreceptor binding site is only available after conformational changes induced by CD4 (12) and since the gp41 ectodomain is only thought to be available after conformational changes induced by coreceptor binding (12), it could be argued that the CD4bs is the most vulnerable region of the viral Env and thus that mutations that shield it from antibodies would provide a fitness advantage to the virus. It has been proposed that the V1V2 and V3 loops interact to form a protective barrier around the CD4 binding site (19, 69). The V3 loop is thought to play the predominant role in physical interactions with the coreceptor, and its strong net positive charge can facilitate binding to the negatively charge CCR5 coreceptor (68). Due to this requirement, the polarity of the V3 loop is relatively constrained. However, if the V1V2 loops adopt a more negative charge, this could influence the packing of the variable loops around the CD4bs (25, 47). In this study, all seven animals with divergent quasispecies had sequences that had evolved a more acidic V1V2. Both the N- and C-terminal amino acids in the V5 loop are known to comprise part of the CD4bs, yet little is known about how the V5 region changes over time. In this study, in five of the seven animals with divergent quasispecies a proline was added in V5; such a change could alter the shape of the V5 loop and may in turn alter the CD4bs. Additional analyses of the binding properties of individual Env proteins bearing these mutations are needed to test this hypothesis directly.
A major goal of this study was to determine how nonsynonymous changes in Env are related to the development of homologous and heterologous NAbs. The calculation of dN/dS ratios suggested that the changes that were observed in these macaques are the result of selective pressure. Irrespective of vaccine treatment, the macaques with higher viremia were typically the ones to develop persistent homologous neutralizing antibodies as well as divergent quasispecies. Two exceptions to this trend were observed, i.e., macaques K98099 and L98152. Both macaques received a vaccine consisting of DNA expressing Env from both 89.6 and 89.6P and vaccinia virus expressing Env from 89.6 prior to challenge, and both animals maintained low viremia. K98099 mounted a strong homologous neutralizing antibody response after challenge yet underwent minimal divergence. We speculate that the limited antigen in this macaque resulted in the rapid decline of homologous NAbs and little to no divergence of the viral Env. In contrast, L98152 failed to develop detectable neutralizing antibodies, yet its virus diverged. If the vaccine primed a response that controlled viremia, it did not prevent divergence. This result suggests that divergence per se does not require high levels of virus in the periphery, nor does it imply a poor clinical outcome. The development of persistent homologous NAbs may be both antigen dependent and require exposure to the diverging quasispecies, while divergence of the quasispecies may be driven partially by viral fitness and partially by immune escape. Evidence for viral replication driving antibody maturation has been observed in another system, namely, influenza virus vaccination (7). Five of the seven divergent macaques received vaccines that were ineffective in generating significant B- or T-cell responses prior to challenge, and after challenge these SHIV-specific responses and control of viremia were indistinguishable from those in mock-vaccinated controls. We thus consider it likely that these vaccines had no effect on the development and selection of viral variants after challenge.
It is interesting that several of the animals maintained steady virus loads and that the homologous NAbs were either maintained or continued to increase despite the changes occurring in the viral Env. However, further work is necessary to determine whether any of the mutations that we have sequenced encode escape variants. In the macaques studied here, the majority of homologous NAbs may be directed against the immunodominant V3 loop, which did not undergo change in the time frame of this study. It is possible that mutations in Env sequences contribute to the development of higher-avidity NAbs or to NAbs effective against heterologous isolates. An investigation of this possibility is currently in progress, utilizing the cloned Envs to make pseudovirions and to determine the neutralization profiles of each variant. This study ended after 1 year of observation, at which point none of the plasma samples showed significant evidence of heterologous neutralization. This finding is consistent with studies in HIV-infected humans, who do not typically develop heterologous NAbs prior to 3 years of infection (43, 51).
There is precedent for convergent patterns of change as HIV evolves within independent hosts. It is well documented that the HIV subtype B viruses often undergo defined changes as the virus switches from an R5 to an X4 phenotype (29, 56). Hirsch et al. have shown with the SIV model that convergent changes in the viral Env can affect viral fitness by leading to a CD4-independent virus (21). These examples highlight patterns in the way that the virus evolves to enhance viral fitness. Data from this work demonstrate additional mechanisms of convergent change in the viral envelope that involve glycosylation, the V1V2 charge, and the V5 structure. These changes correlate with both higher viral loads and higher homologous NAb titers. Further work is under way to define whether these predicted convergent changes are utilized and how they affect viral fitness or viral escape. As we continue to deepen our understanding of the intricacies of HIV evolution, we may be able to better inform the design of effective vaccines capable of generating the elusive broadly reactive NAbs.
Acknowledgments
We thank Madhumita Mahalanabis for a critical review of the manuscript, Julie Overbaugh and Leonidas Stamatatos for helpful discussions, and Pushpa Jayaraman, Dina Lauman, Ruth Hotchkiss, and William Sutton for technical assistance. The viral clone Q23ΔEnv was kindly provided by Julie Overbaugh. Virus stocks and cell lines used in neutralization assays were obtained from the NIH AIDS Research and Reference Reagent Program.
This study was supported by PHS grant P01-AI54564 (N.L.H.), a Center for AIDS Research predoctoral fellowship (P30-AI27757) (W.B.), a grant from the Murdock Charitable Trust (N.L.H.), and donations from the James B. Pendleton Charitable Trust.
REFERENCES
- 1.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 1997. Lasergene software suite. DNASTAR Inc., Madison, Wis.
- 3.Balzarini, J., S. Hatse, K. Vermeire, K. Princen, S. Aquaro, C.-F. Perno, E. D. Clercq, H. Egberink, G. V. Mooter, W. Peumans, E. V. Damme, and D. Schols. 2004. Mannose-specific plant lectins from the Amaryllidaceae family qualify as efficient microbicides for prevention of human immunodeficiency virus infection. Antimicrob. Agents Chemother. 48:3858-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balzarini, J., K. V. Laethem, S. Hatse, M. Froeyen, E. V. Damme, A. Bolmstedt, W. Peumans, E. D. Clercq, and D. Schols. 2005. Marked depletion of glycosylation sites in HIV-1 gp120 under selection pressure by the mannose-specific plant lectins of Hippeastrum hybrid and Galanthus nivalis. Mol. Pharmacol. 67:1556-1565. [DOI] [PubMed] [Google Scholar]
- 5.Bernardin, F., D. Kong, L. Peddada, L. A. Baxter-Lowe, and E. Delwart. 2005. Human immunodeficiency virus mutations during the first month of infection are preferentially found in known cytotoxic T-lymphocyte epitopes. J. Virol. 79:11523-11528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Botarelli, P., B. A. Houlden, N. L. Haigwood, C. Servis, D. Montagna, and S. Abrignani. 1991. N-glycosylation of HIV-gp120 may constrain recognition by T lymphocytes. J. Immunol. 147:3128-3132. [PubMed] [Google Scholar]
- 7.Brokstad, K. A., R. J. Cox, D. Major, J. M. Wood, and L. R. Haaheim. 1995. Cross-reaction but no avidity change of the serum antibody response after influenza vaccination. Vaccine 13:1522-1528. [DOI] [PubMed] [Google Scholar]
- 8.Burns, D. P. W., C. Collignon, and R. C. Desrosiers. 1993. Simian immunodeficiency virus mutants resistant to serum neutralization arise during persistent infection of rhesus monkeys. J. Virol. 67:4104-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 10.Cao, Y., L. Qin, L. Zhang, J. Safrit, and D. D. Ho. 1995. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N. Engl. J. Med. 332:201-208. [DOI] [PubMed] [Google Scholar]
- 11.Chackerian, B., L. M. Rudensey, and J. Overbaugh. 1997. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J. Virol. 71:7719-7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, B., E. M. Vogan, H. Gong, J. J. Skehel, D. C. Wiley, and S. C. Harrison. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834-841. [DOI] [PubMed] [Google Scholar]
- 13.Cheng-Mayer, C., A. Brown, J. Harouse, P. A. Luciw, and A. J. Mayer. 1999. Selection for neutralization resistance of the simian/human immunodeficiency virus SHIVSF33A variant in vivo by virtue of sequence changes in the extracellular envelope glycoprotein that modify N-linked glycosylation. J. Virol. 73:5294-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chohan, B., D. Lang, M. Sagar, B. Korber, L. Lavreys, B. Richardson, and J. Overbaugh. 2005. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1-V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J. Virol. 79:6528-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choisy, M., C. H. Woelk, J.-F. Guegan, and D. L. Robertson. 2004. Comparative study of adaptive molecular evolution in different human immunodeficiency virus groups and subtypes. J. Virol. 78:1962-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciurea, A., P. Klenerman, L. Hunziker, E. Horvath, B. M. Senn, A. F. Ochsenbein, H. Hengartner, and R. M. Zinkernagel. 2000. Viral persistence in vivo through selection of neutralizing antibody-escape variants. Proc. Natl. Acad. Sci. USA 97:2749-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dacheux, L., A. Moreau, Y. Ataman-Onal, F. Biron, B. Verrier, and F. Barin. 2004. Evolutionary dynamics of the glycan shield of the human immunodeficiency virus envelope during natural infection and implications for exposure of the 2G12 epitope. J. Virol. 78:12625-12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daniels, R., B. Kurowski, A. E. Johnson, and D. N. Hebert. 2003. N-linked glycans direct the cotranslational folding pathway of influenza hemagglutinin.Mol. Cell 11:79-90. [DOI] [PubMed] [Google Scholar]
- 19.D'Costa, S., K. S. Slobod, R. G. Webster, S. W. White, and J. L. Hurwitz. 2001. Structural features of HIV envelope defined by antibody escape mutant analysis. AIDS Res. Hum. Retrovir. 17:1205-1209. [DOI] [PubMed] [Google Scholar]
- 20.Decker, J. M., F. Bibollet-Ruche, X. Wei, S. Wang, D. N. Levy, W. Wang, E. Delaporte, M. Peeters, C. A. Derdeyn, S. Allen, E. Hunter, M. S. Saag, J. A. Hoxie, B. H. Hahn, P. D. Kwong, J. E. Robinson, and G. M. Shaw. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 201:1407-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dehghani, H., B. A. Puffer, R. W. Doms, and V. M. Hirsch. 2003. Unique pattern of convergent envelope evolution in simian immunodeficiency virus-infected rapid progressor macaques: association with CD4-independent usage of CCR5. J. Virol. 77:6405-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Derdeyn, C. A., J. M. Decker, F. Bibollet-Ruche, J. L. Mokili, M. Muldoon, S. A. Denham, M. L. Heil, F. Kasolo, R. Musonda, B. H. Hahn, G. M. Shaw, B. T. Korber, S. Allen, and E. Hunter. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019-2022. [DOI] [PubMed] [Google Scholar]
- 23.Doria-Rose, N. A., C. Ohlen, P. Polacino, C. C. Pierce, M. T. Hensel, L. Kuller, T. Mulvania, D. Anderson, P. D. Greenberg, S. L. Hu, and N. L. Haigwood. 2003. Multigene DNA priming-boosting vaccines protect macaques from acute CD4+-T-cell depletion after simian-human immunodeficiency virus SHIV89.6P mucosal challenge. J. Virol. 77:11563-11577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doria-Rose, N. A., C. C. Pierce, M. T. Hensel, W. F. Sutton, N. Sheikh, P. Polacino, L. Kuller, Y. D. Zhu, S. L. Hu, D. Anderson, and N. L. Haigwood. 2003. Multigene DNA prime-boost vaccines for SHIV89.6P. J. Med. Primatol. 32:218-228. [DOI] [PubMed] [Google Scholar]
- 25.Etemad-Moghadam, B., G. B. Karlsson, M. Halloran, Y. Sun, D. Schenten, M. Fernandes, N. L. Letvin, and J. Sodroski. 1998. Characterization of simian-human immunodeficiency virus envelope glycoprotein epitopes recognized by neutralizing antibodies from infected monkeys. J. Virol. 72:8437-8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geyer, H., C. Holschbach, G. Hunsmann, and J. Schneider. 1988. Carbohydrates of human immunodeficiency virus. J. Biol. Chem. 263:11760-11767. [PubMed] [Google Scholar]
- 27.Hall, T. 1997-2005. BioEdit, version 5.0.9. Ibis Therapeutics, Carlsbad, Calif.
- 28.Hemming, A., A. Bolmstedt, B. Jansson, J. E. Hansen, B. Travis, S. L. Hu, and S. Olofsson. 1994. Identification of three N-linked glycans in the V4-V5 region of HIV-1 gp 120, dispensable for CD4-binding and fusion activity of gp 120. Arch. Virol. 134:335-344. [DOI] [PubMed] [Google Scholar]
- 29.Jensen, M. A., F.-S. Li, A. B. van't Wout, D. C. Nickle, D. Shriner, H.-X. He, S. McLaughlin, R. Shankarappa, J. B. Margolick, and J. I. Mullins. 2003. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 Env V3 loop sequences. J. Virol. 77:13376-13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalish, M. L., B. T. Korber, S. Pillai, K. E. Robbins, Y. S. Leo, A. Saekhou, I. Verghese, P. Gerrish, C. L. Goh, D. Lupo, B. H. Tan, T. M. Brown, and R. Chan. 2002. The sequential introduction of HIV-1 subtype B and CRF01AE in Singapore by sexual transmission: accelerated V3 region evolution in a subpopulation of Asian CRF01 viruses. Virology 304:311-329. [DOI] [PubMed] [Google Scholar]
- 31.Kimata, J. T., L. Kuller, D. B. Anderson, P. Dailey, and J. Overbaugh. 1999. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nat. Med. 5:535-541. [DOI] [PubMed] [Google Scholar]
- 32.Koch, M., M. Pancera, P. D. Kwong, P. Kolchinsky, C. Grundner, L. Wang, W. A. Hendrickson, J. Sodroski, and R. Wyatt. 2003. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology 313:387-400. [DOI] [PubMed] [Google Scholar]
- 33.Korber, B. T., B. T. Foley, C. L. Kuiken, S. K. Pillai, and J. G. Sodroski. 1998. Numbering positions in HIV relative to HXB2CG. Hum. Retrovir. AIDS III:102-111. [Google Scholar]
- 34.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA 3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 35.Kwong, P. D., R. Wyatt, S. Majeed, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Struct. Fold Des. 8:1329-1339. [DOI] [PubMed] [Google Scholar]
- 36.Kwong, P. D., R. Wyatt, J. Robinson, R. Sweet, J. Sodroski, and W. Hendrickson. 1998. Structure of an HIV-1 gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Land, A., and I. Braakman. 2001. Folding of the human immunodeficiency virus type 1 envelope glycoprotein in the endoplasmic reticulum. Biochimie 83:783-790. [DOI] [PubMed] [Google Scholar]
- 38.Leonard, C. K., M. W. Spellman, L. Riddle, R. J. Harris, J. N. Thomas, and T. J. Gregory. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 265:10373-10382. [PubMed] [Google Scholar]
- 39.Li, Y., L. Luo, N. Rasool, and C. Y. Kang. 1993. Glycosylation is necessary for the correct folding of human immunodeficiency virus gp120 in CD4 binding. J. Virol. 67:584-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long, E. M., S. M. J. Rainwater, L. Lavreys, K. Mandaliya, and J. Overbaugh. 2002. HIV type 1 variants transmitted to women in Kenya require the CCR5 coreceptor for entry, regardless of the genetic complexity of the infecting virus. AIDS Res. Hum. Retrovir. 18:567-576. [DOI] [PubMed] [Google Scholar]
- 41.Ly, A., and L. Stamatatos. 2000. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J. Virol. 74:6769-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCaffrey, R. A., C. Saunders, M. Hensel, and L. Stamatatos. 2004. N-linked glycosylation of the V3 loop and the immunologically silent face of gp120 protects human immunodeficiency virus type 1 SF162 from neutralization by anti-gp120 and anti-gp41 antibodies. J. Virol. 78:3279-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moog, C., H. J. A. Fleury, I. Pellegrin, A. Kirn, and A. M. Aubertin. 1997. Autologous and heterologous neutralizing antibody responses following initial seroconversion in human immunodeficiency virus type 1-infected individuals. J. Virol. 71:3734-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogert, R. A., M. K. Lee, W. Ross, A. Buckler-White, M. A. Martin, and M. W. Cho. 2001. N-linked glycosylation sites adjacent to and within the V1/V2 and the V3 loops of dualtropic human immunodeficiency virus type 1 isolate DH12 gp120 affect coreceptor usage and cellular tropism. J. Virol. 75:5998-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohgimoto, S., T. Shioda, K. Mori, E. E. Nakayama, H. Hu, and Y. Nagai. 1998. Location-specific, unequal contribution of the N glycans in simian immunodeficiency virus gp120 to viral infectivity and removal of multiple glycans without disturbing infectivity. J. Virol. 72:8365-8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pearlman, D. 1995. AMBER, 4.1. University of California San Francisco, San Francisco, Calif.
- 47.Poss, M., A. G. Rodrigo, J. J. Gosink, G. H. Learn, D. de Vange Panteleeff, H. L. Martin, Jr., J. Bwayo, J. K. Kreiss, and J. Overbaugh. 1998. Evolution of envelope sequences from the genital tract and peripheral blood of women infected with clade A human immunodeficiency virus type 1. J. Virol. 72:8240-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quinones-Kochs, M. I., L. Buonocore, and J. K. Rose. 2002. Role of N-linked glycans in a human immunodeficiency virus envelope glycoprotein: effects on protein function and the neutralizing antibody response. J. Virol. 76:4199-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reimann, K. A., J. T. Li, R. Veazey, M. Halloran, I. W. Park, G. B. Karlsson, J. Sodroski, and N. L. Letvin. 1996. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate Env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J. Virol. 70:6922-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reimann, K. A., J. T. Li, G. Voss, C. Lekutis, K. Tenner-Racz, P. Racz, W. Lin, D. C. Montefiori, D. E. Lee-Parritz, Y. Lu, R. G. Collman, J. Sodroski, and N. L. Letvin. 1996. An env gene derived from a primary human immunodeficiency virus type 1 isolate confers high in vivo replicative capacity to a chimeric simian/human immunodeficiency virus in rhesus monkeys. J. Virol. 70:3198-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rudensey, L. M., J. T. Kimata, E. M. Long, B. Chackerian, and J. Overbaugh. 1998. Changes in the extracellular envelope glycoprotein of variants that evolve during the course of simian immunodeficiency virus SIVMne infection affect neutralizing antibody recognition, syncytium formation, and macrophage tropism but not replication, cytopathicity, or CCR-5 coreceptor recognition. J. Virol. 72:209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. Ollmann Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steimer, K. S., C. J. Scandella, P. V. Skiles, and N. L. Haigwood. 1991. Neutralization of divergent HIV-1 isolates by conformation-dependent human antibodies to gp120. Science 254:105-108. [DOI] [PubMed] [Google Scholar]
- 56.Strunnikova, N., S. Ray, R. Livingston, E. Rubalcaba, and R. Viscidi. 1995. Convergent evolution within the V3 loop domain of human immunodeficiency virus type 1 in association with disease progression. J. Virol. 69:7548-7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swofford, D. L. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 58.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, L. X., J. Ni, S. Singh, and H. Li. 2004. Binding of high-mannose-type oligosaccharides and synthetic oligomannose clusters to human antibody 2G12: implications for HIV-1 vaccine design. Chem. Biol. 11:127- 134. [DOI] [PubMed] [Google Scholar]
- 60.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 62.Willey, R. L., R. Shibata, E. O. Freed, M. W. Cho, and M. A. Martin. 1996. Differential glycosylation, virion incorporation, and sensitivity to neutralizing antibodies of human immunodeficiency virus type 1 envelope produced from infected primary T-lymphocyte and macrophage cultures. J. Virol. 70:6431-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 64.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 65.Yang, Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13:555-556. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, M., B. Gaschen, W. Blay, B. Foley, N. Haigwood, C. Kuiken, and B. Korber. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14:1229-1246. [DOI] [PubMed] [Google Scholar]
- 67.Zhu, X., C. Borchers, R. J. Bienstock, and K. B. Tomer. 2000. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochemistry 39:11194-11204. [DOI] [PubMed] [Google Scholar]
- 68.Zolla-Pazner, S. 2004. Identifying epitopes of HIV-1 that induce protective antibodies. Nat. Rev. Immunol. 4:199-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zwick, M. B., R. Kelleher, R. Jensen, A. F. Labrijn, M. Wang, G. V. Quinnan, Jr., P. W. H. I. Parren, and D. R. Burton. 2003. A novel human antibody against human immunodeficiency virus type 1 gp120 is V1, V2, and V3 loop dependent and helps delimit the epitope of the broadly neutralizing antibody immunoglobulin G1 b12. J. Virol. 77:6965-6978. [DOI] [PMC free article] [PubMed] [Google Scholar]